Abstract
Therapeutic blockade of the PD-1/PD-L1 axis is recognized as an effective treatment for numerous cancer types. However, only a subset of patients respond to this treatment, warranting a greater understanding of the biological mechanisms driving immune evasion via PD-1/PD-L1 signaling and other T-cell suppressive pathways. We previously identified a head and neck squamous cell carcinoma with human papillomavirus integration in the PD-L1 locus upstream of the transmembrane domain-encoding region, suggesting expression of a truncated form of PD-L1 (Parfenov et al., Proc Natl Acad Sci USA 111(43):15544–15549, 2014). In this study, we extended this observation by performing a computational analysis of 33 other cancer types as well as human cancer cell lines, and identified additional PD-L1 isoforms with an exon 4 enrichment expressed in 20 cancers and human cancer cell lines. We demonstrate that cancer cell lines with high expression levels of exon 4-enriched PD-L1 generate a secreted form of PD-L1. Further biochemical studies of exon 4-enriched PD-L1 demonstrated that this form is secreted and maintains the capacity to bind PD-1 as well as to serve as a negative regulator on T cell function, as measured by inhibition of IL-2 and IFNg secretion. Overall, we have demonstrated that truncated forms of PD-L1 exist in numerous cancer types, and have validated that truncated PD-L1 can be secreted and negatively regulate T cell function.
Electronic supplementary material
The online version of this article (10.1007/s00262-018-2284-z) contains supplementary material, which is available to authorized users.
Keywords: HPV, PD-L1, Secreted PD-L1, Cancer immunology
Introduction
Cancer immunotherapy is now recognized as an effective strategy for the treatment of numerous tumor types [1, 2]. One prominent example has been the clinical efficacy of antibodies targeting the PD-1 receptor and its ligands, agents that have been approved for the treatment of multiple tumor types [1, 2]. Numerous clinical trials are underway to determine the efficacy of PD-1/PD-L1 blockade in additional cancers, as well as combination approaches to build upon the initial success of PD-1/PD-L1 blockade. Although there has been considerable success in treating patients using anti-PD-1/PD-L1 therapies, only a subset of patients respond to this treatment and our knowledge of the diverse mechanisms driving cancer immune evasion is limited.
The PD-1/PD-L1 signaling axis is a major mechanism of immune evasion in cancer [3], specifically by inhibiting T cell function in the immune microenvironment [4]. Both cancer and hematopoietic cells have been shown to express the PD-1 ligands PD-L1 and PD-L2, which bind to the PD-1 receptor expressed by activated T and B cells [3, 4]. The engagement of the PD-1 receptor with its ligands inhibits CD8 + T cell activation, function and survival [4–6], therefore, providing a direct and potent mechanism of immune evasion.
HPV is the etiologic agent for nearly all cervical cancers and an increasing proportion of head and neck squamous cell carcinomas (HNSCCs) [7]. Oncogenic viruses present foreign antigens to the immune system (e.g., HPV E6 and E7) and induce immune tolerance to prevent an anti-tumor T-cell response to viral antigens [8]. One recently reported mechanism governing anti-viral immune escape is PD-L1/PD-L2 locus amplification in Epstein-Barr virus (EBV) associated lymphomas and gastric cancers, as well as PD-L1 overexpression with HPV infection in head and neck cancer [9–12]. We previously reported pan-cancer computational analyses of HPV integration sites in the human genome [13, 14], and while we did not observe amplification of PD-L1/PD-L2 in HPV-associated cancers, we did identify a novel potential mechanism of immune evasion represented by a case in which integration of HPV into the CD274 (PD-L1) locus between exons 4 and 5 drove overexpression of the 5′ exons (exons 1–4) of PD-L1, suggesting the production of a truncated form of PD-L1 [13]. This putative truncated form of PD-L1 differed from those characterized in a recent report highlighting expression of PD-L1 lacking the 3′ UTR as a mechanism of PD-L1 overexpression in diverse cancers [15].
In this study, we characterize this novel PD-L1 integration event and show that it leads to the production of a truncated and alternatively polyadenylated mRNA and a secreted rather than membrane-bound protein. We extend our observation into a large cohort of human cancers [Cancer Cell Line Encyclopedia (CCLE) and The Cancer Genome Atlas (TCGA) databases] and normal tissue (Genotype-Tissue Expression (GTEx) database) and demonstrate that exon 4-enriched isoforms of PD-L1 are expressed in multiple cancer types including bladder, breast, lung, ovarian and other non-HPV driven cancers and cancer cell lines. We further characterize this exon 4-enriched form of PD-L1 using biochemical approaches and cellular assays and show that this form is indeed secreted and negatively affects T cell function. In all, our data demonstrate that a truncated and secreted form of PD-L1 is produced in numerous cancer types and can negatively regulate T cell function. Our data, combined with previous reports of PD-L1 serum detection [16–18], suggest that a genomic basis exists in cancer for production of secreted PD-L1, a finding that may have important implications for cancer patients receiving PD-1/PD-L1 blocking therapies.
Materials and methods
Cancer cell line encyclopedia analysis
After downloading RNA-seq data for 935 cell lines from the National Cancer Institute (NCI) Genomic Data Commons [19], gene abundances and exon counts were quantified using default parameters in StringTie (v1.3.2d) [20, 21]. Kernel density plot representation was used to express the univariate distribution of RNA-seq reads to determine not-detected cutoff of fragments per kilobase of transcript per million mapped reads (FPKM) < 1. Individual cell line expression values are available at 10.5281/zenodo.1413358.
TCGA and GTEx analysis
Gene and exon quantification data was downloaded for cancer, TCGA [22], and corresponding normal tissue, GTEx [23], from the recount2 database [24]. The cancers included in the analysis were: LAML—acute myeloid leukemia, ACC—adrenocortical carcinoma, BLCA—bladder urothelial carcinoma, BRCA—breast invasive carcinoma, CESC—cervical squamous cell carcinoma and endocervical adenocarcinoma, CHOL—cholangiocarcinoma, COAD—colon adenocarcinoma, DLBC—diffuse large B-cell lymphoma, ESCA—esophageal carcinoma, GBM—glioblastoma multiforme, HNSC—head and neck squamous cell carcinoma, KICH—kidney Chromophobe, KIRC—kidney renal clear cell carcinoma, KIRP—kidney renal papillary cell carcinoma, LIHC—liver hepatocellular carcinoma, LUAD—lung adenocarcinoma, LUSC—lung squamous cell carcinoma, MESO—mesothelioma, OV—ovarian serous cystadenocarcinoma, PAAD—pancreatic adenocarcinoma. PCPG—pheochromocytoma and paraganglioma, PRAD—prostate adenocarcinoma, READ—rectum adenocarcinoma, SARC—sarcoma, SKCM—skin cutaneous melanoma, STAD—stomach adenocarcinoma, TGCT—testicular germ cell tumors, THCA—thyroid carcinoma, THYM—thymoma, UCEC—uterine corpus endometrial carcinoma, UCS—uterine carcinosarcoma, UVM—uveal melanoma. Kernel density plot representation was used to express the univariate distribution of RNA-seq reads of all genes to determine not-detected cutoff of RPKM < 1. Individual tumor expression values are available at 10.5281/zenodo.1413358.
Kernel density
Kernel density plot representations were used to express the univariate distribution of RNA-seq reads for all protein-coding genes and the exon 4-enriched PD-L1. The kernel density plots were calculated in R (ver. 3.4.1) using the density function with default parameters.
Pairwise Spearman’s correlation analyses
To determine the correlations between the expression of exon 4-enriched PD-L1 and T cell genes for all cancer types, we performed pairwise Spearman’s correlation analyses. The correlation and heatmap plots were calculated in R (ver. 3.4.1) using the rcorr and corrplot functions, respectively, with default parameters.
Primary cells and cell lines
PBMCs were isolated from healthy donor blood from the Brigham and Women’s Blood Bank, and cultured in complete Iscove's Modified Dulbecco's Medium media [IMDM (Gibco, #12440053), 10% Human AB male serum (Gemini, #100-512), penicillin/streptomycin/glutamine (PSG; Corning, #30-009-CI)]. HEK293T cells and CAL62 cells were grown in Dulbecco's Modified Eagle's medium (Gibco, #11965092), 10% FCS, PSG, and 1 mM sodium pyruvate (Gibco, #11360-70). RKO cells were grown in EMEM [American Type Culture Collection (ATCC), #30-2003], 10% FCS, PSG, and 1 mM sodium pyruvate. RERF-LC-AD1 cells were grown in RPMI 1640 (Gibco, #11875093), 10% FCS, PSG, and sodium pyruvate.
Cloning and constructs
For hemagglutinin (HA) tagged constructs (PD-L1-HA, sec-PD-L1-long-HA and sec-PD-L1-short-HA) PD-L1 sequences were amplified by PCR from full-length PD-L1 (Origene) and simultaneously HA-tagged and cloned into pcDNA3.1 (-) (Thermo, V79520).
Transfections
HEK293T cells were transfected with PD-L1-HA, sec-PD-L1-long-HA and sec-PD-L1-short-HA with XtremeGene 9 (Roche, #06365779001) at a ratio of 1:3 (DNA:transfection reagent) in Opti-MEM (Gibco, #31985070) according to manufacturer’s instructions. The following day, media was changed to serum-minus media, and the next day media was harvested for protein precipitation. 2 days post-transfection, media was harvested for ELISA, cells were used for flow cytometry, or lysate was collected for western blotting.
Flow cytometry
For measuring surface localization of HA-tagged PD-L1, HEK293T cells transfected with HA-tagged PD-L1 constructs (PD-L1-HA, sec-PD-L1-long-HA and sec-PD-L1-short-HA) or empty vector (pcDNA3.1(-)) were stained for PD-L1 48 h post-transfection. Briefly, cells were lifted with Accutase Cell Detachment Solution (Innovative Cell Technologies, Inc, #AT104) and rinsed. Cells were incubated with primary antibody (anti-human PD-L1-PE, clone MIH1, mouse IgG1, κ, 1:20, eBioscience, #12-5983) or isotype control (PE mouse IgG1, κ, clone P3.6.2.8.1, eBioscience, #12-4714-41), in blocking buffer (2% Human AB serum (Gemini, #100-512), 2% Fc-block (Miltenyi Biotech, #130-059-901), 2 mM EDTA in PBS) for 30 min on ice. Cells were washed and incubated with LIVE/DEAD yellow stain (1:1000, Thermo, #L34959) for 10 min on ice. Cells were washed, resuspended in flow buffer, and analyzed with a BD LSRII/Fortessa. 10,000 events were collected. For characterizing T cell blasts, cells were stained for CD3, CD4 and CD8. T cell blasts were incubated with conjugated primary antibodies at 1:50 (anti-human CD3 PerCP-eFluor 710, eBioscience, #46-0036; APC/Cy7 anti-human CD8, Biolegend, #344713; PE/Cy7 anti-human CD4, Biolegend, #300511) in blocking buffer for 30 min on ice. Cells were washed and incubated with LIVE/DEAD yellow stain for 10 min on ice followed by washing, fixation in 4% paraformaldehyde, and analysis on a BD LSRII/Fortessa. For PD-L1 surface staining of cancer cell lines, cells were stained as described with the following antibodies: anti-human CD274-APC (1:100, clone 29E.2A3, mouse IgG2b, κ, Biolegend, #329707) or isotype control (1:100, clone MPC-11, mouse IgG2b, κ, Biolegend, #400319). Analysis was performed on a FACS Canto RUO. Between 20,000 and 50,000 events were recorded. All flow data was analyzed and automatic compensation was performed using FlowJo (FlowJo, LLC). For data analysis, cells were first gated on forward scatter versus side scatter, then single cells were gated on using side scatter height versus side scatter width, followed by forward scatter height versus forward scatter width. Dead cells were then excluded by gating on LIVE/DEAD yellow negative cells.
PD-L1 sandwich ELISA
Soluble PD-L1 was measured by sandwich ELISA adapted from Zhou et al. [25]. For measuring soluble PD-L1 in media from cell lines and transfected cells, cells were plated, and after 2 days, when cells were about 70–80% confluent, media was taken for ELISA. Briefly, flat bottom 96-well plates (eBioscience, #44-2404-21) were coated with 2 µg/ml anti-PD-L1 capture antibody recognizing the IgV domain aa 19-131 (29E.12B1, gift from Dr. Gordon Freeman), blocked with blocking buffer (1% BSA in PBS), washed, and then samples were added. Standard curves were made with human PD-L1-histidine tagged fusion protein (Sino Biological, #10084-H08H). The plate was washed and a detection antibody recognizing a region in the PD-L1 IgV domain non-overlapping with 29E.12B1 [25] was used at 1:2,500 (29E.2A3-Biotin, Biolegend, #329704) in 1% BSA in PBS. The plate was washed, and streptavidin-HRP (Southern Biotech 7100-05) was then added at 1:20,000 in 1% BSA in PBS. The plate was washed and 3,3′,5,5′-tetramethylbenzidine (TMB) peroxidase substrate (Cell Signaling Technology (CST), #7002S) was added for 30 min. TMB stop solution (CST, #7004S) was added, absorbance was read on a Tecan Infinite M200 Pro plate reader at 450 nm, and protein concentrations were determined using GraphPad Prism 6 Software.
PD-1 binding assay
Binding ability of PD-L1 to PD-1 was measured using a functional ELISA with an R&D Systems protocol based on the literature indicating PD-1 receptor as a binding partner for PD-L1 [26]. Briefly, flat bottom 96-well plates were coated with 100 ng/ml Fc-tagged recombinant human PD-L1 (rhPD-L1-Fc) chimera protein (R&D Systems, #1086-PD-050). To account for non-specific binding, wells with no PD-1 were used. Plates were washed, blocked in blocking buffer (1% BSA in PBS), and coated with rhPD-L1-Fc (extracellular region of PD-L1 as positive control, R&D Systems, #156-B7-100), sec-PD-L1-long-Fc (R&D Systems, custom-made product) or rhIgG1-Fc (negative control, R&D Systems, 110-HG-100) in 1% BSA in PBS at varying concentrations in µg/ml. For antibody blocking experiments, rhPD-L1-Fc and sec-PD-L1-long-Fc was used at 1 µg/ml, and an anti-PD-1 blocking antibody (mouse IgG1, clone EH12 (EH12.1), kind gift from Dr. Gordon Freeman), an anti-PD-L1 blocking antibody that detects a region in the CD80 binding domain different than the detection antibody used [27, 28] (mouse IgG1, MIH1 clone, eBioscience, #16-5983-82), and IgG1 isotype control (LEAF Purified Mouse IgG1, κ Isotype control, MOPC-21 clone, Biolegend, #400124) were used at 10 µg/ml. Anti-PD-L1 and the corresponding IgG1 isotype control antibody was incubated with PD-L1 protein prior to addition to the plates. Anti-PD-1 antibody and the corresponding IgG1 Isotype control antibody was incubated directly on plates in 1% BSA in PBS after blocking of the plate, prior to addition of rhPD-L1-Fc and sec-PD-L1-long-Fc protein. Plates were washed, and incubated with detection antibody recognizing the IgV domain within the CD80 binding region [25] (1:2,500; 29E.2A3-Biotin, Biolegend, #329704) in 1% BSA in PBS. The plate was then washed and streptavidin-HRP (1:20,000; Southern Biotech, #7100-05) was added in 1% BSA in PBS. The plate was washed again and TMB peroxidase substrate was added for 30 min. TMB stop solution was added and absorbance of ELISA plates were read on a SpectraMax 190 plate reader at 450 nm. OD values were normalized to maximum binding values of rhPD-L1. GraphPad Prism 6 was used for non-linear fit of data and Kd value calculation with a one-site specific binding model, and calculated Kd and Bmax values were used to plot the Scatchard graph.
PD-L1 functional assays
The PD-L1 functional assay was performed using an R&D Systems protocol adapted from Freeman et al. [5]. PBMCs were isolated using Ficoll-Paque Plus at a ratio of 1:2 (Ficoll-Paque:blood; GE Healthcare, #GE17-1440-02). The buffy coat was washed with PBS and erythrocytes were lysed according to manufacturer’s instructions (Human Erythrocyte Lysing Kit, R&D Systems, #WL1000). PBMCs were washed and either used fresh or frozen for later use. Fresh or thawed PBMCs were cultured in complete IMDM media and T-cell blasts were generated as per R&D Systems protocol with IL-2 and PHA. Day 9 T-cell blasts were washed with IMDM and plated in complete IMDM media at 1E5 cells per well in round bottom 96 well plates which had been pre-coated with anti-CD3 antibody (3 µg/ml; BD, #555336) overnight at 4C followed by a wash and coating with rhPD-L1-Fc (R&D Systems, #156-B7-100), sec-PD-L1-long-Fc (R&D Systems, custom product) or rhIgG1-Fc (R&D Systems, #110-HG-100) at 20 µg/ml or 40 µg/ml for 3–4 h at 37C. C-terminal Fc-tagged sec-PD-L1-long was custom made by R&D Systems. 24 h after plating, media was harvested and frozen at − 80 °C for IL-2 (OptEIATM IL-2 ELISA kit II, BD, #550611) and IFNg (OptEIATM IL-2 ELISA kit II, BD, #550612) ELISA assays. Absorbance of ELISA plates were read on a SpectraMax 190 (Molecular Devices) plate reader at 450 nm, and protein concentrations were determined with SoftMax Pro 5 (Molecular Devices).
Western blotting
Cell lysates were harvested using Radioimmunoprecipitation assay (RIPA) buffer (CST, #9806S) with protease (Complete Mini Protease Inhibitor Cocktail, Roche, #11836153001) and phosphatase inhibitors (Set I & II, EMD Millipore, #524624, 524625). Briefly, RIPA buffer was added to cell culture dishes, and the lysate was scraped off plates using a cell scraper. Lysate was mixed at 4 °C for 20 min, spun down to remove debris, and protein concentration was measured using a Bradford assay (Protein Assay Dye Reagent Concentrate, Bio-Rad, #5000006). Between 30 and 100 µg of total protein was prepped in Laemmli SDS protein loading buffer (Boston Bioproducts, #BP-111R), boiled, and loaded on a 4–12% Bis-Tris gel (NuPAGE Novex, #NP0322BOX). For protein precipitation, trichloroacetic acid (TCA; Sigma, #T0699) was used. Media was changed on cells to serum-free media the day before media harvest (1 day post-transfection or plating of non-transfected cells). Briefly, TCA was added to media at 1:4, and samples were spun down. The protein pellet was washed with acetone, dried, resuspended in Laemmli SDS protein loading buffer, boiled, and directly loaded on a 4–12% Bis-Tris gel for SDS-PAGE. A nitrocellulose membrane was used (Fisher, #EP4H00010) for transfer. Blots were blocked for an hour at room temperature (RT) and incubated with primary antibody overnight at 4C. 10% milk was used for blocking, and primary antibody was resuspended in 5% milk or 2% BSA. Blots were washed and incubated with the appropriate secondary antibodies in 5% milk or 2% BSA. Blots were washed and enhanced chemiluminescence substrate was used according to manufacturer’s instructions (Amersham ECL Prime Western Blotting Detection Reagent, #RPN2232). Blots were then developed on film and scanned or digitized using an ImageQuant LAS 4000 luminescent image analyzer (GE Healthcare Life Sciences), for the appropriate exposure times. The following primary antibodies were used: rabbit anti-CD274 (1:100; Abcam, #ab58810), anti-mouse beta-actin (1:1000, Cell Signaling technology #3700), mouse anti-HA (1:1000; 16B12, Covance, #MMS-101P), mouse anti-vinculin (1:10,000; hVIN-1, Sigma, #V9131). The following Pierce HRP-conjugated secondary antibodies were used at 1:20,000: goat anti-mouse (#31444), goat anti-rabbit (#31460). Goat anti-mouse secondary antibody (1:5000, Jackson Immuno Research Labs#115035146) was used for detecting beta-actin. Densitometry analysis was performed using ImageJ software, where signal (percentage) was normalized to the loading control [29].
Immunofluorescent staining
HEK293T and cancer cells were grown on coverslips overnight, and HEK293T cells were transfected as described. 48 h later, cells were fixed with 4% paraformaldehyde, washed with PBS, and permeabilized with 0.1% Triton X-100. Cells were washed with PBS, and blocked in Antibody Dilution Buffer (Ventana Medical Systems, Inc., #ADB250) for 1 h at RT. Cells were then incubated with primary antibody diluted in Antibody Dilution Buffer at 4 °C overnight at the following dilutions: mouse anti-HA (1:1000; 16B12, Covance, #MMS-101P), rabbit anti-β-catenin (1:1000; CST, #9587S), rabbit anti-PD-L1 (1:500; EPR19759, Abcam, #ab213524). Cells were washed 3× in PBS, and incubated with the following secondary antibodies diluted in Antibody Dilution Buffer at RT for an hour: goat anti-mouse IgG-Alexa Fluor 488 (1:1000; ThermoFisher Scientific, #A-11001), donkey anti-rabbit IgG-Alexa Fluor 568 (1:1000; ThermoFisher Scientific, #A-10042), goat anti-rabbit IgG Alexa Fluor 488 (1:500; Fisher Scientific, #A11034). Cells were then washed 2× in PBS, and counterstained with Hoechst 33342 diluted in PBS (1:1000; Invitrogen, #H3570) for 10 min at RT. Cells were washed 2× in PBS and mounted on microscope slides (Denville Ultra Frost, Denville Scientific Inc., #M1002) with SlowFade Diamond Antifade Mountant (ThermoFisher Scientific, #S36967) or mounting media containing DAPI was applied to cells (Electron Microscopy Sciences, #1798550) and sealed with nail polish. Cells were then imaged on an Olympus BX50 Upright Fluorescent Microscope using a DP73 camera and a 100× oil immersion lens for 293T cells and inverted Zeiss LSM 780 multiphoton laser scanning confocal microscope using a 63X oil immersion lens for the cancer cell lines.
Statistical analysis
GraphPad Prism 6 Software and Microsoft Excel were used to generate graphs, and GraphPad Prism 6 was used to calculate ANOVA, Kd and Bmax values. One-way ANOVA was used with a Dunnett’s test for multiple comparison correction for IL2 and IFNg ELISA comparisons, and a two-way ANOVA was used with a Dunnett’s test for multiple comparison correction for PD-1 binding with neutralizing antibodies. A p value < 0.05 was considered significant.
Results
PD-L1 with a high exon 4/5 expression ratio is expressed in a variety of cancer types
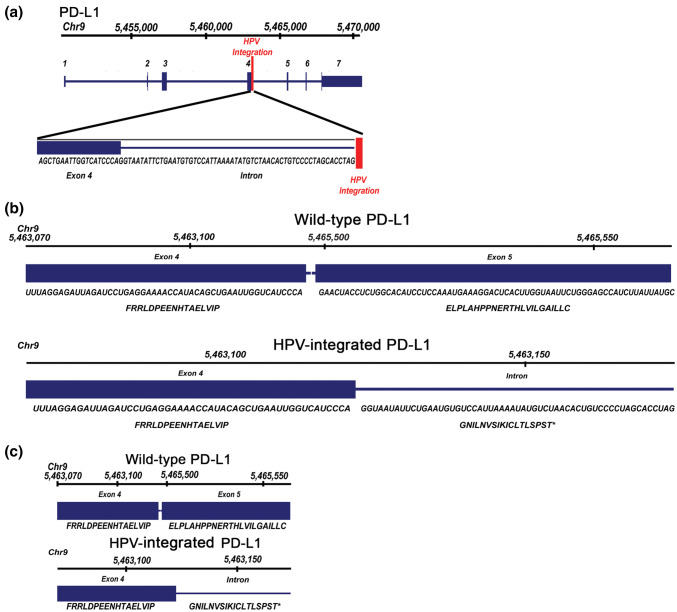
We previously performed a computational analysis of next-generation whole genome and RNA sequencing data from 279 HNSCCs (TCGA), where we investigated genomic sites of HPV integration and identified a case where HPV16 integrated in the intron after exon 4 of the PD-L1 (CD274) locus [13]. We now extended this analysis and performed de novo alignment and re-assembly of PD-L1 transcripts in this case and identified an apparently novel transcript with an alternative 3′ region with a readthrough of 56 nucleotides into the intron after exon 4, an in-frame stop codon and an alternative polyadenylation signal (Fig. 1a,b [13]). There was a predicted difference in amino acid sequence after exon 4 as compared to wild-type PD-L1, specifically, with the inclusion of an alternative 18 amino acid sequence (Fig. 1b,c). We hypothesized that this form of PD-L1 may be secreted, since it lacks the transmembrane domain encoded by exon 5, and prior reports have demonstrated the existence of soluble PD-L1 protein in human serum [16, 30].
Fig. 1.
HPV Integration into the PD-L1 locus in a head and neck squamous cell carcinoma patient. a Genomic track showing a previously published case whereby HPV16 was found to be integrated in the intron between exon 4 and exon 5 of the PD-L1 locus, resulting in expression of a b transcript with an alternative 3′ region including a readthrough of 56 nucleotides into the intron after exon 4, including a stop codon and an alternative polyadenylation signal. The predicted difference in amino acid sequences is depicted b, c, with an alternative 18 amino acid sequence after exon 4 [13]
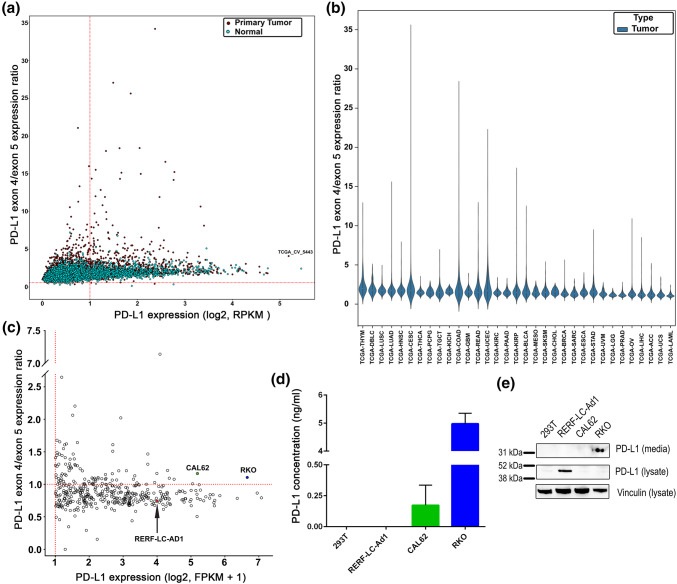
To determine if PD-L1 isoforms predicted to be secreted are unique to the single HNSCC case, we further investigated the presence and prevalence of PD-L1 isoforms with high exon 4 expression relative to the 3′ exons (by measuring the expression ratio of exon 4 to exon 5) in a variety of cancers by analyzing TCGA data from 33 different cancer types as well as corresponding normal tissue (Fig. 2a,b; Figs. S1, S2). The expression of the fourth and fifth exons was measured and plotted as a ratio on the y-axis (PD-L1 exon 4/5 expression ratio), with the expression of total PD-L1 on the x-axis. We observed an enrichment of expression of the fourth exon of PD-L1 as compared to the fifth exon in multiple cancer types, with 116 cases out of a total of 9981 (1.68%) demonstrating a ratio of expression of exon 4–exon 5 at a level equal to or greater than our HNSCC index case. Further examination across tumor types indicated that lung and head and neck cancers expressed the greatest number of cases with elevated ratios of exon 4–exon 5 (adjusted p value < 2e−16 by Wilcoxon rank sum test for both tumor types) and prostate cancer and low grade glioma had the lowest level of enrichment (adjusted p value < 2e−16 by Wilcoxon rank sum test for both tumor types) (Fig. 2b). Additionally, expression of exon 4 versus 5 was significantly enriched when comparing tumor to normal-matched tissue across TCGA (adjusted p value < 2.2e−16 by Kruskal–Wallis test). To determine if enrichment of the fourth exon is unique to cancerous tissues, we analyzed normal tissue PD-L1 mRNA expression from GTEx [23]. The ratio of exon 4/5 of PD-L1 showed an enrichment in multiple tissue types with the greatest expression in normal lung tissue and lowest in the brain (adjusted q value < 2e−16 for both tissue types by Wilcoxon rank sum test) (Fig. S1b).
Fig. 2.
Expression of exon 4-enriched PD-L1 isoforms in various cancer types. a The expression of PD-L1 isoforms with high exon 4/5 expression ratios was measured in 33 different cancer types in the TCGA dataset and corresponding normal tissue from GTEx. The x-axis represents the log2 expression of total PD-L1. The y-axis represents the PD-L1 exon 4/5 expression ratio, which is the expression of the fourth exon of PD-L1 over the expression of the fifth exon of PD-L1. The HNSCC index case is labeled (TCGA_CV_5443). b Violin plot of expression of PD-L1 isoforms with high exon 4/5 expression ratios in cancer types ordered by decreasing median PD-L1 expression. Observed differences are significant as determined by an ANOVA comparison of the means (p value < 0.0001). Cancer type abbreviations are defined in the “Materials and methods”. c Cancer cell lines were analyzed for the expression of exon 4-enriched PD-L1 as described in a), from the CCLE dataset. The y-axis represents the log2 expression. The top two cell lines (RKO, blue; CAL62, green) with the highest PD-L1 exon 4/5 expression ratio as well as with the highest total PD-L1 expression were chosen for protein verification. RERF-LC-Ad1 (red, arrow) was chosen as a negative control, given it had high PD-L1 expression and a low PD-L1 exon 4/5 expression ratio. d, e Media from RKO, RERF-LC-Ad1, CAL62, and HEK293T (negative control) was analyzed for PD-L1 protein expression. d Media was used in an ELISA to measure PD-L1 present in the media. Data are from two separate experiments. The mean and SEM are plotted. e Protein was precipitated from media with TCA precipitation or isolated in cell lysates and analyzed with western blotting for PD-L1 protein expression. Vinculin was used as a loading control
To assess whether overexpression of the 5′ exons of PD-L1 might be associated with specific features of the tumor immune microenvironment, we first confirmed prior reports of a correlation among total PD-L1 expression with expression of T cell activation-associated genes coding for IFNg, CXCL10, PD-1, PRF1, ICOS, CD8, GZMA, and TIGIT [31–33] in 29 tumor types (combined p value < 0.1)(Fig. S4). Similar correlations were not observed when considering tumors with enrichment of exon 4 of PD-L1 as compared to exon 5. Additionally, we investigated the correlation between the ratio of exon 4 versus exon 5 of PD-L1 and patient survival in the TCGA datasets and did not observe an association between a high ratio and overall survival (Fig. S3).
Given that TCGA samples are comprised of tumor cells with stroma, we explored whether this fourth exon-enriched form of PD-L1 could be expressed by pure tumor cells. Utilizing CCLE data [34], we determined exon-level patterns of PD-L1 expression in 935 cancer cell lines in the CCLE (Fig. 2c). We did observe an enrichment of the expression of the fourth exon relative to the fifth exon (PD-L1 4/5 expression ratio) in cancer cell lines, with 4.1% (39/935) of cell lines demonstrating clearly measurable expression of total PD-L1 along with a PD-L1 exon 4/5 expression ratio of > 1. These data demonstrate that numerous different cancer types express PD-L1 isoforms enriched for the 4th exon relative to the 5th exon, and that cancer cells themselves can produce this isoform.
PD-L1 with exon 4 enrichment is secreted
To examine whether a secreted PD-L1 protein could be detected in cell lines with expression of PD-L1 isoforms with a high exon 4/5 ratio, we selected two cell lines (RKO, colon carcinoma, blue; CAL62, thyroid anaplastic carcinoma, green) that had the highest PD-L1 exon 4/5 expression ratios (> 1.1) in conjunction with high total PD-L1 expression (> 5.0 log2 FPKM), for further analysis (Fig. 2c). We also selected a cell line with high full-length PD-L1 expression (3.99 log2 FPKM) but a low exon 4/5 expression ratio (0.74, RERF-LC-Ad1, lung adenocarcinoma, red) as a negative control. To determine if cell lines with enrichment of expression of the 5′ exons of PD-L1 might produce a truncated and secreted form of PD-L1, we performed a PD-L1 ELISA on spun cell culture media from unmanipulated RKO, CAL62, RERF-LC-Ad1 and HEK293T (negative control) cells growing in log phase in standard cell culture conditions. PD-L1 was quantified in the media from cell lines using ELISA (Fig. 2d), where we detected PD-L1 in the media of RKO cells (4.97 ng/ml) and CAL62 cells, albeit at a lower level (0.17 ng/ml). We were unable to detect PD-L1 in the media of HEK293T cells or RERF-LC-Ad1 cells, as predicted from the computational analysis. As an orthogonal approach, we performed trichloroacetic acid (TCA) precipitation of media proteins from the same cell lines and also generated cell lysates for western blotting (Fig. 2e). We detected the presence of a shorter isoform (~ 36 kD) of PD-L1 in media precipitates from RKO cells but not from RERF-LC-Ad1 cells, nor was PD-L1 detected from HEK293T cells. PD-L1 was not detectable from CAL62 cells with this method, possibly explained by the very low level of PD-L1 detected in the media by ELISA. RERF-LC-Ad1 cells were found to have a high level of PD-L1 in the cell lysate, as expected. Flow cytometry was performed to measure the surface expression of PD-L1 (Fig. S5a) with surface PD-L1 detected in RKO, CAL62 and RERF-LC-Ad1 cells by flow. Immunofluorescent microscopy demonstrated principally cytoplasmic staining for RKO, CAL62 and RERF-LC-Ad1 cells (Fig. S5b). These data demonstrate a correlation between truncated PD-L1 isoform RNA and secreted PD-L1 protein confirmed by both ELISA and western blot.
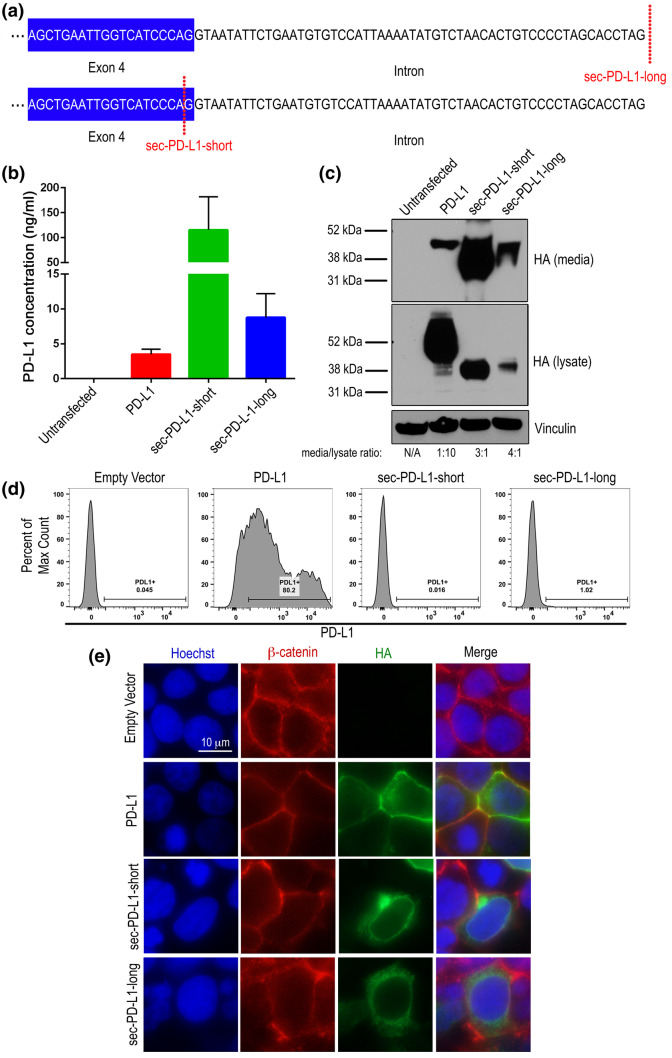
We next sought to further define the localization of truncated PD-L1 using the HEK293T cell line model which does not express detectable baseline PD-L1. We expressed an HA-tagged truncated PD-L1 that we identified in the HNSCC case (sec-PD-L1-long) (Fig. 1) in HEK293T cells, which included the readthrough into the intron of the 56 intronic nucleotides (Fig. 3a). We also overexpressed an HA-tagged short form of sec-PD-L1-long (sec-PD-L1-short) including sequences up until the last codon of exon 4, excluding the intronic readthrough, to determine the effect of this intronic region on the production and localization of the protein. We overexpressed wild-type HA-tagged PD-L1 in HEK293T cells as a control. Media was then subjected to ELISA (Fig. 3b) and TCA precipitation to determine the amount of PD-L1 in media as compared to intracellular PD-L1 (Fig. 3c). Data from the ELISA demonstrated that sec-PD-L1-long was present in the media at higher levels (8.7 ng/ml) as compared to wild-type PD-L1 (3.4 ng/ml), but, interestingly, not as high as the short form of sec-PD-L1-long (sec-PD-L1-short) (115 ng/ml). Total expression of sec-PD-L1-short was higher than sec-PD-L1-long, however (Fig. 3b). Densitometry analysis of the HA signal in the media versus the lysate from the western blot validated that sec-PD-L1-long was indeed secreted more than maintained in the cell (ratio = 4:1), with sec-PD-L1-short being secreted at a slightly lower ratio (3:1) (Fig. 3c). Ratios of HA in media/lysate were higher in both sec-PD-L1-long and sec-PD-L1-short compared to PD-L1 (ratio = 1:10) (Fig. 3c). The higher amount of sec-PD-L1-short detected by ELISA and western blot compared to sec-PD-L1-long is, therefore, likely due to a higher total expression rather than significant impacts of the C-terminal sequence on secretion.
Fig. 3.
Sec-PD-L1-long is secreted and not localized to the membrane. a Diagram depicting the differential sequences of sec-PD-L1-long and sec-PD-L1-short. b, c PD-L1-HA, sec-PD-L1-long-HA, sec-PD-L1-short-HA were expressed in HEK293T cells and PD-L1 protein in media was measured by b ELISA. Data are from three separate experiments. c PD-L1 was measured in media using TCA precipitation and in cell lysates by western blot by probing for HA tag. Vinculin was used as a loading control. Densitometry was performed where the HA signal was normalized to vinculin and the ratio of HA signal in the media versus lysate is depicted. d, e HEK293T cells were transfected with empty vector (pcDNA3.1(1)), PD-L1-HA, sec-PD-L1-long-HA and sec-PD-L1-short-HA, and 2 days later were d stained for PD-L1 (PE), and surface PD-L1 was analyzed with flow cytometry. Duplicates were performed, and 10,000 events were collected. The percent PD-L1 positive cells are shown. Data are representative of multiple experiments. e Cells were immunofluorescently stained for HA tag (green), β-catenin (red), and with Hoechst (blue), and imaged with a fluorescent microscope. 10 µm scale bar is shown
To measure membrane localization of the PD-L1 isoforms, sec-PD-L1-long-HA, sec-PD-L1-short-HA and PD-L1-HA expressing HEK293T cells were stained with an anti-PD-L1 antibody, and the amount of PD-L1 on the surface of cells was measured with flow cytometry (Fig. 3d). High levels of PD-L1 was measured on the surface of PD-L1 cells (80.2%), as expected, but little PD-L1 was measured on the surface of sec-PD-L1-long- (1.02%), and sec-PD-L1-short-expressing (0.016%) cells (Fig. 3d). Immunofluorescent microscopy of HEK293T cells overexpressing sec-PD-L1-long-HA, sec-PD-L1-short-HA and PD-L1-HA corroborated our observations made by flow cytometry, with high membrane staining being observed in cells overexpressing PD-L1, as assessed by co-staining for β-catenin as a membrane marker, but only cytoplasmic staining being observed in cells overexpressing sec-PD-L1-long and sec-PD-L1-short (Fig. 3e). Together, these data demonstrate that sec-PD-L1-long and the short form of sec-PD-L1-long are preferentially secreted by cells as opposed to being membrane-bound.
Sec-PD-L1-long retains the ability to bind PD-1 and inhibits IL-2 and IFNg secretion in primary T Cells
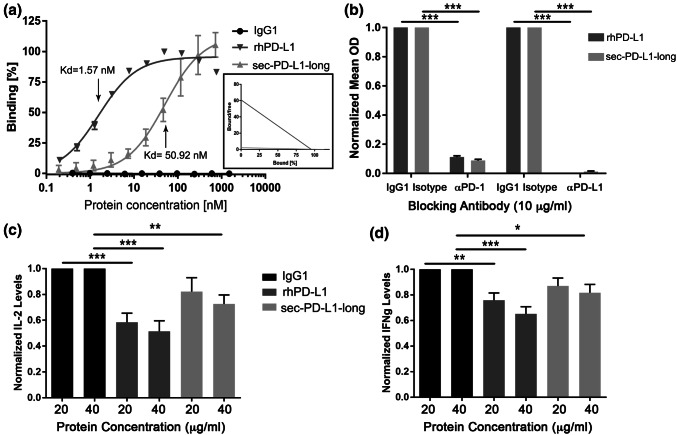
To test whether truncated PD-L1 retains the ability to bind the PD-1 receptor, a PD-1 functional ELISA was performed with an Fc-tagged sec-PD-L1-long (sec-PD-L1-long-Fc), Fc-tagged recombinant human PD-L1 (rhPD-L1-Fc) as a positive control, or IgG1-Fc as a negative control (Fig. 4a). Sec-PD-L1-long retained the ability to bind the PD-1 receptor, albeit at a lower capacity (Kd = 50.92 nM) than rhPD-L1 (Kd = 1.57 nM). Blocking antibodies against PD-1 and PD-L1 reduced the ability of sec-PD-L1-long to bind PD-1 by 11 fold and 80 fold, respectively (Fig. 4b).
Fig. 4.
Sec-PD-L1-long retains the capacity to bind to PD-1 and negatively affects T cells. a PD-1 binding capacity of IgG1-FC (negative control), rhPD-L1-FC (positive control) or sec-PD-L1-long-FC was measured using a functional ELISA, where mean OD was measured at varying concentrations of IgG1-FC, rhPD-L1-FC or sec-PD-L1-long-FC protein. Kd values are displayed. The corresponding Scatchard graph displaying the differences in slope (−1/Kd) of rhPD-L1-FC (medium gray) or sec-PD-L1-long-FC (light gray) is shown as an inset. Data are from two separate experiments. b The ability of PD-L1 and PD-1 neutralizing antibodies (10 µg/ml) to block PD-1 binding to rhPD-L1-FC and sec-PD-L1-long-FC was tested with a functional ELISA. An IgG1 isotype control antibody was used as a negative control. Normalized mean OD values compared to incubation with the appropriate IgG1 isotype control conditions are plotted. Data are from two separate experiments. A two-way ANOVA was used. The mean and SEM are plotted. c, d Primary human T cell blasts were incubated with IgG1-FC (negative control), rhPD-L1-FC (positive control) or sec-PD-L1-long-FC at either 20 µg/ml or 40 µg/ml in the presence of anti-CD3 for 24 h, and media was harvested to quantify c IL-2 and d IFNg with ELISA. Samples were run in duplicate. N = 5. Normalized protein levels compared to the appropriate IgG1-Fc control are plotted. A one-way ANOVA was used. The mean and SEM are plotted. *p < 0.05, **p < 0.01, ***p < 0.001
To determine whether sec-PD-L1-long retains the function of wild-type PD-L1 in terms of T cell suppression, the effect of sec-PD-L1-long on IL-2 and IFNg secretion was measured from cultured primary human T cell blasts. T cell blasts were verified to be mostly CD3+ using flow cytometry, with an average CD3 positivity of 83%, with on average 30% and 64% of CD3+ cells being CD4+ and CD8+, respectively (Fig. S6). T cell blasts were incubated with sec-PD-L1-long-Fc, rhPD-L1-Fc, or IgG1-Fc, in the presence of an anti-CD3 antibody. IL-2 and IFNg levels were measured in the media after 24 h using ELISA (Fig. 4c,d, Fig. S7). IL-2 levels were found to be decreased with sec-PD-L1-long incubation in a dose-dependent manner, with a 1.2 and 1.4 fold decrease in IL2 secretion with 20 µg/ml and 40 µg/ml sec-PD-L1-long incubation, respectively (Fig. 4c). IFNg levels were also found to be decreased with sec-PD-L1-long treatment in a dose-dependent manner, with a decrease of IFNg levels of 1.15 and 1.22 fold with 20 µg/ml and 40 µg/ml sec-PD-L1-long incubation, respectively (Fig. 4d). Interestingly, IL2 and IFNg levels were not inhibited as strongly as with the positive control rhPD-L1, likely due to lower levels of PD-1 binding. Together, these data demonstrate that sec-PD-L1-long acts as a negative regulator of T cells.
Discussion
In this study, we extended observations made in our previous computational analysis of genomic sites of HPV integration, where we identified HPV integration in the PD-L1 locus in a HNSCC tumor, suggesting expression of a truncated form of PD-L1 [13]. We performed computational de novo assembly of this truncated form that led to the identification of a novel expressed PD-L1 variant and identified additional truncated forms of PD-L1 in multiple human cancers and cancer cell lines. We verified the presence of secreted PD-L1 protein in two of the human cancer cell lines that had high expression levels of exon 4 versus exon 5 of PD-L1, including a colon cancer cell line (RKO) and a thyroid anaplastic carcinoma cell line (CAL62), and validated that the truncated PD-L1 isoform from the HNSCC is preferentially secreted and maintains the ability to bind PD-1 as well a negative regulatory effect on T cell function, as measured by inhibition of IL-2 and IFNg secretion.
Our work has provided evidence that the presence of secreted PD-L1 may be due to the expression of novel PD-L1 isoforms, and our data are corroborated by a study by Mahoney et al., co-published together with ours. In their work, titled “A secreted PD-L1 splice variant that covalently dimerizes and mediates immunosuppression”, the authors describe a similarly spliced variant of PD-L1 with a readthrough into the fourth intron, which negatively effects T cell function and is widely expressed in various human cancers as well as normal tissues. Others have also recently demonstrated PD-L1 splice variants being responsible for soluble PD-L1 protein in other cancers, such as melanoma [25]. The soluble PD-L1 proteins found in melanoma are due to splice variants found in either exon 4 (PD-L1-3 and PD-L1-12) or exon 5 (PD-L1-3 and PD-L1-9) which produce a premature stop codon before the transmembrane domain-encoding region (PD-L1-3 and PD-L1-12) or in a loss of the domain (PD-L1-9). However, we propose a truncated PD-L1 due to a readthrough of the intron after fourth exon with a premature stop codon and polyadenylation site.
Interestingly, we and others detected secreted PD-L1 when wild-type PD-L1 was overexpressed [35]. Frigola et al. [36] detected secreted PD-L1 from adherent cells of normal PBMCs, such as dendritic cells, when activated with PHA. Interestingly, Frigola et al. [36] did not detect the presence of secreted PD-L1 in activated T cells, suggesting that increased expression of PD-L1 is not sufficient in itself for the secretion of PD-L1, and other factors likely also help determine whether PD-L1 is membrane-bound or secreted. An additional explanation for secreted PD-L1 protein expression could be active cleavage of wild-type PD-L1 at the cell surface. Chen et al. [35] demonstrated that the levels of secreted PD-L1 can be reduced by treating a PD-L1 overexpressing mouse fibrous cell line with a broad-spectrum matrix metalloprotease inhibitor.
The presence of a secreted form of PD-L1 in the serum of cancer patients has been reported previously, such as in gastric cancer, renal cell carcinoma, non-small cell lung cancer and melanoma [16–18, 25]. Most studies demonstrating the detection of PD-L1 in human blood have not characterized the isoform type. Zhou et al. [25] show the presence of longer as well as truncated PD-L1 forms in human blood. Circulating PD-L1 in patients could be short secreted versions lacking the transmembrane domain similar to that described herein, cleaved extracellular PDL-L1 as described by others [35], or even full-length PD-L1 in exosomes [37]. Nevertheless, there have been mixed reports on the implications of circulating PD-L1 on the prognosis of patients. Zhang et al. [18] report a worse prognosis in pre-treated non-small cell lung cancer patients expressing high levels of circulating PD-L1. Another study demonstrated a better prognosis in gastric cancer patients with higher circulating PD-L1 before treatment [17]. Frigola et al. [16] demonstrated that renal cell carcinoma patients who experienced a doubling of circulating PD-L1 had a 41% increase in the risk of death. The direct link between the secretion of PD-L1 by tumors and the ability to detect levels of circulating PD-L1 has not yet been proven and the diversity and amino acid identity of secreted PD-L1 forms has not yet been comprehensively defined. Clearly, further work needs to be done to better understand how soluble PD-L1 affects conventional treatment response and prognosis.
Our experiments using human T cell blasts to measure the effect of sec-PD-L1-long on T-cell function did support that there is a preservation of a negative regulatory effect of sec-PD-L1-long on T cells. The decreased ability of sec-PD-L1-long to bind to PD-1 as compared to rhPD-L1, which includes only the extracellular region of PD-L1, likely is due to either the absence of amino acids directly proximal to the cell membrane present in rhPD-L1, or the presence of the amino acids encoded for by the intronic read-through of sec-PD-L1-long. This decrease in PD-1 binding capacity likely explains the differences in ability to suppress cytokine secretion by sec-PD-L1-long compared to rhPD-L1.
Our TCGA analysis of wild-type PD-L1 expression correlated with the expression of several T-cell genes in all cancers when combined, highlighting a general adaptive resistance to immune cell response (Fig. S4). However, a similar analysis did not demonstrate a positive correlation between the expression of a truncated form of PD-L1 with T cell genes. Expression of the full-length PD-L1 was higher than that of truncated PD-L1, which could account for this difference in association with T cell gene expression. Further studies are needed to determine the comprehensive effects of a secreted form of PD-L1 on immune response in vivo.
Another unanswered question is whether a high level of secreted PD-L1 detected in the serum of cancer patients predicts a worse response to anti-PD-L1/PD-1 antibody treatment, as it can be imagined that circulating PD-L1, or perhaps higher levels of secreted PD-L1 in the tumor microenvironment, may act as a scavenger of anti-PD-L1 therapeutic antibody or that secreted PD-L1 may outcompete anti-PD-1 antibody, rendering anti-PD-L1/PD-1 treatments less effective. One published paper by Zhou et al. [25] has suggested that in malignant melanoma, patients with the highest pretreatment circulating PD-L1 levels tended to have more rapidly progressing disease after anti-PD-1 treatment, but the sample size was small. More work is needed to understand the implications of circulating PD-L1 levels on immunotherapy efficacy.
In conclusion, our data demonstrate the existence of truncated, secreted PD-L1 isoforms in cancer that retain negative immunomodulatory functions and warrant further investigation into the effect of these PD-L1 isoforms on prognosis and treatment response.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors would like to acknowledge the Dana-Farber Cancer Institute Flow Cytometry Core (Suzan Lazo-Kallanian, John Daley), and UT Southwestern Children’s Research Institute Flow Cytometry Core for help with flow cytometry. The authors would also like to thank Gordon Freeman for PD-L1 antibodies.
Abbreviations
- ATCC
American type culture collection
- CCLE
Cancer cell line encyclopedia
- CST
Cell signaling technology
- EBV
Epstein–Barr virus
- ELISA
Enzyme linked immunosorbent assay
- FPKM
Fragments per kilobase of transcript per million mapped reads
- GTeX
Genotype-Tissue Expression
- HA
Hemagglutinin
- HNSCC
Head and neck squamous cell carcinoma
- HPV
Human papillomavirus
- HRP
Horseradish Peroxidase
- NCI
National Cancer Institute
- PD-L1
Programmed death-ligand-1
- PHA
phytohemagglutinin
- PSG
Penicillin/streptomycin/glutamine
- RIPA
Radioimmunoprecipitation assay
- RT
Room temperature
- TCA
Trichloroacetic acid
- TCGA
The Cancer Genome Atlas
- TMB
3,3′,5,5′-Tetramethylbenzidine
Author contributions
NBH and VSM contributed equally to experimental design, data collection and analysis, and manuscript writing. YH and EMB helped with data collection and analysis. SSF and CSP helped with bioinformatics. SK, SM, NS, K-KW and GD aided with experimental design and provided reagents. PSH and EAA helped with experimental design, data collection, providing reagents, and manuscript writing and editing.
Funding
This work was supported by the NCI R01 CA205150, CA196932, and K08 CA163677 grants, as well as the Starr Consortium for Cancer Research and Stand up to Cancer awards to Peter Hammerman. This work was also supported by the Young Investigator Award from the International Association for the Study of Lung Cancer, Career Enhancement Award (5P50CA070907) through the National Institutes of Health, and Cancer Prevention and Research Institute of Texas Scholar Award (RR160080) to Esra Akbay. Venkat Malladi was supported by the Cancer Prevention and Research Institute of Texas award (RP150596).
Compliance with ethical standards
Conflict of interest
The authors have no relevant conflicts to disclose.
Informed consent
Informed consent was obtained from blood donors from the Brigham and Women’s Blood Bank under the approved IRB protocol 02-180.
Cell line authentication
HEK293T cells were purchased from ATCC, and RKO, CAL62, and RERF-LC-Ad1 cells were obtained from the CCLE [34] through the Broad Institute of Massachusetts Institute of Technology and Harvard. All cells were used within 6 months of initial passage and not further authenticated.
Footnotes
Poster/abstract publication in Cancer Immunology Research, 2017; 5 (3 Suppl); Abstract nr A56; Proceedings of the American Association for Cancer Research Special Conference on Tumor Immunology and Immunotherapy; 2016 Oct 21; Boston, MA, USA.
This paper is published together with the following paper 10.1007/s00262-018-2282-1.
Nadia B. Hassounah and Venkat S. Malladi contributed equally to this study and are joint first authors of this paper.
References
- 1.Mahoney KM, Rennert PD, Freeman GJ. Combination cancer immunotherapy and new immunomodulatory targets. Nat Rev Drug Discov. 2015;14(8):561–584. doi: 10.1038/nrd4591. [DOI] [PubMed] [Google Scholar]
- 2.Postow MA, Callahan MK, Wolchok JD. Immune checkpoint blockade in cancer therapy. J Clin Oncol. 2015;33(17):1974–1982. doi: 10.1200/JCO.2014.59.4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen Z, et al. Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer. 2014;14(8):535–546. doi: 10.1038/nrc3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12(4):252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freeman GJ, et al. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J Exp Med. 2000;192(7):1027–1034. doi: 10.1084/jem.192.7.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keir ME, et al. PD-1 and its ligands in tolerance and immunity. Annu Rev Immunol. 2008;26:677–704. doi: 10.1146/annurev.immunol.26.021607.090331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gillison ML, et al. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J Natl Cancer Inst. 2000;92(9):709–720. doi: 10.1093/jnci/92.9.709. [DOI] [PubMed] [Google Scholar]
- 8.Yim EK, Park JS. The role of HPV E6 and E7 oncoproteins in HPV-associated cervical carcinogenesis. Cancer Res Treat. 2005;37(6):319–324. doi: 10.4143/crt.2005.37.6.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gulley ML. Genomic assays for Epstein–Barr virus-positive gastric adenocarcinoma. Exp Mol Med. 2015;47:e134. doi: 10.1038/emm.2014.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Network CGAR. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513(7517):202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Green MR, et al. Constitutive AP-1 activity and EBV infection induce PD-L1 in Hodgkin lymphomas and posttransplant lymphoproliferative disorders: implications for targeted therapy. Clin Cancer Res. 2012;18(6):1611–1618. doi: 10.1158/1078-0432.CCR-11-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lyford-Pike S, et al. Evidence for a role of the PD-1:PD-L1 pathway in immune resistance of HPV-associated head and neck squamous cell carcinoma. Cancer Res. 2013;73(6):1733–1741. doi: 10.1158/0008-5472.CAN-12-2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parfenov M, et al. Characterization of HPV and host genome interactions in primary head and neck cancers. Proc Natl Acad Sci USA. 2014;111(43):15544–15549. doi: 10.1073/pnas.1416074111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ojesina AI, et al. Landscape of genomic alterations in cervical carcinomas. Nature. 2014;506(7488):371–375. doi: 10.1038/nature12881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kataoka K, Shiraishi Y, Takeda Y, Sakata S, Matsumoto M, Nagano S, Maeda T, Nagata Y. Aberrant PD-L1 expression through 3′-UTR disruption in multiple cancers. Nature. 2016;537:402. doi: 10.1038/nature18294. [DOI] [PubMed] [Google Scholar]
- 16.Frigola X, et al. Identification of a soluble form of B7-H1 that retains immunosuppressive activity and is associated with aggressive renal cell carcinoma. Clin Cancer Res. 2011;17(7):1915–1923. doi: 10.1158/1078-0432.CCR-10-0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng Z, et al. Level of circulating PD-L1 expression in patients with advanced gastric cancer and its clinical implications. Chin J Cancer Res. 2014;26(1):104–111. doi: 10.3978/j.issn.1000-9604.2014.02.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang J, et al. Circulating PD-L1 in NSCLC patients and the correlation between the level of PD-L1 expression and the clinical characteristics. Thorac Cancer. 2015;6(4):534–538. doi: 10.1111/1759-7714.12247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grossman RL, et al. Toward a shared vision for cancer genomic data. N Engl J Med. 2016;375(12):1109–1112. doi: 10.1056/NEJMp1607591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pertea M, et al. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 2016;11(9):1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pertea M, et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33(3):290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weinstein JN, et al. The cancer genome atlas pan-cancer analysis project. Nat Genet. 2013;45(10):1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Consortium G. The genotype-tissue expression (GTEx) project. Nat Genet. 2013;45(6):580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Collado-Torres L, et al. Reproducible RNA-seq analysis using recount2. Nat Biotechnol. 2017;35(4):319–321. doi: 10.1038/nbt.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou J, et al. Soluble PD-L1 as a biomarker in malignant melanoma treated with checkpoint blockade. Cancer Immunol Res. 2017;5(6):480–492. doi: 10.1158/2326-6066.CIR-16-0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keir ME, Francisco LM, Sharpe AH. PD-1 and its ligands in T-cell immunity. Curr Opin Immunol. 2007;19(3):309–314. doi: 10.1016/j.coi.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 27.Butte MJ, et al. Interaction of human PD-L1 and B7-1. Mol Immunol. 2008;45(13):3567–3572. doi: 10.1016/j.molimm.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haile ST, et al. Soluble CD80 restores T cell activation and overcomes tumor cell programmed death ligand 1-mediated immune suppression. J Immunol. 2013;191(5):2829–2836. doi: 10.4049/jimmunol.1202777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng S, et al. PD-L1 gene polymorphism and high level of plasma soluble PD-L1 protein may be associated with non-small cell lung cancer. Int J Biol Markers. 2015;30(4):e364–e368. doi: 10.5301/jbm.5000170. [DOI] [PubMed] [Google Scholar]
- 31.Hendrickx W, et al. Identification of genetic determinants of breast cancer immune phenotypes by integrative genome-scale analysis. Oncoimmunology. 2017;6(2):e1253654. doi: 10.1080/2162402X.2016.1253654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Balar AV, et al. Atezolizumab as first-line treatment in cisplatin-ineligible patients with locally advanced and metastatic urothelial carcinoma: a single-arm, multicentre, phase 2 trial. Lancet. 2017;389(10064):67–76. doi: 10.1016/S0140-6736(16)32455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosenberg JE, et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet. 2016;387(10031):1909–1920. doi: 10.1016/S0140-6736(16)00561-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barretina J, et al. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Y, et al. Development of a sandwich ELISA for evaluating soluble PD-L1 (CD274) in human sera of different ages as well as supernatants of PD-L1 + cell lines. Cytokine. 2011;56(2):231–238. doi: 10.1016/j.cyto.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 36.Frigola X, et al. Soluble B7-H1: differences in production between dendritic cells and T cells. Immunol Lett. 2012;142(1–2):78–82. doi: 10.1016/j.imlet.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Theodoraki MN, et al. Clinical significance of PD-L1. Clin Cancer Res. 2018;24(4):896–905. doi: 10.1158/1078-0432.CCR-17-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.