Key Points
Association of del17p CCF threshold and poor prognosis in NDMM is established via fluorescence in situ hybridization and sequencing methods.
TP53 mutations are enriched in a high-risk patient segment characterized by high del17p CCF.
Abstract
Deletions of chromosome 17p (del17p) that span the TP53 gene are associated with poor outcome in multiple myeloma (MM), but the prognostic value of del17p cancer clonal fraction (CCF) remains unclear. We applied uniform cytogenetic assessments in a large cohort of newly diagnosed MM (NDMM) patients carrying varying levels of del17p. Incremental CCF change was associated with shorter survival, and a robust CCF threshold of 0.55 was established in discovery and replication data sets. After stratification on the 0.55-CCF threshold, high-risk patients had statistically significantly poorer outcomes compared with low-risk patients (median progression-free survival [PFS] and overall survival [OS], 14 and 32 vs 23.1 and 76.2 months, respectively). Analyses of a third data set comprising whole-exome sequencing data from NDMM patients identified presence of TP53 deletions/mutations as a necessary requirement for high-risk stratification in addition to exceeding the del17p CCF threshold. Meta-analysis conducted across 3 data sets confirmed the robustness of the CCF threshold for PFS and OS. Our analyses demonstrate the feasibility of fluorescence in situ hybridization– and sequencing-based methods to identify TP53 deletions, estimate CCF, and establish that both CCF threshold of 0.55 and presence of TP53 deletion are necessary to identify del17p-carrying NDMM patients with poor prognosis.
Visual Abstract
Introduction
The role of tumor protein 53 (TP53) as a tumor suppressor in multiple myeloma (MM) is well known,1,2 and cytogenetic analysis of chromosome 17p deletion (del17p), which spans the TP53 gene, by fluorescence in situ hybridization (FISH) is part of the recommended risk assessment in newly diagnosed MM (NDMM).3-5
Currently, the minimum percentage of del17p cancer clonal fraction (CCF) indicative of poor prognosis is not known, with most studies reporting arbitrary threshold levels ranging from a single FISH+ cell to exceeding 60% FISH+ MM cells.6-9 The lack of uniform analytical methods for cytogenetic assessment of del17p by FISH adds to discordant claims about CCF threshold for poor prognosis.4,6,8,9 Genetic defects in 17p are complex and include either a deletion only or a mutation only in 1 allele of the TP53 gene or both (biallelic inactivation). Biallelic inactivation in the TP53 gene (ie, double hit) is associated with very poor prognosis.10-12
Here, large NDMM patient discovery and replication data sets with uniform del17p assessment by interphase FISH were used to establish the cytogenetic CCF threshold for poor prognosis patients. Computational analyses were applied to genomic Myeloma Genome Project (MGP) data10 to estimate CCF in tumors characterized by varying extent of 17p deletions, including those in the TP53 region.
Study design
The MGP data set comprised 1766 NDMM patients with available whole-exome sequencing, whole-genome sequencing, RNA sequencing, and expression array data, derived from the Myeloma XI trial, Dana-Farber Cancer Institute/Intergroupe Francophone du Myélome (IFM), University of Arkansas for Medical Sciences Myeloma Institute, and Multiple Myeloma Research Foundation (IA1-IA9). Data from 1273 patients were used for analysis.11
Bone marrow plasma cells were obtained at diagnosis using CD138 selection. FISH analysis was performed on samples with ≥70% plasma cells using TP53 gene probes in discovery cohort and independent replication cohort. Copy number calls alongside CCF calls were calculated using SClust.13 Other experimental details are described in supplemental Materials, available on the Blood Web site.
Results and discussion
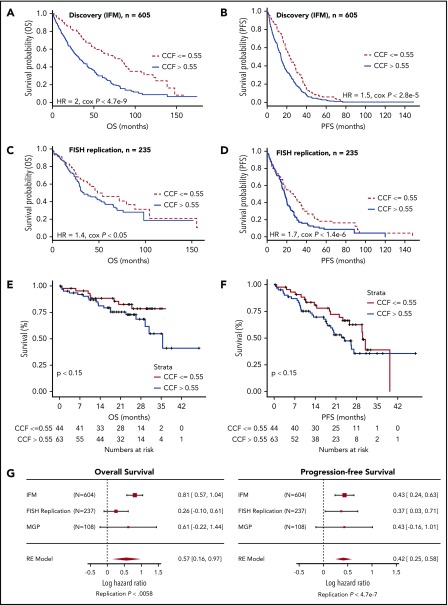
Discovery and FISH replication cohorts (IFM) included NDMM patients enrolled in clinical studies from 1999 to 2015 (supplemental Table 1). To understand the relationship between incremental CCF change and outcome, patients were segmented into 10% increments in CCF range of 0.3 to 0.8. Patients with CCF >0.8 or <0.3 were excluded from this analysis because of inadequate numbers. The optimal CCF value range was 51% to 69%, with 2 optimal cuts identified at 55% and 64% thresholds (supplemental Figure 1). The lowest CCF threshold was selected for further analysis. When patients in the discovery data set were stratified by the CCF threshold of 0.55, patients with lower CCF had significantly longer overall survival (OS; median, 36 vs 84.1 months) and progression-free survival (PFS; median, 14.3 vs 23.9 months) compared with those with higher CCF (Figure 1A-B; supplemental Table 2).
Figure 1.
Identification of 0.55 CCF threshold for del17p/TP53 deletion predictive of survival. Kaplan-Meier plot showing OS (A,C) and PFS (B,D) for patients from discovery cohort (A-B) and replication (RE) cohort (C-D) stratified by high or low CCF value. (E-F) Kaplan-Meier plots were generated for OS (A) and PFS (B) characteristics of patients in MGP del17p cohort stratified by 0.55 CCF value (hazard ratio [HR], 1.8; Cox P < .15 and HR, 1.5; Cox P < .15 for OS and PFS, respectively). Baseline OS and PFS data were not available for 1 patient in the CCF >0.55 group. (G) Meta-analysis shows replication of 55% CCF threshold to be an indicator of poor prognosis based on OS and PFS end points. Numbers on the right of each graph indicate HR and corresponding 95% confidence interval.
CCF threshold robustness was confirmed in a replication cohort with similar demographics (supplemental Table 1). After stratification on CCF of 0.55 (supplemental Table 2), the higher CCF subgroup had inferior OS and PFS (median PFS, 17 months; median OS, 32 months) compared with those with CCF ≤0.55 (median PFS, 29 months; median OS, 54 months; Figure 1C-D). Additional analyses were conducted using a CCF threshold of 0.64 (supplemental Figures 2-5), and results were comparable to those of the 0.55 CCF threshold.
To investigate the effect of high-throughput sequencing–derived del17p CCF on prognosis, we applied the threshold established from cytogenetic data to the available genomic data from MGP. Kaplan-Meier analyses of OS and PFS in the 2 groups are shown in Figure 1E-F. Patients with CCF >0.55 had lower median PFS and OS, as compared with the lower CCF group (supplemental Table 3). These results demonstrate that analysis of del17p by molecular genomic approaches separates poor outcome patients.
We next applied meta-analysis of IFM, replication, and MGP data using the same selection criteria across cohorts (Figure 1G). Summary statistics were combined using a meta-analysis framework that used a random-effect model to integrate the variation across the different populations and derive a single point estimate for the outcome difference in CCF-stratified patients across all 3 cohorts. On the basis of this framework, we derived a replication P value denoting the overall agreement in the summary statistics across the 3 studies.
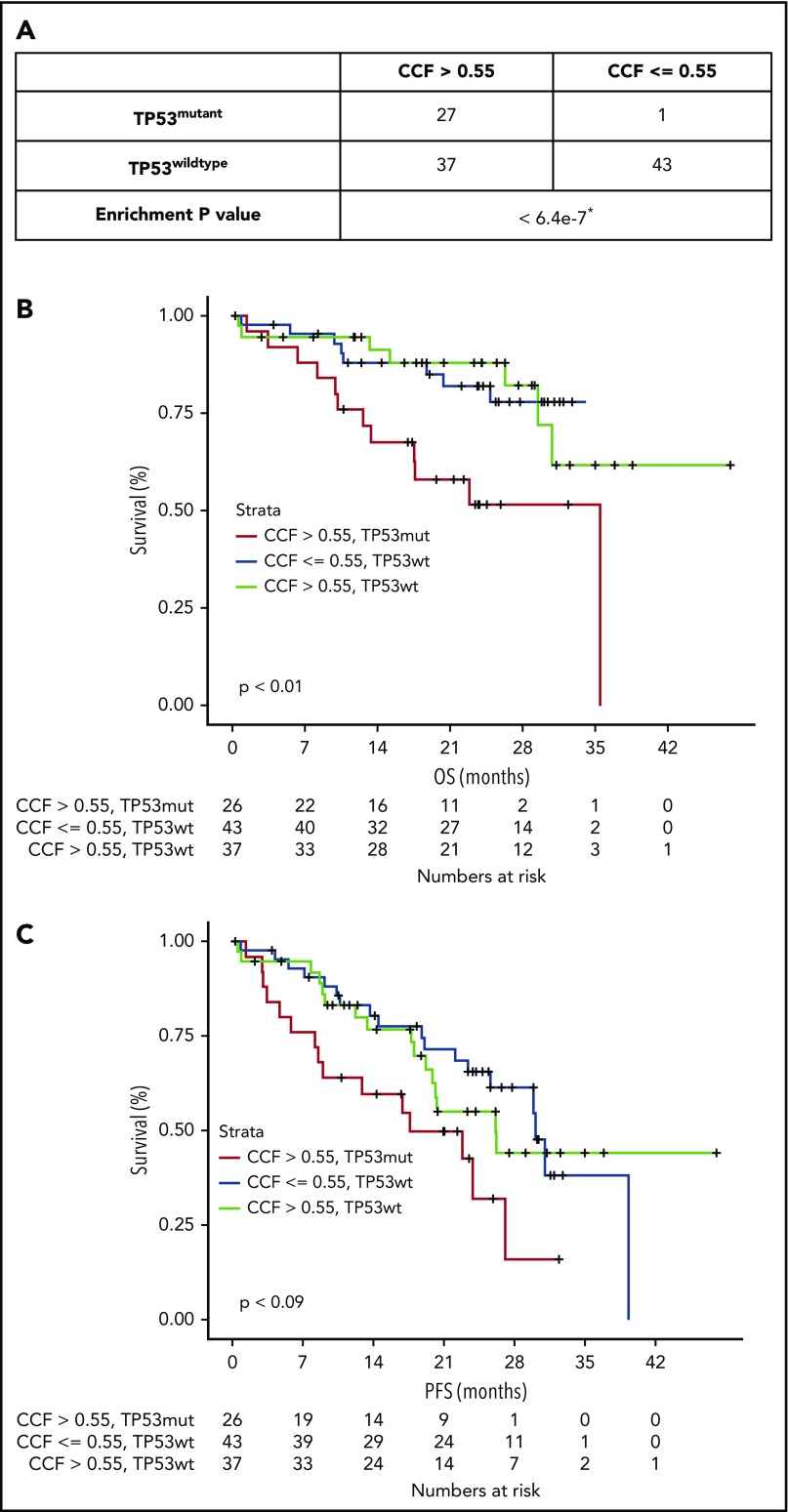
The proposed CCF threshold was further studied in MGP double-hit patients and patients with deletions in 1 allele only. In total, 108 patients had deletions spanning TP53 in the 17p locus, with 28 patients harboring a mutation in the TP53 gene. When separated by CCF level, only 1 patient with TP53 mutation had CCF ≤0.55 (Figure 2A). In contrast, the number of patients with wild-type TP53 was well distributed between low and high CCF groups. We next compared double-hit patients with either patients with only del17p in the CCF >0.55 group or with patients with only del17p in the CCF ≤0.55 group. Double-hit patients had significantly shorter PFS compared with those with deletion only, regardless of CCF category (Figure 2B-C). No significant differences in PFS or OS were observed in the MGP data set between del17p only patients with higher CCF and those with lower CCF. However, this could have been due to the small numbers of patients and events (supplemental Figure 6).
Figure 2.
Nonsynonymous mutations in TP53 are significantly enriched in population of patients with confirmed TP53 deletion. (A) Presence of TP53 mutations was examined in delTP53 patients from MGP data set (n = 108) stratified by CCF value >0.55 or <0.55. Kaplan-Meier plots were generated for OS (B) and PFS (C) for patients with TP53 mutation (mut) with CCF >0.55 (red line), patients with wild-type (wt) TP53 with CCF ≤0.55 (blue line), and patients with TP53wt with CCF >0.55 (green line). *One-sided Fisher’s exact test.
Although the prognostic role of del17p is widely recognized in MM,4,5 there has not been a systematic examination of the role of the extent of deletions spanning TP53 in terms of CCF level. Application of arbitrary CCF cutoffs2 and double-hit phenomena10,12,14 add further complexity to the prognostic impact of subclonal deletions in TP53. In addition, interaction with tumor (eg, 1q amplification) and nontumor (patient and immune) parameters may further influence the prognostic impact of TP53 deletion. Here, using a large cohort of uniformly analyzed NDMM patients, we demonstrate that patients with CCF >0.55 have significantly poorer outcomes as compared with other patients (Figure 1). Additional analyses using MGP non-del17p patients showed clinical outcomes comparable to those of del17 patients with CCF ≤0.55 (supplemental Figure 7). In addition, using high-throughput sequencing–based genomic data, we demonstrate that subclonal TP53 deletions recapitulated the prognostic significance at the same cut point, suggesting consistency across different platforms. The CCF cutoff of 0.55 agrees with a recent study where a multiplex ligation-dependent probe amplification–based assay was used to assess the prognostic relevance of del17p CCF.15 Several key aspects differentiate our study from those previously published. We applied an integrative clinical and genomic approach using an extensive data set of NDMM patients for whom FISH and/or genomic data were available. Our data were derived from 3 large data repositories, lowering the probability of a study-related bias. Our analyses are further strengthened by use of discovery and replication data sets and application of a uniform genomic analysis strategy and confirmatory meta-analysis approach.
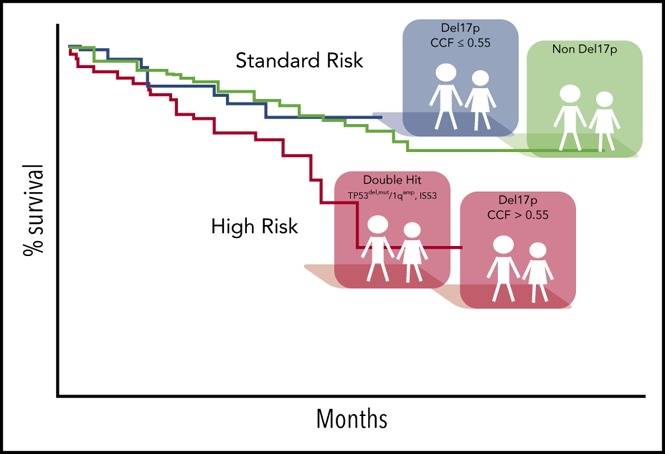
In summary, CCF may be an important parameter in prognostic evaluation of del17p in NDMM patients. Here, we establish a robust threshold of 0.55 for the best separation of patients with poor prognosis. Exceeding this threshold or cooccurrence of TP53 mutation indicates poor prognosis in double-hit patients. Our work suggests the feasibility of applying genomic methods for subclonal analysis of del17p for improved risk stratification of patients, along with or eventually as a substitute for FISH-based analyses. We suggest assessment of a del17p CCF threshold of 0.55 be included in risk assessment for patients enrolled in clinical trials and used for diagnostic testing in NDMM.
Supplementary Material
The online version of this article contains a data supplement.
Acknowledgments
The authors acknowledge the laboratory members of the Avet-Loiseau and Thakurta laboratories and other colleagues for their comments as well as Konstantinos Mavrommatis and Piotr Kozbial for running SClust.
This work was supported by the Myeloma Disease Team in Celgene (A.T.), by the Association Recherche Cancer (IUC-Oncopole Laboratory), and by Celgene, which provided data processing and storage.
Footnotes
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: A.T. and H.A.-L. conceived and designed the project; A.T. acquired funding; M.O., P.B., F.T., Z. Yu, and Z. Yang provided methodology development and computational/statistical analyses; F.T. and E.F. provided project administration; M.O., E.F., and F.T. provided oversight and management of resources (data generation, collection, transfer, infrastructure, data processing); all authors performed analyses and interpretation; M.O., P.B., and F.T. performed data visualization; and A.T., M.O., P.B., F.T., J.C., N.V.S., E.F., and H.A.-L. wrote the manuscript.
Conflict-of-interest disclosure: F.T., E.F., N.V.S., M.O., Z. Yu, Z. Yang, and A.T. are employed by and have equity ownership in Celgene. P.S. received honoraria and research support from Celgene. The remaining authors declare no competing financial interests.
Correspondence: Anjan Thakurta, Celgene Corporation, 566 Morris Ave, Summit, NJ 07901; e-mail: athakurta@celgene.com; and Herve Avet-Loiseau, Unite de Genomique du Myelome, CHU Rangueil, Toulouse, France; e-mail: avet-loiseau.h@chu-toulouse.fr.
REFERENCES
- 1.Tessoulin B, Eveillard M, Lok A, et al. . p53 dysregulation in B-cell malignancies: more than a single gene in the pathway to hell. Blood Rev. 2017;31(4):251-259. [DOI] [PubMed] [Google Scholar]
- 2.Kastenhuber ER, Lowe SW. Putting p53 in context. Cell. 2017;170(6):1062-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kumar SK, Callander NS, Alsina M, et al. . Multiple myeloma, version 3.2017, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2017;15(2):230-269. [DOI] [PubMed] [Google Scholar]
- 4.Chng WJ, Dispenzieri A, Chim CS, et al. ; International Myeloma Working Group. IMWG consensus on risk stratification in multiple myeloma. Leukemia. 2014;28(2):269-277. [DOI] [PubMed] [Google Scholar]
- 5.Palumbo A, Avet-Loiseau H, Oliva S, et al. . Revised International Staging System for multiple myeloma: a report from International Myeloma Working Group. J Clin Oncol. 2015;33(26):2863-2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Avet-Loiseau H, Fonseca R, Siegel D, et al. . Carfilzomib significantly improves the progression-free survival of high-risk patients in multiple myeloma. Blood. 2016;128(9):1174-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen YC, Saranga A, Gatt ME, et al. . Treatment patterns and clinical outcomes in high-risk newly diagnosed multiple myeloma patients carrying the 17p deletion: an observational multi-center retrospective study. Am J Hematol. 2018;93(6):810-815. [DOI] [PubMed] [Google Scholar]
- 8.Lonial S, Dimopoulos M, Palumbo A, et al. ; ELOQUENT-2 Investigators. Elotuzumab therapy for relapsed or refractory multiple myeloma. N Engl J Med. 2015;373(7):621-631. [DOI] [PubMed] [Google Scholar]
- 9.Thanendrarajan S, Tian E, Qu P, et al. . The level of deletion 17p and bi-allelic inactivation of TP53 has a significant impact on clinical outcome in multiple myeloma. Haematologica. 2017;102(9):e364-e367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walker BA, Mavrommatis K, Wardell CP, et al. . A high-risk, double-hit, group of newly diagnosed myeloma identified by genomic analysis. Leukemia. 2018;33(1):159-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walker BA, Mavrommatis K, Wardell CP, et al. . Identification of novel mutational drivers reveals oncogene dependencies in multiple myeloma. Blood. 2018;132(6):587-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weinhold N, Ashby C, Rasche L, et al. . Clonal selection and double-hit events involving tumor suppressor genes underlie relapse in myeloma. Blood. 2016;128(13):1735-1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cun Y, Yang TP, Achter V, Lang U, Peifer M. Copy-number analysis and inference of subclonal populations in cancer genomes using Sclust. Nat Protoc. 2018;13(6):1488-1501. [DOI] [PubMed] [Google Scholar]
- 14.Shah V, Sherborne AL, Walker BA, et al. . Prediction of outcome in newly diagnosed myeloma: a meta-analysis of the molecular profiles of 1905 trial patients. Leukemia. 2018;32(1):102-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shah V, Johnson DC, Sherborne AL, et al. . Subclonal TP53 copy number is associated with prognosis in multiple myeloma. Blood. 2018;132(23):2465-2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.