Abstract
Synaptotagmin-1 (Syt-1) and synaptotagmin-7 (Syt-7) contain analogous tandem C2 domains, C2A and C2B, which together sense Ca2+ to bind membranes and promote the stabilization of exocytotic fusion pores. Syt-1 triggers fast release of neurotransmitters, whereas Syt-7 functions in processes that involve lower Ca2+ concentrations such as hormone secretion. Syt-1 C2 domains are reported to bind membranes cooperatively, based on the observation that they penetrate farther into membranes as the C2AB tandem than as individual C2 domains. In contrast, we previously suggested that the two C2 domains of Syt-7 bind membranes independently, based in part on measurements of their liposome dissociation kinetics. Here, we investigated C2A-C2B interdomain cooperativity with Syt-1 and Syt-7 using directly comparable measurements. Equilibrium Ca2+ titrations demonstrate that the Syt-7 C2AB tandem binds liposomes lacking phosphatidylinositol-4,5-bisphosphate (PIP2) with greater Ca2+ sensitivity than either of its individual domains and binds to membranes containing PIP2 even in the absence of Ca2+. Stopped-flow kinetic measurements show differences in cooperativity between Syt-1 and Syt-7: Syt-1 C2AB dissociates from PIP2-free liposomes much more slowly than either of its individual C2 domains, indicating cooperativity, whereas the major population of Syt-7 C2AB has a dissociation rate comparable to its C2A domain, suggesting a lack of cooperativity. A minor subpopulation of Syt-7 C2AB dissociates at a slower rate, which could be due to a small cooperative component and/or liposome clustering. Measurements using an environment-sensitive fluorescent probe indicate that the Syt-7 C2B domain inserts deeply into membranes as part of the C2AB tandem, similar to the coinsertion previously reported for Syt-1. Overall, coinsertion of C2A and C2B domains is coupled to cooperative energetic effects in Syt-1 to a much greater extent than in Syt-7. The difference can be understood in terms of the relative contributions of C2A and C2B domains toward membrane binding in the two proteins.
Introduction
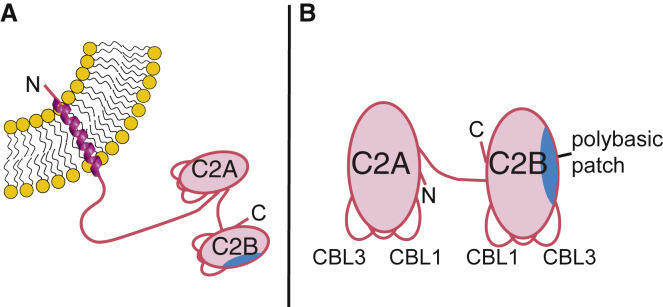
Synaptotagmins are a family of proteins that trigger and regulate fusion of secretory vesicles with the plasma membrane during exocytosis (1, 2, 3). Structurally, synaptotagmins contain an N-terminal transmembrane region, a cytoplasmic juxtamembrane linker region, and tandem C-terminal C2 domains, termed C2A and C2B (Fig. 1 A). In eight of the 17 mammalian isoforms, the C2 domains together function as a Ca2+ sensor to trigger SNARE-mediated membrane fusion (3, 4, 5). Although the synaptotagmins are structurally homologous, their Ca2+ and membrane-binding sensitivities vary (6).
Figure 1.
Schematic structure of synaptotagmin. (A) Full-length synaptotagmins contain an N-terminal transmembrane helix anchored to either the secretory vesicle or plasma membrane (65, 66) and two C-terminal C2 domains. (B) The Syt-1 and Syt-7 C2AB protein fragments used in this study consist of the C-terminal C2 domains, which coordinate Ca2+ ions and insert into membranes primarily via Ca2+-binding loops (CBL1 and CBL3). The C2B domains additionally contain a polybasic patch (blue shading) that can interact with anionic lipids in a Ca2+-independent manner. To see this figure in color, go online.
This study focuses on C2 domains from synaptotagmin-1 (Syt-1) to synaptotagmin-7 (Syt-7), two isoforms that serve as models for high-speed and high-sensitivity Ca2+-dependent vesicle fusion, respectively (7). Syt-1 serves as a primary Ca2+ sensor in the fast, synchronous release of neurotransmitters, whereas Syt-7 is involved in asynchronous neurotransmitter release, vesicle replenishment, synaptic facilitation, and endocrine and neuroendocrine secretion processes that occur at relatively low Ca2+ concentrations (8, 9, 10, 11, 12, 13). Correspondingly, C2 domains of Syt-7 were found to be 400-fold more sensitive to Ca2+ than Syt-1 and have the slowest membrane-dissociation kinetics among the synaptotagmins (14, 15). Both of the individual C2 domains from Syt-7 bind more tightly to membranes in the presence of Ca2+ than their respective counterparts in Syt-1 (16, 17, 18, 19).
Ca2+-dependent membrane binding of synaptotagmin C2 domains is mediated by conserved aspartate residues in each domain’s Ca2+-binding loops (CBLs), of which CBL1 and CBL3 insert into membranes upon binding Ca2+ (Fig. 1 B) (20, 21, 22, 23, 24, 25). The structural origins of membrane-binding differences between Syt-1 and Syt-7 are not yet completely clear, although some features that contribute to the strong membrane binding of Syt-7 have been identified (17, 18, 26). In addition to their Ca2+-dependent membrane binding, the C2B domains of both isoforms contain a polybasic lysine-rich patch centered on the β-4 strand, which has been shown to bind preferentially to polyanionic phospholipids such as phosphatidylinositol-(4,5)-bisphosphate (PIP2) (18, 27, 28, 29, 30). PIP2 binding includes a mechanism that is independent of Ca2+; however, its affinity is modest such that only a fraction of the protein population is typically observed to be membrane bound in the absence of Ca2+ (31). The stronger affinity of Syt-1 C2AB for membranes containing PIP2 manifests primarily through slower dissociation kinetics (koff), whereas association rates (kon) are essentially independent of lipid composition (30).
Mutational studies of Syt-1 and Syt-7 have revealed an interesting difference between the two proteins in the relative functional importance of the two C2 domains. The ability of Syt-1 to trigger fusion appears to depend more on Ca2+ binding by its C2B domain, whereas Syt-7 appears to be dominated by its C2A domain (9, 11, 32, 33, 34). In particular, neutralization of Ca2+-binding aspartate residues in the C2B domain of Syt-1 attenuates synchronous neurotransmitter release much more severely than corresponding mutations in the C2A domain (32, 33). Fusion events mediated by Syt-7 display the opposite pattern: they are more sensitive to mutations in the Ca2+-coordinating residues of the Syt-7 C2A domain (9, 11, 34). In vitro membrane-binding measurements also indicate that C2B dominates membrane binding in Syt-1, whereas C2A dominates in Syt-7 (18). In principle, differences between how these two sets of C2 domains work together could reflect functional specialization in the synaptotagmin family.
In light of these findings, it has become essential to understand how the C2A and C2B domains of Syt-1 and Syt-7 work together to bind and insert into membranes. Several studies have addressed this question for Syt-1 and have found that its C2 domains can either bind to the same target membrane or bridge between two opposing membrane surfaces (35, 36, 37). A combination of these modalities can exist simultaneously depending on the protein fragment used, the lipid composition, and the protein/lipid ratio (29, 38). It has also been reported that Syt-1 C2AB membrane binding is cooperative on the basis of structural observations that both C2 domains copenetrate a target membrane more deeply when present as the C2AB tandem than as isolated individual domains (23, 39). Biophysically, cooperativity implies that additional free-energy stabilization is achieved by the combination of the C2A and C2B domains beyond that which would be expected from simply linking any two membrane-binding protein domains together, i.e., from avidity effects (40). This cooperative energetic model has been supported by single-molecule force measurements on Syt-1, for which C2AB domain dissociation from a target membrane was detected as single events that required a greater pulling force than the individual domains (41, 42). Another study reported that replacement of the C2A-C2B linker region of Syt-1 with a rigid polyproline helix resulted in orientation-dependent effects on both membrane binding in vitro and secretion from neurons, further suggesting that the relative orientation of the two domains may be functionally important for coinsertion into the target membrane (43).
In contrast to this model of cooperative membrane binding by the C2 domains of Syt-1, we have previously reported that the C2 domains of Syt-7 bind a target membrane independently (44). This assessment was made on the basis of both single-molecule diffusion measurements on planar supported lipid bilayers and dissociation kinetics from liposomes lacking PIP2. In this model of independent membrane binding, the C2AB tandem domain is still predicted to have a stronger membrane affinity and Ca2+ sensitivity than the individual domains because the binding strength of a multivalent protein-ligand interaction is enhanced because of the presence of multiple binding sites (40). However, the two binding sites are not predicted to strengthen one another’s affinity, i.e., binding affinities are expected to be additive rather than synergistic.
In this study, we sought to compare more directly the extent of coinsertion and cooperativity in C2AB membrane binding between Syt-1 and Syt-7. First, we measured Ca2+ sensitivity for C2A, C2B, and C2AB domains toward liposomes of physiological lipid compositions, with and without PIP2—the first time, to our knowledge, such a measurement has been reported for the Syt-7 C2B and C2AB domains. Second, we performed direct side-by-side comparisons of liposome dissociation kinetics between Syt-1 and Syt-7 C2A, C2B, and C2AB domains, using the approach developed in our previous study (44). Finally, we applied an environment-sensitive fluorescent reporter assay with Syt-7 that was previously used to show coinsertion of the tandem C2 domains from Syt-1 (23). The results show similarities in the membrane coinsertion of Syt-1 and Syt-7 C2 domains despite significant differences in their kinetics of membrane dissociation. We suggest that coinsertion and energetic cooperativity can be decoupled in Syt-7 because of the relative importance of the Syt-7 C2A domain in governing this protein’s high-affinity membrane binding.
Materials and Methods
Materials
1,2-Dioleoyl-sn-glycero-3-phosphocholine (DOPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 1,2-dioleoyl-sn-glycero-3-phospho-L-serine (sodium salt) (DOPS), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine, liver phosphatidylinositol (PI), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine, brain PIP2, cholesterol, and brain sphingomyelin were from Avanti Polar Lipids (Alabaster, AL). 1,2-Dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(5-dimethylamino-1-naphthalenesulfonyl) (dansyl-PE) was from NOF America (White Plains, NY). 3-[(3-Cholamidopropyl)dimethylammonio]-1-propanesulfonate was from BioVision (Milpitas, CA). 5-({2-[(iodoacetyl)amino]ethyl}amino)naphthalene-1-sulfonic acid (IAEDANS) was from Molecular Probes (Eugene, OR). 2-Mercaptoethanol (βME) and EDTA were from Fisher Scientific (Hampton, NH) (≥99% purity). Nitrilotriacetic acid (NTA) was from Alfa Aesar (Ward Hill, MA). All reagents were American Chemical Society grade or higher.
Protein cloning, expression, and purification
Plasmid DNA encoding human Syt-7 (GenBank ID: BC125170.1) and Syt-1 (GenBank ID: BC058917.1) were obtained from American Type Culture Collection (Manassas, VA). Sequences encoding the Syt-7 C2A domain (residues N135–S266), Syt-7 C2B domain (S261–A403), Syt-7 C2AB domain (N135–A403), Syt-1 C2A domain (K141–E272), Syt-1 C2B domain (S265–K422), and Syt-1 C2AB domain (K141–K422) were subcloned into a glutathione S-transferase-fusion vector developed previously (16, 45). All DNA sequences were verified using primer-extension sequencing (Eton Biosciences, San Diego, CA). Plasmids were transformed into Escherichia coli BL-21 DE3 for protein expression.
All proteins were purified using glutathione affinity chromatography. Cells were lysed in lysis buffer (50 mM Tris, 400 mM NaCl, 1% Triton X-100, 1 mM βME (pH 7.5) with protease inhibitors) using a Sonics VibraCell sonicator (Sonics, Newtown, CT) with a 6-mm probe. For single domains, lysates were treated with DNase (2 U/mL) obtained from Sigma-Aldrich (St. Louis, MO) for 30 min. Lysates were centrifuged to remove insoluble matter, and supernatants were incubated with glutathione sepharose 4B beads (GE Healthcare, Chicago, IL) for 3 h at 4°C. The beads were washed extensively with 50 mM Tris, 400 mM NaCl, 1 mM βME (pH 7.5) and subsequently with 50 mM Tris, 1.1 M NaCl, 5 mM EDTA, 1 mM βME (pH 7.5). Beads were then exchanged into 50 mM Tris, 150 mM NaCl, 0.05 mM EDTA, 1 mM βME (pH 7.7) for cleavage with restriction-grade thrombin (Novagen; Millipore Sigma, Billerica, MA), and eluted using the thrombin cleavage buffer or buffer A (25 mM HEPES, 15 mM NaCl, 140 mM KCl, 0.5 mM MgCl2 (pH 7.4)) plus 1–10 mM βME. βME was omitted during purification of wild-type Syt-1 C2A. For Syt-7 C2AB, 1–3 mM 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate was added to all wash and elution buffers.
Proteins for Ca2+ dependence and kinetic experiments were immediately purified further via gel filtration (Syt-1 C2A) or cation exchange (all other C2 fragments) chromatography using an Akta Purifier fast protein liquid chromatography system (GE Healthcare). Gel filtration was performed using a Superdex G75 10/300 GL column (GE Healthcare) in buffer A. Cation exchange was performed using a HiTrap SP HP 5 mL column (GE Healthcare) in buffer A plus 1 mM βME, and protein was eluted with a gradient of NaCl. Representative chromatograms are shown in Fig. S1. Protein integrity was confirmed using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) (Fig. S2), and masses were further verified using matrix-assisted laser desorption ionization mass spectrometry (MALDI). Absorbance spectra were measured using a Nanodrop 2000 (Thermo Fisher Scientific, Waltham, MA) to assess removal of nucleic acids. All purified proteins had A260/A280 ratios ≤ 0.54, indicating the absence of nucleic acid contamination (46). Finally, protein concentrations were measured using ultraviolet (UV) absorbance (PerkinElmer, Waltham, MA) based on predicted extinction coefficients at 280 nm (http://protcalc.sourceforge.net). The purified proteins were aliquoted, flash-frozen, and stored at −80°C. Before use, aliquots were thawed and centrifuged at 17,000 × g for 2 min to remove any debris, and the UV absorbance spectrum was rechecked to verify protein concentration and lack of nucleic acid.
Liposome preparation
Phospholipids in chloroform were combined at the desired molar ratio for each experiment (Table 1). After the evaporation of chloroform, the lipid films were dried under vacuum for ≥2 h and rehydrated in buffer A containing 10 mM βME to a final concentration of 3 mM total lipid. Small unilamellar vesicles were prepared by sonication to clarity on ice using a Sonics VibraCell sonicator with a 3-mm tip. Liposomes were stored at 4°C for at least 8 h after preparation and were used within 1 week. Lipid concentrations are reported as total accessible lipid, which is approximated as one-half of the total lipid present.
Table 1.
Lipid Compositions Used in This Study
| Target Membrane Lipid Compositions (mol %) |
||||||||
|---|---|---|---|---|---|---|---|---|
| Name | PE | PCa | PSa | PI | PIP2 | Sphingomyelin | Cholesterol | Dansyl-PEb |
| PM | 27.9 | 10.7 | 21.3 | 3.6c | 2.0c | 4.5 | 25 | 5.0 |
| PM(−)PIP2 | 27.9 | 10.7 | 21.3 | 5.6 | – | 4.5 | 25 | 5.0 |
| 1:1 DOPC/DOPS | – | 47.5 | 47.5 | – | – | – | – | 5.0 |
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine were used for the physiological membranes, whereas DOPC and DOPS were used for the simplified lipid composition for consistency with prior experiments (44).
For AEDANS fluorescence measurements, dansyl-PE was omitted and replaced with PC.
For Ca2+ titrations, PM liposomes contained 4.6% PI and 1.0% PIP2.
Equilibrium measurement of Ca2+-dependent protein-to-membrane FRET
C2 domain liposome binding was assessed using a protein-to-membrane fluorescence resonance energy transfer (FRET) assay in which protein Trp residues serve as the donor and dansyl-modified lipids are the acceptor (47). Buffers were prepared using Chelex-treated Ca2+-free water. Liposomes were incubated with 10% (v/v) Chelex beads (Bio-Rad, Hercules, CA) overnight at 4°C to remove residual Ca2+. Protein stocks were dialyzed into Ca2+-free buffer A (plus 1–10 mM βME for cysteine-containing proteins). Quartz cuvettes were soaked in 100 mM EDTA, then rinsed extensively with Ca2+-free water before use. Steady-state fluorescence experiments were performed using a Photon Technology International (Birmingham, NJ) QM-2000-6SE fluorescence spectrometer at 25°C. Excitation slit width was 2.4 nm for Syt-7 C2B on PM liposomes (see Table 1 for lipid composition) and 1 nm for all other samples; emission slit width was 8 nm. CaCl2 was titrated into an initially Ca2+-free solution containing protein (2 μM for Syt-1 C2A and C2B, 0.3–1.0 μM for others) and liposomes (75 μM accessible lipid). Because of the extreme Ca2+ sensitivity of Syt-7 C2 domains, a Ca2+ buffering system containing 1.5 mM NTA was used for most titrations to maintain total [Ca2+] in excess of protein, as described previously (16). For some titrations (Syt-1 C2B and C2AB, as well as titrations indicated in the Supporting Material), a more aggressive Ca2+ buffering system consisting of 1.5 mM NTA and 0.2 mM EDTA was used. Concentrations of free Ca2+ and Mg2+ (the latter held constant at 0.5 mM) were calculated using MaxChelator (https://somapp.ucdmc.ucdavis.edu/pharmacology/bers/maxchelator/webmaxc/webmaxcE.htm). No Ca2+ buffering system was used for Syt-1 C2A. For each titration, FRET was measured (λexcitation = 284 nm, λemission = 510 nm) over a 10-s integration time for each of three replicate samples. Each intensity value was corrected for dilution, and the intensity of a blank sample containing only buffer, lipid, and Ca2+ was subtracted. Reversibility was tested by adding excess EDTA after titrations. Fluorescence emission scans before and after titration are shown in Fig. S3. Normalized data were fitted to the Hill equation,
| (1) |
where ΔF is the fluorescence increase, Ca1/2 is the Ca2+ concentration at which half of the initially unbound protein becomes membrane bound, H is the Hill coefficient, ΔF0 is the fluorescence change in the absence of Ca2+, and ΔFmax is the calculated maximal fluorescence change. Fitting was performed using Kaleidagraph 4.5 (Synergy Software). Data in figures are shown after normalization of ΔFmax to unity for each titration.
Stopped-flow spectroscopy
Stopped-flow fluorescence kinetic measurements were performed using a BioLogic SFM3000 spectrophotometer (Knoxville, TN) using 284-nm excitation and a 455-nm long-pass emission filter. Unless otherwise noted, protein concentrations used were 1 μM for Syt-7 C2 domains and Syt-1 C2AB and 5 μM for individual Syt-1 C2 domains (all concentrations listed are before mixing). Protein-to-membrane FRET (dansyl-PE emission) was monitored after rapid mixing of equal volumes of protein-bound liposomes (200 μM total accessible lipid, 200 μM CaCl2) and 2 mM EDTA in buffer A. Dead time is estimated to be 1.4 ms. Data sets for each sample were calculated as the average of eight or more time courses and were fitted to a single- or double-exponential function (Eqs. 2 or 3, respectively):
| (2) |
| (3) |
where the koff are dissociation rate constants and C is an offset. C was subtracted and ΔFmax (or ΔFmax 1 + ΔFmax 2) normalized to unity in the figures shown. Rate constants listed are average ± SD of ≥3 independent replicate measurements.
Dynamic light scattering
The Z-average diameter of liposome suspensions (1:1 DOPC/DOPS, 200 μM accessible lipids, 100 μM Ca2+) was determined using a Zetasizer Nano S90 (Malvern Instruments, Malvern, UK) before and after addition of protein. The samples were removed from the instrument and used for stopped-flow kinetic measurements as described above.
Purification of AEDANS-labeled proteins
To facilitate labeling, native cysteine residues were removed from the proteins, and unique cysteines were inserted using site-directed mutagenesis (Quik-Change II XL; Agilent, Santa Clara, CA). A cysteine to alanine substitution was performed at C275 in Syt-7 C2AB and C2B. (Note: the C275A mutant of Syt-7 C2B was also used for all Syt-7 C2B kinetic experiments because this mutation greatly simplified purification of the protein, which otherwise had a propensity to form disulfide-linked dimers.) C260 was mutated to serine in Syt-7 C2A and C2AB; we have previously reported that this mutation has only minor effects on the C2A domain (17). For AEDANS attachment, a library of mutants was generated with one of the following residues mutated to cysteine in Syt-7 single and tandem domains: T170 in CBL1 of C2A (single and tandem domains denoted C2A1 and C2A1B, respectively), N232 in CBL3 of C2A (denoted C2A3 and C2A3B), T301 in CBL1 of C2B (denoted C2B1 and C2AB1), and N364 in CBL3 of C2B (denoted C2B3 and C2AB3). Mutations were confirmed using primer-extension sequencing (Eton Biosciences). Proteins were expressed and purified using glutathione affinity chromatography as described above, except that 0.5 mM tris(2-carboxyethyl)phosphine was used in place of 1 mM βME for all domains, and 2.5 mM CaCl2 was included in the thrombin cleavage buffer for C2B. After thrombin cleavage and elution, protein concentrations were estimated from absorbance at 280 nm, and the proteins were incubated with IAEDANS (4:1 molar ratio of IAEDANS/protein). For single domains, proteins were labeled in 140 mM KCl, 0.5 mM MgCl2, 25 mM HEPES, 50 mM glutamic acid, 50 mM arginine (pH 7.1) overnight at 4°C (48). For C2AB tandems, proteins were labeled in thrombin cleavage buffer for 1 h at room temperature. The reaction was quenched by adding 1 mM βME. To remove free dye and anionic contaminants, the proteins were further purified using cation exchange chromatography as described above, including 20 mM CaCl2 in the chromatography buffers. After chromatography, proteins were exchanged into buffer A plus 1 mM βME. Protein mass and labeling was verified using SDS-PAGE gel electrophoresis with visualization under UV light, and concentration was quantified based on AEDANS absorbance.
AEDANS fluorescence assays
AEDANS fluorescence was measured using a Photon Technology International QM-2000-6SE fluorescence spectrometer (Birmingham, NJ) at 25°C. Fluorescence emission was measured from 450 to 600 nm, with λexcitation = 337 nm. Excitation slit widths were 1 nm except for the following samples: C2A3B, 1.6 nm; C2AB1, 1.6 nm; and C2AB3, 2.4 nm. Emission slit widths were 8 nm for all measurements. All measurements were carried out with 0.5–1 μM protein in buffer A with 0.2–1.0 mM Ca2+ before and after addition of liposomes (33 μM total lipid).
Results
Strategy and protein purification
To study C2A-C2B interdomain cooperativity, we expressed and purified individual domains and C2AB tandems of both Syt-1 and Syt-7 (Figs. 1, S1, and S2). Initial purification was accomplished using affinity chromatography with cleavable glutathione S-transferase tags. Because the Syt-1 C2B domain and both Syt-7 C2 domains are cationic and tend to copurify with anionic contaminants, these individual domains and both C2AB tandem domains were additionally purified using cation exchange chromatography (Fig. S1) (16, 18, 49). Syt-1 C2A was purified using glutathione affinity chromatography followed by gel filtration (Fig. S1). Purity of all six protein domains was excellent as assessed using SDS-PAGE, UV absorbance, and mass spectrometry (see Materials and Methods).
Ca2+ sensitivity of Syt-1 and Syt-7 C2 domains
The Ca2+ sensitivities of membrane binding were compared among individual and tandem C2 domains of Syt-7 and Syt-1 (Fig. 2; Table 2). Titration with Ca2+ in the presence of liposome membranes produces sigmoidal binding curves with a characteristic Ca1/2 ([Ca2+] for binding half the Ca2+-sensitive population) and a Hill coefficient typically >1 arising from cooperative binding of multiple Ca2+ ions and membrane to each C2 domain (50). To compare these parameters for Syt-7 C2A, C2B, and C2AB domains, Ca2+ was titrated into solutions containing protein and liposomes with lipid compositions approximating the interior leaflet of the plasma membrane, with or without PIP2 (Table 1), and protein-to-membrane FRET was monitored (47). Because of the strong Ca2+ sensitivity of Syt-7, solutions were treated to remove as much free Ca2+ as possible, and a Ca2+ buffering system was used to enable reliable measurement of binding even at free Ca2+ concentrations less than the protein concentration (16). Consistent with a previous report, the Syt-7 C2A domain bound liposomes containing a physiological membrane composition without PIP2 (PM(−)PIP2, Table 1) with a Ca1/2 around 5 μM and a Hill coefficient of 2.2 (Table 2) (16). Inclusion of PIP2 in the membrane had little impact on the Ca1/2 and Hill coefficient for this domain (Table 2).
Figure 2.
Ca2+ sensitivity of Syt-7 (A and B) and Syt-1 (C) C2 domains. CaCl2 was titrated into solutions containing the indicated C2A (triangles), C2B (squares), or C2AB (circles) domains and liposomes. Data were normalized based on best fits to Eq. 1; the parameters from these fits are given in Table 2. Points and error bars shown are mean ± SD of three replicate titrations; where not visible, error bars are smaller than the symbol. Right panels show the extent of reversibility upon addition of 2.5 mM EDTA.
Table 2.
Equilibrium Ca2+ Titration Parameters of Syt-1 and Syt-7 C2 Domains
| Liposome Composition (Table 1) | Domain | Amount Bound before Ca2+ Addition (%) | Ca1/2 (μM)a | Hill Coefficienta |
|---|---|---|---|---|
| PM | Syt-1 C2A | <5 | 101 ± 2 | 1.7 ± 0.1 |
| PM | Syt-1 C2B | <5 | 14.6 ± 0.6 | 1.3 ± 0.1 |
| PM | Syt-1 C2AB | <5 | 4.5 ± 0.2 | 1.8 ± 0.1 |
| PM(−)PIP2 | Syt-7 C2A | <5 | 4.8 ± 0.4 | 2.2 ± 0.1 |
| PM(−)PIP2 | Syt-7 C2B | 5 ± 1 | 16.0 ± 0.5 | 2.5 ± 0.1 |
| PM(−)PIP2 | Syt-7 C2B C275A | 51 ± 1 | 15.1 ± 0.6 | 2.7 ± 0.2 |
| PM(−)PIP2 | Syt-7 C2AB | 53 ± 3 | 1.2 ± 0.1 | 2.2 ± 0.1 |
| PM | Syt-7 C2A | 6 ± 2 | 4.0 ± 1.0 | 2.2 ± 0.2 |
| PM | Syt-7 C2B | 37 ± 1 | 3.5 ± 0.6 | 0.8 ± 0.1 |
| PM | Syt-7 C2AB | >85 | NDb | NDb |
Uncertainties represent SD from ≥ 3 separate experiments.
Ca1/2 and Hill coefficients were obtained from fitting to Eq. 1. Note that these parameters correspond to the population of protein not initially bound to membrane. Parameters could not be determined accurately for Syt-7 C2AB on PM liposomes because of its largely Ca2+-independent profile.
ND, no data.
In contrast, the Syt-7 C2B domain showed a pronounced PIP2 dependence, consistent with a previous report (18). In the absence of PIP2, Syt-7 C2B bound liposome membranes with somewhat weaker Ca2+ sensitivity than the C2A domain (Fig. 2 A), but the trend was reversed upon inclusion of 1% PIP2 in the membrane because the protein was partially membrane bound even before Ca2+ addition (Fig. 2 B; Table 2). We estimate the level of contaminating free Ca2+ in these measurements to be <0.1 μM based on measurements using the Ca2+-sensitive dye Quin-2 (Fig. S4). The ability of Syt-7 C2B to bind PM liposomes before Ca2+ addition persisted in experiments using a more aggressive Ca2+ buffering system that included 0.2 mM EDTA (Fig. S5); thus, we conclude that Syt-7 C2B has a binding mode to PIP2 that is Ca2+-independent. Its binding was mostly reversed upon addition of 2.5 mM EDTA (Fig. 2 B; Figs. S3 and S5), which might arise from ionic interactions between EDTA and the polybasic region of the Syt-7 C2B domain.
Mutation of Cys275 to alanine amplified a Ca2+-independent component of Syt-7 C2B binding PM(−)PIP2 liposomes, which was not reversible with EDTA (Fig. S5; Table 2). However, the Ca1/2 and Hill coefficient were only modestly affected by this mutation, suggesting that the mutation alters Ca2+-independent but not Ca2+-dependent binding (Table 2). This mutation was required for the AEDANS measurements and was also used for the kinetic experiments with Syt-7 C2B described below.
The Syt-7 C2AB tandem had a lower Ca1/2 toward PM(−)PIP2 liposomes than either individual domain and was ∼50% bound to these liposomes before Ca2+ addition (Fig. 2 A; Fig. S5). Inclusion of 1% PIP2 further enhanced membrane binding because the C2AB tandem was > 85% bound to PM liposomes before Ca2+ addition (Fig. 2 B). Binding of Syt-7 C2AB to PM(−)PIP2, but not PM, liposomes was reversed upon EDTA addition (Fig. 2; Fig. S3). In contrast, C2 domains of Syt-1 did not exhibit measurable Ca2+-independent binding toward PM liposomes under these experimental conditions (Fig. 2 C). Overall, the combination of the two domains increases the Ca2+ sensitivity of membrane binding for both Syt-1 and Syt-7 (Fig. 2, A–C) and enhances a Ca2+-independent binding mode for Syt-7 (Fig. 2, A and B). However, it is not clear from these data whether the enhancements reflect cooperativity or simply additive contributions of two connected membrane-binding domains.
Assessing cooperativity in Syt-1 and Syt-7 C2AB domains from dissociation kinetics
To probe whether the energetics of Syt-1 and Syt-7 C2AB membrane binding are cooperative, we measured the kinetics of protein-membrane dissociation upon addition of the Ca2+ chelator EDTA. In this experiment, protein-to-membrane FRET decreases after the addition of EDTA to protein-liposome complexes. The EDTA coordinates Ca2+ ions as each C2 domain leaves the membrane surface, rendering each domain’s dissociation irreversible. Thus, if the two C2 domains bind and release from membranes independently, then the dissociation kinetic profile of the C2AB tandem should be closely similar to that of the rate-limiting domain. We have previously used this method to report independent binding for Syt-7 C2AB using liposomes composed of 3:1 DOPC/DOPS (44). On the other hand, if the two C2 domains of a C2AB tandem bind membranes cooperatively, then C2AB dissociation is expected to be significantly slower than both of the individual domains.
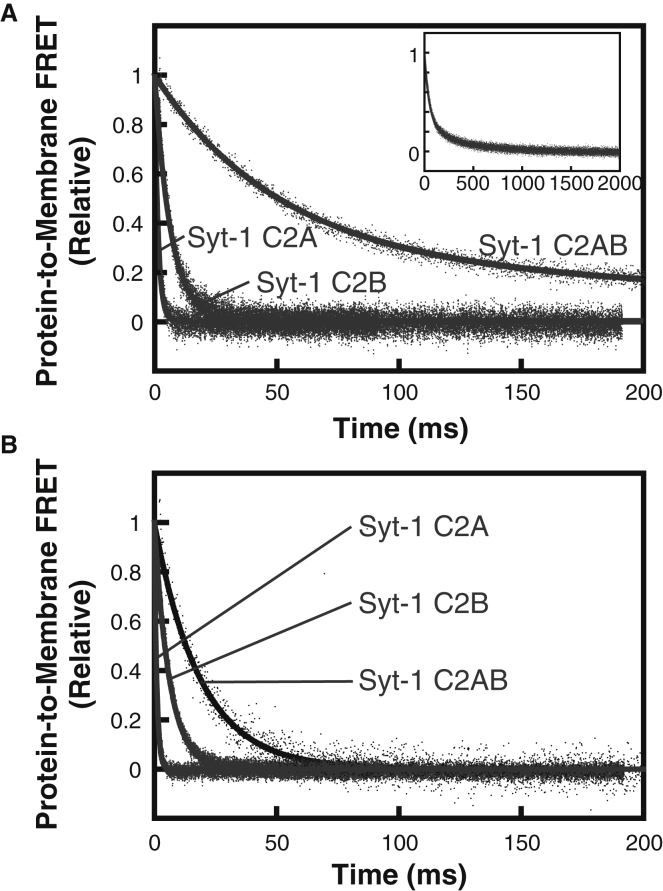
Using this approach, the tandem C2 domains of Syt-1 are clearly observed to bind membranes cooperatively. Although the Syt-1 C2A domain dissociated from 3:1 DOPC/DOPS too quickly to measure, we quantified dissociation kinetics of Syt-1 C2A, C2B, and C2AB using 1:1 DOPC/DOPS (Fig. 3 A) and PM(−)PIP2 (Fig. 3 B). The individual Syt-1 C2 domains’ dissociation time courses fit well to single exponential profiles, with the C2A rate constant similar to previous reports (16, 51). Dissociation of the C2B domain was slower than the C2A domain, consistent with its stronger membrane binding (18, 30, 42).
Figure 3.
Dissociation kinetics of Syt-1 C2 domains on (A) 1:1 DOPC/DOPS liposomes and (B) PM(−)PIP2 liposomes. Dansyl-PE fluorescence was monitored as solutions containing 5 μM (single domains) or 1 μM (tandem domain) protein, 200 μM CaCl2, and liposomes (200 μM accessible lipid) were rapidly mixed with an equal volume of 2 mM EDTA (all concentrations listed are before mixing). Kinetic data were fitted to Eqs. 2 or 3, with rate constants shown in Table 3. Inset shows the full timescale of measurement of Syt-1 C2AB. Data shown are representative of ≥3 independent measurements.
Syt-1 C2AB dissociation from 1:1 DOPC/DOPS fit best to a double-exponential profile, of which the faster rate constant was ∼7-fold slower than the isolated C2B domain and >50-fold slower than the isolated C2A domain (Table 3). A similar trend was seen with PM(−)PIP2 liposomes (Fig. 3 B). All three Syt-1 protein fragments had monoexponential profiles with the latter lipid composition, suggesting a single membrane-bound population of each individual or tandem domain. Although Syt-1 C2AB dissociated from these liposomes faster than from 1:1 DOPC/DOPS, the tandem domain was still >3-fold and 20-fold slower than the respective C2B and C2A individual domains (Table 3). Thus, the energy barrier for membrane release upon EDTA addition is significantly greater for the Syt-1 C2AB tandem than for both of its individual domains, consistent with a model of cooperative membrane binding.
Table 3.
Dissociation Kinetics of Syt-1 and Syt-7 C2 Domains
| Domain | koff, 1:1 DOPC/DOPS (s−1) | koff, PM(−)PIP2 (s−1) |
|---|---|---|
| Syt-1 C2A | 710 ± 30 | 1020 ± 90 |
| Syt-1 C2B | 140 ± 10 | 170 ± 10 |
| Syt-1 C2AB | 21 ± 2 (72% amp) 2.6 ± 0.1 (28% amp) | 48 ± 5 |
| Syt-7 C2A | 6.39 ± 0.04 | 7.4 ± 0.3 |
| Syt-7 C2B | 80 ± 9 (88% amp) 13 ± 2 (12% amp) | 100 ± 20 |
| Syt-7 C2AB | 4.6 ± 0.3 (74% amp) 0.16 ± 0.04 (26% amp) | 5.9 ± 0.6 (77% amp) 0.64 ± 0.09 (23% amp) |
Uncertainties represent SD from ≥ 3 separate experiments. Double-exponential rate constants are reported with their respective percent amplitudes from the best-fit curve.
Dissociation kinetics of Syt-7 C2 domains (Fig. 4) show marked differences from Syt-1. Dissociation of each individual Syt-7 domain was slower than that of the corresponding Syt-1 domain (Table 3), consistent with separate previous reports of Syt-7 C2A and C2AB (15, 16). The difference is particularly pronounced between the C2A domains, for which Syt-7 C2A dissociated >100-fold slower from both membrane compositions tested. Notably, for Syt-7, the C2A domain dissociated slower than the C2B domain, in contrast to Syt-1, for which the isolated C2B domain dissociates slower. This result is consistent with prior reports that the C2A domain dominates membrane binding and fusion properties for Syt-7 (9, 11, 18, 34).
Figure 4.
Dissociation kinetics of Syt-7 C2 domains on (A) 1:1 DOPC/DOPS liposomes and (B) PM(−)PIP2 liposomes. Dansyl-PE fluorescence was monitored as solutions containing 1 μM protein (in A) or 0.25 μM protein (in B), 200 μM CaCl2, and liposomes (200 μM accessible lipid) were rapidly mixed with an equal volume of 2 mM EDTA (all concentrations listed are before mixing). Kinetic data were best fitted to Eqs. 2 or 3, with rate constants listed in Table 3. Data shown are representative of ≥3 independent measurements.
Dissociation of Syt-7 C2AB from both 1:1 DOPC/DOPS and PM(−)PIP2 was biphasic, with a major (faster) rate constant that differed by no more than 30% from the isolated C2A domain on each liposome composition tested (Fig. 4; Table 3). The double-exponential character suggests the presence of multiple populations; one interpretation is that a subpopulation (20–30%) of Syt-7 C2AB is bound to membranes in a cooperative fashion analogous to Syt-1, although other explanations are also possible (see Discussion). Notably, the larger population of Syt-7 C2AB has dissociation kinetics that are nearly the same as the isolated C2A domain (Table 3). This similarity is evident from comparing the initial portions of portions of the dissociation time courses, during which the fast component dominates (Fig. S6). For this population, the kinetics are more consistent with the two C2 domains of Syt-7 having noncooperative membrane-binding energies.
Liposome clustering by Syt-7 C2B
Some Syt C2 domains, including C2AB tandems, are known to promote clustering of liposomes in a Ca2+-dependent manner (18, 36, 37, 52, 53). This effect could conceivably produce slower dissociation rates, e.g., if a subpopulation of protein is initially inaccessible to the added EDTA. To probe whether liposome clustering occurs under the conditions used in the measurements above, we performed dynamic light scattering measurements of 1:1 DOPC/DOPS liposomes in the presence of different concentrations of Syt-7 C2B. Addition of 0.5 μM protein to liposomes resulted in a significant increase in particle size from an average diameter of ∼100 nm to ∼500 nm (Fig. S7 A). Further addition of protein to 2 μM increased the size distribution further, to ∼800 nm (Fig. S7 B). These samples became visibly cloudy, but not flocculated, upon addition of 2 μM protein. Thus, Syt-7 C2B induces liposome clustering but not large-scale aggregation under conditions comparable to those used in the kinetic measurements reported above.
This degree of clustering appears to have no effect on the major dissociation rate constants. Kinetic profiles were measured using 0.5, 1, and 2 μM Syt-7 C2B on the same concentration of liposomes (Fig. S8). At the highest protein concentration, the dissociation profile was clearly double exponential, suggesting the presence of multiple populations. Kinetic profiles at the lower protein concentrations could also be fitted to a double-exponential profile, although the slower component was greatly decreased in intensity (Table S1). The slower phase was only a minor contributor (<20%) to the overall kinetic profile at protein concentrations ≤ 1 μM. Importantly, the rate constants of the fast and slow components showed no trend among measurements using various concentrations of protein (Table S1). Thus, we conclude that liposome clustering and aggregation result in a subpopulation of protein with a significantly slower dissociation; however, at the protein concentrations used in the experiments above, the major, faster dissociation rate constant remains readily measurable. It is possible that liposome clustering may give rise to some of the minor, slower component(s) that we observe with Syt-1 and Syt-7 C2AB dissociation. Importantly, these results support a high level of confidence that the major rate constants of the kinetic profiles correspond to straightforward protein-membrane dissociation that is not influenced by liposome aggregation effects.
Membrane insertion of Syt-7 C2 domains
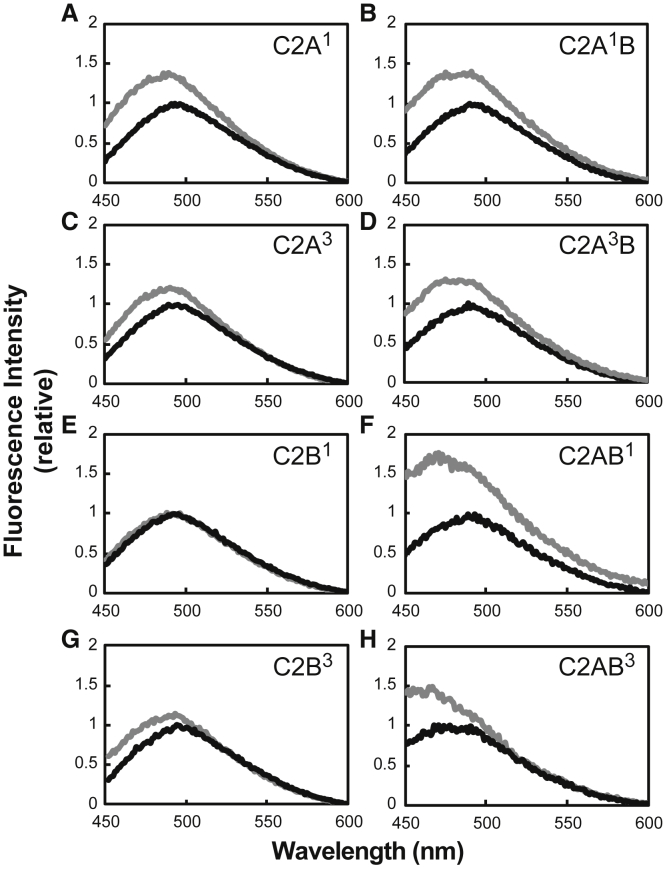
To test whether the tandem structure of Syt-7 C2AB affects its C2 domains’ membrane insertion, we used the environment-sensitive fluorescent label AEDANS. This approach has been used previously to demonstrate membrane insertion of Syt-1 C2 domains (23, 24), but to our knowledge has not been previously applied to other synaptotagmins. AEDANS was attached to a unique engineered cysteine residue on the tip of CBL1 or CBL3 on each Syt-7 C2 domain both individually and in the C2AB tandem. These are the regions that penetrate membranes in many C2 domains, including synaptotagmins (17, 22, 54, 55, 56, 57). We denote these constructs based on the domain and loop in which the AEDANS label was placed; for example, C2A1B has the AEDANS group on CBL1 of C2A within the C2AB tandem (Fig. S9). Upon addition of physiological liposomes lacking PIP2 (PM(−)PIP2, Table 1) to protein in the presence of excess Ca2+, the AEDANS fluorescence emission increased and blueshifted when the label was in either CBL1 or CBL3 of the individual C2A domain (C2A1 or C2A3, Fig. 5, A and C) or CBL3 of the individual C2B domain (C2B3, Fig. 5 G). For all labeling positions, the increase in fluorescence emission was amplified when measured in the C2AB tandem (Fig. 5, B, D, F, and H). The difference between individual and tandem domains is especially striking when comparing constructs labeled on CBL1 of the C2B domain, as the AEDANS fluorescence increased dramatically on addition of liposomes to C2AB1 but not C2B1 (Fig. 5, E and F).
Figure 5.
Membrane insertion of Syt-7 C2A and C2B domain loops in the absence of PIP2. Fluorescence emission spectra are shown of Syt-7 single domains (A, C, E, and G) or the C2AB tandem (B, D, F, and H) labeled with AEDANS on CBL1 (A, B, E, and F) or CBL3 (C, D, G, and H) (see Fig. S9 for schematic of labeling sites and Materials and Methods for residue numbers). Data are of protein in solution alone (black) and after addition of PM(−)PIP2 liposomes (gray). All spectra are normalized to the maximal intensity in the absence of lipid.
Inclusion of 2% PIP2 in the liposome composition resulted in a similar pattern, except that the amount of fluorescence increase for the individual domains was somewhat greater than in the absence of PIP2 (Fig. S10). Overall, the C2 domains of Syt-7 penetrate into membranes when present as single or tandem domains. However, C2B in particular penetrates significantly more deeply when it is part of the C2AB tandem structure. This result is qualitatively similar to what was previously reported for Syt-1 (23).
Discussion
In this study, we have employed FRET-based equilibrium Ca2+ titrations, stopped-flow kinetic measurements, and fluorescence-based membrane insertion assays to explore how the two C2 domains of Syt-7 work together in comparison to those of Syt-1. Four main conclusions can be drawn from the results: 1) membrane-dissociation kinetics indicate the tandem C2 domains of Syt-1 bind membranes with cooperative energetics; 2) Syt-7 C2AB dissociation kinetics from membranes lacking PIP2 reveal two populations, the larger of which has a profile consistent with noncooperative membrane binding (44); 3) the Syt-7 C2AB fragment binds membranes through a combination of Ca2+-independent and highly Ca2+-sensitive mechanisms; and 4) the CBLs of Syt-7 C2B insert more deeply into membranes in the context of the C2AB tandem, similarly to Syt-1 (23). Thus, coinsertion and energetic cooperativity appear to be decoupled in Syt-7 because the linkage of the C2A and C2B domains appears to induce significantly deeper insertion of the C2B domain but has only minor effects on dissociation kinetics relative to the individual C2A domain.
Decoupling of coinsertion and cooperativity in Syt-7
Liposome dissociation kinetics reveal a clear distinction between the extent of cooperative membrane binding in Syt-1 and Syt-7 C2AB tandems. For Syt-1, the C2AB tandem dissociates much slower than either of the C2A or C2B individual domains (Fig. 3; Table 3), consistent with cooperative energetics of insertion. In contrast, Syt-7 C2AB dissociation kinetics are biphasic, indicating multiple populations, of which the larger population dissociates at a rate closely similar to its individual C2A domain (Fig. 4; Table 3). Importantly, these trends among the various domains are the same using two lipid compositions from which dissociation kinetics were measurable for all species, 1:1 DOPC/DOPS and PM(−)PIP2.
The minor, slower phase in Syt-7 C2AB dissociation may arise from a population (∼25%) of the protein in which the C2A and C2B domains bind membranes cooperatively; alternatively, it could arise from liposome clustering. Clustering of liposomes induced by Syt-7 C2AB has been reported previously (18). We observe that liposome clustering gives rise to a minor, slow phase in Syt-7 C2B dissociation kinetics (Figs. S7 and S8; Table S1), and it is likely that a similar phenomenon occurs for the C2AB tandem. Liposome clustering and cooperativity are not mutually exclusive; conceivably, these two phenomena could account for the double-exponential profile of Syt-1 C2AB dissociation from 1:1 DOPC/DOPS liposomes, which has two rate constants that are both significantly slower than the individual domains (Table 3). Syt-7 C2AB dissociation profiles are adequately fitted by a double exponential, but we cannot rule out the possibility of more than two populations. Regardless of the origin of the slow phase(s) in the Syt-7 C2AB dissociation data, the largest population of this protein has dissociation rate constants that are closely similar to the individual C2A domain.
The AEDANS-based membrane insertion results indicate commonalities between Syt-1 and Syt-7 that provide structural context for understanding the differences in kinetics. We show that CBL1 and CBL3 of Syt-7 C2A and C2B insert into membranes in the C2AB tandem (Fig. 5, B, D, F, and H). The trends are the same in the presence of PIP2 (Fig. S10). In particular, the Syt-7 C2B domain penetrates into membranes more deeply as part of the C2AB tandem than as an individual domain (Fig. 5, E–H; Fig. S10, E–H). These results align with previous reports of comparable measurements with Syt-1 (15, 23), suggesting that these two synaptotagmin isoforms bind membranes with similar structural mechanisms despite differences in the extent of interdomain cooperativity that are evident from their dissociation kinetics.
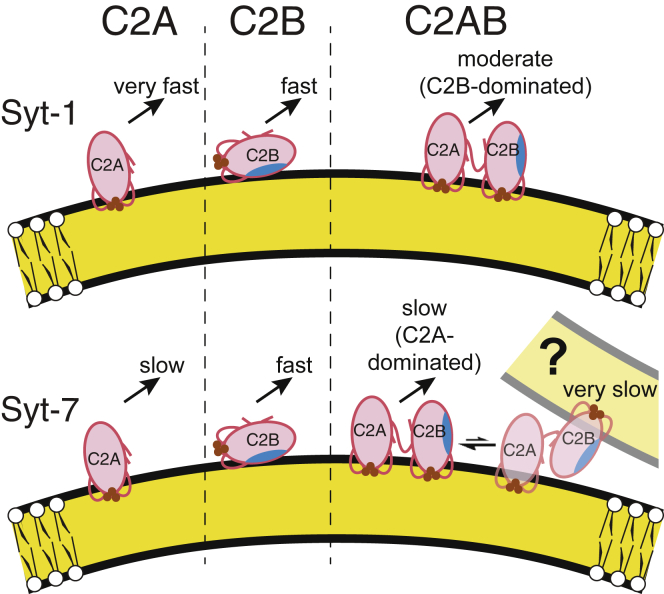
Our results, along with previously reported data, suggest a model in which insertion of the C2B domain is strongly impacted by the C2AB linkage and the relative energetics of membrane binding by C2A and C2B domains differ between the two isoforms (Fig. 6). For Syt-1, the C2B domain has been reported to bind membranes more tightly than the C2A domain does (18, 30). This difference is reflected in our measurements both of Ca2+ dependence in the presence of PIP2 (Fig. 2 C) and dissociation rates in the absence of PIP2 (Fig. 3). In contrast, Syt-7 C2A binds membranes more tightly than Syt-7 C2B in the absence of PIP2 (Figs. 2 A and 4) (18). We suggest that the deeper insertion of the C2B domain significantly slows the membrane-dissociation kinetics of Syt-1 C2AB because that protein’s membrane-binding strength is dominated by its C2B domain (Fig. 6, top). For Syt-7, membrane binding is dominated by the C2A domain, which binds tightly and penetrates deeply as an individual domain (17) and only slightly deeper in the C2AB tandem (Fig. 5, A–D). Thus, C2A-C2B linkage does not significantly enhance the free energy of membrane binding for the C2A-dominated Syt-7 (Fig. 6, bottom).
Figure 6.
Structural model of linkage effects on C2AB insertion and dissociation kinetics. (Left) The individual C2A domains of Syt-1 (top) and Syt-7 (bottom) penetrate membranes primarily via their CBLs (20, 21, 22, 23, 24) (Fig. 5, A and C); Syt-1 C2A dissociates much faster than Syt-7 C2A upon addition of EDTA (16) (Table 3). (Center) The individual C2B domains bind membranes relatively superficially through interaction of the polybasic region (blue shading) with lipid headgroups (30) along with modest insertion of CBL3 (23) (Fig. 5, E and G). (Right) In the C2AB tandem, linkage to C2A affects the binding geometry of the C2B domain, leading to deeper insertion of its CBL1 and CBL3 (23, 24) (Fig. 5, F–H). For Syt-1, this deeper insertion leads to much slower dissociation kinetics compared to the individual domains, i.e., interdomain cooperativity. For Syt-7, dissociation kinetics are dominated by the C2A domain and relatively unaffected by C2A-C2B linkage (Fig. 4). (Bottom right) A minor, very slow phase in Syt-7 C2AB dissociation kinetics may be explained by liposome clustering, which could include a subpopulation of C2AB fragments bound in trans to opposing liposomes. Brown circles represent Ca2+. Curvature is approximately to scale of 100-nm-diameter liposomes (Fig. S7). Kinetics upon EDTA addition are as follows: very fast, >500 s−1; fast, 80–200 s−1; moderate, 10–50 s−1; slow, 1–10 s−1; very slow, <1 s−1 (Table 3). To see this figure in color, go online.
Ca2+ independence of Syt-7 C2 domains toward PIP2
Syt-7 C2AB binding was mostly Ca2+-independent toward membranes containing the physiologically relevant target lipid PIP2 (Fig. 2 B; Table 2). On average, PIP2 comprises 1–2% of plasma membrane lipids, and it is reported to diffuse into fusion pore and secretory vesicle membranes within a few seconds during fusion events; therefore, PIP2 is very likely a target lipid for Syt-1 and Syt-7 in vivo (29, 58, 59, 60). Because Syt-7 C2AB membrane binding to PIP2-containing liposomes was hardly reversed by EDTA under our conditions (Fig. 2 B; Fig. S3 F), we were unable to measure its dissociation kinetics. Thus, further studies are needed to elucidate whether Syt-7 membrane binding becomes cooperative in the presence of PIP2. We observe that Ca2+ sensitivity of the Syt-7 C2B domain is comparable to or stronger than the C2A domain in the presence of PIP2 (Fig. 2 B; Table 2), and the Ca2+ independence of the C2AB tandem suggests that the two domains’ binding energies might be more than additive toward physiological membranes containing PIP2. However, the modest cellular effects reported to arise from mutations in the C2B domain would suggest that the C2A domain continues to dominate Syt-7 Ca2+ sensing under physiological conditions (9, 11, 34). Given the observed Ca2+ independence of Syt-7 C2AB and the high local lipid concentration at sites of fusion, other factors would likely be needed to prevent Syt-7 C2 domains from inserting into the plasma membrane before Ca2+ influx and to bring them off after fusion. Possibly, competing interactions exist, e.g., with SNARE proteins (61, 62), that modulate Ca2+-independent Syt-7 C2AB membrane insertion in vivo.
Liposome clustering and aggregation
Previously, Syt-1 C2B and C2AB as well as Syt-7 C2A and C2AB have been found to induce liposome aggregation (18, 36, 52, 53). In this study, we report that Syt-7 C2B (C275A) shares the same property using 1:1 DOPC/DOPS liposomes. Both of the polybasic regions that have been suggested to impact bilayer bridging by Syt-1 C2B are present in Syt-7 C2B as well (7, 29, 63). As expected, liposome clustering depends on protein concentration (Fig. S7) (28, 64). In these experiments, liposome clustering arising from bilayer bridging may reflect a separate structural state from the coinsertion events whose extent of cooperativity was the main focus in this study, and therefore we sought to minimize aggregation by choosing conditions that favor coinsertion, such as low protein/lipid ratios (38). At higher protein/lipid ratios, liposome aggregation appeared to produce or exaggerate a slow component of the kinetic time course (Fig. S8; Table S1). However, the rate constant of the major component of the kinetic time course remained unaffected; therefore, we conclude that this rate constant represents dissociation of the proteins from the liposome surface.
Implications for evolution of synaptotagmin function
Despite their different Ca2+ sensitivities and kinetics, both Syt-1 and Syt-7 insert their C2B domain CBLs, particularly CBL1, more deeply into membranes as C2AB tandems than as individual domains. The conservation of this structural feature suggests that coinsertion of C2 domains into the same membrane may be a common feature among synaptotagmins, even under conditions in which some degree of liposome clustering is also present (38). We recently posited that synaptotagmins in the three-dimensional context of a fusion pore could assume a geometry in which CBL1 and CBL3 of both C2A and C2B copenetrate the fusion pore neck while the polybasic regions bridge between opposing membranes (7). In other words, cooperative membrane insertion and bilayer bridging are not necessarily contradictory functions. In principle, cooperativity between C2 domains could facilitate rapid and efficient coinsertion to stabilize the fusion pore during a Ca2+ signaling event. Future studies comparing structural and energetic properties of more synaptotagmin isoforms could provide key insights into the evolution of synaptotagmin function in fusion pore formation and stabilization.
Author Contributions
H.T.T., L.H.A., and J.D.K. designed research, performed research, and analyzed data. H.T.T. and J.D.K. wrote the manuscript.
Acknowledgments
The authors thank Arun Anantharam, Skyler Jackman, and Joseph Vasquez for helpful comments on the manuscript.
This work was supported by National Institutes of Health grant R15GM102866 to J.D.K. and University of Colorado Denver Undergraduate Research Opportunity Program funds to H.T.T.
Editor: Arne Gericke.
Footnotes
10 figures and one table are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(19)30105-5.
Supporting Material
References
- 1.Brose N., Petrenko A.G., Jahn R. Synaptotagmin: a calcium sensor on the synaptic vesicle surface. Science. 1992;256:1021–1025. doi: 10.1126/science.1589771. [DOI] [PubMed] [Google Scholar]
- 2.Chapman E.R. How does synaptotagmin trigger neurotransmitter release? Annu. Rev. Biochem. 2008;77:615–641. doi: 10.1146/annurev.biochem.77.062005.101135. [DOI] [PubMed] [Google Scholar]
- 3.Gustavsson N., Han W. Calcium-sensing beyond neurotransmitters: functions of synaptotagmins in neuroendocrine and endocrine secretion. Biosci. Rep. 2009;29:245–259. doi: 10.1042/BSR20090031. [DOI] [PubMed] [Google Scholar]
- 4.Bhalla A., Chicka M.C., Chapman E.R. Analysis of the synaptotagmin family during reconstituted membrane fusion. Uncovering a class of inhibitory isoforms. J. Biol. Chem. 2008;283:21799–21807. doi: 10.1074/jbc.M709628200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Craxton M. A manual collection of Syt, Esyt, Rph3a, Rph3al, Doc2, and Dblc2 genes from 46 metazoan genomes--an open access resource for neuroscience and evolutionary biology. BMC Genomics. 2010;11:37. doi: 10.1186/1471-2164-11-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moghadam P.K., Jackson M.B. The functional significance of synaptotagmin diversity in neuroendocrine secretion. Front. Endocrinol. (Lausanne) 2013;4:124. doi: 10.3389/fendo.2013.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.MacDougall D.D., Lin Z., Anantharam A. The high-affinity calcium sensor synaptotagmin-7 serves multiple roles in regulated exocytosis. J. Gen. Physiol. 2018;150:783–807. doi: 10.1085/jgp.201711944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernández-Chacón R., Königstorfer A., Südhof T.C. Synaptotagmin I functions as a calcium regulator of release probability. Nature. 2001;410:41–49. doi: 10.1038/35065004. [DOI] [PubMed] [Google Scholar]
- 9.Bacaj T., Wu D., Südhof T.C. Synaptotagmin-1 and synaptotagmin-7 trigger synchronous and asynchronous phases of neurotransmitter release. Neuron. 2013;80:947–959. doi: 10.1016/j.neuron.2013.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu H., Bai H., Chapman E.R. Synaptotagmin 7 functions as a Ca2+-sensor for synaptic vesicle replenishment. eLife. 2014;3:e01524. doi: 10.7554/eLife.01524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jackman S.L., Turecek J., Regehr W.G. The calcium sensor synaptotagmin 7 is required for synaptic facilitation. Nature. 2016;529:88–91. doi: 10.1038/nature16507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y., Wang P., Desir G.V. Regulation of insulin secretion and GLUT4 trafficking by the calcium sensor synaptotagmin VII. Biochem. Biophys. Res. Commun. 2007;362:658–664. doi: 10.1016/j.bbrc.2007.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Volynski K.E., Krishnakumar S.S. Synergistic control of neurotransmitter release by different members of the synaptotagmin family. Curr. Opin. Neurobiol. 2018;51:154–162. doi: 10.1016/j.conb.2018.05.006. [DOI] [PubMed] [Google Scholar]
- 14.Bhalla A., Tucker W.C., Chapman E.R. Synaptotagmin isoforms couple distinct ranges of Ca2+, Ba2+, and Sr2+ concentration to SNARE-mediated membrane fusion. Mol. Biol. Cell. 2005;16:4755–4764. doi: 10.1091/mbc.E05-04-0277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hui E., Bai J., Chapman E.R. Three distinct kinetic groupings of the synaptotagmin family: candidate sensors for rapid and delayed exocytosis. Proc. Natl. Acad. Sci. USA. 2005;102:5210–5214. doi: 10.1073/pnas.0500941102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brandt D.S., Coffman M.D., Knight J.D. Hydrophobic contributions to the membrane docking of synaptotagmin 7 C2A domain: mechanistic contrast between isoforms 1 and 7. Biochemistry. 2012;51:7654–7664. doi: 10.1021/bi3007115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Osterberg J.R., Chon N.L., Knight J.D. Membrane docking of the synaptotagmin 7 C2A domain: electron paramagnetic resonance measurements show contributions from two membrane binding loops. Biochemistry. 2015;54:5684–5695. doi: 10.1021/acs.biochem.5b00421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Voleti R., Tomchick D.R., Rizo J. Exceptionally tight membrane-binding may explain the key role of the synaptotagmin-7 C2A domain in asynchronous neurotransmitter release. Proc. Natl. Acad. Sci. USA. 2017;114:E8518–E8527. doi: 10.1073/pnas.1710708114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vermaas J.V., Tajkhorshid E. Differential membrane binding mechanics of synaptotagmin isoforms observed in atomic detail. Biochemistry. 2017;56:281–293. doi: 10.1021/acs.biochem.6b00468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sutton R.B., Davletov B.A., Sprang S.R. Structure of the first C2 domain of synaptotagmin I: a novel Ca2+/phospholipid-binding fold. Cell. 1995;80:929–938. doi: 10.1016/0092-8674(95)90296-1. [DOI] [PubMed] [Google Scholar]
- 21.Fernandez I., Araç D., Rizo J. Three-dimensional structure of the synaptotagmin 1 C2B-domain: synaptotagmin 1 as a phospholipid binding machine. Neuron. 2001;32:1057–1069. doi: 10.1016/s0896-6273(01)00548-7. [DOI] [PubMed] [Google Scholar]
- 22.Zhang X., Rizo J., Südhof T.C. Mechanism of phospholipid binding by the C2A-domain of synaptotagmin I. Biochemistry. 1998;37:12395–12403. doi: 10.1021/bi9807512. [DOI] [PubMed] [Google Scholar]
- 23.Bai J., Wang P., Chapman E.R. C2A activates a cryptic Ca2+-triggered membrane penetration activity within the C2B domain of synaptotagmin I. Proc. Natl. Acad. Sci. USA. 2002;99:1665–1670. doi: 10.1073/pnas.032541099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hui E., Bai J., Chapman E.R. Ca2+-triggered simultaneous membrane penetration of the tandem C2-domains of synaptotagmin I. Biophys. J. 2006;91:1767–1777. doi: 10.1529/biophysj.105.080325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chon N.L., Osterberg J.R., Lin H. Membrane docking of the synaptotagmin 7 C2A domain: computation reveals interplay between electrostatic and hydrophobic contributions. Biochemistry. 2015;54:5696–5711. doi: 10.1021/acs.biochem.5b00422. [DOI] [PubMed] [Google Scholar]
- 26.Bendahmane M., Bohannon K.P., Anantharam A. The synaptotagmin C2B domain calcium-binding loops modulate the rate of fusion pore expansion. Mol. Biol. Cell. 2018;29:773–880. doi: 10.1091/mbc.E17-11-0623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bai J., Tucker W.C., Chapman E.R. PIP2 increases the speed of response of synaptotagmin and steers its membrane-penetration activity toward the plasma membrane. Nat. Struct. Mol. Biol. 2004;11:36–44. doi: 10.1038/nsmb709. [DOI] [PubMed] [Google Scholar]
- 28.Radhakrishnan A., Stein A., Fasshauer D. The Ca2+ affinity of synaptotagmin 1 is markedly increased by a specific interaction of its C2B domain with phosphatidylinositol 4,5-bisphosphate. J. Biol. Chem. 2009;284:25749–25760. doi: 10.1074/jbc.M109.042499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Honigmann A., van den Bogaart G., Jahn R. Phosphatidylinositol 4,5-bisphosphate clusters act as molecular beacons for vesicle recruitment. Nat. Struct. Mol. Biol. 2013;20:679–686. doi: 10.1038/nsmb.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pérez-Lara Á., Thapa A., Jahn R. PtdInsP2 and PtdSer cooperate to trap synaptotagmin-1 to the plasma membrane in the presence of calcium. eLife. 2016;5:e15886. doi: 10.7554/eLife.15886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park Y., Seo J.B., Jahn R. Synaptotagmin-1 binds to PIP2-containing membrane but not to SNAREs at physiological ionic strength. Nat. Struct. Mol. Biol. 2015;22:815–823. doi: 10.1038/nsmb.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mackler J.M., Drummond J.A., Reist N.E. The C2B Ca2+-binding motif of synaptotagmin is required for synaptic transmission in vivo. Nature. 2002;418:340–344. doi: 10.1038/nature00846. [DOI] [PubMed] [Google Scholar]
- 33.Fernández-Chacón R., Shin O.H., Rosenmund C. Structure/function analysis of Ca2+ binding to the C2A domain of synaptotagmin 1. J. Neurosci. 2002;22:8438–8446. doi: 10.1523/JNEUROSCI.22-19-08438.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Segovia M., Alés E., Alvarez de Toledo G. Push-and-pull regulation of the fusion pore by synaptotagmin-7. Proc. Natl. Acad. Sci. USA. 2010;107:19032–19037. doi: 10.1073/pnas.1014070107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herrick D.Z., Kuo W., Cafiso D.S. Solution and membrane-bound conformations of the tandem C2A and C2B domains of synaptotagmin 1: evidence for bilayer bridging. J. Mol. Biol. 2009;390:913–923. doi: 10.1016/j.jmb.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hui E., Gaffaney J.D., Chapman E.R. Mechanism and function of synaptotagmin-mediated membrane apposition. Nat. Struct. Mol. Biol. 2011;18:813–821. doi: 10.1038/nsmb.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seven A.B., Brewer K.D., Rizo J. Prevalent mechanism of membrane bridging by synaptotagmin-1. Proc. Natl. Acad. Sci. USA. 2013;110:E3243–E3252. doi: 10.1073/pnas.1310327110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vennekate W., Schröder S., Walla P.J. Cis- and trans-membrane interactions of synaptotagmin-1. Proc. Natl. Acad. Sci. USA. 2012;109:11037–11042. doi: 10.1073/pnas.1116326109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Herrick D.Z., Sterbling S., Cafiso D.S. Position of synaptotagmin I at the membrane interface: cooperative interactions of tandem C2 domains. Biochemistry. 2006;45:9668–9674. doi: 10.1021/bi060874j. [DOI] [PubMed] [Google Scholar]
- 40.Krishnamurthy V.M., Estroff L.A., Whitesides G.M. Multivalency in ligand design. In: Jahnke W., Erlanson D.A., editors. Fragment-based Approaches in Drug Discovery. Wiley; 2006. pp. 11–54. [Google Scholar]
- 41.Takahashi H., Shahin V., Edwardson J.M. Interaction of synaptotagmin with lipid bilayers, analyzed by single-molecule force spectroscopy. Biophys. J. 2010;99:2550–2558. doi: 10.1016/j.bpj.2010.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma L., Cai Y., Zhang Y. Single-molecule force spectroscopy of protein-membrane interactions. eLife. 2017;6:e30493. doi: 10.7554/eLife.30493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bai H., Xue R., Chapman E.R. Different states of synaptotagmin regulate evoked versus spontaneous release. Nat. Commun. 2016;7:10971. doi: 10.1038/ncomms10971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vasquez J.K., Chantranuvatana K., Knight J.D. Lateral diffusion of proteins on supported lipid bilayers: additive friction of synaptotagmin 7 C2A-C2B tandem domains. Biochemistry. 2014;53:7904–7913. doi: 10.1021/bi5012223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Corbin J.A., Dirkx R.A., Falke J.J. GRP1 pleckstrin homology domain: activation parameters and novel search mechanism for rare target lipid. Biochemistry. 2004;43:16161–16173. doi: 10.1021/bi049017a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Joint Proteomics Laboratory of the Ludwig Institute for Cancer Research. Walter and Eliza Hall Institute of Medical Research, Melbourne, Australia Measuring protein concentration in the presence of nucleic acids by A280/A260: the method of Warburg and Christian. CSH Protoc. 2006;2006 doi: 10.1101/pdb.prot4252. pdb.prot4252. [DOI] [PubMed] [Google Scholar]
- 47.Nalefski E.A., Falke J.J. Use of fluorescence resonance energy transfer to monitor Ca2+-triggered membrane docking of C2 domains. Methods Mol. Biol. 2002;172:295–303. doi: 10.1385/1-59259-183-3:295. [DOI] [PubMed] [Google Scholar]
- 48.Golovanov A.P., Hautbergue G.M., Lian L.Y. A simple method for improving protein solubility and long-term stability. J. Am. Chem. Soc. 2004;126:8933–8939. doi: 10.1021/ja049297h. [DOI] [PubMed] [Google Scholar]
- 49.Ubach J., Lao Y., Rizo J. The C2B domain of synaptotagmin I is a Ca2+-binding module. Biochemistry. 2001;40:5854–5860. doi: 10.1021/bi010340c. [DOI] [PubMed] [Google Scholar]
- 50.Nalefski E.A., Slazas M.M., Falke J.J. Ca2+-signaling cycle of a membrane-docking C2 domain. Biochemistry. 1997;36:12011–12018. doi: 10.1021/bi9717340. [DOI] [PubMed] [Google Scholar]
- 51.Nalefski E.A., Wisner M.A., Falke J.J. C2 domains from different Ca2+ signaling pathways display functional and mechanistic diversity. Biochemistry. 2001;40:3089–3100. doi: 10.1021/bi001968a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Araç D., Chen X., Rizo J. Close membrane-membrane proximity induced by Ca2+-dependent multivalent binding of synaptotagmin-1 to phospholipids. Nat. Struct. Mol. Biol. 2006;13:209–217. doi: 10.1038/nsmb1056. [DOI] [PubMed] [Google Scholar]
- 53.Hamilton D.J., Coffman M.D., Reed S.M. Lipid-coated gold nanoparticles and FRET allow sensitive monitoring of liposome clustering mediated by the synaptotagmin-7 C2A domain. Langmuir. 2017;33:9222–9230. doi: 10.1021/acs.langmuir.7b01397. [DOI] [PubMed] [Google Scholar]
- 54.Frazier A.A., Wisner M.A., Cafiso D.S. Membrane orientation and position of the C2 domain from cPLA2 by site-directed spin labeling. Biochemistry. 2002;41:6282–6292. doi: 10.1021/bi0160821. [DOI] [PubMed] [Google Scholar]
- 55.Kohout S.C., Corbalán-García S., Falke J.J. C2 domain of protein kinase C alpha: elucidation of the membrane docking surface by site-directed fluorescence and spin labeling. Biochemistry. 2003;42:1254–1265. doi: 10.1021/bi026596f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Frazier A.A., Roller C.R., Cafiso D.S. Membrane-bound orientation and position of the synaptotagmin I C2A domain by site-directed spin labeling. Biochemistry. 2003;42:96–105. doi: 10.1021/bi0268145. [DOI] [PubMed] [Google Scholar]
- 57.Rufener E., Frazier A.A., Cafiso D.S. Membrane-bound orientation and position of the synaptotagmin C2B domain determined by site-directed spin labeling. Biochemistry. 2005;44:18–28. doi: 10.1021/bi048370d. [DOI] [PubMed] [Google Scholar]
- 58.Di Paolo G., De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 59.Zhao W.D., Hamid E., Wu L.G. Hemi-fused structure mediates and controls fusion and fission in live cells. Nature. 2016;534:548–552. doi: 10.1038/nature18598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Abbineni P.S., Axelrod D., Holz R.W. Visualization of expanding fusion pores in secretory cells. J. Gen. Physiol. 2018;150:1640–1646. doi: 10.1085/jgp.201812186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rickman C., Craxton M., Davletov B. Comparative analysis of tandem C2 domains from the mammalian synaptotagmin family. Biochem. J. 2004;378:681–686. doi: 10.1042/BJ20031407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chieregatti E., Chicka M.C., Baldini G. SNAP-23 functions in docking/fusion of granules at low Ca2+ Mol. Biol. Cell. 2004;15:1918–1930. doi: 10.1091/mbc.E03-09-0684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xue M., Ma C., Rizo J. The Janus-faced nature of the C2B domain is fundamental for synaptotagmin-1 function. Nat. Struct. Mol. Biol. 2008;15:1160–1168. doi: 10.1038/nsmb.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tucker W.C., Edwardson J.M., Chapman E.R. Identification of synaptotagmin effectors via acute inhibition of secretion from cracked PC12 cells. J. Cell Biol. 2003;162:199–209. doi: 10.1083/jcb.200302060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rao T.C., Passmore D.R., Anantharam A. Distinct fusion properties of synaptotagmin-1 and synaptotagmin-7 bearing dense core granules. Mol. Biol. Cell. 2014;25:2416–2427. doi: 10.1091/mbc.E14-02-0702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sugita S., Han W., Südhof T.C. Synaptotagmin VII as a plasma membrane Ca2+ sensor in exocytosis. Neuron. 2001;30:459–473. doi: 10.1016/s0896-6273(01)00290-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.