Abstract
Deregulation of KRAS4b signaling pathway has been implicated in 30% of all cancers. Membrane localization of KRAS4b is an essential step for the initiation of the downstream signaling cascades that guide various cellular mechanisms. KRAS4b plasma membrane (PM) binding is mediated by the insertion of a prenylated moiety that is attached to the terminal carboxy-methylated cysteine, in addition to electrostatic interactions of its positively charged hypervariable region with anionic lipids. Calmodulin (CaM) has been suggested to selectively bind KRAS4b to act as a negative regulator of the RAS/mitogen-activated protein kinase (MAPK) signaling pathway by displacing KRAS4b from the membrane. However, the mechanism by which CaM can recognize and displace KRAS4b from the membrane is not well understood. In this study, we employed biophysical and structural techniques to characterize this mechanism in detail. We show that KRAS4b prenylation is required for binding to CaM and that the hydrophobic pockets of CaM can accommodate the prenylated region of KRAS4b, which might represent a novel CaM-binding motif. Remarkably, prenylated KRAS4b forms a 2:1 stoichiometric complex with CaM in a nucleotide-independent manner. The interaction between prenylated KRAS4b and CaM is enthalpically driven, and electrostatic interactions also contribute to the formation of the complex. The prenylated KRAS4b terminal KSKTKC-farnesylation and carboxy-methylation is sufficient for binding and defines the minimal CaM-binding motif. This is the same region implicated in membrane and phosphodiesterase6-δ binding. Finally, we provide a structure-based docking model by which CaM binds to prenylated KRAS4b. Our data provide new insights into the KRAS4b-CaM interaction and suggest a possible mechanism whereby CaM can regulate KRAS4b membrane localization.
Introduction
RAS GTPases belong to a family of guanine-nucleotide-binding proteins that are regulators of cell function and play a key role in cell signaling. RAS proteins are regulated by guanine nucleotide exchange factors and GTPase activating proteins, enabling them to function as molecular switches, cycling between an inactive guanine diphosphate (GDP)-bound and active GTP-bound forms. In their active form, RAS proteins bind to various effectors such as RAF and phosphoinositide 3-kinase (PI3K) and leads to downstream activation of the PI3K/MAPK pathways.
The four isoforms of RAS, HRAS, NRAS, and the splice variants KRAS4a and KRAS4b, share a high degree of sequence and structural homology in their N-terminal globular domain (G-domain). However, they exhibit a wide variation in their C-terminal hypervariable region (HVR). Membrane targeting of RAS proteins is essential for initiation of the downstream signaling cascade. Membrane targeting of RAS occurs by post-translational prenylation of the C-terminal cysteine (Cys185) via farnesylation and carboxy-methylation (FMe), allowing for insertion of this lipidated tail into the plasma membrane (PM). Aside from farnesylation, additional modifications in the HVR region of the other RAS isoforms have been shown to play a role in targeting RAS proteins into different PM microdomains. Specifically, a single palmitoyl chain is added for NRAS and KRAS4a, whereas two palmitoyl chains are added to HRAS on cysteines in their respective HVR, which allows them to be associated with ordered lipid microdomains (2, 3, 4). In contrast, KRAS4b does not undergo additional lipid modification and thus is targeted to disordered lipid microdomains, in which its positively charged HVR binds to anionic lipids (5, 6).
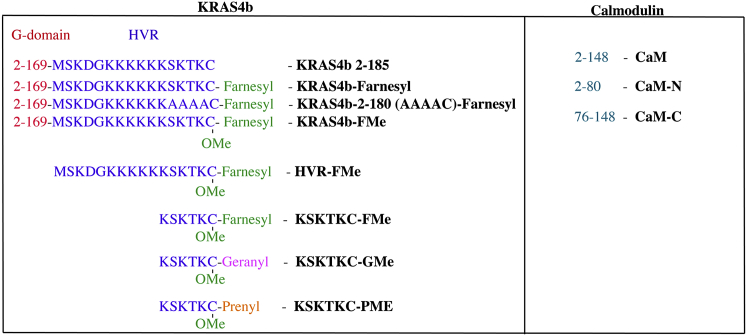
In addition to binding to anionic lipids, the HVR of KRAS4b has also been proposed to selectively bind to calmodulin (CaM) (1) and regulate KRAS activity in cancer cells (1, 7). CaM is a small ubiquitous calcium (Ca2+)-binding protein and acts as a negative regulator of the KRAS/MAPK signaling pathway by sequestering KRAS from the membrane (8). The inhibition of CaM with an antagonist in serum-starved NIH-3T3 and normal rat kidney eptithelial cells restored activation of the RAS/MAPK pathway (1). Among the four RAS isoforms, CaM was shown to selectively bind to KRAS4b in a nucleotide-dependent manner in vivo (1, 7). The mechanism by which CaM can selectively recognize only KRAS4b but not the other RAS isoforms remains unclear given their high degree of sequence and structural similarity in the G-domain. Previous studies show that CaM binds to the G-domain of KRAS4b in a nucleotide-dependent manner (9), whereas another study reveals that the HVR contains the CaM-binding domain motif (10, 11, 12). Structurally, CaM is composed of two pseudosymmetrical EF-hand domains at its N- and C-termini that are joined together by a flexible central linker (13). Binding of Ca2+ to CaM triggers a conformational change that exposes a methionine-rich hydrophobic pocket on each EF domain, enabling it to bind to a target for a specific response (14, 15). CaM-binding motifs are typically composed of short helical hydrophobic regions (16, 17), a motif that is not present in unprenylated KRAS4b (Fig. 1), suggesting that CaM binds to KRAS4b in a noncanonical manner.
Figure 1.
A schematic representation of KRAS4b and CaM domains.
To gain insight into this binding mechanism, we utilized an array of techniques to characterize the interaction between KRAS4b and CaM. Here, we demonstrate that the KRAS4b-CaM interaction is driven primarily by the FMe modification of KRAS4b and the hydrophobic pockets of CaM at the N- and C-termini. Favorable electrostatic interactions between the last five residues of KRAS4b (KSKTK) and the central linker of CaM also contribute to the binding. Interestingly, prenylated KRAS4b CaM forms a tight 2:1 stoichiometric complex with CaM in a nucleotide-independent fashion, with no detectable interaction of the G-domain. Finally, we show that CaM outcompetes a membrane mimetic for binding to KRAS4b because of its higher affinity for KRAS4b and, hence, can displace KRAS4b from the membrane.
Materials and Methods
Cloning of recombinant proteins
The expression construct for pMSP1D1 (#20061; Addgene, Cambridge, MA) was generously provided by Steve Sligar (University of Illinois at Urbana-Champaign, Champaign, IL). Gateway entry clones for human CaM were generated by standard cloning methods and incorporated an upstream tobacco etch virus 18 protease cleavage site (ENLYFQG) to permit tag removal. Entry clones include amino acids 2–148 (full-length CaM), 2–80 (N-terminal domain), or 76–148 (C-terminal domain). An avi-tagged full-length CaM clone was generated by introducing an avi sequence (LNDIFEAQKIEWG) between the tobacco etch virus (TEV) sequence and amino acids 2–148 of CaM. Sequence-validated gateway entry clones were subcloned into PDest-527 (His6 fusion) gateway destination vectors (#11518; Addgene) to produce the final Escherichia coli expression clones (19). Baculovirus expression vectors for production of fully prenylated KRAS4b have been previously described (20).
Escherichia coli expression of recombinant proteins
The BL21 STAR (rne131) E. coli strain containing the DE3 lysogen and rare transfer RNAs (pRare plasmid, CmR) was transformed with the expression plasmids (His6-TEV-POI, AmpR) for the CaM constructs and expressed as in Taylor et al. (19). For proper biotinylation of avi-Hs.CaM, a BL21 STAR DE3 (rne131) strain harboring a KanR version of pRare and a plasmid expressing biotin ligase (pBirA, CamR) were used, and 200 μM D-biotin was added 1 h before induction. pMSP1D1 was expressed as in Travers et al (48). 15N CaM was expressed as in (16) through seed growth, and then 2% of the seed culture was collected and centrifuged at 3000 × g for 10 min at 25°C. The supernatant was decanted, and the pellet was resuspended with 50 mL of T-20052 medium (25 mM Na2HPO4, 25 mM KH2PO4, 50 mM 15NH4Cl, 5 mM Na2SO4, 2 mM MgSO4, 50 μM FeCl3, 20 μM CaCl2, 10 μM MnCl2-4H2O, 10 μM ZnSO4-7H2O, 2 μM CoCl2-6H2O, 2 μM CuCl2-2H2O, 2 μM NiCl2-6H2O, 2 μM Na2MoO4-2H2O, 2 μM Na2SeO3-5H2O, 2 mM H3BO3, 216 mM glycerol, 2.8 mM glucose, and 5.6 mM α-lactose) by gentle mixing and used to inoculate 2 L of T-20052 medium in a 3-L BioFlo 110 bioreactor (Eppendorf/New Brunswick Scientific, Edison, NJ). The culture was grown at 37°C with the airflow set at 2.0 LPM while the agitation was maintained at 481 rotations per minute (rpm). When the OD600 reached 1.0–1.5 (∼7–8 h), the culture was chilled to 20°C and grown overnight (∼16 additional hours), and the cells were collected by centrifugation (5000 × g for 10 min at 4°C). The cell pellet was immediately frozen at −80°C.
Insect expression of recombinant proteins
Throughout, insect cells were grown in appropriately sized Optimum Growth Flasks (Thomson Instrument, Oceanside, CA) using serum free-900 III medium (Thermo Fisher Scientific, Carlsbad, CA). Baculovirus stocks were created by transfecting serum free-9 cultures with bacmid DNA using Cellfectin II transfection reagent (Thermo Fisher Scientific). Titers of baculovirus stocks were determined using a ViroCyt 3100 (Sartorius, Göttingen, Germany). 24 h before infection, Tni-FNL cells were set at 8 × 105 cells/mL and set to shake at 100–110 rpm at 27°C for 24 h. On the day of the infection, the cell count was determined, the virus was added to a multiplicity of infection of 3, and the infected cultures were shaken at 100–110 rpm at 21°C for 72 h. After 72 h, the cultures were centrifuged at 1100 × g for 15 min at 4°C, the supernatants discarded, and the pellets stored at −80°C.
Expression of 15N-labeled KRAS-FMe
15N culture media was prepared as per the manufacturer’s recommendations with modifications. Specifically, after bringing the solution to the correct final volume, the solution was stirred at 37°C for an additional hour, pH adjusted, filter sterilized in a biosafety cabinet hood using a 0.45–0.2 μM Sartopore 2 300 cm2 (Sartorius) inline filter, and collected in a sterile 1-L bottle. 24 h before expression, a Tni-FNL seed culture was split 1:2 with fresh nonlabeled media (CGM-2000-N) from Cambridge Isotope Laboratories (Tewksbury, MA) to keep cells in log phase growth. On the day of infection, prepared 15N-labeling insect media was prewarmed at 27°C for 2–3 h. The cell count was determined, the amount of culture necessary for expression was centrifuged at 400 × g for 6 min, the supernatant was discarded, the cell pellet was suspended in the desired volume of prewarmed 15N-labeling insect media, and the cells were shaken for 30 min at 27°C. After 30 min, the cell count was determined, the virus was added to a multiplicity of infection of 3, and the infected cultures were shaken at 100–110 rpm at 21°C for 72 h. After 72 h, the cultures were centrifuged at 1100 × g for 15 min at 4°C, the supernatants were discarded, and the pellets were stored at −80°C.
Protein purification
All proteins were purified on next generation chromatography systems (Bio-Rad Laboratories, Hercules, CA). Purification of KRAS-FMe and 15N-KRAS-FMe were as in Gillette et al. (20). avi-CaM, CaM, CaM-N (1–80), and CaM-C (76–148) were expressed and purified as noted for proteins of the form His6-MBP-TEV-POI in (21), with the addition of an ion exchange step. Specifically, after the second immobilized metal affinity chromatography column, the proteins were dialyzed overnight against 20 mM Tris-HCl (pH 8.5), 10 mM EDTA, and 1 mM tris(2-carboxyethyl) phosphinehydrochloride (TCEP) (buffer A), loaded onto a HiTrap Q HP column (GE Healthcare, Chicago, IL) equilibrated in buffer A, and eluted using a 10 column volume gradient from 0 to 350 mM NaCl in buffer A. Positive fractions were pooled, and the pools were concentrated to an appropriate volume for injection onto a 16/60 Superdex S-75 (GE Healthcare) column equilibrated and run in 20 mM HEPES (pH 7.3), 150 mM NaCl, and 5 mM CaCl2.
Nucleotide exchange
Nucleotide loading was performed by first exchanging GDP-bound KRAS4b into ammonium phosphatase buffer containing 40 mM Tris-HCl (pH 8.0), 200 mM (NH4)2SO4, and 0.1 mM ZnCl2. Next, 20 μL of alkaline phosphatase beads and a 10:1 molar ratio of nonhydrolysable analog of GTP, GMPPNP (referred to here as GNP), were then added and incubated at 25°C for 3 h. Alkaline phosphatase beads were removed by filtering it out with a 0.22-micron centrifugal filter tube and spun at 14,000 rpm for 5 min. 30 mM MgCl2 and a 5:1 molar ratio of GNP were then added and incubated at 25°C for an additional 2 h. GNP-bound KRAS4b was further purified by HiPrep 26/10 desalting column (GE Healthcare) to remove excess nucleotide. The efficiency of nucleotide exchange of KRAS was determined using high-performance liquid chromatography (HPLC). Specifically, KRAS samples were diluted into 0.1 M K2HPO4 and 1 mM tetrabutyl ammonium hydrogen sulfate (buffer A) and injected onto an Ultrasphere 5 ODS, 250 × 4.6 mm column (Hichrom, Berkshire, UK) on a Waters e2695 Alliance HPLC System (Waters, Milford, MA). Bound nucleotides were eluted using a linear gradient of buffer A containing 30% acetonitrile at a flow rate of 0.6 mL/min. Standards of GDP and GppNHp were included to validate the identity of nucleotide bound to KRAS. GNP-bound KRAS4b was routinely >95% pure as measured by this assay.
Sedimentation velocity analytical ultracentrifugation
Sedimentation velocity experiments were carried out at 50,000 rpm and 20°C on a Beckman Coulter ProteomeLab XL-I analytical ultracentrifuge (Beckman Coulter, Brea, CA) and An50-Ti rotor following standard protocols (22). Samples, as described below, were loaded in 12- or 3-mm two-channel centerpiece cells, and sedimentation data were collected using the absorbance (280 nm) and interference (655 nm) optical detection systems. Time-corrected data (23) were analyzed in SEDFIT 15.01c (24) in terms of a continuous c(s) distribution of sedimenting species with a maximal entropy regularization of 0.68. The solution density, solution viscosity, and protein partial specific volumes were calculated based on the composition in SEDNTERP (25). Protein extinction coefficients at 280 nm and refractive index increments, which were used to determine the interference signal increments, were calculated based on their composition (26). In the case of KRAS, the calculated extinction coefficients do not account for the bound nucleotide. A partial specific volume of 0.981 cm3/g and refractive index of 0.146 cm3/g was used for the lipid portion of the nanodisc (27, 28). Samples of CaM, CaM-C, CaM-N, GDP-bound KRAS4b-FMe 2-185, GNP-bound KRAS4b-FMe 2-185, GDP-bound KRAS4b 2-185, GNP-bound KRAS4b 2-185, GDP-bound KRAS4b 2-169, and GNP-bound KRAS4b 2-169 were studied in 20 mM HEPES (pH 7.2), 150 mM NaCl, 1 mM CaCl2, 1 mM MgCl2, and 1 mM TCEP. KRAS samples were analyzed individually at 5 or 10 μM, and CaM, CaM-C, and CaM-N samples were studied at concentrations in the range of 30–70 μM. Interference signals for the individual species were utilized to determine concentrations of the stock solutions as well as the corresponding absorbance extinction coefficients at 280 nm. Samples of MSP1D1 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC)/1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine (POPS) 70:30 empty nanodiscs, also in 20 mM HEPES (pH 7.2), 150 mM NaCl, 1 mM CaCl2, 1 mM MgCl2, and 1 mM TCEP, were characterized at 5 μM. Mixtures of CaM/KRAS4b-FMe, CaM-N/KRAS4b-FMe, CaM-C/KRAS4b-FMe, nanodiscs/KRAS4b-FMe, and CaM/nanodiscs:KRAS4b-FMe as described in the manuscript were prepared by mixing the appropriate quantities of stock solutions of the various components. Fresh stock solutions prepared for each experiment were first characterized by sedimentation velocity to verify purity and determine concentration. Sedimentation experiments to determine the affinity of CaM-C and CaM-N for GDP-bound KRAS4b-FMe were carried out by preparing samples of 3 μM GDP-bound KRAS4b-FMe with increasing concentrations of CaM-N or CaM-C. The c(s) distributions obtained were integrated to provide the weight-average sedimentation coefficient Sw. The absorbance and interference Sw isotherms were analyzed in terms of an A + B = AB heteroassociation model in SEDPHAT 13.0a (29) to obtain the interaction affinity. Similar experiments were carried out for CaM. Here, samples of 3 and 6 μM KRAS-4b-FMe were prepared with increasing concentrations of CaM, and samples of 6 μM CaM were prepared with increasing concentrations of GDP KRAS4b-FMe. The Sw isotherms obtained were analyzed globally in SEDPHAT 13.0a in terms of a two-site equilibrium model A + B + B = AB + B = BA + B = BAB with two nonsymmetric sites and microscopic K. In this model, sedimentation coefficients for the free species and 1:1 complexes were fixed to experimentally observed values. Affinities for the formation of the 1:1 complex with the N- and C-terminus of CaM were fixed to values determined with CaM-C and CaM-N, above. The sedimentation coefficient for the 2:1 KRAS4b-FMe/CaM complex and co-operativity parameter, reflecting the effects of bound KRAS4b-FMe on the binding of second KRAS-4b-FMe, was refined.
Analysis of the weighted-average sedimentation coefficient isotherms
The weighted-average (or signal-average) sedimentation coefficient, Sw, is a powerful tool for the study of interacting systems. It is obtained by the integration of the continuous c(s) distribution and reflects the average transport contributions from all species in solution as described in Zhao et al. (30):
| (1) |
where ck,tot is the total concentration of each of the components k, εi,λ is the absorption coefficient at wavelength λ for species i or the corresponding interference signal increment, si is the sedimentation coefficient of species i, and ci is the molar concentration of species i for the given total concentrations of the components. In the case of the A + B = AB heteroassociation between CaM-N or CaM-C with GDP-bound KRAS4b-FMe, weighted-average sedimentation coefficients were obtained at different loading concentrations and loaded into SEDPHAT 13.0a for nonlinear regression to determine the interaction affinity. To utilize SEDPHAT and combine Sw isotherms from different experiments, all sedimentation coefficients are corrected to standard conditions:
| (2) |
where ρ, η, and are the solvent density, solvent viscosity, and protein partial specific volume, with the subscripts exp and 20,w denoting the experimental and standard conditions in water at 20°C, respectively. Mass action law determines the molar concentration of each species, and in the case of the A + B = AB heteroassociation,
| (3) |
where subscripts A, B, and AB refer to CaM-C and CaM-N, GDP-bound KRAS4b-FMe, and their 1:1 complex, respectively, and KAB is the equilibrium association constant. Absorbance and interference Sw binding isotherms were globally fitted to separate forms of Eq. 3, depending on the optical detection system, to obtain KAB. Sedimentation and absorbance extinction coefficients and interference signal increments for CaM-C, CaM-N, and GDP-bound KRAS4b-FMe were kept fixed in the analysis, and the sedimentation coefficients for the 1:1 complexes converged to expected values. The formation of the 1:2 CaM/GDP KRAS4b-FMe complex was modeled in a similar manner using a two-site equilibrium model A + B + B = AB + B = BA + B = BAB with two nonsymmetric sites and microscopic K:
| (4) |
Here, distinction is made between GDP-bound KRAS4b-FMe binding to the C- (complex AB, affinity KAB) and N- (complex BA, affinity KBA) termini of full-length CaM. As above, absorbance and interference Sw binding isotherms were globally fitted to Eq. 4, this time to obtain the macroscopic binding affinity K2 describing the formation of the 1:2 complex from both 1:1 complexes. In addition to the sedimentation and absorbance extinction coefficients and interference signal increments for CaM-C, CaM-N, and GDP-bound KRAS4b-FMe, the values of microscopic association constants KAB and KBA and sedimentation coefficients sAB and sBA were kept fixed in the analysis, with the latter values obtained from the A + B = AB heteroassociation analyses of GDP-bound KRAS4b-FMe with CaM-N or CaM-C. The sedimentation coefficients for the 1:2 complex converged to expected values.
In the absence of any co-operativity, the macroscopic constant K2,
| (5) |
and differences in the refined value of K2 are expressed in terms of a cooperativity parameter. Both the A + B = AB heteroassociation and two-site equilibrium A + B + B = AB + B = BA + B = BAB with two nonsymmetric sites and microscopic K models are used as implemented in SEDPHAT.
Surface plasmon resonance
Surface plasmon resonance (SPR) binding experiments were performed on a Biacore T200 Instrument (GE Healthcare). The binding of GNP- and GDP-bound KRAS4b-FMe to CaM, CaM-C, and CaM-N were carried out as follows. Neutravidin was amine coupled to the carboxymethylated dextran surface of a CM5 sensor chip (GE Healthcare) using standard amine coupling chemistry. The surface was activated with 0.1 M N-hydroxysuccinimide and 0.4 M N-ethyl-N′-(3-dimethylaminopropyl) carbodiimide at a flow rate of 20 μL/min. Neutravidin was diluted to 20 μg/mL in 10 mM sodium acetate (pH 4.5) and injected on all four flow cells until a density of ∼5000 response units was attached. Activated amine groups were quenched with an injection of 1 M ethanolamine (pH 8.0). CaM, CaM-C, and CaM-N with an avi-tag was capture on flow cells 2, 3, and 4, respectively. Flow cell 1 was used for referencing purposes. After protein capture, a series of buffer injections were performed in the running buffer 20 mM HEPES (pH 7.2), 50,150, or 500 mM NaCl, 1 mM MgCl2, 1 mM CaCl2, and 1 mM TCEP to establish a stable baseline. KRAS4b-FMe was diluted in running buffer from 10 to 0.08 μM. At the end of the experiments, bound KRAS4b-FMe was removed by a 30-s injection of 1 M NaCl. The data were fitted using the Biacore T200 evaluation software using a single-site binding model as described below:
| (6) |
where Req is the response units measured at a specific concentration, Rmax is the maximal binding capacity of the surface, and RI is the bulk refractive index contribution in the sample. For the membrane competition experiments, anti-His6 monoclonal antibody (Abcam, Cambridge, MA) was coupled to the CM5 sensor chip and activated as described above. Approximately 2500 RU of nanodisc composed of 50% POPC and 50% POPS was captured onto flow cell 2, and flow cell 1 was used for referencing purposes. After nanodisc capture, KRAS4b-FMe at 10 μM was injected followed by 10 μM of either CaM, CaM-C, CaM-N, apo-CaM, and buffer. At the end of the injection, bound KRAS4b-FMe was removed by a 60-s injection of 100 mM phosphoric acid.
Size exclusion chromatography with multiangle light scattering
Size exclusion chromatography with multiangle light scattering (SEC-MALS) data were collected using an Agilent 1260 HPLC system (Agilent Technologies, Santa Clara, CA), attached to a WYATT DAWN HELEOS-II MALS instrument and a Wyatt Optilab rEX differential refractometer (Wyatt Technology, Santa Barbara, CA). For separation, the KRAS4b-FMe/CaM complex was loaded onto a superdex 200 10/300 column (GE Healthcare) in a buffer containing 20 mM Hepes (pH 7.2), 150 mM NaCl, 1 mM CaCl2, 1 mM MgCl2, and 1 mM TCEP. Data were analyzed using the ASTRA VI software (Wyatt Technology).
Membrane nanodisc preparation
An MSP1D1 nanodisc with a diameter of ∼100 Å was prepared as described (32). POPC and POPS (Avanti Polar Lipids, Alabaster, AL) were stored as stock chloroform solutions. After evaporation, POPC and POPS (at a molar ration of 70:30 or 50:50) were resuspended in a buffer containing 10 mM Tris (pH 7.4), 100 mM NaCl, 100 mM sodium cholate, and 0.01% sodium azide. Several freeze-thaw cycles were used to properly redissolve the phospholipids into buffer. Sodium cholate was removed from the nanodisc using Bio-Beads (Bio-Rad Laboratories). Nanodiscs were further purified by gel filtration on a superdex 200 10/300 gL column (GE Healthcare).
Circular dichroism
CaM and KSKTKC-FMe were exchanged into buffer containing 5 mM CaCl2 (pH 7.0). Proteins were quantified by ultraviolet absorbance using ε280 = 19,685 M−1cm−1. Circular dichroism (CD) spectra were collected on a Chirascan Plus (Applied Photophysics, Arlington, VA) at 25°C from 250 to 190 nm with a 1 nm step and bandwidth, time per point 1 s, 1 mm pathlength, and 0.1 mg/mL protein. Buffer blank was autosubtracted from each spectrum, and three were averaged, from which mean residue ellipticity was calculated using the concentration, pathlength, and number of amino acid residues in the polypeptide chain with the program software.
Peptide synthesis
KSKTKC-FMe, AAAAC-FMe, KSKTKC-geranyl methylated (GMe), and KSKTKC-prenyl methylated (PMe) were purchased from AnaSpec (Fremont, CA). Peptide sequences and molecular weight were confirmed via mass spectrometry.
NMR spectroscopy
NMR data were collected on an Agilent 800 MHz and Bruker Avance II 500, 600, and 900 MHz spectrometer at 35°C, processed with NMRPIPE (33), and analyzed via CCPN Analysis (18) or NMR-FAM Sparky (35). Proton, carbon and nitrogen NMR chemical shifts for CaM are reported elsewhere (36, 37). Proton, carbon, and nitrogen chemical shifts for KRAS4b are reported here (31). The backbone resonances of CaM bound to KRAS4b-FMe were assigned using standard triple resonance data (HNCACB and CBCACONH), and 15N-edited HSQC-NOESY data were collected at 35°C on 250 μM samples in a buffer containing 20 mM HEPES (pH 6.5), 100 mM NaCl, 5 mM CaCl2, 1 mM MgCl2, and 1 mM TCEP-HCl. Combined 1H-15N chemical shift changes were calculated as ΔδHN (ppm) = [(ΔδH)2 + (ΔδN/5)2]1/2, where ΔδH and ΔδN are the 1H and 15N chemical shift changes, respectively. The criterion for the selection of significantly affected residues was an average plus one SD of all ΔδHN values.
Isothermal titration calorimetry
The binding affinities of KRAS4b-FMe, HVR-FMe, KSKTKC-FMe, AAAAC-FMe, KSKTKC-GMe, and KSKTKC-PMe were determined using a MicroCal PEAQ-isothermal titration calorimetry (ITC) (Malvern Panalytical, Westborough, MA). All ITC experiments were performed in a buffer containing 20 mM HEPES (pH 7.2), 150 mM NaCl, 1 mM MgCl2, 1 mM CaCl2, and 1 mM TCEP-HCL. CaM at either 200 or 500 μM or 1 mM was titrated into 50 μM of KRAS4b-FMe, HVR-FMe, and KSKTKC-FMe, 30 μM of KSKTKC-GMe, and 25 μM of AAAAC-FMe and KSKTKC-PMe. The heat of the reaction was measured at 25°C for 19 injections, and the heats of dilution were measured by titrating CaM into buffer. Data analysis was performed using a nonlinear least square algorithm incorporated into the MicroCal PEAQ-ITC analysis software applying the “single set of identical sites” fitting model (Malvern Panalytical).
Structural docking
The complex structure was manually modeled in Coot (38) and rendered in Pymol (Schrödinger, New York, NY).
Results and Discussion
Prenylation is required for the binding of KRAS4b to CaM, and the interaction is nucleotide independent
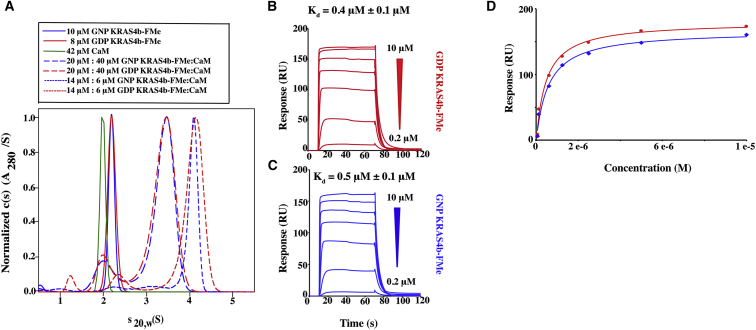
To survey the interaction of CaM with KRAS4b, 30 μM CaM and 20 μM of GDP-bound and GNP-bound unprenylated and preynlated KRAS4b (Fig. 1; Fig. S1) were mixed and analyzed by sedimentation velocity analytical ultracentrifugation (SV-AUC). Each batch of protein used was individually characterized and found to be monodisperse and monomeric with sedimentation coefficients and estimated molar masses shown in Table S1. In contrast to previously published data, which show that CaM binds to unprenylated forms of KRAS4b (10, 11, 12), our studies detect no binding between CaM and unprenylated KRAS4b (Fig. S1). Evidence for complex formation was found only with GDP-bound and GNP-bound prenylated KRAS4b-FMe (Fig. 2 A; Fig. S1 B). Furthermore, the equivalent sedimenting c(s) values of both GDP- and GNP-bound KRAS4b-FMe/CaM complexes suggest that the interaction is nucleotide independent (Fig. 2 A). To investigate the nucleotide-independent interaction, binding affinities of GDP-bound KRAS4b-FMe and GNP-bound KRAS4b-FMe to CaM were obtained via SPR. As shown in Fig. 2, B–D, GDP-bound KRAS4b-FMe and GNP-bound KRAS4b-FMe loaded onto avi-CaM yielded Kd values of 0.4 and 0.5 μM, respectively. Because these binding affinities are similar, the SPR data support that CaM binds to KRAS4b-FMe in a nucleotide-independent manner.
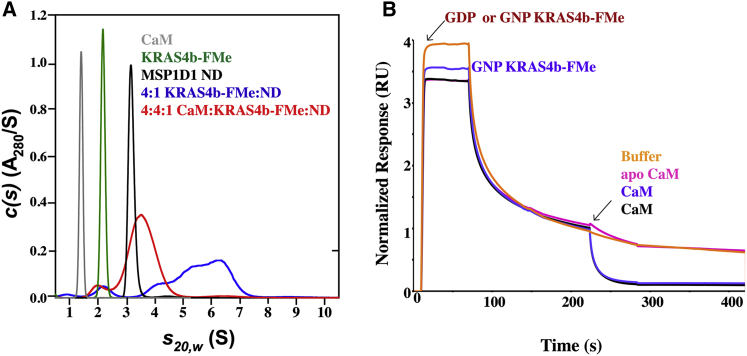
Figure 2.
GDP- and GNP-bound KRAS4b-FMe binds to CaM in a nucleotide-independent manner. (A) Shown are the sedimentation velocity and normalized absorbance c(s) profiles for solutions containing 42 μM CaM (green), 8 μM GDP-bound KRAS4b-FMe (red), and 10 μM GNP-bound KRAS4b-FMe (blue). C(s) distributions for mixtures of 20 μM KRAS4b-FMe and 40 μM CaM as well as 14 μM KRAS4b-FMe and 6 μM CaM are shown as long dash and short dash plots, respectively. Red plots are data for GDP-bound KRAS4b-FMe, and blue plots are for GNP-bound KRAS4b-FMe. (B and C) Shown are SPR binding kinetics of 10–0.2 μM GDP- and GNP-bound KRAS4b-FMe to avi-CaM. Data were fit and yielded Kd values of 0.4 and 0.5 μM, respectively. (D) Fits of the steady-state binding isotherms derived from the SPR data are shown.
KRAS4b-FMe binds to CaM with a 2:1 stoichiometry
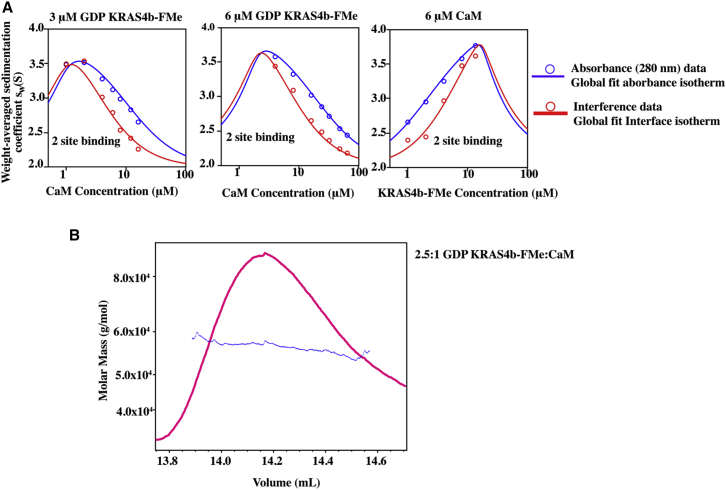
Previous reports indicate that unprenylated KRAS4b binds to CaM with a 1:1 stoichiometry (10, 11, 12). Yet, as demonstrated in Fig. 2 A, the sedimentation properties of the faster sedimenting species observed correlate with the CaM and KRAS4b loading concentrations and their ratios, suggesting that the KRAS4b-FMe-CaM stoichiometry is not 1:1. We therefore employed SV-AUC and SEC-MALS to characterize the stoichiometric states of the KRAS4b-FMe-CaM complex in solution. SV-AUC experiments carried out with mixtures of 20 μM KRAS4b-FMe and 40 μM CaM show a predominant species at 4.06 S, with an estimated mass of 59.3 kDa consistent with a 2:1 KRAS4b-FMe-CaM complex (Fig. S2 A). Various titrations were carried out with full-length CaM (Fig. S2 A), all of which supported the binding of the 2:1 stoichiometry. The combined Sw isotherms were fit to a two-site binding model, in which the sites are not equivalent, corresponding to the two domains of CaM. The microscopic binding constants for these sites were fixed to values determined individually with CaM-N and CaM-C (see below) to obtain a favorable cooperative term of −1.9 kcal/mol (Fig. 3 A), suggesting either a KRAS4b-FMe/KRAS4b-FMe interaction within the complex or a CaM conformational change that favors the binding of a second KRAS4b-FMe. Poor fits were obtained when these isotherms were modeled for a 1:1 interaction.
Figure 3.
KRAS4b-FMe forms a 2:1 complex with CaM. (A) Shown are the absorbance (blue) and interference (red) Sw isotherms for the titration of CaM into 3 μM GDP-bound KRAS4b-FMe (left panel) and 6 μM GDP-bound KRAS4b-FMe (center panel) as well as the titration of GDP-bound KRAS4b-FMe into 6 μM CaM (right panel). Experimental data (circles) were fitted globally to a two-site equilibrium model with two nonsymmetric sites and microscopic K as described in the text to obtain the isotherms shown (solid lines). (B) MALS analysis was performed on GDP-bound KRAS4b-FMe-CaM complex eluted from an SEC column. The absolute molar mass versus elution time displays a monodisperse peak at 57 kDa.
SEC-MALS is a complementary method to SV-AUC for accurate molar mass determination that is independent of the shape and elution time from the column. Therefore, to validate the mass of the complex, a mixture of 40 μM CaM and 100 μM KRAS4b-FMe was characterized by SEC-MALS and yielded an experimental molar mass of 57 kDa for the monodisperse complex, consistent with a 2:1 KRAS4b-FMe/CaM complex as observed in the SV-AUC experiments (Fig. 3 B).
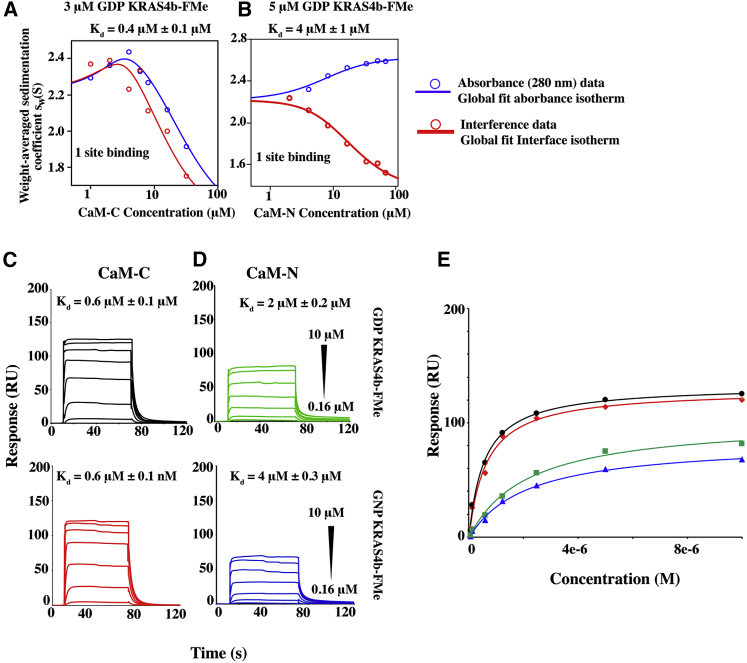
Conclusively, to establish whether CaM binds to two species of KRAS4b-FMe, we truncated CaM into two individual subdomains, CaM-N and CaM-C, and tested their binding to KRAS4b-FMe. SV-AUC experiments in which increasing amounts of CaM-C or CaM-N were added to 3 or 5 μM GDP-bound KRAS4b-FMe, respectively, led to c(s) profiles consistent with the formation of a 1:1 complex (Fig. S2 B). Fitting the Sw isotherms using a 1:1 binding model returned an affinity (Kd) of 0.4 μM for CaM-C binding to GDP-bound KRAS4b-FMe (Fig. 4 A). Similar studies with CaM-N (Fig. S2 C) indicated a weaker binding affinity of 4 μM (Fig. 4 B). Comparable affinities of CaM-C and CaM-N binding to GDP-bound and GNP-bound KRAS4b-FMe were also obtained via SPR, as seen in Fig. 4, C–E and Table S2. No differences in binding affinity were observed between CaM-C and CaM-N with GDP-bound and GNP-bound KRAS4b-FMe, further supporting nucleotide-independent binding. Collectively, our data demonstrate that KRAS4b-FMe binds to CaM with a 2:1 stoichiometry.
Figure 4.
KRAS4b-FMe forms a 1:1 complex with CaM-N and CaM-C. (A) Shown are the absorbance (blue) and interference (red) Sw isotherms for the titration of CaM-C into 3 μM GDP-bound KRAS4b-FMe. Experimental data (circles) were fitted to an A + B = AB heteroassociation model to obtain the best-fit isotherms shown (solid lines). (B) Shown are the absorbance (blue) and interference (red) Sw isotherms for the titration of CaM-N into 3 μM GDP-bound KRAS4b-FMe. Experimental data (circles) were fitted to an A + B = AB heteroassociation model to obtain the best-fit isotherms shown (solid lines). Sedimentation velocity c(s) profiles for these isotherms are shown in Supporting Materials and Methods. (C) Shown are SPR binding kinetics of 10–0.2 μM GDP and GNP-bound KRAS4b-FMe to avi-CaM-C. Data were fit using a one-site model and yielded Kd values of ∼0.6 μM for both GDP- and GNP-bound KRAS4b-FMe to CaM-C. (D) Shown are SPR binding kinetics of 10–0.2 μM GDP- and GNP-bound KRAS4b-FMe to avi-CaM-N. Data were fit using a one-site model and yielded Kd values of ∼2 μM for both GDP- and GNP-bound KRAS4b-FMe to CaM-N. (E) Fits of the steady-state binding isotherms derived from the SPR data are shown.
Electrostatic interactions also contribute to KRAS4b-FMe/CaM interaction
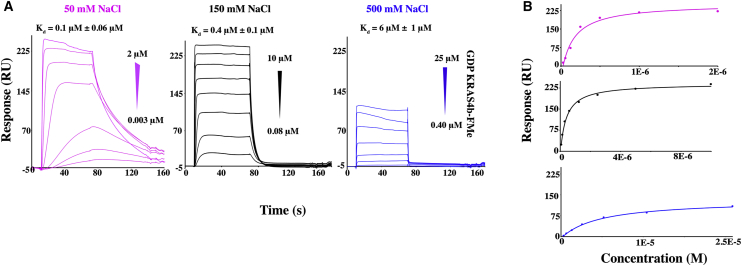
In addition to the confirmed interaction between the hydrophobic farnesyl group of KRAS4b and the hydrophobic pockets of CaM, the acidic central linker of CaM has been shown to play an essential role in the stabilization of CaM-protein complexes (39, 40). Furthermore, KRAS4b contains a highly basic HVR region (Fig. 1), which may bind to the central acidic linker of CaM. Although our AUC and ITC data so far indicate that CaM does not bind to unprenylated KRAS4b (Fig. S1), we wanted to determine what role, if any, additional HVR electrostatic interactions may contribute to CaM binding of KRAS4b. To modulate the strength of electrostatic interaction of this, we injected KRAS4b-FMe onto avi-CaM at 50, 150, and 500 mM NaCl (Fig. 5 A) and show that the Kd values decrease by ∼30-fold over this salt concentration (Fig. 5 B). Therefore, although the FMe is required for binding, our data indicate that favorable electrostatic interactions also contribute to the stabilization of the KRAS4b-FMe/CaM complex.
Figure 5.
Electrostatic interactions contribute to KRAS4b-FMe/CaM binding. (A) Steady-state binding of GDP-bound KRAS4b-FMe to avi-CaM at 50, 150, and 500 mM NaCl is shown. (B) Data were fit using a one-site model and yielded Kd values of 0.1, 0.4, and 6 μM, respectively.
HVR-FMe contains the CaM-binding motif
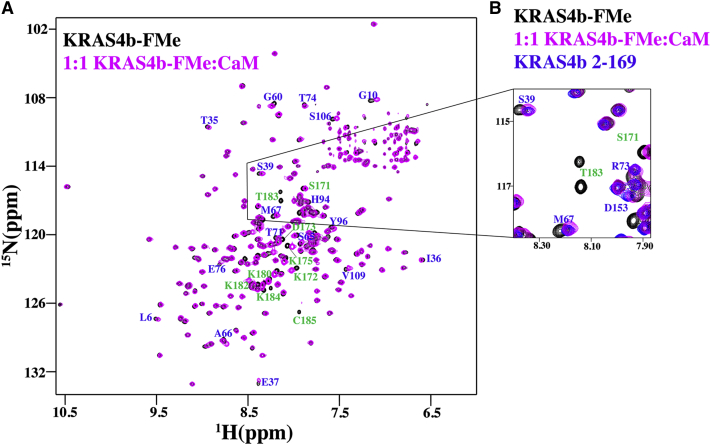
To identify the binding interface between KRAS4b-FMe and CaM binding, we have utilized NMR chemical shift perturbations (CSPs) detected by two-dimensional (2D) 1H-15N heteronuclear single quantum coherence (HSQC) experiments. 2D HSQC experiments can be used to determine residues that are involved in the binding interface in addition to any conformational changes that might occur upon binding. Consistent with our AUC and ITC data (Fig. S1), no CSP or line broadening was observed when 500 μM CaM was added to 50 μM 15N-labeled unprenylated GDP- and GNP-bound KRAS4b (Fig. S3). Because of the molecular weight of the KRAS4b-FMe/CaM complex (∼57 kDa), we collected 2D 1H-15N transverse relaxation optimized spectroscopy (TROSY)-HSQC tailored toward protein(s) with relatively high molecular weights. NMR data were collected only for 15N GDP-bound KRAS4b-FMe titrated with CaM because our earlier data demonstrated that CaM binds to KRAS4b-FMe in a nucleotide-independent manner (Figs. 2 and 4). The 1H-15N TROSY-HSQC data obtained upon titration of CaM into 15N GDP-bound KRAS4b-FMe are shown in Fig. 6 A. Peaks arising from the FMe region of KRAS4b are not observed in these HSQC experiments; however, modest CSPs (>0.05) are observed for subsets of signals corresponding to the peaks within the HVR (green). The most perturbed region (K180, S181, K182, T183, K184, C185) (Fig. 6 A) corresponds to the residues proximal to the FMe moiety.
Figure 6.
CaM disrupts KRAS4b G-domain-HVR interaction. (A) 2D 1H-15N TROSY-HSQC spectra were obtained from GDP-bound 15N-labeled KRAS4b-FMe (70 μM) in the free (black) and CaM-bound (magenta) states. Amide signals that exhibited chemical shift changes in the G-domain of KRAS4b (blue) and HVR (green) are highlighted. (B) Selected region of 2D 1H-15N TROSY-HSQC spectra was obtained of a GDP-bound 15N-labeled KRAS4b-FMe in the free (black) overlaid with CaM-bound (magenta) states and free GDP-bound 15N-labeled KRAS 2-169 (blue).
Although our data infer that CaM primarily binds to the HVR-FMe region of KRAS4b, an additional subset of peaks within the G-domain (blue) are also perturbed upon CaM binding. These residues (L6, G10, T35, I36, E37, S39, G60, Y64, S65, A66, M67, Q70, E76, T71, T74, H94, Y96, S106, and V109), which correspond to switch I, switch II, and helix 2 and 3 regions of KRAS4b, have been proposed to be within the interface of the KRAS4b G-domain/HVR interaction (31). Therefore, the perturbations observed within these regions might be attributed to some disruption of KRAS4b G-domain-HVR interaction by the binding of CaM to the farnesyl group and a part of the HVR. To evaluate this, 1H-15N HSQC spectra was collected for free 15N KRAS4b 2-169 (blue) and overlaid with the spectra of free 15N KRAS4b 2-185-FMe (black) and 1:1 15N KRAS4b-2-185-FMe/CaM spectra (magenta) (Fig. 6 B). The perturbed G-domain peaks in the 15N KRAS4b-FMe-CaM complex (magenta) matches with those of free 15N GDP-bound KRAS4b 2-169 (blue), indicating that the perturbations observed within the G-domain are caused by CaM sequestering the HVR-FMe away from the G-domain.
KSKTKC-FMe is the minimal CaM-binding motif
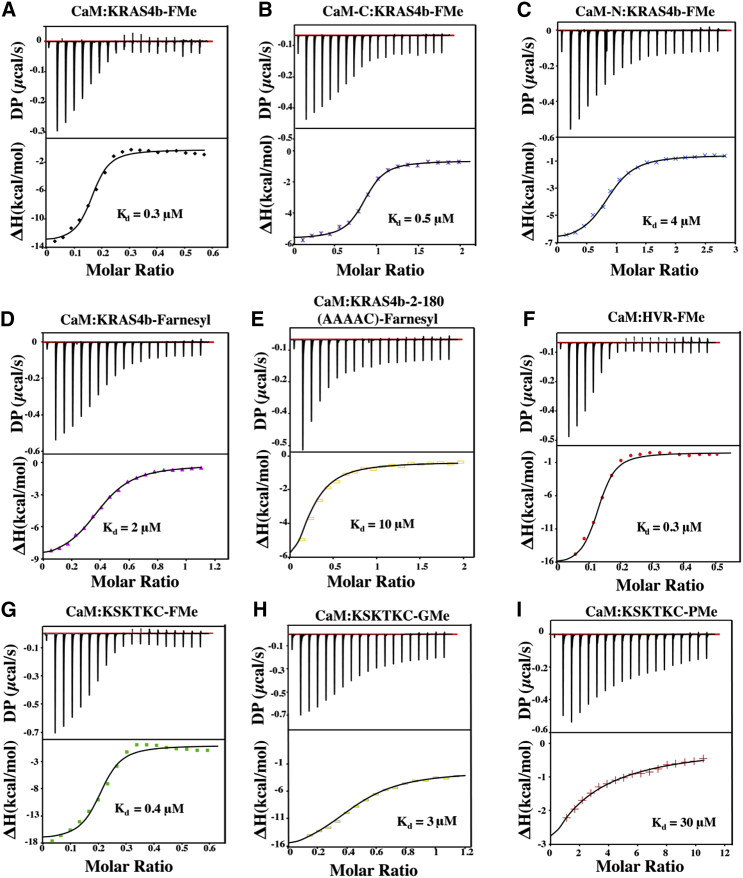
Although we do not observe the binding of CaM to unprenylated KRAS4b, our NMR data show that the HVR is perturbed upon CaM binding (Fig. 6 A). This observation might be ascribed to two factors: favorable electrostatic interactions that occur upon CaM binding to the FMe and/or CaM-induced conformational changes within the HVR upon binding to FMe. We have shown that electrostatic interactions contribute to the stabilization of the complex (Fig. 5). To gain insight into which part of the HVR is involved in CaM binding, we carried out ITC experiments in which CaM was titrated into various KRAS4b constructs, including a peptide representing the most perturbed region near the farnesyl group KSKTKC-FMe based on the NMR measurements. In addition to providing binding affinities, ITC also measures thermodynamic properties, which provide insight into the noncovalent forces governing the interaction. ITC data collected for the titration of CaM into KRAS4b-FMe clearly support the binding of more than one KRAS4b-FMe to CaM (Fig. 7 A). Unfortunately, we were unable to model our data in terms of two nonsymmetric sites. We therefore took advantage of the pseudosymmetrical nature of the CaM-N and the CaM-C lobe and were able to fit our ITC data of CaM binding to KRAS4b variants using the PEAQ-ITC single set of identical sites fitting the model to obtain apparent binding affinities (Table S3). As demonstrated in Fig. 7 A, the binding of CaM to KRAS4b-FMe is exothermic with an effective Kd of 0.3 μM. The binding affinity measured using ITC is consistent with that obtained from AUC and SPR (Kd values ranging from ∼0.3 to 0.4 μM for KRAS4b-FMe binding to CaM). To further establish the affinities obtained from the AUC and SPR that show that CaM-C binds better than CaM-N to KRAS4b-FMe, we collected data upon titration of CaM-C or CaM-N into GDP-bound KRAS4b-FMe. As shown in Fig. 7, B and C and Table S2, binding affinities obtained from ITC are consistent with what we obtained from AUC and SPR. To investigate whether carboxymethylation also played a role in the KRAS4b/CaM interaction, we titrated CaM into KRAS-farnesyl and obtained a weaker binding affinity of 1.5 μM, a fourfold decrease form KRAS4b-FMe, indicating that the carboxymethylation is required for the interaction of KRAS4b and CaM (Fig. 7 D). To determine whether the penultimate five residues of the HVR (KSKTK) were crucial for binding, we mutated those residues to alanine in full-length KRAS4b-farnesyl and tested binding via ITC. Titration of CaM into KRAS4b-2-180 (AAAAC)-farnesyl led to a weaker binding affinity of 10 μM, demonstrating that the last five residues of HVR are critical for CaM binding (Fig. 7 E). Titration of CaM into HVR-FMe and KSKTKC-FMe are also exothermic and enthalpically driven with binding affinities of 0.3 and 0.4 μM, respectively (Fig. 7, F and G). The same binding affinity is measured for KSKTKC-FMe, HVR-FMe, and KRAS4b-FMe binding to CaM, consistent with our data indicating no role of the G-domain in binding to CaM.
Figure 7.
KSKTKC-FMe is the minimal CaM-binding domain. (A) ITC data obtained for GDP-bound KRAS4b-FMe binding to CaM. (B) ITC data obtained for GDP-bound KRAS4b-FMe binding to CaM-C. (C) ITC data obtained for GDP-bound KRAS4b-FMe binding to CaM-N. ITC data obtained for (D) GDP-bound KRAS4b-farnesyl, (E) GDP-bound KRAS4b 2-180 (AAAAC)-farnesyl, (F) HVR-FMe, (G) KSKTKC-FMe, (H) KSKTKC-GMe, (I) and KSTKTC-PMe binding to CaM. Fit data yielded Kd values of 0.3, 0.5, 4, 2, 10, 0.3, 0.4, 3, and 30 μM, respectively.
Lastly, to determine whether CaM could accommodate the whole farnesyl group, we have generated two different constructs, KSKTKC-PMe, which contains a prenyl group, 10 hydrocarbon units shorter than the farnesyl group, and KSKTC-GMe, which contains a geranyl group, five hydrocarbon units shorter than the farnesyl group. As shown in Fig. 7, H and I, the decrease in affinity from FMe-GMe-PMe (0.4 μM − 3 μM − 30 μM) demonstrates that CaM binds most tightly to the farnesyl group, indicating that it accommodates the entire farnesyl moiety. Collectively, our data show that CaM specifically binds the farnesyl group, and the primary binding determinant in KRAS4b appears to be KSKTKC-FMe.
KRAS4b-FMe induces major changes in CaM
To identify the CaM-interacting interface, we collected NMR data of the reverse HSQC experiments with 15N-labeled CaM constructs titrated with unlabeled KRAS4b. To confirm our data that demonstrate that KRAS4b-FMe binds to both lobes of CaM, we collected NMR data upon the titration of GDP-bound KRAS-FMe into CaM-C and CaM-N (Fig. S4). Comparison of the CSP obtained from the titration experiment suggests that CaM-C contains a larger interface than CaM-N (Fig. S4, C–F). This could likely explain why KRAS4b-FMe has a higher affinity to CaM-C over CaM-N. Next, to further confirm whether prenylation is required for KRAS4b to bind to full-length CaM, we titrated both unprenylated and prenylated GDP-bound and GNP-bound KRAS4b into full-length CaM. As expected, titration of 500 μM of unprenylated GDP-bound KRAS4b and GNP-bound KRAS4b into 50 μM 15N CaM did not show any perturbations in the HSQC spectra (Fig. S5).
Because of the relatively large size of the complex (∼57 kDa), titration of KRAS4b-FMe into 15N-CaM led to severe line broadening and the loss of most of the signals (data not shown). Therefore, the HSQC data were collected with the minimal CaM-binding motif of KRAS4b and KSKTKC-FMe to probe into CaM/KSKTKC-FMe interaction.
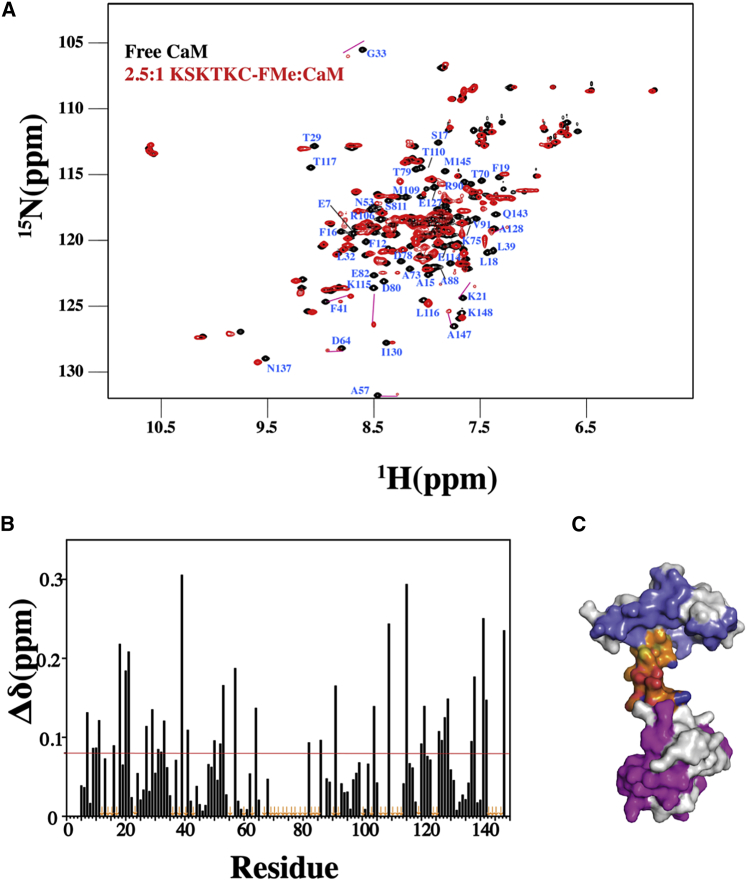
As shown in Fig. 8, most CaM peaks exhibit extensive CSP upon titration with KSKTKC-FMe. The affected regions correspond to residues located throughout the CaM structure (Fig. 8, B and C). Also, severe line broadening led to the disappearance of most the signals of the central linker of CaM (orange) and regions around the hydrophobic pockets of both CaM-N and CaM-C (Fig. 8 B). Based on the relatively small size of the KSTKC-FMe/CaM complex (∼17 kDa), the loss of signals may be accounted for by some of the following factors: an intermediate and/or slow exchange between the free and bound states on an NMR timescale, a large conformational change reflecting secondary-tertiary structural rearrangement, or a global allosteric change in the conformation of CaM upon binding to KSKTKC-FMe. To determine which factor contributes to the severe broadening, we first titrated KSKTKC-FMe up to 10:1 into 15N CaM. However, no further changes in the spectra were observed with the titration of additional KSKTKC-FMe. Furthermore, varying the temperature from 16 to 50°C did not lead to clear improvement of the signal intensity (data not shown), suggesting that the line broadening is not due to the complex being in an intermediate and/or slow exchange form.
Figure 8.
KRAS4b-FMe induces major changes in CaM. (A) 2D 1H-15N TROSY-HSQC spectra obtained of a 15N-labeled CaM (200 μM) in the free (black) and KSKTKC-FMe bound (red states) are shown. The amide signals that exhibited significant chemical shift changes correspond to residues throughout the N- and C-terminal lobes as well as the central linker of CaM. (B) Shown is a histogram of normalized 1H-15N chemical shift changes versus residue number calculated from the HSQC spectra for CaM upon the addition of KSKTKC-FMe. (C) Shown is the surface representation of CaM structure (Protein Data Bank (PDB): 3CLN) highlighting residues that exhibited substantial (>0.1 ppm) chemical shift changes. Residues that were significantly perturbed in CaM-N (blue), central linker (orange), and CaM-C (magenta) regions have been highlighted.
Next, to find out whether binding of KSKTKC-FMe to CaM leads to secondary structural rearrangement of CaM, we collected CD data of CaM and the KSKTKC-FMe/CaM complex. As shown in Fig. S6, CaM maintains its helical structure upon interacting with KSTKTC-FMe. This indicates that KSKTKC-FMe does not induce secondary structural rearrangements in CaM, and therefore, we postulate that the effect we observed in the HSQC spectra is most likely due to a global conformational change in CaM.
CaM outcompetes membrane mimetics for binding to KRAS4b-FMe
The CaM-binding motif, KSKTKC-FMe, is also part of the HVR that interacts with membranes (5, 6). Therefore, we wanted to assess whether CaM could compete with the membrane for KRAS4b-FMe binding. Competition experiments between CaM and membranes for KRAS4b-FMe binding were carried out using AUC and SPR. First, SV-AUC experiments on mixtures of KRAS4b-FMe and nanodiscs composed of MSP1D1 70:30 POPC/POPS at a 4:1 molar ratio resulted in c(s) profiles that show fast sedimenting complexes at 4–7 S (Fig. 9 A). These appear to be 1:1, 2:1, and higher-order complexes of KRAS4b-FMe bound to the nanodisc. The addition of 1:1 equivalents of CaM (per KRAS4b-FMe) appeared to abrogate the interaction of KRAS4b-FMe with the nanodisc, based on the loss of the faster sedimenting material, suggesting that CaM can compete with the membrane for bound KRAS4b-FMe. The loss of the faster sedimenting species implies that CaM can remove bound KRAS4b-FMe from the membrane and/or sequesters the FMe away from rebinding.
Figure 9.
CaM displaces membrane-bound GDP- and GNP-bound KRAS4b-FMe. (A) Left: shown are the sedimentation velocity absorbance c(s) profiles for 42 μM CaM (gray), 9 μM GDP-bound KRAS4b-FMe (green), 4.5 μM MSP1D1 70:30 POPC/POPS nanodisc (black), and mixtures of 2.5 μM nanodisc and 10 μM GDP-bound KRAS4b-FMe without (blue) and with 11 μM CaM (red). Shown are the sedimentation velocity absorbance c(s) profiles for mixtures of 10 μM nanodisc and 40 μM GDP-bound KRAS4b-FMe without (purple) and with 44 μM CaM (red). (B) Shown is a normalized SPR sensogram of single injection of GDP- and GNP-bound KRAS4b-FMe (20 μM) onto immobilized MSP1D1: 50:50 POPC/POPS nanodisc followed by injection of buffer (orange), apo-CaM (magenta), CaM (black) onto GDP-bound nanodisc, and CaM (blue) onto GNP-bound nanodisc. Notice that CaM can displace KRAS4b-FMe from the membrane, whereas buffer and apo-CaM do not.
To further examine this, we used SPR to determine if CaM could extract KRAS4b-FMe that was prebound to a nanodisc. Previous work by Sperlich et al. showed that CaM could extract GDP-bound KRAS4b-FMe but not GNP-bound KRAS4b-FMe from liposomes (41). Based on our data that show the nucleotide-independent binding of CaM to KRAS, this result may be due to the large size and heterogeneity of the liposome preparations. The advantage of using nanodiscs over liposomes is that nanodiscs are homogenous, and the lipid ratio can be precisely controlled (32). To validate this experimental design, we first injected buffer and apo-CaM onto KRAS4b-FMe bound to nanodiscs (Fig. 9 B). No change in the dissociation rate of the KRAS-FMe from the nanodisc was observed, implying that apo-CaM are unable to disrupt the KRAS4b-FMe/nanodisc complex. However, the addition of CaM onto GDP- and GNP-bound forms of KRAS4b-FMe prebound to immobilized nanodiscs led to a significant increase in the dissociation rate of KRAS-FMe, resulting in an almost complete loss of KRAS-FMe from the nanodisc, suggesting that CaM can displace KRAS4b-FMe in a nucleotide-independent manner.
Conclusions
Membrane localization of KRAS4b is required for its initiation of the downstream signaling pathways. The farnesyl chain, in addition to the positively charged HVR of KRAS4b, is essential for the KRAS4b-membrane interaction (5, 6). Among all the RAS isoforms, CaM was shown to exclusively bind to KRAS4b and displace it from the membrane (5, 6, 7). Before this study, the mechanism for this process was not well established. Until the recent high-yield production of fully prenylated KRAS4b-FMe (20), previous studies attempting to characterize KRAS4b-CaM interaction used only the G-domain or the unprenylated form of KRAS4b (7, 8, 9, 10). In other methods for producing prenylated KRAS4b protein, S-farnesylation of the C-terminal cysteine was synthesized by a combination of expressed protein ligation and lipopeptide synthesis (9, 33). However, KRAS proteins produced by these methods may not accurately represent the prenylated form found in mammalian cells.
In this work, we believe that we present the first study that characterizes the interaction between fully prenylated KRAS4b-FMe and CaM using multiple biochemical, biophysical, and structural techniques. We have shown that CaM binds tightly to KRAS4b-FMe in a nucleotide-independent manner. Our observation of the nucleotide-independent binding mode was unexpected based on the previous publications that reported a nucleotide-dependent binding event between KRAS4b and CaM (7, 8, 9, 10). However, given the high degree of sequence and structural homology in the G-domain of all RAS isoforms, the ability for CaM to selectively bind to KRAS4b suggests that the recognition motif is likely to be within the HVR. To support this hypothesis, we present data that demonstrate that the G-domain is not involved in the KRAS4b-FMe-CaM interaction but rather the farnesyl moiety is the primary determinant for KRAS4b binding to CaM (Fig. 2).
A noteworthy finding obtained in our current study was that KRAS4b-FMe forms a 2:1 complex with CaM (Figs. 2 and 3). This finding was unpredicted, but considering the pseudosymmetrical nature of the N- and C-termini of CaM, the hydrophobic pockets of CaM-N and CaM-C subdomain could each accommodate one farnesyl group of KRAS4b. In many CaM-binding motifs, the CaM-C lobe has been shown to bind tighter to targets than the CaM-N lobe (39, 40, 42). Here, we also demonstrate that the affinity for KRAS4b-FMe to CaM-C is much tighter than CaM-N (Fig. 4). Based on the cooperative factors obtained for CaM binding to KRAS4b-FMe from our AUC data, we postulate that KRAS4b-FMe binds to the CaM-C lobe first and induces conformational changes in CaM, which then enhances the binding of a second KRAS4b-FMe to the CaM-N lobe.
It has been widely reported that CaM-binding targets tend to be short helical and hydrophobic peptides, but emerging studies show that CaM binds to other targets in noncanonical ways (16, 17). It was previously shown that CaM binds to the lipidated myristoyl peptide corresponding to the N-terminal domain of CAP-23/NAP (22, 43). Recently, it was reported that CaM interacts with two loops on Akt pleckstrin homology domain adjacent to its lipid binding site (39, 44). Here, we believe that CaM can accommodate the entire farnesyl moiety (Fig. 7), which represents another novel CaM-binding mode. In addition to the binding of the farnesyl moiety to the hydrophobic pockets of CaM, the last five residues of KRAS4b (KSKTK) were shown to be crucial for the KRAS4b-FMe-CaM interaction because mutation of these residues to alanine attenuated CaM binding (Fig. 7). These mostly basic residues that are replaced by neutral residues in HRAS (CMSCK) and NRAS (CMGLP) also contain an additional palmitoyl moiety on another Cys residue located in that region. The placement of this extra palmitoyl group close to the CaM-binding site could potentially sterically hinder and prevent CaM from binding to these proteins. A combination of the neutral amino acids and the possible steric clash with the palmitoyl group could likely explain why CaM binds to KRAS4b but not the other RAS isoforms.
Interestingly, the CaM-binding domain KSKTKC-FMe represents the same motif shown in the structure of KRAS4b-FMe binding to phosphodiesterase6-δ (PDE6δ) (21). PDE6δ has been shown to play a primary role in KRAS trafficking and membrane localization in cells by binding the prenylated HVR and sequestering it away from the cytosol (45). There is evidence that KRAS still gets transported to the PM when PDE6δ is depleted in cells (46). This infers that other proteins bind and transport KRAS from the cytoplasm to the PM in the absence of PDE6δ. In support of this, CaM was shown to translocate KRAS but not HRAS from the PM to the Golgi complex and then to early-recycling endosomes in response to neuronal activity in hippocampal neuronal cells (Fig. 7). The recent crystal structure of KRAS4b-FMe in complex with PDE6δ revealed a large hydrophobic pocket, which binds to the entire farnesyl moiety and thus is able to sequester it away from the cytosol (21).
This implies that KRAS trafficking partners must contain a hydrophobic pocket that can accommodate the farnesyl chain to traffic it from the endoplasmic reticulum to the PM and vice versa. Our ITC data show that the binding affinity of CaM to KRAS4b is tightest with the farnesyl compared to the 10-carbon geranyl and five-carbon prenyl group, respectively (Fig. 7). This indicates that CaM is capable of binding to the entire farnesyl group (Fig. 10) to sequester it away from the cytoplasm just like PDE6δ. Our docking model suggests that CaM can indeed accommodate the entire farnesyl group. In addition, the last five residues of KRAS4b could make a favorable electrostatic interaction with the acidic regions of CaM (Fig. 10). Based on this, we hypothesize that CaM may play an additional role in translocating KRAS4b from and to the membrane.
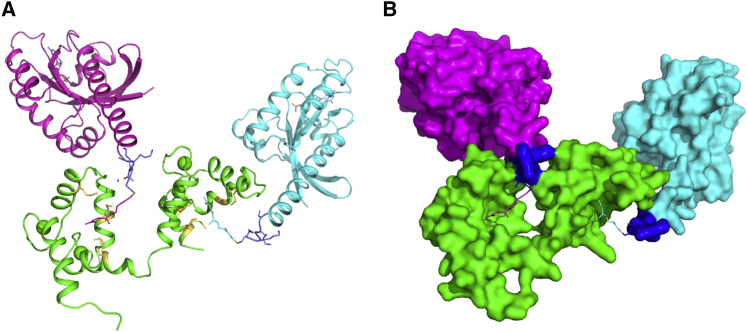
Figure 10.
Proposed structural docking model of CaM-KRAS4b interaction. (A) Cartoon showing two methylated and farnesylated KRAS4b subunits (PDB: 5TAR), color-coded magenta and cyan, modeled into two hydrophobic pockets of the pseudosymmetrical N- and C-terminal domains of calmodulin structure, color-coded green (PDB: 2MGU). The last five residues of KRAS4b (181–185) critical for binding to calmodulin and the hydrophobic residues in the hydrophobic pockets of calmodulin are shown in blue and yellow sticks, respectively. The farnesyl group covalently linked to the last residue of KRAS4b is also shown in sticks, having the same color as its respective KRAS protein subunit. (B) A surface representation of the structural model shown in (A) with the last residue of KRAS4b (185) and the farnesyl group displayed as sticks is given.
Previous studies have shown that CaM is able to dissociate KRAS from membranes. However, the mechanism by which this occurs is not well understood. Based on previous data, two different mechanisms have been proposed: 1) CaM binds to KRAS4b-FMe that has been spontaneously released from the membrane and sequesters it from rebinding to the membrane, and 2) CaM is able to actively bind to KRAS4b-FMe, which is still bound to the membrane. Our data show that CaM binds tightly to the prenylated HVR with a Kd of ∼0.3 μM. The prenylated HVR is the same region of KRAS4b required for membrane binding (5, 6). Therefore, we wanted to assess whether CaM could compete with the membrane for binding to KRAS4b. Using AUC and SPR data (Fig. 9), we show that CaM can disrupt membrane-bound KRAS4b-FMe and prevent it from rebinding to the membrane (Fig. 9).
The ability of CaM to bind to a membrane-bound KRAS4b-FMe suggests that there is a CaM recognition motif of KRAS4b-FMe available even when KRAS4b-FMe is inserted into the membrane. Because CaM does not bind to membranes (data not shown) or the unprenylated form of KRAS4b, this implies that a portion of the farnesyl tail must be solvent exposed to allow CaM to interact with it. A similar hypothesis was proposed for a myristoylated Src peptide in which they show that only 10 carbons of the myristoyl chain was inserted into the bilayers, with the first four carbons being solvent exposed to interact with the lipid headgroups (47). Based on this observation, we propose that CaM binds to a solvent-exposed portion of the membrane-bound KRAS4b-FMe group, perhaps the first five-carbon prenyl group (Fig. 7), which leads to the disruption of KRAS4b-FMe with the membrane and the subsequent release of KRAS4b-FMe into the hydrophobic pockets of CaM. Because of a tighter affinity of CaM to KRAS4b-FMe over the nanodiscs (Kd ∼2 μM) (20), CaM is then able to sequester it away and prevent rebinding to the membrane.
Collectively, our data provide new insights into the KRAS4b-CaM interaction and show that membrane localization of KRAS4b via CaM is important in understanding the regulation of KRAS4b.
Author Contributions
C.A., R.G., and A.G.S. designed the experiments. C.A., R.G., and L.B. performed the experiments and analyzed the data. M.T. collected NMR data. T.T. and S.M. expressed and purified the proteins. F.M. and D.V.N. provided scientific guidance and discussion. T.H.T. helped generate the docking model and reviewed the manuscript. C.A., R.G., and A.G.S. wrote the manuscript.
Acknowledgments
We thank Cammi Bittner, John-Paul Denson, Matthew Drew, Dom Esposito, Peter Frank, Bill Gillette, Clarissa Gorge, Jennifer Mehalko, Shelley Perkins, Rosemilia Reyes, Mukul Sherekar, Kelly Snead, and Vanessa Wall from the Protein Expression Laboratory Frederick National Laboratory for Cancer Research (FNL) for their help in cloning, expression, and purification of recombinant proteins. Timothy Waybright performed nucleotide loading analysis. We thank Patrick Alexander (FNL) for helping with the CD experiments. We would like to thank Hiroshi Matsuo of the FNL for useful discussions about the data.
We also thank David J. Aceti of the National Magnetic Resonance Facility at Madison, which is supported by National Institutes of Health (NIH) grant P41GM103399 (National Institute of General Medical Sciences), old number: P41RR002301. Equipment was purchased with funds from the University of Wisconsin-Madison, the NIH (P41GM103399, S10RR02781, S10RR08438, S10RR023438, S10RR025062, S10RR029220), the National Science Foundation (DMB-8415048, OIA-9977486, BIR-9214394), and the U.S. Department of Agriculture. This project was funded in whole or in part with federal funds from National Cancer Institute, NIH Contract HHSN261200800001E, and by the Intramural Research Program of the NIH, The National Institute of Diabetes and Digestive and Kidney Diseases.
Editor: Kalina Hristova.
Footnotes
Six figures and three tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(19)30131-6.
Supporting Material
References
- 1.Villalonga P., López-Alcalá C., Agell N. Calmodulin binds to K-Ras, but not to H- or N-Ras, and modulates its downstream signaling. Mol. Cell. Biol. 2001;21:7345–7354. doi: 10.1128/MCB.21.21.7345-7354.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roy S., Luetterforst R., Parton R.G. Dominant-negative caveolin inhibits H-Ras function by disrupting cholesterol-rich plasma membrane domains. Nat. Cell Biol. 1999;1:98–105. doi: 10.1038/10067. [DOI] [PubMed] [Google Scholar]
- 3.Prior I.A., Harding A., Hancock J.F. GTP-dependent segregation of H-ras from lipid rafts is required for biological activity. Nat. Cell Biol. 2001;3:368–375. doi: 10.1038/35070050. [DOI] [PubMed] [Google Scholar]
- 4.Parker J.A., Mattos C. The Ras-membrane interface: isoform-specific differences in the catalytic domain. Mol. Cancer Res. 2015;13:595–603. doi: 10.1158/1541-7786.MCR-14-0535. [DOI] [PubMed] [Google Scholar]
- 5.Hancock J.F., Paterson H., Marshall C.J. A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell. 1990;63:133–139. doi: 10.1016/0092-8674(90)90294-o. [DOI] [PubMed] [Google Scholar]
- 6.Hancock J.F. Ras proteins: different signals from different locations. Nat. Rev. Mol. Cell Biol. 2003;4:373–384. doi: 10.1038/nrm1105. [DOI] [PubMed] [Google Scholar]
- 7.Fivaz M., Meyer T. Reversible intracellular translocation of KRas but not HRas in hippocampal neurons regulated by Ca2+/calmodulin. J. Cell Biol. 2005;170:429–441. doi: 10.1083/jcb.200409157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sidhu R.S., Clough R.R., Bhullar R.P. Ca2+/calmodulin binds and dissociates K-RasB from membrane. Biochem. Biophys. Res. Commun. 2003;304:655–660. doi: 10.1016/s0006-291x(03)00635-1. [DOI] [PubMed] [Google Scholar]
- 9.Lopez-Alcalá C., Alvarez-Moya B., Agell N. Identification of essential interacting elements in K-Ras/calmodulin binding and its role in K-Ras localization. J. Biol. Chem. 2008;283:10621–10631. doi: 10.1074/jbc.M706238200. [DOI] [PubMed] [Google Scholar]
- 10.Abraham S.J., Nolet R.P., Gaponenko V. The hypervariable region of K-Ras4B is responsible for its specific interactions with calmodulin. Biochemistry. 2009;48:7575–7583. doi: 10.1021/bi900769j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu L.J., Xu L.R., Liang Y. Both the C-terminal polylysine region and the farnesylation of K-RasB are important for its specific interaction with calmodulin. PLoS One. 2011;6:e21929. doi: 10.1371/journal.pone.0021929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jang H., Banerjee A., Nussinov R. Flexible-body motions of calmodulin and the farnesylated hypervariable region yield a high-affinity interaction enabling K-Ras4B membrane extraction. J. Biol. Chem. 2017;292:12544–12559. doi: 10.1074/jbc.M117.785063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Babu Y.S., Bugg C.E., Cook W.J. Structure of calmodulin refined at 2.2 A resolution. J. Mol. Biol. 1988;204:191–204. doi: 10.1016/0022-2836(88)90608-0. [DOI] [PubMed] [Google Scholar]
- 14.Ishida H., Vogel H.J. Protein-peptide interaction studies demonstrate the versatility of calmodulin target protein binding. Protein Pept. Lett. 2006;13:455–465. doi: 10.2174/092986606776819600. [DOI] [PubMed] [Google Scholar]
- 15.Yamniuk A.P., Vogel H.J. Calmodulin’s flexibility allows for promiscuity in its interactions with target proteins and peptides. Mol. Biotechnol. 2004;27:33–57. doi: 10.1385/MB:27:1:33. [DOI] [PubMed] [Google Scholar]
- 16.Vetter S.W., Leclerc E. Novel aspects of calmodulin target recognition and activation. Eur. J. Biochem. 2003;270:404–414. doi: 10.1046/j.1432-1033.2003.03414.x. [DOI] [PubMed] [Google Scholar]
- 17.Villarroel A., Taglialatela M., Areso P. The ever changing moods of calmodulin: how structural plasticity entails transductional adaptability. J. Mol. Biol. 2014;426:2717–2735. doi: 10.1016/j.jmb.2014.05.016. [DOI] [PubMed] [Google Scholar]
- 18.Vranken W.F., Boucher W., Laue E.D. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 19.Taylor T., Denson J.P., Esposito D. Optimizing expression and solubility of proteins in E. coli using modified media and induction parameters. Methods Mol. Biol. 2017;1586:65–82. doi: 10.1007/978-1-4939-6887-9_5. [DOI] [PubMed] [Google Scholar]
- 20.Gillette W.K., Esposito D., Stephen A.G. Farnesylated and methylated KRAS4b: high yield production of protein suitable for biophysical studies of prenylated protein-lipid interactions. Sci. Rep. 2015;5:15916. doi: 10.1038/srep15916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dharmaiah S., Bindu L., Simanshu D.K. Structural basis of recognition of farnesylated and methylated KRAS4b by PDEδ. Proc. Natl. Acad. Sci. USA. 2016;113:E6766–E6775. doi: 10.1073/pnas.1615316113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao H., Brautigam C.A., Schuck P. Overview of current methods in sedimentation velocity and sedimentation equilibrium analytical ultracentrifugation. Curr. Protoc. Protein Sci. 2013;Chapter 20:Unit20.12. doi: 10.1002/0471140864.ps2012s71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghirlando R., Balbo A., Zhao H. Improving the thermal, radial, and temporal accuracy of the analytical ultracentrifuge through external references. Anal. Biochem. 2013;440:81–95. doi: 10.1016/j.ab.2013.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cole J.L., Lary J.W., Laue T.M. Analytical ultracentrifugation: sedimentation velocity and sedimentation equilibrium. Methods Cell Biol. 2008;84:143–179. doi: 10.1016/S0091-679X(07)84006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao H., Brown P.H., Schuck P. On the distribution of protein refractive index increments. Biophys. J. 2011;100:2309–2317. doi: 10.1016/j.bpj.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Inagaki S., Ghirlando R. Nanodisc characterization by analytical ultracentrifugation. Nanotechnol. Rev. 2017;6:3–14. [Google Scholar]
- 28.Inagaki S., Ghirlando R., Grisshammer R. Biophysical characterization of membrane proteins in nanodiscs. Methods. 2013;59:287–300. doi: 10.1016/j.ymeth.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dam J., Velikovsky C.A., Schuck P. Sedimentation velocity analysis of heterogeneous protein-protein interactions: Lamm equation modeling and sedimentation coefficient distributions c(s) Biophys. J. 2005;89:619–634. doi: 10.1529/biophysj.105.059568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao H., Balbo A., Schuck P. The boundary structure in the analysis of reversibly interacting systems by sedimentation velocity. Methods. 2011;54:16–30. doi: 10.1016/j.ymeth.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chavan T.S., Jang H., Tarasova N.I. High-affinity interaction of the K-Ras4B hypervariable region with the Ras active site. Biophys. J. 2015;109:2602–2613. doi: 10.1016/j.bpj.2015.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bayburt T.H., Sligar S.G. Membrane protein assembly into Nanodiscs. FEBS Lett. 2010;584:1721–1727. doi: 10.1016/j.febslet.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Delaglio F., Grzesiek S., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 35.Lee W., Tonelli M., Markley J.L. NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics. 2015;31:1325–1327. doi: 10.1093/bioinformatics/btu830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghanam R.H., Fernandez T.F., Saad J.S. Binding of calmodulin to the HIV-1 matrix protein triggers myristate exposure. J. Biol. Chem. 2010;285:41911–41920. doi: 10.1074/jbc.M110.179093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ikura M., Kay L.E., Bax A. A novel approach for sequential assignment of 1H, 13C, and 15N spectra of proteins: heteronuclear triple-resonance three-dimensional NMR spectroscopy. Application to calmodulin. Biochemistry. 1990;29:4659–4667. doi: 10.1021/bi00471a022. [DOI] [PubMed] [Google Scholar]
- 38.Emsley P., Lohkamp B., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Agamasu C., Ghanam R.H., Saad J.S. Structural and biophysical characterization of the interactions between calmodulin and the pleckstrin homology domain of Akt. J. Biol. Chem. 2015;290:27403–27413. doi: 10.1074/jbc.M115.673939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fernandez T.F., Samal A.B., Saad J.S. Structural and biophysical characterization of the interactions between the death domain of Fas receptor and calmodulin. J. Biol. Chem. 2013;288:21898–21908. doi: 10.1074/jbc.M113.471821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sperlich B., Kapoor S., Weise K. Regulation of K-Ras4B membrane binding by calmodulin. Biophys. J. 2016;111:113–122. doi: 10.1016/j.bpj.2016.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang X., Boyken S.E., Huang Y.H. Calmodulin and PI(3,4,5)P3 cooperatively bind to the Itk pleckstrin homology domain to promote efficient calcium signaling and IL-17A production. Sci. Signal. 2014;7:ra74. doi: 10.1126/scisignal.2005147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matsubara M., Nakatsu T., Taniguchi H. Crystal structure of a myristoylated CAP-23/NAP-22 N-terminal domain complexed with Ca2+/calmodulin. EMBO J. 2004;23:712–718. doi: 10.1038/sj.emboj.7600093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Agamasu C., Ghanam R.H., Saad J.S. The interplay between calmodulin and membrane interactions with the pleckstrin homology domain of Akt. J. Biol. Chem. 2017;292:251–263. doi: 10.1074/jbc.M116.752816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chandra A., Grecco H.E., Bastiaens P.I. The GDI-like solubilizing factor PDEδ sustains the spatial organization and signalling of Ras family proteins. Nat. Cell Biol. 2011;14:148–158. doi: 10.1038/ncb2394. [DOI] [PubMed] [Google Scholar]
- 46.Bhagatji P., Leventis R., Silvius J.R. Multiple cellular proteins modulate the dynamics of K-ras association with the plasma membrane. Biophys. J. 2010;99:3327–3335. doi: 10.1016/j.bpj.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McLaughlin S., Aderem A. The myristoyl-electrostatic switch: a modulator of reversible protein-membrane interactions. Trends Biochem. Sci. 1995;20:272–276. doi: 10.1016/s0968-0004(00)89042-8. [DOI] [PubMed] [Google Scholar]
- 48.Travers T., Lopez C.A., Gnanakaran S. Molecular recognition of RAS/RAF complex at the membrane: Role of RAF cysteine-rich domain. Sci Rep. 2018;8:8461. doi: 10.1038/s41598-018-26832-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.