Highlights
-
•
Deep learning has become widely used in different fields of computer science such as face recognition, but also in biology, for example to detect malignant skin cancers based on images.
-
•
Deep learning applied to microscopy images of biofilm colonization patterns accurately characterized its bacterial composition.
-
•
Deep learning applications only rarely outperform human experts in classification tasks. Here however, deep learning reached an accuracy of 90%, therefore clearly outperforming human experts (50% accurate).
-
•
Our method provides an accurate alternative to standard, time-consuming biochemical methods, using visual information only.
Keywords: Deep learning, Convolutional neural networks, Biomining, Acidophiles, Bacterial biofilm, Microscopy imaging
Abstract
Background
Deep neural networks have been successfully applied to diverse fields of computer vision. However, they only outperform human capacities in a few cases.
Methods
The ability of deep neural networks versus human experts to classify microscopy images was tested on biofilm colonization patterns formed on sulfide minerals composed of up to three different bioleaching bacterial species attached to chalcopyrite sample particles.
Results
A low number of microscopy images per category (<600) was sufficient for highly efficient computational analysis of the biofilm's bacterial composition. The use of deep neural networks reached an accuracy of classification of ∼90% compared to ∼50% for human experts.
Conclusions
Deep neural networks outperform human experts’ capacity in characterizing bacterial biofilm composition involved in the degradation of chalcopyrite. This approach provides an alternative to standard, time-consuming biochemical methods.
1. Introduction
“Biomining” is an industrial process that employs microorganisms for the recovery of valuable metals from sulfidic ores [1], [2]. Dissolution of metal sulfides, such as the copper mineral chalcopyrite, is catalyzed by microbial oxidation of ferrous iron that provides ferric ions for the chemical oxidation of metal sulfides. This regenerates ferrous ions and a cycle between chemical and biological reactions occurs. In addition, sulfur-oxidizing acidophiles assist the mineral degradation process by producing sulfuric acid from inorganic sulfur compounds. Biomining is less harmful to the environment than conventional metal recovery processes [3] and therefore, it is important to further optimize this method.
Biofilms are communities of microorganisms embedded in a self-generated matrix of extracellular polymeric substances (EPS). This microbial lifestyle confers several advantages compared to free-living planktonic cells, such as water retention, protection against stresses, providing nutritional requirements, etc. [4]. Biofilm-forming microorganisms are crucial in commercial heap biomining operations, in which they partly determine the initial metal sulfide dissolution rate [1], [5], [6]. Acidophilic microbes have differing abilities to generate energy from the conversion of the mineral components under moderately thermophilic temperatures. Acidithiobacillus caldus is an obligate chemolithoautotrophic sulfur oxidizer that thrives at pH 2.5 [7], [8]. Leptospirillum ferriphilum is a ferrous iron oxidizing autotroph that is often the dominant iron-oxidizer in biomining environments at extremely low pH (1.3–1.6) and high redox potential conditions [9]. Sulfobacillus thermosulfidooxidans is a mixotroph that primarily oxidizes iron but is also capable of oxidizing sulfur compounds at higher pH conditions compared to other acidophiles [10], [11]. The interplay of species in mixed community biofilms affects metal dissolution rates and therefore, biofilms consisting of acidophilic microbial consortia are important to understand and optimize during metal dissolution.
Epifluorescence microscopy (EFM) can be used in biomining applications to study biofilm structure and spatial distribution of cells on mineral sulfides [12], [13], [14]. In combination with nucleic acid dyes that label bacterial species, it enables the detection of specific groups of microorganisms attached to metal sulfide surfaces [15], [16], [17]. This allows to evaluate the extent of bacterial colonization on chalcopyrite mineral grains and to visualize the biofilm morphology [18], [17].
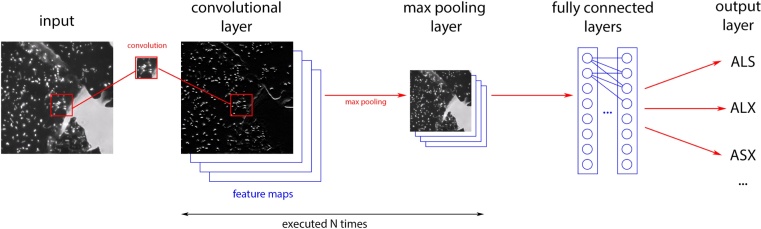
Machine learning is a field of computer science that enables computers to process and learn from data without being explicitly programmed. Convolutional neural networks (CNNs) are a category of deep neural networks that are able to make predictions in areas such as image recognition and classification. A CNN consists of an input and an output layer, as well as multiple hidden layers that carry out the following tasks: (i) Convolutional layers emulate the response of an individual neuron to visual stimuli and applies a convolution operation to the input, passing the result to the next layer. (ii) Pooling layers combine the outputs of neuron clusters at one layer into a single neuron in the next layer. (iii) Fully connected layers connect every neuron in one layer to every neuron in another layer (Fig. 1). Altogether, this workflow allows fitting multiple parameters of a CNN upon training with a set of images and then the trained CNN is used to classify new images and infer the category they belong to. These neural networks have proven successful in many different real-life case studies and applications, including identifying faces [19], detecting objects [20], and assisting self-driving cars [21]. However, while deep learning is commonly applied to diverse fields of computer vision and easing biological image processing in different fields of biology [22], it only rarely outperforms humans in image classification [23], [24].
Fig. 1.
CNN workflow showing how an input image is analyzed by a CNN where image features are detected in the convolutional layer followed by processing of maximum pooling and finally resulting into classification (output layer) of the different microbial species in the biofilm.
In this study, we used CNNs trained with a low number of EFM images representing biofilms of different bacterial compositions, and compared the performance of CNNs versus human experts in correctly classifying new images.
2. Materials and methods
2.1. Microbial species cultivation
EFM pictures were taken of biofilm grown strains of A. caldus DSM 8584 [25], L. ferriphilum DSM 14647 [26], and S. thermosulfidooxidans DSM 9293 [27]. Bacteria were grown in sterile Mackintosh basal salt (MAC) medium [28] with soluble electron donors for inoculation of chalcopyrite cultures. For L. ferriphilum, 4 g/L iron(II)-ions were provided as FeSO4·7H2O. Precipitation of ferric salts was prevented by addition of sulfuric acid to maintain the pH in the range of 1.6–1.8. A. caldus and S. thermosulfidooxidans were pre-cultured using 0.9 g/L potassium tetrathionate (K2S4O6). The medium for S. thermosulfidooxidans was supplemented with 0.02% yeast extract (YE) and 0.1 g/L iron(II)-ions [29]. Cells were harvested by centrifugation at 11,270g for 10 min, washed in sterile medium, and inoculated at an initial cell density of 107 cells/mL to chalcopyrite cultures in 300-mL-Erlenmeyer flasks with 150 mL MAC medium and 2% (wt/vol) chalcopyrite grains (50–100 μm grain size). Equal proportions of cells of each species were used in mixed cultures. All strains were cultivated on a rotary shaker at 37 °C and 150 rpm.
2.2. Microscopy sample preparation
About 25 mg of mineral grain particle samples were withdrawn from mineral cultures using a flame-sterilized spatula and incubated in 1 mL sterile MAC medium (pH 1.8) with 4% formaldehyde at room temperature for 1 hour for fixation of mineral-attached cells. This was followed by two washing steps with water and subsequently with 1 mL phosphate-buffered saline (PBS). Samples were stored at −20 °C in 50% ethanol in PBS. Mineral particles were incubated for 10 min in 200 μL of an aqueous solution of 0.01% 4′,6-diamidine-2′-phenylindole dihydrochloride (DAPI) in 2% formaldehyde. Before and after staining of attached cells, mineral grains were washed with 1 mL PBS. Mineral particles were mounted on 10-well diagnostic glass slides (10-well, 6.7 mm; Thermo Scientific) using a glycerol-based mounting medium (CitiFluor AF2) and 22- by 50-mm cover glasses [30].
2.3. High-throughput epifluorescence microscopy
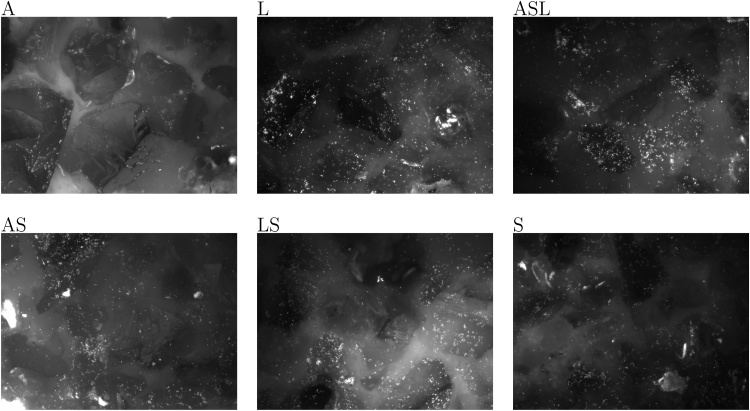
Microscopy images were obtained using the EFM platform AxioImager M2m (Zeiss) equipped with a motorized microscopy stage (IM SCAN 130 × 85 – DC 1 mm, Märzhäuser Wetzlar) and a AxioCam MRm camera. This setup was used to generate sets of images for different acidophile microbial mixtures. Each image represented the bacteria stained on mineral particles in an area of 450 × 335 μm (see Fig. 2 and images in the Supplementary Material section “Test For Humans”). Automated acquisition allowed to image between 180 and 504 images per category composed of a different combination of bacterial species that were then used to train deep neural networks [30]. For all categories, data augmentation was used by simple random image duplication to obtain the same number of images (504) for each category used for deep neural networks training.
Fig. 2.
Example of EFM images representing the different biofilm categories. The leaching mixtures were composed of A. caldus (A), L. ferriphilum (L), and S. thermosulfidooxidans (S) that were used as pure or mixed cultures, resulting in the following categories: A, L, S, AS, LS, and ASL.
2.4. Deep learning application
TensorFlow [31], [32], developed by Google, is one of the most recent deep learning frameworks and provides state-of-the-art implementations to build CNNs. Briefly, the software is written in C++ and offers interfaces to Python. A suite of visualization tools, called TensorBoard, is included within TensorFlow that allows visualization of networks in a web browser and to monitor the training progress. The CNN training was carried out with TensorFlow v1.6 in <10 h on an Intel Core i5 2.0 GHz computer (with 4 CPU cores) but the procedure can be shortened by parallelizing the network training on GPUs (code and images are available in Supplementary Material section “TensorFlow code and microscopy images”). In order to account for variation in accuracy of the algorithm and the effect of over-fitting, resampling was carried out where the same procedure was applied to a different division of the images into training and testing sets (see Supplementary Material section “Deep Learning Resampling”).
2.5. Deep learning versus human expert performance
TensorFlow-based deep neural networks were trained on less than 600 microscopy images (after data augmentation) per experimental category (each category represented by a different microbial composition of the leaching mixture, each grown on the same mineral substrate and under same conditions). The leaching mixtures were composed of A. caldus (A), L. ferriphilum (L), and S. thermosulfidooxidans (S) that were used as pure or mixed cultures, resulting in the following categories: A, L, S, AS, LS, and ASL. The category AL (see examples in Supplementary Material section “Example of AL Images”) was not processed because of the insufficient images at disposition for CNN training compared to the other categories and therefore was omitted in the present results. Deep neural networks were subsequently trained on the different categories and their ability to correctly assign new images was tested on a subset of 100 images per category that were not included in the training set. The training performance progress is shown in Supplementary Material section “Deep Learning Performance vs. Amount of Training Data”. Further, the same approach was applied to human subjects in order to compare the performance of neural networks versus humans in their ability to distinguish biofilms of different bacterial composition, and classify them according to their category based on visual features. Human subjects (n = 20; all experienced in work related to microbial bioleaching) were questioned once by a custom-built double-blind test (see Supplementary Material section “Test For Humans”). The test included a training set of images (70 images per category), followed by a test section where subjects were asked to classify 10 unknown images belonging to the same categories as the training set, which corresponded to the same image dataset used for the deep neural network training. The “AS” category was not selected by the random procedure generating the double-blind test for human experts and therefore, it was omitted in the present results (the accuracy of deep learning for identifying “AS” was 81%).
3. Results and discussion
3.1. Deep neural networks can identify characteristic biofilm patterns
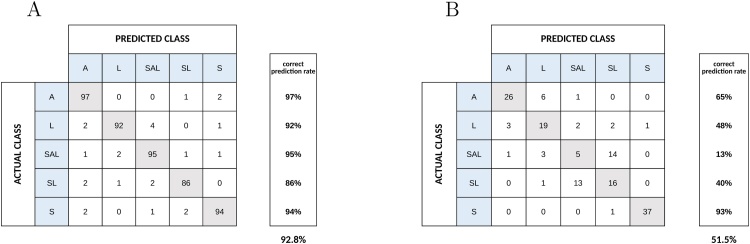
Deep neural networks applied to our bioleaching experiments correctly deduced the species or combination of species in a biofilm of unknown composition. Moreover, the neural networks achieved an accuracy of 92.8% compared to 51.5% by the human experts consensus (Fig. 3; expected accuracy by random guessing was approximately 17%, and the best human expert performance was of 80% (see Supplementary Material section “Human Expert's Performance”), deep neural networks performance with samples devoid of bacteria was 16.67%, stdev = 2.87%, CV = 0.17% (see Supplementary Material section “Negative Control”)). For example, the deep neural network correctly classified 97 images out of 100 belonging to the category “A”, while misclassifying three of them to either “SL” or “S” (Fig. 3). Surprisingly, the “SAL” category was poorly classified by humans (13% accurate), but was the second-best guessed category by neural networks (95% accurate). In comparison, the best performance of deep neural networks for image analysis in another biological area was approximately 72% correct for classification of skin cancers compared to approximately 66% for expert dermatologists [23]. This does not imply superiority of the method presented in this study, but might be related to the nature of the analyzed images.
Fig. 3.
Deep neural networks (A) versus human experts’ (B) ability in predicting the species composition of bacterial biofilms. The matrices indicate the share of images correctly deduced in the diagonal line (shaded grey) and categories the misclassified images were assigned are shown in the horizontal plane.
While it is difficult to determine the image features used by deep neural networks that lead to a correct decision [22], the following features were mentioned by human experts as decisive factors for classification (see Fig. 2 and images in the Supplementary Material section “Test For Humans”): (i) Cellular shape, in particular for the distinction between “S” having longer-shaped cells compared to “A” and “L” plus “A” was identified based on a slightly smaller size than “L”. (ii) Abundance of “L” cells imaged compared to the other categories. (iii) Increased “A” cell clusters in the colonization pattern compared to “L” and “S” being more sparsely distributed. (iv) Poor attachment of “A” cells compared to the other species. (v) The brightness of images representing “L”. The reasons listed above partially account for the outcome of human experts’ best performances, in particular concerning the classification of “S”. However, it does not explain the major source of confusion between the categories “SAL” and “SL” among human experts that was not encountered in the deep learning evaluation.
3.2. Potential of CNNs for the characterization of biofilm colonization pattern images
CNNs were trained on microscopy images of different bacterial compositions and their ability to correctly classify new images was tested and compared to human expert performance. This application demonstrated that deep learning enabled image classification based on recognition of attachment patterns, which are cryptic to human perception. Moreover, our results were achieved with a small training set (between 180 and 504 images per category, on 504 images per category after data augmentation) compared to applications of equivalent performance (e.g., 129,000 images in Ref. [23], 2 millions in Ref. [33]). Finally, the proposed methodology is not limited to a specific experimental setup and therefore represents a method of choice for microscopy imaging-based quantification in environmental microbiology beyond the field of biomining.
3.3. Limitations
One limitation of this study was represented by the low number of human testing subjects that were not stratified into different categories (e.g., experts, non-experts, etc.). Therefore, a reduced number of subjects familiar to the field (termed “experts”, whose expertise relied on their working experience related to microscopy imaging of bacterial biofilms and bioleaching research for at least eight months) were included, assuming that they would perform better than subjects external to the field and that inclusion of the non-experts would not improve the information. A further limitation is the unclear character of the determining features that the CNNs used to classify images. However, this common issue relates to all applications of deep neural networks processing a large amount of data [22]. Finally, the bioleaching bacteria of our experimental setup form simpler biofilms in comparison to other environments [4] and the presented method might have limited performances when applied to other environmental samples with a higher species diversity than typical, low-diversity bioleaching environments.
4. Conclusions
Deep neural networks were trained on a reduced set of microscopy images of microbial biofilms composed of different bacterial bioleaching species colonizing chalcopyrite particles. The performance of deep neural networks in correctly classifying new images was compared to human experts’ ability in performing the same task based on the same training set. Deep neural networks showed superior performance compared to human experts and were able to predict the presence of microbial species or combination thereof in a biofilm of unknown composition. This allowed to measure important features of biofilm development under different experimental conditions by imaging only. Therefore, this provides an efficient alternative to standard and time-consuming biochemical methods, such as qPCR, which may be biased by nonhomogeneous cell lysis during mineral sulfide samples preparation [30]. Biofilms are important in biomining due to ferric attack on the mineral and high concentrations of ferric accumulate in the EPS. Therefore, studies of biofilm can help optimize the bioleaching process. This methodology opens the way to efficient evaluation of high-throughput microscopy imaging in the field of mineral biofilm leaching, and is applicable beyond the presented experimental setup.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgments
This project was supported by Bundesministerium für Bildung und Forschung (BMBF, 031A600A and B), Vetenskapsrådet (contract 2014-6545), the Luxembourg National Research Fund (FNR) (INTER/SYSAPP/14/05), and the Swiss Initiative in Systems Biology (SystemsX.ch, SysMetEx) under the frame of ERASysAPP. MV acknowledges support from Fondecyt1161007 grant. ABD and IVP acknowledge support from the Swiss National Science Foundation grant 205321_173020. We also thank the persons who took the test that allowed us to complete the present study.
Footnotes
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.btre.2019.e00321.
Appendix B. Supplementary data
The following are the supplementary data to this article:
References
- 1.Vera M., Schippers A., Sand W. Progress in bioleaching: fundamentals and mechanisms of bacterial metal sulfide oxidation – Part A. Appl. Microbiol. Biotechnol. 2013;97(17):7529–7541. doi: 10.1007/s00253-013-4954-2. (PubMed:23720034) [DOI] [PubMed] [Google Scholar]
- 2.Brierley C.L., Brierley J.A. Progress in bioleaching: part B: applications of microbial processes by the minerals industries. Appl. Microbiol. Biotechnol. 2013;97(17):7529–7541. doi: 10.1007/s00253-013-5095-3. (PubMed:23877580) [DOI] [PubMed] [Google Scholar]
- 3.Jerez C.A. Biomining of metals: how to access and exploit natural resource sustainably. Microbiol. Biotechnol. 2017;10(5):1191–1193. doi: 10.1111/1751-7915.12792. (PubMed Central:PMC5609284, PubMed:28771998) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flemming H.C., Wingender J. The biofilm matrix. Nat. Rev. Microbiol. 2010;8(9):623–633. doi: 10.1038/nrmicro2415. (PubMed:20676145) [DOI] [PubMed] [Google Scholar]
- 5.Sand W., Gehrke T., Jozsa P.-G., Schippers A. (Bio)chemistry of bacterial leaching-direct vs. indirect bioleaching. Hydrometallurgy. 2001;59:159–175. [Google Scholar]
- 6.Rui-Yong Zhang, Sören Bellenberg, Neu Thomas R., Wolfgang Sand, Mario Vera The biofilm lifestyle of acidophilic metal/sulfur-oxidizing microorganisms. Biotechnol. Extremophiles: Adv. Challenges. 2016:177–213. [Google Scholar]
- 7.Mangold S., Valdes J., Holmes D.S., Dopson M. Sulfur metabolism in the extreme acidophile acidithiobacillus caldus. Front. Microbiol. 2011;2:17. doi: 10.3389/fmicb.2011.00017. (PubMed Central:PMC3109338, PubMed:21687411) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Valdes J., Quatrini R., Hallberg K., Dopson M., Valenzuela P.D., Holmes D.S. Draft genome sequence of the extremely acidophilic bacterium Acidithiobacillus caldus ATCC 51756 reveals metabolic versatility in the genus Acidithiobacillus. J. Bacteriol. 2009;191(18):5877–5878. doi: 10.1128/JB.00843-09. (PubMed Central:PMC2737959, PubMed:19617360) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christel S., Herold M., Bellenberg S., El Hajjami M., Buetti-Dinh A., Pivkin I.V., Sand W., Wilmes P., Poetsch A., Dopson M. Multi-omics reveal the lifestyle of the acidophilic, mineral-oxidizing model species Leptospirillum ferriphilum T. Appl. Environ. Microbiol. 2017 doi: 10.1128/AEM.02091-17. (PubMed Central:PMC5772234, PubMed:29150517) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Justice N.B., Norman A., Brown C.T., Singh A., Thomas B.C., Banfield J.F. Comparison of environmental and isolate Sulfobacillus genomes reveals diverse carbon, sulfur, nitrogen, and hydrogen metabolisms. BMC Genomics. 2014;15:1107. doi: 10.1186/1471-2164-15-1107. (PubMed Central:PMC4378227, PubMed:25511286) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Janosch C., Remonsellez F., Sand W., Vera M. Sulfur oxygenase reductase (Sor) in the moderately thermoacidophilic leaching bacteria: studies in Sulfobacillus thermosulfidooxidans and Acidithiobacillus caldus. Microorganisms. 2015;3(4):707–724. doi: 10.3390/microorganisms3040707. (PubMed Central:PMC5023260, PubMed:27682113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Rui Yong, Zhang Yu Tong, Neu Thomas R., Li Qian, Bellenberg Sören, Sand Wolfgang, Vera Mario. Biotechnologies in Mining Industry and Environmental Engineering, volume 1130 of Advanced Materials Research. Trans Tech Publications; 2015. Initial attachment and biofilm formation of a novel crenarchaeote on mineral sulfides; pp. 127–130. [Google Scholar]
- 13.Florian Bianca M., Noël Nanni, Bellenberg Soeren, Huergo J., Rohwerder Thore, Sand Wolfgang. Biohydrometallurgy 2009 volume 71 of Advanced Materials Research. Trans Tech Publications; 2009. Attachment behavior of leaching bacteria to metal sulfides elucidated by combined atomic force and epifluorescence microscopy; pp. 337–340. [Google Scholar]
- 14.Rui Yong Zhang, Vera Mario, Bellenberg Sören, Sand Wolfgang. Integration of Scientific and Industrial Knowledge on Biohydrometallurgy volume 825 of Advanced Materials Research. Trans Tech Publications; 2013. Attachment to minerals and biofilm development of extremely acidophilic archaea; pp. 103–106. [Google Scholar]
- 15.Noël N., Florian B., Sand W. AFM and EFM study on attachment of acidophilic leaching organisms. Hydrometallurgy. 2010;104(3):370–375. [Google Scholar]
- 16.Florian Bianca, Noël Nanni, Sand Wolfgang. Visualization of initial attachment of bioleaching bacteria using combined atomic force and epifluorescence microscopy. Miner. Eng. 2010;23(05):532–535. [Google Scholar]
- 17.Bellenberg S., Diaz M., Noel N., Sand W., Poetsch A., Guiliani N., Vera M. Biofilm formation communication and interactions of leaching bacteria during colonization of pyrite and sulfur surfaces. Res. Microbiol. 2014;165(9):773–781. doi: 10.1016/j.resmic.2014.08.006. (PubMed:25172572) [DOI] [PubMed] [Google Scholar]
- 18.Bellenberg S., Barthen R., Boretska M., Zhang R., Sand W., Vera M. Manipulation of pyrite colonization and leaching by iron-oxidizing Acidithiobacillus species. Appl. Microbiol. Biotechnol. 2015;99(3):1435–1449. doi: 10.1007/s00253-014-6180-y. (PubMed:25381488) [DOI] [PubMed] [Google Scholar]
- 19.Parkhi O.M., Vedaldi A., Zisserman A. Deep face recognition. British Machine Vision Conference. 2015 [Google Scholar]
- 20.Krizhevsky A., Sutskever I., Hinton G.E. Imagenet classification with deep convolutional neural networks. Advances in Neural Information Processing Systems. 2012:1097–1105. [Google Scholar]
- 21.Bojarski M., Del Testa D., Dworakowski, Firner B., Flepp B., Goyal P., Jackel L.D., Monfort M., Muller U., Zhang J. End to end learning for self-driving cars. arXiv Preprint. 2016 arXiv:1604.07316. [Google Scholar]
- 22.Angermueller C., Parnamaa T., Parts L., Stegle O. Deep learning for computational biology. Mol. Syst. Biol. 2016;12(7):878. doi: 10.15252/msb.20156651. (PubMed Central:PMC4965871, PubMed:27474269) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Esteva A., Kuprel B., Novoa R.A., Ko J., Swetter S.M., Blau H.M., Thrun S. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542(7639):115–118. doi: 10.1038/nature21056. (PubMed:28117445) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gulshan V., Peng L., Coram M., Stumpe M.C., Wu D., Narayanaswamy A., Venugopalan S., Widner K., Madams T., Cuadros J., Kim R., Raman R., Nelson P.C., Mega J.L., Webster D.R. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. 2016;316(22):2402–2410. doi: 10.1001/jama.2016.17216. (PubMed:27898976) [DOI] [PubMed] [Google Scholar]
- 25.Hallberg K.B., Lindstrom E.B. Characterization of Thiobacillus caldus sp. nov., a moderately thermophilic acidophile. Microbiology (Reading Engl.) 1994;140(Pt 12):3451–3456. doi: 10.1099/13500872-140-12-3451. (PubMed:7533596) [DOI] [PubMed] [Google Scholar]
- 26.Coram N.J., Rawlings D.E. Molecular relationship between two groups of the genus Leptospirillum and the finding that Leptospirillum ferriphilum sp. nov. dominates South African commercial biooxidation tanks that operate at 40 degrees C. Appl. Environ. Microbiol. 2002;68(2):838–845. doi: 10.1128/AEM.68.2.838-845.2002. (PubMed Central:PMC126727, PubMed:11823226) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Golovacheva R.S., Karavaiko G.I. Sulfobacillus, a new genus of thermophilic sporulating bacteria. Mikrobiologiia. 1978;47(5):815–822. (PubMed:101742) [PubMed] [Google Scholar]
- 28.Mackintosh M.E. Nitrogen fixation by Thiobacillus ferrooxidans. Microbiology. 1978;105(2):215–218. [Google Scholar]
- 29.Christel S., Herold M., Bellenberg S., Buetti-Dinh A., El Hajjami M., Pivkin I.V., Sand W., Wilmes P., Poetsch A., Vera M., Dopson M. Weak iron oxidation by Sulfobacillus thermosulfidooxidans maintains a favorable redox potential for chalcopyrite bioleaching. Front. Microbiol. 2018;9:3059. doi: 10.3389/fmicb.2018.03059. (PubMed Central:PMC6315122, PubMed:30631311) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bellenberg S., Buetti-Dinh A., Galli V., Ilie O., Herold M., Christel S., Boretska M., Pivkin I.V., Wilmes P., Sand W., Vera M., Dopson M. Automated microscopic analysis of metal sulfide colonization by acidophilic microorganisms. Appl. Environ. Microbiol. 2018;84(20) doi: 10.1128/AEM.01835-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abadi M., Barham P., Chen J., Chen Z., Davis A., Dean J., Devin M., Ghemawat S., Irving G., Isard M. Tensorflow: a system for large-scale machine learning. OSDI, vol. 16. 2016:265–283. [Google Scholar]
- 32.TensorFlow Official Website. www.tensorflow.org.
- 33.Buggenthin F., Buettner F., Hoppe P.S., Endele M., Kroiss M., Strasser M., Schwarzfischer M., Loeffler D., Kokkaliaris K.D., Hilsenbeck O., Schroeder T., Theis F.J., Marr C. Prospective identification of hematopoietic lineage choice by deep learning. Nat. Methods. 2017;14(4):403–406. doi: 10.1038/nmeth.4182. (PubMed Central:PMC5376497, PubMed:28218899) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.