Abstract
The prevalence of nonalcoholic fatty liver disease (NAFLD) has increased with the incidence of obesity; however, the underlying mechanisms are unknown. In this study, high-resolution metabolomics (HRM) along with transcriptomics were applied on animal models to draw a mechanistic insight of NAFLD. Wild type (WT) and catalase knockout (CKO) mice were fed with normal fat diet (NFD) or high fat diet (HFD) to identify the changes in metabolic and transcriptomic profiles caused by catalase gene deletion in correspondence with HFD. Integrated omics analysis revealed that cholic acid and 3β, 7α-dihydroxy-5-cholestenoate along with cyp7b1 gene involved in primary bile acid biosynthesis were strongly affected by HFD. The analysis also showed that CKO significantly changed all-trans-5,6-epoxy-retinoic acid or all-trans-4-hydroxy-retinoic acid and all-trans-4-oxo-retinoic acid along with cyp3a41b gene in retinol metabolism, and α/γ-linolenic acid, eicosapentaenoic acid and thromboxane A2 along with ptgs1 and tbxas1 genes in linolenic acid metabolism. Our results suggest that dysregulated primary bile acid biosynthesis may contribute to liver steatohepatitis, while up-regulated retinol metabolism and linolenic acid metabolism may have contributed to oxidative stress and inflammatory phenomena in our NAFLD model created using CKO mice fed with HFD.
Keywords: Catalase, Nonalcoholic fatty liver disease, Liver metabolism, Inflammation, Metabolomics, Transcriptomics
INTRODUCTION
Nonalcoholic fatty liver disease (NAFLD) is the most common liver disease representing over 75% of chronic liver disease cases (Younossi et al., 2011). The prevalence of NAFLD in the United States rose from 18% to 31% in the last three decades (Ruhl and Everhart, 2015). NAFLD is mainly associated with obesity, type 2 diabetes, dyslipidemia, and cardiovascular diseases (Lin et al., 2000; Cani et al., 2008; Hariri and Thibault, 2010; Panchal et al., 2011; Piao et al., 2017). NAFLD is characterized by the excessive accumulation of triglycerides in the form of lipid droplets in more than 5–10% of hepatocytes in the absence of excessive alcohol consumption. The disease spectrum ranges from simple fatty liver through nonalcoholic steatohepatitis (NASH), characterized by liver cell injury through inflammation, oxidative stress, and increased risk for liver fibrosis and cancer (Singh et al., 2015).
Given the increasing prevalence of NAFLD, understanding the initiation and progression of NAFLD has become crucial for developing therapeutic options (Hassan et al., 2014). Several mechanisms of NAFLD/NASH have been proposed but the exact mechanisms remain unknown. To understand the complex mechanisms of NAFLD in humans, efficient, inexpensive, and relevant animal models are needed. Although there are several diet-induced models of NAFLD/NASH in small animals, the most widely used diet to induce NAFLD/NASH is a high fat diet (HFD) to create the accumulation of hepatic triglycerides (Machado et al., 2015). Furthermore, oxidative stress has been implicated in the pathogenesis of NAFLD. Catalase, one of the antioxidant enzymes, showed its decreased activity in NAFLD (Videla et al., 2004). Recently, catalase in peroxisomes was shown to play a protective role in HFD induced liver injury in a CKO mouse model (Piao et al., 2017). These studies suggested that oxidative stress occurs upon reduced catalase activity and plays an important role in NAFLD/NASH.
Catalase is a common enzyme present in most aerobic cells. This enzyme protects cells from oxidative stress by catalyzing the rapid decomposition of hydrogen peroxide (H2O2). In our previous study, we reported the importance of catalase in protecting against kidney injury under diabetic stress (Hwang et al., 2012). Further, we investigated the role of catalase in HFD induced liver injury (Piao et al., 2017). Our data demonstrated that catalase deficiency accelerated liver injury through increased liver lipid accumulation, inflammation, and oxidative stress, which highlighted the beneficial effects of endogenous catalase in protecting against liver injury through maintaining liver redox balance. Therefore, we applied a metabolomics approach combined with transcriptomics to elucidate the metabolic alterations caused by catalase deficiency in order to understand the molecular mechanism by which endogenous catalase conferred a hepatoprotective effect.
Metabolomics is one of several emerging fields of “omics” research, which focuses on a comprehensive qualitative and quantitative analysis of all small molecules present in a cell, a tissue, or an organism to understand the function of biological systems. Metabolomic analyses determine the slight changes of metabolic profiles related to diseases, disease progression, therapeutic intervention, environmental modification, and genetic modification (Stewart and Bolt, 2011; Gao et al., 2014; Park et al., 2015, 2016). In addition, we extended our research by measuring transcriptomic alteration. Integrative omics, a combined study of more than one omics out of four (genomics, transcriptomics, proteomics, metabolomics), has become a rising field because of this strategy’s remarkable accuracy and integrity (Horgan and Kenny, 2011; Gomez-Cabrero et al., 2014; Cavill et al., 2016; Rajasundaram and Selbig, 2016). In this study, we aimed to identify alterations in metabolic and transcriptomic profiles involved in liver damage caused by HFD in mice and finally to verify if deleting the gene encoding catalase may worsen the liver injury caused by high fat diet by attenuating the level of identified metabolites and the levels of mRNAs. The results of this study suggested that an important association among diet, oxidative stress and inflammation would increase the risk in the development of NAFLD/NASH.
MATERIALS AND METHODS
Mouse preparation and Sampling
CKO mice were supplied by Ewha Womans University. Animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) at Ewha Womans University (No. 2010-30-3). Homozygous CKO male mice were bred with wild type (WT) C57BL/6J female mice to obtain heterozygous CKO mice. Interbreeding of the heterozygous mice was performed to generate homozygous CKO mice. Homozygous CKO mice and WTC57BL/6J mice were fed with normal fat diet (NFD) or high fat diet (HFD). A total of 24 mice were divided into four groups: wild type normal fat diet (WTNF), catalase knockout normal fat diet (CKONF), wild type high fat diet (WTHF), and catalase knockout high fat diet (CKOHF) as shown in Fig. 1. NFD was composed of 24% protein-derived calories, 58% carbohydrate-derived calories, and 18% fat derived calories while HFD had 18.4% protein-derived calories, 21.3% carbohydrate-derived calories, and 60.3% fat derived calories (Harlan TD 06414). All mice were bred with NFD for 29 weeks, following NFD or HFD for the next 11 weeks. Each diet was scheduled twice a week before collection of samples. Liver samples from mice were isolated, frozen in liquid nitrogen, and stored at −80°C for further analysis. Blood was centrifuged at 3,000 rpm for 15 min at 4°C, serum in the supernatant was collected, and alanine aminotransferase (ALT) was measured using EnzyChromTM assay kit (BioAssay Systems, Hayward, CA, USA) following manufacturer instructions. The experiments were performed in duplicate. Hepatic triglyceride concentration was measured as previously described (Norris et al., 2003). In brief, 50 mg of liver tissue was minced to small pieces and incubated in 0.3 ml of ethanolic KOH containing EtOH-30% potassium hydroxide (KOH) (2:1) at 55°C for overnight. After saponification, 0.6 ml of 50% EtOH was added, and centrifuged to get supernatant. Supernatant was neutralized by the addition of 0.215 ml of 1M MgCl2. Hepatic triglyceride content was measured with Free Glycerol Reagent followed by manufacturer’s instruction (F6428, Sigma-Aldrich, MO, USA).
Fig. 1.
Study design of the nonalcoholic fatty liver disease (NAFLD) model. Fig. 1 represents the overall flow of the study. A total of 24 mice were classified into 2 groups, normal fat diet (NFD, green-bordered) and high fat diet (HFD, yellow-bordered). Each group consisted of 6 wild type mice (WTNF or WTHF, light blue-colored) and 6 catalase knockout mice (CKONF or CKOHF, light orange-colored). To observe the effect of HFD (liver fat accumulation) and CKO (oxidative stress and inflammation), statistical analysis and pathway analysis were employed. The comparison between WTNF vs. CKOHF was excluded after preanalysis.
DNA Microarray
For transcriptomic analysis, microarray was performed as follows as recommended by E-Biogen (Seoul, Korea): Total RNA quality was controlled using an Agilent 2100 Bioanalyzer instrument (Agilent, CA, USA). Amplification and labeling of RNA was carried out with Agilent’s Low RNA Input linear amplification kit PLUS (Agilent). Microarray hybridization and sample washing were conducted using Agilent’s Gene Expression Hybridization Kit (Agilent) and Agilent’s Gene Expression Wash Buffer Kit (Agilent), respectively. Following this pre-process, samples were analyzed with Agilent’s DNA microarray scanner and Feature Extraction software (Agilent).
Liquid chromatography with tandem mass spectrometry
Samples were treated with 100 μL acetonitrile at a 1:2 v/v ratio and were centrifuged at 16,000 g for 10 min at 4°C to remove proteins (Want et al., 2006). An Agilent High Performance LC-MS/MS 6530 Q-TOF system (Agilent) was run in positive-ion mode for metabolic profiling. The LC system was equipped with a TARGA (Higgins, Mountain View, CA, USA) C18 column (100 mm×2.1 mm i.d.; 5 μm particle size). The LC parameters were set using an autosampler at a temperature of 4°C, injection volume of 5 μL, column temperature of 40°C, and flow rate of 0.4 mL/min. The compositions of mobile phases were as follows. A: 0.1% formic acid in water and B: 0.1% formic acid in ACN. The sample was injected in triplicate to ensure reliability though repetition. Total run time was 30 minutes with five minutes of post-run gap. Metabolites were identified with a mass/charge range of 50–1000 m/z, and nitrogen was chosen as the collision gas. Mass Hunter Acquisition software (Agilent) was used to collect data from LC-MS.
Metabolic profiling with statistical analysis
Data extraction: The LC-MS data were processed using apLCMS, an R package, to retrieve features of the samples (Yu et al., 2009). The m/z of ions were detected from 50 to 1,000 with a resolution of 20,000. A total of 5208 features were extracted. The triplicated data were then averaged to be used in further analysis.
Statistical analysis with PCA, HCA, and Manhattan plot: Bioinformatics using principal component analysis (PCA) and hierarchical cluster analysis (HCA) was conducted to determine if metabolic phenotypes were different among groups. The extracted features from apLCMS were filtered with interquantile range (IQR), log-transformed and pareto scaled using Metaboanalyst (http://www.metaboanalyst.ca/). Then the pairwise analyses between WTNF vs WTHF and WTHF vs CKOHF were performed. Manhattan plots were applied with a threshold of false discovery rate (FDR), q=0.1 to determine the significant metabolites among total features in pairwise comparisons using Limma, an R package.
Metabolite database search using Metlin database: The significant features were further annotated by their m/z values using the Metlin database (https://metlin.scripps.edu/) to identify the compound names. The positive adducts were used with a confidence limit of 10 ppm (Wolf et al., 2010). KEGG IDs provided by Metlin database (http://metlin.scripps.edu/) were used further used to determine the affected metabolic pathways in the Kyoto Encyclopedia of Genes and Genomes (KEGG) mouse metabolic pathway database (http://www.genome.jp/KEGG/) between WTNF vs WTHF and WTHF vs CKOHF.
Gene data extraction for transcriptomic study: To select altered genes by fold change, the threshold of fold change (FC) was set as 1.5. In this way, a total of 1,038 out of 28,299 genes with FC>1.5 in WTNF vs WTHF, and 1,191 out of 28,299 genes with FC>1.5 in WTHF vs CKOHF were chosen for further pathway analysis using KEGG database.
Pathway analysis with KEGG by integrative omics
KEGG database can visualize interactions between molecular and biological networks, which enables user to link metabolite information with gene data. Significantly altered metabolites (q<0.1) and genes (FC>1.5) were inserted as a single input file in KEGG database. The affected pathways hits were constructed as cumulative bars with the number metabolites and genes hits. Bar graphs of significant metabolites affected in those pathways were analyzed using the GraphPad Prism v 5.03 software (GraphPad Software, Inc., CA, USA) to measure their relative intensities among groups.
Receiver operating characteristic (ROC) curves and prediction of the 5 metabolites
The prediction ability of the five significant metabolites was assessed using receiver operating characteristic (ROC) curve from Metaboanalyst. ROC curve showed the optimal number of variables, sensitivity, specificity and area under curve (AUC) of each metabolite that can differentiate between two group (Obuchowski and Bullen, 2018). Then, the samples were analyzed to investigate how those variables separate between groups using predicted class probabilities.
Correlation analysis based on metabolites and genes
To support our data, we employed correlation study among metabolites and genes. The correlation analysis was visualized using heatmap with clustering based on pearson r correlation method. Then, correlation coefficients of metabolites and genes were shown as bars.
RESULTS
Physiological characteristics after feeding NFD or HFD on wild type
In order to confirm the validity of the NAFLD animal model in mice fed NFD or HFD for 11 weeks, we first measured physiological changes like body weight among NFD or HFD fed WT and CKO mice. Both the WT and CKO mice fed with HFD exhibited increased body weight, as shown in Table 1. WTHF and CKOHF mice weighed 45.4 ± 1.9 and 46.9 ± 2.1 g, respectively, while WTNF mice weighed 33.5 ± 0.9 g. The body weight, alanine aminotransferase (ALT) and liver triglyceride (TG) of HFD mice were significantly elevated compared to NFD mice (p<0.05). Characteristics of WT or CKO mice fed with HFD are described in Table 1. In addition, the lipid accumulation in mice liver was observed by H&E staining (data not shown).
Table 1.
Physiological characteristics of experimental mice after feeding
| WTNF | WTHF | CKONF | CKOHF | |
|---|---|---|---|---|
| Body weight (g) | 33.5 ± 0.9 | 45.4 ± 1.9* | 39.0 ± 2.0* | 46.9 ± 2.1*,† |
| Liver weight (g) | 1.12 ± 0.20 | 1.26 ± 0.16 | 1.32 ± 0.26 | 1.74 ± 0.35 |
| ALT (IU/L) | 34 ± 4 | 86 ± 11* | 60 ± 7* | 193 ± 23*,†,# |
| Liver TG (mg/mg protein) | 0.15 ± 0.02 | 2.97 ± 0.49* | 2.10 ± 0.27* | 4.46 ± 0.47*,†,# |
Data are presented as means ± SE of 6 mice per group.
p<0.05 compared to WTNF.
p<0.05 compared to CKONF.
p<0.05 compared to WTHF. ALT represents Alanine aminotransferase. TG represents Triglyceride.
Metabolic alterations in NAFLD
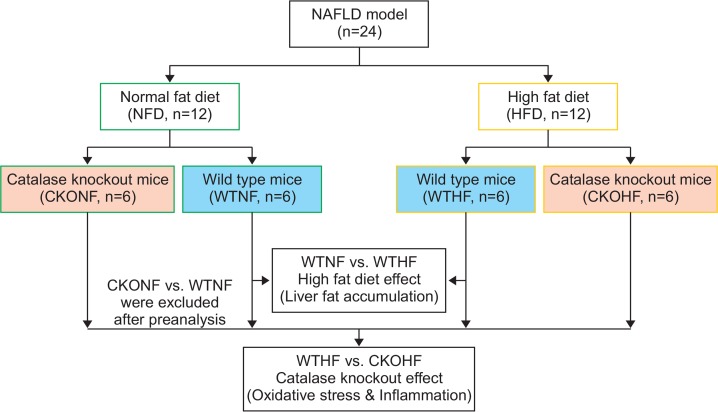
The validity of the established NAFLD mouse model was further ensured by analyzing the metabolic alterations caused by NFD and HFD between the WT or CKO mice. The study scheme was designed as shown in Fig. 2. The analysis was performed by inserting the apLCMS feature table containing 5,208 features in Metaboanalyst 3.0. Unsupervised multivariate statistical PCA and HCA procedures using Metaboanalyst 3.0. were employed to observe metabolic differences among the 4 groups. In Fig. 2A, the PCA score plot showed a tendency of gathering in individual circles: WTNF (sky blue); WTHF (blue); CKONF (green); and CKOHF (red). Additionally, to further define the differential metabolic phenotypes between NFD and HFD fed animals, a two-way HCA was performed among the WT and CKO mice. As shown in Fig. 2B, the upper clustering bar of HCA showed a clear separation among the 4 groups, suggesting that the metabolites were significantly different among groups. These results ensured the metabolic-associated alteration caused by NFD and HFD, as the WT mice fed with NFD or HFD were significantly distinguishable by either PCA or HCA. Similarly, a perturbed metabolic phenotype upon catalase gene knockout was also observed in the PCA and HCA analysis, as CKOHF mice were well-separated from WTHF mice (Fig. 2A, 2B). In order to specifically focus on HFD effect on WT or CKO mice, we excluded the CKONF group in further analysis.
Fig. 2.
PCA and HCA based on discrimination of HFD or NFD fed WT and CKO mice. Fig. 2 shows the result of PCA (A) and HCA (B) (http://www.metaboanalyst.ca/). In Fig. 2A, each group showed a tendency to gather in their respective colored circle. Fig. 2B shows HCA a clear separation of clustering among all 4 groups (WTNF, WTHF, CKONF, and CKOHF) with color keys (upper clustering bar).
Investigating significant metabolites
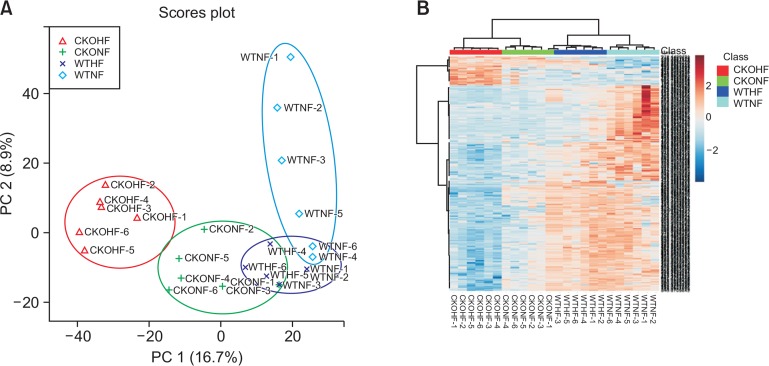
After observing distinct metabolomics-based profiles among HFD and NFD fed WT or CKO mice, we investigated the metabolic features that were responsible for these distinctions. To identify the metabolites responsible for the alterations caused by HFD and CKO, we compared WTNF vs WTHF and WTHF vs CKOHF using Manhattan plots (Fig. 3). Manhattan plots portray metabolites as dots (Clarke et al., 2011), where the dashed line signifies FDR q=0.1 and dots above the black dashed line represent the significant features. As shown in Fig. 3, 871 features were significantly different between WTNF and WTHF (Fig. 3A), while 1,172 features were significantly different in WTHF and CKOHF (Fig. 3B). The significant features annotated with Metlin database were mapped to KEGG pathway analysis to find affected pathways in CKO mice fed with HFD. These pathways were further investigated for their up- or down-regulation by CKO mice fed with HFD.
Fig. 3.
Manhattan plots of (A) WTNF vs WTHF and (B) WTHF vs CKOHF. Fig. 3 represents a Manhattan plot with the black dashed line corresponding to q=0.1. Dots above the dashed line represent 871 significant features (q<0.1) upon comparing WTNF and WTHF (A). Dots above the dashed line represent 1172 significant (q<0.1) upon comparing WTHF and CKOHF (B).
Identification of affected genes by transcriptomic analysis
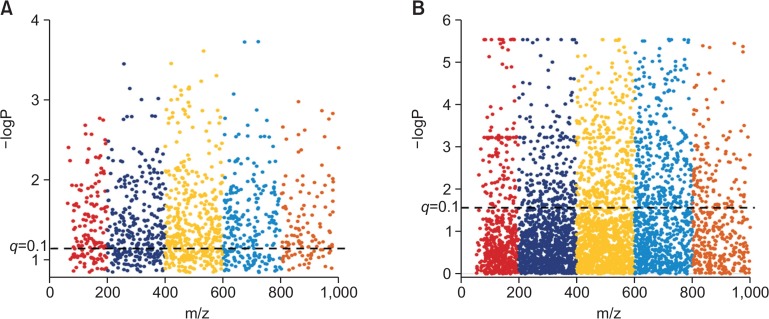
Determining the genes affected due to HFD or catalase deficiency might have an important correlation with our significant features among the top affected metabolic pathways. Therefore, we performed univariate analysis on gene alteration based on their fold change (FC). As mentioned in method section, we observed that 1,038 out of 28,299 genes with threshold of FC 1.5 were affected between WTNF and WTHF possibly due to high fat diet as shown in Fig. 4A. While 1191 out of 28299 genes with threshold of 1.5 between WTHF and CKOHF as shown in Fig. 4B were possibly altered due to catalase deficiency, since both groups were HFD fed. These up- or down-regulated genes based on fold changes were further analyzed for the metabolite–gene integrative analysis. The genes which showed up in gene-metabolite integrated altered pathways were considered important.
Fig. 4.
Fold change analysis of WTNF vs WTHF and WTHF vs CKOHF. This figure represents log2 transformed fold change of each gene. (A) Total 1,038 out of 28,299 genes altered with fold change threshold of 1.5 shown as purple dots. (B) Total 1,191 out of 28,299 genes altered with fold change threshold of 1.5 shown as purple dots.
Integrative pathway analysis with KEGG
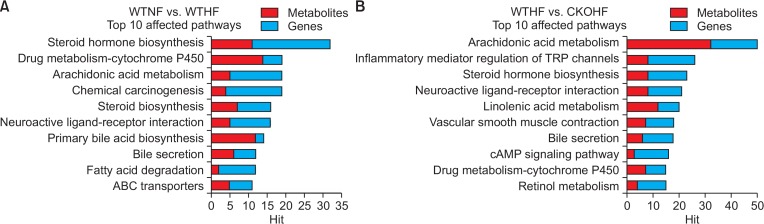
As the major aim of this study was to observe metabolic and transcriptomic alterations caused by HFD and/or catalase deficiency, we first investigated HFD effect between WTNF and WTHF. To do so, based on the 871 significant metabolites from the manhattan plot (Fig. 3A) and the 1,038 altered genes (Fig. 4A), we performed integrative pathway analysis between WTNF and WTHF. The integrative pathway analysis showed highly affected pathways and top 10 pathways were listed with the number of metabolites and genes as shown in Fig. 5A. While, to understand the metabolic and transcriptomic changes induced by catalase deficiency, 1,172 significant metabolites from manhattan plot (Fig. 3B) and 1,191 altered genes (Fig. 4B) among WTHF and CKOHF groups were chosen for KEGG input. The resulting top ten highly affected pathways are shown in Fig. 5B.
Fig. 5.
Integrative pathway analysis of top 10 affected pathways. Fig. 5 represents top 10 affected pathways by both metabolites and genes. (A) The top 10 pathways affected by high fat diet was shown as bar graphs using the number of hit metabolites (red bar) and genes (blue bar) for each pathways. (B) The top 10 pathways affected by catalase knockout was shown as bar graphs using the number of hit metabolites (red bar) and genes (blue bar) for each pathways.
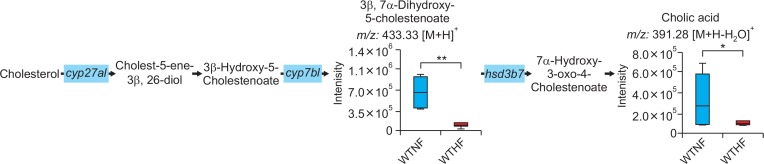
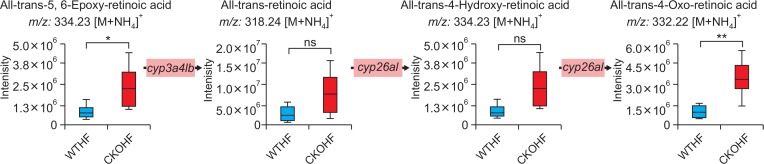
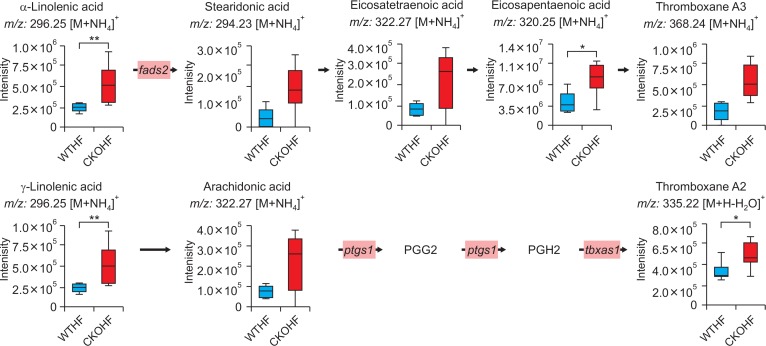
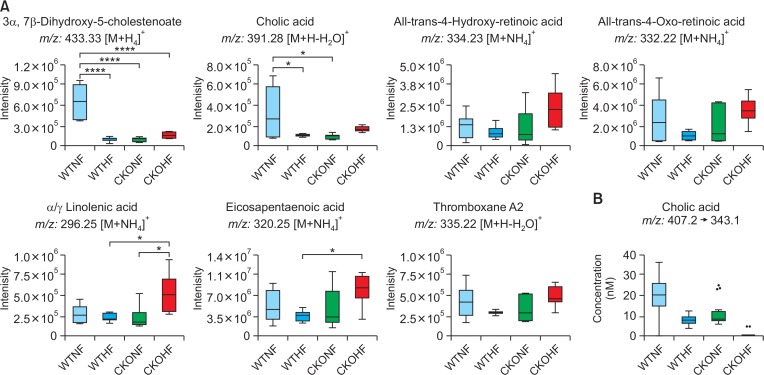
Among top 10 pathways from WTNF vs WTHF, primary bile acid biosynthesis which is related to liver was chosen. As shown in Fig. 6, significant metabolites annotated as cholic acid (391.28 m/z [M+H-H2O]+, q=0.091) and 3β, 7α-dihydroxy-5-cholestenoate (433.3327 m/z [M+H]+, q=0.047) and the gene named cyp7b1 (FC=0.5040) in primary bile acid biosynthesis were highly affected by HFD among WT mice. In addition to cyp7b1, cyp27a1 (FC=0.7754) and hsd3b7 (0.8893) were also affected in primary bile acid biosynthesis but the FC of these genes didn’t meet the threshold 1.5. All metabolites and genes shown in Fig. 6 were found to be consistently down-regulated. Among top 10 pathways from WTHF vs CKOHF, retinol metabolism and linolenic acid metabolism were considered as important because these pathways were related to oxidative stress and immune system, respectively. As shown in Fig. 7, the following significant metabolites; all-trans-5,6-epoxy-retinoic acid or all-trans-4-hydroxy-retinoic acid (334.23 m/z [M+NH4]+, q=0.071), and all-trans-4-oxoretinoic acid (332.22 m/z [M+NH4]+, q=0.027) between WTHF and CKOHF belonged to retinol metabolism. all-trans-Retinoic acid (318.24 m/z [M+NH4]+, q=0.124) was not significantly different but was slightly up-regulated in CKOHF (Fig. 7). The gene named cyp3a41b was found to be highly up-regulated (FC>1.5) in retinol metabolism. cyp26a1 (FC=1.2385) in retinol metabolism was up-regulated but not within the criteria of FC>1.5, however the associated metabolites were significantly increased in CKOHF. While, we previously showed that catalase deficiency elevated macrophage infiltration and accelerated HFD-induced liver inflammation in CKO mice (Piao et al., 2017). Here, in support of our previous results, we found linolenic acid metabolism among the top 10 affected pathways in KEGG analysis between WTHF and CKOHF. Significant metabolites and altered genes that can contribute in liver inflammatory response in NAFLD were as following; α-linolenic acid or γ-linolenic acid (296.25 m/z [M+NH4]+, p=0.006, q=0.039), eicosapentaenoic acid (320.25 m/z [M+NH4]+, q=0.076), and thromboxane A2 (335.22 m/z [M+H-H2O]+, q=0.089), ptgs1 (FC=1.5893) and tbxas1 (FC=2.0721) as shown in Fig. 8. Additionally, fads2 (FC=1.1801) was also increased in CKOHF compared to WTHF but not within the selection threshold 1.5. All significant metabolites and altered genes shown in Fig. 8 were consistently up-regulated. The fold change of all significant metabolites and altered genes shown in each pathway are summarized in Table 2. Further, as shown in Fig. 9A, we separately analyzed the overall tendency of increase or decrease of selected metabolites among 4 groups. In addition, cholic acid was quantified and showed decreased concentration (nM) in WTHF, CKONF and CKOHF compared to WTNF (Fig. 9B). This result concretes our previous analysis that metabolic alteration in retinol metabolism and linolenic acid metabolism were induced not only by single factor but the HFD combined with catalase deficiency. Five out of seven metabolites from our study were further validated with authentic standards as shown in Supplementary Fig. 1–5.
Fig. 6.
Primary bile acid biosynthesis pathway affected by HFD. The bar graphs represent two metabolites, 3β, 7α-dihydroxy-5-cholestenoate and cholic acid, altered significantly between WTNF and WTHF (*q<0.1, **q<0.05). The gene cyp7b1 was found severely down-regulated (FC threshold=1.5) in WTHF shown in light blue boxes. cyp27a1 and hsd3b7 were down-regulated as well but not included in the criteria (FC threshold=1.5).
Fig. 7.
Retinol metabolism pathway altered in CKOHF compared to WTHF. The bar graphs represent that all-trans-5,6-epoxy-retinoic acid, all-trans-4-hydroxy-retinoic acid and all-trans-4-oxo-retinoic acid were significantly different between WTHF and CKOHF (q<0.1). all-trans-Retinoic acid was not significant but increased. Gene expression shown in pink boxes represent cyp3a41b was found highly up-regulated (FC threshold=1.5) in CKOHF compared to WTHF. cyp26a1 was also up-regulated but not within the criteria (FC threshold=1.5). m/z values of all-trans-5,6-epoxy-retinoic acid is identical to all-trans-4-hydroxy-retinoic acid. *q<0.1, **q<0.05, ns=not significant.
Fig. 8.
Linolenic acid metabolism pathway affected in CKOHF compared to WTHF. The bar graphs represent that α/γ-linolenic acid, eicosapentaenoic acid and thromboxane A2 were significantly different in CKOHF compared to WTHF (q<0.1). Gene expression of ptgs1 and tbxas1 in pink boxes were highly up-regulated (FC threshold=1.5) in CKOHF compared to WTHF. FADS2 was up-regulated but not within the criteria (FC threshold=1.5). Stearidonic acid, eicosatetraenoic acid, thromboxane A3 and arachidonic acid showed the tendency of increase in CKOHF compared to WTHF but not significant. m/z values of α-linolenic acid and arachidonic acid were identical to γ-linolenic acid and eicosatetraenoic acid, respectively. *q<0.05 **q<0.01.
Table 2.
Fold change of metabolites and genes for each pathway and group, respectively
| Pathway (Group) | Phenotype | Name | Fold Change |
|---|---|---|---|
| Primary bile acid biosynthesis (WTNF vs WTHF) | Metabolite (m/z) | Cholic acid (391.28) | 0.3071 |
| 3β, 7α-Dihydroxy-5-cholestenoate (433.33) | 0.1501 | ||
| Gene | cyp7b1 | 0.5040 | |
| Retinoic acid metabolism (WTHF vs CKOHF) | Metabolite (m/z) | all-trans-4-Hydroxy-retinoic acid (334.23) | 2.4388 |
| all-trans-4-Oxo-retinoic acid (332.22) | 2.8720 | ||
| Gene | cyp3a41b | 1.5344 | |
| Linolenic acid metabolism(WTHF vs CKOHF) | Metabolite (m/z) | α/γ-Linolenic acid (296.25) | 2.1972 |
| Eicosapentaenoic acid (320.25) | 2.0151 | ||
| Thromboxane A2 (335.22) | 3.3668 | ||
| Gene | ptgs1 | 1.5893 | |
| tbxas1 | 2.0721 |
Fig. 9.
Overall observation of HFD and CKO effect on pathways. (A) This figure represents relative intensities of metabolites related to 3 pathways, primary bile acid biosynthesis, retinol metabolism, and linolenic acid metabolism, in all 4 groups in form of box and whisker plot. Two metabolites from primary bile acid biosynthesis are decreased in WTHF, CKONF and CKOHF compared to WTNF. Five metabolites belonged to retinol metabolism and linolenic acid metabolism showed same tendency increased most in CKOHF among 4 groups. The metabolites were tested with one-way ANOVA with Tukey’s HSD. *p<0.05, ****p<0.001. (B) This figure shows the quantified concentration of cholic acid, a representative composition of bile acid. Cholic acid showed the highest value in WTNF compared to the other groups.
Receiver operating characteristic (ROC) curve of significant metabolites
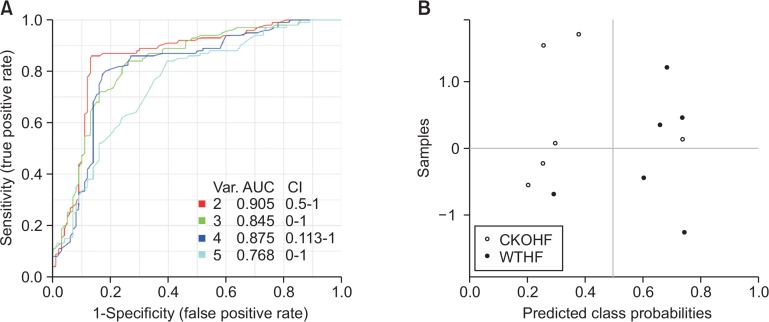
The receiver operating characteristic (ROC) curve with five significant metabolites from retinol metabolism and linolenic acid metabolism between WTHF and CKOHF was performed to investigate AUC, as well as sensitivity and specificity, in order to support our hypothesis of these effects caused by catalase knockout. All metabolites were predicted with AUC>0.78, and showed a good sensitivity (>0.8) and specificity (>0.7) in discrimination of the WTHF and CKOHF (Table 3). With five metabolites, ROC curve showed the highest AUC value in two variable model (AUC=0.905) as shown in Fig. 10A. In addition, the samples were analyzed using two variable model and showed separated by predicted class probability values except one samples from each group (Fig. 10B). The two variable model including all-trans-4-oxo-retinoic acid and thromboxane A2 showed predict accuracy of 84.5% and this value was the highest among the other values from models.
Table 3.
ROC details of retinoic acid and linolenic acid metabolism’s significant metabolites
| m/z | Annotation | AUC | Sensitivity | Specificity |
|---|---|---|---|---|
| 334.23 | all-trans-4-Hydroxy-retinoic acid | 0.89 | 1 | 0.7 |
| 332.22 | all-trans-4-Oxo-retinoic acid | 0.94 | 0.8 | 1 |
| 296.25 | α/γ-Linolenic acid | 0.92 | 0.8 | 1 |
| 320.25 | Eicosapentaenoic acid | 0.89 | 0.8 | 1 |
| 335.22 | Thromboxane A2 | 0.78 | 0.8 | 0.8 |
Fig. 10.
Receiver operating characteristic (ROC) curves of the 5 metabolites in comparison of WTHF and CKOHF. ROC curves were created with 5 significant metabolites. (A) Among five metabolites, two metabolites model showed highest prognostic value of AUC=0.905 with 95% confidence interval. (B) The values of predicted class probabilities of each sample separate the CKOHF (white) from WTHF (black) shown as dots. The observed error rate of 1/6 in CKOHF (n=6) and 1/6 in WTHF (n=6).
Correlation analysis among metabolites and genes
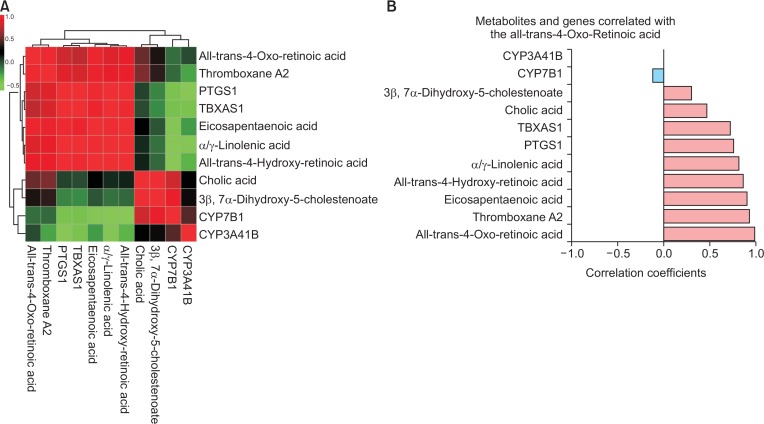
To consolidate our data, we performed correlation analysis with all affected of metabolites with genes from selected pathways. Interestingly, our result showed that metabolites and genes belonging to primary bile acid biosynthesis were clustered together and while those belonging to retinol metabolism and linolenic acid metabolism except cyp3a41b, were clustered together as well (Fig. 11A). Further, we investigated correlation coefficient value of each metabolite or gene against the others. Among them, all-trans-4-oxo-retinoic acid showed positive correlation with others except cyp3a41b and cyp7b1 (Fig. 11B). The other correlation coefficient bar graphs were not shown in this manuscript.
Fig. 11.
Correlation analysis of all metabolites and genes from selected pathways. (A) This figure represents clustered heatmap of correlation with selected metabolites and genes. The red colored metabolites and genes were positively correlated, while negatively correlated metabolites and genes are shown as green colored. (B) Correlation coefficients of each metabolite and gene against all-trans-4-oxo-retinoic acid were shown as horizontal bars. The light blue bar represents negative correlation and the light red bar represents positively correlated metabolites or genes.
DISCUSSION
NAFLD is rapidly growing as one of the most prevalent liver diseases linked to metabolic, cardiovascular, and hepatocellular carcinoma without approved treatment. Since the prevalence of NAFLD in the United States is over 75% of the chronic liver disease cases, this disease represents a major burden to both morbidity and mortality in the US (Hassan et al., 2014). However, NAFLD diagnosis in population studies is usually obtained by ultrasonography, which is known to underestimate the prevalence of fatty liver. NAFLD refers to a spectrum ranging from noninflammatory isolated steatosis to NASH, which is characterized by steatosis, necroinflammatory changes, and varying degrees of liver fibrosis (Arguello et al., 2015). In addition, the pathogenesis of NAFLD remains unknown. In this study, we applied integrative omics to investigate the combined effect of HFD and oxidative stress through catalase deficiency compared to WT in a mouse model to study NAFLD/NASH.
For this study, NAFLD animal model in mice fed NFD or HFD was established. Before investigating metabolic alterations, we measured physiological changes such as body weight, liver weight, ALT and liver TG among NFD or HFD fed WT and CKO mice. Interestingly, CKONF group, although less than WTHF and CKOHF, showed significantly elevated body weight compared to WTNF. CKO affected the body weight of mice, however, it is apparent that HFD combined with CKO significantly contributed to increase in body weight of mice compared to CKONF. Apart from significant elevations in body weight, ALT and TG showed significant increase in CKOHF compared to other groups, indicating that a good NAFLD model was created among the WT or CKO mice fed with HFD.
Based on integrative pathway analysis, we focused on pathway named primary bile acid biosynthesis. The main function of bile acids is not only to emulsify and digest dietary fats and oils into micelles, but also to regulate cholesterol homeostasis (Li et al., 2013). Bile acids are synthesized exclusively in the liver from cholesterol through two major pathways, namely the classic pathway and acidic pathway (Staels and Fonseca, 2009). The enzyme, cyp7b1, is known to catalyze the reaction in the cholesterol catabolic pathway, which converts cholesterol to bile acids (Stiles et al., 2009). The acidic bile acid synthesis pathway is initiated by sterol 27-hydroxylase (cyp27a1) and produces oxysterols, notably 25-hydroxycholesterol and 27-hydroxycholesterol, which are important ligands in regulating inflammation, lipid metabolism, and cell proliferation (Yuan and Bambha, 2015). In our previous study, plasma TG and TC levels were found to be significantly increased only in CKOHF mice compared to those in WTHF mice. However, no difference in body weight, systemic glucose tolerance, and liver insulin signaling was detected between the WTHF and CKOHF mice (Piao et al., 2017). Interestingly, using a metabolomics approach in association with transcriptomics, this study determined that 3 genes (cyp27a1, cyp7b1, and hsd3b7) were down-regulated in WT mice fed with a HFD. Such decreased expression of rate-limiting genes may have caused the decreased production of downstream metabolites. This may imply that an exogenous HFD may have terminated the endogenous bile acid biosynthesis pathway to obstruct excessive fat intake from the intestine affected by bile acids (Yamato et al., 2012). In a recently published study by Mouzaki et al. (2016), it was demonstrated that the level of fecal bile acid including cholic acid was higher in patients with NAFLD/NASH compared to healthy control. Though the median value of BA was increased in feces of NAFL and NASH patients, the maximum values of each group were decreased. On the other hand, our study was performed using mice liver sample and we observed integrative metabolomic and transcriptomic alterations. In comparison to our previous study showing the negligible effect of HFD on WT animals’ TG and TC levels, our result observing altered bile acid biosynthesis among WT mice fed with HFD or NFD, highlights the importance of metabolomics-transcriptomics approaches in investigating a pathological condition at the level of metabolic regulation or variation.
Our previous study confirmed a variety of phenotypes representing the induction of liver injury caused by HFD in CKO mice (Piao et al., 2017). An elevated liver injury phenomenon was promoted by increased levels of hepatic nitrotyrosine (Piao et al., 2017), which was previously shown to be a marker of oxidative stress (Darwish et al., 2007). In this study, we further investigated the metabolic effects caused by oxidative stress in HFD fed CKO mice. Furthermore, Pasquali et al. (Pasquali et al., 2008) previously reported the toxic effects of retinol and its major biologically active metabolite, all-trans retinoic acid (RA), related to pro-oxidant properties. Interestingly, in support of results from Pasquali et al. (2008) and our previous study of oxidative stress in CKO mice, we detected alterations in the retinoic acid pathway of CKO mice. Retinol and retinoic acid enhance intracellular reactive species production and increase catalase activity, which is a major mechanism of defense against cytotoxic effects of H2O2 (Pasquali et al., 2008). Our study observed that the levels of retinoic acids belonging to the pathway was increased significantly in CKOHF compared to that in WTHF mice. In addition, previous studies observed that the mRNA expression levels of monocyte chemoattractant protein-1 (MCP-1) and cyclooxygenase (COX2) was significantly increased in CKOHF compared to those in WTHF mice, as determined in the immunohistochemically stained F4/80 liver homogenate. In accordance, this study showed significantly altered effects of CKO on linolenic acid metabolism. Metabolites in this pathway, such as arachidonic acid and thromboxane A2, are well-known important mediators during the inflammatory response (Kuehl and Egan, 1980; Nakahata, 2008). Our results suggest that a severe inflammatory response occurred due to catalase deficiency in mice fed a HFD.
In essence of unmatched pathways between transcriptomics and metabolomics, we observed retinol metabolism affected by genes among WTNF vs WTHF. This may indicate that retinol metabolism was affected by catalase knockout with high fat diet. While, cancer related pathways showed increased number of genes in WTHF vs CKOHF compared to WTNF vs WTHF as following pathway name and changes of hit genes: cytokine-cytokine receptor interaction (18→31), PI3K-Akt signaling pathway (13→30) and MAPK signaling pathway (15→27). This is interesting, as it imply that transcriptomic alterations caused by catalase knockout with high fat diet does not only propose the hepatic disorder but also contributes in the occurrence of cancer. However, further studies should confirm these relationships.
In spite of such findings, our study has some limitations. First, the number of mice used in this study was six per group, hence, the performance of our statistical analysis needs to be validated in a large scale of in vivo/in vitro study. Second, for the investigation of metabolic alteration induced by nonalcoholic fatty liver disease, extracted liver samples were used in this study. The result from this study further need to be validated in serum or plasma so that liver biopsy accompanied with histological observation can be replaced with serological test for identification of nonalcoholic fatty liver disease.
In summary, our study highlights the explanation of unknown mechanism of NAFLD induced by HFD and CKO using integrative omics combined with physiological changes in body weight, liver weight, ALT and liver triglyceride. Our results provide a better understanding of the mechanisms of NAFLD and a basis of further research or treatment strategy by considering catalase deficiency as an important factor of liver injury and modification of high fat diet to normal fat diet to prevent the NAFLD prevalence.
Acknowledgments
This work was carried out with the support of “Cooperative Research Program for Agriculture Science and Technology Development (Project No. PJ01345402)” Rural Development Administration, Republic of Korea; National Research Foundation of Korea under grant NRF-2017R1A2B4003890 and NRF-2016R1A2B4006575.
Footnotes
CONFLICT OF INTEREST
The authors report no conflict of interest.
REFERENCES
- Arguello G, Balboa E, Arrese M, Zanlungo S. Recent insights on the role of cholesterol in non-alcoholic fatty liver disease. Biochim. Biophys. Acta. 2015;1852:1765–1778. doi: 10.1016/j.bbadis.2015.05.015. [DOI] [PubMed] [Google Scholar]
- Cani PD, Bibiloni R, Knauf C, Waget A, Neyrinck AM, Delzenne NM, Burcelin R. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet–induced obesity and diabetes in mice. Diabetes. 2008;57:1470–1481. doi: 10.2337/db07-1403. [DOI] [PubMed] [Google Scholar]
- Cavill R, Jennen D, Kleinjans J, Briede JJ. Transcriptomic and metabolomic data integration. Brief. Bioinform. 2016;17:891–901. doi: 10.1093/bib/bbv090. [DOI] [PubMed] [Google Scholar]
- Clarke GM, Anderson CA, Pettersson FH, Cardon LR, Morris AP, Zondervan KT. Basic statistical analysis in genetic case-control studies. Nat. Protoc. 2011;6:121–133. doi: 10.1038/nprot.2010.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwish RS, Amiridze N, Aarabi B. Nitrotyrosine as an oxidative stress marker: evidence for involvement in neurologic outcome in human traumatic brain injury. J. Trauma. 2007;63:439–442. doi: 10.1097/TA.0b013e318069178a. [DOI] [PubMed] [Google Scholar]
- Gao Y, Lu Y, Huang S, Gao L, Liang X, Wu Y, Wang J, Huang Q, Tang L, Wang G, Yang F, Hu S, Chen Z, Wang P, Jiang Q, Huang R, Xu Y, Yang X, Ong CN. Identifying early urinary metabolic changes with long-term environmental exposure to cadmium by mass-spectrometry-based metabolomics. Environ. Sci. Technol. 2014;48:6409–6418. doi: 10.1021/es500750w. [DOI] [PubMed] [Google Scholar]
- Gomez-Cabrero D, Abugessaisa I, Maier D, Teschendorff A, Merkenschlager M, Gisel A, Ballestar E, Bongcam-Rudloff E, Conesa A, Tegner J. Data integration in the era of omics: current and future challenges. BMC Syst. Biol. 2014;8(Suppl 2):I1. doi: 10.1186/1752-0509-8-S2-I1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariri N, Thibault L. High-fat diet-induced obesity in animal models. Nutr. Res. Rev. 2010;23:270–299. doi: 10.1017/S0954422410000168. [DOI] [PubMed] [Google Scholar]
- Hassan K, Bhalla V, El Regal ME, A-Kader HH. Nonalcoholic fatty liver disease: a comprehensive review of a growing epidemic. World J. Gastroenterol. 2014;20:12082–12101. doi: 10.3748/wjg.v20.i34.12082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horgan RP, Kenny LC. ‘Omic’ technologies: genomics, transcriptomics, proteomics and metabolomics. TOG. 2011;13:189–195. doi: 10.1576/toag.13.3.189.27672. [DOI] [Google Scholar]
- Hwang I, Lee J, Huh JY, Park J, Lee HB, Ho YS, Ha H. Catalase deficiency accelerates diabetic renal injury through peroxisomal dysfunction. Diabetes. 2012;61:728–738. doi: 10.2337/db11-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehl FA, Jr, Egan RW. Prostaglandins, arachidonic acid, and inflammation. Science. 1980;210:978–984. doi: 10.1126/science.6254151. [DOI] [PubMed] [Google Scholar]
- Li T, Francl JM, Boehme S, Chiang JY. Regulation of cholesterol and bile acid homeostasis by the cholesterol 7alpha-hydroxylase/steroid response element-binding protein 2/microR-NA-33a axis in mice. Hepatology. 2013;58:1111–1121. doi: 10.1002/hep.26427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Thomas T, Storlien L, Huang X. Development of high fat diet-induced obesity and leptin resistance in C 57 Bl/6 J mice. Int. J. Obes. Relat. Metab. Disord. 2000;24:639–646. doi: 10.1038/sj.ijo.0801209. [DOI] [PubMed] [Google Scholar]
- Machado MV, Michelotti GA, Xie G, Almeida Pereira T, Boursier J, Bohnic B, Guy CD, Diehl AM. Mouse models of diet-induced nonalcoholic steatohepatitis reproduce the heterogeneity of the human disease. PLoS ONE. 2015;10:e0127991. doi: 10.1371/journal.pone.0127991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouzaki M, Wang AY, Bandsma R, Comelli EM, Arendt BM, Zhang L, Fung S, Fischer SE, McGilvray IG, Allard JP. Bile acids and dysbiosis in non-alcoholic fatty liver disease. PLoS ONE. 2016;11:e0151829. doi: 10.1371/journal.pone.0151829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata N. Thromboxane A2: physiology/pathophysiology, cellular signal transduction and pharmacology. Pharmacol. Ther. 2008;118:18–35. doi: 10.1016/j.pharmthera.2008.01.001. [DOI] [PubMed] [Google Scholar]
- Norris AW, Chen L, Fisher SJ, Szanto I, Ristow M, Jozsi AC, Hirshman MF, Rosen ED, Goodyear LJ, Gonzalez FJ, Spiegelman BM, Kahn CR. Muscle-specific PPARgamma-deficient mice develop increased adiposity and insulin resistance but respond to thiazolidinediones. J. Clin. Invest. 2003;112:608–618. doi: 10.1172/JCI17305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obuchowski NA, Bullen JA. Receiver operating characteristic (ROC) curves: review of methods with applications in diagnostic medicine. Phys. Med. Biol. 2018;63:07TR01. doi: 10.1088/1361-6560/aab4b1. [DOI] [PubMed] [Google Scholar]
- Panchal SK, Poudyal H, Iyer A, Nazer R, Alam A, Diwan V, Kauter K, Sernia C, Campbell F, Ward L. High-carbohydrate, high-fat diet-induced metabolic syndrome and cardiovascular remodeling in rats. J. Cardiovasc. Pharmacol. 2011;57:611–624. doi: 10.1097/FJC.0b013e3181feb90a. [DOI] [PubMed] [Google Scholar]
- Park YH, Fitzpatrick AM, Medriano CA, Jones DP. High-resolution metabolomics to identify urine biomarkers in corticosteroid-resistant asthmatic children. J. Allergy Clin. Immunol. 2016;139:1518–1524.e4. doi: 10.1016/j.jaci.2016.08.018. [DOI] [PubMed] [Google Scholar]
- Park YH, Shi YP, Liang B, Medriano CAD, Jeon YH, Torres E, Uppal K, Slutsker L, Jones DP. High-resolution metabolomics to discover potential parasite-specific biomarkers in a Plasmodium falciparum erythrocytic stage culture system. Malar. J. 2015;14:122. doi: 10.1186/s12936-015-0651-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquali MA, Gelain DP, Zanotto-Filho A, de Souza LF, de Oliveira RB, Klamt F, Moreira JC. Retinol and retinoic acid modulate catalase activity in Sertoli cells by distinct and gene expression-independent mechanisms. ToxicolIn Vitro. 2008;22:1177–1183. doi: 10.1016/j.tiv.2008.03.007. [DOI] [PubMed] [Google Scholar]
- Piao L, Choi J, Kwon G, Ha H. Endogenous catalase delays high-fat diet-induced liver injury in mice. Korean J. Physiol. Pharmacol. 2017;21:317–325. doi: 10.4196/kjpp.2017.21.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasundaram D, Selbig J. More effort - more results: recent advances in integrative ‘omics’ data analysis. Curr. Opin. Plant Biol. 2016;30:57–61. doi: 10.1016/j.pbi.2015.12.010. [DOI] [PubMed] [Google Scholar]
- Ruhl CE, Everhart JE. Fatty liver indices in the multiethnic United States National Health and Nutrition Examination Survey. Aliment. Pharmacol. Ther. 2015;41:65–76. doi: 10.1111/apt.13012. [DOI] [PubMed] [Google Scholar]
- Singh S, Allen AM, Wang Z, Prokop LJ, Murad MH, Loomba R. Fibrosis progression in nonalcoholic fatty liver vs nonalcoholic steatohepatitis: a systematic review and meta-analysis of paired-biopsy studies. Clin. Gastroenterol. Hepatol. 2015;13:643–54.e1–9. doi: 10.1016/j.cgh.2014.04.014. quiz e39–e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staels B, Fonseca VA. Bile acids and metabolic regulation: mechanisms and clinical responses to bile acid sequestration. Diabetes Care. 2009;32(Suppl 2):S237–S245. doi: 10.2337/dc09-S355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart JD, Bolt HM. Metabolomics: biomarkers of disease and drug toxicity. Arch. Toxicol. 2011;85:3–4. doi: 10.1007/s00204-010-0635-4. [DOI] [PubMed] [Google Scholar]
- Stiles AR, McDonald JG, Bauman DR, Russell DW. CYP7B1: one cytochrome P450, two human genetic diseases, and multiple physiological functions. J. Biol. Chem. 2009;284:28485–28489. doi: 10.1074/jbc.R109.042168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Videla LA, Rodrigo R, Orellana M, Fernandez V, Tapia G, Quinones L, Varela N, Contreras J, Lazarte R, Csendes A, Rojas J, Maluenda F, Burdiles P, Diaz JC, Smok G, Thielemann L, Poniachik J. Oxidative stress-related parameters in the liver of non-alcoholic fatty liver disease patients. Clin. Sci. (Lond) 2004;106:261–268. doi: 10.1042/CS20030285. [DOI] [PubMed] [Google Scholar]
- Want EJ, O’Maille G, Smith CA, Brandon TR, Uritboonthai W, Qin C, Trauger SA, Siuzdak G. Solvent-dependent metabolite distribution, clustering, and protein extraction for serum profiling with mass spectrometry. Anal. Chem. 2006;78:743–752. doi: 10.1021/ac051312t. [DOI] [PubMed] [Google Scholar]
- Wolf S, Schmidt S, Muller-Hannemann M, Neumann S. In silico fragmentation for computer assisted identification of metabolite mass spectra. BMC Bioinformatics. 2010;11:148. doi: 10.1186/1471-2105-11-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamato M, Shiba T, Ide T, Seri N, Kudo W, Ando M, Yamada K, Kinugawa S, Tsutsui H. High-fat diet-induced obesity and insulin resistance were ameliorated via enhanced fecal bile acid excretion in tumor necrosis factor-alpha receptor knockout mice. Mol. Cell. Biochem. 2012;359:161–167. doi: 10.1007/s11010-011-1010-3. [DOI] [PubMed] [Google Scholar]
- Younossi ZM, Stepanova M, Afendy M, Fang Y, Younossi Y, Mir H, Srishord M. Changes in the prevalence of the most common causes of chronic liver diseases in the United States from 1988 to 2008. Clin. Gastroenterol. Hepatol. 2011;9:524–530.e1. doi: 10.1016/j.cgh.2011.03.020. quiz e60. [DOI] [PubMed] [Google Scholar]
- Yu T, Park Y, Johnson JM, Jones DP. apLCMS—adaptive processing of high-resolution LC/MS data. Bioinformatics. 2009;25:1930–1936. doi: 10.1093/bioinformatics/btp291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L, Bambha K. Bile acid receptors and nonalcoholic fatty liver disease. World J. Hepatol. 2015;7:2811–2818. doi: 10.4254/wjh.v7.i28.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.