Abstract
Background
The association between gene polymorphisms and the risk of primary nephrotic syndrome (PNS) is uncovering recently. This study aims to investigate the relationship between single nucleotide polymorphisms (SNPs) on HLA‐DQA1 gene and the risk of PNS.
Methods
In this study, we genotyped eight single nucleotide polymorphisms (SNPs) in the HLA‐DQA1 gene in 501 PNS patients and 532 healthy people in Chinese population. Then we analyzed associations of these SNPs with the clinical features in primary nephrotic syndrome of children in Chinese population.
Results
Significant associations with PNS were found on missense SNP rs1129740 (GG vs AA, odds ratio (OR) = 1.987, 95% confidence interval (CI) = 1.468‐2.652, P = 0.00177049) and rs1047992 (AA vs GG, OR = 1.857, 95% CI = 1.325‐2.391, P = 1.1073E‐10) of the HLA‐DQA1 gene.
Conclusions
This work suggests SNPs of HLA‐DQA1 are risk factors for PNS in Chinese population, which implies roles of immune response in the pathogenesis of PNS.
Keywords: children primary nephrotic syndrome, HLA‐DQA1, polymorphism
1. INTRODUCTION
Primary nephrotic syndrome (PNS) is a kidney disorder that featured by edema, hypoalbuminemia, hyperlipidemia, and massive proteinuria and suffered during childhood.1, 2 The approximate prevalence of PNS is about 16 cases per 100 000 children, which makes it the most frequent glomerular disorder of children.3, 4, 5 The risk factors and pathogenesis underlying PNS have not been well elucidated. Recent studies suggested that genetic factors might justify, a minimum of partially, such risk of children PNS.6, 7
The human leukocyte antigen (HLA) is a gene family encoding the major histocompatibility complex (MHC) proteins, which are liable for the regulation of the immune system in humans.8 HLA genes area unit extremely polymorphic, which suggests that they need many alternative alleles, permitting them to fine‐tune the immune response system. HLA genes play important roles in various diseases.8, 9, 10, 11 Major histocompatibility complex, class II, DQ alpha 1 and DQ beta 1, conjointly referred to as HLA‐DQA1 and HLA‐DQB1, locate on short arm of chromosome 6 (6p21.3) and encode MHC class II present antigens.12 Recent study suggested that single nucleotide polymorphisms (SNPs) (rs1129740, rs1140342, rs1071630, and rs1140343) in HLA‐DQA1 and HLA‐DQB1 were significantly associated with childhood‐onset steroid‐sensitive nephrotic syndrome (SSNS).13 However, it's still unclear whether SNPs in HLA‐DQA1 and HLA‐DQB1 are associated with PNS.
To investigate whether gene polymorphisms of HLA‐DQA1 and HLA‐DQB1 are associated with risk of PNS, we genotyped and analyzed eight SNPs (rs1129740, rs1140342, rs1047992, rs12722051, rs1071630, rs1130432, rs9273651, and rs1140343) in the HLA‐DQA1 and HLA‐DQB1 gene in 501 PNS patients and 532 healthy children, which are from the Chinese Han population. The results indicated gene polymorphisms (rs1129740 and rs1047992) of HLA‐DQA1 but not that of HLA‐DQB1 are significantly associated with the risk of PNS. The results of this study suggest a potential contribution of HLA‐DQA1 gene in the pathogenesis of PNS.
2. MATERIALS AND METHODS
2.1. Patients
All participants (including PNS patients and healthy controls) were enrolled in the study between January 1, 2015, and December 31, 2017, in Xuzhou Children's Hospital and in Chinese Han population. Written consent was obtained from legal guardians of all participants with a full rationalization of the procedure. The look of this study was approved by the ethics panel of Xuzhou Children's Hospital in accordance with the declaration of national capital. PNS patients were diagnosed by PNS specialists through clinical examinations. Criteria for enrollment as an impact enclosed no history of nephrosis, traditional diagnosis, and no puffiness.
2.2. DNA genotyping
DNA of patients and control population was obtained from blood samples using the QIAamp DNA Blood Mini Kit (QIAGEN, Valencia, CA, USA). PCR system was used to capture selected SNPs. The PCR products were sequenced by ABI‐PRISM 3730 genetic analyzer (Sequenom, Inc., San Diego, CA, USA). We genotyped four SNPs in the HLA‐DQA1 gene and four SNPs in the HLA‐DQB1 gene. All the eight SNPs with minor allele frequencies (MAF) > 0.1 in the Chinese population were selected from the dbSNP database (www.ncbi.nlm.nih.gov/SNP). All the eight SNPs were missense SNPs.
2.3. PCR
PCR system for 50 μL was as follows: the reaction mixture contained 2 μL DNA, 2 μL of every primer, 4 μL dNTP, 5 μL PCR buffer, 0.25 μL of Taq enzyme (Takara, Japan), and double water to a volume of 50 μL. The sport conditions were as follows: the initial denaturation at 94°C for 5 minutes, thirty‐nine amplification cycles were administrated per the subsequent temperature profile: 30 second at 94°C, 45 second at 53.7°C, 50 second at 72. The ultimate extension lasted for 5 minutes at 72°C.
2.4. Statistical analysis
Statistical analysis was performed with R (version 3.4.1; RStudio, Boston, MA, USA). The discrepancies in genotype frequencies and allele at single loci between PNS patients and healthy controls were determined by Chi‐square test. Hardy–Weinberg equilibrium (HWE) test was performed using SPSS. The relative risk was determined by the odds ratio (OR), and a 95% confidence interval (CI) was estimated for each OR. Linkage disequilibrium (LD) was determined by R 2. P‐values are two‐tailed, and the cut line of statistical significance was 0.05.
3. RESULTS
3.1. rs1129740 and rs1047992 are associated with PNS
The case–control study consisted of 501 PNS patients (297 boys and 207 girls with a mean age ± SD of 4.8 ± 3.76 years) and 532 healthy controls (289 boys and 243 girls with a mean age ± SD of 5.16 ± 4.26 years) (Table 1). The distribution of the genotypes of the eight SNPs was in HWE in all the controls and in most of the patients, except for rs1129740 and rs1047992 in PNS patients. There were no significant differences in allele frequencies or genotype frequencies between PNS patients (n = 501) and controls (n = 532) for six SNPs (Table 2). The allele frequencies and genotype frequencies of rs1129740 and rs1047992 are significantly different between PNS patients and controls (P < 0.05). The G allele frequency of rs1129740 was higher in PNS patients than in controls (P = 0. 00177049, odds ratio (OR) = 1.987, 95% confidence interval (CI) = 1.468‐2.652). The A allele frequency of rs1047992 was higher in PNS patients than in controls (OR = 1.857, 95% CI = 1.325‐2.391, P = 1.1073E‐10). The genotype frequencies of rs1129740 (P = 0.00014265) and rs1047992 (P = 0.7.916E‐10) are significantly different between PNS patients and controls, respectively. There are no significant differences between PNS patients and controls in either allele frequencies or genotype frequencies in the other six SNPs.
Table 1.
Characteristics of samples
| Patients (n = 501) | Control (n = 532) | |
|---|---|---|
| Age (years) | 4.8 ± 3.76 | 5.16 ± 4.26 |
| Male | 297 | 289 |
| Female | 204 | 243 |
| Body weight (kg) | 39 ± 15 | 41 ± 19 |
| Disease duration (years) | 2.1 ± 2.9 | 2.9 ± 3.5 |
Table 2.
Allele and genotype frequencies of SNPs
| Group | Allele frequency | P | Genotype frequency | P | H–W | |||
|---|---|---|---|---|---|---|---|---|
| rs1129740 | A | G | AA | AG | GG | |||
| Patients | 469 | 533 | 0.00177049 | 95 | 279 | 127 | 0.00014265 | 0.029963 |
| Control | 541 | 523 | 142 | 257 | 133 | 0.741149 | ||
| rs1140342 | C | T | CC | TC | TT | |||
| Patients | 516 | 486 | 0.05720511 | 127 | 262 | 112 | 0.11829002 | 0.577095 |
| Control | 549 | 515 | 145 | 259 | 128 | 0.843209 | ||
| rs1047992 | A | G | AA | AG | GG | |||
| Patients | 309 | 693 | 1.1073E‐10 | 46 | 217 | 238 | 7.916E‐10 | 0.942346 |
| Control | 239 | 825 | 25 | 189 | 318 | 0.900154 | ||
| rs12722051 | A | T | AA | AT | TT | |||
| Patients | 677 | 325 | 0.056872 | 231 | 215 | 55 | 0.2671501 | 0.896503 |
| Control | 717 | 347 | 249 | 219 | 64 | 0.342857 | ||
| rs1071630 | A | T | AA | AT | TT | |||
| Patients | 529 | 473 | 0.05298636 | 139 | 251 | 111 | 0.27128101 | 0.993415 |
| Control | 568 | 496 | 146 | 276 | 110 | 0.620356 | ||
| rs1130432 | C | T | CC | TC | TT | |||
| Patients | 415 | 587 | 0.05409631 | 75 | 265 | 161 | 0.18676304 | 0.131447 |
| Control | 446 | 618 | 89 | 268 | 175 | 0.727897 | ||
| rs9273651 | C | T | CC | TC | TT | |||
| Patients | 191 | 811 | 0.05125835 | 12 | 167 | 322 | 0.05302142 | 0.199128 |
| Control | 197 | 867 | 19 | 159 | 354 | 0.976268 | ||
| rs1140343 | T | G | TT | TG | GG | |||
| Patients | 537 | 465 | 0.04134722 | 134 | 269 | 98 | 0.2753932 | 0.20592 |
| Control | 583 | 481 | 152 | 279 | 101 | 0.401135 | ||
SNP, single nucleotide polymorphism.
3.2. Linkage disequilibrium analysis
The result of the linkage disequilibrium (LD) analysis between the eight SNPs indicated that no SNP pairs exhibited strong LD (R 2 > 0.8) (Figure 1). These SNPs pairs in this study only showed modest or weak LD (Table 3). It revealed that these SNPs play their roles independently in the studied population. Therefore, no haplotypes were built in this study.
Figure 1.
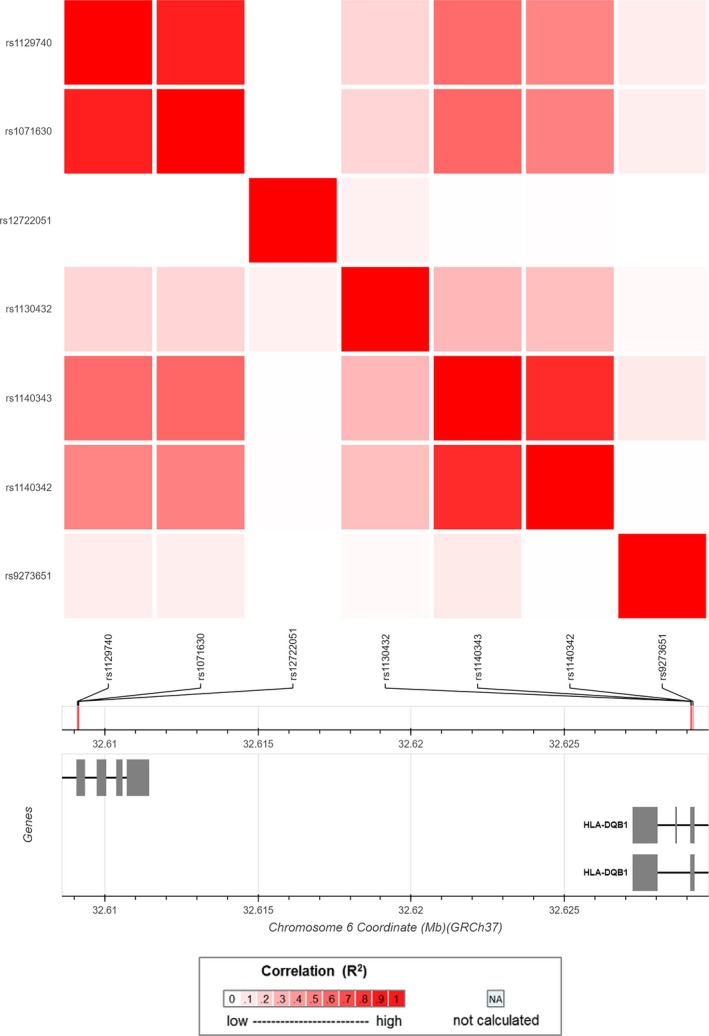
The linkage disequilibrium (LD) analysis between the SNPs
Table 3.
The linkage disequilibrium (R 2) among the SNPs
| R 2 | rs1129740 | rs1071630 | rs12722051 | rs1130432 | rs1140343 | rs1140342 | rs9273651 | rs1047992 |
|---|---|---|---|---|---|---|---|---|
| rs1129740 | 1 | 0.719 | 0.011 | 0.172 | 0.597 | 0.469 | 0.083 | 0.098 |
| rs1071630 | 0.719 | 1 | 0.009 | 0.186 | 0.656 | 0.524 | 0.069 | 0.007 |
| rs12722051 | 0.011 | 0.009 | 1 | 0.069 | 0.011 | 0.012 | 0.001 | 0.086 |
| rs1130432 | 0.172 | 0.186 | 0.069 | 1 | 0.329 | 0.277 | 0.04 | 0.189 |
| rs1140343 | 0.597 | 0.656 | 0.011 | 0.329 | 1 | 0.807 | 0.078 | 0.019 |
| rs1140342 | 0.469 | 0.524 | 0.012 | 0.277 | 0.807 | 1 | 0.001 | 0.075 |
| rs9273651 | 0.083 | 0.069 | 0.001 | 0.04 | 0.078 | 0.001 | 1 | 0.061 |
| rs1047992 | 0.098 | 0.007 | 0.086 | 0.189 | 0.019 | 0.075 | 0.061 | 1 |
SNP, single nucleotide polymorphism.
4. DISCUSSION
Primary nephrotic syndrome (PNS) is a childhood kidney disorder that featured by edema, hypoalbuminemia, hyperlipidemia, and massive proteinuria and suffered.1, 2 Recent studies suggested that genetic factors, such as single nucleotide polymorphisms (SNPs) of genes, might justify, a minimum of partially, the risk of many types of diseases,14, 15, 16 as well as children PNS.6, 7 In this study, we found significant associations between missense SNP rs1129740 (odds ratio (OR) = 1.987, 95% confidence interval (CI) = 1.468‐2.652, P = 0.00177049) and rs1047992 (OR = 1.857, 95% CI = 1.325‐2.391, P = 1.1073E‐10) of the HLA‐DQA1 gene and the risk of children PNS.
The human leukocyte antigen (HLA) is a gene family encoding the major histocompatibility complex (MHC) proteins, which are liable for the regulation of the immune system in humans.8 HLA genes area unit extremely polymorphic and play important roles in various diseases.17, 18, 19 Studies of smaller sample size from totally different populations have antecedently reportable association between SNPs in HLA‐DQA1 and HLA‐DQB1 and childhood‐onset steroid‐sensitive nephrotic syndrome13, which was limited by sample sizes and was restricted to different population.20, 21, 22 This study aims to investigate whether SNPs of HLA‐DQA1 and HLA‐DQB1 are associated with risk of PNS in 501 PNS patients and 532 healthy controls of Chinese Han population. The results indicated that SNPs of HLA‐DQA1 but not that of HLA‐DQB1 are associated with the risk of PNS in Chinese Han population.
The lack of LD between rs1129740 and rs1047992, which are the SNPs associated with PNS, in this study suggests that the loci found in this study could represent independent mechanism. The rs1129740 is the risk locus for childhood‐onset steroid‐sensitive and steroid‐resistant nephrotic syndrome.13, 23 The further finding of HLA loci related to the PNS suggests that all of them share an underlying immune‐mediated mechanism in illness pathological process.24, 25 Our ability to duplicate this locus in a totally different population implied the fact that the risk factor rs1129740 might not be population specific. Meanwhile, the newly identified rs1047992 might be a new risk factor for PNS in Chinese population.
Generally, this study discovered the association of rs1129740 and rs1047992 on HLA‐DQA1 with the risk of PNS in Chinese Han population. It extends our knowledge on the pathogenesis of PNS and may promote diagnostic and therapeutic strategies for PNS.
AUTHORS CONTRIBUTION
Bingbing Zhu, Lijun Tian, and Huandan Yang performed the experiments. Tingting Yuan, Juan Lv, and Qianqian Peng analyzed the data. Ruifeng Zhang wrote and edited the manuscript.
Zhu B, Zhang R, Yang H, et al. Association of HLA‐DQA1 gene polymorphisms with the risk of children primary nephrotic syndrome in Chinese population. J Clin Lab Anal. 2019;33:e22623 10.1002/jcla.22623
REFERENCES
- 1. Sinha A, Bagga A. Nephrotic syndrome. Indian J Pediatr. 2012;79(8):1045‐1055. [DOI] [PubMed] [Google Scholar]
- 2. He B, Lu C, Zheng G, et al. Combination therapeutics in complex diseases. J Cell Mol Med. 2016a;20(12):2231‐2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Eddy AA, Symons JM. Nephrotic syndrome in childhood. Lancet. 2003;362(9384):629‐639. [DOI] [PubMed] [Google Scholar]
- 4. Greenbaum LA, Benndorf R, Smoyer WE. Childhood nephrotic syndrome–current and future therapies. Nat Rev Nephrol. 2012;8(8):445‐458. [DOI] [PubMed] [Google Scholar]
- 5. He B, Lu C, Wang ML, et al. Drug discovery in traditional Chinese medicine: from herbal fufang to combinatory drugs. Science. 2015;350(6262):S74‐S76. [Google Scholar]
- 6. Yang F, Lai X, Deng L, et al. Association of endothelin‐1 gene polymorphisms with the clinical phenotype in primary nephrotic syndrome of children. Life Sci. 2014;118(2):446‐450. [DOI] [PubMed] [Google Scholar]
- 7. Jin J, Ding G, Bao H, et al. Correlation between PPAR Gene polymorphisms and primary nephrotic syndrome in children. PPAR Res. 2013;2013:927915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Howell WM. HLA and disease: guilt by association. Int J Immunogenet. 2014;41(1):1‐12. [DOI] [PubMed] [Google Scholar]
- 9. He B, Zhang H, Shi T. A comprehensive analysis of the dynamic biological networks in HCV induced hepatocarcinogenesis. PLoS ONE. 2011;6(4):e18516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dias FC, Castelli EC, Collares CV, Moreau P, Donadi EA. The role of HLA‐G molecule and HLA‐G gene polymorphisms in tumors, viral hepatitis, and parasitic diseases. Front Immunol. 2015;6:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. He B, Qiu X, Li P, Wang L, Lv Q, Shi T. HCCNet: an integrated network database of hepatocellular carcinoma. Cell Res. 2010;20(6):732‐734. [DOI] [PubMed] [Google Scholar]
- 12. Ruggeri A, Paviglianiti A, Gluckman E, Rocha V. Impact of HLA in cord blood transplantation outcomes. HLA. 2016;87(6):413‐421. [DOI] [PubMed] [Google Scholar]
- 13. Gbadegesin RA, Adeyemo A, Webb NJ, et al. HLA‐DQA1 and PLCG2 are candidate risk loci for childhood‐onset steroid‐sensitive nephrotic syndrome. J Am Soc Nephrol. 2015;26(7):1701‐1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. He B, Lu P, Guan L, et al. Identifying key regulating miRNAs in hepatocellular carcinomas by an omics' method. Oncotarget. 2017;8(61):103919‐103930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ramirez‐Bello J, Vargas‐Alarcon G, Tovilla‐Zarate C, Fragoso JM. Single nucleotide polymorphisms (SNPs): functional implications of regulatory‐SNP (rSNP) and structural RNA (srSNPs) in complex diseases. Gac Med Mex. 2013;149(2):220‐228. [PubMed] [Google Scholar]
- 16. He B, Li T, Guan L, et al. CTNNA3 is a tumor suppressor in hepatocellular carcinomas and is inhibited by miR‐425. Oncotarget. 2016b;7(7):8078‐8089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhu QG, Zhang SM, Ding XX, He B, Zhang HQ. Driver genes in non‐small cell lung cancer: characteristics, detection methods, and targeted therapies. Oncotarget. 2017;8(34):57680‐57692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ding XX, Zhu QG, Zhang SM, et al. Precision medicine for hepatocellular carcinoma: driver mutations and targeted therapy. Oncotarget. 2017;8(33):55715‐55730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dyer P, McGilvray R, Robertson V, Turner D. Status report from ‘double agent HLA’: health and disease. Mol Immunol. 2013;55(1):2‐7. [DOI] [PubMed] [Google Scholar]
- 20. Manju MA, Candel MJ, Berger MP. Optimal and maximin sample sizes for multicentre cost‐effectiveness trials. Stat Methods Med Res. 2015;24(5):513‐539. [DOI] [PubMed] [Google Scholar]
- 21. Huang JH, Su QM, Yang J, et al. Sample sizes in dosage investigational clinical trials: a systematic evaluation. Drug Des Devel Ther. 2015;9:305‐312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. He B, Zhang ZK, Liu J, et al. Bioinformatics and microarray analysis of miRNAs in aged female mice model implied new molecular mechanisms for impaired fracture healing. Int J Mol Sci. 2016c;17(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Adeyemo A, Esezobor C, Solarin A, et al. HLA‐DQA1 and APOL1 as risk loci for childhood‐onset steroid‐sensitive and steroid‐resistant nephrotic syndrome. Am J Kidney Dis. 2018;71(3):399‐406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Drachman R, Schlesinger M, Drukker A. The immune system in primary nephrotic syndrome in childhood. Harefuah. 1987;112(8):394‐397. [PubMed] [Google Scholar]
- 25. Aydin Z, Gursu M, Ozturk S, Kilicaslan I, Kazancioglu R. A case of primary immune deficiency presenting with nephrotic syndrome. NDT Plus. 2010;3(5):456‐458. [DOI] [PMC free article] [PubMed] [Google Scholar]


