Abstract
Biomolecular monitoring in the gastrointestinal tract could offer rapid, precise disease detection and management but is impeded by access to the remote and complex environment. Here, we present an ingestible micro-bio-electronic device (IMBED) for in situ biomolecular detection based on environmentally resilient biosensor bacteria and miniaturized luminescence readout electronics that wirelessly communicate with an external device. As a proof of concept, we engineer heme-sensitive probiotic biosensors and demonstrate accurate diagnosis of gastrointestinal bleeding in swine. Additionally, we integrate alternative biosensors to demonstrate modularity and extensibility of the detection platform. IMBEDs enable new opportunities for gastrointestinal biomarker discovery and could transform the management and diagnosis of gastrointestinal disease.
Microorganisms living on and in the human body constantly interrogate their biochemical surroundings and alter gene expression to adapt to changing environments. Synthetic biology enables the robust engineering of living cells with increasingly complex genetic circuits to sense biological inputs and control gene expression (1). Whole-cell biosensors harness this sensing ability to detect analytes associated with human health (2) or environmental contamination (3). Owing to their innate robust functionality in complex physiological environments, biosensors have been developed to sense clinically relevant biomarkers in serum or urine ex vivo (4), as well as gut biomolecules supplemented in diet (5–7) or generated during disease (8–10). However, despite their promise as noninvasive diagnostics, biosensors have yet to be employed for clinically compatible testing in an unobtrusive, real-time, and user-friendly way. Current research applications of ingestible biosensors in animal models rely on cumbersome analysis of bacterial gene expression or DNA in stool samples (5–10), rather than real-time reporting from within the body.
By contrast, the impressive scaling of semiconductor microelectronics over the past few decades has delivered sophisticated, highly miniaturized platforms for ultra–low-power sensing, computation, and wireless communication (11–13). In turn, these have enabled the recording of patient compliance and the evaluation of the gastrointestinal tract by using optical images, gases, temperature, and pH (14–17). However, the ability of electronics to directly and selectively sense biomolecules in vivo is limited by the availability of labile biochemical transducers and the size of the power-demanding circuits required to sense them.
Here, we describe an ingestible micro-bio-electronic device (IMBED) that combines engineered probiotic sensor bacteria together with ultra–low-power microelectronics to enable in situ detection of gastrointestinal biomolecules associated with health or disease (fig. S1). By partitioning sensing to biological systems and computation and communication to electrical devices, IMBEDs leverage the natural advantages of each approach to enable ingestible gastrointestinal diagnostics. In an IMBED, biosensor probiotics lie adjacent to readout electronics in individual wells separated from the outside environment by a semipermeable membrane that confines cells in the device and allows for diffusion of small molecules. Sensing of target biomarkers by the bacteria generates light, which is detected by photodetectors embedded in the electronics. These electrical signals are processed by an integrated bioluminescence detection circuit (18) and are transmitted wirelessly from the device to an external radio or cellular phone for convenient readout.
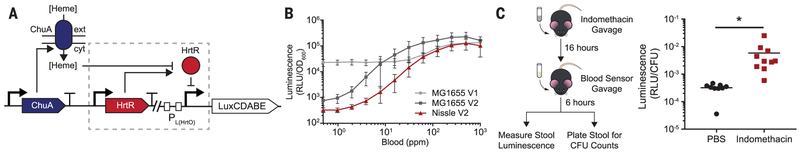
As a proof-of-concept IMBED for a clinically relevant biomarker, we developed a biosensor for gastrointestinal bleeding events via heme liberated from lysed red blood cells. Although cost-effective fecal occult-blood testing is available (19), diagnosis of acute bleeding in the upper gastrointestinal tract often requires endoscopic observation (20), and IMBEDs could offer a rapid, minimally invasive, and cost-effective means of detection. The heme biosensor was based on a synthetic promoter (PL(HrtO)) (fig. S2A), regulated by the Lactococcus lactis heme-responsive transcriptional repressor, HrtR (21), and ChuA, an outer-membrane transporter from Escherichia coli O157:H7 that allows for the transit of extracellular heme through the cell envelope (Fig. 1A) (22). Photorhabdus luminescens luxCDABE was used as the output of the genetic circuit as it functions at body temperature and encodes all components necessary for intracellular substrate production (23). The resultant prototype biosensor in laboratory E. coli (MG1655 V1) responded to increasing heme input with luminescence output with a signal-to-noise ratio (SNR) of 5.9 and a Hill function threshold (KD) of 1 μM heme (fig. S2B). Luminescence production was also induced by whole horse blood (fig. S2C), and lysis of red blood cells in simulated gastric fluid greatly improved sensitivity by liberating heme [KD = 115 parts per million (ppm) blood] (Fig. 1B and fig. S2D). Next, the prototype genetic circuit was iteratively optimized with the goal of improving SNR without compromising maximum luminescence output (MG1655 V2; Fig. 1B and fig. S3). The final gene circuit was transferred to the probiotic E. coli Nissle 1917 (Nissle V2), and the resultant strain responded rapidly to lysed horse blood (SNR = 310; KD = 95 ppm; t1/2 = 45 min) (Fig. 1B and fig. S4) as well as human blood (fig. S5).
Fig. 1. Probiotic E. coli can be engineered to sense blood in vitro and in vivo.
(A) Schematic of the blood sensor gene circuit. Extracellular heme is internalized through the outer-membrane transporter ChuA and interacts with the transcriptional repressor HtrR to allow for expression of the bacterial luciferase operon luxCDABE. (B) Dose-response curves of prototype (V1) and optimized (V2) heme-sensing genetic circuits in laboratory (MG1655) and probiotic (Nissle) strains of E. coli. Error bars represent SEM of three independent biological replicates. (C) In vivo blood sensor performance. C57BL/6J mice were administered indomethacin (10 mg/kg) to induce gastrointestinal bleeding or vehicle (PBS, phosphate-buffered saline) and inoculated with blood sensor E. coli Nissle cells the following day. Normalized luminescence values of fecal pellets were significantly higher in mice administered indomethacin compared to control animals (*P = 0.04; Student’s t test; N = 10 mice). CFU, colony-forming units; RLU, relative luminescence units.
To examine functionality of the bacterial blood sensor in vivo, we evaluated whether bacterial blood sensors passing through the gut could detect upper gastrointestinal bleeding elicited by oral indomethacin administration (Fig. 1C) (24). At baseline, administration of the biosensors did not lead to detectable luminescence activity in stool, indicating that basal heme concentrations in the murine gut are insufficient to activate the gene circuit (fig. S6). After oral administration of indomethacin, blood sensor bacteria demonstrated 18-fold higher luminescence values in fecal pellets as compared to controls (Fig. 1C). Our designed biosensor can thus effectively detect the presence of gastrointestinal bleeding in vivo.
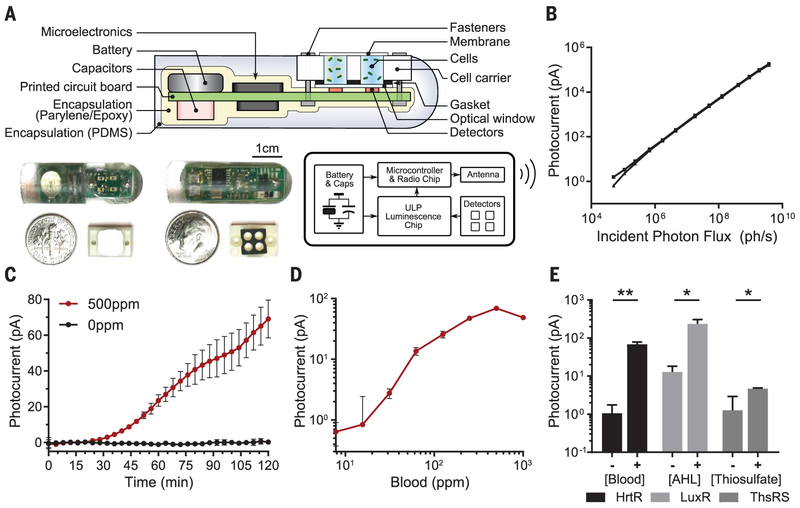
We next sought to integrate our bacterial biosensor with an electronic sensor and wireless transmission platform. Prior demonstrations of sensitive bioluminescence detection electronics have required external wiring and have been limited to bench-top assays (18, 25, 26), whereas a low-power wireless solution could enable convenient in vivo detection. We developed a miniaturized, fully integrated, wireless readout capsule for targeted sensing of small molecules in the gastrointestinal tract (Fig. 2A). The device combined our prior nanowatt-level time-based luminometer chip (18) with a microprocessor, wireless transmitter, and a set of phototransistors inside a molded capsule. Bioluminescence from activated cells was detected by phototransistors located below each cavity. The detected luminescence was converted to a digital code by the low-power luminometer chip and transmitted wirelessly outside the body for calibration, display, and recording. The small button-cell battery inside the capsule (5 mAh) powered the device, and the measured power consumption (table S1) suggests a nominal device shelf-life of >9 months and active operation time of 1.5 months on a full charge. The electronic system was highly sensitive and captured photon flux as low as 5 × 104 photons/s incident on the 0.29-mm2 area of the detectors [white-noise–limited coefficient of variation (CV) 13%, Fig. 2B and fig. S8A]. The mean channel mismatch was less than 6% (CV) (fig. S7A), and mean temperature-induced drift across 5°C variation was less than 2 pA (fig. S7B). Additionally, the electronic system was stable in simulated gastric fluid for up to 36 hours (fig. S7C), providing sufficient time to perform an ingestible measurement during gastrointestinal transit.
Fig. 2. Design and in vitro evaluation of IMBED for miniaturized wireless sensing with cellular biosensors.
(A) Cross section, electrical system diagram, and front- and back-side photos of the device. PDMS, polydi-methylsiloxane. (B) System photocurrent response measured without cells. The incident photon flux was supplied by green light-emitting diode (wavelength λ = 525 nm) and calibrated with an optical power meter (individual traces shown for N = 3 devices). (C) Kinetic response of blood sensor IMBED in bacterial growth media supplemented with 0 and 500 ppm blood. (D) Dose-response of blood sensor IMBEDs in bacterial growth media containing different blood concentrations 2 hours after exposure. The leftmost data point represents the background response in the absence of blood. (E) Detection of multiple gut-relevant small molecules with IMBEDs. HrtR-, LuxR- and ThsRS-containing E. coli Nissle strains in IMBEDs were exposed to 500 ppm blood, 100 nM acyl-homoserine lactone (AHL), or 10 mM thiosulfate for 2 hours. In (C) to (E), error bars denote the SEM for three independent biological replicates conducted with different IMBEDs. *P < 0.05, **P < 0.01, Student’s t test.
To demonstrate integration of the ingestible luminometer capsule and engineered biosensors, we tested the probiotic blood sensor strains in an IMBED in vitro. Upon exposure to 500 ppm blood, induced bioluminescence could be observed in as soon as 30 min (Fig. 2C). The dose-response curve of blood sensor IMBEDs was similar to that of plate-reader measurements (SNR = 76; KD = 135 ppm; compare Fig. 1B and 2D), with saturation achieved at 250 ppm and significant detection as low as 32.5 ppm blood (fig. S8) (Student’s t test; P = 0.03). By combining cellular sensors with ultra–low-power electronic readout, IMBEDs serve as a flexible platform for sensitive detection of bleeding in fluidic environments.
In addition to blood sensing, we adapted IMBEDs to sense alternative biomarkers, thiosulfate and acyl-homoserine lactone (AHL). Thiosulfate could serve as a biomarker of gut inflammation as it is elevated in murine models of colitis (8). AHLs are molecular signatures of particular bacteria, and their detection could indicate the presence of commensal or infectious agents in the gut microbiota (27, 28). Thiosulfate-and AHL-inducible genetic circuits were introduced into E. coli Nissle, and exposure to inducer elicited a dose-dependent bioluminescence response (fig. S9). These alternative biosensors were integrated with IMBEDs, and different analytes were readily detectable in a fluidic environment (Fig. 2E). As additional biosensors of clinically relevant gut biomarkers continue to be developed, we anticipate that the breadth of potential analytes of the IMBED platform will continue to expand.
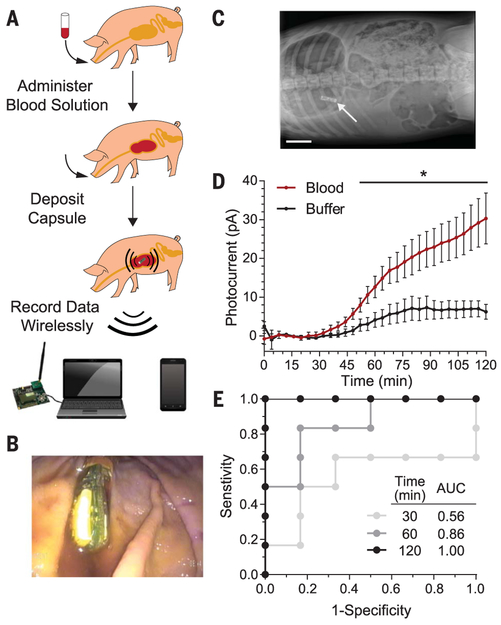
To examine wireless in situ detection of biomolecules with biosensors, we deployed blood sensor IMBEDs in a porcine model of gastrointestinal bleeding. Prior to device deposition, pigs were administered a bicarbonate-glucose neutralization solution with or without 0.25 ml of blood (Fig. 3A). The neutralization solution helped buffer the low pH of the porcine gastric fluid as acidic environments degrade the functionality of the biosensor (fig. S10). The blood sensor IMBED was subsequently deposited into the stomach via orogastric tube and remained resident and stable in the gastric cavity for the entire duration of the experiment (Fig. 3, B and C). Photocurrent data were wirelessly transmitted from the stomach over the course of 2 hours and logged by both a laptop computer and an Android phone equipped with a custom application for real-time data processing and visualization (figs. S11 and S12). The presence of blood in the porcine gastric environment could be observed as early as 52 min (Student’s t test; P < 0.05) and led to a fivefold increase in photocurrent after 120 min as compared to animals given buffer alone (Fig. 3D and fig. S13). Luminescence production was not detected in biosensors lacking the ChuA heme transporter or the luciferase operon, indicating that observed light production was dependent on a functional genetic circuit activated in the presence of heme (fig. S14). The receiver operating characteristic of the bloodsensing IMBED improved over time, with a sensitivity and specificity of 83.3% at 60 min and 100% at 120 min (Fig. 3E). IMBEDs can thus detect small amounts of analyte in the harsh gastric environmental with high specificity and sensitivity.
Fig. 3. IMBEDs can rapidly detect porcine gastric bleeding.
(A) Schematic of the experimental flow, which consisted of blood administration in neutralization solution, capsule deposition, and wireless transmission to a commercial receiver connected to a laptop or a cellular phone. Representative endoscopic (B) and x-ray (C) images illustrate the location of the device in the stomach at the conclusion of our 2-hour experiments, just before device removal [scale bar (C), 5 cm]. (D) Kinetic response of blood sensor IMBED in a porcine model of gastric bleeding. IMBEDs deposited in gastric cavity can rapidly discriminate between pigs administered blood versus buffer control. Error bars denote SEM for six IMBED experiments (three animals on different days, two capsules per animal). *P < 0.05, Student’s t test. (E) Receiver operating characteristic (ROC) of IMBED sensing over time. Perfect detection is achieved at t = 120 min. AUC, area under the curve.
By combining the environmental resilience and natural sensing properties of bacterial cells with the complex data processing and wireless transmission afforded by ultra–low-power microelectronics, we developed a device capable of in vivo biosensing in harsh, difficult-to-access environments. Using gastrointestinal bleeding as a proof-of-concept model system, we demonstrate strategies for genetic circuit design and optimization, fabrication of an ingestible low-power, wireless luminometer, and validation of integrated system functionality both in vitro and in a large animal model. As the field of wholecell biosensors matures, newly developed sensors of clinically relevant biomarkers could be rapidly integrated into an IMBED to perform minimally invasive detection in the gastrointestinal tract. With a test panel of candidate biomolecules, IMBEDs could enable studies of the biochemistry of anatomical regions that are traditionally difficult to access and could lead to the discovery of new clinical biomarkers associated with health or disease. The in situ detection afforded by IMBEDs could also allow sensing of labile gut or microbiota-derived biomolecules that would otherwise be degraded before excretion in stool. Further integration of electronic modules, such as photodetectors, microprocessor, and transmitter, in a single integrated circuit could allow for further miniaturization of IMBEDs as well as lower power consumption. Additional measurement channels would also enable more precise biochemical readings, as the response of replicate biosensors within the same device could be averaged to mitigate the inherent variance of biological sensors as well as the heterogeneity of the complex gastrointestinal environment. Improved preparation of bacterial cultures for long-term storage, such as lyophilization, could be implemented to extend the shelf-life of fully assembled IMBEDs. Furthermore, the devices could be equipped with new orally delivered encapsulation technologies to enable long-term residency, monitoring, and anatomic localization in the gastrointestinal tract (29, 30). This integration of biological engineering and semiconductor electronics offers opportunities to transform diagnosis, management, and monitoring of health and disease.
Supplementary Material
ACKNOWLEDGMENTS
The thiosulfate sensor plasmids were a gift from J. Tabor (Rice University). The luxCDABE genes were obtained from pAKlux2, a gift from A. Karsi (Addgene plasmid no. 14080). We thank D. Glettig for help with pilot experiments in rodents and M. Goulamaly for preparing initial parylene samples.
Funding: Supported by Texas Instruments (A.P.C.); the Hong Kong Innovation and Technology Fund (ITS/195/14FP) (A.P.C. and T.K.L.); Office of Naval Research (N00014-13-1-0424) (T.K.L.); NSF Biological Computing (1522074) (T.K.L.); the Center for Microbiome Informatics and Therapeutics (15127713) (T.K.L.); Division of Gastroenterology, Brigham and Women’s Hospital (G.T.); NIH (EB-000244) (R.L.); the Qualcomm Innovation Fellowship (M.M. and P.N.); the HHMI International Student Fellowship (M.M.); and the Natural Sciences and Engineering Council of Canada Fellowship (P.N.). We also thank the TSMC University Shuttle Program for chip fabrication.
Footnotes
Competing interests: M.M. and T.K.L. have filed a patent application based on the microbial heme sensor (US20170058282A1) with the U.S. Patent and Trademark Office. M.M., P.N., A.P.C., and T.K.L have filed a patent application based on the integrated device (PCT/US2018/027904) with the U.S. Patent and Trademark Office.
Data and materials availability: Genetic constructs are available in Addgene.
SUPPLEMENTARY MATERIALS
REFERENCES AND NOTES
- 1.Brophy JAN, Voigt CA, Nat. Methods 11, 508–520 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Slomovic S, Pardee K, Collins JJ, Proc. Natl. Acad. Sci. U.S.A 112, 14429–14435 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roggo C, van der Meer JR, Curr. Opin. Biotechnol 45, 24–33 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Courbet A, Endy D, Renard E, Molina F, Bonnet J, Sci. Transl. Med 7, 289ra83 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Kotula JW et al. , Proc. Natl. Acad. Sci. U.S.A 111, 4838–4843 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mimee M, Tucker AC, Voigt CA, Lu TK, Cell Syst. 1, 62–71 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lim B, Zimmermann M, Barry NA, Goodman AL, Cell 169, 547–558.e15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daeffler KN-M et al. , Mol. Syst. Biol 13, 923 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riglar DT et al. , Nat. Biotechnol 35, 653–658 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pickard JM et al. , Nature 514, 638–641 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Otis B, Parviz B, Introducing our smart contact lens project. Google Off. Blog (2014); available at https://googleblog.blogspot.com/2014/01/introducing-our-smart-contact-lens.html. [Google Scholar]
- 12.Wang H, IEEE Microw. Mag 14, 110–130 (2013). [Google Scholar]
- 13.Norian H, Field RM, Kymissis I, Shepard KL, Lab Chip 14, 4076–4084 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Iddan G, Meron G, Glukhovsky A, Swain P, Nature 405, 417–417 (2000). [DOI] [PubMed] [Google Scholar]
- 15.Kalantar-Zadeh K et al. , Nat. Electron. 2017 11. 1, 79 (2018). [Google Scholar]
- 16.van der Schaar PJ et al. , Gastrointest. Endosc 78, 520–528 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Hafezi H et al. , IEEE Trans. Biomed. Eng 62, 99–109 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Nadeau P, Mimee M, Carim S, Lu TK, Chandrakasan AP, Nanowatt circuit interface to whole-cell bacterial sensors. 2017 IEEE International Solid-State Circuits Conference (ISSCC), San Francisco, CA, 2017, pp. 352–353. [Google Scholar]
- 19.Rockey DC, Koch J, Cello JP, Sanders LL, McQuaid K, N. Engl. J. Med 339, 153–159 (1998). [DOI] [PubMed] [Google Scholar]
- 20.Barkun A, Bardou M, Marshall JK, Ann. Intern. Med 139, 843–857 (2003). [DOI] [PubMed] [Google Scholar]
- 21.Lechardeur D et al. , J. Biol. Chem 287, 4752–4758 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nobles CL, Clark JR, Green SI, Maresso AW, J. Microbiol. Methods 118, 7–17 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Close D et al. , Sensors (Basel) 12, 732–752 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lanas A, Chan FKL, Lancet 390, 613–624 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Eltoukhy H, Salama K, El Gamal A, IEEEJ. Solid-State Circuits 41, 651–662 (2006). [Google Scholar]
- 26.Singh RR, Leng L, Guenther A, Genov R, IEEE J. Solid-State Circuits 47, 2822–2833 (2012). [Google Scholar]
- 27.Hwang IY et al. , Nat. Commun 8, 15028 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schuster M, Sexton DJ, Diggle SP, Greenberg EP, Annu. Rev. Microbiol 67, 43–63 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Bellinger AM et al. , Sci. Transl. Med 8, 365ra157 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kirtane AR et al. , Nat. Commun 9, 2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.