Abstract
The chemical functionalities within biopolymers determine their physical properties and biological activities. The relationship between the side-chains available to a biopolymer population and the potential functions of the resulting polymers, however, has proven difficult to study experimentally. Using seven sets of chemically diverse charged, polar, and nonpolar side-chains, we performed cycles of artificial translation, in vitro selections for binding to either PCSK9 or IL-6 protein, and replication on libraries of random side-chain-functionalized nucleic acid polymers. Polymer sequence convergence, bulk population target binding, affinity of individual polymers, and head-to-head competition among post-selection libraries collectively indicate that polymer libraries with nonpolar side-chains outperformed libraries lacking these side-chains. The presence of nonpolar groups, resembling functionality present in proteins but missing from natural nucleic acids, thus may be strong determinants of binding activity. This factor may contribute to the apparent evolutionary advantage of proteins over their nucleic acid precursors for some molecular recognition tasks.
Introduction
The chemical structures of nucleic acid and protein building blocks determine the properties of biopolymers and thus many of the functions of living systems. What chemical functionalities are most likely to enable the evolution of polymers with activities essential to living systems, such as the ability to bind other molecules, or to catalyze chemical reactions? Insights into this fundamental question would enhance our understanding of life’s molecular requirements, and inform our ability to navigate chemical space to discover non-natural polymers with useful properties. Many studies that have probed these questions have searched for function using limited sets of building blocks from naturally derived monomers of proteins1–5 and nucleic acids6–9, such as using just three DNA bases instead of four, or using a subset of amino acids out of the canonical 20. While these studies have shown the adequacy or limitations of smaller subsets of natural building blocks, the fundamental relationship between the availability of diverse combinations of many different side-chains in a biopolymer’s genetic code and the evolutionary potential of the resulting polymers has not been experimentally illuminated.
Nucleic acids are well-suited biopolymers for exploring these questions because functional members can be rapidly enriched from large, diverse libraries of sequences using SELEX10–12, iterated cycles of Darwinian selection and replication. Using SELEX and related processes, researchers have previously discovered and isolated many DNA-based receptors13–15 and catalysts16–21. Researchers have also performed SELEX using side-chain-modified DNA, and have demonstrated that the replacement of one or two nucleotides with side-chain-appended nucleotides can enable target binding22–24 or catalysis25–28 that may be more difficult without the additional side-chain or pair of side-chains29–31. One hypothesis emerging from these studies is that replacing all cytosines (Cs) or thymines (Ts) with nucleotides containing aromatic side-chains may lead to polymer libraries with better affinity to protein targets31,32. To illuminate the relationship between side-chain access and biopolymer functional potential more comprehensively, however, requires generating and comparing polymer libraries with access to larger sets of multiple, diverse side-chains, beyond one or two types of chemical modification in an otherwise unmodified nucleic acid.
We and others previously described an artificial translation system that uses DNA ligase to mediate the DNA-templated synthesis of long, sequence-defined highly functionalized nucleic acid polymers (HFNAPs)33–361. In our system, each HFNAP building block contains a YNN trinucleotide backbone (where Y = T or C, and N = A, T, C, or G) that is covalently linked to one of up to 32 chemically diverse side-chains on the 5’ pyrimidine base. In recent applications of the HFNAP system34, eight different side-chains are encoded by 32 trinucleotide codons, such that four trinucleotide sequences provide access to each side-chain group. HFNAP libraries in which each polymer contains 15 consecutive building blocks can be generated in this system from corresponding libraries of DNA templates (Fig. 1)33,34. The resulting libraries contain sequence-defined 45-mers with a theoretical library diversity of ~4 × 1022 different polymers with a variable sequence region of ~15 kD. These libraries can be selected in vitro for target binding, and surviving polymers can be “reverse translated” back into DNA using Q5 polymerase and PCR, resulting in DNA templates suitable for a subsequent round of translation and selection34. We recently used this system to discover HFNAPs that bind two disparate proteins of biomedical interest, PCSK9 and IL-6, with high affinity (Kd ~1–10 nM)34.
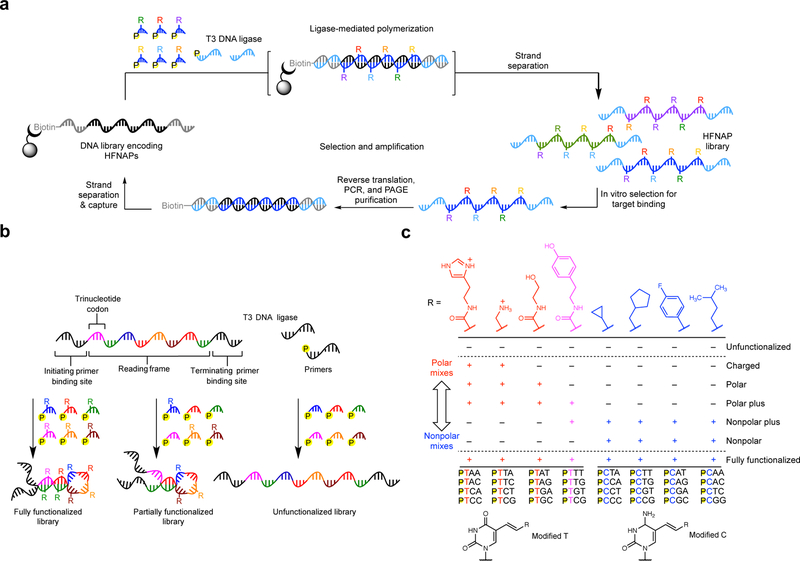
Figure 1: HFNAP translation, selection, and library design.
a, Overview of a complete cycle of HFNAP translation from DNA templates, selection for target affinity, reverse translation back to DNA templates, and template replication by PCR. b, Pool design showing how combinations of monomers are combined to make fully side-chain functionalized, partially functionalized, or unfunctionalized libraries. c, The seven sets of side-chain availability in each “genetic code” (top) and the 32 possible trinucleotide codons for each HFNAP.
Since researchers can choose the chemical structure of the side-chain in every building block, the HFNAP system provides an ideal opportunity to experimentally test how the side-chain repertoire in a sequence-defined polymer library containing many different side-chains influences the functional potential of the resulting polymers. Here we performed in parallel iterated rounds of translation, selection, and replication of HFNAPs for binding to PCSK9 or IL-6 using seven different “genetic codes”, each of which allow polymers to access a different subset of side-chain functionalities. Using several metrics to evaluate both bulk populations of HFNAPs and individual polymers post-selection, we observed that access to hydrophobic side-chains strongly promoted the emergence of polymers that bind with high affinity to their protein targets for both selections. Our findings illuminate the relationship between side-chain composition and polymer binding activities, and experimentally demonstrate how the availability of nonpolar side-chains could provide biopolymers such as proteins with evolutionary advantages over biopolymers such as natural DNA and RNA that lack such groups.
Results
HFNAP library and selection design
The HFNAP translation system (Fig. 1a) uses a DNA template containing a 45-base coding region of 15 consecutive codons flanked by two primer binding sites. Trinucleotide building blocks hybridize to complementary codons on the template33,34, and T3 DNA ligase catalyzes the covalent coupling of adjacent building blocks on a template in sequence-defined order37. From the resulting heteroduplex HFNAP:DNA double-stranded product, the desired polymer strands are isolated through alkaline denaturation (Fig. 1a).
To study how side-chain composition in a polymer library influences selection outcomes, we prepared seven HFNAP libraries of random sequence from aliquots of the same DNA template library containing 15 consecutive NNR codons (where N = A, T, C, or G; and R = A or G). Each of the seven libraries differed in the presence or absence of different side-chains during translation (Fig. 1b). The library templates were drawn from the same random sequence mixture, although statistically each library shares a negligible fraction of identical template sequences with the other six libraries. For libraries designated to omit specific side-chains, we replaced the building blocks that would normally contain that side-chain with sequence-matched unfunctionalized DNA trinucleotide building blocks lacking any side-chain.
The seven different “genetic codes” included a fully functionalized code in which all eight side-chains including amine, alcohol, aliphatic, fluorinated, and aryl groups were introduced, and a completely unfunctionalized code, in which all building blocks were unfunctionalized DNA trinucleotides lacking side-chains (Fig. 1c). In addition, we used five genetic codes in which different subsets of side-chains were included based on their polarity. The “charged” code included building blocks with primary amine or imidazole side-chains that can be protonated in equilibrium with their neutral, non-protonated forms. The positively charged side-chains complement the negatively charged backbone of DNA. The “polar” code added an alcohol side-chain along with the charged side-chains. The “nonpolar” code contained only hydrocarbon and fluorocarbon side-chains, including cyclopropyl, cyclopentyl, p-fluorophenyl, and isopentyl side-chains. Since the phenolic side-chain possesses both polar and nonpolar characteristics38 and is frequently involved in protein-protein interactions39,40, we investigated how the addition of this side-chain influenced selection outcomes using two additional codes in which the phenolic side-chain was added to either the polar code (resulting in the “polar plus” code), or to the nonpolar code (resulting in the “nonpolar plus” code).
Following DNA-templated polymerization (translation) of each of the seven polymer libraries, we performed on each library iterated cycles of target binding selection, “reverse translation” from HFNAP back to DNA, PCR replication of surviving DNA templates, and re-translation into HFNAPs to enrich the population of HFNAP binders34. Briefly, translated polymers were incubated with the agarose bead-immobilized protein target. The non-binding flow-through was removed and the beads were washed three times with buffer. Bound polymers were eluted by heating in buffered detergent, then reverse translated and amplified by PCR with Q5 DNA polymerase. Full-length amplicons were purified by PAGE, and the template DNA strands were isolated by streptavidin capture and alkaline denaturation to enter the next round of HFNAP library translation. The sequence composition of each of the seven parallel populations was analyzed after each PCR step by high-throughput DNA sequencing (HTS).
Outcomes of PCSK9 selections
We chose PCSK9 and interleukin 6 (IL-6), two disparate proteins of biomedical interest41–43 for which we34 and others31,44,45 have previously isolated nucleic acid receptors, as the in vitro selection targets. We examined how the seven libraries matured when iteratively challenged to bind PCSK9 and IL-6 (Fig. 2). In the first selection campaign for PCSK9 binding, we translated 7.5 × 1013 DNA template molecules (125 pmol) for each of the seven genetic codes and challenged the resulting polymer libraries to bind PCSK9 using the selection method described above. After each round of selection, the quantity of the HFNAP pool retained by the PCSK9-linked beads was assessed by qPCR (Supplementary Fig. 1). The stringency of selections was increased gradually as the rounds progressed by decreasing the amount of target PCSK9 protein provided, or by decreasing the incubation time. All seven HFNAP libraries were subjected to the same selection conditions throughout all rounds of selection (Supplementary Fig. 1). Starting in round 7, we added a counter-selection at the end of each round to remove polymers that bound beads lacking PCSK9.
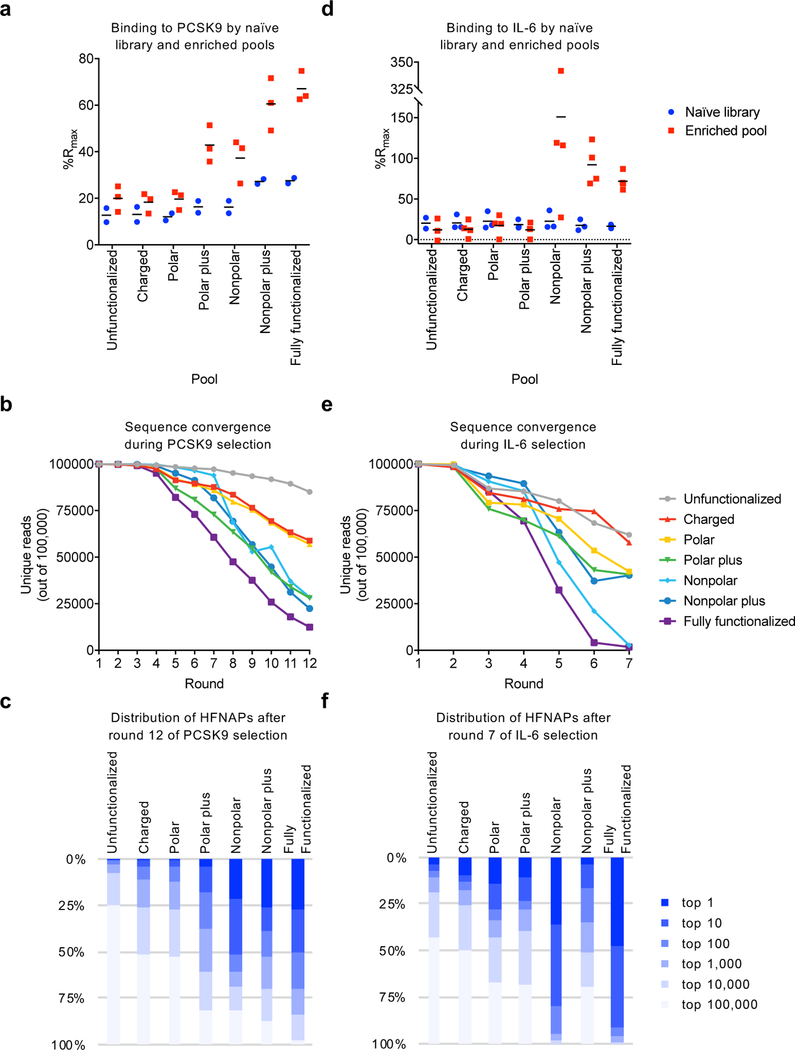
Figure 2: Protein target binding activity, sequence convergence, and sequence distribution of HFNAPs selected for PCSK9 or IL-6 affinity.
a, Normalized SPR response (%Rmax) observed when immobilizing biotinylated naïve libraries or enriched pools on an SPR chip and flowing PCSK9. b, Side-chain-dependent convergence of libraries in PCSK9 selections as determined by high-throughput sequencing (HTS). c, Polymer density in the final round of PCSK9 selection represented as the fraction of 100,000 HTS reads captured by the top 1, 10, 100, etc. HFNAPs. d, e, and f, analogous data to a, b, and c for the IL-6 selections. In a and d, horizontal bars denote mean values of three or more replicates; the value from each replicate is shown.
Over 12 rounds of iterated selection, reverse translation, template replication, and translation, qPCR analysis revealed that the polymer pools with access to the nonpolar or phenolic side-chains became enriched in PCSK9-binding polymers earlier in the iterated selection process, and to a greater extent, than the pools with access only to the unfunctionalized or polar side-chains (Supplementary Fig. 1). Using the templates from the final round of selection, we prepared 3’-biotinylated polymer pools and assessed the relative performance of these HFNAP populations by surface plasmon resonance (SPR), a direct binding assay that is distinct from bead retention. We examined the SPR response to PCSK9 using surfaces coated with each polymer pool and normalized the observed response by the maximum response expected from a 1:1 binding interaction (Rmax) calculated from each polymer pool’s immobilization level. Consistent with the qPCR data, the enriched pools with access to the full side-chain set and those containing nonpolar or the phenolic side-chains demonstrated the highest levels of polymer binding (%Rmax) to PCSK9 (Fig. 2a and Supplementary Fig. 2). Naïve libraries, prepared and assayed similarly, demonstrated much lower binding responses (Fig. 2a), suggesting that stronger binding to PCSK9 was a property enriched during the selection process. Together, these results suggest that the polymer libraries with highest side-chain diversity or access to nonpolar side-chains more strongly support the enrichment of PCSK9-binding HFNAPs.
To further characterize the progress of each HFNAP population as it matured through the iterated translation, selection, and replication process, we subjected DNA templates emerging from each round of selection to HTS. The data revealed that the two populations containing mostly nonpolar side-chains (the nonpolar and nonpolar plus pools), the polar plus pool, as well the population with access to the full set of side-chains, converged much more quickly to a smaller number of unique HFNAPs compared to the populations having access to mostly polar side-chains (Fig. 2b). Moreover, the most abundant HFNAPs from the pools with access to the nonpolar side-chains represented a higher percentage of the total pool than in the pools lacking nonpolar side-chains, suggesting that these strongly enriched polymers were indeed of high fitness in these populations (Fig. 2c). For example, by the end of round 12, the ten most abundant HFNAPs from the fully functionalized and nonpolar plus pools, represented 50% and 39%, respectively, of all HFNAPs in the pool. In contrast, in the unfunctionalized DNA pool and the pool accessing only polar side-chains, the most abundant 10 HFNAPs represented only 0.72% and 3.7% of the total population, respectively (Fig. 2c).
To illuminate the relationship between side-chain abundance in a polymer and enrichment during protein-binding selection, we examined the prevalence of each type of side-chain in the post-selection sequences. The most enriched sequences from the nonpolar pool were not predominantly composed of building blocks containing side-chains (Supplementary Table 1), even though the starting library contained 50% side-chain-containing building blocks, further suggesting that the benefit conferred by the availability of nonpolar side-chains is not simply to maximize the abundance of groups that participate in non-specific hydrophobic interactions with the target protein.
To gain additional insight into the functional potential of the different pools that emerged after the final rounds of selection, we performed a single-solution, head-to-head affinity competition (Fig. 3). Such a competition offers a direct comparison of polymer library performance as a function of side-chain availability. Polymers emerging from each of the seven pools’ final round of selection for PCSK9 binding were mixed together in equimolar ratio (~160 fmol of each post-round 12 polymer library). The resulting mixture was subjected to an additional selection for binding to PCSK9 (Supplementary Fig. 3a). PCSK9-bound polymers were eluted, reverse-translated, sequenced by HTS, and assigned back to one of the seven pools based on alignment to sequences from previous rounds. The vast majority (94%) of the polymers surviving the head-to-head selection could be unambiguously assigned to their original pools (Supplementary Fig. 3b). The changes in relative abundance of each pool’s polymers when combined and reselected for PCSK9 binding reflected the relative fitness of each population’s polymers in the competition.
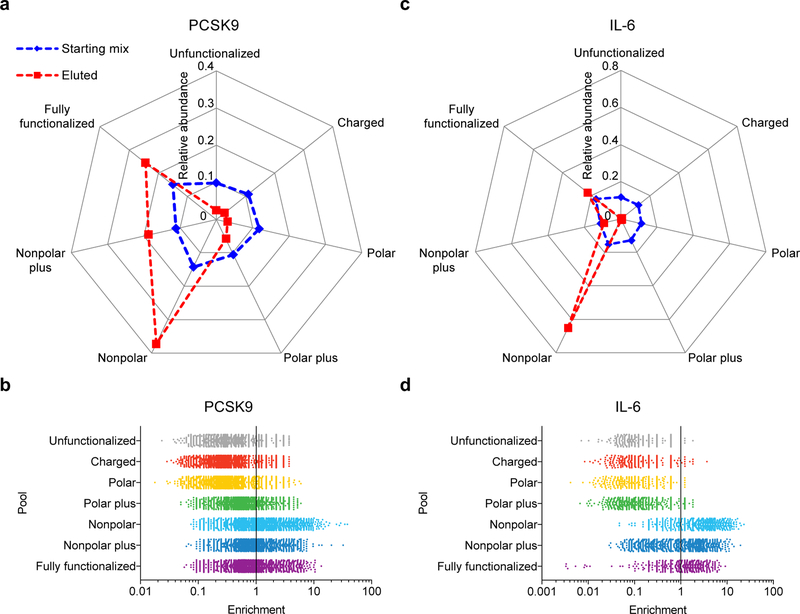
Figure 3: Head-to-head competition of seven pools following iterated selections for binding to PCSK9 or IL-6.
a, Plot showing the distribution of assigned polymers before head-to-head competition selection (blue) and after head-to-head selection (red) for PCSK9 binding. The values represent relative abundances of HFNAPs from each pool (total abundance = 1.0, including unassigned; see Supplementary Fig. 3 for a detailed breakdown). b, The distribution of individual polymer enrichment values is shown for each pool in the PCSK9 competition experiment. Enrichment for each polymer was determined from the ratio of its abundance in the eluted material divided by its abundance in the initial combined pool. c, Distribution of assigned polymers before head-to-head competition selection (blue) and after head-to-head selection (red) for IL-6 binding (see Supplementary Fig. 10 for a detailed breakdown). d, Distribution of individual polymer enrichment values for each pool in the IL-6 competition experiment.
Consistent with the above findings, the ability of each polymer pool to survive the head-to-head PCSK9 binding competition correlated with the availability of nonpolar side-chains. The HFNAPs from the fully functionalized, nonpolar plus, and nonpolar populations enriched at the expense of polymers from the unfunctionalized, charged, polar, and polar plus populations (Fig. 3a, b). This conclusion was not dependent on how the 5.7% of unassigned sequences were categorized, as the unfunctionalized, charged, and polar pools were depleted to a much larger extent than could be accounted for by those unassigned sequences (Supplementary Fig. 3b). On an individual polymer level, we also observed clear advantages for polymers that accessed nonpolar side-chains. For example, all of the 100 assignable polymers undergoing the greatest enrichment from the head-to-head competition were from the three pools (nonpolar, nonpolar plus, and fully functionalized) containing mostly nonpolar side-chains (Fig 3b and Supplementary Fig. 4). Importantly, the simple number of side-chains present in each polymer was not sufficient to explain these performance differences, as those 100 most strongly enriched polymers averaged only 7.4 out of 15 possible side-chains per polymer. Together, the results of this head-to-head competition experiment further support the finding that the populations containing nonpolar side-chains yielded polymers that outperform populations lacking nonpolar side-chains in a target protein-binding task.
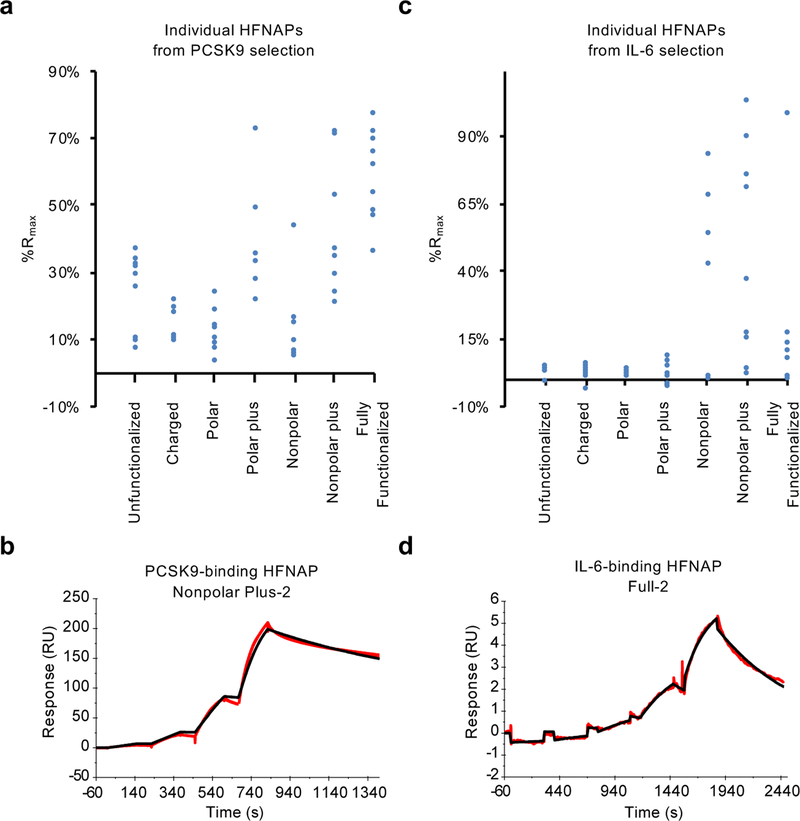
Next, we sought to validate and compare the binding of individual polymers identified during the selection (Fig. 4). We synthesized biotinylated versions of the most abundant polymers at the end of the selection and also those HFNAPs undergoing the greatest enrichment during any stage of the selection. We subjected the resulting polymers to PCSK9-binding analysis by SPR using a streptavidin-linked chip and a buffer similar to that used in selection (Fig. 4a, Supplementary Fig. 5, and Supplementary Table 2). Most of the individual polymers tested exhibited detectable binding to PCSK9, consistent with apparent bulk library affinity (Fig. 2a), with Kd values as low as 11±14 nM (Fig. 4b), consistent with our previous studies34. Some polymers did not demonstrate simple 1:1 binding kinetics to PCSK9, complicating the determination of Kd values and polymer comparison. We therefore injected a fixed concentration of PCSK9 onto surfaces coated with each polymer and normalized the observed response to Rmax, calculated from each polymer’s immobilization level. Individual HFNAPs from the polar plus, nonpolar plus, and fully functionalized pools generally exhibited higher normalized SPR response (%Rmax) than the less functionalized and more polar pools (Fig. 4a). Collectively these data on individual polymer abundance, enrichment, and binding characteristics suggest that individual polymers from the best-performing libraries indeed exhibit greater overall PCSK9 binding activity.
Figure 4: Target-binding activity of individual HFNAPs by SPR.
a, Observed %Rmax for individual polymers emerging from iterated cycles of translation and selection for binding to PCSK9, calculated from SPR response following a single injection of 60 nM PCSK9. b, Background-subtracted sensorgram (red) of PCSK9-binding HFNAP Nonpolar Plus-2 binding to 10, 30, 100, and 300 nM PCSK9 and curve fitting with a 1:1 binding model (black) results in an apparent Kd of 28 nM (shown); the average among three replicates is Kd = 11±14 nM (see Methods). c, Observed %Rmax for individual polymers emerging from iterated cycles of translation and selection for binding to IL-6, calculated from SPR response following the third of four injections of IL-6 in a single-cycle kinetics assay (8.9, 27, 80, and 240 nM IL-6; see Methods for details). d, Background-subtracted sensorgram (red) of IL-6-binding polymer Full-2 binding to 0.3, 1, 3, 10, and 30 nM IL-6 and fitting with a 1:1 binding model (black) results in an apparent Kd of 12 nM (shown); the average among three replicates is Kd = 5.3±5.9 nM, although we note that this polymer exhibits complex binding behavior at concentrations above 30 nM. Kd values represent mean ± standard deviation (n = 3).
Investigation of potential sources of bias
To examine if the trends emerging from the enriched pools following the PCSK9 selections were due to target binding and not artifacts of other steps in the selection cycle, we also examined potential bias in the HFNAP translation, reverse translation and amplification steps of the selection process starting with aliquots of naïve template libraries used in the PCSK9 selection. We found that the overall translation and amplification efficiency of libraries containing side-chain functionalized building blocks is lower, rather than higher, compared to the translation and amplification efficiency of unfunctionalized DNA, up to a factor of approximately six for the fully functionalized library (Supplementary Fig. 6). Given that the fully functionalized library was among the best-performing polymer libraries, this observation suggests that the advantage of functionalization with side-chains overcomes any overall bias against incorporating side-chains in the polymer translation and amplification process.
To examine the potential bias of the system toward individual building blocks, we performed a neutral selection46 in which we collected HTS data after two cycles of translation, dilution (without selection for target binding), reverse translation, and PCR for each genetic code set. Analysis of the distribution of all 32 codons revealed that over the course of two neutral selection rounds, none of the building blocks enriched more than approximately 2-fold or de-enriched more than approximately 4-fold (Supplementary Fig. 7). Importantly, the most prevalent building blocks in the most enriched pools from the PCSK9 selection were different from the most prevalent building blocks in the neutral selection (Supplementary Fig. 7). Moreover, none of the building blocks were completely eliminated during the course of any of the selections described above. Taken together, this analysis suggests that the changes to the HFNAP populations during target-binding selections are not primarily due to codon or side-chain biases separate from those that contribute to binding activity.
Outcomes of IL-6 selections
To test if the above findings also apply to protein targets other than PCSK9, we repeated the HFNAP library translation and iterated selections, as well as all other experiments with enriched polymers described above, with IL-6, an unrelated protein involved in inflammation41. Over seven rounds of translation, selection, and replication, qPCR analysis once again suggested that the HFNAP libraries with access to the nonpolar side-chains generally exhibited superior binding to immobilized IL-6 than libraries lacking access to nonpolar side-chains (Supplementary Fig. 8). With substantial differences already evident among the enriched polymer pools, we stopped IL-6 selections after round 7. Following the seventh selection round, we immobilized the naïve polymer libraries and the enriched polymer pools to an SPR chip as described above to study their binding to IL-6 in a distinct assay modality. Similar to our findings for PCSK9 binding, all of the enriched polymer pools with access to the nonpolar set of side-chains (nonpolar, nonpolar plus, and the fully functionalized set) exhibited higher SPR responses to IL-6, suggesting greater binding activity (Fig. 2d and Supplementary Fig. 9). These experiments further support that the bulk binding affinity of the enriched polymer libraries with nonpolar side-chains to IL-6 is greater than those without non-polar side-chains, consistent with the results from PCSK9-binding selections.
HTS data indicated that all of the libraries in the IL-6 selection campaign converged more quickly than those in the PCSK9 selection campaign (Fig. 2e). Nonetheless, during IL-6 binding selections the nonpolar and fully functionalized populations converged more quickly to a smaller number of HFNAPs than the other libraries, again suggesting the benefit of nonpolar side-chains to HFNAP enrichment in IL-6 binding. Moreover, the 10 most abundant HFNAPs from the fully functionalized and nonpolar pools following the seventh selection round represented 91% and 80%, respectively, of all HFNAPs in the pool (Fig. 2f). The other side-chain-containing libraries (charged, polar, polar plus, and nonpolar plus) enriched much less strongly, with the ten most abundant HFNAPs representing only 13–28% of the total population of each pool.
We performed a head-to-head IL-6-binding competition of an equimolar mixture of the IL-6 post-selection polymer pools. The nonpolar pool outperformed all others, while the fully functionalized and nonpolar plus pools approximately maintained their relative abundance before and after competitive selection (Fig. 3c, d and Supplementary Figs 10–11). In contrast, the pools without side-chains or containing only polar side-chains were depleted in the IL-6 binding competition. As 99.4% of the sequences were assignable based on their similarity to sequences observed during selections on isolated pools, these results did not depend on the 0.6% unassigned sequences. In addition, all of the top 100 most enriched polymers from the head-to-head competition originated from pools with access to nonpolar side-chains. Furthermore, among this top set of polymers, the average number of side-chains present was 6.2 out of 15 possible, suggesting that the total number of nonpolar side-chains is not a primary determinant of IL-6 binding. Collectively, these results are consistent with the findings from the PCSK9 selections.
We prepared individual biotinylated polymers that exhibited the greatest enrichment during the IL-6 selection process for analysis, and immobilized them on the surface of a streptavidin-linked SPR chip to assess their binding to varying concentrations of IL-6. Individual polymers derived from the nonpolar, nonpolar plus, and fully functionalized pools exhibited substantially higher %Rmax values than polymers from the unfunctionalized and exclusively polar side-chain sets (Fig. 4c, Supplementary Fig. 12, and Supplementary Table 4). Individual Kd values were as low as 5.3±5.9 nM (Fig. 4d). These data confirm that individual polymers identified from the selections indeed bind IL-6 with high affinity.
Discussion
This work demonstrates that in both separate and competitive Darwinian selections for binding to two unrelated proteins, HFNAP libraries with access to nonpolar side-chains outperformed those without access to nonpolar side-chains. Despite some differences between the outcomes of PCSK9 and IL-6 selections, which may reflect differences in the rarity of polymers that bind to the target proteins, in selection stringency or other parameters during selection, or other factors that influence HFNAP fitness, the overall trends in polymer pool performance between the two selection campaigns were remarkably similar. In both selections, the libraries with access to nonpolar side-chains generally converged more quickly, yielded polymers with higher target protein binding activity, and competed more effectively compared with populations with access to only polar side-chains, or to those with no side-chains. Furthermore, the enriched pools and individual polymers containing nonpolar side-chains demonstrated higher apparent binding activity to their protein targets in an orthogonal SPR assay when compared to those from other pools.
These findings mirror observations during the development of some bioactive small-molecule probes and therapeutics47, in which the addition of lipophilic groups is known to contribute to target-binding activity. While the addition of too many nonpolar groups in small-molecule drugs is known to lead to promiscuous protein binding48, the data from our study suggest that nonpolar side-chains among the HFNAPs characterized in this work do not simply serve to increase non-specific protein affinity. We previously found that the specific structure, location, and sequence context of key nonpolar side-chains among PCSK9-binding and IL-6-binding HFNAPs, and not simply their abundance, were critical determinants of binding activity34. Indeed, our analysis of the top sequences from the head-to-head competition in this study indicate that the number of non-polar side-chains among top-performing sequences did not approach the maximum number allowed, further suggesting that the position of the nonpolar side-chains in the context of the surrounding building clocks, rather than the absolute number of nonpolar side-chains, contributes strongly to target-binding activity. Consistent with these conclusions, the nonpolar side-chains in these DNA-based polymers represent only a minor fraction of groups among the extensive and fairly polar architecture of HFNAPs.
This study experimentally illuminates how nonpolar side-chains offer benefits when multiple side-chains are available, complementing previous findings that nonpolar groups in singly- or doubly-modified DNA-based polymers are beneficial31 compared to unmodified DNA for generating protein-binding aptamers. It is tempting to speculate about the applicability of these findings to other protein targets and to other types of biopolymers. While the systems and experiments reported by others working with functionalized DNA are distinct from those in this work, our findings together with previous observations that DNA modified with nonpolar side chains has a higher propensity to provide functional receptors to a variety of protein targets22,30 suggest that these principles may apply more broadly than to HFNAPs, PCSK9, and IL-6.
In addition to directly demonstrating how functional competition between classes of evolving polymers could have favored polymers with access to nonpolar side-chains similar to those found in proteins but absent from natural nucleic acids, these findings also inform future synthetic polymer evolution efforts. Of all known biopolymers, proteins arguably have access to the most diverse repertoire of chemical functionality—one that is further augmented by post-translational modifications. By providing exquisite control over the structure and position of diverse chemical groups including those unavailable to native biopolymers, the HFNAP translation system may eventually provide polymers with binding, catalytic, and other activities that complement—and perhaps even exceed—those in the impressive repertoire of natural proteins.
Methods
General Methods.
Oligonucleotides were purchased from IDT, and are listed in the Supplementary Information. Monomer building blocks were synthesized as described in ref 34. T3 DNA ligase (3,000 U/µL), ATP, T4 RNA ligase buffer were purchased from New England Biolabs. PCSK9 (PC9-H5223) and IL-6 (IL6-H4218) were purchased from AcroBiosystems and reconstituted according to the manufacturers recommendations. Aliquots were flash frozen in liquid N2 and stored at −80 ºC until ready to use. The proteins were prepared for SPR by dialyzing into the indicated buffer and concentration was determined by absorbance at 280 nm. Streptavidin-coated magnetic beads (Dynabeads MyOne C1) were purchased from Life Technologies. AminoLink Plus Micro Immobilization Kits (ThermoFisher Scientific) were used for protein immobilization and for blank beads. HBS-P+ was purchased from GE Healthcare Life Sciences.
Preparation of initial DNA libraries prior to selection.
Starting template libraries were prepared through a primer extension of the appropriate naïve library (purchased as a hand mixed library from IDT; e.g., naïve library AZ) using a biotinylated primer (e.g., BtBt_EPrimerA, see Supporting Information). Binding the resulting dsDNA to magnetic beads, washing, and strand separation with 20 mM NaOH furnished the biotinylated complement after gel purification and cleanup with Omega Bio-tek HiBind DNA Midi columns.
DNA template libraries that were used to synthesize polymers in the start of the selections were from different aliquots from the same starting template library mix. Due to the high diversity of naïve libraries, a negligible fraction of template sequences in each of the pools are identical to each other.
General comments on library comparison studies.
A 2’,3’-dideoxy 3’-primer was used in the translation of the libraries to limit propagation of cheaters arising from incorporation of a primer-binding sequence in the polymer-coding region, as we previously described (ref. 34).
First round of HFNAP library synthesis.
DNA template (125 pmol), initiation primer (188 pmol), termination primer (188 pmol), monomer mix containing 1.25 nmol each monomer, 12.5 µL 10x T4 RNA ligase buffer, and enough water to bring the volume to approximately 112 µL were added to tubes of a PCR strip, then mixed. The strips were placed in a thermal cycler and run with the following program: heat to 95°C for 10 seconds, cool to 65°C, hold at 65 ºC for 4 min, ramp down at 0.1 °C per 10 seconds to 4 °C. While still at 4 ºC, 6.25 µL of 10 mM ATP and 6.25 µL T3 DNA ligase (3,000 U/µL) were added to each reaction without removing the PCR strip from thermal cycler. The reactions were kept at 4 ºC for 12 h, then warmed to 16 ºC until ready for work up.
Each 125 µL reaction was mixed with a suspension of streptavidin-coated beads (250 µL from bottle, 2.5 mg beads; which had been prewashed with B&W buffer) that were suspended in 125 µL 2x B&W buffer (20 mM Tris•HCl, 2 M NaCl, 2 mM EDTA, pH 7.5). The bead suspensions were mixed on a rotary mixer for 1 h before removing the supernatant on a magnetic tube rack. The non-biotinylated DNA strands from the translations were separated from the bead-bound DNA through two ~10 min washes with freshly prepared 20 mM NaOH, which were combined. The combined supernatants from each reaction were mixed with a solution of 2.5 mL (5x volume) of 2:3 sat. aq. guanidine•HCl:isopropanol with 1% v/v 3 M NaOAc and 2.5 µL pH indicator. The resulting material was cleaned up with a Qiaprep 2.0 column and eluted into 52 µL buffer EB (Qiagen). A 1-µL aliquot was removed from each sample for analysis.
Protocol for second-round and later translations for HFNAP pools.
An aliquot of double-stranded template in an amount based upon the table in Supplementary Figs 1 or 8 was added to a 10-µL aliquot of streptavidin-coated magnetic beads in 1x B&W buffer and mixed for 1 h. The non-biotinylated strand was removed from the template strand by washing twice (10 min each) with 20 mM NaOH (aq.) and discarding the flow through. The beads were washed twice with 1x B&W buffer and once with 1x T4 RNA ligase buffer. The supernatant was removed.
The beads were then mixed with a solution containing initiation primer (1.5 equiv), termination primer (1.5 equiv), prepared monomer mix containing 10 equiv each of 32 monomers, 1 µL 10x T4 RNA ligase buffer, and enough water to bring the volume to approximately 8 µL. The strips were placed in a thermal cycler and run with the following program: heat to 95°C for 10 seconds, cool to 65°C, hold at 65 ºC for 4 min, ramp down at 0.1 °C per 10 seconds to 4 °C. While still at 4 ºC, 1 µL of 10 mM ATP and 1 µL T3 DNA ligase (3,000 U/µL) were added to each reaction. The reactions were kept at 4 ºC for 12 h, then warmed to 16 ºC until ready for work up.
Synthesis of individual HFNAPs.
A mixture of template (1 µL, 100 µM, 100 pmol), primers (e.g., pp1A and BtBt-pp1Z, 1.5 equiv, 150 pmol), 10x RNA ligase buffer (10 µL), 10 equiv of each of 15 monomers (either functionalized or unfunctionalized) found in HFNAP sequence of interest, and enough water to bring the volume to 80 µL was prepared. The mixture was distributed in 16-µL aliquots to five tubes of a strip, placed in a thermal cycler, and run with the following program: heat to 95°C for 10 seconds, cool to 65°C, hold at 65 ºC for 4 min, then ramp down at 0.1 °C per 10 seconds to 4 °C. While still at 4 ºC, 10 µL of 10 mM ATP and 10 µL T3 DNA ligase (3,000 U/µL) were distributed equally across the tubes. The reactions were kept at 4 ºC for at least 12 h.
The materials were pooled and mixed with a suspension of streptavidin-coated beads (100 µL from bottle, 1.0 mg beads; which had been prewashed with B&W buffer) that were suspended in 100 µL 2x B&W buffer. The bead suspensions were mixed on a rotary mixer for 1 h before removing the supernatant on a magnetic tube rack. The translations were washed first with 100 µL 1x B&W buffer and then the non-biotinylated DNA strands from the translations were separated from the bead-bound DNA through two ~10-min washes with freshly prepared 20 mM NaOH. The beads were washed twice with 100 µL B&W buffer. The beads were mixed with a solution of 95% formamide/5% EDTA (500 mM, aqueous). The beads were heated at 95 ºC for 30 min, then cooled to room temperature. The supernatants were removed, mixed with ~1 µL Gel Pilot loading buffer, and added to a 10% TBE-Urea gel, which was run for 1 h at 200 V.
The gel was visualized by UV shadowing, identifying bands that appear when shining UV light over a gel placed over a TLC plate with F254 indicator. The gel bands containing the fully-translated HFNAPs were excised from the gel and the HFNAPs were extracted from the gel pieces into 300 µL TE buffer (10 mM Tris, 0.1 mM EDTA, pH 8) by shaking overnight at 37 ºC. The resulting material was mixed with 1.5 mL “Dmitry Buffer” (2:3 guanidine•HCl:IPA with 1% (v/v) aq. NaOAc (3 M)) and concentrated with a MinElute column.
Preparation of protein beads for in vitro selection.
Protein aliquots were prepared for coupling to agarose beads in the PBS by first diluting in water and then concentrating using Amicon Ultra 0.5 mL 10k centrifugal filters. The concentrated material was dissolved in PBS spun down to concentrate. This dilution-concentration protocol was repeated again before removing the protein solution from the filter and diluting into PBS buffer. The resulting proteins were bound to agarose beads using an AminoLink Plus Micro Immobilization Kit (ThermoFisher Scientific), using 100 µg per 100 µL beads initially, and reducing the loading in later rounds, down to 20 µg PCSK9 or 10 µg IL-6 per 100 µL beads for the PCSK9 and IL-6 selections, respectively.
To make the blank beads for the negative selection, the same agarose beads were prepared without the addition of protein.
In vitro selections for protein binding.
As a general note, the selection protocol used in each one of the rounds was similar, but the selection stringency applied at the different rounds varied. The same conditions were applied equally to all of the different pools, but these conditions varied between selections and rounds.
Protein-bound beads were transferred as a suspension, mixing thoroughly until homogeneous prior to transfer, to a spin column with filter. The supernatant was removed by spinning down and the protein beads were washed with selection buffer (0.1 mg/mL BSA, 0.05% Tween-20, in 0.5x DPBS with Ca and Mg ions (Lonza)). The translated materials were mixed with a 2x selection buffer and added to column containing protein-bound beads, then mixed for specified time.
For rounds in which a negative selection was implemented, the translated polymer solution in selection buffer was first mixed with blank beads for an hour and the supernatant was collected. A 1-µL aliquot was removed for analysis and the bulk of the material was mixed with protein-bound beads in the positive selection, mixing for the specified time.
The flow-through from the positive selection was collected and the beads from each of the selection pools were washed thrice with selection buffer. The beads were collected into a tube, mixed with LDS buffer, and incubated at 95 ºC for 15 min. The resulting material was purified by spin column (Qiagen MinElute) and eluted with buffer EB. Generally, the eluted material was mixed with additional buffer, making this material of equal volume to the washes (in the first round, this material was brought to only half the volume of the washes).
A 1-µL aliquot of all of the samples from each selection were analyzed by qPCR using Q5 Hot Start High Fidelity 2x Master Mix (New England Biolabs) and 0.5x SYBR Green I (Life Technologies). The data from the qPCR was used to compare the progress of each of the pools at each round and to determine the number of PCR cycles necessary to amplify the eluted material to late-exponential growth phase.
The preparative PCR was also performed with the same general conditions as the qPCR, except that a biotinylated version of one of the primers was introduced such that the template strand would carry a biotin. The 600-µL reactions were prepared and distributed among a PCR strip in aliquots of 100 µL. The number of PCR cycles from each selection pool was adjusted based upon the qPCR data such that each pool may be amplified with a differing number of cycles.
In the first round, the eluted material PCR-amplified with Q5 Hot Start High-Fidelity 2x Master Mix and purified in two portions such that all of the eluted material was carried forward. In subsequent rounds only about one-third of the material was amplified in future selection rounds.
Following amplification, the materials were cleaned up and concentrated by addition to 3 mL Dmitry Buffer (defined above) and purification using MinElute columns (Qiagen). The resulting material was further purified by PAGE gel using a non-denaturing 10% TBE gel (Bio-Rad). The gel was visualized by UV shadowing, identifying bands that appear when shining UV light over a gel placed over a TLC plate with F254 indicator. The gel bands were excised from the gel with special care to reduce potential contamination between the pools (e.g., a designated gel box was used for these studies, and gels were wrapped in plastic). The resulting double-stranded templates were extracted from the gel pieces into 300 µL TE buffer (10 mM Tris, 0.1 mM EDTA, pH 8) by shaking overnight at 37 ºC. The resulting material was mixed with 1.5 mL Dmitry Buffer and concentrated using a MinElute column. The resulting double-stranded DNA was quantified by Nanodrop.
HTS analysis of libraries.
Aliquots collected over the course of the selection were amplified by PCR using Q5 Hot Start High-Fidelity 2x Master Mix with primers installing flanking sequences (see Oligonucleotide sequences: Sequences used for Illumina MiSeq analysis). To ensure adequate sample diversity, a portion of this PCR reaction was analyzed by qPCR to determine the number of cycles to amplify the bulk of the material to a sub-saturation number of cycles. The resulting amplicons were PAGE-purified and amplified again with Illumina barcoding primers, again to sub-saturation level determined by separate qPCR analysis. The amplicons were PAGE purified and interrogated using an Illumina MiSeq.
When performing the HTS experiments, several barcoded samples were pooled together. Since all of the libraries from each selection campaign had identical primer binding sites, four different sets of tiled adapter primers were used to ensure sequence diversity. The adapter primers were designed to all yield amplicons of the same size. Furthermore, all of the samples from the same set (e.g., all samples from the round 5 eluted libraries) were amplified with the same set of adapters.
The FASTQ files were parsed to first furnish a list of sequences that contain both primer-binding sites with a coding region of correct length. Next, the quality of the resulting sequences was evaluated such that only reads with all base calls of Q32 or higher in the coding region were kept.
The resulting filtered list of reads was processed to computationally translate into the appropriate nucleic acid and/or functionalized polymer sequences. A list of the unique sequences was compiled and the number of instances of each of those unique sequences was recorded. For some of the analyses, the first 100,000 of the filtered reads were analyzed (e.g., for convergence data). In other analyses, like the overall ranking of sequences (which was used to select sequences to synthesize), the entire list of reads was processed.
Analysis of head-to-head competition experiments.
Mixed samples prior to the head-to-head competition and those following the elution from the competition were prepared for HTS analysis as described above. Lists of reads were also processed as above, removing improperly sized and low-quality reads. We compiled a list of all of the unique sequences and associated read counts for each sample.
Each of the unique sequences from the head-to-head sample were compared against each of the seven sequence lists resulting from HTS analysis of the enriched pools in the final round of selection (e.g., round 12 for PCSK9). If a sequence from the head-to-head sample only appeared in a single pool, then it was assigned as originating from that pool. If a sequence appeared in multiple pools, then we looked at the ranking of the sequence in the originating pools. Some of the most abundant sequences in the final rounds of selection appeared in multiple pools, likely due to contamination facilitated by the higher copy number of template or polymers. If the sequence found in multiple parent pools was present in the top 1,000 most abundant sequences in one of the pools, that sequence was assigned to the pool in which it was most abundant. If the sequence did not make the top 1,000 threshold, it was considered an unassignable sequence and flagged as a duplicate (see Supplementary Figs 3 and 10).
Some of the sequences from the head-to-head competition samples were not matched to a parent pool after the initial assignment. In these cases, the same matching protocol was applied using larger lists of sequences that contained all of the unique sequences identified at any round. For instance, the larger list from the unfunctionalized pool contained all of the unique sequences found in rounds 1 through 11. Sequences that failed to match after this more extensive search were considered unassignable.
General protocols for SPR Analysis.
SPR analysis was conducted using a Biacore T200 or S200 (GE Healthcare Life Sciences). Analyses were conducted at 25 ºC with a flow rate of 30 µL/min in a buffer of HBS-P+ containing 130 mg/L CaCl2•H2O, 100 mg/L MgCl2•H2O, and 200 mg/L KCl (unless otherwise specified). During the SPR experiments, reagents were stored at 4–10 ºC in the reagent compartment.
The choice of which report point to use in single-cycle kinetics runs to evaluate binding differences between different polymers or polymer mixtures was made such that reference binding of the analyte solution was minimal.
Unless otherwise specified, biotinylated HFNAPs or HFNAP libraries were immobilized to an SPR chip surface using a Biotin CAPture kit (GE Life Sciences 28–9202-34). Prior to use, the chip was conditioned three times for 1 min at 30 µL/min using a 3:1 solution of 8 M guanadine•HCl:1 M NaOH. Next, three startup cycles with buffer were performed.
Prior to immobilization on the surface, the active and reference surfaces of the chip were treated with the Biotin CAPture reagent according to the manufacturers recommendations and the ligands were captured onto the active cell. In a single cycle kinetics run, four or five concentrations of protein analyte were flowed over the active and reference surfaces, each for 150 s at 30 µL/min, followed by a dissociation phase of 600 s (unless otherwise specified). The chip surface was regenerated with a 30-s treatment with a 3:1 mixture of 8 M guanadine•HCl and 1 M NaOH at 30 µL/min.
The analysis of each sample ligand in binding to protein was accompanied by a blank run without protein. Sensorgrams were double reference subtracted. Specified response points were used with the following equation to calculate %Rmax, where s, the stoichiometry, is taken as 1:
| (eq. 1) |
| (eq. 2) |
Assessment of pool binding by SPR.
To prepare naïve libraries or enriched polymer pools for SPR analysis, the naïve library or the biotinylated templates following the final selection rounds were each amplified by PCR so as to furnish non-biotinylated template strands. PCR reactions were monitored by qPCR to ensure that the amplification was stopped prior to reaching saturation. The non-biotinylated templates of the enriched libraries were translated with their respective monomer sets to furnish un-/partially-/fully-functionalized nucleic acid polymer pools.
The relative binding of the enriched polymer pools was studied by SPR by using a CAP chip, functionalizing with the CAPture reagent, and treating the chip surfaces with enriched polymer pools for an 1,800 s contact time at 1 µL/min or 900 s at 2 µL/min, followed by a 30 s stabilization period.
The naïve and enriched polymer pools from the PCKS9 selections were immobilized to achieve loading of 56–161 RU for all polymer pools. Concentrations of 7.3, 22, 67, and then 200 nM PCSK9, each with 0.05 mg/mL BSA, were flowed over the surface in the single-cycle kinetics mode. Example reference subtracted sensorgrams from the enriched library samples are shown in Supplementary Fig. 2. The binding report points from the end of the 200 nM injections were used to calculate the %Rmax values shown in Fig. 2a, according to eq. 1 and 2. Enriched polymer pool data in Fig. 2a represents %Rmax determinations from different Biacore machines, CAP chips, batches of enriched polymer pools, and PCSK9 batches.
The libraries derived from the IL-6 selection were immobilized to the surface with a loading between 7 and 132 RU. Concentrations of 8.9, 27, 80, and 240 nM IL-6 were flowed over the surface in the single-cycle kinetics mode. Example reference subtracted sensorgrams from the enriched library samples are shown in Supplementary Fig. 9. The values from Fig. 2d were derived from calculation of the %Rmax achieved following the 80 nM IL-6 injection in the single cycle kinetics run according to eq. 1 and 2. Enriched polymer pool data in Fig. 2d represents %Rmax determinations from different Biacore instruments, CAP chips, batches of enriched polymer pools, and IL-6 batches.
Since the identity and composition of the libraries determines the total mass, the mass used in the calculation of each of the polymer libraries was approximated based upon the average of the calculated masses for each of the most abundant individual polymers synthesized.
Assessment of individual HFNAP binding by SPR.
The %Rmax for each polymer was determined using the instrument response from the double reference subtracted sensorgram. For the polymers resulting from the PCSK9 selections, the %Rmax was calculated using the SPR response at the binding point after a single injection of 60 nM PCSK9 at 30 µL/min for 600 s. Measurements were taken using a Biacore S200 with the loading of polymers on the surface ranged from 9 to 119 RU. Polymers exhibited a range of behavior, with some representative background subtracted sensorgrams appearing in Supplementary Fig. 5. Some values are calculated as the average of multiple measurements.
For assaying affinity of HFNAPs derived from the IL-6 selections, the buffer used was HBS-P+ with added CaCl2, MgCl2, and KCl. Injections of 8.8, 27, 80, and then 240 nM IL-6 were performed in single-cycle kinetics experiments. SPR response was measured at the binding point at the end of the final, 240 nM IL-6 injection and used to calculate the %Rmax. The amount captured on the surface varied between 46 and 176 RU. Example sensorgrams are shown in Supplementary Fig. 12. Some values are calculated as the average of multiple measurements.
Kd values of individual polymers were determined from single-cycle kinetics analysis immobilizing biotinylated polymers. The affinity of HFNAP Nonpolar Plus-2 to PCSK9 was determined by immobilizing biotinylated HFNAP on the surface of an SA chip. Data were collected from runs with 10, 30, 100, and 300 nM PCSK9 and 0.3, 1, 3, 10, and 30 nM PCSK9 concentrations. Binding of HFNAP Full-2 from the IL-6 selection to IL-6 was studied by immobilizing biotinylated Full-2 to a CAP chip. Data were collected from runs with 0.3, 1, 3, 10, and 30 nM IL-6. All assessments were made using HBS-P+ buffer.
Supplementary Material
Acknowledgments
This work was supported by DARPA N66001-14-2-4053, NIH R35 GM118062, and the Howard Hughes Medical Institute. Z.C. was partially supported by the Y. Kishi Graduate Prize in Chemistry and Chemical Biology sponsored by the Eisai Corporation. The authors thank Dr. Alix Chan, Dr. Juan Pablo Maianti, Dr. Michael Packer, Dr. Dmitry L. Usanov, Dr. Ben Thuronyi, and Dr. Chris Wilson for helpful comments and discussions. We also thank Dr. Kelly Arnett and Robyn Stoller for guidance with SPR experiments and analysis.
Footnotes
Competing interests
The authors declare competing financial interests: Z.C., P.A.L., and D.R.L. have filed patent applications on DNA-templated polymerization. The authors declare no competing non-financial interests.
Code availability. The scripts used to analyze data from the head-to-head competition experiments are described and included in Supplementary Information, Supplementary Notes 2–4.
Data availability. High-throughput sequencing data will be available from the NCBI Sequence Read Archive under accession code SRP153119.
References
- 1.Kamtekar S, Schiffer JM & Xiong H Protein design by binary patterning of polar and nonpolar amino acids. Science 262, 1680–1685 (1993). [DOI] [PubMed] [Google Scholar]
- 2.Davidson AR & Sauer RT Folded proteins occur frequently in libraries of random amino acid sequences. Proc. Natl. Acad. Sci. U.S.A 91, 2146–2150 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heim EN et al. Biologically active LIL proteins built with minimal chemical diversity. Proc. Natl. Acad. Sci. U.S.A 112, E4717–E4725 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taylor SV, Walter KU, Kast P & Hilvert D Searching sequence space for protein catalysts. Proc. Natl. Acad. Sci. U.S.A 98, 10596–10601 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beasley JR & Hecht MH Protein Design: The Choice of de Novo Sequences. J. Biol. Chem 272, 2031–2034 (1997). [DOI] [PubMed] [Google Scholar]
- 6.Reader JS & Joyce GF A ribozyme composed of only two different nucleotides. Nature 420, 841–844 (2002). [DOI] [PubMed] [Google Scholar]
- 7.Ruff KM, Snyder TM & Liu DR Enhanced functional potential of nucleic acid aptamer libraries patterned to increase secondary structure. J. Am. Chem. Soc 132, 9453–9464 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rogers J & Joyce GF A ribozyme that lacks cytidine. Nature 402, 323–325 (1999). [DOI] [PubMed] [Google Scholar]
- 9.Schlosser K & Li Y DNAzyme-mediated catalysis with only guanosine and cytidine nucleotides. Nucleic Acids Res 37, 413–420 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tuerk C & Gold L Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249, 505–510 (1990). [DOI] [PubMed] [Google Scholar]
- 11.Ellington AD & Szostak JW In vitro selection of RNA molecules that bind specific ligands. Nature 346, 818–822 (1990). [DOI] [PubMed] [Google Scholar]
- 12.Robertson DL & Joyce GF Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature 344, 467–468 (1990). [DOI] [PubMed] [Google Scholar]
- 13.Bock LC, Griffin LC, Latham JA, Vermaas EH & Toole JJ Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature 355, 564 (1992). [DOI] [PubMed] [Google Scholar]
- 14.Ellington AD & Szostak JW Selection in vitro of single-stranded DNA molecules that fold into specific ligand-binding structures. Nature 355, 850–852 (1992). [DOI] [PubMed] [Google Scholar]
- 15.Keefe AD, Pai S & Ellington A Aptamers as therapeutics. Nat. Rev. Drug Discov 9, 537–550 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Breaker RR & Joyce GF A DNA enzyme that cleaves RNA. Chem. Biol 1, 223–229 (1994). [DOI] [PubMed] [Google Scholar]
- 17.Li YF, Liu Y & Breaker RR Capping DNA with DNA. Biochemistry 39, 3106–3114 (2000). [DOI] [PubMed] [Google Scholar]
- 18.Silverman SK In vitro selection, characterization, and application of deoxyribozymes that cleave RNA. Nucleic Acids Res 33, 6151–6163 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schlosser K & Li Y Biologically Inspired Synthetic Enzymes Made from DNA. Chem. Biol 16, 311–322 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Walsh SM, Sachdeva A & Silverman SK DNA catalysts with tyrosine kinase activity. J. Am. Chem. Soc 135, 14928–14931 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Silverman SK Catalytic DNA: Scope, Applications, and Biochemistry of Deoxyribozymes. Trends Biochem. Sci 41, 595–609 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gold L et al. Aptamer-Based Multiplexed Proteomic Technology for Biomarker Discovery. PLoS ONE 5, e15004–17 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ren X, Gelinas AD, Carlowitz von I, Janjic N & Pyle AM Structural basis for IL-1α recognition by a modified DNA aptamer that specifically inhibits IL-1α signaling. Nat. Commun 8, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ochsner UA, Katilius E & Janjic N Detection of Clostridium difficile toxins A, B and binary toxin with slow off-rate modified aptamers. Diagn. Microbiol. Infect. Dis 76, 278–285 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Zhou C et al. DNA-Catalyzed Amide Hydrolysis. J. Am. Chem. Soc 138, 2106–2109 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hollenstein M, Hipolito CJ, Lam CH & Perrin DM A DNAzyme with three protein-like functional groups: enhancing catalytic efficiency of M2+-independent RNA cleavage. ChemBioChem 10, 1988–1992 (2009). [DOI] [PubMed] [Google Scholar]
- 27.Sidorov AV, Grasby JA & Williams DM Sequence-specific cleavage of RNA in the absence of divalent metal ions by a DNAzyme incorporating imidazolyl and amino functionalities. Nucleic Acids Res 32, 1591–1601 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Santoro SW, Joyce GF, Sakthivel K, Gramatikova S & Barbas CF RNA cleavage by a DNA enzyme with extended chemical functionality. J. Am. Chem. Soc 122, 2433–2439 (2000). [DOI] [PubMed] [Google Scholar]
- 29.Thomas JM, Yoon J-K & Perrin DM Investigation of the Catalytic Mechanism of a Synthetic DNAzyme with Protein-like Functionality: An RNaseA Mimic? J. Am. Chem. Soc 131, 5648–5658 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Rohloff JC et al. Nucleic Acid Ligands With Protein-like Side Chains: Modified Aptamers and Their Use as Diagnostic and Therapeutic Agents. Mol. Ther. Nucleic Acids 3, e201 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gawande BN et al. Selection of DNA aptamers with two modified bases. Proc. Natl. Acad. Sci. U.S.A 114, 2898–2903 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davies DR et al. Unique motifs and hydrophobic interactions shape the binding of modified DNA ligands to protein targets. Proc. Natl. Acad. Sci. U.S.A 109, 19971–19976 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hili R, Niu J & Liu DR DNA Ligase-Mediated Translation of DNA Into Densely Functionalized Nucleic Acid Polymers. J. Am. Chem. Soc 135, 98–101 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Z, Lichtor PA, Berliner AP, Chen JC & Liu DR Evolution of sequence-defined highly functionalized nucleic acid polymers. Nat. Chem 346, 818 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kong D, Lei Y, Yeung W & Hili R Enzymatic Synthesis of Sequence-Defined Synthetic Nucleic Acid Polymers with Diverse Functional Groups. Angew. Chem. Int. Ed 55, 13164–13168 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kong D, Yeung W & Hili R In Vitro Selection of Diversely Functionalized Aptamers. J. Am. Chem. Soc 139, 13977–13980 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Lei Y, Washington J & Hili R Efficiency and fidelity of T3 DNA ligase in ligase-catalysed oligonucleotide polymerisations. Org. Biomol. Chem 1, 0076 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rose GD, Geselowitz AR, Lesser GJ, Lee RH & Zehfus MH Hydrophobicity of amino acid residues in globular proteins. Science 229, 834–838 (1985). [DOI] [PubMed] [Google Scholar]
- 39.Koide S & Sidhu SS The importance of being tyrosine: lessons in molecular recognition from minimalist synthetic binding proteins. ACS Chem. Biol 4, 325–334 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramaraj T, Angel T, Dratz EA, Jesaitis AJ & Mumey B Antigen-antibody interface properties: composition, residue interactions, and features of 53 non-redundant structures. Biochim. Biophys. Acta 1824, 520–532 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hunter CA & Jones SA IL-6 as a keystone cytokine in health and disease. Nature Immunology 16, 448–457 (2015). [DOI] [PubMed] [Google Scholar]
- 42.Halford B Cholesterol-Lowering PCSK9 Inhibitors Near Market Entry. Chemical and Engineering News 93, 10–14 (2015). [Google Scholar]
- 43.Sabatine MS et al. Evolocumab and Clinical Outcomes in Patients with Cardiovascular Disease. N Engl J Med 376, 1713–1722 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Gupta S et al. Chemically Modified DNA Aptamers Bind Interleukin-6 with High Affinity and Inhibit Signaling by Blocking Its Interaction with Interleukin-6 Receptor. J. Biol. Chem 289, 8706–8719 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gelinas AD et al. Crystal structure of interleukin-6 in complex with a modified nucleic acid ligand. J. Biol. Chem 289, 8720–8734 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zimmermann B, Gesell T, Chen D, Lorenz C & Schroeder R Monitoring genomic sequences during SELEX using high-throughput sequencing: neutral SELEX. PLoS ONE 5, e9169 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lipinski CA, Lombardo F, Dominy BW & Feeney PJ Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev 23, 3–25 (1997). [DOI] [PubMed] [Google Scholar]
- 48.Hughes JD et al. Physiochemical drug properties associated with in vivo toxicological outcomes. Bioorganic Med. Chem. Lett 18, 4872–4875 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.