Abstract
Background
Mal de Río Cuarto virus (MRCV) infects several monocotyledonous species including maize and wheat. Infected plants show shortened internodes, partial sterility, increased tillering and reduced root length. To better understand the molecular basis of the plant-virus interactions leading to these symptoms, we combined RNA sequencing with metabolite and hormone measurements.
Results
More than 3000 differentially accumulated transcripts (DATs) were detected in MRCV-infected wheat plants at 21 days post inoculation compared to mock-inoculated plants. Infected plants exhibited decreased levels of TaSWEET13 transcripts, which are involved in sucrose phloem loading. Soluble sugars, starch, trehalose 6-phosphate (Tre6P), and organic and amino acids were all higher in MRCV-infected plants. In addition, several transcripts related to plant hormone metabolism, transport and signalling were increased upon MRCV infection. Transcripts coding for GA20ox, D14, MAX2 and SMAX1-like proteins involved in gibberellin biosynthesis and strigolactone signalling, were reduced. Transcripts involved in jasmonic acid, ethylene and brassinosteroid biosynthesis, perception and signalling and in auxin transport were also altered. Hormone measurements showed that jasmonic acid, brassinosteroids, abscisic acid and indole-3-acetic acid were significantly higher in infected leaves.
Conclusions
Our results indicate that MRCV causes a profound hormonal imbalance that, together with alterations in sugar partitioning, could account for the symptoms observed in MRCV-infected plants.
Electronic supplementary material
The online version of this article (10.1186/s12870-019-1709-y) contains supplementary material, which is available to authorized users.
Keywords: Disease symptoms, Fijivirus, MRCV, Plant hormones, Reoviridae, Sucrose metabolism, SWEET transporters, Trehalose 6-phosphate, Wheat
Background
Mal de Río Cuarto virus (MRCV, genus Fijivirus, family Reoviridae) causes the most important viral maize disease in Argentina [1], the world’s second largest maize exporter (https://atlas.media.mit.edu/en/visualize/tree_map/hs92/export/show/all/1005/2016/). Other viruses in this genus cause severe rice and maize diseases in Asia [2, 3]. Besides maize, MRCV infects several monocotyledonous crops such as wheat (Triticum aestivum), oat (Avena sativa), rye (Secale cereale) and diverse species of weeds that act as winter reservoirs [4, 5]. The virus is transmitted by a number of species of delphacid planthoppers in a persistent propagative manner [6–9]. Viral replication is phloem-limited and causes severe symptoms in plants, whereas replication in planthoppers is asymptomatic [10].
Its high infection rate [10], the growing and handling advantages over maize and the availability of genetic and genomic resources [11–13] makes wheat an excellent model species to study Mal de Río Cuarto disease [14].
One of the earliest disease symptoms in wheat (appearing around 20 days post inoculation, dpi) is a darker coloration in basal leaves. Then, leaves become shorter, erect and coriaceous. Curled leaves with cross-cut edges and thickened ribs are commonly observed (Additional file 1: Figure S1). Sporadically, hypertrophy of the phloem leads to the presence of white waxy tumors called enations that are located along the veins [4, 5]. In addition, infected plants exhibit severe shortening of internodes and roots, a stunted appearance and an increased number of tillers in the vegetative stage. In the reproductive stage, plants show deformed ears that are partially or totally sterile [5]. Interestingly, symptoms occasionally appear only in a single tiller [4].
Viral infection triggers a cascade of events that profoundly disrupt host physiology, leading to virus-induced disease symptoms [15–17]. This process is systemically coordinated by a fine-tuned system in which phytohormones play a master role. Moreover, viral symptoms often resemble the phenotypes of mutants with compromised hormone biosynthesis, signalling or transport [18]. In general, salicylic acid (SA), cytokinins (CKs) and brassinosteroids (BRs) exert positive effects on plant defence against viruses, whereas auxins, ethylene (ET) and jasmonic acid (JA) have negative effects. While abscisic acid (ABA) can have both positive and negative effects in response to bacteria and fungi, it predominantly enhances plant defences upon virus infections [19].
Sugar availability is one of the major determinants of plant growth and is influenced by a large number of abiotic and biotic external factors. The impact of biotic interactions on sugar availability has been extensively studied for bacteria and fungi that colonize the apoplast, but much less is known regarding the impact of virus infections that exclusively replicate in the symplast [20–22]. Plant viral infections were reported to alter carbohydrate allocation and signalling. Soluble sugars and starch accumulate in the leaves where the virus is actively replicating, photosynthesis is reduced and respiration increased [20, 23]. The underlying mechanisms are specific to each plant-virus pathosystem and can involve the expression of viral movement proteins that interfere with plasmodesmata function and/or callose deposition, thus altering their function [24–26].
The vast majority of studies on the molecular basis of plant-virus interactions were conducted in eudicotyledonous plants, such as Arabidopsis thaliana (hereafter Arabidopsis) and Nicotiana benthamiana. Far less is known about the physiological responses of grasses to viral infections [16]. In the present study we performed an exhaustive analysis of the transcriptome, metabolite and hormone profiles of systemic MRCV-infected wheat leaves at two early time points of infection. We found that MRCV infection causes a strong alteration in sugar partitioning, characterized by high levels of sucrose, starch and the signal metabolite Tre6P. At the transcriptomic level, we found 3233 DATs at 21 dpi, including reduced transcript levels of nine genes encoding sucrose transporters from the SWEET (Sugars Will Eventually be Exported Transporters) family and for class II Tre6P synthases (TPS). In addition, our results implicated several phytohormones in the production of viral symptoms.
Results
Effect of MRCV-infection on the wheat transcriptome
Controlled infection experiments were performed to compare the wheat transcriptome between MRCV-infected and mock-inoculated plants. We were particularly interested in understanding the changes that may contribute to the establishment of a systemic infection rather than the late, pleiotropic effects of the infection. Therefore, the youngest fully expanded systemic leaf of each plant was sampled at 12 and 21 dpi. Plants at 12 dpi are asymptomatic, whereas the latter time point coincides with the appearance of macroscopic symptoms (Additional file 1: Figure S1). For each time point, four infected plants with similar viral loads and four mock-inoculated plants treated with non-viruliferous planthoppers were selected. We extracted total RNA from the individual leaf samples, constructed RNA-seq libraries and sequenced them using Illumina technology. On average, 50.0 million reads per library were obtained, 98.2% of which were retained after adapter and quality trimming (Additional file 1: Figure S2). Reads were mapped to the wheat genome (assembly version TGACv1) and, using only uniquely mapped reads (~ 43% of the total), we identified DATs between infected and control plants at each time point. At 12 dpi, before the appearance of disease symptoms, we identified just two DATs (FDR-adjusted P-value< 0.05, Additional file 2: Table S1). This result indicates that MRCV infection has almost no impact on the wheat transcriptome at this early time point. In contrast, at 21 dpi, following the onset of visual disease symptoms, we identified 3233 DATs, 42% with lower and 58% with higher transcript levels in infected plants compared to mock-inoculated samples (Additional file 2: Table S1). Twelve DATs at 21 dpi were randomly selected to validate the RNA-seq by qRT-PCR. We found consistent results using both technologies (Student’s t Test, P < 0.05), indicating that RNA-seq data is reliable (Additional file 1: Figure S3).
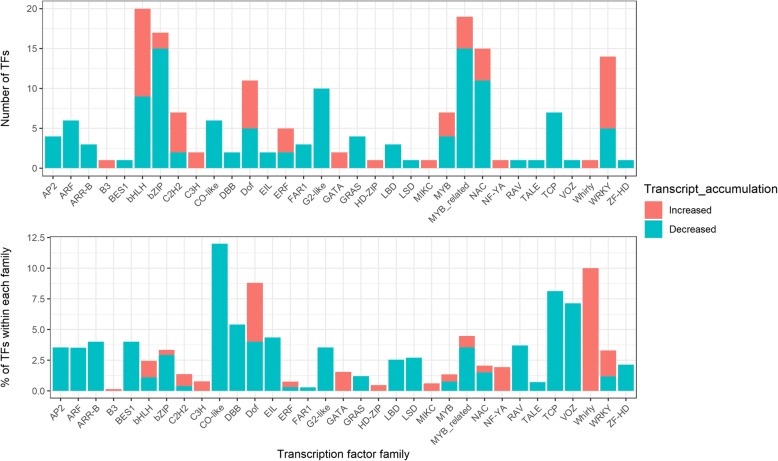
Using a set of annotated wheat transcription factors (TFs) [27], we identified 180 TFs from 33 different families among DATs at 21 dpi (Fig. 1 and Additional file 2: Table S2). Although 58% of the total DATs showed increased accumulation in infected plants, 68.9% of the transcription factor DATs showed reduced levels upon the infection. TFs from the basic helix-loop-helix (bHLH), MYB-related, basic leucine zipper (bZIP), NAC and WRKY families were the most represented. When analyzing the proportion of altered TFs within each family, CONSTANS-like (CO-like), WHIRLY, DNA BINDING WITH ONE FINGER (Dof), TEOSINTE-BRANCHED1/CYCLOIDEA/PCF (TCP) and VASCULAR PLANT ONE-ZINC FINGER (VOZ) families were most affected by MRCV infection (Fig. 1). Some of the TFs found to be altered upon MRCV infection are involved in hormone responses and are discussed below.
Fig. 1.
Effect of MRCV infection on transcripts coding for TFs of different families. Absolute number (upper panel) and proportion within each family (lower panel) of DATs coding for TF upon the infection. Red and blue colours represent increased and decreased transcripts, respectively
We next performed a gene ontology (GO) analysis to identify classes of enriched genes within DATs at 21 dpi (Table 1). Over-represented GO categories for the 1874 DATs with higher levels in infected plants included translation, cellulose biosynthesis and oxylipin biosynthetic process (Table 1). Among the 1359 DATs with reduced levels in infected plants, enriched GO categories included carbohydrate transport, trehalose biosynthesis, CK biosynthesis and strigolactone (SL) signalling.
Table 1.
Over-represented GO categories of DATs during infection for “Biological Process” ontology (Fisher’s exact test with P < 0.05)
| GO ID | Term | Annotated | Significant | Expected | classicFisher |
|---|---|---|---|---|---|
| GO categories of increased DATs | |||||
| GO:0006412 | Translation | 1624 | 257 | 42.09 | < 1e-30 |
| GO:0030244 | Cellulose biosynthetic process | 117 | 22 | 3.03 | 3.4e-13 |
| GO:0006414 | Translational elongation | 121 | 20 | 3.14 | 4.7e-11 |
| GO:0034975 | Protein folding in endoplasmic reticulum | 6 | 6 | 0.16 | 3.0e-10 |
| GO:0007017 | Microtubule-based process | 366 | 30 | 9.49 | 1.1e-08 |
| GO:0006457 | Protein folding | 346 | 42 | 8.97 | 3.6e-08 |
| GO:0007010 | Cytoskeleton organization | 322 | 21 | 8.35 | 5.3e-08 |
| GO:0042026 | Protein refolding | 32 | 9 | 0.83 | 8.4e-08 |
| GO:0030245 | Cellulose catabolic process | 60 | 11 | 1.56 | 3.6e-07 |
| GO:0031408 | Oxylipin biosynthetic process | 47 | 9 | 1.22 | 2.9e-06 |
| GO:0006564 | L-serine biosynthetic process | 17 | 6 | 0.44 | 2.9e-06 |
| GO:0006165 | Nucleoside diphosphate phosphorylation | 186 | 12 | 4.82 | 1.9e-05 |
| GO:0006183 | GTP biosynthetic process | 14 | 5 | 0.36 | 1.9e-05 |
| GO:0006228 | UTP biosynthetic process | 14 | 5 | 0.36 | 1.9e-05 |
| GO:0006167 | AMP biosynthetic process | 14 | 4 | 0.36 | 6.8e-05 |
| GO:0042254 | Ribosome biogenesis | 269 | 22 | 6.97 | 7.8e-05 |
| GO:0006166 | Purine ribonucleoside salvage | 5 | 3 | 0.13 | 0.00017 |
| GO:0006555 | Methionine metabolic process | 46 | 8 | 1.19 | 0.00032 |
| GO:0000028 | Ribosomal small subunit assembly | 6 | 3 | 0.16 | 0.00033 |
| GO categories of decreased DATs | |||||
| GO:0045454 | Cell redox homeostasis | 379 | 37 | 5.96 | 1.4e-18 |
| GO:0005992 | Trehalose biosynthetic process | 56 | 11 | 0.88 | 1.1e-09 |
| GO:0019538 | Protein metabolic process | 11,517 | 136 | 181.11 | 1.1e-07 |
| GO:0006809 | Nitric oxide biosynthetic process | 9 | 5 | 0.14 | 1.1e-07 |
| GO:1904143 | Positive regulation of carotenoid biosynthesis | 9 | 5 | 0.14 | 1.1e-07 |
| GO:0048577 | Negative regulation of short-day photoperiodism, flowering | 9 | 5 | 0.14 | 1.1e-07 |
| GO:0015798 | Myo-inositol transport | 6 | 4 | 0.09 | 8.9e-07 |
| GO:0009691 | Cytokinin biosynthetic process | 37 | 7 | 0.58 | 1.6e-06 |
| GO:1902348 | Cellular response to strigolactone | 3 | 3 | 0.05 | 3.9e-06 |
| GO:0042128 | Nitrate assimilation | 17 | 5 | 0.27 | 5.0e-06 |
| GO:0016573 | Histone acetylation | 61 | 8 | 0.96 | 5.1e-06 |
| GO:0008643 | Carbohydrate transport | 151 | 12 | 2.37 | 5.3e-06 |
| GO:0008654 | Phospholipid biosynthetic process | 148 | 9 | 2.33 | 2.2e-05 |
| GO:1901562 | Response to paraquat | 5 | 3 | 0.08 | 3.8e-05 |
| GO:0019375 | Galactolipid biosynthetic process | 5 | 3 | 0.08 | 3.8e-05 |
| GO:0071492 | Cellular response to UV-A | 6 | 3 | 0.09 | 7.5e-05 |
| GO:0006021 | Inositol biosynthetic process | 6 | 3 | 0.09 | 7.5e-05 |
| GO:0009585 | Red, far-red light phototransduction | 17 | 4 | 0.27 | 0.00012 |
| GO:0009584 | Detection of visible light | 8 | 3 | 0.13 | 0.00020 |
| GO:0006821 | Chloride transport | 35 | 5 | 0.55 | 0.00021 |
Effect of MRCV infection on primary metabolism and sugar allocation
Viral replication demands the biosynthesis of a great amount of new molecules in infected tissues and thus has a strong impact on primary metabolism [23]. At the transcript level, we identified 34 DATs involved in carbohydrate synthesis, degradation and transport, according to their associated GO terms and Ensembl annotations (Additional file 2: Table S3). Four sucrose synthase transcripts were significantly increased, including four transcripts with homology to Arabidopsis sucrose synthase 5 and 1. In addition, 12 transcripts coding for invertases and/or fructosyltransferases (evolutionarily related to vacuolar invertases) were altered at 21 dpi (Additional file 2: Table S3). Vacuolar invertases and fructosyltransferases directly affect the levels of soluble sugars and may thus contribute to the generation of turgor, which further drives cell expansion [28]. Since wheat invertases and fructosyltransferases are not fully annotated in the TGACv1 assembly version of the wheat genome, we built a phylogenetic tree including sequences of previously reported homologs to better classify them [29, 30]. We identified four significantly decreased DATs coding for vacuolar invertases and eight significantly increased DATs encoding fructosyltransferases (1 sucrose:sucrose 1-fructosyltransferase, 1-SST; 4 fructan:fructan 1-fructosyltransferases, 1-FFT; and 3 sucrose:fructan 6-fructosyltransferases, 6-SFT). In line with a possible accumulation of fructans in the infected plants, a fructan 6-exohydrolase involved in fructan degradation was decreased upon the infection (Additional file 1: Figure S4, Additional file 2: Table S3). In addition, 12 transcripts encoding homologs of enzymes involved in starch metabolism (2 glucoamylases, 2 granule-bound starch synthases, 2 branching enzymes, and 6 alpha-glucan phosphorylases) were significantly increased, while four β-amylase transcripts were significantly decreased and one was increased at 21 dpi (Additional file 2: Table S3). We also found evidence of alterations in cell wall remodeling transcripts. The GO categories of cellulose biosynthetic and catabolic processes were over-represented among transcripts with increased levels at 21 dpi (Table 1). These categories include several cellulose synthase, endoglucanase and xyloglucan endotransglucosylase/hydrolase transcripts (Additional file 2: Table S3).
The changes observed in transcripts coding for enzymes involved in primary metabolism prompted us to quantify soluble sugars and starch. MRCV-infected plants showed a significant increase in glucose, fructose, sucrose and starch at 21 dpi (Fig. 2, P < 0.05). In agreement with the gene expression analysis, we found no significant differences for any of these metabolites between MRCV-infected and control plants at 12 dpi. To further investigate the metabolic status of MRCV-infected plants, we used liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) to determine the levels of phosphorylated intermediates and organic acids. No differences were observed for any of these metabolites at 12 dpi; conversely, 10 out of 23 metabolites were significantly accumulated in MRCV-infected leaves at 21 dpi (Fig. 3, P < 0.05).
Fig. 2.
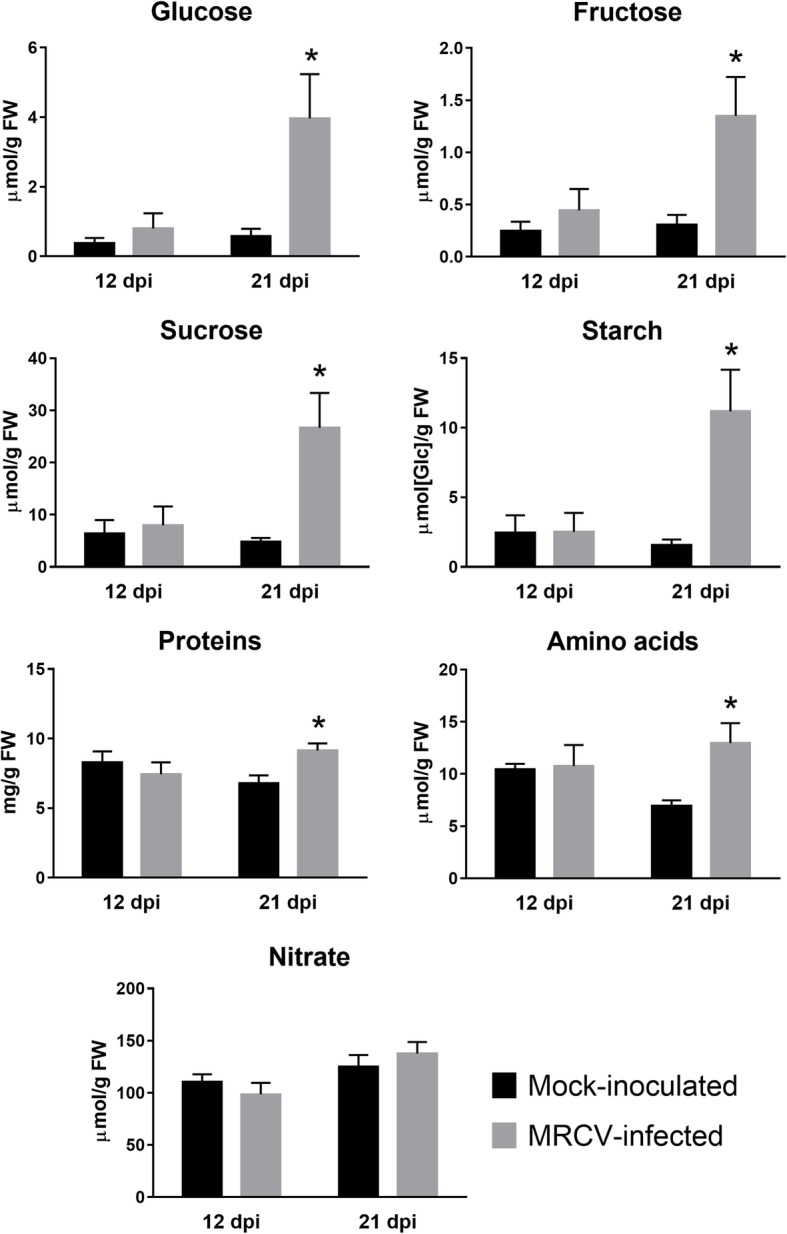
Effect of MRCV infection on the accumulation of sugars, starch, total amino acids, proteins and nitrate in MRCV-infected wheat leaves at 12 and 21 dpi. Student’s t test: *P < 0.05; n = 5. Error bars: standard error
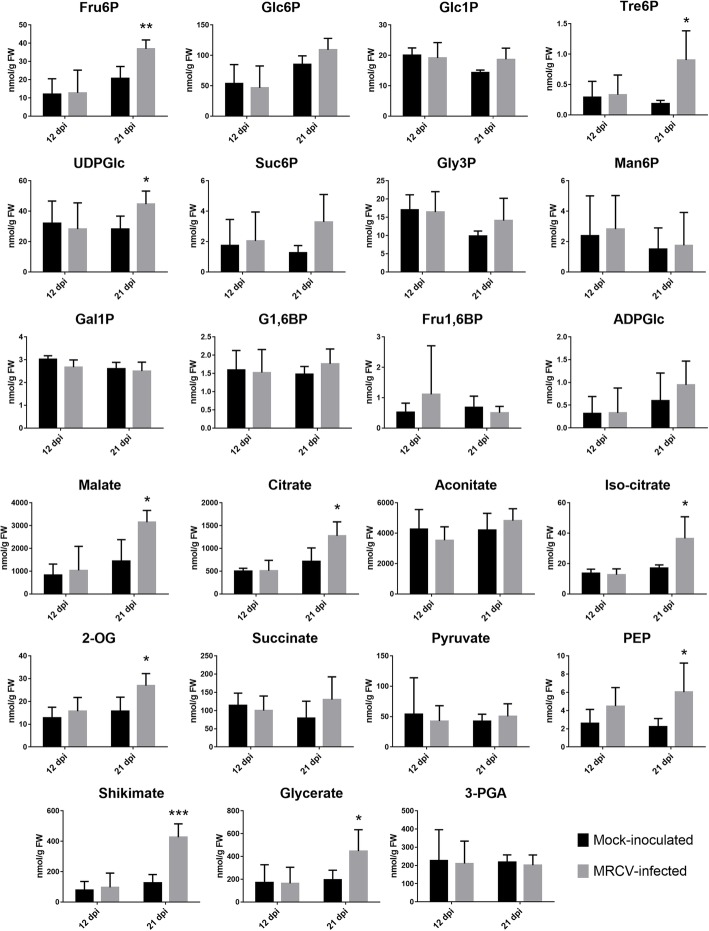
Fig. 3.
Accumulation of primary metabolites during MRCV infection at 12 and 21 dpi. Metabolites were analyzed by LC-MS/MS. Student’s t test: *P < 0.05, **P < 0.01, ***P < 0.001; n = 5. Error bars: standard error
Interestingly, the signal metabolite Tre6P was 5-fold more abundant in MRCV-infected leaves at 21 dpi than in controls (Fig. 3). Tre6P is both a signal and a negative regulator of sucrose levels in plants, thus linking growth and development to carbon status [31, 32]. In illuminated Arabidopsis leaves, Tre6P promotes the synthesis of organic and amino acids by post-translational activation of phosphoenolpyruvate (PEP) carboxylase and nitrate reductase [33]. Similarly, the levels of Tre6P and certain amino acids concomitantly increased in axillary buds from decapitated pea plants [34]. In line with these findings, we observed accumulation of several tricarboxylic acid (TCA) cycle intermediates (citrate, iso-citrate, 2-oxoglutarate and malate), PEP, shikimate and total amino acids in MRCV-infected plants at 21 dpi (Figs. 2 and 3). Total protein levels were also increased, whereas nitrate content was not significantly impacted by MRCV infection (Fig. 2).
The GO category trehalose biosynthesis was over-represented among transcripts with decreased levels at 21 dpi (Table 1). Previous phylogenetic analyses of plant TPS proteins showed that they cluster in two distinct sub-families [35, 36]. Class I TPS proteins are catalytically active, whereas the function of class II TPS proteins remains unknown [32, 37, 38]. Because wheat TPS genes are not fully annotated in the TGACv1 assembly version of the wheat genome, we built a phylogenetic tree including 21 putative wheat TPS proteins (Additional file 2: Table S4) and previously reported TPS sequences from Arabidopsis, rice, poplar and common bean [35, 36, 39]. Additional file 1: Figure S5 shows that 11 DATs coding for class II TPSs (TaTPS2, TaTPS5, TaTPS6 and TaTPS10) were significantly decreased, whereas two transcripts encoding TaTPS4 (another class II TPS) were increased.
The GO category carbohydrate transport was over-represented among DATs with decreased levels at 21 dpi (Table 1). We considered this interesting since it has been shown that bacterial and fungal plant pathogens are able to hijack sucrose transporters [22, 40, 41]. In Arabidopsis leaves, sucrose efflux from phloem parenchyma cells to the apoplast is driven by AtSWEET11 and 12 [42, 43]. Interestingly, we found 13 altered SWEET transcripts in MRCV-infected leaves at 21 dpi (Additional file 2: Table S3). To further characterize these transcripts, we searched for sequences coding for SWEET transporters in the wheat genome and found 90 putative members for this family (Additional file 2: Table S4). Next, we built a phylogenetic tree including sequences from Arabidopsis and rice [40, 44]. The levels of nine transcripts coding for TaSWEET13 were decreased in MRCV-infected plants. The proteins encoded by these transcripts cluster with AtSWEET11/12 and OsSWEET14 (Additional file 1: Figure S6), which are sucrose exporters located in phloem parenchyma cells [42].
Amino acid metabolism is also modulated by viral infections [23]. Interestingly, we observed a significant accumulation of amino acids and total proteins upon MRCV infection (Fig. 2, P < 0.05). The levels of several transcripts encoding enzymes involved in amino acid synthesis were altered at 21 dpi (22 increased and 6 decreased; Additional file 2: Table S5). The recent identification of wheat amino acid transporters [45] allowed us to detect 18 transcripts significantly altered upon MRCV infection (15 decreased and 3 increased; Additional file 2: Table S5), suggesting that amino acid transport is also affected by MRCV infection. Overall, our results show that MRCV infection has a profound effect on carbon partitioning and amino acid metabolism.
Effect of MRCV infection on phytohormone metabolism and transport
Changes in hormone metabolism, transport and signalling are frequently associated with virus symptoms. Thus, we analyzed transcripts involved in phytohormone synthesis, catabolism, transport and signalling that were significantly altered in MRCV-infected plants at 21 dpi. We also quantified several hormones including their biosynthetic precursors and metabolites by ultra-high performance liquid chromatography–electrospray tandem mass spectrometry (UHPLC–MS/MS). The transcripts associated with the different hormones are listed and classified in Additional file 2: Table S6, while Fig. 4 shows the fold change of hormone levels between infected and mock-inoculated plants. Each hormone is discussed separately in the sections below.
Fig. 4.
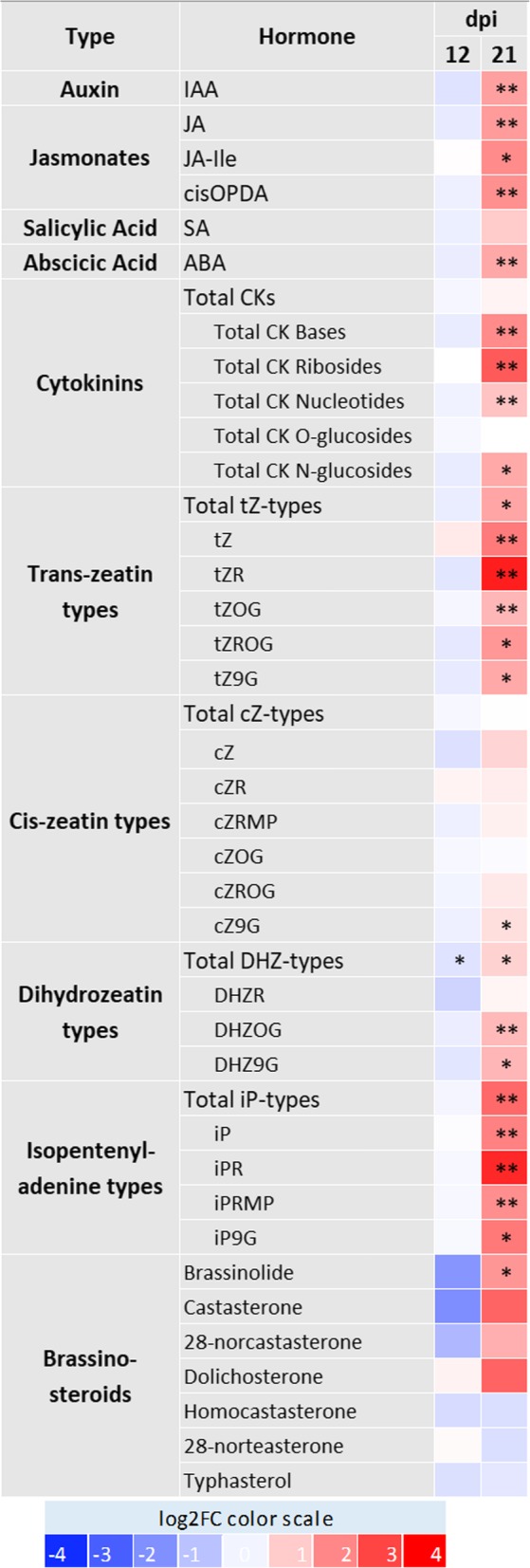
Heat-map visualization of hormone accumulation in MRCV-infected plants at 12 and 21 dpi. The log2FC color-scale is indicated. Asterisks indicate statistically significant difference between infected and mock-inoculated plants. Student’s t tests; * and ** correspond to P < 0.01 and P < 0.001, respectively; n = 5
Cytokinins
The levels of eight transcripts of LOG (LONELY GUY) genes were reduced in MRCV-infected plants. These genes encode cytokinin riboside 5′-monophosphate phosphoribohydrolase, an enzyme that converts inactive cytokinin nucleotides to the biologically active free-base forms [46]. In addition, transcript levels of three type-B ARR (AUTHENTIC RESPONSE REGULATOR) family members (Fig. 1), which are associated with cytokinin signal transduction [47], were also decreased. These results suggest that MRCV infection may be associated with a reduction in CK activity. However, when we measured different CK forms, total CK bases (bioactive forms), CK ribosides (transport forms), CK nucleotides (precursor forms) and CK N-glucosides (irreversible catabolites) were all significantly increased upon MRCV infection.
Brassinosteroids
A wheat transcript homologous to Arabidopsis and rice DIMINUTO/DWARF1 (DIM/DWF1) genes was significantly more abundant in MRCV-infected plants. These proteins are involved in the synthesis of the BR precursor campesterol [48, 49]. By contrast, DWF5, a gene encoding a protein involved in BR biosynthesis [50] was significantly decreased in MRCV-infected plants. Remarkably, the abundance of a transcript encoding the BR receptor BRASSINOSTEROID INSENSITIVE1 (BRI1) [51] was 19-fold higher in infected plants than in controls. Finally, a transcript coding for the transcription factor BES1/BZR1 and three transcripts coding for 14-3-3-like proteins identified as BZR1- and BZR2/BES1-interacting proteins [52] were significantly reduced in MRCV-infected plants. These transcriptional changes are consistent with a reduction in the levels of bioactive BRs. However, when we measured the endogenous levels of these compounds, the most active form of BRs, brassinolide (BL), was 3.4 times more abundant in MRCV-infected plants (Fig. 4).
Auxin
WALLS ARE THIN1 (WAT1) is a vacuolar auxin transport facilitator required for auxin homeostasis [53]. Six WAT1 transcripts were decreased while eight transcripts were significantly increased upon infection. In addition, three transcripts coding for the F-box TRANSPORT INHIBITOR RESPONSE 1 (TIR1) auxin receptor [54], two transcripts coding for the auxin carrier PIN5 (PIN-FORMED5) [55] and two others coding for PIN-LIKES [56] were decreased. Moreover, the levels of six transcripts coding for auxin response factors (ARFs, Fig. 1) [57] were reduced. Consistent with these expression profiles, we found that the levels of IAA, the most abundant of the four native forms of auxins, were 3-fold higher in MRCV-infected leaves (Fig. 4).
Abscisic acid
The levels of an abscisic aldehyde oxidase AAO2 transcript involved in ABA and AUX biosynthesis [58] rose upon MRCV infection, while the levels of an ABA 8′-hydroxylase transcript involved in ABA catabolism [59] were reduced. In addition, seven transcripts coding for ABA receptors PYL1 and PYL5 [60], were decreased and three transcripts coding for SKP1-like protein involved in ABA signalling [61] and the levels of TANDEM ZINC FINGER PROTEIN 1 (TZF1) (Fig. 1), which confers hypersensitivity to ABA seedlings in rice [62], were increased. Consistent with these expression profiles, we found that the levels of ABA were approximately three times higher in infected plants (P < 0.001) (Fig. 4).
Ethylene
The levels of a transcript coding for 1-aminocyclopropane-1-carboxylate synthase (ACS), which catalyses the generally rate-limiting step in ethylene biosynthesis [63], and two transcripts coding for ethylene-responsive transcription factors [64] were 40-, 6.5- and 2.2-fold higher in infected plants, respectively.
Jasmonic acid
The levels of 5 transcripts that code for lipoxygenase enzymes LOX2 involved in JA biosynthesis [65] were elevated in MRCV-infected plants. In addition, 14 WRKY TFs, some of them known to be induced by MeJA treatment in wheat and rice [66], showed altered expression in MRCV-infected plants (Fig. 1). The levels of transcripts coding for JA-responsive proteins involved in defence such as pathogenesis-related and germin-like proteins were elevated (Additional file 2: Table S1). We found that the endogenous levels of jasmonates were approximately 3.5-fold higher in MRCV-infected plants than control plants (Fig. 4).
Salicylic acid
SA synthesis depends on the activity of methylesterases [67]. The levels of three transcripts coding for methylesterases were increased while the levels of one of these transcripts was decreased upon infection. However, the levels of SA were not significantly affected by MRCV infection (Fig. 4).
Gibberellin
Bioactive GA levels strongly depend on 2-oxoglutarate-dependent dioxygenases (2-ODDs: GA20ox, GA3ox, GA2ox), which catalyze the latter steps of the biosynthetic pathway [68]. Transcript levels of two homeologous copies of GA20ox2 (GA20ox-A2 and GA20ox-D2), were decreased. These expression profiles are consistent with MRCV-infection being associated with a reduction in GA biosynthesis. In addition, two transcripts coding for gibberellin receptor GID1 were increased upon the infection [69].
Strigolactones
SLs are hormones that inhibit tillering and shoot branching through the MORE AXILLARY BRANCHING (MAX)-dependent pathway [70–72]. Remarkably, in MRCV-infected plants we found a significant decrease in transcript levels of the genes encoding the putative SL receptor strigolactone esterase D14 [73, 74]; the second component of the pathway, F-box/LRR-repeat MAX2 from Arabidopsis [75] and rice [76]; and SMAX1-LIKE proteins with homology to Arabidopsis SMXL 6, 7 and 8, which act redundantly in response to SL [77, 78].
Collectively, these results indicate that MRCV infection causes a profound disruption in hormone accumulation. Some of these changes could be linked to variations in the levels of transcripts coding for transcription factors and proteins responsible for hormone biosynthesis, transport or degradation.
Discussion
Grasses are the primary source of calories for humans and livestock [79]. Understanding the molecular basis underlying their growth, development and defence is particularly relevant for the foundation of breeding strategies for increasing and stabilizing yields. As plant viruses induce profound disruptions in host development and physiology, it will be important to dissect these mechanisms to identify key elements that can aid the improvement of agricultural traits.
To study MRCV infection in wheat, we chose two time points that precede (12 dpi) or coincide with (21 dpi) the appearance of macroscopic symptoms. We found only 2 DATs and no significant changes in metabolite levels in MRCV-infected plants at 12 dpi, whereas we detected large-scale alterations at 21 dpi (Figs. 2 and 3 and Additional file 2: Table S1). Importantly, MRCV non-structural proteins P5–2, P7–1 and P7–2 are localized in the nucleus in insect and plant cells [80, 81] where they could affect the regulation of host target genes, as has been shown for rice black streaked dwarf virus (RBSDV) P8 non-structural protein [82].
We found that both sucrose and Tre6P accumulated in fully expanded wheat leaves during MRCV infection at 21 dpi (Figs. 2 and 3). Sucrose accumulation in leaves has been repeatedly reported after virus infections [83, 84]. Soluble sugars and starch accumulate in MRCV-infected wheat leaves during late stages of infection [85], while several proteins involved in carbohydrate metabolism and photosynthesis were differentially accumulated in maize leaf tumours caused by RBSDV [86]. However, to the best of our knowledge, Tre6P accumulation has never been reported in virus-infected plants. According to the Tre6P-sucrose nexus model, the Tre6P:sucrose ratio is part of a homeostatic mechanism that ensures the maintenance of sucrose levels within an appropriate range for each cell type and/or developmental stage [31]. We observed that Tre6P and sucrose proportionally increased in MRCV-infected plants at 21 dpi compared to controls (Figs. 2 and 3); i.e. the Tre6P:sucrose ratio was not affected by the viral infection (0.040 and 0.037 nmol μmol− 1 for infected and control plants, respectively). These values are similar to those reported for Arabidopsis rosettes and maize leaves [32], suggesting that the Tre6P-sucrose nexus is still functional in these plants. In Arabidopsis, Tre6P regulates starch turnover [87, 88] and coordinates organic and amino acid metabolism with carbon availability [33]. Accordingly, we observed accumulation of starch, organic acids (including several TCA intermediates) and total amino acids in infected plants at 21 dpi (Figs. 2 and 3). Accumulation of Tre6P has been linked to reproductive failure [89, 90]; interestingly, seed abortion is one of MRCV symptoms [5]. We speculate that reduced sucrose export from source tissues due to MRCV infection is perceived as sugar starvation by developing spikes, thus leading to seed abortion.
We found 13 class II TPS transcripts with altered levels in MRCV-infected plants at 21 dpi (Additional file 1: Figure S5 and Additional file 2: Tables S3 and S4). From these, 11 DATs were decreased (TaTPS2, TaTPS5, TaTPS6 and TaTPS10) and two DATs were increased (TaTPS4) upon MRCV infection. This is consistent with previous reports showing divergent transcriptional regulation of different TPS genes [31]. Transcripts coding for TaTPS2 and TaTPS6 cluster with AtTPS7, while those encoding TaTPS4 and TaTPS10 are in the same branch as AtTPS6 (Additional file 1: Figure S5). These results are consistent with those observed for AtTPS5–7 transcripts in C-starved Arabidopsis seedlings 3 h after sucrose re-addition; i.e. high Tre6P and sucrose levels and concomitant, opposite changes in AtTPS5 and AtTPS6–7 transcripts [31]. It has been suggested that all Arabidopsis class II TPS (AtTPS5–11) lack catalytic activity, as they fail to complement the tps1Δ and tps1Δtps2Δ yeast mutants [37, 38]. As the functions of the class II TPSs are largely unknown, it is unclear whether the increased Tre6P levels are linked to the changes in class II TPS expression. One possible hypothesis is that Tre6P and TPS transcript levels reflect the high sucrose content of these plants [31]. Nevertheless, class II TPS proteins could play a key role in sugar sensing and Tre6P homeostasis [91, 92]. The use of novel reverse genetics resources for wheat [11, 93] will be invaluable to study the specific functions of the different members of this clade.
It has previously been shown that transcripts coding for the sucrose transporters AtSWEET11 and 12 are induced by fungal and bacterial pathogens that colonize the apoplast. As a consequence, sugar efflux is increased at the site of infection, affecting both pathogen growth and plant immunity [22, 94]. We observed a significant decrease in the levels of nine paralogous and homeologous TaSWEET13 transcripts and a concomitant increase of TaSWEET15 transcripts in MRCV-infected plants at 21 dpi (Additional file 1: Figure S6 and Additional file 2: Table S4). Likewise, the homologous genes in rice [95], sorghum [96] and Arabidopsis [97] showed opposed expression patterns in different physiological situations. We speculate that these changes may lead to reduced sucrose phloem loading, which in turn would contribute to the observed sucrose accumulation in source leaves (Fig. 2).
Interestingly, maize SWEET13 paralogs (a, b and c) are highly expressed in the leaf vasculature and phloem loading is impaired in the triple zmsweet13a, b, c knock-out mutant [98]. In MRCV-infected wheat, impaired sucrose phloem loading could reduce carbon partitioning to the roots (and other sink tissues), negatively affecting their growth, which is a well-known symptom of MRCV. Strikingly, AtSWEET13 and 14 mediate cellular GA uptake when expressed in yeast and Xenopus oocytes and defects of double atsweet13/14 mutants (such as delayed anther dehiscence) are reversed by GA treatment [99]. It is tempting to speculate that reduction in TaSWEET13 levels caused by MRCV infection could modulate GA responses, thus contributing to disease symptoms in wheat. It would be interesting to characterize the phenotype of wheat sweet13 loss-of-function mutants to test this hypothesis.
High sucrose levels in MRCV-infected leaves might have multiple consequences. From a plant physiology perspective, it could downregulate photosynthesis and promote starch and fructan accumulation [100–102]. In addition, the increase of the cell osmotic potential caused by sucrose accumulation could be partially relieved by cell wall expansion and/or by a decrease in the activity or expression of vacuolar invertases. Consistent with this hypothesis, we identified 39 DATs involved in cell wall synthesis, remodeling and expansion and 4 transcripts coding for vacuolar invertases with reduced accumulation at 21 dpi.
From a viral life-cycle perspective, high sucrose levels in infected tissues could contribute to increased carbon availability to sustain viral replication [23] and to attract planthopper vectors for feeding on infected plants or modify their feeding behaviors. Interestingly, non-viruliferous planthoppers prefer fijivirus-infected rice plants than healthy ones, whereas persistently-infected planthoppers prefer to feed on healthy plants [103]. The attractiveness of fijivirus-infected rice plants to planthoppers is related to virus titer and varies along the virus infection cycle [104].
Sugar, hormone signalling and defence networks are profoundly intertwined since hormone-driven processes need to be energetically compatible with the carbon status of the plant. Numerous mechanistic links between sugar and hormone-mediated processes have been described, and were recently reviewed. Moreover, several studies connect sucrose with auxin production and transport [105]. Accordingly, we detected high levels of auxin along with a reduction of transcripts coding for ARFs and for PIN5, PIN-likes and WATs auxin transporters along with high sucrose levels in MRCV-infected leaves (Additional file 2: Table S6).
Dwarfism is the most conspicuous symptom caused by fijivirus infection. It was recently shown that RBSDV P7–2 non-structural protein binds both rice S-PHASE KINASE-ASSOCIATED PROTEIN 1 (SKP1) and GIBBERLLIN INSENSITIVE DWARF2 (GID2). Since GID2 is essential for regulating the GA signalling pathway [106], these results suggest that GA signalling may play an important role during fijivirus infection [107]. In wheat, we found that TaGA20ox-A2 and –D2 transcript levels were significantly decreased in MRCV-infected plants (Additional file 2: Table S6). GA20ox2 encodes a protein which catalyzes the rate-limiting step during GA biosynthesis [108]. In rice, OsGA20ox2 loss-of-function mutations are responsible for the semi-dwarf phenotype of “green revolution” varieties [109] and in barley, reduced expression of HvGA20ox2 is also associated with a semi-dwarf phenotype [110]. We hypothesize that decreases in GA20ox2 transcript levels contribute to the dwarfed phenotype characteristic of MRCV infection in wheat. It will be important to assay bioactive GA levels in MRCV-infected plants to test this hypothesis.
We also found that the levels of transcripts encoding key SL signalling components, such as D14, MAX2 and SMAX1-like proteins SMXL 6, 7 and 8, were significantly reduced in MRCV-infected plants (Additional file 2: Table S6). In Arabidopsis, a MAX2 homolog is regulated by sugar, and both MAX2 and SMXL 6, 7 and 8 proteins promote auxin transport in the stem and growth of leaf length and width [77]. Although we were unable to quantify SL levels in our samples, the key role of these components suggests that MRCV infection may partially impair SL signalling causing excessive tillering and contributing to the reduced root and altered leaf morphology observed in MRCV-infected plants. The participation of SL in fijivirus symptoms has not been previously proposed.
Intriguingly, MRCV infection decreased the transcript levels of several LOGs involved in CK synthesis and ARR transcription factors involved in CK signaling. Yet, CK bioactive, transport and precursor forms were increased upon infection. This is in agreement with the result that ahk loss-of-function mutants have increased cytokinin content [111] and thus underpins the existence of homeostatic control mechanisms. It was further supported by analysis of endogenous cytokinin concentrations in the rock2 and rock3 mutants with enhanced cytokinin signalling. Taken together, the gain-of-function alleles of AHK2 and AHK3 have an impact on cytokinin homeostasis and lower the cytokinin content supporting a feedback regulation of cytokinin metabolism by the cytokinin signalling pathway [112]. Part of these control mechanisms is an influence of cytokinin signalling on the transcript level of cytokinin metabolism genes [113].
MRCV infection increased both bioactive BRs and transcript levels of the BR receptor BRI1 (Fig. 4 and Additional file 2: Table S6). Interestingly, TaBRI1 overexpression in Arabidopsis increased BR sensitivity and resulted in root length inhibition in a concentration-dependent manner [114]. Therefore, enhanced BR signalling could contribute to the reduced root length phenotype caused by MRCV infection.
Finally, MRCV-infected plants also exhibited increased ABA content. The antiviral roles of ABA were recently reviewed [19]. In other viral infections, ABA accumulation increases callose deposition on plasmodesmata, thus limiting viral spread [19, 115]. Interestingly, two CalS transcripts with homology to OsCal7 and AtCal7 that are responsible for specific callose deposition in the phloem [116] were significantly accumulated.
Conclusions
Fijiviruses cause severe rice and maize diseases that threaten crop production worldwide [1–3, 117] while wheat and other winter species act as virus reservoirs. Our work provides novel insights into the mechanisms underlying fijivirus symptom development in wheat highlighting for the first time the participation of the signal metabolite Tre6P, TaSWEET13 transcripts and the SL and GA signaling pathways in this process. The recent establishment of CRISPR/Cas9 genome editing protocols [11, 118, 119], along with the availability of sequenced TILLING populations both in tetraploid and hexaploid wheat [11, 93], will contribute to further deepen into these mechanisms.
Methods
Plant inoculation and RNA sequencing
Six-day-old wheat (T. aestivum cv. ProINTA Federal) seedlings were infected with MRCV as previously described [14]. This cultivar was selected because it is susceptible to MRCV infection [4]. Briefly, second instar Delphacodes kuscheli (delphacid planthopper vector) nymphs fed on MRCV-infected wheat plants were used for 1:1 transmission assays. Nymphs fed on non-infected wheat were used as control treatment. Plants were grown in 750 cm3 pots with soil sterilized by solarization in greenhouses with temperature-controlled conditions (24 ± 3 °C) in 16h light / 8h dark photoperiod and daily irrigation with no fertilization. The plants were rotated regularly within the greenhouse to reduce any positional effects. The youngest fully-expanded leaf was collected at 12 and 21 dpi, frozen in liquid nitrogen and kept at − 80 °C until use. Wheat plants were identified as MRCV infected or non-infected by DAS-ELISA at 50 dpi [4]. Absolute viral load was quantified as previously described [14, 120] and plants with similar viral load (log10(MRCV-S3 molecules)/(μg of total RNA) = 20.60 ± 0.96) for each time point were selected. To avoid leaf oxidative stress caused by ovipositions, only plants infected by male delphacids were used. Total RNA samples from mock-inoculated and MRCV-infected plants were extracted at 12 and 21 dpi (n = 4, a total of 16 individual samples) using a mirVana kit (Thermo Fisher Scientific Inc., Waltham, MA, USA) following the total RNA extraction protocol. cDNA libraries were synthesized using an Illumina TruSeq RNA sample prep kit (Illumina, San Diego, CA, USA). Library quality was checked using a High Sensitivity DNA chip in a Bioanalyzer 2100 kit (Agilent Technologies, Santa Clara, CA, USA). Libraries were sequenced on the HiSeq 3000 platform (Illumina, San Diego, CA, USA) at the UC Davis Genome Center using a single-end 50 bp (SE 50) module. Raw sequences of the 16 libraries were deposited in NCBI Sequence Read Archive (SRA) under the accession SRP160433.
Sequence data analysis
Raw reads were processed as previously described [121]. Briefly, “Scythe” (https://github.com/vsbuffalo/scythe) was used to remove adapter contamination (default options) and “Sickle” (https://github.com/najoshi/sickle) to remove low-quality reads (default options except –l 25 –q 25). Trimmed reads were mapped to the Chinese Spring (CS42) TGACv1 genome assembly [12] using GSNAPl (version 2017-09-11, default parameters except -m 2 -n 1 -N 1 -A sam) [122]. Raw counts for gene features were obtained using ht-seq count (default parameters except -m union -a 30) [123] using the GFF file available in Ensembl plants release 37. A threshold for mapping quality was selected so that only uniquely-mapped reads were retained, ensuring homoeologue-specific expression profiles [124]. DESeq2 version 1.17.39 [125] and R version 3.4.0 were used for the normalization and classification of differentially accumulated transcripts (DATs) between infected and mock-inoculated samples (FDR-adjusted P-value ≤0.05, Additional file 2: Table S1).
Annotation data were retrieved from Ensembl plants release 37 using BioMart [126, 127]. GO annotation was used for functional enrichment analysis using the ‘R’ package TopGO version 2.29.0. “Biological Process” ontology was used and the significance values were calculated using ‘classic’ Fishers’ exact test (P < 0.01). When detailed annotation data were not available for T. aestivum, data from the closest homolog of Aegilops tauschii (goatgrass, the diploid progenitor of the D-genome of hexaploid wheat) was used. A reference-quality genome sequence was recently reported with more than 76% of genes annotated [128].
qRT-PCR validations
One μg of DNase-treated total RNA, SuperScript III Reverse transcriptase (Invitrogen, Carlsbad, CA, USA) and random primers were used to synthesize the first-strand of cDNA, according to the manufacturer’s instructions. Twelve DATs were randomly selected and the respective oligonucleotides (listed in Additional file 2: Table S7) were designed using Primer3 [129]. SYBR green PCR reactions were carried out in 20 μl final volume containing: 3 mM MgCl2, 200 nM each primer, 0.2 mM dNTPs, 1 U Platinum Taq DNA Polymerase (Invitrogen, Carlsbad, CA, USA), 20 mM Tris-HCl (pH 8.4), 50 mM KCl. Cycling conditions were as follows: 95 °C for 5 min, 40 cycles at 95 °C for 15 s, 60 °C for 60 s and a final extension at 72 °C for 30 s, in an Applied Biosystems Step One Plus Real Time PCR system. qRT-PCR data analysis and primer efficiencies were obtained using LinRegPCR software [130]. The genes coding for GTP-BINDING PROTEIN (GTPB, Genbank ID: JQ673330.1) and ELONGATION FACTOR 1 ALPHA (EF1α, Genbank ID: AK455225.1) were used as reference [131]. Relative expression ratios and statistical analysis were performed using fgStatistics software interface [132]. Six biological replicates (the same four as in the RNA-seq plus two additional replicates) and two technical replicates were performed. Statistical significance between RNA-seq and qPCR Fold Change values was assessed by Two-tailed Student’s T tests.
Extraction and analysis of metabolites
Soluble sugars (glucose, fructose and sucrose) were extracted from the same previously described leaf samples of five individual plants (the same four as in the RNA-seq plus one additional replicate) using ethanol and enzymatically quantified [133]. Total amino acids and nitrate were determined using the same extract, while starch and proteins were assayed in the insoluble material obtained after the ethanolic extraction, following the protocols described by Cross et al. [134]. Tre6P, phosphorylated intermediates and organic acids were extracted with chloroform/methanol, and measured by high-performance anion-exchange LC-MS/MS as described by Lunn et al. [135], with the modifications introduced by Figueroa et al. [33].
Phylogenetic analysis
Protein sequences for TPS, SWEET, invertases and fructosyltransferases from Triticum aestivum (Ta), the progenitors of the A and D subgenomes Triticum urartu (Tu) and Aegilops tauschii (AEGt), Triticum turgidum (Tt), Aegilops searsii (As), Oryza sativa (Os), Arabidopsis thaliana (At), Solanum lycopersicum (Sl) and Populus trichocarpa (Pt) were downloaded from the NCBI database (https://www.ncbi.nlm.nih.gov/). Protein sequences from wheat were selected using the BLASTP algorithm in Ensembl Plants (http://plants.ensembl.org/index.html). Briefly, representative sequences from rice (for TPS, SWEET and invertases) or wheat (for fructosyltransferases) were used to search for homologs encoded in the T. aestivum genome (assembly TGACv1). The resulting sequences were downloaded from UniProt (http://www.uniprot.org/), manually curated and automatically aligned with those obtained for other species using the ClustalW program in GenomeNet (http://www.genome.jp/). Phylogenetic trees were built using the neighbor-joining algorithm (bootstrap of 1000) in Seaview 4.3.0 [136] and figures were prepared with the FigTree 1.3.1 program (http://tree.bio.ed.ac.uk/). Sequences used in this study are listed in Additional file 2: Table S4.
Quantitative analysis of plant hormones
The endogenous phytohormones concentrations were determined in leaf samples by an ultra-high performance liquid chromatography–electrospray tandem mass spectrometry (UHPLC–MS/MS) using stable isotope-labelled internal standards as a reference. The isolation of CKs was performed according to Antoniadi et al. [137] and their levels were quantified by UHPLC–MS/MS [138]. Briefly, samples (20 mg FW) were extracted in 1 ml of modified Bieleski buffer [139] together with a cocktail of stable isotope-labeled internal standards used as a reference (0.25 pmol of CK bases, ribosides, N-glucosides, and 0.5 pmol of CK O-glucosides, nucleotides per sample added). The extracts were purified using the Oasis MCX column (30 mg/1 ml, Waters) and cytokinin levels were determined using the LC-MS/MS system consisting of an ACQUITY UPLC System and a Xevo TQ-S triple quadrupole mass spectrometer (Waters). The acidic phytohormones (JA; jasmonoyl-L-isoleucine, JA-Ile; cis-12-oxo-phytodienoic acid, cis-OPDA; indole-3-acetic acid, IAA; ABA; and SA) were extracted using an aqueous solution of methanol (10% MeOH/H2O, v/v) [140]. A cocktail of stable isotope-labelled standards was added as follows: 5 pmol of [13C6]IAA, 10 pmol of [2H6]JA, [2H2]JA-Ile, and [2H6]ABA, and 20 pmol of [2H4]SA and [2H5]OPDA (all from Olchemim Ltd., Czech Republic) per sample. The extracts were purified using Oasis HLB columns (30 mg/1 ml, Waters), targeted analytes were eluted using 80% MeOH and then analyzed by LC-MS/MS method [140]. The brassinosteroid content in the samples was determined using the previously described UHPLC–MS/MS method [141]. All experiments were performed in five independent biological replicates (the same four as in the RNA-seq plus one additional replicate).
Additional files
Figure S1. Representative pictures of MRCV symptoms in wheat at 21 dpi. Insets highlighting curled leaves and cross-cut edges are shown. Figure S2. Library mapping statistics. For each library, total reads, clean reads, mapped reads, and uniquely mapped reads that fall within a gene feature are indicated. T stands for treated (MRCV-infected) and C for control (mock-inoculated). R1 to R4 stand for each of the four plants sequenced from each treatment. Figure S3. qPCR validation of DATs at 21 dpi. For qPCR experiments, n = 6. Error bars: standard error. Statistical significance between RNA-seq and qPCR Fold Change values was assessed by Two-tailed Student’s T tests (*P < 0.05). Figure S4. Phylogenetic relationships of wheat vacuolar invertases and fructosyltransferases with other plant sequences. Protein sequences from Triticum aestivum (Ta), the progenitors of the A and D subgenomes Triticum urartu (Tu) and Aegilops tauschii (AEGt), Triticum turgidum (Tt), Aegilops searsii (As), Oryza sativa (Os), Arabidopsis thaliana (At), Solanum lycopersicum (Sl) and Populus trichocarpa (Pt) were used to build a phylogenetic tree using the neighbor-joining algorithm with a bootstrap of 1000. Figure S5. Phylogenetic relationships of wheat TPS with other plant TPS. Protein sequences of class II TPS from Triticum aestivum (Ta), Oryza sativa (Os), Arabidopsis thaliana (At), Phaseolus vulgaris (Pv) and Populus trichocarpa (Pt) were used to build a phylogenetic tree using the neighbor-joining algorithm with a bootstrap of 1000. Figure S6. Phylogenetic relationships of wheat SWEET with other plant SWEET. Sequences from Triticum aestivum (Ta), Oryza sativa (Os) and Arabidopsis thaliana (At) were used to build a phylogenetic tree using the neighbor-joining algorithm with a bootstrap of 1000. Wheat protein sequences whose transcripts were significantly increased or decreased in the RNA-seq analysis at 21 dpi are coloured in red and blue, respectively. (DOCX 2662 kb)
Table S1. Differentially accumulated transcripts (DATs) upon MRCV infection at 12 and 21 dpi. For each gene, the normalized mean count for mock-inoculated and MRCV-infected samples, the fold change and the FDR-adjusted P-value (padj) are indicated. The annotations from Ensembl plants Release 37 or inferred in this work are included. Table S2. DATs classified as wheat Transcription factors according to reference [27]. Table S3. DATs classified as Carbohydrate-related according to their functional annotation. Table S4. Putative wheat invertases and fructosyltransferases, Tre6P synthases (TPS) and Sugars Will Eventually be Exported Transporters (SWEET) used to build the phylogenetic trees shown on Additional file 1: Figures S4-S6. Sequences encoded by DATs at 21 dpi are highlighted in red (increased upon the infection) and blue (decreased). A/N Inv, Vac Inv and CW Inv stand for acid and/or neutral, vacuolar and cell wall invertases respectively. Fructosyltransferases included sucrose:sucrose 1-fructosyltransferase (1-SST), fructan:fructan 1-fructosyltransferases (1-FFT) and sucrose:fructan 6-fructosyltransferases (6-SFT). Table S5. DATs classified as Amino acid-related according to their functional annotation. Amino acid transporters were extracted from reference [45]. Table S6. DATs classified as Hormone-related according to their functional annotation. Table S7. Primer sequences used for qRT-PCR validations. (XLSX 717 kb)
Acknowledgements
The authors would like to thank Jorge Dubcovsky for supervising the wheat RNA-seq analysis and providing hardware resources. Advice given by Hans Vasquez-Gross and Tyson Howell for bioinformatic analyses is greatly appreciated. Special thanks to Laura Inés Ramos for technical assistance.
Funding
The maintenance of planthopper populations, plant infection experiments, sample collection, construction and sequencing of RNAseq libraries, qPCR validations, and sugar and protein measurements were supported by Research project PNBIO 1131022 from the Instituto National de Tecnología Agropecuaria (INTA) and by PICT 2012 N 0391 from the Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT). Metabolite analysis was supported by the Max Planck Society (R.F., and J.E.L.). CMF is funded by the Max Planck Society (Partner Group for Plant Biochemistry). The quantification of hormones was supported by the Ministry of Education, Youth and Sports of Czech Republic from European Regional Development Fund-Project “Centre for Experimental Plant Biology”: No. CZ.02.1.01/0.0/0.0/16_019/0000738. LdH was granted with a CONICET Doctorate fellowship and a Fulbright-BEC.AR short term fellowship.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files. Raw sequences of RNA-seq libraries were deposited in NCBI Sequence Read Archive (SRA) (https://www.ncbi.nlm.nih.gov/sra) under the accession number SRP160433.
Abbreviations
- ABA
Abscisic acid
- ARF
Auxin response factor
- AUX
Auxin
- BR
Brassinosteroid
- cis-OPDA
Cis-12-oxo-phytodienoic acid
- CK
Cytokinin
- DAS-ELISA
Double Antibody Sandwich ELISA
- DAT
Differentially accumulated transcript
- dpi
Days post inoculation
- ELISA
Enzyme-linked immunosorbent assay
- ET
Ethylene
- FDR
False discovery rate
- GO
Gene ontology
- IAA
Indole-3-acetic acid
- JA
Jasmonic acid
- JA-Ile
Jasmonoyl-L-isoleucine
- LC-MS/MS
Liquid chromatography coupled to tandem mass spectrometry
- MeJA
Methyl jasmonate
- MRCV
Mal de Río Cuarto virus
- PEP
Phosphoenolpyruvate
- qRT-PCR
Quantitative reverse transcription PCR
- RBSDV
Rice black streaked dwarf virus
- SA
Salicylic acid
- SL
Strigolactone
- SRA
Sequence read archive
- SWEET
Sugars Will Eventually be Exported Transporters
- TCA
Tricarboxylic acid
- TF
Transcription factor
- TPS
Tre6P synthase
- Tre6P
Trehalose 6 Phosphate
- UHPLC–MS/MS
Ultra-high performance liquid chromatography coupled to tandem mass spectrometry
Authors’ contributions
LAdH followed the infection experiment, collected the samples, constructed RNA-seq libraries, performed qPCR validations, performed all bioinformatic analyses, analyzed the results and wrote the manuscript. SMA performed qPCR validations, measured sugars and proteins, prepared samples for mass spec analysis. ON and DT quantified plant hormones and discussed the manuscript. RF performed mass spec analysis of metabolites. ADD and MFM raised planthoppers, maintained MRCV inocula, performed plant infections and ELISA tests. GL followed infection and collected samples. MS and JEL discussed the results and revised the manuscript. SP contributed with bioinformatics assistance and training, analyzed GA genes, discussed the results and wrote the manuscript. CMF measured sugars, proteins and nitrate, prepared samples for mass spec analysis, performed phylogenetic analyses, discussed the results and wrote the manuscript. MdV conceived the study, discussed the results and wrote the manuscript. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
T. aestivum cv. ProINTA Federal seeds were obtained from the germplasm bank of EEA Marcos Juárez, INTA, WIEWS instcode ARG1345. Details available at http://www.fao.org/wiews/data/organizations/en/?instcode=ARG1345.
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Luis Alejandro de Haro, Email: deharo.luis@inta.gob.ar.
Sofía Maité Arellano, Email: arellano.sofia@inta.gob.ar.
Ondrej Novák, Email: ondrej.novak@upol.cz.
Regina Feil, Email: Feil@mpimp-golm.mpg.de.
Analía Delina Dumón, Email: dumon.analia@inta.gob.ar.
María Fernanda Mattio, Email: mattio.fernanda@inta.gob.ar.
Danuše Tarkowská, Email: tarkowska@ueb.cas.cz.
Gabriela Llauger, Email: llauger.gabriela@inta.gob.ar.
Miroslav Strnad, Email: miroslav.strnad@upol.cz.
John Edward Lunn, Email: Lunn@mpimp-golm.mpg.de.
Stephen Pearce, Email: Stephen.Pearce@colostate.edu.
Carlos María Figueroa, Email: carfigue@fbcb.unl.edu.ar.
Mariana del Vas, Phone: (0054) 114621-1676, Email: delvas.mariana@inta.gob.ar.
References
- 1.Lenardón SL, March GJ, Nome SF, Ornaghi JA. Recent outbreak of “Mal de Río Cuarto” virus on corn in Argentina. Plant Dis. 1998;82(4):448. doi: 10.1094/PDIS.1998.82.4.448C. [DOI] [PubMed] [Google Scholar]
- 2.Wang HD, Chen JP, Wang AG, Jiang XH, Adams MJ. Studies on the epidemiology and yield losses from rice black-streaked dwarf disease in a recent epidemic in Zhejiang province. China Plant Pathol. 2009;58(5):815–825. doi: 10.1111/j.1365-3059.2009.02091.x. [DOI] [Google Scholar]
- 3.Zhou G, Xu D, Xu D, Zhang M. Southern rice black-streaked dwarf virus: A white-backed planthopper-transmitted fijivirus threatening rice production in Asia. Front Microbiol. 2013;4(SEP):1–9. doi: 10.3389/fmicb.2013.00270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Truol GA, Usugi T, Hirao J, Arneodo JD, Gimenez Pecci M, Laguna IG. Transmisión experimental del virus del Mal de Río Cuarto por Delphacodes kuscheli. Phytopatologia Bras. 2001;26(1):39–44. doi: 10.1590/S0100-41582001000100007. [DOI] [Google Scholar]
- 5.Rodriguez Pardina PE, Gimenez Pecci MP, Laguna IG, Dagoberto E, Truol G. Wheat: a new natural host for the Mal de Río Cuarto virus in the endemic disease area, Río Cuarto, Córdoba Province, Argentina. Plant Dis. 1998;82:149–152. doi: 10.1094/PDIS.1998.82.2.149. [DOI] [PubMed] [Google Scholar]
- 6.Mattio MF, Cassol A, Marino de Remes Lenicov AN, Truol G. Tagosodes orizicolus: nuevo vector potencial del Mal de Río Cuarto virus. Trop Plant Pathol. 2008;33(3):237–240. doi: 10.1590/S1982-56762008000300010. [DOI] [Google Scholar]
- 7.Velázquez PD, Arneodo JD, Guzmán FA, Conci LR, Truol GA. Delphacodes haywardi muir, a new natural vector of Mal de Río Cuarto virus in Argentina. J Phytopathol. 2003;151:669–672. doi: 10.1046/j.1439-0434.2003.00786.x. [DOI] [Google Scholar]
- 8.Velázquez PD, De Remes Lenicov AMM, Truol GA. Caenodelphax teapae (fowler) (hemiptera: delphacidae): a new natural vector of the Mal de Río Cuarto virus (MRCV, fijivirus) in Argentina. AgriScientia. 2017;34:39. doi: 10.31047/1668.298x.v34.n1.17357. [DOI] [Google Scholar]
- 9.Marino de Remes Lenicov AM, Tesón A, Dagoberto E, Huguet N. Hallazgo de uno de los vectores del Mal de Río Cuarto virus en maíz. Gac Agronómica. 1985;25:251–258. [Google Scholar]
- 10.Arneodo JD, Guzmán FA, Conci LR, Laguna IG, Truol GA. Transmission features of Mal de Río Cuarto virus in wheat by its planthopper vector Delphacodes kuscheli. Ann Appl Biol. 2002;141:195–200. doi: 10.1111/j.1744-7348.2002.tb00212.x. [DOI] [Google Scholar]
- 11.Uauy C, Wulff BBH, Dubcovsky J. Combining Traditional Mutagenesis with New High-Throughput Sequencing and Genome Editing to Reveal Hidden Variation in Polyploid Wheat. Annu Rev Genet. 2017;51(September):435–454. doi: 10.1146/annurev-genet-120116-024533. [DOI] [PubMed] [Google Scholar]
- 12.Clavijo BJ, Venturini L, Schudoma C, Accinelli GG, Kaithakottil G, Wright J, et al. An improved assembly and annotation of the allohexaploid wheat genome identifies complete families of agronomic genes and provides genomic evidence for chromosomal translocations. Genome Res. 2017;27(5):885–896. doi: 10.1101/gr.217117.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Appels R, Eversole K, Feuillet C, Keller B, Rogers J, Stein N, et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science. 2018;361(6403):eaar7191. doi: 10.1126/science.aar7191. [DOI] [PubMed] [Google Scholar]
- 14.de Haro LA, Dumón AD, Mattio MF, Argüello Caro EB, Llauger G, Zavallo D, et al. Mal de Río Cuarto virus infection triggers the production of distinctive viral-derived siRNA profiles in wheat and its planthopper vector. Front Plant Sci. 2017;8(May):1–11. doi: 10.3389/fpls.2017.00766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Culver JN, Padmanabhan MS. Virus-induced disease: altering host physiology one interaction at a time. Annu Rev Phytopathol. 2007;45(1):221–243. doi: 10.1146/annurev.phyto.45.062806.094422. [DOI] [PubMed] [Google Scholar]
- 16.Mandadi KK, Scholthof K-BG. Plant immune responses against viruses: how does a virus cause disease? Plant Cell. 2013;25(5):1489–1505. doi: 10.1105/tpc.113.111658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang A. Dissecting the molecular network of virus-plant interactions: the complex roles of host factors. Annu Rev Phytopathol. 2015;53:45. doi: 10.1146/annurev-phyto-080614-120001. [DOI] [PubMed] [Google Scholar]
- 18.Alazem M, Lin NS. Roles of plant hormones in the regulation of host-virus interactions. Mol Plant Pathol. 2015;16(5):529–540. doi: 10.1111/mpp.12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alazem M, Lin N-S. Antiviral roles of abscisic acid in plants. Front Plant Sci. 2017;8(October):1–10. doi: 10.3389/fpls.2017.01760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Biemelt S, Sonnewald U. Plant-microbe interactions to probe regulation of plant carbon metabolism. J Plant Physiol. 2006;163:307. doi: 10.1016/j.jplph.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 21.Lemoine R, La Camera S, Atanassova R, Dédaldéchamp F, Allario T, Pourtau N, et al. Source-to-sink transport of sugar and regulation by environmental factors. Front Plant Sci. 2013;4:272. doi: 10.3389/fpls.2013.00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bezrutczyk M, Yang J, Eom J-SS, Prior M, Sosso D, Hartwig T, et al. Sugar flux and signaling in plant-microbe interactions. Plant J. 2018;93(4):675–85. [DOI] [PubMed]
- 23.Llave C. Dynamic cross-talk between host primary metabolism and viruses during infections in plants. Curr Opin Virol. 2016;19:50–55. doi: 10.1016/j.coviro.2016.06.013. [DOI] [PubMed] [Google Scholar]
- 24.Tccsi L, Smith AM, Maule AJ, Leegood RC. A spatial analysis of physiological changes associated with infection of cotyledons of marrow plants with cucumber mosaic virus. Plant Physiol. 1996;11(1):975–985. doi: 10.1104/pp.111.4.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Balachandran S, Hurry VM, Kelley SE, Osmond CB, Robinson SA, Rohozinski J, et al. Concepts of plant biotic stress. Some insights into the stress physiology of virus-infected plants, from the perspective of photosynthesis. Physiol Plant. 1997;100(2):203–213. doi: 10.1111/j.1399-3054.1997.tb04776.x. [DOI] [Google Scholar]
- 26.Herbers K, Takahata Y, Melzer M, Mock H-P, Hajirezaei M, Sonnewald U. Regulation of carbohydrate partitioning during the interaction of potato virus Y with tobacco. Mol Plant Pathol. 2000;1:51. doi: 10.1046/j.1364-3703.2000.00007.x. [DOI] [PubMed] [Google Scholar]
- 27.Borrill P, Harrington SA, Uauy C. Genome-Wide Sequence and Expression Analysis of the NAC Transcription Factor Family in Polyploid Wheat. G3. 2017;7(September). 10.1534/g3.117.043679. [DOI] [PMC free article] [PubMed]
- 28.Lunn JE. Sucrose Metabolism. eLS. 2016;(2008):1–9.
- 29.Huynh BL, Mather DE, Schreiber AW, Toubia J, Baumann U, Shoaei Z, et al. Clusters of genes encoding fructan biosynthesizing enzymes in wheat and barley. Plant Mol Biol. 2012;80(3):299–314. doi: 10.1007/s11103-012-9949-3. [DOI] [PubMed] [Google Scholar]
- 30.Wan H, Wu L, Yang Y, Zhou G, Ruan YL. Evolution of sucrose metabolism: the dichotomy of Invertases and beyond. Trends Plant Sci. 2018;23(2):163–177. doi: 10.1016/j.tplants.2017.11.001. [DOI] [PubMed] [Google Scholar]
- 31.Yadav UP, Ivakov A, Feil R, Duan GY, Walther D, Giavalisco P, et al. The sucrose-trehalose 6-phosphate (Tre6P) nexus: specificity and mechanisms of sucrose signalling by. J Exp Bot. 2014;65(4):1051–1068. doi: 10.1093/jxb/ert457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lunn JE, Delorge I, Figueroa CM, Van Dijck P, Stitt M. Trehalose metabolism in plants. Plant J. 2014;79:544. doi: 10.1111/tpj.12509. [DOI] [PubMed] [Google Scholar]
- 33.Figueroa CM, Feil R, Ishihara H, Watanabe M, Kölling K, Krause U, et al. Trehalose 6-phosphate coordinates organic and amino acid metabolism with carbon availability. Plant J. 2016;85(3):410–423. doi: 10.1111/tpj.13114. [DOI] [PubMed] [Google Scholar]
- 34.Fichtner F, Barbier FF, Feil R, Watanabe M, Annunziata MG, Chabikwa TG, et al. Trehalose 6-phosphate is involved in triggering axillary bud outgrowth in garden pea (Pisum sativum L.) Plant J. 2017;92(4):611–623. doi: 10.1111/tpj.13705. [DOI] [PubMed] [Google Scholar]
- 35.Lunn JE. Gene families and evolution of trehalose metabolism in plants. Funct Plant Biol. 2007;34(6):550–563. doi: 10.1071/FP06315. [DOI] [PubMed] [Google Scholar]
- 36.Yang HL, Liu YJ, Wang CL, Zeng QY. Molecular evolution of Trehalose-6-phosphate synthase (TPS) gene family in populus, arabidopsis and rice. PLoS One. 2012;7(8):e42438. doi: 10.1371/journal.pone.0042438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramon M, De Smet I, Vandesteene L, Naudts M, Leyman B, Van Dijck P, et al. Extensive expression regulation and lack of heterologous enzymatic activity of the class II trehalose metabolism proteins from Arabidopsis thaliana. Plant Cell Environ. 2009;32(8):1015–1032. doi: 10.1111/j.1365-3040.2009.01985.x. [DOI] [PubMed] [Google Scholar]
- 38.Delorge I, Figueroa CM, Feil R, Lunn JE, Van Dijck P. Trehalose-6-phosphate synthase 1 is not the only active TPS in Arabidopsis thaliana. Biochem J. 2015;466:283. doi: 10.1042/BJ20141322. [DOI] [PubMed] [Google Scholar]
- 39.Barraza A, Contreras-Cubas C, Estrada-Navarrete G, Reyes JL, Juárez-Verdayes MA, Avonce N, et al. The class II trehalose 6-phosphate synthase gene PvTPS9 modulates trehalose metabolism in Phaseolus vulgaris nodules. Front Plant Sci. 2016;7:1589. doi: 10.3389/fpls.2016.01589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen LQ, Hou BH, Lalonde S, Takanaga H, Hartung ML, Qu XQ, et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature. 2010;468(7323):527–532. doi: 10.1038/nature09606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen LQ. SWEET sugar transporters for phloem transport and pathogen nutrition. New Phytol. 2014;201(4):1150–1155. doi: 10.1111/nph.12445. [DOI] [PubMed] [Google Scholar]
- 42.Chen L-Q, Qu X-Q, Hou B-H, Sosso D, Osorio S, Fernie AR, et al. Sucrose Efflux Mediated by SWEET Proteins as a Key Step for Phloem Transport. Science. 2012;335(6065):207–211. doi: 10.1126/science.1213351. [DOI] [PubMed] [Google Scholar]
- 43.Chen L-Q, Cheung LS, Feng L, Tanner W, Frommer WB. Transport of sugars. Annu Rev Biochem. 2015;84(1):865–894. doi: 10.1146/annurev-biochem-060614-033904. [DOI] [PubMed] [Google Scholar]
- 44.Lin IW, Sosso D, Chen LQ, Gase K, Kim SG, Kessler D, et al. Nectar secretion requires sucrose phosphate synthases and the sugar transporter SWEET9. Nature. 2014;508(7497):546–549. doi: 10.1038/nature13082. [DOI] [PubMed] [Google Scholar]
- 45.Wan Y, King R, Mitchell RAC, Hassani-Pak K, Hawkesford MJ. Spatiotemporal expression patterns of wheat amino acid transporters reveal their putative roles in nitrogen transport and responses to abiotic stress. Sci Rep. 2017;7(1):1–13. doi: 10.1038/s41598-016-0028-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kurakawa T, Ueda N, Maekawa M, Kobayashi K, Kojima M, Nagato Y, et al. Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature. 2007;445(7128):652–655. doi: 10.1038/nature05504. [DOI] [PubMed] [Google Scholar]
- 47.Mason MG, Mathews DE, Argyros DA, Maxwell BB, Kieber JJ, Alonso JM, et al. Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell. 2005;17(11):3007–3018. doi: 10.1105/tpc.105.035451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Choe S, Dilkes BP, Gregory BD, Ross AS, Yuan H, Noguchi T, et al. The Arabidopsis dwarf1 Mutant Is Defective in the Conversion of 24-Methylenecholesterol to Campesterol in Brassinosteroid Biosynthesis Published by: American Society of Plant Biologists (ASPB). 2016; 10.1104/pp.119.3.897. [DOI] [PMC free article] [PubMed]
- 49.Hong Z, Ueguchi-Tanaka M, Fujioka S, Takatsuto S, Yoshida S, Hasegawa Y, et al. The Rice brassinosteroid-deficient dwarf2 mutant, defective in the rice homolog of Arabidopsis DIMINUTO/DWARF1, is rescued by the endogenously accumulated alternative bioactive brassinosteroid, dolichosterone. Plant Cell. 2005;17(8):2243–2254. doi: 10.1105/tpc.105.030973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Choe S, Tanaka A, Noguchi T, Fujioka S, Takatsuto S, Ross AS, et al. Lesions in the sterol D 7 reductase gene of Arabidopsis cause dwar ® sm due to a block in brassinosteroid biosynthesis. 2000. p. 21. [DOI] [PubMed] [Google Scholar]
- 51.Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–938. doi: 10.1016/S0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- 52.Bai M-Y, Zhang L-Y, Gampala SS, Zhu S-W, Song W-Y, Chong K, et al. Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc Natl Acad Sci U S A. 2007;104(34):13839–13844. doi: 10.1073/pnas.0706386104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ranocha P, Dima O, Nagy R, Felten J, Corratge-Faillie C, Novak O, et al. Arabidopsis WAT1 is a vacuolar auxin transport facilitator required for auxin homoeostasis. Nat Commun. 2013;4:2625. doi: 10.1038/ncomms3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kepinski S, Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 2005;435(7041):446–451. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- 55.Mravec J, Skůpa P, Bailly A, Hoyerová K, Křeček P, Bielach A, et al. Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature. 2009;459(7250):1136–1140. doi: 10.1038/nature08066. [DOI] [PubMed] [Google Scholar]
- 56.Barbez E, Kubeš M, Rolčík J, Béziat C, Pěnčík A, Wang B, et al. A novel putative auxin carrier family regulates intracellular auxin homeostasis in plants. Nature. 2012;485(7396):119–122. doi: 10.1038/nature11001. [DOI] [PubMed] [Google Scholar]
- 57.Lavy M, Estelle M. Mechanisms of auxin signaling. Development. 2016;143:3226. doi: 10.1242/dev.131870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mano Y, Nemoto K. The pathway of auxin biosynthesis in plants. J Exp Bot. 2012;63(8):2853–2872. doi: 10.1093/jxb/ers091. [DOI] [PubMed] [Google Scholar]
- 59.Xu Z, Yoo Y, Hwang I. Abscisic Acid: Metabolism, Transport and Signaling. 2014. pp. 77–87. [Google Scholar]
- 60.Park S, Fung P, Nishimura N, Jensen DR, Zhao Y, Lumba S, et al. Abscisic acid inhibits PP2Cs via the PYR/PYL family of ABA- binding START proteins. Science. 2010;324(5930):1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li C, Liu Z, Zhang Q, Wang R, Xiao L, Ma H, et al. SKP1 is involved in abscisic acid signalling to regulate seed germination, stomatal opening and root growth in Arabidopsis thaliana. Plant Cell Environ. 2012;35:952. doi: 10.1111/j.1365-3040.2011.02464.x. [DOI] [PubMed] [Google Scholar]
- 62.Zhang C, Zhang F, Zhou J, Fan Z, Chen F, Ma H, et al. Overexpression of a phytochrome-regulated tandem zinc finger protein gene, OsTZF1, confers hypersensitivity to ABA and hyposensitivity to red light and far-red light in rice seedlings. Plant Cell Rep. 2012;31(7):1333–1343. doi: 10.1007/s00299-012-1252-x. [DOI] [PubMed] [Google Scholar]
- 63.Rzewuski G, Sauter M. Ethylene biosynthesis and signaling in rice. Plant Sci. 2008;175(1–2):32–42. doi: 10.1016/j.plantsci.2008.01.012. [DOI] [Google Scholar]
- 64.Rashid M, Guangyuan H, Guangxiao Y, Hussain J, Xu Y. AP2/ERF transcription factor in rice: genome-wide anvas and yntenic relationships between monocots and udicots. Evol Bioinforma. 2012;2012(8):321–355. doi: 10.4137/EBO.S9369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wasternack C, Hause B. Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. Ann Bot. 2013;6:1021–1058. doi: 10.1093/aob/mct067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu H, Carvalhais LC, Kazan K, Schenk PM. Development of marker genes for jasmonic acid signaling in shoots and roots of wheat. Plant Signal Behav. 2016;11(5):1–8. doi: 10.1080/15592324.2016.1176654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Seo HS, Song JT, Cheong J-J, Lee Y-H, Lee Y-W, Hwang I, et al. Jasmonic acid carboxyl methyltransferase: a key enzyme for jasmonate-regulated plant responses. Proc Natl Acad Sci. 2001;98(8):4788–4793. doi: 10.1073/pnas.081557298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pearce S, Huttly AK, Prosser IM, Li Y, Vaughan SP, Gallova B, et al. Heterologous expression and transcript analysis of gibberellin biosynthetic genes of grasses reveals novel functionality in the GA3ox family. BMC Plant Biol. 2015;15:130. doi: 10.1186/s12870-015-0520-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ueguchi-Tanaka M, Ashikari M, Nakajima M, Itoh H, Katoh E, Kobayashi M, et al. GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature. 2005;437(7059):693–698. doi: 10.1038/nature04028. [DOI] [PubMed] [Google Scholar]
- 70.Gomez-Roldan V, Fermas S, Brewer PB, Puech-Pagès V, Dun EA, Pillot JP, et al. Strigolactone inhibition of shoot branching. Nature. 2008;455:189. doi: 10.1038/nature07271. [DOI] [PubMed] [Google Scholar]
- 71.Bennett T, Leyser O. Strigolactone signalling: standing on the shoulders of DWARFs. Curr Opin Plant Biol. 2014;22:7. doi: 10.1016/j.pbi.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 72.Teichmann T, Muhr M. Shaping plant architecture. Front Plant Sci. 2015;6:233. doi: 10.3389/fpls.2015.00233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Arite T, Umehara M, Ishikawa S, Hanada A, Maekawa M, Yamaguchi S, et al. D14, a strigolactone-Insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol. 2009;50(8):1416–1424. doi: 10.1093/pcp/pcp091. [DOI] [PubMed] [Google Scholar]
- 74.Zhou F, Lin Q, Zhu L, Ren Y, Zhou K, Shabek N, et al. D14-SCF D3 -dependent degradation of D53 regulates strigolactone signalling. Nature. 2013;504:406. doi: 10.1038/nature12878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stirnberg P, van De Sande K, Leyser HMO. MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development. 2002;129(5):1131–1141. doi: 10.1242/dev.129.5.1131. [DOI] [PubMed] [Google Scholar]
- 76.Ishikawa S, Maekawa M, Arite T, Onishi K, Takamure I, Kyozuka J. Suppression of tiller bud activity in tillering dwarf mutants of rice. Plant Cell Physiol. 2005;46(1):79–86. doi: 10.1093/pcp/pci022. [DOI] [PubMed] [Google Scholar]
- 77.Soundappan I, Bennett T, Morffy N, Liang Y, Stanga JP, Abbas A, et al. SMAX1-LIKE/D53 family members enable distinct MAX2-dependent responses to Strigolactones and Karrikins in Arabidopsis. Plant Cell. 2015;27(11):3143–3159. doi: 10.1105/tpc.15.00562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang L, Wang B, Jiang L, Liu X, Li X, Lu Z, et al. Strigolactone signaling in Arabidopsis regulates shoot development by targeting D53-like SMXL repressor proteins for ubiquitination and degradation. Plant Cell. 2015;27(11):3128–3142. doi: 10.1105/tpc.15.00605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Awika JM. Major cereal grains production and use around the world. Adv Cereal Sci Implic to food Process Heal Promot. 2011;1089:1–13. doi: 10.1021/bk-2011-1089.ch001. [DOI] [Google Scholar]
- 80.Maroniche G, Mongelli VC, Llauger G, Alfonso V, Taboga O, del Vas M. In vivo subcellular localization of Mal de Río Cuarto virus (MRCV) non-structural proteins in insect cells reveals their putative functions. Virology. 2012;430(2):81–89. doi: 10.1016/j.virol.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 81.Pons ABS. Inmunodetección y localización subcelular in vivo de las proteínas codificadas por el segmento bicistrónico S7 del Mal de Río Cuarto virus (MRCV) en hospedantes vegetales. Córdoba: Universidad Nacional de Córdoba; 2012. [Google Scholar]
- 82.Liu H, Wei C, Zhong Y, Li Y. Rice black-streaked dwarf virus minor core protein P8 is a nuclear dimeric protein and represses transcription in tobacco protoplasts. FEBS Lett. 2007;581(13):2534–2540. doi: 10.1016/j.febslet.2007.04.071. [DOI] [PubMed] [Google Scholar]
- 83.Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444(7117):323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 84.Dodds PN, Rathjen JP. Plant immunity: towards an integrated view of plant–pathogen interactions. Nat Rev Genet. 2010;11(8):539–548. doi: 10.1038/nrg2812. [DOI] [PubMed] [Google Scholar]
- 85.Di Feo L, Laguna IG, Biderbost EB. Alteraciones fisiológicas asociadas a la infección con Mal de Río Cuarto virus (MRCV) y a fitotoxicidad provocada por su insecto vector (Delphacodes kuscheli Fennah) en trigo. Trop Plant Pathol. 2010;35(2):79–87. doi: 10.1590/S1982-56762010000200002. [DOI] [Google Scholar]
- 86.Shen J, Chen X, Chen J, Sun L. A phloem-limited fijivirus induces the formation of neoplastic phloem tissues that house virus multiplication in the host plant. Sci Rep. 2016;6(May):29848. doi: 10.1038/srep29848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Martins MCM, Hejazi M, Fettke J, Steup M, Feil R, Krause U, et al. Feedback inhibition of starch degradation in Arabidopsis leaves mediated by Trehalose 6-phosphate. Plant Physiol. 2013;163(3):1142–63. doi: 10.1104/pp.113.226787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.dos Anjos L, Pandey PK, Moraes TA, Feil R, Lunn JE, Stitt M. Feedback regulation by trehalose 6-phosphate slows down starch mobilization below the rate that would exhaust starch reserves at dawn in Arabidopsis leaves. Plant Direct. 2018;2(8):e00078. doi: 10.1002/pld3.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bledsoe SW, Henry C, Griffiths CA, Paul MJ, Feil R, Lunn JE, et al. The role of Tre6P and SnRK1 in maize early kernel development and events leading to stress-induced kernel abortion. BMC Plant Biol. 2017;17:74. doi: 10.1186/s12870-017-1018-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lauxmann MA, Annunziata MG, Brunoud G, Wahl V, Koczut A, Burgos A, et al. Reproductive failure in Arabidopsis thaliana under transient carbohydrate limitation: flowers and very young siliques are jettisoned and the meristem is maintained to allow successful resumption of reproductive growth. Plant Cell Environ. 2016;39:745. doi: 10.1111/pce.12634. [DOI] [PubMed] [Google Scholar]
- 91.Harthill JE, Meek SEM, Morrice N, Peggie MW, Borch J, Wong BHC, et al. Phosphorylation and 14-3-3 binding of Arabidopsis trehalose-phosphate synthase 5 in response to 2-deoxyglucose. Plant J. 2006;47(2):211–223. doi: 10.1111/j.1365-313X.2006.02780.x. [DOI] [PubMed] [Google Scholar]
- 92.Vandesteene L, Ramon M, Le Roy K, Van Dijck P, Rolland F. A single active trehalose-6-P synthase (TPS) and a family of putative regulatory TPS-like proteins in arabidopsis. Mol Plant. 2010;3(2):406–419. doi: 10.1093/mp/ssp114. [DOI] [PubMed] [Google Scholar]
- 93.Krasileva KV, Vasquez-Gross HA, Howell T, Bailey P, Paraiso F, Clissold L, et al. Uncovering hidden variation in polyploid wheat. Proc Natl Acad Sci. 2017:201619268 Available from: http://www.pnas.org/lookup/doi/10.1073/pnas.1619268114. [DOI] [PMC free article] [PubMed]
- 94.Chandran D. Co-option of developmentally regulated plant SWEET transporters for pathogen nutrition and abiotic stress tolerance. IUBMB Life. 2015;67(7):461–471. doi: 10.1002/iub.1394. [DOI] [PubMed] [Google Scholar]
- 95.Yuan M, Wang S. Rice MtN3/saliva/SWEET family genes and their homologs in cellular organisms. Mol Plant. 2013;6(3):665–674. doi: 10.1093/mp/sst035. [DOI] [PubMed] [Google Scholar]
- 96.Mizuno H, Kasuga S, Kawahigashi H. The sorghum SWEET gene family: stem sucrose accumulation as revealed through transcriptome profiling. Biotechnol Biofuels. 2016;9(1):127. doi: 10.1186/s13068-016-0546-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gebauer P, Korn M, Engelsdorf T, Sonnewald U, Koch C, Voll LM. Sugar accumulation in leaves of Arabidopsis sweet11/sweet12 double mutants enhances priming of the salicylic acid-mediated defense response. Front Plant Sci. 2017;8(August):1–13. doi: 10.3389/fpls.2017.01378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bezrutczyk M, Hartwig T, Horschman M, Char SN, Yang J, Yang B, et al. Impaired phloem loading in zmsweet13a, b, c sucrose transporter triple knock-out mutants in Zea mays. New Phytol. 2018;218(2):594–603. doi: 10.1111/nph.15021. [DOI] [PubMed] [Google Scholar]
- 99.Kanno Y, Oikawa T, Chiba Y, Ishimaru Y, Shimizu T, Sano N, et al. AtSWEET13 and AtSWEET14 regulate gibberellin-mediated physiological processes. Nat Commun. 2016;7:1–11. doi: 10.1038/ncomms13245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Noël G, Tognetti JA, Pontis HG. Protein kinase and phosphatase activities are involved in fructan synthesis initiation mediated by sugars. Planta. 2001;213(4):640–646. doi: 10.1007/s004250100550. [DOI] [PubMed] [Google Scholar]
- 101.Blasing OE. Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell Online. 2005;17(12):3257–3281. doi: 10.1105/tpc.105.035261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bolouri Moghaddam MR, Van Den Ende W. Sweet immunity in the plant circadian regulatory network. J Exp Bot. 2013;64(6):1439–1449. doi: 10.1093/jxb/ert046. [DOI] [PubMed] [Google Scholar]
- 103.Wang H, Xu D, Pu L, Zhou G. Southern rice black-streaked dwarf virus alters insect vectors’ host orientation preferences to enhance spread and increase Rice ragged stunt virus co-infection. Phytopathology. 2014;104(2):196–201. doi: 10.1094/PHYTO-08-13-0227-R. [DOI] [PubMed] [Google Scholar]
- 104.Lu G, Zhang T, He Y, Zhou G. Virus altered rice attractiveness to planthoppers is mediated by volatiles and related to virus titre and expression of defence and volatile-biosynthesis genes. Sci Rep. 2016;6(November):1–10. doi: 10.1038/srep38581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ljung K, Nemhauser JL, Perata P. New mechanistic links between sugar and hormone signalling networks. Curr Opin Plant Biol. 2015;25:130–7. [DOI] [PubMed]
- 106.Ueguchi-Tanaka M, Nakajima M, Motoyuki A, Matsuoka M. Gibberellin receptor and its role in gibberellin signaling in plants. Annu Rev Plant Biol. 2007;58:183–198. doi: 10.1146/annurev.arplant.58.032806.103830. [DOI] [PubMed] [Google Scholar]
- 107.Tao T, Zhou CJ, Wang Q, Chen XR, Sun Q, Zhao TY, et al. Rice black streaked dwarf virus P7-2 forms a SCF complex through binding to Oryza sativa SKP1-like proteins, and interacts with GID2 involved in the gibberellin pathway. PLoS One. 2017;12(5):1–16. doi: 10.1371/journal.pone.0177518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yamaguchi S. Gibberellin metabolism and its regulation. Annu Rev Plant Biol. 2008;59(1):225–251. doi: 10.1146/annurev.arplant.59.032607.092804. [DOI] [PubMed] [Google Scholar]
- 109.Spielmeyer W, Ellis MH, Chandler PM. Semidwarf (sd-1), “green revolution” rice, contains a defective gibberellin 20-oxidase gene. Proc Natl Acad Sci. 2002;99(13):9043–9048. doi: 10.1073/pnas.132266399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jia QJ, Zhang JJ, Westcott S, Zhang XQ, Bellgard M, Lance R, et al. GA-20 oxidase as a candidate for the semidwarf gene sdw1/denso in barley. Funct Integr Genomics. 2009;9(2):255–262. doi: 10.1007/s10142-009-0120-4. [DOI] [PubMed] [Google Scholar]
- 111.Riefler M. Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell Online. 2006;18(1):40–54. doi: 10.1105/tpc.105.037796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bartrina I, Jensen H, Novák O, Strnad M, Werner T, Schmülling T. Gain-of-function mutants of the Cytokinin receptors AHK2 and AHK3 regulate plant organ size, flowering time and plant longevity. Plant Physiol. 2017;173(3):1783–1797. doi: 10.1104/pp.16.01903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Brenner WG, Ramireddy E, Heyl A, Schmülling T. Gene regulation by cytokinin in Arabidopsis. Front Plant Sci. 2012;3(January):1–22. doi: 10.3389/fpls.2012.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Singh A, Breja P, Khurana JP, Khurana P. Wheat Brassinosteroid-Insensitive1 (TaBRI1) interacts with members of TaSERK gene family and cause early flowering and seed yield enhancement in arabidopsis. PLoS One. 2016;11(6):1–22. doi: 10.1371/journal.pone.0153273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mauch-Mani B, Mauch F. The role of abscisic acid in plant-pathogen interactions. Curr Opin Plant Biol. 2005;8(4):409–414. doi: 10.1016/j.pbi.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 116.Xie B, Wang X, Zhu M, Zhang Z, Hong Z. CalS7 encodes a callose synthase responsible for callose deposition in the phloem. Plant J. 2011;65(1):1–14. doi: 10.1111/j.1365-313X.2010.04399.x. [DOI] [PubMed] [Google Scholar]
- 117.Achon MA, Serrano L, Sabate J, Porta C. Understanding the epidemiological factors that intensify the incidence of maize rough dwarf disease in Spain. Ann Appl Biol. 2015;166(2):311–320. doi: 10.1111/aab.12184. [DOI] [Google Scholar]
- 118.Wang Y, Cheng X, Shan Q, Zhang Y, Liu J, Gao C, et al. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotechnol. 2014;32(9):947–951. doi: 10.1038/nbt.2969. [DOI] [PubMed] [Google Scholar]
- 119.Liang Z, Chen K, Li T, Zhang Y, Wang Y, Zhao Q, et al. Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat Commun. 2017;8:1–5. doi: 10.1038/s41467-016-0009-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Argüello Caro EB, Maroniche GA, Dumón AD, Sagadín MB, del Vas M, Truol G. High viral load in the Planthopper vector Delphacodes kuscheli (Hemiptera: Delphacidae) is associated with successful transmission of Mal de Río Cuarto virus. Ann Entomol Soc Am. 2013;106(1):93–99. doi: 10.1603/AN12076. [DOI] [Google Scholar]
- 121.Pearce S, Kippes N, Chen A, Debernardi JM, Dubcovsky J. RNA-seq studies using wheat PHYTOCHROME B and PHYTOCHROME C mutants reveal shared and specific functions in the regulation of flowering and shade-avoidance pathways. BMC Plant Biol. 2016;16(1):1–19. doi: 10.1186/s12870-016-0831-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26(7):873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Anders S, Pyl PT, Huber W. HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pearce S, Vazquez-Gross H, Herin SY, Hane D, Wang Y, Gu YQ, et al. WheatExp: an RNA-seq expression database for polyploid wheat. BMC Plant Biol. 2015;15(1):1–8. doi: 10.1186/s12870-014-0410-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):1–21. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kinsella RJ, Kähäri A, Haider S, Zamora J, Proctor G, Spudich G, et al. Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database. 2011;2011(February):1–9. doi: 10.1093/database/bar030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Durinck S, Spellman PT, Birney E, Huber W. Mapping identifiers for the integration of genomic datasets with the R/ Bioconductor package biomaRt. Nat Protoc. 2009;4(8):1184–1191. doi: 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Luo MC, Gu YQ, Puiu D, Wang H, Twardziok SO, Deal KR, et al. Genome sequence of the progenitor of the wheat D genome Aegilops tauschii. Nature. 2017;551(7681):498–502. doi: 10.1038/nature24486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, et al. Primer3-new capabilities and interfaces. Nucleic Acids Res. 2012;40(15):1–12. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ramakers C, Ruijter JM, Lekanne Deprez RH, Moorman AFM. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett. 2003;339(1):62–66. doi: 10.1016/S0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- 131.Zhang K, Niu S, Di D, Shi L, Liu D, Cao X, et al. Selection of reference genes for gene expression studies in virus-infected monocots using quantitative real-time PCR. J Biotechnol. 2013;168(1):7–14. doi: 10.1016/j.jbiotec.2013.08.008. [DOI] [PubMed] [Google Scholar]
- 132.Di Rienzo JA. fgStatistics [Internet] 2009. [Google Scholar]
- 133.Stitt M, Lilley RMC, Gerhardt R, Heldt HW. Metabolite levels in specific cells and subcellular compartments of plant leaves. Methods Enzymol. 1989;174(C):518–552. doi: 10.1016/0076-6879(89)74035-0. [DOI] [Google Scholar]
- 134.Cross JM, von Korff M, Altmann T, Bartzetko L, Sulpice R, Gibon Y, et al. Variation of enzyme activities and metabolite levels in 24 Arabidopsis accessions growing in carbon-limited conditions. Plant Physiol. 2006;142(4):1574–1588. doi: 10.1104/pp.106.086629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lunn JE, Feil R, Hendriks JHM, Gibon Y, Morcuende R, Osuna D, et al. Sugar-induced increases in trehalose 6-phosphate are correlated with redox activation of ADPglucose pyrophosphorylase and higher rates of starch synthesis in Arabidopsis thaliana. Biochem J. 2006;397(1):139–148. doi: 10.1042/BJ20060083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gouy M, Guindon S, Gascuel O. Sea view version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27(2):221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 137.Antoniadi I, Plačková L, Simonovik B, Doležal K, Turnbull C, Ljung K, et al. Cell-type-specific Cytokinin distribution within the Arabidopsis primary root apex. Plant Cell. 2015;27(7):1955–1967. doi: 10.1105/tpc.15.00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Svačinová J, Novák O, Plačková L, Lenobel R, Holík J, Strnad M, et al. A new approach for cytokinin isolation from Arabidopsis tissues using miniaturized purification: pipette tip solid-phase extraction. Plant Methods. 2012;8(1):1–14. doi: 10.1186/1746-4811-8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hoyerová K, Gaudinová A, Malbeck J, Dobrev PI, Kocábek T, Šolcová B, et al. Efficiency of different methods of extraction and purification of cytokinins. Phytochemistry. 2006;67(11):1151–1159. doi: 10.1016/j.phytochem.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 140.Floková K, Tarkowská D, Miersch O, Strnad M, Wasternack C, Novák O. UHPLC-MS/MS based target profiling of stress-induced phytohormones. Phytochemistry. 2014;105:147–157. doi: 10.1016/j.phytochem.2014.05.015. [DOI] [PubMed] [Google Scholar]
- 141.Tarkowská D, Novák O, Oklestkova J, Strnad M. The determination of 22 natural brassinosteroids in a minute sample of plant tissue by UHPLC–ESI–MS/MS. Anal Bioanal Chem. 2016;408(24):6799–6812. doi: 10.1007/s00216-016-9807-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Representative pictures of MRCV symptoms in wheat at 21 dpi. Insets highlighting curled leaves and cross-cut edges are shown. Figure S2. Library mapping statistics. For each library, total reads, clean reads, mapped reads, and uniquely mapped reads that fall within a gene feature are indicated. T stands for treated (MRCV-infected) and C for control (mock-inoculated). R1 to R4 stand for each of the four plants sequenced from each treatment. Figure S3. qPCR validation of DATs at 21 dpi. For qPCR experiments, n = 6. Error bars: standard error. Statistical significance between RNA-seq and qPCR Fold Change values was assessed by Two-tailed Student’s T tests (*P < 0.05). Figure S4. Phylogenetic relationships of wheat vacuolar invertases and fructosyltransferases with other plant sequences. Protein sequences from Triticum aestivum (Ta), the progenitors of the A and D subgenomes Triticum urartu (Tu) and Aegilops tauschii (AEGt), Triticum turgidum (Tt), Aegilops searsii (As), Oryza sativa (Os), Arabidopsis thaliana (At), Solanum lycopersicum (Sl) and Populus trichocarpa (Pt) were used to build a phylogenetic tree using the neighbor-joining algorithm with a bootstrap of 1000. Figure S5. Phylogenetic relationships of wheat TPS with other plant TPS. Protein sequences of class II TPS from Triticum aestivum (Ta), Oryza sativa (Os), Arabidopsis thaliana (At), Phaseolus vulgaris (Pv) and Populus trichocarpa (Pt) were used to build a phylogenetic tree using the neighbor-joining algorithm with a bootstrap of 1000. Figure S6. Phylogenetic relationships of wheat SWEET with other plant SWEET. Sequences from Triticum aestivum (Ta), Oryza sativa (Os) and Arabidopsis thaliana (At) were used to build a phylogenetic tree using the neighbor-joining algorithm with a bootstrap of 1000. Wheat protein sequences whose transcripts were significantly increased or decreased in the RNA-seq analysis at 21 dpi are coloured in red and blue, respectively. (DOCX 2662 kb)
Table S1. Differentially accumulated transcripts (DATs) upon MRCV infection at 12 and 21 dpi. For each gene, the normalized mean count for mock-inoculated and MRCV-infected samples, the fold change and the FDR-adjusted P-value (padj) are indicated. The annotations from Ensembl plants Release 37 or inferred in this work are included. Table S2. DATs classified as wheat Transcription factors according to reference [27]. Table S3. DATs classified as Carbohydrate-related according to their functional annotation. Table S4. Putative wheat invertases and fructosyltransferases, Tre6P synthases (TPS) and Sugars Will Eventually be Exported Transporters (SWEET) used to build the phylogenetic trees shown on Additional file 1: Figures S4-S6. Sequences encoded by DATs at 21 dpi are highlighted in red (increased upon the infection) and blue (decreased). A/N Inv, Vac Inv and CW Inv stand for acid and/or neutral, vacuolar and cell wall invertases respectively. Fructosyltransferases included sucrose:sucrose 1-fructosyltransferase (1-SST), fructan:fructan 1-fructosyltransferases (1-FFT) and sucrose:fructan 6-fructosyltransferases (6-SFT). Table S5. DATs classified as Amino acid-related according to their functional annotation. Amino acid transporters were extracted from reference [45]. Table S6. DATs classified as Hormone-related according to their functional annotation. Table S7. Primer sequences used for qRT-PCR validations. (XLSX 717 kb)
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files. Raw sequences of RNA-seq libraries were deposited in NCBI Sequence Read Archive (SRA) (https://www.ncbi.nlm.nih.gov/sra) under the accession number SRP160433.