Summary
Schizophrenia and bipolar disorder are two distinct diagnoses that share symptomology. Understanding the genetic factors contributing to the shared and disorder-specific symptoms will be crucial for improving diagnosis and treatment. In genetic data consisting of 53,555 cases (20,129 BD, 33,426 SCZ) and 54,065 controls, we identified 114 genome-wide significant loci implicating synaptic and neuronal pathways shared between disorders. Comparing SCZ to BD (23,585 SCZ, 15,270 BD) identified four genomic regions including one with disorder-independent causal variants and potassium ion response genes as contributing to differences in biology between the disorders. Polygenic risk score (PRS) analyses identified several significant correlations within case-only phenotypes including SCZ PRS with psychotic features and age of onset in BD. For the first time, we discover specific loci that distinguish between BD and SCZ and identify polygenic components underlying multiple symptom dimensions. These results point to the utility of genetics to inform symptomology and potentially treatment.
Introduction
Bipolar disorder (BD) and schizophrenia (SCZ) are severe psychiatric disorders and among the leading causes of disability worldwide(Whiteford et al., 2013). Both disorders have significant genetic components with heritability estimates ranging from 60–80%(Nöthen et al., 2010). Recent genetic and epidemiological studies have demonstrated substantial overlap between these two disorders with a genetic correlation from common variation near 0.6–0.7(Cross-Disorder Group of the Psychiatric Genomics Consortium, 2013) and high relative risks (RR) among relatives of both BD and SCZ patients (RRs for parent/offspring: BD/BD: 6.4, BD/SCZ: 2.4; SCZ/BD: 5.2, SCZ/SCZ: 9.9)(Lichtenstein et al., 2009). Despite shared genetics and symptomology, the current diagnostic systems(“Diagnostic and Statistical Manual of Mental Disorders | DSM Library,” n.d.),(“WHO | International Classification of Diseases,” n.d.) adhere to historical distinctions from the late 19th century and represent BD and SCZ as independent categorical entities differentiated on the basis of their clinical presentation, with BD characterized by predominant mood symptoms, mood-congruent delusions and an episodic disease course and SCZ considered a prototypical psychotic disorder. Identifying genetic components contributing to both disorders provides insight into the biology underlying the shared symptoms of the disorders.
While the shared genetic component is substantial, studies to date have also implicated genetic architecture differences between these two disorders(Curtis et al., 2011; Ruderfer et al., 2014). A polygenic risk score created from a case only SCZ vs BD genome-wide association study (GWAS) significantly correlated with SCZ or BD diagnosis in an independent sample(Ruderfer et al., 2014), providing the first evidence that differences between the disorders also have a genetic basis. An enrichment of rare, moderate to highly penetrant copy number variants (CNVs) and de novo CNVs are seen in SCZ patients(CNV and Schizophrenia Working Groups of the Psychiatric Genomics Consortium, 2017; Gulsuner and McClellan, 2015; Kirov et al., 2012; Stone et al., 2008; Szatkiewicz et al., 2014), while, the involvement of CNVs in BD is less clear(Green et al., 2016). Although the role of de novo single nucleotide variants in BD and SCZ has been investigated in only a handful of studies, enrichment in pathways associated with the postsynaptic density has been reported for SCZ, but not BD(Fromer et al., 2014; Kataoka et al., 2016). Identifying disorder-specific variants and quantifying the contribution of genetic variation to specific symptom dimensions remain important open questions. Characterizing these genetic differences will facilitate an understanding of the dimensions of the disorders instead of the dichotomous diagnosis. For example, we have shown that SCZ patients with greater manic symptoms have higher polygenic risk for BD(Ruderfer et al., 2014). These findings demonstrate shared genetic underpinnings for symptoms across disorders and may enable us to characterize patients by genetic liability to symptom dimensions thereby informing disease course and treatment.
Here, we utilize large collections of genotyped samples for BD and SCZ along with clinically-relevant measures identifying 28 subphenotypes to address three questions: 1) Are there specific variants, genes or pathways that are either shared by, or differentiate BD and SCZ? 2) Are the shared symptoms between these disorders driven by the same underlying genetic profiles? and 3) Can we demonstrate independent genetic signatures for subphenotypes within these disorders?
Results
Shared genetic contribution to BD and SCZ
We performed association analysis of BD and SCZ combined into a single phenotype, totaling 53,555 cases (20,129 BD, 33,426 SCZ) and 54,065 controls on 15.5 million SNP allele dosages imputed from 1000 genomes phase 3(The 1000 Genomes Project Consortium, 2015). Logistic regression was performed controlling for 13 principal components of ancestry, study sites and genotyping platform. We identified 11,231 SNPs with p-value below our genome-wide significance (GWS) threshold of 5×10−8. After grouping SNPs in linkage disequilibrium with each other (r2 > 0.2), 114 genomic risk loci remained. For the most significant variant in each of the 114 GWS loci, we performed conditional analysis with any GWS hit within 1Mb of the extent of the locus from the previously performed single disease GWAS of SCZ(Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014) and BD(Stahl et al., 2017) and identified 32 loci that were independently significant defined strictly as no single disease locus within 1Mb or a GWS p-value after conditional analysis (Supplementary Table 1). We further performed gene-set based tests using MAGMA(Leeuw et al., 2015) across 10,891 curated pathways(Watanabe et al., 2017) and identified 8 pathways surpassing Bonferroni correction (p < 4.6×10−6) with all but one pathway implicating synaptic and neuronal biology (Supplementary Table 2a). Establishing independent controls (see Methods) allowed us to perform disorder-specific GWAS in 20,129 BD cases vs 21,524 BD controls and 33,426 SCZ cases and 32,541 SCZ controls. Using these results, we compared effect sizes of these 114 loci across each disorder independently showing the subsets of variants that had larger effects in SCZ compared to BD and vice versa (Figure 1a).
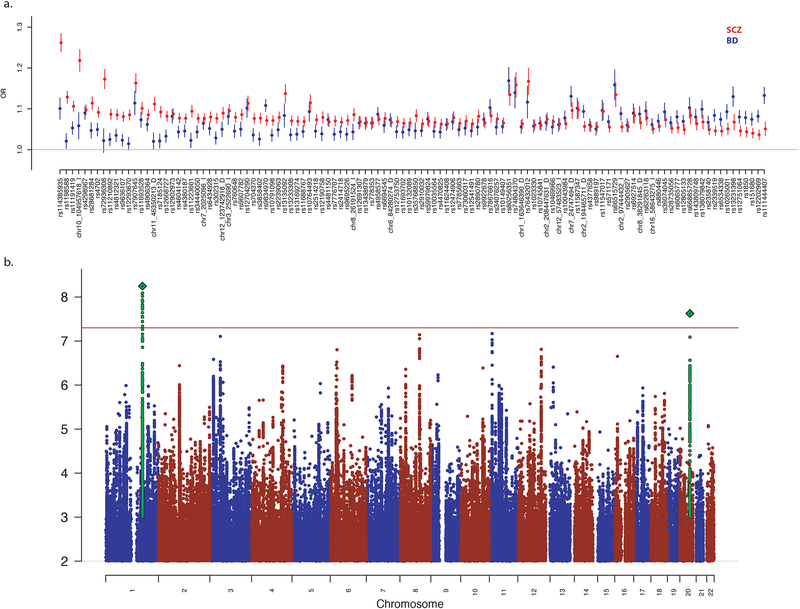
Figure 1. Associated Genomic Loci Shared and Divergent Between BD and SCZ.
a) Odds ratios (OR) from independent data sets of BD (blue) and SCZ (red) for each of the 114 genome-wide significant variants in the BD and SCZ vs controls GWAS. b) Manhattan plot for SCZ vs BD GWAS.
Differentiating genetic contribution to BD and SCZ
To identify loci with divergent effects on BD and SCZ, we performed an association analysis comparing 23,585 SCZ cases with 15,270 BD cases matched for shared ancestry and genotyping platform (see Methods, Figure 1b, Table 1). Two genome-wide significant loci were identified, the most significant of which was rs56355601 located on chromosome 1 at position 173,811,455 within an intron of DARS2 (Supplementary Figure 1). The second most significant locus was rs200005157, a four base-pair insertion/deletion, on chromosome 20 at position 47638976 in an intron of ARFGEF2 (Supplementary Figure 2). For both variants, the minor allele frequency was higher in BD cases than SCZ cases and disease-specific GWAS showed opposite directions of effect when compared to controls. We sought to identify additional disease-specific loci by comprehensively incorporating expression information with association results to perform fine-mapping and identify novel variants(Gamazon et al., 2015; Giambartolomei et al., 2014; Gusev et al., 2016; He et al., 2013). Here, we applied the summary-data-based Mendelian randomization (SMR) method(Zhu et al., 2016) (see Methods) utilizing the cis-QTLs derived from peripheral blood(Westra et al., 2013), human dorsolateral prefrontal cortex (DLPFC)(Fromer et al., 2016) from the Common Mind Consortium and 11 brain regions from the GTEx consortium(Consortium, 2015). We identified one SNP-probe combination that surpassed the threshold for genome-wide significance in blood but was also the most significant finding in brain. We found that SNP rs4793172 in gene DCAKD is associated with SCZ vs BD analysis (pGWAS = 2.8×10−6) and is an eQTL for probe ILMN 1811648 (peQTL = 2.9×10−168), resulting in pSMR = 4.1×10−6 in blood (peQTL = 2.9×10−25, pSMR = 2.0×10−5 in DLFC, and peQTL = 4.6×10−15, pSMR = 6.0×10−5 in GTEx cerebellar hemisphere) (Supplementary Table 3, Supplementary Figure 3) and shows no evidence of heterogeneity (pHET =0.66) which implies only a single causal variant in the locus.
Table 1.
Most Significant Associated Loci from SCZ vs BD GWAS
| SCZ vs BD (23,584 vs 15,270) | SCZ vs controls (33,426 vs 32,541) | BD vs controls (20,129 vs 21,524) | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | CHR | BP | A1 | SCZ freq | BD freq | P | OR | BD freq | control freq | P | OR | SCZ freq | control freq | P | OR | Het p-value | ||||
| rs56355601 | 1 | 173811455 | T | 0.7410 | 0.7230 | 5.7E-09 | 1.12 | 0.7340 | 0.7250 | 0.012 | 1.04 | 0.7220 | 0.7290 | 0.0449 | 0.96 | 1.74E-03 | ||||
| rs200005157 | 20 | 47638976 | D | 0.5300 | 0.5110 | 2.4E-08 | 1.10 | 0.5290 | 0.5190 | 0.004 | 1.04 | 0.5130 | 0.5220 | 0.0012 | 0.95 | 1.51E-05 | ||||
| rs10832027 | 11 | 13357183 | A | 0.6630 | 0.6840 | 6.8E-08 | 0.91 | 0.6610 | 0.6580 | 0.050 | 0.98 | 0.6840 | 0.6710 | 0.0004 | 1.06 | 6.86E-05 | ||||
| rs7823741 | 8 | 92904254 | C | 0.3720 | 0.3930 | 7.2E-08 | 0.91 | 0.3800 | 0.3880 | 0.115 | 0.98 | 0.3910 | 0.3840 | 0.0476 | 1.03 | 1.12E-02 | ||||
| rs7645466 | 3 | 50049958 | T | 0.4930 | 0.4760 | 7.8E-08 | 1.10 | 0.4990 | 0.5010 | 0.547 | 1.01 | 0.4800 | 0.4890 | 0.0018 | 0.95 | 5.01E-03 | ||||
Association results for the five most significant variants in the SCZ vs BD GWAS with the top two being genome-wide significant. Each variant includes results from the independent BD vs controls and SCZ vs controls GWAS and the comparable p-value from a heterogeneity test when performing a two cohort meta-analysis of SCZ and BD.
In an effort to prioritize genes for the two GWS loci from the GWAS, we performed fine-mapping(Benner et al., 2016) using an LD map derived from a majority of the control samples. We then performed SMR on each of the variants with causal probability greater than 1% using all eQTLs from the CommonMind Consortium DLPFC reference. All the most likely causal variants were shown to most significantly regulate the same gene suggesting CSE1L is the most likely relevant gene on chromosome 20 (rs200005157: causal probability=0.21, pGWAS=2.4×10−8, peQTL 3×10−8, pSMR=8.5×10−5, pHET=0.34). For the locus on chromosome 1, SLC9C2 is the most significantly regulated gene. However, a highly significant heterogeneity test indicates a complex genetic architecture making it difficult to infer a causal role for the associated SNP. Therefore, DARS2 presents as the most likely relevant gene on chromosome 1 (rs56355601: pGWAS=5.6×10−9, causal probability=0.079, peQTL 7.4×10−13, pSMR=6.17×10−6, pHET=0.03). We note however, that in both cases there are less associated variants that are stronger eQTLs for these genes complicating a straightforward causal interpretation. Finally, using the same gene-set test used for the combined analysis GO biological process “response to potassium ion” (p=1.6×10−6) was the only pathway surpassing our Bonferroni corrected significance threshold (Supplementary Table 2b).
Regional joint association
We expanded our efforts to identify disorder-specific genomic regions by jointly analyzing independent GWAS results from BD and SCZ(Pickrell et al., 2016). The genome was split into 1,703 previously defined approximately LD independent regions(Berisa and Pickrell, 2015). Thirteen percent, or 223 regions, had a posterior probability greater than 0.5 of having a causal variant for at least one disorder. Of these, 132 best fit the model of a shared causal variant influencing both BD and SCZ, 88 were most likely specific to SCZ, 3 demonstrated evidence of two independent variants (with one impacting each of the two disorders) and none were BD-specific. Of note, this approach calculates a prior probability that any given region is disease-specific and from these data the probability of having a BD specific region was 0.1% compared to 15% for SCZ, likely a result of increased power from the larger SCZ sample size and/or a difference in genetic architecture between these disorders.
The 114 GWS SNPs from the combined BD and SCZ GWAS localized into 99 independent regions (13 regions had multiple GWS SNPs), of which 78 (79%) were shared with a posterior probability of greater than 0.5. Sixty regions had at least one GWS SNP in the independent SCZ GWAS, of which 30 (50%) are shared and 8 regions contained a GWS SNP in the independent BD GWAS, of which 6 (75%) are shared using the same definition. For the three regions showing evidence for independent variants, two had highly non-overlapping association signals in the same region stemming from independent variants. The third, on chromosome 19 presented a different scenario where association signals were overlapping. The most significant variant in BD was rs111444407 (chr19:19358207, p = 8.67×10−10) and for SCZ was rs2315283 (chr19:19480575, p=4.41×10−7). After conditioning on the most significant variant in the other disorder, the association signals of the most significant variant in BD and SCZ were largely unchanged (BD rs111444407 =1.3×10−9, SCZ rs2315283 p=6.7×10−5). We further calculated the probability of each variant in the region being causal for both BD and SCZ(Benner et al., 2016) and found no correlation (r= −0.00016). The most significant variants had the highest posterior probability of being causal (SCZ: rs2315283, prob = 0.02, BD: rs111444407, prob = 0.16). Both variants most significantly regulate the expression of GATAD2A in brain(Fromer et al., 2016) but in opposite directions (rs111444407 peQTL = 6×10−15, beta = 0.105; rs2315283 peQTL = 1.5×10−28, beta = −0.11).
Regional SNP-heritability estimation
Across the genome, regional SNP-heritabilities (h2snp) were estimated separately for SCZ and BD(Shi et al., 2016) and were found to be moderately correlated (r=0.25). We next defined risk regions as those containing the most associated SNP for each GWS locus. In total, there were 101 SCZ risk regions from the 105 autosomal GWS loci reported previously(Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014) and 29 BD risk regions from 30 GWS loci reported previously(Stahl et al., 2017). Ten regions were risk regions for both BD and SCZ comprising 33% of BD risk regions and 10% of SCZ risk regions. We further stratified regional h2snp by whether a region was a risk region in one disorder, none or both (Supplementary Figure 4). Since the discovery data for the regions overlapped with the data used for the heritability estimation, we expected within-disorder analyses to show significant results. In risk regions specific to SCZ (n=91) there was a significant increase in regional h2snp in SCZ, as expected (p = 1.1×10−22), but also in BD (p = 1.2×10−6). In risk regions specific to BD (n=19), significantly increased regional h2snp was observed in BD, as expected (p = 0.0007), but not in SCZ (p = 0.89). Risk regions shared by both disorders had significantly higher h2snp in both disorders, as expected (BD p = 5.3×10−5, SCZ p = 0.006), compared to non-risk regions. However, we observed a significant increase in BD h2snp in shared risk regions compared to BD risk regions (BD p = 0.003) but not SCZ h2snp for shared risk regions compared to SCZ risk regions (p = 0.62). Using a less stringent p-value threshold for defining risk regions (p < 5×10−6), thereby substantially increasing the number of regions, resulted in similar results. Seven regions contributed to substantially higher h2snp in SCZ compared to BD but no region showed the inverse pattern. Of these regions, all but one was in the major histocompatibility region (MHC), the sole novel region was chr10:104380410–106695047 with regional h2snp= 0.0019 in SCZ and h2snp=0.00063 in BD.
Polygenic dissection of subphenotypes
Subphenotypes were collected for a subset of patients with either BD or SCZ (see Methods). For SCZ, we had clinical quantitative measurements of manic, depressive, positive and negative symptoms generated from factor analysis of multiple instruments as described previously(Ruderfer et al., 2014) but in larger sample sizes (n=6908, 6907, 8259, 8355 respectively). For BD, 24 subphenotypes were collected among nearly 13,000 cases in distinct categories including comorbidities, clinical information such as rapid cycling and psychotic features as well as additional disease course data such as age of onset and number of hospitalizations. For each BD or SCZ patient, we calculated a polygenic risk score (PRS) using all SNPs, from each of the four main GWAS analyses (BD+SCZ, BD, SCZ and SCZvsBD). We then used regression analysis including principal components and site to assess the relationship between each subphenotype and the 4 PRS. Specifically, we tested whether polygenic risk scores of BD+SCZ, BD, SCZ or SCZvsBD were correlated with each of these subphenotypes separately within BD and SCZ cases. When testing if the variance explained by the PRS was different from zero, we applied a significance cutoff of p < 0.0004 based on Bonferroni correction for 112 tests. In total, we identified 6 significant results after correction (Figure 2, Table 2).
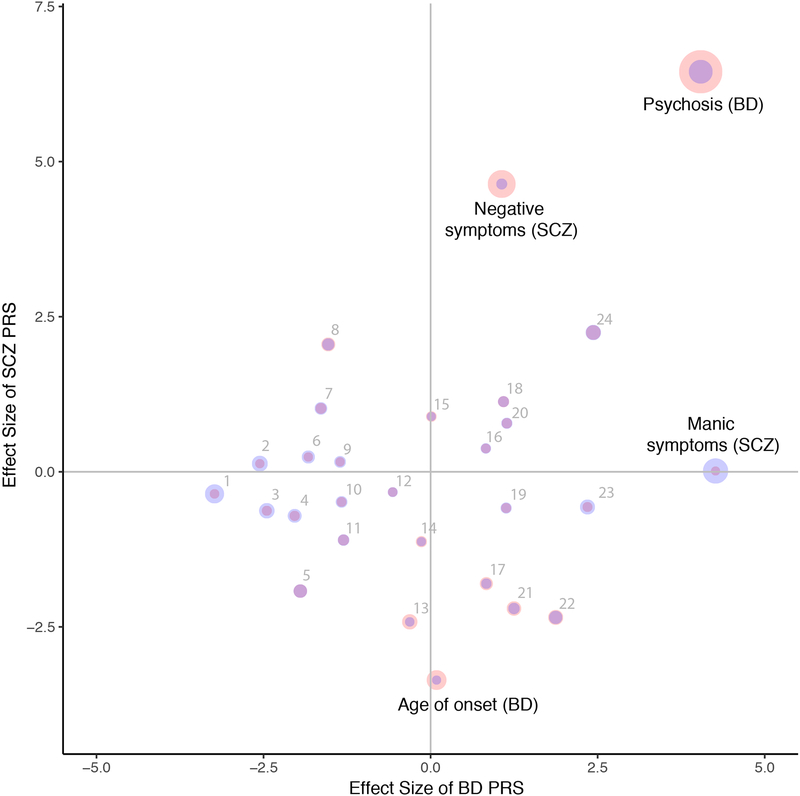
Figure 2. Polygenic Risk Score Dissection of Clinical Symptom Dimensions.
Effect size (calculated by dividing regression estimate by standard error) from regression analysis including ancestry covariates for each subphenotype and PRS for BD (x-axis) and SCZ (y-axis). Point size represents −log10(p-value) with SCZ (red) and BD (blue). Numbered subphenotypes are 1) comorbid migraine, 2) panic attacks 3) suicide attempt 4) mixed states 5) rapid cycling 6) comorbid eating disorder 7) comorbid OCD 8) year of birth 9) suicide ideation 10) panic disorder 11) number of suicide attempts 12) depressive symptoms (SCZ) 13) episodes depressive 14) episodes total 15) positive symptoms (SCZ) 16) irritable mania 17) age of onset depression 18) family history 19) episodes mixed mania 20) unipolar mania 21) alcohol substance dependence 22) age of onset mania 23) age at interview 24) number of hospitalizations. All subphenotypes are in BD except those labeled (SCZ).
Table 2.
Complete Results of Polygenic Risk Score Dissection Analysis
| P-value | Effect | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Disorder | Subphenotype | N | Cases | Controls | BPSCZ | BP | SCZ | BAS | BPSCZ | BP | SCZ | BAS | ||
| BD | psychosis | 8131 | 4632 | 3499 | 7.9E-13 | 5.3E-05 | 1.2E-10 | 5.8E-01 | 7.17 | 4.04 | 6.45 | 0.55 | ||
| suicide ideation | 5399 | 3801 | 1598 | 7.8E-01 | 1.8E-01 | 8.7E-01 | 1.7E-01 | −0.28 | −1.35 | 0.16 | 1.37 | |||
| family history | 4971 | 2730 | 2241 | 6.1E-02 | 2.8E-01 | 2.6E-01 | 6.9E-01 | 1.87 | 1.09 | 1.13 | −0.39 | |||
| irritable mania | 4230 | 2401 | 1829 | 3.8E-01 | 4.1E-01 | 7.1E-01 | 1.0E-01 | 0.88 | 0.83 | 0.38 | −1.63 | |||
| rapid cycling | 5214 | 1744 | 3470 | 7.9E-03 | 5.1E-02 | 5.5E-02 | 3.1E-01 | −2.66 | −1.95 | −1.92 | 1.01 | |||
| alcohol substance dependence | 5440 | 1494 | 3946 | 4.5E-01 | 2.1E-01 | 2.8E-02 | 1.7E-01 | −0.75 | 1.25 | −2.20 | −1.36 | |||
| panic disorder | 4647 | 863 | 3784 | 2.8E-01 | 1.8E-01 | 6.3E-01 | 4.0E-01 | −1.07 | −1.33 | −0.49 | 0.83 | |||
| panic attacks | 3976 | 851 | 3125 | 1.3E-01 | 1.1E-02 | 9.0E-01 | 4.7E-02 | −1.50 | −2.56 | 0.13 | 1.98 | |||
| mixed states | 4044 | 826 | 3218 | 1.0E-01 | 4.2E-02 | 4.8E-01 | 6.0E-02 | −1.64 | −2.03 | −0.71 | 1.88 | |||
| unipolar mania | 4863 | 461 | 4402 | 2.4E-02 | 2.5E-01 | 4.3E-01 | 6.1E-01 | 2.26 | 1.14 | 0.78 | 0.51 | |||
| comorbid migraine | 2652 | 410 | 2242 | 1.3E-02 | 1.2E-03 | 7.2E-01 | 4.4E-01 | −2.48 | −3.23 | −0.36 | 0.77 | |||
| comorbid OCD | 4215 | 386 | 3829 | 9.7E-01 | 1.0E-01 | 3.1E-01 | 1.9E-01 | −0.04 | −1.64 | 1.02 | 1.30 | |||
| comorbid eating disorder | 3839 | 331 | 3508 | 2.1E-01 | 6.7E-02 | 8.1E-01 | 6.3E-01 | −1.25 | −1.83 | 0.24 | 0.48 | |||
| suicide attempt | 6308 | 2412 | 3896 | 1.2E-01 | 1.4E-02 | 5.3E-01 | 2.8E-01 | −1.54 | −2.45 | −0.63 | 1.09 | |||
| age of onset | 8610 | 6.2E-03 | 9.3E-01 | 7.9E-04 | 6.2E-01 | −2.74 | 0.09 | −3.36 | −0.50 | |||||
| age at interview | 8062 | 5.9E-01 | 1.9E-02 | 5.7E-01 | 4.4E-01 | 0.54 | 2.35 | −0.57 | −0.78 | |||||
| episodes mixed mania | 6587 | 6.3E-01 | 2.6E-01 | 5.6E-01 | 3.2E-01 | −0.48 | 1.13 | −0.58 | −1.00 | |||||
| episodes depressive | 6252 | 7.4E-03 | 7.6E-01 | 1.6E-02 | 9.6E-01 | −2.68 | −0.31 | −2.42 | −0.05 | |||||
| episodes total | 5958 | 1.3E-01 | 8.9E-01 | 2.6E-01 | 3.9E-01 | −1.51 | −0.14 | −1.13 | −0.87 | |||||
| year of birth | 5317 | 1.7E-01 | 1.3E-01 | 4.0E-02 | 3.6E-02 | 1.39 | −1.53 | 2.05 | 2.10 | |||||
| number of suicide attempts | 5015 | 6.2E-02 | 1.9E-01 | 2.7E-01 | 4.9E-01 | −1.87 | −1.30 | −1.10 | −0.69 | |||||
| number of hospitalizations | 3944 | 4.2E-04 | 1.5E-02 | 2.5E-02 | 7.4E-01 | 3.53 | 2.43 | 2.25 | −0.33 | |||||
| age of onset depression | 3467 | 2.3E-01 | 4.0E-01 | 7.2E-02 | 2.2E-01 | −1.19 | 0.83 | −1.80 | 1.24 | |||||
| age of onset mania | 3395 | 2.5E-01 | 6.1E-02 | 1.9E-02 | 2.2E-01 | −1.14 | 1.87 | −2.35 | −1.23 | |||||
| SCZ | Manic | 6908 | 2.4E-02 | 2.0E-05 | 9.9E-01 | 3.5E-02 | 2.26 | 4.26 | 0.01 | −2.10 | ||||
| Depressive | 6907 | 9.0E-01 | 5.7E-01 | 7.4E-01 | 1.8E-01 | 0.13 | −0.57 | −0.33 | −1.36 | |||||
| Negative | 8355 | 1.5E-05 | 2.9E-01 | 3.6E-06 | 2.1E-02 | 4.33 | 1.06 | 4.64 | 2.31 | |||||
| Positive | 8259 | 4.1E-01 | 9.9E-01 | 3.7E-01 | 5.1E-01 | 0.82 | 0.01 | 0.89 | 0.65 | |||||
Polygenic scoring results of all four GWAS phenotypes (BD+SCZ vs controls, BD vs controls, SCZ vs controls and SCZ vs BD) and 24 subphenotypes from BD and 4 subphenotypes from SCZ, rows without case/control counts are quantitative measures. Significance and effects are from regression analysis of subphenotype on PRS including principal components of ancestry and site as covariates. Effect is the regression estimate divided by the standard error.
A significant positive correlation existed between BD PRS and manic symptoms in SCZ cases as seen previously(Ruderfer et al., 2014) (p=2×10−5, t=4.26) and BD PRS and psychotic features in BD patients (p=5.3×10−5, t=4.04). A significant increase in SCZ PRS was seen for BD cases with versus without psychotic features (p=1.2×10−10, t=6.45) and patients with increased negative symptoms in SCZ patients (p=3.60×10−6, t=4.64). The BD+SCZ vs controls PRS was significantly associated with psychotic features in BD (p=7.9×10−13, t=7.17) and negative symptoms in SCZ (p=1.5×10−5, t=4.33). The next two most significant results which did not survive our conservative correction were both indicative of a more severe course in BD: increased BD+SCZ PRS with increased numbers of hospitalizations in BD cases (p=4.2×10−4, t=3.53) and increased SCZ PRS with earlier onset of BD (p=7.9×10−4, t=−3.36). We assessed the role of BD subtype on the correlation between SCZ PRS and psychotic features and identified a significant correlation when restricted to only BD type I cases indicating the result was not likely driven by BD patients with a schizoaffective subtype (BDI: 3,763 with psychosis, 2,629 without, p=1.55×10−5, Supplementary Table 4).
We performed a GWAS for all 8 quantitative subphenotypes and 9 binary subphenotypes with at least 1,000 cases and calculated heritability and genetic correlation with BD and SCZ. Only two subphenotypes had significant h2snp estimates using LD-score regression(Bulik-Sullivan et al., 2015) both in BD: psychotic features in BD (h2snp=0.15, SE=0.06) and suicide attempt (h2snp=0.25, SE=0.1). Only psychotic features demonstrated a significant genetic correlation with SCZ (rg=0.34, SE=0.13, p=0.009). The significant genetic correlation demonstrates a genome-wide relationship between common variants contributing to SCZ risk and those contributing to psychotic features in BD cases. We tested whether the most significantly associated SCZ loci contributed directly to psychotic features in BD. One hundred of the 105 autosomal genome-wide significant SCZ SNPs previously published(Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014) were in our dataset after QC and 60 were in the same direction of effect for risk of psychotic features in BD (p=0.028, one-sided binomial-test).
Discussion
Here we present a genetic dissection of bipolar disorder and schizophrenia from over 100,000 genotyped subjects. Consistent with earlier results(Cross-Disorder Group of the Psychiatric Genomics Consortium, 2013), we found extensive genetic sharing between these two disorders, identifying 114 genome-wide significant loci contributing to both disorders of which 32 are novel. These findings point to the relevance of neuronal and synaptic biology for the shared genetic substrate of these disorders. However, despite this degree of sharing, we identified several loci that significantly differentiated between the two disorders, having opposite directions of effect. We also found polygenic components that significantly correlated from one disorder to symptoms of the other.
Two GWS loci were identified from the case only SCZ versus BD analysis providing opportunities to inform the underlying biological distinctions between BD and SCZ. The most significant locus implicates DARS2 (coding for the mitochondrial Aspartate-tRNA ligase) which is highly expressed in the brain and significantly regulated by the most significant SNP rs56355601 (peQTL=2.5×10−11). Homozygous mutations in DARS2 are responsible for leukoencephalopathy with brainstem and spinal cord involvement and lactate elevation (LBSL), which was characterized by neurological symptoms such as psychomotor developmental delay, cerebellar ataxia and delayed mental development(Yamashita et al., 2013, p. 2). Based on methylation analysis from the prefrontal cortex of stress models (rats and monkeys) and from peripheral samples (in monkeys and human newborns), DARS2, among others, has been suggested as a potential molecular marker of early-life stress and vulnerability to psychiatric disorders(Luoni et al., 2016). The second most significant locus implicates CSE1L, a nuclear transport factor that plays a role in cellular proliferation as well as in apoptosis(Bera et al., 2001). Intronic SNPs in CSE1L have been associated with subjective well-being(Okbay et al., 2016) and, nominally to antidepressant response(Li et al., 2016). More interestingly, CSE1L is a potential target gene of miR-137, one of the well-known schizophrenia risk loci(Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014), which is able to negatively regulate CSE1L by interacting with complementary sequences in the 3’ UTR of CSE1L(Li et al., 2013). Although falling short of genome-wide significance, the third most significant locus implicates ARNTL (Aryl Hydrocarbon Receptor Nuclear Translocator Like), which is a core component of the circadian clock. ARNTL has been previously hypothesized for relevance in bipolar disorder,(Yang et al., 2008) although human genetic evidence is currently limited(Byrne et al., 2014).
The ability to generate transcriptional data on multiple tissues across many individuals using RNA-sequencing has provided detailed information on the role common variants play in regulating expression of specific genes in specific tissues. These eQTLs can be integrated with the genetic association data from GWAS to inform on the relationship between variant association and variant regulation of expression for each gene. Performing this integration, we identified a third genome-wide significant finding in DCAKD. The gene codes for Dephospho-CoA Kinase Domain Containing protein, a member of the human postsynaptic density proteome from human neocortex(Bayés et al., 2011). In the mouse cortical synaptoproteome DCAKD is among the proteins with the highest changes between juvenile postnatal days and adult stage, suggesting a putative role in brain development(Gonzalez-Lozano et al., 2016; Moczulska et al., 2014). Discerning between pleiotropy (variant independently regulates expression and alters risk to disease) from causality (variant regulates expression which thereby alters risk to disease) through statistical analysis alone is difficult, this analytical approach is stringent in excluding loci where colocalised SNP-phenotype and SNP-expression associations may reflect confounding driven by linkage disequilibrium (LD) (one variant regulates expression and a different variant alters risk but the variants in the region are in LD). Hence, this approach utilizes currently available data to prioritize genes, including direction of effect, for functional follow-up. These analyses will become more powered with increased sample sizes for both phenotype and eQTL data sets.
Performing pathway analysis based on the full association results shows enrichment of genes involved in response to potassium ions, including potassium voltage-gated channel subfamily members and a number of genes regulated by cellular potassium concentration. This is in line with previous genetic evidence pointing to a key etiologic role of potassium channels, in particular, in BD(Judy and Zandi, 2013), which could be explained by their role in multiple neurobiological mechanisms involved in the development of psychiatric disorders such as regulation of the dopaminergic circuits, synaptic plasticity, and myelination(Balaraman et al., 2015).
We further assessed the contribution of regions of the genome to each disorder through joint regional association and heritability estimation. These results point to an additional locus that may contribute differentially to liability to BD and SCZ. The region on chr19 shows overlapping association peaks that are driven by independent causal variants for each disorder. Both variants significantly regulate the same gene GATAD2A but in opposite directions. GATAD2A is a transcriptional repressor, which is targeted by MBD2 and is involved in methylation-dependent gene silencing. The protein is part of the large NuRD (nucleosome remodeling and deacetylase) complex, for which also HDAC1/2 are essential components. NurD complex proteins have been associated with autism(Li et al., 2015). Their members, including GATAD2A, display preferential expression in fetal brain development(Li et al., 2015) and in recent work has been implicated in SCZ through open chromatin(Fullard et al., n.d.). Further, p66α (mouse GATAD2A) was recently shown to participate in memory preservation through long-lasting histone modification in hippocampal memory-activated neurons(Ding et al., 2017). SNP-heritability appears to be consistently shared across regions and chromosomes between these two disorders. Regions with GWS loci often explain higher proportions of heritability as expected. When looking at the effect on heritability of the presence of a GWS locus in the other disorder, we identified a significant increase in BD heritability for regions containing a GWS locus for SCZ but no significant increase in SCZ heritability in regions having a BD one. This result suggests a directionality to the genetic sharing of these disorders with a larger proportion of BD loci being specific to BD. However, we cannot exclude that the asymmetry of results may reflect less power of discovery for BD than SCZ. The degree to which power and subphenotypes contribute to this result requires further examination.
We note that as with nearly all GWAS findings, the calculated population-based effect sizes of the variants identified here are small and independently explain only a modest fraction to the heritability of these disorders. The identification of these variants is dependent on the ability to have highly accurate allele frequency estimates that can only be ascertained from large sample sizes. As sample sizes get larger the power to identify variants of smaller effect increases meaning that increasing sample size results in the identification of variants of smaller effect. However, a small population effect size does not exclude the possibility of a substantially larger effect on molecular phenotypes nor does it preclude the utility of association regions in understanding biology or having a clinical impact. Efforts following up GWAS results to date have demonstrated the value of these findings in pointing to genes that can aid in understanding the underlying biology of the trait(Claussnitzer et al., 2015; Mohanan et al., 2018; Sekar et al., 2016). Further, there is a clear relationship between GWAS results of a phenotype and gene targets of drugs that treat that phenotype pointing to the potential for improved therapeutic understanding(Nelson et al., 2015; Ruderfer et al., 2016). A major challenge of GWAS is the sheer number of findings and the substantial time/cost required for functional follow up of these findings in the classical paradigms used for genes causal for monogenic disorders. In silico bioinformatic analyses (such as SMR used here) that integrate GWAS results with ‘omics data (transcription, protein, epigenetic, etc.) have the potential to put a clearer biological focus on GWAS results. Such analyses can become more complex as more reference omics data sets (with genome-wide genotyping) become available. Additional analytical efforts will be required to facilitate the transition from GWAS to biology but substantial data has shown there is much to be learned from these variants despite their small effects(Visscher et al., 2017).
We have now identified multiple genomic signatures that correlate between one disorder and a clinical symptom in the other disorder, illustrating genetic components underlying particular symptom dimensions within these disorders. Medical symptoms, including those seen in psychiatric disorders, can manifest through a multitude of causes. The classic example often used is headache for which many different paths lead to the same symptom. Psychiatric symptoms also have many potential causes. For example, symptoms of psychosis can be the result of highly heritable diseases such as BD and SCZ but also infectious and neurodegenerative diseases, sleep/sensory deprivation or psychedelic drugs. Demonstrating a shared biological underpinning to these symptoms suggests they could be treated through modulating the same pathway. As previously shown, we find a significant positive correlation between the PRS of BD and manic symptoms in SCZ. We also demonstrate that BD cases with psychotic features carry a significantly higher SCZ PRS than BD cases without psychotic features and this result is not driven by the schizoaffective BD subtype. Further, we show that increased PRS is associated with more severe illness. This is true for BD with psychotic features having increased SCZ PRS, earlier onset BD having higher SCZ PRS and cases with higher BD+SCZ PRS having a larger number of hospitalizations. We demonstrated that psychotic features within BD is a heritable trait and GWS loci for SCZ have a consistent direction of effect in psychotic features in BD, demonstrating the potential to study psychosis more directly to identify variants contributing to that symptom dimension.
This work illustrates the utility of genetic data, in aggregate, at dissecting symptom heterogeneity among related disorders and suggests that further work could aid in characterizing patients for more personalized treatment. Genetic risk scores have demonstrated their ability to inform and predict pathology(Cleynen et al., 2016) and more recently have been shown to be able to identify patients with risk equivalent to monogenic variants(Khera et al., 2017). In psychiatry, we lack objective biological measurements (biomarkers) with which to assess the ability of a genetic signature to predict or inform. Lacking diagnostic pathology for psychiatric disorders leaves a genuine opportunity for the genetics to drive diagnosis and treatment to a much larger degree than in other domains. One potential model assumes that each individual has a quantitative loading of a series of symptom dimensions (i.e. manic, psychotic, cognitive, etc.) and that these symptoms can be assessed at the genetic level to characterize a patient’s dysfunction and used to inform disease course and optimal treatment. Making this a reality will require more detailed information on disease course and outcomes. For example, if treatment response data existed for these samples one could ask whether a genetic loading for psychosis was correlated with response to treatment. Initial work has already shown the potential of this approach using a SCZ PRS to inform lithium response in BD(Amare et al., 2018). Ultimately, the goal will be to quantify multiple genetic loadings of each individual’s illness and use those measures to inform treatment based on the outcomes of previous individuals with similar profiles.
In conclusion, we present a detailed genetic dissection of BD and SCZ pointing to substantial shared genetic risk but also demonstrating that specific loci contribute to the phenotypic differences of these disorders. We show that genetic risk scores can correspond to symptoms within and across disorders. Finally, we present data that points to these disorders being neither independent nor the same but sharing particular symptom dimensions that can be captured from the genetics and used to characterize patients to ultimately inform diagnosis and treatment.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Genotype and phenotype data use is restricted and governed by the Psychiatric Genetics Consortium. Further information and requests for analytical results or additional information should be directed to and will be fulfilled by the Lead Contact, Douglas Ruderfer (douglas.ruderfer@vanderbilt.edu).
SUBJECT DETAILS
Genotyped Sample Description
SCZ samples are a substantial subset of those analyzed previously(Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014). BD samples are the newest collection from Psychiatric Genomics Consortium Bipolar Disorder Working Group(Stahl et al., 2017).
Below we provide information on the individual samples used here as provided by the original PGC disorder publications. Additionally, most studies have been described in detail in the citations provided. The boldfaced first line for each sample is study PI, PubMed ID, country (study name), and the PGC internal tag or study identifier.
European ancestry, case-control design
Schizophrenia
Adolfsson, R | NP | Umeå, Sweden | scz_umeb_eur
Adolfsson, R | NP | Umeå, Sweden | scz_umes_eur
Cases of European ancestry were ascertained from multiple different studies of schizophrenia (1992–2009). The diagnostic processes were similar between studies, and the final diagnosis is a best-estimate consensus lifetime diagnosis based on multiple sources of information such as clinical evaluation by research psychiatrists, different types of semi-structured interviews made by trained research nurses and research psychiatrists, medical records, course of the disease and data from multiple informants. Diagnosis was made in accordance with the Diagnostic and Statistical Manual of Mental Disorders-Version IV (DSM-IV) or International Classification of Diseases, 10th Revision (ICD-10) criteria. Controls were recruited from the Betula study, an ongoing longitudinal, prospective, population-based study from the same geographic area (North Sweden) that is studying aging, health, and cognition in adults. All subjects (cases and controls) participated after giving written informed consent and the regional Ethical Review Board at the University of Umeå approved all original studies and participation in the PGC. GWAS genotyping was performed at Broad Institute.
Andreassen, O | 19571808 | Norway (TOP) | scz_top8_eur
In the TOP study (Tematisk omrade psykoser), cases of European ancestry, born in Norway, were recruited from psychiatric hospitals in the Oslo region. Patients were diagnosed according to SCID and further ascertainment details have been reported. Healthy control subjects were randomly selected from statistical records of persons from the same catchment area as the patient groups. All participants provided written informed consent and the human subjects protocol was approved by the Norwegian Scientific-Ethical Committee and the Norwegian Data Protection Agency.
Blackwood, D | 19571811 | Edinburgh, UK | scz_edin_eur
Cases and controls were recruited from the southeast of Scotland, and ascertainment has been previously described as part of the International Schizophrenia Consortium studies. All participating subjects gave written, informed consent and the human subjects protocol was approved by the Scotland A Research Ethics Committee. DNA samples were genotyped at the Broad Institute.
Børglum, A | 19571808 | Denmark | scz_aarh_eur
DNA samples for all subjects were collected from blood spots systematically collected by the Danish Newborn Screening Biobank), with case/control status established using the Danish Psychiatric Central Register. Cases were diagnosed clinically according to ICD-10 criteria. Controls were selected to match the cases by birth cohort. The Danish Data Protection Agency and the ethics committees in Denmark approved the human subjects protocol.
Bramon | 23871474 | Seven countries (PEIC, WTCCC2) | scz_pewb_eur
Bramon | 23871474 | Spain (PEIC, WTCCC2) | scz_pewb_eur
The Psychosis Endophenotypes International Consortium (PEIC) was part of WTCCC2. Samples were collected through seven centers in Europe and Australia (the Institute of Psychiatry, King’s College London, London; GROUP (consisting of the University of Amsterdam, Amsterdam; the University of Groningen, Groningen; Maastricht University Medical Centre, Maastricht; and the University of Utrecht, Utrecht); the University of Western Australia, Perth; the Universidad de Cantabria, Santander; the University of Edinburgh, Edinburgh; Heidelberg University, Heidelberg and Ludwig-Maximilians-Universität München, Munich). To allow for a DSM-IV diagnosis to be ascertained or ruled out, all participants (including controls and unaffected family members) underwent a structured clinical interview with the Schedule for Affective Disorders and Schizophrenia (SADS), the Structured Clinical Interview for DSM Disorders (SCID), or the Schedules for Clinical Assessment in Neuropsychiatry (SCAN). We included cases with schizophrenia and schizoaffective disorder. Participants in all groups were excluded if they had a history of neurological disease or head injury resulting in loss of consciousness.
Buxbaum, J | 20489179 | New York, US & Israel | scz_msaf_eur
Samples contributed by Mount Sinai were derived from three cohorts. In all cohorts, ethical approval was obtained from all participating sites, and all subjects provided informed consent. Two of the cohorts were in a prior paper on copy number variation. One of the cohorts was from the Mount Sinai brain bank, where DNA was extracted from postmortem samples, and another comprised of patients ascertained in Israel. The third cohort included subjects more recently recruited through the Mount Sinai Conte Center.
Corvin, A | 19571811 | Ireland | scz_dubl_eur
The case sample was collected primarily in the Dublin area and the ascertainment procedure has been previously described. The controls were recruited, from the same region through the Irish Blood Transfusion Services. All participants gave written, informed consent and the collections were approved through the Federated Dublin Hospitals and Irish Blood Transfusion Services Research Ethics Committees, respectively. DNA samples were genotyped at the Broad Institute.
Corvin, A; Riley, B | 22883433 | Ireland (WTCCC2) | scz_irwt_eur
The case sample was recruited from the Republic of Ireland and Northern Ireland. All cases had four Irish grandparents and ascertainment details have been reported elsewhere. Ethics approval was obtained from all participating hospitals and centers. Controls were blood donors from the Irish Blood Transfusion Service, whose Ethics Committee approved the human subjects protocol. All participants gave written informed consent. Samples were genotyped at Affymetrix (Santa Clara, California, US) laboratory as part of the WTCCC2 genotyping pipeline.
Ehrenreich, H | 20819981 | Germany (GRAS) | scz_gras
The Gottingen Research Association for Schizophrenia (GRAS) collection included cases recruited across 23 German hospitals. Controls were unscreened blood donors recruited at the Georg-August-University according to national blood donation guidelines. Cases completed a structured clinical interview and were diagnosed with DSM-IV schizophrenia or schizoaffective disorder. The study was approved by the Georg-August-University ethics committee and local internal review boards of the participating centers. All participants gave written informed consent.
Esko, T | 15133739 | Estonia (EGCUT) | scz_egcu_eur
The Estonian cohort comes from the population-based biobank of the Estonian Genome Project of University of Tartu (EGCUT). The project was conducted according to the Estonian Gene Research Act and all participants provided informed consent (www.biobank.ee). In total, 52,000 individuals aged 18 years or older participated in this cohort (33% men, 67% women). The population distributions of the cohort reflect those of the Estonian population (83% Estonians, 14% Russians and 3% other). General practitioners (GP) and physicians in the hospitals randomly recruited the participants. A Computer-Assisted Personal interview was conducted over 1–2 ours at doctors’ offices. Data on demographics, genealogy, educational and occupational history, lifestyle and anthropometric and physiological data were assessed. Schizophrenia was diagnosed prior to the recruitment by a psychiatrist according to ICD-10 criteria and identified from the Estonian Biobank phenotype database. Controls were drawn from a larger pool of genotyped biobank samples by matching on gender, age and genetic ancestry. All the controls were population-based and have not been sampled for any specific disease.
Esko, T; Li, Q; Dominici E | 15133739, 24166486 | J&J and Roche cases, EGCUT controls | scz_jr3a_eur
Esko, T; Li, Q; Domenici E | 15133739, 24166486 | J&J and Roche cases, EGCUT controls | scz_jr3b_eur
Esko, T; Li, Q; Domenici E | 15133739, 24166486 | J&J and Roche cases, EGCUT controls | scz_jri6_eur
Esko, T; Li, Q; Dominici E | 15133739, 24166486 | J&J and Roche cases cases, EGCUT controls | scz_jrsa_eur
Cases were collected by Johnson and Johnson (J&J) and Roche as part of clinical collaborations with hospitals and outpatient centers. Cases were diagnosed according to DSMIV criteria, with medical record review by a trained psychiatrist. There were reliability trials across centers for the J&J studies. The J& J cases were mostly collected in Eastern Europe, with most coming from Estonian and Russia (>100); intermediate numbers from Austria, the Czech Republic, Latvia, Lithuania, and Spain (50–100); and smaller collections from Bulgaria, Hungary, and Poland (<50). The Roche cases were assessed with a structured psychiatric assessment by trained interviewers. Most of the Eastern European controls were from the Estonian Biobank project (EGCUT) and were ancestrally matched with cases from the J&J sample.
Gejman, P | 19571809 | US, Australia (MGS) | scz_mgs2_eur
European ancestry case samples were collected by the Molecular Genetics of Schizophrenia (MGS) collaboration across multiple sites in the USA and Australia as described in detail elsewhere. Cases gave written informed consent, and IRBs at each collecting site approved the human subjects protocol. A survey company (Knowledge Networks, under MGS guidance) collected the European ancestry control sample and ascertainment is described in detail elsewhere. DNA samples were genotyped at the Broad Institute.
Gurling, H | 19571811 | London, UK | scz_uclo_eur
All cases and controls were collected by University College London and had both parents from England, Scotland or Wales. All participants gave written informed consent and the U.K. National Health Service multicenter and local research ethics committee approved the human subjects protocol. Further details on ascertainment are available elsewhere. The samples were genotyped at the Broad Institute.
Jönsson, E | 19571808 | Sweden (Hubin) | scz_ersw_eur
Cases were recruited from northwestern Stockholm County and ascertainment has been described previously. Cases gave informed consent and the human subjects protocol was approved by the ethical committees of the Karolinska Hospital and the Stockholm Regional Ethical Committee. Controls were recruited either among subjects previously participating in biological research at the Karolinska Institute or drawn from a representative register of the population of Stockholm County. All participants provided informed consent.
Kirov, G | Not published | Bulgaria | scz_buls_eur
All cases were recruited from Bulgaria and had a history of hospitalization for treatment of schizophrenia. Controls were recruited from the two largest cities in Bulgaria as previously described. All participants gave written informed consent and the study was approved by local ethics committees at the participating centers.
Knight, J; Collier DA; Nisenbaum L| Not published | Canada (Toronto)-US(Lilly)-US (MIGen)| scz_lktu_eur
Toronto cases were recruited by referral and advertisement. Diagnoses were made according to DSM-III or DSM-IV criteria following interview and medical record review. US cases were recruited from schizophrenia clinical trials in a range of settings as part of a trial with Eli Lilly. Diagnoses were made according to DSM-III or DSM-IV criteria following interview by psychiatrist and medical record review. No controls were sampled as part of the study, and ancestrally-matched controls were chosen from the Myocardial Infarction Genetics Consortium (MIGen, dbGaP ID phs000294.v1.p1) that was genotyped with the same SNP array.
Lencz, T; Darvasi A | 23325106 | Israel | scz_ajsz_eur
Cases and controls were sampled from an Ashkenazi Jewish repository (Hebrew University Genetic Resource, http://hugr.huji.ac.il). Patients were recruited from hospitalized inpatients at 7 medical centers in Israel and were diagnosed with DSM-IV schizophrenia or schizoaffective disorder. Controls were sampled through the Israeli Blood Bank and did not report any chronic disease or regularly prescribed medication at the time of assessment. Full ascertainment details have previously been reported. Local ethics committees and the National Genetic Committee of the Israeli Ministry of Health approved the studies and all participants gave informed, written consent.
Levinson, D | 22885689 | Six countries, WTCCC controls | scz_lacw_eur
Cases collected as part of a larger pedigree-based study were partitioned into two subsamples. Cases with two genotyped parents were analyzed as trios (see PI Levinson, ms.scz_lemu_eur in the Trio section below). Unrelated cases who could not be used as part of a trio were included as a separate case-control analysis, using independent controls, matched by ancestry and genotyping array, from the Wellcome Trust Case Control Consortium. Cases were identified from different clinical settings (e.g. inpatients, outpatients and community facilities) in six countries (Australia, France, Germany, Ireland, UK, and the US). Diagnoses were established using semi-structured interviews, psychiatric records and informant reports. Case subjects were diagnosed with schizophrenia or schizoaffective disorder according to DSM-III-R criteria. All protocols were approved by loci IRBs, and all cases provided written informed consent.
Malhotra, A | 17522711 | New York, US | scz_zhh1_eur
The case and control subjects were recruited in the New York metropolitan area and ascertainment methods have been described previously. All participants gave written, informed consent and the IRB of the North Shore-Long Island Jewish Health System approved the human subjects protocols. DNA was genotyped at Zucker Hillside.
Mowry, B | 21034186 | Australia | scz_asrb_eur
These subjects were part of the Australian Schizophrenia Research Bank. The case sample was recruited in four Australian States (New South Wales, Queensland, Western Australia and Victoria) through hospital inpatient units, community mental health services, outpatient clinics and rehabilitation services, non-government mental illness support organizations, and, in the initial stages, through a large-scale, national, multi-media advertising campaign. This sample is comprised of 509 cases from larger metropolitan centers of Brisbane, Newcastle, Sydney, Melbourne, and Perth. Cases gave written informed consent, and the human subjects protocol was initially approved by the Hunter New England Area Health Research Committee and subsequently approved by relevant Institutional Ethics Committees in Brisbane, Sydney, Melbourne and Perth. Healthy controls were recruited through multi-media advertisements, and other sources. Controls were from the metropolitan centers of Brisbane, Newcastle, Sydney, Melbourne, and Perth. Controls gave written informed consent, and the human subjects protocol was approved by the Hunter New England Area Health Research Committee and Institutional Ethics Committees in Brisbane, Sydney, Melbourne and Perth. The samples were genotyped in two stages at the Hunter Medical Research Institute, University of Newcastle, Newcastle, Australia.
O’Donovan, M: Owen, M | 19571811 | Cardiff, UK | scz_caws_eur
The case sample included European ancestry schizophrenia cases recruited in the British Isles and described previously. All cases gave written informed consent to. The study was approved by the Multicentre Research Ethics Committee in Wales and Local Research Ethics Committees from all participating sites. The control sample used the Wellcome Trust CaseControl Consortium (WTCCC) sample described elsewhere, but included similar numbers of individuals from the 1958 British Birth Cohort and a panel of consenting blood donors (UK Blood Service). Samples were genotyped at Affymetrix service lab (San Francisco, USA).
O’Donovan, M: Owen, M: Walters, J | 22614287 | UK (CLOZUK) | scz_clm2_eur
O’Donovan, M: Owen, M: Walters, J | 22614287 | UK (CLOZUK) | scz_clo3_eur
CLOZUK cases were taking the antipsychotic clozapine and had received a clinical diagnosis of treatment-resistant schizophrenia. Patients taking clozapine provide blood samples to allow detection of adverse drug-effects. Through collaboration with Novartis (the manufacturer of a proprietary form of clozapine, Clozaril), we acquired blood from people with treatment-resistant schizophrenia according to the clozapine registration forms completed by treating psychiatrists as previously reported. The samples were genotyped at the Broad Institute. The UK Multicentre Research Ethics Committee (MREC) approved the study. The controls were drawn from the WTCCC2 control samples (~3,000 from the 1958 British Birth Cohort and ~3,000 samples from the UK Blood Service Control Group). An additional 900 controls, held by Cardiff University, were recruited from the UK National Blood Transfusion Service. They were not specifically screened for psychiatric illness. All control samples were from participants who provided informed consent.
Ophoff, R | 19571808 | Netherlands | scz_ucla_eur
The case sample consisted of inpatients and outpatients recruited through psychiatric hospitals and institutions throughout the Netherlands. Cases with DSM-IV schizophrenia were included in the analysis. Further details on ascertainment are provided elsewhere. Controls came from the University Medical Centre Utrecht and were volunteers with no psychiatric history. Ethical approval was provided by local ethics committees and all participants gave written informed consent.
Palotie, A | 19571808 | Finland | scz_fi3m_eur
Palotie, A | Not published | Finnish | scz_fii6_eur
Finnish cases were drawn from a nationwide collection of families with schizophrenia spectrum disorders. The control sample was derived from the Finnish Health 2000 survey. All participants provided written informed consent and approval was obtained from the ethics committees at each location.
Pato, C | 19571811 | Portugal | scz_port_eur
Cases and controls lived in Portugal, the Azorean and Madeiran islands, or were the direct (firstor second-generation) Portugese immigrant population in the US, as previously described. Controls were not biologically related to cases. All participants gave written informed consent and the IRB of SUNY Upstate Medical University approved the protocol. The samples were genotyped at the Broad Institute.
Petryshen, T | 24424392| Boston, US (CIDAR) | scz_cims_eur
Cases were recruited from inpatient and outpatient settings in the Boston area by clinician referral, through review of medical records, or through advertisements in local media. Cases were diagnosed with DSM-IV schizophrenia through a structured clinical interview (SCID) by trained interviewers with review of medical records and a best estimate diagnostic procedure including reliability trials across interviewers. A psychiatrist or a PhD-level mental health professional made the final diagnostic determination. Controls were ascertained through local advertisements from the same geographical area. Ethical approval was provided by local ethics committees and all participants gave written informed consent.
Rietschel/Rujescu/Nöthen | 19571808 | Bonn/Mannheim, Germany | scz_boco_eur
These German samples were collected by separate groups within the MooDS Consortium in Mannheim, Bonn, Munich and Jena. For the PGC analyses, the samples were combined by chip and ancestry. In Bonn/Mannheim, cases were ascertained as previously described. Controls were drawn from three population-based epidemiological studies (PopGen), the Cooperative Health Research in the Region of Augsburg (KORA) study, and the Heinz Nixdorf Recall (HNR) study. All participants gave written informed consent and the local ethics committees approved the human subjects protocols. Additional controls were randomly selected from a Munich-based community sample and screened for the presence of anxiety and affective disorders using the Composite International Diagnostic Screener. Only individuals negative for the above mentioned disorders were included in the sample.
Rujescu, D | 19571808 | Munich, Germany | scz_munc_eur
For the Munich sample, cases were ascertained from the Munich area of Germany, as described previously. The controls were unrelated volunteers randomly selected from the general population of Munich. All were screened to exclude a history of psychosis/central neurological disease either personally or in a first-degree relative. All participants gave written informed consent and the local ethics committees approved the human subjects protocols.
St Clair, D | 19571811 | Aberdeen, UK | scz_aber_eur
Ascertainment and inclusion/exclusion criteria for cases and controls have been previously described. All participating subjects were born in the UK (95% Scotland) and gave written informed consent. Both local and multiregional academic ethical committee approved the human subjects protocol. The samples were genotyped at the Broad Institute.
Sullivan, PF | 18347602 | US (CATIE) | scz_cati_eur
Cases were collected as part of the Clinical Antipsychotics Trials of Intervention Effectiveness (CATIE) project and ascertainment was previously described. Participants were recruited from multiple sites in the USA with informed written consent and approval from the IRBs at each CATIE site and the University of North Carolina (Chapel Hill). The control subjects were collected by MGS (described above) and gave online informed consent and were fully anonymized. There was no overlap with controls included in the MGS collaboration sample.
Sullivan, PF; Sklar P; Hultman C | 23974872 | Sweden | scz_swe1_eur
Sullivan, PF; Sklar P; Hultman C | 23974872 | Sweden | scz_s234_eur
Sullivan, PF; Sklar P; Hultman C | 23974872 | Sweden | scz_swe5_eur
Sullivan, PF; Sklar P; Hultman C | 23974872 | Sweden | scz_swe6_eur
Samples from the Swedish Schizophrenia Study were collected in a multi-year project and genotypes in six batches (sw1–6). All procedures were approved by ethical committees at the Karolinska Institutet and the University of North Carolina, and all subjects provided written informed consent (or legal guardian consent and subject assent). All samples were genotyped at the Broad Institute. Cases with schizophrenia were identified via the Swedish Hospital Discharge Register which captures all public and private inpatient hospitalizations. The register is complete from 1987 and is augmented by psychiatric data from 1973–1986. The register contains International Classification of Disease discharge diagnoses made by attending physicians for each hospitalization. Case inclusion criteria included ≥2 hospitalizations with a discharge diagnosis of schizophrenia, both parents born in Scandinavia and age ≥18 years. Case exclusion criteria included hospital register diagnosis of any medical or psychiatric disorder mitigating a confident diagnosis of schizophrenia as determined by expert review. The validity of this case definition of schizophrenia was strongly supported by clinical, epidemiological, genetic epidemiological and genetic evidence. Controls were selected at random from Swedish population registers, with the goal of obtaining an appropriate control group and avoiding ‘super-normal’ controls. Control inclusion criteria included never being hospitalized for schizophrenia or bipolar disorder (given evidence of genetic overlap with schizophrenia), both parents born in Scandinavia and age of ≥18 years.
Walters, J | 21850710 | Cardiff, UK (CogUK) | scz_cou3_eur
Cases were recruited from community mental health teams in Wales and England on the basis of a clinical diagnosis of schizophrenia or schizoaffective disorder (depressed sub-type) as described previously. 35 Diagnosis was confirmed following a SCAN interview and review of case notes followed by consensus diagnosis according to DSM-IV criteria. The samples were genotyped at the Broad Institute. The UK Multicentre Research Ethics Committee (MREC) approved the study and all participants provided valid informed consent.
Weinberger, D | 11381111 | NIMH CBDB | scz_lie2_eur
Weinberger, D | 11381111 | NIMH CBDB | scz_lie5_eur
Subjects were recruited from the Clinical Brain Disorders Branch of the NIMH ‘Sibling Study’ as previously described. In brief, cases and controls gave informed consent and only participants of European ancestry were included in the current analysis. Cases completed a structured clinical interview and were diagnosed with schizophrenia-spectrum disorders. Samples were genotyped at the NIMH.
Wendland/Schubert | Pfizer | Not Published | Multiple countries | scz_pfla_eur
Pfizer contributed anonymized individual genotypes for cases from seven multi-center randomized, double-blind efficacy and safety clinical trials (A1281063, A1281134, A1281148, A245–102, NRA7500001, NRA7500002, NRA7500003, and NRA7500004) as well as a set of purchased samples (NRA9000099). Trial samples were collected for antipsychotic medications across outpatient and inpatient treatment settings. All participating cases had a diagnosis of schizophrenia and were assessed using a structural clinical interview by trained interviewers, with systematic procedures to quality-control diagnostic accuracy and reliability trials across participating sites in the United States and internationally. Purchased blood samples were obtained from PrecisionMed International by Pharmacia and Upjohn Corporation, and were collected from diagnosed subjects with schizophrenia and schizoaffective disorder. All studies were reviewed by both central and local institutional review boards, depending on the study site, before recruitment of subjects started. Protocol amendments were approved while the study was in progress and before the data were unblinded. The studies were conducted in conformity with the U.S. Food and Drug Administration Code of Federal Regulations (21CFR, Part 50) and the Declaration of Helsinki and its amendments, and were consistent with Good Clinical Practice and the applicable regulatory requirements. Participants provided written informed consent before enrollment. An optional blood sample was collected from clinical trial subjects for pharmacogenetic analysis to investigate potential associations between genetic variant drug response and general characteristics of schizophrenia and related disorders. Sample collection was not required for participation in the original clinical trials. The controls (A9011027) were recruited in a multi-site, cross-sectional, non-treatment prospective trial to collect data, including DNA, from cognitive normal and free of psychiatric diseases elderly subjects in the US. Subjects were specifically recruited to match the gender, age, and ethnicity information from the LEADe and UCSD MCI studies. The study described here is within the scope of patient consent.
Werge, T | 19571808 | Denmark | scz_denm_eur
Cases were ascertained through psychiatric departments and twin pair studies, and were of Danish parentage for at least the prior three generations. The controls were collected at the University of Aarhus, and included 500 medical students, all of Danish parentage for at least three generations. All subjects gave written informed consent and the Danish Data Protection Agency and the ethics committees of Denmark approved the human subjects protocol.
Bipolar Disorder
Adolfsson, R | Not published | Umeå, Sweden | bip_ume4_eur
Clinical characterization of the patients included the Mini-International Neuropsychiatric Interview (MINI), the Diagnostic Interview for Genetic Studies (DIGS), the Family Interview for Genetic Studies (FIGS) and the Schedules for Clinical Assessment in Neuropsychiatry (SCAN). The final diagnoses were made according to the DSM-IV-TR and determined by consensus of 2 research psychiatrists. The unrelated Swedish control individuals, consisting of a large population-based sample representative of the general population of the region, were randomly selected from the ‘Betula study’.
Alda, M; Smoller, J | Not published | Nova Scotia, Canada; I2B2 controls | bip_hal2_eur
The case samples were recruited from patients longitudinally followed at specialty mood disorders clinics in Halifax and Ottawa (Canada). Cases were interviewed in a blind fashion with the Schedule of Affective Disorders and Schizophrenia-Lifetime version (SADS-L) and consensus diagnoses were made according to DSM-IV and Research Diagnostic Criteria (RDC). Protocols and procedures were approved by the local Ethics Committees and written informed consent was obtained from all patients before participation in the study. Control subjects were drawn from the I2B2 (Informatics for Integrating Biology and the Bedside) project. The study consists of de-identified healthy individuals recruited from a healthcare system in the Boston, MA, US area. The de-identification process meant that the Massachusetts General Hospital Institutional Review Board elected to waive the requirement of seeking informed consent as detailed by US Code of Federal Regulations, Title 45, Part 46, Section 116 (46.116).
Andreassen, OA | PMID:21926972 [PGC1], PMID:20451256 | Norway (TOP) | bip_top7_eur
In the TOP study (Tematisk omrade psykoser), cases of European ancestry, born in Norway, were recruited from psychiatric hospitals in the Oslo region. Patients were diagnosed according to the SCID and further ascertainment details have been reported. Healthy control subjects were randomly selected from statistical records of persons from the same catchment area as the patient groups. The control subjects were screened by interview and with the Primary Care Evaluation of Mental Disorders (PRIME-MD). None of the control subjects had a history of moderate/severe head injury, neurological disorder, mental retardation or an age outside the age range of 18–60 years. Healthy subjects were excluded if they or any of their close relatives had a lifetime history of a severe psychiatric disorder. All participants provided written informed consent and the human subjects protocol was approved by the Norwegian Scientific-Ethical Committee and the Norwegian Data Protection Agency.
Andreassen, OA | Not published | Norway (TOP) | bip_top8_eur
The TOP8 bipolar disorder cases and controls were ascertained in the same way as the bip_top7_eur (TOP7) samples described above, and recruited from hospitals across Norway.
Biernacka, JM; Frye, MA | 27769005 | Mayo Clinic, USA | bip_may1_eur
Bipolar cases were drawn from the Mayo Clinic Bipolar Biobank. Enrolment sites included Mayo Clinic, Rochester, Minnesota; Lindner Center of HOPE/University of Cincinnati College of Medicine, Cincinnati, Ohio; and the University of Minnesota, Minneapolis, Minnesota. Enrolment at each site was approved by the local Institutional Review Board approval, and all participants consented to use of their data for future genetic studies. Participants were identified through routine clinical appointments, from in-patients admitted in mood disorder units, and recruitment advertising. Participants were required to be between 18 and 80 years old and be able to speak English, provide informed consent, and have DSM-IV-TR diagnostic confirmation of type 1 or 2 bipolar disorder or schizoaffective bipolar disorder as determined using the SCID. Controls were selected from the Mayo Clinic Biobank. Potential controls with ICD9 codes for bipolar disorder, schizophrenia or related diagnoses in their electronic medical record were excluded.
Blackwood, D | 18711365 [PGC1] | Edinburgh, UK | bip_edi1_eur
This sample comprised Caucasian individuals contacted through the inpatient and outpatient services of hospitals in South East Scotland. A BD-I diagnosis was based on an interview with the patient using the SADS-L supplemented by case note review and frequently by information from medical staff, relatives and caregivers. Final diagnoses, based on DSM-IV criteria were reached by consensus between two trained psychiatrists. Ethnically-matched controls from the same region were recruited through the South of Scotland Blood Transfusion Service. Controls were not directly screened to exclude those with a personal or family history of psychiatric illness. The study was approved by the Multi-Centre Research Ethics Committee for Scotland and patients gave written informed consent for the collection of DNA samples for use in genetic studies.
Breen, G; Vincent, JB | 24387768; 19416921; 21926972 [PGC1] |London, UK; Toronto, Canada [BACC] | bip_bac1_eur
The total case/control cohort (N=1922) includes 871 subjects from Toronto, Canada (N=431 cases (160 male; 271 female); N=440 controls (176 male; 264 female)), 1051 subjects from London, UK (N=538 cases (180 male; 358 female); N=513 controls (192 male; 321 female)). A summary of mean and median age at interview, age of onset (AOO), diagnostic subtypes (BD 1 versus BD 2), presence of psychotic symptoms, suicide attempt and family history of psychiatric disorders has been provided previously for both the Toronto and London cohorts. From the Toronto site (Centre for Addiction & Mental Health (CAMH)), BD individuals and unrelated healthy controls matched for age, gender and ethnicity were recruited. Inclusion criteria for patients: a) diagnosed with DSMIV/ICD 10 BD 1 or 2; b) 18 years old or over; c) Caucasian, of Northern and Western European origin, and three out of four grandparents also N.W. European Caucasian. Exclusion criteria include: a) Use of intravenous drugs; b) Evidence of intellectual disability; c) Related to an individual already in the study; d) Manias that only ever occurred in relation to or resulting from alcohol or substance abuse/dependence, or medical illness; e) Manias resulting from non-psychotropic substance usage. The SCAN interview (Schedule for Clinical Assessments in Neuropsychiatry) was used for subject assessment. Using the SCAN interview along with case note review, each case was assigned DSM-IV and ICD 10 diagnoses by two independent diagnosticians, according to lifetime consensus best-estimate diagnosis. Lifetime occurrence of psychiatric symptoms was also recorded using the OPCRIT checklist, modified for use with mood disorders. Similar methods and criteria were also used to collect a sample of 538 BD cases and 513 controls for the London cohort (King’s College London; KCL). Both studies were approved by respective institutional research ethics committees (the CAMH Research Ethics Board (REB) in Toronto, and the College Research Ethics Committee (CREC) at KCL), and informed written consent was obtained from all participants. GWAS results have previously been published for the entire KCL/CAMH cohort.
Corvin, A | 18711365 [PGC1] | Ireland | bip_dub1_eur
Samples were collected as part of a larger study of the genetics of psychotic disorders in the Republic of Ireland, under protocols approved by the relevant IRBs and with written informed consent that permitted repository use. Cases were recruited from Hospitals and Community psychiatric facilities in Ireland by a psychiatrist or psychiatric nurse trained to use the SCID. Diagnosis was based on the structured interview supplemented by case note review and collateral history where available. All diagnoses were reviewed by an independent reviewer. Controls were ascertained with informed consent from the Irish GeneBank and represented blood donors who met the same ethnicity criteria as cases. Controls were not specifically screened for psychiatric illness.
Rietschel, M; Nöthen, MM, Cichon, S | 21926972 [PGC1] | BOMA-Germany I | bip_bonn_eur
Cases for the BOMA-Bipolar Study were ascertained from consecutive admissions to the inpatient units of the Department of Psychiatry and Psychotherapy at the University of Bonn and at the Central Institute for Mental Health in Mannheim, University of Heidelberg, Germany. DSM-IV lifetime diagnoses of bipolar I disorder were assigned using a consensus best-estimate procedure, based on all available information, including a structured interview with the SCID and SADS-L, medical records, and the family history method. In addition, the OPCRIT checklist was used for the detailed polydiagnostic documentation of symptoms. Controls were ascertained from three population-based studies in Germany (PopGen, KORA, and Heinz-Nixdorf-Recall Study). The control subjects were not screened for mental illness. Study protocols were reviewed and approved in advance by Institutional Review Boards of the participating institutions. All subjects provided written informed consent.
Rietschel, M; Nöthen, MM; Schulze, TG; Reif, A; Forstner, AJ | 24618891 | BOMA-Germany II | bip_bmg2_eur
Cases were recruited from consecutive admissions to psychiatric in-patient units at the University Hospital Würzburg. All cases received a lifetime diagnosis of BD according to the DSM-IV criteria using a consensus best-estimate procedure based on all available information, including semi-structured diagnostic interviews using the Association for Methodology and Documentation in Psychiatry, medical records and the family history method. In addition, the OPCRIT system was used for the detailed polydiagnostic documentation of symptoms.
Control subjects were ascertained from the population-based Heinz Nixdorf Recall (HNR) Study. The controls were not screened for a history of mental illness. Study protocols were reviewed and approved in advance by Institutional Review Boards of the participating institutions. All subjects provided written informed consent.
Rietschel, M; Nöthen, MM; Schulze, TG; Bauer, M; Forstner, AJ; Müller-Myhsok, B | 24618891 | BOMA-Germany III | bip_bmg3_eur
Cases were recruited at the Central Institute of Mental Health in Mannheim, University of Heidelberg, and other collaborating psychiatric hospitals in Germany. All cases received a lifetime diagnosis of BD according to the DSM-IV criteria using a consensus best-estimate procedure based on all available information including structured diagnostic interviews using the AMDP, Composite International Diagnostic Screener (CID-S), SADS-L and/or SCID, medical records, and the family history method. In addition, the OPCRIT system was used for the detailed polydiagnostic documentation of symptoms. Controls were selected randomly from a Munich-based community sample and recruited at the Max-Planck Institute of Psychiatry. They were screened for the presence of anxiety and mood disorders using the CID-S. Only individuals without mood and anxiety disorders were collected as controls. Study protocols were reviewed and approved in advance by Institutional Review Boards of the participating institutions. All subjects provided written informed consent.
Hauser, J; Lissowska, J; Forstner, AJ | 24618891 | BOMA-Poland | bip_bmpo_eur
Cases were recruited at the Department of Psychiatry, Poznan University of Medical Sciences, Poznan, Poland. All cases received a lifetime diagnosis of BD according to the DSM-IV criteria on the basis of a consensus best-estimate procedure and structured diagnostic interviews using the SCID. Controls were drawn from a population-based case-control sample recruited by the Cancer-Center and Institute of Oncology, Warsaw, Poland and a hospital-based case-control sample recruited by the Nofer Institute of Occupational Medicine, Lodz, Poland. The Polish controls were produced by the International Agency for Research on Cancer (IARC) and the Centre National de Génotypage (CNG) GWAS Initiative for a study of upper aerodigestive tract cancers. The controls were not screened for a history of mental illness. Study protocols were reviewed and approved in advance by Institutional Review Boards of the participating institutions. All subjects provided written informed consent.
Rietschel, M; Nöthen, MM; Rivas, F; Mayoral, F; Kogevinas, M; others | 24618891 | BOMA-Spain | bip_bmsp_eur
Cases were recruited at the mental health departments of the following five centers in Andalusia, Spain: University Hospital Reina Sofia of Córdoba, Provincial Hospital of Jaen; Hospital of Jerez de la Frontera (Cádiz); Hospital of Puerto Real (Cádiz); Hospital Punta Europa of Algeciras (Cádiz); and Hospital Universitario San Cecilio (Granada). Diagnostic assessment was performed using the SADS-L; the OPCRIT; a review of medical records; and interviews with first and/or second degree family members using the Family Informant Schedule and Criteria (FISC). Consensus best estimate BD diagnoses were assigned by two or more independent senior psychiatrists and/or psychologists, and according to the RDC, and the DSM-IV. Controls were Spanish subjects drawn from a cohort of individuals recruited in the framework of the European Community Respiratory Health Survey (ECRHS, http://www.ecrhs.org/). The controls were not screened for a history of mental illness. Study protocols were reviewed and approved in advance by Institutional Review Boards of the participating institutions. All subjects provided written informed consent.
Fullerton, J.M.; Mitchell, P.B.; Schofield, P.R.; Martin N.G.; Cichon, S. | 24618891 | BOMA-Australia | bip_bmau_eur
Cases were recruited at the Mood Disorder Unit, Prince of Wales Hospital in Sydney. All cases received a lifetime diagnosis of BD according to the DSM-IV criteria on the basis of a consensus best-estimate procedure and structured diagnostic interviews using the DIGS, FIGS, and the SCID. Controls were parents of unselected adolescent twins from the Brisbane Longitudinal Twin Study. The controls were not screened for a history of mental illness. Study protocols were reviewed and approved in advance by Institutional Review Boards of the participating institutions. All subjects provided written informed consent.
Grigoroiu-Serbanescu, M; Nöthen, MM | 21353194 | BOMA-Romania | bip_rom3_eur
Cases were recruited from consecutive admissions to the Obregia Clinical Psychiatric Hospital, Bucharest. Patients were administered the DIGS and FIGS interviews. Information was also obtained from medical records and close relatives. The diagnosis of BP-I was assigned according to DSM-IV criteria using the best estimate procedure. All patients had at least two hospitalized illness episodes. Population-based controls were evaluated using the DIGS to exclude a lifetime history of major affective disorders, schizophrenia, schizoaffective disorders, and other psychoses, obsessive-compulsive disorder, eating disorders, and alcohol or drug addiction.
Craddock, N, Jones, I, Jones, L | 17554300 | WTCCC | bip_wtcc_eur_sr-qc
Cases were all over the age of 17 yr, living in the UK and of European descent. Recruitment was undertaken throughout the UK and included individuals who had been in contact with mental health services and had a lifetime history of high mood. After providing written informed consent, participants were interviewed by a trained psychologist or psychiatrist using a semi-structured lifetime diagnostic psychiatric interview (Schedules for Clinical Assessment in Neuropsychiatry) and available psychiatric medical records were reviewed. Using all available data, best-estimate life-time diagnoses were made according to the RDC. In the current study we included cases with a lifetime diagnosis of RDC bipolar 1 disorder, bipolar 2 disorder or schizo-affective disorder, bipolar type. Controls were recruited from two sources: the 1958 Birth Cohort study and the UK Blood Service (blood donors) and were not screened for history of mental illness. All cases and controls were recruited under protocols approved by the appropriate IRBs. All subjects gave written informed consent.
Kelsoe, J | 21926972 [PGC1] | USA (GAIN) | bip_gain_eur
Genetic Association Information Network (GAIN)/The Bipolar Genome Study (BiGS) The BD sample was collected under the auspices of the NIMH Genetics Initiative for BD (http://zork.wustl.edu/nimh/), genotyped as part of GAIN and analyzed as part of a larger GWAS conducted by the BiGS consortium. Approximately half of the GAIN sample was collected as multiplex families or sib pair families (waves 1–4), the remainder were collected as individual cases (wave 5). Subjects were ascertained at 11 sites: Indiana University, John Hopkins University, the NIMH Intramural Research Program, Washington University at St. Louis, University of Pennsylvania, University of Chicago, Rush Medical School, University of Iowa, University of California, San Diego, University of California, San Francisco, and University of Michigan. All investigations were carried out after the review of protocols by the IRB at each participating institution. At all sites, potential cases were identified from screening admissions to local treatment facilities and through publicity programs or advocacy groups. Potential cases were evaluated using the DIGS, FIGS, and information from relatives and medical records. All information was reviewed through a best estimate diagnostic procedure by two independent and non-interviewing clinicians and a consensus best-estimate diagnosis was reached. In the event of a disagreement, a third review was done to break the tie. Controls were from the NIMH Genetic Repository sample obtained by Dr. P. Gejman through a contract to Knowledge Networks, Inc. Only individuals with complete or near-complete psychiatric questionnaire data who did not fulfill diagnostic criteria for major depression and denied a history of psychosis or BD were included as controls for BiGS analyses. Controls were matched for gender and ethnicity to the cases.
Kelsoe, J; Sklar, P; Smoller, J | [PGC1 Replication] | USA (FAT2; FaST, BiGS, TGEN) | bip_fat2_eur
Cases were collected from individuals at the 11 U.S. sites described for the GAIN sample. Eligible participants were age 18 or older meeting DSM-IV criteria for BD-I or BD-II by consensus diagnosis based on interviews with the Affective Disorders Evaluation (ADE) and MINI. All participants provided written informed consent and the study protocol was approved by IRBs at each site. Collection of phenotypic data and DNA samples were supported by NIMH grants MH063445 (JW Smoller); MH067288 (PI: P Sklar), and MH63420 (PI: V Nimgaonkar). The control samples were NIMH controls that were using the methods described in that section. The case and control samples were independent of those included in the GAIN sample.
Kirov, G | 25055870 | Bulgarian trios | bip_butr_eur
All cases were recruited in Bulgaria from psychiatric inpatient and outpatient services. Each proband had a history of hospitalisation and was interviewed with an abbreviated version of the SCAN. Consensus best-estimate diagnoses were made according to DSM-IV criteria by two researchers. All participants gave written informed consent and the study was approved by local ethics committees at the participating centers.
Kirov, G | 25055870 | UK trios | bip_uktr_eur
The BD subjects were recruited from lithium clinics and interviewed in person by a senior psychiatrist, using abbreviated version of the SCAN. Consensus best-estimate diagnoses were made based on the interview and hospital notes. Ethics committee approval for the study was obtained from the relevant research ethics committees and all individuals provided written informed consent for participation.
Landén, M; Sullivan, PF; Sklar, P | [ICCBD] | Sweden (ICCBD) | bip_swa2_eur
The BD subjects were identified using the Swedish National Quality Register for Bipolar Disorders (BipoläR) and the Swedish National Patient Register (using a validated algorithm requiring at least two hospitalizations with a BD diagnosis). A confirmatory telephone interview with a diagnostic review was conducted. Additional subjects were recruited from the St. Göran Bipolar Project (Affective Center at Northern Stockholm Psychiatry Clinic, Sweden), enrolling new and ongoing patients diagnosed with BD using structured clinical interviews. Diagnoses were made according to the DSM-IV criteria (BipoläR and St. Göran Bipolar Project) and ICD-10 (National Patient Register). The control subjects used were the same as for the SCZ analyses described above. All ascertainment procedures were approved by the Regional Ethical Committees in Sweden.
Landén, M; Sullivan, PF; Sklar, P | [ICCBD] | Sweden (ICCBD) | bip_swei_eur
The cases and controls in the bip_swei_eur sample were recruited using the same ascertainment methods described for the bip_swa2_eur sample.
Leboyer, M | [PGC1 replication] | France | bip_fran_eur
Cases with BD1 or BD2 and control samples were recruited as part of a large study of genetics of BD in France (Paris-Creteil, Bordeaux, Nancy) with a protocol approved by relevant IRBs and with written informed consent. Cases were of French descent for more than 3 generations were assessed by a trained psychiatrist or psychologist using structured interviews supplemented by medical case notes, mood scales and self-rating questionnaire assessing dimensions.
Li, Q | 24166486; 27769005 | USA (Janssen), SAGE controls | bip_jst5_eur
The study included unrelated patients with bipolar 1 disorder from 6 clinical trials (IDs: NCT00253162, NCT00257075, NCT00076115, NCT00299715, NCT00309699, and NCT00309686). Participant recruitment was conducted by Janssen Research & Development, LLC (formerly known as Johnson & Johnson Pharmaceutical Research & Development, LLC) to assess the efficacy and safety of risperidone. Bipolar cases were diagnosed according to DSM-IV-TR criteria. The diagnosis of bipolar disorder was confirmed by the Schedule for Affective Disorders and Schizophrenia for School-Age Children-Present and Lifetime Version (K-SADS-PL) in NCT00076115, by the SCID in NCT00257075 and NCT00253162, or by the MINI in NCT00299715 and NCT00309699, and NCT00309686, respectively. Additional detailed descriptions of these clinical trials can be found at ClinicalTrials.gov. Only patients of European ancestry with matching controls were included in the current analysis. Controls subjects were drawn from the Study of Addiction: Genetics and Environment (SAGE, dbGaP Study Accession: phs000092.v1.p1). Control subjects did not have alcohol dependence or drug dependence diagnoses; however, mood disorders were not an exclusion criterion.
McQuillin, A; Gurling, H | 18317468 [PGC1] | UCL (University College London), London, UK | bip_uclo_eur
The UCL sample comprised Caucasian individuals who were ascertained and received clinical diagnoses of bipolar 1 disorder according to UK National Health Service (NHS) psychiatrists at interview using the categories of the International Classification of Disease version 10. In addition bipolar subjects were included only if both parents were of English, Irish, Welsh or Scottish descent and if three out of four grandparents were of the same descent. All volunteers read an information sheet approved by the Metropolitan Medical Research Ethics Committee who also approved the project for all NHS hospitals. Written informed consent was obtained from each volunteer. The UCL control subjects were recruited from London branches of the National Blood Service, from local NHS family doctor clinics and from university student volunteers. All control subjects were interviewed with the SADS-L to exclude all psychiatric disorders.
Craddock, N; Jones, I; Jones, L | [ICCBD] | Cardiff and Worcester, UK (ICCBD-BDRN) | bip_icuk_eur
Cases were all over the age of 17 yr, living in the UK and of European descent. Cases were recruited via systematic and not systematic methods as part of the Bipolar Disorder Research Network project (www.bdrn.org), provided written informed consent and were interviewed using a semi-structured diagnostic interview, the Schedules for Clinical Assessment in Neuropsychiatry. Based on the information gathered from the interview and case notes review, best-estimate lifetime diagnosis was made according to DSM-IV. Inter-rater reliability was formally assessed using 20 randomly selected cases (mean ĸ Statistic = 0.85). In the current study we included cases with a lifetime diagnosis of DSM-IV bipolar disorder or schizo-affective disorder, bipolar type. The BDRN study has UK National Health Service (NHS) Research Ethics Committee approval and local Research and Development approval in all participating NHS Trusts/Health Boards.Controls were part of the Wellcome Trust Case Control Consortium common control set, which comprised healthy blood donors recruited from the UK Blood Service and samples from the 1958 British Birth Cohort. Controls were not screened for a history of mental illness. All cases and controls were recruited under protocols approved by the appropriate IRBs. All subjects gave written informed consent.
Ophoff, RA | Not Published | Netherlands | bip_ucla_eur
The case sample consisted of inpatients and outpatients recruited through psychiatric hospitals and institutions throughout the Netherlands. Cases with DSM-IV bipolar disorder, determined after interview with the SCID, were included in the analysis. Controls were collected in parallel at different sites in the Netherlands and were volunteers with no psychiatric history after screening with the (MINI). Ethical approval was provided by UCLA and local ethics committees and all participants gave written informed consent.
Paciga, S | [PGC1] | USA (Pfizer) | bip_pf1e_eur
This sample comprised Caucasian individuals recruited into one of three Geodon (ziprasidone) clinical trials (NCT00141271, NCT00282464, NCT00483548). Subjects were diagnosed by a clinician with a primary diagnosis of Bipolar 1 Disorder, most recent episode depressed, with or without rapid cycling, without psychotic features, as defined in the DSM-IV-TR (296.5x) and confirmed by the MINI (version 5.0.0). Subjects also were assessed as having a HAM-D-17 total score of >20 at the screening visit. The trials were conducted in accordance with the protocols, International Conference on Harmonization of Good Clinical Practice Guidelines, and applicable local regulatory requirements and laws. Patients gave written informed consent for the collection of blood samples for DNA for use in genetic studies.
Pato, C | [ICCBD] | Los Angeles, USA (ICCBD-GPC)| bip_usc2_eur
Genomic Psychiatry Consortium (GPC) cases and controls were collected via the University of Southern California healthcare system, as previously described. Using a combination of focused, direct interviews and data extraction from medical records, diagnoses were established using the OPCRIT and were based on DSM-IV-TR criteria. Age and gender-matched controls were ascertained from the University of Southern California health system and assessed using a validated screening instrument and medical records.
Scott, L; Myer, RM; Boehnke, M | 19416921 [PGC1] | Michigan, USA (Pritzker and NIMH) | bip_mich_eur
The Pritzker Neuropsychiatric Disorders Research Consortium (NIMH/Pritzker) case and controls samples were from the NIMH Genetics Initiative Genetics Initiative Repository. Cases were diagnosed according to DMS-III or DSM-IV criteria using diagnostic interviews and/or medical record review. Cases with low confidence diagnoses were excluded. From each wave 1–5 available non-Ashkenazi European-origin family, two BD1 siblings were included when possible and the proband was preferentially included if available (n=946 individuals in 473 sibling pairs); otherwise a single BD1 case was included (n=184). The bipolar sibling pairs were retained within the NIMH/Pritzker sample when individuals in more than one study were uniquely assigned to a study set. Controls had non-Ashkenazi European-origin, were aged 20–70 years and reported no diagnosis with or treatment for BD or schizophrenia, and that they had not heard voices that others could not hear. Individuals with suspected major depression were excluded based on answers to questions related to depressive mood. NIMH controls were further selected as the best match(es) to NIMH cases based on self-reported ancestry.
Sklar, P; Smoller, J | 18317468 [PGC1] | USA (STEP1) | bip_stp1_eur
The Systematic Treatment Enhancement Program for Bipolar Disorder (STEP-BD) was a seven-site, national U.S., longitudinal cohort study designed to examine the effectiveness of treatments and their impact on the course of BD that enrolled 4,361 participants who met DSM-IV criteria for BD1, BD2, bipolar not otherwise specified (NOS), schizoaffective manic or bipolar type, or cyclothymic disorder based on diagnostic interviews. From the parent study, 2,089 individuals who were over 18 years of age with BD1 and BD2 diagnoses consented to the collection of blood samples for DNA. BD samples with a consensus diagnosis of BD1 were selected for inclusion in STEP1. Two groups of controls samples from the NIMH repository were used. One comprised DNA samples derived from US Caucasian anonymous cord blood donors. The second were controls who completed the online self-administered psychiatric screen and were ascertained as described above, by Knowledge Networks Inc. For the second sample of controls only those without history of schizophrenia, psychosis, BD or major depression with functional impairment were used.
Sklar, P; Smoller, J | 18711365 [PGC1] | USA (STEP2) | bip_stp2_eur
The STEP2 sample included BD-1 and BD-2 samples from the STEP-BD study described above along with BD-2 subjects from UCL study also described above. The controls samples for this study were from the NIMH repository as described above for the STEP1 study.
European ancestry, trio design
Schizophrenia
Kirov, G: Owen M | 22083728| Bulgaria | ms.scz_butr_eur
Families from Bulgaria were recruited if a proband had schizophrenia or schizoaffective disorder, both parents were available, and all members of the trio agreed to participate in the study. Recruitment took place between 1999 and 2004 in several psychiatric hospitals in Bulgaria. Ethical Committee approval was obtained from each of these hospitals. All probands and all parents received an Information Sheet and signed Informed Consent Forms. All participants had attended mainstream schools, which at the time in Bulgaria, excluded people with mental retardation. Probands were either in- or out-patients at the time of the study but each had a history of hospitalization. A team of psychiatrists was trained in using the rating scales and methods of the study. We used the SCAN instrument to perform an interview for psychotic and mood symptoms. This instrument has been translated into Bulgarian and validated by one of its authors (A. Jablensky). Consensus diagnoses were made according to DSM-IV criteria on the basis of an interview and inspection of hospital notes by two clinicians. If consensus was not attained, the patient was re-interviewed by a research interview trained clinician and was excluded if consensus could still not be reached. In addition, approximately 23% of the sample was selected at random and re-interviewed by a research interview trained clinician. Hospital notes were also collected for affected relatives in order to confirm diagnoses.
Levinson, D | 22885689 | Six countries | ms.scz_lemu_eur
Schizophrenia cases were included from the family sample of European-ancestry pedigrees described by Levinson et al. Participants and their families in this trio study, probands were ascertained and recruited from different clinical settings (e.g. inpatients, outpatients and community facilities) in six countries (Australia, France, Germany, Ireland, UK, and the US). (Unrelated individuals were included as part of a case-control design, see Levinson, D, scz_lacw_eur above.) Diagnoses were established using semi-structured interviews, psychiatric records and informant reports. Case probands were diagnosed with schizophrenia or schizoaffective disorder according to DSM-III-R criteria. The trio-based analysis included families where there was at least one affected proband and two available parents. Each affected sibling in such families was included, with the parents, as an independent trio. All protocols were approved by loci IRBs, and all cases provided written informed consent.
Kirov, G: Owen, M | Not Published | Bulgaria | ms.scz_uktr_eur
All cases and parents were recruited from UK and had a history of hospitalization for treatment of schizophrenia. Diagnosis was confirmed following a SCAN interview and review of case notes followed by consensus diagnosis according to DSM-IV criteria. The samples were genotyped at the Broad Institute. All participants gave written informed consent and the study was approved by local ethics committees at the participating centers. The samples were genotyped at the Broad Institute.
Genotype Quality Control
To ensure independence of the data sets, individuals were excluded until no individual showed a relatedness (pihat) value greater than 0.2 to any other individual in the collection, while preferentially keeping the case over the control for case-control related pairs. In total 1,795 BD cases, 1,165 SCZ cases and 27,274 controls were removed (most of which were previously known), leaving 20,129 BD cases 33,426 SCZ cases and 54,065 controls for the final meta-analysis.
For analyses directly comparing BD and SCZ, we matched cases from both phenotypes on genotyping platform and ancestry, resulting in 15,270 BD cases versus 23,585 SCZ cases. Hence, we were able to match 76% of BD cases and 71% of SCZ cases for this case vs case analysis.
Among our entire dataset, 44% of the sample was female, 51% was male and 5% were unreported by the collection site. This work focused explicitly on the autosomes and sought maximal power across the analyses, sex was not used except for during quality control and sex-specific analyses were not performed in this effort. Individual ages were not provided. For a subset of cases, we had information for age of onset which were used in subphenotype specific analyses only.
Sub-phenotype Description
BD sub-phenotypes were collected by each study site using a combination of diagnostic instruments, case records and participant interviews. Ascertainment details for each study site are described in the supplementary data of the PGC Bipolar Working Group paper(Stahl et al., 2017). The selection of phenotypes for collection by this group was determined by literature searches in order to determine phenotypes with prior evidence for heritability. It was further refined dependent on the availability of phenotype data across a range of study sites and the consistency by which the phenotypes were defined. Schizophrenia subphenotypes represent quantitative traits extracted using factor analysis from a set of standard psychiatric assessments and represent four symptom dimensions (manic, depressive, positive and negative). These subphenotypes were used previously(Ruderfer et al., 2014) but in this work we have increased the sample size with additional cohorts being added.
METHOD DETAILS
QUANTIFICATION AND STATISTICAL ANALYSIS
Quality Control, Imputation, Association Analysis and Polygenic Risk Score Testing
Quality control and imputation were performed on each of the study cohort datasets (n=81), according to standards established by the Psychiatric Genomics Consortium (PGC). The quality control parameters for retaining SNPs and subjects were: SNP missingness < 0.05 (before sample removal); subject missingness (p < 0.02); autosomal heterozygosity deviation (| Fhet | < 0.2); SNP missingness < 0.02 (after sample removal); difference in SNP missingness between cases and controls < 0.02; and SNP Hardy-Weinberg equilibrium (p > 10−6 in controls or p > 10−10 in cases). Genotype imputation was performed using the pre-phasing/imputation stepwise approach implemented in IMPUTE2(Howie et al., 2011) / SHAPEIT(Delaneau et al., 2013) (chunk size of 3 Mb and default parameters). The imputation reference set consisted of 2,186 phased haplotypes from the full 1000 Genomes Project dataset (August 2012, 30,069,288 variants, release “v3.macGT1”), all variants align to human genome build 19 (hg19). After imputation, we used the best guess genotypes (genotype probability > 0.8), for further robust relatedness testing and population structure analysis. Here we required very high imputation quality (INFO > 0.8) and low missingness (<1%) for further quality control. After linkage disequilibrium (LD) pruning (r2 < 0.02) and frequency filtering (MAF > 0.05), there were 14,473 autosomal SNPs in the data set. Principal component estimation was done with the same collection of autosomal SNPs. We tested the first 20 principal components for phenotype association (using logistic regression with study indicator variables included as covariates) and evaluated their impact on the genome-wide test statistics using λ. Thirteen principal components namely 1,2,3,4,5,6,7,8,10,12,15,18,20 were included in all association analyses (λ=1.45). Analytical steps were repeated for SCZ vs BD analysis.
We performed four main association analyses (Figure 1), i.e. (i) GWAS of BD and SCZ as a single combined case phenotype, as well as disorder-specific GWAS using independent control sets in (ii) BD cases vs BD controls and (iii) SCZ cases vs SCZ controls, and (iv) association analysis of SCZ cases vs BD cases. For all GWS loci from the GWAS of BD and SCZ vs controls we identified any GWS loci within 1Mb from the extent of the locus in the previously published PGC SCZ vs controls(Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014) and the most recent PGC GWAS of BD vs controls(Stahl et al., 2017) and performed conditional analysis. Specifically, we transformed the genotype probabilities of the disease variant into dosages and used it as an additional covariate for the association analysis for the BD+SCZ vs controls index SNP. This was done within each cohort and an OR based inverse SE weighted meta-analysis was performed for the final result. All datasets were included except for those with trios.
Summary-data-based Mendelian Randomization (SMR)
SMR(Zhu et al., 2016) is a method that integrates summary level GWAS data with gene expression quantitative trait loci (eQTL) identified in independent data sets. This integration aims to identify variants that have pleotropic effects on expression of a given gene and the phenotype. While significant findings may indeed reflect a causal path from variant to phenotype through expression, it is impossible to discern statistically between pleiotropy and causality. However, the method can remove linkage as driving the result, and uses the data available to prioritise amongst genes in genomic regions that show association with disease. We used SMR as a statistical fine-mapping tool applied to the SCZ vs BD GWAS results to identify loci with strong evidence of causality via gene expression. SMR analysis is limited to significant (FDR < 0.05) cis SNP-expression quantitative trait loci (eQTLs) with MAF > 0.01. eQTLs passing these thresholds were combined with GWAS results in the SMR test, with significance (pSMR) reported at a Bonferroni-corrected threshold for each eQTL data set. The eQTL architecture may differ between genes. For example, through LD, many SNPs can generate significant associations with the same gene, but in some instances multiple SNPs may be independently associated with the expression of a gene. After identification of significant SNP-expression-trait association through the SMR test, a follow-up heterogeneity test aims to prioritize variants by excluding regions for which there is conservative evidence for multiple causal loci (pHET < 0.05). SMR analyses were conducted using eQTL data from whole peripheral blood(Westra et al., 2013), dorsolateral prefrontal cortex generated by the CommonMind Consortium8, and 11 brain sub-regions from the GTEx consortium(Consortium, 2015).
Regional joint GWAS
Summary statistic Z-scores were calculated for each marker in each of the four main GWAS results, using the logistic regression coefficient and its standard error. Rare SNPs (MAF < 0.01), and SNPs with a low INFO score (< 0.3) in either dataset were removed. The causal variant relationships between SCZ and BD were investigated using the Bayesian method software pw-gwas (v0.2.1), with quasi-independent regions determined by estimate LD blocks in an analysis of European individuals (n=1,703)(Berisa and Pickrell, 2015; Pickrell et al., 2016). Briefly, pw-gwas takes a Bayesian approach to determine the probability of five independent models of association. (1) There is no causal variant in BD or SCZ; (2) a causal variant in BD, but not SCZ (3); a causal variant in SCZ, but not BD; (4) a shared causal variant influencing both BD and SCZ; (5) two causal variants where one influences BD, and one influences SCZ (Figure 2). The posterior probability of each model is calculated using model priors, estimated empirically within pw-gwas. Regions were considered to support a particular model when the posterior probability of the model was greater than 0.5.
Regional SNP-heritability estimation
We calculated local SNP-heritability independently for SCZ and BD using the Heritability Estimator from Summary Statistics (HESS) software(Shi et al., 2016) for each of the independent regions defined above. The sum of these regional estimates is the total SNP-heritability of the trait. To calculate local SNP-heritability HESS requires reference LD matrices representative of the population from which the GWAS samples were drawn. We utilized the 1000 genomes European individuals as the reference panel(The 1000 Genomes Project Consortium, 2015). Unlike pw-gwas(Pickrell et al., 2016), HESS does not assume that only one causal variant can be present in each region.
DATA AND SOFTWARE AVAILABILITY
Summary statistics from GWAS are publically available at https://www.med.unc.edu/pgc/results-and-downloads/downloads.
Supplementary Material
Figure S1. Related to Figure 1b. Regional Association Plot and Forest Plot for the First Genome-wide Significant Hit in the SCZ vs BD GWAS.
Figure S2. Related to Figure 1b. Regional Association Plot and Forest Plot for the Second Genome-wide Significant Hit in the SCZ vs BD GWAS.
Figure S3. Related to Summary-data-based Mendelian Randomization. Detailed Association of DCAKD from SMR. Results at the DCAKD locus from SMR analysis of SCZ vs BD. Top plot, brown dots represent the P values for SNPs from SCZ vs BD GWAS, diamonds represent the P values for probes from the SMR test. Bottom plot, the eQTL P values of SNPs from the Westra study for the ILMN_1811648 probe tagging DCAKD. The top and bottom plots include all the SNPs available in the region in the GWAS and eQTL summary data, respectively, rather than only the SNPs common to both data sets. Highlighted in red is the gene (DCAKD) that passed the SMR and HEIDI tests.
Figure S4. Related to Regional SNP-heritability estimation. Heritability Estimates for BD and SCZ in Genome-wide Significant Regions of BD and SCZ. Regional SNP-heritability estimates for SCZ and BD stratified by whether the region contains the most significant variant in a genome-wide significant locus in BD, SCZ, neither or both.
Acknowledgements
The work of the contributing groups was supported by numerous grants from governmental and charitable bodies as well as philanthropic donation. Specifically, DMR (R01MH111776), NRW (NHMRC 1078901, 1087889).
Footnotes
Declaration of Interests
The authors declare no competing interests.
References
- Amare AT, Schubert KO, Hou L, Clark SR, Papiol S, Heilbronner U, Degenhardt F, Tekola-Ayele F, Hsu Y-H, Shekhtman T, Adli M, Akula N, Akiyama K, Ardau R, Arias B, Aubry J-M, Backlund L, Bhattacharjee AK, Bellivier F, Benabarre A, Bengesser S, Biernacka JM, Birner A, Brichant-Petitjean C, Cervantes P, Chen H-C, Chillotti C, Cichon S, Cruceanu C, Czerski PM, Dalkner N, Dayer A, Zompo MD, DePaulo JR, Étain B, Falkai P, Forstner AJ, Frisen L, Frye MA, Fullerton JM, Gard S, Garnham JS, Goes FS, Grigoroiu-Serbanescu M, Grof P, Hashimoto R, Hauser J, Herms S, Hoffmann P, Hofmann A, Jamain S, Jiménez E, Kahn J-P, Kassem L, Kuo P-H, Kato T, Kelsoe J, Kittel-Schneider S, Kliwicki S, König B, Kusumi I, Laje G, Landén M, Lavebratt C, Leboyer M, Leckband SG, Tortorella A, Manchia M, Martinsson L, McCarthy MJ, McElroy S, Colom F, Mitjans M, Mondimore FM, Monteleone P, Nievergelt CM, Nöthen MM, Novák T, O’Donovan C, Ozaki N, Ösby U, Pfennig A, Potash JB, Reif A, Reininghaus E, Rouleau GA, Rybakowski JK, Schalling M, Schofield PR, Schweizer BW, Severino G, Shilling PD, Shimoda K, Simhandl C, Slaney CM, Squassina A, Stamm T, Stopkova P, Maj M, Turecki G, Vieta E, Volkert J, Witt S, Wright A, Zandi PP, Mitchell PB, Bauer M, Alda M, Rietschel M, McMahon FJ, Schulze TG, Baune BT, 2018. Association of Polygenic Score for Schizophrenia and HLA Antigen and Inflammation Genes With Response to Lithium in Bipolar Affective Disorder: A Genome-Wide Association Study. JAMA Psychiatry 75, 65–74. 10.1001/jamapsychiatry.2017.3433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaraman Y, Lahiri DK, Nurnberger JI, 2015. Variants in Ion Channel Genes Link Phenotypic Features of Bipolar Illness to Specific Neurobiological Process Domains. Mol. Neuropsychiatry 1, 23–35. 10.1159/000371886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayés À, van de Lagemaat LN, Collins MO, Croning MDR, Whittle IR, Choudhary JS, Grant SGN, 2011. Characterization of the proteome, diseases and evolution of the human postsynaptic density. Nat. Neurosci 14, 19–21. 10.1038/nn.2719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benner C, Spencer CCA, Havulinna AS, Salomaa V, Ripatti S, Pirinen M, 2016. FINEMAP: efficient variable selection using summary data from genome-wide association studies. Bioinformatics 32, 1493–1501. 10.1093/bioinformatics/btw018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bera TK, Bera J, Brinkmann U, Tessarollo L, Pastan I, 2001. Cse1l Is Essential for Early Embryonic Growth and Development. Mol. Cell. Biol 21, 7020–7024. 10.1128/MCB.21.20.7020-7024.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berisa T, Pickrell JK, 2015. Approximately independent linkage disequilibrium blocks in human populations. Bioinformatics 32, 283–285. 10.1093/bioinformatics/btv546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik-Sullivan BK, Loh P-R, Finucane HK, Ripke S, Yang J, Schizophrenia Working Group of the Psychiatric Genomics Consortium, Patterson N, Daly MJ, Price AL, Neale BM, 2015. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet 47, 291–295. 10.1038/ng.3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne EM, Heath AC, Madden PA, Pergadia ML, Hickie IB, Montgomery GW, Martin NG, Wray NR, 2014. Testing the role of circadian genes in conferring risk for psychiatric disorders. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. Off. Publ. Int. Soc. Psychiatr. Genet 0, 254–260. 10.1002/ajmg.b.32230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claussnitzer M, Dankel SN, Kim K-H, Quon G, Meuleman W, Haugen C, Glunk V, Sousa IS, Beaudry JL, Puviindran V, Abdennur NA, Liu J, Svensson P-A, Hsu Y-H, Drucker DJ, Mellgren G, Hui C-C, Hauner H, Kellis M, 2015. FTO Obesity Variant Circuitry and Adipocyte Browning in Humans. N. Engl. J. Med 373, 895–907. 10.1056/NEJMoa1502214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleynen I, Boucher G, Jostins L, Schumm LP, Zeissig S, Ahmad T, Andersen V, Andrews JM, Annese V, Brand S, Brant SR, Cho JH, Daly MJ, Dubinsky M, Duerr RH, Ferguson LR, Franke A, Gearry RB, Goyette P, Hakonarson H, Halfvarson J, Hov JR, Huang H, Kennedy NA, Kupcinskas L, Lawrance IC, Lee JC, Satsangi J, Schreiber S, Théâtre E, van der Meulen-de Jong AE, Weersma RK, Wilson DC, Parkes M, Vermeire S, Rioux JD, Mansfield J, Silverberg MS, Radford-Smith G, McGovern DPB, Barrett JC, Lees CW, 2016. Inherited determinants of Crohn’s disease and ulcerative colitis phenotypes: a genetic association study. Lancet Lond. Engl 387, 156–167. 10.1016/S0140-6736(15)00465-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CNV and Schizophrenia Working Groups of the Psychiatric Genomics Consortium, 2017. Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat. Genet 49, 27–35. 10.1038/ng.3725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium, T.Gte., 2015. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 348, 648–660. 10.1126/science.1262110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross-Disorder Group of the Psychiatric Genomics Consortium, 2013. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat. Genet 45, 984–994. 10.1038/ng.2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis D, Vine A, McQuillin A, Bass N, Pereira A, Kandaswamy R, Lawrence J, Anjorin A, Choudhury K, Datta S, Puri V, Krasucki R, Pimm J, Thirumalai S, Quested D, Gurling H, 2011. Case-case genome wide association analysis reveals markers differentially associated with schizophrenia and bipolar disorder and implicates calcium channel genes. Psychiatr. Genet 21, 1–4. 10.1097/YPG.0b013e3283413382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaneau O, Zagury J-F, Marchini J, 2013. Improved whole-chromosome phasing for disease and population genetic studies. Nat. Methods 10, 5–6. 10.1038/nmeth.2307 [DOI] [PubMed] [Google Scholar]
- Diagnostic and Statistical Manual of Mental Disorders | DSM Library [WWW Document], n.d. URL http://dsm.psychiatryonline.org/doi/book/10.1176/appi.books.9780890425596 (accessed 3.14.17).
- Ding X, Liu S, Tian M, Zhang W, Zhu T, Li D, Wu J, Deng H, Jia Y, Xie W, Xie H, Guan J-S, 2017. Activity-induced histone modifications govern Neurexin-1 mRNA splicing and memory preservation. Nat. Neurosci. advance online publication. 10.1038/nn.4536 [DOI] [PubMed] [Google Scholar]
- Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P, Georgieva L, Rees E, Palta P, Ruderfer DM, Carrera N, Humphreys I, Johnson JS, Roussos P, Barker DD, Banks E, Milanova V, Grant SG, Hannon E, Rose SA, Chambert K, Mahajan M, Scolnick EM, Moran JL, Kirov G, Palotie A, McCarroll SA, Holmans P, Sklar P, Owen MJ, Purcell SM, O’Donovan MC, 2014. De novo mutations in schizophrenia implicate synaptic networks. Nature 506, 179–184. 10.1038/nature12929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromer M, Roussos P, Sieberts SK, Johnson JS, Kavanagh DH, Perumal TM, Ruderfer DM, Oh EC, Topol A, Shah HR, Klei LL, Kramer R, Pinto D, Gümüş ZH, Cicek AE, Dang KK, Browne A, Lu C, Xie L, Readhead B, Stahl EA, Xiao J, Parvizi M, Hamamsy T, Fullard JF, Wang Y-C, Mahajan MC, Derry JMJ, Dudley JT, Hemby SE, Logsdon BA, Talbot K, Raj T, Bennett DA, De Jager PL, Zhu J, Zhang B, Sullivan PF, Chess A, Purcell SM, Shinobu LA, Mangravite LM, Toyoshiba H, Gur RE, Hahn C-G, Lewis DA, Haroutunian V, Peters MA, Lipska BK, Buxbaum JD, Schadt EE, Hirai K, Roeder K, Brennand KJ, Katsanis N, Domenici E, Devlin B, Sklar P, 2016. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat. Neurosci 19, 1442–1453. 10.1038/nn.4399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullard JF, Giambartolomei C, Hauberg ME, Xu K, Voloudakis G, Shao Z, Bare C, Dudley JT, Mattheisen M, Robakis NK, Haroutunian V, Roussos P, n.d. Open chromatin profiling of human postmortem brain infers functional roles for non-coding schizophrenia loci. Hum. Mol. Genet 10.1093/hmg/ddx103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamazon ER, Wheeler HE, Shah KP, Mozaffari SV, Aquino-Michaels K, Carroll RJ, Eyler AE, Denny JC, GTEx Consortium, Nicolae DL, Cox NJ, Im HK, 2015. A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet 47, 1091–1098. 10.1038/ng.3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giambartolomei C, Vukcevic D, Schadt EE, Franke L, Hingorani AD, Wallace C, Plagnol V, 2014. Bayesian Test for Colocalisation between Pairs of Genetic Association Studies Using Summary Statistics. PLOS Genet. 10, e1004383 10.1371/journal.pgen.1004383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Lozano MA, Klemmer P, Gebuis T, Hassan C, van Nierop P, van Kesteren RE, Smit AB, Li KW, 2016. Dynamics of the mouse brain cortical synaptic proteome during postnatal brain development. Sci. Rep 6 10.1038/srep35456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green EK, Rees E, Walters JTR, Smith K-G, Forty L, Grozeva D, Moran JL, Sklar P, Ripke S, Chambert KD, Genovese G, McCarroll SA, Jones I, Jones L, Owen MJ, O’Donovan MC, Craddock N, Kirov G, 2016. Copy number variation in bipolar disorder. Mol. Psychiatry 21, 89–93. 10.1038/mp.2014.174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulsuner S, McClellan JM, 2015. Copy Number Variation in Schizophrenia. Neuropsychopharmacology 40, 252–254. 10.1038/npp.2014.216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusev A, Ko A, Shi H, Bhatia G, Chung W, Penninx BWJH, Jansen R, de Geus EJC, Boomsma DI, Wright FA, Sullivan PF, Nikkola E, Alvarez M, Civelek M, Lusis AJ, Lehtimäki T, Raitoharju E, Kähönen M, Seppälä I, Raitakari OT, Kuusisto J, Laakso M, Price AL, Pajukanta P, Pasaniuc B, 2016. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet 48, 245–252. 10.1038/ng.3506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Fuller CK, Song Y, Meng Q, Zhang B, Yang X, Li H, 2013. Sherlock: Detecting Gene-Disease Associations by Matching Patterns of Expression QTL and GWAS. Am. J. Hum. Genet 92, 667–680. 10.1016/j.ajhg.2013.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie B, Marchini J, Stephens M, 2011. Genotype Imputation with Thousands of Genomes. G3 GenesGenomesGenetics 1, 457–470. 10.1534/g3.111.001198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judy JT, Zandi PP, 2013. A review of potassium channels in bipolar disorder. Front. Genet 4 10.3389/fgene.2013.00105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka M, Matoba N, Sawada T, Kazuno A-A, Ishiwata M, Fujii K, Matsuo K, Takata A, Kato T, 2016. Exome sequencing for bipolar disorder points to roles of de novo loss-of-function and protein-altering mutations. Mol. Psychiatry 21, 885–893. 10.1038/mp.2016.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khera AV, Chaffin M, Aragam K, Emdin CA, Klarin D, Haas M, Roselli C, Natarajan P, Kathiresan S, 2017. Genome-wide polygenic score to identify a monogenic risk-equivalent for coronary disease. bioRxiv 218388 10.1101/218388 [DOI] [Google Scholar]
- Kirov G, Pocklington AJ, Holmans P, Ivanov D, Ikeda M, Ruderfer D, Moran J, Chambert K, Toncheva D, Georgieva L, Grozeva D, Fjodorova M, Wollerton R, Rees E, Nikolov I, van de Lagemaat LN, Bayés À, Fernandez E, Olason PI, Böttcher Y, Komiyama NH, Collins MO, Choudhary J, Stefansson K, Stefansson H, Grant SGN, Purcell S, Sklar P, O’Donovan MC, Owen MJ, 2012. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol. Psychiatry 17, 142–153. 10.1038/mp.2011.154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeuw CA, de Mooij JM, Heskes T, Posthuma D, 2015. MAGMA: Generalized Gene-Set Analysis of GWAS Data. PLOS Comput. Biol 11, e1004219 10.1371/journal.pcbi.1004219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Ma Z, Shi M, Malty RH, Aoki H, Minic Z, Phanse S, Jin K, Wall DP, Zhang Z, Urban AE, Hallmayer J, Babu M, Snyder M, 2015. Identification of Human Neuronal Protein Complexes Reveals Biochemical Activities and Convergent Mechanisms of Action in Autism Spectrum Disorders. Cell Syst. 1, 361–374. 10.1016/j.cels.2015.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li KK-W, Yang L, Pang JC-S, Chan AK-Y, Zhou L, Mao Y, Wang Y, Lau K-M, Poon WS, Shi Z, Ng H-K, 2013. MIR-137 Suppresses Growth and Invasion, is Downregulated in Oligodendroglial Tumors and Targets CSE1L. Brain Pathol. 23, 426–439. 10.1111/bpa.12015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QS, Tian C, Seabrook GR, Drevets WC, Narayan VA, 2016. Analysis of 23andMe antidepressant efficacy survey data: implication of circadian rhythm and neuroplasticity in bupropion response. Transl. Psychiatry 6, e889 10.1038/tp.2016.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenstein P, Yip BH, Björk C, Pawitan Y, Cannon TD, Sullivan PF, Hultman CM, 2009. Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: a population-based study. The Lancet 373, 234–239. 10.1016/S0140-6736(09)60072-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luoni A, Massart R, Nieratschker V, Nemoda Z, Blasi G, Gilles M, Witt SH, Suderman MJ, Suomi SJ, Porcelli A, Rizzo G, Fazio L, Torretta S, Rampino A, Berry A, Gass P, Cirulli F, Rietschel M, Bertolino A, Deuschle M, Szyf M, Riva MA, 2016. Ankyrin-3 as a molecular marker of early-life stress and vulnerability to psychiatric disorders. Transl. Psychiatry 6, e943 10.1038/tp.2016.211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moczulska KE, Pichler P, Schutzbier M, Schleiffer A, Rumpel S, Mechtler K, 2014. Deep and Precise Quantification of the Mouse Synaptosomal Proteome Reveals Substantial Remodeling during Postnatal Maturation. J. Proteome Res. 13, 4310–4324. 10.1021/pr500456t [DOI] [PubMed] [Google Scholar]
- Mohanan V, Nakata T, Desch AN, Lévesque C, Boroughs A, Guzman G, Cao Z, Creasey E, Yao J, Boucher G, Charron G, Bhan AK, Schenone M, Carr SA, Reinecker H-C, Daly MJ, Rioux JD, Lassen KG, Xavier RJ, 2018. C1orf106 is a colitis risk gene that regulates stability of epithelial adherens junctions. Science eaan 0814 10.1126/science.aan0814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MR, Tipney H, Painter JL, Shen J, Nicoletti P, Shen Y, Floratos A, Sham PC, Li MJ, Wang J, Cardon LR, Whittaker JC, Sanseau P, 2015. The support of human genetic evidence for approved drug indications. Nat. Genet. advance online publication. 10.1038/ng.3314 [DOI] [PubMed] [Google Scholar]
- Nöthen MM, Nieratschker V, Cichon S, Rietschel M, 2010. New findings in the genetics of major psychoses. Dialogues Clin. Neurosci 12, 85–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okbay A, Baselmans BML, De Neve J-E, Turley P, Nivard MG, Fontana MA, Meddens SFW, Linnér RK, Rietveld CA, Derringer J, Gratten J, Lee JJ, Liu JZ, de Vlaming R, Ahluwalia TS, Buchwald J, Cavadino A, Frazier-Wood AC, Furlotte NA, Garfield V, Geisel MH, Gonzalez JR, Haitjema S, Karlsson R, van der Laan SW, Ladwig K-H, Lahti J, van der Lee SJ, Lind PA, Liu T, Matteson L, Mihailov E, Miller MB, Minica CC, Nolte IM, Mook-Kanamori D, van der Most PJ, Oldmeadow C, Qian Y, Raitakari O, Rawal R, Realo A, Rueedi R, Schmidt B, Smith AV, Stergiakouli E, Tanaka T, Taylor K, Wedenoja J, Wellmann J, Westra H-J, Willems SM, Zhao W, LifeLines Cohort Study, Amin N, Bakshi A, Boyle PA, Cherney S, Cox SR, Davies G, Davis OSP, Ding J, Direk N, Eibich P, Emeny RT, Fatemifar G, Faul JD, Ferrucci L, Forstner A, Gieger C, Gupta R, Harris TB, Harris JM, Holliday EG, Hottenga J-J, De Jager PL, Kaakinen MA, Kajantie E, Karhunen V, Kolcic I, Kumari M, Launer LJ, Franke L, Li-Gao R, Koini M, Loukola A, Marques-Vidal P, Montgomery GW, Mosing MA, Paternoster L, Pattie A, Petrovic KE, Pulkki-Råback L, Quaye L, Räikkönen K, Rudan I, Scott RJ, Smith JA, Sutin AR, Trzaskowski M, Vinkhuyzen AE, Yu L, Zabaneh D, Attia JR, Bennett DA, Berger K, Bertram L, Boomsma DI, Snieder H, Chang S-C, Cucca F, Deary IJ, van Duijn CM, Eriksson JG, Bültmann U, de Geus EJC, Groenen PJF, Gudnason V, Hansen T, Hartman CA, Haworth CMA, Hayward C, Heath AC, Hinds DA, Hyppönen E, Iacono WG, Järvelin M-R, Jöckel K-H, Kaprio J, Kardia SLR, Keltikangas-Järvinen L, Kraft P, Kubzansky LD, Lehtimäki T, Magnusson PKE, Martin NG, McGue M, Metspalu A, Mills M, de Mutsert R, Oldehinkel AJ, Pasterkamp G, Pedersen NL, Plomin R, Polasek O, Power C, Rich SS, Rosendaal FR, den Ruijter HM, Schlessinger D, Schmidt H, Svento R, Schmidt R, Alizadeh BZ, Sørensen TIA, Spector TD, Steptoe A, Terracciano A, Thurik AR, Timpson NJ, Tiemeier H, Uitterlinden AG, Vollenweider P, Wagner GG, Weir DR, Yang J, Conley DC, Smith GD, Hofman A, Johannesson M, Laibson DI, Medland SE, Meyer MN, Pickrell JK, Esko T, Krueger RF, Beauchamp JP, Koellinger PD, Benjamin DJ, Bartels M, Cesarini D, 2016. Genetic variants associated with subjective well-being, depressive symptoms, and neuroticism identified through genome-wide analyses. Nat. Genet. advance online publication. 10.1038/ng.3552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK, Berisa T, Liu JZ, Ségurel L, Tung JY, Hinds DA, 2016. Detection and interpretation of shared genetic influences on 42 human traits. Nat. Genet 48, 709–717. 10.1038/ng.3570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruderfer DM, Charney AW, Readhead B, Kidd BA, Kähler AK, Kenny PJ, Keiser MJ, Moran JL, Hultman CM, Scott SA, Sullivan PF, Purcell SM, Dudley JT, Sklar P, 2016. Polygenic overlap between schizophrenia risk and antipsychotic response: a genomic medicine approach. Lancet Psychiatry 3, 350–357. 10.1016/S2215-0366(15)00553-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruderfer DM, Fanous AH, Ripke S, McQuillin A, Amdur RL, Gejman PV, O’Donovan MC, Andreassen OA, Djurovic S, Hultman CM, Kelsoe JR, Jamain S, Landén M, Leboyer M, Nimgaonkar V, Nurnberger J, Smoller JW, Craddock N, Corvin A, Sullivan PF, Holmans P, Sklar P, Kendler KS, 2014. Polygenic dissection of diagnosis and clinical dimensions of bipolar disorder and schizophrenia. Mol. Psychiatry 19, 1017–1024. 10.1038/mp.2013.138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427. 10.1038/nature13595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekar A, Bialas AR, de Rivera H, Davis A, Hammond TR, Kamitaki N, Tooley K, Presumey J, Baum M, Van Doren V, Genovese G, Rose SA, Handsaker RE, Schizophrenia Working Group of the Psychiatric Genomics Consortium, Daly MJ, Carroll MC, Stevens B, McCarroll SA, 2016. Schizophrenia risk from complex variation of complement component 4. Nature 530, 177–183. 10.1038/nature16549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Kichaev G, Pasaniuc B, 2016. Contrasting the Genetic Architecture of 30 Complex Traits from Summary Association Data. Am. J. Hum. Genet 99, 139–153. 10.1016/j.ajhg.2016.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl E, Forstner A, McQuillin A, Ripke S, Pgc BDWG of the, Ophoff R, Scott L, Cichon S, Andreassen OA, Sklar P, Kelsoe J, Breen G, 2017. Genomewide association study identifies 30 loci associated with bipolar disorder. bioRxiv 173062 10.1101/173062 [DOI] [Google Scholar]
- Stone JL, O’Donovan MC, Gurling H, Kirov GK, Blackwood DHR, Corvin A, Craddock NJ, Gill M, Hultman CM, Lichtenstein P, McQuillin A, Pato CN, Ruderfer DM, Owen MJ, Clair DS, Sullivan PF, Sklar P (Leader), Korn SMP, Macgregor J, Morris S, O’Dushlaine DW, Daly CT, Visscher MJ, Holmans PM, Purcell PA, Scolnick SM, M. E (Leader), Williams PS, Georgieva NM, Nikolov L, Norton I, Williams N, Toncheva H, Milanova D, Thelander V, Sullivan EF, Kenny P, Waddington E, Choudhury JL, Datta K, Pimm S, Thirumalai J, Puri S, Krasucki V, Lawrence R, Quested J, Bass D, Curtis N, Crombie D, Fraser C, Kwan G, Walker SL, Muir N, McGhee WJ, Pickard KA, Malloy B, Maclean P, Beck AW, Pato MV, Medeiros MT, Middleton H, Carvalho F, Morley C, Fanous C, Conti A, Knowles D, Ferreira JA, Macedo CP, Azevedo A, McCarroll MH, Daly SA, Chambert M, Gates K, Gabriel C, Mahon SB, Ardlie, K. S, 2008. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature 455, 237–241. 10.1038/nature07239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szatkiewicz JP, O’Dushlaine C, Chen G, Chambert K, Moran JL, Neale BM, Fromer M, Ruderfer D, Akterin S, Bergen SE, Kähler A, Magnusson PKE, Kim Y, Crowley JJ, Rees E, Kirov G, O’Donovan MC, Owen MJ, Walters J, Scolnick E, Sklar P, Purcell S, Hultman CM, McCarroll SA, Sullivan PF, 2014. Copy number variation in schizophrenia in Sweden. Mol. Psychiatry 19, 762–773. 10.1038/mp.2014.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The 1000 Genomes Project Consortium, 2015. A global reference for human genetic variation. Nature 526, 68–74. 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher PM, Wray NR, Zhang Q, Sklar P, McCarthy MI, Brown MA, Yang J, 2017. 10 Years of GWAS Discovery: Biology, Function, and Translation. Am. J. Hum. Genet 101, 5–22. 10.1016/j.ajhg.2017.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Taskesen E, Bochoven A van, Posthuma D, 2017. FUMA: Functional mapping and annotation of genetic associations. bioRxiv 110023 10.1101/110023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westra H-J, Peters MJ, Esko T, Yaghootkar H, Schurmann C, Kettunen J, Christiansen MW, Fairfax BP, Schramm K, Powell JE, Zhernakova A, Zhernakova DV, Veldink JH, Van den Berg LH, Karjalainen J, Withoff S, Uitterlinden AG, Hofman A, Rivadeneira F, ‘t Hoen PAC, Reinmaa E, Fischer K, Nelis M, Milani L, Melzer D, Ferrucci L, Singleton AB, Hernandez DG, Nalls MA, Homuth G, Nauck M, Radke D, Völker U, Perola M, Salomaa V, Brody J, Suchy-Dicey A, Gharib SA, Enquobahrie DA, Lumley T, Montgomery GW, Makino S, Prokisch H, Herder C, Roden M, Grallert H, Meitinger T, Strauch K, Li Y, Jansen RC, Visscher PM, Knight JC, Psaty BM, Ripatti S, Teumer A, Frayling TM, Metspalu A, van Meurs JBJ, Franke L, 2013. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet 45, 1238–1243. 10.1038/ng.2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteford HA, Degenhardt L, Rehm J, Baxter AJ, Ferrari AJ, Erskine HE, Charlson FJ, Norman RE, Flaxman AD, Johns N, Burstein R, Murray CJ, Vos T, 2013. Global burden of disease attributable to mental and substance use disorders: findings from the Global Burden of Disease Study 2010. The Lancet 382, 1575–1586. 10.1016/S0140-6736(13)61611-6 [DOI] [PubMed] [Google Scholar]
- WHO | International Classification of Diseases [WWW Document], n.d.. WHO. URL http://www.who.int/classifications/icd/en/ (accessed 3.14.17).
- Yamashita S, Miyake N, Matsumoto N, Osaka H, Iai M, Aida N, Tanaka Y, 2013. Neuropathology of leukoencephalopathy with brainstem and spinal cord involvement and high lactate caused by a homozygous mutation of DARS2. Brain Dev. 35, 312–316. 10.1016/j.braindev.2012.05.007 [DOI] [PubMed] [Google Scholar]
- Yang S, Van Dongen HPA, Wang K, Berrettini W, Bućan M, 2008. Assessment of circadian function in fibroblasts of patients with bipolar disorder. Mol. Psychiatry 14, 143–155. 10.1038/mp.2008.10 [DOI] [PubMed] [Google Scholar]
- Zhu Z, Zhang F, Hu H, Bakshi A, Robinson MR, Powell JE, Montgomery GW, Goddard ME, Wray NR, Visscher PM, Yang J, 2016. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet 48, 481–487. 10.1038/ng.3538 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Related to Figure 1b. Regional Association Plot and Forest Plot for the First Genome-wide Significant Hit in the SCZ vs BD GWAS.
Figure S2. Related to Figure 1b. Regional Association Plot and Forest Plot for the Second Genome-wide Significant Hit in the SCZ vs BD GWAS.
Figure S3. Related to Summary-data-based Mendelian Randomization. Detailed Association of DCAKD from SMR. Results at the DCAKD locus from SMR analysis of SCZ vs BD. Top plot, brown dots represent the P values for SNPs from SCZ vs BD GWAS, diamonds represent the P values for probes from the SMR test. Bottom plot, the eQTL P values of SNPs from the Westra study for the ILMN_1811648 probe tagging DCAKD. The top and bottom plots include all the SNPs available in the region in the GWAS and eQTL summary data, respectively, rather than only the SNPs common to both data sets. Highlighted in red is the gene (DCAKD) that passed the SMR and HEIDI tests.
Figure S4. Related to Regional SNP-heritability estimation. Heritability Estimates for BD and SCZ in Genome-wide Significant Regions of BD and SCZ. Regional SNP-heritability estimates for SCZ and BD stratified by whether the region contains the most significant variant in a genome-wide significant locus in BD, SCZ, neither or both.
Data Availability Statement
Summary statistics from GWAS are publically available at https://www.med.unc.edu/pgc/results-and-downloads/downloads.