Abstract
Na+-H+ exchanger regulatory factor-1 (NHERF1) is a PDZ protein that scaffolds membrane proteins, including sodium–phosphate co-transport protein 2A (NPT2A) at the plasma membrane. NHERF1 is a phosphoprotein with 40 Ser and Thr residues. Here, using tandem MS analysis, we characterized the sites of parathyroid hormone (PTH)-induced NHERF1 phosphorylation and identified 10 high-confidence phosphorylation sites. Ala replacement at Ser46, Ser162, Ser181, Ser269, Ser280, Ser291, Thr293, Ser299, and Ser302 did not affect phosphate uptake, but S290A substitution abolished PTH-dependent phosphate transport. Unexpectedly, Ser290 was rapidly dephosphorylated and rephosphorylated after PTH stimulation, and we found that protein phosphatase 1α (PP1α), which binds NHERF1 through a conserved VxF/W PP1 motif, dephosphorylates Ser290. Mutating 257VPF259 eliminated PP1 binding and blunted dephosphorylation. Tautomycetin blocked PP1 activity and abrogated PTH-sensitive phosphate transport. Using fluorescence lifetime imaging (FLIM), we observed that PTH paradoxically and transiently elevates intracellular phosphate. Added phosphate blocked PP1α-mediated Ser290 dephosphorylation of recombinant NHERF1. Hydrogen–deuterium exchange MS revealed that β-sheets in NHERF1's PDZ2 domain display lower deuterium uptake than those in the structurally similar PDZ1, implying that PDZ1 is more cloistered. Dephosphorylated NHERF1 exhibited faster exchange at C-terminal residues suggesting that NHERF1 dephosphorylation precedes Ser290 rephosphorylation. Our results show that PP1α and NHERF1 form a holoenzyme and that a multiprotein kinase cascade involving G protein–coupled receptor kinase 6A controls the Ser290 phosphorylation status of NHERF1 and regulates PTH-sensitive, NPT2A-mediated phosphate uptake. These findings reveal how reversible phosphorylation modifies protein conformation and function and the biochemical mechanisms underlying PTH control of phosphate transport.
Keywords: PDZ domain, protein–protein interaction, protein phosphorylation, phosphoprotein phosphatase 1 (PP1), hydrogen–deuterium exchange, structural biology, NHERF1, NPT2A, phosphate transport
Introduction
Extracellular phosphate homeostasis in vertebrates is controlled largely by the kidneys, where parathyroid hormone (PTH)4 and fibroblast growth factor 23 (FGF23) regulate phosphate absorption mediated principally by the NPT2A sodium–phosphate transporter (SLC34A1) in a manner that requires the adapter protein Na+-H+ exchanger regulatory factor-1 (NHERF1, SLC9A3R1, known also as the 50-kDa ezrin–binding protein EBP50). NHERF1 is a widely expressed multifunctional protein that scaffolds integral membrane proteins with cytoplasmic proteins (1–4). It was first identified as a regulator of Na+-H+ exchange and as a binding partner for active ezrin (3, 5). Notable NHERF1 structural features include the presence of two tandem PDZ (Postsynaptic density 95/Disk large/Zonula occludens) domains and an ezrin-binding domain (EBD) (Fig. 1). PDZ domains are among the most common protein interaction modules in the human proteome (6). Class I PDZ proteins such as NHERF1 bind to proteins harboring a C-terminal motif having an ideal profile of the form (Asp/Glu)-(Ser/Thr)-Xaa-Φ, where Xaa is promiscuous, and Φ is a hydrophobic residue, generally Leu, Ile, or Val (7, 8).
Figure 1.
Domain structure of NHERF1. Linear representation showing tandem arrangement of PDZ1 and PDZ2, and EBD domain. Start and stop sites for each domain are shown along with the location of the 257VxF/W259 PP1 motif and 288SASSDTS294 Ser-rich cluster in the disordered linker region between PDZ2 and the EBD.
The NPT2A sodium–phosphate co-transporter possesses a canonical Class I type PDZ ligand (–Thr–Arg–Leu). Mutating the PDZ ligand disrupts binding to NHERF1 (9). PTH and FGF23 down-regulate NPT2A expression and function by distinct signaling pathways that converge at NHERF1 (10). Mice lacking NHERF1 display characteristic mineral-ion wasting and osteopenia (11, 12). Humans with NHERF1 inactivating mutations exhibit elevated phosphate excretion and a prominent bone phenotype with fractures (13, 14). Likewise, knockout of Npt2a5 (15) or SLC34A1 mutations disrupt phosphate metabolism with a constellation of mineral-ion and skeletal disorders (16, 17).
NHERF1 is a phosphoprotein possessing 31 Ser and 9 Thr residues. Although these sites are dispersed throughout the protein, there is a conspicuous Ser-rich cluster located in the linker region between PDZ2 and the EBD (Fig. 1). NHERF1 displays a combination of structurally defined and undefined regions. The two PDZ segments are highly organized, which permitted their structures to be solved by X-ray diffraction and solution NMR (18, 19). The linker regions separating the two PDZ domains and between PDZ2 and the EBD are intrinsically disordered (ID). This flexibility permits NHERF1 to assume open and closed conformations, wherein the C-terminal tail of NHERF1, itself a PDZ ligand (–Ser–Asn–Leu), engages the core-binding segment of PDZ2 in an intramolecular manner (20–22). The ID region between PDZ2 and the EBD may be unavailable for binding in the closed conformation. Previous work established that some human NHERF1 mutations, viz. R153Q, stabilize the closed NHERF1 conformation, preventing access of PKA regulatory subunits to bound ezrin that in turn interferes with hormone action and leads to renal phosphate wasting (13, 23). Introducing a second compensatory mutation of the NHERF1 PDZ ligand (−SNA) prevented formation of the closed conformation and overcame the nominally deleterious action of the inherited mutation on signaling and function. These observations regarding the Ser-rich cluster within the linker domain, combined with the finding that the closed NHERF1 conformation prevents engagement of PKA regulatory subunit binding and function, suggest that phosphorylation within this cluster may regulate hormone action by controlling access to these critical binding sites.
Compared with structurally determined rigid protein domains, ID regions contain a higher density of phosphorylation sites (24). Site-specific phosphorylation within ID regions, in turn, promotes structurally relevant conformational transitions that affect protein function (24, 25). Phosphorylation elicits diverse effects on the biological functions of proteins harboring ID regions by altering the conformational landscape and by stabilizing secondary structural elements (26). In silico analysis of NHERF1 predicts 22 putative phosphorylation sites (27). Constitutive and ligand-induced phosphorylation has been reported at Ser77 (28, 29), Thr95 (29), Thr156 (30, 31), Ser162 (32), Ser279/Ser301 (33, 34), Ser290 (35), and Ser339/340 (Table 1)6 (36, 37). Phosphorylation of these residues depends on an activating kinase such as PKC or CDK1 (Cdc2 kinase), which have been implicated in altered conformation and function (37, 38). In addition to PKC, PKA, GRK6A, and SGK1 are AGC family kinases involved in constitutive or PTH-mediated NHERF1 phosphorylation (28, 29, 35, 39). The described phosphorylation sites are associated with an array of actions ranging from intramolecular structural reorganization to physiological functions, including renal phosphate transport, cell division, and ion channel activity (Table 1). Notably, the majority of described phosphorylation sites are located in structurally defined NHERF1 PDZ domains.
Table 1.
Reported NHERF1 phosphorylation sites
Amino acid residue numbering corresponds to human NHERF1. CFTR is cystic fibrosis transmembrane conductance regulator.
| Site | Kinase | Proposed function | Refs. |
|---|---|---|---|
| Ser77, Thr95 | PKC | Renal phosphate transport | 28, 29, 102 |
| Thr156 | Akt | Cell division | 30 |
| Thr156 | RSK1 | Nuclear localization | 31 |
| Ser162 | PKC | CFTR gating | 32 |
| Ser280/Ser302 | Cdc2 | Cell division | 33, 34, 103 |
| CFTR expression | |||
| Ser290 | GRK6a | Unknown | 35 |
| Ser339/Ser340 | PKC | NHERF1 oligomerization and CFTR macromolecular assembly | 36, 37 |
Early reports showed that phosphorylation of Ser77 and Thr95 is essential for hormone-regulated phosphate transport (28, 29). Moreover, the diverse functional events regulated by NHERF1 phosphorylation raise the question whether a well-established phosphorylation pattern exists, where individual or arrays of phosphorylated residues are uniquely associated with the distinct cellular activities. However, the dynamic, time-dependent pattern of ligand-induced phosphorylation and dephosphorylation and its relation to the signaling, trafficking, and functional actions of NHERF1 are essentially unknown. Adding (phosphorylation) or removing (dephosphorylation) a phosphate group can impact protein structure at either a local or global level (40). In the absence of a full-length NHERF1 structure, such conformational changes upon phosphorylation can be probed by a single or combined biophysical and biochemical methods, including hydrogen–deuterium exchange MS (HDX-MS).
The unifying hypothesis of the present work is that phosphorylation regulates NHERF1 interactions with NPT2A to promote PTH-sensitive activity. To test this theory, we focused herein on identifying PTH-induced NHERF1 phosphorylation sites using an unbiased approach that combined MS and 32P labeling. SILAC-based MS revealed three different phosphorylation patterns of NHERF1. Site-directed mutagenesis of identified phosphorylation sites by Ala replacement showed that only endogenous confirmed Ser290 is relevant to PTH-inhibited phosphate uptake. Rapid dephosphorylation of this site is highly time-coincident with an acute cessation of phosphate uptake in the presence of high concentrations of extracellular phosphate, as observed by real-time measurement of phosphate in living cells using fluorescence lifetime imaging (FLIM). HDX-MS was applied to probe the conformational changes upon Ser290 dephosphorylation. We demonstrate that Ser/Thr-protein phosphoprotein phosphatase 1α (PPP1CA, PP1α) acts as a holoenzyme with NHERF1 and is responsible for dephosphorylating Ser290. Our data suggest that PTH-induced reversible phosphorylation at Ser290 serves as an on/off switch to regulate NPT2A-mediated phosphate uptake. Obtaining sufficient phosphorylation-dependent structural dynamics will help with understanding how protein phosphorylation functions in the regulation of cellular signaling and functioning in general and the biochemical mechanisms underlying PTH regulation of phosphate transport in particular.
Results
Identification of NHERF1 phosphorylation sites
HEK293 GnTI− cells were stably transfected with TAP–NHERF1 and FLAG–PTHR to generate sufficient NHERF1 protein to map phosphorylation sites by MS. Because they produce higher protein yields than other cell lines and their ability to be grown in suspension permitting operational scale-up, GnTI− cells are a convenient tool for overexpressing membrane proteins for biochemical and related analyses (41). They lack N-acetylglucosaminyltransferase I (GnTI) that is required for processing complex N-glycans. PTHR possesses four N-glycosylation sites, and the absence of GnTI restricts PTHR glycosylation to a single Man5GlcNAc2. This residual glycosylation, however, is sufficient for PTHR function (42). PTH(1–34) elicited increases of intracellular cAMP (Fig. 2A) and calcium (Fig. 2B), two PTHR core signaling events, in the NHERF1–PTHR double stable cell line. The results validate the use of FLAG–PTHR–TAP–NHERF1 GnTI− cells to investigate PTH-induced NHERF1 phosphorylation.
Figure 2.
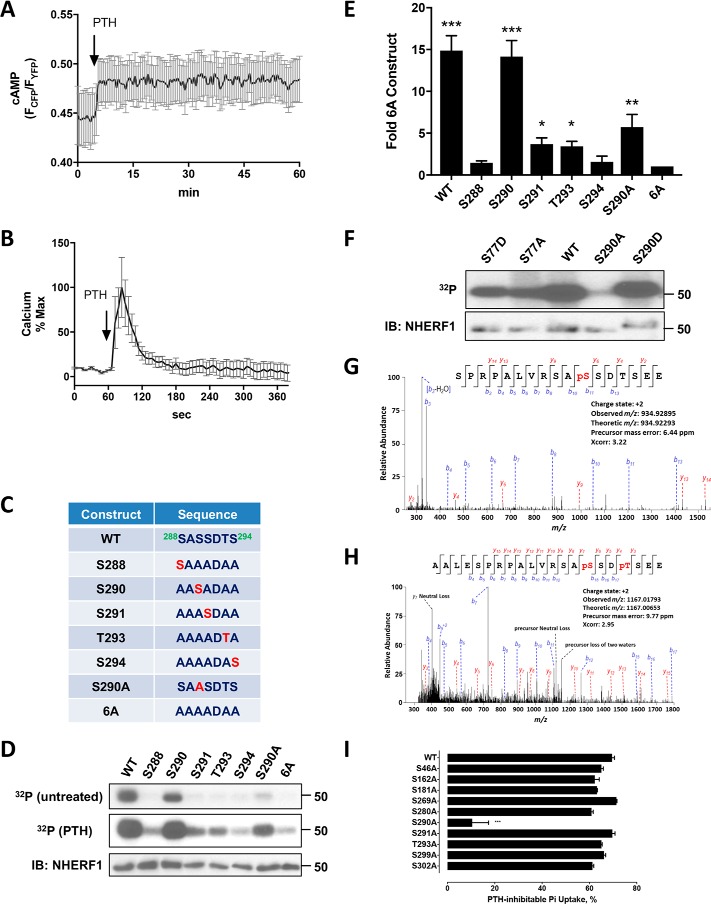
Identification and function of NHERF1 phosphorylation sites. A, cAMP in double-stable FLAG–PTHR–TAP–NHERF1 GnTI− cells transiently transfected with the cAMP FRET sensor EPACCFP/YFP (104). 100 nm PTH(1–34) was added at the time indicated by the arrow. Data represent the mean ± S.E. of three independent experiments. B, calcium changes in double-stable GnTI− cells transiently transfected with the pCMV-R-GECO1.2 calcium sensor (105). 100 nm PTH(1–34) was added at the indicated time. Epifluorescence was monitored at 585 nm emission following 488 nm excitation as detailed under “Experimental procedures.” Results are the mean ± S.E. of three independent experiments. C, WT and engineered mutant constructs of the 288SASSDTS294 Ser-rich cluster. D, 32P incorporation in specified FLAG–NHERF1 constructs (top, middle rows) individually transfected into TAP–PTHR-stable GnTI− cells. FLAG–NHERF1 was purified as described under “Experimental procedures” and resolved by SDS-PAGE followed by autoradiography. In untreated cells (upper row), only Ser290 exhibited appreciable 32P incorporation, similar to wildtype (WT) NHERF1. Upon PTH treatment (middle row), Ser290, Ser291, and Thr293 exhibited noticeable 32P incorporation compared with the 6A construct. Total NHERF1 levels are shown in the lower row. Results represent three independent experiments. E, quantitative analysis of PTH-treated samples in D. Densitometry signals for the various constructs were normalized to the amount of WT–NHERF1. In the presence of 100 nm PTH, WT–NHERF1 (WT), Ser290 (S290), Ser291 (S291), Thr293 (T293), and S290A exhibited significant phosphorylation compared with the 6A construct. Results are shown as means ± S.D. (n = 3; *, p < 0.05; **, p < 0.01; ***, p < 0.001). IB, immunoblot. F, 32P metabolic labeling of WT and targeted Ser77 and Ser290 mutants. Labeling and purification were as described above and detailed under “Experimental procedures.” The illustrated Western blotting is representative of three separate experiments. G, MS/MS fragmentation spectrum showing constitutive Ser290 phosphorylation in human RPTEC cells with endogenous NHERF1 expression. Peak heights show the relative abundance of the corresponding fragmentation ions, with the annotation of the identified matched N terminus containing b ions in blue and the C terminus containing y ions in red. H, Ser290 and Thr293 phosphorylation in NHERF1 from RPTEC cells treated for 5 min with 100 nm PTH(1–34). I, PTH-inhibitable phosphate uptake in OKH cells transiently expressing the specified NHERF1 construct. S290A reduced uptake by 90%. Other tested NHERF1 constructs exhibited less than a 10% inhibitory effect. Results report the mean ± S.E. (n = 3; ***, p < 0.001, ANOVA).
NHERF1 expressed in double-stable GnTI− cells was purified and subjected to in-solution digestion by trypsin or GluC as detailed under “Experimental procedures.” GluC cleaves peptide bonds C-terminal to Glu and was used to enhance and complement coverage of potential phosphorylation sites, including Ser339/Ser340 that are not covered by trypsin digestion, which cleaves peptide bonds at the C-terminal side of Lys or Arg, except when followed by Pro. Digesting protein samples with multiple proteases improves sequence coverage compared with single protease cleavage (43). The resulting phosphopeptide mixtures were enriched with titanium dioxide (TiO2) beads and analyzed by MS. The 10 most frequently phosphorylated Ser and Thr residues assigned by phosphoRS (44) with confidences greater than 75% site possibility are listed in Table 2. The results confirm several previously reported phosphorylation events at Ser162, Ser280, Ser290, and Ser302 (Table 1) and reveal six functionally undefined phosphorylated residues at Ser46, Ser181, Ser269, Ser291, Thr293, and Ser299, consistent with the sites deposited in the PhosphoSitePlus database (45). Notably, the majority of these newly identified phosphorylation sites map to the linker region between PDZ2 and the EBD (Fig. 1).
Table 2.
Identified NHERF1 phosphorylation sites
Overexpressed TAP–NHERF1 was purified as described under “Experimental procedures” and digested by GluC (cleavage site, Asp or Glu) or trypsin (cleavage site, Arg or Lys). Samples were loaded on an LTQ Orbitrap XL mass spectrometer. Data were searched using Sequest (99) against a human sequence database. Xcorr values indicate the proximity of the observed spectrum to an ideal spectrum for the matched peptide. Parts/million (ppm) indicate the difference between the observed peptide mass and the expected peptide mass calculated from the peptide sequence. The confidence of phosphorylation site localization was evaluated with phosphoRS 2.0 (44) implemented in Proteome Discoverer (ThermoFisher Scientific). The phosphoRS score is based on the cumulative binomial probability (0–100%) that the observed match is a random event. Values >75% are confidence scores, good evidence of true phosphorylation sites.
| Position | Identified peptide | Protease | Charge | Xcorr | ppm | PhosphoRS site probability |
|---|---|---|---|---|---|---|
| 46 | R.LVEPGSPAEK.A | Trypsin | 2 | 1.972 | 1.13 | 100 |
| 162 | K.KGPSGYGFNLHSDK.S | Trypsin | 2 | 2.057 | 9.69 | 92.0 |
| 181 | R.SVDPDSPAEASGLR.A | Trypsin | 2 | 2.192 | 0.72 | 100 |
| 269 | E.NSREALAE.A | GluC | 2 | 2.446 | −4.64 | 100 |
| 280 | R.EALAEAALESPRPALVR.S | Trypsin | 2 | 2.565 | −2.66 | 100 |
| 290 | R.SASSDTSEELNSQDSPPK.Q | Trypsin | 2 | 4.616 | 3.73 | 92.4 |
| 291 | R.SASSDTSEELNSQDSPPK.Q | Trypsin | 2 | 4.529 | −0.06 | 92.9 |
| 293 | R.SASSDTSEELNSQDSPPK.Q | GluC | 2 | 4.711 | −1.30 | 86.4 |
| 299 | E.LNSQDSPPKQDSTAPSSTSSSDPILD.F | GluC | 2 | 4.052 | −3.21 | 87.2 |
| 302 | E.LNSQDSPPKQD.S | GluC | 2 | 3.453 | 0.12 | 100 |
We metabolically labeled NHERF1 with 32P to validate the phosphorylation sites identified by MS in the upstream Ser-rich cluster: 288Ser-Ala-Ser-Ser-Asp-Thr-Ser294. To exclude the uncertainty arising from the presence of neighboring Ser or Thr residues, we mutated all Ser/Thr in the cluster to Ala and then singly reverted to them to their original form. These constructs were designated Ser288 (S288), Ser290 (S290), Ser291 (S291), Thr293 (T293), and Ser294 (S294) to indicate the restored native residue (Fig. 2C). These constructs, tagged with FLAG, were individually transfected into GnTI− cells and metabolically labeled with 32P. Under control conditions, WT and Ser290 constructs were phosphorylated. Upon PTH(1–34) treatment, WT and Ser290 constructs displayed greater phosphorylation, and the other constructs showed varying levels of phosphorylation. Ser288, Ser291, Thr293, and Ser294 constructs exhibited limited phosphorylation compared with the WT construct or to Ser290 (Fig. 2D). Mutating all Ser/Thr residues in this cluster to Ala (6A) substantially reduced phosphorylation. The Ser290 construct displayed 32P incorporation comparable with WT–NHERF1 reinforcing the view that Ser290 is the primary phosphorylation site within this cluster and in the apoprotein (35). S290A displayed reduced 32P incorporation compared with WT. This difference in phosphorylation may arise from phosphorylation of other sites such as Ser291 or Thr293 in the WT sequence, as shown in Fig. 2, D and E. Thus, 32P incorporation at Ser290, Ser291, and Thr293 is consistent with the MS analysis. Several previously identified phosphorylation sites (Ser77, Thr95, Thr156, and Ser339,340) (Table 1) were not detected here. Ser77 and Thr95 phosphorylation have not been consistently demonstrated by MS (36). The inability to identify post-translationally modified sites, and phosphorylation in particular, by MS may arise, at least in part, by incomplete coverage following GluC or trypsin digestion or by rapid dephosphorylation with attendant loss of the transient phosphorylation event. This explanation is supported by in vivo metabolic 32P labeling coupled with mutagenesis. As shown in Fig. 2F, Ala or Asp replacement at Ser77 caused equivalently modest reductions in phosphorylation compared with WT–NHERF1, whereas Ala substitution at Ser290 greatly reduced phosphorylation. Asp mutants displayed somewhat slower electrophoretic mobility, likely due to replacing Ser/Thr with negatively charged Asp (46). Unexpectedly, the phosphomimic S290D construct exhibited phosphorylation comparable with WT–NHERF1 (Fig. 2F) suggesting that Ser290 phosphorylation might be important for preserving a favorable conformation to facilitate phosphorylation of other residues, likely including Ser77, a site required for PTH action (28).
To ensure that NHERF1 phosphorylation observed in GnTI−–FLAG–PTHR–TAP–NHERF1 cells did not arise from overexpression, we examined the pattern of NHERF1 phosphorylation in human renal proximal tubule cells (RPTEC), which constitutively express endogenous NHERF1 and PTHR (10). We developed a procedure to purify NHERF1 from these cells by preparing FERM-agarose beads as described under “Experimental procedures.” Notably, Ser290 was phosphorylated both in the absence (Fig. 2G) and presence (Fig. 2H) of PTH treatment, suggesting that Ser290 is constitutively phosphorylated (35). PTH exposure promoted Thr293 phosphorylation (Fig. 2H). Most of phosphorylation sites identified in GnTI−–FLAG–PTHR–TAP–NHERF1 cells could not be verified at endogenous expression levels in RPTEC, presumably due to insufficient mass of phosphopeptides available from cultures of these slow-growing native cells.
PTH inhibits phosphate absorption mediated by the sodium–phosphate co-transporter NPT2A/Npt2a. NHERF1 is essential for this process (10, 39, 47, 48). To determine which of the identified NHERF1 phosphorylation sites were involved in PTH action on phosphate transport, we generated phosphoresistant mutants of the identified sites: S46A, S162A, S181A, S269A, S280A, S290A, S291A, T293A, S299A, and S302A, and we evaluated their participation in hormone-sensitive phosphate transport. For these experiments, we used opossum kidney cells, an accepted model for hormone-regulated phosphate transport (49). The OKH cell strain lacks NHERF1 and expressing exogenous NHERF1 rescues PTH-sensitive phosphate transport (50, 51). Use of NHERF1-deficient OKH cells permitted testing the phosphoresistant NHERF1 constructs. The various constructs were individually transfected into OKH cells. Basal and PTH-sensitive phosphate uptake was measured as described under “Experimental procedures.” The results (Fig. 2I) showed that Ala substitution at Ser46, Ser162, Ser181, Ser269, Ser280, Ser291, Thr293, Ser299, or Ser302 did not interfere with PTH-sensitive phosphate uptake compared with WT–NHERF1. In contrast, replacing Ala at Ser290 (S290A) decreased PTH-sensitive phosphate uptake by 90%. Hence, phosphorylation of Ser290 plays a critical role in regulating NPT2A-mediated phosphate uptake.
PTH promotes dynamic NHERF1 phosphorylation
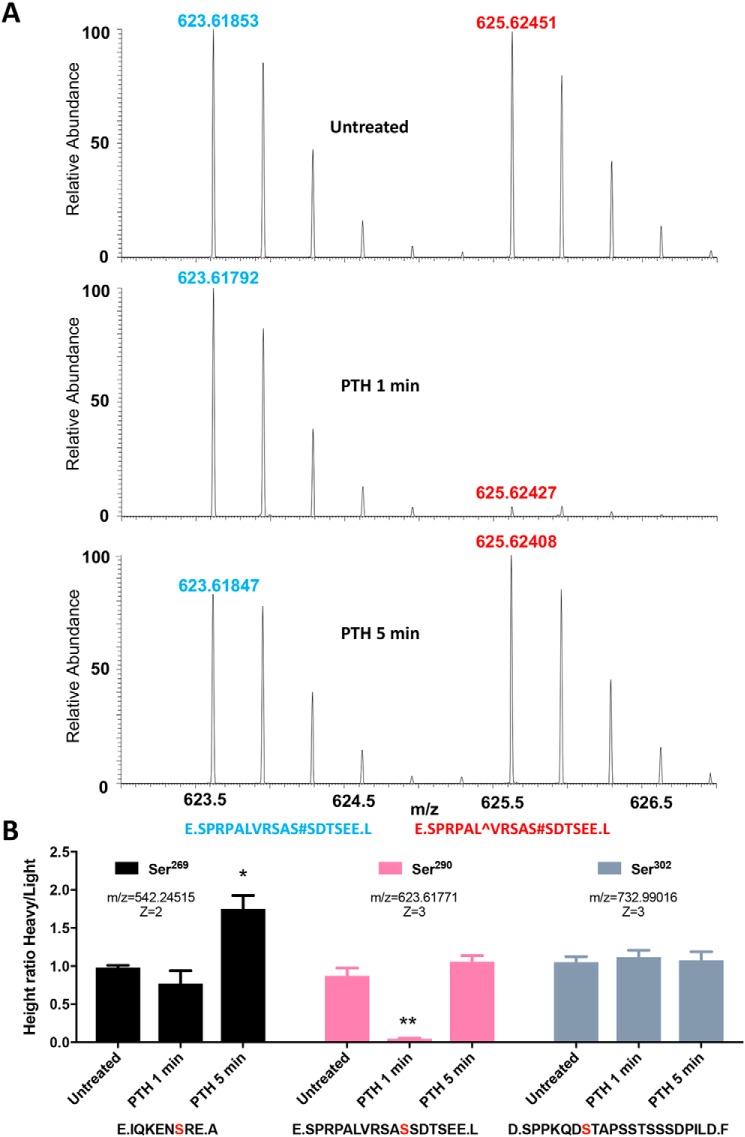
Mass spectrometry analysis using stable isotope labeling by amino acids in cell culture (SILAC) was performed to quantify the magnitude of PTH stimulation of site-specific NHERF1 phosphorylation. HEK293 GnTI− cells stably expressing FLAG–PTHR and TAP–NHERF1 were grown in parallel in SILAC medium containing l-Lys-12C6 and l-Leu-12C6 (light) or l-Lys-13C6 and l-Leu-13C6 (heavy). After labeling to isotopic equilibrium, cells grown in heavy medium were treated with 100 nm PTH for 0, 1, or 5 min, whereas cells propagated in light medium were not exposed to PTH and served as controls. The treatment time was chosen because maximal PTH stimulation of cAMP and Ca2+ occurred within 0.5–5 min (Fig. 2, A and B). Equal amounts of lysates prepared from cells grown in light and heavy media were combined. NHERF1 was purified, digested, and analyzed by MS. The resulting spectra appeared as a series of peptide pairs (Fig. 3A) allowing quantification of relative peptide abundance under control and PTH-treated conditions. The spectrum for a representative peptide, 280SPRPALVRSASSDTSEE296, harboring phosphorylated Ser290 (pSer290) is shown in Fig. 3A without treatment and following 1 or 5 min exposure to PTH. The ratio of the MS signal intensity of the paired light (Fig. 3A, blue) and heavy (red) phosphopeptides corresponds to the difference in their relative abundance and the change in phosphorylation. Using this SILAC-based quantitative analysis, we found Ser269 phosphorylation increased at 5 min (Fig. 3B, left panel). Among the 10 phosphorylation sites identified by MS, Ser290 displayed an unexpected and conspicuous dynamic time-dependent dephosphorylation at 1 min of PTH treatment. Ser290 phosphorylation decreased by 90% at 1 min (p < 0.01) and was fully rephosphorylated at 5 min (Fig. 3B, center panel). These results were qualitatively and quantitatively verified independently using an anti-pSer290 antibody (Fig. S1, A–C). Ser302 phosphorylation was unchanged in response to PTH (Fig. 3B, right panel). Additionally, no discernible phosphorylation pattern was found for other identified sites, suggesting these residues are not involved in early PTH responses. We next sought to determine the origin and functionality of the cyclical pattern of pSer290 dephosphorylation and rephosphorylation.
Figure 3.
Quantitative analysis of PTH-stimulated NHERF1 phosphorylation. A, representative MS spectra generated using SILAC. FLAG–PTHR–TAP–NHERF1 GnTI− cells grown in medium containing heavy l-Leu-13C6 and l-Lys-13C6 and treated with vehicle or 100 nm PTH(1–34) for 1 or 5 min. Control cells were cultured in normal l-Leu-12C6- and l-Lys-12C6-containing media without PTH exposure. Cell lysates were prepared as outlined under “Experimental procedures” and were digested by either trypsin or GluC. Light- and heavy-labeled peptides are shown for the E.SPRPALVRSAS#SDTSEE.L fragment in untreated cells or after 1 or 5 min of PTH exposure. Identified phosphorylation sites are marked with “#”, and heavy l-Leu-13C6 is indicated as “[caret]”. Mass spectra of heavy peptides containing l-Leu-13C6 have an increased mass of 6 Da and are shifted to the right, compared with the corresponding light peptide spectrum, by m/z of 2 caused by a +3 ionization. The ratio of labeled heavy (red)/light (blue) for each sample was calculated by comparing the differences in the respective mass spectra peaks. B, summary of dynamic phosphorylation changes of the identified sites. The phosphorylation pattern for Ser269, Ser290, and Ser302 is shown in untreated cells and at 1 and 5 min following PTH. The phosphorylation sites in each peptide are highlighted in bold red. Results are shown as means ± S.E. (n = 3–4; *, p < 0.05; **, p < 0.01, ANOVA).
Protein phosphatase 1α (PP1α) binds NHERF1 and mediates Ser290 dephosphorylation
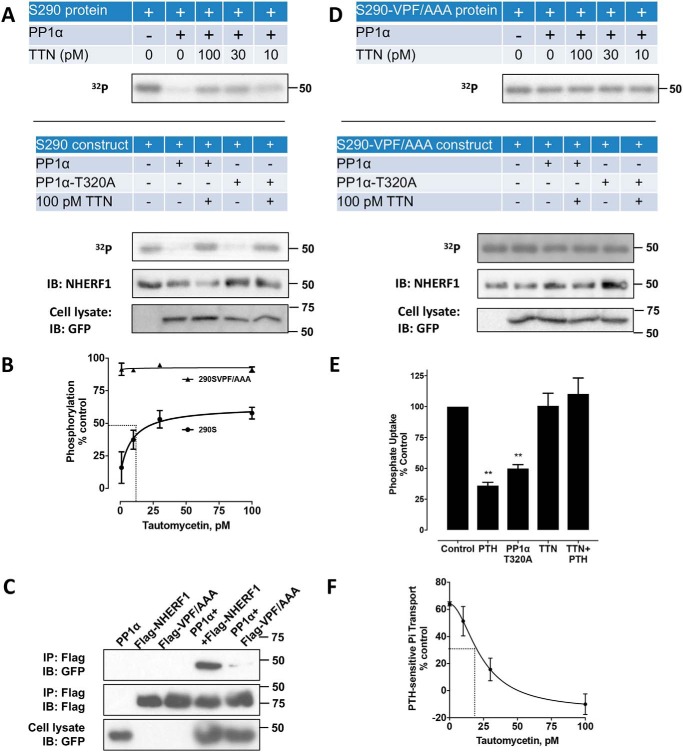
We first directed our attention at identifying the phosphatase responsible for the rapid Ser290 dephosphorylation. Our interest centered on PP1α because it was shown to bind NHERF1 (52) and was found here to be among the most frequent interacting NHERF1 partners (Table S1). Mass spectrometry analysis of pulldown samples revealed 10 unique PP1α peptides with greater than 45% sequence coverage (Table S2). We tested the hypothesis that PP1α dephosphorylates pSer290 in vivo by co-transfecting PP1α and NHERF1 constructs into cultured GnTI− cells or in vitro using recombinant PP1α and NHERF1. For the in vitro studies FLAG–NHERF1–Ser290 protein (288AASADAA294) expressed in HEK293 GnTI− cells was metabolically labeled with 32P, purified with anti-FLAG beads, and eluted with 3× FLAG peptide. The resulting protein was incubated with or without PP1α in the presence of various concentrations of TTN, a specific PP1 inhibitor (53). As shown in Fig. 4A (top panel), PP1α virtually abolished Ser290 phosphorylation. With increasing concentrations of TTN (0–100 pm), dephosphorylation by PP1α was inhibited with an IC50 of ∼10 pm (Fig. 4B). The results were confirmed in situ (Fig. 4A, bottom panel) by transfecting the Ser290 construct alone or with WT PP1α or with constitutively active PP1α-T320A into GnTI− cells that were then labeled with 32P. Compared with control, pSer290 was extensively dephosphorylated upon exposure to either WT PP1α or PP1α-T320A. Consistent with the in vitro dephosphorylation, 100 pm TTN virtually abolished dephosphorylation by PP1α. Inhibition of pSer290 dephosphorylation by TTN was further confirmed in vivo using an anti-pSer290 antibody (Fig. S1D). These findings are compatible with the conclusion that Ser290 is the primary NHERF1 phosphorylation site (Fig. 2D) and that PP1α is responsible for the decreased 32P incorporation due to dephosphorylation.
Figure 4.
PP1α-catalyzed NHERF1 dephosphorylation and effects on phosphate transport. A, Ser290 dephosphorylation by PP1α in vitro (top panel) and in vivo (lower panel). For in vitro dephosphorylation, equal amounts of purified 32P-labeled FLAG–Ser290 were incubated with or without PP1α in the presence of the indicated concentration of the PP1α inhibitor TTN. For in vivo dephosphorylation, the FLAG–Ser290 construct alone or together with native GFP–PP1α or constitutively active GFP–PP1α–T320A was transfected into HEK293 GnTI− cells. 48 h after transfection, cells were incubated with 100 pm TTN for 30 min where indicated and labeled with 32P. Purified protein was separated on SDS-PAGE. Assay details are described under “Experimental procedures.” Results represent three independent experiments. B, quantitative analysis of Fig. 4A (top panel). At an optimum concentration of PP1α (∼30 nm), TTN blocked PP1α dephosphorylation of recombinant FLAG–Ser290 in a concentration-dependent manner with an IC50 of about 10 pm. C, PP1α binds to the 257VPF259 motif in NHERF1. FLAG–WT–NHERF1 or the mutant VPF/AAA NHERF1 construct was transfected alone or together with GFP–PP1α into GnTI− cells expressing TAP–PTHR. Similar findings were obtained in three independent experiments. D, dephosphorylation of mutant NHERF1 by PP1α in vitro (top panel) or in vivo (lower panel). The experiments were performed using a similar protocol to A except with the VPF/AAA construct (Ser290-VPF/AAA). All blots or dried gels are illustrative of three separate experiments. Notably, PP1α had no discernable effect on Ser290 dephosphorylation in the presence or absence of TTN. Quantitative analysis for in vitro dephosphorylation (top panel) using recombinant Ser290-VPF/AAA protein is shown in B. E, PTH-inhibitable phosphate uptake in OK cells. Overexpressing constitutively active PP1α-T320A inhibited phosphate uptake nearly as extensively as did PTH treatment. Blocking phosphatase activity with 100 pm TTN in the presence of 100 nm PTH(1–34) abolished PTH action. Results are the means ± S.D. (n = 3; **, p < 0.01, ANOVA). F, concentration/response of TTN on PTH-inhibitable phosphate transport assayed in OK cells. TTN exhibited an IC50 of 20 pm.
Solid-phase binding assays excluded the possibility that PP1, which lacks a canonical PDZ-recognition sequence, engaged NHERF1 through PDZ1 or PDZ2 (Fig. S2). Careful examination revealed that NHERF1 possesses a previously unrecognized VxF (257VPF259) PP1-binding motif (54, 55) located within the linker region between the PDZ2 and the EBD domain (Fig. 1). Co-immunoprecipitation experiments illustrated in Fig. 4C show that PP1α extensively bound NHERF1, and this interaction was effectively eliminated upon mutating 257VPF259 to 257AAA259. Furthermore, the binding-defective VPF/AAA NHERF1 mutant was refractory to pSer290 dephosphorylation by PP1α in vitro (Fig. 4, B and D, top panel) or in vivo (Fig. 4D, lower panel) either in the presence or absence of TTN. These results support the view that PP1α engages NHERF1 through the 257VPF259 motif and that binding to NHERF1 is required for PP1α phosphatase activity at Ser290.
The findings thus far demonstrate that Ser290 phosphorylation is required for the inhibitory action of PTH on NPT2A (Fig. 2H). We next sought to determine whether pSer290 dephosphorylation is likewise necessary for PTH action. As expected, TTN, which blocked pSer290 dephosphorylation (Fig. 4A), likewise abolished PTH-sensitive phosphate transport (Fig. 4E) with an IC50 of 20 pm (Fig. 4F), similar to the inhibitory action on dephosphorylation (Fig. 4B).
We reasoned that if the inhibitory action of PTH on phosphate transport required PP1α-mediated pSer290 dephosphorylation, then constitutively active PP1α should mirror the biological effect of PTH. Overexpressing constitutively active PP1α-T320A (52, 56) inhibited phosphate uptake almost as much as did PTH (Fig. 4E). These results support the view that pSer290 dephosphorylation is a necessary step in hormone-dependent phosphate transport and that dephosphorylation is mediated by PP1α. Thus, PTH initiates a phospho-transfer cycle at Ser290 in NHERF1 that is essential for hormone action.
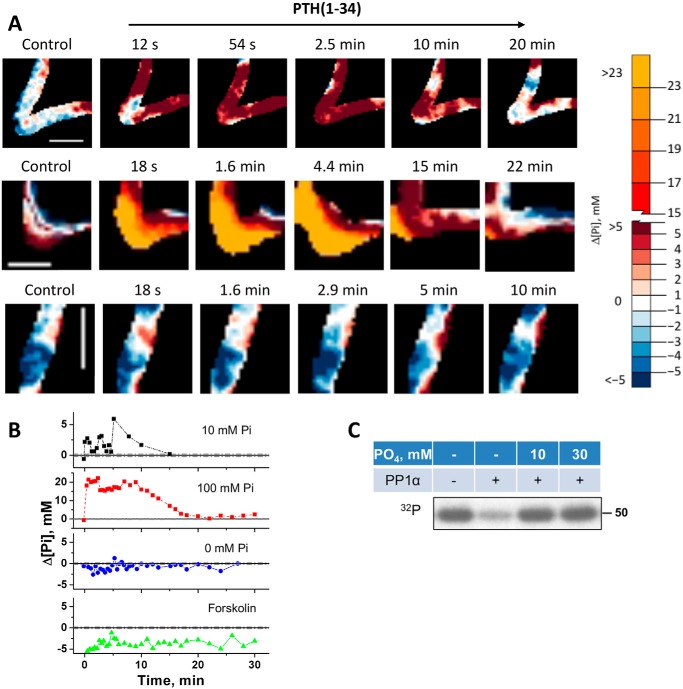
The rapid dephosphorylation of Ser290 and its requirement for PTH action led us to question whether this effect stemmed from a signaling event or was associated with phosphate transport. To examine the relationship between Ser290 phosphorylation status and NPT2A-mediated phosphate uptake, we measured changes of intracellular phosphate in live cells using FLIM. Here, we used mouse MC3T3-E1 preosteoblasts, which constitutively express PTHR, Npt2a, and NHERF1 (57, 58), and are well-established for live-cell imaging of phosphate measurement (59) using a reporter sensitive to total phosphate concentration (Figs. S3 and S4). Unexpectedly, addition of 100 nm PTH(1–34) at normal or high extracellular phosphate concentrations in the presence of sodium rapidly but transiently increased intracellular phosphate (Fig. 5A and Figs. S3–S6). In PBS (∼12 mm phosphate), intracellular phosphate increased by 3–6 mm, whereas in 100 mm extracellular phosphate, the local intracellular concentration increased by 20 mm. This action was sustained for some 10 min and was followed by a remarkable decline to resting levels thereafter (Fig. 5, A and B; Figs. S3–S6). In the absence of sodium, no increase of intracellular phosphate was observed (data not shown), consistent with the view that the rise of intracellular phosphate had its origin in sodium-dependent phosphate entry. This is consistent with the fact that phosphate uptake mediated by NPT2A is sodium-dependent (23, 60). The possibility that the rise of intracellular phosphate stemmed from a signaling event was excluded by the finding that phosphate liberated by ATP hydrolysis upon forskolin-activated adenylyl cyclase did not detectably increase intracellular phosphate (Fig. 5B). These results suggest that extracellular phosphate is the primary source of Npt2a-mediated intracellular phosphate. The prompt decrease of phosphate within 1–5 min in the presence of 10 or 100 mm phosphate (Fig. 5, A and B) may be associated with the rapid dephosphorylation of Ser290 at 1 min.
Figure 5.
Intracellular phosphate in living cells and autoregulatory inhibition. A, representative FLIM images of MC3T3-E1 preosteoblast cells loaded with the fluorescent dye 2Me-4OMe-TM. Results are shown before and after treatment with 100 nm PTH(1–34) for the time indicated. Results show changes when the extracellular buffer contained 10 mm phosphate (PBS, upper row), 100 mm phosphate (center row), or was phosphate-free (bottom row). Scale bars, 5 μm. The color scale indicates changes of the intracellular phosphate concentration (Δ[Pi]). B, quantitative summary of changes of intracellular phosphate (Δ[Pi], mm) from A. The findings indicate a rapid spike in the presence of phosphate followed by small (10 mm Pi; black) or large (100 mm Pi; red) sustained increase of ΔPi at 1–5 min followed by decreased intracellular phosphate after 10 min. No burst of phosphate uptake was evident in the absence of extracellular phosphate (blue). Directly activating adenylyl cyclase with forskolin did not change ΔPi (green). Experiments were performed in triplicate with equivalent results (see supporting information Fig. S6). C, phosphate inhibits PP1α phosphatase activity. Purified 32P-labeled FLAG–Ser290 was incubated with PP1α for 30 min as described in Fig. 4. 10 mm (PBS) or 30 mm phosphate inhibited Ser290 dephosphorylation.
The unanticipated transient elevation of intracellular phosphate raised the question whether it served as an autoregulatory mechanism to terminate phosphatase activity. We tested this theory by measuring the effect of phosphate on PP1α-mediated Ser290 dephosphorylation using purified 32P-labeled FLAG–Ser290, harboring Ser290 but with the surrounding Ser/Thr residues replaced by Ala (Fig. 2C) as described earlier. The results illustrated in Fig. 5C show that exogenous phosphate added at concentrations comparable with those measured in cells (Fig. 5, A and B) inhibited pSer290 dephosphorylation supporting the conclusion that PP1α action is auto-inhibited by intracellular phosphate, then allowing rephosphorylation of Ser290.
pSer290 dephosphorylation reduces NHERF1 binding to NPT2A
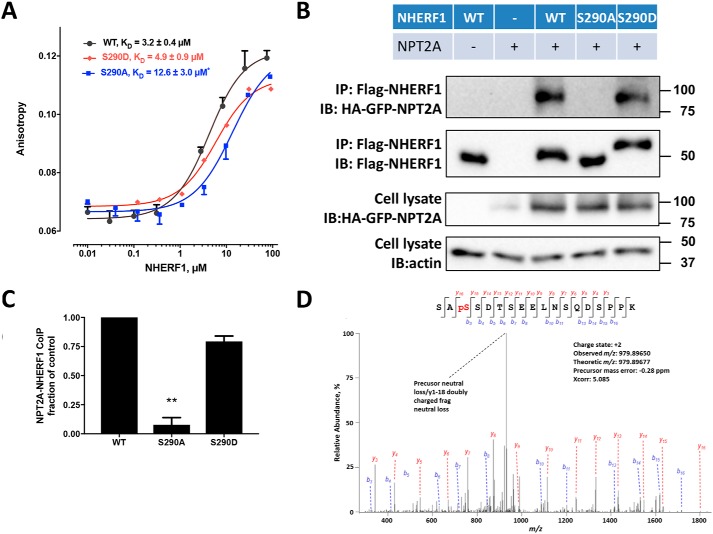
Direct interaction between NHERF1 and NPT2A is required for phosphate absorption (50). We therefore wondered whether NHERF1 Ser290 phosphorylation affects NPT2A binding. To address this question, we prepared S290D phosphomimic and S290A phosphomutant recombinant proteins by site-directed mutagenesis. Fluorescence anisotropy was used to measure the affinity of WT–NHERF1, S290A, and S290D constructs for NPT2A. For these measurements, we employed a 22-amino acid, human C-terminal NPT2A peptide labeled with carboxytetramethylrhodamine (TAMRA) as outlined under “Experimental procedures” and detailed previously (61). As shown in Fig. 6A, purified WT–NHERF1 had a KD = 3.2 μm for NPT2A, which was not significantly different from the S290D phosphomimic (KD = 4.9 μm). The S290A phosphomutant, however, exhibited significantly weaker binding to NPT2A (KD = 12.6 μm). This observation was independently confirmed by co-immunoprecipitation assays (Fig. 6, B and C), where the NHERF1 S290A phosphomutant exhibited diminished interaction with NPT2A compared with the NHERF1 S290D phosphomimic.
Figure 6.
NHERF1 Ser290 dephosphorylation disrupts NPT2A binding. A, binding of NPT2A to wildtype (WT), S290D, or S290A NHERF1. Fluorescence anisotropy measurements were performed as detailed under “Experimental procedures” and elsewhere (61). TAMRA-labeled 22-residue NPT2A C-terminal peptide was incubated with the indicated form of NHERF1 for 15 min at room temperature. Affinities are shown as means ± S.E. (n = 3; *, p < 0.05 versus WT). B, NPT2A co-immunoprecipitation (IP) with WT–NHERF1 (WT), S290A, or S290D mutants. HEK293 GnTI− cells were transfected with the indicated FLAG-tagged NHERF1 construct and HA-GFP-NPT2A. Cell lysates were prepared 48 h later and were analyzed by immunoblot (IB). WT and S290D NHERF1 exhibited stronger binding to NPT2A than S290A, consistent with the fluorescence anisotropy experiments in A. Western blottings are illustrative of three independent experiments. C, quantitative summary of NPT2A binding to the indicated NHERF1 construct. Results are means ± S.E. (n = 3; **, p < 0.01, ANOVA). D, MS/MS fragmentation spectrum for the Ser288–Lys305 NHERF1 peptide showing phosphorylated Ser290. Protein was prepared from E. coli as described under “Experimental procedures.”
In the absence of NHERF1, GnTI− cells express relatively little NPT2A (Fig. 6B) due to constitutive NPT2A internalization and down-regulation (11), as described previously (42). Remarkably, in both fluorescence anisotropy and co-immunoprecipitation experiments, NPT2A displayed somewhat higher binding to WT–NHERF1, essentially equivalent to the phosphomimic S290D, and much stronger than to the phosphoresistant S290A as reflected by the higher KD of 3.2 μm in fluorescence anisotropy measurements (Fig. 6A) and by more extensive pulldown of HA–NPT2A (Fig. 6B). Additional MS analysis disclosed that Ser290 from overexpressed WT–NHERF1 is phosphorylated (Fig. 6D), which explains the greater binding affinity.
GRK6A-mediated Ser290 rephosphorylation is required for NPT2A binding to NHERF1 and hormone-regulated phosphate transport
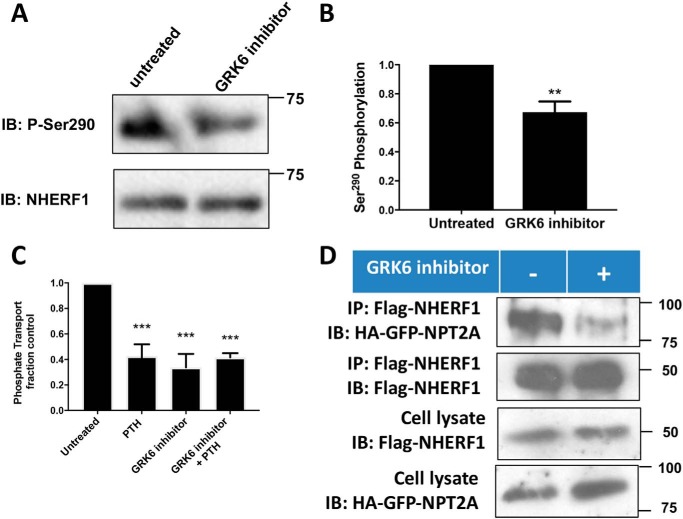
Having identified the mechanism of Ser290 dephosphorylation and its role in hormone-sensitive phosphate transport, we turned our attention to Ser290 phosphorylation. Grk6a constitutively phosphorylates Ser290 (35), but the biological relevance of this activity was unknown. Using a pSer290 antibody, we confirmed that NHERF1 is phosphorylated in the absence of prior treatment (Fig. 7, A and B). Constitutive phosphorylation of overexpressed NHERF1 was reduced by application of a cell-permeable small molecule GRK6 kinase inhibitor. Notably, disruption of GRK6A-mediated Ser290 phosphorylation decreased basal phosphate uptake and abolished PTH action (Fig. 7C). GRK6 inhibition of Ser290 phosphorylation likewise decreased NHERF1 binding to NPT2A (Fig. 7D), similar to the effect of the phosphoresistant S290A NHERF1 analog (Fig. 6B). Thus, both dephosphorylation of Ser290 by PP1α and its phosphorylation by GRK6A are required for NPT2A binding and regulatory control of phosphate transport. These findings motivated us to examine the structural features of NHERF1 binding to NPT2A and NHERF1 conformational changes associated with phosphorylation cycling at Ser290.
Figure 7.
Ser290 phosphorylation and effect on basal and PTH-inhibitable phosphate uptake. Double-stable GnTI− cells expressing TAP–NHERF1 and FLAG–PTHR were grown on 6-cm dishes for 48 h. Cells were treated with vehicle or 10 μm GRK6 inhibitor OICR0009944A02 (K3) for 30 min. NHERF1 protein was purified using streptavidin beads and analyzed by immunoblotting (IB). A, representative blot from three independent experiments. B, quantitative summary of A. GRK6 inhibitor treatment decreased Ser290 phosphorylation. Results are means ± S.D. (n = 3; **, p < 0.01). C, GRK6 inhibition abolished phosphate uptake by OK cells. Phosphate transport was measured as described under “Experimental procedures.” Results are means ± S.D. (n = 3–4; **, p < 0.01, ANOVA). D, GRK6 inhibitor impairs NHERF1 binding to NPT2A. HEK293 cells were transfected with FLAG–NHERF1 and HA–GFP–NPT2A. IP, immunoprecipitation. 48 h post-transfection, cells were serum-starved and followed by 10 μm GRK6 inhibitor treatment for 1 h. Immunoblot analysis was performed using protein purified with anti-FLAG beads.
Structural determinants of NHERF1 binding to NPT2A and conformational effects of phosphorylation
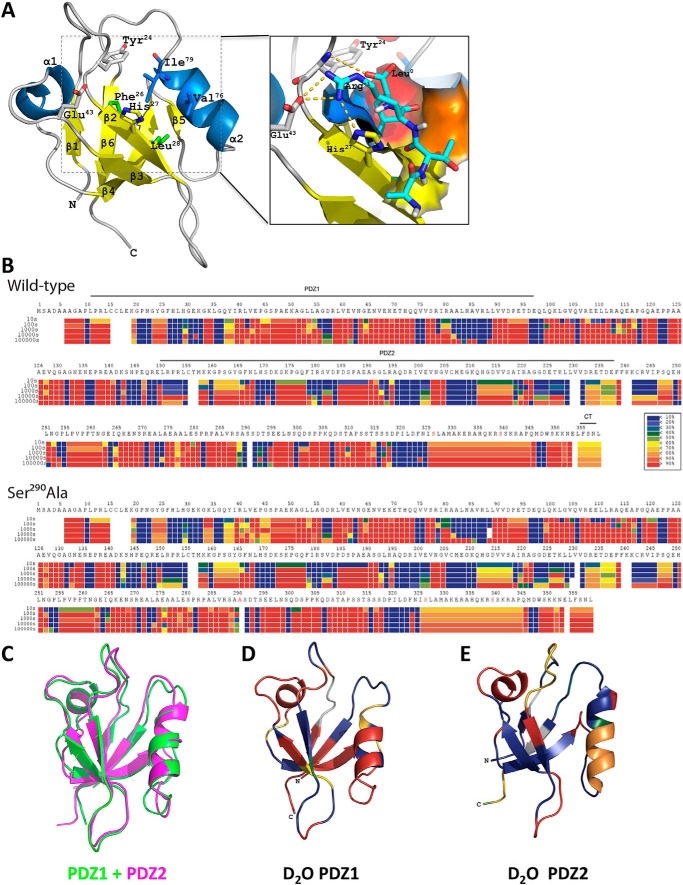
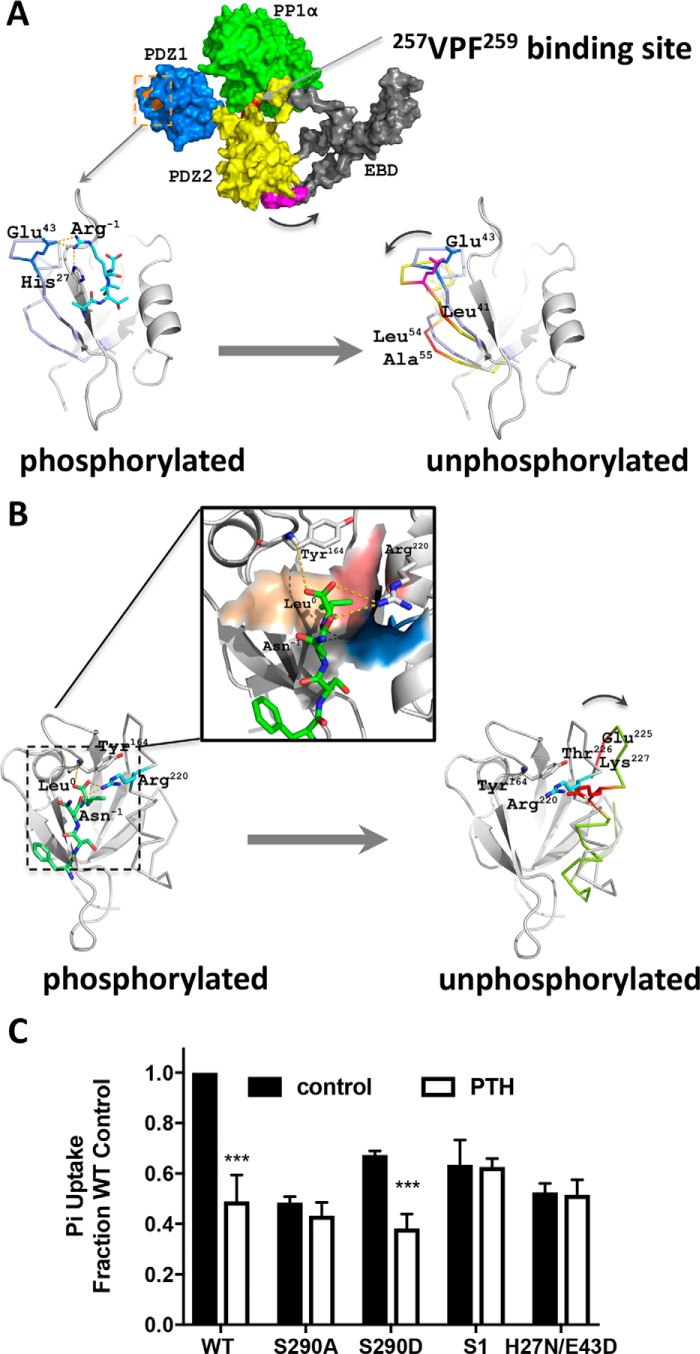
The observed phosphocycling at Ser290 suggested that this activity in the disordered region may induce conformational changes of NHERF1. Key positions within the 23GYGF26 core-binding motif, as well as at His27 and Glu43, which are critical for NPT2A interactions (62), may be altered upon phosphorylation (Fig. 8A). To explore this possibility, we applied HDX-MS to monitor structural changes by comparing time-dependent deuterium incorporation between WT-phosphorylated NHERF1 and dephosphorylated NHERF1, for which we used phosphoresistant S290A as a surrogate. Pepsin digestion yielded 521 peptides covering >98% of the protein across phosphorylated and phosphoresistant conditions for each time point. The high sequence coverage of NHERF1 allowed analysis of the structural characteristics of the entire protein, including two segments not observed by X-ray crystallography (spanning residues 95–149 and 236–358). Deuterium exchange heat maps for WT–NHERF1 shown in Fig. 8B (top panel) reveal many “hot regions” (red) indicating rapid deuterium uptake, consistent with the characteristic elongated NHERF1 open conformation allowing greater exposure for deuterium incorporation. Locations with slow deuterium uptake (Fig. 8B, blue) are more ordered and less prone to hydrogen–deuterium exchange. NHERF1 contains three domains: PDZ1, PDZ2, and EBD (Fig. 1). Superimposition of two available PDZ domain structures determined by X-ray crystallography suggests that they share high overall structural resemblance as indicated by a relatively low root-mean-square deviation of 0.769 for 81 α-carbons (Fig. 8C). However, the structural resemblance does not promote similar deuterium uptake. Discrete peptides from equivalent positions in PDZ1 and PDZ2 display distinct HDX kinetics as shown in Fig. 8B (top panel). A clear deuterium incorporation pattern was observed, for instance, when mapping HDX amplitude of WT–NHERF1 PDZ structures at 100 s (Fig. 8, D and E). Surface residues on both PDZ domains displayed higher deuteration on a short time scale, consistent with the view that solvent accessibility in the crystal structure correlates with the relative deuterium uptake (63). Strikingly, the β-sheets in PDZ2 (Fig. 8E) show lower deuterium uptake than those in PDZ1 (Fig. 8D) suggesting that PDZ2 is more cloistered and protected from HDX, likely due to steric shielding imparted by the EBD.
Figure 8.
Phosphorylation changes NHERF1 conformation. A, molecular ribbon model of PDZ1 (PDB code 1I92) complexed with NPT2A ligand after 100 ns of MD simulation (62). The 23GYGF26 core-binding motif along with His27 and Glu43 are critical structural determinants for binding NPT2A. The enlarged inset shows the four NPT2A C-terminal residues −3ATRL0 (cyan) located in a deep cavity formed by four hydrophobic residues (Phe26 (blue), Leu28 (yellow), Val76 (orange), and Ile79 (red)). A hydrogen bond links the carboxyl group of Leu0 and the amide group of Phe24 in PDZ1 (gold dashed lines). NPT2A Arg−1 forms a salt bridge with His27 and hydrogen bonds with Glu43 in PDZ1. The nomenclature for secondary structural elements is adapted from Ref. 18. B, representative heat maps of deuterium incorporation in wildtype (WT, upper panel) and phosphoresistant S290A (lower panel) NHERF1 peptides. Deuterium uptake over 10, 100, 1000, 10,000, and 100,000 s is indicated by sets of horizontal bars below the amino acid sequence. The color coding indicates the percentage of deuterium incorporation (black box below the WT heat map). C, superimposition of PDZ1 (green, PDB code 1I92) and PDZ2 (magenta, PDB code 4Q3H) indicating their structural similarity based on the low root-mean-square deviation for 81 α-carbons of 0.769 Å. D and E, mapping deuterium uptake of WT–NHERF1 at 100 s onto the structures of PDZ1 (D) and PDZ2 (E), respectively. Although the structures of PDZ1 and PDZ2 are similar, their structural dynamic profiles differ. PDZ1 is more dynamic than PDZ2. The most noticeable differences in HDX rates are in the β-sheets of PDZ1 (red) and PDZ2 (blue).
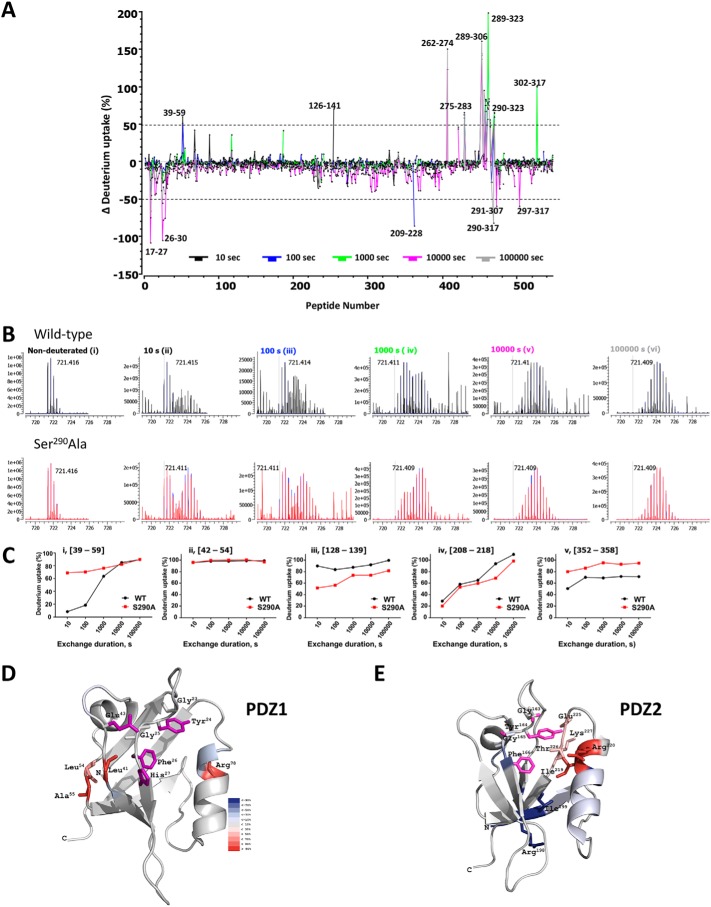
HDX-MS data obtained under the same conditions for the phosphoresistant mutant S290A–NHERF1 used here as a surrogate for the dephosphorylated Ser290 condition (Fig. 8B, lower panel) enabled mapping the regions undergoing conformational changes when Ser290 is dephosphorylated. As shown in Fig. 9A, the butterfly plot of differential deuterium uptake between phosphorylated and dephosphorylated states indicates that multiple sites exhibited structural changes as reflected by deuterium uptakes greater than 50%. Compared with structurally well-defined PDZ domains that exhibited comparable deuterium uptake in phosphorylated WT–NHERF1 and the phosphoresistant S290A protein, extensive differential deuterium incorporation was found within the unstructured PDZ2–EBD linker region ranging from residue 289 to 323 that harbors Ser290 (Fig. 9A). Large differences in deuterium incorporation may reflect global conformational changes, making this region more available to HDX. In contrast, only a few peptides in PDZ1(17–59) or PDZ2(126–141) display different deuterium incorporation (Fig. 9A) suggesting that the PDZ domains may undergo only local conformational changes upon pSer290 dephosphorylation. Consistent with previous observations in other proteins (64, 65), and as illustrated in Fig. 9B (panels ii and iv), the regions exhibiting conformational changes upon pSer290 dephosphorylation display a typical bimodal distribution of deuterium uptake as typified by the NHERF1(39–59) peptide. This peptide exhibited 60% more deuterium incorporation even at 10 s in the S290A–NHERF1 construct (Fig. 9C (panel i)) than in WT–NHERF1, suggesting that this region is not protected from HDX and likely undergoes localized conformational changes in the relatively rigid PDZ1.
Figure 9.
Quantitative HDX analysis and NHERF1 conformation. A, differential deuterium uptake for WT–NHERF1 and S290A–NHERF1 and at 10 s (black), 100 s (blue), 1000 s (green), 10000 s (pink), and 100,000 s (gray). Peptides exhibiting significant differential deuterium uptake greater than 50% are indicated; others were omitted for clarity. Of note, there are large peaks of differential deuterium uptake surrounding Ser290 from Ala289 to Phe323 in the S290A phosphoresistant construct. The (39–58)-peptide, where Glu43 is adjacent to the PDZ1 23GYGF26 core-binding motif as shown in Fig. 8A, displays high differential deuterium uptake. B, representative isotope profile of the (39–58)-peptide at the indicated times for WT (black) and S290A–NHERF1 (red). Nondeuterated controls for WT and S290A peptides are indistinguishable. A bimodal distribution at 10, 100, and 1000 s (panels ii–iv) reflects conformational change between phosphorylated and unphosphorylated conditions. C, dynamic exchange profiles for the indicated peptides from WT (black) and S290A–NHERF1 (red). Peptides with unique, time-dependent HDX behavior are shown. C-terminal residues in S290A–NHERF1 display greater deuterium uptake. D, model showing residues exhibiting differential deuterium uptake between WT and S290A–NHERF1 at 100 s mapped on the structure of PDZ1 (PDB code 1I92). The critical residues His27, Glu43, and the GYGF core-binding motif for PDZ ligand binding are colored in purple. Residues undergoing increased deuteration of 30% or more are shown. E, differential deuterium uptake between WT and S290A–NHERF1 at 100 s mapped on PDZ2 (PDB code 4Q3H). Critical binding residues based on the same criteria as in D are colored purple. The differential deuteration levels are color-coded as in D. The high HDX rate at Ile219, Arg220, Glu225, Thr226, and Lys227 suggests that these residues undergo significant structural movement in the opposite direction from what occurs in PDZ1.
As summarized in Fig. 9C, three distinct patterns of deuteration were found across the entire protein resulting in increased (Fig. 9C, panels i and v), unchanged (Fig. 9C, panel ii), or decreased (Fig. 9C, panels iii and iv) incorporation, reflecting various conformational fluctuations within discrete regions in response to pSer290 dephosphorylation. As shown in Fig. 9C, panel v, the C-terminal (352–358) peptide exhibited faster HDX exchange in the Ser290 phosphoresistant construct (S290A) than in phosphorylated WT–NHERF1 indicating greater exposure to HDX. This region has been proposed to be critical for intramolecular “head-to-tail” interactions between the EBD and PDZ2 and for the switch between open and closed NHERF1 conformations (20). The greater deuteration seen here likely reflects release from the closed conformation and may serve to signal Ser290 rephosphorylation by GRK6A.
We next mapped differential deuterium uptake on PDZ1 and PDZ2 X-ray crystal structures to uncover how pSer290 dephosphorylation influences local conformations. The results showed that Leu41, Leu54, Ala55, and Arg78 in PDZ1 (Fig. 9D), Ile219 and Arg220 of the α2 helix, and Glu225, Thr226, and Lys227 within the PDZ2 loop region displayed faster exchange (Fig. 9E) when pSer290 was dephosphorylated. These residues are vicinal to the GYGF PDZ-binding motif and other critical docking sites located between the sides of the PDZ1 ligand-binding pocket, in one case, and on one side of the PDZ2 cavity, in the other case. Faster deuterium incorporation rates suggest that restricted conformational changes may occur that permit greater solvent access for deuterium incorporation. Specifically, for PDZ1, the movement of Leu41, Leu54, and Ala55 located in the flexible loop may be toward to the left side of the pocket. In the case of PDZ2, the direction of Glu225, Thr226, Lys227, Ile219, and Arg220 may be toward to the right side of the pocket, thereby forming a more compact structure at the pocket base as indicated by less deuterated Arg198 and Ile199 in the phosphoresistant S290A–NHERF1 construct.
We then sought to define how pSer290 dephosphorylation alters the conformational landscape to develop a mechanistic explanation of NHERF1 function through coordinated PDZ ligand binding and release. The structural details of the human PP1α VxF/W motif (PDB code 5IOH) have been disclosed by X-ray crystallography (66). Likewise, PDZ1 (PDB codes 1I92 and 4LMM) and PDZ2–EBD (PDB code 2KRG) structures have been solved by X-ray crystallography (18, 67, 68) and by NMR (67), respectively. Using these structures as a starting point, we developed a working model for NHERF1 binding to PP1α and for PP1α-dependent pSer290 dephosphorylation, along with an experimentally testable regulatory mechanism for PTH-sensitive phosphate transport. As shown in Fig. 10A (top panel), PDZ1 together with PDZ2–EBD form an elongated molecule as measured by small-angle X-ray scattering (69). A head-to-tail interaction connects PDZ2 with the EBD (19, 20), where the NHERF1 C-terminal tail (355FSNL358) is inserted in the ligand-binding pocket of PDZ2 (Fig. 10, A and B, top panels). According to this model, PP1α engages NHERF1 through the 257VPF259-binding site with neighboring spatial contacts in both PDZ1 and PDZ2. Upon PP1α-mediated pSer290 dephosphorylation, outward movement of Leu41, Leu54, and Ala55 within the (39–58)-peptide region at the periphery of the β2-sheet allows greater surface exposure for deuterium exchange (Fig. 10A, lower panel), leading to weaker NPT2A binding because Glu43 in NHERF1 is displaced and unable to bind Arg−1 in NPT2A. Likewise, the interaction network between PDZ2 and the NHERF1 C-terminal tail through residues Tyr164 and Arg220 (Fig. 10B, top panel) is dissembled because of outward movement of Arg220, Glu225, Thr226, and Lys227 on the side of the α2-helix (Fig. 10B, lower panel). According to this model, His27 and Glu43, together with the 23GYGF26 core-binding motif, are critical structural determinants for NPT2A binding to PDZ1 (61). Indeed, disrupting the 23GYGF26 PDZ1 core-binding motif (23) or double mutation at H27N/E43D reduces binding to NPT2A (61, 62). We tested the functional consequences of these mutations by analyzing their effect on basal and PTH-dependent phosphate uptake. As shown in Fig. 10C, the phosphoresistant S290A construct displayed reduced baseline phosphate uptake and was resistant to PTH. The S290D phosphomimic exhibited behavior similar to WT–NHERF1, although basal uptake was somewhat lower. Mutating the PDZ1 23GYGF26 core-binding motif depressed basal phosphate uptake and was refractory to PTH. Finally, the double H27N/E43D mutant that was predicted to weaken binding to NPT2A was essentially inert.
Figure 10.
Ser290 dephosphorylation induced PDZ1 conformational modifications and associated changes in phosphate transport. A, NHERF1 (PDB code 1I92 for PDZ1; PDB code 2KRG for PDZ2–EBD) complexed with PP1α (PDB code 5IOH). PDZ1 is colored in marine, PDZ2 in yellow, and EBD in gray. The PDZ1 GYGF core-binding (orange) is the site of NPT2A interaction. Bound PP1α (green) is assembled with NHERF1 at the 257VPF259 motif (red). The C-terminal −3FSNL0 EBD ligand (purple) inserts in the PDZ2-binding pocket. The lower panel showing the outward movement against the PDZ-binding ligand, as implied from Fig. 9D, may dissociate NPT2A from NHERF1. B, possible conformational changes in PDZ2 and EBD upon Ser290 dephosphorylation by PP1α. Top panel shows a modeled structure by MD simulation, based on a similar protocol as described (61), of the four EBD C-terminal residues, FSNL inserted in the PDZ2-binding pocket formed by hydrophobic residues Phe166 (orange), Leu168 (gray), Val216 (blue), and Ile219 (red). The interacting network formed by hydrogen bonds (gold dashed lines) further stabilizes the liganded EBD C-terminal tail. As implied in Fig. 9E, an outward movement against the PDZ-binding ligand may release the EBD C-terminal tail from PDZ2 (lower panel). This action would follow kinase-induced rephosphorylation and dissociate PP1α. C, PTH-sensitive phosphate uptake in cells expressing constructs with residues critical for NPT2A binding. Wildtype (WT), S290A, S290D, mutated PDZ1 core-binding motif (S1), or double H27N/E43D NHERF1 constructs are shown. Compared with WT–NHERF1, all mutant constructs display decreased basal phosphate uptake. PTH inhibited phosphate uptake only in cells transfected with WT–NHERF1 and the S290D phosphomimic. Results are means ± S.D. (n = 3; **, p < 0.01).
Discussion
In this study, we set out to characterize the sites and function of NHERF1 phosphorylation on PTH-regulated phosphate transport mediated by NPT2A. Several key findings were made. 1) Ser290 was rapidly and reversibly dephosphorylated. 2) Dephosphorylation was mediated by PP1, which binds NHERF1. Mutating the VxF/W motif abolished PP1 binding and dephosphorylation. At picomolar concentrations, TTN blocked PP1 activity and abolished PTH-sensitive phosphate transport. 3) Ser290 phosphorylation was mediated by GRK6A. Pharmacologic inhibition of GRK6A decreased NHERF1 rephosphorylation and inhibited hormone-dependent phosphate transport. 4) Phosphocycling at Ser290 induces conformation changes consistent with reversible binding and release of NPT2A.
Protein phosphorylation is the most common reversible post-translational modification (70). Catalyzed by kinases, a phosphate moiety is covalently attached by a phosphoester bond to Ser, Thr, or Tyr. Other linkages are possible that permit phosphorylation of His, Lys, Arg, or Asp and Glu. In most cases, phosphorylation may be reversed by a protein phosphatase. Activity-dependent, reversible phosphorylation can modify protein structure and stability, control protein–protein interactions, and affect enzyme activity and subcellular localization (71). The resulting changes provide a dynamic phospho-regulatory mechanism that mediates many aspects of cellular function. The scaffolding phosphoprotein NHERF1 exemplifies the diverse roles of protein phosphorylation (Table 1). These distinctive cellular processes mediated by NHERF1 may arise from the multiple Ser and Thr sites that singly or collectively govern cellular responses elicited by extra- or intracellular stimuli (72).
Mass spectrometry permitted identification of 10 phosphorylated Ser and Thr residues in NHERF1. Some sites, including Ser162, Ser280, Ser290, and Ser302, were previously identified (Table 1) by in vitro 32P phosphorylation assays or by in vivo metabolic labeling of full-length or truncated NHERF1. It should be pointed out that NHERF1 phosphorylation sites compiled in PhosphoSitePlus (45) have not been independently verified, nor has a confidence level been established for their identification. Also, the functionality of these sites, if any, remains to be determined. SILAC approaches do not augment sensitivity. Finally, it should be borne in mind that PTH-induced phosphorylation may alter the phospho-map of NHERF1 upon PTH induction.
Based on top-down tandem MS and 32P metabolic labeling, we confirmed phosphorylation at Ser290, Ser291, and Thr293 within the Ser-rich cluster surrounding Ser290 (Fig. 2, D and E). Importantly, Ser290 and Thr293 were verified by MS at endogenous expression levels in RPTEC cells. In addition, other functionally undefined sites, including Ser46, Ser181, Ser269, Ser291, Thr293, and Ser299, were found, although their existence in native cells and at constitutive expression needs to be validated. The corresponding exploration of biological functions associated with these sites will doubtlessly lead to new findings. Certain phosphorylation sites reported previously like Ser77, Thr95, Thr156, and Ser339/Ser340 were not detected by MS. The unidentified sites do not rule out phosphorylation events at the specific position that occurred in cells as shown in Fig. 2F for Ser77. Low abundance and variable fragmentation of these sites containing peptides presumably limit the sensitivity of MS detection of phosphorylation events. Although major phosphorylation sites are readily detected, the limited dynamic range makes minor sites more difficult to identify. Also, enzymatic digestion can produce multiple basic residues with higher-charge–state ions making identification of phosphorylated residues by standard fragmentation methods difficult (73). Ser77 is located in an α-helix, and modeling predicts that it could be involved in hydrogen bonding with Gln73. Failure of trypsin digestion to disrupt the α-helix could preclude its detection by MS.
The described phosphorylation sites are associated with distinct NHERF1 physiological functions (74). We systematically examined quantitative, time-dependent changes in PTH-induced NHERF1 phosphorylation by SILAC labeling. The results revealed three patterns of NHERF1 phosphorylation in response to PTH. In the first scheme (Ser269), phosphorylation increased over 5 min. The second pattern (Ser290) displayed dynamic, cyclical dephosphorylation followed by full rephosphorylation (Fig. 3, A and B). These results were independently verified using an anti-pSer290–NHERF1 antibody (Fig. S1, A–C). In the third example (Ser302), no detectable change of phosphorylation occurred over the first 5 min after PTH addition. Among the 10 identified phosphorylation sites, only Ser290 was required for PTH-inhibitable, NPT2A-mediated phosphate transport (Fig. 2I).
The initial dephosphorylation of Ser290 was not anticipated. As noted earlier, we directed our attention to PP1α as the candidate phosphatase responsible for this activity because it co-purified (Table S2) and immunoprecipitated with NHERF1 (Fig. 4C). Prior work first identified NHERF1 binding to PP1α but did not disclose a binding site (52). PP1 lacks a C-terminal PDZ sequence, suggesting that it did not bind NHERF1 through a canonical PDZ–ligand recognition motif. When this supposition was confirmed (Fig. S2), we considered alternative motifs, which revealed a heretofore unrecognized VxF/W-binding motif located in the intrinsically disordered region between PDZ2 and the EBD (Fig. 1).
The free PP1 catalytic subunit lacks substrate specificity (75). Targeted activity is conferred by assembly as a holoenzyme thereby imparting distinct substrate specificity (76). Three lines of evidence now support the view that NHERF1 is a novel PP1 binding partner. 1) PP1α binds to the putative VxF/W motif to dephosphorylate NHERF1. 2) Mutating the motif abolished both PP1α binding and dephosphorylation. 3) TTN, a PP1-specific inhibitor with a 1000-fold selectivity over PP2 (53), blocked PP1 activity and abolished PTH-sensitive phosphate transport at picomolar concentrations. Although not required, the location of the VxF/W locus in a structurally undefined region is consistent with PP1 forming a holoenzyme with NHERF1. As noted by others, binding of PP1 to a RVxF motif does not affect the conformation of the catalytic subunit but, rather, increases the resident concentration of the regulatory partner, thereby promoting secondary interactions that augment PP1 activity and substrate specificity (77).
Applying an established constitutively active form of PP1α (52) verified that Ser290 dephosphorylation is required for PTH inhibition of Npt2a-mediated phosphate transport. The findings (Fig. 4E) further indicate that Ser290 dephosphorylation not only is necessary for phosphate transport but also is effectively sufficient because TTN inhibited phosphate transport in the presence of PP1α–T320A nearly as completely as observed with PTH.
Upon PTH treatment, we detected a transient burst of intracellular phosphate. The increased phosphate was attributable to entry from the extracellular solution and was mediated by a sodium-dependent process inasmuch as sodium removal abolished this action. Because the time frame of this effect coincided with that of Ser290 dephosphorylation and rephosphorylation, we theorized that elevated phosphate inhibited PP1α, thereby terminating its phosphatase activity. Remarkably, PP1α was inhibited over the same range (10 mm) of added phosphate as the magnitude of the rise of intracellular phosphate. The catalytic action of PP1-mediated phosphate monoester hydrolysis has a Kd value in this range, 2–10 mm (78, 79). Thus, we conclude that PTH initiates a cascade of events that includes PP1α activation.
Targeted disruption of murine Slc9a3r1 promotes Npt2a internalization with attendant renal phosphate wasting (11). NHERF1 expression is required for PTH-controlled Npt2a endocytosis and down-regulation (80, 81). By extension, it is plausible to assume that the diminished NPT2A binding caused by Ser290 dephosphorylation (Fig. 6, A–C) facilitates NPT2A internalization or even degradation, as implicated previously (82–84). Preventing Ser290 phosphorylation with a phosphoresistant mutant or pharmacologically inhibiting GRK6 eliminated NPT2A binding to NHERF1 and reduced PTH-sensitive phosphate transport. We conclude that GRK6A-mediated Ser290 phosphorylation supports strong binding of NHERF1 to NPT2A that retains the transporter at the cell surface. Upon pSer290 dephosphorylation, or replacement of Ser290 with Ala, binding of NPT2A is disrupted, which may initiate NPT2A internalization and cessation of phosphate transport. The temporal coincidence between the prompt rise of intracellular phosphate (Fig. 5, A and B) with that of pSer290 dephosphorylation at 1–5 min (Fig. 3, A and B) is compatible with the view that pSer290 dephosphorylation is responsible for the acute modulation of cellular phosphate uptake.
The signal activating PP1α here is unknown. PTH occupancy of its cognate PTHR elicits prompt activation of PKA (85, 86) and of PKC (87), either of which could be an upstream signal for PP1α (88) and Ser290 dephosphorylation. As shown here, PTH action on NPT2A also requires Ser290 rephosphorylation. GRK6A specifically mediates this event. GRK6 possesses three isoforms. Notably, GRK6A harbors a C-terminal PDZ-binding sequence (−TRL), and it is the only GRK6 isoform that binds and phosphorylates NHERF1 (35). We reasoned that if GRK6A played a central role in PTH control of NHERF1 phosphorylation, then its inhibition should interfere with phosphate transport, and indeed, this was the case (Fig. 7). The extent to which the GRK6 inhibitor reduced Ser290 phosphorylation (Fig. 7B) was conspicuously less than the degree of suppression of phosphate transport (Fig. 7C) or the blockade of NHERF1 binding to NPT2A (Fig. 7D). The origin of this disparity is unclear and may suggest that other kinases contribute indirectly to Ser290 phosphorylation or that even small changes of Ser290 phosphorylation have large effects on NHERF1 protein–protein interaction and phosphate uptake.
Phosphorylation-dependent structural changes are well-known to alter protein-binding affinity along with other biological actions (71). Such dynamic conformational changes theoretically could be detected by X-ray crystallography (78), solid-state nuclear magnetic resonance (NMR) (14), single-pair FRET (spFRET) (79), or more recently by HDX-MS (80). By comparing constitutively phosphorylated WT–NHERF1 with recombinant, phosphoresistant S290A NHERF1, we observed phosphorylation-dependent structural changes at positions critical for binding NPT2A (Figs. 8 and 9). The regional flexibility at near-residue resolution, indicated by deuterium labeling from the solvent, suggests that the most notable structural differences occur in the linker region between the PDZ2 and the EBD. Small but significant conformational changes were also found in PDZ1 around Glu43, in PDZ2 around Arg220, and in the EBD at the C-terminal tail, where the internal PDZ-binding ligand resides. When combined with available PDZ structures, the observed conformational changes permit developing a functional mechanism for NHERF1 despite the absence of a full-length structure.
Only structures of truncated PDZ1 (18) and extended PDZ2 (19) are available, limiting insight into the mechanisms of NHERF1 structural plasticity and functional activity. In addition to simple tethering, long-range allosteric communications between the two PDZ domains have been described (19). Understanding of the molecular mechanism of long-range allosteric communication requires resolving the structure of full-length NHREF1. We applied HDX-MS technology to characterize the static conformations and the conformational dynamics of full-length NHERF1 protein under conditions where the critical Ser290 was phosphorylated. The high-protein coverage (>98%) reveals the conformational changes and dynamics of different regions in NHERF1 and also the global structural characteristics of the apoprotein. As indicated by the red bars throughout the protein (Fig. 8B), rapid deuterium uptake confirms that NHERF1 is highly flexible. The linker regions between PDZ1–PDZ2 (residues 98–150) and PDZ2–EBD (residues 238–329) displayed high deuterium incorporation rates. These observations provide direct experimental evidence for the molecular and structural mechanisms of tethering and long-range allosteric communication.
We previously analyzed isolated PDZ1 and PDZ2 by HDX-MS and compared them with full-length NHERF1 (89). Deuterium incorporation in that study was lower than reported here. Deuterium back-exchange occurs while digesting samples and during LC/MS. If not corrected, this leads to underestimating the extent of deuterium incorporation because HDX values are the sum of those collected in time-dependent mode plus back-exchange. Fully deuterated (FD, or equilibrium deuterated) samples can be used to determine the extent of back-exchange by calculating the experimental maximum deuterium incorporation (usually lower than the theoretical maximum deuterium incorporation, MaxD) under the same experimental conditions as for time-dependent on-exchange measurements (90). This explains why the earlier HDX values (89) are lower than those reported here. Indeed, analysis of the current collected HDX spectra without FD correction produced heat maps similar to those previously reported (Fig. S7).
Although an improvement, inclusion of FD samples only permits more precise measurement of authentic deuterium incorporation; it does not affect the results for protein conformational dynamics. Thus, the disparity in deuterium incorporation notwithstanding, the two studies lead to similar conclusions that PDZ1 is more solvent-accessible than PDZ2 under resting conditions and provide a structural framework for the distinct ligand selectivity of each NHERF1 PDZ domain despite their harboring identical GYGF core-binding motifs.
To reveal the structural changes in NHERF1 induced by phospho-cycling at Ser290, we compared the HDX-MS profiles of WT (phosphorylated) and S290A (phosphoresistant) NHERF1 as a surrogate for unphosphorylated Ser290. The most notable difference was in the PDZ2–EBD linker region harboring Ser290, where deuterium incorporation significantly increased in the S290A mutant compared with WT–NHERF1. This observation suggests that a substantial and global conformational change occurs during Ser290 dephosphorylation, leading to a more accessible linker region. Notable local conformational dynamics were also found in the 39–58-residue region in PDZ1 that displayed faster deuterium incorporation in S290A NHERF1, suggesting substantial conformational change accompanying Ser290 dephosphorylation in this restricted region. The 39–58-region included Glu43, a critical structural determinant for NPT2a binding to PDZ1 (62). This local conformational change may affect the NHERF1-NPT2a interaction.
The region surrounding Arg220 in PDZ2 and the C-terminal 352–358-residue peptide simultaneously exhibited faster deuterium incorporation rates in the phosphoresistant S290A construct compared with phosphorylated WT–NHERF1. This observation suggests that the Arg220 region in PDZ2 interacts with the EBD C-terminal tail in phosphorylated NHERF1 and that pSer290 dephosphorylation disrupts this interaction to release the self-inhibited C-terminal tail from PDZ2 and promote the open NHERF1 conformation.
The HDX-MS data advance a partial model for conformational changes in NHERF1 during phospho-cycling at Ser290. In the phosphorylated state, we propose that NPT2A is bound to PDZ1. Upon pSer290 dephosphorylation by PP1α, outward movement of Leu41, Leu54, and Ala55 at the periphery of the PDZ1 β2-sheet weakens NPT2A binding because Glu43 in NHERF1 is displaced and unable to bind Arg−1. Evidently, changes in phosphorylation at Ser290 work in concert with other coordinated sites (50). In the presence of phosphorylated Ser290, phosphorylation cycling at Ser77 regulates PTH-sensitive phosphate transport. This may also result from conformational changes affecting NPT2A binding.
Normal phosphate homeostasis involving the intestines, kidneys, and bone is required for extracellular mineral-ion homeostasis and skeletal mineralization. Acquired and inherited disorders of the proteins involved in these functions often cause hypophosphatemia and abnormal bone growth and deformities (91, 92).
We advance here the view that rapid dynamic phosphorylation cycling and associated NHERF1 conformational changes play a central role in coordinating the hormonal response to PTH. Ser290 dephosphorylation disrupts binding of NPT2A to NHERF1 leading to cessation of hormone-regulated phosphate transport by initiating or facilitating NPT2A internalization. Dephosphorylation is rapidly extinguished by terminating PP1α activity by a transient increase of intracellular phosphate. In this manner, Ser290 undergoes a regulated cycle of phosphorylation and dephosphorylation. Such an acute regulatory strategy that requires NHERF1 involves a rapid on/off switch between phosphorylation and dephosphorylation at Ser290 to limit cellular phosphate uptake. This mechanism helps explain the biochemical, molecular, and cellular events involved in phosphate homeostasis and may provide new targets for controlling NPT2A abundance and could serve as a therapeutic alternative for managing clinical disorders of phosphate balance.
Experimental procedures
Chemical reagents, plasmids, and antibodies
All chemicals were analytical grade from various companies as follows: TTN (Tocris Bioscience); protease inhibitor mixture Set I (EMD-Millipore); phosphatase inhibitor mixture PhosSTOP (Roche Applied Science). The 22-residue C-terminal NPT2A peptide was synthesized using standard procedures (62). The fluorescent dye 7-hydroxy-5,5-dimethyl-10-(4-methoxy-2-methyl-phenyl)-dibenzo-[b,e]-silin-3(5H)-one (2Me-4OMe-TM) was synthesized as described (93). All other reagents were of the highest purity available and were obtained from Sigma unless otherwise stated.
Plasmids were prepared by PCR; human-derived NHERF1 was inserted into pIRESpuro-Glue-N1 (94) to overexpress TAP–NHERF1 (TAP contains a streptavidin-binding protein tag, a tobacco etch virus cleavage site, an HA epitope, and a calmodulin-binding protein tag) (kindly provided by Drs. Jean-Luc Parent and Terence Herbert); FLAG–NHERF1 or PTHR was cloned in pcDNA3.1(+). All NHERF1 mutants were generated using QuikChange site-directed mutagenesis kit. Sequences of all constructs were confirmed by DNA sequencing.
Antibodies were purchased from the following suppliers: rabbit anti-NHERF1 (Alomone Labs, APZ-006, lot 6AN0302); anti-HA (Cell Signaling Technology, 3724, lot 9); anti-GFP (Invitrogen, A6455, lot 1853896); anti-FLAG (Sigma, F7425, lot 085M4774V); anti-actin (Sigma, A1978, lot 076M4786V); and anti-phospho-Ser290 NHERF1 (pSer290) (GenScript, U5674CJ090, lot A318030013) generated against 284ALVRSApSSDTSEEL297).
Cell culture and transfection
HEK293 GnTI− cells were grown in high-glucose Dulbecco's modified Eagle's medium (DMEM; Mediatech, 10-013-CV) supplemented with 10% FBS and 1% penicillin and streptomycin (pen/strep). Opossum kidney (OK and OKH) cells (23) were grown in DMEM/Ham's F-12 50:50 medium (Mediatech, 10-090-CV) supplemented with 10% FBS and 1% pen/strep. Telomerase-immortalized human RPTEC (10, 95) were obtained from ATCC under license from Geron Corp. Cells were cultured in defined medium (DMEM/F-12 (Mediatech, 10-090-CV) supplemented with 5 pm triiodo-l-thyronine, 10 ng/ml recombinant human epidermal growth factor, 25 ng/ml prostaglandin E1, 3.5 μg/ml ascorbic acid, 1 mg/ml insulin, 0.55 mg/ml transferrin, 0.5 μg/ml sodium selenite, 25 ng/ml hydrocortisone plus 1% pen/strep, and 0.1 mg/ml G418. MC3T3-E1 preosteoblast (ECACC 99072810, Cell Culture Facility, University of Granada) were grown in α-minimum essential medium (10-012-CV) containing 10% FBS and 1% pen/strep.
Cells were transfected with the indicated plasmids using FuGENE 6 (Promega) or Lipofectamine 3000 (Invitrogen) according to the manufacturer's instructions. Stable cells expressing TAP–NHERF1, FLAG–PTHR, or TAP–PTHR were prepared by screening with puromycin or G418.
Intracellular cAMP and calcium
GnTI− cells stably expressing TAP–NHERF1 and FLAG–PTHR were seeded on 35-mm coverslips coated with poly-d-lysine and grown on 6-well plates cultured in 2 ml of high-glucose DMEM. Cells were transiently transfected with pcDNA3 CFP–Epac1–YFP using FuGENE 6 transfection reagent. Experiments were performed in a culture chamber in a FRET buffer (137 mm NaCl, 5 mm KCl, 20 mm HEPES, pH 7.5, 1 mm CaCl2, and 1 mm MgCl2) supplemented with 0.1% BSA. Coverslips were mounted on the stage of an inverted Nikon Ti-E microscope and imaged at ×63 magnification and excited at 444 nm, and emission at 535 nm was monitored. Data were digitized by NIS-Elements (Nikon), and FRET was expressed as the normalized ratio of YFP/CFP signals.
Changes of intracellular calcium were monitored in double-stable GnTI− cells transfected with the calcium sensor pCMV-R-GECO1.2 (Addgene). Approximately 24 h later, cells were washed once in FRET buffer and then in FRET buffer plus 0.1% BSA. Cells were imaged on a Nikon A1 confocal microscope at ×63 and excited at 561 nm, and emission at 585 nm was monitored at 6-s intervals. Image data were processed using NIS-Elements.
Preparation of endogenous and overexpressed NHERF1
The N-terminal 4.1 FERM domain (1–299) from human ezrin (kindly provided by Dr. Zimei Bu) was prepared, as described previously (96), and 10 mg/ml of this protein was coupled to 1 ml of CNBr-activated agarose by following the supplier's instructions. The conjugated FERM beads were used to isolate endogenous NHERF1 from RPTECs. Briefly, confluent RPTEC cells from 20 15-cm dishes were harvested by centrifugation and lysed in a modified NP40 buffer containing 50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 5 mm EDTA, and 0.5% NP40 supplemented with 0.1 mm phenylmethylsulfonyl fluoride, protease inhibitors (Millipore), and phosphatase inhibitors (Roche Applied Science). After high-speed centrifugation at 12,000 rpm for 30 min at 4 °C, the supernatant was incubated with the FERM beads for 2 h in the cold room. Beads were then washed extensively with a wash buffer containing 50 mm Tris-HCl, pH 8.0, and 150 mm NaCl. Protein was eluted using 0.1 m glycine, pH 2.5, and immediately neutralized by 1 m Tris-HCl, pH 8.0.
The overexpressed TAP–NHERF1 protein generated from double-stable FLAG–PTHR–TAP–NHERF1 GnTI− cells was purified by a similar protocol as for endogenous NHERF1. In brief, the clear supernatant after centrifugation was incubated with streptavidin–agarose beads, and TAP–NHERF1 was eluted using a buffer containing 50 mm Tris-HCl, pH 8.0, 150 mm NaCl, and 10 mm biotin.
Transiently expressed FLAG–NHERF1 in HEK293 GnTI− cells was purified using anti-FLAG beads (Sigma). Briefly, WT or mutant NHERF1 tagged with FLAG epitope was transfected using Lipofectamine 3000 (Invitrogen). 36–48 h later, following 32P labeling, where indicated, cells were rinsed with an ice-cold buffer containing 50 mm Tris-HCl, pH 8.0, and 150 mm NaCl. Cells were then lysed in the modified NP40 buffer supplemented with protease inhibitors (Millipore). The clarified supernatant by centrifugation was incubated with anti-FLAG beads for 2 h in the cold room. FLAG–NHERF1 was eluted using the 3× FLAG peptide (Sigma) in a buffer containing 50 mm Tris-HCl, pH 8.0, and 150 mm NaCl. Protein concentrations were determined by Bradford assay.
SILAC
Double-stable GnTI− cells expressing TAP–NHERF1 and FLAG–PTHR were grown in SILAC DMEM supplemented with 10% dialyzed FBS and antibiotics. The “light” (l-Lys-12C6 and l-Leu-12C6) and “heavy” (l-Lys-13C6 and l-Leu-13C6) SILAC DMEM was prepared as per manufacturer's instructions. Cells were grown for at least 7 doublings in the labeling medium to achieve >95% isotope incorporation as determined by MS. 100 nm PTH was added to cells grown in heavy isotope-containing media for the indicated times. The EC50 values for PTH binding and receptor activation are about 10 nm (97), and the lowest PTH concentration eliciting maximal effects is 100 nm (98).
For semiquantitative SILAC-based MS/MS, the protein concentration was measured using the BCA assay. Equal amounts of light and heavy samples were combined and subjected to purification by streptavidin affinity chromatography as outlined above.
In-solution protein digestion and mass spectrometry
Purified NHERF1 samples (∼1–5 μg) were mixed with 0.02% ProteaseMax (Promega) and 25 mm DTT. Disulfide reduction was performed by incubating at 99 °C for 10 min. After cooling to room temperature for 10 min, the sample was mixed with 45 mm indoacetamide and incubated in the dark for 1 h. Overnight digestion with 0.2 μg/μl GluC or trypsin was carried out at 37 °C. The next day, digestion was stopped by adding 1% TFA to the sample, and digested peptides were desalted using Pierce C18 Spin Columns. After desalting, each digested peptide sample was divided into two fractions (20 and 80%). The 20% fraction sample was analyzed directly on the LC-MS system to identify the NHERF1-interacting proteins. The collected data were also used to calculate the ratio of “heavily-labeled” and “lightly-labeled” proteins in SILAC experiments. The resulting ratios were further used as normalization factors to the phosphopeptides from those heavily- and lightly-labeled NHERF1. The remaining 80% of digested and desalted peptides were subjected to phosphopeptide-enrichment procedures using Titansphere Phos-TiO kit (GL Sciences) as per the manufacturer's instructions. After lyophilizing in SpeedVac centrifuge, the peptides were reconstituted in 0.1% formic acid. Samples were analyzed on a Velos Orbitrap mass spectrometer (ThermoFisher Scientific). MS data analysis was performed using SEQUEST (99) referencing SwissProt Homo sapiens forward and reverse protein sequences unless otherwise stated. Initial peptide mass tolerance was set to 10 ppm, and fragment ion tolerance was 0.8 Da. Enzyme specificity was set to full trypsin or GluC digestion allowing for up to two missed cleavages. Carbamidomethylation of cysteine was set as a fixed modification (57.02146 Da); oxidation of Met (+15.99491 Da) and phosphorylation of Ser, Thr, and Tyr (+79.96633 Da) were included as variable modifications. For SILAC data, Lys (+6.02012 Da) and Leu (+6.02012 Da) were set as variable modifications.
Immunoprecipitation and Western blotting
Transfected cells were incubated at 37 °C for 48 h, and cells were then harvested and lysed in a lysis buffer (50 mm Tris, pH 8.0, 150 mm NaCl, 1% NP40, and protease inhibitors). The supernatant after high-speed centrifugation was incubated with corresponding agarose beads while rotating at 4 °C for 4 h. The resin was washed three times with the lysis buffer and mixed with 2× Laemmli sample buffer. The protein samples were then resolved on SDS-polyacrylamide gel and transferred to Immobilon-P polyvinylidene difluoride membrane for Western blotting (EMD Millipore). Proteins were detected by using ECL Western blotting detection reagent (GE Healthcare).
Metabolic 32P labeling
TAP–PTHR-stable HEK293 GnTI− cells were grown in 6- or 10-cm dishes and transfected with WT or mutant FLAG–NHERF1 constructs. 36–48 h post-transfection, cells were serum-starved by phosphate-free DMEM (REF: 11971-025; Lot: 1668852; Gibco) for 4 h. Subsequently, cells were incubated with the same phosphate-free DMEM supplemented with [32P]orthophosphate (0.1 mCi/ml) for 10 min followed by a 5-min pretreatment with the Ser/Thr phosphatase inhibitor calyculin A (50 nm). Cells were then exposed to PTH (100 nm) or vehicle for 15 min at 37 °C, rinsed with ice-cold PBS, and lysed in the lysis buffer containing phosphatase and protease inhibitors. The cell lysates were immunoprecipitated with FLAG–agarose beads overnight at 4 °C. After washing, the beads were resuspended in 2× SDS sample buffer. The NHERF1 samples were resolved on 12% SDS-polyacrylamide gel. The gel was dried, autoradiographed, and quantified with ImageJ software (100).
Phosphate uptake
OKH cells grown on 12-well plates were transfected with WT or mutant FLAG–NHERF1. 36–48 h after transfection, cells were serum-starved for 4 h and treated with 100 nm PTH(1–34) for 2 h. After aspirating the media, cells were washed at room temperature with sodium-containing buffer (140 mm NaCl, 4.8 mm KCl, 1.2 mm MgSO4, 0.1 mm KH2PO4, 10 mm HEPES, pH 7.4) three times. Cells were subsequently incubated with sodium-containing buffer supplemented with 32P (1 μCi/ml) for 10 min at 37 °C. Phosphate uptake was stopped by washing the cells with ice-cold sodium-free buffer (140 mm N-methyl-d-glucamine, 4.8 mm KCl, 1.2 mm MgSO4, 0.1 mm KH2PO4, 10 mm HEPES, pH 7.4). Cells were lysed overnight in 0.5 ml of 1% Triton X-100 at 4 °C. The following day, 0.25 ml of cell lysate was mixed with 5 ml of scintillation fluid and β emission was counted on an LS 6500 multipurpose counter (Beckman Coulter).
Dephosphorylation
For in vitro dephosphorylation, purified 32P-labeled Ser290 or Ser290-VPF/AAA was used as substrates. Briefly, FLAG–Ser290-NHERF1 or FLAG–Ser290-VPF/AAA was transfected in GnTI− cells grown on 10-cm dishes. 48 h later, cells were metabolically labeled with 32P as described above. Cells were then lysed in the lysis buffer, and the resulting lysate was incubated with anti-FLAG beads for 2 h at 4 °C. Protein was eluted with a buffer containing 50 mm HEPES, pH 7.4, and 150 mm NaCl supplemented with 3× FLAG peptide. 1 μg of protein was incubated with or without PP1α (provided by Dr. Meng Choy) in the presence or absence of TTN in a buffer containing 50 mm HEPES, pH 7.4, 100 mm NaCl, 4 mm MnCl2, 0.1 mm EGTA, 5 mm DTT, and 0.025% Tween 20 at 30 °C for 30 min. To determine the effect of phosphate on PP1α activity, PBS (∼10 mm Na2HPO4) or 30 mm Na2HPO4 was added to the assay buffer. The dephosphorylation reaction was terminated by addition of SDS sample buffer and boiling. Protein samples were further resolved in 12% SDS-polyacrylamide gel, dried and exposed to X-ray films.
For in vivo dephosphorylation, FLAG–Ser290 or FLAG–Ser290-VPF/AAA was transfected in GnTI− cells alone or together with GFP–PP1α or constitutively active GFP–PP1α–T320A (kindly provided by Dr. Karen E. Hedin, Mayo Clinic College of Medicine). 48 h after transfection, cells were serum-starved in phosphate-free DMEM for 4 h followed by a 30-min treatment with 100 pm TTN as indicated. Cells were then metabolically labeled for 30 min in phosphate-free DMEM with 0.1 mCi/ml [32P]phosphate. After labeling, cells were lysed in the same lysis buffer as above. Protein was purified with anti-FLAG beads and resolved by 12% SDS-PAGE. 32P incorporation was examined by autoradiography.
Real-time intracellular phosphate
MC3T3-E1 preosteoblasts were seeded on 25-mm coverslips at a density of 11,250 cells/cm2 as described (59). Stock solutions of 2Me-4OMe-TM, at a concentration of 8.3 × 10−5 m, dissolved in 0.1 m NaOH were kept at 4 °C and in the dark before use. Stock solutions of sodium phosphate were prepared using NaH2PO4 and Na2HPO4 (Fluka) in appropriate amounts to obtain the required pH. Commercially available PBS was also used. Phosphate-free buffer contained 12 mm sodium tetramethylammonium sulfate, 130 mm NaCl, 4 mm KCl, and 1 mm CaCl2. High-phosphate buffer contained 100 mm NaH2PO4, 4 mm KCl, and 1 mm CaCl2. Aliquots of forskolin (1 mm) dissolved in DMSO and aqueous solutions of α-toxin from Staphylococcus aureus was prepared and stored at −20 °C until use. Protein-free aqueous solutions were prepared using Milli-Q water and filtered through 0.02-μm filters (Whatman) before use. Coverslips were washed twice with the working buffer before placing in the microscope holder and adding the working buffer.
Fluorescence lifetime images were obtained with a MicroTime 200 (PicoQuant GmbH, Berlin) based on an IX-71 Olympus inverted confocal microscope equipped with a 1.4 NA, ×100 oil immersion objective. The excitation source was a 532-nm pulsed laser (LDH-P-FA-530B, PicoQuant) controlled with a PDL-800 driver (PicoQuant) operating at 20 MHz. The excitation light beam crossed a quarter-wave plate (ThorLabs) and was directed into the specimen through the microscope objective after being reflected in the excitation dichroic mirror (Z532RDC, Chroma). The fluorescence emission was collected back through the microscope objective, filtered by a 550LP (AHF/Chroma) cutoff filter, and focused on a 75-μm pinhole. After the confocal filtering, the fluorescence light crossed through a 630/60 bandpass filter (ThorLabs) and was refocused onto a single photon avalanche diode SPCM-AQR 14 (PerkinElmer Life Sciences). The photon event detection signal was tagged and processed in a TimeHarp 200 single photon timing module (PicoQuant), working in time-tagging time-resolved mode. The photons of each pixel were temporally sorted with respect to the excitation pulse in the histograms with a time resolution of 116 ps/channel.
For FLIM image collection and analysis, the coverslip with cells in the desired buffer was set on the microscope stage, and 4-MeO-2-Me-TM was added to a final concentration of 8 × 10−8 m. An initial pre-scan of an 80 × 80 μm2 was performed to locate a single cell. Then, a small region (between 6 × 6 μm and 12.8 × 12.8 μm) containing the cell membrane was selected. This area was imaged in a fast mode, with 50 × 50 pixel resolution, in bidirectional mode, and a collection time of 3.84 ms/pixel. This allows scanning the selected area near the membrane in 9.6 s. The fluorescence lifetime of the dye, τ, in unaltered cells was collected over 5 min, as an average of at least five images. Once the control τ was established, PTH was added to the sample at a final concentration of 100 nm. Then, images of the selected area were collected over 30 min to follow the changes in the phosphate levels in this area, as evidenced by changes in fluorescence lifetime. Experiments with forskolin were performed by adding 1 μm reagent to the cell culture media during 15 min before imaging. Calibration experiments to establish the response of the dye's τ to total phosphate concentration were performed by treating the cells with 2 μg/ml α-toxin for 30 min (93) before imaging following the same procedure described above (for calibration procedure, see supporting information).
FLIM image data were analyzed using SymphoTime 32 (PicoQuant) and FiJi (100). The fluorescence decay trace within each pixel of the FLIM image was fitted to a single exponential decay function, using a reconstructed instrument response function for the deconvolution analysis. A maximum likelihood estimator was employed as the fitting criterion to improve parameter estimation in decays with low count rates (101). Once the FLIM images were obtained, the τ-based images, where τ is the fluorescence lifetime, were converted to changes of phosphate (Pi) using the calibration information obtained from measurements with α-toxin (Figs. S3 and S4) as described (59).
HDX mass spectrometry
HDX-MS experiments were performed as described (90). In brief, purified NHERF1 protein (WT or S290A) was diluted in working buffer containing 150 mm NaCl and 8.3 mm Tris-HCl, pH 7.2, for HDX-MS analysis. For each protein sample, three sets of samples were prepared: 1) nondeuterated (ND); 2) fully deuterated (FD); and 3) time-dependent on-exchange. A protein/buffer/quench solution ratio of 1:3:6 (volume) was used for all samples. The FD sample sets were prepared by mixing samples with D2O buffer (0.8% formic acid in 100% D2O) and incubated at room temperature for 12 h before quenching. The ND sample sets were prepared using a similar procedure with H2O buffer (150 mm NaCl, 8.3 mm Tris-HCl, pH 7.2 in H2O) without the incubation step. The on-exchange sample sets were prepared by adding 3 volumes of D2O buffer (150 mm NaCl, 8.3 mm Tris-HCl, pH 7.2, in D2O) at 0 °C and incubating for 10, 100, 1000, 10,000, or 100,000 s. Six volumes of ice-cold quench solution was then added to each sample, followed by snap-freezing on dry ice and storage at −80 °C. Samples were thawed immediately before on-line pepsin digestion at 0 °C using a cryogenic autosampler and passed at once over an immobilized porcine pepsin column (16 μl bed volume). Peptide fragments were collected contemporaneously on a C18 trap column, desalted, and separated over 30-min on a Michrom Magic C18AQ (New Objective, Woburn, MA) column using a linear 6.4–38.4% acetonitrile gradient, followed by LC/MS analysis using an Orbitrap Elite mass spectrometer (ThermoFisher Scientific). Both MS1 and MS2 spectra were collected in data-dependent acquisition mode. Peptide identification was performed from LC/MS data sets collected from ND samples using Proteome Discoverer (ThermoFisher Scientific) and the SEQUEST database search engine. The SEQUEST database search results were submitted to HDExaminer (Sierra Analytics Inc.), filtered using several threshold parameters to create an initial peptide pool. The quality of the MS1 data for each filtered peptide was then checked by assigning an initial quality score by HDExaminer software, followed by a quality control process that included manual investigation of peak isotopic envelope and adjusting/improving the quality score. Only high-quality peptide MS1 spectra (mass tolerance <10 ppm) were included in the final peptide pool. The retention times and m/z ranges of each peptide from the final peptide pool were manually verified and adjusted across all LC/MS data sets from on-exchange samples and FD samples to ensure that HDExaminer had selected the correct peptide for all experiments. Results from FD samples were used to monitor the back-exchange rates during on-line pepsin digestion and LC/MS analysis. The centroids of isotopic envelopes of nondeuterated, partially deuterated, and fully deuterated peptides were measured using HDExaminer and then converted to deuteration level with corrections for back-exchange. A deuterium accumulation plot was created for each peptide as a further quality check and data refinement process.
Data analysis
Results were analyzed using Prism 7 software (GraphPad, La Jolla, CA). Results represent the mean ± S.D. or S.E. of n ≥ 3 independent experiments, unless indicated otherwise, and were compared by analysis of variance (ANOVA) or Sidak's multiple comparisons test with post hoc analysis using the Bonferonni procedure. p values < 0.05 were considered statistically significant.
Author contributions
Q. Z. and B. J. resources; Q. Z., K. X., J. M. P., T. M., W. B. S., H. L., D. W., S. L., J. C. M., F. J.-A., and A. O. investigation; Q. Z., K. X., J. M. P., T. M., and B. J. methodology; Q. Z., T. M., A. O., and P. A. F. writing-original draft; Q. Z., D. U., A. O., and P. A. F. writing-review and editing; T. M., S. L., A. O., and P. A. F. formal analysis; T. M. visualization; D. U., R. A.-a., and P. A. F. conceptualization; A. O. and P. A. F. funding acquisition; P. A. F. supervision; P. A. F. project administration.
Supplementary Material
Acknowledgments
We thank Dr. Karen Hedin (Mayo Clinic College of Medicine, Rochester, MN), Dr. Zimei Bu (CCNY, New York), Drs. Jean-Luc Parent (Université de Sherbrooke), and Terence Herbert (McGill University) for providing plasmid DNAs used in this study. We are especially grateful to Dr. Meng S. Choy (Department of Chemistry and Biochemistry, University of Arizona) for generously supplying the PP1α used here.
This work was supported by National Institutes of Health Awards DK105811 and DK111427 (to P. A. F.), Spanish Ministry of Economy and Competitiveness Grant CTQ2014-56370-R, the Agencia Estatal de Investigacion (AEI), and the European Regional Development Fund (ERDF) (to A. O.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S7, Tables S1–S2 and supporting Ref. 1.
Human proteins are indicated by three-letter uppercase convention; genes are in italics. Only the first letter is uppercase for the corresponding mouse protein or gene.
Residue numbering refers to human NHERF1. Where results from other species were reported, the corresponding human residue is used.
- PTH
- parathyroid hormone
- PTHR
- PTH receptor
- NHERF1
- Na+/H+-exchanger regulatory factor 1
- GnTI−
- N-acetylglucosaminyltransferase-deficient HEK-293S cells
- TAP–NHERF1
- tandem affinity purification-tagged NHERF1
- FLIM
- fluorescence lifetime imaging
- HDX
- hydrogen/deuterium exchange mass spectrometry
- pSer290
- phosphorylated Ser290
- NP40
- Nonidet P-40
- TAMRA
- carboxytetramethylrhodamine
- SILAC
- stable isotope labeling of amino acids in cell culture
- DMEM
- Dulbecco's modified Eagle's medium
- FBS
- fetal bovine serum
- ND
- nondeuterated
- FD
- fully deuterated
- EBD
- ezrin-binding domain
- PDB
- Protein Data Bank
- 2Me-4OMe-TM
- 7-hydroxy-5,5-dimethyl-10-(4-methoxy-2-methyl-phenyl)-dibenzo-[b,e]-silin-3(5H)-one
- ANOVA
- analysis of variance
- ID
- intrinsically disordered
- TTN
- tautomycetin
- RPTEC
- renal proximal tubule epithelial cell
- pen/strep
- penicillin and streptomycin
- CFP
- cyan fluorescent protein
- FERM
- Ezrin-Radixin-Moesin
- MD
- molecular dynamics.
References
- 1. Broadbent D., Ahmadzai M. M., Kammala A. K., Yang C., Occhiuto C., Das R., and Subramanian H. (2017) Roles of NHERF family of PDZ-binding proteins in regulating GPCR functions. Adv. Immunol. 136, 353–385 [DOI] [PubMed] [Google Scholar]
- 2. Vaquero J., Nguyen Ho-Bouldoires T. H., Clapéron A., and Fouassier L. (2017) Role of the PDZ-scaffold protein NHERF1/EBP50 in cancer biology: from signaling regulation to clinical relevance. Oncogene 36, 3067–3079 10.1038/onc.2016.462 [DOI] [PubMed] [Google Scholar]
- 3. Reczek D., Berryman M., and Bretscher A. (1997) Identification of EBP50: a PDZ-containing phosphoprotein that associates with members of the ezrin-radixin-moesin family. J. Cell Biol. 139, 169–179 10.1083/jcb.139.1.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Centonze M., Saponaro C., and Mangia A. (2018) NHERF1 between promises and hopes: overview on cancer and prospective openings. Transl. Oncol. 11, 374–390 10.1016/j.tranon.2018.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Weinman E. J., Steplock D., and Shenolikar S. (1993) cAMP-mediated inhibition of the renal brush border membrane Na+-H+ exchanger requires a dissociable phosphoprotein cofactor. J. Clin. Invest. 92, 1781–1786 10.1172/JCI116767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hung A. Y., and Sheng M. (2002) PDZ domains: structural modules for protein complex assembly. J. Biol. Chem. 277, 5699–5702 10.1074/jbc.R100065200 [DOI] [PubMed] [Google Scholar]
- 7. Songyang Z., Fanning A. S., Fu C., Xu J., Marfatia S. M., Chishti A. H., Crompton A., Chan A. C., Anderson J. M., and Cantley L. C. (1997) Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science 275, 73–77 10.1126/science.275.5296.73 [DOI] [PubMed] [Google Scholar]
- 8. Romero G., von Zastrow M., and Friedman P. A. (2011) Role of PDZ proteins in regulating trafficking, signaling, and function of GPCRs. Means, motif, and opportunity. Adv. Pharmacol. 62, 279–314 10.1016/B978-0-12-385952-5.00003-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Weinman E. J., Steplock D., Cha B., Kovbasnjuk O., Frost N. A., Cunningham R., Shenolikar S., Blanpied T. A., and Donowitz M. (2009) PTH transiently increases the percent mobile fraction of Npt2a in OK cells as determined by FRAP. Am. J. Physiol. Renal Physiol. 297, F1560–F1565 10.1152/ajprenal.90657.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sneddon W. B., Ruiz G. W., Gallo L. I., Xiao K., Zhang Q., Rbaibi Y., Weisz O. A., Apodaca G. L., and Friedman P. A. (2016) Convergent signaling pathways regulate parathyroid hormone and fibroblast growth factor-23 action on NPT2A-mediated phosphate transport. J. Biol. Chem. 291, 18632–18642 10.1074/jbc.M116.744052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shenolikar S., Voltz J. W., Minkoff C. M., Wade J. B., and Weinman E. J. (2002) Targeted disruption of the mouse NHERF-1 gene promotes internalization of proximal tubule sodium–phosphate co-transporter type IIa and renal phosphate wasting. Proc. Natl. Acad. Sci. U.S.A. 99, 11470–11475 10.1073/pnas.162232699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Morales F. C., Takahashi Y., Kreimann E. L., and Georgescu M. M. (2004) Ezrin-radixin-moesin (ERM)-binding phosphoprotein 50 organizes ERM proteins at the apical membrane of polarized epithelia. Proc. Natl. Acad. Sci. U.S.A. 101, 17705–17710 10.1073/pnas.0407974101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Karim Z., Gérard B., Bakouh N., Alili R., Leroy C., Beck L., Silve C., Planelles G., Urena-Torres P., Grandchamp B., Friedlander G., and Prié D. (2008) NHERF1 mutations and responsiveness of renal parathyroid hormone. N. Engl. J. Med. 359, 1128–1135 10.1056/NEJMoa0802836 [DOI] [PubMed] [Google Scholar]
- 14. Courbebaisse M., Leroy C., Bakouh N., Salaün C., Beck L., Grandchamp B., Planelles G., Hall R. A., Friedlander G., and Prié D. (2012) A new human NHERF1 mutation decreases renal phosphate transporter NPT2a expression by a PTH-independent mechanism. PLoS ONE 7, e34764 10.1371/journal.pone.0034764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Beck L., Karaplis A. C., Amizuka N., Hewson A. S., Ozawa H., and Tenenhouse H. S. (1998) Targeted inactivation of Npt2 in mice leads to severe renal phosphate wasting, hypercalciuria, and skeletal abnormalities. Proc. Natl. Acad. Sci. U.S.A. 95, 5372–5377 10.1073/pnas.95.9.5372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Prié D., Huart V., Bakouh N., Planelles G., Dellis O., Gérard B., Hulin P., Benqué-Blanchet F., Silve C., Grandchamp B., and Friedlander G. (2002) Nephrolithiasis and osteoporosis associated with hypophosphatemia caused by mutations in the type 2a sodium–phosphate co-transporter. N. Engl. J. Med. 347, 983–991 10.1056/NEJMoa020028 [DOI] [PubMed] [Google Scholar]
- 17. Lederer E., and Wagner C. A. (2019) Clinical aspects of the phosphate transporters NaPi-IIa and NaPi-IIb: mutations and disease associations. Pflugers Arch. 471, 137–148 [DOI] [PubMed] [Google Scholar]
- 18. Karthikeyan S., Leung T., and Ladias J. A. (2001) Structural basis of the Na+/H+ exchanger regulatory factor PDZ1 interaction with the carboxyl-terminal region of the cystic fibrosis transmembrane conductance regulator. J. Biol. Chem. 276, 19683–19686 10.1074/jbc.C100154200 [DOI] [PubMed] [Google Scholar]
- 19. Bhattacharya S., Dai Z., Li J., Baxter S., Callaway D. J., Cowburn D., and Bu Z. (2010) A conformational switch in the scaffolding protein NHERF1 controls autoinhibition and complex formation. J. Biol. Chem. 285, 9981–9994 10.1074/jbc.M109.074005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Morales F. C., Takahashi Y., Momin S., Adams H., Chen X., and Georgescu M. M. (2007) NHERF1/EBP50 head-to-tail intramolecular interaction masks association with PDZ domain ligands. Mol. Cell. Biol. 27, 2527–2537 10.1128/MCB.01372-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cheng H., Li J., Fazlieva R., Dai Z., Bu Z., and Roder H. (2009) Autoinhibitory interactions between the PDZ2 and C-terminal domains in the scaffolding protein NHERF1. Structure 17, 660–669 10.1016/j.str.2009.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ali Khajeh J., Ju J. H., Atchiba M., Allaire M., Stanley C., Heller W. T., Callaway D. J., and Bu Z. (2014) Molecular conformation of the full-length tumor suppressor NF2/Merlin-A small-angle neutron scattering study. J. Mol. Biol. 426, 2755–2768 10.1016/j.jmb.2014.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang B., Means C. K., Yang Y., Mamonova T., Bisello A., Altschuler D. L., Scott J. D., and Friedman P. A. (2012) Ezrin-anchored PKA coordinates phosphorylation-dependent disassembly of a NHERF1 ternary complex to regulate hormone-sensitive phosphate transport. J. Biol. Chem. 287, 24148–24163 10.1074/jbc.M112.369405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Iakoucheva L. M., Radivojac P., Brown C. J., O'Connor T. R., Sikes J. G., Obradovic Z., and Dunker A. K. (2004) The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 32, 1037–1049 10.1093/nar/gkh253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Darling A. L., and Uversky V. N. (2018) Intrinsic disorder and posttranslational modifications: the darker side of the biological dark matter. Front. Genet. 9, 158 10.3389/fgene.2018.00158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bah A., and Forman-Kay J. D. (2016) Modulation of intrinsically disordered protein function by post-translational modifications. J. Biol. Chem. 291, 6696–6705 10.1074/jbc.R115.695056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Blom N., Sicheritz-Pontén T., Gupta R., Gammeltoft S., and Brunak S. (2004) Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 4, 1633–1649 10.1002/pmic.200300771 [DOI] [PubMed] [Google Scholar]
- 28. Weinman E. J., Biswas R. S., Peng G., Peng Q., Shen L., Turner C. L., E X Steplock D., Shenolikar S., and Cunningham R. (2007) Parathyroid hormone inhibits renal phosphate transport by phosphorylation of serine 77 of sodium-hydrogen exchanger regulatory factor-1. J. Clin. Invest. 117, 3412–3420 10.1172/JCI32738 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29. Weinman E. J., Steplock D., Zhang Y., Biswas R., Bloch R. J., and Shenolikar S. (2010) Cooperativity between the phosphorylation of Thr95 and Ser77 of NHERF-1 in the hormonal regulation of renal phosphate transport. J. Biol. Chem. 285, 25134–25138 10.1074/jbc.M110.132423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Song G. J., Leslie K. L., Barrick S., Mamonova T., Fitzpatrick J. M., Drombosky K. W., Peyser N., Wang B., Pellegrini M., Bauer P. M., Friedman P. A., Mierke D. F., and Bisello A. (2015) Phosphorylation of ezrin-radixin-moesin-binding phosphoprotein 50 (EBP50) by Akt promotes stability and mitogenic function of S-phase kinase associated protein-2 (Skp2). J. Biol. Chem. 290, 2879–2887 10.1074/jbc.M114.609768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lim H. C., and Jou T. S. (2016) Ras-activated RSK1 phosphorylates EBP50 to regulate its nuclear localization and promote cell proliferation. Oncotarget 7, 10283–10296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Raghuram V., Hormuth H., and Foskett J. K. (2003) A kinase-regulated mechanism controls CFTR channel gating by disrupting bivalent PDZ domain interactions. Proc. Natl. Acad. Sci. U.S.A. 100, 9620–9625 10.1073/pnas.1633250100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sun C., Zheng J., Cheng S., Feng D., and He J. (2013) EBP50 phosphorylation by Cdc2/cyclin B kinase affects actin cytoskeleton reorganization and regulates functions of human breast cancer cell line MDA-MB-231. Mol. Cells 36, 47–54 10.1007/s10059-013-0014-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. He J., Lau A. G., Yaffe M. B., and Hall R. A. (2001) Phosphorylation and cell cycle-dependent regulation of Na+/H+ exchanger regulatory factor-1 by Cdc2 kinase. J. Biol. Chem. 276, 41559–41565 10.1074/jbc.M106859200 [DOI] [PubMed] [Google Scholar]
- 35. Hall R. A., Spurney R. F., Premont R. T., Rahman N., Blitzer J. T., Pitcher J. A., and Lefkowitz R. J. (1999) G protein-coupled receptor kinase 6A phosphorylates the Na+/H+ exchanger regulatory factor via a PDZ domain-mediated interaction. J. Biol. Chem. 274, 24328–24334 10.1074/jbc.274.34.24328 [DOI] [PubMed] [Google Scholar]
- 36. Fouassier L., Nichols M. T., Gidey E., McWilliams R. R., Robin H., Finnigan C., Howell K. E., Housset C., and Doctor R. B. (2005) Protein kinase C regulates the phosphorylation and oligomerization of ERM binding phosphoprotein 50. Exp. Cell Res. 306, 264–273 10.1016/j.yexcr.2005.02.011 [DOI] [PubMed] [Google Scholar]
- 37. Li J., Poulikakos P. I., Dai Z., Testa J. R., Callaway D. J., and Bu Z. (2007) Protein kinase C phosphorylation disrupts Na+/H+ exchanger regulatory factor 1 autoinhibition and promotes cystic fibrosis transmembrane conductance regulator macromolecular assembly. J. Biol. Chem. 282, 27086–27099 10.1074/jbc.M702019200 [DOI] [PubMed] [Google Scholar]
- 38. Garbett D., LaLonde D. P., and Bretscher A. (2010) The scaffolding protein EBP50 regulates microvillar assembly in a phosphorylation-dependent manner. J. Cell Biol. 191, 397–413 10.1083/jcb.201004115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Andrukhova O., Zeitz U., Goetz R., Mohammadi M., Lanske B., and Erben R. G. (2012) FGF23 acts directly on renal proximal tubules to induce phosphaturia through activation of the ERK1/2-SGK1 signaling pathway. Bone 51, 621–628 10.1016/j.bone.2012.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xin F., and Radivojac P. (2012) Post-translational modifications induce significant yet not extreme changes to protein structure. Bioinformatics 28, 2905–2913 10.1093/bioinformatics/bts541 [DOI] [PubMed] [Google Scholar]
- 41. Chaudhary S., Pak J. E., Gruswitz F., Sharma V., and Stroud R. M. (2012) Overexpressing human membrane proteins in stably transfected and clonal human embryonic kidney 293S cells. Nat. Protoc. 7, 453–466 10.1038/nprot.2011.453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang Q., Xiao K., Liu H., Song L., McGarvey J. C., Sneddon W. B., Bisello A., and Friedman P. A. (2018) Site-specific polyubiquitination differentially regulates parathyroid hormone receptor-initiated MAPK signaling and cell proliferation. J. Biol. Chem. 293, 5556–5571 10.1074/jbc.RA118.001737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Swaney D. L., Wenger C. D., and Coon J. J. (2010) Value of using multiple proteases for large-scale mass spectrometry-based proteomics. J. Proteome Res. 9, 1323–1329 10.1021/pr900863u [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Taus T., Köcher T., Pichler P., Paschke C., Schmidt A., Henrich C., and Mechtler K. (2011) Universal and confident phosphorylation site localization using phosphoRS. J. Proteome Res. 10, 5354–5362 10.1021/pr200611n [DOI] [PubMed] [Google Scholar]
- 45. Hornbeck P. V., Kornhauser J. M., Tkachev S., Zhang B., Skrzypek E., Murray B., Latham V., and Sullivan M. (2012) PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 40, D261–D270 10.1093/nar/gkr1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Normanno D., Négrel A., de Melo A. J., Betzi S., Meek K., and Modesti M. (2017) Mutational phospho-mimicry reveals a regulatory role for the XRCC4 and XLF C-terminal tails in modulating DNA bridging during classical non-homologous end joining. Elife 6, e22900 10.7554/eLife.22900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cunningham R., X E, Steplock D., Shenolikar S., and Weinman E. J. (2005) Defective PTH regulation of sodium-dependent phosphate transport in NHERF-1−/− renal proximal tubule cells and wildtype cells adapted to low phosphate media. Am. J. Physiol. Renal Physiol. 289, F933–F938 10.1152/ajprenal.00005.2005 [DOI] [PubMed] [Google Scholar]
- 48. Weinman E. J., Steplock D., Shenolikar S., and Biswas R. (2011) Fibroblast growth factor-23-mediated inhibition of renal phosphate transport in mice requires sodium-hydrogen exchanger regulatory factor-1 (NHERF-1) and synergizes with parathyroid hormone. J. Biol. Chem. 286, 37216–37221 10.1074/jbc.M111.288357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Biber J., Malmström K., Reshkin S., and Murer H. (1990) Phosphate transport in established renal epithelial cell lines. Methods Enzymol. 191, 494–505 10.1016/0076-6879(90)91032-2 [DOI] [PubMed] [Google Scholar]
- 50. Mahon M. J., Cole J. A., Lederer E. D., and Segre G. V. (2003) Na+/H+ exchanger-regulatory factor 1 mediates Inhibition of phosphate transport by parathyroid hormone and second messengers by acting at multiple sites in opossum kidney cells. Mol. Endocrinol. 17, 2355–2364 10.1210/me.2003-0043 [DOI] [PubMed] [Google Scholar]
- 51. Wang B., Ardura J. A., Romero G., Yang Y., Hall R. A., and Friedman P. A. (2010) Na/H exchanger regulatory factors control PTH receptor signaling by differential activation of Gα protein subunits. J. Biol. Chem. 285, 26976–26986 10.1074/jbc.M110.147785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kremer K. N., Dudakovic A., Hess A. D., Smith B. D., Karp J. E., Kaufmann S. H., Westendorf J. J., van Wijnen A. J., and Hedin K. E. (2015) Histone deacetylase inhibitors target the leukemic microenvironment by enhancing a Nherf1-protein phosphatase 1α-TAZ signaling pathway in osteoblasts. J. Biol. Chem. 290, 29478–29492 10.1074/jbc.M115.668160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Choy M. S., Swingle M., D'Arcy B., Abney K., Rusin S. F., Kettenbach A. N., Page R., Honkanen R. E., and Peti W. (2017) PP1:tautomycetin complex reveals a path toward the development of PP1-specific inhibitors. J. Am. Chem. Soc. 139, 17703–17706 10.1021/jacs.7b09368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhao S., and Lee E. Y. (1997) A protein phosphatase-1-binding motif identified by the panning of a random peptide display library. J. Biol. Chem. 272, 28368–268372 10.1074/jbc.272.45.28368 [DOI] [PubMed] [Google Scholar]
- 55. Shi Y. (2009) Serine/threonine phosphatases: mechanism through structure. Cell 139, 468–484 10.1016/j.cell.2009.10.006 [DOI] [PubMed] [Google Scholar]
- 56. Dohadwala M., da Cruz e Silva E. F., Hall F. L., Williams R. T., Carbonaro-Hall D. A., Nairn A. C., Greengard P., and Berndt N. (1994) Phosphorylation and inactivation of protein phosphatase 1 by cyclin-dependent kinases. Proc. Natl. Acad. Sci. U.S.A. 91, 6408–6412 10.1073/pnas.91.14.6408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wang B., Yang Y., Liu L., Blair H. C., and Friedman P. A. (2013) NHERF1 regulation of PTH-dependent bimodal Pi transport in osteoblasts. Bone 52, 268–277 10.1016/j.bone.2012.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Nagai S., Okazaki M., Segawa H., Bergwitz C., Dean T., Potts J. T. Jr., Mahon M. J., Gardella T. J., and Jüppner H. (2011) Acute down-regulation of sodium-dependent phosphate transporter NPT2a involves predominantly the cAMP/PKA pathway as revealed by signaling-selective parathyroid hormone analogs. J. Biol. Chem. 286, 1618–1626 10.1074/jbc.M110.198416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Paredes J. M., Giron M. D., Ruedas-Rama M. J., Orte A., Crovetto L., Talavera E. M., Salto R., and Alvarez-Pez J. M. (2013) Real-time phosphate sensing in living cells using fluorescence lifetime imaging microscopy (FLIM). J. Phys. Chem. B 117, 8143–8149 10.1021/jp405041c [DOI] [PubMed] [Google Scholar]
- 60. Ba J., Brown D., and Friedman P. A. (2003) CaSR regulation of PTH-inhibitable proximal tubule phosphate transport. Am. J. Physiol. Renal Physiol. 285, F1233–F1243 10.1152/ajprenal.00249.2003 [DOI] [PubMed] [Google Scholar]
- 61. Mamonova T., Kurnikova M., and Friedman P. A. (2012) Structural basis for NHERF1 PDZ domain binding affinity. Biochemistry 51, 3110–3120 10.1021/bi201213w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mamonova T., Zhang Q., Khajeh J. A., Bu Z., Bisello A., and Friedman P. A. (2015) Canonical and noncanonical sites determine NPT2A binding selectivity to NHERF1 PDZ1. PLoS ONE 10, e0129554 10.1371/journal.pone.0129554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Eisinger M. L., Dörrbaum A. R., Michel H., Padan E., and Langer J. D. (2017) Ligand-induced conformational dynamics of the Escherichia coli Na+/H+ antiporter NhaA revealed by hydrogen/deuterium exchange mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 114, 11691–11696 10.1073/pnas.1703422114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhang Q., Chen J., Kuwajima K., Zhang H. M., Xian F., Young N. L., and Marshall A. G. (2013) Nucleotide-induced conformational changes of tetradecameric GroEL mapped by H/D exchange monitored by FT-ICR mass spectrometry. Sci. Rep. 3, 1247 10.1038/srep01247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhou J., Yang L., DeColli A., Freel Meyers C., Nemeria N. S., and Jordan F. (2017) Conformational dynamics of 1-deoxy-d-xylulose 5-phosphate synthase on ligand binding revealed by H/D exchange MS. Proc. Natl. Acad. Sci. U.S.A. 114, 9355–9360 10.1073/pnas.1619981114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kumar G. S., Gokhan E., De Munter S., Bollen M., Vagnarelli P., Peti W., and Page R. (2016) The Ki-67 and RepoMan mitotic phosphatases assemble via an identical, yet novel mechanism. eLife 5, e16539 10.7554/eLife.16539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Karthikeyan S., Leung T., and Ladias J. A. (2002) Structural determinants of the Na+/H+ exchanger regulatory factor interaction with the β2 adrenergic and platelet-derived growth factor receptors. J. Biol. Chem. 277, 18973–18978 10.1074/jbc.M201507200 [DOI] [PubMed] [Google Scholar]
- 68. Jiang Y., Lu G., Trescott L. R., Hou Y., Guan X., Wang S., Stamenkovich A., Brunzelle J., Sirinupong N., Li C., and Yang Z. (2013) New conformational state of NHERF1-CXCR2 signaling complex captured by crystal lattice trapping. PLoS ONE 8, e81904 10.1371/journal.pone.0081904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Li J., Callaway D. J., and Bu Z. (2009) Ezrin induces long-range interdomain allostery in the scaffolding protein NHERF1. J. Mol. Biol. 392, 166–180 10.1016/j.jmb.2009.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Khoury G. A., Baliban R. C., and Floudas C. A. (2011) Proteome-wide post-translational modification statistics: frequency analysis and curation of the Swiss-Prot database. Sci. Rep. 1, srep00090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Nishi H., Hashimoto K., and Panchenko A. R. (2011) Phosphorylation in protein-protein binding: effect on stability and function. Structure 19, 1807–1815 10.1016/j.str.2011.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Pawson T., and Scott J. D. (2005) Protein phosphorylation in signaling–50 years and counting. Trends Biochem. Sci. 30, 286–290 10.1016/j.tibs.2005.04.013 [DOI] [PubMed] [Google Scholar]
- 73. Dephoure N., Gould K. L., Gygi S. P., and Kellogg D. R. (2013) Mapping and analysis of phosphorylation sites: a quick guide for cell biologists. Mol. Biol. Cell 24, 535–542 10.1091/mbc.e12-09-0677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ardura J. A., and Friedman P. A. (2011) Regulation of G protein-coupled receptor function by Na+/H+ exchange regulatory factors. Pharmacol. Rev. 63, 882–900 10.1124/pr.110.004176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Bollen M., Peti W., Ragusa M. J., and Beullens M. (2010) The extended PP1 toolkit: designed to create specificity. Trends Biochem. Sci. 35, 450–458 10.1016/j.tibs.2010.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Bertolotti A. (2018) The split protein phosphatase system. Biochem. J. 475, 3707–3723 10.1042/BCJ20170726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Hendrickx A., Beullens M., Ceulemans H., Den Abt T., Van Eynde A., Nicolaescu E., Lesage B., and Bollen M. (2009) Docking motif-guided mapping of the interactome of protein phosphatase-1. Chem. Biol. 16, 365–371 10.1016/j.chembiol.2009.02.012 [DOI] [PubMed] [Google Scholar]
- 78. McWhirter C., Lund E. A., Tanifum E. A., Feng G., Sheikh Q. I., Hengge A. C., and Williams N. H. (2008) Mechanistic study of protein phosphatase-1 (PP1), a catalytically promiscuous enzyme. J. Am. Chem. Soc. 130, 13673–13682 10.1021/ja803612z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Andrews L. D., Zalatan J. G., and Herschlag D. (2014) Probing the origins of catalytic discrimination between phosphate and sulfate monoester hydrolysis: comparative analysis of alkaline phosphatase and protein tyrosine phosphatases. Biochemistry 53, 6811–6819 10.1021/bi500765p [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Hernando N., Déliot N., Gisler S. M., Lederer E., Weinman E. J., Biber J., and Murer H. (2002) PDZ-domain interactions and apical expression of type IIa Na/Pi co-transporters. Proc. Natl. Acad. Sci. U.S.A. 99, 11957–11962 10.1073/pnas.182412699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Gisler S. M., Stagljar I., Traebert M., Bacic D., Biber J., and Murer H. (2001) Interaction of the type IIa Na/Pi co-transporter with PDZ proteins. J. Biol. Chem. 276, 9206–9213 10.1074/jbc.M008745200 [DOI] [PubMed] [Google Scholar]
- 82. Bacic D., Wagner C. A., Hernando N., Kaissling B., Biber J., and Murer H. (2004) Novel aspects in regulated expression of the renal type IIa Na/Pi-co-transporter. Kidney Int. Suppl. 2004, S5–S12 10.1111/j.1523-1755.2004.09102.x [DOI] [PubMed] [Google Scholar]
- 83. Pfister M. F., Lederer E., Forgo J., Ziegler U., Lötscher M., Quabius E. S., Biber J., and Murer H. (1997) Parathyroid hormone-dependent degradation of type II Na+/Pi co-transporters. J. Biol. Chem. 272, 20125–20130 10.1074/jbc.272.32.20125 [DOI] [PubMed] [Google Scholar]
- 84. Pfister M. F., Ruf I., Stange G., Ziegler U., Lederer E., Biber J., and Murer H. (1998) Parathyroid hormone leads to the lysosomal degradation of the renal type II Na/Pi co-transporter. Proc. Natl. Acad. Sci. U.S.A. 95, 1909–1914 10.1073/pnas.95.4.1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Castro M., Nikolaev V. O., Palm D., Lohse M. J., and Vilardaga J. P. (2005) Turn-on switch in parathyroid hormone receptor by a two-step parathyroid hormone binding mechanism. Proc. Natl. Acad. Sci. U.S.A. 102, 16084–16089 10.1073/pnas.0503942102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Ferrandon S., Feinstein T. N., Castro M., Wang B., Bouley R., Potts J. T., Gardella T. J., and Vilardaga J. P. (2009) Sustained cyclic AMP production by parathyroid hormone receptor endocytosis. Nat. Chem. Biol. 5, 734–742 10.1038/nchembio.206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Weinman E. J., Steplock D., Shenolikar S., and Blanpied T. A. (2011) Dynamics of PTH-induced disassembly of Npt2a/NHERF-1 complexes in living OK cells. Am. J. Physiol. Renal Physiol. 300, F231–F235 10.1152/ajprenal.00532.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Sim A. T., and Scott J. D. (1999) Targeting of PKA, PKC and protein phosphatases to cellular microdomains. Cell Calcium 26, 209–217 10.1054/ceca.1999.0072 [DOI] [PubMed] [Google Scholar]
- 89. Park J. Y., Duc N. M., Kim D. K., Lee S. Y., Li S., Seo M. D., Woods V. L., and Chung K. Y. (2015) Different conformational dynamics of PDZ1 and PDZ2 in full-length EBP50 analyzed by hydrogen/deuterium exchange mass spectrometry. Biochem. Cell Biol. 93, 290–297 10.1139/bcb-2014-0145 [DOI] [PubMed] [Google Scholar]
- 90. Xiao K., Zhao Y., Choi M., Liu H., Blanc A., Qian J., Cahill T. J. 3rd, Li X., Xiao Y., Clark L. J., and Li S. (2018) Revealing the architecture of protein complexes by an orthogonal approach combining HDXMS, CXMS, and disulfide trapping. Nat. Protoc. 13, 1403–1428 10.1038/nprot.2018.037 [DOI] [PubMed] [Google Scholar]
- 91. Alizadeh Naderi A. S., and Reilly R. F. (2010) Hereditary disorders of renal phosphate wasting. Nat. Rev. Nephrol. 6, 657–665 10.1038/nrneph.2010.121 [DOI] [PubMed] [Google Scholar]
- 92. Clinkenbeard E. L., and White K. E. (2017) Heritable and acquired disorders of phosphate metabolism: Etiologies involving FGF23 and current therapeutics. Bone 102, 31–39 10.1016/j.bone.2017.01.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Herrero-Foncubierta P., Paredes J. M., Giron M. D., Salto R., Cuerva J. M., Miguel D., and Orte A. (2018) A red-emitting, multidimensional sensor for the simultaneous cellular imaging of biothiols and phosphate ions. Sensors 18, 161 10.3390/s18010161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Roy S. J., Glazkova I., Fréchette L., Iorio-Morin C., Binda C., Pétrin D., Trieu P., Robitaille M., Angers S., Hébert T. E., and Parent J. L. (2013) Novel, gel-free proteomics approach identifies RNF5 and JAMP as modulators of GPCR stability. Mol. Endocrinol. 27, 1245–1266 10.1210/me.2013-1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Wieser M., Stadler G., Jennings P., Streubel B., Pfaller W., Ambros P., Riedl C., Katinger H., Grillari J., and Grillari-Voglauer R. (2008) hTERT alone immortalizes epithelial cells of renal proximal tubules without changing their functional characteristics. Am. J. Physiol. Renal Physiol. 295, F1365–F1375 10.1152/ajprenal.90405.2008 [DOI] [PubMed] [Google Scholar]
- 96. Li J., Dai Z., Jana D., Callaway D. J., and Bu Z. (2005) Ezrin controls the macromolecular complexes formed between an adapter protein Na+/H+ exchanger regulatory factor and the cystic fibrosis transmembrane conductance regulator. J. Biol. Chem. 280, 37634–37643 10.1074/jbc.M502305200 [DOI] [PubMed] [Google Scholar]
- 97. Jüppner H., Abou-Samra A. B., Freeman M., Kong X. F., Schipani E., Richards J., Kolakowski L. F. Jr., Hock J., Potts J. T. Jr., and Kronenberg H. M. (1991) A G protein-linked receptor for parathyroid hormone and parathyroid hormone-related peptide. Science 254, 1024–1026 10.1126/science.1658941 [DOI] [PubMed] [Google Scholar]
- 98. Gesek F. A., and Friedman P. A. (1992) On the mechanism of parathyroid hormone stimulation of calcium uptake by mouse distal convoluted tubule cells. J. Clin. Invest. 90, 749–758 10.1172/JCI115947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Eng J. K., McCormack A. L., and Yates J. R. (1994) An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989 10.1016/1044-0305(94)80016-2 [DOI] [PubMed] [Google Scholar]
- 100. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J. Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., and Cardona A. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Maus M., Cotlet M., Hofkens J., Gensch T., De Schryver F. C., Schaffer J., and Seidel C. A. (2001) An experimental comparison of the maximum likelihood estimation and nonlinear least-squares fluorescence lifetime analysis of single molecules. Anal. Chem. 73, 2078–2086 10.1021/ac000877g [DOI] [PubMed] [Google Scholar]
- 102. Voltz J. W., Brush M., Sikes S., Steplock D., Weinman E. J., and Shenolikar S. (2007) Phosphorylation of PDZ1 domain attenuates NHERF-1 binding to cellular targets. J. Biol. Chem. 282, 33879–33887 10.1074/jbc.M703481200 [DOI] [PubMed] [Google Scholar]
- 103. Rubino R., Bezzerri V., Favia M., Facchini M., Tebon M., Singh A. K., Riederer B., Seidler U., Iannucci A., Bragonzi A., Cabrini G., Reshkin S. J., and Tamanini A. (2014) Pseudomonas aeruginosa reduces the expression of CFTR via post-translational modification of NHERF1. Pflugers Arch. 466, 2269–2278 10.1007/s00424-014-1474-6 [DOI] [PubMed] [Google Scholar]
- 104. Nikolaev V. O., Bunemann M., Hein L., Hannawacker A., and Lohse M. J. (2004) Novel single chain cAMP sensors for receptor-induced signal propagation. J. Biol. Chem. 279, 37215–37218 10.1074/jbc.C400302200 [DOI] [PubMed] [Google Scholar]
- 105. Wu J., Liu L., Matsuda T., Zhao Y., Rebane A., Drobizhev M., Chang Y. F., Araki S., Arai Y., March K., Hughes T. E., Sagou K., Miyata T., Nagai T., Li W. H., and Campbell R. E. (2013) Improved orange and red Ca2+ indicators and photophysical considerations for optogenetic applications. ACS Chem. Neurosci. 4, 963–972 10.1021/cn400012b [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.