Abstract
Tests to detect the presence and activity of hepatitis B virus (HBV) are the cornerstones of diagnosis and management. Assays that detect or measure serum levels of HB surface antigen, HB surface antibody, and HB core antibody are used to identify patients with exposure to HBV, whereas other tests provide information on the level of virus replication, the presence of specific variants, and presence of virus reservoirs. Newer diagnostic tests, used only in research settings so far, aim to quantify levels of intrahepatic HBV replication. Other tests have been developed to detect HBV infection in resource-limited settings. We review point of care tests (essential in global screening efforts), standard diagnostic tests used in routine clinical management, and newer tests that might be used in clinical trials of agents designed to cure HBV infection.
Keywords: HBsAg, point-of-care, quantitative, HBV RNA
Assays that detect or measure serum levels of HB surface antigen (HBsAg), HB surface antibody (anti-HBs), and HB core antibody (anti-HBc) are used to identify patients who have been exposed to HBV, whereas other tests provide information on the level of virus replication, the presence of specific variants, and presence of virus reservoirs. Tests are being developed to quantify levels of intrahepatic HBV replication. These biomarkers are used to identify patients with HBV infection, follow disease progression and response to therapy, and determine efficacy of new agents in clinical trials.
The goal of HBV treatment is sterilizing cure, defined as a sustained loss of HB surface antigen HBsAg from serum, loss of HBV DNA from serum and liver, and loss of closed circular DNA (cccDNA) and integration of HBV DNA into genome. This may or may not be achievable. A more immediately feasible goal is functional cure, defined as loss of HBsAg, with or without loss of anti-HBs. Access to HBV tests varies worldwide; resource-constrained areas are less likely to have access to tests that measure virus replication or detect variants and have greater reliance on serologic tests. In difficult-to-access populations, point of care tests are important and significant advances have been made in the past few years.
Point of Care and Dried-blood Spot Tests
An estimated 292 million persons have chronic HBV infection worldwide, but only 10% have been diagnosed 1. Acute or chronic HBV infection is established based on detection of HBsAg in serum using an enzyme immunoassay (EIA) or chemiluminescence immunoassay. However, these laboratory-based immunoassays may not be readily accessible or affordable, particularly in resource-constrained countries. Point of care (POC) tests provide an alternative means of diagnosis (Figure 1). In some high-income counties, rapid diagnostic tests offered at the point of care are needed for populations unable or unwilling to access regular medical care, such as injection drug users or homeless or uninsured individuals. Ideal rapid diagnostic tests are inexpensive, easy to use, and placed within a closed system to avoid cross-contamination. They need a long shelf life for tropical climates and should not require cold chain transportation and storage. Currently available POC tests are small devices that use blood or saliva to detect or measure of viral antibodies and/or antigens 2. The World Health Organization (WHO) has endorsed the use of rapid diagnostic tests for diagnosis of chronic HBV infection, 3 but the American Association for the Study of Liver Diseases (AASLD) and European Association for the Study of the Liver (EASL) guidelines do not. The WHO recommends point of care tests to improve access and linkage to care and treatment. Only a few rapid diagnostic tests for HBsAg have met WHO qualification criteria (Vikia HBsAg, Biomérieux, France; BIOLINE HBsAg, Standard Diagnostics).
Figure 1. Point of Care Tests.
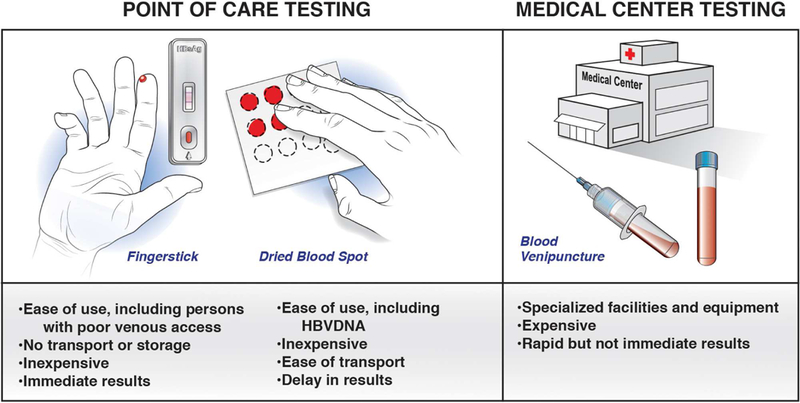
POC tests are important for populations unable or unwilling to access regular medical care, such as among injection drug users, homeless, or the uninsured. POC tests use either fingersticks or dried blood spots. They are easy to use and inexpensive. Fingerstick POC tests provide immediate results whereas dry blood spots must be mailed for central testing.
Recent meta-analyses have shown the performance characteristics of rapid diagnostic tests for HBsAg, using EIA and nucleic-acid tests as reference standards. An analysis of 30 studies, in 23,716 individuals from 23 countries, assessed the diagnostic accuracy of 33 brands of rapid diagnostic tests against a reference standard of enzyme immunoassays. These tests identified patients with chronic HBV infection with a pooled sensitivity of 90.0% and a pooled specificity of 99.5% (ref 4). The accuracy of these tests did not differ with use of serum, plasma, or venous or capillary whole blood. The brand of rapid diagnostic test associated with accuracy. The presence of HIV coinfection significantly reduced sensitivity, to 72%, but the high specificity was preserved 4. False-negative results have been linked with low levels of HBsAg, HBV DNA, mutations in HBsAg, and different HBV genotypes or subtypes 5. However, newer rapid tests have overcome these limitations 6, 7. A study from Gambia compared 3 different POC tests (fingerstick and dried blood spot [DBS]) with a serum EIA, and found these POC tests to identify patient with chronic HBV infection with 89%–94% sensitivity and 95%–100% specificity; almost all false-negative results were from inactive carriers with low levels of HBsAg 8. Although lower levels of sensitivity may be an acceptable in areas in which access to any testing is limited, the application of POC tests requires consideration of benefits (broader access) vs risks (missed cases). One study found increased liver stiffness (above 7.2 kPa) in 17% of patients with negative results from POC tests for HBsAg, raising concerns about missed cases with significant liver disease 8. Certainly, POC or DBS tests are useful for epidemiology studies to define disease burden and effects of vaccination or other interventions.
Capillary DBS tests are cheap and accessible, and unlike rapid diagnostic tests, can measure levels of HBsAg and HBV DNA (Figure 1). In a meta-analysis of DBS for detection of HBsAg, the DBS identified patients with chronic HBV infection with a weighted sensitivity value of 99% and specificity value of 100% (compared to venous blood tests) 9. When DBS are used to detect HBV DNA, the amount of blood analyzed affects sensitivity. In a meta-analysis of DBS (compared to venous blood tests), DBS detected HBV DNA with a pooled estimated sensitivity value of 95% and specificity of 99% 10. Additionally, DBS might not always detect lower levels of virus—this might not be a major issue, because pati ents with low-level viremia are typically not candidates for antiviral therapy. No DBS tests have been approved for use by the Food and Drug Administration.
Rapid diagnostic tests for anti-HBs are available but have been less-extensively studied than HBsAg tests. These tests detect anti-HBs with 96%–98% specificity but only 60%–70% sensitivity; false-negative results are largely related to low titers of anti-HBs 11, 12. Levels of sensitivity are above 90% when levels of anti-HBs are greater than 150 U/L12. In resource-constrained countries, rapid diagnostic tests for HBsAg followed by vaccination could be a better approach than additional tests for anti-HBs by rapid diagnostic tests. It would be helpful to have rapid tests to measure levels of HB e antigen (HBeAg), to select patients for treatment when tests for HBV DNA are not available. This is particularly relevant for identification of pregnant women who may benefit from antiviral therapy in the last trimester as a means of reducing mother to child transmission of HBV. Limited data indicate the high specificity and sensitivity of POC tests for HBeAg. In a study comparing POC tests to EIA, as a reference standard, in 942 patients (303 HBeAg positive), the serum POC identified patients with chronic HBV infection with 96.4% sensitivity and 99.4% specificity, with comparable or higher performance for whole blood samples13. No rapid diagnostic test for HBeAg has been approved by the WHO.
Tests Used in Routine Clinical Practice
Tests to detect antigens, antibodies, and viral nucleic acids are used routinely for diagnosis and monitoring of HBV infection (Table 1). Progression of chronic HBV infection involves interactions between the virus and the immune response. Whether the infection persists for months or a lifetime, the serial evaluation of serologic markers, levels of alanine aminotransferase (ALT), and HBV DNA guides management.
Table 1.
Tests Used in Management of Patients with HBV Infection
| Diagnostic Test | Clinical Interpretation | Typical Scenarios for its Use |
|---|---|---|
| HBsAg | Marker of acute and chronic HBV infection |
Initial presentation; Annually in patients with inactive infection; Concern for sero-reversion (in immune- suppressed patients) |
|
Quantification of HBsAg |
Aids in defining phase of infection, identifying patients most likely to respond to interferon, and determining likelihood of HBV reactivation |
Aids in distinguishing patients with HBeAg-negative chronic HBV infection from inactive carriers; Prediction of mother to child transmission Determine frequency of measurement of ALT and elastography tests for patients with inactive infections; To identify patients for withdrawal of NA therapy; To identify patients unlikely to respond to continue peginterferon therapy |
| Anti-HBs | Marker of immunity (natural or with vaccination) |
Upon presentation and when HBsAg loss has been documented |
| Anti-HBc | Marker of HBV exposure | Initial presentation |
| HBeAg | Associated with high levels of HBV DNA, marker of infectivity |
Initial presentation Every 6 months in HBeAg-positive patients on treatment; HBV flares (changes in levels of ALT and HBV DNA) |
| Anti-HBe | Associated with lower levels of HBV DNA |
Initial presentation; Every 6 months in HBeAg-positive patients receiving treatment; HBV flares (changes in levels of ALT and HBV DNA) |
| HBV DNA | Detects HBV infection, used to define phase of infection and need for HBV therapy |
Initial presentation; Every 6–12 months in untreated patients; Every 3 months for patients receiving treatment until undetectable, then every 6 months; For patients with ALT flares; Anti-HBc positive persons at risk for HBV reactivation (such as immunosuppressed patients) |
| HBV genotype | Genotypes A–H, determined by detection of specific sequences |
When considering peginterferon therapy; Useful for epidemiology studies and to broadly define disease progression |
|
Precore or basal core promoter mutations |
Detects presence of mutations in the precore and BCP regions |
None established; Useful for epidemiology studies and to broadly define disease progression |
|
HBV resistance tests |
Detects presence (more than 10%) of specific HBV substitutions associated with resistance to NAs |
In patients with virologic breakthrough on NAs |
In each HBsAg-positive person, tests are used to define the phase of infection (Figure 2), determine viral coinfections, and stage liver disease severity. Establishing whether a patient has advanced fibrosis or cirrhosis is important for surveillance procedures (for liver cancer and varices) and strongly influences antiviral treatment recommendations. The EASL and AASLD each recommend antiviral therapy for all patients with cirrhosis, regardless of levels of HBV DNA and/or ALT 14, 15. For patients without cirrhosis, AASLD and Asian Pacific Association for the Study of the Liver treatment guidelines recommend treatment for individuals with levels of HBV DNA above 2000 IU/mL, if they are HBeAg-negative, and above 20,000 IU/mL if they are HBeAg-positive and have levels of ALT twice upper limit of normal 16, 17. EASL treatment guidelines endorse a lower ALT threshold15. Clearly, accurate tools to measure HBV DNA are essential for management.
Figure 2. Phases of Chronic HBV Infection.
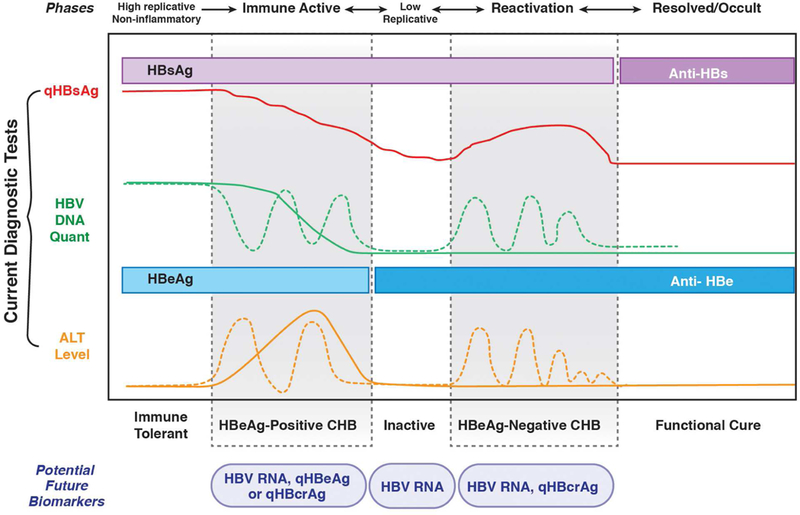
Tests recommended for persons with chronic HBV infection include detection and quantification (q) of HBsAg, HBV DNA, HBeAg, and ALT, which indicate changes of the course of chronic HBV infection. There are 5 phase of infection: immune tolerance (also referred to as high-replicative, non-inflammatory phase), HBeAg-positive and HBeAg-negative chronic infection, inactive chronic infection (inactive carrier), and resolved chronic infection (functional cure). Transitions between phases are not unidirectional. Patients can revert back to an earlier phase or move back and forth between 2 phases over the course of their chronic infection.
Tests to measure HBeAg and the antibody against HBeAg define phase of infection, determine HBV DNA thresholds for initiation of treatment, and serve as an intermediate treatment endpoint for some patients. Tests to quantify the level of HBsAg (qHBsAg) have been widely used in Europe, Canada, and Asia but have been available in the United States only since 2017. These tests have an increasingly important role in management of patients with chronic HBV infection. HBV genotypes and mutations in precore, and basal core promoter (BCP) promoters are analyzed in epidemiology and disease progression studies but this information is rarely used in diagnosis or management.
HBV DNA quantification
Accurate methods to detect and quantify HBV DNA are essential to diagnose acute and chronic infections, guide treatment decisions, assess responses to treatment, and determine risk of HBV-related complications. Historically, hybridization assays were used to estimate HBV DNA levels in blood samples but their level of sensitivity was suboptimal. Amplification-based assays detect HBV DNA with high specificity (99%) and sensitivity (≥95%), with limits of quantitation ranging from 10–20 IU/mL (approximately 50–100 virus genome copies/mL). The WHO established the first international standard for HBV DNA, IU, for calibration of reference reagents used in HBV nucleic acid amplifications techniques. The establishment of this international standard allowed comparison of HBV DNA among different laboratories and assays. There are 3 FDA-approved assays for HBV DNA quantification (Supplemental Table 1). The high cost and need for specialized equipment limit accessibility of these assays in resource-limited settings.
The sensitivity of transcription-mediated and real-time PCR-based quantitative assays for HBV DNA obviates the need for qualitative HBV DNA tests. During acute HBV infection, HBV DNA is the only marker of infection during the window-phase of the infection. Detection of window-phase infections is most relevant to tests for blood and organ donors; use of sensitive nucleic acid tests reduces time to detection of HBV infection after exposure from an average of approximately 32 days (HBsAg detected) to 15 days (HBV DNA detected) 18, 19. A second window for testing can occur after HBsAg clearance and before anti-HBs is measurable—in this period, anti-HBc is the marker of HBV infection, but HBV DNA can be measured to determine infectivity. Additionally, HBV DNA tests can confirm viremia in patients with chronic infection but with HBsAg mutations that are not detected. The frequency of such HBsAg mutants in the general population is unknown. Next-generation HBsAg tests, which include monoclonal and polyclonal antibodies directed against epitopes within and outside the a determinant of HBsAg, have increased capacity to detect these HBsAg mutants (such as the Elecsys HBsAg II assay; Roche Diagnostics) 20.
HBV DNA tests are important for management of patients with chronic HBV infection (Supplemental Table 2) 14–16. Serial monitoring of levels of HBV DNA and ALT is used to determine the need for and response to anti-HBV therapy. Recommended minimum thresholds of HBV DNA for which initiation of antiviral therapy is recommended in patients with increased levels of ALT are 2000 IU/mL or more for HBeAg-negative patients and 20,000 IU/mL or more for HBeAg-positive patients 14–16. The goal of therapy is to achieve undetectable HBV DNA level with a sensitive PCR assay. Serial monitoring of HBV DNA is used to detect the emergence of viral resistance and medication adherence 21.
Measurement of HBsAg
HBsAg originates from the episomal mini-chromosome (cccDNA) and is translated from pre-S1 and S2 mRNAs that are transcribed from the S gene. An additional source of HBsAg comes from randomly integrated locations in the host genome. The contributions of HBsAg from cccDNA and integrated sources can differ among individuals (Figure 3). For example, the correlation between level of HBsAg and serum level of HBV DNA, intrahepatic cccDNA, and total HBV DNA was found to be high in HBeAg-positive patients (r = 0.69, 0.71, 0.76). Among HBeAg-negative individuals, there was low correlation between level of HBsAg with HBV DNA (r = 0.28) and no correlation between level of intrahepatic cccDNA and total HBV DNA, possibly due to increasing integration of HBV with longer duration of infection22. HBsAg exists as 3 protein subtypes (small, middle, and large) that are not currently differentiated by commercial assays. Additionally, there are 2 non-infectious subviral particles secreted, in spherical and filamentous forms, with 100-fold to 100,000-fold higher levels than mature virions. The production of HBsAg from these subviral particles and integrated HBV DNA within the host genome have been proposed to contribute to HBV’s capacity to evade immune surveillance23.
Figure 3. New Serum Markers of HBV.
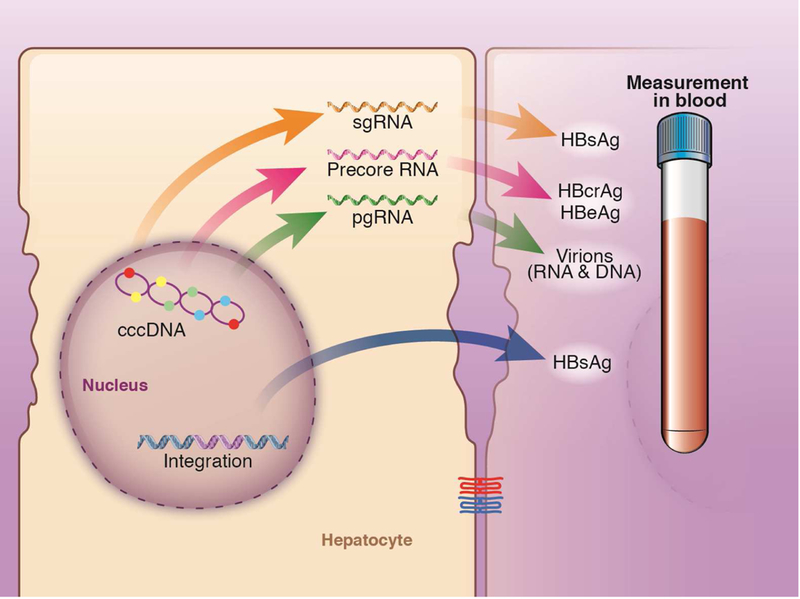
HBV cccDNA is the template for all known HBV RNA transcripts. These include (i) HBV full-length pregenomic RNA, which is eventually packaged to form progeny virions, (ii) shorter subgenomic RNAs, which are translated into HBsAg, and (iii) pre-core RNA, which encodes the secreted HBeAg and HBcrAg. HBsAg can also originate from noninfectious integrated viral DNA.
Commercially available assays for HBsAg include Architect HBsAg QT (Abbott Diagnostics), Elecsys II (Roche Diagnostics), and Liaison XL Murex HBsAg quant (DiaSorin)—all have lower limits of detection of 0.05 IU/mL (upper limit above 50,000 IU/mL with dilution). There is a high level of agreement between results from the Architect HBsAg QT and Elecsys II assays24. Additionally, a linearized HBsAg assay is being tested, with increased sensitivity at lower limits of detection (0.005 IU/mL) 25. New monoclonal antibody tests to quantify HBsAg protein composition have identified different patterns of proteins with phase of disease—lower proportions of large and middle protein subtypes are detected in inactive carriers compared to patients with acute or chronic HBV infections 26.
Measurement of qHBsAg and disease progression
Levels of HBsAg differ with phase of infection as well as genotype. The highest levels (above 4 log10 IU/mL) are seen in the immune-tolerant (non-inflammatory, high replication) phase and lowest in inactive carriers (approximately 2 log10 IU/mL). Within the same phase, levels of HBsAg are higher in patients with HBV genotype A infection than other genotypes, and lower if pre-S escape mutants are dominant 27. Although level of HBsAg is not associated with likelihood of HBeAg seroconversion, its greatest utility may lie in its ability to differentiate patients with HBeAg-negative, immune-active hepatitis from inactive carriers among indeterminate or grey zone patients—a frequently encountered group in cli nical practice 28. Baseline level of HBsAg below 1000 IU/mL with levels of HBV DNA below 2000 IU/mL identifies patients with inactive chronic HBV genotype D infection with a 90% PPV 29. When these cut-off values were applied to patients with HBV genotype B or C patients infection in the HBV-REVEAL community-based cohort, they identified inactive carriers with a PPV of 83% 30. In patients with inactive chronic HBV infection, levels of HBsAg below 100 IU/mL have been associated with and incorporated into simple scoring systems to predict spontaneous loss of HBsAg 31. A more prominent decrease in qHBsAg (such as a reduction of 1 log IU/mL or more) in patients with inactive chronic HBV infection heralds spontaneous HBsAg loss 29. Therefore, baseline, and possibly longitudinal, measurement of HBsAg, in conjunction with measurement of HBV DNA and ALT, might be used to identify patients with HBeAg-negative infection (Table 1). Under EASL guidelines, follow-up intervals for HBeAg-negative patients with HBV DNA levels below 2000 IU/mL differ based on qHBsAg, with yearly measurements of ALT and HBV DNA and assessment of fibrosis every 3 years, if baseline levels of HBsAg are below 1000 IU/mL vs every 6 months and every 2 years if levels of HBsAg are above 1000 IU/mL 15. The AASLD does not incorporate quantification of HBsAg in management algorithms 17.
Levels of HBsAg have been used to predict vertical transmission; in resource-constrained settings where HBV DNA testing is complex and costly, this may be particularly useful. Level of HBsAg above 4.1 log10 IU/mL identified mothers who had transmitted the virus to their newborn with 100% sensitivity and 71% specificity 32. Quantification of HBsAg has also been added to prediction rules for incident HCC. In a study of HBeAg-negative individuals with low viral load, the risk of HCC was 14-fold higher when levels of HBsAg were above 1000 vs below 1000 IU/mL 33. Studies are needed to replicate these findings larger and more diverse cohorts.
Measurement of qHBsAg and treatment
Levels of HBsAg decrease slowly in patients receiving nucleos(t)ide analogue (NA) treatment, compared to levels of HBV DNA 34. Although baseline qHBsAg and rapid decrease in qHBsAg have been linked to likelihood of HBeAg seroconversion and virus suppression in HBeAg-positive patients receiving NAs, no correlation was observed in HBeAg-negative patients 35. In addition, the lower the level of HBsAg, the greater the likelihood for HBsAg loss, although HBsAg loss occurs in only 1%–2% of patients receiving treatment per year, regardless of NA type 36. A decrease in qHBsAg of 1 log IU/mL or more at week 24 of treatment is associated with loss of HBsAg in HBeAg-positive patients receiving NA treatment 37, 38. The role of clinical monitoring of HBsAg levels in patients receiving NA therapy is evolving, with a potential use in identifying individuals with low levels of HBsAg (below 100 IU/mL), who may be candidates for treatment discontinuation39.
In contrast, qHBsAg can be a valuable tool in management of patients receiving peginterferon therapy 40. Peginterferon can induce a strong decrease in level of HBsAg, attributed to immune-modulatory mechanisms, with the most- and least-striking decreases seen in patients with HBV genotypes A or D, respectively. In patients with HBV and HIV co-infection, decreases in qHBsAg on antiviral therapy are associated with increasing numbers of CD4+ T cells, indicating that immune control is necessary for HBsAg clearance 41.
Given the toxicities and side effects of peginterferon, early identification of non-responders is important. In HBeAg-positive patients with genotype B or C HBV infection, a level of HBsAg below 1500 IU/mL at week 12 of peginterferon treatment was associated with subsequent HBeAg seroconversion in 57% of patients and HBsAg loss in 18%. Conversely, no patient with HBV genotype B or C infection with level of HBsAg above 20,000 IU/mL at weeks 12 and 24 achieved a response to treatment (negative predictive value, 100%), defined as HBeAg-seroconversion and HBV DNA level below 2000 IU/mL after 6 months and 3 years of follow up 42. In patients with HBV genotype D, a less than 2 log decrease in level of HBsAg at 12 weeks identified patients without a response to therapy with a negative predictive value of 100%. Among HBV genotypes, level of HBsAg below 1500 IU/mL at weeks 12 and 24 was associated with higher rates of response to treatment. Based on these data, genotype-specific stopping rules for treatment have been proposed 15. Specifically, in HBeAg-positive patients, levels of HBsAg above 20,000 IU/mL in patients with HBV genotypes B or C, or no decrease in HBsAg in patients with HBV genotypes A or D at 12 weeks, are indications to stop therapy (Table 1). In HBeAg-negative patients with genotype D HBV infection, those without any decrease in level of HBsAg by week 12 and at least a 2-log10 decrease in HBV DNA at 12 weeks, are recommended to stop treatment—this profile identifies patients who do not respond to therapy with a 100% negative predictive value 43. Fewer HBeAg-negative patients who do not have HBV genotype D infections have been studied and no stopping rules have been endorsed.
It would be ideal to identify patients who will maintain or attain inactive status or develop HBsAg loss if they discontinue long-term NA therapy. A trial from Greece initially found that a lower level of HBsAg prior to NA withdrawal increased the odds of subsequent HBsAg loss (median 1733 IU/mL vs 4905 IU/mL in patients with vs without loss, respectively) 44. A meta-analysis of mostly retrospective studies of patients who stopped NA therapy found that a level of HBsAg below 100–200 IU/mL prior to withdrawal indicated a high chance of sustained response as well as HBsAg loss in HBeAg-negative individuals who had at least 2 years of consolidation therapy 45. Conversely, higher end of treatment level of HBsAg was an independent factor for relapse (HBV DNA level above 200–1000 IU/mL) 46–49, although studies vary in definitions of relapse 50. Ongoing prospective studies will be helpful in validating these cut-offs. Changes in qHBsAg are increaingly included as primary or secondary endpoints of studies of new anti-viral agents. For example, among patients given ARB-1467 (a small interfering RNA), 71% achieved qHBsAg below 50 IU/mL, although none had HBsAg loss 51. .
HBV Genotypes
Phylogenetic analyses have parsed 10 distinct HBV genotypes (A–J) with genetic divergence of more than 8% and more than 30 subtypes with divergence greater than 4%. These genotype and subtypes have specific geographic patterns (Supplemental Figure 1), routes of transmission, and progression52. Outcomes have been mainly compared between genotypes B and C, in Asia, and a few studies have compared genotypes A and D in Europe and India. Individuals with HBV genotype C infection are more often HBeAg-positive and, based on analysis of an Alaskan cohort, have a 2-decade delay in HBeAg seroconversion compared to persons with HBV genotypes A, B, D, or F 53. HBeAg seroconversion and the inactive carrier state are associated with better outcomes, so higher proportions of patients with HBV genotype C infection, who have longer periods of intermittent active inflammation, develop cirrhosis. Patients with HBV genotype C are also at higher risk of HCC compared to patients with HBV genotypes A, B or D, based on a large meta-analysis of observational studies 54. Patients with HBV genotype B are more likely to develop HCC without cirrhosis at a younger age (less than 35 years) 55. No difference has been observed in HCC incidence between patients with HBV genotypes A vs D. However, there may be a higher incidence of HCC in patients with HBV genotype A1 (Africa) compared to HBV genotype A2 (US/Europe). Higher rates of spontaneous loss of HBsAg have been reported in patients with HBV genotype A or B56, 57.
The AASLD and EASL recommend routine testing of HBV genotype only if peginterferon therapy is being considered 15, 17; the value of selecting patients for peginterferon based on HBV genotype has been increasingly recognized 58. Available data from clinical trials indicated higher odds of HBeAg seroconversion in HBeAg-positive patients infected with HBV genotypes A or B, and loss of HBsAg in patients infected with HBV genotypes C or D—these f indings should be considered in selecting candidates for peginterferon therapy 17. The highest probability of sustained response (HBeAg loss and HBV DNA below 2000 IU/ml at 6 months) was observed in patients with genotype A infection and high baseline levels of ALT or low levels of HBV DNA. Patients with HBV genotype D infection have the lowest odds of sustained response, regardless of level of ALT or HBV DNA 59. Among HBeAg-negative patients, those with HBV genotypes B or C have better odds of ALT normalization and reduction of HBV DNA to below 2000 IU/mL than those with HBV genotype D 60. Genotype-specific stopping rules, based on levels of HBsAg at 12 and 24 weeks of treatment, are recommended by EASL 15. Notably, stopping rules for HBeAg-negative patients exist only for patients with HBV genotype D—there are few reliable data on other genotypes. Data on treatment response is also sparse for patients with HBV genotypes E–J or mixed-genotype infections.
HBV genotype does not impact patient selection for NA treatment 58 or monitoring intervals. A meta-analysis of genotype effect on response (HBeAg seroconversion in HBeAg-positive and undetectable HBV DNA in HBeAg-negative) to all NA therapies (including entecavir and tenofovir) showed no significant difference 61. In addition, drug resistance, reductions in HBV DNA, and ALT normalization were similar across all studied genotypes 62. Genotype may, however, influence likelihood of on-treatment HBsAg loss. Among tenofovir-treated HBeAg positive patients with HBsAg loss, 61% were genotype A and 30% genotype D, as compared to 4% and 0% with genotypes B and C, respectively 38.
HBeAg and precore or basal core mutations
The HBV core gene encodes HBeAg, which is secreted from infected hepatocytes and is a marker of active virus replication. HBeAg and antibody against it (anti-HBe) are measured by immunoassays. HBeAg is detected 6–12 weeks after exposure to HBV and indicates high level of HBV DNA and high infectivity. Seroconversion from HBeAg to anti-HBe is an important hallmark of disease progression, and is typically associated with a transition to an inactive infection, with a low level HBV DNA. However, acquisition of precore and/or basal core promoter (BCP) mutations can prevent or reduce HBeAg production, leading to an HBeAg-negative phenotype of patients with high levels of HBV DNA and ALT (Figure 2).
The precore mutation G1896A (at codon 28) inhibits translation by causing a frameshift and insertion of a premature stop codon (Supplemental Figure 2A). Mutations in BCP at nt 1742–1849 prevent transcription of precore mRNA, resulting in defective synthesis of HBeAg (Supplemental Figure 2B). The A1762T and G1746A mutations in BCP are frequently detected together and prevent HBeAg production. Mutations in BCP are detected alone or in conjunction with mutations in the precore protein. The G1896 mutations is rarely detected in HBV genotypes A or H, or subgenotypes C1, F2, or F3 because this change would impair Watson-Crick base pairing within the epsilon structure and thereby compromise encapsidation of pregenomic RNA (pgRNA).
Precore and BCP mutations and effects on disease progression and treatment
Tests to detect precore or BCP mutations are not recommended for routine clinical care but many studies have examined associations with responses to treatment (such as peginterferon and NAs), HBeAg and HBsAg seroclearance, and risk of fibrosis progression and HCC. The prevalence of precore or BCP mutations increases with duration of chronic infection and age, and most notably in association with HBeAg seroconversion 63. In all cases, increases in percentages of mutations are preceded or accompanied by increases in serum levels of ALT63. A longitudinal study with a mean follow-up time of 14 years found that children with spontaneous or treatment-induced HBeAg seroconversion who subsequently developed HBeAg-negative chronic HBV infections (high viremia, increased level of ALT), underwent seroconversion at older ages and more frequently had BCP mutations 64. These studies indicated a correlation between acquisition of BCP or precore mutations and risk of HBeAg-negative HBV infection. A double mutation in BCP (A1762T and G1764A) has been associated with advanced liver disease and HCC 65, 66. In adults with HBV genotype C infection, this double mutation was associated with a 3.5-fold higher risk of HCC compared to infected adults without the mutation 65. These mutations are believed to cause upregulation of pregenomic RNA (higher virus levels) and/or the overlap with the HBx protein-encoding region, which has oncogenic properties. Mutations in the precore region alone have been less consistently linked with increased risk of HCC 65, 67.
In HBeAg-positive positive patients with HBV genotypes B or C treated with interferon, higher numbers of precore and BCP mutations at baseline are associated with HBeAg seroconversion and HBV DNA below 2000 IU/mL at 6 months after treatment48, 49. In contrast, in patients with HBV genotypes A or D infections, the presence of precore and BCP mutants (vs only wild type virus) before peginterferon therapy had lower rates of sustained response 48. In treatment studies that used loss of HBsAg as endpoint, a higher prevalence of BCP and precore mutations was associated with lower likelihood of loss among tenofovir-treated patients with HBV genotypes A or D 68, but not among patients treated with antiviral agents for HBV genotype B or C infection 69. Interpretation of study results therefore requires attention to the methods they used to quantify precore and BCP variants, as well as HBV genotypes and length of follow up (risk period for outcomes of interest).
New Diagnostic Tests
It is important to discover new biomarkers that can be used to monitor response to therapy (with NAs and peginterferon) and test the efficacy of therapeutic agents in development (Table 2).
Table 2.
New Biomarkers for HBV Infection
| New biomarkers (References) |
Detection target | Diagnostic test | Potential roles in management of patients with chronic HBV infection |
|---|---|---|---|
| Measurement of serum HBV RNA84, 87, 88 |
HBV pre-genomic RNA and total serum HBV RNA (variants that cause defects in RNA splicing). Serum levels of pre- genomic indicate transcription of cccDNA |
RACE and real- time PCR (Abbott m2000 RNA RUO assay) |
Defining stopping rules for patients on NA therapy; Identifying which patients with cirrhosis may safely discontinue HBV therapy; Endpoint for testing therapeutic agents that cccDNA |
| Measurement of HBcrAg104, 105 |
Core antigen associated with circulating virus particles, denatured HBeAg, and p22 core related protein. Surrogate marker of intrahepatic cccDNA |
ELISA (Lumipulse G HBcrAg, Fujirebio, Gent, Belgium) |
Diagnostic tool for differentiating disease states; Identifying which patients may safely discontinue NA therapy; Might identify patients at risk for reactivation of HBV during therapy; Might be used to determine risk for HCC |
| Measurement of HBeAg 93, 95, 96, 106 |
Soluble protein produced during acute or chronic infection, encoded by core gene, indicates active viral replication |
ELISA (Abbott, Chicago,IL, USA) |
To establish spontaneous HBeAg seroconversion; Might be used to monitor response to treatment with NAs or new antiviral therapies |
| Measurement of anti-HBc 98, 102 |
Antibody against the hepatitis B core antigen; indicates prior HBV exposure, marker of immune response |
ELISA (Beijing Wantai Biological Pharmacy, Beijing, China) |
Assess immune response against HBV; Identify patients with HBeAg seroconversion Might be used with immune- modulatory therapies, independent of HBV genotype |
cccDNA
Intrahepatic cccDNA is the transcriptional template for HBV70. The existence of cccDNA allows for reactivation of HBV in persons who are negative for HBsAg but positive for anti-HBc who receive immune suppressive medications; cccDNA is also the reason for frequent relapse after withdrawal of long-term NA therapy 71, 72. HBV cccDNA is maintained within the nucleus of infected cells, and eradication is required to permanently cure infection, which this is not achievable with current therapeutics. Agents to target cccDNA or disrupt its formation are therefore in development 73. Barriers to measurement of cccDNA, as a biomarker for viral replication and a therapeutic endpoint include the need for liver biopsy collection and analysis and lack of standardized methods for quantification, such as PCR or in situ hybridization 74. The standard for quantification of cccDNA detection, Southern blot analysis, does not always detect low levels and is too complex and time-consuming for routine use.
HBV cccDNA can be detected at low levels in patients with occult HBV infection (lack of detectable HBsAg in serum), including core-positive donor livers and HBsAg-negative HCC tumor cells 70. During chronic HBV infection, levels of cccDNA correlate with intrahepatic and circulating levels of HBV DNA. The highest levels of cccDNA are observed in HBeAg-positive individuals, followed by HBeAg-negative patients with immune-active chronic HBV infection, inactive carriers, and last, patients with loss of HBsAg 75. In cross-sectional studies, lower levels of cccDNA were associated with inactive chronic HBV infection, HBsAg loss, and lower levels of inflammation (detected by histology) and ALT. However there have been no prospective studies of the relationship between cccDNA levels and changes in phases or outcomes of chronic HBV infection. A lower ratio of cccDNA to intrahepatic HBV DNA was associated with more severe fibrosis (determined by histology and survival of patients with HCC who underwent hepatectomy. So, relative proportions of intracellular DNA associate with disease progression 76.
There have been few studies of the kinetics of cccDNA in patients receiving treatment, due to challenges in accurate quantification. NA therapy has no direct effect on cccDNA, because reverse transcription is downstream step of HBV replication. However, a reduction of about 1 log10 during 1 year of therapy has been reported, likely mediated by insufficient recycling of nucleocapsids to the nucleus with inhibition of cytoplasmic DNA synthesis 70. Studies peginterferon combination therapies reported greater decreases in cccDNA (1.4–2.4 log10) 71. This could result from a decrease in transcription, via epigenetic regulation and nonhepatotoxic degradation of the cccDNA pool 77. In either case, incomplete elimination of cccDNA invariably leads to relapse in most patients who stop therapy. In patients with chronic HBV infection who received liver transplants, undetectable intrahepatic cccDNA has been associated with success in weaning from HBV prophylaxis 78.
As we look to the future, cccDNA is unlikely to be routine biomarker because liver biopsies are required for its analysis. Researchers reported detection of cccDNA in the serum, presumed from lysed hepatocytes79, but these findings must be validated. Levels of HBV RNA and HBcrAg correlate with level of cccDNA and might be used as markers.
Measurement of serum HBV RNA
HBV pgRNA and total RNA (such as from variants with defects in RNA splicing) are in virions, released into the hepatocyte cytoplasm, and can be detected in serum 80, 81. Serum pgRNA is constitutively transcribed from cccDNA, so levels of serum RNA can indicate the presence of cccDNA virus transcriptional activity 82. Application of rapid amplification of cDNA ends (RACE) to HBV research, 83 along with real-time PCR 84, has allowed HBV RNA to be used as a biomarker. A commercial high through-put HBV RNA test was recently developed (Abbott m2000 RNA Research Use Only (RUO) assay, LLOD 45 U/L, where 1 U HBV RNA = 1 IU HBV DNA); it detects HBV RNA with greater sensitivity than RACE analysis 84.
Levels of HBV RNA correlate with levels of HBV DNA and associate with serum levels of HBsAg and ALT, HBV genotype, and presence of basal core promotor mutations in untreated patients 85. HBV RNA levels vary with phase of infection—they are highest in the immune-tolerant phase and lowest in patients with inactive HBV infection 86. In patients receiving treatment with NAs, levels of HBV RNA are higher than levels of HBV DNA 87. In HBeAg-negative patients off treatment with peginterferon and adefovir, lower baseline serum levels of RNA were associated with maintained response (normal level of ALT and HBV DNA below 2000 IU/mL) 87. HBeAg-positive patients with significant reductions in serum levels of HBV RNA while receiving NA therapy were more likely to achieve HBeAg seroconversion 84. In a cross-sectional study of 47 patients with reductions in HBV DNA while receiving entecavir treatment, serum HBV RNA was detected in 35/47 (74.4%), with levels ranging from 2.3–4.8 log10 copies/mL; levels correlated with intrahepatic HBV RNA and cccDNA as well as liver histologic scores 80. Large scale studies are required to determine whether measurement of HBV RNA in serum can be used to assess intrahepatic HBV cccDNA activity, because levels vary with peginterferon and NA therapies87 and with HBeAg status 88.
Overall, serum levels of HBV RNA indicate cccDNA transcriptional activity and reservoir size. This biomarker (in combination with HBV DNA and level of HBsAg) might be used to monitor response to treatment in patients receiving NA therapy. It might also be used identify patients that can safely discontinue NA and/or achieve HBsAg loss (a functional cure). Serial assessment of intrahepatic HBV cccDNA is not feasible, so measurement of HBV RNA along with assays for viral activity could be used to determine cccDNA activity—especially with therapies in development that aim for sterilizing and/or functional cure.
Measurement of HBcrAg
HBcrAg has been proposed to be a marker of intrahepatic HBV cccDNA and its transcription. A chemiluminescence enzyme immunoassay (Lumipulse G HBcrAg, Fujirebio, Gent, Belgium) is available that measures a combination of HBcrAg, HBeAg, and the 22 kDa truncated precore protein in serum samples 89. In a retrospective study of 118 HBeAg-positive adults (genotypes A–G) treated with NAs, combined levels of HBsAg and HBcrAg had the greatest predictive value for HBeAg seroconversion at 12 months; the cut-offs for HBsAg 3.8 log10 IU/mL and HBcrAg 5.5 log10 U/mL identified patients with HBeAg seroconversion with a negative predictive value of 86.1% and an area under the curve of 0.807 90. In a study of 121 Thai patients with HBeAg-negative chronic HBV infection treated with peginterferon alone or in combination with entecavir, baseline level of HBcrAg correlated with HBV DNA and cccDNA but not HBsAg 91. At week 12, a decrease of less than 0.5 log10 in levels of HBsAg, HBcrAg, or both antigens combined identified patients with off-treatment virologic remission with negative predictive values of 90%, 82%, and 96%, respectively. A systematic review of published studies proposed HBcrAg as biomarker to identify patients most likely to undergo HBeAg seroconversion, determine the phase of HBV infection, determine risks of HCC and liver disease progression, and identify patients who can safely discontinue NA therapy and /or achieve HBsAg loss92. HBcrAg can be detected even in HBsAg-negative patients and appears to be a reliable marker of intrahepatic cccDNA, so assays to measure it in serum might be used in development of therapeutic agents that target cccDNA and to achieve a sterilizing cure. Quantification of HBcrAg alone or in combination with measurement of HBsAg and HBV RNA might be used to monitor response to current therapies and evaluate therapeutic agents in development.
Measurement of HBeAg
HBeAg is measured in an immunoassay, although commercial quantitative tests for HBeAg are not available. There have been studies of the association between levels of HBeAg and HBeAg seroconversion in patients with chronic HBV infection. In a retrospective study of 76 HBeAg-positive patients who received 96 weeks of NA therapy (lamivudine and adefovir), baseline level of HBeAg and decreases at 24-weeks were independently associated with HBeAg seroconversion (area under the curve values of 0.828 and 0.814, respectively) 93. In a study of treatment of patients with HBV and HIV co-infection, decreased levels of HBeAg of 0.5 log10 at week 12 and 1.0 log10 at week 24 were associated with seroconversion to anti-HBe seroconversion and loss of HBsAg 94. Other studies have sought to determine if measurement of HBeAg could be an alternative to quantification of HBV DNA, which is a challenge in some regions. In a study of 82 patients in Malaysia, levels of HBeAg correlated with HBV DNA levels above 300 IU/mL (r = 0.893) whereas levels of HBsAg did not 95. In a large community-based study of 788 HBeAg-positive subjects in China with normal levels of ALT, levels of HBeAg correlated with HBV DNA levels, particularly at high HBV DNA levels 96.
In 2013, the WHO proposed the 1st International Standard for HBeAg with an assigned unit of 100 IU/mL 97. Heterogeneity among immunoassays used and reference standards make it difficult to compare findings among studies—an issu e that could be improved by standardization. There are limited data published on the role for quantification of HBeAg, although similar to other serum markers, levels might be used to follow disease progression, predict HBeAg seroconversion, and determine response to approved and novel anti-HBV therapies.
Measurement of anti-HBc
Anti-HBc is a marker of HBV core antigen-specific immune activity. A sandwich-based ELISA assay is commercially available in China for quantification of anti-HBc (Beijing Wantai Biological Pharmacy Beijing, China)98. Higher levels of anti-HBc might indicate a strong immune response against HBV and therefore be used to identify patients most likely to respond to anti-HBV therapies 98. Levels of anti-HBc in serum parallel levels of ALT and hepatic inflammation in patients with HBeAg-positive and HBeAg-negative active HBV infection 99,100. In a study of 397 patients in the REVEAL-HBV cohort, followed for 6.8 years, baseline level of anti-HBc were independently associated with HBeAg seroclearance in untreated individuals, although no association was found with HBcrAg101. The authors concluded that measurements of anti-HBc might be used to determine patient risk and identify patients most likely to undergo spontaneous HBeAg seroconversion.
Several studies have investigated whether baseline measurements of anti-HBc can identify patients who respond to NA and peginterferon treatment. Peginterferon-treated patients who underwent seroconversion to HBeAg and anti-HBe after the end of treatment or during a follow-up period had significantly higher levels of anti-HBc at baseline 98. Further, level of anti-HBc at baseline (4.4–4.5 log10 IU/mL or more) was the best independent predictor of HBeAg seroconversion following either peginterferon or NA treatment, compared to level of ALT or HBV DNA 98, 102. In a study of 100 patients (71% HBeAg-positive at start of NA therapy) followed for 4 years after NA discontinuation, higher level of anti-HBc at the time of NA discontinuation was associated with a reduced risk of relapse (defined as HBV DNA above 2000 IU/mL and level of ALT more than 2-fold the upper limit of normal). Among patients who had stopped taking NAs for 4 years, 85% of those with levels of anti-HBc below 100 IU/mL had a relapse compared to 21% of patients with levels of anti-HBc above 1000 IU/mL 103. Level of anti-HBc might therefore be a marker of an antiviral immune response that can be used to identify patients most likely to respond to therapy (independent of HBV genotype and unlike level of HBsAg) 98. Anti-HBc also has a longer half-life in serum than ALT.
Future Directions
Tests to measure biomarkers such as HBsAg, HBeAg, and HBV DNA are used in management of patients with chronic HBV infection, as specified in association guidelines. Markers of virus activity have been identified—following these could increase our understanding HBV pathogenesis and patient disease progression and response to treatment. With many new therapies for HBV infection in development, new diagnostic tools are needed to measure response and establish cure. Tests to detect and measure cccDNA, HBV RNA, anti-HBc, and HBcrAg are in development, but methods to quantify virus activity and anti-HBV immune response, and to identify genetic variants, are needed.
Supplementary Material
Acknowledgements:
The authors would like to thank Dr. S. Joshi, PhD for graphic design of Fig. 3.
NT has received institutional grant support from Gilead, AbbVie, Merck and BMS. CSC has received Investigator Initiated institutional research Grant support and/or research materials from GSK, Gilead Sciences, Arbutus Biopharma and Bristol-Myers Squibb. KZ is supported by T32 5T32DK060414-14 from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- 1.Polaris Observatory C Global prevalence, treatment, and prevention of hepatitis B virus infection in 2016: a modelling study. Lancet Gastroenterol Hepatol 2018;3:383–403. [DOI] [PubMed] [Google Scholar]
- 2.Chevaliez S, Pawlotsky JM. New virological tools for screening, diagnosis and monitoring of hepatitis B and C in resource-limited settings. J Hepatol 2018. [DOI] [PubMed]
- 3.Organization WH. Guidelines on Hepatitis B and C Testing, 2017.
- 4.Amini A, Varsaneux O, Kelly H, et al. Diagnostic accuracy of tests to detect hepatitis B surface antigen: a systematic review of the literature and meta-analysis. BMC Infect Dis 2017;17:698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scheiblauer H, El-Nageh M, Diaz S, et al. Performance evaluation of 70 hepatitis B virus (HBV) surface antigen (HBsAg) assays from around the world by a geographically diverse panel with an array of HBV genotypes and HBsAg subtypes. Vox Sang 2010;98:403–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chevaliez S, Challine D, Naija H, et al. Performance of a new rapid test for the detection of hepatitis B surface antigen in various patient populations. J Clin Virol 2014;59:89–93. [DOI] [PubMed] [Google Scholar]
- 7.Gish RG, Gutierrez JA, Navarro-Cazarez N, et al. A simple and inexpensive point-of-care test for hepatitis B surface antigen detection: serological and molecular evaluation. J Viral Hepat 2014;21:905–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Njai HF, Shimakawa Y, Sanneh B, et al. Validation of rapid point-of-care (POC) tests for detection of hepatitis B surface antigen in field and laboratory settings in the Gambia, Western Africa. J Clin Microbiol 2015;53:1156–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lange B, Cohn J, Roberts T, et al. Diagnostic accuracy of serological diagnosis of hepatitis C and B using dried blood spot samples (DBS): two systematic reviews and meta-analyses. BMC Infect Dis 2017;17:700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lange B, Roberts T, Cohn J, et al. Diagnostic accuracy of detection and quantification of HBV-DNA and HCV-RNA using dried blood spot (DBS) samples - a systematic review and meta-analysis. BMC Infect Dis 2017;17:693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bottero J, Boyd A, Gozlan J, et al. Performance of rapid tests for detection of HBsAg and anti-HBsAb in a large cohort, France. J Hepatol 2013;58:473–8. [DOI] [PubMed] [Google Scholar]
- 12.Poiteau L, Soulier A, Roudot-Thoraval F, et al. Performance of rapid diagnostic tests for the detection of anti-HBs in various patient populations. J Clin Virol 2017;96:64–66. [DOI] [PubMed] [Google Scholar]
- 13.Clement F, Dewint P, Leroux-Roels G. Evaluation of a new rapid test for the combined detection of hepatitis B virus surface antigen and hepatitis B virus e antigen. J Clin Microbiol 2002;40:4603–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Terrault NA, Lok ASF, McMahon BJ, et al. Update on prevention, diagnosis, and treatment of chronic hepatitis B: AASLD 2018 hepatitis B guidance. Hepatology 2018;67:1560–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.European Association for the Study of the Liver. Electronic address eee, European Association for the Study of the L. EASL 2017 Clinical Practice Guidelines on the management of hepatitis B virus infection. J Hepatol 2017. [DOI] [PubMed]
- 16.Sarin SK, Kumar M, Lau GK, et al. Asian-Pacific clinical practice guidelines on the management of hepatitis B: a 2015 update. Hepatol Int 2016;10:1–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Terrault NA, Lok AS, McMahon BJ, et al. Update on Prevention, Diagnosis, and Treatment and of Chronic Hepatitis B: AASLD 2018 Hepatitis B Guidance. Hepatology 2018;67:1560–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Biswas R, Tabor E, Hsia CC, et al. Comparative sensitivity of HBV NATs and HBsAg assays for detection of acute HBV infection. Transfusion 2003;43:788–98. [DOI] [PubMed] [Google Scholar]
- 19.Candotti D, Laperche S. Hepatitis B Virus Blood Screening: Need for Reappraisal of Blood Safety Measures? Front Med (Lausanne) 2018;5:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Louisirirotchanakul S, Khupulsup K, Akraekthalin S, et al. Comparison of the technical and clinical performance of the Elecsys HBsAg II assay with the Architect, AxSym, and Advia Centaur HBsAg screening assays. J Med Virol 2010;82:755–62. [DOI] [PubMed] [Google Scholar]
- 21.Hongthanakorn C, Chotiyaputta W, Oberhelman K, et al. Virological breakthrough and resistance in patients with chronic hepatitis B receiving nucleos(t)ide analogues in clinical practice. Hepatology 2011;53:1854–63. [DOI] [PubMed] [Google Scholar]
- 22.Thompson AJ, Nguyen T, Iser D, et al. Serum hepatitis B surface antigen and hepatitis B e antigen titers: disease phase influences correlation with viral load and intrahepatic hepatitis B virus markers. Hepatology 2010;51:1933–44. [DOI] [PubMed] [Google Scholar]
- 23.Wooddell CI, Yuen MF, Chan HL, et al. RNAi-based treatment of chronically infected patients and chimpanzees reveals that integrated hepatitis B virus DNA is a source of HBsAg. Sci Transl Med 2017;9. [DOI] [PMC free article] [PubMed]
- 24.Wursthorn K, Jaroszewicz J, Zacher BJ, et al. Correlation between the Elecsys HBsAg II assay and the Architect assay for the quantification of hepatitis B surface antigen (HBsAg) in the serum. J Clin Virol 2011;50:292–6. [DOI] [PubMed] [Google Scholar]
- 25.Seto WK, Wong DK, Fung J, et al. Linearized hepatitis B surface antigen and hepatitis B core-related antigen in the natural history of chronic hepatitis B. Clin Microbiol Infect 2014;20:1173–80. [DOI] [PubMed] [Google Scholar]
- 26.Pfefferkorn M, Bohm S, Schott T, et al. Quantification of large and middle proteins of hepatitis B virus surface antigen (HBsAg) as a novel tool for the identification of inactive HBV carriers. Gut 2018;67:2045–2053. [DOI] [PubMed] [Google Scholar]
- 27.Cornberg M, Wong VW, Locarnini S, et al. The role of quantitative hepatitis B surface antigen revisited. J Hepatol 2017;66:398–411. [DOI] [PubMed] [Google Scholar]
- 28.Di Bisceglie AM, Lombardero M, Teckman J, et al. Determination of hepatitis B phenotype using biochemical and serological markers. J Viral Hepat 2017;24:320–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brunetto MR, Oliveri F, Colombatto P, et al. Hepatitis B surface antigen serum levels help to distinguish active from inactive hepatitis B virus genotype D carriers. Gastroenterology 2010;139:483–90. [DOI] [PubMed] [Google Scholar]
- 30.Liu J, Yang HI, Lee MH, et al. Serum Levels of Hepatitis B Surface Antigen and DNA Can Predict Inactive Carriers With Low Risk of Disease Progression. Hepatology 2016;64:381–9. [DOI] [PubMed] [Google Scholar]
- 31.Liu J, Lee MH, Batrla-Utermann R, et al. A predictive scoring system for the seroclearance of HBsAg in HBeAg-seronegative chronic hepatitis B patients with genotype B or C infection. J Hepatol 2013;58:853–60. [DOI] [PubMed] [Google Scholar]
- 32.Wen WH, Huang CW, Chie WC, et al. Quantitative maternal hepatitis B surface antigen predicts maternally transmitted hepatitis B virus infection. Hepatology 2016;64:1451–1461. [DOI] [PubMed] [Google Scholar]
- 33.Tseng TC, Liu CJ, Yang HC, et al. High levels of hepatitis B surface antigen increase risk of hepatocellular carcinoma in patients with low HBV load. Gastroenterology 2012;142:1140–1149 e3; quiz e13–4. [DOI] [PubMed] [Google Scholar]
- 34.Chevaliez S, Hezode C, Bahrami S, et al. Long-term hepatitis B surface antigen (HBsAg) kinetics during nucleoside/nucleotide analogue therapy: finite treatment duration unlikely. J Hepatol 2013;58:676–83. [DOI] [PubMed] [Google Scholar]
- 35.Chen CH, Chiu YC, Lu SN, et al. Serum hepatitis B surface antigen levels predict treatment response to nucleos(t)ide analogues. World J Gastroenterol 2014;20:7686–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tseng TC, Liu CJ, Yang HC, et al. Determinants of spontaneous surface antigen loss in hepatitis B e antigen-negative patients with a low viral load. Hepatology 2012;55:68–76. [DOI] [PubMed] [Google Scholar]
- 37.Heathcote EJ, Marcellin P, Buti M, et al. Three-year efficacy and safety of tenofovir disoproxil fumarate treatment for chronic hepatitis B. Gastroenterology 2011;140:132–43. [DOI] [PubMed] [Google Scholar]
- 38.Marcellin P, Buti M, Krastev Z, et al. Kinetics of hepatitis B surface antigen loss in patients with HBeAg-positive chronic hepatitis B treated with tenofovir disoproxil fumarate. J Hepatol 2014;61:1228–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen CH, Lu SN, Hung CH, et al. The role of hepatitis B surface antigen quantification in predicting HBsAg loss and HBV relapse after discontinuation of lamivudine treatment. J Hepatol 2014;61:515–22. [DOI] [PubMed] [Google Scholar]
- 40.Martinot-Peignoux M, Asselah T, Marcellin P. HBsAg quantification to optimize treatment monitoring in chronic hepatitis B patients. Liver Int 2015;35 Suppl 1:82–90. [DOI] [PubMed] [Google Scholar]
- 41.Jaroszewicz J, Reiberger T, Meyer-Olson D, et al. Hepatitis B surface antigen concentrations in patients with HIV/HBV co-infection. PLoS One 2012;7:e43143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sonneveld MJ, Hansen BE, Piratvisuth T, et al. Response-guided peginterferon therapy in hepatitis B e antigen-positive chronic hepatitis B using serum hepatitis B surface antigen levels. Hepatology 2013;58:872–80. [DOI] [PubMed] [Google Scholar]
- 43.Rijckborst V, Hansen BE, Ferenci P, et al. Validation of a stopping rule at week 12 using HBsAg and HBV DNA for HBeAg-negative patients treated with peginterferon alfa-2a. J Hepatol 2012;56:1006–11. [DOI] [PubMed] [Google Scholar]
- 44.Hadziyannis SJ, Sevastianos V, Rapti I, et al. Sustained responses and loss of HBsAg in HBeAg-negative patients with chronic hepatitis B who stop long-term treatment with adefovir. Gastroenterology 2012;143:629–636 e1. [DOI] [PubMed] [Google Scholar]
- 45.Chang ML, Liaw YF, Hadziyannis SJ. Systematic review: cessation of long-term nucleos(t)ide analogue therapy in patients with hepatitis B e antigen-negative chronic hepatitis B. Aliment Pharmacol Ther 2015;42:243–57. [DOI] [PubMed] [Google Scholar]
- 46.Wang CC, Tseng KC, Hsieh TY, et al. Assessing the Durability of Entecavir-Treated Hepatitis B Using Quantitative HBsAg. Am J Gastroenterol 2016;111:1286–94. [DOI] [PubMed] [Google Scholar]
- 47.Chen CH, Hsu YC, Lu SN, et al. The incidence and predictors of HBV relapse after cessation of tenofovir therapy in chronic hepatitis B patients. J Viral Hepat 2018;25:590–597. [DOI] [PubMed] [Google Scholar]
- 48.Su TH, Yang HC, Tseng TC, et al. Distinct Relapse Rates and Risk Predictors After Discontinuing Tenofovir and Entecavir Therapy. J Infect Dis 2018;217:1193–1201. [DOI] [PubMed] [Google Scholar]
- 49.Liang Y, Jiang J, Su M, et al. Predictors of relapse in chronic hepatitis B after discontinuation of anti-viral therapy. Aliment Pharmacol Ther 2011;34:344–52. [DOI] [PubMed] [Google Scholar]
- 50.Papatheodoridis GV, Manolakopoulos S, Su TH, et al. Significance of definitions of relapse after discontinuation of oral antivirals in HBeAg-negative chronic hepatitis B. Hepatology 2017. [DOI] [PubMed]
- 51.Agarwal K, Gane EJ, Cheng W, et al. Bi-weekly dosing of ARB-1467 LNP siRNA in HBeAg negative, virally suppressed patients with chronic HBV infection leads to deeper declines in HBSAG and potential association with IL28B. Hepatology 2017:2017.
- 52.Sunbul M Hepatitis B virus genotypes: global distribution and clinical importance. World J Gastroenterol 2014;20:5427–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Livingston SE, Simonetti JP, Bulkow LR, et al. Clearance of hepatitis B e antigen in patients with chronic hepatitis B and genotypes A, B, C, D, and F. Gastroenterology 2007;133:1452–7. [DOI] [PubMed] [Google Scholar]
- 54.Wong GL, Chan HL, Yiu KK, et al. Meta-analysis: The association of hepatitis B virus genotypes and hepatocellular carcinoma. Aliment Pharmacol Ther 2013;37:517–26. [DOI] [PubMed] [Google Scholar]
- 55.Kao J, Chen P, Lai M, et al. Hepatitis B genotypes correlate with clinical outcomes in patients with chronic hepatitis B. Gastroenterology 2000;118:554–9. [DOI] [PubMed] [Google Scholar]
- 56.Sanchez-Tapias JM, Costa J, Mas A, et al. Influence of hepatitis B virus genotype on the long-term outcome of chronic hepatitis B in western patients. Gastroenterology 2002;123:1848–56. [DOI] [PubMed] [Google Scholar]
- 57.Yuen MF, Wong DK, Sablon E, et al. HBsAg seroclearance in chronic hepatitis B in the Chinese: virological, histological, and clinical aspects. Hepatology 2004;39:1694–701. [DOI] [PubMed] [Google Scholar]
- 58.Rajoriya N, Combet C, Zoulim F, et al. How viral genetic variants and genotypes influence disease and treatment outcome of chronic hepatitis B. Time for an individualised approach? J Hepatol 2017;67:1281–1297. [DOI] [PubMed] [Google Scholar]
- 59.Buster EH, Hansen BE, Lau GK, et al. Factors that predict response of patients with hepatitis B e antigen-positive chronic hepatitis B to peginterferon-alfa. Gastroenterology 2009;137:2002–9. [DOI] [PubMed] [Google Scholar]
- 60.Bonino F, Marcellin P, Lau GK, et al. Predicting response to peginterferon alpha-2a, lamivudine and the two combined for HBeAg-negative chronic hepatitis B. Gut 2007;56:699–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wiegand J, Hasenclever D, Tillmann HL. Should treatment of hepatitis B depend on hepatitis B virus genotypes? A hypothesis generated from an explorative analysis of published evidence. Antivir Ther 2008;13:211–20. [PubMed] [Google Scholar]
- 62.Raimondi S, Maisonneuve P, Bruno S, et al. Is response to antiviral treatment influenced by hepatitis B virus genotype? J Hepatol 2010;52:441–9. [DOI] [PubMed] [Google Scholar]
- 63.Nie H, Evans AA, London WT, et al. Quantitative dynamics of hepatitis B basal core promoter and precore mutants before and after HBeAg seroconversion. J Hepatol 2012;56:795–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu JF, Chiu YC, Chang KC, et al. Predictors of hepatitis B e antigen-negative hepatitis in chronic hepatitis B virus-infected patients from childhood to adulthood. Hepatology 2016;63:74–82. [DOI] [PubMed] [Google Scholar]
- 65.Yang Z, Zhuang L, Lu Y, et al. Naturally occurring basal core promoter A1762T/G1764A dual mutations increase the risk of HBV-related hepatocellular carcinoma: a meta-analysis. Oncotarget 2016;7:12525–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang HI, Yeh SH, Chen PJ, et al. Associations between hepatitis B virus genotype and mutants and the risk of hepatocellular carcinoma. J Natl Cancer Inst 2008;100:1134–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liao Y, Hu X, Chen J, et al. Precore mutation of hepatitis B virus may contribute to hepatocellular carcinoma risk: evidence from an updated meta-analysis. PLoS One 2012;7:e38394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bayliss J, Yuen L, Rosenberg G, et al. Deep sequencing shows that HBV basal core promoter and precore variants reduce the likelihood of HBsAg loss following tenofovir disoproxil fumarate therapy in HBeAg-positive chronic hepatitis B. Gut 2017;66:2013–2023. [DOI] [PubMed] [Google Scholar]
- 69.Tseng TC, Liu CJ, Chen CL, et al. Higher lifetime chance of spontaneous surface antigen loss in hepatitis B carriers with genotype C infection. Aliment Pharmacol Ther 2015;41:949–60. [DOI] [PubMed] [Google Scholar]
- 70.Allweiss L, Dandri M. The Role of cccDNA in HBV Maintenance. Viruses 2017;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang HC, Kao JH. Persistence of hepatitis B virus covalently closed circular DNA in hepatocytes: molecular mechanisms and clinical significance. Emerg Microbes Infect 2014;3:e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kumar R, Perez-Del-Pulgar S, Testoni B, et al. Clinical relevance of the study of hepatitis B virus covalently closed circular DNA. Liver Int 2016;36 Suppl 1:72–7. [DOI] [PubMed] [Google Scholar]
- 73.Yang HC, Chen PJ. The potential and challenges of CRISPR-Cas in eradication of hepatitis B virus covalently closed circular DNA. Virus Res 2018;244:304–310. [DOI] [PubMed] [Google Scholar]
- 74.Li X, Zhao J, Yuan Q, et al. Detection of HBV Covalently Closed Circular DNA. Viruses 2017;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Werle-Lapostolle B, Bowden S, Locarnini S, et al. Persistence of cccDNA during the natural history of chronic hepatitis B and decline during adefovir dipivoxil therapy. Gastroenterology 2004;126:1750–8. [DOI] [PubMed] [Google Scholar]
- 76.Wang Q, Fiel MI, Luan W, et al. Impact of intrahepatic hepatitis B DNA and covalently closed circular DNA on survival after hepatectomy in HBV-associated hepatocellular carcinoma patients. Ann Surg Oncol 2013;20:3761–70. [DOI] [PubMed] [Google Scholar]
- 77.Lucifora J, Xia Y, Reisinger F, et al. Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA. Science 2014;343:1221–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lenci I, Tisone G, Di Paolo D, et al. Safety of complete and sustained prophylaxis withdrawal in patients liver-transplanted for HBV-related cirrhosis at low risk of HBV recurrence. J Hepatol 2011;55:587–93. [DOI] [PubMed] [Google Scholar]
- 79.Huang JT, Yang Y, Hu YM, et al. A Highly Sensitive and Robust Method for Hepatitis B Virus Covalently Closed Circular DNA Detection in Single Cells and Serum. J Mol Diagn 2018;20:334–343. [DOI] [PubMed] [Google Scholar]
- 80.Wang J, Shen T, Huang X, et al. Serum hepatitis B virus RNA is encapsidated pregenome RNA that may be associated with persistence of viral infection and rebound. J Hepatol 2016;65:700–710. [DOI] [PubMed] [Google Scholar]
- 81.Volz T, Lutgehetmann M, Wachtler P, et al. Impaired intrahepatic hepatitis B virus productivity contributes to low viremia in most HBeAg-negative patients. Gastroenterology 2007;133:843–52. [DOI] [PubMed] [Google Scholar]
- 82.Giersch K, Allweiss L, Volz T, et al. Serum HBV pgRNA as a clinical marker for cccDNA activity. J Hepatol 2017;66:460–462. [DOI] [PubMed] [Google Scholar]
- 83.Kock J, Theilmann L, Galle P, et al. Hepatitis B virus nucleic acids associated with human peripheral blood mononuclear cells do not originate from replicating virus. Hepatology 1996;23:405–13. [DOI] [PubMed] [Google Scholar]
- 84.van Bommel F, Bartens A, Mysickova A, et al. Serum hepatitis B virus RNA levels as an early predictor of hepatitis B envelope antigen seroconversion during treatment with polymerase inhibitors. Hepatology 2015;61:66–76. [DOI] [PubMed] [Google Scholar]
- 85.van Campenhout MJH, van Bommel F, Pfefferkorn M, et al. Host and viral factors associated with serum hepatitis B virus RNA levels among patients in need for treatment. Hepatology 2018;68:839–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang J, Yu Y, Li G, et al. Natural history of serum HBV-RNA in chronic HBV infection. J Viral Hepat 2018;25:1038–1047. [DOI] [PubMed] [Google Scholar]
- 87.Jansen L, Kootstra NA, van Dort KA, et al. Hepatitis B Virus Pregenomic RNA Is Present in Virions in Plasma and Is Associated With a Response to Pegylated Interferon Alfa-2a and Nucleos(t)ide Analogues. J Infect Dis 2016;213:224–32. [DOI] [PubMed] [Google Scholar]
- 88.Butler EK, Gersch J, McNamara A, et al. Hepatitis B Virus Serum DNA and RNA Levels in Nucleos(t)ide Analog-Treated or Untreated Patients During Chronic and Acute Infection. Hepatology 2018. [DOI] [PubMed]
- 89.Park Y, Hong DJ, Shin S, et al. Performance evaluation of new automated hepatitis B viral markers in the clinical laboratory: two quantitative hepatitis B surface antigen assays and an HBV core-related antigen assay. Am J Clin Pathol 2012;137:770–7. [DOI] [PubMed] [Google Scholar]
- 90.Wang B, Carey I, Bruce M, et al. HBsAg and HBcrAg as predictors of HBeAg seroconversion in HBeAg-positive patients treated with nucleos(t)ide analogues. J Viral Hepat 2018;25:886–893. [DOI] [PubMed] [Google Scholar]
- 91.Chuaypen N, Posuwan N, Chittmittraprap S, et al. Predictive role of serum HBsAg and HBcrAg kinetics in patients with HBeAg-negative chronic hepatitis B receiving pegylated interferon-based therapy. Clin Microbiol Infect 2018;24:306 e7–306 e13. [DOI] [PubMed] [Google Scholar]
- 92.Mak LY, Wong DK, Cheung KS, et al. Review article: hepatitis B core-related antigen (HBcrAg): an emerging marker for chronic hepatitis B virus infection. Aliment Pharmacol Ther 2018;47:43–54. [DOI] [PubMed] [Google Scholar]
- 93.Gao YH, Meng QH, Zhang ZQ, et al. On-treatment quantitative hepatitis B e antigen predicted response to nucleos(t)ide analogues in chronic hepatitis B. World J Hepatol 2016;8:1511–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Matthews GV, Ali RJ, Avihingsanon A, et al. Quantitative HBsAg and HBeAg predict hepatitis B seroconversion after initiation of HAART in HIV-HBV coinfected individuals. PLoS One 2013;8:e61297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hudu SA, Niazlin MT, Nordin SA, et al. Quantitative Hepatitis B e Antigen: A Better Predictor of Hepatitis B Virus DNA than Quantitative Hepatitis B Surface Antigen. Clin Lab 2018;64:443–449. [DOI] [PubMed] [Google Scholar]
- 96.Chen P, Xie Q, Lu X, et al. Serum HBeAg and HBV DNA levels are not always proportional and only high levels of HBeAg most likely correlate with high levels of HBV DNA: A community-based study. Medicine (Baltimore) 2017;96:e7766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Organization WH. Collaborative Study to Establish a World Health Organization International Standard for Hepatitis B e Antigen (HBeAg) Geneva, 2013. [Google Scholar]
- 98.Yuan Q, Song LW, Liu CJ, et al. Quantitative hepatitis B core antibody level may help predict treatment response in chronic hepatitis B patients. Gut 2013;62:182–4. [DOI] [PubMed] [Google Scholar]
- 99.Song LW, Liu PG, Liu CJ, et al. Quantitative hepatitis B core antibody levels in the natural history of hepatitis B virus infection. Clin Microbiol Infect 2015;21:197–203. [DOI] [PubMed] [Google Scholar]
- 100.Li J, Zhang TY, Song LW, et al. Role of quantitative hepatitis B core antibody levels in predicting significant liver inflammation in chronic hepatitis B patients with normal or near-normal alanine aminotransferase levels. Hepatol Res 2018;48:E133–E145. [DOI] [PubMed] [Google Scholar]
- 101.Liu J, Hu HH, Chang CL, et al. Association Between High Levels of Hepatitis B Core Antibody and Seroclearance of Hepatitis B e Antigen in Individuals With Chronic Hepatitis B Virus Infection. Clin Gastroenterol Hepatol 2018. [DOI] [PubMed]
- 102.Fan R, Sun J, Yuan Q, et al. Baseline quantitative hepatitis B core antibody titre alone strongly predicts HBeAg seroconversion across chronic hepatitis B patients treated with peginterferon or nucleos(t)ide analogues. Gut 2016;65:313–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chi H, Li Z, Hansen BE, et al. Serum Level of Antibodies Against Hepatitis B Core Protein Is Associated With Clinical Relapse After Discontinuation of Nucleos(t)ide Analogue Therapy. Clin Gastroenterol Hepatol 2018. [DOI] [PubMed]
- 104.Wang ML, Liao J, Wei B, et al. Comparison of hepatitis B virus core-related antigen and hepatitis B surface antigen for predicting HBeAg seroconversion in chronic hepatitis B patients with pegylated interferon therapy. Infect Dis (Lond) 2018;50:522–530. [DOI] [PubMed] [Google Scholar]
- 105.Martinot-Peignoux M, Lapalus M, Maylin S, et al. Baseline HBsAg and HBcrAg titres allow peginterferon-based ‘precision medicine’ in HBeAg-negative chronic hepatitis B patients. J Viral Hepat 2016;23:905–911. [DOI] [PubMed] [Google Scholar]
- 106.Li Y, Xie J, Wang H, et al. Elevated pre-treatment IL-18 level is associated with HBeAg seroconversion in HIV-HBV coinfection. Antivir Ther 2017;22:523–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


