Plasmodium falciparum erythrocyte-binding antigen 140 (EBA-140) plays a role in tight junction formation during parasite invasion of red blood cells and is a potential vaccine candidate for malaria. Individuals in areas where malaria is endemic possess EBA-140-specific antibodies, and individuals with high antibody titers to this protein have a lower rate of reinfection by parasites.
KEYWORDS: Plasmodium falciparum, antibody function, epitope, immunogen design, malaria, structural vaccinology, three-dimensional structure, vaccines
ABSTRACT
Plasmodium falciparum erythrocyte-binding antigen 140 (EBA-140) plays a role in tight junction formation during parasite invasion of red blood cells and is a potential vaccine candidate for malaria. Individuals in areas where malaria is endemic possess EBA-140-specific antibodies, and individuals with high antibody titers to this protein have a lower rate of reinfection by parasites. The red blood cell binding segment of EBA-140 is comprised of two Duffy-binding-like domains, called F1 and F2, that together create region II. The sialic acid-binding pocket of F1 is essential for binding, whereas the sialic acid-binding pocket in F2 appears dispensable. Here, we show that immunization of mice with the complete region II results in poorly neutralizing antibodies. In contrast, immunization of mice with the functionally relevant F1 domain of region II results in antibodies that confer a 2-fold increase in parasite neutralization compared to that of the F2 domain. Epitope mapping of diverse F1 and F2 monoclonal antibodies revealed that the functionally relevant F1 sialic acid-binding pocket is a privileged site inaccessible to antibodies, that the F2 sialic acid-binding pocket contains a nonneutralizing epitope, and that two additional epitopes reside in F1 on the opposite face from the sialic acid-binding pocket. These studies indicate that focusing the immune response to the functionally important F1 sialic acid binding pocket improves the protective immune response of EBA-140. These results have implications for improving future vaccine designs and emphasize the importance of structural vaccinology for malaria.
INTRODUCTION
Malaria affects one-third of the world’s population, with over 200 million cases annually worldwide and an estimated 445,000 deaths, primarily in children under the age of five (www.who.int). Malaria is caused by Plasmodium parasites, and of the five species that infect humans, the majority of deaths are attributed to Plasmodium falciparum. The Plasmodium parasite has a complex life cycle that includes a human host and a mosquito vector (1). The clinical symptoms of malaria arise from the asexual replicative red blood cell (RBC) cycle in the human host (1). The red blood cell cycle is characterized by multiple rounds of invasion of, replication within, and lysis of RBCs (1). RBC invasion can be broken down into 5 steps: (i) low-affinity interactions between the merozoite stage parasite and the RBCs, (ii) apical reorientations of the merozoite, (iii) tight junction formation, (iv) active invasion utilizing an actin-myosin motor, and (v) shedding of the parasite surface proteins and formation of the parasitophorous vacuole (1). Multiple distinct families of parasite proteins are involved in the various steps of invasion; several of these proteins are vaccine candidates (1).
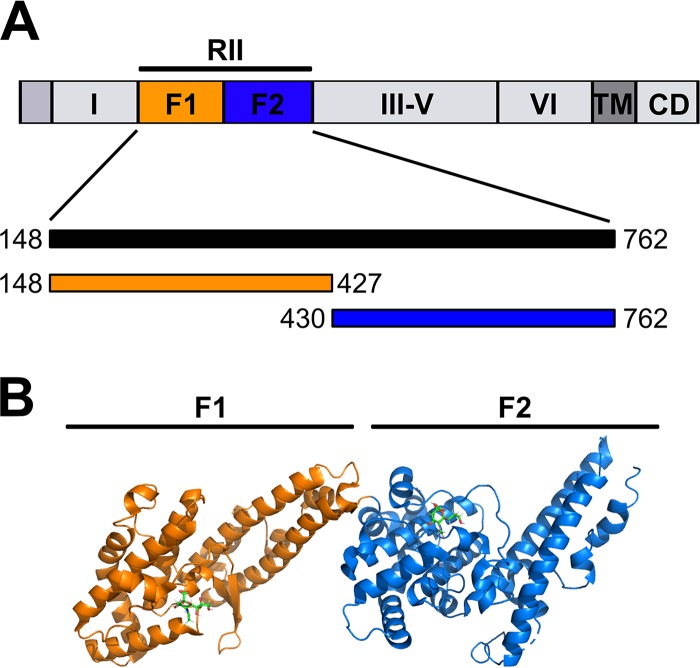
One such family of proteins is the erythrocyte binding-like (EBL) family of proteins that are involved in tight junction formation, including erythrocyte-binding antigen 140 (EBA-140) (2–14), EBA-175 (15–22), EBA-181 (8, 10, 23–26), EBL1 (27), and Duffy-binding protein (DBP) (28–38). EBL family proteins share a similar domain structure (Fig. 1) that includes a region II (RII), consisting of either a single or double Duffy binding-like (DBL) domain, regions RIII to RV, which include coiled-coil like repetitions, a cysteine-rich domain in region VI, a transmembrane domain, and a cytoplasmic domain (6, 7, 31, 34, 35, 39–41). In all cases examined, RII comprises the minimal segment necessary to bind host cell receptors.
FIG 1.
Domain structure of the full-length PfEBA-140 and the details of the two DBL domains in region II. (A) EBA-140 is comprised of regions I to VI: RII consists of two DBL domains, RIII to RV contain coiled-coil like repetitions, and RVI is a conserved cysteine-rich domain. A transmembrane (TM) domain and a cytoplasmic domain (CD) are located at the carboxyl terminus. The amino acid boundaries for the RII domain construct, as well as the F1 domain and F2 domain, are shown below in the domain structure model. (B) Crystal structure of RII in complex with sialyllactose adapted from the crystal structure 4JNO. Orange, F1 domain; blue, F2 domain. Sialyllactose is represented by a stick model colored green.
P. falciparum EBA-140 contains two DBL domains in region II, termed F1 and F2 (2–4, 6, 7). The multimeric states of EBA-175 and DBP have profound effects on receptor binding (20, 34, 35). In contrast, EBA-140 has only been observed as a monomer, and it is unclear if multimeric assembly occurs upon receptor binding (6, 7). EBA-140 binds to glycophorin C on the surface of RBCs in a sialic acid-dependent manner (2, 3, 5–7). Structural and functional studies have shown that both F1 and F2 harbor structurally conserved sialic acid-binding pockets (6, 7). However, receptor binding appears to be dependent on sialic acid binding to F1, while the sialic acid-binding pocket in F2 appears to be dispensable (7).
Antibodies to EBA-140 are correlated with acquired immunity and protection in individuals in regions where malaria is endemic (8, 9, 11, 14). This acquired immunity is age and antibody response dependent (8, 9, 11, 14). Individuals with higher antibody titers to EBA-140 show greater protection against infection than those with low titers (9). IgG responses are higher for the RII domain of EBA-140 and EBA-175, although responses to RIII to RV are also correlated with protection (9, 14). Polysera from individuals from different continents and countries show reactivity to EBA-140, suggesting EBA-140 is a conserved target of protective immunity and therefore a viable vaccine candidate (8, 9, 11, 13, 14). Previous animal studies for EBA-140 have focused on the F2 domain of RII and the RIII to RV domains (10, 12). However, immunization with the F2 domain resulted in minimal neutralization of parasites in culture (10).
Here, we examine the neutralizing antibody response via immunization with RII, F1, and F2 of EBA-140, characterize the ability of individual monoclonal antibodies (MAbs) to neutralize parasites, and map the epitopes of antibodies with diverse neutralizing potential. We show that focusing the immune response to the F1 domain results in more effective neutralizing antibodies for blocking parasite growth. Immunization with the full-length RII reduces neutralizing antibody generation compared to that of F1 alone, potentially by diluting the immune response to target the F2 sialic acid-binding pocket. Finally, we find that immunodominant epitopes to nonfunctional regions exist in the F1 domain, sheltering the F1 sialic acid-binding pocket from the immune response.
RESULTS
Immunizing mice with RII-140 resulted in poorly neutralizing monoclonal antibodies that all map to the F2 domain.
Three BALB/c mice were immunized with RII from EBA-140, and antibody titers were measured after three rounds of boosting. All three animals showed similar levels of reactivity to RII-140 (see Fig. S1A in the supplemental material). Splenocytes from mouse 2 were fused with a murine myeloma to generate hybridoma lines. Hybridomas secreting RII-140-specific antibodies were subcloned by limiting dilution to establish stable cell lines secreting unique antibodies, resulting in four monoclonal antibodies, which were further characterized.
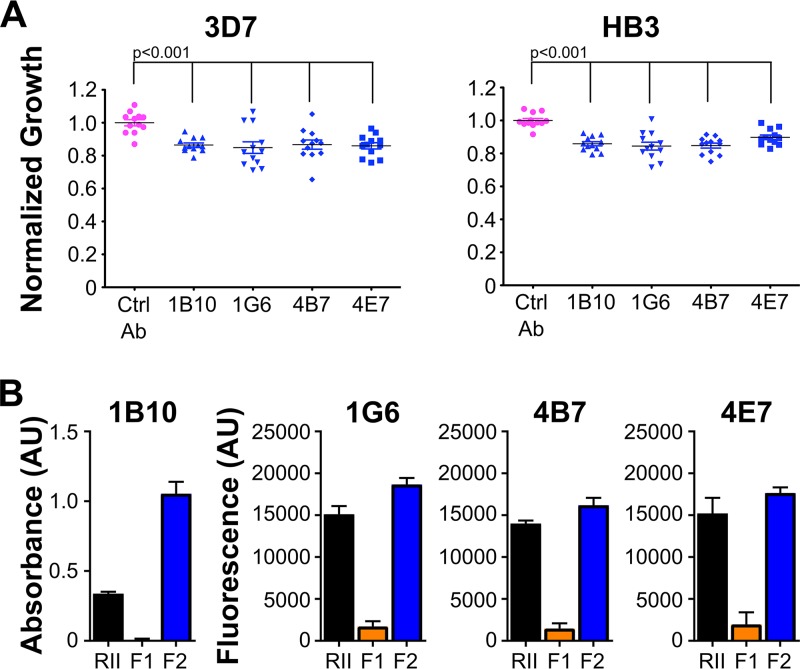
We tested the four MAbs 1G6, 1B10, 4B7, and 4E7 for their ability to neutralize P. falciparum 3D7 and HB3 strains (Fig. 2A). 3D7 and HB3 represent two strains from distinct geographical locations and two of the five polymorphic groups for the RII domain of EBA-140. All four anti-RII-140 antibodies tested were only slightly neutralizing, 10% to 15%, on both 3D7 and HB3 culture growth (Fig. 2A), compared to the control antibody, an IgG against West Nile virus. This level of parasite growth neutralization by RII-140 antibodies is similar in range to that reported by others who examined EBA-140 antibodies (10).
FIG 2.
Four monoclonal antibodies derived from immunization with RII have minimal neutralization effects and map to F2 domain of RII-140. (A) Neutralization analysis of schizont-stage P. falciparum 3D7 and HB3 parasites cultured in the absence (PBS) or presence of 3 mg/ml control antibody (purple) or anti-RII antibody (blue; 1B10, 1G6, 4B7, or 4E7). Parasitemia was assessed after two cycles by microscopy and growth normalized to that of PBS-treated cultures. The experiment was done four independent times in triplicate, and all data are shown. Significant values were determined by one-way ANOVA with Dunnett’s multiple-comparison tests. (B) ELISA for MAbs 1B10, 1G6, 4E7, and 4B7 utilizing RII (black bars), F1 (orange bars), and F2 (blue bars) construct-coated plates.
To determine the mechanism for minimal neutralization of parasite growth, we examined whether these antibodies preferentially mapped to particular domains in RII of EBA-140. A domain fragment enzyme-linked immunosorbent assay (ELISA) showed that all four antibodies bind to the F2 domain of RII-140 and full-length RII-140 but not to the F1 domain (Fig. 2B). F2 appears functionally dispensable for receptor binding, and it is plausible these antibodies show limited neutralization due to their domain specificity. Strikingly, no F1-specific MAbs were obtained by this immunization and hybridoma protocol, suggesting immunization with RII of EBA-140 dilutes the antibody response to the F2 domain.
Immunization with F1 generates a greater protective response than immunization with F2 or RII.
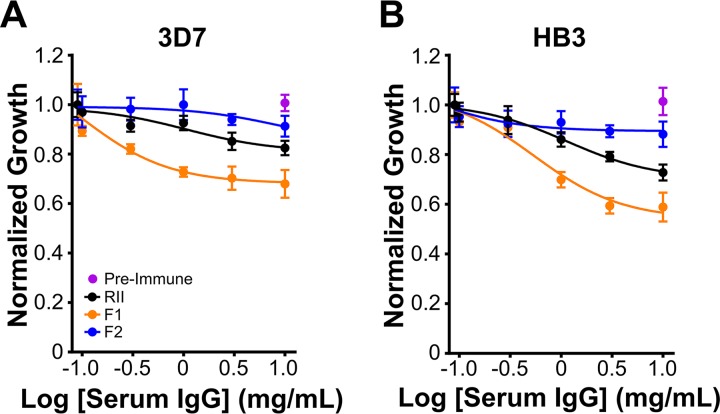
To understand the immune response to individual regions of the receptor binding domain, we immunized mice with F1, F2, or RII, isolated the total IgGs from mouse polysera, and performed growth neutralization assays. Compared to the preimmune sera, purified IgGs from F1-immunized mice neutralized the growth of 3D7 and HB3 cultures by 35% and 40%, respectively (Fig. 3). In contrast, sera from F2-140-immunized mice neutralized growth by ∼10%. The sera from RII-140-immunized mouse neutralized parasite growth by 20% on 3D7 and 30% on HB3. All immunizations showed a similar level of response to their respective antigens, indicating total IgG levels are similar and cannot account for the difference in neutralization (Fig. S1B). Together, the results indicate that immunization with F1 results in more potent neutralizing antibodies and that the addition of F2 in RII dilutes the antibody response toward nonneutralizing antibodies.
FIG 3.
Immunization with F1 results in a greater protective response than with F2 or RII. Growth of 3D7 (A) or HB3 (B) schizont-stage parasites treated with PBS or with various concentrations (0.1 to 10 mg/ml) of purified serum IgGs from preimmune or RII-, F1-, or F2-immunized mice. Serum IgGs from preimmune mice (10 mg/ml) served as a control. Parasitemia was determined by microscopy after two cycles. Parasite growth in serum IgG-treated cultures was normalized to that in PBS-treated cultures. Data shown are representative of parasite neutralization by serum IgGs from three sets of mice assayed in triplicate.
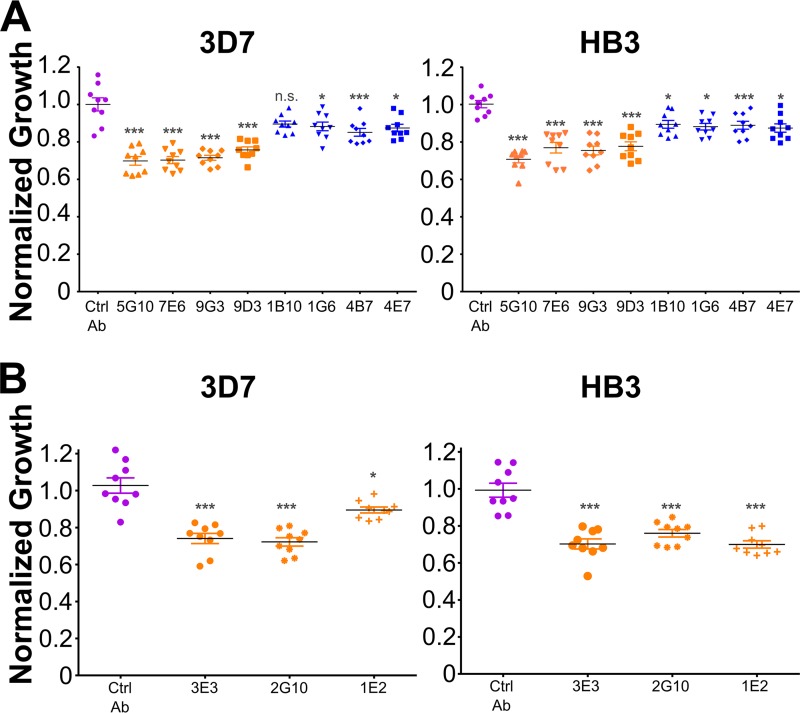
Monoclonal antibodies derived from F1-140-immunized mice are F1 specific and are more effective at neutralizing parasites.
A hybridoma fusion with the spleen from an F1-140-immunized mouse and myeloma cells resulted in a total of seven F1-specific monoclonal antibodies. All seven MAbs (1E2, 2G10, 9G3, 3E3, 5G10, 7E6, and 9D3) recognized both the F1 domain and the intact RII of EBA-140 but not the F2 domain (Fig. S2). To compare the effectiveness of F1- and F2-specific antibodies in parasite neutralization, we incubated 3D7 and HB3 cultures with 3 mg/ml of the control monoclonal antibody or the various domain-specific MAbs for two cycles. All the F1-specific MAbs neutralized parasite growth by 20 to 30% for both 3D7 and HB3 (Fig. 4), with the exception of 1E2, which showed preferential neutralization of HB3 over 3D7 (Fig. 4B). In contrast, all the F2-specific MAbs neutralized parasite growth by 10% to 15% (Fig. 4A). The F1-specific MAbs are significantly better at parasite neutralization than the F2-specific MAbs, with a 2-fold increase in neutralization (Table S1). Taken together, all F1-specific MAbs neutralized parasites to a greater extent than F2-specific MAbs.
FIG 4.
Monoclonal antibodies derived from F1-immunized mice are more effective at neutralizing parasites. (A) Growth of 3D7 and HB3 parasite cultures incubated with 3 mg/ml of the control Ab (purple) or with F1-specific (orange) or F2-specific (blue) antibody for two cycles. The experiment was done three independent times in triplicate, and all data are shown. Significant differences (Table S1) were determined by one-way ANOVA with Dunnett’s multiple-comparison tests. P values were <0.001 (***) and <0.05 (*) for comparisons made to the control. The extents of antibody inhibition for 3D7 and HB3 are 30% and 30% (5G10), 30% and 23% (7E6), 29% and 25% (9G3), 24% and 22% (9D3), 10% and 11% (1B10), 12% and 12% (1G6), 15% and 11% (4B7), and 13% and 13% (4E7), respectively. (B) Additional F1-specific MAbs (3 mg/ml) tested for neutralization of 3D7 and HB3 parasites. The extents of F1 antibody inhibition for 3D7 and HB3 are 26% and 30% (3E3), 28% and 24% (2G10), and 10% and 30% (1E2), respectively. Growth in all antibody-treated cultures (inclusive of control antibody culture) was normalized to that of PBS-treated cultures, since PBS was the vehicle for all antibodies. Significance was determined as described above. P values were <0.001 (***) and <0.05 (*) for comparisons made to the control.
HDX-MS revealed five distinct epitopes in the RII domain.
We then turned to hydrogen deuterium exchange mass spectrometry (HDX-MS) experiments to determine the epitopes of F1-specific MAbs 1E2, 2G10, 9G3, 3E3, 5G10, 7E6, and 9D3 and the F2-specific MAb 4E7. With optimized conditions for HDX quenching and for liquid chromatography (LC) gradient separation (see Materials and Methods), we acquired 98% sequence coverage of RII (Fig. S3) (a color gradient is shown in the figure legend for different levels of deuterium uptake). The deuterium uptake behavior is consistent with the secondary structure elements of RII (Fig. S3), that is, structured regions show less HDX.
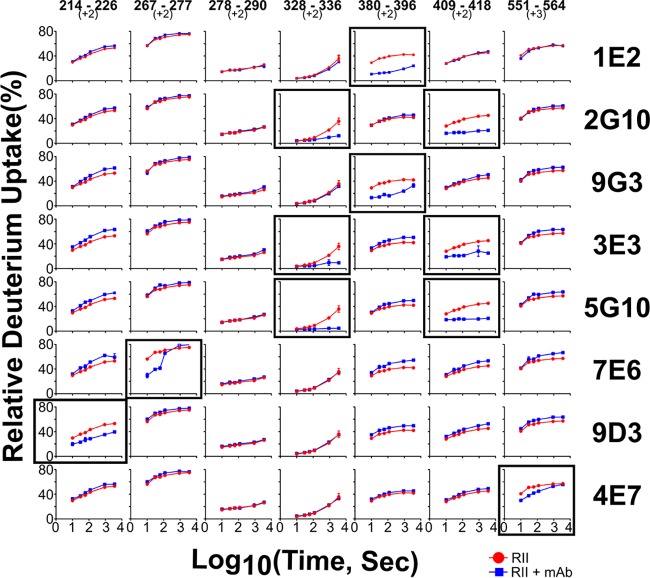
We measured the extent of HDX in the presence or absence of the MAbs and displayed differences in the average HDX extent across all time points. This data processing revealed four distinct epitopes in F1 (Fig. 5 and Table S2). The F1 MAb 9D3 had a distinct epitope, indicated by moderate protection for the region of residues 205 to 213, and pronounced protection from HDX in peptides covering the region of residues 184 to 189 and residues 214 to 226. A separate, distinct epitope in F1 was seen for MAb 7E6, with a marked difference in deuterium uptake for the region of residues 267 to 277. A third epitope in the F1 domain appears to be shared by MAbs 9G3 and 1E2. 9G3-bound F1 showed reduction in deuterium uptake for the region of residues 366 to 407, with the biggest difference in residues 380 to 396. Similarly, MAb 1E2 showed reduced deuterium uptake for the region of residues 374 to 407, with the biggest difference occurring in the same region as that with MAb 9G3 (residues 380 to 396). Finally, a fourth epitope in the F1 domain appeared to be shared by MAbs 2G10, 5G10, and 3E3. Modest protection revealed by HDX occurs in the regions of residues 328 to 351 and 352 to 361 for 2G10, with the biggest difference for the region of residues 408 to 420 (residues 409 to 418 showed the largest change for three peptides representing this region). 5G10 and 3E3 showed similar binding behavior with reduced HDX for regions of residues 327 to 336 and 408 to 420 compared to EBA-140 alone, with the greatest protection for the region of residues 409 to 418.
FIG 5.
HDX uptake plots for different regions of PfEBA-140. The kinetic plots for seven peptides from PfEBA-140 in the presence of various MAbs (PfEBA-140 plus MAbs, depicted in blue) and in the absence of the MAb (PfEBA-140 alone, depicted in red). Each region (column) is represented by a peptic peptide and its charge state, as measured by mass spectrometry. Each row represents a state bound with a MAb; the antibody is listed on the right. Those regions showing reduced rates or extents of exchange for the sample of PfEBA-140 with MAbs (blue) are considered to contain the epitopes. Those regions with no difference are examples of regions that do not contain the epitopes and can be viewed as controls. Boxed plots are those that showed a difference upon addition of antibody for each MAb tested.
Most other peptides identified in these experiments exhibited little or no difference in HDX upon complex formation with MAbs, as represented by peptide 278-290 (Fig. 5 and Table S2). The HDX-MS mapping of the epitopes indicated that the epitopes were excluded from the F1 sialic acid-binding pocket. All of the F1 MAb epitopes are, in fact, distant from the binding pocket, and many were on the opposite face of the domain from the sialic acid-binding pocket.
In addition to the F1-specific MAbs, we also mapped the F2-specific MAb 4E7 by HDX-MS. The only region that showed protection upon binding by 4E7 comprised region of residues 551 to 564 (Fig. 5 and Table S2). This epitope region overlaps one side of the F2 sialic acid-binding pocket. This result suggests that antibody binding to and blocking of the F2 sialic acid-binding pocket results in limited neutralization, emphasizing that the F2 sialic acid-binding pocket is dispensable for receptor binding (6, 7). The F2 sialic acid-binding pocket is also targetable by the immune response, in contrast to the F1 sialic acid-binding pocket that appears sheltered from the antibody response.
RII mutants reveal the residues important for MAb binding in all five epitopes.
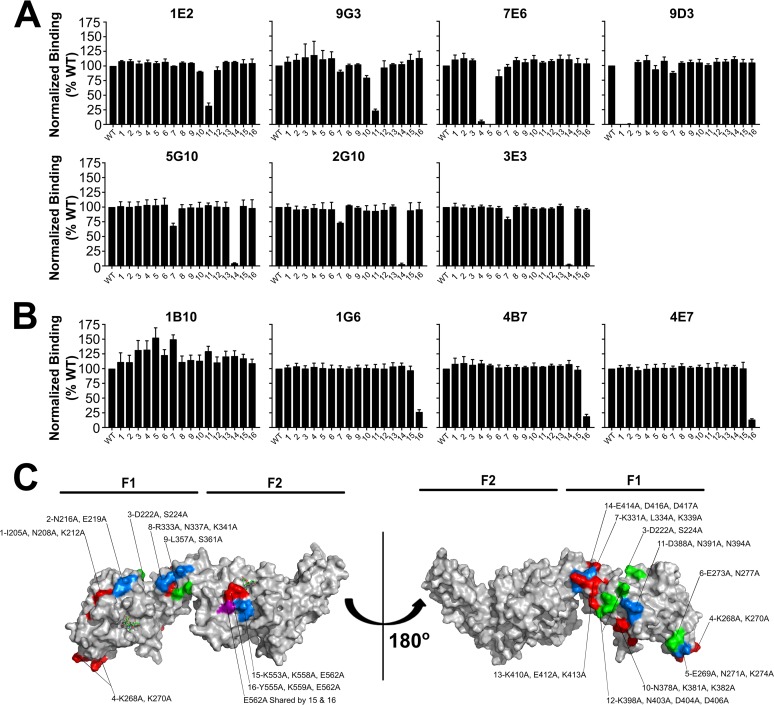
On the basis of the epitopes determined by HDX-MS, we generated 16 mutants in RII (Fig. S4 and Table S3). Fourteen of the mutants had residues in the F1 domain that were replaced with alanine, and two of the mutants had residues in the F2 domain replaced with alanine (Table S3). The mutants were numbered in order from the N terminus to the C terminus of RII. All RII mutant proteins expressed well and were properly folded, as evidenced by size exclusion chromatography and Coomassie-stained protein gels (Fig. S4).
We tested all MAbs for binding to the 16 mutants by ELISA, normalized the results to those of binding to wild-type RII, and determined significance by using one-way analysis of variance (ANOVA). The F1-specific MAbs all showed loss of binding to one or more mutants with substitutions located in the F1 domain of RII. 9D3 showed loss of binding to mutants 1 and 2 (P ≤ 0.0001) and to mutant 7 (P = 0.0142) (Fig. 6A and C). The substituted residues for mutants 1 and 2 are within the epitope region determined by HDX for 9D3, whereas the substituted residue in mutant 7 is distant from that determined by HDX. 7E6 showed loss of binding to mutants 4 and 5 (P ≤ 0.0001) and to mutant 6 (P = 0.0073) (Fig. 6A and C). All substitutions for mutants that showed loss of binding for 7E6 are within the HDX-MS identified epitope. 9G3 and 1E2 showed loss of binding to mutant 11 (P ≤ 0.0001), and 1E2 also showed loss of binding to mutant 10 (P = 0.0217) (Fig. 6A and C). All substitutions for mutants that showed loss of binding for 9G3 and 1E2 are within the HDX-MS epitope. 2G10, 5G10, and 3E3 all showed loss of binding to mutant 14 (P ≤ 0.0001) (Fig. 6A and C). 2G10, 5G10, and 3E3 also showed a loss of binding to mutant 7 (P = 0.0014, P ≤ 0.0001, and P ≤ 0.0001, respectively) (Fig. 6A and C). All residue substitutions that showed loss of binding for 2G10, 5G10, and 3E3 are within the epitope region identified by HDX-MS.
FIG 6.
Mutations within the epitopes of MAbs result in loss of binding. (A) ELISA for MAbs 1E2, 9G3, 7E6, 9D3, 5G10, 2G10, and 3E3 utilizing RII wild-type (WT) and mutant construct-coated plates (residues mutated are listed in Table S3). 1E2, mutant 10, P = 0.0217; mutant 11, P ≤ 0.001; 9G3, mutant 11, P ≤ 0.001; 7E6, mutants 4 and 5, P ≤ 0.001; mutant 6, P = 0.0073; 5G10, mutants 7 and 14, P ≤ 0.001; 2G10, mutant 7, P = 0.0014; mutant 14, P ≤ 0.001; 3E3, mutants 7 and 14, P ≤ 0.001. (B) ELISA for MAbs 1B10, 1G6, 4E7, and 4B7 utilizing RII wild-type and mutant construct-coated plates (residues mutated are listed in Table S3). 1B10, no significant loss of binding; 1G6, 4B7, and 4E7, lane 16, P ≤ 0.001. (C) Residues substituted for each mutant mapped on the structure of RII. Sialyllactose is represented by green sticks.
We also tested the F2-specific MAbs for loss of binding to mutants by ELISA. Although only 4E7 was mapped by HDX-MS, the ELISAs revealed that 4E7, 4B7, and 1G6 all share an epitope. 4E7, 4B7, and 1G6 all showed a loss of binding to mutant 16 (P ≤ 0.0001) for all three MAbs (Fig. 6B and C). The mutant that showed loss of binding for 4E7 has a substitution within the epitope determined by HDX-MS. This epitope is highly immunogenic, as 75% of the MAbs derived from this immunization target these residues of the F2-binding pocket. The F2-specific MAb 1B10 was also tested for binding to all mutants and showed no significant loss of binding to any mutant (Fig. 6B and C).
DISCUSSION
The EBA family of proteins is under selective pressure to evade the immune system. This selective pressure results in decoy epitopes that prevent targeting of functional regions of the receptor binding domains and the emergence of polymorphisms. Decoy epitopes are immunodominant and found in nonfunctional regions of proteins. Antibodies that bind to the functionally important regions are more neutralizing than those binding to nonfunctional regions. One example of this phenomenon is the pair of MAbs (R217 and R218) derived from an immunization with the RII domain of the related EBL family protein EBA-175. MAb R217 targets the dimer interface and receptor binding pocket of RII-175 and has a 50% inhibitory concentration (IC50) of 10 to 100 μg/ml; MAb R218 targets a nonfunctional region of RII-175 and has an IC50 of >1 mg/ml (42). Similarly, this work shows that immunizing with the whole RII domain of EBA-140 results in the isolation of four F2-specific MAbs with limited (10% to 15%) parasite neutralization activity and no F1-specific MAbs (Fig. 2). Three of the F2-specific MAbs (4E7, 4B7, and 1G6) target the receptor-binding pocket of the F2 domain (Fig. 4, 5, and 7), demonstrating that this binding pocket is dispensable for parasite viability. The fact that most F2-specific antibodies bind to a single epitope with little neutralizing ability suggests that this epitope is a decoy in the RII domain.
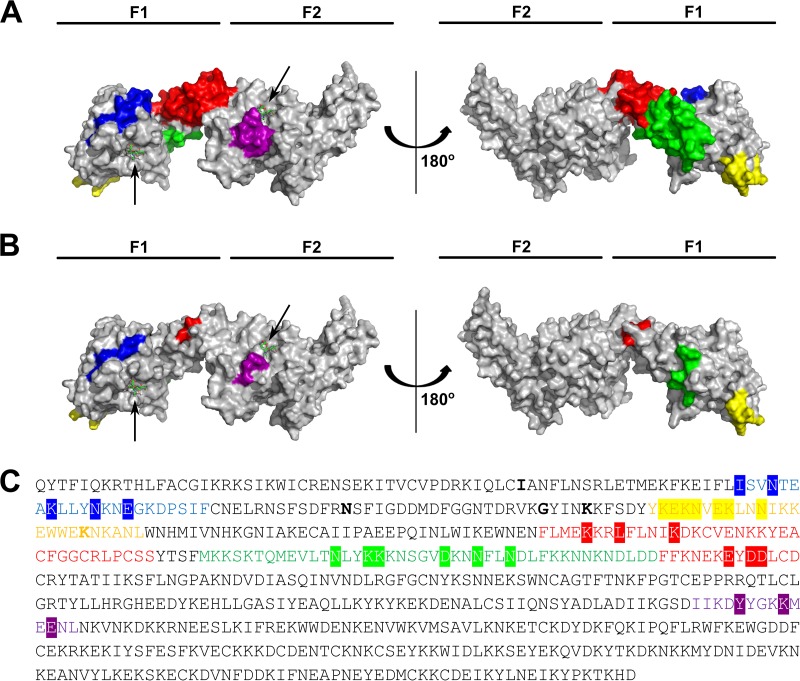
FIG 7.
Two immunodominant regions exist in RII. (A) Surface representation of HDX-MS epitopes in a structure adapted from 4JNO. (B) Surface representation of epitopes based on ELISAs of the RII mutants in a structure adapted from 4JNO. (C) HDX-MS and mutant ELISA epitopes mapped on the amino acid sequence of RII. Colored residues, HDX-MS epitopes; highlighted residues, mutant ELISA epitopes; boldface residues, polymorphisms. Blue, 9D3; yellow, 7E6; red, 5G10, 2G10, and 3E3; green, 9G3 and 1E2; purple, 4E7, 4B7, and 1G6.
Immunization with the F1 domain alone resulted in isolation of antibodies that are 2-fold more neutralizing than the F2-specific MAbs at 3 mg/ml (Fig. 4). This concentration reflects the fact that laboratory strains have adapted to be less dependent on glycophorin C for invasion than field strains (43). It is anticipated that the concentration of EBA-140 MAbs required to neutralize field strains will be lower, as field strains showed a greater range of dependence on glycophorin C for invasion than laboratory-adapted strains, up to a 47% reduction of invasion for field strains in glycophorin C knockdown RBCs compared to the 0% to 10% reduction seen in laboratory strains (43). In addition to EBA-140, the merozoite antigens EBA-175 (42, 44, 45), AMA-1 (46–51), RON2 (52), and RH5 (53–56) all have demonstrated the ability to elicit antibodies that potently neutralize growth in vitro. The concentrations of EBA-140 antibodies required here for neutralization are higher than those reported for some of these alternative antigens, and this is likely due to immunization with EBA-140 eliciting antibodies to nonneutralizing or moderately neutralizing epitopes away from the sialic acid-binding pocket of the F1 domain. Although the F1 immunization produced antibodies with greater neutralizing potential, it still failed to produce an antibody that targets the F1 receptor-binding pocket (Fig. 5 and 7). The results from HDX-MS and ELISAs of mutated proteins located the epitopes of the seven F1-specific antibodies to regions that excluded the F1 receptor-binding pocket (Fig. 7). All epitopes mapped were on the opposite face of F1 from the sialic acid-binding pocket. HDX-MS and mutant ELISA mapping also revealed that the two most distant epitopes from the F1 sialic acid-binding pocket are also the two most immunodominant epitopes in the F1 domain, as evidenced by three MAbs targeting one epitope and two MAbs targeting the other epitope (Fig. 5 and 7).
In addition to decoy epitopes, EBA-140 contains four polymorphisms located in the F1 domain of RII. In contrast, EBA-175 contains 14 polymorphisms in both DBL domains that comprise its RII domain. The lack of polymorphisms in the F2 domain of EBA-140 may be due to the minimal role F2 plays in receptor binding and may be a mechanism by which the parasite attempts to evade the immune system. The development of polymorphisms as a method for immune evasion has been shown for the related EBA family DBP of Plasmodium vivax (57, 58). Surprisingly, all of the epitopes determined by mutant ELISA exclude the four known polymorphisms in the F1 domain. Although the MAbs from the F1 immunization are only 20% to 30% neutralizing in culture, they are broadly neutralizing because of the exclusion of the polymorphisms from the epitopes.
Previous animal immunization studies and analysis of patient antibody response against EBA-140 focused on the F2 domain of RII-140. Total IgG purified from rabbits immunized with F2 resulted in a 17.6% neutralization of parasite growth, which is similar to the 15% maximum neutralization observed here with F2-specific MAbs (10). Immunization with the functional F1 domain described here results in a 2-fold increase in neutralization of parasite growth. In contrast, immunization of rabbits with regions III to V of EBA-140 resulted in a 50% to 80% neutralization of parasites. This neutralization, however, was strain specific, as demonstrated by a greater than 6-fold difference in IC50 for the diverse Plasmodium strains tested (12). Immunization with F1 here showed little difference in neutralization of the Plasmodium strains tested, likely due to the exclusion of polymorphisms from the epitopes defined, in contrast to the strain specificity seen for immunization with regions III to V.
The current paradigm in the field is to immunize with either whole proteins or domains for vaccine candidates. We have shown that this type of immunization results in an antibody response that is skewed toward antibodies targeting nonfunctional immunodominant epitopes in Plasmodium antigens. Our study here showed that even within functional domains, functional regions were protected from that antibody response. By combining immunization studies with structural and functional data, more effective vaccines can be rationally designed to produce the greatest protection against pathogens by immunizing with only functional regions of the antigens. This study shows that targeting a functional domain increases the neutralization seen both in polysera and in MAb on parasites. While we were able to produce neutralizing antibodies to the F1 domain, immunodominant epitopes within this domain draw the antibody response away from the receptor-binding pocket. These results suggest the functionally important F1 sialic acid-binding pocket is immunologically privileged and protected from antibody recognition. By using the epitopes defined in this study along with structural data, it may be possible to design a better antigen that targets the F1 receptor-binding pocket. Focusing the immune response away from the immunodominant epitopes in F1 and toward the sialic acid-binding pocket through immunogen engineering may lead to novel EBA-140-based immunogens that improve the protective antibody response.
MATERIALS AND METHODS
Recombinant protein expression and purification.
RII-140 was expressed and purified as described previously (6, 7). Briefly, the recombinant proteins were expressed as insoluble proteins in Escherichia coli and refolded by rapid dilution in 400 mM l-arginine, 50 mM Tris, pH 8.0, 10 mM ethylenediaminetetraacetic acid, 0.1 mM phenylmethanesulfonyl fluoride, 2 mM reduced glutathione, and 0.2 mM oxidized glutathione and refolded for 48 h at 4°C. The refolded protein was purified by ion exchange resin and gel filtration using a HiLoad Superdex 200 16/60 column (GE Healthcare) and phosphate-buffered saline (PBS). F1, F2, and RII mutants were expressed and purified as described above for RII-140.
Immunization of mice.
The Hybridoma Center at the Washington University School of Medicine provided experimental support in accordance with the Washington University Institutional Animal Care and Use Committee (IACUC) protocol number 20160232. Three BALB/c mice were each immunized with 10 μg of antigen. For the first round with RII-140, mice were boosted with additional antigen twice intravenously on days 22 and 44 and once intraperitoneally on day 70 prior to fusion. For the second round of immunization with RII-140, F1-140, or F2-140, boostings were done on days 16 and 34 and prefusion boost was on day 81. Polyserum from each mouse was tested by ELISA for reactivity toward its specific antigen. The ELISA plates were coated with 0.02 mg/ml antigen overnight at 4°C and blocked with 2% bovine serum albumin in PBS with 0.05% Tween 20 for 1 h at room temperature. A 10-fold serial dilution series of polysera was added to the plates and incubated for 1 h at room temperature. An anti-mouse IgG secondary antibody conjugated to horseradish peroxidase (HRP; Jackson ImmunoResearch) was then added to plates at a 1:5,000 dilution and incubated for 1 h at room temperature. Plates were developed using TMB substrate (Sigma), and the absorbance at 450 nm was measured with a BMG POLARStar plate reader. At day 73 or 85, the mice were sacrificed and terminal bleeds collected, along with the spleens, for later use in generation of monoclonal antibodies.
Generation of monoclonal antibodies.
For those mice immunized with either RII-140 or F1-140, the spleen from mouse with the highest reactivity to antigen was used for the generation of monoclonal antibodies. B cells from the mice were fused with myeloma cells to generate hybridoma cell lines. Two rounds of screening on the polyclonal cell lines were done to analyze reactivity toward their respective antigen by ELISA. The ELISA plates were coated and blocked as described above. Supernatant from the polyclonal cell lines was then added to the plates and incubated for 1 h at room temperature. An anti-mouse IgG-HRP secondary antibody was added, and absorbance at 450 nm was measured. Those lines with reactivity of ≥0.5 arbitrary units (AU) were chosen for subcloning. These lines were plated by limited dilution at 100 cells/well, 10 cells/well, and 1 cell/well and allowed to reach confluence, at which time the supernatants from each well were tested for reactivity by ELISA as described above. For each cell line, wells that initially contained 1 cell/well and retained reactivity toward antigen were chosen for an additional round of subcloning, as described above. After two rounds of subcloning, the individual hybridoma cell lines were declared monoclonal. At this point, each line was isotyped, expanded, and put into production flasks (Cell Line 1000 flasks; ThermoFisher Scientific) for antibody expression and purification.
Purification of IgGs from polysera and monoclonal IgG from hybridoma lines.
Preimmune sera or pooled sera from the same animal collected at days 9, 28, and 41 were diluted 1:1.5 (vol/vol) with IgG binding buffer (ThermoFisher Pierce) and incubated with a 0.5-ml bed volume of protein A resin (Goldbio Technologies) for 30 min at room temperature. The resin was washed 3 times with 3 ml of IgG binding buffer, and the IgGs were eluted with 2 ml of IgG elution buffer (ThermoFisher Pierce). The eluted IgGs were buffer exchanged 3 times with PBS by using Amicon spin concentrators (10-kDa-molecular-mass cutoff), filter sterilized, and added to parasite cultures at 10 mg/ml.
For monoclonal antibody purification, supernatant from monoclonal hybridoma cell lines was diluted 1:3 with IgG binding buffer and passed over a 3-ml-bed-volume protein A resin column. The resin was washed with 15 ml of IgG binding buffer. IgG was eluted from the column using 15 ml of IgG elution buffer into 15 ml of PBS. The eluted antibody was buffer exchanged 3 times with PBS using Amicon spin concentrators with a molecular mass cutoff of 10 kDa. For those experiments using parasite cultures, the antibody was filter sterilized after buffer exchanging into PBS.
Domain fragment mapping by ELISA.
For monoclonal antibody domain mapping, the ELISA plates were coated with antigen and blocked. A solution of 250 ng/ml of each monoclonal antibody in PBS was then added to the plates and incubated for 1 h at room temperature. An anti-mouse IgG secondary antibody conjugated to Alexa Fluor 488 (Life Technologies) was then added to plates at a 1:1,000 dilution and incubated for 1 h at room temperature. Fluorescence (in AU) was measured with a BMG POLARStar plate reader. For antibody 1B10 not recognized by the Alexa Fluor-conjugated antibody, an anti-mouse IgG secondary antibody conjugated to HRP was added to plates at a 1:5,000 dilution, incubated for 1 h at room temperature, and developed using TMB substrate. Absorbance at 450 nm was measured with a BMG POLARStar plate reader.
Parasite neutralization assay.
The P. falciparum strains 3D7 and HB3 cultured in human O+ RBCs in RPMI 1640 medium with 0.5% AlbuMAX were synchronized with successive rounds of 5% sorbitol treatment, as described previously (59). Schizont-stage parasites enriched with magnetic bead columns (MACSQuant columns; Miltenyi Biotec) were mixed with fresh RBCs to adjust the parasitemia to 0.1% to 0.5% at the start of the invasion assay. The cultures at 2% hematocrit (100 μl/well, triplicate wells per condition) were then incubated in the absence or presence of 3 mg/ml of various EBA-140 monoclonal antibodies or isotype-matched control monoclonal antibodies (anti-West Nile virus and anti-P. vivax CelTOS 8C4) for two rounds of invasion. Giemsa-stained thin smears were then prepared from each well, and 10 random field images per smear were acquired using a Zeiss Axioskop microscope equipped with a 100× oil immersion lens (1.3 numerical aperture [NA]) and an AxioCam MRm camera with AxioVision v3.1 software (Carl Zeiss). The number of infected erythrocytes in each image was visually counted, and the total erythrocytes in each image were analyzed using Volocity 6.3 cellular imaging software (Perkin Elmer). Parasitemia was calculated as infected erythrocytes divided by the total number of erythrocytes per field. Parasite growth in antibody-treated cultures was normalized to that of nontreated cultures. All data groups were subjected to normality testing, followed by one-way ANOVA. Bonferroni’s multiple-comparison test was used to analyze the significance level between control and antibody-treated groups.
HDX-MS.
Continuous HDX labeling of P. falciparum EBA-140 (PfEBA-140) was at 25°C for 0, 10, 30, 60, 120, 900, and 3,600 s, as previously described but with modifications (60). Briefly, stock solutions of PfEBA-140 with or without the antibodies were prepared in 1× PBS, pH 7.4, and incubated on ice for 1 h. Continuous HDX was initiated by diluting stock samples 25-fold in D2O and 1× PBS buffer. Quenching and LC-MS analysis were as previously described (60), except the peptic fragments were separated with a 15-min gradient of 5% to 100% acetonitrile in 0.1% formic acid at 100 μl/min. The linear part of the gradient from 0.3 min to 11 min raised the acetonitrile content from 10% to 50%. Duplicate measurements were taken at each time point. Acquired spectra were searched using HDX workbench (61). Peptic peptides were identified in a data-dependent mode on a Thermo LTQ-FT mass spectrometer (Waltham, MA) by submitting product-ion spectra from MassMatrix (version 2.4.2) for identification (62), followed by manual validation for HDX runs. The HDX was adjusted for 96% D2O. Deuterium oxide was from Cambridge Isotope Laboratories (Tewksbury, MA); other reagents and proteases were from Sigma-Aldrich (St. Louis, MO).
RII mutant ELISAs.
The ELISA plates were coated with antigen and blocked as described above. A solution of 250 ng/ml of each monoclonal antibody in PBS was added to the plates and incubated for 1 h at room temperature. An anti-mouse IgG secondary antibody conjugated to HRP was added to plates at a 1:5,000 dilution, incubated for 1 h at room temperature, and developed using TMB substrate. Absorbance at 450 nm was measured with a BMG POLARStar plate reader.
Data availability.
All epitopes have been deposited in the IEDB under reference ID 1034379.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health. We thank the Hybridoma Center and Protein Production and Purification Facility of the Rheumatic Diseases Core Center (P30AR048335) at the Washington University School of Medicine for experimental support. This work was supported by NIH grant R56 AI080792 (to N.T.), NIH contract HHSN272201400018C (to N.T., J.A., and M.L.G.), and Burroughs Wellcome Fund grant 1013514 (to N.T.). We also thank the Goldberg, Sibley, and Odom laboratories for discussions, reagents, and equipment sharing, as well as J. Patrick Gorres for assistance in editing the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00716-18.
REFERENCES
- 1.Cowman AF, Tonkin CJ, Tham WH, Duraisingh MT. 2017. The molecular basis of erythrocyte invasion by malaria parasites. Cell Host Microbe 22:232–245. doi: 10.1016/j.chom.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thompson JK, Triglia T, Reed MB, Cowman AF. 2001. A novel ligand from Plasmodium falciparum that binds to a sialic acid-containing receptor on the surface of human erythrocytes. Mol Microbiol 41:47–58. doi: 10.1046/j.1365-2958.2001.02484.x. [DOI] [PubMed] [Google Scholar]
- 3.Mayer DC, Kaneko O, Hudson-Taylor DE, Reid ME, Miller LH. 2001. Characterization of a Plasmodium falciparum erythrocyte-binding protein paralogous to EBA-175. Proc Natl Acad Sci U S A 98:5222–5227. doi: 10.1073/pnas.081075398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Narum DL, Fuhrmann SR, Luu T, Sim BK. 2002. A novel Plasmodium falciparum erythrocyte binding protein-2 (EBP2/BAEBL) involved in erythrocyte receptor binding. Mol Biochem Parasitol 119:159–168. doi: 10.1016/S0166-6851(01)00428-5. [DOI] [PubMed] [Google Scholar]
- 5.Lobo CA, Rodriguez M, Reid M, Lustigman S. 2003. Glycophorin C is the receptor for the Plasmodium falciparum erythrocyte binding ligand PfEBP-2 (baebl). Blood 101:4628–4631. doi: 10.1182/blood-2002-10-3076. [DOI] [PubMed] [Google Scholar]
- 6.Lin DH, Malpede BM, Batchelor JD, Tolia NH. 2012. Crystal and solution structures of Plasmodium falciparum erythrocyte-binding antigen 140 reveal determinants of receptor specificity during erythrocyte invasion. J Biol Chem 287:36830–36836. doi: 10.1074/jbc.M112.409276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malpede BM, Lin DH, Tolia NH. 2013. Molecular basis for sialic acid-dependent receptor recognition by the Plasmodium falciparum invasion protein erythrocyte-binding antigen-140/BAEBL. J Biol Chem 288:12406–12415. doi: 10.1074/jbc.M113.450643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ford L, Lobo CA, Rodriguez M, Zalis MG, Machado RL, Rossit AR, Cavasini CE, Couto AA, Enyong PA, Lustigman S. 2007. Differential antibody responses to Plasmodium falciparum invasion ligand proteins in individuals living in malaria-endemic areas in Brazil and Cameroon. Am J Trop Med Hyg 77:977–983. [PubMed] [Google Scholar]
- 9.Richards JS, Stanisic DI, Fowkes FJ, Tavul L, Dabod E, Thompson JK, Kumar S, Chitnis CE, Narum DL, Michon P, Siba PM, Cowman AF, Mueller I, Beeson JG. 2010. Association between naturally acquired antibodies to erythrocyte-binding antigens of Plasmodium falciparum and protection from malaria and high-density parasitemia. Clin Infect Dis 51:e50–e60. doi: 10.1086/656413. [DOI] [PubMed] [Google Scholar]
- 10.Lopaticki S, Maier AG, Thompson J, Wilson DW, Tham WH, Triglia T, Gout A, Speed TP, Beeson JG, Healer J, Cowman AF. 2011. Reticulocyte and erythrocyte binding-like proteins function cooperatively in invasion of human erythrocytes by malaria parasites. Infect Immun 79:1107–1117. doi: 10.1128/IAI.01021-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Persson KE, Fowkes FJ, McCallum FJ, Gicheru N, Reiling L, Richards JS, Wilson DW, Lopaticki S, Cowman AF, Marsh K, Beeson JG. 2013. Erythrocyte-binding antigens of Plasmodium falciparum are targets of human inhibitory antibodies and function to evade naturally acquired immunity. J Immunol 191:785–794. doi: 10.4049/jimmunol.1300444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Healer J, Thompson JK, Riglar DT, Wilson DW, Chiu YH, Miura K, Chen L, Hodder AN, Long CA, Hansen DS, Baum J, Cowman AF. 2013. Vaccination with conserved regions of erythrocyte-binding antigens induces neutralizing antibodies against multiple strains of Plasmodium falciparum. PLoS One 8:e72504. doi: 10.1371/journal.pone.0072504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zerka A, Rydzak J, Lass A, Szostakowska B, Nahorski W, Wroczyńska A, Myjak P, Krotkiewski H, Jaskiewicz E. 2016. Studies on immunogenicity and antigenicity of baculovirus-expressed binding region of Plasmodium falciparum EBA-140 merozoite ligand. Arch Immunol Ther Exp (Warsz) 64:149–156. doi: 10.1007/s00005-015-0367-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCallum FJ, Persson KE, Fowkes FJ, Reiling L, Mugyenyi CK, Richards JS, Simpson JA, Williams TN, Gilson PR, Hodder AN, Sanders PR, Anders RF, Narum DL, Chitnis C, Crabb BS, Marsh K, Beeson JG. 2017. Differing rates of antibody acquisition to merozoite antigens in malaria: implications for immunity and surveillance. J Leukoc Biol 101:913–925. doi: 10.1189/jlb.5MA0716-294R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Orlandi PA, Sim BK, Chulay JD, Haynes JD. 1990. Characterization of the 175-kilodalton erythrocyte binding antigen of Plasmodium falciparum. Mol Biochem Parasitol 40:285–294. [DOI] [PubMed] [Google Scholar]
- 16.Sim BK, Toyoshima T, Haynes JD, Aikawa M. 1992. Localization of the 175-kilodalton erythrocyte binding antigen in micronemes of Plasmodium falciparum merozoites. Mol Biochem Parasitol 51:157–159. [DOI] [PubMed] [Google Scholar]
- 17.Orlandi PA, Klotz FW, Haynes JD. 1992. A malaria invasion receptor, the 175-kilodalton erythrocyte binding antigen of Plasmodium falciparum recognizes the terminal Neu5Ac(alpha 2-3)Gal- sequences of glycophorin A. J Cell Biol 116:901–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klotz FW, Orlandi PA, Reuter G, Cohen SJ, Haynes JD, Schauer R, Howard RJ, Palese P, Miller LH. 1992. Binding of Plasmodium falciparum 175-kilodalton erythrocyte binding antigen and invasion of murine erythrocytes requires N-acetylneuraminic acid but not its O-acetylated form. Mol Biochem Parasitol 51:49–54. doi: 10.1016/0166-6851(92)90199-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sim BK, Chitnis CE, Wasniowska K, Hadley TJ, Miller LH. 1994. Receptor and ligand domains for invasion of erythrocytes by Plasmodium falciparum. Science 264:1941–1944. [DOI] [PubMed] [Google Scholar]
- 20.Tolia NH, Enemark EJ, Sim BK, Joshua-Tor L. 2005. Structural basis for the EBA-175 erythrocyte invasion pathway of the malaria parasite Plasmodium falciparum. Cell 122:183–193. doi: 10.1016/j.cell.2005.05.033. [DOI] [PubMed] [Google Scholar]
- 21.Salinas ND, Tolia NH. 2014. A quantitative assay for binding and inhibition of Plasmodium falciparum erythrocyte binding antigen 175 reveals high affinity binding depends on both DBL domains. Protein Expr Purif 95:188–194. doi: 10.1016/j.pep.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salinas ND, Paing MM, Tolia NH. 2014. Critical glycosylated residues in exon three of erythrocyte glycophorin A engage Plasmodium falciparum EBA-175 and define receptor specificity. mBio 5:e01606-14. doi: 10.1128/mBio.01606-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilberger TW, Thompson JK, Triglia T, Good RT, Duraisingh MT, Cowman AF. 2003. A novel erythrocyte binding antigen-175 paralogue from Plasmodium falciparum defines a new trypsin-resistant receptor on human erythrocytes. J Biol Chem 278:14480–14486. doi: 10.1074/jbc.M211446200. [DOI] [PubMed] [Google Scholar]
- 24.Mayer DC, Mu JB, Kaneko O, Duan J, Su XZ, Miller LH. 2004. Polymorphism in the Plasmodium falciparum erythrocyte-binding ligand JESEBL/EBA-181 alters its receptor specificity. Proc Natl Acad Sci U S A 101:2518–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Treeck M, Struck NS, Haase S, Langer C, Herrmann S, Healer J, Cowman AF, Gilberger TW. 2006. A conserved region in the EBL proteins is implicated in microneme targeting of the malaria parasite Plasmodium falciparum. J Biol Chem 281:31995–32003. doi: 10.1074/jbc.M606717200. [DOI] [PubMed] [Google Scholar]
- 26.Maier AG, Baum J, Smith B, Conway DJ, Cowman AF. 2009. Polymorphisms in erythrocyte binding antigens 140 and 181 affect function and binding but not receptor specificity in Plasmodium falciparum. Infect Immun 77:1689–1699. doi: 10.1128/IAI.01331-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mayer DC, Cofie J, Jiang L, Hartl DL, Tracy E, Kabat J, Mendoza LH, Miller LH. 2009. Glycophorin B is the erythrocyte receptor of Plasmodium falciparum erythrocyte-binding ligand, EBL-1. Proc Natl Acad Sci U S A 106:5348–5352. doi: 10.1073/pnas.0900878106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller LH, Mason SJ, Dvorak JA, McGinniss MH, Rothman IK. 1975. Erythrocyte receptors for (Plasmodium knowlesi) malaria: Duffy blood group determinants. Science 189:561–563. [DOI] [PubMed] [Google Scholar]
- 29.Miller LH, Mason SJ, Clyde DF, McGinniss MH. 1976. The resistance factor to Plasmodium vivax in blacks. The Duffy-blood-group genotype, FyFy. N Engl J Med 295:302–304. doi: 10.1056/NEJM197608052950602. [DOI] [PubMed] [Google Scholar]
- 30.Adams JH, Hudson DE, Torii M, Ward GE, Wellems TE, Aikawa M, Miller LH. 1990. The Duffy receptor family of Plasmodium knowlesi is located within the micronemes of invasive malaria merozoites. Cell 63:141–153. [DOI] [PubMed] [Google Scholar]
- 31.Adams JH, Sim BK, Dolan SA, Fang X, Kaslow DC, Miller LH. 1992. A family of erythrocyte binding proteins of malaria parasites. Proc Natl Acad Sci U S A 89:7085–7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chitnis CE, Miller LH. 1994. Identification of the erythrocyte binding domains of Plasmodium vivax and Plasmodium knowlesi proteins involved in erythrocyte invasion. J Exp Med 180:497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chitnis CE, Chaudhuri A, Horuk R, Pogo AO, Miller LH. 1996. The domain on the Duffy blood group antigen for binding Plasmodium vivax and P. knowlesi malarial parasites to erythrocytes. J Exp Med 184:1531–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Batchelor JD, Zahm JA, Tolia NH. 2011. Dimerization of Plasmodium vivax DBP is induced upon receptor binding and drives recognition of DARC. Nat Struct Mol Biol 18:908–967. doi: 10.1038/nsmb.2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Batchelor JD, Malpede BM, Omattage NS, DeKoster GT, Henzler-Wildman KA, Tolia NH. 2014. Red blood cell invasion by Plasmodium vivax: structural basis for DBP engagement of DARC. PLoS Pathog 10:e1003869. doi: 10.1371/journal.ppat.1003869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen E, Salinas ND, Ntumngia FB, Adams JH, Tolia NH. 2015. Structural analysis of the synthetic duffy binding protein (DBP) antigen DEKnull relevant for Plasmodium vivax malaria vaccine design. PLoS Negl Trop Dis 9:e0003644. doi: 10.1371/journal.pntd.0003644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen E, Salinas ND, Huang Y, Ntumngia F, Plasencia MD, Gross ML, Adams JH, Tolia NH. 2016. Broadly neutralizing epitopes in the Plasmodium vivax vaccine candidate duffy binding protein. Proc Natl Acad Sci U S A 113:6277–6282. doi: 10.1073/pnas.1600488113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ntumngia FB, Pires CV, Barnes SJ, George MT, Thomson-Luque R, Kano FS, Alves JRS, Urusova D, Pereira DB, Tolia NH, King CL, Carvalho LH, Adams JH. 2017. An engineered vaccine of the Plasmodium vivax Duffy binding protein enhances induction of broadly neutralizing antibodies. Sci Rep 7:13779. doi: 10.1038/s41598-017-13891-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paing MM, Tolia NH. 2014. Multimeric assembly of host-pathogen adhesion complexes involved in apicomplexan invasion. PLoS Pathog 10:e1004120. doi: 10.1371/journal.ppat.1004120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malpede BM, Tolia NH. 2014. Malaria adhesins: structure and function. Cell Microbiol 16:621–631. doi: 10.1111/cmi.12276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tolia N, Enemark E, Sim B, Joshua-Tor L. 2005. Structural basis for the EBA-175 erythrocyte invasion pathway of the malaria parasite Plasmodium falciparum. Cell 122:485. [DOI] [PubMed] [Google Scholar]
- 42.Sim BK, Narum DL, Chattopadhyay R, Ahumada A, Haynes JD, Fuhrmann SR, Wingard JN, Liang H, Moch JK, Hoffman SL. 2011. Delineation of stage specific expression of Plasmodium falciparum EBA-175 by biologically functional region II monoclonal antibodies. PLoS One 6:e18393. doi: 10.1371/journal.pone.0018393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dankwa S, Chaand M, Kanjee U, Jiang RHY, Nobre LV, Goldberg JM, Bei AK, Moechtar MA, Grüring C, Ahouidi AD, Ndiaye D, Dieye TN, Mboup S, Weekes MP, Duraisingh MT. 2017. Genetic evidence for erythrocyte receptor glycophorin B expression levels defining a dominant Plasmodium falciparum invasion pathway into human erythrocytes. Infect Immun 85:e00074-17. doi: 10.1128/IAI.00074-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen E, Paing MM, Salinas N, Sim BKL, Tolia NH. 2013. Structural and functional basis for inhibition of erythrocyte invasion by antibodies that target Plasmodium falciparum EBA-175. PLoS Pathog 9:e1003390. doi: 10.1371/journal.ppat.1003390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ambroggio X, Jiang L, Aebig J, Obiakor H, Lukszo J, Narum DL. 2013. The epitope of monoclonal antibodies blocking erythrocyte invasion by Plasmodium falciparum map to the dimerization and receptor glycan binding sites of EBA-175. PLoS One 8:e56326. doi: 10.1371/journal.pone.0056326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Coley AM, Gupta A, Murphy VJ, Bai T, Kim H, Foley M, Anders RF, Batchelor AH. 2007. Structure of the malaria antigen AMA1 in complex with a growth-inhibitory antibody. PLoS Pathog 3:1308–1319. doi: 10.1371/journal.ppat.0030138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Collins CR, Withers-Martinez C, Hackett F, Blackman MJ. 2009. An inhibitory antibody blocks interactions between components of the malarial invasion machinery. PLoS Pathog 5:e1000273. doi: 10.1371/journal.ppat.1000273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Collins CR, Withers-Martinez C, Bentley GA, Batchelor AH, Thomas AW, Blackman MJ. 2007. Fine mapping of an epitope recognized by an invasion-inhibitory monoclonal antibody on the malaria vaccine candidate apical membrane antigen 1. J Biol Chem 282:7431–7441. doi: 10.1074/jbc.M610562200. [DOI] [PubMed] [Google Scholar]
- 49.Coley AM, Parisi K, Masciantonio R, Hoeck J, Casey JL, Murphy VJ, Harris KS, Batchelor AH, Anders RF, Foley M. 2006. The most polymorphic residue on Plasmodium falciparum apical membrane antigen 1 determines binding of an invasion-inhibitory antibody. Infect Immun 74:2628–2636. doi: 10.1128/IAI.74.5.2628-2636.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maskus DJ, Królik M, Bethke S, Spiegel H, Kapelski S, Seidel M, Addai-Mensah O, Reimann A, Klockenbring T, Barth S, Fischer R, Fendel R. 2016. Characterization of a novel inhibitory human monoclonal antibody directed against Plasmodium falciparum apical membrane antigen 1. Sci Rep 6:39462. doi: 10.1038/srep39462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mugyenyi CK, Elliott SR, McCallum FJ, Anders RF, Marsh K, Beeson JG. 2013. Antibodies to polymorphic invasion-inhibitory and non-inhibitory epitopes of Plasmodium falciparum apical membrane antigen 1 in human malaria. PLoS One 8:e68304. doi: 10.1371/journal.pone.0068304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Srinivasan P, Ekanem E, Diouf A, Tonkin ML, Miura K, Boulanger MJ, Long CA, Narum DL, Miller LH. 2014. Immunization with a functional protein complex required for erythrocyte invasion protects against lethal malaria. Proc Natl Acad Sci U S A 111:10311–10316. doi: 10.1073/pnas.1409928111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wright KE, Hjerrild KA, Bartlett J, Douglas AD, Jin J, Brown RE, Illingworth JJ, Ashfield R, Clemmensen SB, de Jongh WA, Draper SJ, Higgins MK. 2014. Structure of malaria invasion protein RH5 with erythrocyte basigin and blocking antibodies. Nature 515:427–430. doi: 10.1038/nature13715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chiu CY, Healer J, Thompson JK, Chen L, Kaul A, Savergave L, Raghuwanshi A, Li Wai Suen CS, Siba PM, Schofield L, Mueller I, Cowman AF, Hansen DS. 2014. Association of antibodies to Plasmodium falciparum reticulocyte binding protein homolog 5 with protection from clinical malaria. Front Microbiol 5:314. doi: 10.3389/fmicb.2014.00314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Douglas AD, Williams AR, Knuepfer E, Illingworth JJ, Furze JM, Crosnier C, Choudhary P, Bustamante LY, Zakutansky SE, Awuah DK, Alanine DG, Theron M, Worth A, Shimkets R, Rayner JC, Holder AA, Wright GJ, Draper SJ. 2014. Neutralization of Plasmodium falciparum merozoites by antibodies against PfRH5. J Immunol 192:245–258. doi: 10.4049/jimmunol.1302045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ord RL, Caldeira JC, Rodriguez M, Noe A, Chackerian B, Peabody DS, Gutierrez G, Lobo CA. 2014. A malaria vaccine candidate based on an epitope of the Plasmodium falciparum RH5 protein. Malar J 13:326. doi: 10.1186/1475-2875-13-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tsuboi T, Kappe SH, Al-Yaman F, Prickett MD, Alpers M, Adams JH. 1994. Natural variation within the principal adhesion domain of the Plasmodium vivax duffy binding protein. Infect Immun 62:5581–5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cole-Tobian JL, Michon P, Biasor M, Richards JS, Beeson JG, Mueller I, King CL. 2009. Strain-specific duffy binding protein antibodies correlate with protection against infection with homologous compared to heterologous plasmodium vivax strains in Papua New Guinean children. Infect Immun 77:4009–4017. doi: 10.1128/IAI.00158-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Drew ME, Banerjee R, Uffman EW, Gilbertson S, Rosenthal PJ, Goldberg DE. 2008. Plasmodium food vacuole plasmepsins are activated by falcipains. J Biol Chem 283:12870–12876. doi: 10.1074/jbc.M708949200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fernandez E, Kose N, Edeling MA, Adhikari J, Sapparapu G, Lazarte SM, Nelson CA, Govero J, Gross ML, Fremont DH, Crowe JE Jr, Diamond MS. 2018. Mouse and human monoclonal antibodies protect against infection by multiple genotypes of Japanese encephalitis virus. mBio 9:e00008-18. doi: 10.1128/mBio.00008-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pascal BD, Willis S, Lauer JL, Landgraf RR, West GM, Marciano D, Novick S, Goswami D, Chalmers MJ, Griffin PR. 2012. HDX workbench: software for the analysis of H/D exchange MS data. J Am Soc Mass Spectrom 23:1512–1521. doi: 10.1007/s13361-012-0419-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu H, Freitas MA. 2009. MassMatrix: a database search program for rapid characterization of proteins and peptides from tandem mass spectrometry data. Proteomics 9:1548–1555. doi: 10.1002/pmic.200700322. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All epitopes have been deposited in the IEDB under reference ID 1034379.