Abstract
An increasing number of publications had reported the association between single‐nucleotide polymorphisms (SNPs) and esophageal cancer (EC) risk in the past decades. Results from these publications were controversial. We used PubMed, Medline, and Web of Science to identify meta‐analysis articles published before 30 July 2018, that summarize a comprehensive investigation for cumulative evidence of genetic polymorphisms of EC and its subtype risk. Two methods, Venice criteria and false‐positive report probability (FPRP) tests, were used to assess cumulative evidence of significant associations. At last, 107 meta‐analyses were considered to be in conformity with the inclusion criteria, yielding 51 variants associated with EC or esophageal squamous cell carcinoma (ESCC). Thirty‐eight variants were considered to be nominally significant associated with risk of EC or ESCC, whereas the rest showed non‐association. In additional, five variants on five genes were rated as strong cumulative epidemiological evidence for a nominally significant association with EC and ESCC risk, including CYP1A1 rs1048943, EGF rs444903, HOTAIR rs920778, MMP2 rs243865, and PLCE1 rs2274223, 10 variants were rated as moderate, and 18 variants were rated as weak. Additionally, 17 SNPs were verified noteworthy in six genomewide association studies (GWAS) using FPRP methods. Collectively, this review offered a comprehensively referenced information with cumulative evidence of associations between genetic polymorphisms and EC and ESCC risk.
Keywords: esophageal cancer, genetic polymorphisms, genome-wide association study, meta‐analyses, susceptibility
1. INTRODUCTION
Esophageal cancer (EC) is a digestive tract carcinoma and remains the sixth leading cause of a carcinoma‐associated deaths worldwide.1, 2 In 2015 in the USA, there had been an approximate 16 980 new patients had EC and 15 590 patients died of it,3 and its overall five‐year survival rate was only about 20%.4, 5, 6, 7 Esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EADC) were two different common pathological types of EC, and they were caused by different risk factors. For ESCC, the risk factors were smoking, alcoholic beverages, socioeconomic status (SES), polycyclic aromatic hydrocarbons (PAHs), betel quid, diet quality, low fruit and vegetable intake, micronutrients, pickled vegetables, hot food and beverages and so on. While gastroesophageal reflux disease, Barrett's esophagus and smoking tobacco have been verified as risk factors for EADC. Although these environmental factors were considered to be risk factors for EC, epidemiological and etiological studies have shown that the role of genetic variants was also needed to be considered.8
Over the past few decades, a large number of candidate genes association studies9, 10, 11, 12 were performed to explore the relationship between gene polymorphisms and EC risk. However, due to the small sample size and inadequate statistical power, the results were instability. Meta‐analysis could present more credible results and stronger statistical power through integrating individual study findings,13, 14 and more than 100 meta‐analyses had been performed in recent years. Most of the results for same variant, however, were inconsistent. For EGF rs4444903, Xu et al15 performed a meta‐analysis and found that the variant rs4444903 could decrease the risk of EC (OR = 0.73, 95% CI = 0.61‐0.86), whereas Li et al16 found the variant rs4444903 could increase the risk of EC (OR = 1.17, 95% CI = 1.09‐1.25). For TP53 rs1042522, Zhao et al performed a meta‐analysis and found that the variant rs1042522 could increase the risk of EC (OR = 1.20, 95% CI = 1.06‐1.36), whereas Jiang et al found that the variant rs1042522 had an opposite association with EC risk (OR = 0.73, 95% CI = 0.57‐0.94).17 GWAS could screen the sequence variation in the human genome and identify SNPs related to human diseases,18 and they extended our understanding of associations between genetic variations and cancer risk.19 To date, GWAS that have two stages (discovery and replication) have identified to be a commonly powerful and successful tool in the identification of genetic variants associated with susceptibility to complex human diseases or phenotypes.20, 21
As early as 2008, Dong et al22 reported that three variants were significantly related to EC by assessing the FPRP of meta‐analysis. In recent years, more than 100 meta‐analyses had been performed. When GWAS method is applied, gene mutations or susceptibility loci were identified to have relationship with many diseases.23 In 2010, of the 18 SNPs, Wang LD et al24 summarized two identified susceptibility loci (10q23 and 20p13) associated with ESCC risk. Meanwhile, Abnet et al25 found variants on PLCE1 gene associated with ESCC risk and Wang et al24 found that the gene, C20orf54, had significant association with ESCC risk in Chinese population. Later, Jin et al26 found consistent associations two loci (6p21.1 and 7p15.3) with EC risk in both GWAS and replication stages. In 2013, Levine et al27 added three new susceptibility loci (3p13, 9q22, and 19p13) for EADC. In following years, Chang et al28 found another two variants on 13q22.1.
Although more than 100 meta‐analyses and several GWAS with association between genetic variants and EC susceptibility had been performed, these results of different studies for same variant were inconsistent, indicating the possibility of false‐positive associations. Ioannidis et al indicated that mechanisms for summarizing and assessing genetic epidemiological evidence require periodic updates of all appropriate association studies based on widely accepted assessment criteria.29 However, the current literature still lacks an updated comprehensive assessment report covering all possible variants of multiple genes with EC risk.
Therefore, we attempt to collect cumulative evidence of associations between genetic variants and EC risk from published meta‐analyses and GWAS, and evaluate these associations, which may offer referenced information for further investigation of genetic risk factors for EC and its subtype.
2. METHOD
2.1. Literature search strategy and criteria for inclusion
The following items were used in search process: (“esophageal”) and (“cancer” or “adenocarcinoma” or “carcinoma” or “tumor” or “squamous cell carcinoma”) and (“meta‐analysis” or “Meta‐analysis” or “systematic review” or “literature review”) and (“genetic association” or “Genetic” or “SNP” or “polymorphism” or “single nucleotide polymorphism” or “genotype” or “variant” or “variation” or “mutation” or “susceptibility”). Additionally, we also checked all the relevant references to find other potential meta‐analyses that could offer relevant data.
Meta‐analysis articles met to the following criteria: (a) The publications were in English; (b) cancer type was EC or including subtypes; (c) the patients with EC were diagnosed by pathological or histological examination; (d) sample size was not fewer than 1000; (e) they were studied of EC incidence/susceptibility (rather than mortality or survival rate). The following criteria should be met for screening the SNPs in GWAS on PubMed: (a) The publications were in English; (b) cancer type was EC, which includes all the subtype of EC; (c) the patients with EC were diagnosed by pathological or histological examination; (d) the studies included two phases (discovery and replication); (e) OR and 95% CI were provided and the less than cutoff of 1 × 10‐8 of P value was considered statistically significant; (f) they were studied of EC incidence/susceptibility (rather than mortality or survival rate).
2.2. Data extraction
Data were extracted by J.T and checked by two authors (C.L and GL). Information extracted from each eligible publication in GWAS included PMID of article, first author, publishing year, gene name, genetic variant, ethnicity of participants, the amount of subjects (cases and controls), minor allele frequency (MAF), OR, 95% CI, P value. In meta‐analysis, the following data were collected: first author, publishing year, gene name, genetic variant, OR and 95% CI,the number of studies, the number of subjects (cases and controls), ethnicity, I‐square, test for heterogeneity (Q test) between studies,30 and the test for publication bias (Egger's test).31 I‐square refers to the percentage of variation across studies due to heterogeneity. We referenced the Cochran's Q test32 to evaluate heterogeneity between studies. Generally, the cutoff P value used for between‐study test of heterogeneity (Q‐test) was 0.10. P value >0.10 represents little heterogeneity while P value <0.10 indicates the presence of heterogeneity. We attempted to extract information of EC, including ESCC and EADC; however, the results showed that almost all of the articles were studied for EC and ESCC, not for EADC. Therefore, we were unable to conduct assessment of EADC, and then, we just evaluated cumulative epidemiological evidence for EC and ESCC. In addition, the eligible studies reported two major ethnicities, Asian and Caucasian. Majority of the meta‐analyses included a combination of two or more ethnicities, which were defined as “diverse populations.” Therefore, we extracted the information of diverse population, when applicable, Asian and Caucasian population were also extracted. Multiple studies concerning the same SNP reported conflicting results due to varied sample sizes and the selection of different association models. Therefore, given study quality and result credibility, we selected the most recently published study with the greatest number of and most integrated participants and the standardized report of the genetic association study based on guidelines of the Human Genome Epidemiology Network for systematic review of genetic association studies33 and Preferred Reporting Items for Systematic Reviews and Meta‐Analyses (PRISMA).34 In addition, articles usually offered different genetic models; therefore, the additive model (see Table S1) was considered as the priority model for data extraction and evaluation in order to reduce selection bias. Specifically, the rest models were also used when additive model was not usable. For variant name, the most recent gene names were used to identify the different variants. An association was considered to be statistically significant if the 95% CI excluded 1.0 or if the reported P value was <0.05.
2.3. Assessment of cumulative evidence
Venice criteria were applied to assess the epidemiological credibility of significant associations identified by meta‐analysis.29, 35 Credibility was rated as strong, moderate, or weak (grade A, B, or C) according to three elements: amount of evidence, replication of association, and protection from bias. The first element was evaluated by the total of alleles or genotypes among cases and controls which was divided into three groups: >1000, 100‐1000, and fewer than 100 (representing A, B and C, respectively). Although certain test allele or genotype amounts were not provided, we could obtain the MAF from database of SNP on NCBI and further calculate the amounts. Association replication was calculated using heterogeneity statistics assigned one of three grades, as follows: grade A (I 2 < 25%), grade B (25% < I 2 < 50%), or grade C (I 2 > 50%). Protection from bias was mainly determined by sensitivity analysis and a series of bias tests including publication bias, small‐study bias, as well as an excess of significant findings (see Table S2). Briefly, protection from bias was graded as A if there was no observable bias, and bias was unlikely to explain the presence of the association, B if bias could be present, or C if bias was evident or was likely to explain the presence of the association. Assessment of protection from bias also considered the magnitude of association; a score of C was assigned to an association with a summary OR < 1.15, unless the association had been replicated prospectively by several studies with no evidence of publication bias (ie, GWAS or GWAS meta‐analysis from collaborative studies). Nevertheless, the associations did not participant in grading if the information was insufficient for assessment. For cumulative epidemiological evidence, all three elements were A would be considered as strong evidence, a C for any grade were weak, the rest of combinations were moderate.
We performed a false‐positive report probability (FPRP) assay with a prior probability of 0.001 and an FPRP cutoff value of 0.2 to uncover potential false‐positive results among significant associations and evaluate whether these associations should be omitted, and we used the statistical power to detect an odds ratio of 1.5 for alleles with an elevated risk in FPRP calculations, as suggested by Wacholder et al.36 Statistical power and FPRP values were calculated by the Excel spreadsheet which was offered on Wacholder's website. If the calculated FPRP value was below the prespecified noteworthiness value of 0.2, we would consider the association noteworthy, indicating the association might be true. Value for FPRP was assigned to three groups: <0.05, 0.05‐0.2, >0.2 (representing strong, moderate and weak, respectively). Cumulative evidence was upgraded from moderate to strong or from weak to moderate base on FPRP <0.05. Conversely, cumulative evidence was downgraded from strong to moderate or from moderate to weak base on FPRP >0.2.
3. RESULTS
3.1. Description of search results and characteristics of the studies
As presented in Figure 1, our search yielded a total of 918 publications. Of these, 179 publications were excluded due to overlaps, 157 irrelevant articles were excluded for reading the title or abstract, 21 articles were excluded due to not meta‐analysis, genetic polymorphism, or esophageal cancer, 19 articles were excluded due to not latest meta‐analysis. In addition, 15 additional articles identified through relevant reference publications. At last, 107 meta‐analyses were eligible for criteria in our review. On the basis of the data that extracted from the method mentioned above, yielding 51 SNPs associated with EC or ESCC susceptibility (45 SNPs in 41 genes, six SNPs in miRNA). We used the PubMed to identify the SNPs in GWAS. As a result, 17 SNPs were identified in six GWAS studies (15 SNPs are in 10 genes and two SNPs are near one gene or between genes).
Figure 1.
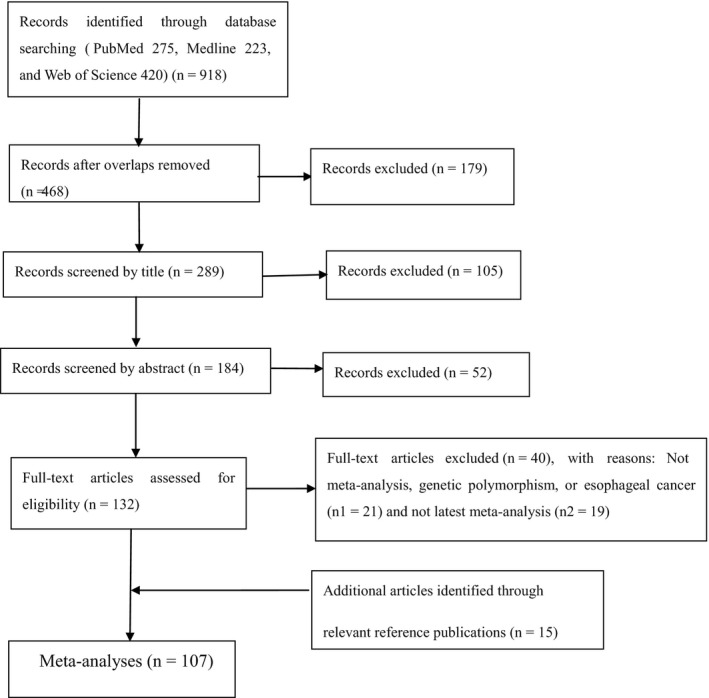
Provides selection of studies
3.2. Significant association in meta‐analyses and GWAS
For 51 SNPs identified by meta‐analysis, 38 SNPs had statistically significant association with EC or ESCC susceptibility.37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74 Meta‐analysis results showed that there were 27 SNPs associated with increased EC or ESCC risk, whereas 11 SNPs decreased the risk of EC or ESCC risk. In addition, 17 SNPs were evaluated using the additive model. Ten SNPs were evaluated using the dominant model and another 11 SNPs were assessed using recessive or homozygous model because the additive model was not available.
As presented in Table 1, cumulative epidemiological evidence was graded for 38 significant associations among the main meta‐analyses. Venice criteria were firstly used to assess these associations. Strong, moderate, and weak evidence were assigned to four, seven, and 17 SNPs for EC, and were assigned to one, three, and two SNPs for ESCC, respectively. Next, cumulative evidence were upgraded from moderate to strong for CYP1A1 rs1048943, PLCE1 rs2274223, MMP2 rs243865 in EC and HOTAIR rs920778 in ESCC, from weak to moderate for ADH1B rs1229984 and COX‐2 rs20417 in EC, based on FPRP <0.05. Cumulative evidence were downgraded from strong to moderate for IL‐18 −607C>A, MMP1 rs1799750, SLC52A3 rs13042395 and MDM2 rs2279744 in EC, and TNF‐α rs1800629, C20orf54 rs13042395, microRNA124 rs531564 in ESCC, from moderate to weak for GSTT1 null/present, XRCC1 rs1799782, Hsa‐mir rs3746444, hOGG1 rs1052133, STK15 rs2273535 in EC, and microRNA‐34b/c rs4938723, NAT2 rapid/slow in ESCC, based on FPRP >0.2. Finally, five SNPs on five genes were rated as strong for cumulative epidemiological evidence of association by combining Venice criteria and FPRP results, including CYP1A1 rs1048943, EGF rs444903, MMP2 rs243865, PLCE1 rs2274223 for EC and HOTAIR rs920778 for ESCC. Seven SNPs with EC and three with EC were rated as moderate. Sixteen with EC and two with ESCC were rated as weak. Great discrepancy between the calculated amount and the true amount makes the grade arduous to determine. Therefore, calculated amounts of less than 3000 were not included for assessment in MAFs obtained from the dbSNP.
Table 1.
Statistically significant variants from meta‐analyses, false‐positive report probabilities (FPRP), and cumulative epidemiological evidence
| PMID | Gene (variant) | Cancer type | Year | Comparison | Ethnicity | OR (95% CI) | Publication bias/heterogeneity | I 2 (%) | No. of studies | Cases/control | Number of test allele or genotype (calculated value according to MAF) | Maf | Venice Criteriaa | Venice Grade | Power Or of 1.5 | FPRP values at prior probability of 0.001 at power OR of 1.5 | Cumulative epidemiological evidenceb | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CYP1A1 | ||||||||||||||||||
| 25048966 | CYP1A1 exon7 (rs1048943) | EC | 2014 | Dominant | Diverse | 1.49 (1.33‐1.66) | P = 0.815/0.036 | 38.30% | 18 | 6165 (2552/3613) | 2863 | 0.2281 | ABA | Moderate | 0.548 | 0.000 | Strong | 37 |
| EC | 2014 | Dominant | Asian | 1.48 (1.33‐1.66) | P = 0.925/0.026 | 44.20% | 15 | 5431 (2381/3050) | 2703 | 0.2598 | ABA | Moderate | 0.591 | 0.000 | Strong | 37 | ||
| EC | 2014 | Dominant | Caucasian | 1.50 (0.87‐2.59) | P = 0.537/0.254 | 25.20% | 3 | 734 (171/563) | Na | Na | Na | Na | Na | Na | Na | 37 | ||
| CYP1A1 | ||||||||||||||||||
| 25886559 | CYP1A1 (rs4646903) | EC | 2015 | Additive | Asian | 1.25 (1.04‐1.51) | P = 0.550/0.000 | 67.10% | 12 | 3161 (1359/1802) | 2645 | 0.4001 | ACA | Weak | 0.971 | 0.955 | Weak | 38 |
| ESCC | 2015 | Additive | Asian | 1.17 (1.04‐1.32) | 0.544c/0.055c | 47.4%c | 9 | 2384 (1027/1357) | 1897 | 0.3868 | ABA | Moderate | 1.000 | 0.915 | Weak | 38 | ||
| ERCC2 751 | ||||||||||||||||||
| 25748732 | ERCC2 (rs13181) | EC | 2015 | Dominant | Diverse | 1.30 (1.07‐1.57) | P = No/<0.05 | 80.00% | 21 | 14 832 (6581/8251) | 5570 | 0.2424 | ACA | Weak | 0.931 | 0.873 | Weak | 39 |
| EC | 2015 | Dominant | Asian | 1.27 (1.04‐1.56) | P = No/<0.05 | 66.7%c | 12 | 7265 (3338/3927) | 1778 | 0.1179 | ACA | Weak | 0.944 | 0.960 | Weak | 39 | ||
| ESCC | 2015 | Dominant | diveres | 1.27 (1.04‐1.55) | P = 0.229/0.000c | 66.7%c | 13 | 8111 (3351/4760) | 2385 | 0.1673 | ACA | Weak | 0.949 | 0.952 | Weak | 39 | ||
| EADC | 2015 | Dominant | diveres | 1.16 (0.87‐1.55) | Na | Na | 7 | 5122 (1726/3396) | Na | Na | Na | Na | Na | Na | Na | 39 | ||
| 21667112 | ERCC2 (rs1052559) | EC | 2012 | Homozygous | Asian | 2.45 (1.10‐5.44) | P = 0.83/0.355 | 7.7%c | 4 | 2352 (1093/1259) | 28 | 0.0436 | CAA | Weak | 0.114 | 0.996 | Weak | 40 |
| EADC | 2012 | Homozygous | diveres | 1.26 (1.02‐1.56) | P = 0.277/0.054 | 54%c | 6 | 4341 (1281/3060) | 593 | 0.3565 | BCA | Weak | 0.945 | 0.973 | Weak | 40 | ||
| ESCC | 2012 | Homozygous | diveres | 1.32 (0.85‐2.06) | Na | Na | 6 | 3185 (1294/1281) | Na | Na | Na | Na | Na | Na | Na | 40 | ||
| ERCC2 | ||||||||||||||||||
| 25209371 | ERCC2 (rs238406) | ESCC | 2014 | Dominant | Asian | 1.24 (1.04‐1.49) | P = No/0.017 | Na | 1 | 2257 (1126/1131) | 1534 | 0.424 | AXA | Na | 0.979 | 0.957 | Na | 41 |
| Fas | ||||||||||||||||||
| 24598538 | Fas (rs2234767) | EC | 2014 | Recessive | Diverse | 1.58 (1.16‐2.13) | P=>0.05/0.089 | 58.70% | 3 | 2660 (1126/1534) | Na (0‐490) | 0.1841d | XCA | Na | 0.367 | 0.880 | Na | 42 |
| GSTP1 | ||||||||||||||||||
| 25280543 | GSTP1 (rs1695) | EC | 2015 | Additive | Caucasian | 1.146 (1.031‐1.275) | P = 0.901/0.175 | 30.40% | 9 | 3289 (1198/2019) | 2333 | 0.3479 | ABCe | Weak | 1.000 | 0.925 | Weak | 43 |
| ESCC | 2015 | Additive | Caucasian | 1.041 (0.956‐1.134) | Na | Na | 15 | Na | Na | Na | Na | Na | Na | Na | Na | 43 | ||
| EADC | 2015 | Additive | Caucasian | 1.096 (0.971‐1.237) | Na | Na | 10 | Na | Na | Na | Na | Na | Na | Na | Na | 43 | ||
| HOTAIR | ||||||||||||||||||
| 27791260 | HOTAIR (rs920778) | ESCC | 2017 | Recessive | Asian | 2.525 (1.921‐3.320) | P=>0.396/0.368 | 0.1%c | 3 | 4221 (2071/2150) | 259 | 0.2105 | BAA | Moderate | 0.000 | 0.000 | Strong | 44 |
| IL‐18 | ||||||||||||||||||
| 26214646 | IL‐18 (−607C>A) | EC | 2015 | Dominant | Diverse | 1.29 (1.00‐1.66) | P = 0.088/0.70 | 0.00%c | 2 | 1749 (1305/444) | 1175 | 0.4561 | AAA | Strong | 0.879 | 0.982 | Moderate | 45 |
| MMP1 | ||||||||||||||||||
| 23644699 | MMP1 (rs1799750) | EC | 2013 | Dominant | Diverse | 1.47 (1.18‐1.82) | P = 0.127/0.78 | 0.00% | 3 | 1936 (856/1080) | 1457 | 0.5181 | AAA | Strong | 0.574 | 0.415 | Moderate | 46 |
| MnSOD | ||||||||||||||||||
| 23679296 | MnSOD (rs4880) | EC | 2013 | Dominant | Na | 1.74 (1.36‐2.22) | P = 0.61/0.64 | 0.00% | 4 | 1529 (620/909) | Na (628‐1256) | 0.4107d | XAA | Na | 0.116 | 0.067 | Na | 47 |
| MTHFR C677T | ||||||||||||||||||
| 24606463 | MTHFR (rs1801133) | EC | 2014 | Additive | Asian | 1.19 (1.06‐1.34) | P = 0.667/<0.001 | Na | 14 | 6633 (2808/3825) | 6491 | 0.4567 | AXA | Na | 1.000 | 0.803 | Na | 48 |
| NAT2 | ||||||||||||||||||
| 24595082 | NAT2 (rapid/slow) | ESCC | 2013 | slow vs rapid | Asian | 1.35 (1.03‐1.77) | P = 0.805/0.093 | 49.70% | 5 | 1534 (441/1093) | 402 | 0.118 | BBA | Moderate | 0.777 | 0.975 | Weak | 49 |
| hOGG1 | ||||||||||||||||||
| 23909557 | hOGG1 (rs1052133) | EC | 2013 | Recessive | diveres | 1.40 (1.12‐1.74) | P = 0.140/0.176 | 27.40% | 12 | 5984 (2363/3621) | 586 | 0.2813 | BBA | Moderate | 0.733 | 0.767 | Weak | 50 |
| EC | 2013 | Recessive | Asian | 1.51 (1.15‐1.96) | P = 0.140/0.22 | 29.00% | 6 | 2461 (1123/1338) | 408 | 0.3961 | BBA | Moderate | 0.48 | 0.803 | Weak | 50 | ||
| ESCC | 2013 | Recessive | diveres | 1.86 (1.36‐2.53) | P = 0.140/0.73 | 0.00% | 3 | 1271 (589/682) | 200 | 0.3512 | BAA | Moderate | 0.085 | 0.474 | Weak | 50 | ||
| EADC | 2013 | Recessive | Caucasian | 1.08 (0.69‐1.67) | Na | Na | 3 | 2611 (1189/1422) | Na | Na | Na | Na | Na | Na | Na | 50 | ||
| TNF‐α | ||||||||||||||||||
| 27821804 | TNF‐α (rs1800629) | ESCC | 2016 | Dominant | Diverse | 1.19 (1.00‐1.41) | P = No/0.405 | 3.30% | 8 | 4469 (1144/3325) | 1297 | 0.1582 | AAA | Strong | 0.996 | 0.978 | Moderate | 51 |
| PLCE1 | ||||||||||||||||||
| 25422186 | PLCE1 (rs2274223) | EC | 2014 | Dominant | Diverse | 1.30 (1.16‐1.46) | P = No/0.0003 | 68.00% | 12 | 22 935 (9912/13 023) | Na (6846‐13 692) | 0.2985d | ABA | Moderate | 0.992 | 0.009 | Strong | 52 |
| EC | 2014 | Dominant | Asian | 1.39 (1.24‐1.57) | P = No/0.009 | 61.00% | 10 | 19 263 (8737/10 526) | Na (5750‐11 500) | 0.2985d | ABA | Moderate | 0.89 | 0.000 | Strong | 52 | ||
| STK15 T>A | ||||||||||||||||||
| 25452806 | STK15 (rs2273535) | EC | 2015 | Recessive | Asian | 1.19 (1.03‐1.38) | P = 0.835/0.24 | 26.00% | 6 | 3725 (1767/1958) | 1260 | 0.5523 | ABA | Moderate | 0.999 | 0.955 | Weak | 53 |
| C20orf54 | ||||||||||||||||||
| 26154995 | C20orf54 (rs13042395) | ESCC | 2015 | Additive | Diverse | 0.95 (0.90‐0.99) | P = 0.604/0.33 | 13.00% | 7 | 88 324 (29 922/58 402) | Na (19 855) | 0.1124d | AAA | Strong | 1.000 | 0.937 | Moderate | 54 |
| CASP8 −652 6N | ||||||||||||||||||
| 28915630 | CASP8 −652 6 N (rs3834129) | EC | 2017 | Additive | Asian | 0.81 (0.72‐0.92) | P = 0.002/0.712 | 0.00%c | 3 | 2608 (1412/1196) | 1338 | 0.3020 | AAC | Weak | 0.999 | 0.542 | Weak | 55 |
| CYP2E1 | ||||||||||||||||||
| 23226753 | CYP2E1 (RSqI/PstI) | EC | 2012 | Additive | Asian | 0.64 (0.50‐0.81) | P = Na/<0.01 | 80.00% | 17 | 4226 (1663/4266) | 2279 | 0.3016 | ACX | Na | 0.367 | 0.358 | Na | 56 |
| Hsa‐mir | ||||||||||||||||||
| 25433484 | Hsa‐mir‐499 (rs3746444) | EC | 2014 | Additive | Asian | 0.80 (0.66‐0.98) | P = >0.05/0.75 | 0.00%c | 2 | 1358 (669/689) | 498 | 0.2017 | BAA | Moderate | 0.961 | 0.97 | Weak | 57 |
| MicroRNA 34 | ||||||||||||||||||
| 28415817 | MicroRNA (rs4938723) | ESCC | 2017 | Homozygous | Asian | 0.787 (0.638‐0.972) | P = 0.622/0.345 | 9.50% | 4 | 4650 (2226/2424) | 433 | 0.3245 | BAA | Moderate | 0.938 | 0.965 | Weak | 58 |
| MicroRNA 124 | ||||||||||||||||||
| 26171202 | MicroRNA‐124 (rs531564) | ESCC | 2015 | Additive | Asian | 0.87 (0.77‐0.98) | P = No/0.69 | 0.00% | 3 | 4077 (1964/2113) | 1257 | 0.1621 | AAA | Strong | 1.000 | 0.956 | Moderate | 59 |
| MMP2 | ||||||||||||||||||
| 23644699 | MMP2 (rs243865) | EC | 2013 | Dominant | Asian | 0.67 (0.55‐0.80) | P = 0.072/0.59 | 0.00% | 3 | 2781 (1050/1731) | 668 | 0.1416 | BAA | Moderate | 0.522 | 0.018 | Strong | 60 |
| SLC52A3 | ||||||||||||||||||
| 27600099 | SLC52A3 (rs13042395) | EC | 2016 | Recessive | Diverse | 0.84 (0.76‐0.93) | P = 0.357/0.738 | 0.00% | 11 | 26 956 (10 027/16 929) | 2009 | 0.3592 | AAA | Strong | 1.000 | 0.44 | Moderate | 61 |
| ADH1B | ||||||||||||||||||
| 27450204 | ADH1B (rs1229984) | EC | 2016 | Additive | Diverse | 0.67 (0.59‐0.76) | P = No/<0.00001 | 87.00% | 20 | 23 148 (9158/13 990) | 30 676 | 0.6983 | ACA | Weak | 0.531 | 0.000 | Moderate | 62 |
| EC | 2016 | Additive | Asian | 0.66 (0.57‐0.75) | P = No/<0.00001 | 88.00% | 18 | 22 038 (8687/13 351) | 30 595 | 0.7299 | ACA | Weak | 0.439 | 0.000 | Moderate | 62 | ||
| ALDH2 | ||||||||||||||||||
| 25848305 | ALDH2 (rs671) | EC | 2015 | Homozygous | Diverse | 0.69 (0.48‐0.98) | P = 0.682/<0.001 | 74.80% | 31 | 24 707 (8510/16 197) | 1439 | 0.2446 | ACA | Weak | 0.576 | 0.985 | Weak | 63 |
| EC | 2015 | Homozygous | Asian | 0.68 (0.60‐0.79)c | P = 0.682/0.000c | 74.3%c | 30 | 23 481 (8130/15 351) | 1429 | 0.2573 | ACA | Weak | 0.602 | 0.001 | Moderate | 63 | ||
| CCND1 | ||||||||||||||||||
| 24944806 | CCND1 (rs603965) | EC | 2014 | Recessive | Diverse | 1.33 (1.03‐1.73) | P < 0.001/0.000 | 72.10% | 11 | 5343 (2111/3232) | 1471 | 0.5203 | ACC | Weak | 0.815 | 0.976 | Weak | 64 |
| EC | 2014 | Recessive | Asian | 1.22 (0.93‐1.60) | P < 0.001/0.0228 | 30.70% | 4 | Na | Na | Na | Na | Na | Na | Na | Na | 64 | ||
| EC | 2014 | Recessive | Caucasian | 1.44 (0.97‐2.14) | P < 0.001/0.000 | 80.70% | 7 | Na | Na | Na | Na | Na | Na | Na | Na | 64 | ||
| ESCC | 2014 | Recessive | diveres | 1.28 (0.93‐1.75) | Na | Na | Na | Na | Na | Na | Na | Na | Na | Na | Na | 64 | ||
| EADC | 2014 | Recessive | Caucasian | 1.59 (0.69‐3.70) | Na | Na | Na | Na | Na | Na | Na | Na | Na | Na | Na | 64 | ||
| COX‐2 | ||||||||||||||||||
| 21304218 | COX‐2 (rs20417) | EC | 2011 | Additive | Diverse | 1.45 (1.23‐1.71) | P = 0.922/0.003 | 76.00% | 4 | 3779 (1562/2217 ) | 638 | 0.0753 | BCA | Weak | 0.656 | 0.015 | Moderate | 65 |
| EC | 2011 | Additive | Asian | 1.71 (1.37‐2.17) | P = 0.922/Na | Na | 2 | 2686 (1200/1486 ) | 307 | 0.0451 | BXA | Na | 0.141 | 0.067 | Na | 65 | ||
| EGF | ||||||||||||||||||
| 23403233 | EGF (rs4444903) | EC | 2013 | Additive | Diverse | 1.38 (1.20‐1.59) | P = 0.476/0.997 | 0.00% | 3 | 1713 (779/934) | 1659 | 0.4534 | AAA | Strong | 0.876 | 0.009 | Strong | 66 |
| ERCC2 Asp312Asn | ||||||||||||||||||
| 25356096 | ERCC2 (rs1799793) | EC | 2014 | Dominant | Diverse | 1.14 (1.03‐1.27) | P = 0.096/1.00 | 0.00% | 15 | 9940 (3928/6012) | 2981 | 0.1865 | AACe | Weak | 1.000 | 0.946 | Weak | 67 |
| EC | 2014 | Dominant | diveres | 1.12 (0.99‐1.27) | Na | Na | Na | Na | Na | Na | Na | Na | Na | Na | Na | 67 | ||
| EC | 2014 | Dominant | diveres | 1.20 (0.99‐1.47) | Na | Na | Na | Na | Na | Na | Na | Na | Na | Na | Na | 67 | ||
| GSTM1 | ||||||||||||||||||
| 26855551 | GSTM1 (null/present) | EC | 2016 | Additive | Diverse | 1.33 (1.12‐1.57) | P = 0.0873/<0.000001 | 77.00% | 37 | 11 949 (4572/7377) | 5478 | 0.2197 | ACA | Weak | 0.922 | 0.449 | Weak | 68 |
| EC | 2016 | Additive | Asian | 1.53 (1.26‐1.86) | P = 0.0873/0.000001 | 77.2%c | 27 | 8406 (3336/5070) | 3814 | 0.2099 | ACA | Weak | 0.421 | 0.045 | Moderate | 68 | ||
| GSTT1 | ||||||||||||||||||
| 23244092 | GSTT1 (null/present) | EC | 2012 | Additive | Asian | 1.26 (1.05‐1.52) | P = 0.270/0.04 | 42.70% | 15 | 3842 (1626/2216) | 1686 | 0.2128 | ABA | Moderate | 0.966 | 0.942 | Weak | 69 |
| MDM2 | ||||||||||||||||||
| 24844868 | MDM2 (rs2279744) | EC | 2015 | Additive | Diverse | 0.88 (0.81‐0.96) | P = 0.83/0.263 | 22.80% | 6 | 4915 (1899/3016) | 4762 | 0.4741 | AAA | Strong | 1.000 | 0.799 | Moderate | 70 |
| EC | 2015 | Homozygous | Asian | 0.7 (0.58‐0.84) | P = 0.94/0.539 | 0.00% | 5 | 4150 (1059/2562) | 1076 | 0.4881 | AAA | Strong | 0.7 | 0.152 | Strong | 70 | ||
| MTHFR A1298C | ||||||||||||||||||
| 23679298 | MTHFR (rs1801131) | EC | 2013 | Recessive | Diverse | 1.843 (1.414‐2.402) | P = 0.801/0.435 | 0.00% | 6 | 3693 (1302/2391) | 246 | 0.2388 | BAA | Moderate | 0.064 | 0.087 | Moderate | 71 |
| EC | 2013 | Recessive | Asian | 3.997 (1.614‐9.900) | P = 0.801/0.409 | 0.00% | 4 | 1652 (598/1054) | 20 | 0.1456 | CAA | Weak | 0.017 | 0.994 | Weak | 71 | ||
| EC | 2013 | Recessive | Caucasian | 1.693 (1.280‐2.240) | P = 0.801/0.936 | 0.00% | 2 | 2037 (704/1333) | 226 | 0.3132 | BAA | Moderate | 0.198 | 0.534 | Weak | 71 | ||
| ESCC | 2013 | Recessive | diveres | 2.57 (1.76‐3.76) | Na | Na | Na | Na | Na | Na | Na | Na | Na | Na | Na | 71 | ||
| EADC | 2013 | Recessive | diveres | 1.47 (1.07‐2.01) | Na | Na | Na | Na | Na | Na | Na | Na | Na | Na | Na | 71 | ||
| NQO1 607C>T | ||||||||||||||||||
| 28203294 | NQO1 (rs1800566) | EC | 2017 | Additive | Diverse | 1.13 (1.01‐1.26) | P = >0.05/0.000 | 74.40% | 13 | 5385 (2357/3028) | Na (3110) | 0.2889d | ACCe | Weak | 1.000 | 0.965 | Weak | 72 |
| TP53 | ||||||||||||||||||
| 23844939 | TP53 (rs1042522) | EC | 2013 | Additive | Diverse | 1.146 (1.106‐1.293) | P = 0.481/<0.001 | 70.10% | 14 | 11 492 (4184/7308) | 11 550 | 0.4698 | ACCe | Weak | 1.000 | 0.964 | Weak | 73 |
| EC | 2013 | Additive | Asian | 1.194 (1.031‐1.384) | P = 0.499/<0.001 | 70.40% | 11 | 7614 (2317/5297) | 6714 | 0.4188 | ACA | Weak | 0.999 | 0.949 | Weak | 73 | ||
| XRCC1 Arg194Trp | ||||||||||||||||||
| 23543084 | XRCC1 (rs1799782) | EC | 2013 | Recessive | Asian | 1.332 (1.093‐1.624) | P = 0.902/0.074c | 42.30% | 10 | 5290 (1946/3344) | 457 | 0.2929 | BBA | Moderate | 0.88 | 0.839 | Weak | 74 |
| ESCC | 2013 | Recessive | Asian | 1.43 (1.16‐1.75)c | P = 0.872/0.315c | 14.3%c | 9 | 5068 (1840/3228) | 429 | 0.2893 | BAA | Moderate | 0.679 | 0.432 | Weak | 74 | ||
Na, Not available; No significant publication bias/heterogeneity was not found, diversee: two or more ethnicities were reported in the meta‐analysis.
Venice Criteria grades are evidence of amount, replication of the association, and protection from bias.
Cumulative epidemiological evidence as graded by combination of results from Venice Creteria and FPRP.
The information is calculated according to the data provided in the article since the article did not present (such as I 2,OR, publication bias and heterogeneity).
The MAF is obtained from dbSNP database.
The grade of C is given because the OR value is less than 1.15 and the association is not replicated by GWAS or GWAS meta‐analysis.
The associations including 17 SNPs identified in six GWAS studies were presented in Table 2.24, 26, 27, 28, 75, 76, 77 Eleven SNPs listed in chart had associations with increased EC risk. Opposite association was found in six SNPs,26, 27, 28, 75 all of which were regarded as noteworthy based on FPRP method. The way of Venice Criteria was not applicable to GWAS which has not enough datasets even regarding the two‐step GWAS including discovery and replication phases as individual studies,29 we did not further evaluate these results. In addition, two variant (rs13042395 on C20orf54 and rs2274223 on PLCE1) were performed both in meta‐analysis and in GWAS.
Table 2.
Statistically significant variants from GWAS
| PMID | Gene | Year | Variant | ethnicity | OR (95% CI) | MAFb | P‐valuec | Cases/control | Power OR of 1.5 | FPRP values at prior probability of 0.001 at power OR of 1.5 | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 20729852 | PLCE1 | 2010 | rs3765524 | Asian | 1.35 (1.22‐1.49) | 0.258/0.207 | 1.74E‐09 | 5417 (2115/3302) | 0.982 | 0.000 | 75 |
| PLCE1 | 2010 | rs2274223 | Asian | 1.43 (1.37‐1.49) | Na | 7.46E‐56 | 5417 (2115/3302) | 0.842 | 0.000 | 75 | |
| 20729853 | C20orf54 | 2010 | rs13042395 | Asian | 0.86 (0.82‐0.90) | Na | 1.21E‐11 | 22 336 (9053/13 283) | 1.000 | 0.000 | 24 |
| HEATR | 2012 | rs4785204 | Asian | 1.24 (1.18‐1.29) | Na/0.26 | 2.24E‐20 | 20 787 (10 123/10 664) | 1.000 | 0.000 | 24 | |
| 22960999 | HEATR | 2012 | rs7206735 | Asian | 1.20 (1.15‐1.26) | Na/0.28 | 1.97E‐16 | 20 787 (10 123/10 664) | 1.000 | 0.000 | 76 |
| HAP1 | 2012 | rs6503659 | Asian | 1.27 (1.20‐1.34) | Na/0.13 | 2.73E‐10 | 20 787 (10 123/10 664) | 1.000 | 0.000 | 76 | |
| XBP1 | 2012 | rs2239815 | Asian | 1.18 (1.13‐1.23) | Na/0.37 | 3.88E‐15 | 20 787 (10 123/10 664) | 1.000 | 0.000 | 76 | |
| ST6GAL1 | 2012 | rs2239612 | Asian | 1.21 (1.15‐1.27) | Na/0.19 | 5.74E‐15 | 20 787 (10 123/10 664) | 1.000 | 0.000 | 76 | |
| SMG6 | 2012 | rs17761864 | Asian | 1.21 (1.14‐1.28) | Na/0.14 | 2.21E‐11 | 20 787 (10 123/10 664) | 1.000 | 0.000 | 76 | |
| PTPN2 | 2012 | rs2847281 | Asian | 1.20 (1.14‐1.26) | Na/0.16 | 2.49E‐11 | 20 787 (10 123/10 664) | 1.000 | 0.000 | 76 | |
| CHEK2 | 2012 | rs4800983 | Asian | 1.27 (1.21‐1.34) | Na/0.20 | 1.94E‐22 | 20 787 (10 123/10 664) | 1.000 | 0.000 | 76 | |
| CHEK2 | 2012 | rs1033667 | Asian | 1.25 (1.19‐1.30) | Na/0.25 | 4.85E‐22 | 20 787 (10 123/10 664) | 1.000 | 0.000 | 76 | |
| 24121790 | KLF5 | 2015 | rs1924966 | Asian | 0.84 (0.80‐0.89) | 0.35/0.40 | 1.37E‐10 | 12 356 (6177/6179) | 1.000 | 0.000 | 27 |
| 26315552 | KLF5 | 2015 | rs115797771 | Asian | 0.69 (0.62‐0.78) | 0.05/0.06 | 2.32E‐10 | 12 356 (6177/6179) | 0.709 | 0.000 | 28 |
| KLF5 | 2015 | rs58090485 | Asian | 0.69 (0.62‐0.77) | 0.05/0.07 | 1.23E‐10 | 12 356 (6177/6179) | 0.731 | 0.000 | 28 | |
| TMEM173a | 2014 | rs7447927 | Asian | 0.85 (0.82‐0.88) | Na | 7.72E‐20 | 30 286 (15 667/14 619) | 1.000 | 0.000 | 28 | |
| 25129146 | ATP1B2a | 2014 | rs1642764 | Asian | 0.88 (0.85‐0.91) | Na | 3.10E‐13 | 30 281 (15 474/14 807) | 1.000 | 0.000 | 77 |
Na, Not available.
The SNP is near the genes in brackets.
Minor Allele Frequency (MAF) in Case/Control.
The P values are all <1.00E‐08.
3.3. Nonsignificant association in meta‐analyses
We performed statistical power analyses to determine the stability of the associations. In our meta‐analysis results, 13 variants were not significantly associated with EC or ESCC risk.37, 42, 60, 78, 79 The variants (Arg399Gln on XRCC1) with sample sizes >10 000 were also not significantly associated with EC; further investigations for this variant may not be fruitful. Certain variants presented with relatively small sample sizes; as such, the evidence for nonsignificant (see Table S3) was considered unstable.
3.4. Inconsistency among meta‐analyses
Twenty variants presented inconsistent results in our review (see Table S4). All in all, 13 SNPs were deemed to have significant association, as follows: ADH1B rs1229984, ALDH2 rs25848305, MDM2 rs2279744, XRCC1 rs1799782, TP53 rs1042522, CCND1 rs603965, COX‐2 rs20417, EGF rs4444903, ERCC2 rs1799793, GSTM1 null/present, GSTT1 null/present, MTHFR rs1801131 and NQO1 rs1800566. Seven SNPs were deemed to have nonassociation: EPHX1 (rs2234922 and rs1051740), Fas 1800682, XPA rs1800975, microRNA146 rs2910164, microRNA196 rs11614913, XRCC1 rs25487. The results of two variants, EPHX1 (Try113His and His139Arg)81, 83, 84 and XRCC1 (Arg399Gln and Arg194Trp),74, 86, 87 should be prudently interpreted due to similar sample size.
4. DISCUSSION
This review collates a comprehensive investigation for cumulative evidence of genetic polymorphisms of EC and its subtype risk. We extracted relevant useful information from meta‐analyses and GWAS to support a comprehensive assessment for further evaluation. Using FPRP tests and Venice criteria, we scored strong, moderate, or weak cumulative evidence as credibility and strength of an association with cancer susceptibility. Five SNPs on five genes with strong evidence of association were identified, including CYP1A1 rs1048943, EGF rs444903, MMP2 rs243865, PLCE1 rs2274223 for assessing risk of EC and HOTAIR rs920778 for ESCC. Ten variants were found to have moderate evidence of association with EC or ESCC risk, and 18 variants weak evidence.
CYP1A1, located on chromosome 15‐q22, is an isozyme of cytochrome P450 and encods aryl hydrocarbon hydroxylase (AHH) which may combine with DNA to form adducts via a series of biochemical reactions. The ultimate carcinogens converted from the DNA adducts were considered to be associated with the development of EC.37, 90 SNP (rs1048943) was rated as strong evidence of association with a 1.49‐fold increased risk of EC in overall population based on over 6000 sample size. This SNP triggers an increase in the enzymatic activity and increases the activation of enzyme induction, thus may accelerate cancer development.37, 91 In our subgroup analysis, this SNP increased EC risk based on 5431 sample size in Asians, whereas nonassociation was found in Caucasians based on 734 sample size. More studies of this variant in Caucasians or other ethnic groups should be performed.
EGF, located on chromosome 4q25‐q27,92, 93 participates in the process of proliferation and differentiation of cells94 and promotes gene transcription when EGF binds to its receptor.95 Quiet a few studies have identified the G allele promoted the EGF protein expression when EGF binds to its receptor which could interfering DNA folding and further increased susceptibility of a range of human cancers.96 Our review showed that the G allele of EGF +61A>G (rs4444903) polymorphism was rated as strong evidence of association with 1.38‐fold increased risk of EC based on 1713 sample size.
MMP‐2 is a sort of zinc‐dependent endopeptidases, which can regulate various cell behaviors such as tumor initiation and growth by modulating cell proliferation, apoptosis and angiogenesis.97, 98 The SNP (rs243865), located in the promoter region of the MMP‐2, disrupts an Sp1‐type promoter site (CCACC box) and then affects MMP‐2 expression or activity, which was considered to be associated with development of cancer condition.99 Our review showed that the SNP (rs243865) was rates as strong evidence of association with EC risk (OR = 0.67, 95% CI = 0.55‐0.80) under dominant model. All studies were performed on a single ethnic group (Asian), and we recommend expanding studies on this variant to other ethnic groups.
PLCE1, located on chromosome 10q23, participate in cell growth, differentiation, gene expression and oncogenesis.100, 101 Our review showed that this SNP (rs2274223) was rated as strong evidence of association with increased risk of EC for a common A to G transition of PLCE1 that may increase expression of PLCE1 protein102, 103 in a diverse population based on a sample size of over 20 000. In our subgroup analysis, this SNP was found significant association with EC risk in Asians, whereas nonassociation was found in Caucasians. Although the mechanism for ethnic differences is still unclear, one possible reason is due to differences in genetic backgrounds and in the environmental and lifestyle context.
HOTAIR, located on chromosome 12q13.13, might transform normal cells to malignant.44 As Zhang et al105 suggested, the TT genotype was nominally significant related to ESCC susceptibility among Chinese population when compared with the rs920778 CC genotype.44 In our review, the SNP (rs920778) was rated as strong evidence of true association with ESCC risk for the T allele of HOTAIR that may induce genome‐wide retargeting of polycomb‐repressive complex 2, trimethylates histone H3 lysine‐27 (H3K27me3) and deregulation of multiple downstream genes which participated in development and progression of ESCC105 based on 4221 sample size (OR = 2.525, 95% CI = 1.921‐3.320). Again, all studies were performed on a single ethnic group (Asian), and we again recommend expanding studies on this polymorphism to other ethnic groups.
There are six variants showing moderate evidence of association in our review, all of which downgraded from strong to moderate: IL‐8 −607C>A, MMP‐1 rs1799750, TNF‐α rs1800629, MicroRNA124 rs531564, SLC52A3 rs13042395 and MDM2 rs2279744 based on a high FPRP (> 0.2). The FPRP method considers the P value, prior probability, and statistical power of the test; as we calculated FPRP at prior probability of 0.001 and used the statistical power to detect an odds ratio of 1.5 for alleles with an elevated risk in FPRP calculations, certain otherwise significant associations may have been excluded. Previous studies using different prior probabilities have classified their results as more noteworthy. Further investigations on these six variants may be necessary to analyze their associations in greater depth. Additionally, SNP (rs13042395) on C20orf54 also showing moderate evidence of association with ESCC susceptibility, which downgraded from strong to moderate. This SNP was both verified in GWAS and assessed in meta‐analysis. The association in GWAS, however, may be more statistically significant and convincing when related meta‐analysis results are inconsistent or the association evidences are not strong by combination Venice criteria and FPRP method. The Venice criteria could assess multiple reasons of potential bias such as genotype error or misclassification and ethnicity stratification, which were difficult to perform in meta‐analysis. Therefore, results might be more convincing if different weights in the Venice Criteria were reset.
Cumulative evidence of two variants (ADH1B rs1229984 and COX‐2 rs20417) with risk of EC was upgraded from weak to moderate based on FPRP <0.05. Additional assessment of two variants were necessary, particularly the variant (COX‐2 rs20417) since sample size of study for this variant are relatively small (a total of 3779 sample size).65, 106
Thirteen variants were found not to be significantly associated with EC risk, to include nine variants on seven genes and two mRNAs in a sample of approximately 4000 patients, at approximately 85% power to detect an OR of 1.15 under different model for a variant with MAF of 20%. The MAFs of the aforementioned seven variants were almost all over 0.3 despite sample sizes >4000. We can safely conclude, therefore, that these seven variants are unlikely to be associated with EC (see Table S5). Further investigations evaluating these seven variants will probably not yield meaningful results with regards to EC.
Certain limitations do apply to this report. Although we did a comprehensive literature search, some articles may have been missed. A variability in sample size presented among different studies; the smaller sizes may have impacted the credibility of the data. The evaluated data extracted from only one source which may be main cause of bias. Finally, only the susceptibility/incidence between genetic variants and EC risk were evaluated; however, other roles of genetic polymorphisms such as tumor progression, metastasis, drug resistance for EC were not be assessed due to lack of data or information. Despite the limitations of our method, we believe that our study, as an updated summary and evaluation of the existing literature reporting genetic predisposition of EC, is of value for further genetic studies.
This review evaluated the cumulative epidemiological evidence of significant associations by combining the Venice criteria and FPRP results which identified five SNPs having strong evidence of true association. Collectively, our review provides referenced information for further investigation into genetic susceptibility of EC and ESCC.
CONFLICT OF INTEREST
None declared.
Supporting information
Tian J, Liu C, Liu G, Zuo C, Chen H. Cumulative evidence for association between genetic polymorphisms and esophageal cancer susceptibility: A review with evidence from meta‐analysis and genome‐wide association studies. Cancer Med. 2019;8:1289–1305. 10.1002/cam4.1972
Jie Tian and Caiyang Liu are contributed equally.
REFERENCES
- 1. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN2012. Int J Cancer. 2015;136:E359‐386. [DOI] [PubMed] [Google Scholar]
- 2. Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87‐108. [DOI] [PubMed] [Google Scholar]
- 3. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5‐29. [DOI] [PubMed] [Google Scholar]
- 4. Merkow RP, Bilimoria KY, Keswani RN, et al. Treatment trends, risk of lymph node metastasis, and outcomes for localized esophageal cancer. J Natl Cancer Inst. 2014;106(7):pii:dju133. [DOI] [PubMed] [Google Scholar]
- 5. Lao‐Sirieix P, Fitzgerald RC. Screening for oesophageal cancer. Nat Rev Clin Oncol. 2012;9:278‐287. [DOI] [PubMed] [Google Scholar]
- 6. Kaz AM, Grady WM. Epigenetic biomarkers in esophageal cancer. Cancer Lett. 2014;28(342):193‐199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pennathur A, Gibson MK, Jobe BA, et al. Oesophageal carcinoma. Lancet. 2013;381:400‐412. [DOI] [PubMed] [Google Scholar]
- 8. Diao J, Bao J, Peng J, et al. Correlation between NAD(P)H: quinone oxidoreductase 1 C609T polymorphism and increasedrisk of esophageal cancer: evidence from a meta‐analysis. Ther Adv Med Oncol. 2017;9:13‐21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wu C, Kraft P, Zhai K, et al. Genome‐wide association analyses of esophageal squamous cell carcinoma in Chinese identify multiple susceptibility loci and gene‐environment interactions. Nat Genet. 2012;44:1090‐1097. [DOI] [PubMed] [Google Scholar]
- 10. Wu C, Hu Z, He Z, et al. Genome‐wide association study identifies three new susceptibility loci for esophageal squamous‐cell carcinoma in Chinese populations. Nat Genet. 2011;43:679‐684. [DOI] [PubMed] [Google Scholar]
- 11. Wang LD, Zhou FY, Li XM, et al. Genome‐wide association study of esophageal squamous cell carcinoma in Chinese subjects identifies susceptibility loci at PLCE1 and C20orf54. Nat Genet. 2010;42:759‐763. [DOI] [PubMed] [Google Scholar]
- 12. Abnet CC, Freedman ND, Hu N, et al. A shared susceptibility locus in PLCE1 at 10q23 for gastric adenocarcinoma and esophageal squamous cell carcinoma. Nat Genet. 2010;42:764‐767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Glass GV. Primary, secondary and meta‐analysis of research. Educ. Res. 1976;10:3‐8. [Google Scholar]
- 14. Lau J, Ioannidis JP, Schmid CH. Summing up evidence: one answer is not always enough. Lancet. 1998;351:123‐127. [DOI] [PubMed] [Google Scholar]
- 15. Xu W, Li Y, Wang X, et al. Association between EGF promoter polymorphisms and cancer risk: a meta‐analysis. Med Oncol. 2010;27:1389‐1397. [DOI] [PubMed] [Google Scholar]
- 16. Li TF, Ren KW, Liu PF. Meta‐analysis of epidermal growth factor polymorphisms and cancer risk: involving 9,779 cases and 15,932 controls. DNA Cell Biol. 2012;31:568‐574. [DOI] [PubMed] [Google Scholar]
- 17. Zhao Y, Wang F, Shan S, et al. Genetic polymorphism of p53, but not GSTP1, is association with susceptibility to esophageal cancer risk ‐ a meta‐analysis. Int J Med Sci. 2010;7:300‐308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chang J, Wei L, Miao X, et al. Two Novel Variants on 13q22.1 Are Associated with Risk of Esophageal Squamous Cell Carcinoma. Cancer Epidemiol Biomarkers Prev. 2015;24:1774‐1780. [DOI] [PubMed] [Google Scholar]
- 19. Chung CC, Chanock SJ. Current status of genome‐wide association studies in cancer. Hum Genet. 2011;130:59‐78. [DOI] [PubMed] [Google Scholar]
- 20. Hardy J, Singleton A. Genomewide association studies and human disease. N Engl J Med. 2009;360:1759‐1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bush WS, Moore JH. Chapter 11: Genome‐wide association studies. PLoS Comput Biol. 2012;8:e1002822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dong LM, Potter JD, White E, et al. Genetic susceptibility to cancer: the role of polymorphisms in candidate genes. JAMA. 2008;299:2423‐2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xue W, Zhu M, Wang Y, et al. Association between PLCE1 rs2274223 A > G polymorphism and cancer risk: proof from a meta‐analysis. Sci Rep. 2015;5:7986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang LD, Zhou FY, Li XM, et al. Genome‐wide association study of esophageal squamous cell carcinoma in Chinese subjects identifies susceptibility loci at PLCE1 and C20orf54. Nat Genet. 2010;42:759‐763. [DOI] [PubMed] [Google Scholar]
- 25. Abnet CC, Freedman ND, Hu N, et al. A shared susceptibility locus in PLCE1 at 10q23 for gastric adenocarcinoma and esophagealsquamous cell carcinoma. Nat Genet. 2010;42:764‐767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jin G, Ma H, Wu C, et al. Genetic variants at 6p21.1 and 7p15.3 are associated with risk of multiple cancers in Han Chinese. Am J Hum Genet. 2012;91:928‐934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Levine DM, Ek WE, Zhang R, et al. A genome‐wide association study identifies new susceptibility loci for esophageal adenocarcinoma and barrett’s esophagus. Nat Genet. 2013;45:1487‐1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chang J, Wei L, Miao X, et al. Two novel variants on 13q22.1 associated with risk of esophageal squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2015;24:1774‐1780. [DOI] [PubMed] [Google Scholar]
- 29. Ioannidis JP, Boffetta P, Little J, et al. Assessment of cumulative evidence on genetic associations: interim guidelines. Int J Epidemiol. 2008;37:120‐132. [DOI] [PubMed] [Google Scholar]
- 30. Cochran WG. The combination of estimates from different experiments. Bio metrics.1954;1;101‐129. [Google Scholar]
- 31. Egger M, Davey Smith G, Schneider M, et al. Bias in meta‐analysis detected by a simple, graphical test. BMJ. 1997;315:629‐634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lau J, Ioannidis JP, Schmid CH. Quantitative synthesis in systematic reviews. Ann Intern Med. 1997;127:820‐826. [DOI] [PubMed] [Google Scholar]
- 33. Sagoo GS, Little J, Higgins JP. Systematic reviews of genetic association studies. Human Genome Epidemiology Network. PLoS Med.. 2009;6:e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Moher D, Liberati A, Tetzlaff J, et al. Preferred reporting items for systematic reviews and metaanalyses: the PRISMA statement. PLoS Med.. 2009;151:264‐269. [DOI] [PubMed] [Google Scholar]
- 35. Khoury MJ, Bertram L, Boffetta P, et al. Genome‐wide association studies, field synopses, and the development of the knowledge baseon genetic variation and human diseases. Am J Epidemiol. 2009;170:269‐279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wacholder S, Chanock S, Garcia‐Closas M, et al. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J Natl Cancer Inst. 2004;96:434‐442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gong FF, Lu SS, Hu CY, et al. Cytochrome P450 1A1 (CYP1A1) polymorphism and susceptibility to esophageal cancer: anupdated meta‐analysis of 27 studies. Tumour Biol. 2014;35:10351‐10361. [DOI] [PubMed] [Google Scholar]
- 38. Zheng H, Zhao Y. Association of CYP1A1 MspI polymorphism in the esophageal cancer risk: a meta‐analysis in theChinese population. Eur J Med Res. 2015;20:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Guo XF, Wang J, Lei XF, et al. Lys751Gln polymorphisms and the risk of esophageal cancer: an updated meta‐analysis. Intern Med. 2015;54:251‐259. [DOI] [PubMed] [Google Scholar]
- 40. Ding DP, Ma WL, He XF, et al. XPD Lys751Gln polymorphism and esophageal cancer susceptibility: a meta‐analysis of case‐control studies. Mol Biol Rep. 2012;39:2533‐2540. [DOI] [PubMed] [Google Scholar]
- 41. Zhu ML, He J, Wang M, et al. Potentially functional polymorphisms in the ERCC2 gene and risk of esophageal squamous cellcarcinoma in Chinese populations. Sci Rep. 2014;4:6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xu Y, He B, Li R, et al. Association of the polymorphisms in the Fas/FasL promoter regions with cancer susceptibility: asystematic review and meta‐analysis of 52 studies. PLoS ONE. 2014;9:e90090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tan X, Chen M. Association between glutathione S‐transferases P1 Ile 105 Val polymorphism and susceptibility to esophageal cancer: evidence from 20 case ‐ control studies. Mol Biol Rep. 2015;42:399‐408. [DOI] [PubMed] [Google Scholar]
- 44. Ge Y, Jiang R, Zhang M, et al. Analyzing 37,900 samples shows significant association between HOTAIR polymorphisms and cancer susceptibility: a meta‐analysis. Int J Biol Markers. 2017;32:e231‐e242. [DOI] [PubMed] [Google Scholar]
- 45. Li X, Ren D, Li Y, et al. Increased cancer risk associated with the ‐607C/A polymorphism in interleukin‐18 gene promoter: an updated meta‐analysis including 12,502 subjects. J BUON. 2015;20:902‐917. [PubMed] [Google Scholar]
- 46. Li X, Qu L, Zhong Y, et al. Association between promoters polymorphisms of matrix metalloproteinases and risk ofdigestive cancers: a meta‐analysis. J Cancer Res Clin Oncol. 2013;139:1433‐1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sun GG, Wang YD, Lu YF, et al. Different association of manganese superoxide dismutase gene polymorphisms with risk of prostate, esophageal, and lung cancers: evidence from a meta‐analysis of 20,025 subjects. Asian Pac J Cancer Prev. 2013;14:1937‐1943. [DOI] [PubMed] [Google Scholar]
- 48. Yang YB, Shang YH, Tan YL, et al. Methylenetetrahydrofolate reductase polymorphisms and susceptibility to esophageal cancer in Chinese populations: a meta‐analysis. Asian Pac J Cancer Prev. 2014;15:1345‐1349. [DOI] [PubMed] [Google Scholar]
- 49. Ma RL, Min L, Chen D, et al. N‐acetyltransferase 2 phenotype and risk of esophageal cancer: a meta analysis. Cancer Biomark. 2013;13:447‐455. [DOI] [PubMed] [Google Scholar]
- 50. Wang Z, Gan L, Nie W, et al. The OGG1 Ser326Cys polymorphism and the risk of esophageal cancer: a meta‐analysis. Genet Test Mol Biomarkers. 2013;17:780‐785. [DOI] [PubMed] [Google Scholar]
- 51. Luo M, Yang Y, Luo D, et al. Tumor necrosis factor‐alpha promoter polymorphism 308 G/A is not significantly associated with esophageal cancer risk: a meta‐analysis. Oncotarget. 2016;7:79901‐79913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Guo LY, Yang N, Hu D, et al. PLCE1 rs2274223 polymorphism and susceptibility to esophageal cancer: a meta‐analysis. Asian Pac J Cancer Prev. 2014;15:9107‐9112. [DOI] [PubMed] [Google Scholar]
- 53. Luo J, Yan R, Zou L. Serine/threonine kinase 15 gene polymorphism and risk of digestive system cancers: A meta‐analysis. Exp Ther Med. 2015;9:219‐226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Duan F, Cui S, Song C, et al. Esophageal squamous cell carcinoma and gastric cardia adenocarcinoma shared susceptibility locus in C20orf54: evidence from published studies. Sci Rep. 2015;5:11961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cai J, Ye Q, Luo S, et al. CASP8 ‐652 6N insertion/deletion polymorphism and overall cancer risk: evidence from 49 studies. Oncotarget. 2017;8:56780‐56790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Leng WD, Zeng XT, Chen YJ, et al. Cytochrome P450 2E1 RsaI/PstI polymorphism and risk of esophageal cancer: A meta‐analysis of 17 case‐control studies. Exp Ther Med. 2012;4:938‐948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chen C, Yang S, Chaugai S, et al. Meta‐analysis of Hsa‐mir‐499 polymorphism (rs3746444) for cancer risk: evidence from 31 case‐control studies. BMC Med Genet. 2014;15:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Li H, Diao S, Li J, et al. An updated meta‐analysis of 23 case‐control studies on the association between miR‐34b/c polymorphism and cancer risk. Oncotarget. 2017;8:28888‐28896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Fang C, Zeng H, Li A, et al. Association of the pri‐miR‐124‐1 rs531564 polymorphism with cancer risk: A meta‐analysis. Mol Clin Oncol. 2015;3:892‐896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Li X, Qu L, Zhong Y, et al. Association between promoters polymorphisms of matrix metalloproteinases and risk of digestive cancers: a meta‐analysis. J Cancer Res Clin Oncol. 2013;139:1433‐1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Li J, Wang S, Li M, et al. Decreased risk of developing cancer in subjects carrying SLC52A3 rs13042395 polymorphism: proof from a meta‐analysis. Biomark Med. 2016;10:1105‐1118. [DOI] [PubMed] [Google Scholar]
- 62. Mao N, Nie S, Hong B, et al. Association between alcohol dehydrogenase‐2 gene polymorphism and esophageal cancer risk: a meta‐analysis. World J Surg Oncol. 2016;14:191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhao T, Wang C, Shen L, et al. Clinical significance of ALDH2 rs671 polymorphism in esophageal cancer: evidence from 31 case‐control studies. Onco Targets Ther. 2015;8:649‐659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wen L, Hu YY, Yang GL, et al. CCND1 G870A polymorphism contributes to the risk of esophageal cancer: An updated systematic review and cumulative meta‐analysis. Biomed Rep. 2014;2:549‐554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Liang Y, Liu JL, Wu Y, et al. Cyclooxygenase‐2 polymorphisms and susceptibility to esophageal cancer: a meta‐analysis. Tohoku J Exp Med. 2011;223:137‐144. [DOI] [PubMed] [Google Scholar]
- 66. Piao Y, Liu Z, Ding Z, et al. EGF +61A>G polymorphism and gastrointestinal cancer risk: a HuGE review and meta‐analysis. Gene. 2013;519:26‐33. [DOI] [PubMed] [Google Scholar]
- 67. Guo XF, Wang J, Lei XF, et al. XPD Asp312Asn polymorphism and esophageal cancer risk: an update meta‐analysis based on 3928 cases and 6012 controls. Int J Clin Exp Med. 2014;7:2443‐2452. [PMC free article] [PubMed] [Google Scholar]
- 68. Lu QJ, Bo YC, Zhao Y, et al. Glutathione S‐transferase M1 polymorphism and esophageal cancer risk: An updated meta‐analysis based on 37 studies. World J Gastroenterol. 2016;22:1911‐1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yi SM, Li GY. Null genotype of GSTT1 contributes to esophageal cancer risk in Asian populations: evidence from a meta‐analysis. Asian Pac J Cancer Prev. 2012;13:4967‐4971. [DOI] [PubMed] [Google Scholar]
- 70. Chen B, Xiong MM, Meng XL. Current evidence on the relationship between murine double minute 2 T309G polymorphism and esophageal cancer susceptibility. Dis Esophagus. 2015;28:593‐601. [DOI] [PubMed] [Google Scholar]
- 71. Tan X, Wang YY, Dai L, et al. Genetic polymorphism of MTHFR A1298C and esophageal cancer susceptibility: a meta‐analysis. Asian Pac J Cancer Prev. 2013;14:1951‐1955. [DOI] [PubMed] [Google Scholar]
- 72. Diao J, Bao J, Peng J, et al. Correlation between NAD(P)H: quinone oxidoreductase 1 C609T polymorphism and increased risk of esophageal cancer: evidence from a meta‐analysis. Ther Adv Med Oncol. 2017;9:13‐21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zhao L, Zhao X, Wu X, et al. Association of p53 Arg72Pro polymorphism with esophageal cancer: a meta‐analysis based on 14 case‐control studies. Genet Test Mol Biomarkers. 2013;17:721‐726. [DOI] [PubMed] [Google Scholar]
- 74. Li S, Deng Y, You JP, et al. XRCC1 Arg399Gln, Arg194Trp, and Arg280His polymorphisms in esophageal cancer risk: a meta‐analysis. Dig Dis Sci. 2013;58:1880‐1890. [DOI] [PubMed] [Google Scholar]
- 75. Abnet CC, Freedman ND, Hu N, et al. A shared susceptibility locus in PLCE1 at 10q23 for gastric adenocarcinoma and esophageal squamous cell carcinoma. Nat Genet. 2010;42:764‐767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wu C, Kraft P, Zhai K, et al. Genome‐wide association analyses of esophageal squamous cell carcinoma in Chinese identify multiple susceptibility loci and gene‐environment interactions. Nat Genet. 2012;44:1090‐1097. [DOI] [PubMed] [Google Scholar]
- 77. Wu C, Wang Z, Song X, et al. Joint analysis of three genome‐wide association studies of esophageal squamous cell carcinoma in Chinese populations. Nat Genet. 2014;46:1001‐1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ji HH, Hong‐Luo H, et al. Association between microRNA‐196a2 rs11614913, microRNA‐146a rs2910164, and microRNA‐423 rs6505162 polymorphisms and esophageal cancer risk: A meta‐analysis. Meta Gene. 2015;3:14‐25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Liu C, Wang Y, Jiang H, et al. Association between Cytotoxic T‐lymphocyte antigen 4 (CTLA‐4) + 49 G > A (rs231775) polymorphism and esophageal cancer: from a case‐control study to a meta‐analysis. Int J Clin Exp Med. 2015;8:17664‐17673. [PMC free article] [PubMed] [Google Scholar]
- 80. Chen XP, Xu DF, Xu WH, et al. Association studies of CYP1A1 Exon7 polymorphism and ‐GSTM1 interaction with esophageal cancer risk: a meta‐analysis in the Chinese population. Clin Lab. 2016;62:1795‐1802. [DOI] [PubMed] [Google Scholar]
- 81. Yan YL, Chen X, Liang HJ, et al. Lack of association between EPHX1 polymorphism and esophageal cancer risk: evidence from meta‐analysis. Dis Esophagus. 2015;28:164‐171. [DOI] [PubMed] [Google Scholar]
- 82. He L, Deng T, Luo H. XPA A23G polymorphism and risk of digestive system cancers: a meta‐analysis. Onco Targets Ther. 2015;8:385‐394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Zhao W, Luo J, Cai X. Association between microsomal epoxide hydrolase 1 polymorphisms and susceptibility to esophageal cancer: a meta‐analysis. Tumour Biol. 2013;34:2383‐2388. [DOI] [PubMed] [Google Scholar]
- 84. Li QT, Kang W, Wang M, et al. Association between esophageal cancer risk and EPHX1 polymorphisms: a meta‐analysis. World J Gastroenterol. 2014;20:5124‐5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Tan X, He WW, Wang YY, et al. EPHX1 Tyr113His and His139Arg polymorphisms in esophageal cancer risk: a meta‐analysis. Genet Mol Res. 2014;13:649‐659. [DOI] [PubMed] [Google Scholar]
- 86. Yin M, Tan D, Wei Q. Genetic variants of the XRCC1 gene and susceptibility to esophageal cancer: a meta‐analysis. Int J Clin Exp Med. 2009;2:26‐35. [PMC free article] [PubMed] [Google Scholar]
- 87. Dai L, Wang K, Zhang J, et al. XRCC1 gene polymorphisms and esophageal squamous cell carcinoma risk in Chinese population: A meta‐analysis of case‐control studies. Int J Cancer. 2009;125:1102‐1109. [DOI] [PubMed] [Google Scholar]
- 88. Zhang ZY, Xuan Y, Jin XY, et al. Meta‐analysis demonstrates association of XRCC1 genetic polymorphism Arg399Gln with esophageal cancer risk in the Chinese population. Genet Mol Res. 2013;12:2567‐2577. [DOI] [PubMed] [Google Scholar]
- 89. Huang J, Zhang J, Zhao Y, et al. The Arg194Trp polymorphism in the XRCC1 gene and cancer risk in Chinese Mainland population: a meta‐analysis. Mol Biol Rep. 2011;38:4565‐4573. [DOI] [PubMed] [Google Scholar]
- 90. Liu C, Jiang Z, Deng QX, et al. Meta‐analysis of association studies of CYP1A1 genetic polymorphisms with digestive tract cancer susceptibility in Chinese. Asian Pac J Cancer Prev. 2014;15:4689‐4695. [DOI] [PubMed] [Google Scholar]
- 91. Cascorbi I, Brockmöller J, Roots I. A C4887A polymorphism in exon 7 of human CYP1A1: population frequency, mutation linkages, and impact on lung cancer susceptibility. Cancer Res. 1996;56:4965‐4969. [PubMed] [Google Scholar]
- 92. Kobayashi D, Nomoto S, Kodera Y, et al. Suppressor of cytokine signaling 4 detected as a novel gastric cancer suppressor gene using double combination array analysis. World J Surg. 2012;36:362‐372. [DOI] [PubMed] [Google Scholar]
- 93. Normanno N, De Luca A, Bianco C, et al. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene. 2006;366:2‐16. [DOI] [PubMed] [Google Scholar]
- 94. Lanuti M, Liu G, Goodwin JM, et al. A functional epidermal growth factor (EGF) polymorphism, EGF serum levels, and esophageal adenocarcinoma risk and outcome. Clin Cancer Res. 2008;14:3216‐3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Araújo AP, Costa BM, Pinto‐Correia AL, et al. Association between EGF +61A/G polymorphism and gastric cancer in Caucasians. World J Gastroenterol. 2011;17:488‐492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Jain M, Upadhyay R, Kumar S, et al. Influence of apoptosis (BCL2, FAS), cell cycle (CCND1) and growth factor (EGF, EGFR) geneticpolymorphisms on survival outcome: an exploratory study in squamous cell esophageal cancer. Cancer Biol Ther. 2007;6:1553‐1558. [DOI] [PubMed] [Google Scholar]
- 97. Bäck M, Ketelhuth DF, Agewall S. Matrix metalloproteinases in atherothrombosis. Prog Cardiovasc Dis. 2010;52:410‐428. [DOI] [PubMed] [Google Scholar]
- 98. González‐Arriaga P, Pascual T, García‐Alvarez A, et al. Genetic polymorphisms in MMP 2, 9 and 3 genes modify lung cancer risk and survival. BMC Cancer. 2012;12:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Gonçalves FM, Martins‐Oliveira A, Lacchini R, et al. Matrix metal‐ loproteinase (MMP)‐2 gene polymorphisms affect circulating MMP‐2 levels in patients with migraine with aura. Gene. 2013;512:35‐40. [DOI] [PubMed] [Google Scholar]
- 100. Wing MR, Bourdon DM, Harden TK. PLC‐epsilon: a shared effector protein in Ras‐, Rho‐, and G alpha beta gamma‐mediatedsignaling. Mol Interv. 2003;3:273‐280. [DOI] [PubMed] [Google Scholar]
- 101. Thatcher JD. The inositol trisphosphate (IP3) signal transduction pathway. Sci Signal. 2010;3:tr3. [DOI] [PubMed] [Google Scholar]
- 102. Wing MR, Bourdon DM, Harden TK. PLC‐epsilon: a shared effector protein in Ras‐, Rho‐, and G alpha beta gamma‐mediated signaling. Mol Interv. 2003;3:273‐280. [DOI] [PubMed] [Google Scholar]
- 103. Bunney TD, Harris R, Gandarillas NL, et al. Structural and mechanistic insights into ras association domains of phospholipase C epsilon. Mol Cell. 2006;21:495‐507. [DOI] [PubMed] [Google Scholar]
- 104. Bunney TD, Katan M. Phospholipase C epsilon: linking second messengers and small GTPases. Trends Cell Biol. 2006;16:640‐648. [DOI] [PubMed] [Google Scholar]
- 105. Zhang X, Zhou L, Fu G, et al. The identification of an ESCC susceptibility SNP rs920778 that regulates the expression of lncRNA HOTAIR via a novel intronic enhancer. Carcinogenesis. 2014;35:2062‐2067. [DOI] [PubMed] [Google Scholar]
- 106. Zhu W, Wei BB, Shan X, et al. 765G > C and 8473 T > C polymorphisms of COX‐2 and cancer risk: a meta‐analysis based on 33 case‐control studies. Mol Biol Rep. 2010;37:277‐288. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


