Abstract
Raw and undercooked meat are regarded as important sources of Toxoplasma gondii infection of people in Europe; however, data concerning this issue in Poland are still insufficient. The aim of this study was to determine the prevalence of T. gondii DNA isolated from raw meat products retailed in Poland. The molecular characteristics of detected DNA were also performed. Samples of cured bacon, raw or smoked sausages, ham, and minced meat were examined for the presence of T. gondii DNA. Samples were digested by pepsin solution, followed by the DNA isolation. Nested and real-time polymerase chain reaction (PCR) was performed based on the amplification of 35-fold-repetitive B1 fragment gene of T. gondii. For selected B1-positive samples, multiplex PCR was performed using SAG1, SAG2 (5′-SAG2 and 3′-SAG2), altSAG2, SAG3, GRA6, BTUB, C29-2, and L358 genetic markers. Amplicons were sequenced and analyzed with NCBI database. Among 3223 examined samples, 175 (5.4%) were PCR positive. The highest percentages of positive results were found for samples originating from south-east regions of Poland—Podkarpackie (17.9%), Małopolskie (12.6%), and Lubelskie (10.8%) (p < 0.001). The percentages of positive results for particular types of meat products—sausages, smoked meat products, ham, and minced meat—ranged from 4.5% to 5.8% and the differences between them were not significant (p > 0.05). Sequence analysis of selected B1-positive samples demonstrated mostly the alleles of clonal type III (49.0%), and less—type II (17.3%), and type I (10.2%) based on nine used genetic markers. The combinations of types I/II or II/III or I/III alleles at different loci were also found in 23.5% of cases. Detection of T. gondii DNA in raw meat products may indicate the potential health threat for consumers in Poland; however, for complete risk assessment of T. gondii infection, the additional studies, including detection of live parasite, are needed.
Keywords: Toxoplasma gondii, prevalence, raw meat products, PCR, genotyping, Poland
Background
Toxoplasmosis, caused by an obligatory intracellular protozoan parasite Toxoplasma gondii, may pose a severe medical problem in a congenital form, as cerebral and ocular damage in newborns, and as an acquired infection in immunocompromised individuals (Dubey and Beattie, 1988). Toxoplasmosis has been also associated with behavioral changes and the development of psychiatric disorders (Flegr, 2007). T. gondii can infect humans and many species of warm blooded animals (Da Silva et al., 2005). The prevalence of T. gondii infection in humans varies depending on age, geographical location, nutritional habits, and keeping of hygienic standards (Tenter et al., 2000).
Toxoplasmosis has been demonstrated as a foodborne infection of global concern, posing the greatest disease risk among all parasitic infections (WHO, 2015; Limon et al., 2017; Bouwknegt et al., 2018). Fresh pork meat and meat products are regarded as one of the important risk factors of T. gondii infection, since viable T. gondii parasites have been isolated thereof (van der Giessen et al., 2007; Kijlstra et al., 2009; Limon et al., 2017). By contrast, little is known about the presence of T. gondii in cured meat products.
T. gondii has a clonal populational structure (Ajzenberg, 2010) and most of the parasite isolates in Europe and North America are classified into three clonal lineages (type I–III) (Howe and Sibley, 1995; Ajzenberg et al., 2002). Other clonal lineages were found in wildlife in North America (Khan et al., 2011). In South America, T. gondii isolates are more genetically diverse, what can be a result of high biodiversity, geographical differences, and the important role of recombination in strain diversification (Lehmann et al., 2006; Dubey et al., 2008b; Pena et al., 2008).
In Poland, the seropositivity of T. gondii in humans is estimated up to 60–70%, depending on a tested group (Nowakowska et al., 2006b). The overall incidence of human toxoplasmosis in Poland is underestimated as only the congenital cases are recorded (17 cases in 2017; NIH Raport, 2017). This represents a small proportion of the total number of clinical (i.e., lymphadenopathy, chorioretinitis, and neurotoxoplasmosis) and asymptomatic cases. The identification of a possible correlation between the severity or type of disease and strain genotype might be important for determining the proper treatment and possible outcome of the disease in human T. gondii infection cases. However, there is no clear opinion so far, on the correlation between T. gondii strain genotype and character of symptoms in infected people (Fuentes et al., 2001). So far, only few studies in Poland applied the genotyping of T. gondii. Nowakowska et al. (2006a) genotyped T. gondii isolated from infants with congenital toxoplasmosis and demonstrated the presence of type II. Dubey et al. (2008a) detected the nonclonal strains of T. gondii in chickens in Poland. In the study by Lass et al. (2012), genotyping of T. gondii oocysts found on fruits and vegetables showed types I and II at SAG2 locus. Recently, our own studies showed high prevalence of type III in goats' milk (Sroka et al., 2017).
Since pork is widely consumed in Europe, determining the frequency of rate of T. gondii infection in pigs and pork is very important for the prognosis of disease risk. The prevalence of T. gondii infection in pigs in Poland varies depending on type of housing and production system. Recent serological studies performed by the National Veterinary Institute in Pulawy (Poland), in 3600 pigs, showed positive results in 11.1–14.3% of examined animals (Sroka et al., 2011; Sroka et al. unpublished data).
Aiming to continue our previous research concerning the prevalence of T. gondii in slaughtered animals in Poland, we chose, as a goal of this study, to determine the prevalence and genetic assortment of T. gondii strains in raw meat products from retail stores in Poland, in aspect of potential threat to human health.
Materials and Methods
Meat products samples
The research was conducted as a part of the surveillance program realized by the National Veterinary Institute in Pulawy (Poland) (2014–2018). Samples were collected from the meat producers and retail meat stores offering raw meat products, randomly selected in each of 13 analyzed regions of Poland (Table 1; Fig. 1). The number of samples obtained in each of the regions depended on the availability of meat plants and retail stores offering raw meats products, as well as on the size of their assortment.
Table 1.
Results of Toxoplasma gondii DNA Detection in Meat Products Retailed in Poland
| Type of meat products | Results for particular regions of Poland | Total | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N of positive/N of examined samples | ||||||||||||||
| % of positive (CI)* | ||||||||||||||
| Mp | Pk | Lb | Pl | W-m | Pm | Wk | Dl | Śl | Św | Zp | M | Kp | ||
| Smoked meat products | 9/74 | 11/61 | 8/95 | 3/77 | 3/85 | 3/23 | 2/42 | 1/72 | 1/61 | 1/87 | 2/48 | 4/79 | 0/52 | 48/856 |
| 12.2% (6.5–21.5) | 18.0% (10.4–29.5) | 8.4% (4.3–15.7) | 3.9% (1.3–10.9) | 3.4% (1.2–9.9) | 13% (4.5–32.1) | 4.8% (1.3–15.8) | 1.4% (0.3–7.5) | 1.6% (0.2–8.7) | 1.1% (0.2–6.2) | 4.2% (1.2–14.0) | 5.1% (2–12.3) | 0.0% (0.0–6.9) | 5.7% (4.3–7.4) | |
| Sausages | 18/101 | 18/93 | 5/65 | 4/112 | 3/201 | 4/66 | 10/98 | 5/92 | 0/77 | 0/156 | 2/70 | 5/143 | 5/81 | 79/1355 |
| 17.8% (11.6–26.4) | 19.4% (12.6–28.5) | 7.7% (3.3–16.8) | 3.6% (1.4–8.8) | 1.5% (0.5–4.3) | 6.1% (2.4–14.6) | 1.0% (5.6–11.8) | 5.4% (5.2–11.6) | 0.0% (0.0–4.8) | 0.0% (0.0–2.4) | 2.9% (0.8–9.8) | 3.5% (1.5–7.9) | 6.2% (2.7–13.7) | 5.8% (4.7–7.2) | |
| Ham | 3/16 | 9/31 | 2/19 | n/e | 0/26 | 0/9 | 0/28 | 0/20 | 0/9 | 0/34 | 0/18 | 0/19 | 0/27 | 14/256 |
| 18.8% (6.6–43.0) | 29.0% (16.1–46.6) | 10.5% (2.9–31.4) | 0.0% (0.0–12.8) | 0.0%/(0.0–29.9) | 0.0% (0.0–12.1) | 0.0% (0.0–16.1) | 0.0% (0.0–29.9) | 0.0% (0.0–10.2) | 0.0% (0.0–17.6) | 0.0% (0.0–16.8) | 0.0% (0.0–12.5) | 5.5% (3.3–9.0) | ||
| Minced meat | 3/70 | 4/50 | 10/53 | 2/55 | 3/102 | 0/30 | 2/45 | 0/100 | 7/94 | 0/100 | 0/6 | n/e | 3/51 | 34/756 |
| 4.3% (1.5–11.9) | 8.0% (3.2–18.8) | 18.9% (10.6–31.4) | 3.6% (0.6–13.6) | 2.9% (1.0–8.3) | 0.0% (0.0–11.4) | 4.4% (1.2–14.8) | 0.0% (3.7–14.6) | 7.4% (3.7–14.6) | 0.0% (0.0–3.7) | 0.0% (0.0–39.0) | 5.9% (2.0–15.9) | 4.5% (3.3–9.0) | ||
| Total | 33/261 | 42/235 | 25/232 | 9/244 | 9/414 | 7/128 | 14/213 | 6/284 | 8/241 | 1/377 | 4/142 | 9/241 | 8/211 | 175/3223 |
| 12.6%* (9.1–17.2) | 17.9%* (13.5–23.3) | 10.8%* (7.4–15.4) | 3.7% (2.0–6.9) | 2.2% (1.2–4.1) | 5.5% (2.7–10.9) | 6.6% (4.0–10.7) | 2.1% (1.0–4.5) | 3.3% (1.7–6.4) | 0.3% (0.0–1.5) | 2.8% (1.1–7.0) | 3.7% (2.0–7.0) | 3.8% (2.0–7.3) | 5.4% (4.7–6.3) | |
Significant differences in percentages of positive results in comparison with other regions of Poland (p < 0.05).
The regions of Poland: Mp, Małopolskie; Pk, Podkarpackie; Lb, Lubelskie; Pl, Podlaskie; W-m, Warmińsko-Mazurskie; Pm, Pomorskie; Wk, Wielkopolskie; Dl, Dolnośląskie; Śl, Śląskie; Św, Świętokrzyskie; Zp, Zachodniopomorskie; M, Mazowieckie; Kp, Kujawsko-pomorskie.
n/e, not examined; CI, 95% confidence interval.
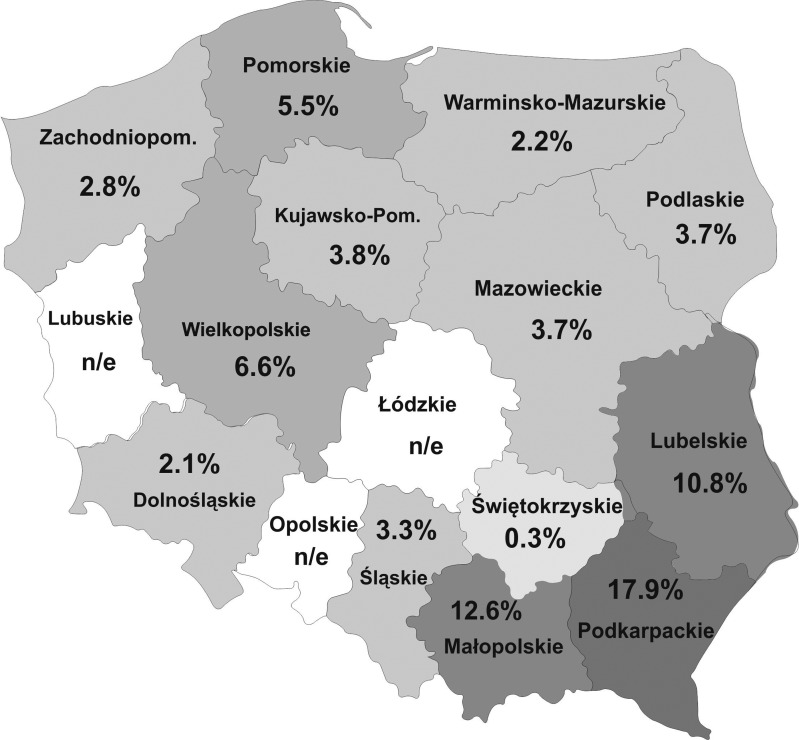
FIG. 1.
Results of Toxoplasma gondii DNA detection in raw meat products from selected regions of Poland.
In total, 3223 meat product samples were tested, including 2467 samples purchased (2015–2017) in retail meat stores and 756 samples collected by Veterinary Inspection in meat plants from 13 regions of Poland. The following kind of meat products were tested: raw sausages (i.e., Polish sausage, white sausage, Frankfurters, minced “Metka” sausage, kindikas, salami, “Cracow” dry sausage, thin dry-smoked pork sausage, juniper dry sausage, and hunter's sausage), raw or smoked bacon, smoked ribs, raw loin or tenderloin, raw (smoked) fermented ham, and minced meat.
Digesting of meat product samples and DNA extraction
Meat product samples were digested by pepsin solution according to the method described by Dubey and Beattie (1988). Briefly, 50 g samples were cut and homogenized in 125 mL of 0.9% NaCl. Next, the homogenate of each sample was mixed with 250 mL of acid-pepsin solution (2.6 g pepsin, 7 mL HCl, and 0.9% NaCl filled up to 500 mL, pH 1.1–1.2), incubated in a shaker bath at 37°C for 90 min., poured out through the gauze, and centrifuged at 1200 × g for 10 min. The collected pellet was resuspended in 35 mL of phosphate-buffered saline (pH 7.4) and centrifuged (1200 × g × 10 min.). Supernatant was removed and pellet was resuspended in 5 mL of 0.9% NaCl, and stored at −20°C until further analysis. Genomic DNA was extracted from the pellet using a commercial kit (QIAmp DNA Mini Kit; Qiagen), according to the manufacturer's instructions.
Nested and real-time polymerase chain reaction
Briefly, T. gondii DNA was detected by amplification of 35-fold-repetitive B1 fragment gene in nested polymerase chain reaction (PCR) (according to the method by Grigg and Boothroyd, 2001) and, as an alternative technique, to confirm B1 PCR-positive results, by real-time PCR (according to the method by Lin et al., 2000). In real-time PCR, the commercial master mix QI Supermix (Bio-Rad, Hercules, CA) was used.
Multilocus PCR
To investigate the clonal type of B1 gene-positive samples, multilocus PCR was carried out using nine markers: SAG1, 3′SAG2, 5′SAG2, SAG3, BTUB, GRA6, altSAG2, C29-2, and L358, based on the method described by Su et al. (2010).
Nested and multilocus PCR were carried out in a C1000 Thermal Cycler (Bio-Rad). Real-time PCR amplification was performed in a thermal cycler CFX-96 (Bio-Rad). As positive controls, RH (type I), ME49 (type II), and C56 (type III) DNA isolates of T. gondii strains, and as a negative control nuclease-free water, were used.
DNA sequencing of amplicons was performed using ABI PRISM 310 Genetic Analyzer (Applied Biosystems, Inc., Foster City, CA), with the use of Abi Prism Big Dye Terminator v. 3.1. Cycle Sequencing Kits and Big Dye XTerminator Purification Kit (Applied Biosystems). Sequences were analyzed using Geneious v. 11.1.4. software (Geneious Co., Wellington, New Zealand) and compared with the sequences deposited in NCBI database using Blast.
Statistical analysis
The results were analyzed with χ2 test, using STATISTICA v. 5.1 package (Statsoft, Tulsa, OK).
Results
PCR results
In total, among 3223 meat products samples examined, 175 (5.4%) were PCR positive, including 48 smoked meat products (27.4% of total positive), 79 sausages (45.1%), 14 hams (8.0%), and 34 minced meat samples (19.4%). All B1 PCR-positive samples were positive in RT PCR; however, for part of samples (62 out of 175 samples), the borderline results (Ct values exceeded 40) were obtained. The highest percentages of positive results were obtained for samples originating from south-east regions of Poland—Podkarpackie (Pk) (17.9%), Małopolskie (Mp) (12.6%), and Lubelskie (Lb) (10.8%). The percentages of positive results for samples originating from other regions of Poland were significantly lower (0.3–6.6%) (p < 0.001) (Table 1). In individual groups of meat products, T. gondii DNA was detected in 5.7% of smoked meat products samples, 5.8% of sausage samples, 5.5% of ham samples, and 4.5% of minced meat samples; the differences between percentages were not significant. In the group of smoked meat products, the highest percentages of positive results were obtained for samples from Pk and Mp regions (18.0% and 12.2%), lower for samples from Pm and Lb regions (13% and 8.4%), and lowest for other regions (0.0–5.1%) (p < 0.05). In the group of sausages, significant differences were found between percentages of positive results obtained for samples from Pk and Mp regions (19.4% and 17.8%) in comparison with other regions (0.0–7.7%) (p < 0.05) The positive results for ham samples were obtained only for samples originating from three regions—Pk (29.0%), Mp (18.8%), and Lb (10.5%). Minced meat samples from Lb (18.9%) were significantly (p < 0.05) more likely to be PCR positive than samples from other regions (0.0–8.0%) (Table 1).
Genotyping results
Among 175 DNA samples positive in PCR (B1), we were able to sequence 61 samples (in RT PCR, for these samples, Ct value did not exceed 39). The remaining samples did not have enough DNA amount to be processed for sequencing, or sequences were not of high quality for analysis.
Among DNA meat product samples, one sample was able to be genotyped with six markers (SAG1, 3′SAG2, SAG3, GRA6, altSAG2, and L358), one sample with four markers (5′SAG2, SAG3, GRA6, and altSAG2), and five samples with three markers. Next, 19 samples were genotyped with two markers. The remaining sequenced samples (35) were positive only with one marker (7 samples with SAG1; 11 samples with SAG3; 9 samples with GRA6; 1 sample with BTUB, 1 sample with 3′SAG2, 4 samples with 5′SAG2, and 2 samples with C358). In total, by using nine markers, 98 amplicons were obtained (Table 2).
Table 2.
Distribution of Toxoplasma gondii Genotypes in Sixty-One Meat Products Samples Included in the Study
| Pattern of possible amplifications | Total number of meat products DNA samples | SAG1a | 3′SAG2 | 5′SAG2 | SAG3 | GRA6 | BTUB | AltSAG2 | C29-2 | L358 |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | n/a | n/a | III | n/a | II | n/a | — | — | — |
| 1 | II | III | n/a | n/a | n/a | |||||
| 2 | 1 | II/III | III | n/a | III | n/a | n/a | — | — | — |
| 3 | 1 | II/III | III | III | n/a | n/a | n/a | — | — | — |
| 4 | 1 | I | n/a | n/a | III | n/a | n/a | — | — | — |
| 2 | II/III | III | ||||||||
| 5 | 1 | I | n/a | III | n/a | n/a | n/a | — | — | — |
| 1 | II/III | I/IIb | ||||||||
| 1 | II/III | n/a | II | n/a | n/a | n/a | ||||
| 6 | 1 | n/a | n/a | III | III | n/a | n/a | — | — | — |
| 7 | 1 | n/a | n/a | n/a | III | I/IIb | n/a | — | — | — |
| 1 | I/IIIb | III | n/a | n/a | n/a | |||||
| 1 | II | III | n/a | n/a | n/a | |||||
| 2 | III | III | — | — | — | |||||
| 8 | 1 | n/a | n/a | n/a | III | n/a | I/IIIb | — | — | — |
| 9 | 6 | II/III | n/a | n/a | n/a | n/a | n/a | -/n/a | -/n/a | -/n/a |
| 1 | II/III | n/a | n/a | n/a | ||||||
| 10 | 6 | n/a | n/a | n/a | III | n/a | n/a | — | — | — |
| 2 | I | |||||||||
| 3 | II | |||||||||
| 11 | 1 | n/a | n/a | n/a | n/a | I/IIb | n/a | — | — | — |
| 3 | II | -/n/a | -/n/a | -/n/a | ||||||
| 5 | ||||||||||
| III | -/n/a | -/n/a | -/n/a | |||||||
| 12 | 2 | n/a | n/a | I/IIb | n/a | n/a | n/a | — | — | — |
| 2 | III | -/n/a | -/n/a | -/n/a | ||||||
| 13 | 1 | n/a | n/a | n/a | n/a | n/a | I/IIIb | — | — | — |
| 14 | 1 | I | II | n/a | II | I | n/a | III | n/a | II |
| 15 | 1 | n/a | n/a | III | III | III | n/a | n/a | n/a | n/a |
| 16 | 1 | n/a | n/a | III | III | III | n/a | III | n/a | n/a |
| 17 | 1 | n/a | n/a | n/a | III | III | n/a | n/a | n/a | II |
| 18 | 1 | n/a | n/a | n/a | II | n/a | n/a | II | n/a | n/a |
| 19 | 1 | n/a | n/a | n/a | I | n/a | n/a | n/a | I | n/a |
| 1 | II | III | ||||||||
| 20 | 1 | n/a | I/IIIb | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| 21 | 2 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | III |
| 22 | 1 | I | n/a | I/IIb | I | n/a | n/a | n/a | n/a | n/a |
| 23 | 1 | n/a | n/a | n/a | III | n/a | n/a | n/a | n/a | III |
GenBank accession numbers for selected sequences: MH429059- MH429071, MH536007- MH536012, and MH606148–MH606184.
At SAG1 locus, types II and III are indistinguishable.
Distinction between these types was not possible, probably a mix of T. gondii strains occurred.
—, not examined; n/a, product not amplified; -/n/a, part of samples not examined, for next part of samples − product not amplified.
In total, type I, type II, and type III T. gondii lineages were determined for 10 (10.2%), 17 (17.3%), and 48 (49.0%) amplicons, respectively. Alleles types I/II, II/III, and I/III had 6 (6.1%), 13 (13.3%), and 4 (4.1%) amplicons, respectively (Table 3). The new single-nucleotide polymorphisms (SNPs) were found in three amplicons.
Table 3.
The Summary of Toxoplasma gondii Genotypes Determined at Individual Markers Used in Study
| Genotype markers | N of positive DNA samples | Type I | Type II | Type III | Type I/IIa | Type II/IIIa | Type I/IIIa |
|---|---|---|---|---|---|---|---|
| SAG1 | 17 | 4 | 0 | 0 | 0 | 13 | 0 |
| 3′SAG2 | 4 | 0 | 1 | 2 | 0 | 0 | 1 |
| 5′SAG2 | 14 | 0 | 2 | 8 | 4 | 0 | 0 |
| SAG3 | 31 | 4 | 7 | 19 | 0 | 0 | 1 |
| GRA6 | 20 | 1 | 4 | 13 | 2 | 0 | 0 |
| BTUB | 2 | 0 | 0 | 0 | 0 | 0 | 2 |
| altSAG2 | 3 | 0 | 1 | 2 | 0 | 0 | 0 |
| C29-2 | 2 | 1 | 0 | 1 | 0 | 0 | 0 |
| L-358 | 5 | 0 | 2 | 3 | 0 | 0 | 0 |
| Total | 98 | 10b (10.2%) | 17b (17.3%) | 48b (49.0%) | 6b (6.1%) | 13b (13.3%) | 4b (4.1) |
Distinction between these types was not possible, probably a mix of T. gondii strains occurred.
N of T. gondii DNA samples (percentage).
In relation to the genotyping results of particular amplicons and the regions of study, the high numbers of T. gondii type III and mix of types II/III were determined for samples from Mazowieckie and Wielkopolskie (Wk, Type III), and Mp (types II/III) regions (p < 0.05), whereas, type I was mostly detected in samples from Mp and Zp regions (Table 4).
Table 4.
The Summary of Genotyping in Relation to the Regions of Study (Poland)
| Regions of study | N of positive DNA samples | Type I | Type II | Type III | Type I/IIa | Type II/IIIa | Type I/IIIa |
|---|---|---|---|---|---|---|---|
| Kujawsko-Pomorskie | 9 | 0 | 4 | 5 | 0 | 0 | 0 |
| Małopolskie | 40 | 5 | 1 | 18 | 3 | 11b | 2 |
| Mazowieckie | 10 | 0 | 2 | 7b | 0 | 0 | 1 |
| Podkarpackie | 13 | 0 | 4 | 6 | 2 | 1 | 0 |
| Wielkopolskie | 14 | 1 | 3 | 7b | 1 | 1 | 1 |
| Zachodniopomorskie | 12 | 4 | 3 | 5 | 0 | 0 | 0 |
| Total | 98 | 10 (10.2%) | 17 (17.3%) | 48 (49.0%) | 6 (6.1%) | 13 (13.3%) | 4 (4.1%) |
Distinction between these types was not possible, probably a mix of T. gondii strains occurred.
Significant differences in the number of genotypes determined for particular amplicons, in comparison with other regions of Poland (p < 0.05).
Taking into account only the results with homogenous genotype obtained for particular meat product samples, finally, type III was determined for 21, type II for 7 samples, and type I for 3 samples. The highest number of meat product samples with T. gondii type III genotypes was detected in Mp region; however, this difference did not attain a level of statistical significance (Table 5).
Table 5.
The Number of Meat Product Samples with the Homologous Genotypes Obtained at Used Markers, in Relation to the Regions of Study
| Regions of Poland | Type I | Type II | Type III | Total |
|---|---|---|---|---|
| Kujawsko–Pomorskie | 0 | 2 | 1 | 3 |
| Małopolskie | 2 | 0 | 7 | 9 |
| Mazowieckie | 0 | 0 | 3 | 3 |
| Podkarpackie | 0 | 3 | 4 | 7 |
| Wielkopolskie | 0 | 2 | 3 | 5 |
| Zachodniopomorskie | 1 | 0 | 3 | 4 |
| Total | 3 (9.7%) | 7 (22.6%) | 21 (67.7%) | 31 |
The comparison of genotyping results in selected 61 meat products, in relation to the type of meat products, showed the high number of type II and type I in minced meat samples (beef), whereas type III was prevalent in sausages and pork minced meat samples (Table 6).
Table 6.
The Summary of Genotyping in Sixty-One Meat Products Samples, in Relation to Their Types
| Types of meat products | N of positive DNA samples | Type I | Type II | Type III | Type I/IIa | Type II/IIIa | Type I/IIIa |
|---|---|---|---|---|---|---|---|
| Sausages | 37 | 5 | 4 | 20 | 3 | 3 | 2 |
| Smoked meat products | 24 | 0 | 2 | 12 | 0 | 9 | 1 |
| Hams | 7 | 0 | 2 | 2 | 2 | 0 | 1 |
| Minced meat (total), including | 30 | 5 | 9b | 14 | 1 | 1 | 0 |
| Pork | 8 | 0 | 0 | 7b | 0 | 1 | 0 |
| Beef | 22 | 5 | 9 | 7 | 1 | 0 | 0 |
| Total | 98 | 10 (10.2%) | 17 (17.3%) | 48 (49.0%) | 6 (6.1%) | 13 (13.3%) | 4 (4.1%) |
Distinction between these types was not possible, probably a mix of T. gondii strains occurred.
Significant differences in the number of genotypes determined for particular amplicons, in comparison with other types of meat products or in a particular group of meat products (p < 0.05).
Discussion
The foodborne route of human T. gondii infection is mainly linked with consumption of raw or undercooked meat and meat products (Kapperud et al., 1996; Cook et al., 2000; WHO, 2015; Jones et al., 2009). Serological examination in pigs, in general, is useful for indirect diagnosis of T. gondii infection. Seropositivity in pigs can indicate parasite presence in their tissues. Some authors report that the success of live T. gondii isolation increases with antibody titer in the pig (Dubey et al., 1995), whereas the others have not found such correlation. In a recent study, Kuruca et al. (2017) detected T. gondii DNA in diaphragm tissues of eight pigs, of which three were seronegative. In the study by Dubey et al. (2012), live T. gondii strains were isolated from 17 pigs, including one from a seronegative animal. Thus, meat from seronegative pigs may also represent a source of potential infection to consumers.
Recent trends in consumer habits, with a shift toward the consumption of free-range and organic pork, where animals have a higher risk of exposure to T. gondii from the environment, may result in a higher risk of consumer exposure to T. gondii (van der Giessen et al., 2007; Kijlstra et al., 2009). Thus, the genotyping of T. gondii strains isolated from various sources of infection may provide new insights to evaluate the possible interaction between parasite types and severity of the disease in humans (Da Silva et al., 2005).
In Poland, there are still regions with small, family farms with traditional rearing of pigs, which have direct contact with potential sources of T. gondii. Having outdoor access, the presence of cats in farm, and feed stored with the possibility for contamination with cat feces have been previously reported as risk factors for T. gondii infection (Assadi-Rad et al., 1995; Weigel et al., 1995; Kijlstra et al., 2004; Klun et al., 2006; Guo et al., 2016).
In this context, noteworthy are regional differences between the frequency rates of T. gondii in meat samples, stated by us in this study. The highest prevalence of T. gondii DNA was found in two regions (Małopolskie and Podkarpackie) situated in the southeastern part of Poland. These are mountainous regions with numerous small farms, where pigs are often free-range reared and might have direct contact with cats, wild animals, and other potential sources of T. gondii infection.
Basic traditional methods of the production of Polish hams and sausages are based on drying (hanging until mature), followed by smoking. Especially, Śląskie, Podlaskie and Wielkopolskie are the regions in Poland where raw sausage, ham, and smoked meat products are produced according to traditional, Polish recipes. There are preserved with salt only, then smoked, and dried.
In this study, a total of 5.4% tested raw meat products were detected, T. gondii positive, mainly sausage (45.1% of total positive), less smoked meat products (27.4%), and least ham (8.0%). The other reports from European countries may confirm our results; Aspinall et al. (2002) in a study conducted in United Kingdom showed 38% of meat and meat product samples contaminated with T. gondii. Vergara et al. (2018) detected T. gondii DNA in carcasses of 14 out of 103 pigs (13.6%) at an abattoir in Italy. In a study performed in Turkey (Ergin et al., 2009), T. gondii DNA was detected in 19% of fermented sausages. The possibility of live T. gondii persistence in meat products was reported by Warnekulasuriya et al. (1998), who detected viable T. gondii organisms in one of 67 cured meat samples from United Kingdom, including dried and semidried sausages and hams. In the studies performed in the New World, the prevalence of T. gondii in meat samples ranged in fairly wide limits from 0.5% to 43% (Dias et al., 2005; Dubey et al., 2005; Galvan-Ramırez et al., 2010; Franco-Hernandez et al., 2016).
There is little knowledge concerning the effects of several processing conditions during preparation of meat products on inactivation of parasite, such as salt, nitrites, nitrates, and organic acid concentration, time, and temperature processing. In opinions of some researchers, tissue cysts of T. gondii are killed during commercial curing procedures with salt (Dubey, 1997; Hill et al., 2004, 2006; Genchi et al., 2017). In contrast, other studies have indicated the potential failure of curing to inactivate T. gondii (Warnekulasuriya et al., 1998; Herrero et al., 2017).
In this study, the clonal T. gondii type III was the most prevalent, while types I and II were less common. This finding is concordant with the report by Zia-Ali et al. (2007) from Iran, who found T. gondii type III (SAG2) in the majority of tested tissues samples of chicken and sheep. Similarly, Lehmann et al. (2003) found 25 T. gondii isolates from market-age pigs in the United States of America, 20 belonging to type III and 5 to type II. In the study performed in Italy, Vergara et al. (2018) identified in pigs types III (3.9%), I (3.9%), and II (5.8%). However, in other European studies, T. gondii type II isolates appeared to be most common (Richomme et al., 2009; Aubert et al., 2010). By contrast, in some genotypic studies performed in United Kingdom and South America, prevalence of type I was stated among T. gondii strains isolated from meat samples (Aspinall et al., 2001; Da Silva et al., 2005).
Since molecular methods for detecting T. gondii DNA were used, we were unable to distinguish viable from nonviable parasites. Because condiments (potential PCR inhibitors) are present in cured meat products, the efficiency of the amplification of T. gondii DNA from cured meat products may be problematic. Amplification failure with some markers might depend on the quality of tissue/DNA samples (Franco-Hernandez et al., 2016). To gain good efficiency of marker amplification, a good quality DNA is needed; however, this is not always possible in epidemiological studies.
In this study, some of B1 PCR-positive samples failed to be completely genotyped and only partial data were obtained due to a low DNA concentration. Considering this limitation, a cautious presumption could be made that genotype III seems to be the most prevalent in the analyzed area followed by genotype II and I, while alleles types I/II, II/III, and I/III had lower prevalence. For three DNA samples, new SNPs were also found. These results seem to be different from those obtained in other European countries, in which T. gondii genotype II was the most widespread. The prevalence of T. gondii genotype II ranged in Europe from 50% in Spain (Calero-Bernal et al., 2015) to 100% in France (Richomme et al., 2009; Aubert et al., 2010). However, in some regions of Europe (i.e., Italy, Spain, Switzerland, Slovakia, and Portugal), the genetic variability can be higher than previously stated and type III, type I, and mixed or atypical strains of T. gondii may be more frequent (Fuentes et al., 2001; de Sousa et al., 2006; Berger-Schoch et al., 2011; Mancianti et al., 2013; Turčeková et al., 2013; Verin et al., 2013; Bacci et al., 2015; Formenti et al., 2016; Battisti et al., 2018). In Poland, the results of our own, previous research, where types III and I, as well as mixed or atypical were detected (i.e., in goat milk, ticks from vegetative stage, and wildlife), may confirm this statement (Sroka et al., 2016, 2017; Cisak et al., 2017; Zając et al., 2017). According to some authors' opinions (Xiao and Yolken, 2015), T. gondii type I and atypical strains can cause more severe disease symptoms, especially in immunocompromised persons.
Conclusion
The results of this study showed that raw meat products seem to be potentially important source of T. gondii infection for humans in Poland, which may be important for persons with immunodeficiency and pregnant women, who should avoid eating raw meat product.
However, for a complete risk assessment, the additional studies, including detection of live parasites, are needed.
Acknowledgment
The investigation was supported by the Polish Ministry of Agriculture and Rural Development (in the frame of the Multiannual Programme “Protection of Animal and Public Health”).
Disclosure Statement
No competing financial interests exist.
References
- Ajzenberg D. Type I strains in human toxoplasmosis: Myth or reality? Future Microbiol 2010;5:841–843 [DOI] [PubMed] [Google Scholar]
- Ajzenberg D, Bañuls AL, Tibayrenc M, Dardé ML. Microsatellite analysis of Toxoplasma gondii shows considerable polymorphism structured into two main clonal groups. Int J Parasitol 2002;32:27–38 [DOI] [PubMed] [Google Scholar]
- Aspinall TV, Marlee D, Hyde JE, Sims PF. Prevalence of Toxoplasma gondii in commercial meat products as monitored by polymerase chain reaction food for thought? Int J Parasitol 2002;32:1193–1199 [DOI] [PubMed] [Google Scholar]
- Assadi-Rad AM, New JC, Patton S. Risk factors associated with transmission of Toxoplasma gondii to sows kept in different management systems in Tennessee. Vet Parasitol 1995;57:289–297 [DOI] [PubMed] [Google Scholar]
- Aubert D, Ajzenberg D, Richomme C, Gilot-Fromont E, Terrier ME, de Gevigney C, Game Y, Maillard D, Gibert P, Dardé ML, Villena I. Molecular and biological characteristics of Toxoplasma gondii isolates from wildlife in France. Vet Parasitol 2010;171:346–349 [DOI] [PubMed] [Google Scholar]
- Bacci C, Vismarra A, Mangia C, Bonardi S, Bruini I, Genchi M, Kramer L, Brindani F. Detection of Toxoplasma gondii in free-range, organic pigs in Italy using serological and molecular methods. Int J Food Microbiol 2015;202:54–56 [DOI] [PubMed] [Google Scholar]
- Battisti E, Zanet S, Trisciuoglio A, Bruno S, Ferroglio E. Circulating genotypes of Toxoplasma gondii in Northwestern Italy. Vet Parasitol 2018;253:43–47 [DOI] [PubMed] [Google Scholar]
- Berger-Schoch AE, Herrmann DC, Schares G, Müller N, Bernet D, Gottstein B, Frey CF. Prevalence and genotypes of Toxoplasma gondii in feline faeces (oocysts) and meat from sheep, cattle and pigs in Switzerland. Vet Parasitol 2011;177:290–297 [DOI] [PubMed] [Google Scholar]
- Bouwknegt M, Devleesschauwer B, Graham H, Robertson L, van der Giessen JWB, The Euro-FBP workshop participants. Prioritisation of food-borne parasites in Europe, 2016. Euro Surveill 2018;23 [Epub ahead of print]; DOI: 10.2807/1560-7917.ES.2018.23.9.17-00161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calero-Bernal R, Saugar JM, Frontera E, Perez-Martin JE, Habela MA, Serrano FJ, Reina D, Fuentes I. Prevalence and genotype identification of Toxoplasma gondii in wild animals from Southwestern Spain. J Wildl Dis 2015;51:233–238 [DOI] [PubMed] [Google Scholar]
- Cisak E, Zając V, Sroka J, Sawczyn A, Kloc A, Dutkiewicz J, Wójcik-Fatla A. Presence of pathogenic Rickettsiae and protozoan in samples of raw milk from cows, goats, and sheep. Foodborne Pathog Dis 2017;14:189–194 [DOI] [PubMed] [Google Scholar]
- Cook AJC, Gilbert RE, Buffolano W, Zufferey J, Petersen E, Jenum PA, Foulon W, Semprini AE, Dunn DT. Sources of Toxoplasma infection in pregnant women: European multicenter case control study. BMJ 2000;321:142–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Silva AV, De Oliveira Mendonça A, Bergamaschi Pezerico S, Domingues PF, Langoni H. Genotyping of Toxoplasma gondii strains detected in pork sausage. Parasitol Latino Am 2005;60:65–68 [Google Scholar]
- de Sousa S, Ajzenberg D, Canada N, Freire L, Da Costa JM, Dardé ML, Thulliez P, Dubey JP. Biologic and molecular characterization of Toxoplasma gondii isolates from pigs from Portugal. Vet Parasitol 2006;135:133–136 [DOI] [PubMed] [Google Scholar]
- Dias RA, Navarro IT, Ruffolo BB, Bugni FM, Castro MV, Freire RL. Toxoplasma gondii in fresh pork sausage and seroprevalence in butchers from factories in Londrina, Parana' State, Brazil. Rev Inst Med Trop Sao Paulo 2005;47:185–189 [DOI] [PubMed] [Google Scholar]
- Dubey JP. Survival of Toxoplasma gondii tissue cysts in 0.85–86% NaCl solutions at 4–20°C. J Parasitol 1997;83:946–949 [PubMed] [Google Scholar]
- Dubey JP, Beattie CP. Toxoplasmosis of Animals and Man. 1st edition. Boca Raton, FL: CRC Press, 1988 [Google Scholar]
- Dubey JP, Hill DE, Jones JL, Hightower AW, Kirkland E, Roberts JM, Marcet PL, Lehmann T, Vianna MCB, Miska K, Sreekumar C, Kwok OCH, Shen SK, Gamble HR. Prevalence of viable Toxoplasma gondii in beef, chicken, and pork from retail meat stores in the United States: Risk assessment to consumers. J Parasitol 2005;91:1082–1093 [DOI] [PubMed] [Google Scholar]
- Dubey JP, Hill DE, Rozeboom DW, Rajendran C, Choudhary S, Ferreira LR, Kwok OC, Su C, et al. High prevalence and genotypes of Toxoplasma gondii isolated from organic pigs in northern USA. Vet Parasitol 2012;188:14–18 [DOI] [PubMed] [Google Scholar]
- Dubey JP, Huong LT, Lawson BW, Subekti DT, Tassi P, Cabaj W, Sundar N, Velmurugan GV, Kwok OC, Su C. Seroprevalence and isolation of Toxoplasma gondii from free-range chickens in Ghana, Indonesia, Italy, Poland, and Vietnam. J Parasitol 2008a;94:68–71 [DOI] [PubMed] [Google Scholar]
- Dubey JP, Sundar N, Hill D, Velmurugan GV, Bandini LA, Kwok OC, Majumdar D, Su C. High prevalence and abundant atypical genotypes of Toxoplasma gondii isolated from lambs destined for human consumption in the USA. Int J Parasitol 2008b;38:999–1006 [DOI] [PubMed] [Google Scholar]
- Dubey JP, Thulliez P, Powell EC. Toxoplasma gondii in Iowa sows: Comparison of antibody titers to isolation of T. gondii by bioassays in mice and cats. J Parasitol 1995;81:48–53 [PubMed] [Google Scholar]
- Ergin S, Ciftcioglu G, Midilli K, Issa G, Gargili A. Detection of Toxoplasma gondii from meat and meat products by the nested PCR method and its relationship with seroprevalence in slaughtered animals. Bull Vet Inst Pulawy 2009;53:657–661 [Google Scholar]
- Flegr J. Effects of Toxoplasma on human behavior. Schizophr Bull 2007;33:757–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formenti N, Gaffuri A, Trogu T, Viganò R, Ferrari N, Lanfranchi P. Spread and genotype of Toxoplasma gondii in naturally infected alpine chamois (Rupicapra r. rupicapra). Parasitol Res 2016;115:2115–2120 [DOI] [PubMed] [Google Scholar]
- Franco-Hernandez EN, Acosta A, Cortés-Vecino J, Gómez-Marín JE. Survey for Toxoplasma gondii by PCR detection in meat for human consumption in Colombia. Parasitol Res 2016;115:691–695 [DOI] [PubMed] [Google Scholar]
- Fuentes I, Rubio JM, Ramírez C, Alvar J. Genotypic characterization of Toxoplasma gondii strains associated with human toxoplasmosis in Spain: Direct analysis from clinical samples. J Clin Microbiol 2001;39:1566–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galván-Ramirez ML, Madriz Elisondo AL, Rico Torres CP, Luna-Pastén H, Rodríguez Pérez LR, Rincón-Sánchez AR, Franco R, Salazar-Montes A, Correa D. Frequency of Toxoplasma gondii in pork meat in Ocotlán, Jalisco, Mexico. J Food Prot 2010;73:1121–1123 [DOI] [PubMed] [Google Scholar]
- Genchi M, Vismarra A, Mangia, Faccini S, Vicari N, Rigamonti S, Prati P, Marino AM, Kramer L, Fabbi M. Lack of viable parasites in cured “Parma Ham” (PDO), following experimental Toxoplasma gondii infection of pigs. Food Microbiol 2017;66:157–164 [DOI] [PubMed] [Google Scholar]
- Grigg ME, Boothroyd JC. Rapid identification of virulent type I strains of the protozoan pathogen Toxoplasma gondii by PCR restriction fragment length polymorphism analysis at the B1 gene. J Clin Microbiol 2001;39:398–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Mishra A, Buchanan RL, Dubey JP, Hill DE, Gamble HR, Jones JL, Pradhan AK. A systematic meta-analysis of Toxoplasma gondii prevalence in food animals in the United States. Foodborne Pathog Dis 2016;13:109–118 [DOI] [PubMed] [Google Scholar]
- Herrero L, Gracia MJ, Pérez-Arquillué C, Lázaro R, Herrera A, Bayarri S. Toxoplasma gondii in raw and dry-cured ham: The influence of the curing proces. Food Microbiol 2017;65:213–220 [DOI] [PubMed] [Google Scholar]
- Hill DE, Benedetto SM, Coss C, McCrary JL, Fournet VM, Dubey JP. Effects of time and temperature on the viability of Toxoplasma gondii tissue cysts in enhanced pork loin. J Food Prot 2006;69:1961–1965 [DOI] [PubMed] [Google Scholar]
- Hill DE, Sreekumar C, Gamble HR, Dubey JP. Effect of commonly used enhancement solutions on the viability of Toxoplasma gondii tissue cysts in pork loin. J Food Prot 2004;67:2230–2233 [DOI] [PubMed] [Google Scholar]
- Howe DK, Sibley LD. Toxoplasma gondii comprises three clonal lineages: Correlation of parasite genotype with human disease. J Infect Dis 1995;172:1561–1566 [DOI] [PubMed] [Google Scholar]
- Jones JL, Dargelas V, Roberts J, Press C, Remington JS, Montoya JG. Risk factors for Toxoplasma gondii infection in the United States. Clin Infect Dis 2009;15:878–884 [DOI] [PubMed] [Google Scholar]
- Kapperud G, Jenum PA, Stray-Pedersen B, Melby KK, Eskild A, Eng J. Risk factors for Toxoplasma gondii infection in pregnancy. Results of a prospective case-control study in Norway. Am J Epidemiol 1996;144:405–412 [DOI] [PubMed] [Google Scholar]
- Khan A, Dubey JP, Su C, Ajioka JW, Rosenthal BM, Sibley LD. Genetic analyses of atypical Toxoplasma gondii strains reveal a fourth clonal lineage in North America. Int J Parasitol 2011;41:645–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kijlstra A, Eissen OA, Cornelissen J, Munniksma K, Eijck I, Kortbeek T. Toxoplasma gondii infection in animal-friendly pig production systems. Invest Ophthalmol Vis Sci 2004;45: 165–169 [DOI] [PubMed] [Google Scholar]
- Kijlstra A, Meerburg BG, Bos AP. Food safety in free-range and organic livestock systems: Risk management and responsibility. J Food Prot 2009;12:2448–2681 [DOI] [PubMed] [Google Scholar]
- Klun I, Djurkovic-Djakovic O, Katic-Radivojevic S, Nikolic A. Cross-sectional survey on Toxoplasma gondii infection in cattle, sheep and pigs in Serbia: Seroprevalence and risk factors. Vet Parasitol 2006;135:121–131 [DOI] [PubMed] [Google Scholar]
- Kuruca L, Klun I, Uzelac A, Nikolić A, Bobić B, Simin S, Lalošević V, Lalošević D, Djurković-Djaković O. Detection of Toxoplasma gondii in naturally infected domestic pigs in Northern Serbia. Parasitol Res 2017;116:3117–3123 [DOI] [PubMed] [Google Scholar]
- Lass A, Pietkiewicz H, Szostakowska B, Myjak P. The first detection of Toxoplasma gondii DNA in environmental fruits and vegetables samples. Eur J Clin Microbiol Infect Dis 2012;31:1101–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann T, Graham DH, Dahl E. Transmission dynamics of Toxoplasma gondii on a pig farm. Infect Gen Evol 2003;3:135–141 [DOI] [PubMed] [Google Scholar]
- Lehmann T, Marcet PL, Graham DH, Dahl ER, Dubey JP. Globalization and the population structure of Toxoplasma gondii. Proc Natl Acad Sci U S A 2006;103:11423–11428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limon G, Beauvais W, Dadios N, Villena I, Cockle C, Blaga R, Guitian J. Cross-sectional study of Toxoplasma gondii infection in pig farms in England. Foodborne Pathog Dis 2017;14:269–281 [DOI] [PubMed] [Google Scholar]
- Lin MH, Chen TC, Kuo TT, Tseng CC, Tseng CP. Real-time PCR for quantitative detection of Toxoplasma gondii. J Clin Microbiol 2000;38:4121–4125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancianti F, Nardoni S, D'Ascenzi C, Pedonese F, Mugnaini L, Franco F, Papini R. Seroprevalence, detection of DNA in blood and milk, and genotyping of Toxoplasma gondii in a goat population in Italy. BioMed Res Int 2013;2013: Article ID:905326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annual Report of Infections with Selected Infectious Diseases in Poland from 1 January to 31 December 2017 and in the Comparable Period of 2016. National Institute of Public Health, Department of Epidemiology and Surveillance of Infectious Diseases, Laboratory of Monitoring and Epidemiological Analysis, Warsaw, Poland. http://wwwold.pzh.gov.pl/oldpage/epimeld/2017/INF_17_12B.pdf Accessed October29, 2018
- Nowakowska D, Colón I, Remington JS, Grigg M, Golab E, Wilczynski J, Sibley LD. Genotyping of Toxoplasma gondii by multiplex PCR and peptide-based serological testing of samples from infants in Poland diagnosed with congenital toxoplasmosis. J Clin Microbiol 2006a;44:1382–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowakowska D, Stray-Pedersen B, Spiewak E, Sobala W, Małafiej E, Wilczyński J. Prevalence and estimated incidence of Toxoplasma infection among pregnant women in Poland: A decreesing trend in the younger population. Clin Microbiol Infect 2006b;12:913–917 [DOI] [PubMed] [Google Scholar]
- Pena HFJ, Gennari SM, Dubey JP, Su C. Population structure and mouse-virulence of Toxoplasma gondii in Brazil. Int J Parasitol 2008;38:561–569 [DOI] [PubMed] [Google Scholar]
- Richomme C, Aubert D, Gilot-Fromont E, Ajzenberg D, Mercier A, Ducrot C, Ferté H, Delorme D, Villena I. Genetic characterization of Toxoplasma gondii from wild boar (Sus scrofa) in France. Vet Parasitol 2009;164:296–300 [DOI] [PubMed] [Google Scholar]
- Sroka J, Karamon J, Bilska-Zając E, Stojecki K, Różycki M, Kusyk P, Wójcik-Fatla A, Zając V, Cencek T. Study on prevalence of Toxoplasma gondii infection in free-living animals in Poland. Ann Parasitol 2016;(62 Suppl):36 [Google Scholar]
- Sroka J, Karamon J, Cencek T, Dutkiewicz J. Preliminary assessment of usefulness of cELISA test for screening pig and cattle populations for presence of antibodies against Toxoplasma gondii. Ann Agric Environ Med 2011;18:335–339 [PubMed] [Google Scholar]
- Sroka J, Kusyk P, Bilska-Zając E, Karamon J, Dutkiewicz J, Wójcik-Fatla A, Zając V, Stojecki K, Różycki M, Cencek T. Seroprevalence of Toxoplasma gondii infection in goats from the south-west region of Poland and the detection of T. gondii DNA in goat milk. Folia Parasitol 2017;64:023. [DOI] [PubMed] [Google Scholar]
- Su C, Shwab EK, Zhou P, Zhu XQ, Dubey JP. Moving towards an integrated approach to molecular detection and identification of Toxoplasma gondii. Parasitology 2010;137:1–11 [DOI] [PubMed] [Google Scholar]
- Tenter AM, Heckeroth AR, Weiss LM. Toxoplasma gondii: From animals to humans. Int J Parasitol 2000;30:1217–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turčeková L, Antolová D, Reiterová K, Spišák F. Occurrence and genetic characterization of Toxoplasma gondii in naturally infected pigs. Acta Parasitol 2013;58:361–366 [DOI] [PubMed] [Google Scholar]
- van der Giessen J, Fonville M, Bouwknegt M, Langelaar M, Vollema A. Seroprevalence of Trichinella spiralis and Toxoplasma gondii in pigs from different housing systems in The Netherlands. Vet Parasitol 2007;148:371–374 [DOI] [PubMed] [Google Scholar]
- Verin R, Mugnaini L, Nardoni S, Papini R, Ariti G, Poli A, Mancianti F. Serologic, molecular and pathologic survey of Toxoplasma gondii infection in freeranging red foxes (Vulpes vulpes) in Central Italy. J Wildl Dis 2013;49:545–551 [DOI] [PubMed] [Google Scholar]
- Vergara A, Marangi M, Caradonna T, Pennisi L, Paludi D, Papini R, Ianieri A, Giangaspero A, Normanno G. Toxoplasma gondii lineages circulating in slaughtered industrial pigs and potential risk for consumers. J Food Prot 2018;81:1373–1378 [DOI] [PubMed] [Google Scholar]
- Warnekulasuriya MR, Johnson JD, Holliman RE. Detection of Toxoplasma gondii in cured meats. Int J Food Microbiol 1998;45:211–215 [DOI] [PubMed] [Google Scholar]
- Weigel RM, Dubey JP, Siegel AM, Kitron UD, Manelli A, Mitchell MA, Mateus-Pinilla NE, Thulliez P, Shen SK, Kwok OC, Todd KS. Risk factors for transmission of Toxoplasma gondii on swine farms in Illinois. J Parasitol 1995;81:723–729 [PubMed] [Google Scholar]
- WHO. WHO Estimates of the Global Burden of Foodborne Disease: Foodborne Disease Burden Epidemiology Reference Group 2007–2015. Geneva, Switzerland: World Health Organization, 2015 [Google Scholar]
- Xiao J, Yolken RH. Strain hypothesis of Toxoplasma gondii infection on the outcome of human disease. Acta Physiol 2015;213:828–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zając V, Wójcik-Fatla A, Sawczyn A, Cisak E, Sroka J, Kloc A, Zając Z, Buczek A, Dutkiewicz J, Bartosik K. Prevalence of infections and co-infections with 6 pathogens in Dermacentor reticulatus ticks collected in eastern Poland. Ann Agric Environ Med 2017;24:26–32 [DOI] [PubMed] [Google Scholar]
- Zia-Ali N, Fazaeli A, Khoramizadeh M, Ajzenberg D, Dardé M, Keshavarz-Valian H. Isolation and molecular characterization of Toxoplasma gondii strains from different hosts in Iran. Parasitol Res 2007;101:111–115 [DOI] [PubMed] [Google Scholar]