Abstract
Transport of small molecules across the cell membrane is a crucial biological mechanism for the maintenance of the cell activity. ABC transporter family is a huge group in the transporter membrane proteins and actively transports the substrates using the energy derived from ATP hydrolysis. In humans, there are 48 distinct genes for ABC transporters. A variation of a single amino acid in the amino acid sequence of ABC transporter has been known to be linked with certain disease. The mechanism of the onset of the disease by the variation is, however, still unclear. Recent progress in the method to measure the structures of huge membrane proteins has enabled determination of the 3D structures of ABC transporters and the accumulation of coordinate data of ABC transporter has enabled us to obtain clues for the onset of the disease caused by a single variation of amino acid residue. We compared the structures of ABC transporter in apo and ATP-binding forms and found a possible conformation shift around pivot-like residues in the transmembrane domains. When this conformation change in ABC transporter and the location of pathogenic variation were compared, we found a reasonable match between the two, explaining the onset of the disease by the variation. They likely cause impairment of the pivot-like movement, weakening of ATP binding and weakening of membrane surface interactions. These findings will give a new interpretation of the variations on ABC transporter genes and pave a way to analyse the effect of variation on protein structure and function.
Keywords: coupling helix, differential map, pathogenic variation, pivot-like residue, protein 3D structure
Significance.
ABC transporter is known to be involved in multidrug transport and its function relates to drug resistance. Nucleotide variations in ABC transporter genes found in human genomes may relate to different responses to drugs, but only a few studies have been conducted on the relationship between the atomic function of the protein and the variations. We took a database-driven approach to address this relationship and found that variations at pivot-like, ATP-binding and membrane-surface regions tend to be annotated as pathogenic variation in the databases.
Transport of small molecules across the cell membrane is a fundamental biological mechanism for the maintenance of the cell activity [1]. The transport is mainly handled by a group of membrane proteins including channels, solute carrier (SLC) transporters and ATP-binding cassette (ABC) transporters [2]. Channel proteins passively transport ions and water molecules by diffusion [3], SLC transporters carry small molecules along with conformational changes in the transporter without ATP hydrolysis [4], and ABC transporter proteins actively transport the substrates using the energy derived from ATP hydrolysis [5]. A protein family of ABC transporters consists of homologous proteins and they share a nucleotide-binding domain (NBD) where ATP molecule binds and is hydrolysed [6]. Based on the sequence similarity, ABC transporter family is divided into seven sub-families, named ABCA, ABCB, ABCC, ABCD, ABCE, ABCF, and ABCG (Table 1). Of them, ABCA, ABCB, ABCC, ABCD and ABCG have similar transmembrane domains (TMDs), but ABCE and ABCF do not have TMDs and are not involved in membrane transport [7]. The evolution of the ABC transporter family appears to be complicated, because the number of domains amongst the sub-families as well as in each sub-family is different [8]. In humans, there are 48 distinct ABC transporters, which belong to one of the sub-families [9].
Table 1.
All known human ABC transporters
| Family | Name | ENSEMBL gene | UniProt | Length | Function | 3D structure in use in this study |
|---|---|---|---|---|---|---|
| ABCA | ABCA1 | ENSG00000165029 | Q9UN09 | 2261 | sulfonylurea-sensitive anion transporter | 5xjy (apo form[human ABCA1])[35] |
| ABCA2 | ENSG00000107331 | Q9BZC7 | 2435 | putative transporter | ||
| ABCA3 | ENSG00000167972 | Q99758 | 1704 | putative cholesterol transporter | ||
| ABCA4 | ENSG00000198691 | P78363 | 2273 | inward-directed retinoid flipase in the visual cycle | ||
| ABCA5 | ENSG00000154265 | Q8WWZ7 | 1642 | autolysosome | ||
| ABCA6 | ENSG00000154262 | Q8N139 | 1617 | macrophage lipid homeostasis | ||
| ABCA7 | ENSG00000064687 | Q8IZY2 | 2146 | mcrophage phagocytosis | ||
| ABCA8 | ENSG00000141338 | O94911 | 1581 | lipophilic drug transporter | ||
| ABCA9 | ENSG00000154258 | Q8IUA7 | 1624 | monocyte differentiation | ||
| ABCA10 | ENSG00000154263 | Q8WWZ4 | 1543 | macrophage lipid homeostasis | ||
| ABCA12 | ENSG00000144452 | Q86UK0 | 2595 | lipid homeostasis | ||
| ABCA13 | ENSG00000179869 | Q8N248 | 5058 | lipid homeostasis | ||
|
| ||||||
| ABCB | ABCB1 | ENSG00000085563 | P08183 | 1280 | efflux pump in multidrug-resistant cells | 5ko2 (apo form[mouse ABCB4])[36] 4xwk (inhibitor bound[mouse ABCB4])[37] 5och (ADP bound[human ABCB8]) 6c0v (ATP bound[human ABCB1])[38] |
| TAP1 | ENSG00000168394 | Q03518 | 808 | (heterodimer with TAP2) antigens transporter to ER | ||
| TAP2 | ENSG00000204267 | Q03519 | 686 | (heterodimer with TAP1) antigens transporter to ER | ||
| ABCB4 | ENSG00000005471 | P21439 | 1286 | phospholipid efflux translocator. function as floppase | ||
| ABCB5 | ENSG00000004846 | Q2M3I5 | 1257 | drug efflux transporter | ||
| ABCB6 | ENSG00000115657 | Q9NP58 | 842 | (homodimer) heme and porphyrin uptake to mitochondrion | ||
| ABCB7 | ENSG00000131269 | O75027 | 752 | (homodimer) heme transport from mitochondrion to cytosol | ||
| ABCB8 | ENSG00000197150 | Q9NUT2 | 735 | (homodimer) multidrug resistance exporter at mitochondrion | ||
| ABCB9 | ENSG00000150967 | Q9NP78 | 766 | (homodimer) low-affinity peptide transporter | ||
| ABCB10 | ENSG00000135776 | Q9NRK6 | 738 | (homodimer) mitochondrial transport for heme biosynthesis | ||
| ABCB11 | ENSG00000073734 | O95342 | 1321 | secretion of bile salts | ||
|
| ||||||
| ABCC | ABCC1 | ENSG00000103222 | Q9UQ99 | 1531 | export of organic anions and drugs from the cytoplasm | 5wua (apo form[hamster ABCC8])[26] 5ywc (ADP bound[hamster ABCC8])[39] 5w81 (ATP bound[zebrafish CFTR])[40] |
| ABCC2 | ENSG00000023839 | Q92887 | 1545 | hepatobiliary excretion of numerous organic anions | ||
| ABCC3 | ENSG00000108846 | O95290 | 1527 | transporter in the intestinal excretion of anions | ||
| ABCC4 | ENSG00000125257 | O15439 | 1325 | organic anion pump | ||
| ABCC5 | ENSG00000114770 | Q9UNP5 | 1437 | multispecific organic anion pump | ||
| ABCC6 | ENSG00000091262 | O95255 | 1503 | transport of drugs into subcellular organelles | ||
| CFTR | ENSG00000001626 | P13569 | 1480 | epithelial ion channel | ||
| ABCC8 | ENSG00000006071 | Q09428 | 1581 | subunit of ATP-sensitive potassium channel | ||
| ABCC9 | ENSG00000069431 | O60706 | 1549 | subunit of ATP-sensitive potassium channel | ||
| ABCC10 | ENSG00000124574 | Q5T3U5 | 1492 | lipophilic anion extrusion | ||
| ABCC11 | ENSG00000121270 | Q96J66 | 1382 | enhance the cellular extrusion of cAMP and cGMP | ||
| ABCC12 | ENSG00000140798 | Q96J65 | 1359 | putative transporter | ||
|
| ||||||
| ABCD | ABCD1 | ENSG00000101986 | P33897 | 745 | putative transporter | |
| ABCD2 | ENSG00000173208 | Q9UBJ2 | 740 | putative transporter | ||
| ABCD3 | ENSG00000117528 | P28288 | 659 | branched-chain fatty acid transporter | ||
| ABCD4 | ENSG00000119688 | O14678 | 606 | intracellular processing of vitamin D | ||
|
| ||||||
| ABCE | ABCE1 | ENSG00000164163 | P61221 | 599 | RNase L inhibitor (no TMD) | |
|
| ||||||
| ABCF | ABCF1 | ENSG00000204574 | Q8NE71 | 845 | mRNA translation initiation (no TMD) | |
| ABCF2 | ENSG00000033050 | Q9UG63 | 623 | unknown (no TMD) | ||
| ABCF3 | ENSG00000161204 | Q9NUQ8 | 709 | antiviral effect against flaviviruses (no TMD) | ||
|
| ||||||
| ABCG | ABCG1 | ENSG00000160179 | Q9BXL1 | 678 | macrophage lipid homeostasis | 5do7 (apo form[human ABCG5/ABCG8])[27] 6hbu (ATP bound[human ABCG2])[41] |
| ABCG2 | ENSG00000118777 | Q86V64 | 655 | High-capacity urate exporter | ||
| ABCG4 | ENSG00000172350 | Q9H172 | 646 | macrophage lipid homeostasis | ||
| ABCG5 | ENSG00000138075 | Q9H222 | 651 | sterol transporter | ||
| ABCG8 | ENSG00000143921 | Q9H221 | 673 | sterol transporter | ||
ABC transporters are known as causative agents of inherited diseases. A variation of a single amino acid type in ABC transporter is annotated as a cause of a certain disease in ClinVar database [10]. ABC transporters are also known as an obstacle for drug therapy, because the transporters pump a broad range of chemical substances out of the cell including cancer chemotherapeutics [11]. Therefore, there is a high demand in the community studying ABC transporters for the combinations of the mechanisms of function/malfunction and disease to understand the molecular and atomic mechanisms causing diseases [12]. The mechanical detail of the function and the variations in the disease has not been combined, because the three-dimensional (3D) structures of ABC transporters have been difficult to obtain.
Recent progress in the method to determine the protein structures, especially the structure of membrane proteins, by electron microscopy (EM) has enabled determination of the 3D structures of ABC transporters [13]. EM measurement of protein structures is pushing up the number of 3D structures in Protein Data Bank (PDB) [14] including the number of ABC transporters. Hence the recent increase in the structural data of ABC transporters has enabled us to compare the structures, especially in different states. The structures of ABC transporter are now determined in a number of different conditions, namely apo form, ATP-binding form, ADP-binding form and inhibitor-binding form.
In this study, we compared the structure of ABC transporters in different states and deduced the conformation shift upon ATP or inhibitor binding. The locations of key residues for conformation shift and for biological function were compared with the positions of known variations on the amino acid sequence. We tried to elucidate a possible cause-and-effect relationship between the variations and the onset of diseases.
Methods
Search for human ABC transporter from ENSEMBL, UniProt, gnomAD and PDB
The DNA and amino acid sequence data including the chromosome location of human ABC transporters were extracted from ENSEMBL and UniProt databases [15,16]. Based on the chromosome location, all the variations were extracted from gnomAD database [17]. The variation data in gnomAD including the relationship between the variations and diseases had been synchronized with the data in ClinVar [10]. Hence the variation data in this study were originated in ClinVar. The 3D structures of ABC transporters were extracted from PDB [14]. To obtain the whole entries of ABC transporters, we used all the amino acid sequences of ABC transporters obtained from ENSEMBL and performed homology search by BLAST [18] locally. In addition, we performed an online homology search on PDBj [19], so as not to miss the recent deposition of the coordinate in PDB.
Detecting ATP-binding residues
ATP-binding residue was detected by the method employed by Kobayashi et al. [20]. Briefly, accessible surface area of each atom of the ATP-bound ABC transporter was calculated by in-house program. The ATP molecule was, then, eliminated from the coordinate set and the accessible surface area was calculated again. An atom with two different values was, then, defined as an ATP-binding atom and the residue with the atom was defined as ATP-binding residue.
Differential map
Two different coordinates of ABC transporter were compared using differential map, which corresponds to the subtraction of two distance maps of the ABC transporter. The similar map was previously used in identifying domain movement under the name of difference map [21]. A distance map was an isosceles right triangular map with the amino acid residue numbers on two right sides. The distance between amino acid residue i and amino acid residue j, which was defined as the distance between two Cα atoms, was mapped on the intersection of i and j inside the triangle [22]. The distance map based on conformation A of ABC transporter was subtracted from the distance map based on conformation B in the following manner;
where dij is the distance between residues i and j, and the correspondence of amino acid residues i and j in different conformations of ATP transporter was taken based on the amino acid sequence alignment. Ideally conformations of the same sequence should be used for the comparison, but the real data often had variations in the sequences including proteins from different species, hence sequence alignment was required and a gap may exist in the map. As far as unambiguous correspondence could be obtained, we used the 3D structures derived from species other than human. The range of ΔD was limited between −10 Å and 10 Å and depicted in the map using blue to white gradation in minus values and white to red in plus values.
Differential map can identify a change in conformation by measuring the relative locations of the residues. Superposition of two proteins are often used for comparing the structures, but conformation changes determined by the super-position method depend on the sites of being superimposed. Superposition with different regions ends in different results which may miss the real conformation change. Differential map has no ambiguity in this sense, but the results tend to be qualitative. Therefore, differential map and superposition method can be complementary. Differential map can identify constant regions in two proteins and superposition method can be performed based on the constant regions to quantify conformation changes. We, however, introduce a differential plot, another quantification method in the following section.
Differential plot
The dimension of the differential map was reduced in the following manner to obtain the differential plot;
where N is the number of ΔD from residue i. Ideally when ΔWi is zero, the relative location of residue i remains the same in the protein during the conformational change. The residue is then a candidate for a pivot in the conformational change (possibly rotation) in the internal dynamics of the protein, if there is such dynamics. If ΔWi is in the local minimum, then residue i is a candidate of the local centre of the conformational change. For the real data, identification of the minimum site was not straight forward. We visually located the low value region from the differential plot and found the least value within the range.
Results and Discussion
ABC transporters of human
The results of database search were summarized in Table 1. As already known, there were 48 independent ABC transporter genes in human genome and they were classified into seven sub-families [7]. ABCE and ABCF did not have TMD and were highly diverged from other sub-families. Therefore, we omitted these sub-families from further analyses. Sub-families ABCA and ABCC were monomer proteins with two TMDs and NBDs, whereas ABCD and ABCG were homo- or hetero-dimer proteins and each subunit had one TMD and one NBD. ABCB contained both monomer type and dimer type transporters. The order of domains along the amino acid sequence of ABCG differed from the others. Majority of the proteins had the order of TMD-NBD-TMD-NBD, whereas ABCG alone had the order of NBD-TMD. At the time of the submission of this manuscript, 3D structure of only a single state was registered in ABCA, structures of multiple states were known in ABCB, ABCC and ABCG, and no structure of ABCD was available.
Benign and pathogenic variations in ABC transporters
We extracted variation information of human ABC transporters from ClinVar and found 30,384 missense variations. Out of these variations, 201 were annotated as benign, 407 were as pathogenic and the rest as unknown. We used the known variations (benign and pathogenic variations) here-after. Frequently observed types of variations in those benign and pathogenic variations were summarized in Table 2, in descending order in the form of major amino acid to variant. One of the differences between the benign and pathogenic variations was the frequent appearance of arginine in pathogenic variation as an original amino acid (bold font in the table). In benign variations, 16% of the variations was originated in arginine, whereas in pathogenic variations, 28% of the variations was from arginine to different amino acid types. Log2-odds values of the pathogenic variations indicated that except for Arg-Gln variation, most of the other variations were twice as frequent as the one in benign case. The alteration from arginine to other amino acid types seemed to have an impact on the function of ABC transporter to a great extent. We previously found the similar trend in SLC transporter proteins and discussed that they were frequently found close to the surface of the membrane [23]. In the case of ABC transporter, however, these arginine residues were not necessarily located close to the presumptive surface of the membrane as we discuss below. The mechanisms leading to the disease by the same type of variations seemed to vary in different proteins.
Table 2.
Variation types in ABC transporters
| benign | pathogenic | |||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| variation | # | % | variation | # | % | log2-odds* |
| Arg-Gln | 13 | 6.5 | Arg-Trp | 23 | 5.7 | 0.7 |
| Val-Ile | 13 | 6.5 | Arg-His | 22 | 5.4 | 1.44 |
| Ala-Thr | 8 | 4.0 | Arg-Gln | 22 | 5.4 | −0.25 |
| Arg-Trp | 7 | 3.5 | Arg-Cys | 20 | 4.9 | 1.3 |
| Ile-Val | 7 | 3.5 | Gly-Arg | 15 | 3.7 | 1.3 |
| Ala-Val | 6 | 3.0 | Glu-Lys | 15 | 3.7 | 0.89 |
| Ile-Met | 5 | 2.5 | Leu-Pro | 11 | 2.7 | 2.44 |
| Val-Met | 5 | 2.5 | Arg-Gly | 10 | 2.5 | 1.3 |
| Val-Ala | 5 | 2.5 | Ala-Val | 10 | 2.5 | −0.29 |
| Arg-Leu | 4 | 2.0 | Arg-Pro | 9 | 2.2 | 2.15 |
| Arg-Cys | 4 | 2.0 | Trp-Cys | 9 | 2.2 | 2.15 |
| Arg-His | 4 | 2.0 | Arg-Leu | 9 | 2.2 | 0.15 |
| Glu-Lys | 4 | 2.0 | ||||
| Leu-Phe | 4 | 2.0 | ||||
| Pro-Leu | 4 | 2.0 | ||||
| Thr-Met | 4 | 2.0 | ||||
| Val-Leu | 4 | 2.0 | ||||
log2-odds was calculated by the logarithm (base 2) of the percentage of pathogenic variation over the percentage of benign variation in the same variation type.
Location of variations on ABCA
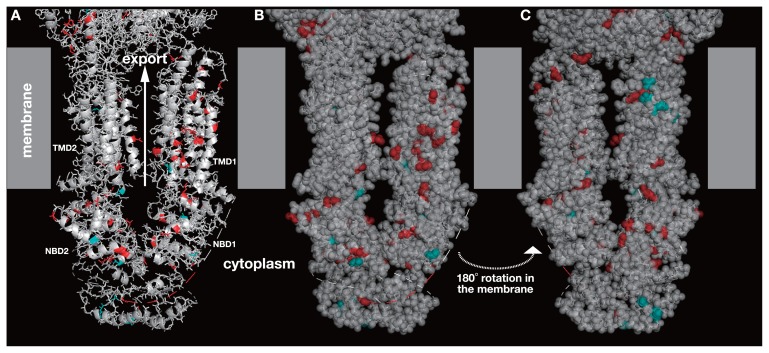
All the missense variations on human ABCA genes were mapped on to the 3D structure of ABCA protein (Fig. 1). Residues in red were the locations of pathogenic variations and residues in cyan were the locations of benign variations. The pathogenic variations were seemingly located all over the proteins except for TMD2 and benign variations were located mostly on two NBDs. The variation mapping, however, did not show any difference between pathogenic and benign variations leading to a different effect in protein function.
Figure 1.
3D structure of human ABCA1 (PDB ID: 5xyj). Grey boxes are the presumptive location of the membrane. The top side is the extracellular and the bottom side is the cytosolic spaces. ABCA1 has a huge extracellular domain, but the domain is only partially shown. Amino acid position in red is the location of pathogenic variation and in cyan is that of benign variation. The structure is shown by ribbon model in A, by ribbon model with translucent space-filling model in B, and the other side of B in C. TMD stands for transmembrane domain and NBD for nucleotide-binding domain. TMD1 and NBD1 are the N-terminal domains, and TMD2 and NBD2 are the C-terminal domains.
Conformation shift and location of variations on ABCB
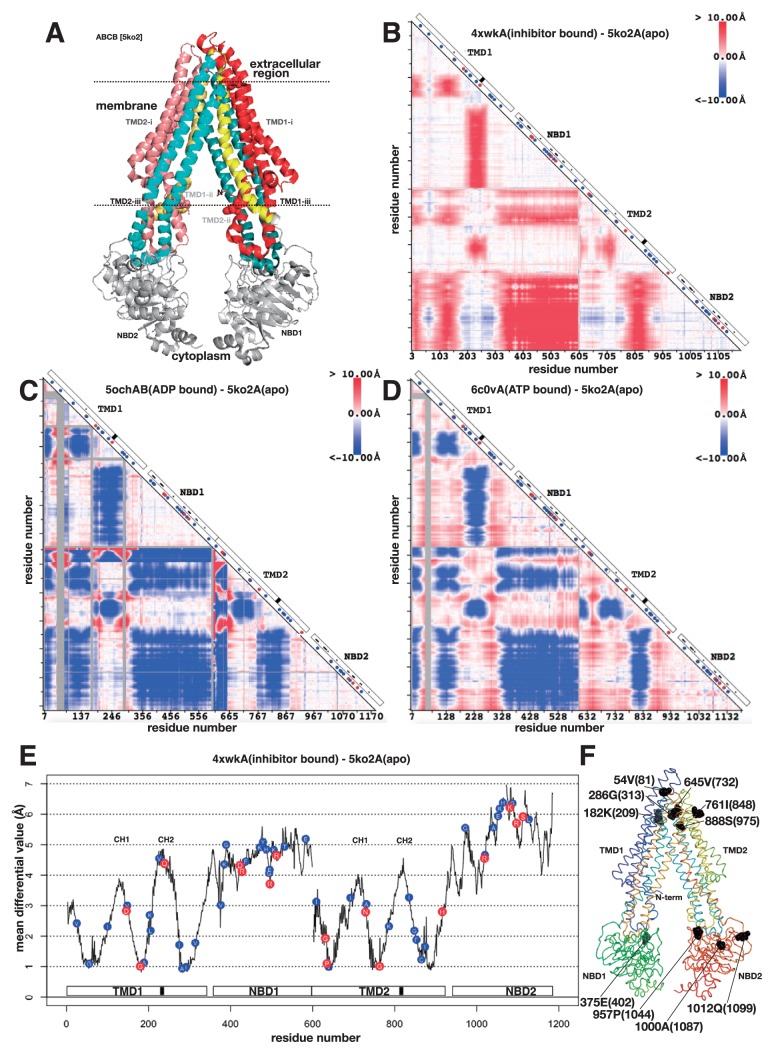
Quite a number of 3D structures have been determined in ABCB and we selected four structures in different states derived from humans or closely related species in relatively high resolution (Table 1). One of the structures is shown in Figure 2A. The overall structure of ABC transporter subfamily B was similar to that of sub-family A, but the structure shown in Figure 2A was in the pose that two NBDs were separated. NBDs of sub-family A in Figure 1 were in contact. Comparison of two structures from sub-family B is shown in Figure 2B using the differential map. The structure of mouse P-glycoprotein (ABCB4) with an inhibitor (PDB ID: 4xwk) was compared against the same protein in apo form (PDB ID: 5ko2). The numbering in the axes is irregular, because of some missing residues in the structure data. This differential map was derived from the subtraction of two distance maps drawn on 4xwk and 5ko2. Ideally the correspondence of two distance maps should be complete, because both coordinate data were derived from the same protein, however there were some residues missing in the data, and hence there were some gaps in the differential map. In the case of Figure 2B, the first two residues (residues 1 and 2) and two residues in the middle (residues 600 and 601) were missing in 4xwk, hence the map has two gaps at the corresponding positions. In this study, the numbering of the residues in the 3D structure was based on the observed Cα atoms. The residue with the first Cα atom from the N-terminal side was annotated as residue 1, the one with the second Cα atom was annotated as residue 2 and the same thereafter.
Figure 2.
3D structure, differential map and differential plot of sub-family B of ABC transporters. A. 3D structure of mouse P-glycoprotein (ABCB4) (PDB ID: 5ko2). See text for the colouring of the protein. Dotted lines are the putative surfaces of the membrane. Seven residues are in space-filling model. They are the locations of certain pathogenic variations. B. Differential map between mouse P-glycoprotein with an inhibitor (PDB ID: 4xwk) and apo form of mouse P-glycoprotein (PDB ID: 5ko2). The horizontal axis from left to right and the vertical axis from top to bottom are the residue number of P-glycoprotein. The difference of the distances between residues i and j (measured between Cα atoms) in two 3D structures of P-glycoproteins is colour-coded at the intersection in the triangular map. Red colour means that the distance of the pair of residues became longer, and blue colour means shorter. On the diagonal axis, a white box indicates domain organization, a black box indicates a location of the second coupling helix (see text), a small dot in the white box indicates ATP-binding residue, a blue circle indicates a location of benign variation, a red circle indicates a location of pathogenic variation, and a white circle indicates a location of conflicting variation. C. The same as B, the differential map between human ABCB8 with ADP molecule (PDB ID: 5ocha) and mouse P-glycoprotein (PDB ID: 5ko2). Grey colour in the triangle indicates an insertion region in human ABCB8 compared with mouse P-glycoprotein. D. The differential map between human P-glycoprotein in ATP-binding outward-facing conformation (PDB ID: 6c0v) and mouse P-glycoprotein (PDB ID: 5ko2). E. Differential plot based on B. Horizontal axis is the residue number and the vertical axis is the mean differential value (). A white box on the horizontal axis shows domain organization and a black box is the location of the second coupling helix. A blue circle with amino acid one letter code on the plot is the position of benign variation and red circle is the position of pathogenic variation. Those variations were derived from the ones on ABCB genes. CH1 and CH2 indicate the locations of the first and the second coupling helices, respectively, defined in refs. 24 and 25. F. Ribbon model of mouse P-glycoprotein protein, the flip side of A. The chain trace is shown in rainbow colour. Ten residues in space-filling model are located in the local minima of the differential plot in E. The residue is identified by the sequential number (equal to the horizontal axis of E), amino acid one letter code and the residue number (the number from the N-terminus of the contiguous sequence).
One of the outstanding patterns in Figure 2B was a clear separation around 600, which separated the N-terminal domains (TMD1 plus NBD1) and C-terminal domains (TMD2 plus NBD2). The N-terminal two domains moved collectively against the C-terminal two domains. The rhombus region on the diagonal axis around 600 are mostly red, indicating that the sets of domains were separated by inhibitor binding. In Figure 2A, this motion corresponds to the horizontal separation between TMD1+NBD1 and TMD2+NBD2. The two sets of domains are, however, connected at the extracellular region. Hence the separation motion is conjectured to be a rotation motion around a pivot located in the extracellular region.
The second outstanding pattern was found in each set of domains. Both TMD1+NBD1 and TMD2+NBD2 were separated into three sections based on the colour pattern in the differential map; the first part of TMD (from 1 to 182, and from 600 to 764 in 5ko2), the middle of TMD (from 183 to 291, and from 765 to 876 in 5ko2), and the last part of TMD (from 292 to 341, and from 877 to 927 in 5ko2) plus NBDs (Numbering method here was explained above). This separation of TMD was mapped in Figure 2A; TMD-i, the first part of TMD is in redish colour, TMD-ii, the middle of TMD is in blueish colour, and the last part of TMD is in yellowish colour (Fig. 2A). Note that TMD-ii performed a domain swapping. TMD-ii conducted a motion separating from TMD-i and from TMD-iii, and reduced the distance to NBDs, which was confirmed by a red belt in TMD+NBD in Figure 2B. By inhibitor binding to P-glycoprotein, TMD-ii seemed to be squeezed out from the bundle of TMD-i and TMD-iii to some extent and located closer to NBD. TMD-ii contained one of the short cytoplasmic helices that is almost parallel to the membrane surface. The helix seemed to play an interface role between TMD and NBD, hence the helix has been termed coupling helix [24]. Dawson, R. J. P. and Locher, K. P. found two coupling helices in each TMD. The one that we found here was the second coupling helix in each TMD. The location of the second coupling helix in each TMD is shown by a black box in the domain diagram on the diagonal axis of the map. TMD-ii apparently played the role of transferring the information of conformation shift in TMD to NBD.
The conformation shift observed in the comparison between the apo and inhibitor-binding forms of P-glycoprotein was also observed in the comparison between the apo and ADP-binding form, but in the opposite direction (Fig. 2C). The ADP-bound form was derived from human ABCB8 (PDB ID: 5och). Human ABCB8 is a homodimer transporter, of which each subunit has one TMD and one NBD. For drawing the differential map in Figure 2C, we therefore concatenated the coordinate files of the two subunits and compared with apo form of mouse P-glycoprotein. The overall sequence identity was about 32%. The locations of the colour regions appearing on the differential map were very similar to the ones in Figure 2B, except for the colour, which means that the direction of the conformation shift was opposite between Figures 2B and 2C. In the case of ADP binding, the transporter was closed, namely, the distance between TMD1+NBD1 and TMD2+NBD2 was closer and the coupling helices were closer to NBDs. The same conformation shift was observed in the comparison between the apo and ATP-binding form (Fig. 2D). The ATP-binding form was derived from human ABCB1 (PDB ID: 6c0v). The overall sequence identity was about 89%. The locations of the colour regions as well as the colour spectra appearing in Figures 2C and 2D are very similar (Pearson’s correlation coefficient = 0.39). In the case of ADP/ATP binding, both molecules bound to NBD, and hence conformation change in NBDs should be transmitted to TMDs and the transporter should assume a closed conformation. Hence TMD-ii likely plays the role of information transfer and the coupling helices play the roles of interfaces between NBD and TMD.
In all the cases, the differential maps present the following two conformation shifts. 1) Increase/Decrease in the distance between TMD1+NBD1 and TMD2+NBD2. And 2) Increase/Decrease in the distance between TMD-ii and NBD. These two types of conformation shift in ABC transporter sub-family B were conjectured to be a rotation movement as discussed above. To clarify this conjecture, we further drew a differential plot (Fig. 2E and Supplementary Fig. 1). As explained in the method section, the differential plot is the mean of the absolute differential values in each residue plotted against the residue number. Figure 2E shows that TMD has lower values than NBD on average, indicating that conformation shift of NBD is greater than the shift of TMD. Though the magnitude of the shift was not so different in ATP/ADP-binding forms (Supplementary Fig. 1). Each TMD had three local minima. When the sum of absolute differential values is zero, then the relative location of the residue does not shift during the conformational change, hence the residue is a candidate of pivot of the conformation shift. We cannot expect such an ideal situation in the real proteins, but when the value is in a local minimum, the residue is the candidate of the pivot for the conformation shift and the existence of such low value suggests that the conformation shift is the rotation-like movement. Finding six low values in TMDs strongly suggests that the conformation shift in ABCB should be a hinge motion. The locations of these six residues in the 3D structure are shown in Figure 2F. Figure 2F showed the flip side of Figure 2A. Six residues on the top side in Figure 2F corresponds to the six minima of TMDs in Figure 2E. In NBDs, there are a number of local minima. At the N-terminal side of NBDs, there locate a couple of deep minima with the value around 3.0 Å. These residues are the candidates for pivot of local conformational change in NBD. A pivot for local conformational change may have great overall shift in the location, but small shift against the local group of residues. When the deep minima in NBDs (402, 1044, 1087 and 1099 in Fig. 2E) were mapped on the 3D structure, they were located at the neck of NBDs (Fig. 2F). These residues likely play the roles of the pivots for hinge-bending motion between TMD and NBD. In Figure 2E, the second coupling helices (black boxes in Fig. 2E) mark the greatest values in TMD. The coupling helices defined by Dawson R. J. P. and Locher, K. P. [24] were noted as CH1 and CH2 in Figure 2E. All of them mark the local maxima in TMD.
One may speculate that the mean differential values of the residues correlates with accessibility of the residues, but we found no correlation (correlation coefficient was about 0.15) between the two. Another reservation one may have is on the comparison of proteins derived from different species. Current data set could not allow the comparison of the different forms derived from the same species due to the paucity of data. But when ABC transporters from mouse and that from Staphylococcus (sequence identity was about 32%) in ADP-binding form were compared, the mean differential value was about 3.2 Å. The corresponding value for Figure 2C was 5.6 Å, much greater than the value of the same form from different species. This result suggests that the values we obtained above were mainly derived from the difference in the states rather than from the difference of the species.
We plotted a position of residue variation in humans on Figures 2B to 2E. A red dot indicates a location of variation that causes pathogenic condition and a blue dot indicates the location of variation that is benign. Blue dots seem to be located throughout the protein. The locations of red dots, however, can be classified into three regions, namely, TMD-ii, NBD, and others. The ones appearing in TMD-ii may hamper the information transfer between NBD and TMD. The ones appearing in NBDs were in fact close to ATP-binding residues that are shown by a small black dot on the diagonal axis in the diagram of Figures, 2B, C, and D, but only one of the variations was on the ATP-binding residue. The variation on ATP-binding residue may impair ATP binding to some extent leading to the impairment of the function of the transporter. If the impairment were too severe, then the function of the transporter should have been lost, hence the variation would have a lethal effect. But the effect of variation should be mild, because there was only one exact match to the ATP-binding residues and ATP binding is a collective function involving many residues (there are 53 dots in the figure). A single variation likely has a limited effect that mildly impairs the function of the transporter that does deprive the residue of the function. The ones appearing in other regions were in fact found either close to the pivots or the interface of the membrane surfaces. These residues with the variations had the mean differential value less than 3.0 Å (Fig. 2E). Three of them had the mean differential value close to 1.0 Å, hence they were likely the pivots for the hinge movement of the TMDs. Mild impairment of the pivot may have impact on the hinge movement and reduce the efficacy of the conformation shift in the transporter protein. The remaining four of them were located close to one of the presumptive surfaces of the membrane (Fig. 2A). These variations likely impair the stable interactions between the protein and the membrane.
Furuta, T. et al. analysed MsbA, ABC transporter of Escherichia coli, and found that the second coupling helix was significantly important for nodding-like motion of NBD based on wet-lab experiments and molecular dynamics simulation [25]. These coupling helices are shown as CH1 and CH2 in Figure 2E. The second coupling helix of MsbA corresponds to the coupling helix in the current analyses. In Figure 2E, the second coupling helix (CH2), undergoes greater conformation shift than the first coupling helix (CH1) and one of the second coupling helices had a pathogenic variation. The results from Figure 2E well corresponded with their results.
Conformation shift and location of variations on ABCC
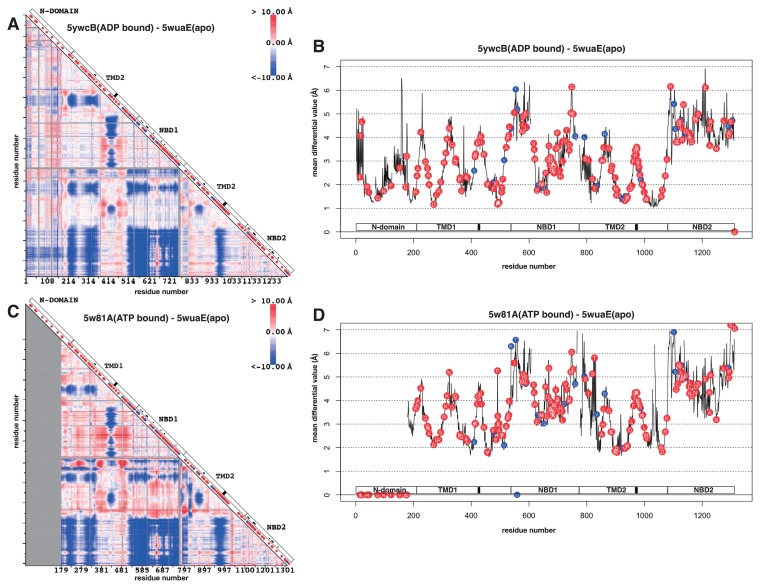
The conformation shift in ABCC was analysed between mouse sulfonylurea receptor SUR1 in apo form (PDB ID: 5wua) and the same protein in ADP-binding form (PDB ID: 5ywc), and between mouse SUR1 in apo form and zebrafish cystic fibrosis transmembrane conductance regulator (CFTR) in ATP-binding form (PDB ID: 5w81). These three structures were all determined by electron microscopy. The differential maps and the differential plots are shown in Figure 3. The regions in colour and the colour spectra of the differential maps (Figs. 3A and 3C) were surprisingly similar to the ones in ABCB (Figs. 2C and 2D). The conformation shifts found in ABCC were, 1) Decrease in the distance between TMD1+NBD1 and TMD2+NBD2, and 2) Decrease in the distance between the coupling helices and NBD. SUR1 had additional N-domain and the domain tended to behave concomitant to TMD1+NBD1. The differential plots for both differential maps (Figs. 3B and 3D) behaved similar to the plot of ABCB (Fig. 2E).
Figure 3.
Differential map and differential plot of sub-family C of ABC transporters. A. Differential map between mouse sulfonylurea receptor SUR1 in apo form (PDB ID: 5wua) and the same protein in ADP-binding form (PDB ID: 5ywc). See the figure caption of Figure 2B for the detail of the map. B. Differential plot based on A. See the figure caption of Figure 2E for the detail of the plot. C. Differential map between mouse sulfonylurea receptor SUR1 in apo form (PDB ID: 5wua) and zebrafish cystic fibrosis transmembrane conductance regulator (CFTR) in ATP-binding form (PDB ID: 5w81). D. Differential plot based on C. The circles located on the horizontal axis indicate that these residues were located on the gap of the alignment amongst human ABCCs, mouse SUR1 and zebrafish CFTR.
In ABCC, a huge number of variations in humans have been identified and most of the variations were pathogenic, especially on multi-specific organic anion transporter E (ABCC6) and cystic fibrosis transmembrane conductance regulator (ABCC7) [12]. When these variations were mapped on to the differential maps and plots, pathogenic variations appeared everywhere including the regions identified in ABCB (Fig. 3). This scattered distribution of the pathogenic variations on ABCC suggests that the way the ABCC realises function is quite different from the way of ABCB. The intolerance of variations in ABCC suggests that there should be many protein-protein interactions involved in ABCC. In fact, SUR1 works with potassium channel. SUR1 surrounds the channel and stabilises a closed conformation of the channel [26].
Conformation shift and location of variations on ABCG
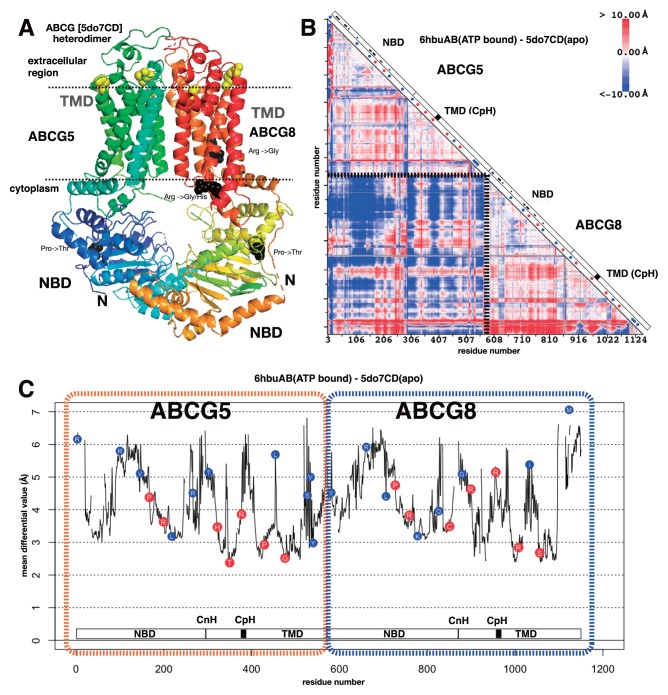
The overall structure of ABCG is different from the members in other ABC sub-families. ABCG is a dimer protein and each subunit correspond to the half of other ABC sub-family proteins. The sequential order of the domains in ABCG is also different from other ABC sub-families. ABCG starts with NBD followed by TMD, whereas other ABC proteins start with TMD followed by NBD. These differences are evidently reflected to the differences in static and dynamic conformations of ABCG from other ABC subfamilies. Figure 4A shows the static structure of human sterol transporter ABCG5/ABCG8 heterodimer. The TMDs are buried in relatively parallel against each other compared with TMDs in other ABC transporters (Fig. 2A). In addition, the length of helices in TMDs is shorter. The conformation shift in ABCG was analysed between human sterol transporter ABCG5/ABCG8 heterodimer in apo form (PDB ID: 5do7) and human ABCG2 in ATP-binding form (PDB ID: 6hbu). The sequence identity was about 27%. The subunits of each protein were concatenated so that shift in subunit location can also be detected in the map. As expected from the difference in the structures between ABCG and other ABC transporters, the coloured regions as well as the colour spectra of the differential map of ABCG are quite different from other ABC transporters (Fig. 4B). The rhombus region on the diagonal axis around the junction of two subunits (two edges are depicted by dotted lines) is mainly blue, which means that upon ATP binding, the distance between two subunits became closer. The regions corresponding to the coupling helix in other ABC transporters are indicated by black boxes in Figure 4B, which is named CpH by Lee, J.-Y. et al. [27], but no characteristic conformation shift was observed on the region in the differential map. Instead the characteristic conformation shift was found close to the junction of NBD and TMD, especially in ABCG5. This region corresponded to a helix connecting the domains and the helix lay parallel to the presumptive membrane surface. Lee, J.-Y. et al. [27] named this helix CnH. The mean differential values of the residues in CpH and CnH were high (Fig. 4C). This observation suggests that both CpH and CnH likely play the role of coupling helix in ABCG, as suggested by Lee, J.-Y. et al. [27]. Ferreira R. J. et al. performed a molecular dynamics calculation and showed that the loop between transmembrane helices 2 and 3 should substitute for the function of the coupling helix (thus they named ‘coupling loop’) [28]. The corresponding loop did not show a conformation shift characteristic to the coupling helix in our study. In either case, ABCG seems to have different information transmission pathway between NBD and TMD.
Figure 4.
3D structure, differential map and differential plot of sub-family G of ABC transporters. A. 3D structure of human sterol transporter ABCG5/ABCG8 heterodimer (PDB ID: 5do7). Dotted lines are the presumptive surfaces of the membrane. Four residues are in black space-filling model. They are located at the positions of pathogenic variation with mean differential value >4.0 Å. Five residues are in yellow space-filling model. They are located at the positions of pathogenic variation with mean differential value <3.0 Å. B. Differential map between human sterol transporter ABCG5/ABCG8 heterodimer in apo form (PDB ID: 5do7) and human ABCG2 in ATP-binding form (PDB ID: 6hbu). The dotted line on the map indicates the separation of two subunits. See the figure caption of Figure 2B for the detail of the map. CpH indicates the location of the coupling helix defined in ref. 27. C. Differential plot based on B. Two subunits (ABCG5 and ABCG8) were concatenated and shown in a single graph. See the figure caption of Figure 2E for the detail of the plot. CnH and CpH indicate the locations of the connecting helix and coupling helix, respectively, defined in ref. 27.
The differential plot of ABCG is shown in Figure 4C. Quite different from the differential plots of other subfamilies, the minimum values did not reach around 1.0 Å and hence there existed no global pivot residues in the conformation shift of ABCG upon ATP binding. However, five pathogenic variations were still located close to the local minimum of the plot. These positions are visualized in Figure 4A with a space-filling model in yellow. All of them are on the tip of the transmembrane helices on the side of extracellular region. This result suggests that the TMDs of ABCG should undergo a hinge motion upon ATP binding as in other ABC transporters, yet the extent of the motion is limited to TMDs at most, and that the mild impairment of the hinge motion by variation at or close to the pivot residue should lead to a decline in the activity of ABCG, which onsets the disease. Ferreira R. J. et al. [28] performed a molecular dynamics simulation on ABCG protein to gain insights for the mechanisms of the pump and they mainly analysed the dynamics of NBD where ATP bound, but the result in their Figure 8A showed a clear conformation shift motion at the extracellular regions where we identified pivot-like residues for hinge motions and pathogenic variations. Pathogenic variations located at the position with mean differential value >4.0 Å is also mapped in Figure 4A with a black space-filling model. These pathogenic variations were located either in NBD or close to the presumptive cytoplasmic interface of the membrane. Especially the two variations, changing Arg to other types of amino acid on the surface of the membrane, likely destabilise the interactions between the TMD and the membrane, and hence likely destabilise the position or the direction of ABCG in the membrane [23]. Figure 4C shows an apparent skewed distribution of pathogenic variations to the positions with lower mean differential value, especially <4.0 Å. The P-value of this skewed distribution can be derived from Fisher’s exact test as 5.0×10−3, suggesting that pathogenic variations tends to be found close to the local pivot-like position in ABCG.
Conclusion
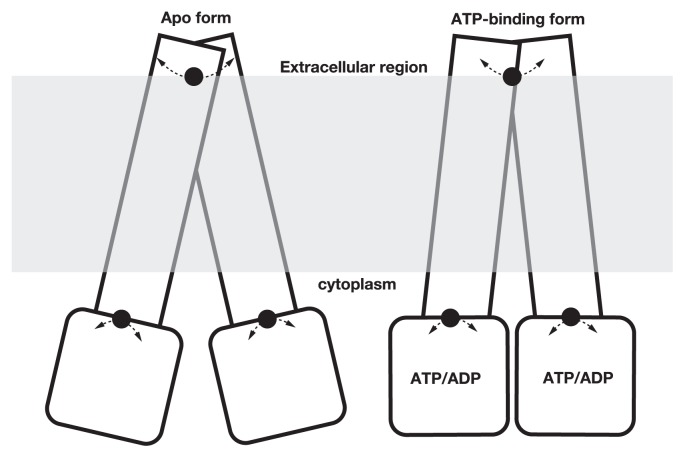
In this study, we found several pivot-like residues using differential maps and plots for the conformation shift deduced by ATP binding in ABC transporters by comparing different 3D structures registered in PDB [14]. The locations of pivot-like residues differed in different sub-families of ABC transporters, but they were often found on the residues in extracellular region (Fig. 5). The importance of the coupling helix has been reported in many previous studies and we also identified the conformation shift in the helix.
Figure 5.
A schematic view of the conformation shift identified by differential map and plot.
The similar, but not exactly the same, studies have been deployed using molecular dynamics simulation [29–33]. Molecular dynamics simulation is a powerful method to elucidate the atomic movement of the protein, yet due to the huge size of ABC transporters, a sufficient phase space search has been difficult to attain. Condic-Jurkic, K. et al. cast a doubt on the results of molecular dynamics simulation of big protein including ABC transporters [34]. Our analyses can be an alternative to address the ambiguity that molecular dynamics simulation has.
Making distinction between benign and pathogenic variations from the location of the variations is still considered to be a difficult task, but accurate prediction of the effect of the variation to the protein function will solve the issue. In this study, we combined the information of conformation shift and the locations of variations and found that amino acid variations on pivot-like residues tend to have pathogenic results. This trend could be found in ABCB, subtle but statistically significant in ABCG, and hard to be applied in ABCC because of the scattered distribution of pathogenic variations over the entire protein. The information of pivot-like residue obtained by the comparison of different 3D structures of ABC transporters, in addition to the information of ATP-binding sites and sites close to the surface of membrane, will help to improve the annotation of variations found in ABC transporter genes in human genome sequences.
Supplementary Information
Acknowledgement
The authors would like to thank the Genome Aggregation Database (gnomAD) and the groups that provided exome and genome variant data to this resource. This research is partly supported by Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS) [JP18am0101065] from Japan Agency for Medical Research and Development (AMED) and by Tokyo Interdisciplinary Life Science Consortium. The authors would also like to thank the anonymous reviewers for quite valuable comments that eliminated all the ambiguous and misinterpreted points that the original manuscript has had.
Footnotes
Conflicts of interest
M. S., H. S. and K. Y. declare that they have no conflict of interest.
Author Contribution
M. S. and K. Y. directed the entire project, H. S. gathered protein structure data, M. S. gathered sequence variation data, M. S., H. S., and K. Y. performed analyses, and M. S., H. S. and K. Y. co-wrote the manuscript.
References
- 1.Alberts B, Johnson A, Lewis J, Morgan D, Raff M, Roberts K, et al. Molecular Biology of The Cell. Sixth Edition. Garland Science; New York: 2015. [Google Scholar]
- 2.Vasiliou V, Vasiliou K, Nebert DW. Human ATP-binding cassette (ABC) transporter family. Human Genomics. 2009;3:281–290. doi: 10.1186/1479-7364-3-3-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gouaux E, Mackinnon R. Principles of selective ion transport in channels and pumps. Science. 2005;310:1461–1465. doi: 10.1126/science.1113666. [DOI] [PubMed] [Google Scholar]
- 4.Schlessinger A, Khuri N, Giacomini KM, Sali A. Molecular modeling and ligand docking for Solute Carrier (SLC) transporters. Curr Top Med Chem. 2013;13:843–856. doi: 10.2174/1568026611313070007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tanaka KJ, Song S, Mason K, Pinkett HW. Selective substrate uptake: The role of ATP-binding cassette (ABC) importers in pathogenesis. Biochim Biophys Acta. 2017;S0005-2736:30261–30264. doi: 10.1016/j.bbamem.2017.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dean M, Annilo T. Evolution of the ATP-binding cassette (ABC) transporter superfamily in vertebrates. Annu Rev Genomics Hum Genet. 2005;6:123–142. doi: 10.1146/annurev.genom.6.080604.162122. [DOI] [PubMed] [Google Scholar]
- 7.Glavinas H, Krajcsi P, Cserepes J, Sarkadi B. The role of ABC transporters in drug resistance, metabolism and toxicity. Curr Drug Deliv. 2004;1:27–42. doi: 10.2174/1567201043480036. [DOI] [PubMed] [Google Scholar]
- 8.Xiong J, Feng J, Yuan D, Zhou J, Miao W. Tracing the structural evolution of eukaryotic ATP binding cassette transporter family. Sci Rep. 2015;5:16724. doi: 10.1038/srep16724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Locher KP. Mechanistic diversity in ATP-binding cassette (ABC) transporters. Nat Struct Mol Biol. 2016;23:487–493. doi: 10.1038/nsmb.3216. [DOI] [PubMed] [Google Scholar]
- 10.Landrum MJ, Lee JM, Benson M, Brown GR, Chao C, Chitipiralla S, et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46:D1062–D1067. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Higgins CF. Multiple molecular mechanisms for multidrug resistance transporters. Nature. 2007;446:749–757. doi: 10.1038/nature05630. [DOI] [PubMed] [Google Scholar]
- 12.Theodoulou FL, Kerr ID. ABC transporter research; going strong 40 years on. Biochem Soc Trans. 2015;43:1033–1040. doi: 10.1042/BST20150139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim JM, Wu S, Tomasiak TM, Mergel C, Winer MB, Stiller SB, et al. Subnanometre-resolution electron cryo-microscopy structure of a heterodimeric ABC exporter. Nature. 2015;517:396–400. doi: 10.1038/nature13872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berman H, Henrick K, Nakamura H. Announcing the worldwide Protein Data Bank. Nat Struct Biol. 2003;10:980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- 15.Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, et al. ENSEMBL. Nucleic Acids Res. 2018;2018;46:D754–D761. doi: 10.1093/nar/gkx1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.The UniProt Consortium. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017;45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 19.Kinjo AR, Bekker G-J, Wako H, Endo S, Tsuchiya Y, Sato H, et al. New tools and functions in Data-out activities at Protein Data Bank Japan (PDBj) Protein Sci. 2018;27:95–102. doi: 10.1002/pro.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kobayashi E, Yura K, Nagai Y. Distinct Conformation of ATP Molecule in Solution and on Protein. Biophysics. 2013;9:1–12. doi: 10.2142/biophysics.9.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suzuki Y, Yura K. Conformational shift in the closed state of GroEL induced by ATP-binding triggers a transition to the open state. Biophys Physicobiol. 2016;13:127–134. doi: 10.2142/biophysico.13.0_127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nishikawa K, Ooi T, Isogai Y, Saitô N. Tertiary structure of proteins. I. representation and computation of the conformations. J Phys Soc Jpn. 1972;32:1331–1337. [Google Scholar]
- 23.Higuchi A, Nonaka N, Yura K. iMusta4SLC: Database for the structural property and variations of solute carrier transporters. Biophys Physicobiol. 2018;15:94–103. doi: 10.2142/biophysico.15.0_94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dawson RJP, Locher KP. Structure of a bacterial multi-drug ABC transporter. Nature. 2006;443:180–185. doi: 10.1038/nature05155. [DOI] [PubMed] [Google Scholar]
- 25.Furuta T, Yamaguchi T, Kato H, Sakurai M. Analysis of the structural and functional roles of coupling helices in the ATP-binding cassette transporter MsbA through enzyme assays and molecular dynamics simulations. Biochemistry. 2014;53:4261–4272. doi: 10.1021/bi500255j. [DOI] [PubMed] [Google Scholar]
- 26.Li N, Wu JX, Ding D, Cheng J, Gao N, Chen L. Structure of a pancreatic ATP-sensitive potassium channel. Cell. 2017;168:101–110. doi: 10.1016/j.cell.2016.12.028. [DOI] [PubMed] [Google Scholar]
- 27.Lee JY, Kinch LN, Borek DM, Wang J, Wang J, et al. Crystal structure of the human sterol transporter ABCG5/ABCG8. Nature. 2016;533:561–564. doi: 10.1038/nature17666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferreira RJ, Bonito CA, Cordeiro MNDS, Ferreira M-JU, dos Santos DJVA. Structure-function relationships in ABCG2: insights from molecular dynamics simulations and molecular docking studies. Sci Rep. 2017;7:15534. doi: 10.1038/s41598-017-15452-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones PM, George AM. Mechanism of ABC transporters: A molecular dynamics simulation of a well characterized nucleotide-binding subunit. Proc Natl Acad Sci USA. 2002;99:12639–12644. doi: 10.1073/pnas.152439599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones PM, George AM. Nucleotide-dependent allostery within the ABC transporter ATP-binding cassette: A computational study of the Mj0796 dimer. J Biol Chem. 2007;282:22793–22803. doi: 10.1074/jbc.M700809200. [DOI] [PubMed] [Google Scholar]
- 31.Damas JM, Oliveira SF, Baptista AM, Soares CM. Structural consequences of ATP hydrolysis on the ABC transporter NBD dimer: Molecular dynamics studies of HlyB. Protein Sci. 2011;20:1220–1230. doi: 10.1002/pro.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones PM, George AM. Molecular-Dynamics simulations of the ATP/apo state of a multidrug ATP-binding cassette transporter provide a structural and mechanistic basis for the asymmetric occluded state. Biophys J. 2011;100:3025–3034. doi: 10.1016/j.bpj.2011.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.St-Pierre J-F, Bunker A, Rog T, Karttunen M, Mousseau N. Molecular dynamics simulations of the bacterial ABC transporter SAV1866 in the close form. J Phys Chem B. 2012;116:2934–2942. doi: 10.1021/jp209126c. [DOI] [PubMed] [Google Scholar]
- 34.Condic-Jurkic K, Subramanian N, Mark AE, O’Mara ML. The reliability of molecular dynamics simulations of the multidrug transporter P-glycoprotein in a membrane environment. PLoS ONE. 2018;13:e0191882. doi: 10.1371/journal.pone.0191882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qian H, Zhao X, Cao P, Lei J, Yan N, Gong X. Structure of the human lipid exporter ABCA1. Cell. 2017;169:1228–1239. doi: 10.1016/j.cell.2017.05.020. [DOI] [PubMed] [Google Scholar]
- 36.Esser L, Zhou F, Pluchino KM, Shiloach J, Ma J, Tang WK, et al. Structures of the Multidrug Transporter P-glycoprotein Reveal Asymmetric ATP Binding and the Mechanism of Polyspecificity. J Biol Chem. 2017;292:446–461. doi: 10.1074/jbc.M116.755884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nicklisch SC, Rees SD, McGrath AP, Gokirmak T, Bonito LT, Vermeer LM, et al. Global marine pollutants inhibit P-glycoprotein: Environmental levels, inhibitory effects, and cocrystal structure. Sci Adv. 2016;2:e1600001. doi: 10.1126/sciadv.1600001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim Y, Chen J. Molecular structure of human P-glycoprotein in the ATP-bound, outward-facing conformation. Science. 2018;359:915–919. doi: 10.1126/science.aar7389. [DOI] [PubMed] [Google Scholar]
- 39.Wu JX, Ding D, Wang M, Kang Y, Zeng X, Chen L. Ligand binding and conformational changes of SUR1 subunit in pancreatic ATP-sensitive potassium channels. Protein Cell. 2018;9:553–567. doi: 10.1007/s13238-018-0530-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Z, Liu F, Chen J. Conformational changes of CFTR upon phosphorylation and ATP binding. Cell. 2017;170:483–491. doi: 10.1016/j.cell.2017.06.041. [DOI] [PubMed] [Google Scholar]
- 41.Manolaridis I, Jackson SM, Taylor NMI, Kowal J, Stahlberg H, Locher KP. Cryo-EM structures of a human ABCG2 mutant trapped in ATP-bound and substrate-bound states. Nature. 2018;563:426–430. doi: 10.1038/s41586-018-0680-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.