Abstract
The Endoplasmic Reticulum (ER), a network of membranous sheets and pipes, supports functions encompassing biogenesis of secretory proteins and delivery of functional solutes throughout the cell1,2. Molecular mobility through the ER network enables these functionalities, but diffusion alone is not sufficient to explain luminal transport across supramicron distances. Understanding the ER structure-function relationship is critical in light of mutations in ER morphology regulating proteins that give rise to neurodegenerative disorders3,4. Here, super-resolution microscopy and analysis of single particle trajectories of ER luminal proteins revealed that the topological organization of the ER correlates with distinct trafficking modes of its luminal content: with a dominant diffusive component in tubular junctions and a fast flow component in tubules. Particle trajectory orientations resolved over time revealed an alternating current of the ER contents, whilst fast ER super-resolution identified energy-dependent tubule contraction events at specific points as a plausible mechanism for generating active ER luminal flow. The discovery of active flow in the ER has implications for timely ER content distribution throughout the cell, particularly important for cells with extensive ER-containing projections such as neurons.
The Endoplasmic Reticulum (ER) is a contiguous network of membranous sheet-like reservoirs and tubes extending throughout the cell. Maintained by membrane-shaping proteins1,2, this morphology supports the distribution of ER luminal content to distant sites. ER-content distribution rate affects the efficiency of ER-mediated intracellular connectivity. Perturbation of this fundamental process may contribute to diseases caused by mutations in ER-shaping proteins3,4. Measurements of ER-luminal protein mobility using fluorescence recovery after photo-bleaching (FRAP) have previously uncovered an energy dependence that is difficult to reconcile with passive diffusion5,6. An increase in luminal crowding due to the incapacitation of ATP-dependent ER chaperones has been suggested as a plausible explanation for this effect (direct crowding measurements here do not detect such an effect). Addressing this paradox remained challenging since FRAP measurements report on bulk mobility, and do not inform as to the nature of forces driving mobility at a molecular level. A passive diffusion model for luminal transport is also challenged by the notion that traversal time of random walking molecules increases exponentially with distance. This poses kinetic limits for material exchange in an expanded ER network. Recent advances in super-resolution microscopy afford a basis for development of a single particle tracking approach to provide a detailed description of molecular motion in the ER lumen, with potential to generate a wealth of information regarding directionality and velocity from a large number of simultaneous single-molecule displacement events. Using live cell super-resolution microscopy, we visualised and analysed single molecule trajectories traversing tubular ER, and the organelle’s real-time morphological dynamics.
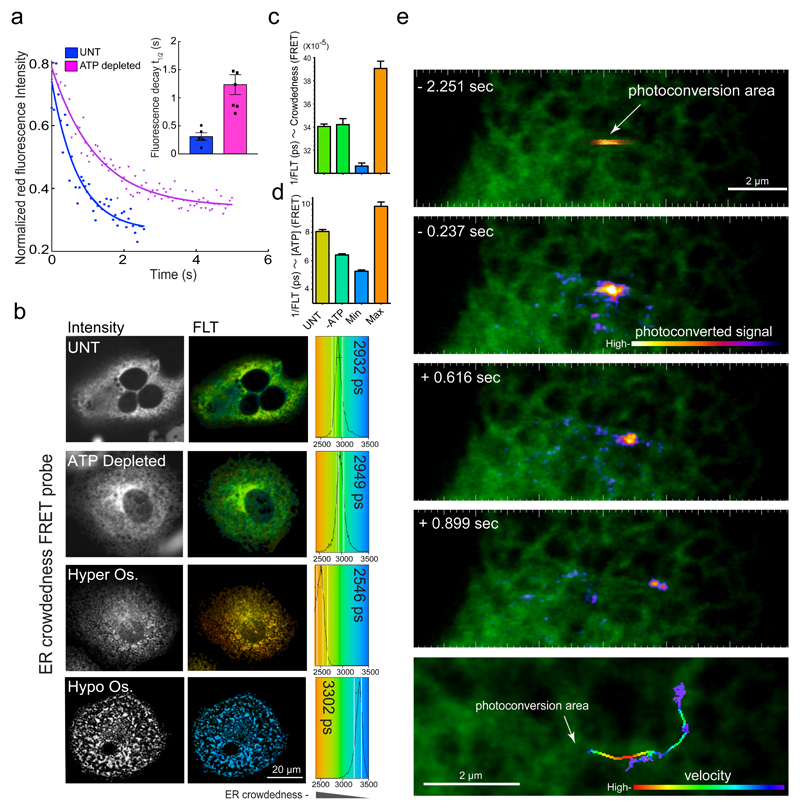
First we examined, in our experimental system, the energy dependence of luminal protein mobility, previously demonstrated for green fluorescent protein (GFP) using FRAP6. Escape-rate of photo-convertible fluorescent protein (Dendra2-ER) from a region of activation was attenuated by ATP depletion (Fig. 1a). This may reflect either an increase in resistance to motion or a decrease in active transport of proteins upon energy starvation. The former is contradicted by measurements of ER crowdedness, using a sensitive FRET-based probe7, that, over a broad range of expression levels, remained unaltered by ATP-depletion (Fig. 1b – d, Supplementary Fig. 1a). Furthermore, displacement of Dendra2 proteins (initially localized as a “packet” in a small volume of the tubular ER that had been subjected to a colour-photoswitching laser pulse) occurred with variable speed and had a conspicuous unidirectional component (Fig. 1e, Supplementary Video1). These features are inconsistent with Brownian motion and suggest, instead, active transport.
Fig. 1. ATP depletion affects ER mobility without altering luminal crowdedness.
(a) Trace of time-dependent decay in the intensity of the fluorescence signal from an ER-localised photoconvertible protein, Dendra2-ER, after a pulse of photo-converting illumination delivered to a small patch of untreated or ATP-depleted COS7 cells. Inset denotes mean ± SEM, (n=5 traces per condition) of fluorescence decay half time, reflecting the probe’s escape from the photoconversion area.
(b) Fluorescence intensity (left) and colour-coded fluorescence lifetime (FLT) images (right) of COS7 cells transfected with an ER-localised molecular crowding probe7. FLT distribution within imaged cells is displayed in colour-coded histograms with mean FLT noted (in picoseconds, ps, right). Cells were left untreated (UNT), ATP depleted, or treated with hyper-osmotic (Hyper Os.) or hypo-osmotic buffers that induce cell shrinking or swelling to obtain maximal and minimal crowding values, respectively. Shown are characteristic images observed in three independent repeats.
(c) Bar diagram of FRET-donor FLT values measured as in (b) (mean values ± SD, n=22 independently sampled cells).
(d) Bar diagram of relative intracellular ATP concentration measured with FRET-based ATP-probe (A-Team)23 in cells untreated or ATP depleted as in (a & b). Minimum and maximum values represent the probe readings in ATP depleted or saturating conditions respectively, imposed in semi-permeabilised cells. Shown are mean values ± SD, n=10 independently sampled cells.
(e) Images of COS7 cell expressing Dendra2. A brief pulse of illumination photoconverted Dendra2 from green to red in a restricted region of the ER. The progression of the photoconverted packet of proteins is revealed by the time series and summated in the bottom panel with its velocity colour coded.
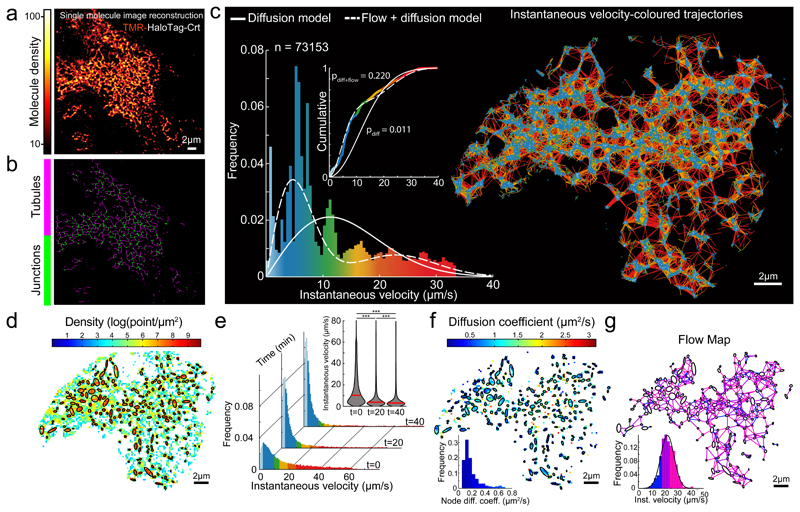
To characterize motion in the ER lumen we performed real-time single particle tracking (SPT) in live cells, acquiring trajectories at optical super-resolution. Imaging at 56 Hz, we recorded single molecule fluorescent signals from functional (Halo-tagged ER luminal chaperone, Calreticulin, Crt) and inert (ER targeted HaloTag) proteins, both sparsely labelled with a fluorescent ligand (chloroalkane-Tetramethylrhodamine, TMR). Brightness and photostability of the TMR ligand enabled tracking single particles over longer trajectories than genetically-encoded fluorophores (Supplementary Fig. 1b), thus establishing spatio-temporal correlations in motion patterns over extended periods of time. This approach offered information on directionality and instantaneous velocity at a single molecule level, not available in bulk methods such as FRAP. By implementing a single molecule localisation algorithm, images reconstructed from the SPT series revealed a pattern typical of the ER network, confirming ER-localisation of the HaloTag-Crt and washout of the unbound dye (Fig. 2a & b, Supplementary Videos 2–4).
Fig. 2. Characteristics of single particle displacement tracked in the tubular ER lumen.
(a) Image reconstructed from single molecule localizations of TMR-labeled Halo-tagged Calreticulin (Crt), in HEK-293 cells, rendered with a molecular density colour code.
(b) Skeletonization view of image in (a). Shown are representatives of n=3 independent experiments.
(c) Single molecule trajectories generated using particle-tracking algorithm from time series of image (a)), color-coded according to instantaneous velocity distribution shown in histogram. Overlaid traces: velocity distribution simulated assuming exclusively diffusion-driven motion (solid line, using apparent D5 from (h)), or combination of diffusion and flow (using D and flow rate from f & g). Inset: cumulative distribution, Kolmogorov-Smirnov test of observed vs. expected distributions.
(d) Density map computed for grid of square bins (sides of 0.2 μm) imposed on particle displacement map. Ellipses mark boundaries of higher density regions (correspond to tube-connecting reservoirs/junctions).
(e) Histograms of instantaneous velocity frequency distributions of SPT from a cell before/after ATP depletion (as in Fig. 1 a-c). Inset: violin plot presenting the medians (red bars) and density (grey) of the distributions. A two-sided Mann-Whitney U-test was used to compare median of each pair of distributions (*** p-value < 1e-3), p(0-20 min)=1e-80, p(20-40 min)=9.889e-64, p(0-40 min)=1e-80; n=20526, n=14591 and n=10108 trajectory displacements respectively.
(f) Diffusion map extracted from the empirical estimator of the Langevin equation (1, Suppl. Note 1) and computed from a square grid as in d. Inset: distributions of the diffusion coefficients inside nodes (AVG +/- SD=0.19 +/- 0.13, n=226 nodes).
(g) Flow map computed by averaging non-Brownian velocity jumps of particles moving between pairs of neighbouring nodes identified in (d) and color-coded according to the inset histogram. Inset: distribution of average instantaneous velocity between pairs of neighbouring nodes (n=705 node-pairs; AVG +/- SD=22.90 +/- 6.92).
Raw source single molecule time series and image-reconstruction are shown in Supplementary Video 2.
Trajectories were generated from single-molecule time series by sequentially implementing spot detection and tracking algorithms8,9. ER geometry constrains particle movement to a narrow tubular network, which limits overlapping trajectories, contributing to the tracking algorithm’s ability to faithfully trace many molecules simultaneously. Resulting trajectories reconstruct a map recognizable as a pattern of ER tubes and their connecting reservoirs (Fig. 2c), reflecting tracking fidelity. Notably, spatio-temporal particle distributions were non-uniform, with higher time-integrated abundance in the tube-connecting reservoirs (Fig. 2d). This heterogeneity correlated with spatial distribution of instantaneous velocities, revealing distinct subgroups. Relatively slow-moving particles predominant in segments of trajectories mapped to the nodes and particles with relatively high and variable velocities to node-connecting tubes (Fig. 2c). These characteristics could be observed for two different markers (Calreticulin and HalotagER) and three different cell types (HEK 293T, COS-7, SH-SY5Y, Supplementary Table 1). The long, single-tailed velocity distribution observed (histogram, Fig. 2c) is incompatible with Brownian motion (Fig. 2c, modelled by solid line). Furthermore, ATP depletion led to selective loss of the fast-moving population (Fig. 2e). These observations suggest that diffusive motion is manifest by the slow-moving particles in the tube-connecting reservoirs (nodes), while the rapidly-moving particles in the tubes are subjected to an ATP-dependent propulsive force, resulting in an ER luminal flow. The displacement profile observed fits well a bi-modal distribution of instantaneous velocities (Fig. 2c, modelled by dashed line).
Next we quantified temporal coordinate changes of HaloTag-Crt by analysing SPT data using the overdamped limit of the Langevin model (where velocity is described as the sum of diffusional and drift forces, and motion parameters are estimated from local statistics of the displacement, see Methods)10–13. Motion is described by the stochastic model (a sum of directed and diffusional motion terms): where w is Brownian motion. The source of the noise w is the ambient thermal agitation, while the drift term b represents transport in tubules and D is the effective local diffusion. This analysis (see Supplementary Note 1) allows estimating D and b from large numbers (~ 104 per cell acquisition) of single particle trajectories.
The global nature of this statistical approach considers not only particle displacement speed but also direction patterns, extracted from a large number of trajectories repeatedly traversing the same regions, thereby unmixing the contribution of Brownian and deterministic forces. This computation identifies a slow diffusional (D=0.19+/-0.13 μm/s2) component that maps predominantly onto the nodes (Fig. 2f). The relatively fast movement of internode particles required an additional component to account for their directionality and persistence, consistent with a propulsive force with normal distribution of velocity (22.9+/-6.92 μm/s, Fig. 2g). The super-diffusive nature of particle motion in ER tubules was further confirmed by analysis of time-averaged mean squared displacement (MSD) of SPT. MSD described as ~ tα, where the anomalous exponent α defines motion as sub-diffusive if α<1, Brownian/diffusive if α=1 and super-diffusive if α>1. Conducted on the entire ensemble of trajectories, the MSD analysis revealed a broad range of particle behaviours (0<α<1.5, Fig. 2b), whereas the same analysis restricted to trajectory fragments located in nodes revealed clearly confined diffusion dynamics (α<0.8, Supplementary Fig. 2c). Trajectories of particles moving outside the nodes exhibited super-diffusive dynamics (α>1, Supplementary Fig. 2d, Video 3). These results are consistent with the active motion mode identified in analysis of Figure 2.
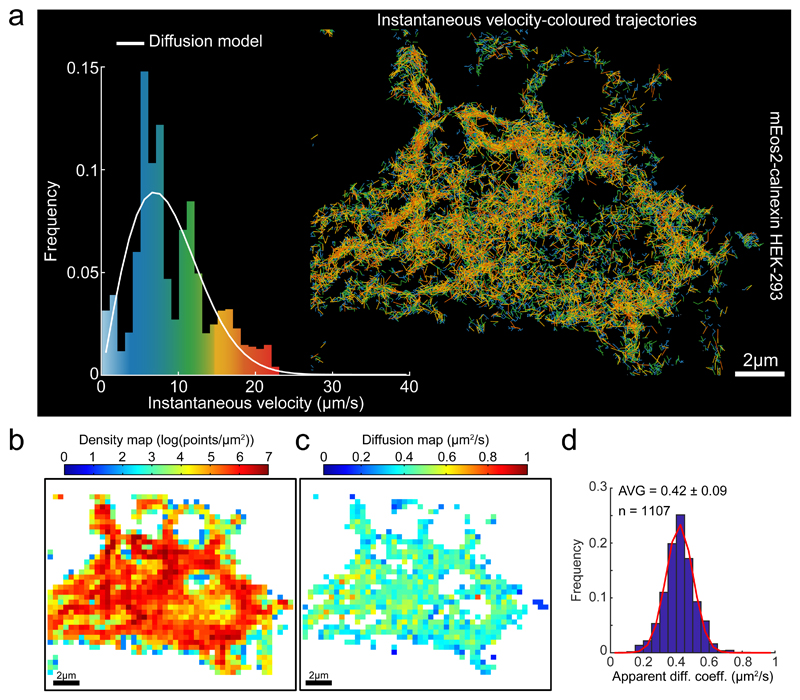
Similar observations were made in Green Monkey kidney (COS-7) and Human neuroblastoma (SH-SY5Y) cell lines, attesting to their broad validity in describing ER flow dynamics and its spatial organization (Supplementary Fig. 3, Table1, Video 2). Measurements of the motion parameters of a lower mass ER-localised protein, HaloTag-KDEL, showed similar values to those observed with the tagged Crt (Supplementary Fig. 3, Table1). Flow velocity was slightly higher in COS cells than in HEK-293T and SH-SY5Y cells. The behaviour of the luminal ER markers, HaloTag-KDEL and Calreticulin, contrasted with that of a membrane-associated analogue of the latter, mEOS2-Calnexin: its velocities were distributed relatively homogeneously through the ER network (Fig. 3a, b), lacked the thick tail of high values in distribution of instantaneous velocities and fit well to a purely diffusional model (Fig. 3a-d).
Fig. 3. Statistics of Single Particle trajectories recorded from the ER membrane.
(a) Single molecule trajectories of mEOS2-calnexin expressed in HEK-293 cells generated as in Figure 2, color-coded according to instantaneous velocity distribution Inset: Instantaneous velocity distribution histograms computed from the displacements extracted from the trajectories and overlaid by the expected distribution for a purely diffusive motion with the diffusion coefficient extracted from (d).
(b) Density map computed for a grid of square bins (sides of 0.4 μm) imposed on the particle displacement map.
(c) Diffusion map extracted from the empirical estimator of the Langevin equation (1, methods) and computed from the same square grid as in (b).
(d) Histograms of diffusion coefficients computed from individual square bins, pooled from two cells, for the entire domain as presented in (c). The red curve on top of the diffusion histogram corresponds to a fit (Trust Region Reflecting algorithm) to a Gaussian distribution with μD = 0.42 μm2/s, σD = 0.12 μm2/s and a determination coefficient R2 = 0.986. Descriptive statistics given as AVG +/- SD.
To establish whether recently reported ER macrostructure motion characteristics14 are reflected in the SPT analysis, we focused on their numerical parameters. Motion of ER tubules characterized by their relatively slow transverse oscillation (4Hz, with an amplitude < 50 nm, which translates to velocity <0.2 μm/s14), does not significantly contribute to the relatively fast velocities of flow-assisted marker particles moving along the tubules (27-42 μm/s, Supplementary Table 1). Junction fluctuations contribute a similarly insignificant component to the diffusional motion inside the junction since the diffusion coefficient calculated from tracking of whole junctions was 69 times slower than the mean diffusion coefficient computed for single molecules (14, Supplementary Fig. 4, and Video 5). Furthermore, contribution of tubule growth to single particle trajectories was found to be negligible, with a mean percentage of tubules growing at any given time of 0.14 +/- 0.04 % (Supplementary Videos 5-7).
Consistently, considering trajectory motion as a purely diffusive process yielded an apparent diffusion coefficient of 1.13 μm2/s (Fig. S2a) similar to that previously estimated by FRAP5, 15, 16.
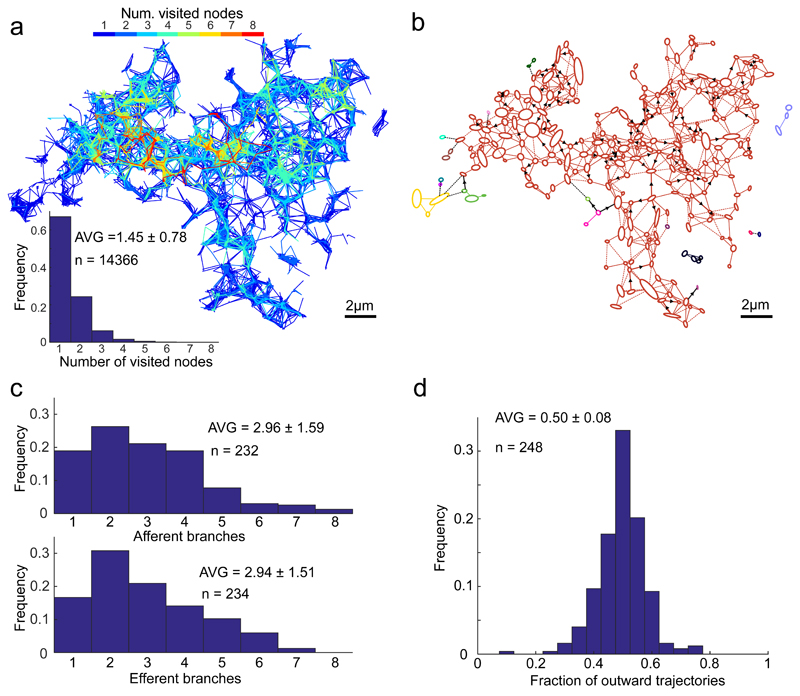
Though most individual trajectories visited only a limited number of nodes (Fig. 4a), an Oriented Network Graph analysis, which identifies directly or proxy interconnected junctions through trajectory directions17 revealed that, regardless of their starting point, particles have the potential to visit almost the entire ER network (Fig. 4b; the disconnected periphery is likely contributed by signals from neighbouring cell(s)). This analysis is consistent with the notion that the ER network maintains a luminal continuum. The ER appeared to be in a state of equilibrium, with nodes, on average, connected by an equal number of inward and outward trajectories (Fig. 4c & d). These findings are consistent with an interconnected system of flows that preserves the content across the ER.
Fig. 4. Properties of ER luminal trajectories’ directionality.
(a) Number of nodes visited by individual particles. Trajectories map, as in Figure 2, colour-coded according to the number of nodes visited by a particle; and the distribution of the number of nodes visited by each individual trajectory (excluding trajectories visiting 0 node).
(b) Vectorial representation of the ER network from Figure 2 analysed using Oriented Network Graph analysis, to assess the direct or proxy, uni/bi-lateral trajectory-connectivity of the nodes, assigning single colour for each interconnected area. Note a strong connected component resulting in a monochromatic appearance of almost the entire network. Arrows denote prevalent displacement directionality (detected in 18% of tubes), defined as such if steady-state ratio of flow in one direction vs. the total flow exceed 0.75. Dashed links represent flows whose directionality could not be determined due to insufficient number of displacement events.
(c) Distribution histogram of the number of outward (efferent) and inward (afferent) directed branching for individual nodes. Efferent branches were defined as the number of nodes, reached by the outward trajectories originating in the examined node, in the time-integrated map; accordingly, afferent branches reflect the number of nodes-of-origin for the trajectories arriving at the examined node.
(d) Distribution of the fraction of exiting trajectories for each node.
All values are given as AVG ± SD.
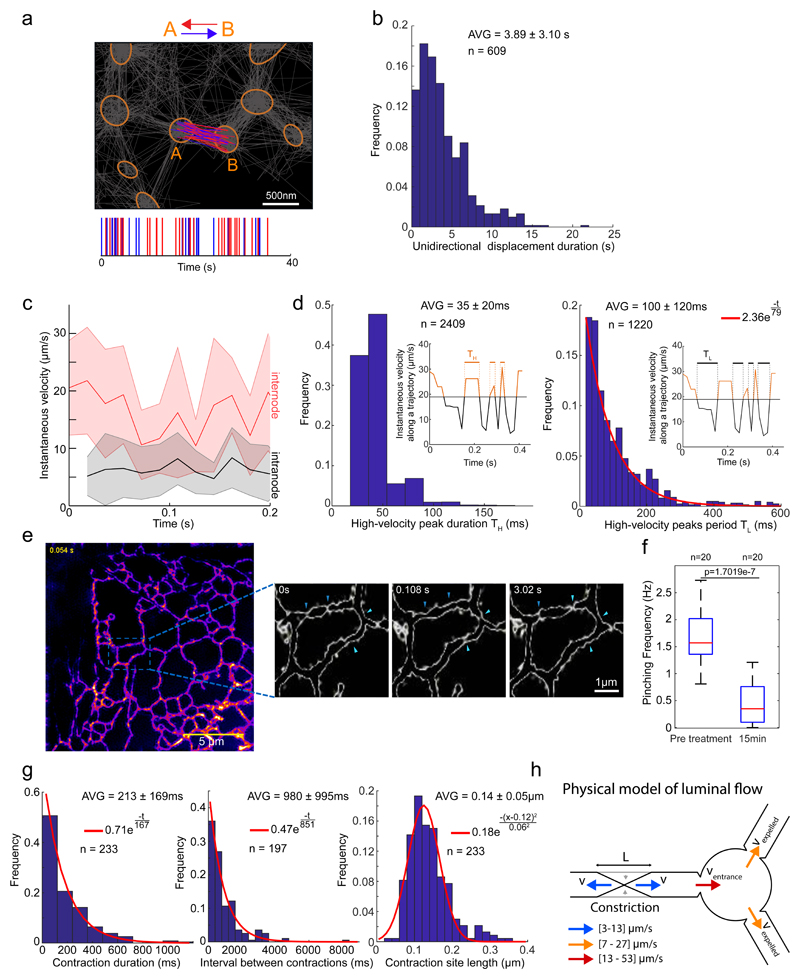
Closer scrutiny of the directionality of individual tube-traversing particles suggested a pattern whereby the direction of visible flow alternates with variable frequency (switching on average every 4 seconds, maintained for up to 14 seconds Fig. 5a & b); and particles accelerated periodically following their exit from a node, reaching brief velocity peaks that lasted up to 120 ms (Fig. 5c & d). Intervals between velocity peaks and flow-directionality alternations were distributed stochastically (Fig. 5d, inset), suggesting that flow-inducing events (e.g. transient tubule contractions, discussed below) are not produced by synchronized oscillators and are therefore not centrally coordinated. However, we cannot exclude the contribution of synchronization processes whose phase is lost, as trajectories are recorded asynchronously. Note that temporal profiles of directionality are not available using low spatio-temporal resolution approaches (e.g. FRAP or photoconversion pulse-chase, Fig. 1e).
Fig. 5. Dynamics of ER luminal flow correlated with tubule contractions.
a. Analysis of particle trajectories’ directionality. Tubular junctions/nodes denoted by orange ellipses; grey lines denote all particle trajectories. Trajectories connecting two nodes indicated as A and B are colour coded according to their direction either in red, denoting travel from A to B, or blue for travel from B to A. Lower graph represents the temporal pattern of traversal-directionality. Shown is representative of n=108 node-pairs.
b. Distribution of time periods of unidirectional inter-node displacement.
c. Plots of instantaneous particle velocities fluctuations. Velocities of particles following departure from nodes and traveling along tubules (between nodes, red), and those of particles residing within nodes (black). Solid lines represent mean values for all trajectories, shaded regions represent SD of total sample size: n=111 internode and n=140 intranode trajectory displacements.
d. Analysis of time duration TH of high-velocity (v > 20 μm/s) peaks (left) and time interval TL between high-velocity peaks. Red line represents an exponential fit (R2 = 0.998).
e. High-speed Structured Illumination Microscopy (SIM) super-resolved images of the tubular ER in live COS7 cells stained with an ER membrane dye (ER Tracker-Green). Images were acquired in 54 ms intervals and processed as described in Methods. The resulting SIM reconstructions were colour-coded according to intensity. The magnified area shows the contours of ER tubules at higher magnification. Arrows denote positions where transient contraction events occur repeatedly. Shown are frames from a time series measurement presented in full in supplementary Video S2. Tubule contractions are better visualized in COS7 cells, but detectable in HEK-293 cells too (Fig. S5). Shown is representative of n=5 independent experiments.
f. Box plot of tubule contraction frequencies extracted from high-speed SIM time series as shown in (e) before and after ATP depletion. Red line – median, boxes’ bottom/top edges – the 25th and 75th percentiles respectively, whiskers – extreme data points. ***: Two-sided Mann-Whitney U-test p=1.7019e-7, n=20 ER tubules.
g. Distributions of contraction duration, intervals and lengths from SIM videos as in (e) and Fig. S5. Red curves: exponential (left and middle) and Gaussian (right) fits (R2 = 0.988, R2 = 0.969, R2 = 0.937 respectively).
h. Schematic representation of the model for estimating tubule contraction-induced particle velocity.
All values are given as AVG ± SD for noted n.
The oscillatory luminal motion suggested the possibility of nanoperistalsis-like18 propulsion, attainable by tubule contractions. To test this, we obtained high-resolution time-series images of the ER tube structure of live cells by fast Structured Illumination Microscopy (SIM)14,19,20. These revealed transient, asynchronous constriction of the tubes at specific locations (Fig. 5e, Supplementary Fig. 5a, b; Video8 & 9), consistent with a role for tube constriction in generating flow. Constriction-driven propulsion is also consistent with the observed velocity values and variation of packets of photo-converted Dendra2 during their deterministic traversal of the tubular ER (Vmax = 19.9 μm/s, Fig. 1e). Furthermore, frequency of contraction events decreased four-fold upon ATP depletion (Fig. 5f, Supplementary Video 10). It is expected that following an individual contraction event (with a frequency of hundreds of milliseconds), both deterministic and acceleratory displacement of multiple particles would be detected, as SPT acquisition operates at approximately ten times the contraction frequency. Assuming uncoordinated contractions throughout the tubular network, consecutive contractions have the same probability to preserve or invert the direction of the next set of detectable SPT events (consistent with observed distribution of directionality preservation time, Fig. 5b).
The notion that tubule contractions generate high-velocity peaks in luminal particles is supported by the fact that their temporal distributions are both Poissonian (Fig. 5d & g), indicating compatible physical processes. The larger time constant of contractions (~900ms) compared to that of high-velocity peaks (~80ms) is expected since several contraction points may contribute to the particles’ acceleration incidence.
Furthermore, a physical model simulating forces resulting from tubule contraction, and based on their empirical characteristics (Fig. 5h and Supplementary Note1), predicts flow velocities of 10-40 μm/s, in agreement with the high-velocities observed in SPT (Fig. 5c). Notably, the contraction frequency is low enough to avoid coinciding proximal contractions that may cancel the local flows (probability of simultaneous contractions of two points = 0.022, calculations in Supplementary Note 1). Whilst the existence of a mechanism for spatio-temporal coordination of the contraction events cannot be ruled out, our findings indicate that an uncoordinated system, inducing fast local currents with alternating directionality inside the tubular network, is sufficient to ensure a rapid luminal content homogenization/distribution and thereby overcome a critical kinetic limitation of passive diffusion as a mechanism for ER content mixing in large cells.
Localised contraction of ER tubules, leading to ER deformation, was observed during calcium manipulation21 (Supplementary Fig. 5c), or phototoxicity (Supplementary Fig. 5e and Video 11), both reversible processes affecting ER morphology (Supplementary Fig. 5e). These super-resolved images of the ER’s structural dynamics under severe experimental perturbation highlight a potential for ER tubes to contract, revealing that fragments of the perturbed ER that had lost their tubular structure displayed characteristically slow-velocity diffusional motion (Supplementary Fig. 5d). Other physiological membrane dynamic processes involving molecular motors, vesicular fusion and budding, network oscillation14 and even tube elongation/withdrawal may also contribute to flow and warrant further investigation.
Regardless of its origin(s), the alternating luminal currents described here are well suited to serve as a mixing device, enhancing distribution of ER content throughout the cell. Given that diffusion-driven connectivity (matter exchange rate) decreases exponentially with distance, it is expected that the active process described here would be especially important in cells with extensive ER projections, such as motor neurones. It is therefore tempting to speculate that perturbed luminal flow might contribute to diseases such as hereditary spastic paraplegia, associated with defective ER membrane-shaping proteins3,4,22.
Methods
1. Intra-vital and fixation cell microscopy
1.1. Cell culture, transfections and expression constructs
COS7 (RRID:CVCL _0224), HEK 293 cells were cultured in Dulbecco’s Modified Eagle’s medium (DMEM) supplemented with 10% foetal calf serum and 1X non-essential amino acids (M7145 -SIGMA, Gillingham, Dorset, UK). SHSY-5Y cells were cultured in MEM/F12 medium supplemented with 15% foetal calf serum. Transfections were performed using the Neon Transfection System (Invitrogen, Paisley, UK) applying 1.5g of ATeam or crowding probe DNA to 3.105 cells. An expression vector encoding a cytosol localised crowding probe7 was modified to encode an endoplasmic reticulum localised probe by N-terminal addition of mouse preprotrypsin signal sequence and C-terminal addition of a KDEL motif by Gibson assembly. Description of plasmids used in this study are presented in Supplementary Table 2.
1.2. Photo-conversion microscopy
Photo-conversion pulse chase experiments were performed using Leica SP8 confocal microscope. Images were acquired using frame size of 256 × 256 pixels to allow imaging at 9 frames/sec rate, in Green (488 nm excitation, 510 - 530 nm emission) and Red (561 nm excitation, 590 - 620 nm emission) channels. Photo-conversion illumination (405 nm) was introduced at frame 20 in a region of interest for duration of 20 frames, using On The Fly FRAP acquisition mode (enabling image recording during the photoconverting illumination). Normalized intensity of the Red channel at the region of photo-conversion was plotted as a function of time post-photoconversion, and fitted to a mono-exponential decay function, to extract the decay t1/2, that was used as an estimator of Dendra2-ER mobility.
1.3. Cell manipulations and Fluorescence Lifetime Imaging Microscopy (FLIM)
FLIM was carried out as previously described25,26, using a pulsed (sub10ps, 20-40MHz) supercontinuum (430-2000nm) light source (SC 450, Fianium Ltd., Southampton, UK). An acousto-optic tunable filter (AA Optoelectronic AOTFnC-VIS) was used to define the 438 nm excitation wavelength for both the ER-crowding probe and ATeam ATP sensor. Emitted light was collected using a 470/20nm emission filter and detected by a fast photomultiplier tube (PMC-100, Becker & Hickl GmbH, Berlin, Germany). Data were processed using SPCImage (Becker & Hickl GmbH), fitting a monoexponential decay function. Osmosis-driven changes in cell volume were induced by addition of 0 mM NaCl (hypo-osmotic) or 500 mM NaCl (hyperosmotic) supplemented Hanks Balanced Salt solution (2.5mM KCl, 1.2 mM CaCl2, 0.5 mM MgCl2, 5 mM glucose, 10 mM HEPES pH 7.5) to a final ratio of 1:1 with DMEM (final osmolalities of 0.152 osmol/kg and 0.609 osmol/kg respectively). ATP depletion was achieved by incubating the cells in the presence of NaN3 (0.05% w/v) and 2-deoxi-glucose (20 mM) for 2 hours prior to imaging; cells preserving the tubular ER pattern were selected upon treatments.
1.4. Single particle localisation and tracking (SPT)
Cells were transiently transfected with vectors encoding HaloTag (Promega) targeted to the ER by N-terminal the preprotrypsin signal sequence and a C-terminal retrieval signal (SS-HaloTag-KDEL); or Halo-Tag Calreticulin. 24 hours after transfection, cells were labelled with 0.5 nM cell-permeant HaloTag TMR ligand (tetra-methyl-rhodamine, Promega G8251) for 10 minutes, followed by 3 washes with label free medium; and imaged with 18 ms exposure on Elyra Super Resolution microscope (Zeiss) using α-Plan Apochromat X100 oil, 1.46C na objective; at 561/570-620 nm excitation/emission, in HiLo (pseudo-TIRF) mode using EMCCD iXON DU897 camera (Andor). The obtained single particle image series (at least 2000 frames) were processed using the PALM image reconstruction module or particle-tracking module at the instruments software (Elyra Zen edition, Zeiss). Trajectory generation fidelity was verified using the ICY software27 version 1.9.5 and Imaris software version 8.4.1 (Bitplane).
The tracking algorithms was set to identify diffraction-limited spots as particles if their signal to noise ratio was > 4 and the spot does not exceed 9 pixels in diameter; then to identify the centers of spots to refine the positioning of the particle beyond the diffraction limit. The algorithm terminated trajectories if the signal disappears for 1 frame (linking is not permitted if particles leave the focal plane, blink etc.).
The tracking stage produces Nt two-dimensional trajectories X1 … XNt each possessing Mi points: Trajectories containing less than three points were discarded from the analysis. For each pair of successive points (tj+1, tj) of a trajectory i, we defined the displacement as: ΔXi (t) = Xi (tj+1) − Xi (tj). See Supplementary Note 1 for further details of SPT mathematical analysis and modeling.
1.5. High-speed Structured Illumination microscopy
Live 2D-SIM (light modulation/grazing incidence illumination microscopy, GI-SIM) images of cells stained for 20 minutes with ER Tracker Green (ThermoFisher scientific, E34251) were acquired with a custom-built highspeed SIM microscope19,20, using a spatial light modulator (SLM). ER Tracker Green was imaged using a 100X/1.49NA TIRF oil-immersion objective (Olympus) with a 488 nm diode laser (Toptica) at an irradiance of 50W/cm2, with emission imaged via a notch filter (FF01-525/30, Semrock) onto an sCMOS camera (ORCA Flash 4.0, Hamamatsu). 2D-SIM gratings displayed on the SLM resulted in a line spacing of 228 nm at the sample, corresponding to an angle of incidence of 44.6°. Each super-resolved frame was obtained from the reconstruction24,28 of nine raw frames acquired at 6 ms/exposure (54 ms/SIM frame). 3D-SIM (23 slices, 2.4 µm) was performed using Elyra microscope (Zeiss), with Plan Apochromat X63/1.4NA oil objective and sCMOS PCO Edge camera (Andor) on Paraforamdehyde 2%, Gluteraldehyde 2%, 100mM sodium cacodylate, 2mM CaCl2, pH7.4 1 hour room temperature. See Supplementary Note 1 for further details of SIM image analysis.
Mathematical modelling and analysis
Details of the computational analyses of single particle tracking and structured illumination microscopy, and their mathematical modelling are described in Supplementary Note 1.
Statistics and Reproducibility
Statistical analyses and visualisation were performed using Matlab 9. Error bars, P values and statistical tests and sample sizes are reported in the figure legends. Statistical tests: differences between probability distribution were assessed using two-way Kolmogorov-Smirnov tests and differences between distribution medians were assessed using two-sided Mann-Whitney U-tests. All experiments were performed independently at least three times.
Supplementary Material
Acknowledgements
We are grateful to Matthew Gratian and Mark Bowen (CIMR, Cambridge) and Gregory Strachan (Institute of Metabolic Science, Cambridge) for assistance in establishing microscopy data acquisition approaches; Steffen Preissler (CIMR, Cambridge) for discussion.
This study was supported by grants from Wellcome Trust to DR (Wellcome 200848/Z/16/Z, WT: UNS18966); Wellcome Trust Strategic Award for core facilities to the Cambridge Institute for Medical Research (Wellcome 100140); EPSRC (EP/L015889/1 and EP/H018301/1), MRC (MR/K015850/1 and MR/K02292X/1) and Wellcome Trust (3-3249/Z/16/Z and 089703/Z/09/Z) to CFK; FRM team research grant to DH; a DIM fellowship from Ile-de-France to PP. DR is a Wellcome Trust Principal Research Fellow. EA is a United Kingdom Dementia Research Institute Fellow.
Footnotes
Code availability
Custom code generated for single particle tracking analysis and visualisation as well as for SIM ER network analysis of SIM images can be obtained from Zenodo database along with experimental raw data examples, DOI: 10.5281/zenodo.1317630 and DOI: 10.5281/zenodo.1318129 respectively.
Data Availability
Source image-series data for Fig. 1e, Supplementary Fig. 2 and Fig 4e, f have been provided as supplementary video1, 2 and 8 - 10 respectively; and statistical information for Fig. 2 and supplementary Fig. 2 have been provided in Supplementary Table 1. Custom code has been deposited in the Zenodo database DOI: 10.5281/zenodo.1317630 with experimental raw data DOI: 10.5281/zenodo.1318129. All other data supporting the findings of this study are available from the corresponding authors on reasonable request.
References
- 1.Powers RE, Wang S, Liu TY, Rapoport TA. Reconstitution of the tubular endoplasmic reticulum network with purified components. Nature. 2017;543:257–260. doi: 10.1038/nature21387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 2006;124:573–586. doi: 10.1016/j.cell.2005.11.047. [DOI] [PubMed] [Google Scholar]
- 3.Hubner CA, Kurth I. Membrane-shaping disorders: a common pathway in axon degeneration. Brain : a journal of neurology. 2014;137:3109–3121. doi: 10.1093/brain/awu287. [DOI] [PubMed] [Google Scholar]
- 4.Blackstone C, O'Kane CJ, Reid E. Hereditary spastic paraplegias: membrane traffic and the motor pathway. Nature reviews. Neuroscience. 2011;12:31–42. doi: 10.1038/nrn2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dayel MJ, Hom EF, Verkman AS. Diffusion of green fluorescent protein in the aqueous-phase lumen of endoplasmic reticulum. Biophysical journal. 1999;76:2843–2851. doi: 10.1016/S0006-3495(99)77438-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nehls S, et al. Dynamics and retention of misfolded proteins in native ER membranes. Nature cell biology. 2000;2:288–295. doi: 10.1038/35010558. [DOI] [PubMed] [Google Scholar]
- 7.Boersma AJ, Zuhorn IS, Poolman B. A sensor for quantification of macromolecular crowding in living cells. Nature methods. 2015;12:227–229. doi: 10.1038/nmeth.3257. 221 p following 229. [DOI] [PubMed] [Google Scholar]
- 8.Cheezum MK, Walker WF, Guilford WH. Quantitative comparison of algorithms for tracking single fluorescent particles. Biophysical journal. 2001;81:2378–2388. doi: 10.1016/S0006-3495(01)75884-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chenouard N, et al. Objective comparison of particle tracking methods. Nature methods. 2014;11:281–289. doi: 10.1038/nmeth.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schuss Z. Theory and Applications of Stochastic Processes. Appl Math Sci. 2010;170 Cover1-U4. [Google Scholar]
- 11.Hoze N, Holcman D. Residence times of receptors in dendritic spines analyzed by stochastic simulations in empirical domains. Biophysical journal. 2014;107:3008–3017. doi: 10.1016/j.bpj.2014.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoze N, et al. Heterogeneity of AMPA receptor trafficking and molecular interactions revealed by superresolution analysis of live cell imaging. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:17052–17057. doi: 10.1073/pnas.1204589109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Langevin P. The theory of brownian movement. Cr Hebd Acad Sci. 1908;146:530–533. [Google Scholar]
- 14.Nixon-Abell J, et al. Increased spatiotemporal resolution reveals highly dynamic dense tubular matrices in the peripheral ER. Science. 2016;354 doi: 10.1126/science.aaf3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lai CW, Aronson DE, Snapp EL. BiP availability distinguishes states of homeostasis and stress in the endoplasmic reticulum of living cells. Molecular biology of the cell. 2010;21:1909–1921. doi: 10.1091/mbc.E09-12-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Snapp EL, Sharma A, Lippincott-Schwartz J, Hegde RS. Monitoring chaperone engagement of substrates in the endoplasmic reticulum of live cells. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:6536–6541. doi: 10.1073/pnas.0510657103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tarjan R. Depth first search and linear graph algorithms. Siam Journal on Computing. 1972;1 [Google Scholar]
- 18.Nadeem S, Maraj EN. The mathematical analysis for peristaltic flow of nano fluid in a curved channel with compliant walls. Applied Nanoscience. 2014;4:85–92. [Google Scholar]
- 19.Young LJ, Strohl F, Kaminski CF. A Guide to Structured Illumination TIRF Microscopy at High Speed with Multiple Colors. Journal of visualized experiments : JoVE. 2016 doi: 10.3791/53988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kner P, Chhun BB, Griffis ER, Winoto L, Gustafsson MG. Super-resolution video microscopy of live cells by structured illumination. Nature methods. 2009;6:339–342. doi: 10.1038/nmeth.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Subramanian K, Meyer T. Calcium-induced restructuring of nuclear envelope and endoplasmic reticulum calcium stores. Cell. 1997;89:963–971. doi: 10.1016/s0092-8674(00)80281-0. [DOI] [PubMed] [Google Scholar]
- 22.Lo Giudice T, Lombardi F, Santorelli FM, Kawarai T, Orlacchio A. Hereditary spastic paraplegia: clinical-genetic characteristics and evolving molecular mechanisms. Experimental neurology. 2014;261:518–539. doi: 10.1016/j.expneurol.2014.06.011. [DOI] [PubMed] [Google Scholar]
- 23.Imamura H, et al. Visualization of ATP levels inside single living cells with fluorescence resonance energy transfer-based genetically encoded indicators. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:15651–15656. doi: 10.1073/pnas.0904764106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gustafsson MG, et al. Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophysical journal. 2008;94:4957–4970. doi: 10.1529/biophysj.107.120345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frank JH, Elder AD, Swartling J, Venkitaraman AR, Jeyasekharan AD, Kaminski CF. A white light confocal microscope for spectrally resolved multidimensional imaging. Journal of Microscopy. 2007;227(3):203–215. doi: 10.1111/j.1365-2818.2007.01803.x. * [DOI] [PubMed] [Google Scholar]
- 26.Avezov E, Cross BC, Schierle GSK, Winters M, Harding HP, Melo EP, Kaminski C, Ron D. Lifetime imaging of a fluorescent protein sensor reveals surprising stability of ER thiol redox. J Cell Biol. 2013;201(2):337–349. doi: 10.1083/jcb.201211155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chenouard N, Bloch I, Olivo-Marin JC. Multiple hypothesis tracking for cluttered biological image sequences. IEEE transactions on pattern analysis and machine intelligence. 2013;35(11):2736–3750. doi: 10.1109/TPAMI.2013.97. [DOI] [PubMed] [Google Scholar]
- 28.Wicker K, Mandula O, Best G, Fiolka R, Heintzmann R. Phase optimisation for structured illumination microscopy. Optics express. 2013;21(2):2032–2049. doi: 10.1364/OE.21.002032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.