Abstract
Tuberculosis caused by Mycobacterium tuberculosis is currently one of the leading causes of death from an infectious agent. The main difficulties encountered in eradicating this bacteria are mainly related to (i) a very complex lipid composition of the bacillus cell wall, (ii) its ability to hide from the immune system inside the granulomas, and (iii) the increasing number of resistant strains. In this context, we were interested in the Rv0646c (lipGMTB) gene located upstream to the mmaA cluster which is described as being crucial for the production of cell wall components and required for the bacilli adaptation and survival in mouse macrophages. Using biochemical experiments combined with the construction of deletion and overexpression mutant strains in Mycobacterium smegmatis, we found that LipGMTB is a cytoplasmic membrane-associated enzyme that displays both phospholipase and thioesterase activities. Overproduction of LipGMTB decreases the glycopeptidolipids (GPL) level concomitantly to an increase in phosphatidylinositol (PI) which is the precursor of the PI mannoside (PIM), an essential lipid component of the bacterial cell wall. Conversely, deletion of the lipGMS gene in M. smegmatis leads to an overproduction of GPL, and subsequently decreases the strain susceptibility to various antibiotics. All these findings demonstrate that LipG is involved in cell envelope biosynthesis/remodeling, and consequently this enzyme may thus play an important role in mycobacterial physiology.
Keywords: antibiotics, cell-envelope, lipolytic enzymes, Mycobacterium tuberculosis, phospholipid homeostasis
Introduction
With more than 10 million new cases and almost 1.6 million deaths in 2017, tuberculosis (TB) caused by the etiologic agent Mycobacterium tuberculosis (M. tuberculosis) still remains the deadliest infectious disease worldwide [1]. The current antibiotherapy, which is a combination of four antibiotics (i.e. isoniazid (INH), rifampicin (RIF), pyrazinamide, and ethambutol), allows to heal 95% of infected people with active disease when administered under directly observed therapy [2]. This treatment is, however, not effective enough to overcome the emergence of multidrug and extensively drug-resistant strains of M. tuberculosis [3,4]. More recently, some totally drug-resistant M. tuberculosis strains have also been detected, which are virtually incurable [5]. Most of the time this antibiotic resistance is directly related to the unique mycobacterial cell wall composition which is an essential factor contributing to virulence, pathogenesis, and survival of the bacilli. Therefore, identifying proteins and mechanisms involved in the biosynthesis and/or remodeling of this complex structure could lead to the development of new therapeutics to control and fight TB.
Indeed, amongst all bacterial phyla, Actinobacteria and more specifically M. tuberculosis, possess an unusual cell envelope with a specific architecture constituted by lipids which may represent up to 40–60% of the dry weight of the cell, as compared with only 5–10% in other bacteria [6,7]. The outer membrane is an extremely hydrophobic barrier mainly composed of long-chain mycolic acids (MAs) and several atypical lipids, which are responsible for the high intrinsic tolerance of the bacteria to various antibiotics [8]. Moreover, it is now acknowledged that most of these lipids play important roles in the viability and/or virulence of the bacteria [9]. As a consequence, the structural characterization of these complex lipids, as well as their biosynthetic pathways have been the subject of many studies in the past decade with the aim of identifying putative therapeutic targets [10–12].
Of particular interest, the synthesis of MAs involves two fatty acid synthase (FAS) enzymatic complexes, FAS-I and FAS-II [13,14]. FAS-I synthesizes de novo medium chain fatty acids (C16–18 and C24–26) as acyl-CoA derivatives, which are further elongated by the FAS-II complex to form the C48–64 meromycolic chains, a major component of the envelope arabinogalactan layer [13,14]. The various methyltransferases closely interact with enzymes belonging to the FAS-II system to functionalize meromycolate chains by adding specific chemical modifications (such as cyclopropane, methyl, hydroxyl, and ketone) [14–18]. Amongst the eight putative methyltransferases identified in M. tuberculosis H37Rv genome, the mmaA gene cluster; i.e. mmaA1 (Rv0645c), mmaA2 (Rv0644c), mmaA3 (Rv0643c), and mmaA4 (Rv0642c); is essential for chemical group introduction and functionalization of Mas, thus participating actively in the envelope integrity/virulence of M. tuberculosis [14,19,20].
Interestingly, these mmaA genes are also clustered with the Rv0646c gene, which is highly conserved amongst mycobacterial species and encodes for LipG (LipGMTB), a putative lipase/esterase which has been recently described as a carboxylesterase [21]. M. tuberculosis H37Rv genome encodes for 36 lipolytic enzymes annotated as putative esterases, lipases, cutinases, or phospholipases based on their sequence homology and the presence of the consensus G-x-S-x-G motif, characteristic of the α/β hydrolase-fold family [19,22–24]. LipGMTB has been classified within the Lip family which includes 24 members [23,25]. This enzyme possesses approximately 50% sequence identity with the EstB protein from Acinetobacter calcoaceticus and 43% with the putative hydrolase PA3586 from Pseudomonas aeruginosa PAO1. Interestingly, the Rv0646c gene has been annotated as essential for M. tuberculosis H37Rv survival within primary murine cells by transposon site hybridization [26]. Due to its high conservation and its peculiar location upstream to the mmaA cluster, which is crucial for the production of cell wall components and so for the mycobacterial envelope integrity, the potential physiological function of LipG deserves to be studied and deciphered.
In this context, Rv0646c gene from M. tuberculosis H37Rv has been cloned, expressed, purified, and fully biochemically characterized. Since its potential important role in mycobacterial lifecycle makes LipGMTB an attractive target for future therapeutic developments against TB, inhibition studies on pure recombinant enzyme were also conducted with two recent families of antitubercular compounds; namely, oxadiazolone-core (OX) derivatives [23,27], and monocyclic analogs of Cyclophostin and Cyclipostins analogs (CyC) [28–30] which affect the growth of M. tuberculosis both in vitro and in infected macrophages.
In order to investigate the physiological function of LipG in mycobacterial species, both deletion and overexpression mutant strains have been generated into the surrogate strain Mycobacterium smegmatis. By using phenotypic assays, subcellular location, in silico modeling, lipid extraction followed by TLC analysis, and susceptibility testing via the resazurin microtiter assay (REMA), we determined that LipGMTB is a cytoplasmic membrane-associated enzyme involved in phospholipids remodeling and cell wall integrity.
Experimental procedures
Bacterial strains and culture conditions
Escherichia coli DH10β and E. coli C41 (DE3)/pLyS strain were cultured in LB broth or Terrific Broth (Invitrogen, France). M. smegmatis mc2 155 and M. smegmatis mc2 155 groEL1ΔC [31] strains were cultured in Middlebrook 7H9 broth (BD Difco, Le Pont-de-Claix, France) supplemented with 0.05% (v/v) Tween-80 and 0.2% (v/v) glycerol (Sigma–Aldrich, Saint-Quentin Fallavier, France) (7H9-S). When needed, ampicillin and kanamycin (Euromedex, Souffelweyersheim, France) were added to the medium at final concentrations of 100 and 50 µg/ml for both E. coli and mycobacterial species, respectively. Hygromycin B (Euromedex) was used at a final concentration of 200 and 50 µg/ml for recombinant E. coli and recombinant mycobacteria, respectively.
In silico analyses
Genomic sequences of lipG and its orthologs were obtained from the NCBI database. M. tuberculosis H37Rv, Mycobacterium leprae TN, M. marinum M, and M. smegmatis genomic clusters were re-adjusted manually based on the genomic annotations provided by the Mycobrowser database [32]. The amino acid sequence of M. tuberculosis H37Rv LipG and its orthologs were aligned using Multalign software [33] and results were displayed using ESPrit 3.0 software [34]. The percentage of identity between the different orthologs was generated with the Clustal Ω multiple sequence alignment program (http://www.ebi.ac.uk/Tools/msa/clustalo/). A 3D model of M. tuberculosis H37Rv LipG (LipGMTB) was also generated, by using the I-TASSER server [35,36] and visualized with PyMOL software (The PyMOL Molecular Graphics System, version 1.8 Schrödinger, LLC). Electrostatic surface potentials of LipGMTB model structure was built using freely available PDB2PQR web service [37,38]. The electrostatic potential data thus generated, were further visualized using the PyMOL APBS Tools 2 plugin [39].
Construction of recombinant plasmids, expression and purification of recombinant LipGMTB protein
Rv0646c ORF encoding LipGMTB protein, was amplified from M. tuberculosis H37Rv genomic DNA by polymerization chain reaction, using primers containing the attB1, Shine–Dalgarno, 6× His, and a TEV (tobacco etch virus) NIa site sequences at the 5′-end and the attB2 recombination site at the 3′-end (Supplementary Table S1, P1MTB and P2MTB). The resulting PCR fragments were cloned using the Gateway® technology (Invitrogen, Carlsbad, CA, U.S.A.) into the pDonR221 entry vector, and then into the pDEST14 expression vector (pDEST14::lipGMTB). A similar strategy was used to clone the MSMEG_1352 ORF encoding LipGMS protein. PCR was performed with P1MS and P2MS primers (Supplementary Table S1), thus giving the pDEST14::lipGMS construct. DNA sequences of recombinant clones were analyzed by GATC Biotech (Ebersberg, Germany). The newly generated pDEST14::lipGMTB was used as a template for generating pDEST14::lipGMTBS123Aby PCR mutagenesis using specific primers (Supplementary Table S1, P3 and P4). The resulting plasmids pDEST14::lipGMTB, pDEST14::lipGMTBS123A, and pDEST14::lipGMS were used to transform E. coli C41 (DE3)/pLyS strain harboring the pRare plasmid (Novagen).
For protein expression, 1 L of Terrific Broth medium was inoculated with the recombinant strains at initial OD600 of 0.05, and was further incubated at 37°C and 200 rpm. When the OD600 reached 0.6, cells were put on ice for 30 min. Then, 0.5 mM isopropyl-d-thiogalactopyranoside was added to the medium and the culture was further incubated at 17°C under shaking. After 18 h of incubation, cells were pelleted, re-suspended in 30 ml of ice-cold lysis buffer (50 mM Tris/HCl pH 8.0, 150 mM NaCl, 1 mM EDTA, 0.1% (v/v) Triton X-100, 0.25 mg/ml lysozyme), and stored at −80°C. To purify recombinant LipGMTB or LipGMS, the supernatant (S1) obtained after cell lysis and centrifugation at 17000×g for 30 min, was stored at 4°C. Pellet was then re-suspended in lysis buffer, sonicated twice for 30 s with 30 s breaks between each cycle, solubilized by stirring at 4°C for 1 h using a magnetic bar, and further centrifuged at 17000×g for 30 min (giving S2 supernatant). Both supernatants (S1 and S2) were pooled and loaded on to a Ni2+-NTA affinity column previously equilibrated with buffer A (10 mM Tris/HCl pH 8.0, 150 mM NaCl). The column was washed with buffer A before performing elution with buffer A and 125 and 250 mM of imidazole. Fraction purity was assessed on SDS/PAGE. The corresponding 35 kDa protein band was further analyzed by tryptic digestion and MS. Pure protein was concentrated to 0.5–1 mg/ml, flash frozen in liquid nitrogen, and stored at −80°C.
Biochemical characterization and inhibition of rLipGMTB/MS
Esterase activity assay
The esterase activity of rLipGMTB or rLipGMS was determined as described previously [23] with slight modifications. Briefly, p-nitrophenyl (pNP) esters with different carbon chain lengths were used as substrates, including pNP acetate (pNP-C2), butyrate (pNP-C4), caproate (pNP-C6), caprylate (pNP-C8), caprate (pNP-C10), laurate (pNP-C12), myristate (pNP-C14), and palmitate (pNP-C16). Stock solution (200 mM) of each substrate was freshly prepared in acetonitrile. Release of pNP was monitored at 410 nm and pH 8 using a 96-well plate spectrophotometer (PowerWave™, Bio-Tek Instruments) and quantitated using a pNP calibration curve (10 µM to 0.5 mM) with (λ = 410 nm) = 6.0 mM−1. Enzymatic reactions were performed at 37°C over a period of 20 min in a 96-well microplate filled with 10 mM Tris buffer (pH 8.0) containing 150 mM NaCl with 0.5% (v/v) Triton X-100 to a final volume of 200 µl. In each well, 2 mM of substrate and 1 µg of rLipGMTB (0.15 µM final concentration) were added. Activities were expressed in international units (U), corresponding to 1 µmol of pNP released per min. Specific activities (S.A) were expressed as U/mg of pure enzyme. Experiments were done in triplicate.
Phospholipase A1 and A2 activities assay
The phospholipase A1 and A2 activities of rLipGMTB and rLipGMS were monitored continuously using BODIPYH dye-labeled phospholipids: PED-A1 (N-((6-(2,4-DNP)Amino)Hexanoyl)-1-(BODIPYH FL C5)-2-hexyl-sn-glycero-3-phosphoethanolamine) and red/green BODIPYH PC-A2(1-O-(6-BODIPYH558/568-aminohexyl)-2-BODIPYH FLC5-sn-glycero-3-phosphocholine), respectively, as previously described [40,41]. Briefly, substrate stock solutions (50 µM) were prepared in ethanol. All enzyme activities were assayed in 10 mM Tris (pH 8.0), 150 mM NaCl. Enzymatic reactions were performed at 25°C for 60 min in a final volume of 200 µl containing 1.5 µg rLipGMTB (0.23 µM final concentration) and 5 µM substrate. The release of BODIPYH (BFCL5) (Life Technologies) was recorded at λexc = 485 nm and λem = 538 nm using a 96-well plate fluorimeter (Fluoroskan Ascent, Thermo Fisher Scientific). Each experiment was done in triplicate. Enzymatic activities were quantitated using a BFCL5 calibration curve (0.08–200 pmoles). S.A were expressed in mU of fatty acid (or BFLC5) released per mg of pure protein (mU.mg−1). One unit correspond to the release of one µmol of product release per min. PLA1 from T. lanuginosus and Bv PLA2 (Sigma–Aldrich) were used as positive standards for the PLA1 and the PLA2 activities, respectively.
Thioesterase activity assay
Hydrolysis of palmitoyl-Coenzyme A (palmitoyl-CoA) (Sigma–Aldrich) was used to measure thioesterase activity. After the hydrolysis of the thioester bond, the free sulphur on CoA is attacked by DTNB (5-5′-Dithio-bis 2-nitrobenzoic acid) (Sigma–Aldrich) which releases a measurable nitrophenyl group, TNB2− (5-Thio-2-nitrobenzoate) ion detectable at 415 nm. Fresh stock solution of palmitoyl-CoA (1 mM) and DTNB (0.4 mM) were prepared in water. Palmitoyl-CoA and DTNB were added in each well of a 96-well plate at final concentration of 10 µM. The thioesterase activity of rLipGMTB or rLipGMS (30 µg) was compared with that of TesA (Rv2928) [42] and LipY [43] used as positive and negative controls, respectively. Triplicate assays were done at 37°C in 10 mM Tris (pH 8.0) 150 mM NaCl buffer. The absorbance at 415 nm was continuously measured and the release of TNB2− was quantitated using a calibration curve (0.5–30 nmoles).
Enzymatic inhibition assays using OX derivatives, CyC analogs, and Orlistat
The six oxadiazolone derivatives (OX); i.e. MmPPOX, iBPOX, iBpPPOX, HPOX, HpPPOX, and BePOX; were synthesized as described previously [23,27]. The five CyC analogs; i.e. CyC17, CyC7α, CyC7β, CyC8α, and CyC8β; were synthesized according to [44–46].
Inhibition experiments were performed using the classical enzyme-inhibitor pre-incubation method, as described previously [30,47,48]. Stock solutions (10 mM) of each CyC and OX inhibitor, as well as Orlistat were prepared in DMSO. rLipGMTB was pre-incubated for 30 min at 25°C with each compound at various inhibitor molar excess (xI), related to 1 mol of enzyme, ranging from 1 to 200. Sample of the incubation medium (1.0 µg rLipGMTB) was then collected for measuring the enzyme residual activity using the colorimetric assay with the substrate pNP-C6, as described above. The variation in the residual activity allowed to determine the inhibitor molar excess which reduced the enzyme activity to 50% of its initial value (xI50) [47,48]. Thereby, a xI50 value of 0.5 is corresponding to a 1:1 stoichiometric ratio between the enzyme and the inhibitor, and is therefore the highest level of inhibitory activity that can be achieved. In each case, control experiments were done in the absence of inhibitor but with the same volume of solvent. It is worth noting that DMSO at a final volume concentration of less than 10% has no effect on the enzyme activity. Inhibition dose–response curves were fitted in Kaleidagraph 4.2 Software (Synergy Software). Results are expressed as mean values ± S.D. of at least three independent assays.
Creation of M. smegmatis MSMEG_1352 disrupted strain
The MSMEG_1352 gene encoding LipGMS was amplified with 500-bp flanking regions by PCR using M. smegmatis mc2 155 genomic DNA as template and specific primers (Supplementary Table S1, P9 and P10). PCR product of ∼2000 bp was cloned into pJET1.2/blunt vector following manufacturer’s instructions giving pJET-lipGMS. Recombinant plasmid was then digested with NruI, and a hygromycin resistance cassette amplified from pUC-Hygro using primer (Supplementary Table S1, P17 and P18). The resulting blunt-ended PCR product was cloned in order to disrupt lipGMS gene. This new plasmid, pJET-lipGMS::Hyg which contains an inactive version of lipGMS gene was used as template for amplifying an allelic exchange substrate by using specific primers (Supplementary Table S1, P9 and P10). Approximately 300 ng of purified PCR product was electroporated into electro-competent M. smegmatis mc2 155 strain harboring pJV53 plasmid, and cells were plated on to Middlebrook 7H10 agar medium supplemented with 50 µg/ml of both kanamycin and hygromycin and incubated at 37°C during 5 days as previously described [49]. Finally, recombinant clones were analyzed by PCR using P7-P8 and P11-P12 primers’ pairs (Supplementary Table S1).
Creation of overexpression/complementation vectors
ORFs Rv0646c and MSMEG_1352 were amplified by PCR with specific primers (Supplementary Table S1, P5–P6 and P7–P8) containing NdeI and BamHI restriction sites in 5′ and 3′ ends, respectively. Digested and purified products were cloned into the pVV16 mycobacterial expression vector in frame with the 6His-tag located in C-terminal position giving the pVV16::lipGMTB and pVV16::lipGMS complementation plasmids allowing a constitutive production of recombinant LipG. Rv0646c ORFs was also cloned within a homemade pMyC Gateway® vector, allowing a strong overexpression following acetamide induction, thus giving pMyC::lipGMTB. Competent M. smegmatis mc2 155, M. smegmatis groEL1ΔC, and lipGMS ::Hyg cells were prepared as previously described [50] and were electroporated with a single pulse of 2.5 kV, 25 µF, and 1000 Ω by using a Gene Pulser Xcell™ Electroporation System (Bio-Rad). Selection was achieved by plating cells on complete Middlebrook agar medium supplemented with the proper antibiotic.
Subcellular fractionation and immunoblotting
Recombinant M. smegmatis groEL1ΔC harboring pVV16::lipGMTB and pVV16::lipGMS were cultured in 7H9-S medium supplemented with 50 µg/ml kanamycin until OD600 was approximately 1–1.5. Cells were harvested for 10 min at 4000×g, then washed twice with PBS (pH 7.4) buffer containing 0.05% (v/v) Tween-20, and re-suspended in lysis buffer (PBS pH 7.4 containing EDTA and 1 mM PMSF). Cells were then disrupted by using a French pressure cell at 1100 psi. Subcellular fractionation was performed as previously described [51] with slight modifications. Briefly, the cell lysate was centrifuged two times at 1000×g at 4°C in order to remove unbroken cells. Supernatant (total lysate) was further centrifuged two times at 27000×g at 4°C to separate cell wall/membrane from the cytoplasmic fraction. The pellet containing insoluble material was washed twice with PBS (pH 7.4) to remove any cytoplasmic contaminant, and supernatant containing the cytoplasmic fraction was further ultracentrifuged at 100000×g for 90 min at 4°C to remove any membrane contaminant. Each fraction was loaded and separated on to 12% SDS/PAGE and transferred on to a nitrocellulose membrane using a Trans-Blot Turbo Transfer System (Bio-Rad). Immunoblotting of 6× His-tagged proteins was performed using anti-HisProbe™ HRP conjugate (Thermo Fisher Scientific). MSMEG_0220 was used as the control for subcellular location, and immunoblotting was performed by using rabbit polyclonal antibodies [52,53] and horseradish peroxidase–conjugated anti-rabbit IgG (Sigma-Aldrich). Revelation was achieved using Pierce™ ECL Western Blotting substrate solution (Thermo Fisher Scientific) and visualized with a ChemiDoc™ MP Imaging System (Bio-Rad).
Genapol® extraction and immunoblotting
Approximately 10 uOD600 of recombinant cells cultured in 7H9-S medium supplemented with 50 µg/ml kanamycin at OD600 ∼1-1.5 were harvested for 10 min at 4000×g. Cells were washed twice in PBS (pH 7.4) buffer containing 0.05% (v/v) Tween-20 and surface-exposed proteins were isolated by incubating cells with PBS containing 0.5% (v/v) Genapol®-X080 (Sigma–Aldrich) at room temperature [54,55]. Control samples were treated with a PBS buffer without Genapol®-X080. After approximately 30 min, all supernatants were collected and precipitated with 12% (v/v) trichloroacetic acid. Finally, samples were separated on to 12% SDS/PAGE and immunoblotting was achieved as described above.
Dynamic subcellular location using fluorescence microscopy
Gene encoding superfolder GFP (sfGFP) was amplified from pKD4-sfGFP plasmid with specific primers (Supplementary Table S1, P13–P15 and P14–P15) which were cloned in pMV261 and pVV16 vectors at either EcoRI/HindIII or only HindIII restriction sites. Thus, creating pMV261::sfGFP and pVV16::sfGFP. Rv0646c and MSMEG_1352 ORFs were amplified by PCR with specific primers (Supplementary Table S1, P5–P6 and P7–P8) containing NdeI and BamHI restriction sites in 5′ and 3′ ends, respectively. Digested and purified products were cloned into the pVV16::sfGFP in frame with the sfGFP gene and thus generating a translational fusion. M. smegmatis was electroporated with 1.5 µg of pMV261::sfGFP and these latter constructs; recombinant clones were selected on to specific medium and overexpression of the two chimeric proteins were checked by immunoblotting as described above. Recombinant strains were cultured until OD600 reaches ∼1-1.5, and then harvested by centrifugation at 4000×g for 10 min. Cells were washed once in PBS (pH 7.4) buffer, and bacterial suspension (5 µl) was spotted between a coverslip of 170 µm thickness and a 1.5% agarose-PBS pad. Bacteria were analyzed by snapshot imaging at room temperature using an Olympus FV1000 confocal microscope and 100× oil-objective. Exposure time was 800 ms for both phase-contrast and fluorescence images (λexc\λem = 490/530 ±10 nm). Images recorded were processed using the open source program ImageJ 1.51K (NIH, U.S.A.).
Lipid extraction and analysis
Mycobacterial strains were grown in 1 l of 7H9-S broth until an OD600 of ∼1 was reached, except when using acetamide inducible strains where cells were collected at specific time points. Cultures were then centrifuged at 4000g for 15 min at 4°C and washed twice with distilled water. Pellets were lyophilized overnight and weighed to determine the exact mass of bacterial dry extract for normalization calculations.
MAs methyl esters (MAMEs) and fatty acids methyl esters (FAMEs) were extracted following previously described protocol [56,57]. Approximately, 10 mg of cellular dry weight were re-suspended in 2 ml TBAH (40% v/v, Sigma–Aldrich) and incubated at 100°C overnight. Then 4 ml of CH2Cl2, 300 µl CH3I, and 2 ml of water were added to the mixture and incubated for 1 h at room temperature under shaking. Sample was centrifuged and the lower organic phase was kept, washed three times with distilled water, dried over MgSO4, and then concentrated under nitrogen stream. Finally, 3 ml of DTT were added to the dried extract, and sonication was performed for 10 min in a water bath. Sample was centrifuged for 10 min at 4000×g to remove any precipitate. The DTT layer was evaporated and the residue was re-suspended in 100 µl of DTT. The same volume of extract containing MAMEs and FAMEs were separated on 1D TLC using hexane/ethyl acetate (19:1, v/v) as eluent, and revealed by using a 10% phosphomolybdic acid solution in absolute ethanol.
Total lipids were extracted as previously described [58] with slight modifications. Briefly, lipids from dry extract were incubated for 16 h with CHCl3-MeOH (1:2; v/v), at room temperature under shaking. Residual lipids were re-extracted for 16 h with the same solvents, but using alternative ratios (1:1 and 2:1 v/v). All the three organic phases were pooled and concentrated under reduced pressure. Samples were re-suspended in a CHCl3-MeOH solution (3:1, v/v), and washed with 0.3% (w/v) NaCl in water. Organic and aqueous phases were separated by centrifugation 4000×g for 10 min, and only the organic phases were conserved and dried over MgSO4. Samples were finally evaporated under a nitrogen stream, weighed, and re-suspended in a CHCl3-MeOH solution (3:1, v/v).
Equal volume of extract containing glycopeptidolipids (GPLs) were separated on TLC (Silica Gel 60, Merck) by using CHCl3-MeOH (90:10, v/v) as eluent and further visualized by vaporization of 20% (v/v) sulphuric acid in ethanol. Phospholipid analyses were carried out by using both 1- or 2D TLC. Mono-dimensional TLC were performed by loading the same volume of extract and using CHCl3-MeOH-H2O (65:25:4; v/v/v) as eluent. Concerning bi-dimensional TLC, the first dimension was resolved by using CHCl3-MeOH-H2O (65:25:4; v/v/v) as solvent system 1, and CHCl3-MeOH-CH3COOH-H2O (80:12:15:3; v/v/v/v) as solvent system 2. Phospholipids were visualized by vaporization of a copper acetate/orthophosphoric acid mixture prepared by mixing a copper acetate saturated solution with a 85% (1:1, v/v) aqueous orthophosphoric acid solution. Phospholipid standards were purchased from Sigma–Aldrich. Finally, each resolved plate was heated at 120°C for 5–10 min, scanned using a ChemiDoc™ MP Imaging System (Bio-Rad), and densitometric analyses was performed using the ImageLab™ software version 5.0 (Bio-Rad) allowing to determine the relative content of each sample.
REMA for MIC determination
Susceptibility testing was performed using the Middlebrook 7H9 broth microdilution method. All assays for each strain were carried out at least in triplicate. MICs against the various M. smegmatis bacterial strains were determined in 96-well flat-bottom Nunclon Delta Surface microplates with lid (Thermo Fisher Scientific) using REMA method [59]. Briefly, log-phase bacteria were diluted to a cell density of 5 × 106 cells/ml in 7H9-S medium. Then 100 µl of the above inoculum (i.e. 5 × 105 cells per well) was added to each well containing 100 µl of 7H9-S medium, serial two-fold dilutions of the selected antibiotics (RIF or INH), CyC17 [28,29], HPOX [27], or Orlistat to a final volume of 200 µl. Growth controls containing no inhibitor (i.e., bacteria only), inhibition controls containing 50 μg/ml kanamycin and sterility controls (i.e. medium only) without inoculation were also included. Plates were incubated at 37°C in a humidity chamber [60] to prevent evaporation for 3–5 days. Then, 20 µl of a 0.02% (w/v) resazurin solution was added to each well, and the plates were incubated at 37°C for color change from blue to pink or violet and for a reading of fluorescence units (FU). Fluorescence corresponding to the resazurin reduction to its metabolite resorufin was quantitated using a Tecan Spark 10M multimode microplate reader (Tecan Group Ltd, France) with excitation at 530 nm and emission at 590 nm. Fluorometric MICs were determined by fitting the RFU% sigmoidal dose–response curves [28,29] in Kaleidagraph 4.2 software (Synergy Software). MIC values determined as the lowest compound concentrations inhibiting 50% of growth were defined as the MIC50.
Results
LipG is a highly conserved mycobacterial protein
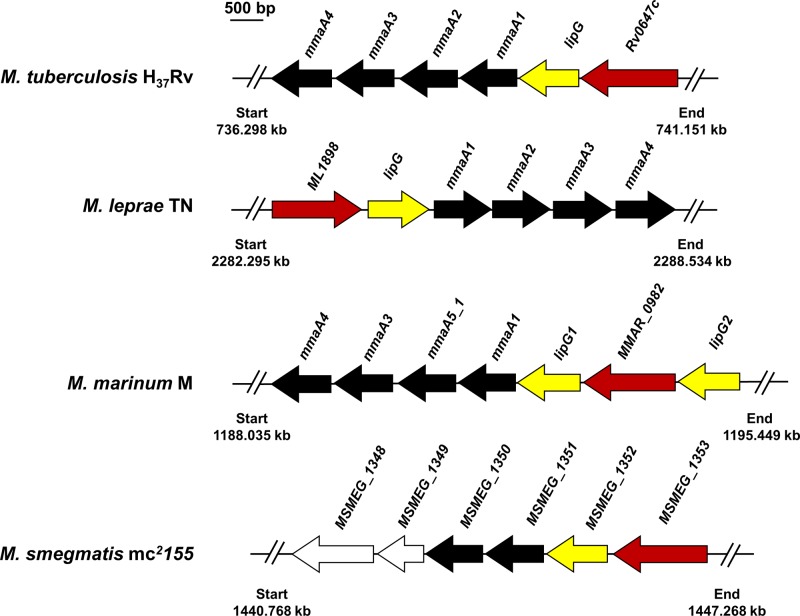
Gene Rv0646c encoding LipGMTB protein was not only found within M. tuberculosis genome. Genomic context containing Rv0646c gene and its orthologs was indeed surprisingly highly conserved within many mycobacterial species. In M. tuberculosis, Rv0646c is surrounded by an essential uncharacterized gene (Rv0647c) and the mmaA cluster composed of four mmaA genes (mmaA1–4) encoding proteins involved in MAs maturation processes (Figure 1). Interestingly, the occurrence and genome organization of lipG clusters are also highly conserved in M. tuberculosis, M. leprae, M. marinum, or M. smegmatis genomes (Figure 1), with two copies of genes encoding for LipG proteins in M. marinum, M. abscessus, or M. ulcerans genomes. Multiple sequence alignment of these various LipG proteins and sequence comparison, revealed a high level of amino-acid conservation, also with some consensus amino-acids blocks between each mycobacterial species [21] (Supplementary Figure S1). The LipG characteristics (i.e. sizes of genes/proteins and sequence identities) from different mycobacterial strains are summarized in Table 1. Each of these proteins possess a molecular weight ∼32 kDa, with the number of amino acids ranging from 301 to 324. Taken together, all these findings suggest that the cluster including the lipG gene may be dedicated to a common mycobacterial feature either in strict pathogenic, opportunistic, or non-pathogenic strains.
Figure 1. Genomic organization of lipG clusters in mycobacterial species.
Schematic organization of the lipG genomic loci from M. tuberculosis, M. leprae, M. marinum, and M. smegmatis were designed based on the Mycobrowser database [32]. lipG genes are represented in yellow whereas genes involved in MAs maturation processes are in black. Furthermore, a conserved ORF encoding a putative ABC transporter was highlighted in red. Co-ordinates of each gene cluster are indicated in kb. Scale represents 500 bp.
Table 1. Conservation and characteristics of mycobacterial LipG proteins.
| Mycobacterial species/strains | LipG and orthologs | ||||
|---|---|---|---|---|---|
| Gene | Gene ID | Protein | Protein size (aa) | Identity1 (%) | |
| M. tuberculosis H37Rv | Rv0646c | 888065 | LipG | 301 | - |
| M. bovis AF2122/97 | Mb_0665c | 1091775 | LipG | 301 | 100 |
| M. bovis BCG Pasteur 1173P2 | BCG_0695c | 4697156 | LipG | 301 | 100 |
| M. canetti CIPT 140010059 | MCAN_06451 | 10986452 | LipG | 301 | 99 |
| M. leprae TN | ML_1899 | 910591 | LipG | 305 | 71 |
| M. marinum M | MMAR_0981 | 6225233 | LipG1 | 315 | 73 |
| MMAR_0983 | 6225235 | LipG2 | 310 | 47 | |
| M. ulcerans Agy99 | MUL_0733 | 4552132 | LipG1 | 314 | 73 |
| MUL_0735 | 4552134 | LipG2 | 303 | 47 | |
| M. abscessus ATCC 19977 | MABS_3878 | 5967421 | LipG2 | 310 | 40 |
| MABS_3880 | 5966346 | LipG1 | 324 | 54 | |
| M. smegmatis mc2155 | MSMEG_1352 | 4534086 | LipG | 305 | 64 |
Amino-acid sequence identity related to M. tuberculosis H37Rv.
Cloning, expression purification, and biochemical characterization of LipGMTB
In an attempt to characterize LipG biochemical activities more finely, to confirm its carboxylesterase properties [21], and also to decipher the physiological role of this protein, the Rv0646c and MSEG_1352 genes have been cloned. The respective recombinant rLipGMTB and rLipGMS were produced in E. coli C41 (DE3)/pLyS strain harboring the pRare plasmid. rLipGMTB and purification was achieved using Ni2+-NTA affinity chromatography, yielding ∼1 mg of pure recombinant protein per culture liter. The purity and the expected molecular weight (∼35 kDa) were confirmed by 12% SDS/PAGE (Figure 2A) and MALDI-TOF analysis.
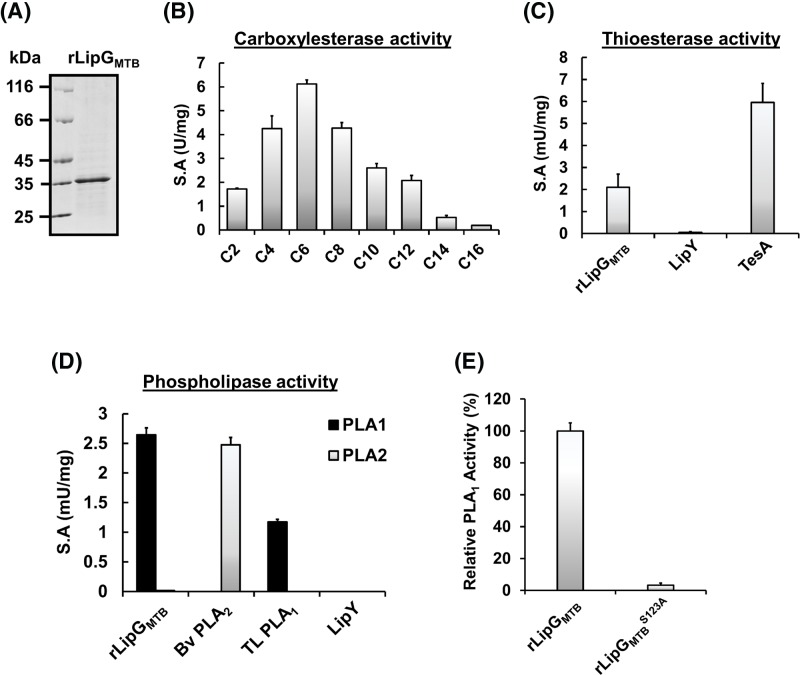
Figure 2. Purification and biochemical characterization of recombinant rLipGMTB.
(A) Purity assessment of rLipGMTB. rLipGMTB (5 µg) was loaded on to 12% polyacrylamide gel. Unstained protein molecular weight marker was used as standard (Euromedex). (B) Carboxylesterase S.A determination on pNP-ester substrates with chain lengths varying from C2 to C16. (C) Thioesterase activity measurements corresponding to palmitoyl-CoA hydrolysis. (D) PLA1 and PLA2 activity using fluorescent phospholipids. TL PLA1, Bv PLA2, and LipY were used as positive and negative controls, respectively. (E) PLA1 enzymatic activity of the rLipGMTBS123A compared with rLipGMTB. Results are expressed as relative activity in %. Each result is expressed as mean values ± S.D. of at least three independent assays. Abbreviation: Bv PLA2, Bee venom PLA2, TL PLA1, T. lanuginosus lipase PLA1.
The esterase activity of rLipGMTB was first assayed using pNP esters with various carbon chain lengths ranging from C2 to C16 (Figure 2B). Whatever their chain length, rLipGMTB was able to hydrolyze all pNP esters, therefore displaying a large substrate specificity as previously reported by Rastogi et al. [21]. While similar S.A were obtained with short pNP-C4 and medium pNP-C8 esters (mean S.A = 4.26 ± 0.11 U/mg). However, rLipGMTB exhibited a clear significant preference for pNP-C6 ester with a S.A value of 6.12 ± 0.17 U/mg. For pNP esters carrying more than eight carbon atoms, this S.A dropped sharply to 2.6 ± 0.18, 2.1 ± 0.21, 0.53 ± 0.08, and 0.19 ± 0.01 U/mg for pNP-C10, pNP-C12, pNP-C14, and pNP-C16, respectively (Figure 2B). Interestingly, amongst the Lip protein family member’s, rLipGMTB substrate specificity using pNP esters was similar to those of the esterase’s LipU and LipR [23].
The lipase activity of rLipGMTB has also been tested using a large panel of triacylglycerol (TAG) substrates, such as trioctanoin and triolein using the pH-stat technique, and pomegranate oil-coated TAG method [48]. In all cases, rLipGMTB was not able to hydrolyze any of these TAGs, even when using highly sensitive fluorogenic TAG substrates [40]. It is well known that lipases differ from esterases by their ability to hydrolyze long-chain acylglycerols at the oil–water interface, whereas esterases can only hydrolyze substrates with short- or medium-fatty acid chains [61]. From these findings, rLipGMTB could be considered as a true carboxylesterase, as recently proposed by Rastogi et al. [21].
Due to its genomic localization in the same cluster as the mmaA genes encoding for methyltransferase proteins, it is tempting to suggest that rLipGMTB might also be able to hydrolyze the thioester bond of acyl-CoA or MA intermediates as previously shown for Cut6 (Rv3802c). Cut6 is an essential bifunctional phospholipase A and thioesterase enzyme belonging to the Cutinase-like family proteins from M. tuberculosis [62]. Thioesterases are a group of enzymes that catalyze the hydrolysis of a thioester bond between a carbonyl group and a sulphur atom. Here, the hydrolysis of palmitoyl-CoA was used to measure the thioesterase activity of rLipGMTB. As shown in Figure 2C, rLipGMTB also exhibited a thioesterase activity with an S.A value of 2.10 ± 0.59 mU/mg. It is noteworthy that this value is of the same order of magnitude as that of TesA, a thioesterase of M. tuberculosis [42] with a S.A of 5.95 ± 0.85 mU/mg.
In order to investigate further rLipGMTB enzyme activity, its phospholipase activity was also investigated. Interestingly, rLipGMTB was able to hydrolyze only fluorescent phospholipids present at the sn−1 stereospecific position with S.A value of 2.75 ± 0.25 mU/mg (Figure 2D) suggesting that rLipGMTB is a strict PLA1. T. lanuginosus lipase PLA1 (TL PLA1) and Bee venom PLA2 (Bv PLA2) were used as positive controls for the PLA1 and PLA2 activities, respectively.
Finally, since Rastogi et al. [21] had proposed that the catalytic machinery was governed by Ser123, we thus generated by site-directed mutagenesis a rLipGMTBS123A variant, in which this Ser123 residue was mutated into an Ala residue. As predicted, no enzyme activity was observed with fluorescent-phospholipids, pNP-substrates or palmitoyl-CoA with this inactive mutant (Figure 2E and data not shown). Taken together, these data provide full evidence that Ser123 contained in the consensus sequence G121ASMG125 is the catalytic residue responsible for the enzyme activity, and consequently, confirms that LipGMTB is a true serine hydrolase. More specifically and given the level of S.A reached with the various substrates investigated, our results suggest that rLipGMTB is a phospholipase A1 enzyme which also displays esterase and thioesterase activities.
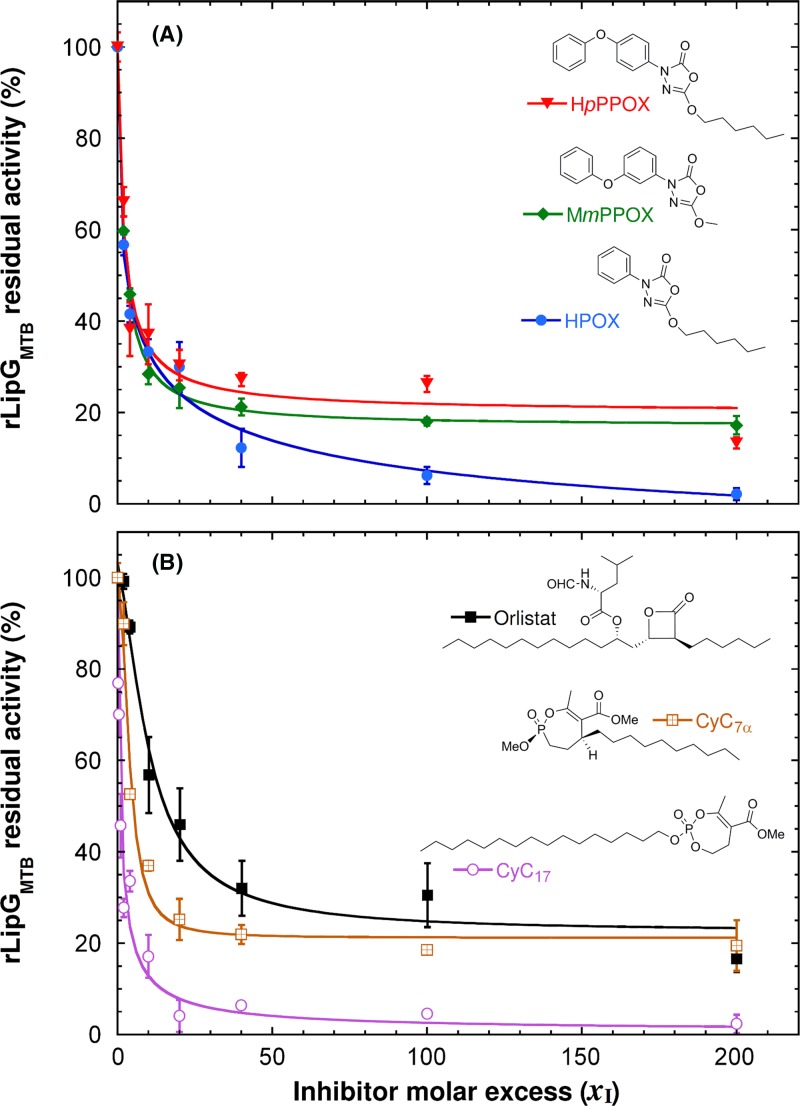
rLipGMTB is inhibited by OX derivatives and CyC analogs
Two series of recently reported antitubercular inhibitors discovered in our laboratory (OX derivatives and CyC analogs), exhibit activity against enzymes by forming a covalent bond with the catalytic serine or cysteine residue in their active sites [27–29]. In order to further investigate the ability of these compounds to efficiently inhibit the lipolytic activity of rLipGMTB, six OX derivatives (i.e. MmPPOX, iBPOX, iBpPPOX, HPOX, HpPPOX, and BePOX) [27]; and five CyC analogs (i.e. the CyC17, CyC7α, CyC7β, CyC8α, and CyC8β) [28,29] were then tested for inhibition of rLipGMTB. In our previous work, whereas iBpPPOX, and CyC7β and CyC8α exhibited strong activity against both extracellular and intramacrophagic M. tuberculosis; iBPOX and HpPPOX, and CyC7α and CyC8β were found active on infected macrophages only [28,42]. In contrast, HPOX and BePOX, and CyC17 efficiently block M. tuberculosis growth in vitro with no activity against intracellular bacilli [28,42]. Orlistat which has been previously reported as an inhibitor of LipG was used as positive control [63]. The chemical structures of the 12 compounds used in this study are provided in Supplementary Figure S2.
rLipGMTB was inactivated by all the six OX derivatives, with medium 60.6% to good 93.8% inhibition levels at xI = 100 (Table 2). The best inhibitors were HPOX, MmPPOX, and HpPPOX, which displayed inhibitor molar excess leading to 50% enzyme inhibition, i.e. xI50 values of ∼3.0 (Table 2 and Figure 3A). Amongst the CyC analogs, CyC7β, CyC8α, and CyC8β exhibited very weak inhibitory effects up to a high molar excess xI = 100. Whereas, a strong dose-dependent inhibition was observed with CyC17 and CyC7α (Table 2 and Figure 3B). In particular, CyC17 was found to react almost stoichiometrically with rLipGMTB, as confirmed by its xI50 value of 0.98. It is noteworthy, that Orlistat, used as reference inhibitor, displayed only a moderate inhibitory activity against pure rLipGMTB with only 69.5% inhibition at xI = 100 and a xI50 value of 15.2, similar to those of iBpPPOX, iBPOX, and BePOX.
Table 2. Inhibition of rLipGMTB after a 30-min incubation period with each inhibitor1.
| Compounds | % inhibition | xI50 | |
|---|---|---|---|
| xI = 4 | xI = 100 | ||
| MmPPOX | 54.1 ± 1.5 | 82.0 ± 3.8 | 3.1 |
| iBpPPOX | 40.5 ± 3.1 | 60.6 ± 0.22 | 14.7 |
| iBPOX | 32.7 ± 0.70 | 80.3 ± 0.90 | 15.0 |
| HpPPOX | 61.8 ± 4.7 | 73.8 ± 1.0 | 3.5 |
| HPOX | 58.5 ± 1.4 | 93.8 ± 5.7 | 2.9 |
| BePOX | 9.8 ± 0.28 | 68.8 ± 1.1 | 12.5 |
| CyC7α | 47.4 ± 0.81 | 81.4 ± 1.6 | 5.0 |
| CyC7β | 7.9 ± 0.72 | 57.6 ± 2.0 | 48.1 |
| CyC8α | 2.5 ± 0.10 | 51.3 ± 2.6 | 58.5 |
| CyC8β | 3.2 ± 0.16 | 32.0 ± 1.5 | >200 |
| CyC17 | 66.4 ± 4.5 | 95.4 ± 2.6 | 0.98 |
| Orlistat | 10.8 ± 0.17 | 69.5 ± 5.3 | 15.2 |
Inhibition data (% of initial activity), at inhibitor molar excess (xI) of 4 and 100 related to 1 mol of enzyme. Results are expressed as mean values ± S.D. of at least three independent assays. The inhibitor molar excess leading to 50% lipase inhibition, xI50, was determined as described in Experimental procedures section.
Figure 3. Inhibition studies of rLipGMTB by OX derivatives and CyC analogs.
Residual activities of LipGMTB measured on pNP-C6 as substrate. Effects of increasing molar excess (xI) of compounds (A) MmPPOX, HpPPOX, HPOX, and (B) CyC7α, CyC17 and Orlistat on the rates of hydrolysis of pNP-C6. LipGMTB was pre-incubated at various inhibitor molar excess (xI) for 30 min at 25°C. Kinetic assays were performed as described in Experimental procedures section. Results are expressed as mean values ± S.D. of three independent assays.
rLipGMTB and rLipGMS are peripheral membrane protein
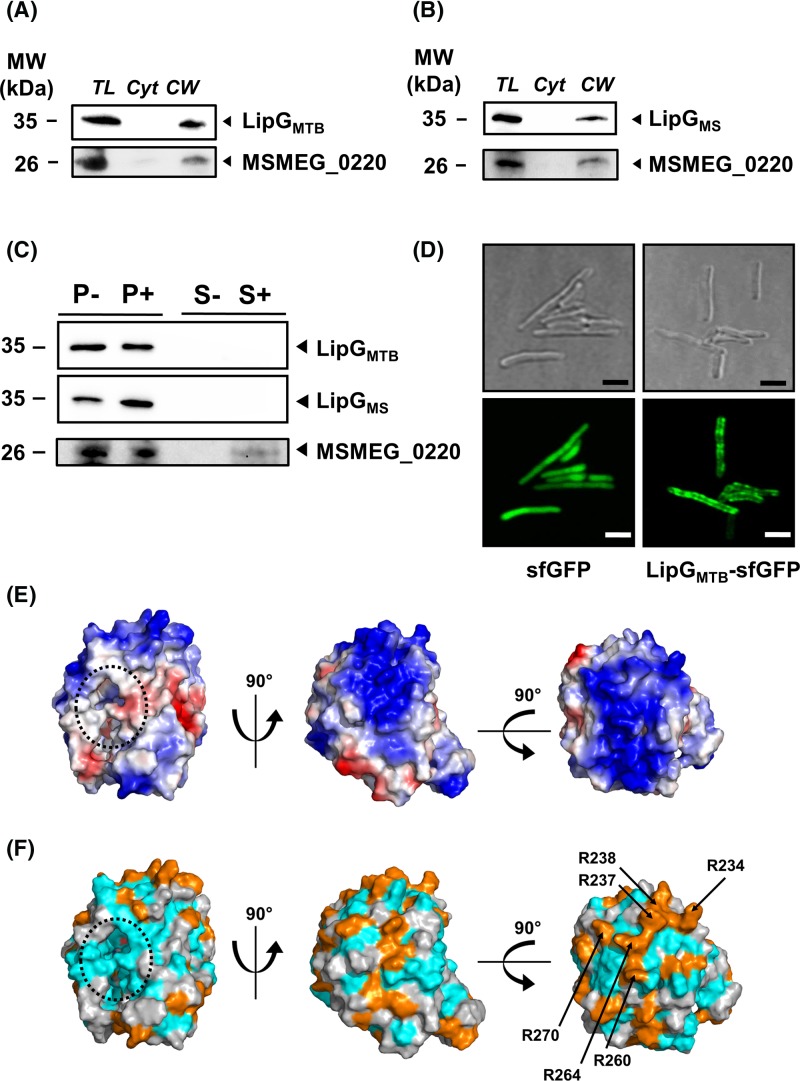
Although LipGMTB has been identified in the membrane fraction of M. tuberculosis H37Rv by using proteomic profiling approach [64], in silico experiments performed using SignalP 4.1 server [65] revealed that neither a peptide signal for the general secretion pathway or for the twin-arginine translocation pathway, nor a Type VII consensus secretion motif YxxxD/E were present in the primary sequence of LipG proteins. In this context, recombinant M. smegmatis strains, overexpressing either rLipGMS (i.e. MSMEG_1352 gene) or rLipGMTB (i.e. Rv0646c gene) cloned under the control of the hsp60 promoter in frame with a C-terminal 6× His-tag, were subjected to subcellular fractionation followed by immunoblotting experiments. The well-characterized cell wall exported monoglyceride lipase, MSMEG_0220, was used as control for cell fractionation [52,53]. In each case, both proteins were found only within the cell wall fraction and not in the soluble cytoplasmic fraction suggesting that LipG is a membrane/cell wall-associated protein (Figure 4A,B). Additional differential detergent extraction of surface-exposed mycobacterial proteins using Genapol®-X080 [54,55], followed by precipitation and immunoblotting (Figure 4C), demonstrated that LipG protein remains associated with the pellet fraction. This results suggest that this enzyme would not be accessible to the detergent and thus is not a surface-exposed protein. In contrast, MSMEG_0220 was found within the supernatant fraction, as expected (Figure 4C). All these data strengthen the fact that mycobacterial LipG are peripheral proteins, interacting with the inner leaflet of the cytoplasmic membrane.
Figure 4. Subcellular location of LipG proteins.
(A,B) Subcellular localization of LipGMS and LipGMTB in M. smegmatis groEL1ΔC determined by ultracentrifugation. Recombinant cells expressing 6× His-tagged LipG were lysed, total lysate (TL), cytoplasm (Cyt), and cell wall (CW) fractions were separated by ultracentrifugation. Samples were loaded on to 12% SDS/PAGE and immunoblotted using HisProbe reagent. The exported MSMEG_0220 was used as control. (C) Determination of cell-surface-exposition of LipGMS and LipGMTB in M. smegmatis groEL1ΔC by detergent extraction. Recombinant cells expressing 6× His-tagged LipG were treated with PBS-buffer or with PBS-buffer containing Genapol®-X080 detergent. Pellet (P) and supernatant (S) fractions containing cytoplasmic and surface-exposed proteins respectively, were separated by centrifugation. Samples were loaded on to 12% SDS/PAGE and immunoblotted using HisProbe reagent. The exported MSMEG_0220 was used as a control. (D) Fluorescence microscopy analysis of M. smegmatis groEL1ΔC strains carrying either WT sfGFP gene or lipGMTB-sfGFP translational fusions. Cells were analyzed in both phase contrast (upper panel) and fluorescent channels (bottom panel). Scale bars represent 1 µm. (E) Electrostatic potential of LipGMTB 3D model. The electrostatic surface potentials were displayed color-coded on to a van der Waals surface using the PyMOL Molecular Graphics System (version 1.8, Schrödinger, LLC). Red and blue colors represent net negative and positive charges, while white color represents overall neutral positions, respectively Black lines are showing the catalytic pocket. Two rotations of 90° were performed in order to provide a better view of the high potential area on top of the protein. (F) Position of positively charged residues in LipGMTB 3D model. Positives residues were highlighted in orange. The two rotations of 90° were conserved in order to provide a better view of the positively charged residues on top of the protein. An arginine-rich patch has been identified and position of the respective residues (R234, R237, R238, R260, R264 and R270) are marked with black arrows.
To go further, the dynamic location of LipG within living cells was investigated by using fluorescence microscopy [66]. Gene fusions between Rv0646c or MSMEG_1352 and the sfGFP gene encoding the sfGFP [67] were generated and used to transform M. smegmatis cells. Recombinant clones harboring stable fusions (Supplementary Figure S3) with a constant and important expression level were grown to mid-log exponential phase (i.e. OD600 = 1) in classical 7H9-S medium and further analyzed by fluorescence microscopy. As shown in Figure 4D (right panel), the strains harboring LipGMTB-sfGFP chimeric construct emitted a strong fluorescence signal mainly localized at the periphery of the inner mycobacterial cell membrane. In contrast, the fluorescence signal of the recombinant strain expressing only a wild-type sfGFP was less intense and diffuse through the cytoplasmic compartment (Figure 4D, left panel). Similar results were obtained when the bacteria was processed either at early exponential phase (i.e. OD600 = 0.2) or at the stationary phase (i.e. OD600 = 3). These findings strongly suggest that the resulting chimeric LipG proteins remain clustered into the bacterial cytoplasmic membrane whatever the phase of bacterial growth.
To get structural insights concerning LipGMTB and its potential interaction within the cytoplasmic membrane, a 3D structural model was built using the I-TASSER server [35,36]. The X-ray structure of Streptomyces lividans methylesterase RdmC (PDB id: 1Q0R), solved at 1.45 Å resolution was used as a template [68]. This generated model revealed that LipGMTB is a serine hydrolase constituted by 11 α-helices and 8 β-strands that are forming a central β-sheet (Supplementary Figure S4) as previously described [21]. A putative catalytic triad composed by Ser123, Asp251, and His279 is located within a highly accessible active site surrounded by hydrophobic residues forming the catalytic pocket (Supplementary Figure S4). In an effort to better characterize the molecular interactions that might govern LipG binding on to the inner cell membrane, the electrostatic surface potential of the previously built 3D model structure of LipGMTB (Supplementary Figure S4) was generated and is displayed in Figure 4E,F. Interestingly, a positive electrostatic patch located opposite to the active site, on top of the model structure, was found. Notably, this specific amino-acid patch is mainly constituted by six arginine residues highly conserved in several LipG orthologs (Figure 4E,F and Supplementary Figure S5). Such arginine residues, which are indeed known to play important role in protein binding to zwitterionic and anionic glycerolipid polar head groups [69,70], might thus contribute to the binding of LipG to the cytoplasmic membrane (Figure 4E,F).
Disruption of MSMEG_1352 triggers cell wall modifications
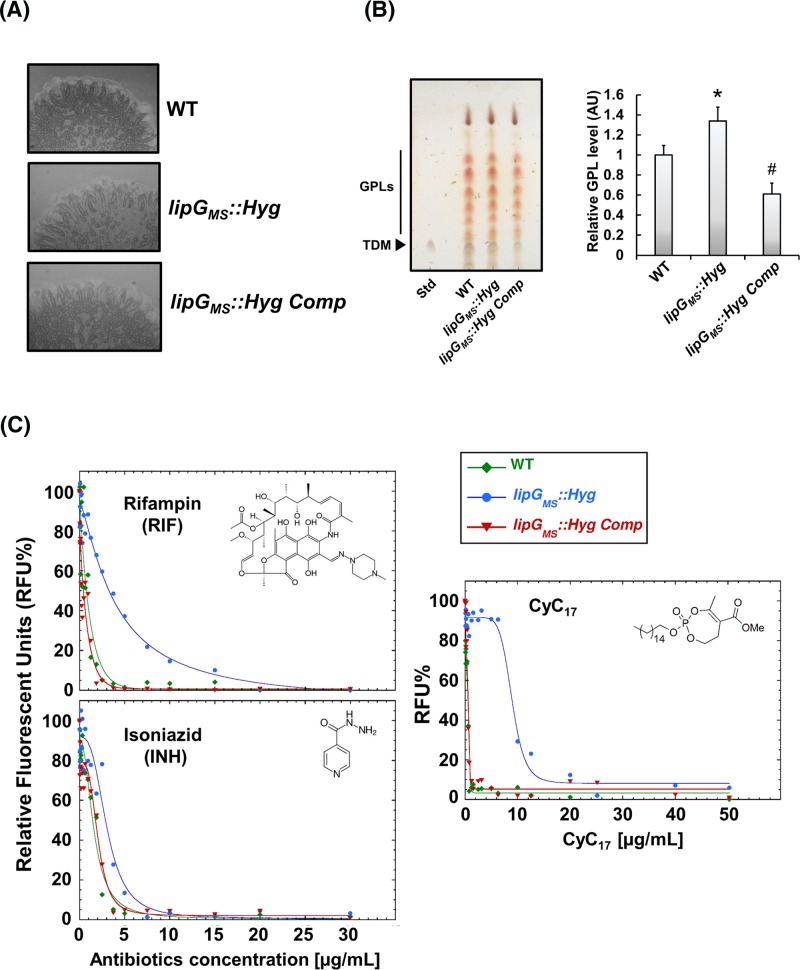
In order to identify the physiological role of the LipG proteins within the mycobacterial lifecycle, a M. smegmatis MSMEG_1352 disrupted strain named lipGMS::Hyg, in which the gene has been interrupted with a hygromycin resistance cassette, as well as a lipGMS::Hyg complemented strain (i.e. lipGMS::Hyg Comp) were produced (Supplementary Figure S6). When grown in 7H9-S medium for 48 h at 37°C, no significant differences in growth rate were observed between the WT, lipGMS::Hyg, and lipGMS::Hyg Comp strains. Interestingly, when the strains were grown on to 7H11TG agar plates (i.e. 7H11 medium supplemented with 0.02% Tween 80 and 0.2% glycerol), no differences in colony morphologies were observed with naked eyes. In contrast, when agar plates were spot-inoculated on to the same medium devoid of detergent and observed under a stereomicroscope, the lipGMS::Hyg mutant strain harbored a slightly smoother and rounder phenotype than the WT and lipGMS::Hyg Comp strains (Figure 5A). This finding would suggest that disruption of lipGMS may trigger some minor cell surface modifications (Figure 5A). TLC analysis of total lipid extract reveals that disruption of lipGMS induces a slight but significant overproduction of GPLs by 1.35-fold compared with WT strain (Figure 5B). In contrast, no detectable changes were observed within FAMEs and MAMEs levels as well as phospholipid content between each strain (Supplementary Figure S7A,B).
Figure 5. Disruption of lipGMS triggers cell-wall modifications and decreases antibiotics susceptibility.
(A) Colony morphology of WT, lipGMS disrupted and complemented strains. Recombinant strains were grown in 7H9-S before being spotted on to Middlebrook 7H11 agar plate devoid of Tween-80. Images are representative of two independent experiments. (B) GPL analysis of WT, lipGMS disrupted and complemented strains. Cells were grown in 7H9-S until OD600 nm ∼1–1.5. Lipid extraction was followed by TLC (left panel) and densitometry analysis (right panel). Results are expressed as mean ± S.D. of total GPL level from three independent experiments, where WT level was arbitrarily adjusted to 1 relative unit. Statistical analysis was performed by using the Student’s t test. Relative GPL level from lipGMS disrupted mutant were compared with the WT where * corresponded to a P-value <0.05. Relative GPL level from lipGMS complemented strain were compared with the lipGMS disrupted mutant where # corresponded to a P-value <0.05. (C) Antibiotic susceptibility of WT, lipGMS disrupted and complemented strains. Susceptibility toward INH, RIF, and CyC17 were determined using the REMA. Fluorometric MIC50s were determined by fitting the RFU% sigmoidal dose–response curves. Graph fitting are representative of three independent assays performed in duplicate.
Although bacterial cell wall remodeling (including misregulation of GPL production) may also affect biofilm/pellicle formation, neither global nor visible modifications where observed amongst the WT, lipGMS::Hyg, and lipGMS::Hyg Comp strains regarding pellicle formation (Supplementary Figure S7C) in 7H9-S medium containing either glycerol or acetate as carbon source.
Since the mycobacterial cell wall is the first effective barrier against bactericidal compounds, we hypothesized that increase in GPLs could also play an important role in cell wall integrity. To answer this question, the WT, lipGMS::Hyg, and lipGMS::Hyg Comp strains were subjected to antibiotic susceptibility testing toward standard antibiotics INH and RIF, using the REMA method [28,59] (Figure 5C and Table 3). The effects of Orlistat, HPOX as well as the CyC17 inhibitor on the growth of each strains were also investigated.
Table 3. Susceptibility testing of M. smegmatis WT and mutant strains.
| MIC50 (µg/ml)1 | Fold change compared with WT | |||||||
|---|---|---|---|---|---|---|---|---|
| INH | RIF | Orlistat | CyC17 | INH | RIF | Orlistat | CyC17 | |
| WT | 1.4 | 0.76 | 0.97 | 0.56 | - | - | - | - |
| lipGMS::Hyg | 2.9 | 3.3 | 6.5 | 9.6 | ×2.0 | ×4.3 | ×6.7 | ×15.7 |
| lipGMS::Hyg Comp | 1.8 | 0.89 | 1.2 | 0.60 | ×1.2 | ×1.2 | ×1.3 | ×0.94 |
| lipGMS::Hyg CompMTB | 1.9 | 0.96 | 1.6 | 0.65 | ×1.3 | ×1.3 | ×1.6 | ×1.1 |
MIC50 values corresponding to the compound minimal concentration leading to 50% growth inhibition have been determined by the REMA method and are expressed as mean values of three independent assays performed in duplicate.
It is noteworthy that the respective MIC50 values obtained with INH (0.76 µg/ml), RIF (1.4 µg/ml), and CyC17 (0.56 µg/ml) against the WT strain are in accordance with previously published studies (Figure 5C and Table 3) [28,29]. Regarding Orlistat, this well-known serine hydrolase inhibitor [23,71] is able to impair M. smegmatis growth with an MIC50 of 0.97 µg/ml. Surprisingly, deletion of lipGMS leads to an increased drug tolerance toward the four compounds. As depicted in Figure 5C and Table 3, MIC50 of INH, RIF, Orlistat, and CyC17 against the lipGMS::Hyg strain amounted to 2.9, 3.3, 6.5, and 9.6 µg/ml, respectively, thus corresponding to a respective fold change compared with the WT strain of ×2.0, ×4.3, ×6.7, and ×15.7. The complementation of the mutant strain with the pVV16::lipGMS restored the drug susceptibility with mean MIC50 fold change compared with WT strain of approximately ×1.2 to ×1.3.
Similar results were obtained with HPOX inhibitor. MIC50 values of 8.4 and 18.4 µg/ml were obtained for the WT and lipGMS::Hyg strains, respectively; leading to a fold change compared with the WT strain of ×2.2. However, with this compound the complementation with pVV16::lipGMS vector only partially restored the drug susceptibility with an intermediate MIC50 value of 12.2 µg/ml (i.e. fold change compared with WT strain of approximately ×1.5).
Finally, we also carried out these experiments by performing complementation of the lipGMS::Hyg strain with the pVV16::lipGMTB vector (lipGMS::Hyg CompMTB). Interestingly, production of LipGMTB protein was perfectly able to restore the drug susceptibility, suggesting that the two proteins may possess similar enzymatic activities and/or physiological functions (Table 3).
To further confirm this result, the recombinant rLipGMS was produced, purified, and biochemically characterized as described above for rLipGMTB (Supplementary Figure S8 and Table S2). Both pure rLipGMS and rLipGMTB proteins thus exhibit the same substrate specificity toward pNP ester substrates, the best S.A being also found with pNP caproate (C6). Moreover, rLipGMS was also able to hydrolyze acyl-CoA and phospholipids as substrates, confirming that the substrate specificity is well conserved between the two proteins. Despite their similar reactivity pattern with various esters, we noticed, however, that the rLipGMS was more active than its ortholog (Supplementary Table S2). Interestingly, these differences where only observed in vitro using pure proteins, but not during trans-complementation with the lipGMTB gene during in vivo experiments as described above.
Overexpression of lipGMTB impacts free fatty acids and phospholipids homeostasis
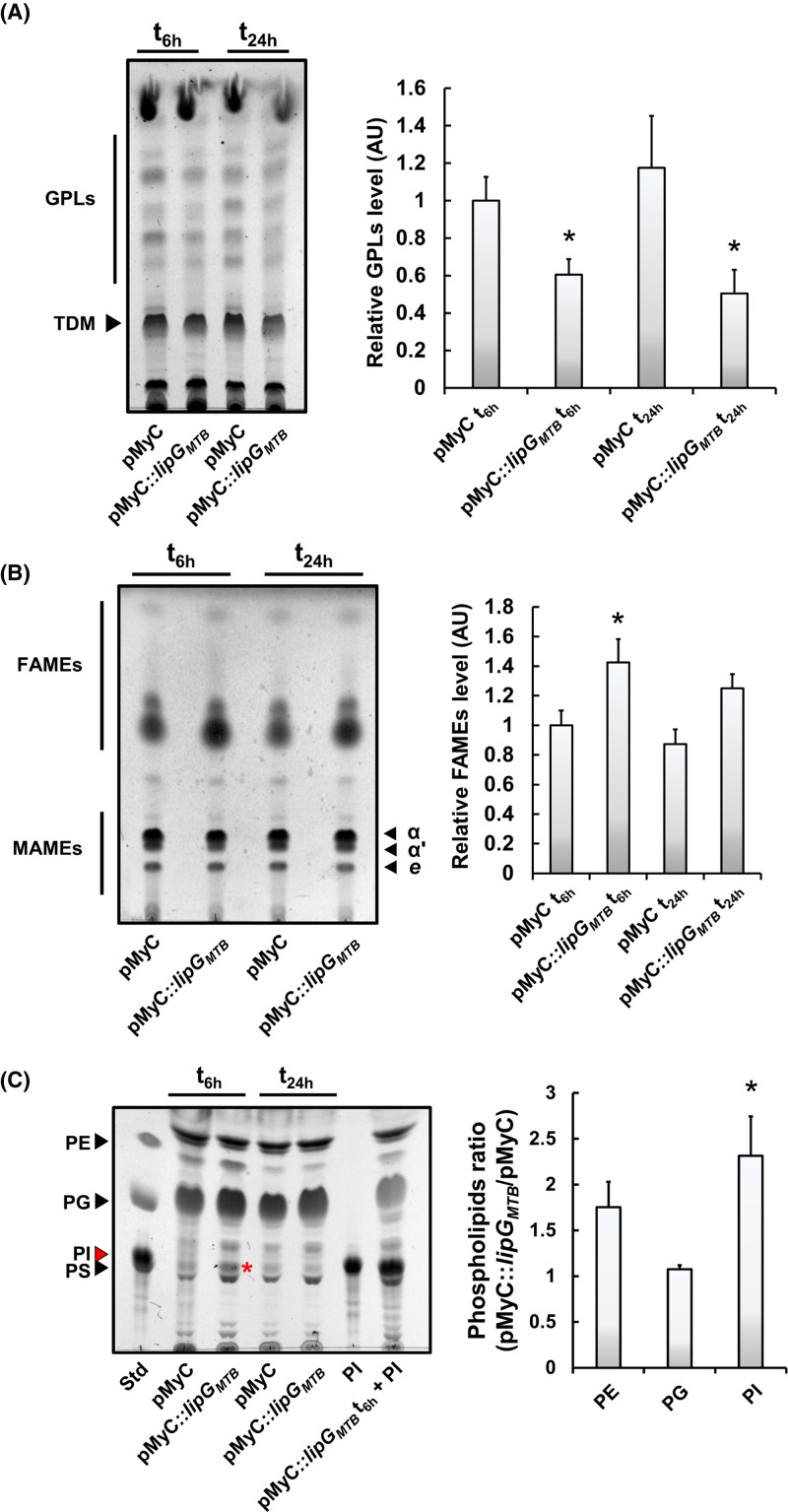
Despite the observed impact of lipGMS gene disruption on to the penetration/diffusion of the latter antitubercular molecules within the mycobacterial envelope, no clear detectable phenotype has been observed. To overcome such issue, a recombinant strain harboring the pMyC::lipGMTB plasmid which allows a strong overproduction of the LipGMTB protein upon acetamide induction, has been constructed. Approximately 6 and 24 h following induction by acetamide, total lipid extraction was performed on cell cultures either in exponential (OD600 ∼1–1.5) or stationary (OD600 ∼3–4) phase, respectively. The effect of LipGMTB overproduction was scrutinized by TLC analysis. As expected from our previous results which showed a slight increase in GPL production in absence of LipGMS (Figure 5A,B), in the strain overproducing LipGMTB (Figure 6A), a clear drop in GPLs production was observed at 6 h (61 ± 8%) and 24 h (51 ± 12%) post-induction when compared with the WT strain harboring the empty pMyC vector as reference. However, in contrast with M. smegmatis, the fact that M. tuberculosis H37Rv does not produce GPLs may suggest a different functional role of LipG in this latter mycobacteria. This prompted us to further analyze other lipid subclasses. Accordingly, a semi-quantitative analysis of M. smegmatis strains harboring pMyC and pMyC::lipGMTB revealed that the overproduction of the protein significantly increases the level of FAMEs by 1.42-fold and 1.25-fold compared with control strain at 6 and 24 h post-induction, respectively (Figure 6B), without affecting MAMEs level or ratio.
Figure 6. LipGMTB impacts free fatty acids and phospholipids biosynthesis.
Mycobacterial cultures harboring pMyC and pMyC::lipGMTB were grown in 7H9-S medium until reaching OD600 nm ∼0.5, then 0.2% of acetamide was added inside the culture medium. Cells were collected at 6 or 24 h post-induction corresponding to exponential or stationary phase, lyophilized and the same amount of dry cell weight was use for lipid extraction. After normalization, the same volume of each sample was loaded on to TLC. (A) Global GPL levels are decreasing in an M. smegmatis pMyC::lipGMTB overexpression strain. Lipid extracts of each samples were analyzed by TLC followed by densitometry analysis. (B) FAMEs levels are increasing in an M. smegmatis pMyC::lipGMTB overexpression strain. Lipid extracts of each samples were analyzed by TLC followed by densitometry analysis. In each case, results are expressed as mean ± S.D. of each respective total lipid species levels from at least two independent experiments, where control strain level at t6 h was arbitrary adjusted to 1 relative unit. (C) Phospholipid biosynthesis is increased in an M. smegmatis pMyC::lipGMTB overexpression strain. Lipid extracts of each samples were analyzed by TLC followed by densitometry analysis. TLC was performed with CHCl3-MeOH-H2O (65:25:4; v/v/v) as eluent and PI overproduction is indicated with a red asterisk (left panel). Lipid sample from M. smegmatis pMyC::lipGMTB at t6 h was also co-injected with pure PI standard as control lane, to clearly identify PI as an overproduced subspecies. Histograms correspond to densitometry analysis at t6 h. Results are displayed as intensity ratio where phospholipids levels from the pMyC::lipGMTB strain were divided by the phospholipids levels from the pMyC strain (right panel). Results are expressed as mean ± S.D. from three independent experiments. Statistical analysis was performed by using the Student’s t test where * correspond to a P-value <0.05. Abbreviation: PI, phosphatidylinositol.
Regarding phospholipid levels, surprisingly overexpression of lipGMTB slightly increases the phospholipid level of phosphatidylethanolamine (PE) and phosphatidylglycerol (PG) subspecies (1.75- and 1.07-fold in comparison with the control strain, respectively), but more importantly, this overproduction drastically impacts phosphatidylinositol (PI) production with a fold-change related to the control strain of ×2.3 (Figure 6C). These results were further confirmed by using 2D-TLC analysis, where a nice and efficient separation of all species of phospholipids allowed to confirm a consequent increase in PI biosynthesis upon LipGMTB overproduction (Supplementary Figure S9).
Altogether, the increase in the levels of both FAMEs and specific subclass of phospholipids suggest that the LipGMTB acyl-CoA-thioesterase activity may be the predominant biochemical activity in vivo, and that the products of the reaction are directly influencing cell wall phospholipids composition.
Discussion
M. tuberculosis is one of the most successful infectious agents worldwide. Therefore, understanding the molecular mechanisms responsible for its pathogenesis remains one of the most important challenges in TB fundamental research. By using highly elaborate genetic screens, several independent teams have indeed tried to identify the genes required for in vitro or in vivo bacterial survival [26,72,73]. These approaches have highlighted the Rv0646c gene of M. tuberculosis which is required for adaptation and survival of the bacilli in mouse macrophages [26]. Interestingly, this gene has been annotated as encoding a putative lipase/esterase protein based on the presence of a G-x-S-x-G consensus pentapeptide motif. More importantly, the Rv0646c gene encoding LipGMTB is widely conserved through the Mycobacterium genus and is associated with a gene cluster involved in MAs maturation processes [19]. Strikingly, bioinformatics analysis demonstrated that this gene is highly conserved in both slow- and fast-growing mycobacteria, and even in the obligate intracellular parasite M. leprae. This latter unusual pathogen is harboring a genome with a large number of inactivated pseudogenes which are not required anymore in vivo [74]. However, amongst the 24 genes encoding lipolytic enzymes from the Lip family in M. tuberculosis, only lipE, lipU, and lipG are conserved in M. leprae, thus reinforcing an essential role played by these enzymes in intracellular survival and proliferation [74,75]. In this study based on biochemical characterization of rLipGMTB protein associated with microbiology experiments, we uncovered the potential role of this enzyme in mycobacterial lipid metabolism.
In accordance with a recent report, we confirmed that recombinant LipGMTB protein is an esterase acting preferentially on medium chain-length substrates [21,75]. More interestingly, we also found that the protein was able to hydrolyze phospholipids at the sn−1 position and cleave the thioester bond from palmitoyl-CoA, showing that rLipGMTB possesses esterase and phospholipase, as well as thioesterase activities (Figure 2). Both phospholipase and thioesterase activities have been already observed by Parker et al. [62], who described that M. tuberculosis Rv3802c gene (cut6) encodes a functional phospholipase/thioesterase protein. Using radiolabeled phospholipids coupled with TLC analysis, the authors showed that Cut6 was not only able to act on PE, PC, and phosphatidylserine (PS) molecules [62], but that it was also able to hydrolyze both palmitoyl- and decanoyl-CoA, suggesting that the protein may act as an acyl donor in vivo.
The fact that LipG was required for adaptation and survival within macrophages, prompted us to investigate the inhibitory properties of two new series of serine hydrolase inhibitors active against M. tuberculosis growth [27–29]. During in vitro inhibition experiments on pure protein (Figure 3 and Table 2), the best inhibitors from the two series of compounds were HPOX (for the OX derivatives) on the one hand, and CyC17 (for the CyC analogs) on the other hand; two molecules that are only active against extracellularly growing M. tuberculosis strain [27,28]. In the case of CyC17, this result was a posteriori predictable. Following activity-based protein profiling experiment coupled with proteomics analyses [28], LipG was indeed found as a potential target of this compound from a lysate of M. tuberculosis (unpublished data). However, its low coverage (21% sequence identity) with only five distinct peptide sequences identified together with a peptide score ∼2.0–3.4 (given by SequestHT algorithm) made that this protein was below our threshold and was not selected. Conversely, LipG was not identified from similar ABPP experiments performed with HPOX directly on living M. tuberculosis cells [27]. We recently demonstrated that both the OX and CyC families of compound are multitarget inhibitors, which are probably displaying pleiotropic and cumulative effects toward mycobacterial Ser- and Cys-containing enzymes involved in lipid metabolism and/or cell wall maintenance, thus leading to M. tuberculosis growth arrest [27,28]. From these findings and despite their strong inhibitory effect in vitro on pure LipG enzyme, we thus proposed that in vivo HPOX and CyC17 preferentially impair other previously identified target enzymes such as the Ag85 complex or the thioesterase TesA rather than LipGMTB.
Based on proteomic analysis of the M. tuberculosis membrane fraction, it has been recently proposed that LipGMTB may be localized to the cell surface and directly interact with the host immune system [21,64]. However, bioinformatics analysis did not reveal the presence of any secretion signals or specific motifs, which may indicate that the protein is surface exposed. Herein, using different cell-fractionation approaches, we demonstrated that LipGMTB is associated with the membrane fraction [54,55,66], but not translocated across the cytoplasmic membrane and remains a cytoplasmic protein which is mainly peripheral (Figure 4). It has been described that such peripheral proteins require either a lipophilic attachment (i.e. acylation) or the presence of basic residues positively charged [69,70] which induce electrostatic interaction with the acidic and negatively charged phospholipids head-groups, thus allowing anchoring of the protein within the cytoplasmic membrane [70]. Based on our 3D structure model, surface electrostatic potential analysis allows identification of a region on top of the active site mainly composed by positively charged residues (Figure 4E,F). This area is essentially constituted by a C-terminal cluster of arginine residues fully conserved in numerous LipG proteins (Supplementary Figure S5). Since this type of electrostatic interaction has already been described for a mycobacterial protein involved in acyl-CoA metabolism [76], one can assume that this cluster of Arg residues would play an important role in LipG binding to the cell membrane, and thus for its physiological function.
In contrast with Cut6, which is known for being secreted and acting outside the mycobacterial cell [62,77], LipGMTB and LipGMS are presumably hydrolyzing cytoplasmic substrates. Two distinct strategies were employed: (i) the generation of a lipGMS disrupted strain which was trans-complemented with either lipGMS or lipGMTB and (ii) the generation of a lipGMTB heterologous overexpression strain which were useful in identifying the potential physiological role of LipG proteins and to define their substrates in vivo. On one hand, the observed increase in GPLs production due to lipG gene disruption (Figure 5) resulted in a significant reduced susceptibility to RIF, INH, Orlistat, HPOX, and above all CyC17 for which the highest fold change compared with WT strain in MIC50 value has been reached (Table 3). Since RIF and INH are inhibiting specifically cytoplasmic targets, we hypothesized that such increase in GPL will affect cell wall fluidity and integrity; which subsequently impacts the penetration of the main two first-line anti-TB drugs [78]. Such results are in agreement with Khoo et al. work [79] who reported that Mycobacterium avium GPLs positive serovar were less sensitive to ethambutol than GPLs negative serovar, and concluded that hydrophobic GPLs production may alter cell wall permeability and correlate with antibiotic tolerance [79]. In other hand, the overproduction of the LipGMTB protein leads to a significant increase in the FAMEs level (1.42-fold and 1.25-fold at 6 and 24 h post-induction, respectively), but more importantly a decrease in GPLs levels within M. smegmatis concomitant to an increase in level of phospholipid species and more particularly, the phosphatidyl inositol (Figure 6). However this is not the first time that such modifications of the cell wall composition have been observed. Similar phenotypes with drastic alteration of FAMEs/MAMEs and phospholipid subspecies levels have already been described in 2009, by Meniche et al. [58]. The authors demonstrated that overexpression of the Corynebacterium glutamicum gene NCgl2775 encoding a functional thioesterase/phospholipase protein leads to an increase in MAs production concomitantly with a decrease in glycerophosphoslipids levels [58]. A similar result was also obtained when the MSMEG_6394 protein (i.e. NCgl2775 and Cut6 orthologs) was overproduced, which prompted the authors to propose that this kind of bifunctional enzyme is may be able to regulate membrane compositions under specific conditions [58].
Since not all the mycobacterial strain are GPLs producers, especially M. tuberculosis H37Rv, we postulated that this phenotype was more an indirect effect, than a direct catabolic process transposable to M. tuberculosis.
From all these findings, it is tempting to propose that LipG is mainly acting as a thioesterase dedicated to phospholipid metabolism in vivo. In a large number of bacteria, the contribution of thioesterase proteins to phospholipid biosynthetic process is however not well-defined. For example, in E. coli, glycerol starvation leads to an increase in acyl-ACP and a drastic diminution of phospholipid levels. Overexpression of TesA, a type I thioesterase, drastically reduced the accumulation of long-chain acyl-ACP species, probably by rerouting free fatty acids to phospholipids [80]. It has also been proposed that the ybgC gene encoding a functional acyl thioesterase is playing a role in phospholipid metabolism in Gram-negative bacteria [81–83]. Astonishingly, this latter enzyme is known for interacting with a large number of proteins dedicated to phospholipid homeostasis, such as the acyl-carrier protein, the glycerol 3-phosphate acyltransferase (PlsB), and the PS synthase (PssA) [82]. Such systems dedicated to phospholipids have not been described so far in mycobacteria, but a similar multicomponent complex is involved in MA biosynthesis [16,17]. Overall, it might be possible that some proteins work together in order to respond to specific stimuli and thus modulate biological membrane composition and fluidity. Such a physiological process has also been reported in eukaryotic cells, where upon inflammatory stimulus, the acyl-CoA thioesterase 7 (ACOT7) converts acyl-CoA molecules into free fatty acids, thus triggering a remodeling of phospholipid containing unsaturated long (≥C20)-acyl chains in macrophages [84].
Finally, PI is known for being an essential and abundant phospholipid species within mycobacterial cytoplasmic membrane [85], but also for being a major precursor for complex glycoconjugated lipids such as phosphatidyl mannosides, lipomannan, or lipoarabinomannan [86,87]. Observation of an important increase in PI subspecies during LipGMTB overproduction may also impact the formation of such glycolipids that are highly contributing to virulence and modulation of the host immune response. The potential role of the LipGMTB protein within these pathways is currently under investigation in our laboratory and may bring new insight concerning the biosynthesis of such complex lipids, but also the role of this highly conserved protein during host–pathogen cross-talk.
Supporting information
Table S1. Primers used in this study. Restrictions sites or mutated codons are indicated in bold.
Table S2. Specific activities (S.A) of rLipG proteins. Experiments were performed with same amount of rLipGMTB or rLipGMS following the procedures described in Material and Methods.
Figure S1. LipG proteins multiple sequence alignment.
Figure S2. Chemical structure of CyC analogs and OX derivatives used in this study.
Figure S3. Western-Blot analysis of recombinant strains expressing LipG or LipG-sfGFP translational fusions.
Figure S4. 3D model of LipGMTB protein and identification of a putative catalytic triad.
Figure S5. Arginine-rich patch conservation and composition.
Figure S6. Schematic representation of lipGMS::Hyg strain construction and validation.
Figure S7. Biofilm phenotypes and lipid analysis of WT, lipGMS::Hyg and complemented strains.
Figure S8. Purification and biochemical characterization of recombinant rLipGMS.
Figure S9. Phosphatidylinositol identification by two-dimension TLC analysis.
Acknowledgments
We thank E. Cascales for providing the pKD4-sfGFP plasmid. Special thanks are addressed to Dr Chantal de Chastellier, Prof Sophie Bleves and Dr Eric Durand for their advice and continuous support. P.S., J.-F.C. and S.C. would like to thank Orane Chabiron, Loic Marcelly and Pauline Théolissat for technical assistance. We also thank Dr Thierry Durand for his precious help and advice in chemical synthesis of oxadiazolone compounds. This work has benefited from the facilities and expertise of the Platform for Microscopy of the Institut de Microbiologie de la Méditerranée FR3479 CNRS.
Abbreviations
- CyC
cyclophostin and cyclipostins analogs
- DTNB
5-5′-Dithio-bis 2-nitrobenzoic acid
- FAME
fatty acid methyl ester
- FAS
fatty acid synthase
- GPL
glycopeptidolipid
- INH
isoniazid
- MA
mycolic acid
- MAME
MA methyl ester
- OX
oxadiazolone-core derivative
- palmitoyl-CoA
palmitoyl-Coenzyme A
- PI
phosphatidylinositol
- pNP
p-nitrophenyl
- PS
phosphatidylserine
- REMA
resazurin microtiter assay
- RIF
rifampicin
- S.A
specific activity
- sfGFP
superfolder GFP
- TAG
triacylglycerol
- TB
tuberculosis
Competing interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was supported by the CNRS; the Aix-Marseille Université; and the Ph.D. fellowship from the Ministère Français de l’Enseignement Supérieur, de la Recherche et de l’Innovation.
Author contribution
P.S., J.-F.C., and S.C. conceived and designed research. P.S., V.P., I.P., C.S.-A., and J.-F.C. performed the experiments. P.S., V.P., C.-S.A., J.-F.C., and S.C. contributed to data analysis and interpretation. C.C., J.M.G., A.G., and C.D.S. provided chemical compounds. P.S., J.-F.C., and S.C. wrote the manuscript. This work was proposed and supervised by S.C.
References
- 1.WHO (2017) Global Tuberculosis Report 2017, http://www.who.int/tb/publications/global_report/gtbr2017_main_text.pdf [Google Scholar]
- 2.Zumla A., Nahid P. and Cole S.T. (2013) Advances in the development of new tuberculosis drugs and treatment regimens. Nat. Rev. Drug Discov. 12, 388–404 10.1038/nrd4001 [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y. and Yew W.W. (2009) Mechanisms of drug resistance in Mycobacterium tuberculosis. Int. J. Tuberc. Lung Dis. 13, 1320–1330 [PubMed] [Google Scholar]
- 4.Zhang Y. and Yew W.W. (2015) Mechanisms of drug resistance in Mycobacterium tuberculosis: update 2015. Int. J. Tuberc. Lung Dis. 19, 1276–1289 10.5588/ijtld.15.0389 [DOI] [PubMed] [Google Scholar]
- 5.Velayati A.A., Masjedi M.R., Farnia P., Tabarsi P., Ghanavi J., ZiaZarifi A.H.. et al. (2009) Emergence of new forms of totally drug-resistant tuberculosis bacilli: super extensively drug-resistant tuberculosis or totally drug-resistant strains in iran. Chest 136, 420–425 10.1378/chest.08-2427 [DOI] [PubMed] [Google Scholar]
- 6.Daffe M. and Draper P. (1998) The envelope layers of mycobacteria with reference to their pathogenicity. Adv. Microb. Physiol. 39, 131–203 10.1016/S0065-2911(08)60016-8 [DOI] [PubMed] [Google Scholar]
- 7.Daffe M. (2015) The cell envelope of tubercle bacilli. Tuberculosis (Edinb.) 95 (Suppl. 1), S155–S158 10.1016/j.tube.2015.02.024 [DOI] [PubMed] [Google Scholar]
- 8.Marrakchi H., Laneelle M.A. and Daffe M. (2014) Mycolic acids: structures, biosynthesis, and beyond. Chem. Biol. 21, 67–85 10.1016/j.chembiol.2013.11.011 [DOI] [PubMed] [Google Scholar]
- 9.Neyrolles O. and Guilhot C. (2011) Recent advances in deciphering the contribution of Mycobacterium tuberculosis lipids to pathogenesis. Tuberculosis (Edinb.) 91, 187–195 10.1016/j.tube.2011.01.002 [DOI] [PubMed] [Google Scholar]
- 10.Banerjee A., Dubnau E., Quemard A., Balasubramanian V., Um K.S., Wilson T.. et al. (1994) inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science 263, 227–230 10.1126/science.8284673 [DOI] [PubMed] [Google Scholar]
- 11.Vilcheze C., Wang F., Arai M., Hazbon M.H., Colangeli R., Kremer L.. et al. (2006) Transfer of a point mutation in Mycobacterium tuberculosis inhA resolves the target of isoniazid. Nat. Med. 12, 1027–1029 10.1038/nm1466 [DOI] [PubMed] [Google Scholar]
- 12.North E.J., Jackson M. and Lee R.E. (2014) New approaches to target the mycolic acid biosynthesis pathway for the development of tuberculosis therapeutics. Curr. Pharm. Des. 20, 4357–4378 10.2174/1381612819666131118203641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takayama K., Wang C. and Besra G.S. (2005) Pathway to synthesis and processing of mycolic acids in Mycobacterium tuberculosis. Clin. Microbiol. Rev. 18, 81–101 10.1128/CMR.18.1.81-101.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pawelczyk J. and Kremer L. (2014) The molecular genetics of mycolic acid biosynthesis. Microbiol. Spectr. 2, MGM2–0003-2013, [DOI] [PubMed] [Google Scholar]
- 15.Brennan P.J. and Nikaido H. (1995) The envelope of mycobacteria. Annu. Rev. Biochem. 64, 29–63 10.1146/annurev.bi.64.070195.000333 [DOI] [PubMed] [Google Scholar]
- 16.Veyron-Churlet R., Guerrini O., Mourey L., Daffe M. and Zerbib D. (2004) Protein-protein interactions within the Fatty Acid Synthase-II system of Mycobacterium tuberculosis are essential for mycobacterial viability. Mol. Microbiol. 54, 1161–1172 10.1111/j.1365-2958.2004.04334.x [DOI] [PubMed] [Google Scholar]
- 17.Veyron-Churlet R., Bigot S., Guerrini O., Verdoux S., Malaga W., Daffe M.. et al. (2005) The biosynthesis of mycolic acids in Mycobacterium tuberculosis relies on multiple specialized elongation complexes interconnected by specific protein-protein interactions. J. Mol. Biol. 353, 847–858 10.1016/j.jmb.2005.09.016 [DOI] [PubMed] [Google Scholar]
- 18.Cantaloube S., Veyron-Churlet R., Haddache N., Daffe M. and Zerbib D. (2011) The Mycobacterium tuberculosis FAS-II dehydratases and methyltransferases define the specificity of the mycolic acid elongation complexes. PLoS ONE 6, e29564 10.1371/journal.pone.0029564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cole S.T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D.. et al. (1998) Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544 10.1038/31159 [DOI] [PubMed] [Google Scholar]
- 20.Dubnau E., Chan J., Raynaud C., Mohan V.P., Laneelle M.A., Yu K.. et al. (2000) Oxygenated mycolic acids are necessary for virulence of Mycobacterium tuberculosis in mice. Mol. Microbiol. 36, 630–637 10.1046/j.1365-2958.2000.01882.x [DOI] [PubMed] [Google Scholar]
- 21.Rastogi R., Kumar A., Kaur J., Saini V. and Bhatnagar A. (2018) Rv0646c, an esterase from M. tuberculosis, up-regulates the host immune response in THP-1 macrophages cells. Mol. Cell. Biochem. 10.1007/s11010-018-3303-2 [DOI] [PubMed] [Google Scholar]
- 22.Camus J.C., Pryor M.J., Medigue C. and Cole S.T. (2002) Re-annotation of the genome sequence of Mycobacterium tuberculosis H37Rv. Microbiology 148, 2967–2973 10.1099/00221287-148-10-2967 [DOI] [PubMed] [Google Scholar]
- 23.Delorme V., Diomande S.V., Dedieu L., Cavalier J.F., Carriere F., Kremer L.. et al. (2012) MmPPOX inhibits Mycobacterium tuberculosis lipolytic enzymes belonging to the hormone-sensitive lipase family and alters mycobacterial growth. PLoS ONE 7, e46493 10.1371/journal.pone.0046493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dedieu L., Serveau-Avesque C., Kremer L. and Canaan S. (2013) Mycobacterial lipolytic enzymes: a gold mine for tuberculosis research. Biochimie 95, 66–73 10.1016/j.biochi.2012.07.008 [DOI] [PubMed] [Google Scholar]
- 25.Deb C., Daniel J., Sirakova T.D., Abomoelak B., Dubey V.S. and Kolattukudy P.E. (2006) A novel lipase belonging to the hormone-sensitive lipase family induced under starvation to utilize stored triacylglycerol in Mycobacterium tuberculosis. J. Biol. Chem. 281, 3866–3875 10.1074/jbc.M505556200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rengarajan J., Bloom B.R. and Rubin E.J. (2005) Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages. Proc. Natl. Acad. Sci. U.S.A. 102, 8327–8332 10.1073/pnas.0503272102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nguyen P.C., Delorme V., Bénarouche A., Guy A., Landry V., Audebert S.. et al. (2018) Oxadiazolone derivatives, new promising multi-target inhibitors against M. tuberculosis. Bioorg. Chem. 81, 414–424 10.1016/j.bioorg.2018.08.025 [DOI] [PubMed] [Google Scholar]
- 28.Nguyen P.C., Delorme V., Benarouche A., Martin B.P., Paudel R., Gnawali G.R.. et al. (2017) Cyclipostins and Cyclophostin analogs as promising compounds in the fight against tuberculosis. Sci. Rep. 7, 11751 10.1038/s41598-017-11843-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nguyen P.C., Madani A., Santucci P., Martin B.P., Paudel R.R., Delattre S.. et al. (2018) Cyclophostin and Cyclipostins analogues, new promising molecules to treat mycobacterial-related diseases. Int. J. Antimicrob. Agents 51, 651–654 10.1016/j.ijantimicag.2017.12.001 [DOI] [PubMed] [Google Scholar]
- 30.Viljoen A., Richard M., Nguyen P.C., Fourquet P., Camoin L., Paudal R.R.. et al. (2018) Cyclipostins and cyclophostin analogs inhibit the antigen 85C from Mycobacterium tuberculosis both in vitro and in vivo. J. Biol. Chem. 293, 2755–2769 10.1074/jbc.RA117.000760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noens E.E., Williams C., Anandhakrishnan M., Poulsen C., Ehebauer M.T. and Wilmanns M. (2011) Improved mycobacterial protein production using a Mycobacterium smegmatis groEL1DeltaC expression strain. BMC Biotechnol. 11, 27 10.1186/1472-6750-11-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kapopoulou A., Lew J.M. and Cole S.T. (2011) The MycoBrowser portal: a comprehensive and manually annotated resource for mycobacterial genomes. Tuberculosis (Edinb.) 91, 8–13 10.1016/j.tube.2010.09.006 [DOI] [PubMed] [Google Scholar]
- 33.Corpet F. (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16, 10881–10890 10.1093/nar/16.22.10881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robert X. and Gouet P. (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–324 10.1093/nar/gku316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y. (2008) I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9, 40 10.1186/1471-2105-9-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roy A., Kucukural A. and Zhang Y. (2010) I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 10.1038/nprot.2010.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dolinsky T.J., Nielsen J.E., McCammon J.A. and Baker N.A. (2004) PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 32, W665–667 10.1093/nar/gkh381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dolinsky T.J., Czodrowski P., Li H., Nielsen J.E., Jensen J.H., Klebe G.. et al. (2007) PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 35, W522–W525 10.1093/nar/gkm276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baker N.A., Sept D., Joseph S., Holst M.J. and McCammon J.A. (2001) Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 10.1073/pnas.181342398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dedieu L., Serveau-Avesque C. and Canaan S. (2013) Identification of residues involved in substrate specificity and cytotoxicity of two closely related cutinases from Mycobacterium tuberculosis. PLoS ONE 8, e66913 10.1371/journal.pone.0066913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Flaugnatti N., Le T.T., Canaan S., Aschtgen M.S., Nguyen V.S., Blangy S.. et al. (2016) A phospholipase A1 antibacterial Type VI secretion effector interacts directly with the C-terminal domain of the VgrG spike protein for delivery. Mol. Microbiol. 99, 1099–1118 10.1111/mmi.13292 [DOI] [PubMed] [Google Scholar]
- 42.Nyguen P.C., Nguyen V.S., Martin B.P., Fourquet P., Camoin L., Spilling C.D.. et al. (2018) Biochemical and Structural Characterization of TesA, a Major Thioesterase Required for Outer-Envelope Lipid Biosynthesis in Mycobacterium tuberculosis. J Mol Biol. 430 (24), 5120–5136 10.1016/j.jmb.2018.09.017 [DOI] [PubMed] [Google Scholar]
- 43.Brust B., Lecoufle M., Tuaillon E., Dedieu L., Canaan S., Valverde V.. et al. (2011) Mycobacterium tuberculosis lipolytic enzymes as potential biomarkers for the diagnosis of active tuberculosis. PLoS ONE 6, e25078 10.1371/journal.pone.0025078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Point V., Malla R.K., Diomande S., Martin B.P., Delorme V., Carriere F.. et al. (2012) Synthesis and kinetic evaluation of cyclophostin and cyclipostins phosphonate analogs as selective and potent inhibitors of microbial lipases. J. Med. Chem. 55, 10204–10219 10.1021/jm301216x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vasilieva E., Dutta S., Malla R.K., Martin B.P., Spilling C.D. and Dupureur C.M. (2015) Rat hormone sensitive lipase inhibition by cyclipostins and their analogs. Bioorg. Med. Chem. 23, 944–952 10.1016/j.bmc.2015.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martin B.P., Vasilieva E., Dupureur C.M. and Spilling C.D. (2015) Synthesis and comparison of the biological activity of monocyclic phosphonate, difluorophosphonate and phosphate analogs of the natural AChE inhibitor cyclophostin. Bioorg. Med. Chem. 23, 7529–7534 10.1016/j.bmc.2015.10.044 [DOI] [PubMed] [Google Scholar]
- 47.Point V., Malla R.K., Diomande S., Martin B.P., Delorme V., Carriere F.. et al. (2012) Synthesis and kinetic evaluation of cyclophostin and cyclipostins phosphonate analogs as selective and potent inhibitors of microbial lipases. J. Med. Chem. 55, 10204–10219 10.1021/jm301216x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ulker S., Placidi C., Point V., Gadenne B., Serveau-Avesque C., Canaan S.. et al. (2016) New lipase assay using Pomegranate oil coating in microtiter plates. Biochimie 120, 110–118 10.1016/j.biochi.2015.09.004 [DOI] [PubMed] [Google Scholar]
- 49.van Kessel J.C. and Hatfull G.F. (2007) Recombineering in Mycobacterium tuberculosis. Nat. Methods 4, 147–152 10.1038/nmeth996 [DOI] [PubMed] [Google Scholar]
- 50.Goude R., Roberts D.M. and Parish T. (2015) Electroporation of mycobacteria. Methods Mol. Biol. 1285, 117–130 10.1007/978-1-4939-2450-9_7 [DOI] [PubMed] [Google Scholar]
- 51.Rezwan M., Laneelle M.A., Sander P. and Daffe M. (2007) Breaking down the wall: fractionation of mycobacteria. J. Microbiol. Methods 68, 32–39 10.1016/j.mimet.2006.05.016 [DOI] [PubMed] [Google Scholar]
- 52.Cotes K., Dhouib R., Douchet I., Chahinian H., de Caro A., Carriere F.. et al. (2007) Characterization of an exported monoglyceride lipase from Mycobacterium tuberculosis possibly involved in the metabolism of host cell membrane lipids. Biochem. J. 408, 417–427 10.1042/BJ20070745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dhouib R., Laval F., Carriere F., Daffe M. and Canaan S. (2010) A monoacylglycerol lipase from Mycobacterium smegmatis Involved in bacterial cell interaction. J. Bacteriol. 192, 4776–4785 10.1128/JB.00261-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cascioferro A., Delogu G., Colone M., Sali M., Stringaro A., Arancia G.. et al. (2007) PE is a functional domain responsible for protein translocation and localization on mycobacterial cell wall. Mol. Microbiol. 66, 1536–1547 [DOI] [PubMed] [Google Scholar]
- 55.Daleke M.H., Cascioferro A., de Punder K., Ummels R., Abdallah A.M., van der Wel N.. et al. (2011) Conserved Pro-Glu (PE) and Pro-Pro-Glu (PPE) protein domains target LipY lipases of pathogenic mycobacteria to the cell surface via the ESX-5 pathway. J. Biol. Chem. 286, 19024–19034 10.1074/jbc.M110.204966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Slayden R.A. and Barry C.E. III (2001) Analysis of the lipids of Mycobacterium tuberculosis. Methods Mol. Med. 54, 229–245 [DOI] [PubMed] [Google Scholar]
- 57.Dupont C.M. and Kremer L. (2014) Extraction and purification of mycobacterial mycolic acids. Bio-Protocol 4 (20), e1265 10.21769/bioProtoc.1265 [DOI] [Google Scholar]
- 58.Meniche X., Labarre C., de Sousa-d’Auria C., Huc E., Laval F., Tropis M.. et al. (2009) Identification of a stress-induced factor of Corynebacterineae that is involved in the regulation of the outer membrane lipid composition. J. Bacteriol. 191, 7323–7332 10.1128/JB.01042-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Palomino J.C., Martin A., Camacho M., Guerra H., Swings J. and Portaels F. (2002) Resazurin microtiter assay plate: simple and inexpensive method for detection of drug resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 46, 2720–2722 10.1128/AAC.46.8.2720-2722.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Walzl A., Kramer N., Mazza M.R., Falkenhagen D., Hengstschläger M., Schwanzer-Pfeiffer D.. et al. (2012) A simple and cost efficient method to avoid unequal evaporation in cellular screening assays, which restores cellular metabolic activity. Int. J. Appl. Sci. Technol. 2, 17–25 [Google Scholar]
- 61.Chahinian H., Nini L., Boitard E., Dubes J.P., Comeau L.C. and Sarda L. (2002) Distinction between esterases and lipases: a kinetic study with vinyl esters and TAG. Lipids 37, 653–662 10.1007/s11745-002-0946-7 [DOI] [PubMed] [Google Scholar]
- 62.Parker S.K., Barkley R.M., Rino J.G. and Vasil M.L. (2009) Mycobacterium tuberculosis Rv3802c encodes a phospholipase/thioesterase and is inhibited by the antimycobacterial agent tetrahydrolipstatin. PLoS ONE 4, e4281 10.1371/journal.pone.0004281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ravindran M.S., Rao S.P., Cheng X., Shukla A., Cazenave-Gassiot A., Yao S.Q.. et al. (2014) Targeting lipid esterases in mycobacteria grown under different physiological conditions using activity-based profiling with tetrahydrolipstatin (THL). Mol. Cell. Proteomics 13, 435–448 10.1074/mcp.M113.029942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gu S., Chen J., Dobos K.M., Bradbury E.M., Belisle J.T. and Chen X. (2003) Comprehensive proteomic profiling of the membrane constituents of a Mycobacterium tuberculosis strain. Mol. Cell. Proteomics 2, 1284–1296 10.1074/mcp.M300060-MCP200 [DOI] [PubMed] [Google Scholar]
- 65.Nielsen H. (2017) Predicting secretory proteins with SignalP. Methods Mol. Biol. 1611, 59–73 [DOI] [PubMed] [Google Scholar]
- 66.Belardinelli J.M. and Jackson M. (2017) Green Fluorescent Protein as a protein localization and topological reporter in mycobacteria. Tuberculosis (Edinb.) 105, 13–17 10.1016/j.tube.2017.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pedelacq J.D., Cabantous S., Tran T., Terwilliger T.C. and Waldo G.S. (2006) Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 24, 79–88 10.1038/nbt1172 [DOI] [PubMed] [Google Scholar]
- 68.Jansson A., Niemi J., Mantsala P. and Schneider G. (2003) Crystal structure of aclacinomycin methylesterase with bound product analogues: implications for anthracycline recognition and mechanism. J. Biol. Chem. 278, 39006–39013 10.1074/jbc.M304008200 [DOI] [PubMed] [Google Scholar]
- 69.Whited A. and Johs A. (2015) The interactions of peripheral membrane proteins with biological membranes. Chem. Phys. Lipids 192, 51–59 10.1016/j.chemphyslip.2015.07.015 [DOI] [PubMed] [Google Scholar]
- 70.Murray D., Arbuzova A., Honig B. and McLaughlint S. (2002) The role of electrostatic and nonpolar interactions in the association of peripheral proteins with membranes. Curr. Top. Membr. 52, 277–307 10.1016/S1063-5823(02)52012-3 [DOI] [Google Scholar]
- 71.Lehmann J., Cheng T.Y., Aggarwal A., Park A.S., Zeiler E., Raju R.M.. et al. (2018) An Antibacterial beta-lactone kills Mycobacterium tuberculosis by disrupting mycolic acid biosynthesis. Angew. Chem. Int. Ed. Engl. 57, 348–353 10.1002/anie.201709365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sassetti C.M., Boyd D.H. and Rubin E.J. (2003) Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48, 77–84 10.1046/j.1365-2958.2003.03425.x [DOI] [PubMed] [Google Scholar]
- 73.Griffin J.E., Gawronski J.D., Dejesus M.A., Ioerger T.R., Akerley B.J. and Sassetti C.M. (2011) High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog. 7, e1002251 10.1371/journal.ppat.1002251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cole S.T., Eiglmeier K., Parkhill J., James K.D., Thomson N.R., Wheeler P.R.. et al. (2001) Massive gene decay in the leprosy bacillus. Nature 409, 1007–1011 10.1038/35059006 [DOI] [PubMed] [Google Scholar]
- 75.Kaur G. and Kaur J. (2017) Multifaceted role of lipids in Mycobacterium leprae. Future Microbiol. 12, 315–335 10.2217/fmb-2016-0173 [DOI] [PubMed] [Google Scholar]
- 76.Andersson C.S., Lundgren C.A., Magnusdottir A., Ge C., Wieslander A., Martinez Molina D.. et al. (2012) The Mycobacterium tuberculosis very-long-chain fatty acyl-CoA synthetase: structural basis for housing lipid substrates longer than the enzyme. Structure 20, 1062–1070 10.1016/j.str.2012.03.012 [DOI] [PubMed] [Google Scholar]
- 77.West N.P., Chow F.M., Randall E.J., Wu J., Chen J., Ribeiro J.M.. et al. (2009) Cutinase-like proteins of Mycobacterium tuberculosis: characterization of their variable enzymatic functions and active site identification. FASEB J. 23, 1694–1704 10.1096/fj.08-114421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sarathy J.P., Dartois V. and Lee E.J. (2012) The role of transport mechanisms in Mycobacterium tuberculosis drug resistance and tolerance. Pharmaceuticals (Basel) 5, 1210–1235 10.3390/ph5111210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Khoo K.H., Jarboe E., Barker A., Torrelles J., Kuo C.W. and Chatterjee D. (1999) Altered expression profile of the surface glycopeptidolipids in drug-resistant clinical isolates of Mycobacterium avium complex. J. Biol. Chem. 274, 9778–9785 10.1074/jbc.274.14.9778 [DOI] [PubMed] [Google Scholar]
- 80.Jiang P. and Cronan J.E. Jr (1994) Inhibition of fatty acid synthesis in Escherichia coli in the absence of phospholipid synthesis and release of inhibition by thioesterase action. J. Bacteriol. 176, 2814–2821 10.1128/jb.176.10.2814-2821.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhuang Z., Song F., Martin B.M. and Dunaway-Mariano D. (2002) The YbgC protein encoded by the ybgC gene of the tol-pal gene cluster of Haemophilus influenzae catalyzes acyl-coenzyme A thioester hydrolysis. FEBS Lett. 516, 161–163 10.1016/S0014-5793(02)02533-4 [DOI] [PubMed] [Google Scholar]
- 82.Gully D. and Bouveret E. (2006) A protein network for phospholipid synthesis uncovered by a variant of the tandem affinity purification method in Escherichia coli. Proteomics 6, 282–293 10.1002/pmic.200500115 [DOI] [PubMed] [Google Scholar]
- 83.Masilamani R., Cian M.B. and Dalebroux Z.D. (2018) Salmonella Tol-Pal reduces outer membrane glycerophospholipid levels for envelope homeostasis and survival during bacteremia. Infect. Immun. 86 pii: e00173–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wall V.Z., Barnhart S., Kramer F., Kanter J.E., Vivekanandan-Giri A., Pennathur S.. et al. (2017) Inflammatory stimuli induce acyl-CoA thioesterase 7 and remodeling of phospholipids containing unsaturated long (>/ = C20)-acyl chains in macrophages. J. Lipid Res. 58, 1174–1185 10.1194/jlr.M076489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jackson M., Crick D.C. and Brennan P.J. (2000) Phosphatidylinositol is an essential phospholipid of mycobacteria. J. Biol. Chem. 275, 30092–30099 10.1074/jbc.M004658200 [DOI] [PubMed] [Google Scholar]
- 86.Haites R.E., Morita Y.S., McConville M.J. and Billman-Jacobe H. (2005) Function of phosphatidylinositol in mycobacteria. J. Biol. Chem. 280, 10981–10987 10.1074/jbc.M413443200 [DOI] [PubMed] [Google Scholar]
- 87.Crellin P.K., Luo C.-Y. and Morita Y.S. (2013) Metabolism of plasma membrane lipids in Mycobacteria and Corynebacteria. In Lipid Metabolism, InTech [Google Scholar]