Abstract Abstract
Correct mosquito species identification is essential for mosquito and disease control programs. However, this is complicated by the difficulties in morphologically identifying some mosquito species. In this study, variation of a partial sequence of the cytochrome c oxidase unit I (COI) gene was used for the molecular identification of British mosquito species and to facilitate the discovery of cryptic diversity, and monitoring invasive species. Three DNA extraction methods were compared to obtain DNA barcodes from adult specimens. In total, we analyzed 42 species belonging to the genera Aedes Meigen, 1818 (21 species), Anopheles Meigen, 1818 (7 species), Coquillettidia Theobald, 1904 (1 species), Culex Linnaeus, 1758 (6 species), Culiseta Felt, 1904 (7 species), and Orthopodomyia Theobald, 1904 (1 species). Intraspecific genetic divergence ranged from 0% to 5.4%, while higher interspecific divergences were identified between Aedesgeminus Peus, 1971/Culisetalitorea (Shute, 1928) (24.6%) and Ae.geminus/An.plumbeus Stephens, 1828 (22.5%). Taxonomic discrepancy was shown between An.daciae Linton, Nicolescu & Harbach, 2004 and An.messeae Falleroni, 1828 indicating the poor resolution of the COI DNA barcoding region in separating these taxa. Other species such as Ae.cantans (Meigen, 1818)/Ae.annulipes (Meigen, 1830) showed similar discrepancies indicating some limitation of this genetic marker to identify certain mosquito species. The combination of morphology and DNA barcoding is an effective approach for the identification of British mosquitoes, for invasive mosquitoes posing a threat to the UK, and for the detection of hidden diversity within species groups.
Keywords: DNA extraction methods, hidden genetic diversity, molecular identification, vector species
Introduction
The family Culicidae includes approximately 112 genera and 3,547 described species (Harbach 2017, 2018). Several species are biting pests playing an important role as vectors of pathogens of humans and livestock (Becker et al. 2010). These include chikungunya, dengue, Japanese encephalitis, yellow fever, West Nile, Rift Valley fever, and Zika viruses, as well as several nematodes and protozoans such as Plasmodium Marchiafava & Celli, 1885 (Becker et al. 2010, Medlock et al. 2007). In addition to their medical and veterinary importance, mosquitoes are significant nuisance biters of humans and within the environment may serve additional roles such as key indicators of landscape degradation (Dorvillé 1996, Guedes and Navarro-Silva 2014, Montagner et al. 2018). As a result, mosquitoes are one of the principal target groups within surveillance and control programs worldwide (Hernández-Triana et al. 2017).
Current approaches to species identification still rely heavily upon morphology-based procedures, which typically require substantial training and may not always provide a good resolution on a specimen’s identity due to homogeneity between life stages of different species and the presence of species complexes (Cywinska et al. 2006, Hernández-Triana et al. 2012, 2014, 2015, Linton et al. 2005, Packer et al. 2009, Versteirt et al. 2015). To overcome this obstacle, a small region (658 bp) of the mitochondrial cytochrome c oxidase unit I (COI) gene was proposed as a standardized DNA marker in support of species identification for animal barcodes, in a process commonly referred to as DNA barcoding (Hebert et al. 2003a, b).
Until recently, thirty-four mosquito species have been recorded in the United Kingdom (UK) (Medlock et al. 2015, Medlock and Vaux 2009, 2015). However, Medlock et al. (2017a) detected the presence of Ae.albopictus (Skuse, 1895) in southern England, and Dallimore et al. (2017) collected a single male Ae.aegypti (Linnaeus, 1762) in Merseyside in north west England, although these two invasive species are not believed to be locally established. Nonetheless, these findings demonstrated that the UK is at risk of introduction by invasive species of Aedes (Dallimore et al., 2017). In addition, Harbach et al. (2017) discovered Ae.nigrinus (Eckstein, 1918) in the New Forest, southern England, which brings the total count of named species to 37 [35 native species plus two records of invasive species]. In addition, the occurrence of certain species has been very sporadic as in the case of Ae.vexans (Meigen, 1830); however, Medlock et al. (2017b) reported a resident population of this species at Marston Marshes, Norwich. Although no mosquito-borne pathogen affecting humans or livestock is presently thought to circulate in the UK, there is potential for future pathogen emergence (Medlock and Leach 2015; Medlock et al. 2017a, b; Vaux et al. 2015) and there remains continuing mosquito nuisance in various parts of the country (Brugman et al. 2017a, b). Collectively, these discoveries highlight the need for continued research on the native mosquito fauna of the UK, taking into account the potential incursion of invasive species.
There is, however, a paucity of data on the utility of molecular methods for species identification of the British mosquito fauna. During the first development of a molecular assay for the identification of hybrids and sibling species within Culexpipiens s.l., Smith and Fonseca (2004) used specimens from England and Scotland. Golding et al. (2012) subsequently employed the COI marker to compare sequences of Cx.modestus Ficalbi, 1890 with other Culex Linnaeus, 1758 species in southeast England, and Danabalan et al. (2012) employed a combination of the internal transcribed spacer gene-2 (ITS-2) and COI markers in their assessment of molecular identification tools to determine the status of Cx.pipiens s.l. The same approach was used by Danabalan et al. (2014) to confirm the occurrence of species within the Anophelesmaculipennis complex Theobald, 1911 in England and Wales. Recently, Hernández-Triana et al. (2015) employed an integrated approach to determine mosquito host feeding preferences (via identification of blood meal origin), as well as the molecular characterization of mosquito species carrying pathogens such as myxoma virus (Brugman et al. 2015, 2017a, b) and Theileriaorientalis Yakimoff & Soudatschenkoff, 1931 within their bloodmeal (de Marco et al. 2016).
In the present paper, we apply the COI DNA barcoding approach in support of the identification of native British mosquitoes and known invasive species in continental Europe. In addition, we assessed the DNA barcode variability using genetic distance methods to detect cryptic diversity across the taxa.
Materials and methods
Collection of specimens
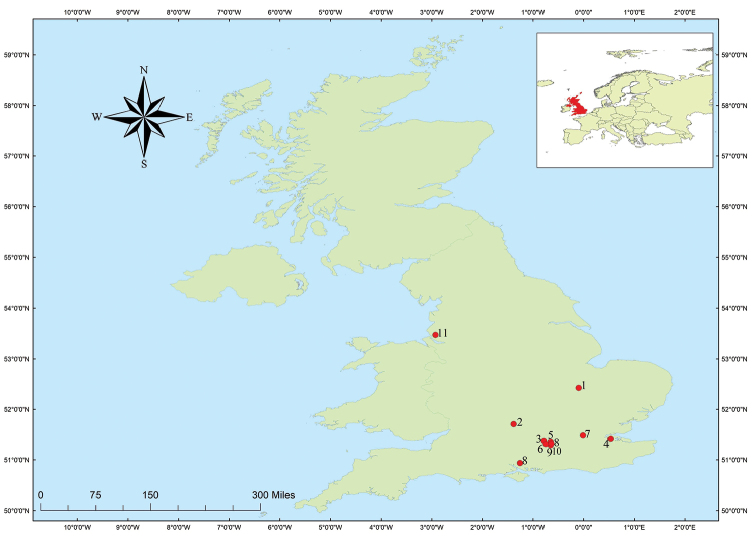
Ten locations were visited between March and October in the years 2012 to 2015 and specimens were collected following the protocols of Brugman et al. (2015, 2017a, b) (see Table 1, Fig. 1). Further samples were obtained by collecting mosquitoes alighting on the collectors and by standard larval dipping procedures followed by laboratory rearing according to Brugman et al. (2015, 2017a, b). All specimens were kept either at -20 °C or dry-pinned, and were morphologically identified using the key of Cranston et al. (1987). We followed the classification of Wilkerson et al. (2015) for taxa in Aedini. The subgeneric placement for all species can be found in Harbach (2017) and Harbach et al. (2017).
Table 1.
Description of key collecting sites with reference to habitat and the main livestock present. Further information can be found in Brugman et al. (2017b).
| Locality/Farms | County | Coordinates | Habitat | Main livestock types present |
|---|---|---|---|---|
| 1. ADAS Arthur Rickwood | Cambridgeshire | 52.422560, -0.098302 | Grazing farm | Sheep |
| 2. Church Farm | Oxfordshire | 51.715807, -1.380813 | Rural area | Cattle, sheep |
| 3. Coombelands Farm | Surrey | 51.360241, -0.652256 | Mixed farm | Cattle, sheep, pigs, horses |
| 4. Elmley Nature Reserve | Kent | 51.377587, 0.783954 | Grazing marsh | Cattle, sheep |
| 5. Glendell Livery, Mill Lane | Surrey | 51.290499, -0.652256 | Mixed woodland | Horses |
| 6. Frimley | Surrey | 51.313037, -0.745237 | Peri-urban | n/a |
| 7. Mudchute Farm | Greater London | 51.491732, -0.009367 | City farm | Cattle, sheep, pigs, horses |
| 8. Northney Farm, Hayling Island | Hampshire | 50.828166, -0.962151 | Arable farm | Cattle |
| 9. White Lodge, Bisley | Surrey | 51.322255, -0.637692 | Mixed woodland | Cattle |
| 10. Bartley Heath | Hampshire | 50.919701, -1.565337 | Woodland | Cattle, horses, deer |
| 11. Dee Marsh | Cheshire | 52.8322, -3.7656 | Salt marsh | n/a |
Figure 1.
Location of study sites in the United Kingdom. Key: 1 ADAS Arthur Rickwood; 2 Church Farm; 3 Coombelands Farms; 4 Elmley Nature Reserve; 5 Glendell Livery, Mill Lane; 6 Frimley; 7 Mudchute Farm; 8 Northney Farm, Hayling Island; 9 White Lodge, Bisley; 10 Bartley Heath; 11 Dee Marsh.
The source of specimens from invasive species is as follows: Ae.albopictus – Luke Alphey, UK (colony from Malaysia); Aleksandra Ignjatović-Ćupina, Serbia (wild caught); Ae.aegypti – Shahida Begum, UK (colony from West Africa); Ae.atropalpus (Coquillet, 1902), Ae.japonicus (Theobald, 1901), Ae.koreicus (Edwards, 1917) – Norbert Becker and Daniel Hoffman, Germany, and Ignacio Justicia-Ibáñez, Holland (all wild caught); Culextritaeniorhynchus Giles, 1901, Filiz Gunay, Turkey (wild caught); Cx.quinquefasciatus Say, 1823 [for sequences from NCBI and further details Suppl. material 1].
DNA extraction methods
Three methods were used for DNA extraction from two mosquito tissue types (Brugman et al. 2015, 2017). Firstly, 1–2 legs of specimens were placed in 100 µl of molecular grade water in a 96-well plate, which was then sonicated at room temperature for 10 min to release DNA (Hunter et al. 2008). Secondly, we employed a modified Hotshot technique (Montero-Pau et al. 2008). In this case, 1–2 legs were placed directly into 50 µl of alkaline lysis buffer in a 96-well plate, which was then sonicated in a water bath for 10 min. The plate was subsequently incubated in a thermocycler for 30 min at 94 °C, cooled for 5 min at 4 °C, and then centrifuged for 3 min at 3000 rpm, after which 50 µl of the neutralizing buffer was added to each sample. The plate was then centrifuged again for 10 min at 3000 rpm, and stored at -80 °C until analysis. Thirdly, engorged female abdomens were processed using Qiagen DNeasy Blood and Tissue kits following the procedures detailed in Brugman et al. (2015, 2017a, b) and the manufacturer’s instructions.
COI DNA barcoding region amplification
For molecular species identification using the COI DNA barcoding region, the protocols of Hernández-Triana et al. (2012, 2014) and Hebert et al. (2003a, b) were followed. We used the primers developed by Folmer et al. (1994), which amplify the 658 bp long target region of the COI gene. PCR products were obtained using a Qiagen PCR system following the reaction mix of Hernández-Triana et al. (2017).
Data analysis
Paired bi-directional sequence traces were combined to produce a single consensus sequence (i.e., the full-length 658 bp barcode sequence). To achieve this, individual forward and reverse traces were oriented, edited, and aligned using the Sequencer (v.4.5; Genes Codes Corporation, Ann Harbour, MI), GenDoc (v. 2.6.02) and ClustalX sequence analysis programs (Hernández-Triana et al. 2017). Full details for each specimen and sequence information can be found at the Barcode of Life Database (BOLD) within the “Human Pathogens and Zoonoses Initiative”, Working Group 1.4. The Digital Object Identifier (DOI) for the publically available projects in BOLD is dx.doi.org/10.5883/DS-MQFWUK and dx.doi.org/10.5883/DS-MQIUV. Accession numbers for all sequences were obtained from NCBI (accession numbers: MK403007-MK403548). For certain species, we used COI barcode sequences deposited at NCBI due to the lack of available material from UK populations (Table 3; Suppl. material 1). The dataset was analyzed in MEGA v.6 (Tamura et al. 2007). The Neighbor Joining (NJ) analysis was performed using the Kimura 2-Parameter distance metric to determine their distribution pattern. The tree robustness was measured by the bootstrap approach using 1000 pseudoreplicates (Hernández-Triana et al. 2012, 2014). To barcode sequences larger than 500 bp, a Barcode Index Number (BIN) was assigned and each BIN was mapped according to species (see Fig. 2). The taxonomic discordance in the dataset (Hernández-Triana et al. 2017) was analyzed using BOLD as detailed in Ratnasingham and Hebert (2013).
Table 3.
List of mosquito species (in alphabetical order), country of collection, and number of specimens with DNA barcodes. Mean (%) intraspecific values of sequence divergence (Kimura2-Parameter distance) are shown with missing entries indicating that less than two specimens were analyzed. Asterisks indicate species complexes (*) and taxa with deep splits (**) in the Neighbor Joining tree; (***) taxa with above 2% genetic divergence. Invasive species in Europe are in Bold.
| Species | Collection Country | n | mean % |
|---|---|---|---|
| Aedes aegypti | West Africa | 10 | 0 |
| Aedes albopictus | Malaysia; Montenegro | 12 | 0.12 |
| Aedes annulipes | Belgium | 12 | 0.89 |
| Aedes atropalpus | Holland, USA, Canada | 11 | 0.69 |
| Aedes cantans | Belgium; UK | 44 | 0.80 |
| Aedes caspius | Belgium; UK | 40 | 0.78 |
| Aedes cinereus | Sweden; UK | 30 | 0.61 |
| Aedes communis | Belgium | 13 | 0.14 |
| Aedes detritus | Belgium; UK | 44 | 0.66 |
| Aedes dorsalis | USA; Canada | 8 | 0.16 |
| Aedes flavescens | UK | 10 | 0.18 |
| Aedes geminus | Germany | 4 | 0.58 |
| Aedes geniculatuss | Belgium | 16 | 0.25 |
| Aedes japonicus | Germany | 14 | 0.32 |
| Aedeskoreicus**;*** | Belgium; Holland; Hungary | 6 | 2.19 |
| Aedes leucomelas | Sweden | 2 | 0.40 |
| Aedes nigrinus | Sweden | 2 | 0.77 |
| Aedes punctor | Belgium; UK | 47 | 0.67 |
| Aedes rusticus | Belgium; UK | 31 | 0.07 |
| Aedes sticticus | Belgium; UK | 10 | 1.29 |
| Aedesvexans** | Belgium; Spain; Holland; Sweden; UK | 38 | 1.46 |
| Anopheles algeriensis | Spain | 6 | 0.41 |
| Anopheles atroparvus | UK; Belgium | 91 | 0.92 |
| Anophelesclaviger s.l. | Belgium; UK | 26 | 0.65 |
| Anopheles daciae | Romania; UK | 28 | 0.76 |
| Anopheles messeae | UK | 35 | 1.01 |
| Anopheles plumbeus | Belgium; UK | 17 | 0 |
| Coquillettidia richiardii | Belgium; UK | 42 | 0.07 |
| Culex modestus | Germany; Romania; Turkey; UK | 49 | 0 |
| Culexpipiens s.l.* | Belgium; UK | 187 | 0.06 |
| Culex quinquefasciatus | Pakistan; Turkey | 12 | 0 |
| Culex territans | Belgium; Germany | 5 | 2.05 |
| Culex torrentium | Belgium; Germany; UK | 66 | 0.43 |
| Culex tritaeniorhynchus | Turkey | 5 | 0.65 |
| Culiseta alaskaensis | Canada | 3 | 1.13 |
| Culiseta annulata | Belgium; UK | 192 | 0.05 |
| Culiseta fumipennis | Belgium | 2 | 0.30 |
| Culisetalitorea*** | Spain; UK | 9 | 5.35 |
| Culiseta longiareolata | Spain | 5 | 0.12 |
| Culiseta morsitans | Belgium; UK | 7 | 0.34 |
| Culiseta subochrea | Spain; UK | 6 | 0.34 |
| Orthopodomyia pulcripalpis | Austria | 1 | n/a |
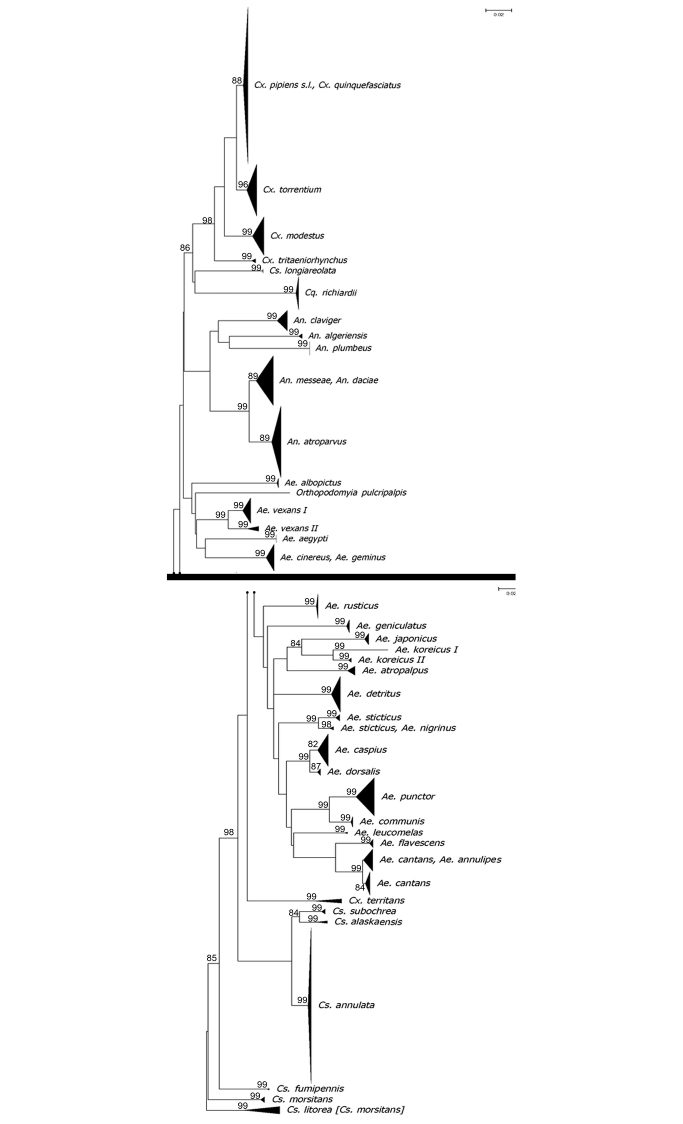
Figure 2.
Neighbor joining tree of COI DNA barcodes (658 bp) for mosquito species. A divergence of > 2% may be indicative of separate operational taxonomic units. Only bootstrap values higher than 70% are shown.
Results
Assessment of DNA extraction methodologies
In general, adding 1–2 legs to molecular grade water and then sonicating them for 10 min proved to be an effective method for obtaining DNA (30 min total time); however, only 41 barcodes (43.1%) yielded sufficient sequence data for inclusion in our analysis (Table 2). The Hotshot technique also proved to be an efficient approach (1 hour per plate) for processing 1–2 legs with high percentages of target DNA amplification and COI DNA barcode sequences (429 sequences, 90.3%). In terms of cost, reagents for the preparation of the Hotshot working stock buffers were only 200 GBP, one purchase of which we estimate can last up to one year. DNA extraction from blood-engorged abdomens processed using the Qiagen DNeasy Blood and Tissue kit also provided barcodes for 306 specimens (64.4%), but this approach was time consuming, with a sample processing rate of 32 specimens per four hour session of DNA extraction. The time for the DNA extraction was also increased by the limitation of the number of wells in the centrifuge available (30 wells). In addition, non-target PCR product was also encountered as vertebrate DNA present was amplified from cows, chicken, sheep, rabbits and birds [169 samples] (see Table 2).
Table 2.
DNA extraction methods and percentage of PCR amplification success in obtaining COI DNA barcodes from mosquitoes.
| Extraction method | No. plates / samples | Time per plate | Amplification success n (%) | Observations |
|---|---|---|---|---|
| 1. Legs directly into molecular grade water and sonicated for 10 min | 1 plate / 95 samples | 30 min | 41 (43.1%) | High sequencing failure (54 samples) |
| 2. Legs directly into alkaline lysis buffer and sonicated for 10 min (Hotshot) | 5 plates / 475 samples | 1hr each plate | 429 (90.3%) | Target length barcodes obtained |
| 3. Abdomen processed using Qiagen kit | 5 plates / 475 samples | Only 32 samples per 4hr session for DNA extraction for each plate | 306 (64.4%) | Target length barcodes obtained. Vertebrate DNA amplified |
Mosquito species identification using DNA barcoding
In total, we analyzed DNA barcode sequences for 42 species belonging to the genera Aedes (21 species), Anopheles (7 species), Coquillettidia (1 species), Culex (6 species), Culiseta (7 species), and Orthopodomyia (1 species) (Table 3). Of these, we analyzed sequences for 23 of the 37 species of mosquito that have been recorded in the UK (62%) (Harbach et al. 2017, Medlock et al. 2007a, b). In addition, we also analyzed representatives of invasive Aedes species (Ae.aegypti, Ae.albopictus, Ae.japonicus, Ae.koreicus, Ae.atropalpus) and two Culex species (Cx.quinquefasciatus, Cx.tritaeniorhynchus), which are of epidemiological relevance in Europe (Medlock et al. 2017a, b). Three or more representatives were available for 38 morphospecies in the dataset (see Table 3). In total, 1198 barcode sequences were analyzed.
Even though in most cases individuals of the same species clustered together, this was not the case for all species. Within the genus Aedes, the first incongruence was identified between Ae.sticticus (Meigen, 1838) and Ae.nigrinus. Although the majority of specimens from Belgium and the two UK specimens identified as Ae.sticticus (voucher number APHA-4-2015G06, APHA-4-2015G07) grouped together in a separate cluster with 100% bootstrap support, the only two available COI sequences of Ae.nigrinus in NCBI (KP942769, KP942770) grouped with the two specimens collected in Belgium, identified as Ae.sticticus (CULBE-833009, CULBE-833008) (Fig. 2). To further support our identification of the two UK specimens as Ae.sticticus, we obtained ITS-2 sequences (data not shown) and searched the NCBI database using the BLAST algorithm; both queries retrieved Ae.sticticus with 96% match (KF535079) [this relative low percentage could be due to the low coverage of the ITS-2 sequences we obtained (338 bp and 369 bp, respectively)]. Similar results have been obtained by Versteirt et al. (2015) (see Fig. 2). Certain specimens grouped only as Ae.cantans or Ae.annulipes, but another group was composed of Ae.cantans and Ae.annulipes (Fig. 1) with 100% bootstraps support values. Similarly, no successful identification was reached between Ae.cinereus (Meigen, 1818) and Ae.geminus, species which are morphologically similar.
Within Anophelesmaculipennis s.l. (Linton et al. 2002, 2005), no accurate identification was achieved between An.messeae [also molecularly identified by ITS-2 in our laboratory; see also Kronefeld et al. 2012] and An.daciae (sensu Nicolescu et al. 2004), although An.atroparvus was clearly separated from the aforementioned species (Fig. 2). This is not surprising as all members of the An.maculipennis complex are phylogenetically related, and cannot be readily identified based on adult morphological traits or only using the COI genetic marker (Linton et al. 2002, Kronefeld et al. 2014, Ruiz-Arrondo et al. 2017). In the genus Culex, COI was not able to separate Culexpipiens s.l. (including both forms pipiens and molestus) and Cx.quinquefasciatus, in agreement with results by Gunay et al. (2015).
Our DNA barcodes dataset from the genus Culiseta separated certain species with high support bootstrap values such as Cs.alaskaensis (Ludlow, 1906), Cs.annulata (Schrank, 1776), Cs.longiareolata (Macquart, 1838) and Cs.subochrea (Edwards, 1921) (Fig. 2). However, we could not achieve the same resolution for Cs.fumipennis (Stephens, 1825), Cs.litorea (Shute, 1928) and Cs.morsitans (Theobald, 1901). The specimens identified as Cs.fumipennis (Versteirt et al. 2015) from Belgium grouped separately from one specimen (KU748471) collected in the UK (de Marco et al. 2016) previously identified as Cs.fumipennis, which clustered with specimens from Belgium identified as Cs.morsitans (CULBE-816017, CULBE-816018, CULBE-997001, CULBE-997002, CULBE-9972101, CULBE-972103) with 99% bootstrap values. Therefore, we now consider the UK specimen to be Cs.morsitans. In addition, seven specimens from the UK identified as Cs.morsitans in de Marco et al. (2016) (KU748440, KU748443, KU748450, KU748453, KU748460, KU748500, KU748488), grouped with 99% bootstraps values with two males recently collected from Spain identified as Cs.litorea. Subsequent dissection of the genitalia of these specimens confirmed their identification as Cs.litorea based on the key of Becker et al. (2010); therefore, we now considered these seven specimens from the UK as Cs.litorea.
The levels of sequence divergence were variable across the taxa, with conspecific individuals collected from a single site often exhibiting zero, or 0.07% to 0.1% divergence values, while other specimens showed higher percentages (see Table 3). The intraspecific genetic divergence measured 1.3%, ranging from 0% to 5.4% (Table 3) (Ae.aegypti, An.plumbeus, Cx.modestus and Cx.quinquefasciatus), while the interspecific divergence ranged between 0.19% to 24.6% (Suppl. material 2). Interspecific genetic divergence values were higher between species from different genera. The pairs Ae.geminus/Cs.litorea (24.6%) and Ae.geminus/An.plumbeus (22.5%) were the most divergent species. As known, the smallest values of genetic divergence were found among species in the same genus, for example Cx.pipiens s.l./Cxquinquefasciatus (0.19%), Ae.cantans/Ae.annulipes (1.2%) and Ae.geminus/Ae.cinereus (0.65%) (Suppl. material 2).
In this study, we analyzed three species which are known, or suspected to be, part of species complexes [species which can only be distinguished either by cytotaxonomy or molecular methods (Danabalan et al. 2012, 2014; Linton et al. 2001)]: An.maculipennis s.l., An.claviger s.l. (Meigen, 1804), and Cx.pipiens s.l. All specimens grouped together in either Cx.pipiens s.l. or An.claviger s.l., and we did not detect high levels of genetic diversity or deep splits in the NJ tree as found in other studies (Gunay et al. 2015, Versteirt et al. 2015). This might be due to specimens originating from localities in relatively close proximity to one another in England (Fig. 1). Specimens of Cx.pipiens s.l. in this study originated from the study of Brugman et al. (2015, 2017), in which the CQ11 PCR assay was conducted to separate the forms molestus and pipiens. Only specimens from the typical pipiens form of Cx.pipiens were detected in the aforementioned studies, with 0.06% genetic diversity in our dataset. Nonetheless, not all morphologically identified species clustered as expected. Certain species exhibited higher levels of divergence above 2% (see Table 1) and other showed deep splits in the NJ tree (Fig. 2). For example, intraspecific genetic divergence averaged 1.46% in Ae.vexans, but the specimens separated into two defined clusters (I and II) (Fig. 2). Similarly, Ae.koreicus showed a deep split in the NJ with 2.19% genetic divergence.
The BIN counts in our dataset in BOLD of 721 full length barcode sequences from 1006 records in BOLD datasets found 21 BINs. The BIN analysis did not include sequences downloaded from the NCBI database. In general, 487 barcodes were assigned a BIN number, which represented 14 concordant BINs, three BINs were singletons (Cs.fumipennisBOLD:AAR2210, Ae.geniculatusBOLD:AAM5898, and Cs.subochreaBOLD:AAV90 75), and only four BINs (231 records) were discordant. The discordant BINs occurred at the species level, mainly because of the discrepancy in taxonomic information assigned to certain specimens, for example Ae.cinereus versus Ae.nr.cinereus, and Ae.caspius versus Ae.nr.caspius. Another discordance was in a single specimen identified as Cx.torrentium, which appears to be close to a BIN within Cx.pipiens s.l. (BOLD:AAA4751; Process ID:MSEMV855-15); however, morphological traits in the male genitalia and other analysis (CQ11 PCR) showed that it does belongs to Cx.torrentium (Manley et al. 2015), and it did cluster with this species when further 66 barcode sequences of Cx.torrentium were added to the dataset.
Discussion
This study assessed minimally destructive approaches that retained a significant part of the sample as a referenced voucher and the development of a COI DNA barcoding library in mosquitoes, and assessed the use of the variability within COI in support of species identification. Overall, the three extraction methods used provided sufficient DNA for subsequent analysis; however the modified Hotshot technique of Montero-Pau et al. (2008) proved to be the most efficient and inexpensive method for obtaining COI DNA barcode sequences. This method has been applied to other groups such as the Hymenoptera (Guzmán-Larralde et al. 2016) with good results as assessed by DNA yield and PCR amplification success. The amplification of vertebrate DNA from engorged abdomens using the Qiagen DNeasy Blood and Tissue extraction kit highlights the need to use insect specific primers, for example LepF/LepR (see www.boldsystems.org/index.php/ Public_Primer_PrimerSeach) instead of the standard Folmer primers (Folmer et al. 1994). In terms of cost, considering that we did not have to buy a DNA extraction kit to perform the DNA extraction for processing the legs, the Hotshot technique represented savings of around 500 GBP per 96-well plate to our laboratory, making it a cost-effective method for performing DNA extractions.
The majority of morphologically identified species in this study formed defined groups in the NJ analysis based on DNA barcodes (Fig. 2), supporting the use of COI DNA barcoding in combination with morphological observation as a suitable approach for species identification. Genetic divergence between morphospecies ranged from 0.19% to 24.6%, whereas intraspecific genetic divergences within distinct species ranged from 0% to 5.4% (average 1.30%; Table 1, Suppl. material 2). Most of the specimens within a morphospecies were resolved in the NJ tree (Fig. 2). However, some individuals in certain taxa such as Ae.annulipes/Ae.cantans, Ae.cinereus/Ae.geminus, Ae.sticticus/Ae.nigrinus, An.daciae/An.messeae, and Cx.quinquefasciatus/Cx.pipiens s.l. clustered together (Fig. 2), indicating some limitations of the COI gene as a marker to separate these species. This finding is not surprising as these taxa are phylogenetically, as it has been highlighted in the literature (Harbach 2017, Harbach et al. 2017).
With regard to Anophelesmaculipennis s.l., although some morphological traits in egg structure provide an effective method to separate some members of this group, there is some dispute regarding the taxonomic status of An.daciae (e.g. Linton et al. 2002, 2005, Kronefeld et al. 2012). This species was described by Nicolescu et al. (2004) based on all life stages collected from the Black Sea region in Romania. The authors stated that An.daciae and An.messeae have been misidentified in the past because of similar morphology. However, they showed that An.daciae eggs are generally smaller than those of An.messeae, with patches of larger deck tubercles that contrast more sharply with patches of smaller tubercles to impart greater definition to the mottled surface of the deck (see Nicolescu et al. 2004; fig. 3 A, C). In contrast, the deck of An.messeae eggs has a more diffuse or weakly mottled appearance (see Nicolescu et al. 2004; fig. 3 B, D). In the same study, molecular analysis of ITS-2 sequences identified single nucleotide polymorphisms and unique haplotypes which, in the authors’ views, confirmed the specific status of An.daciae. However, other authors have queried the specific status of An.daciae, and stated that COI offered poor resolution and advocated for further work to determine the status of An.daciae. Similarly, in their study of Belgian mosquitoes, Versteirt et al. (2015) reported that specimens identified as Ae.annulipes and Ae.cantans grouped together in their NJ tree, and stated that COI cannot separate these two species; the same results have been obtained in our study (Fig. 2).
Moreover, COI DNA barcoding highlighted mis-identifications within the genus Culiseta (Cs.fumipennis, Cs.litorea and Cs.morsitans). These species are placed in the subgenus Culicella, and the females are difficult to identify because of their morphological similarity and absence of reliable diagnostic characteristics (Becker 2010, Cranston et al. 1987, Medlock and Vaux 2010). All of our females were collected in traps and were in relatively poor condition. In addition, no COI sequences of reliably identified material of Cs.litorea were available in the NCBI or BOLD databases to compare with our specimens (Ruiz-Arrondo et al. 2017). Because the sequences of females of Cs.morsitans matched the two males identified as Cs.litorea from Spain, we considered all our specimens to be Cs.litorea. In the literature (Medlock et al. 2007), Cs.morsitans is considered a widespread species in the UK, found in permanent waters, while Cs.litorea has a more restricted distribution in coastal regions of southern England. Both species feed mainly on birds, but can also bite humans, and they are considered bridge vectors of arboviruses (Medlock et al. 2007). Our findings highlight the need for careful examination of material obtained from traps in combination with the application of molecular techniques for a reliable identification of these species. Even though in our dataset Ae.vexans showed a low genetic divergence of approximately 2%, specimens of this species separated into two distinct groups in the NJ tree (Fig. 2), which may be an indication of some genetic differentiation within the population. This agrees with the findings of Lilja et al. (2018) in which the authors reported a distinct genotype of Ae.vexans in Europe.
Regarding non-indigenous mosquito species, although the adults of certain species are easily identified using morphological keys, for example Ae.aegypti and Ae.albopictus (Becker 2010, Cranston et al. 1987), the development of a molecular library for species identification is important, in particular when specimens are found in a poor stage of preservation. This is an essential step for the establishment of control measures (Versteirt et al. 2015) in the event of a recent introduction, as in the case of the detection of Ae.albopictus in the UK (see Medlock et al. 2017a, b).
In our dataset, Ae.koreicus and Cs.litorea showed higher intraspecific genetic divergences (Table 1, Fig. 2), which may indicate the presence of cryptic diversity. For all other species, the variation in intra- and interspecific genetic values reported in this study fall within the range for DNA barcoding studies of European mosquitoes (Engdahl et al. 2014, Gunay et al. 2015, Versteirt et al. 2015) or other zoogeographical regions such as the Nearctic and Oriental Regions (Cywinska et al. 2006, Kumar et al. 2007, Murugan et al. 2016). Nonetheless, we advocate the combination of the COI DNA barcoding with other genetic markers such as the Elongator Complex Protein 1 gene (ECP1) (Low et al. 2016, Pangjanda and Pramual 2016, Senatore et al. 2014) and ITS-2 sequences from a larger number of specimens across the species distribution range in order to resolve some of the taxonomic problems highlighted in this study.
Conclusions
This study provides COI DNA barcoding data to support the molecular identification of mosquito species in the UK as well as invasive mosquito species, many of which are currently expanding their geographical range in continental Europe. We augment the barcoding data for anthropophilic species such as Ae.cinereus, Ae.detritus, Ae.sticticus, Ae.vexans, and Cx.modestus, as well as other species of veterinary importance such as the bridge vector Cs.annulata. Even though the majority of specimens were separated by COI, certain taxa could not be distinguished using this genetic marker within the genera Aedes, Anopheles and Culex. The use of COI also underlined identification problems in Culiseta species (Cs.fumipennis, Cs.litorea and Cs.morsitans) within the BOLD and NCBI databases. This finding supports the need for continuing research combining the use of molecular methodologies with morphological traits for species delineation in the Culicidae.
Acknowledgements
We thank the kind assistance of each of the field site employees without whom the work could not have been completed. Thanks are given to our colleagues at Public Health England (Jolyon Medlock, Alex Vaux, and Ben Cull), The Pirbright Institute (Simon Carpenter) and the Natural History Museum (Ralph Harbach) for discussions on mosquito ecology and distribution. Luis M. Hernández-Triana gives special thanks to Quetzaly Siller (Juarez University, Durango Estate) for the preparation of the distribution map. In addition, we also thank Luke Alphey (The Pirbright Institute, UK), Aleksandra Ignjatović-Ćupina (University of Novi Sad, Faculty of Agriculture, Serbia), Jenny Hesson (Liverpool School of Tropical Medicine, UK), Gale Chapman (School of Veterinary Medicine and Science, University of Nottingham, UK), Shahida Begum (London School of Hygiene and Tropical Medicine, UK), Norbert Becker and Daniel Hoffman (University of Heidelberg, Heidelberg, Germany), Renke Lühken (Bernhard Nocht Institute for Tropical Medicine, WHO Collaborating Centre for Arbovirus and Hemorrhagic Fever Reference and Research, Hamburg, Germany), Adolfo Justicia-Ibáñez (National Centre for Monitoring of Vector, Netherlands Food and Consumer Product Safety Authority, Ministry of Economic Affairs, Wageningen, Holland), and Filiz Gunay (Hacettepe University, Faculty of Science, Department of Biology, Ecology Section, ESRL Laboratories Turkey) for providing material for invasive species analyzed in this study. Funding for LMHT was provided by the UK Department for Environment Food and Rural Affairs (DEFRA), Scottish Government and Welsh Government through grants SV3045, and SE4113, and the EU Framework Horizon 2020 Innovation Grant, European Virus Archive (EVAg, grant no. 653316). Funding for VAB was additionally provided by the Biotechnology and Biological Sciences Research Council (BBSRC), grant BB/F016492/1 and the Pirbright Institute as part of his PhD project. All German collections between 2011 and 2014 were part of projects conducted by the Bernhard Nocht Institute for Tropical Medicine, Hamburg, Germany, and collaborators. This work was funded by the Leibnitz Association, grant no. SAW-2011-BNI-3, and the German Federal Ministry for Environment, Nature Conservation, Building and Nuclear Safety (BMUB) through the Federal Environment Agency (UBA), grant no. FKZ371148404.
Citation
Hernández-Triana LM, Brugman VA, Nikolova NI, Ruiz-Arrondo I, Barrero E, Thorne T, de Marco MF, Krüger A, Lumley S, Johnson N, Fooks AR (2019) DNA barcoding of British mosquitoes (Diptera, Culicidae) to support species identification, discovery of cryptic genetic diversity and monitoring invasive species. ZooKeys 832: 57–76. https://doi.org/10.3897/zookeys.832.32257
Supplementary materials
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Luis M. Hernández-Triana, Victor A. Brugman, Nadya I. Nikolova, Ignacio Ruiz-Arrondo, Elsa Barrero, Leigh Thorne, Mar Fernández de Marco, Andreas Krüger, Sarah Lumley, Nicholas Johnson, Anthony R. Fooks
Accession number(s) of COI DNA barcode sequences used in this study downloaded from the NCBI database or provided by colleagues
Data type: molecular data
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Luis M. Hernández-Triana, Victor A. Brugman, Nadya I. Nikolova, Ignacio Ruiz-Arrondo, Elsa Barrero, Leigh Thorne, Mar Fernández de Marco, Andreas Krüger, Sarah Lumley, Nicholas Johnson, Anthony R. Fooks
Percentage of Interspecific (between groups) pairwise K2P genetic divergence of unique DNA barcodes (658 bp), representing 42 species of mosquitoes
Data type: molecular data
Explanation note: Highest pairwise distances (most divergent taxa) and lowest pairwise distances (most closely related taxa) are highlighted in yellow/bold, and green, respectively.
References
- Becker N, Petrić D, Zgomba M, Boase C, Madon M, Dahl C, Kaiser A. (2010) Mosquitoes and their control. 2nd edn. Springer-Verlag, Berlin Heidelberg, 577 pp 10.1007/978-3-540-92874-4 [DOI] [Google Scholar]
- Brugman VA, England ME, Stoner J, Tugwell L, Harrup LE, Wilson JW, Medlock JM, Logan JG, Fooks AR, Mertens PPC, Johnson N, Carpenter S. (2017a) How often do mosquitoes bite humans in southern England? A standardised summer trial at four sites reveals spatial, temporal and site-related variation in biting rates. Parasites & Vectors 10: 420. 10.1186/s13071-017-2360-9 [DOI] [PMC free article] [PubMed]
- Brugman VA, Hernández-Triana LM, England ME, Medlock JL, Logan JG, Wilson AJ, Fooks AR, Johnson N, Carpenter S. (2017b) Host-feeding patterns and insights into the potential role of native mosquitoes as pathogen vectors in the United Kingdom. Parasites & Vectors 10: 163. 10.1186/s13071-017-2098-4 [DOI] [PMC free article] [PubMed]
- Brugman VA, Hernández-Triana LM, Prosser SW, Weland C, Westcott DG, Fooks AR, Johnson N. (2015) Molecular species identification, host preference and detection of myxoma virus in the Anophelesmaculipennis complex (Diptera: Culicidae) in southern England, UK. Parasites & Vectors 8: 1–8. 10.1186/s13071-015-1034-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cranston PS, Ramsdale CD, Snow KR, White GB. (1987) Keys to the adults, male hypopygia, fourth-instar larvae and pupae of the British mosquitoes (Culicidae) with notes on their ecology and medical importance. Freshwater Biological Association Scientific Publication 48: 1‒152.
- Cywinska A, Hunter FF, Hebert PDN. (2006) Identifying Canadian mosquitoes species through DNA barcodes. Medical and Veterinary Entomology 20: 413–24. 10.1111/j.1365-2915.2006.00653.x [DOI] [PubMed] [Google Scholar]
- Dallimore T, Hunter T, Medlock JM, Vaux GC, Harbach RR, Strode C. (2017) Discovery of a single male Aedesaegypti (L.) in Merseyside, England. Parasites & Vectors 10: 239. [DOI] [PMC free article] [PubMed]
- Danabalan R, Monaghan MT, Ponsonby DJ, Linton Y-M. (2014) Occurrence and host preferences of Anophelesmaculipennis group mosquitoes in England and Wales. Medical and Veterinary Entomology 28: 169–78. 10.1111/mve.12023 [DOI] [PubMed] [Google Scholar]
- Danabalan R, Ponsonby DJ, Linton Y-M. (2012) A critical assessment of available molecular identification tools for determining the status of Culexpipiens s.l. in the United Kingdom. Journal of American Mosquito Control Association 28: 68–74. 10.2987/8756-971X-28.0.68 [DOI] [PubMed] [Google Scholar]
- de Marco MF, Brugman VA, Hernández-Triana LM, Nikolova NI, Fooks AR, Johnson N. (2016) Detection of Theileriaorientalis in the blood meals of Culisetaannulata in southern England, United Kingdom. Veterinary Parasitology 229: 31–36. 10.1016/j.vetpar.2016.09.012 [DOI] [PubMed] [Google Scholar]
- Dorvillé LFM. (1996) Mosquitoes as bioindicators of forest degradation in southeastern Brazil, a statistical evaluation of published data in the literature. Studies Neotropical Environment 31: 68–78. 10.1076/snfe.31.2.68.13331 [DOI] [Google Scholar]
- Engdahl C, Larsson P, Näslund J, Bravo M, Evander M, Lundström JO. (2014) Identification of Swedish mosquitoes based on molecular barcoding of the COI gene and SNP analysis. Molecular Ecology Resources 140: 478–88. 10.1111/1755-0998.12202 [DOI] [PubMed] [Google Scholar]
- Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. (1994) DNA primers for amplification of mitochondrial cytochrome oxidase subunit I from diverse metazoan invertebrates. Molecular Microbiology and Biological Biotechnology 3: 294–299. [PubMed] [Google Scholar]
- Golding N, Nunn MA, Medlock JM, Purse VB, Vaux AGC, Schäfer SM. (2012) West Nile virus vector Culexmodestus established in southern England. Parasites & Vectors 5: 32. 10.1186/1756-3305-5-32 [DOI] [PMC free article] [PubMed]
- Guedes MLP, Navarro-Silva MA. (2014) Mosquito community composition in dynamic landscapes from the Atlantic Forest biome (Diptera, Culicidae). Revista Brasileira de Entomologia 58: 88–94. 10.1590/S0085-56262014000100014 [DOI] [Google Scholar]
- Gunay F, Alten B, Simsek F, Aldemir A, Linton T-M. (2015) Barcoding Turkish Culex mosquitoes to facilitate arbovirus vector incrimination studies reveals hidden diversity and new potential vectors. Acta Tropica 143: 112‒120. 10.1016/j.actatropica.2014.10.013 [DOI] [PubMed]
- Guzmán-Larralde AJ, Suaste-Dzul AP, Gallou A, Peña-Carrillo KI, González-Hernández A. (2016) DNA recovery from microhymenopterans with six non-destructive methodologies and considerations for museum slides preparations. Genome 260: 85–91. 10.1139/gen-2015-0172 [DOI] [PubMed] [Google Scholar]
- Harbach RE. (2017) Mosquito Taxonomic Inventory. http://mosquito-taxonomic-inventory.info [Accessed on 23 January 2018]
- Harbach RE. (2018) Culicipedia: species-group, genus-group and family-group names in Culicidae (Diptera). CABI, United Kingdom, 396 pp 10.1079/9781786399052.0000 [DOI] [Google Scholar]
- Harbach RE, Dallimore T, Briscoe AG, Culverwell CL, Vaux AGC, Medloch JM. (2017) Aedesnigrinus (Eckstein, 1918) (Diptera, Culicidae) a new country record for England, contrasted with Aedessticticus (Meigen, 1838). ZooKeys 671: 119–130. 10.3897/zookeys.671.12447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert PDN, Cywinska A, Ball SL, de Waard JR. (2003a) Biological identifications through DNA barcodes. Proceedings of the Royal Society B: Biological Sciences 270: 313–321. 10.1098/rspb.2002.2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert PDN, Ratnasingham S, de Waard JR. (2003b) Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proceedings of the Royal Society B: Biological Sciences 270: S96–S99. 10.1098/rsbl.2003.0025 [DOI] [PMC free article] [PubMed]
- Hernández-Triana LM, Brugman VA, Prosser SWJ, Weland C, Nikilova N, Thorne L, de Marco MF, Fooks AR, Johnson N. (2017) Molecular approaches for blood meal analysis and species identification of mosquitoes (Insecta: Diptera: Culicidae) in rural locations in southern England, United Kingdom. Zootaxa 4250: 067–076. 10.11646/zootaxa.4250.1.5 [DOI] [PubMed] [Google Scholar]
- Hernández-Triana LM, Chaverri LG, Rodríguez-Pérez MA, Prosser SWJ, Hebert PDN, Ryan-Gregory T, Johnson N. (2015) DNA barcoding of Neotropical black flies (Diptera: Simuliidae): Species identification and discovery of cryptic diversity in Mesoamerica. Zootaxa 3936: 93–114. 10.11646/zootaxa.3936.1.5 [DOI] [PubMed] [Google Scholar]
- Hernández-Triana LM, Crainey JL, Hall A, Fatih F, Mackenzie-Dodds J, Shelley AJ, Zhou X, Post RJ, Gregory RT, Hebert DN. (2012) The utility of DNA barcoding for species identification within the blackfly SubgenusTrichodagmia Enderlein (Diptera: Simuliidae: Simulium) and related taxa in the New World. Zootaxa 3154: 43–69. [Google Scholar]
- Hernández-Triana LM, Prosser SW, Rodríguez-Pérez MA, Chaverri LG, Hebert PDN, Gregory TR. (2014) Recovery of DNA barcodes from blackfly museum specimens (Diptera: Simuliidae) using primer sets that target a variety of sequence lengths. Molecular Ecology Resources 4: 508–18. 10.1111/1755-0998.12208 [DOI] [PubMed] [Google Scholar]
- Hunter SJ, Goodall TI, Walsh KA, Owen R, Day JC. (2008) Nondestructive DNA extraction from blackflies Diptera: Simuliidae: retaining voucher specimens from DNA barcoding projects. Molecular Ecology Resources 8: 56–61. 10.1111/j.1471-8286.2007.01879.x [DOI] [PubMed] [Google Scholar]
- Kronefeld M, Werner D, Kampen H. (2014) PCR identification and distribution of Anophelesdaciae (Diptera, Culicidae) in Germany. Parasitological Resources 113: 2079–2086. 10.1007/s00436-014-3857-1 [DOI] [PubMed] [Google Scholar]
- Kronefeld M, Dittmann M, Zielke D, Werner D, Kampen H. (2012) Molecular confirmation of the occurrence in Germany of Anophelesdaciae (Diptera, Culicidae). Parasites and Vectors 5: 250. 10.1186/1756-3305-5-250 [DOI] [PMC free article] [PubMed]
- Kumar NP, Rajavel AR, Natarajan R, Jambulingam P. (2007) DNA barcodes can distinguish species of Indian mosquitoes (Diptera: Culicidae). Journal of Medical Entomology 44: 1‒7. 10.1093/jmedent/41.5.01 [DOI] [PubMed]
- Lilja T, Troell K, Kirik H, Lindström A. (2018) A distinct group of north European Aedesvexans as determined by mitochondrial and nuclear markers. Journal and Veterinary Entomology 32: 282–289. 10.1111/mve.12294 [DOI] [PubMed] [Google Scholar]
- Linton Y, Lee A, Curtis C. (2005) Discovery of a third member of the Maculipennis group in SW England. European Mosquito Bulletin 2005: 19:5–9.
- Linton Y-M, Samanidou-Voyadjoglou A, Harbach RE. (2002) Ribosomal ITS2 sequence data for Anophelesmaculipennis and An.messeae in northern Greece, with a critical assessment of previously published sequences. Institute of Molecular Biology 11: 379–383. 10.1046/j.1365-2583.2002.00338.x [DOI] [PubMed] [Google Scholar]
- Linton Y-M, Harbach RE, Seng CM, Anthony TG, Matusop A. (2001) Morphological and molecular identity of Anopheles (Cellia) sundaicus (Diptera: Culicidae), the nominotypical member of a malaria vector species complex in Southeast Asia. Systematic Entomology 26: 357‒366.
- Low VL, Pramual P, Adler PH, Ya’cob Z, Huang YT, Da Pham X, Sofian-Azirun M. (2016) Delineating taxonomic boundaries in the largest species complex of black flies (Simuliidae) in the Oriental Region. Scientific Reports 6: 1–8. 10.1038/srep20346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley R, Harrup LE, Veronesi E, Stubbins F, Stoner J, Gubbins S, Gubbins S, Wilson A, Batten C, Koenraadt HM, Barber J, Carpenter S. (2015) Testing of UK populations of Culexpipiens L. for Schmallenberg virus vector competence and their colonization. PLoS ONE 10: 0134453. 10.1371/journal.pone.0134453 [DOI] [PMC free article] [PubMed]
- Medlock JM, Leach SA. (2015) Effect of climate change on vector-borne disease risk in the UK. The Lancet Infectious Diseases 15: 721–730. 10.1016/S1473-3099(15)70091-5 [DOI] [PubMed] [Google Scholar]
- Medlock JM, Vaux AGC. (2009) Aedes (Aedes) geminus Peus (Diptera, Culicidae) – an addition to the British mosquito fauna. Diptera Digest 16: 1–4. [Google Scholar]
- Medlock JM, Vaux AGC. (2010) Morphological separation of the European members of the genus Culiseta (Diptera, Culicidae). Dipterists Digest 17: 1–6. [Google Scholar]
- Medlock JM, Vaux AGC. (2015) Seasonal dynamics and habitat specificity of mosquitoes in an English wetland: implications for UK wetland management and restoration. Journal of Vector Ecology 40: 90–106. 10.1111/jvec.12137 [DOI] [PubMed] [Google Scholar]
- Medlock JM, Snow KR, Leach SA. (2007) Possible ecology and epidemiology of medically important mosquito-borne arboviruses in Great Britain. Epidemiology and Infection 135: 466–82. 10.1017/S0950268806007047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medlock JM, Cull B, Vaux AGC, Irwin AG. (2017b) The mosquito Aedesvexans in England. Veterinary Record 181: 243. 10.1136/vr.j4048 [DOI] [PubMed]
- Medlock JM, Vaux AGC, Cull B, Schaffner F, Gillingham E, Pfluger V, Leach S. (2017a) Detection of the invasive mosquito species Aedesalbopictus in southern England. Lancet Infectious Diseases 2017. 10.1016/S1473-3099(17)30024-5 [DOI] [PubMed]
- Montagner FRG, Silva OS, Jahnke SM. (2018) Mosquito species occurrence in association with landscape composition in green urban areas. Brazilian Journal of Biology 78: 233–239. 10.1590/1519-6984.04416 [DOI] [PubMed] [Google Scholar]
- Montero-Pau J, Gómez A, Muñoz J. (2008) Application of an inexpensive and high throughput genomic DNA extraction method for the molecular ecology of zooplanktonic diapausing eggs. Limnology and Oceaneography: Methods 6: 218–222. 10.4319/lom.2008.6.218 [DOI] [Google Scholar]
- Murugan K, Vadivalagan C, Karthika P, Panneerselvam C, Paulpandi M, Subramaniam J. et al. (2016) DNA barcoding and molecular evolution of mosquito vectors of medical and veterinary importance. Parasitology Resources 115: 107–21. 10.1007/s00436-015-4726-2 [DOI] [PubMed] [Google Scholar]
- Nicolescu G, Linton YM, Vladimirescu A, Howard TM, Harbach RE. (2004) Mosquitoes of the Anophelesmaculipennis group (Diptera: Culicidae) in Romania, with the discovery and formal recognition of a new species based on molecular and morphological evidence. Bulletin of Entomological Research 94: 525–535. 10.1079/BER2004330 [DOI] [PubMed] [Google Scholar]
- Packer L, Gibbs J, Sheffields C, Hanner R. (2009) DNA barcoding and the mediocrity of morphology. Molecular Ecology Resources, Special issue on Barcoding Life, supplement 1: 42–50. 10.1111/j.1755-0998.2009.02631.x [DOI] [PubMed] [Google Scholar]
- Pangjanda S, Pramual P. (2016) Tests of conspecificity for closely related black fly (Diptera: Simuliidae) species of Simuliummultistriatum group in Thailand. Zootaxa 4231(3): 421–430. 10.11646/zootaxa.4231.3.8 [DOI] [PubMed] [Google Scholar]
- Ratnasingham S, Hebert PDN. (2013) A DNA-Based registry for all animal species: The Barcode Index Number (BIN) system. PLoS ONE 8(8): e66213. 10.1371/journal.pone.0066213 [DOI] [PMC free article] [PubMed]
- Ruiz-Arrondo I, Hernández-Triana LM, Oteo J. (2017) Fauna de mosquitos (Diptera, Culicidae) presentes en el humedal de La Grajera (Logroño) y sus implicaciones en salud pública. Zubía, Instituto de Estudios Riojanos 35: 123–140. [Google Scholar]
- Senatore GL, Alexander EA, Adler PH, Moulton JK. (2014) Molecular systematics of the Simuliumjenningsi species group (Diptera: Simuliidae), with three new fast-evolving nuclear genes for phylogenetic inference. Molecular Phylogenetics and Evolution 75: 138–148. 10.1016/j.ympev.2014.02.018 [DOI] [PubMed] [Google Scholar]
- Smith JL, Fonseca DM. (2004) Rapid assays for identification of members of the Culex (Culex) pipiens complex, their hybrids, and other sibling species (Diptera: Culicidae). American Journal of Tropical Medicine and Hygiene 70: 339–345. 10.4269/ajtmh.2004.70.339 [DOI] [PubMed] [Google Scholar]
- Tamura K, Stoecher G, Peterson N, Filipski A, Kumar S. (2007) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Molecular Biology and Evolution 30: 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaux AGC, Gibson G, Hernández-Triana LM, Cheke RA, McCracken F, Jeffries CL, Horton DL, Springate S, Johnson N, Fooks AR, Leach S, Medlock JM. (2015) Enhanced West Nile virus surveillance in the North Kent marshes, UK. Parasites & Vectors 8: 91. 10.1186/s13071-015-0705-9 [DOI] [PMC free article] [PubMed]
- Versteirt V, Nagy ZT, Roelants P, Denis L, Breman FC, Damiens D, Dekoninck W, Backeljau T, Coosemans M, Van Bortel W. (2015) Identification of Belgian mosquito species (Diptera: Culicidae) by DNA barcoding. Molecular Ecology Resources 15: 449‒457. 10.1111/1755-0998.12318 [DOI] [PubMed]
- Wilkerson RC, Linton Y-M, Fonseca DM, Schultz TR, Price DC, Strickman DA. (2015) Making mosquito taxonomy useful: A stable classification of tribe Aedini that balances utility with current knowledge of evolutionary relationships. PloS ONE 10: e0133602. 10.1371/journal.pone.0133602 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Luis M. Hernández-Triana, Victor A. Brugman, Nadya I. Nikolova, Ignacio Ruiz-Arrondo, Elsa Barrero, Leigh Thorne, Mar Fernández de Marco, Andreas Krüger, Sarah Lumley, Nicholas Johnson, Anthony R. Fooks
Accession number(s) of COI DNA barcode sequences used in this study downloaded from the NCBI database or provided by colleagues
Data type: molecular data
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Luis M. Hernández-Triana, Victor A. Brugman, Nadya I. Nikolova, Ignacio Ruiz-Arrondo, Elsa Barrero, Leigh Thorne, Mar Fernández de Marco, Andreas Krüger, Sarah Lumley, Nicholas Johnson, Anthony R. Fooks
Percentage of Interspecific (between groups) pairwise K2P genetic divergence of unique DNA barcodes (658 bp), representing 42 species of mosquitoes
Data type: molecular data
Explanation note: Highest pairwise distances (most divergent taxa) and lowest pairwise distances (most closely related taxa) are highlighted in yellow/bold, and green, respectively.