Abstract
Background:
Gut microbiota may play a role in egg allergy. We sought to examine the association between early-life gut microbiota and egg allergy.
Methods:
We studied 141 children with egg allergy and controls from the multi-center Consortium of Food Allergy Research study. At enrollment (age 3 to 16 months), fecal samples were collected and clinical evaluation, egg specific IgE measurement, and egg skin prick test were performed. Gut microbiome was profiled by 16S rRNA sequencing. Analyses for the primary outcome of egg allergy at enrollment, and the secondary outcomes of egg sensitization at enrollment and resolution of egg allergy by age 8 years, were performed using Quantitative Insights into Microbial Ecology (QIIME), Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt), and Statistical Analysis of Metagenomic Profiles (STAMP).
Results:
Compared to controls, increased alpha diversity and distinct taxa (PERMANOVA P=5.0×10−4) characterized the early-life gut microbiome of children with egg allergy. Genera from the Lachnospiraceae, Streptococcaceae, and Leuconostocaceae families were differentially abundant in children with egg allergy. Predicted metagenome functional analyses showed differential purine metabolism by the gut microbiota of egg allergic subjects (Kruskal Wallis Padj=0.021). Greater gut microbiome diversity and genera from Lachnospiraceae and Ruminococcaceae were associated with egg sensitization (PERMANOVA P = 5.0×10−4). Among those with egg allergy, there was no association between early-life gut microbiota and egg allergy resolution by age 8 years.
Conclusion:
The distinct early-life gut microbiota in egg allergic and egg-sensitized children identified by our study may point to targets for preventive or therapeutic intervention.
Keywords: Egg allergy, egg sensitization, food allergy, microbiome, purine
Introduction
Egg allergy is one of the most common childhood food allergies, affecting 0.5 to 2.5% of young children (1). However, the prevalence of food allergy has increased in recent years (2), with egg allergy reported in up to 8.9% of infants in some populations (3). Although host genetics predispose toward food allergy (4), genetic change does not occur on this time scale. Increasing attention has been paid to environmental factors, including the microbiome, that may be contributing to food allergy risk. Growing evidence points to a role for gut microbiota in the pathogenesis and course of food allergy (5–9).
We hypothesized that gut microbiota play a role in egg allergy. The etiology of food allergy is thought to involve deviation from the default state of mucosal immune tolerance that may be driven by diet, commensal microbiota, and interactions between them (10, 11). Variations in infant gut flora have been associated with allergen sensitization (12), milk allergy (7, 13–15), and food allergy as a general category (16). Microbiota associated with individual food allergies may differ by food allergen, consistent with the distinct natural histories and clinical courses of individual food allergies (6). To date, there has been no dedicated study of the microbiome in egg allergic subjects.
In this multi-center study, we used 16S rRNA sequencing to characterize the gut microbiome of 141 children age 3-16 months with egg allergy and controls (Figure 1). We examined for associations between early-life gut microbiota and our primary outcome of egg allergy, as well as with our secondary outcomes of egg allergen sensitization and egg allergy resolution.
Figure 1. Study design.
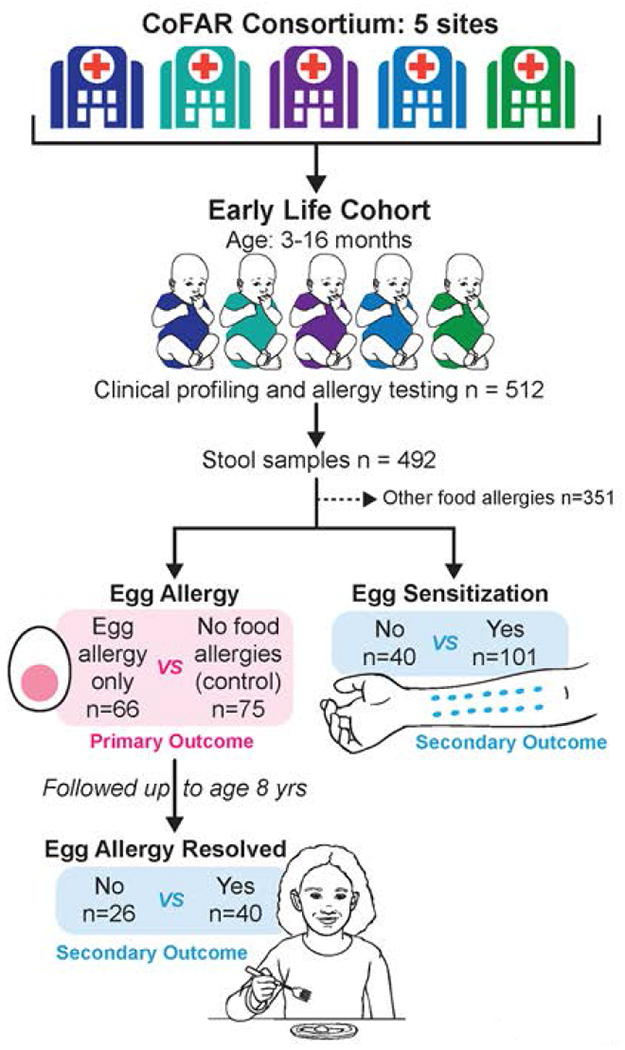
Participants were a subset of the multi-center CoFAR early-life observational cohort of children at risk for allergy. Children were enrolled at age 3-16 months, at which time clinical profiling, allergy testing, and stool collection were performed. Children with non-egg food allergy at enrollment or at any time during the ensuing observational period were excluded from this study. The goal of this study was to examine for associations between early-life gut microbiome and egg allergy at enrollment (primary outcome). We additionally examined for associations between early-life gut microbiome and egg sensitization at enrollment (secondary outcome) and early-life gut microbiome and the longitudinal outcome of egg allergy resolution by age 8 years (secondary outcome).
Methods
Study protocols were approved by the institutional review boards of the participating institutions.
Study design and subjects
The subjects of this study are a subset of a larger observational cohort study by the Consortium of Food Allergy Research (CoFAR) of 512 atopic participants (17). Participants were recruited at age 3-16 months from five study centers across the United States and were observed over time. The study sites included the Icahn School of Medicine at Mount Sinai, New York, New York; Duke University School of Medical Center, Durham, North Carolina; Johns Hopkins University School of Medicine, Baltimore, Maryland; National Jewish Health, Denver, Colorado; and Arkansas Children’s Hospital, Little Rock, Arkansas. The goal of the CoFAR observational study was to identify factors associated with the development of peanut allergy in a high risk cohort (17). Evaluation of egg allergy in this cohort was a secondary objective (18).
Stool samples were collected from CoFAR subjects at or near the time of enrollment (Figure 1). Samples were collected during a study visit or by the parent at home using a stool collection kit provided by CoFAR, transported with ice packs (consistent with protocols used by the Human Microbiome Project (19–21)), and frozen at −80C upon arrival. Because microbiota associated with individual food allergies may differ by food allergen, and the goal of this study was to examine microbiota associated with egg allergy in particular, we removed from the analysis CoFAR subjects with any other food allergies at enrollment or during the CoFAR observational period so that we could optimally compare subjects with egg allergy only to subjects with no food allergy. Specifically, among the 492 CoFAR subjects with available stool samples, we removed 345 samples from subjects with non-egg food allergy at any point during the CoFAR study and 6 samples that failed sequencing or quality control, leaving 141 samples, of which 66 had egg allergy only and the remaining 75 had no food allergy (control group) (Figure 1).
DNA isolation and 16S rRNA sequencing
DNA was isolated with the MoBio Power Soil DNA Isolation kit (Carlsbad, CA). The V4 region of the 16S rRNA gene was amplified with barcoded primers and 16S rRNA sequencing was performed on the Illumina MiSeq platform using 2×250 bp paired-end reads as previously described (7). Samples were submitted for sequencing as a single batch. Quality control on the raw reads was performed using Quantitative Insights into Microbial Ecology (QIIME 1.8.0) as previously described (7). Samples with less than 1000 sequences per sample were removed (6 samples). The median number of reads per sample was 23,786.
Outcomes
Participants were considered to have egg allergy if they had either (1) a positive physician-supervised oral food challenge to egg; (2) a convincing reaction and sensitization to egg by egg-specific IgE level ≥ 0.35 kUA/L and/or skin prick test (SPT) > 3mm; or (3) a flare of atopic dermatitis associated with egg ingestion along with an egg specific IgE level > 2 kUA/L (18). Control subjects had no known food allergies. At entry, dietary, medical, and social histories were obtained by questionnaires administered to the parents of participants. Egg-specific IgE levels and egg skin prick test were performed at entry and atopic dermatitis severity was graded based on criteria previously described (17) (Figure 1). Following their enrollment visit, participants were evaluated at 6 months, 12 months, and then yearly thereafter up until age 8 years for egg allergy status and other clinical information.
The primary outcome of this study was egg allergy at enrollment (Figure 1). Secondary outcomes included (1) egg sensitization at enrollment as evidenced by egg sIgE ≥ 0.35 kUa/L, and (2) among those with egg allergy at enrollment, resolution of egg allergy by age 8 years, defined as a change from egg allergy at enrollment to no egg allergy by the last documented encounter.
Microbiome analyses
Unless noted otherwise, we performed all analyses using QIIME 1.9.1 (22), an open-source bioinformatics pipeline for performing analysis of microbiome sequence data. To map the 16S rRNA reads into operational taxonomic units (OTUs) (23), defined as taxonomic units based on DNA sequences that share high identity, we used closed reference OTU picking as previously described (7). Each read was assigned to an OTU based on sequence similarity of at least 97% to the representative sequence of the OTU collection from the GreenGenes Database (v13.8) (http://greengenes.secondgenome.com). Alpha diversity (the richness and/or evenness) of a sample in terms of the diversity of OTUs observed in it) was calculated based on the Chao1 Index (24), Faith’s phylogenetic diversity (25), and Shannon (H) index (26) by sub-sampling ten times at maximum rarefaction depth of 2,000 reads. To assess between-sample composition differences, beta diversity was measured using unweighted UniFrac distances (27). PERMANOVA (permutational multivariate analysis of variance) (28) was used to test beta diversity based on UniFrac distances. PERMANOVA is a non-parametric test similar to ANOVA but that does not require the data to be normally distributed. PERMANOVA P values were calculated by performing 2000 permutations and were corrected for multiple testing using the Benjamini-Hochberg method (<5% FDR). To identify fecal microbiota differentiating subjects with and without egg allergy, as well as subjects with and without egg sensitization, we used linear discriminant analysis effect size (LEfSe) (29), a method for biomarker discovery, on the normalized relative abundances of taxa. LEfSe scores measure the consistence of differences in relative abundance between taxa in the groups analyzed (e.g. egg allergy vs. controls), with a higher score indicating higher consistency. As LEfSe does not allow for multiple covariate adjustment, we then used logistic regression to identify bacterial genera associated with egg allergy in models adjusted for these potential confounders: age, breast-feeding status, antibiotic usage, and baseline atopic dermatitis score. Solid food intake was considered as a potential confounder but not included in the adjusted model due its colinearity with age based on variancePartition (30) analysis (Figure S1). We used an analogous approach to identify genera associated with egg sensitization (egg sIgE ≥ 0.35 kUA/L).
Prediction of metagenome functional content
We used Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) v1.0.0 (31) to predict functional and metabolic pathways from the 16S rRNA data. OTUs associated with egg allergy after adjustment for covariates were used as input to construct functional content. We then used Statistical Analysis of Metagenomic Profiles (STAMP) (32) to detect and visualize the significant pathways present among samples with egg allergy and controls using the Kruskal-Wallis test, with correction for multiple testing via the Benjamini-Hochberg method.
Significance thresholds
Baseline characteristic comparisons and logistic regression results were considered statistically significant with unadjusted P <0.05, significant results from LEfSe linear discriminant analysis (LDA) required Kruskal Wallis P < 0.05 and LDA score > 2.0, and PERMANOVA and metagenome analyses were considered statistically significant with adjusted P < 0.05 and mean relative sequence difference > 0.1%.
Results
Study population
The baseline characteristics of the 141 participants are shown in Table 1. The population was mostly Caucasian (73%) with more male (67%) than female children. The majority of children had been born vaginally (68%) with 39% breastfeeding and 90% taking solid foods at entry. The median age of participants was 9.5 months (IQR 7.1–12.3). Compared to controls, those with egg allergy were older, less likely to be currently breastfeeding, and had milder atopic dermatitis.
Table 1:
Baseline characteristics of the participating subjects
| Characteristics | All subjects (n=141) | Egg Allergy (n=66) | Controls (n=75) | P value* |
|---|---|---|---|---|
| Sex- Female | 46 (32.6%) | 19 (28.8%) | 27 (36.0%) | 0.38 |
| Age- Months | 9.7 (3.4) | 11.7 (2.8) | 7.9 (2.8) | 4.5×10−13 |
| Race- Caucasian | 103 (73.0%) | 47 (71.2%) | 56 (74.7%) | 0.71 |
| Egg sIgE (kUA/L) | 3.4 (5.9) | 5.3 (7.6) | 1.7 (3.2) | 5.3×10−4 |
| Egg SPT (wheal mm) | 7.0 (4.3) | 8.4 (4.0) | 5.7 (4.2) | 1.8×10−4 |
| Atopic dermatitis | 0.03 | |||
| None | 7 (5.0%) | 6 (9.1%) | 1 (1.3%) | |
| Mild | 25 (17.7%) | 16 (24.2%) | 9 (12.0%) | |
| Moderate | 70 (49.6%) | 27 (40.9%) | 43 (57.3%) | |
| Severe | 39 (27.7%) | 17 (25.8%) | 22 (29.3%) | |
| Currently breastfeeding | 55 (39.0%) | 16 (24.2%) | 39 (52.0%) | 9.7×10−4 |
| Mode of delivery- vaginal | 96 (68.1%) | 44 (66.7%) | 52 (69.3%) | 0.86 |
| Solid food intake | 127 (90.1%) | 65 (98.5%) | 62 (82.7%) | 1.5×10−3 |
| Antibiotics– any during lifetime | 88 (62.4%) | 50 (75.8%) | 38 (50.7%) | 2.9×10−3 |
| Resolution of egg allergy by age 8 yrs | n/a | 40 (60.6%) | n/a | n/a |
Number (%) or mean (SD) reported.
Fisher Exact test for categorical variables, T test for continuous variables
Distinct gut microbiome composition and increased diversity associated with egg allergy
Children with egg allergy had a distinct gut microbiome composition (PERMANOVA R2= 2.7%, P=5.0×10−4). At the phylum level, Firmicutes and Verrucomicrobia were enriched in subjects with egg allergy (Figure 2A). To more specifically identify bacterial genera associated with egg allergy while also accounting for potential confounding by variables associated with gut microbiome as well as egg allergy (Table 1), we used LEfSe analysis and logistic regression models adjusted for age, atopic dermatitis score, breastfeeding, and antibiotic usage. The adjusted models identified 3 genera significantly associated with egg allergy: Ruminococcus, Lactococcus and Leuconostoc (Figure 2B). Ruminococcus (family Lachnospiraceae) and Lactococcus (family Streptococcaceae) were enriched in children with egg allergy, while Leuconostoc (family Leuconostocaceae) was enriched in controls.
Figure 2. Distinct gut microbiome composition in children with egg allergy.
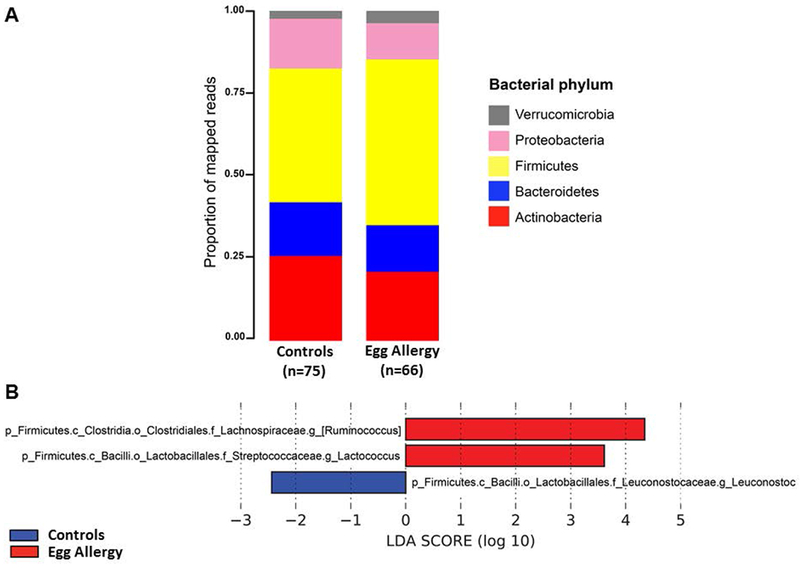
(A) Bacterial phyla in subjects with egg allergy and controls. (B) Bacterial genera associated with egg allergy after adjustment for potential confounders (age, breastfeeding, antibiotic usage and atopic dermatitis score). Genera from the Lachnospiraceae and Streptococcaceae families were enriched in subjects with egg allergy, whereas Leuconostoc was enriched in controls. Only genera with Linear Discriminant Analysis (LDA) scores > 2 and P<0.05 are presented.
Compared to controls, children with egg allergy had increased within-sample bacterial diversity as estimated by multiple alpha diversity indices (Figure 3). In subjects with egg allergy, community richness was greater in those with egg allergy by both species-based (Chao1 Index, Figure 3A; mean 299.3 (SD 97.2) in subjects with egg allergy vs. mean 223.2 (SD 92.0) in controls) and divergence-based (Faith’s phylogenetic diversity, Figure 3B; mean 12.3 (SD 3.3) in egg allergy vs. 9.7 (SD 3.1) in controls) indices. Egg allergic children also had higher diversity when richness and evenness were considered together (Shannon Index, Figure 3C; mean 4.4 (SD 0.9) in egg allergy vs 3.8 (SD 1.1) in controls). In analyses stratified by age, atopic dermatitis, breastfeeding, and antibiotic use, alpha diversity indices were also higher in subjects with egg allergy vs. controls (Figure S2-S5), although significance was not observed in some strata due to smaller sample sizes.
Figure 3: Increased gut bacterial diversity in children with egg allergy.
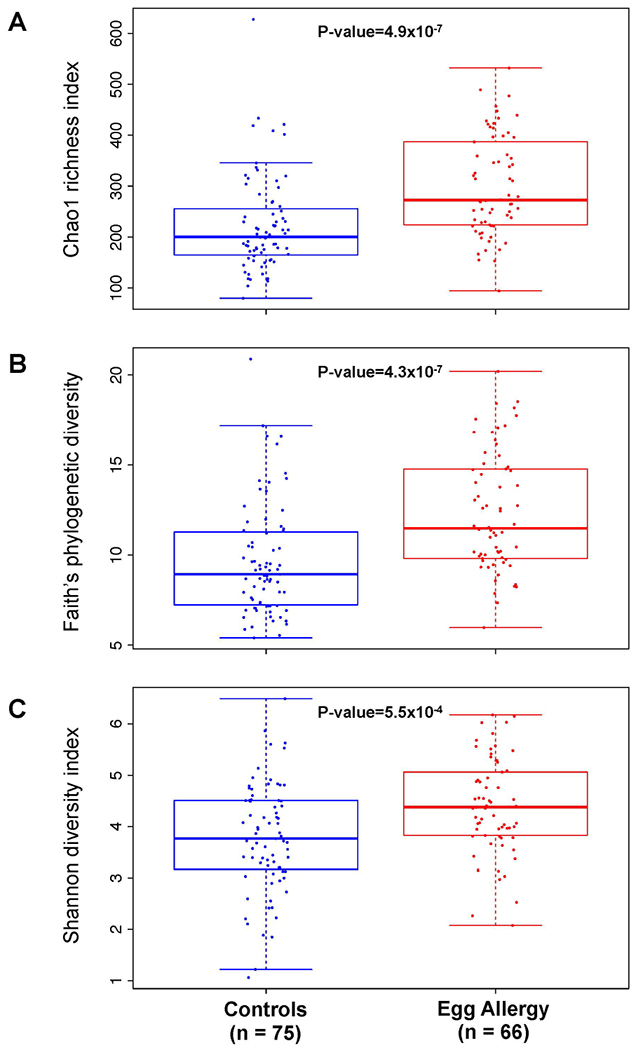
Alpha diversity estimated by (A) Chao1 index (species-based richness), (B) Faith’s phylogenetic diversity (divergence-based richness), and (C) Shannon (H) index (richness and evenness) at rarefaction depth of 2000 reads for subjects with egg allergy and controls. Mean and standard deviation alpha diversity were estimated by subsampling 10 times for each sample at this rarefaction depth. P-values were calculated using the Wilcoxon rank sum test.
Predicted functional pathway of bacterial taxa associated with egg allergy
Predicted metagenome function of OTUs associated with egg allergy revealed a single inferred metabolic pathway that was differentially abundant based on egg allergy status. Compared to controls, purine metabolism was decreased in subjects with egg allergy (K-W test Padj=0.021) (Figure 4).
Figure 4: Differential purine metabolism by gut microbiota of children with egg allergy.
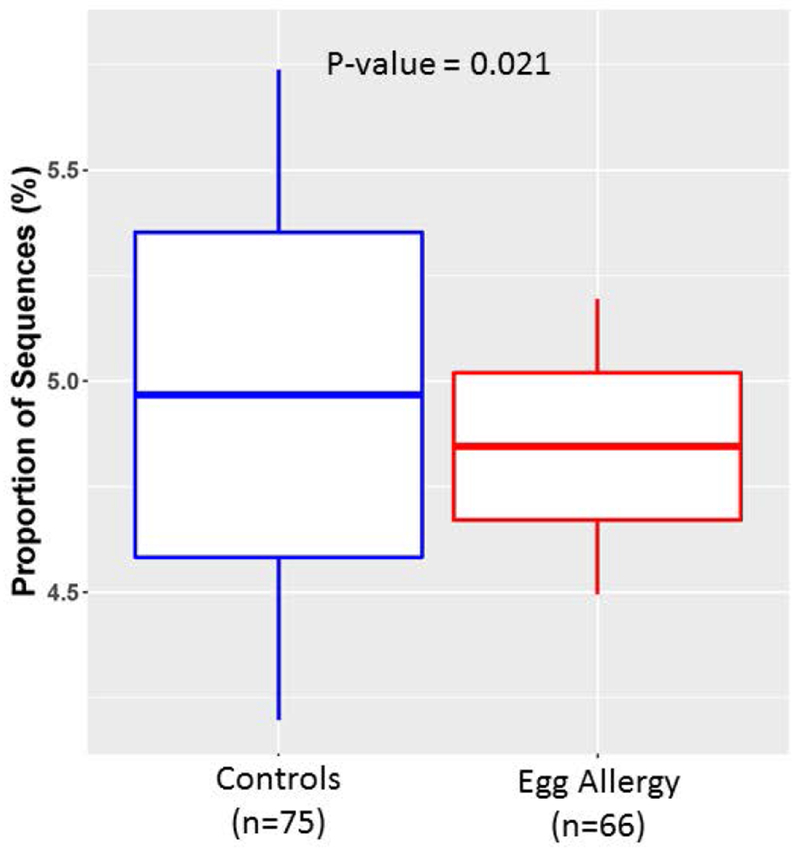
Prediction of metagenome functional content showed differential abundance of purine metabolic pathways based on egg allergy status. The vertical axis indicates proportion of sequences annotated as belonging to purine metabolic pathways based on predicted Kyoto Encyclopedia of Genes and Genomes (KEGG) orthologs.
Distinct gut microbiome composition and increased diversity associated with egg sensitization
Egg sensitization (egg sIgE ≥ 0.35 kUa/L) was associated with a distinct gut microbiome composition (PERMANOVA R2=1.6%, P= 5.0×10−4). Relative abundances at the phylum level based on egg sensitization status are shown in Figure 5A, which showed an enrichment of Firmicutes and Verrucomicrobia in egg sensitized subjects concordant with what was seen for egg allergy. Univariate analyses for potential confounders showed that age and race were associated with egg sensitization (P = 2 × 10−4 and P = 7 × 10−4, respectively), but there were no significant associations between other covariates (sex, atopic dermatitis, breastfeeding, mode of birth, antibiotic use) and egg sensitization. LEfSe analysis and logistic regression models adjusted for age and race identified 2 genera that were enriched in egg sensitized children: Roseburia (family Lachnospiraceae) and Faecalibacterium (family Ruminococcaceae) (Figure 5B).
Figure 5. Distinct gut microbiome composition in children with egg sensitization.
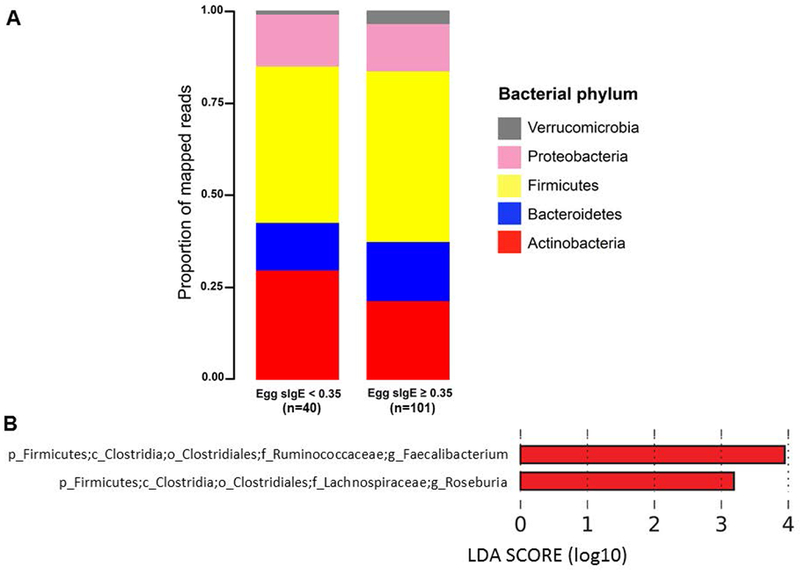
(A) Bacterial phyla in subjects with and without egg sensitization (egg sIgE ≥ 0.35 kUA/L vs. egg sIgE < 0.35 kUA/L, respectively). (B) Bacterial genera significantly associated with egg sensitization after adjustment for relevant confounders (age, race). Genera from Ruminococcaceae and Lachnospiraceae were enriched in egg sensitized children. Only genera with Linear Discriminant Analysis (LDA) scores > 2 and P<0.05 are presented.
Similar to egg allergy, children with egg sensitization had increased alpha diversity of the gut microbiome compared to subjects without egg sensitization (Figure 6). In subjects sensitized to egg, gut bacterial community richness was greater relative to controls (Chao1 Index, Figure 6A and Faith’s phylogenetic diversity, Figure 6B). Egg sensitized children also had higher diversity when richness and evenness were considered together (Shannon Index, Figure 6C). Spearman correlation analyses between egg sIgE and alpha diversity indices demonstrated positive correlation trends that did not reach statistical significance: r2=0.16, P=0.059 for Chao1 Index; r2 = 0.16, P=0.052 for Faith’s phylogenetic diversity; and r2=0.09, P=0.31 for Shannon index. In analyses stratified by age, atopic dermatitis, breastfeeding, and antibiotic use, alpha diversity indices also trended higher in subjects with egg sensitization (Figure S6-S9), except for those not breastfeeding, where diversity levels appeared more equivalent.
Figure 6: Increased gut bacterial diversity in children with egg sensitization.
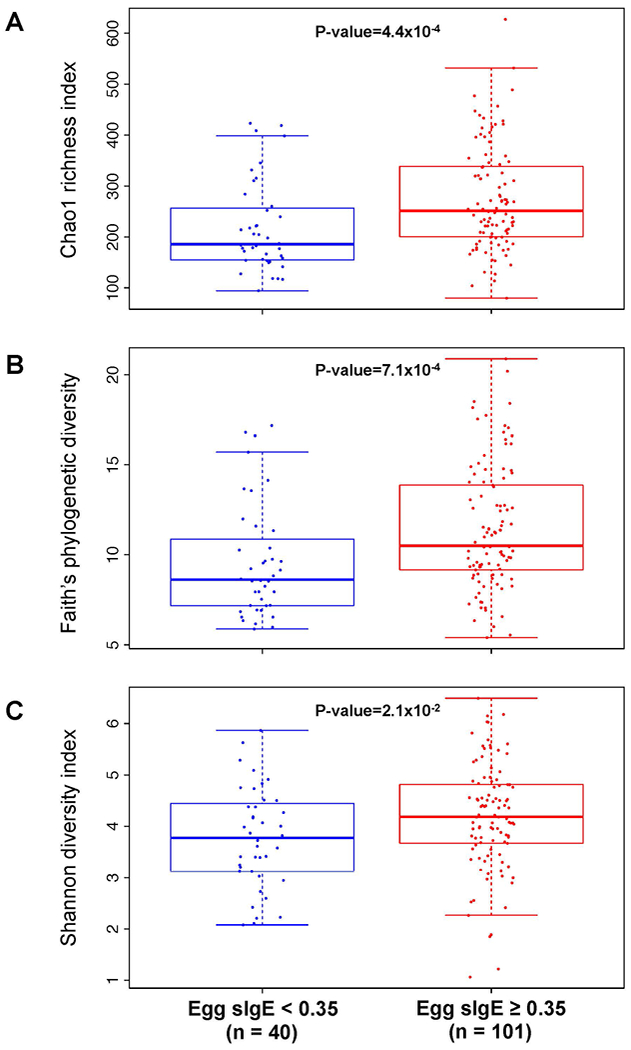
Alpha diversity estimated by (A) Chao1 index (species-based richness), (B) Faith’s phylogenetic diversity (divergence-based richness), and (C) Shannon (H) index (richness and evenness) at rarefaction depth of 2000 reads for subjects with and without egg sensitization. Mean and standard deviation alpha diversity were estimated by subsampling 10 times. P-values were calculated using the Wilcoxon rank sum test.
No difference in early-life bacterial composition or diversity associated with egg allergy resolution by age 8 years
Among the children with egg allergy, those whose egg allergy resolved by age 8 years showed no significant compositional differences in gut microbiota compared to those with persistent egg allergy (PERMANOVA R2=1.6%, P=0.28). There was also no significant difference in gut bacterial diversity based on egg allergy resolution vs. persistence (Figure S10).
Discussion
In this study of 141 children, we found increased diversity and distinct taxa in the early-life gut microbiome of children with egg allergy compared to children without food allergy. Genera belonging to the Lachnospiraceae and Streptococcaceae families were enriched in children with egg allergy, while Leuconostocaceae were enriched in controls. The predicted functional pathway of these bacterial taxa associated with egg allergy was purine metabolism. Similar to egg allergy, increased gut bacterial diversity was associated with egg sensitization, with enrichment of genera from Lachnospiraceae and Ruminococcaceae in egg sensitized children. Among the subjects with egg allergy, there were no associations between early-life gut microbiota and resolution of egg allergy by age 8 years.
To our knowledge, this is the first dedicated study of the microbiome of egg allergic subjects. It is possible that specific microbiota associated with individual food allergies differ depending on the food allergen, in line with the distinct natural histories and clinical courses of particular food allergies (6). There are few studies targeting the gut microbiome of specific food allergies other than milk (5–7, 14, 33). Because egg allergy is one of the most common food allergies in children (1), we found it compelling to study its associated gut microbiome, as this may elucidate pathways for its potential treatment and prevention.
At first glance, our finding of increased gut bacterial diversity in children with egg allergy versus controls may seem counter-intuitive to reasoning based on other diseases, where bacterial diversity is often higher in controls relative to affected individuals. For example, obese individuals have fewer types of gut bacteria than their lean counterparts (34). A beneficial association between bacterial diversity and disease is not always the case, however, as multiple studies of asthma have shown that airway bacterial diversity can be higher in individuals with asthma vs. controls (35, 36). Linked epidemiologically by the atopic march (37), egg allergy and asthma have some shared etiological origins, and observing a consistency in their respective associations with increased bacterial diversity suggests a nuanced relationship between microbial diversity and specific diseases. It is likely that the role of the microbiome in health and disease is not easily captured by any one dimension such as alpha diversity and should be considered with component taxa, their interactions, and their metabolic effects in the context of specific disease phenotypes (6, 38).
We found that genera belonging to the Lachnospiraceae and Streptococcaceae families were enriched in children with egg allergy (Figure 2), a finding that has both similarities and contrasts with prior studies of the microbiome in other food allergies. Consistent with our findings, a murine study of food allergy demonstrated differential abundance of taxa from Lachnospiraceae in ovalbumin-sensitized food allergy-prone mice with a gain-of-function mutation in the IL-4 receptor-a chain (39). Microbial communities dominated by Lachnospiraceae have also been reported in children with milk allergy (14). However, the same study detected lower levels of Streptococcaceae in milk allergic children. We note that the associations from our and others’ studies are from cross-sectional analyses, so causation cannot be inferred. In children without egg allergy, we found enrichment of Leuconostoc. Consistent with potentially protective effects from this genus on allergy diathesis, strains from this genus inhibit serum levels of total IgE, IgG1, IgG2, and increase OVA-specific IFN-gamma production in an ovalbumin-induced allergy mouse model (40). A limitation of cross-study comparisons is that implicated taxa are often reported at varying taxonomic levels (6). Because the biological impacts of microbiota are likely at the strain level, different strains within the same family may have disparate effects. Additionally, the heterogeneity of food allergy and different practices in clinical phenotyping, sample collection, and sample processing can lead to disparate results in microbiome studies (6).
The functional effects of gut microbiota in food allergy likely occur via the metabolites they produce (6, 41). To better understand the implications of the bacterial taxa associated with egg allergy, we used metagenomic functional prediction to infer metabolic pathways impacted by these microbiota. Interestingly, we found differential purine metabolism by the gut microbiota of egg allergic subjects. Metabolomic studies have shown that the purine pathway may be involved in the induction of peanut allergy, with peanut allergic children and mice undergoing peanut sensitization both demonstrating altered levels of uric acid, a product of purine metabolism (42). Depletion of uric acid in mice changes the development of peanut-specific IgE and IgG1 and anaphylaxis, while exogenous manipulation of uric acid can modulate the peanut allergic phenotype (42). It is possible that the altered purine metabolism associated with the early-life gut microbiota of egg allergic children in our study reflects a related role for purine metabolism in egg allergy.
Genera from Lachnospiraceae and Ruminococcaceae (order Clostridia) were enriched in children with egg sensitization (Figure 5). In line with our findings, Kalliomaki and colleagues found that Clostridia were more abundant in fecal samples from infants sensitized to at least one food or environmental allergen (including egg) compared to nonatopic controls (12). In contrast, other studies have reported no association between gut microbiota and food allergen sensitization (7, 43, 44). The studies that found no association between sensitization and gut microbiota focused on sensitization to different foods (e.g. milk (7)) or several foods simultaneously (43, 44). Gut microbiota may be specific to individual food allergens (6).
Although a prior study identified bacterial taxa associated with milk allergy resolution (7), we found no significant differences in early-life gut microbiota based on egg allergy resolution vs. persistence by age 8 years. Power may have been a limitation, as this analysis was limited to the 66 subjects with egg allergy at enrollment. While this study’s sample of egg allergic children was large compared to most studies of the microbiome of food allergic subjects (6), it was smaller than the sample studied for milk allergy resolution (n=226).
We recognize the limitations of our study. It would have been ideal for our controls to have been completely nonatopic, but we were constrained by the inclusion criteria for the overarching CoFAR study (17). That said, our controls had no egg allergy or other food allergy. Some were sensitized (without clinical allergy) and most had atopic dermatitis. Because large population studies have shown that a significant proportion of sensitized individuals have no clinical allergy (45), it was reasonable not to exclude sensitized non-allergic individuals from the control group. Further, we specifically performed a secondary analysis focused on egg sensitization. To address the limitation of high atopic dermatitis prevalence, we adjusted for atopic dermatitis (and other potential confounders including age, breastfeeding, and antibiotic usage) in our analyses. We applied bioinformatic approaches to 16S rRNA data to infer metabolic pathways affected by the gut bacteria differentially abundant in egg allergy. Bias by the 16S region targeted is possible and future work could include direct metabolite profiling to corroborate these inferences. Last, although our sample processing was consistent with well-established protocols (19–21) and was uniformly implemented for all samples, we cannot rule out that results for particular taxa could be influenced by methodological parameters (46–49). By studying children from CoFAR, we examined a well-established and large, multi-center sample of children with and without egg allergy determined by rigorous criteria (18) with contemporaneous stool collections at enrollment. We emphasize that this is one of the largest studies of the microbiome in food allergy (6), and the only to focus on egg allergy.
Conclusions
This multi-center study is the first to characterize the early-life gut microbiome in children with egg allergy. The distinct gut microbiota in egg allergic and egg-sensitized children identified by our study may point to targets for preventive or therapeutic intervention, especially if specific strains within these taxa can be identified via deeper sequencing. Our finding of differential purine metabolism by gut microbiota associated with egg allergy should be independently validated. The results from this study contribute to an evolving picture of egg allergy and food allergy overall, where resident bacteria may have more mechanistic impact on allergy than previously thought.
Supplementary Material
Figure S1. Pairwise correlation between egg allergy and potential covariates. All five variables were significantly associated with egg allergy (Table 1). Based on canonical correlation analyses (30), age, antibiotic use, breastfeeding status, and atopic dermatitis were included as covariates in the adjusted model. Because of colinearity with age, solid food intake was not included in the adjusted model.
Figure S2. Alpha diversity indices in children with and without egg allergy stratified by age. Alpha diversity estimated by (A) Chao1 index (species-based richness), (B) Faith’s phylogenetic diversity (divergence-based richness), and (C) Shannon (H) index (richness and evenness) at rarefaction depth of 2000 reads for subjects with egg allergy and controls, stratified by age. Mean and standard deviation alpha diversity were estimated by subsampling 10 times for each sample at this rarefaction depth. P-values were calculated using the Wilcoxon rank sum test.
Figure S3. Alpha diversity indices in children with and without egg allergy stratified by atopic dermatitis. Alpha diversity estimated by (A) Chao1 index (species-based richness), (B) Faith’s phylogenetic diversity (divergence-based richness), and (C) Shannon (H) index (richness and evenness) at rarefaction depth of 2000 reads for subjects with egg allergy and controls, stratified by atopic dermatitis. Mean and standard deviation alpha diversity were estimated by subsampling 10 times for each sample at this rarefaction depth. P-values were calculated using the Wilcoxon rank sum test.
Figure S4. Alpha diversity indices in children with and without egg allergy stratified by current breastfeeding status. Alpha diversity estimated by (A) Chao1 index (species-based richness), (B) Faith’s phylogenetic diversity (divergence-based richness), and (C) Shannon (H) index (richness and evenness) at rarefaction depth of 2000 reads for subjects with egg allergy and controls, stratified by current breastfeeding status. Mean and standard deviation alpha diversity were estimated by subsampling 10 times for each sample at this rarefaction depth. P-values were calculated using the Wilcoxon rank sum test.
Figure S5. Alpha diversity indices in children with and without egg allergy stratified by antibiotic use (any during lifetime). Alpha diversity estimated by (A) Chao1 index (species-based richness), (B) Faith’s phylogenetic diversity (divergence-based richness), and (C) Shannon (H) index (richness and evenness) at rarefaction depth of 2000 reads for subjects with egg allergy and controls, stratified by lifetime antibiotic use. Mean and standard deviation alpha diversity were estimated by subsampling 10 times for each sample at this rarefaction depth. P-values were calculated using the Wilcoxon rank sum test.
Figure S6. Alpha diversity indices in children with and without egg sensitization stratified by age. Alpha diversity estimated by (A) Chao1 index (species-based richness), (B) Faith’s phylogenetic diversity (divergence-based richness), and (C) Shannon (H) index (richness and evenness) at rarefaction depth of 2000 reads for subjects with and without egg sensitization, stratified by age. Mean and standard deviation alpha diversity were estimated by subsampling 10 times for each sample at this rarefaction depth. P-values were calculated using the Wilcoxon rank sum test.
Figure S7. Alpha diversity indices in children with and without egg sensitization stratified by atopic dermatitis. Alpha diversity estimated by (A) Chao1 index (species-based richness), (B) Faith’s phylogenetic diversity (divergence-based richness), and (C) Shannon (H) index (richness and evenness) at rarefaction depth of 2000 reads for subjects with and without egg sensitization, stratified by atopic dermatitis. Mean and standard deviation alpha diversity were estimated by subsampling 10 times for each sample at this rarefaction depth. P-values were calculated using the Wilcoxon rank sum test.
Figure S8. Alpha diversity indices in children with and without egg sensitization stratified by current breastfeeding status. Alpha diversity estimated by (A) Chao1 index (species-based richness), (B) Faith’s phylogenetic diversity (divergence-based richness), and (C) Shannon (H) index (richness and evenness) at rarefaction depth of 2000 reads for subjects with and without egg sensitization, stratified by current breastfeeding status. Mean and standard deviation alpha diversity were estimated by subsampling 10 times for each sample at this rarefaction depth. P-values were calculated using the Wilcoxon rank sum test.
Figure S9. Alpha diversity indices in children with and without egg sensitization stratified by antibiotic use (any during lifetime). Alpha diversity estimated by (A) Chao1 index (species-based richness), (B) Faith’s phylogenetic diversity (divergence-based richness), and (C) Shannon (H) index (richness and evenness) at rarefaction depth of 2000 reads for subjects with and without egg sensitization, stratified by lifetime antibiotic use. Mean and standard deviation alpha diversity were estimated by subsampling 10 times for each sample at this rarefaction depth. P-values were calculated using the Wilcoxon rank sum test.
Figure S10. No difference in gut bacterial diversity between subjects with persistent vs. resolved egg allergy by age 8 years. Alpha diversity at rarefaction depth of 2000 reads for egg allergic subjects with and without egg allergy resolution by age 8 years. The vertical axis indicates alpha diversity measured using Faith’s phylogenetic diversity. Mean and standard deviation alpha diversity were estimated by subsampling 10 times.
Acknowledgments
We thank Dr. Marshall Plaut, CoFAR scientific and medical officer and chief of the NIAID Food Allergy, Atopic Dermatitis, and Allergic Mechanisms Section. We thank Monica Andrade and Jose Clemente for their assistance with library preparation and OTU picking. We thank the families who kindly participated. We thank the staff of the clinical research units at each institution and the Statistical and Clinical Coordinating Center.
Funding: This study was supported by the National Institutes of Health (U19AI066738, U01AI066560, K08AI093538, R01AI118833), and the Department of Scientific Computing at the Icahn School of Medicine at Mount Sinai. The study was also supported by the National Center for Research Resources (NCRR)/National Institutes of Health (NIH), UL1 TR000154 from the NIH/National Center for Advancing Translational Sciences (National Jewish), UL1 TR000067 (Mount Sinai), UL1 TR000039 (Arkansas), UL 1 RR024128 (North Carolina), and UL1 RR 025005 (Johns Hopkins) from the NCRR.
Abbreviations:
- CoFAR
Consortium for Food Allergy Research
- FDR
false discovery rate
- LDA
linear discriminant analysis
- LEfSe
Linear discriminant analysis effect size
- OTU
operational taxonomic units
- PERMANOVA
permutational multivariate analysis of variance
- PICRUSt
Phylogenetic Investigation of Communities by Reconstruction of Unobserved States
- QIIME
Quantitative Insights into Microbial Ecology
- sIgE
specific IgE
- SPT
skin prick test
- STAMP
Statistical Analysis of Metagenomic Profiles
Footnotes
Conflicts of interest: None.
References
- 1.Caubet JC, Wang J. Current understanding of egg allergy. Pediatr Clin North Am 2011;58(2):427–443, xi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Branum AM, Lukacs SL. Food allergy among U.S. children: trends in prevalence and hospitalizations. NCHS Data Brief 2008(10):1–8. [PubMed] [Google Scholar]
- 3.Osborne NJ, Koplin JJ, Martin PE, Gurrin LC, Lowe AJ, Matheson MC, et al. Prevalence of challenge-proven IgE-mediated food allergy using population-based sampling and predetermined challenge criteria in infants. J Allergy Clin Immunol 2011;127(3):668–676 e662. [DOI] [PubMed] [Google Scholar]
- 4.Sicherer SH, Furlong TJ, Maes HH, Desnick RJ, Sampson HA, Gelb BD. Genetics of peanut allergy: a twin study. J Allergy Clin Immunol 2000;106(1 Pt 1):53–56. [DOI] [PubMed] [Google Scholar]
- 5.Bunyavanich S, Schadt EE. Systems biology of asthma and allergic diseases: a multiscale approach. J Allergy Clin Immunol 2015;135(1):31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang YJ, Marsland BJ, Bunyavanich S, O’Mahony L, Leung DY, Muraro A, et al. The Microbiome in Allergic Disease: Current Understanding and Future Opportunities - 2017 PRACTALL Document of the American Academy of Allergy, Asthma & Immunology and the European Academy of Allergy and Clinical Immunology. J Allergy Clin Immunol 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bunyavanich S, Shen N, Grishin A, Wood R, Burks W, Dawson P, et al. Early-life gut microbiome composition and milk allergy resolution. J Allergy Clin Immunol 2016;138(4):1122–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blazquez AB, Berin MC. Microbiome and food allergy. Transl Res 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stefka AT, Feehley T, Tripathi P, Qiu J, McCoy K, Mazmanian SK, et al. Commensal bacteria protect against food allergen sensitization. Proc Natl Acad Sci U S A 2014;111(36):13145–13150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berin MC, Sampson HA. Mucosal immunology of food allergy. Curr Biol 2013;23(9):R389–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol 2009;9(5):313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalliomaki M, Kirjavainen P, Eerola E, Kero P, Salminen S, Isolauri E. Distinct patterns of neonatal gut microflora in infants in whom atopy was and was not developing. J Allergy Clin Immunol 2001;107(1):129–134. [DOI] [PubMed] [Google Scholar]
- 13.Thompson-Chagoyan OC, Vieites JM, Maldonado J, Edwards C, Gil A. Changes in faecal microbiota of infants with cow’s milk protein allergy--a Spanish prospective case-control 6-month follow-up study. Pediatr Allergy Immunol 2010;21(2 Pt 2):e394–400. [DOI] [PubMed] [Google Scholar]
- 14.Berni Canani R, Sangwan N, Stefka AT, Nocerino R, Paparo L, Aitoro R, et al. Lactobacillus rhamnosus GG-supplemented formula expands butyrate-producing bacterial strains in food allergic infants. ISME J 2016;10(3):742–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berni Canani R, Nocerino R, Terrin G, Coruzzo A, Cosenza L, Leone L, et al. Effect of Lactobacillus GG on tolerance acquisition in infants with cow’s milk allergy: a randomized trial. J Allergy Clin Immunol 2012;129(2):580–582, 582 e581–585. [DOI] [PubMed] [Google Scholar]
- 16.Savage JH, Lee-Sarwar KA, Sordillo J, Bunyavanich S, Zhou Y, O’Connor G, et al. A prospective microbiome-wide association study of food sensitization and food allergy in early childhood. Allergy 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sicherer SH, Wood RA, Stablein D, Burks AW, Liu AH, Jones SM, et al. Immunologic features of infants with milk or egg allergy enrolled in an observational study (Consortium of Food Allergy Research) of food allergy. J Allergy Clin Immunol 2010;125(5):1077–1083 e1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sicherer SH, Wood RA, Vickery BP, Jones SM, Liu AH, Fleischer DM, et al. The natural history of egg allergy in an observational cohort. J Allergy Clin Immunol 2014;133(2):492–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Human Microbiome Project C. Structure, function and diversity of the healthy human microbiome. Nature 2012;486(7402):207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Human Microbiome Project C. A framework for human microbiome research. Nature 2012;486(7402):215–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aagaard K, Petrosino J, Keitel W, Watson M, Katancik J, Garcia N, et al. The Human Microbiome Project strategy for comprehensive sampling of the human microbiome and why it matters. FASEB J 2013;27(3):1012–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010;7(5):335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blaxter M, Mann J, Chapman T, Thomas F, Whitton C, Floyd R, et al. Defining operational taxonomic units using DNA barcode data. Philos Trans R Soc Lond B Biol Sci 2005;360(1462):1935–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chao A Nonparametric estimation of the number of classes in a population. Scand J Statist 1984;11:265–270. [Google Scholar]
- 25.Faith DP. Conservation evaluation and phylogenetic diversity. Biological Conservation 1992;61:1–10. [Google Scholar]
- 26.Shannon C A mathematical theory of communication. Bell Syst Tech J 1948;27:379–423. [Google Scholar]
- 27.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 2005;71(12):8228–8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecology 2001;26:32–46. [Google Scholar]
- 29.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol 2011;12(6):R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoffman GE, Schadt EE. variancePartition: Quantifying and interpreting drivers of variation in complex gene expression studies. BMC Bioinformatics 2016;17(1):483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 2013;31(9):814–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parks DH, Tyson GW, Hugenholtz P, Beiko RG. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 2014;30(2):3123–3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thompson-Chagoyan OC, Fallani M, Maldonado J, Vieites JM, Khanna S, Edwards C, et al. Faecal microbiota and short-chain fatty acid levels in faeces from infants with cow’s milk protein allergy. Int Arch Allergy Immunol 2011;156(3):325–332. [DOI] [PubMed] [Google Scholar]
- 34.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature 2006;444(7122):1022–1023. [DOI] [PubMed] [Google Scholar]
- 35.Huang YJ, Nelson CE, Brodie EL, DeSantis TZ, Baek MS, Liu J, et al. Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. Journal of Allergy and Clinical Immunology 2011;127(2):372–381. e373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marri PR, Stern DA, Wright AL, Billheimer D, Martinez FD. Asthma-associated differences in microbial composition of induced sputum. Journal of Allergy and Clinical Immunology 2013;131(2):346–352. e343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alduraywish SA, Lodge CJ, Campbell B, Allen KJ, Erbas B, Lowe AJ, et al. The march from early life food sensitization to allergic disease: a systematic review and meta-analyses of birth cohort studies. Allergy 2016;71(1):77–89. [DOI] [PubMed] [Google Scholar]
- 38.Bunyavanich S, Schadt EE. Systems biology of asthma and allergic diseases: A multiscale approach. J Allergy Clin Immunol 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Noval Rivas M, Burton OT, Wise P, Zhang YQ, Hobson SA, Garcia Lloret M, et al. A microbiota signature associated with experimental food allergy promotes allergic sensitization and anaphylaxis. J Allergy Clin Immunol 2013;131(1):201–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kang H, Oh YJ, Ahn KS, Eom HJ, Han N, Kim YB, et al. Leuconostoc citreum HJ-P4 (KACC 91035) regulates immunoglobulin E in an ovalbumin-induced allergy model and induces interleukin-12 through nuclear factor-kappa B and p38/c-Jun N-terminal kinases signaling in macrophages. Microbiol Immunol 2009;53(6):331–339. [DOI] [PubMed] [Google Scholar]
- 41.Chinthrajah RS, Hernandez JD, Boyd SD, Galli SJ, Nadeau KC. Molecular and cellular mechanisms of food allergy and food tolerance. J Allergy Clin Immunol 2016;137(4):984–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kong J, Chalcraft K, Mandur TS, Jimenez-Saiz R, Walker TD, Goncharova S, et al. Comprehensive metabolomics identifies the alarmin uric acid as a critical signal for the induction of peanut allergy. Allergy 2015;70(5):495–505. [DOI] [PubMed] [Google Scholar]
- 43.Adlerberth I, Strachan DP, Matricardi PM, Ahrne S, Orfei L, Aberg N, et al. Gut microbiota and development of atopic eczema in 3 European birth cohorts. J Allergy Clin Immunol 2007;120(2):343–350. [DOI] [PubMed] [Google Scholar]
- 44.Kendler M, Uter W, Rueffer A, Shimshoni R, Jecht E. Comparison of fecal microflora in children with atopic eczema/dermatitis syndrome according to IgE sensitization to food. Pediatr Allergy Immunol 2006;17(2):141–147. [DOI] [PubMed] [Google Scholar]
- 45.Salo PM, Arbes SJ Jr., Jaramillo R, Calatroni A, Weir CH, Sever ML, et al. Prevalence of allergic sensitization in the United States: Results from the National Health and Nutrition Examination Survey (NHANES) 2005–2006. J Allergy Clin Immunol 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo Y, Li SH, Kuang YS, He JR, Lu JH, Luo BJ, et al. Effect of short-term room temperature storage on the microbial community in infant fecal samples. Sci Rep 2016;6:26648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bassis CM, Moore NM, Lolans K, Seekatz AM, Weinstein RA, Young VB, et al. Comparison of stool versus rectal swab samples and storage conditions on bacterial community profiles. BMC Microbiol 2017;17(1):78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hill CJ, Brown JR, Lynch DB, Jeffery IB, Ryan CA, Ross RP, et al. Effect of room temperature transport vials on DNA quality and phylogenetic composition of faecal microbiota of elderly adults and infants. Microbiome 2016;4(1):19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fouhy F, Deane J, Rea MC, O’Sullivan O, Ross RP, O’Callaghan G, et al. The effects of freezing on faecal microbiota as determined using MiSeq sequencing and culture-based investigations. PLoS One 2015;10(3):e0119355. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Pairwise correlation between egg allergy and potential covariates. All five variables were significantly associated with egg allergy (Table 1). Based on canonical correlation analyses (30), age, antibiotic use, breastfeeding status, and atopic dermatitis were included as covariates in the adjusted model. Because of colinearity with age, solid food intake was not included in the adjusted model.
Figure S2. Alpha diversity indices in children with and without egg allergy stratified by age. Alpha diversity estimated by (A) Chao1 index (species-based richness), (B) Faith’s phylogenetic diversity (divergence-based richness), and (C) Shannon (H) index (richness and evenness) at rarefaction depth of 2000 reads for subjects with egg allergy and controls, stratified by age. Mean and standard deviation alpha diversity were estimated by subsampling 10 times for each sample at this rarefaction depth. P-values were calculated using the Wilcoxon rank sum test.
Figure S3. Alpha diversity indices in children with and without egg allergy stratified by atopic dermatitis. Alpha diversity estimated by (A) Chao1 index (species-based richness), (B) Faith’s phylogenetic diversity (divergence-based richness), and (C) Shannon (H) index (richness and evenness) at rarefaction depth of 2000 reads for subjects with egg allergy and controls, stratified by atopic dermatitis. Mean and standard deviation alpha diversity were estimated by subsampling 10 times for each sample at this rarefaction depth. P-values were calculated using the Wilcoxon rank sum test.
Figure S4. Alpha diversity indices in children with and without egg allergy stratified by current breastfeeding status. Alpha diversity estimated by (A) Chao1 index (species-based richness), (B) Faith’s phylogenetic diversity (divergence-based richness), and (C) Shannon (H) index (richness and evenness) at rarefaction depth of 2000 reads for subjects with egg allergy and controls, stratified by current breastfeeding status. Mean and standard deviation alpha diversity were estimated by subsampling 10 times for each sample at this rarefaction depth. P-values were calculated using the Wilcoxon rank sum test.
Figure S5. Alpha diversity indices in children with and without egg allergy stratified by antibiotic use (any during lifetime). Alpha diversity estimated by (A) Chao1 index (species-based richness), (B) Faith’s phylogenetic diversity (divergence-based richness), and (C) Shannon (H) index (richness and evenness) at rarefaction depth of 2000 reads for subjects with egg allergy and controls, stratified by lifetime antibiotic use. Mean and standard deviation alpha diversity were estimated by subsampling 10 times for each sample at this rarefaction depth. P-values were calculated using the Wilcoxon rank sum test.
Figure S6. Alpha diversity indices in children with and without egg sensitization stratified by age. Alpha diversity estimated by (A) Chao1 index (species-based richness), (B) Faith’s phylogenetic diversity (divergence-based richness), and (C) Shannon (H) index (richness and evenness) at rarefaction depth of 2000 reads for subjects with and without egg sensitization, stratified by age. Mean and standard deviation alpha diversity were estimated by subsampling 10 times for each sample at this rarefaction depth. P-values were calculated using the Wilcoxon rank sum test.
Figure S7. Alpha diversity indices in children with and without egg sensitization stratified by atopic dermatitis. Alpha diversity estimated by (A) Chao1 index (species-based richness), (B) Faith’s phylogenetic diversity (divergence-based richness), and (C) Shannon (H) index (richness and evenness) at rarefaction depth of 2000 reads for subjects with and without egg sensitization, stratified by atopic dermatitis. Mean and standard deviation alpha diversity were estimated by subsampling 10 times for each sample at this rarefaction depth. P-values were calculated using the Wilcoxon rank sum test.
Figure S8. Alpha diversity indices in children with and without egg sensitization stratified by current breastfeeding status. Alpha diversity estimated by (A) Chao1 index (species-based richness), (B) Faith’s phylogenetic diversity (divergence-based richness), and (C) Shannon (H) index (richness and evenness) at rarefaction depth of 2000 reads for subjects with and without egg sensitization, stratified by current breastfeeding status. Mean and standard deviation alpha diversity were estimated by subsampling 10 times for each sample at this rarefaction depth. P-values were calculated using the Wilcoxon rank sum test.
Figure S9. Alpha diversity indices in children with and without egg sensitization stratified by antibiotic use (any during lifetime). Alpha diversity estimated by (A) Chao1 index (species-based richness), (B) Faith’s phylogenetic diversity (divergence-based richness), and (C) Shannon (H) index (richness and evenness) at rarefaction depth of 2000 reads for subjects with and without egg sensitization, stratified by lifetime antibiotic use. Mean and standard deviation alpha diversity were estimated by subsampling 10 times for each sample at this rarefaction depth. P-values were calculated using the Wilcoxon rank sum test.
Figure S10. No difference in gut bacterial diversity between subjects with persistent vs. resolved egg allergy by age 8 years. Alpha diversity at rarefaction depth of 2000 reads for egg allergic subjects with and without egg allergy resolution by age 8 years. The vertical axis indicates alpha diversity measured using Faith’s phylogenetic diversity. Mean and standard deviation alpha diversity were estimated by subsampling 10 times.


