Abstract
The α-hemolysin nanopore has been studied for applications in DNA sequencing, various single-molecule detections, biomolecular interactions, and biochips. The detection of single molecules in a clinical setting could dramatically improve cancer detection and diagnosis as well as develop personalized medicine practices for patients. This brief review shortly presents the current solid state and protein nanopore platforms and their applications like biosensing and sequencing. We then elaborate on various epigenetic detections (like microRNA, G-quadruplex, DNA damages, DNA modifications) with the most widely used alpha-hemolysin pore from a biomedical diagnosis perspective. In these detections, a nanopore electrical current signature was generated by the interaction of a target with the pore. The signature often was evidenced by the difference in the event duration, current level, or both of them. An ideal signature would provide obvious differences in the nanopore signals between the target and the background molecules. The development of cancer biomarker detection techniques and nanopore devices have the potential to advance clinical research and resolve health problems. However, several challenges arise in applying nanopore devices to clinical studies, including super low physiological concentrations of biomarkers resulting in low sensitivity, complex biological sample contents resulting in false signals, and fast translocating speed through the pore resulting in poor detections. These issues and possible solutions are discussed.
Keywords: α-hemolysin, microRNA, G-quadruplex, DNA damage, guanine oxidation, abasic site, DNA methylation, DNA hydroxymethylation, nanopore device, cancer biomarker
1. Introduction
1.1. Epigenetics and its detections
The term epigenetics was introduced by Conrad Waddington in the early 1940s. Originally, epigenetics referred to the molecular pathways modulating the expression of a genotype into a particular phenotype, but the meaning of the term has gradually narrowed over the decades [1]. Unlike genetics studies based on alterations to the DNA sequence (the genotype), the epigenetics refer to the changes in gene expression or cellular phenotype which have other causes. Epigenetics also refers to the changes to the genome that do not involve a change in the nucleotide sequence. Examples of mechanisms that produce such changes are DNA methylation, loss of imprinting and histone modification, each of which affects how genes are expressed but without altering the underlying DNA sequence.
Epigenetic are affected by many factors like development, environmental chemicals, diet, drugs or pharmaceuticals, aging, which can cause cancer, autoimmune disease, mental disorder, diabetes and so on. Epigenetic modulations are fundamental to the genesis of cancer [2–4]. Detection of epigenetic alterations as biomarkers for cancer detection, diagnosis and prognosis have been studied extensively and were advanced rapidly [5–7].
1.2. Nanopore
A nanopore is a small hole, with the diameter from several nanometers (nm) to hundreds of nanometers. It could be a pore-forming protein, ion channels (biological nanopores), or a hole in synthetic materials such as glass, silicon, silicon nitride or graphene (solid state nanopores). Solid state nanopores are generally made in silicon membranes, the most common being silicon nitride [8–10]. Others like graphene [11–13], glass [14, 15] or glass slides [16] are used, as well. Solid state nanopores can be manufactured with several techniques including ion-beam sculpting [17], the transmission electron microscope (TEM) technique, or by electron beams [18]. Current biological nanopore platforms include the most widely used alpha-hemolysin [19], and others like MspA [20,21], aerolysin [22], OccK channels [23], FhuA [24], β-barrel protein nanopore [25], SP1 [26], Phi29 [27] and Cytolysin [28]. Composing hybrid nanopores is a newly evolved research field that began around 2007. Researchers are working on functionalizing solid state nanopores with biologically compatible polymer coatings and inserting biological-forming pores into the solid state nanopores, which will make the hybrid nanopores available for many different applications [29]. Studies have found that pre-assembled aHL protein pore can be inserted into a silicon nitride membrane with small holes, of diameters from 2.4–3.6 nm [30]. Many research groups are working on coating specific recognition sequences and receptors to nanopores, chemical functionalization of solid nanopores, like DNA Origami Nanopores [31], functionalized Solid Nanopores [32–35], and biofriendly nanochannels in a thin solid membrane [36].
1.3. Nanopore applications
Utilizing the electrophysiology method, i.e., patch clamp single channel recording technology, the ion current through a nanopore is very sensitive to the target molecules when occupying and translocating the pore. The interactions of the molecules and the pore can generate characteristic change in the nanopore current, therefore forming nanopore signatures (current-time relationship) as well as different molecular states which can be electrically identified.
The nanopores have been extensively studied for genetic detections [37–40], epigenetic detections [20,41–43], single-molecule detections [44–49], and biomolecular interactions [50–56] and are now being developed for next-generation sequencing [57–59].
This review will focus on epigenetic detections with the most widely used alpha-hemolysin nanopore (α-HL). Significance regarding biological functions and medicinal research of these epigenetic detections will also be reviewed and discussed.
2. microRNA Detections
In humans, microRNA genes represent approximately 1% of genome but regulate approximately 30% of human genes [60]. MicroRNAs are a group of small non-coding cellular RNAs with important biological functions in regulation of cell development, differentiation, apoptosis, and signaling pathways [61]. By systematic analysis of microRNA expression profiles, microRNA “signatures” have been identified in different types of cancers. Furthermore, cancer cells release certain microRNAs into circulation. These blood microRNAs are present in a very stable format and have thus been considered as potential biomarkers for cancer detection [62–64].
2.1. Detection of circulating microRNAs in cancer patients
It is difficult to distinguish the translocation of different microRNAs in α-HL because the sequences of all microRNAs are short and similar in length. One way to overcome this challenge is to use a signature that can detect target microRNA in the mixture. Studies have identified such a microRNA signature signal in the α-HL using an oligonucleotide probe with a signal tag (Figure 1 a, b). The analysis indicated two important functions performed by the signal tag of the probe: 1) guidance of the microRNA·probe complex entrapment in the pore and 2) inducement of the dissociation of the microRNA·probe complex. The configuration change during the unzipping process gave rise to signature current patterns, which enabled the recognition of single target microRNA molecules. Because of the specificity of the probe, the frequency of the signature signal was independent of the presence of multiple nucleic acid components and could therefore be used to quantify target microRNA in the mixture.
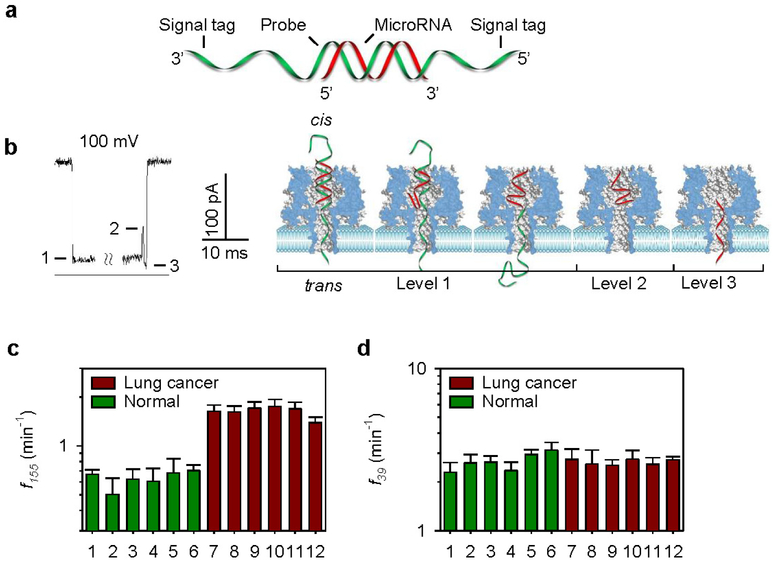
Figure 1. microRNA detections in the lung cancer patients.
a, Molecular diagram of a microRNA (red) bound to a probe (green) with signal tags. b, The unzipping signature (left) and the molecule pathways of the dsDNA unzipping signature (right) in the nanopore. c, The frequencies of miR-155 signature events (f155) from six healthy individuals (#1 to #6) and six patients with lung cancer (#7 to #12) in the presence of spiked-in synthetic miR-39. d The frequencies of spike-in miR-39 signature events detected all the samples that were used in c. The patient conditions were the following: #7, metastatic squamous lung carcinoma; #8, recurrent small-cell cancer; #9, early-stage small-cell carcinoma, status post-chemotherapy and -radiation; #10, early-stage small-cell cancer, status post-chemotherapy; #11, late-stage non-small cell carcinoma, status post-resection and -chemotherapy; #12, late-stage adenocarcinoma, status post-chemotherapy. Reprinted with permission from reference [65].
The level of mir-155 in blood samples from cancer patients was significantly higher than the normal healthy controls, but the level of spiked-in internal control of mir-39 (synthetic C. elegans microRNA, not presented in human) remained unchanged in these samples (Figure 1 c, d). Overall, the signature signal ensured the high selectivity required for microRNA detection in plasma RNA extract [65]. The key component of the nanopore sensor is the probe, the sequence of which is programmable and can be optimized to achieve high sensitivity and selectivity. The nanopore method can be a useful tool for quantitative studies of microRNAs and the discovery of disease markers, which are important for non-invasive screening and the early diagnosis of diseases such as cancer.
2.2. Developing a novel polycationic probe for simultaneous enrichment and detection of MicroRNAs
Generally, the clinical samples used to test for microRNA are RNA extractions from a patient’s biofluids such as plasma. These extractions are a complex collection of various RNA species: miRNAs, mRNAs, tRNAs, etc. When the nanopore is used to detect the target miRNA, any free nucleic acids in the RNA mixture can also nonspecifically interact with the pore. These interactions can result in “contaminative” signals in the α-HL that severely influence the target miRNA determination, and these signals should be eliminated.
By using a polycationic probe as the carrier, the nanopore can selectively capture and detect the target miRNA. The probe comprises a sequence of peptide nucleic acids (PNA) conjugated with a polycationic peptide lead. The PNA is designed to specifically capture the target miRNA. Upon hybridization, the positively charged peptide lead and the negatively charged miRNA together form a dipole (Figure 2). This structure can be driven into the nanopore by a large electric field gradient around the nanopore opening. At the same time, any free nucleic acids without probe hybridization would carry negative charge and migrate away from the pore opening. Consequently, only the signatures for the miRNA·probe complex and probe alone in the nanopore will be identified, and any interference signal originating from free nucleic acids is completely eliminated [66]. This method allows researchers to selectively detect only those nucleic acid sequences that hybridize with the probe, even when many other confounding species are present. If validated in clinical samples, for example, the detection of target miRNA from RNA extractions derived from a patients’ biofluids, this method would have applications in many areas such as early disease diagnosis, cancer metastasis prediction, and the monitoring of a patient’s response to therapy. In conclusion, this novel approach introduces a new method of detecting clinically relevant DNA or RNA fragments in a complex nucleic acid mixture.
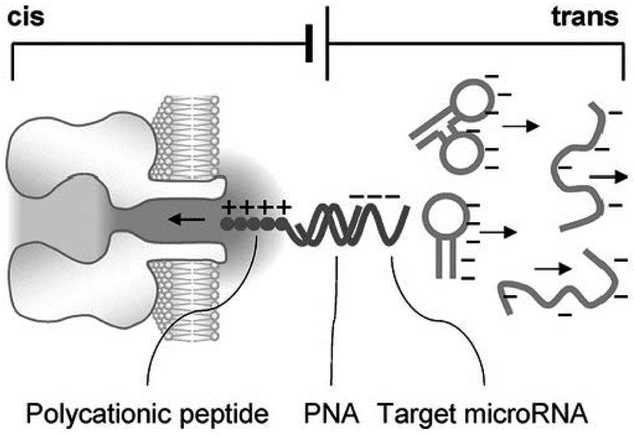
Figure 2. Cationic peptide probe-based interference-free detection of microRNAs in the nanopore.
The probe includes: a polycationic polymer lead (peptide, blue) and a capture domain (PNA, green). The capture domain can hybridize with the target microRNA (red). The miRNA·probe complex is drawn into the nanopore by the applied voltage at the pore (trans) opening, while all other negatively charged free nucleic acids without the probe binding (gray) were electrophoretically expelled away from the pore. Reprinted with permission from reference [66].
2.3. Developing probes for multiplex microRNA detection
Cancer/disease detection and diagnosis require accurate measurement of a biomarker panel, rather than a single miRNA species. The current nanopore technology cannot meet this need because it can analyze only one miRNA per detection. Although nanopore multiplex detection has been reported, their common principle, i.e., different targets (unlabeled or labeled) generating distinct nanopore signatures, is not applicable to miRNA detection. Due to their similar polymer lengths (18–22 bases), miRNAs cannot be distinguished from each other by their signatures. This demands new nanopore strategies for multiplex biomarker detection.
Studies have designed a series of barcode probes that were constructed through click chemistry (Figure 3). Each barcode motif trapped in the nanopore can specifically modulate the nanopore ionic current and therefore can encode different target nucleic acids. These universal barcode probes working together enable simultaneous detection of multiple miRNAs in a biomarker panel [67]. The target was a panel of four lung cancer-derived miRNAs: miR-155, miR-182-5p, miR-210, and miR-21. For each miRNA, four probes were constructed: one without a tag (P0) and the other three tagged with a 3-, 8-, and 24-mer PEG [68] (P3, P8, and P24) on the lead, respectively. The PEG label allows generating a distinct current profile compared with the block using unlabeled probe, and modulation of miRNA·probe blocking level by PEGs of different lengths were successful. Simultaneous observation of multiple miRNA·probe blocking levels in a current trace was also achieved. This method elucidated a biophysical mechanism for modulating nanopore ionic flow through tagging a barcode motif on the nucleic acid duplex. The barcode tag sliding in the pore marked the molecular processes of trapping, unzipping, and translocation. As each barcode tag specifically blocked the nanopore, different barcodes could be used to encode different target sequences, therefore realizing nanopore multiplex detection.
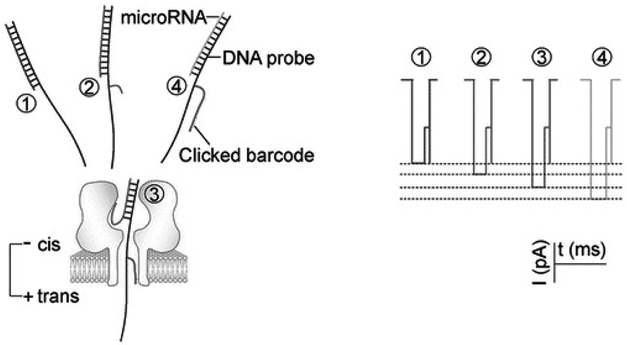
Figure 3. Barcode tagged probe through click chemistry for nanopore ionic current modulation and multiplex detection of mircroRNAs in the nanopore.
Each PEG-labeled DNA probe can target a specific microRNA and generate a specific nanopore current profile. Upon being captured in the nanopore, the PEG on the probe specifically regulates the nanopore current profile, thereby generating a signature for target identification. Reprinted with permission from reference [68].
3. G-quadruplex Folding and Detections
Repetitive DNA sequences (telomere sequence) located at the ends of chromosomes can fold into compact structures called G-quadruplexes (also known as G-tetrads or G4-DNA). They are rich in guanine and are capable of forming a four-stranded structure. The quadruplex structure is further stabilized by the presence of a cation, like potassium. G-quadruplex plays an important role in epigenetics (replication, recombination, DNA repair, maintaining genome stability) and regulation processes, as well as having the potential to serve as the drug target of G-quadruplex binding proteins [69–73].
3.1. Folding and unfolding of G-quadruplex subtypes in a confined space
A particular study demonstrated that various G-quadruplex conformations (hybrids, basket, and propeller folds) adopted by the human telomeric sequence are formed under different physical conditions, and these conformations were identified. These subtype conformations were found to form characteristic current patterns in the nanopore (Figure 4).
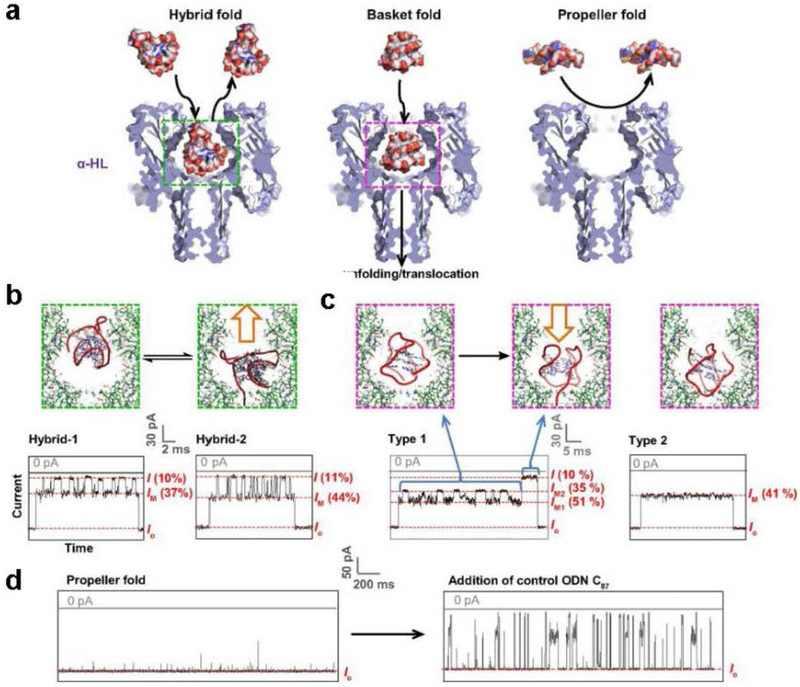
Figure 4. Interactions between G-quadruplex [5’-TAGGG(TTAGGG)3TT-3’] subtypes and the alpha-hemolysin reveal the size-dependent properties of the protein nanopore.
a, Space-filling models of G-quadruplexes interacting with the nanopore constructed using different PBD structures, b-d, Stick models of the proposed interaction mechanisms and current-time traces yielded by (b) the hybrid folds (50 mM KCl, 950 mM LiCl, 25 mM Tris, pH 7.9), (c) the basket folds (1 M NaCl, 25 mM Tris, pH 7.9), and (d) the propeller folds (20 mM KCl, 5 M LiCl, 25 mM Tris, pH 7.9). All current traces were recorded under 120 mV (trans vs. cis). I and IM values are indicated as a percentage of the open-channel current Io. Reprinted with permission from reference [75].
The basket-folded structure was able to unravel within the nanopore, while the hybrid folds had much more difficulty unfolding. In contrast, the propeller fold could not enter the nanopore opening because of its disk-like shape and larger dimensions; however, it could unfold outside the protein vestibule much faster than the basket fold. This study demonstrated the ability of the nanopore to discriminate different G-quadruplex secondary structures based on their specific shapes and sizes, as well as monitoring their unfolding kinetics at different locations (pore opening, nanocavity, constriction site) in the nanopore, thus expanding the applications of nanopore technology [74].
As we described above, the human telomere sequence (repetitive 5’-TTAGGG-3’) can fold into G-quadruplexes with different secondary structures. Hybrid folds with triplex folding intermediates can generate structure-dependent electrical current signatures within the nanocavity of the α-HL [75]. This telomere sequence is hypersensitive to UV-induced thymine dimer (T=T) formation. Studies have demonstrated that the generation of T=T only slightly changes the stabilities of hybrid and basket folds, and the presence of the T=T pair changed the ratio of the hybrid types in the hybrid folds [76].
3.2. Folding and unfolding of thrombin-binding aptamer formed G-quadruplex in electrolytes with different cations
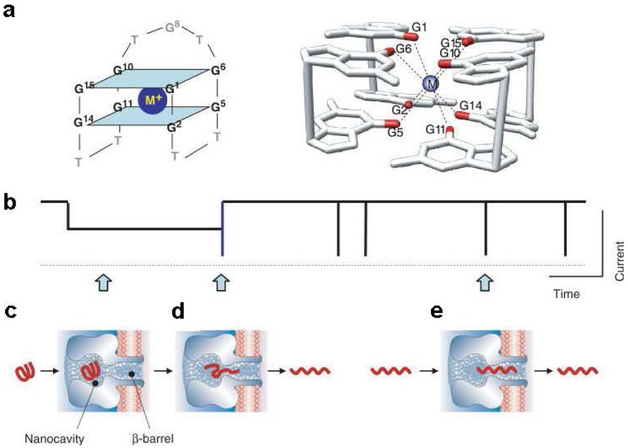
G-quadruplexes play important roles in both gene regulation and nanosensor constructions. Studies found that cations can regulate the folding and unfolding of the G-quadruplex formed by the thrombin-binding aptamer (TBA, GGTTGGTGTGGTTGG). Single G-quadruplexes can be trapped in the nanocavity of the nanopore. The trapped Single G-quadruplexes trapped in the nanocavity of the nanopore specifically blocked the current through the nanopore [77]. The nanopore electrical signatures (Figure 5) revealed that the G-quadruplex formation was regulated by the type of cations. Cations like K+, Ba2+ and NH4+ were more favorable over Cs+, Na+ and Li+ for G-quadruplexes. Meanwhile, cations like Mg2+ and Ca2+ did not induce G-quadruplex formation [78]. The nanopore was also demonstrated to be efficient for the study of interactions with the protein ligand [79].
Figure 5. Detection of thrombin-binding aptamer (TBA, GGTTGGTGTGGTTGG) with metal ions in the nanopore.
a, the sequence and structure of TBA G-quadruplex (left) and the two G-tetrad planes in the TBA G-quadruplex formed by guanines at the position 1, 6, 10 and 15, and the bottom one by guanine 2, 5, 11 and 14 (right). A cation in between is coordinated with eight carbonyls. b, Schemes of the current trace showing characteristic signature blocks. c, Long-lived block for capturing a single G-quadruplex in the nanocavity enclosed by the α-hemolysin nanopore; d, The long block terminal spike produced by translocation of the unfolded G-quadruplex through the β-barrel. The long-lived block with an ending spike served as the nanopore signature for the folded form of TBA. e, Short-lived block formed by translocation of linear form TBA. Reprinted with permission from reference [78].
This study demonstrated that α-HL is a useful single-molecule tool for studying specific molecular processes like the ion-regulated oligonucleotides. The method used in this study may be expanded for the kinetic study of other quadruplexes and their variants. Potential targets include various biologically relevant intramolecular quadruplexes, such as the i-motif (quadruplexes formed by cytidine-rich sequences) and chemically modified quadruplexes with unique functionalities. This research may also be helpful in constructing new molecular species with tunable properties for nano-constructions and the manufacture of biosensors [79].
4. Detection of DNA Damages
DNA damage can happen in a variety of ways, which can ultimately lead to mutations and genomic instability without a means of repair. This might result in the development of a variety of cancers. Damage can be caused by the environment (like excessive exposure to UV radiation in the form of sunlight or tobacco smoke), oxidative damage (like free radicals), error in DNA replication, or the loss of DNA bases known as AP (apurinic/apyrimidinic) sites. Developing different kinds of biosensors for DNA damage detection has been extensively studied recently [80–83]. Below, the alpha-hemolysin based sensors for detecting DNA damages are summarized and discussed.
4.1. Detection of guanine oxidation in the human telomere repeat sequence
8-oxo-7,8-dihydroguanine (OG) produced from guanine (G) oxidation is a biomarker of oxidative stress which could induce telomere shortening and cellular senescence. Firstly, the natural forms of G and OG in the human telomere sequence (hTelo, 24-mer) at different locations (top tetrad, middle tetrad and bottom tetrad) were studied in the nanopore, and the singly oxidized G4 system can be discriminated from the natural form based on the event durations. However, it is limited in the clinical studies since a telomere from a cellular source will possess ~104 5’-TTAGGG-3’ repeats that can fold to series of ~103 G4s, leading to the event durations being very broad. It then will not be able to distinguish the events generated by the G and OG strands in the nanopore [84].
Ideally, the signature for DNA damage detection is that such damage can introduce both event duration and current level changes. By the labelling of aminomethyl-[18-cown-6] (18c6) to the OG sites and optimizing of the electrolyte, detecting and quantification of OG sites in the long a Long hTelo Sequence (120-mer) after exposure to 1O2 was achieved. The labeled OG yielded a pulse-like signal (signature) in the nanopore current profile (current vs time) when the DNA strand was pulled through the α-HL nanopore by the applied voltage (Figure 6). Identification and detection of telomeres for OG utilizing the nanopore is an innovative approach that is envisioned to simultaneously allow quantification of OG and determination of telomere length in one single experiment. The principles described here can be applied to research surrounding the oxidative stress and telomere attrition observed in different diseases including prostate cancer and diabetes [84].
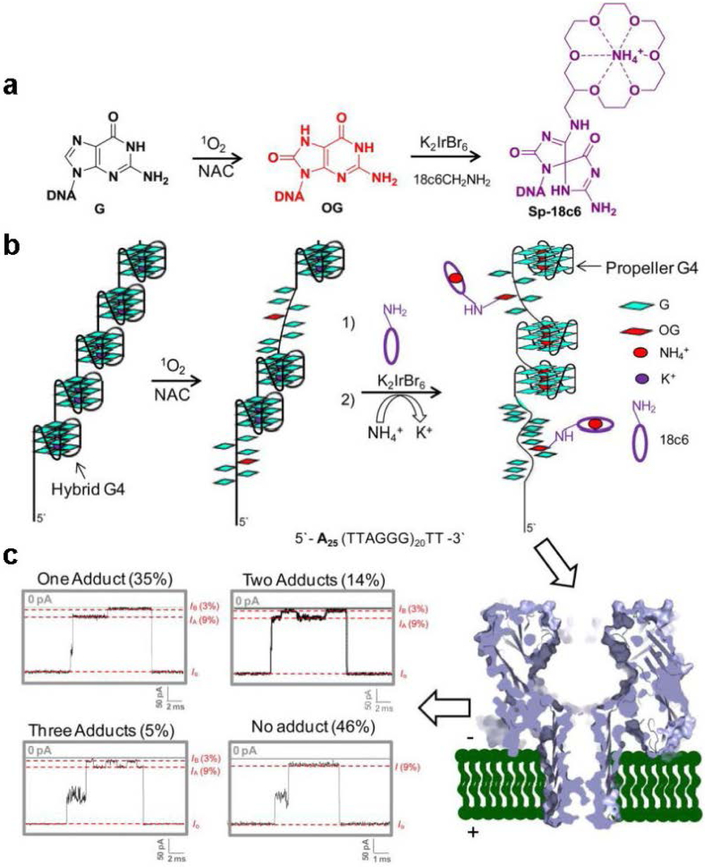
Figure 6. Detection of guanine oxidation in the human telomere repeat sequence.
Oxidation of a 120-mer portion of the human telomere repeat sequence (Q5) with 1O2 to yield 8-oxo-7,8-dihydroguanine (OG) that was labeled with 18c6 followed by nanopore detection and quantification. a, Reaction scheme for oxidation of guanine to yield OG and the labeling reaction of OG by 18c6 in the presence of K2IrBr6. b, Model of the Q5 strand in biologically relevant salts (hybrid G4) followed by oxidation labeling and refolding in NH4Cl (100 mM) and LiCl (2 M) electrolyte to yield the propeller fold. c, More than 50% of the events contained pulse-like current modulations: i.e., ~35% of the events presented one modulation, ~14% two modulations, and ~5% three modulations. Nanopore recordings were conducted in 25 mM Tris, pH 7.9, 100 mM NH4Cl, and 2 M LiCl at 25 °C. Reprinted with permission from reference [84].
4.2. Detection of abasic site in the β-barrel site of the nanopore
DNA abasic (AP) sites are one of the most frequent lesions in the genome derived from either spontaneous hydrolysis or the enzymatic removal of modified bases by glycosylase. AP sites could induce strand breaks and transcriptional mutations resulting in cellular dysfunction [85,86,87,88]. AP site detection has attracted tremendous interest, and many methods have been developed, like fluorescence measurements [89,90], atomic force microscopy (AFM) [91], mass spectrometry [92], ELISA-based assay [93], utilization of aldehyde-reactive probes [94], metalloinsertors [95], and isotope labels [96]. Though, detection methods that can detect multiple sites are still in urgent need [97].
In the α-HL, studies have demonstrated that the 18c6 labeled abasic site (AP-18c6) can be detected and identified in single-stranded DNA in a NaCl electrolyte solution, which generated a diagnostic pulse-like current signature in the nanopore when the AP-18c6 strand interacted with the β-barrel (Figure 7). It is possible that 18c6-Na+ hesitated at the protein constriction and prevented the movement of the DNA molecule. However, after the dissociation of the Na+, the 18c6 adduct passed through the constriction site, and a deeper current blockage was generated. Finally, when the DNA had passed through the nanopore, the current returned to its open current level. Most importantly, this method was able to detect multiple AP sites in one DNA strand [98]. Applying the same chemical strategy combined with the help of lesion-specific glycosylases, researches can envision that a variety of DNA base damages and mismatches can be specifically converted to AP sites and then functionalized to the AP-18c6 adduct, allowing their sequence-specific detection in single molecules. This method has the potential to be used for disease/cancer detections caused by an unrepaired AP site.
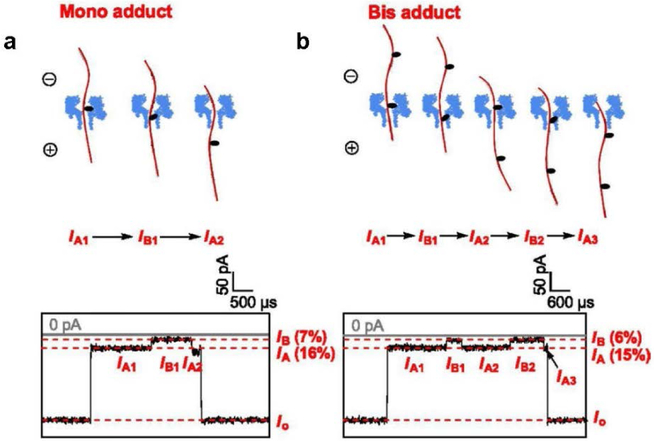
Figure 7. Detection of abasic site in the β-barrel of the nanopore. Individual i-t traces of AP-18c6 in homopolymeric strands.
a, Identification of single AP-18c6 adduct. Sample i-t traces for mono adduct (120 mV trans vs. cis). b, Identification of two AP-18c6 adducts. Sample i-t traces for bis adducts (120 mV trans vs. cis). DNA was captured into the nanopore from 5’ entry. The pulse like nanopore signature for AP-18c6 was generated for the AP detection. Reprinted with permission from reference [98].
4.3. Abasic site detection in the latch zone of the nanopore
The α-HL nanopore has been employed for DNA sequencing, various single-molecule detections, and bimolecular interactions by utilizing its narrowest constriction site, since it has always been considered as the sensing region of the pore. Interestingly, the latch zone, i.e., the vicinity of the vestibule constriction of the pore was recently found to constitute a sensing region as well. This previously unrecognized sensing region was found to be able to detect individual abasic sites in dsDNA in 2013 [99] (Figure 8). Later, further research was carried out which obtained the optimized experimental conditions for identifying the base modification [100].
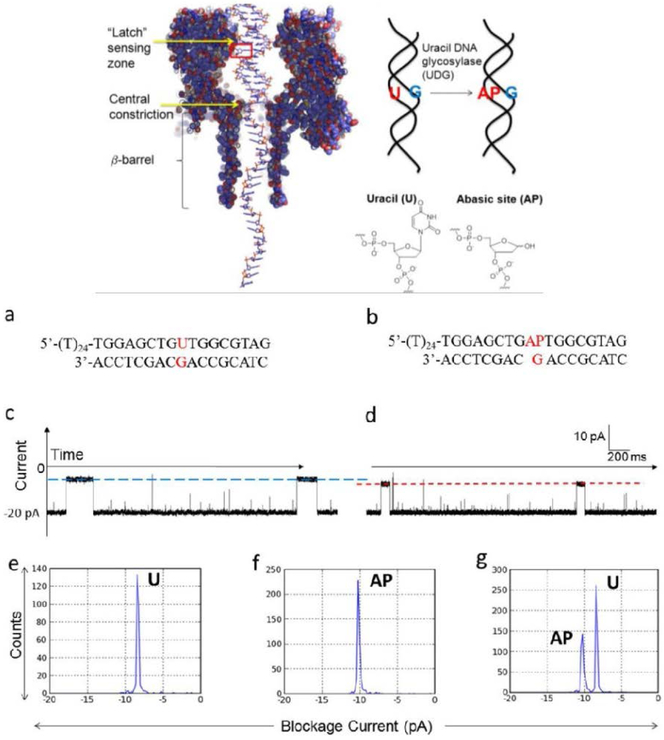
Figure 8. Abasic site detection in the latch zone of the nanopore by monitoring the UDG enzyme activity for dsDNA in the nanopore.
Top scheme: Left: The structure of dsDNA with a 5’- poly(T)24 tail within WT α-HL. The box indicates the location of the uracil (U) base or the abasic site (AP). Right: Scheme of the UDG hydrolysis reaction. The α-HL structure was taken from pdb 7AHL. DNA structure is shown on a 1:1 scale with α-HL. a, Sequence of the starting material formed by a 41-mer U-containing strand hybridized to a 17-mer strand. b, Sequence of the product containing AP. c, d, Sample current-time (i-t) traces for blockages generated by the U duplex (c) or the AP duplex (d) in individual experiments. The blue and red lines indicate the current blockage levels used to determine the duplex identity. e, f, g, Histograms of current blockage levels for the U duplex (e), AP duplex (f) and for a mixture of U and AP duplexes (g, mole ratio ~2:1). Single-nucleotide recognition was achieved between the U-containing duplex (a, c, and e) and the AP-containing duplex (b, d, and f) based on a ~2 pA difference in blockage current levels of the unzipping events in a 14 μM DNA, 150 mM buffered KCl solution at −120 mV. Reprinted with permission from reference [99].
Studies have successfully demonstrated that U and AP sites can be discriminated in the nanopore by monitoring uracil-DNA glycosylase (UDG) enzyme activity. The UDG hydrolysis reaction converts a uracil (U) base to an AP site (Figure 8). When the dsDNA is trapped at the nanocavity, and the U-G or AP-G pairs are placed at the vicinity of the latch zone of the pore, they can generate distinguishable current levels. During the hydrolysis reaction, the two current levels can be observed at the same time due to partial conversion of U to AP (Figure 8g). This study also provides a guideline for developing new approaches for monitoring enzymatic activity on DNA bases [99]. This newly discovered sensing zone at the latch zone suggests the possible development of new approaches to detect site-specific changes in dsDNA structure relevant to genetic, epigenetic and medical diagnostic applications. Protein mutagenesis that could alter the amino acids at the latch region can possibly change the size and dimension of the channel and further change the chemical and physical interactions between the channel wall and DNA. It is therefore potentially useful in the future to enhance detection specificity and sensitivity.
5. Cytosine Modifications Detections
Silver ions specifically interact with C-C mismatches [101–104], while mercury ions specifically interact with T-T mismatches [105–108]. These interactions that strongly stabilize DNA duplexes have been extensively studied recently [109]. Considering that cytosine (C) modifications such as 5-methylcytosine (mC) and 5-hydroxymethylcytosine (hmC) are important epigenetic markers associated with gene expression and tumorigenesis [110–112], we were motivated to explore the interactions of Ag+with a DNA duplex containing a single C-C, C-mC, or C-hmC mismatch in the α-HL nanopore. The α-HL has a nanocavity (2.6nm opening with a 1.4nm constriction site) that can capture and hold the DNA duplex, providing an ideal platform for studying both the cations/C-C interaction and how cytosine modifications change the interaction.
5.1. Direct and label-free discrimination of cytosine and cytosine modifications utilizing silver ions in the nanopore
In previous studies [113–116], researchers have found that C, mC, or hmC can be recognized by immobilizing the DNA with streptavidin [115] or by chemical modifications [113] in α-HL. When in a solid-state nanopore, it was found that DNA duplexes containing mC and hmC could be discriminated [116], and by using methylated CpG binding proteins [43,114] and chemical modifications via sequencing, the hmC, mC, and C bases could be discriminated as well [117–119].
Recently, studies have shown that cytosine modifications can change the current profiles when the modifications containing dsDNA were trapped in the nanocavity. With the help of silver ions, these DNA modifications could modify DNA stability and lead to changes in event durations. Secondly, these modifications also could alter the DNA space resulting in changes of current levels. This was because mC has only an extra methyl group, while hmC has an extra methyl group and a hydroxy group, which blocked more ion current when these modifications were placed at the latch zone of the nanopore [120].
The results were supported by the melting temperature measurement and molecule dynamic simulations. These interactions that strongly stabilize DNA duplexes have been extensively studied recently [109], but the nature of coordination of Ag+ with C-C mismatches is still not clearly understood [104,121–123]. The simulations suggest that the paring, via a hydrogen bond, of a C-C mismatch results in a binding site for cations, such as K+ and Ag+. It is a dynamic coordination between N3A and O2B, or N3B and O2A for C-Ag-C interactions [120,124]. The interactions suggest that the coordination of Ag+ in C-Ag-C complexes may have a different mechanism (Figure 9). This approach might be expanded to investigate other cytosine modifications like 5-formylcytosine (5fC) and other metallo-pair interactions.
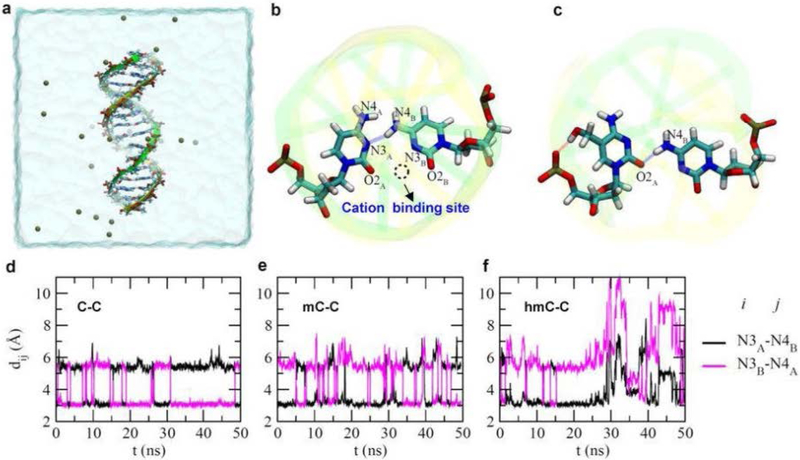
Figure 9. Molecule dynamic simulations revealed a cation binding site in the C-C mismatch and unstable hmc-C pairing. The DNA duplex is in the “stick” presentation, and the two backbones are illustrated as yellow and green belts respectively.
Potassium ions that neutralize the entire simulation system are shown as tan balls. Water in a cubic box (78.5 × 78.5 × 78.5 Å3) is shown transparently. (b) A snap-shot of pairing between two cytosine bases. The dashed circle highlights the binding site for a cation. (c) A snap-shot of hmC-C pairing before the pairing was broken. (d-f) Time-dependent distances between the N3 atom of one base and the N4 atom of the other base, in C-C(d), mC-C(e) and hmC-C(f) mismatches. Reprinted with permission from reference [120].
5.2. Methylation detection by designing a mercury interstrand lock
DNA methylation is one of the most commonly occurring epigenetic events in the human genome. It is a covalent addition of a methyl group to the cytosine ring by DNA methyltransferases [125,126]. Before nanopore detection, a uracil is converted from unmethylated cytosine by bisulfite treatment. The α-HL nanopore studies demonstrated that mercury ions can bind both T-T mismatches and U-T mismatches. The Hg2+ binding produces a reversible interstrand lock, called MercuLock, which can enhance the hybridization strength by two orders of magnitude (evidenced by the prolonged event durations). Such MercuLock cannot be formed in a 5-methylcytosine-thymine mismatch (mC-T). Therefore the nanopore can be used to distinguish single bases between uracil and 5-methylcytosine in a sequence [127].
More importantly, the study demonstrated the methylation analysis of multiple CpGs in a p16 gene CpG island. Different numbers and distributions of methylated cytosines in the segments of p16 gene can be successfully detected (Figure 10). This work provides a powerful biophysical tool to explore metal ion-nucleic acid interactions in living organisms and in humans. For example, whether Hg2+ binds to T-T or a U-T mismatched pair which can compromise the DNA repair process in humans, especially during tumorigenesis, needs further study. This work opens an avenue to the application of metal ion-nucleic acid interactions in rapid detection of single nucleotide alteration in gene sequences, such as pathological point mutations, single nucleotide polymorphism (SNPs), and DNA methylation in variety of disease states including cancer.
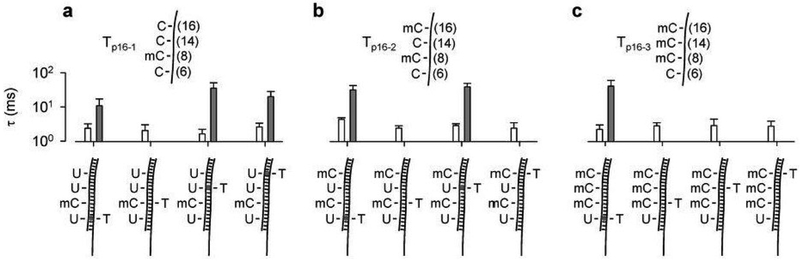
Figure 10. Methylation detection by designing a mercury interstrand lock.
(a), (b) and (c) compared the duration of short and long signature blocks for targets Tp16-1 (a), Tpl6-2 (b) and Tpl6-3 (c) detected by four probes PC6, PC8, PC14 and PC16. The duration of signature blocks allowed determining of the methylation status for each of four CpG cytosines. The DNA sequences of the three p16 fragments containined bases 1, 2 and 3 mC in the DNA strand. C can be converted to U by bisulfite treatment and then form a U-T pair which can be stabilized by a mercury ion evidenced by the prolonged duration (gray bar). However, mC can not be converted, so it forms a mC-T pair which can not be stabilized by a mercury ion, therefore only short blocks were observed (white bar). All traces were recorded at +130 mV in 1 M KCl and 10 mM Tris (pH 7.4). Reprinted with permission from reference [127].
6. Nanopore Devices
The successful development of a nanopore device platform will rapidly advance both basic and clinical research. With minimum sample preparation, an easy learning and handling curve, and real time testing and analyzation of experimental data, the nanopore device can be used by a doctor for early diagnosis, staging and monitoring of cancer and for observing effectiveness of disease and cancer related drug treatments. This is due to the many disease/cancer biomarkers that exist, like circulating nucleic acids (such as microRNA, DNA) [128–130], peptides [131–133], and proteins [134–137], DNA damages and DNA mutations which can all be detected by the nanopore. Lots of effort is being put in to create a portable and robust nanopore device using solid state nanopores [138–141], protein nanopores [142–144], and α-HL nanopores [145–148] for vast breadth of applications. Through the detection and identification of cancer biomarkers, many academic research and clinical application problems can be solved, which would help to advance modern health care and economy.
The Minion (Oxford Nanopore Technologies) is a commercial portable genome sequencing instrument containing nanopores embedded within a synthetic membrane [149]. It has been used to generate a bacterial genome dataset [150], identify the chromosomal insertion site of a composite antibiotic resistance island [151], and assemble a genome [152,153]. This device is capable of sequencing long DNA fragments (>10 kb) without amplification.
7. Conclusion
Biomedical diagnosis for cancer research have attracted great interests for both scientists and physicians, utilizing different kinds of techniques like biophotonics [154], nanoparticles [155], computational intelligence [156], confocal Raman and quantitative phase microscopy [157], nanowire field-effect transistor [158], optical imaging [159], metabolic fingerprinting [160], fluorescence spectroscopy [161] and so on. A steady stream of money flows into endeavors to apply whole genome and sequencing to the development of patient-specific treatment strategies and personalized medicine.
With newly emerged nanopore sequencing technologies, devices like Ion Proton (California-based Life Technologies), HiSeq (San Diego-based Illumina), and Swiss giant Roche, a frontrunner in personalized healthcare, have made huge contributions to the sequencing technology field. Competition between companies has been able to push fast, accurate, and low-cost sequencing into the clinical space and market. Another growing trend in the field of nanopore research is the development of sensors for biodetection of intramolecular and intermolecular interactions as we discussed above. The detection of these targets using the nanopore has advanced considerably over the past ten years [46], and this has led to promising developments of new bioanalytic and diagnostic tools [58]. Unfortunately, the low physiological concentrations of many of these biomarkers [162–165] can dramatically lower the rate at which a target molecule enters the nanopore. This requires the enrichment of the target and limits the practical applications of nanopore-based sensors [165]. All nanopore sensors are facing two common challenges which are low detection efficiency and fast translocation speeds [166].
Enhancing the capture rate and slowing down the translocation speed at the same time are seemingly contradictory actions, but promisingly, the transmembrane salt gradient has proven to be an effective approach for enhancing the capture of double-stranded DNA (dsDNA) and reducing their translocation speed in a synthetic nanopore [167]. Several theoretical studies [168–170] were performed to explore the principles behind this phenomenon, and they suggest that osmotic water flow, electro-osmotic flow (EOF), and electrophoresis may each play their own role. However, these calculations without experimental data were focused on a synthetic pore system, and their results were still being debated. More efforts are urgently needed in this direction of study for developing versatile and accurate nanopore sensors. Another challenge for the application of a nanopore in clinical detections is identifying the target in a mixture environment and discriminating the target from its background molecules. Reliable nanopore signatures are required for this accurate detection. These nanopore signatures have both a specific event duration and a current level change compared to the background signals.
Footnotes
Conflict of Interest
All authors declare no conflict of interest.
References
- 1.Dupont C, Armant DR, Brenner CA (2009) Epigenetics: Definition, mechanisms and clinical perspective. Semin Reprod Med 27: 351–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dawson MA, Kouzarides T (2012) Cancer epigenetics: From mechanism to therapy. Cell 150: 12–27. [DOI] [PubMed] [Google Scholar]
- 3.Esteller M (2008) Molecular origins of cancer: Epigenetics in cancer. New Engl J Med 358: 1148–1159+1096. [DOI] [PubMed] [Google Scholar]
- 4.Laird PW (2005) Cancer epigenetics. Hum Mol Genet 14: R65–R76. [DOI] [PubMed] [Google Scholar]
- 5.Okugawa Y, Grady WM, Goel A (2015) Epigenetic Alterations in Colorectal Cancer: Emerging Biomarkers. Gastroenterology 149: 1204–1225e1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deng D, Liu Z, Du Y (2010) Epigenetic Alterations as Cancer Diagnostic, Prognostic, and Predictive Biomarkers. Adv Genet 71: 126–176. [DOI] [PubMed] [Google Scholar]
- 7.Herceg Z, Hainaut P (2007) Genetic and epigenetic alterations as biomarkers for cancer detection, diagnosis and prognosis. Mol Oncol 1: 26–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dekker C (2007) Solid-state nanopores. Nat Nanotechnol 2: 209–215. [DOI] [PubMed] [Google Scholar]
- 9.Keyser UF (2011) Controlling molecular transport through nanopores. JR Soc Interface 8: 1369–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siwy ZS, Howorka S (2010) Engineered voltage-responsive nanopores. Chem Soc Rev 39: 1115–1132. [DOI] [PubMed] [Google Scholar]
- 11.Garaj S, Hubbard W, Reina A, et al. (2010) Graphene as a subnanometre trans-electrode membrane. Nature 467: 190–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siwy ZS, Davenport M (2010) Nanopores: Graphene opens up to DNA. Nat Nanotechnol 5: 697–698. [DOI] [PubMed] [Google Scholar]
- 13.Tada K, Haruyama J, Yang HX, et al. (2011) Graphene magnet realized by hydrogenated graphene nanopore arrays. ApplPhys Lett 99: 183111. [DOI] [PubMed] [Google Scholar]
- 14.Ding S, Gao C, Gu LQ (2009) Capturing single molecules of immunoglobulin and ricin with an aptamer-encoded glass nanopore. Anal Chem 81: 6649–6655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu LQ, Shim JW (2010) Single molecule sensing by nanopores and nanopore devices. Analyst 135:441–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fertig N, Meyer C, Blick RH, et al. (2001) Microstructured glass chip for ion-channel electrophysiology. Phys Rev E Stat Nonlin Soft Matter Phys 64: 409011–409014. [DOI] [PubMed] [Google Scholar]
- 17.Li J, Stein D, McMullan C, et al. (2001) Ion-beam sculpting at nanometre length scales. Nature 412:166–169. [DOI] [PubMed] [Google Scholar]
- 18.Storm AJ, Chen JH, Ling XS, et al. (2003) Fabrication of solid-state nanopores with single-nanometre precision. Nat Mater 2: 537–540. [DOI] [PubMed] [Google Scholar]
- 19.Kasianowicz JJ, Brandin E, Branton D, et al. (1996) Characterization of individual polynucleotide molecules using a membrane channel. Proc Natl Acad Sci USA 93: 13770–13773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laszlo AH, Derrington IM, Brinkerhoff H, et al. (2013) Detection and mapping of 5- methylcytosine and 5-hydroxymethylcytosine with nanopore MspA. Proc Natl Acad Sci USA 110:18904–18909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laszlo AH, Derrington IM, Ross BC, et al. (2014) Decoding long nanopore sequencing reads of natural DNA. Nat Biotechnol 32: 829–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Montana V, Grubišić V, et al. (2015) Nanopore sensing of botulinum toxin type B by discriminating an enzymatically cleaved peptide from a synaptic protein synaptobrevin 2 derivative. ACS ApplMater Inter 7: 184–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J, Eren E, Vijayaraghavan J, et al. (2012) OccK channels from Pseudomonas aeruginosa exhibit diverse single-channel electrical signatures but conserved anion selectivity. Biochemistry 51: 2319–2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mohammad MM, Iyer R, Howard KR, et al. (2012) Engineering a rigid protein tunnel for biomolecular detection. J Am Chem Soc 134: 9521–9531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheneke BR, van den Berg B, Movileanu L (2015) Quasithermodynamic contributions to the fluctuations of a protein nanopore. ACS Chem Biol 10: 784–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang HY, Li Y, Qin LX, et al. (2013) Single-molecule DNA detection using a novel SP1 protein nanopore. Chem Commun (Camb) 49: 1741–1743. [DOI] [PubMed] [Google Scholar]
- 27.Wang S, Haque F, Rychahou PG, et al. (2013) Engineered nanopore of phi29 dna-packaging motor for real-time detection of single colon cancer specific antibody in serum. ACS Nano 7: 9814–9822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu D, Bi S, Zhang L, et al. (2014) Single-molecule study of proteins by biological nanopore sensors. Sensors (Basel) 14: 18211–18222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Venkatesan BM, Bashir R (2011) Nanopore sensors for nucleic acid analysis. Nat Nanotechnol 6: 615–624. [DOI] [PubMed] [Google Scholar]
- 30.Hall AR, Scott A, Rotem D, et al. (2010) Hybrid pore formation by directed insertion of +Ý-haemolysin into solid-state nanopores. Nat Nanotechnol 5: 874–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bell NA, Engst CR, Ablay M, et al. (2012) DNA origami nanopores. Nano Lett 12: 512–517. [DOI] [PubMed] [Google Scholar]
- 32.Iqbal SM, Akin D, Bashir R (2007) Solid-state nanopore channels with DNA selectivity. Nat Nanotechnol 2: 243–248. [DOI] [PubMed] [Google Scholar]
- 33.Tinazli A, Tang J, Valiokas R, et al. (2005) High-affinity chelator thiols for switchable and oriented immobilization of histidine-tagged proteins: a generic platform for protein chip technologies. Chemistry 11: 5249–5259. [DOI] [PubMed] [Google Scholar]
- 34.Wei R, Pedone D, Zurner A, et al. (2010) Fabrication of metallized nanopores in silicon nitride membranes for single-molecule sensing. Small 6: 1406–1414. [DOI] [PubMed] [Google Scholar]
- 35.Yusko EC, Johnson JM, Majd S, et al. (2011) Controlling protein translocation through nanopores with bio-inspired fluid walls. Nat Nanotechnol 6: 253–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang D, Harrer S, Luan B, et al. (2014) Regulating the transport of DNA through Biofriendly Nanochannels in a thin solid membrane. Sci Rep 4: 3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Branton D, Deamer DW, Marziali A, et al. (2008) The potential and challenges of nanopore sequencing. Nat Nanotechnol 26: 1146–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cherf GM, Lieberman KR, Rashid H, et al. (2012) Automated forward and reverse ratcheting of DNA in a nanopore at 5-A precision. Nat Biotechnol 30: 344–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kasianowicz JJ, Brandin E, Branton D, et al. (1996) Characterization of individual polynucleotide molecules using a membrane channel. Proc Natl Acad Sci US A 93: 13770–13773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manrao EA, Derrington IM, Laszlo AH, et al. (2012) Reading DNA at single-nucleotide resolution with a mutant MspA nanopore and phi29 DNA polymerase. Nat Biotechnol 30: 349–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.An N, Fleming AM, White HS, et al. (2012) Crown ether-electrolyte interactions permit nanopore detection of individual DNA abasic sites in single molecules. Proc Natl Acad Sci U S A 109:11504–11509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wallace EV, Stoddart D, Heron AJ, et al. (2010) Identification of epigenetic DNA modifications with a protein nanopore. Chem Commun(Camb) 46: 8195–8197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shim J, Kim Y, Humphreys GI, et al. (2015) Nanopore-based assay for detection of methylation in double-stranded DNA fragments. ACS Nano 9: 290–300. [DOI] [PubMed] [Google Scholar]
- 44.Bayley H, Cremer PS (2001) Stochastic sensors inspired by biology. Nature 413: 226–230. [DOI] [PubMed] [Google Scholar]
- 45.Gu LQ, Shim JW (2010) Single molecule sensing by nanopores and nanopore devices. Analyst 135:441–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Howorka S, Siwy Z (2009) Nanopore analytics: sensing of single molecules. Chem Soc Rev 38: 2360–2384. [DOI] [PubMed] [Google Scholar]
- 47.Luan B, Peng H, Polonsky S, et al. (2010) Base-by-base ratcheting of single stranded DNA through a solid-state nanopore. Phys Rev Lett 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Asandei A, Chinappi M, Lee JK, et al. (2015) Placement of oppositely charged aminoacids at a polypeptide termini determines the voltage-controlled braking of polymer transport through nanometer-scale pores. Sci Rep 5: 10419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mereuta L, Roy M, Asandei A, et al. (2014) Slowing down single-molecule trafficking through a protein nanopore reveals intermediates for peptide translocation. Sci Rep 4: 3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deamer D (2010) Nanopore analysis of nucleic acids bound to exonucleases and polymerases. Annu RevBiophys 39: 79–90. [DOI] [PubMed] [Google Scholar]
- 51.Olasagasti F, Lieberman KR, Benner S, et al. (2010) Replication of individual DNA molecules under electronic control using a protein nanopore. Nat Nanotechnol 5: 798–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chu J, Gonzalez-Lopez M, Cockroft SL, et al. (2010) Real-time monitoring of DNA polymerase function and stepwise single-nucleotide DNA strand translocation through a protein nanopore. Angew Chem Int Ed Engl 49: 10106–10109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Benner S, Chen RJ, Wilson NA, et al. (2007) Sequence-specific detection of individual DNA polymerase complexes in real time using a nanopore. Nat Nanotechnol 2: 718–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Asandei A, Iftemi S, Mereuta L, et al. (2014) Probing of various physiologically relevant metals: Amyloid-β peptide interactions with a lipid membrane-immobilized protein nanopore. J Membrane Biol 247: 523–530. [DOI] [PubMed] [Google Scholar]
- 55.Asandei A, Schiopu I, Iftemi S, et al. (2013) Investigation of Cu2+ binding to human and rat amyloid fragments Aβ (1–16) with a protein nanopore. Langmuir 29: 15634–15642. [DOI] [PubMed] [Google Scholar]
- 56.Ying YL, Wang HY, Sutherland TC, et al. (2011) Monitoring of an ATP-binding aptamer and its conformational changes using an α-hemolysin nanopore. Small 7: 87–94. [DOI] [PubMed] [Google Scholar]
- 57.Clarke J, Wu HC, Jayasinghe L, et al. (2009) Continuous base identification for single-molecule nanopore DNA sequencing. Nat Nanotechnol 4: 265–270. [DOI] [PubMed] [Google Scholar]
- 58.Branton D, Deamer DW, Marziali A, et al. (2008) The potential and challenges of nanopore sequencing. Nat Biotechnol 26: 1146–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bayley H (2006) Sequencing single molecules of DNA. Curr Opin Chem Biol 10: 628–637. [DOI] [PubMed] [Google Scholar]
- 60.Bartel DP (2004) MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- 61.Carthew RW, Sontheimer EJ (2009) Origins and Mechanisms of miRNAs and siRNAs. Cell 136: 642–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Calin GA, Croce CM (2006) MicroRNA signatures in human cancers. Nat Rev Cancer 6: 857–866. [DOI] [PubMed] [Google Scholar]
- 63.Chen X, Ba Y, Ma L, et al. (2008) Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 18: 997–1006. [DOI] [PubMed] [Google Scholar]
- 64.Mitchell PS, Parkin RK, Kroh EM, et al. (2008) Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA 105: 10513–10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang Y, Zheng D, Tan Q, et al. (2011) Nanopore-based detection of circulating microRNAs in lung cancer patients. Nat Nanotechnol 6: 668–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tian K, He Z, Wang Y, et al. (2013) Designing a polycationic probe for simultaneous enrichment and detection of microRNAs in a nanopore. ACS Nano 7: 3962–3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang X, Wang Y, Fricke BL, et al. (2014) Programming nanopore ion flow for encoded multiplex microRNA detection. ACS Nano 8: 3444–3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cao C, Ying YL, Gu Z, et al. (2014) Enhanced resolution of low molecular weight poly(ethylene glycol) in nanopore analysis. Anal Chem 86: 11946–11950. [DOI] [PubMed] [Google Scholar]
- 69.Baral A, Kumar P, Pathak R, et al. (2013) Emerging trends in G-quadruplex biology-role in epigenetic and evolutionary events. MolBioSyst 9: 1568–1575. [DOI] [PubMed] [Google Scholar]
- 70.Frees S, Menendez C, Crum M, et al. (2014) QGRS-Conserve: A computational method for discovering evolutionarily conserved G-quadruplex motifs. Human Genomics 8: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Paeschke K, Bochman ML, Daniela Garcia P, et al. (2013) Pifl family helicases suppress genome instability at G-quadruplex motifs. Nature 497: 458–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Qiu J, Wang M, Zhang Y, et al. (2015) Biological function and medicinal research significance of g-quadruplex interactive proteins. Curr Top Med Chem 15: 1971–1987. [DOI] [PubMed] [Google Scholar]
- 73.Fleming AM, Burrows CJ (2013) G-quadruplex folds of the human telomere sequence alter the site reactivity and reaction pathway of guanine oxidation compared to duplex DNA. Chem Res Toxicol 26: 593–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.An N, Fleming AM, Middleton EG, et al. (2014) Single-molecule investigation of G-quadruplex folds of the human telomere sequence in a protein nanocavity. Proc Natl Acad Sci USA 111: 14325–14331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.An N, Fleming AM, Burrows CJ (2013) Interactions of the human telomere sequence with the nanocavity of the α-hemolysin ion channel reveal structure-dependent electrical signatures for hybrid folds. J Am Chem Soc 135: 8562–8570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wolna AH, Fleming AM, Burrows CJ (2014) Single-molecule analysis of thymine dimer-containing G-quadruplexes formed from the human telomere sequence. Biochemistry 53: 7484–7493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shim JW, Gu LQ (2008) Encapsulating a single G-quadruplex aptamer in a protein nanocavity. J PhysChemB 112: 8354–8360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shim JW, Tan Q, Gu LQ (2009) Single-molecule detection of folding and unfolding of the G-quadruplex aptamer in a nanopore nanocavity. Nucleic Acids Res 37: 972–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shim J, Gu LQ (2012) Single-molecule investigation of G-quadruplex using a nanopore sensor. Methods 57: 40–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yang J, Xu ZP, Huang Y, et al. (2004) ATM and ATR: Sensing DNA damage. World J Gastroentero 10: 155–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Diculescu VC, Paquim AMC, Brett AMO (2005) Electrochemical DNA sensors for detection of DNA damage. Sensors 5: 377–393. [Google Scholar]
- 82.Fojta M (2002) Electrochemical sensors for DNA interactions and damage. Electroanalysis 14: 1449–1463. [Google Scholar]
- 83.Paleček E Bartošík M (2012) Electrochemistry of nucleic acids. Chem Rev 112: 3427–3481. [DOI] [PubMed] [Google Scholar]
- 84.An N, Fleming AM, White HS, et al. (2015) Nanopore detection of 8-oxoguanine in the human telomere repeat sequence. ACS Nano 9: 4296–4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Boiteux S, Guillet M (2004) Abasic sites in DNA: Repair and biological consequences in Saccharomyces cerevisiae. DNA Repair 3: 1–12. [DOI] [PubMed] [Google Scholar]
- 86.Wilson Hi DM, Barsky D (2001) The major human abasic endonuclease: Formation, consequences and repair of abasic lesions in DNA. Mutat Res-DNA Repair 485: 283–307. [DOI] [PubMed] [Google Scholar]
- 87.Reardon JT, Sancar A (2005) Nucleotide Excision Repair In: Moldave K, editor. Progress in Nucleic Acid Research and Molecular Biology, 183–235. [DOI] [PubMed] [Google Scholar]
- 88.Gates KS (2009) An overview of chemical processes that damage cellular DNA: Spontaneous hydrolysis, alkylation, and reactions with radicals. Chem Res Toxicol 22: 1747–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Greco NJ, Tor Y (2005) Simple fluorescent pyrimidine analogues detect the presence of DNA abasic sites. J Am Chem Soc 127: 10784–10785. [DOI] [PubMed] [Google Scholar]
- 90.Lucotti A, Tommasini M, Pezzoli D, et al. (2014) Molecular interactions of DNA with transfectants: A study based on infrared spectroscopy and quantum chemistry as aids to fluorescence spectroscopy and dynamic light scattering analyses. RSCAdv 4: 49620–49627. [Google Scholar]
- 91.Sun HB, Qian L, Yokota H (2001) Detection of abasic sites on individual DNA molecules using atomic force microscopy. Anal Chem 73: 2229–2232. [DOI] [PubMed] [Google Scholar]
- 92.Bowler FR, Diaz-Mochon JJ, Swift MD, et al. (2010) DNA analysis by dynamic chemistry. Angew Chem Int Edit 49: 1809–1812. [DOI] [PubMed] [Google Scholar]
- 93.Atamna H, Cheung I, Ames BN (2000) A method for detecting abasic sites in living cells: Age-dependent changes in base excision repair. Proc Natl Acad Sci USA 97: 686–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kojima N, Takebayashi T, Mikami A, et al. (2009) Construction of highly reactive probes for abasic site detection by introduction of an aromatic and a guanidine residue into an aminooxy group. J Am Chem Soc 131: 13208–13209. [DOI] [PubMed] [Google Scholar]
- 95.Zeglis BM, Boland JA, Barton JK (2008) Targeting abasic sites and single base bulges in DNA with metalloinsertors. J Am Chem Soc 130: 7530–7531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang Y, Liu L, Wu C, et al. (2009) Direct detection and quantification of abasic sites for in vivo studies of DNA damage and repair. Nucl Med Biol 36: 975–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hada M, Sutherland BM (2006) Spectrum of complex DNA damages depends on the incident radiation. Radiat Res 165: 223–230. [DOI] [PubMed] [Google Scholar]
- 98.An N, Fleming AM, White HS, et al. (2012) Crown ether-electrolyte interactions permit nanopore detection of individual DNA abasic sites in single molecules. Proc Natl Acad Sci USA 109:11504–11509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jin Q, Fleming AM, Johnson RP, et al. (2013) Base-excision repair activity of uracil-DNA glycosylase monitored using the latch zone of α-hemolysin. J Am Chem Soc 135: 19347–19353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Johnson RP, Fleming AM, Jin Q, et al. (2014) Temperature and electrolyte optimization of the α-hemolysin latch sensing zone for detection of base modification in double-stranded DNA. BiophysJ 107: 924–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lin YH, Tseng WL (2009) Highly sensitive and selective detection of silver ions and silver nanoparticles in aqueous solution using an oligonucleotide-based fluorogenic probe. Chem Commun (Camb): 6619–6621. [DOI] [PubMed] [Google Scholar]
- 102.Ono A, Cao S, Togashi H, et al. (2008) Specific interactions between silver(I) ions and cytosine-cytosine pairs in DNA duplexes. Chem Commun (Camb): 4825–4827. [DOI] [PubMed] [Google Scholar]
- 103.Wen Y, Xing F, He S, et al. (2010) A graphene-based fluorescent nanoprobe for silver(I) ions detection by using graphene oxide and a silver-specific oligonucleotide. Chem Commun (Camb) 46:2596–2598. [DOI] [PubMed] [Google Scholar]
- 104.Torigoe H, Okamoto I, Dairaku T, et al. (2012) Thermodynamic and structural properties of the specific binding between Ag(+) ion and C:C mismatched base pair in duplex DNA to form C-Ag-C metal-mediated base pair. Biochimie 94: 2431–2440. [DOI] [PubMed] [Google Scholar]
- 105.Miyake Y, Togashi H, Tashiro M, et al. (2006) MercuryII-mediated formation of thymine-Hgll-thymine base pairs in DNA duplexes. J Am Chem Soc 128: 2172–2173. [DOI] [PubMed] [Google Scholar]
- 106.Tanaka Y, Oda S, Yamaguchi H, et al. (2007) 15N-15N J-coupling across Hg(II): direct observation of Hg(II)-mediated T-T base pairs in a DNA duplex. J Am Chem Soc 129: 244–245. [DOI] [PubMed] [Google Scholar]
- 107.Torigoe H, Ono A, Kozasa T (2010) Hg(II) ion specifically binds with T:T mismatched base pair in duplex DNA. Chemistry 16: 13218–13225. [DOI] [PubMed] [Google Scholar]
- 108.Wen S, Zeng T, Liu L, et al. (2011) Highly sensitive and selective DNA-based detection of mercury(II) with alpha-hemolysin nanopore. J Am Chem Soc 133: 18312–18317. [DOI] [PubMed] [Google Scholar]
- 109.Ono A, Torigoe H, Tanaka Y, et al. (2011) Binding of metal ions by pyrimidine base pairs in DNA duplexes. Chem Soc Rev 40: 5855–5866. [DOI] [PubMed] [Google Scholar]
- 110.Brena RM, Huang TH, Plass C (2006) Quantitative assessment of DNA methylation: Potential applications for disease diagnosis, classification, and prognosis in clinical settings. J Mol Med (Berl) 84: 365–377. [DOI] [PubMed] [Google Scholar]
- 111.Kriaucionis S, Heintz N (2009) The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324: 929–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lister R, Pelizzola M, Dowen RH, et al. (2009) Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462: 315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li WW, Gong L, Bayley H (2013) Single-molecule detection of 5-hydroxymethylcytosine in DNA through chemical modification and nanopore analysis. Angew Chem Int Ed 52: 4350–4355. [DOI] [PubMed] [Google Scholar]
- 114.Shim J, Humphreys GI, Venkatesan BM, et al. (2013) Detection and quantification of methylation in DNA using solid-state nanopores. Sci Rep 3: 1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wallace EV, Stoddart D, Heron AJ, et al. (2010) Identification of epigenetic DNA modifications with a protein nanopore. Chem Commun (Camb) 46: 8195–8197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wanunu M, Cohen-Karni D, Johnson RR, et al. (2011) Discrimination of methylcytosine from hydroxymethylcytosine in DNA molecules. J Am Chem Soc 133: 486–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yu M, Hon GC, Szulwach KE, et al. (2012) Tet-assisted bisulfite sequencing of 5- hydroxymethylcytosine. Nat Protoc 7: 2159–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yu M, Hon GC, Szulwach KE, et al. (2012) Base-resolution analysis of 5- hydroxymethylcytosine in the mammalian genome. Cell 149: 1368–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Booth MJ, Branco MR, Ficz G, et al. (2012) Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science 336: 934–937. [DOI] [PubMed] [Google Scholar]
- 120.Wang Y, Luan BQ, Yang Z, et al. (2014) Single Molecule Investigation of Ag+ Interactions with Single Cytosine-, Methylcytosine-and Hydroxymethylcytosine-Cytosine Mismatches in a Nanopore. Sci Rep 4: 5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Torigoe H, Miyakawa Y, Ono A, et al. (2011) Thermodynamic properties of the specific binding between Ag+ ions and C:C mismatched base pairs in duplex DNA. Nucleos Nucleot Nucl 30: 149–167. [DOI] [PubMed] [Google Scholar]
- 122.Kistenmacher TJ, Rossi M, Marzilli LG (1979) Crystal and molecular structure of (nitrato)(1- methylcytosine)silver(I): An unusual cross-linked polymer containing a heavy metal and a modified nucleic acid constituent. Inorg Chem 18: 240–244. [Google Scholar]
- 123.Urata H, Yamaguchi E, Nakamura Y, et al. (2011) Pyrimidine-pyrimidine base pairs stabilized by silver(I) ions. Chem Commun (Camb) 47: 941–943. [DOI] [PubMed] [Google Scholar]
- 124.Wang Y, Ritzo B, Gu L-Q (2015) Silver (i) ions modulate the stability of DNA duplexes containing cytosine, methylcytosine and hydroxymethylcytosine at different salt concentrations. RSC Adv 5: 2655–2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Egger G, Liang G, Aparicio A, et al. (2004) Epigenetics in human disease and prospects for epigenetic therapy. Nature 429: 457–463. [DOI] [PubMed] [Google Scholar]
- 126.Bestor TH (2000) The DNA methyltransferases of mammals. Hum Mol Genet 9: 2395–2402. [DOI] [PubMed] [Google Scholar]
- 127.Kang I, Wang Y, Reagan C, et al. (2013) Designing DNA interstrand lock for locus-specific methylation detection in a nanopore. Sci Rep 3: 2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Calin GA, Croce CM (2006) MicroRNA signatures in human cancers. Nat Rev Cancer 6: 857–866. [DOI] [PubMed] [Google Scholar]
- 129.Vlassov VV, Laktionov PP, Rykova EY (2010) Circulating nucleic acids as a potential source for cancer biomarkers. Curr Mol Med 10: 142–165. [DOI] [PubMed] [Google Scholar]
- 130.Mitchell PS, Parkin RK, Kroh EM, et al. (2008) Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 105: 10513–10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Oremek GM, Sapoutzis N (2003) Pro-gastrin-releasing peptide (Pro-GRP), a tumor marker for small cell lung cancer. Anticancer Res 23: 895–898. [PubMed] [Google Scholar]
- 132.Molina R, Auge JM, Bosch X, et al. (2009) Usefulness of serum tumor markers, including progastrin-releasing peptide, in patients with lung cancer: correlation with histology. TumourBiol 30: 121–129. [DOI] [PubMed] [Google Scholar]
- 133.Albrethsen J, Moller CH, Olsen J, et al. (2006) Human neutrophil peptides 1, 2 and 3 are biochemical markers for metastatic colorectal cancer. Eur J Cancer 42: 3057–3064. [DOI] [PubMed] [Google Scholar]
- 134.Bassani-Sternberg M, Barnea E, Beer I, et al. (2010) Soluble plasma HLA peptidome as a potential source for cancer biomarkers. Proc Natl Acad Sci USA 107: 18769–18776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Stella Tsai CS, Chen HC, Tung IN, et al. (2010) Serum cellular apoptosis susceptibility protein is a potential prognostic marker for metastatic colorectal cancer. Am J Pathol 176: 1619–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Stoeva SI, Lee JS, Smith JE, et al. (2006) Multiplexed detection of protein cancer markers with biobarcoded nanoparticle probes. J Am Chem Soc 128: 8378–8379. [DOI] [PubMed] [Google Scholar]
- 137.Zheng W, He L (2010) Multiplexed detection of protein cancer markers on Au/Ag-barcoded nanorods using fluorescent-conjugated polymers. AnalBioanal Chem 397: 2261–2270. [DOI] [PubMed] [Google Scholar]
- 138.Jin H, Liu GL (2012) Fabrication and optical characterization of light trapping silicon nanopore and nanoscrew devices.Nanotechnology 23. [DOI] [PubMed] [Google Scholar]
- 139.Merchant CA, Drndić M (2012) Graphene nanopore devices for DNA sensing In: Gracheva ME, editor. Methods in Molecular Biology, 211–226. [DOI] [PubMed] [Google Scholar]
- 140.Park S, Lim J, Pak YE, et al. (2013) A solid state nanopore device for investigating the magnetic properties of magnetic nanoparticles. Sensors (Basel, Switzerland) 13: 6900–6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wu GS, Zhang Y, Si W, et al. (2014) Integrated solid-state nanopore devices for third generation DNA sequencing. Sci China Technol Sc 57: 1925–1935. [Google Scholar]
- 142.Inoue K, Kawano R, Yasuga H, et al. Logic operation in DNA nano device: Electrical input/output through biological nanopore; 2013; Freiburg: Chemical and Biological Microsystems Society. 1881–1883. [Google Scholar]
- 143.Lim MC, Lee MH, Kim KB, et al. (2015) A mask-free passivation process for low noise nanopore devices. JNanosci Nanotechno 15: 5971–5977. [DOI] [PubMed] [Google Scholar]
- 144.Mulero R, Prabhu AS, Freedman KJ, et al. (2010) Nanopore-Based Devices for Bioanalytical Applications. J Assoc Lab Automat 15: 243–252. [Google Scholar]
- 145.Cockroft SL, Chu J, Amorin M, et al. (2008) A single-molecule nanopore device detects DNA polymerase activity with single-nucleotide resolution. J Am Chem Soc 130: 818–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kawano R, Osaki T, Sasaki H, et al. (2010) A polymer-based nanopore-integrated microfluidic device for generating stable bilayer lipid membranes. Small 6: 2100–2104. [DOI] [PubMed] [Google Scholar]
- 147.Shim JW, Gu LQ (2007) Stochastic sensing on a modular chip containing a single-ion channel. Anal Chem 79: 2207–2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kang XF, Cheley S, Rice-Ficht AC, et al. (2007) A storable encapsulated bilayer chip containing a single protein nanopore. J Am Chem Soc 129: 4701–4705. [DOI] [PubMed] [Google Scholar]
- 149.Eisenstein M (2012) Oxford Nanopore announcement sets sequencing sector abuzz. Nat Biotechnol 30: 295–296. [DOI] [PubMed] [Google Scholar]
- 150.Quick J, Quinlan AR, Loman NJ (2014) A reference bacterial genome dataset generated on the MinlON™ portable single-molecule nanopore sequencer. GigaScience 3: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ashton PM, Nair S, Dallman T, et al. (2015) MinlON nanopore sequencing identifies the position and structure of a bacterial antibiotic resistance island. Nat Biotechnol 33: 296–302. [DOI] [PubMed] [Google Scholar]
- 152.Madoui MA, Engelen S, Cruaud C, et al. (2015) Genome assembly using Nanopore-guided long and error-free DNA reads. BMC Genomics 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Loman NJ, Quick J, Simpson JT (2015) A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat Methods 12: 733–735. [DOI] [PubMed] [Google Scholar]
- 154.Palshikar P, Sharma A, Chauhan CS, et al. (2013) Biophotonics: A novel approach biomedical diagnosis. Int JPharm Sci Rev Res 21: 350–354. [Google Scholar]
- 155.Liu A, Ye B (2013) Application of gold nanoparticles in biomedical researches and diagnosis. Clin Lab 59: 23–36. [PubMed] [Google Scholar]
- 156.Nahar J, Imam T, Tickle KS, et al. (2012) Computational intelligence for microarray data and biomedical image analysis for the early diagnosis of breast cancer. Expert Syst Appl 39: 12371–12377. [Google Scholar]
- 157.Kang JW, Lue N, Kong CR, et al. (2011) Combined confocal Raman and quantitative phase microscopy system for biomedical diagnosis. Biomed Opt Express 2: 2484–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Chen KI, Li BR, Chen YT (2011) Silicon nanowire field-effect transistor-based biosensors for biomedical diagnosis and cellular recording investigation. Nano Today 6: 131–154. [Google Scholar]
- 159.Balas C (2009) Review of biomedical optical imaging - A powerful, non-invasive, non-ionizing technology for improving in vivo diagnosis. Meas Sci Technol 20: 104020. [Google Scholar]
- 160.Ellis DI, Goodacre R (2006) Metabolic fingerprinting in disease diagnosis: Biomedical applications of infrared and Raman spectroscopy. Analyst 131: 875–885. [DOI] [PubMed] [Google Scholar]
- 161.Bottiroli G, Croce AC (2004) Autofluorescence spectroscopy of cells and tissues as a tool for biomedical diagnosis. Photoch Photobio Sci 3: 189–210. [PubMed] [Google Scholar]
- 162.Urbanova M, Plzak J, Strnad H, et al. (2010) Circulating nucleic acids as a new diagnostic tool. Cell Mol Biol Lett 15: 242–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Liu KJ, Brock MV, Shih Ie M, et al. (2010) Decoding circulating nucleic acids in human serum using microfluidic single molecule spectroscopy. J Am Chem Soc 132: 5793–5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Thompson L, Turko I, Murad F (2006) Mass spectrometry-based relative quantification of human neutrophil peptides 1, 2, and 3 from biological samples. Mol Immunol 43: 1485–1489. [DOI] [PubMed] [Google Scholar]
- 165.Wanunu M, Dadosh T, Ray V, et al. (2010) Rapid electronic detection of probe-specific microRNAs using thin nanopore sensors. Nat Nanotechnol 5: 807–814. [DOI] [PubMed] [Google Scholar]
- 166.Squires A, Meller A (2013) DNA Capture and Translocation through Nanoscale Pores-a Fine Balance of Electrophoresis and Electroosmosis. Biophys J 105: 543–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wanunu M, Morrison W, Rabin Y, et al. (2010) Electrostatic focusing of unlabelled DNA into nanoscale pores using a salt gradient. Nat Nanotechnol 5: 160–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hatlo MM, Panja D, van Roij R (2011) Translocation of DNA molecules through nanopores with salt gradients: the role of osmotic flow. Phys Rev Lett 107: 068101. [DOI] [PubMed] [Google Scholar]
- 169.He Y, Tsutsui M, Scheicher RH, et al. (2013) Mechanism of How Salt-Gradient-Induced Charges Affect the Translocation of DNA Molecules through a Nanopore. Biophys J 105: 776–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Chou T (2009) Enhancement of charged macromolecule capture by nanopores in a salt gradient. J Chem Phys 131: 034703. [DOI] [PMC free article] [PubMed] [Google Scholar]