Abstract
A rapidly developing paradigm for modern health care is a proactive and individualized response to patients’ symptoms, combining precision diagnosis and personalized treatment. Precision medicine is becoming an overarching medical discipline that will require a better understanding of biomarkers, phenotypes, endotypes, genotypes, regiotypes, and theratypes of diseases. The 100-year-old personalized allergen-specific management of allergic diseases has particularly contributed to early awareness in precision medicine. Polyomics, big data, and systems biology have demonstrated a profound complexity and dynamic variability in allergic disease between individuals, as well as between regions. Escalating health care costs together with questionable efficacy of the current management of allergic diseases facilitated the emergence of the endotype-driven approach. We describe here a precision medicine approach that stratifies patients based on disease mechanisms to optimize management of allergic diseases.
Introduction and general concepts
Allergic diseases remain a worldwide problem, with almost one billion cases, causing significant morbidity and mortality and accounting for a considerable portion of health care expenditure. The heterogeneous, dynamic combination of dysregulated immune response, chronic inflammation, tissue remodeling, and hyperresponsiveness in affected tissues defines the complexity of asthma, anaphylaxis, food allergy, allergic rhinitis, chronic rhinosinusitis (CRS), and atopic dermatitis (AD).
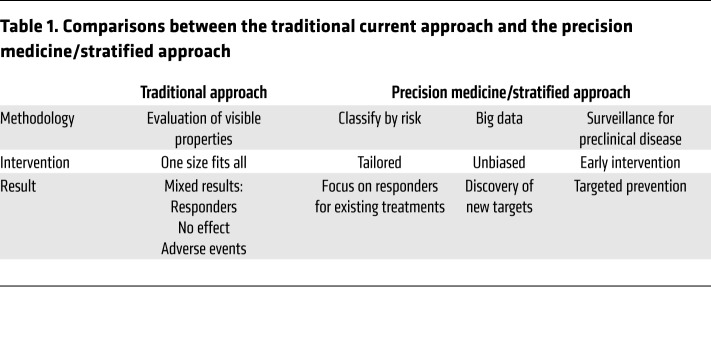
At present, management guidelines for allergic diseases are based on evaluation of symptoms, target organ function, exacerbations, need for rescue medication, and limitation of quality of life, all the while reinforcing the importance of achieving disease control and reducing future risk (1). Nonetheless, the treatments available today merely offer long-term relief of symptoms and do not cure the disease, making it increasingly clear that new approaches for the management of allergic diseases are required. Accordingly, the current understanding of precision medicine has developed and is continuously improving (Table 1).
Table 1. Comparisons between the traditional current approach and the precision medicine/stratified approach.
Disease phenotypes cluster together relevant visible properties such as age at onset, triggers, comorbidities, physiological traits, remodeling, inflammation type (eosinophilic and non-eosinophilic), and treatment response (2, 3). Asthma phenotypes defined based on the predominant inflammatory cell identified in induced sputum (eosinophilic, neutrophilic, mixed granulocytic, and paucigranulocytic phenotypes) or in blood (eosinophilic asthma) are currently being used for a stratified approach to severe asthma (4, 5). Phenotypes do not necessarily relate to or give insights into the underlying pathogenetic mechanism (2, 3). In addition, they frequently overlap and are subject to change over time (2, 6). Thus, defining disease endotypes based on key pathogenetic mechanisms has become a rational development (2, 7–9). The development of precision medicine brought together its own taxonomy, summarized in Table 2.
Table 2. Nomenclature and key terms for the precision medicine/stratified approach.
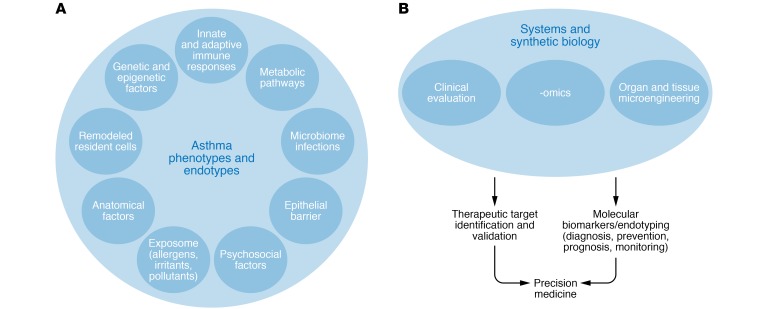
The difficulty of the endotype-driven approach derives from the fact that even though endotypes linked to a single molecular mechanism can be defined, most endotypes share etiological and pathogenic pathways with nonlinear dynamic interactions that may or may not be present in all patients, or in each patient at all time points (Figure 1). Thus, the concept of dynamic complex endotypes (e.g., the complex type 2 endotype), which consist of several subendotypes longitudinally, exists for allergic diseases (10–12). A complex type 2 endotype involves Th2 cells, type 2 innate lymphoid cells (ILC2s), type 2 subsets of B cells, type 2 subset of NKT cells, eosinophils, mast cells, basophils, and their cytokines (e.g., IL-4, IL-5, IL-13); IgE-isotype antibodies; surface molecules such as CRTH2; and soluble mediators such as histamine. To confirm the validity of an endotype, longitudinal follow-up studies of clinical and molecular profiles need to be performed.
Figure 1. Factors influencing disease endotypes and precision medicine.
(A) A multitude of factors can induce or suppress certain genes or pathways and may play a role in the development of certain phenotypes and endotypes as well as control of asthma. (B) Methodologies addressing the dynamic and complex interaction between risk factors, disease phenotype and endotypes, and expression modulators in allergic diseases in the context of precision medicine.
Biological markers (biomarkers) represent measurable indicators linking an endotype with a phenotype (10, 13). At present, biomarkers have been advanced that predict response to treatment in the endotype-driven approach in asthma, CRS, and AD. Regrettably, current biomarkers are not precise in selecting the specific endotype that will respond to a targeted treatment. A good example is the observation that blood eosinophilia predicts therapeutic response to all currently available or future targeted interventions in severe asthma (i.e., anti–IL-5, anti–IL-4/IL-13, and anti-IgE targeted treatment, CRTH2 antagonists) (13, 14).
The endotype-driven approach offers a way to better diagnose, monitor, and stratify patients (Figure 2). Biomarkers and combinations of biomarkers help tailor the management of allergic diseases. Presently, the most salient obstacles to ubiquitous biomarker usage are its feasibility and the cost of measuring samples. Work is under way to create rapid point-of-care tests that are both user-friendly and low-cost. The advent of these new methods combined with insights into biomarker combination strategies will likely yield robust information that will improve diagnosis and management of allergic diseases.
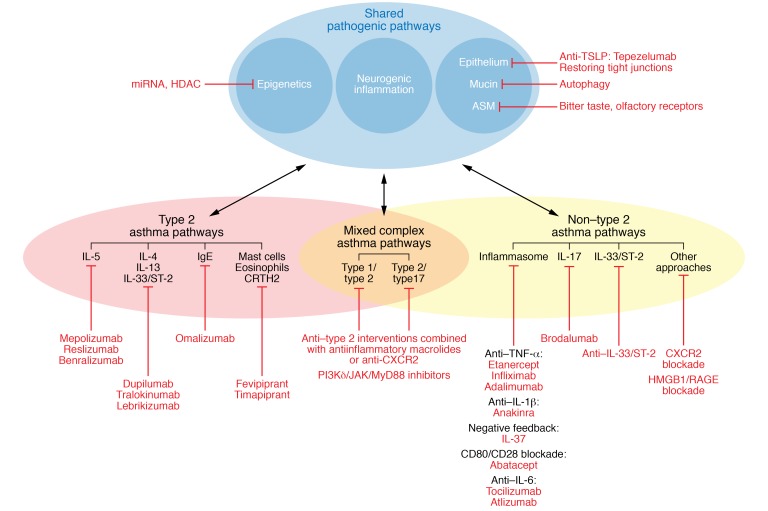
Figure 2. The dynamic and complex interaction between risk factors, disease phenotypes and endotypes, and expression modulators in allergic diseases in the context of precision medicine.
In type 2 asthma, four main pathways can be targeted: the IgE pathway (omalizumab), the IL-5 pathway (mepolizumab, reslizumab, benralizumab), the IL-4/IL-13 pathway (dupilumab, tralokinumab, lebrikizumab), and the CRTH2 receptor (fevipiprant and timapiprant). In non–type 2 asthma, three main pathways can be targeted: the inflammasome pathway (anti–TNF-α interventions such as etanercept, infliximab, and adalimumab; anti–IL-1β interventions such as anakinra; anti–IL-6 targeting with tocilizumab or atlizumab; negative feedback with IL-37; or CD80-CD28 blockade with abatacept), the IL-17 pathway (brodalumab), or the anti–IL-33 pathway, with CXCR2 and HMGB1/RAGE blockade as additional approaches. For the mixed complex endotypes, a combination of anti–type 2 interventions with antiinflammatory macrolides or anti-CXCR2 can be indicated. As an alternative, PI3Kδ/JAK/MyD88 inhibitors might block the upstream effects of type 1, type 2, and type 17 cytokines.
Physiopathology of allergic diseases and endotypes
MicroRNA- and gene signature–based endotypes.
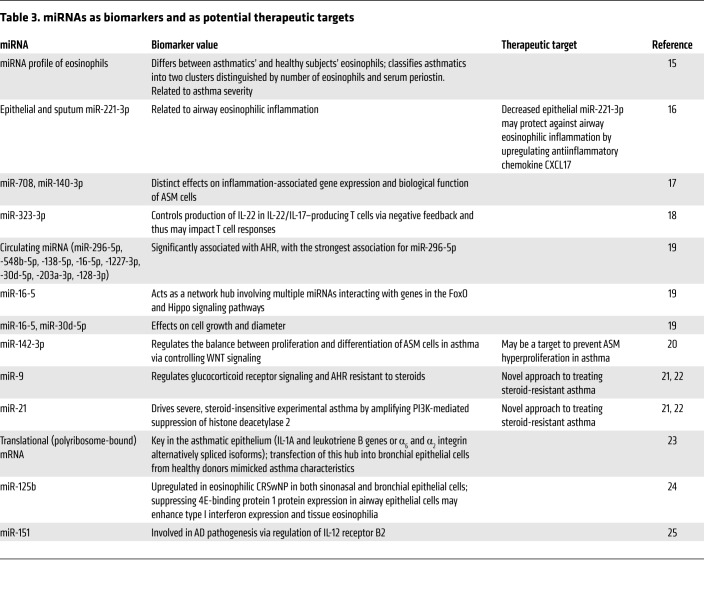
There is extensive posttranscriptional gene dysregulation in allergic diseases, in which microRNAs (miRNAs) play a central role (Table 3). High-throughput profiling studies of well-characterized patients, including analyses of gene expression (e.g., microarrays and RNA sequencing transcriptomics), can help to identify combined signatures for type 2 and non–type 2 endotypes. They are enriching the available data for the identification of new potential targets. As critical components in the regulation of tissue homeostasis, miRNAs represent a very attractive field of precision medicine research in asthma and allergic diseases, as they have been linked to many biological mechanisms underlying Th2 cell and macrophage polarization, regulation of ILC2 biology, steroid-resistant asthma phenotype, airway smooth muscle dysfunction, and impaired antiviral innate immunity (Table 3 and refs. 15–25). Thus, miRNAs represent a modifiable downstream target that will expand the precision medicine approach beyond the type 2/non–type 2 paradigm.
Table 3. miRNAs as biomarkers and as potential therapeutic targets.
In one notable example of this approach, the use of gene expression microarrays on bronchial epithelial cells obtained from patients with asthma led to the identification of periostin, an IL-13–responsive biomarker (26–30). However, expression signatures are not highly specific for type 2 asthma: a recent study showed type 2 airway gene expression in a subset of patients with chronic obstructive pulmonary disease (COPD) (31).
Recently reported gene expression profiles support the concept of complex endotypes. Specific gene expression profiles from induced sputum, nasal, and endobronchial brushings show severe, adult-onset asthma gene signatures associated both with eosinophilic airway inflammation and mast cells and with type 3 ILCs (ILC3s), while childhood-onset asthma has signatures associated with induced lung injury (32). In patients with recurrent wheeze, orosomucoid 1–like protein 3 (ORMDL3) gene expression shows a linear correlation with the stimulator of interferon gene (STING), which controls TLR-independent cytosolic responses to viruses. STING promotes the transcriptional activity of ORMDL3 through the TANK-binding kinase 1–interferon regulatory factor 3–signal transducer and activator of transcription 6 (TBK1-IRF3-STAT6) complex (33). In CRS with nasal polyps (CRSwNP), polyp tissue gene expression analyzed by hierarchical clustering and principal component analysis showed the presence of a complex endotype for both eosinophilic and non-eosinophilic phenotypes. Type 2 cytokines (IL-4, IL-5, CSF2) and cell cycle regulators (cyclin-dependent kinase inhibitor 1A and cyclin D1) were identified in association with tissue eosinophilia, while mainly interferons and acute inflammatory cytokines (IL-1 and IL-6) were demonstrated as upstream regulators in the non-eosinophilic phenotype (34).
NSAID-exacerbated allergic disease.
NSAID-exacerbated respiratory disease (N-ERD) is a chronic eosinophilic, inflammatory disorder occurring in patients with asthma and/or CRSwNP (35). It affects 1 in 10 patients with asthma and CRSwNP. Newly described subphenotypes and emerging subendotypes of N-ERD are potentially relevant to the development of new treatments, such as biologics. Novel mechanisms that can be subendotypes are continually being identified (36, 37).
Regiotypes: a new concept in precision medicine.
The triggers of allergy and disease appearance vary widely between regions of the world. The predominant type of allergens is determined by a region’s source of allergens — for example, a high prevalence of allergy to birch and birch pollen in Nordic countries, ragweed in North America and Eastern Europe, and olive and cypress in the Mediterranean, Middle East, southern regions of North America, and Japan (38–40). A good example of heterogeneous disease appearance is in CRSwNP: in Chinese populations, polyps appear with a tendency toward neutrophil predominance, whereas polyps in Western populations are mostly eosinophilic (41–43).
Biomarkers are part of the regiotype concept that we are proposing. For example, serum periostin did not discriminate between asthmatics and nonasthmatics in a Chinese population, while in Japan and in white populations it is considered a good biomarker for type 2 asthma. Serum periostin levels were higher in the Chinese than in the Caucasian nonasthma group, and were also sex-dependent in the Chinese participants. This suggests that ethnicity should be considered in the interpretation of periostin levels in asthma patients, and sex is an additional consideration in Chinese patients (44). In addition, filaggrin mutations seem to play a less pathogenic role in patients of African origin than in individuals of European or Asian ancestry (45). In inner-city African American children, CXCL1, IL-5, IL-8, and IL-17A were positively associated with difficult-to-control asthma, while IL-4 and IL-13 were positively associated with easy-to-control asthma (46).
Complex molecular pathobiology defines new endotypes and subendotypes
At present, two main subtypes of immune responses driving the allergic disease endotypes have been defined: type 2–high and type 2–low (refs. 10, 11, 26, 47, and Figure 2). Several biomarkers were linked to the type 2–high endotype: total and specific serum IgE, serum periostin and dipeptidyl peptidase 4 (DPP4), blood eosinophils, and, for asthma, sputum eosinophils and exhaled nitric oxide. Most of the new biologic drugs target the type 2–high pathway: IgE, IL-5, IL-4/IL-13, chemoattractant receptor–homologous molecule expressed on Th2 cells (CRTH2), thymic stromal lymphopoietin (TSLP).
Mechanisms of the type 2–high endotype.
Eosinophils have been associated with asthma, CRSwNP, and AD in many studies. Anti–IL-5 interventions have been shown to have benefit in all these diseases, suggesting that eosinophils, which are activated by IL-5, play a pathophysiological role. Mast cells and basophils are also present. Anti-IgE antibody interventions have shown efficacy in asthma and some efficacy in CRS, suggesting that mast cells and/or basophils are involved as well. Eosinophils, mast cells, and basophils express sialoglycan-binding proteins (Siglecs) such as Siglec-8 and Siglec-9, which inhibit cell function and survival. Targeting Siglec-8 or Siglec-9 with antibodies causes eosinophil and neutrophil death, especially in cytokine-primed cells. Siglec-8 targeting in mast cells does not induce cell death, but inhibits the activation of high-affinity IgE receptors (FcεRI), triggering the release of mediators such as histamine and prostaglandin D2. Siglec-9 has also been shown to bind broadly to many different TLR agonists (48, 49). In the lung, mucin 5B (MUC5B) and MUC4 are carriers of sialylated glycan ligands for Siglec-F, and MUC1 is the ligand for Siglec-9 (50). Purified mucin preparations carrying sialylated and sulfated glycans have been shown to bind to mouse eosinophils and induce their death. Mice deficient in MUC5b displayed exaggerated eosinophilic inflammation in response to intratracheal instillation of IL-13. These data define an innate antiinflammatory property of airway mucins by which their glycan composition can control lung eosinophilia through engagement of Siglec-F. Evaluation of turbinate tissue sections from patients with CRS revealed a marked increase in the level of staining of both Siglec-8 and Siglec-9 ligands, suggesting that inflammatory cells or mediators involved in the disease activate the submucosal gland cells to increase ligand expression (51). In type 2 nasal polyps, MUC5AC, MUC5B, and IL-4 receptor-α are overexpressed, supporting the role of IL-4 and IL-13 in upregulating mucin expression (42). In the future, the endotype-driven approach should be expanded beyond the type 2/non–type 2 paradigm, and interventions should target specific molecular pathways such as IL-5–, IL-4/IL-13–, or IgE-driven inflammation as subendotypes of the complex type 2 endotype.
Steroid-unresponsive subtypes.
Over time, chronic inflamed airways can lose tolerance to immunogenic entities that are released following frequent eosinophil degranulation. A recent study reported a “polyclonal” autoimmune event in the airways of prednisone-dependent asthmatic patients with increased eosinophil activity, recurrent pulmonary infections, or both, based on evidence of sputum anti–eosinophil peroxidase and anti-nuclear antibodies of the IgG subtype. In addition to displaying a type 2 microenvironment, extensive profiling of sputum cytokines (including B cell–activating factor and B cell–attracting chemokine 1) indicated the formation of ectopic lymphoid structures. Immunoprecipitated sputum immunoglobulins from patients with autoantibodies triggered eosinophil degranulation, with release of extensive histone-rich extracellular traps, an event unsuppressed by dexamethasone and possibly contributing to the patients’ steroid-unresponsiveness (52).
Impairment of epithelial barrier: the leaky barrier endotype.
Impaired tight junction (TJ) structures are observed in patients with asthma, AD, and CRS. Protease-containing allergens, environmental pollutants, viruses, and type 2 cytokines enhance paracellular transport through disruption of epithelial barrier function (53, 54). The ability of protease allergens to cleave TJs directly and to affect their function via cytokine expression provides a powerful mechanism to compromise epithelial defense: IL-4 and IL-13, released by Th2 cells and ILC2s, open the TJ barrier to cause a chronic epithelial barrier leakiness (55–57). Lung-specific claudin-18 is downregulated by IL-13, whose release is augmented by peptidase allergens (58). These mechanisms are accompanied by protease-driven inflammatory events with intracellular generation of reactive signaling molecules and TLR4 activation. An analogous mechanism operates in skin, wherein house dust mite (HDM) protease allergens cause the epidermis to become leaky, then impede its restitution, while at the same time promoting cytokine and chemokine production (59). Downregulation of phospholipase C-δ1 (PLC-δ1), which is normally abundantly expressed in the epidermis, impairs the barrier functions of the stratum corneum and the localization of TJ proteins (60). Physiological levels of the cytokine oncostatin M impair barrier function in differentiated airway epithelium. Oncostatin M levels are increased in nasal polyps of CRS patients, in bronchoalveolar lavage fluid after segmental allergen challenge in allergic asthmatic patients, and in sputum (61).
Immunometabolic changes associated with type 2 endotypes.
Changes in metabolic functions of various cells have an important impact in asthma development in models. Models of abnormal NADPH oxidase 2 (Nox2) function suffer from hyperinflammatory responses in lung as well as bacterial and fungal infection. Nox2 deficiency (Nox2-null mice) results in exaggerated experimental asthma, which is caused by enhanced Th2 effector function in a T cell–intrinsic manner (62).
Variations in ILC2s underlie allergic subtypes.
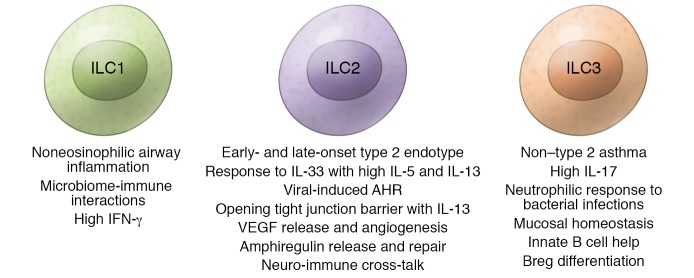
ILC2s play a role in both early- and late-onset type 2 endotypes (63–66). They also play a role in viral-induced airway hyperresponsiveness (AHR). ILC2s are induced by IL-33, which is implicated in the pathophysiology of asthma and allergic diseases (67). Influenza A virus can rapidly release IL-33 from alveolar macrophages and NKT cells, activating ILC2s and causing subsequent production of the type 2 cytokines IL-13 and IL-5 (68). However, through amphiregulin, ILC2s also promote repair of wounded lung tissue after viral infection (69). ILC2s were shown to produce VEGFA, which drives AHR and facilitates autocrine regulation of IL-13 (70). Further, ILC2s support neuro-immune crosstalk: murine ILC2s express NMUR1, the receptor for the neuropeptide NMU, and signaling by NMU through this receptor amplifies ILC2-mediated immune responses (71). Sex hormones also play a role in controlling ILC2 numbers. Female asthma patients have higher levels of ILC2s in their peripheral blood than male patients. Female mice also have more lung ILC2s than males, an apparent result of testosterone metabolite–mediated inhibition of ILC2 numbers and activation (72, 73). ILC3s producing IL-17A might be important drivers in non–type 2 asthma, as IL-17A and Th17 cells play a role in neutrophilic inflammation, and glucocorticoid resistance has been associated with Th17-driven inflammation (74). HDM-induced airway inflammation, remodeling, and Th2/Th17-type cell accumulation involve STAT3 activation that can be prevented by specific inhibition of STAT3 (75). They are also involved in maintenance of mucosal homeostasis: human CD40L+ ILC3s provide innate B cell help and are involved in an innate immunoregulatory mechanism through induction of regulatory B cell differentiation (ref. 76 and Figure 3).
Figure 3. The role of innate lymphoid cells in allergic diseases.
Type 1, 2, and 3 ILCs play different roles in different endotypes of asthma. The findings mostly come from mouse models and detection of cells in body fluids.
Additional subtypes involving dysregulation of cytokines.
TSLP, IL-25, and IL-33 are implicated in the dysregulation of Th2 immune responses in severe allergic asthma (77, 78). TSLP, a cytokine secreted by epithelial and innate immune cells, stimulates type 2 cell responses. Dual stimulation of Th2 cells by TSLP and IL-4 produces higher amounts of IL-5 and IL-13 and other proinflammatory cytokines compared with stimulation with IL-4 alone. Children with asthma showed enhancement of TSLP-driven T cell responses in peripheral blood. These data support sequential cytokine activation for pathogenic Th2 cell differentiation and provide a mechanistic basis for the therapeutic targeting of TSLP signaling in human allergic diseases (79). Aryl hydrocarbon receptor (AhR) activation aggravates allergic inflammation, and AhR activation by diesel exhaust particles mediates upregulation of IL-33, IL-25, and TSLP with Th2 activation, potentially linking environmental pollution and allergic severe asthma (80).
IL-37b, a member of the IL-1 regulatory cytokine family, can suppress innate immunity and inflammatory activity. IL-37–mediated regulation of intracellular inflammation mechanisms links bacterial infection with the activation of human eosinophils. In a coculture of human eosinophils and bronchial epithelial cells, IL-37b showed remarkable suppression of TNF-α, IL-1β, IL-6, CCL2, and CXCL8 production that was evoked by stimulation with the bacterial TLR2 ligand peptidoglycan. In contrast, IL-37b blocked the activation of intracellular NF-κB, the PI3K/Akt pathway, and ERKs 1 and 2 and suppressed gene transcription of allergic inflammation–related PYCARD, S100A9, and CAMP (81).
Bitter taste receptors in allergy and precision medicine.
Following chronic inflammation in asthma, airway structural changes occur, including subepithelial fibrosis, increased airway smooth muscle (ASM) mass, gland enlargement, neovascularization, and epithelial alterations. Increased ASM mass is a hallmark feature of remodeling in asthma. Bitter taste receptors (T2Rs) are expressed on human ASM cells. Six subtypes (T2R10, 14, 31, 5, 4, and 19) were found at levels greater than β2-adrenergic receptor (β2AR) (82). Unlike β2AR-mediated bronchodilation, T2R function is not impaired in asthma and shows little tachyphylaxis. When stimulated by the bacterial acyl-homoserine lactones, T2Rs on ASM may act to open airways and, together with T2Rs of cilia, maintain patency and promote clearance of pathogenic bacteria and debris in the lung during infection. T2R agonists (e.g., chloroquine and quinine) promote ASM relaxation and bronchodilation and inhibit mitogen-induced ASM growth by modulating mitochondrial structure and function, resulting in ASM cell death. The mitochondrial protein BCL2/Bnip3 plays a central role in T2R agonist–induced autophagy (83). T2Rs have recently been identified as important players in sinonasal innate immunity and contribute to CRS. T2R38, T2R4, and T2R16 are ubiquitously expressed in the ciliated cells of the human sinonasal epithelium. The T2R isoform T2R38 (family 2 isoform 38) regulates sinonasal defense against Gram-negative organisms through detection of quorum-sensing molecules secreted by pathogens (84). This activation triggers the production of nitric oxide (NO), a potent antimicrobial defense molecule, in a calcium-dependent manner, which results in increased ciliary beat frequency and acceleration of sinonasal mucociliary clearance. NO also diffuses into the mucus and directly targets bacteria and viruses (85). Polymorphisms in T2R38 that confer decreased receptor functionality lead to an increased susceptibility to Gram-negative infection and can predict surgical outcomes of patients with CRS without nasal polyps and biofilm formation (86, 87). T2R38’s ability to modulate the antimicrobial NO pathway makes it an attractive therapeutic target to promote endogenous immune responses in patients with upper respiratory infections. However, patients with homozygous AVI polymorphisms (i.e., individuals expressing AVI/AVI), which render T2R38 nonfunctional, would be predicted to be less responsive to the treatments with T2R38 agonists. It therefore remains important to identify other T2Rs that activate similar mechanisms.
T2R4 and T2R16 signal through the same NO pathway, independent of T2R38, and thus may be additional targets for novel therapeutic interventions. Keratinocytes express T2R1 and T2R38, which play a role in keratinocyte differentiation by inducing keratin 10, involucrin, and transglutaminase expression, as proven by stimulation of keratinocytes with their agonist, amarogentin (88). Amarogentin inhibited substance P–induced production of newly synthesized TNF-α in mast cells, but their degranulation and release of stored histamine were not affected. In keratinocytes, amarogentin reduced histamine- and TNF-α–induced IL-8 and MMP-1 expression to an extent similar to that achieved by the histamine H1 receptor antagonist azelastine (89). In a proof-of-concept study, Gentiana lutea extract (containing amarogentin) significantly increased the lipid content of the treated skin areas. Enhancing lipid synthesis in human keratinocytes is essential for building an intact epidermal barrier, and therefore this therapeutic approach might be used to improve skin disorders like AD that are characterized by an impaired epidermal barrier (90).
How T2Rs relate to precision medicine is an important question. T2Rs are a novel target for asthma, CRS, and AD treatment independent of the inflammatory profile (type 2 or non–type 2) and could be the perfect example of a smart drug design starting from the endotype and theratype and extending through the drug development program. They are also related to disease outcomes (asthma control, CRS severity) and display significant heterogeneity, which affects their function. For example, genetic variations were associated with lower asthma control and better bronchodilator response (91). T2R38 genetic polymorphism predisposes to infection in upper airways via decreased NO production from epithelial cells in response to bacterial products (84). Allele expression studies in patients with CRS show that T2R38 is not the only genetic determinant of disease severity. Several other loci, such as that of TAS2R14 and TAS2R49, show an allele frequency difference of more than 10% between CRS patients and controls (92). Human skin expressed T2Rs, and the relative amounts differ markedly among individuals based on sun exposure, sex, and age (93).
Endocannabinoids in the type 2 allergic response.
Endogenous cannabinoids acting via cannabinoid 2 (CB2) receptors play important roles in both homeostatic and inflammatory processes in the lungs, including eosinophilic inflammation and the suppression of ILC2s by NK cells. CB2 activation controls NK cell activity and induces a predominant type 2 microenvironment and eosinophilia (94, 95).
Olfactory receptors and the gut-lung axis.
ASM olfactory receptors (ORs) do not bronchodilate, but rather modulate cytoskeletal remodeling and hyperplasia, two cardinal features of asthma (96). Short-chain fatty acids (SCFAs), the by-products of fermentation of polysaccharides by the gut microbiome, activate these receptors, thus supporting a nonimmune gut-lung axis that may affect asthma susceptibility (96). Given the link between obesity and asthma, this potential interaction requires further exploration. Agonists directed to ORs might mitigate increases in ASM in asthma. Alternatively, dietary modification to promote microbial communities (and the appropriate substrates) to generate SCFAs as natural agonists for ORs might represent a nonpharmacological approach.
Corticosteroid responsiveness theratype.
It was recently hypothesized that treatment response in lung function, bronchodilator response, airway responsiveness, symptoms, need for oral steroids, and frequency of emergency department visits and hospitalizations are modulated by a single quantitative corticosteroid responsiveness endotype or, perhaps more accurately, a theratype. A composite corticosteroid responsiveness theratype was tested in four replication populations. This composite theratype measured the endophenotype with higher accuracy, higher stability across populations, and higher robustness to missing data than any clinical phenotype (97). Accordingly, several SNPs have been described that associate with response to common asthma controllers (98–103) or with response to treatment of asthma exacerbations (104, 105). There are also SNPs that predict one of the adverse effects of inhaled corticosteroids (ICS), which is adrenal suppression (106, 107).
In asthma clinical trials, surrogates such as blood or sputum eosinophils and exhaled NO have been used. For example, blood or sputum eosinophil counts have been used to titrate inhaled corticosteroids, and this intervention yielded better results compared with guidelines-driven asthma management in decreasing exacerbation (108).
Fibrotic mechanisms contribute to asthmatic responses.
Activin A induces lung fibroblast proliferation and differentiation into myofibroblasts, and stimulates the production of collagen (81). Follistatin is a protein that binds activin with high affinity and low reversibility and antagonizes the pleiotropic actions of activin. Follistatin-like 3 (FSTL3), a glycoprotein secreted by bronchial epithelial cells that binds to and inhibits activin A, is diminished in children with asthma, especially when airway obstruction is present. Decreased FSTL3 expression in the asthmatic bronchial epithelium led to subepithelial fibrosis (109).
The microbiome in allergic endotyping.
High microbial diversity in the environment has been associated with lower asthma risk, particularly in children living in farm environments and exposed to farm milk. This effect operates through alterations in the microbiome of airway mucosal surfaces (110). Extensive molecular analysis of host and commensal microbe interactions continues, and has thus far generated several major molecular findings relevant to asthma pathogenesis that together open a new concept for disease endotyping related to microbiome (111).
Metabolites of microbes regulate immune responses in several aspects. Decreased fatty acid desaturase activity is observed in patients with asthma, and experimental models in mice and human epithelial cells suggest that inhibition of desaturase activity leads to AHR and reduced antiviral defense (112). Bacterial production of histamine and other monoamines was recently revealed as a possible concept in the pathogenesis of several diseases and should be further explored (113–115). Bacteria-derived SCFAs, such as butyric acid, propionic acid, and acetic acid, have also been shown to be active in many inflammatory diseases, in particular butyrate in asthma models. Particular gut commensals, such as Bifidobacterium longum, are known to show protection from inflammatory diseases. Recently, a molecular mechanism of action was ascribed to exopolysaccharide produced by the commensal bacteria (116, 117).
Treatable traits and asthma management.
A new taxonomic and management approach, termed treatable traits, has been proposed for airway diseases including severe asthma, and eosinophilia is included as a treatable trait, indicating potential response to intervention with corticosteroids. Treatable traits can be assessed using asthma registries. Patients with severe asthma express more treatable traits than patients with nonsevere asthma associated with risk of future asthma exacerbation, demonstrating the clinical utility of assessing treatable traits. In Australasia, allergic sensitization, upper airway disease, airflow limitation, eosinophilic inflammation, and frequent exacerbations, as assessed using a severe asthma registry, were common in severe asthma. Among the traits that most strongly predict exacerbation risk in asthma are proneness to exacerbations, depression, inhaler device polypharmacy, vocal cord dysfunction, and obstructive sleep apnea. The treatable traits concept develops in parallel to endotypes and phenotypes and will probably find a niche in personalized medicine (118).
Tissue responses and disease endotypes
Using microarray and PCR analyses of airway epithelial brushings, Woodruff was the first to classify asthma based on high or low expression of IL-13–inducible genes (periostin, chloride channel regulator 1, and serpin peptidase inhibitor, clade B, member 2). This classification was validated through analyses of cytokine expression in bronchial biopsies, reproducibility on repeated examination, and relation with clinical endpoints such as lung function and responsiveness to inhaled corticosteroids (26).
Inflammatory signatures of CRS vary around the world, with less eosinophilic and more neutrophilic inflammation found in Asia compared with Europe and North America. These differences are associated with a lower asthma comorbidity and risk of disease recurrence after surgery in the Asian population. Interestingly, the percentage of type 2 signature disease in patients with CRS is dramatically increasing in several Asian countries over the last 20 years (this phenomenon has been termed eosinophilic shift). As a hallmark of severe type 2 inflammation, eosinophils attacking Staphylococcus aureus at the epithelial barrier have been described recently; they can also be found in a subgroup of Asian patients with nasal polyps (119).
Expression profiling of skin biopsy specimens has established molecular features of the skin in patients with AD. The minimally invasive skin tape strip transcriptome analysis has been evaluated for its value to describe molecular disease endotypes. Skin tape expression profiles were deemed better at evaluating epidermal differentiation complex genes. Both immune- and epidermis-related genes distinguish patients with AD from healthy subjects. Fifty percent of AD patients exhibit a type 2–high endotype that is characterized by more severe disease (120).
The “lung- and small airway-on-a-chip” technologies will rapidly contribute to filling the current translational gap in lung research and advance drug development, biomarker identification, and personalized therapy. Despite the higher cost and operational complexity of these technologies, they enable real-time analysis of dynamic cell-cell interactions together with the ability to stretch the cells, mimicking expansion and retraction, as well as the ability to study inhalation exposure under physiological breathing. These methods also provide a high-resolution kinetic analysis of biological responses (e.g., time-course release of secreted factors in response to challenge) (121). The small airways disease asthma phenotype, which is not easily accessible through classic biopsies, is particularly amenable to the small airway-on-a-chip technology. Bioelectronics has enabled development of biosensors that can detect biological signals in a living microenvironment using micro- or nanofabricated probes. Thus the formation and disruption of epithelium barrier function can be investigated noninvasively in real time (122). In a recent study using this technology, a significant increase in the number of mucin-producing cells and secretion of proinflammatory cytokines, as well as a decrease in ciliary beat frequency, was observed in IL-13–treated mucociliated airway epithelial cells. The same study showed the ability of tofacitinib to reverse the observed asthmatic phenotypes to normal levels by inhibiting Janus kinase signaling (123).
Conclusions
Precision endotyping that links pathobiological mechanisms with visible properties via specific biomarkers and can be translated into pathway-specific diagnostic tests opens a pathway to accurate disease classification and individualized targeted treatments. It is expected to change the diagnostic and therapeutic landscape of all medical disciplines. Endotype-driven treatment still faces multiple challenges before its implementation in daily practice in allergic diseases and asthma. Currently, most of the endotype research is conducted in severe forms of allergic diseases; however, endotypes should be considered as a disease entity on their own. Early endotyping of milder or at-risk patients might provide more insight into disease mechanisms. At the early stages of the disease, putative endotypes may be easier to differentiate, while as the disease progresses, other cell types and structures would be affected, making the distinction of each endotype more difficult. Sustainability of an endotype is one of the major questions. Although a stable endotype has been observed in the absence of disease exacerbation, changes in an endotype can be observed during exacerbations, in smokers, during development of COPD, or during a shift from asthma to asthma-COPD overlapping syndrome.
A huge amount of omics data has been developing during recent years that can only be rationally analyzed in depth with cooperation between biologists, data scientists, and health care professionals. Translational bioinformatics had been proposed and developed as a new field only recently. The emergence of deep learning and artificial intelligence has impacted numerous machine learning–based applications and much research. The reason for their success lies in two main advantages: they provide the ability to learn very complex nonlinear relationships between features, and they allow one to leverage information from unlabeled data that do not belong to the problem being handled. The deep learning–based molecular disease classification can be used to guide decisions made on the diagnosis and treatment of allergic diseases, and therefore may have important applications in precision medicine.
It is becoming increasingly obvious that a revised taxonomy of allergic diseases based on precision medicine and endotype profiling is necessary to stimulate targeted research and identify biomarkers that can predict patient response to treatment. Type 2 and non–type 2 allergic disease profiling should involve the interconnected concepts of phenotype-biomarker-endotype, but also expand toward new targets such as ASM, epithelial components, and epigenetic modifications. In the future, many new endotypes and theratypes particularly defined by treatment response will be introduced. It is important to emphasize here that there are many clinical studies ongoing, with biologicals that target single cytokines in the pipeline. Combination therapies and dual-specific antibodies are also ahead.
The endotype-driven strategy should be modeled per race, per age group, and per sex, because disease endotypes are also modulated by unmodifiable factors, such as race/ethnicity, sex, and age. For example, neutrophilic inflammation is more frequently encountered in women, teenagers, or the elderly, or in the Chinese population. 17q21 gene variants are the strongest known genetic determinants for childhood asthma; however, they do not substantially influence asthma risk in adulthood or susceptibility to detrimental effects of active smoking. This contrasts with the findings in children and suggests that this locus is associated with a childhood-specific asthma endotype (124). The transition between age groups is also interesting. The ALL Age Asthma Cohort (ALLIANCE) offers a unique, integrative, and interdisciplinary framework with a comprehensive molecular approach in a prospective and identical fashion across ages in order to identify biomarkers and predictors for distinct childhood wheeze and asthma trajectories as well as their further course during adulthood (125). Taken together, the implementation of precision medicine in clinical practice should consider insight into high-risk groups indicated by public health studies, including those linked to age, sex, and ethnicity.
Finally, collaboration of health care providers with pharmaceutical and biotechnical companies, scientific organizations, and governmental regulatory agencies will play a crucial role in better control and even cure of allergic diseases within the framework set forth in this Review.
Acknowledgments
Ioana Agache developed this review as part of the research project PN-II-RU-TE-2014-4-2303. The Lab of Cezmi Akdis is supported by Grants from Swiss National Science Foundation.
Version 1. 03/11/2019
Electronic publication
Version 2. 04/01/2019
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019 American Society for Clinical Investigation
Reference information: J Clin Invest. 2019;129(4):1493–1503.https://doi.org/10.1172/JCI124611.
Contributor Information
Ioana Agache, Email: ibrumaru@unitbv.ro.
Cezmi A. Akdis, Email: akdisac@siaf.uzh.ch.
References
- 1.Bousquet J, et al. Allergic Rhinitis and its Impact on Asthma (ARIA) 2008 update (in collaboration with the World Health Organization, GA(2)LEN and AllerGen) Allergy. 2008;63(suppl 86):8–160. doi: 10.1111/j.1398-9995.2007.01620.x. [DOI] [PubMed] [Google Scholar]
- 2.Agache I, Akdis C, Jutel M, Virchow JC. Untangling asthma phenotypes and endotypes. Allergy. 2012;67(7):835–846. doi: 10.1111/j.1398-9995.2012.02832.x. [DOI] [PubMed] [Google Scholar]
- 3.Agusti A, et al. Treatable traits: toward precision medicine of chronic airway diseases. Eur Respir J. 2016;47(2):410–419. doi: 10.1183/13993003.01359-2015. [DOI] [PubMed] [Google Scholar]
- 4.Simpson JL, Scott R, Boyle MJ, Gibson PG. Inflammatory subtypes in asthma: assessment and identification using induced sputum. Respirology. 2006;11(1):54–61. doi: 10.1111/j.1440-1843.2006.00784.x. [DOI] [PubMed] [Google Scholar]
- 5.Green RH, Brightling CE, Woltmann G, Parker D, Wardlaw AJ, Pavord ID. Analysis of induced sputum in adults with asthma: identification of subgroup with isolated sputum neutrophilia and poor response to inhaled corticosteroids. Thorax. 2002;57(10):875–879. doi: 10.1136/thorax.57.10.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Al-Samri MT, et al. Variability of sputum inflammatory cells in asthmatic patients receiving corticosteroid therapy: a prospective study using multiple samples. J Allergy Clin Immunol. 2010;125(5):1161–1163.e4. doi: 10.1016/j.jaci.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 7.Lötvall J, et al. Asthma endotypes: a new approach to classification of disease entities within the asthma syndrome. J Allergy Clin Immunol. 2011;127(2):355–360. doi: 10.1016/j.jaci.2010.11.037. [DOI] [PubMed] [Google Scholar]
- 8.Anderson GP. Endotyping asthma: new insights into key pathogenic mechanisms in a complex, heterogeneous disease. Lancet. 2008;372(9643):1107–1119. doi: 10.1016/S0140-6736(08)61452-X. [DOI] [PubMed] [Google Scholar]
- 9.Agache IO. From phenotypes to endotypes to asthma treatment. Curr Opin Allergy Clin Immunol. 2013;13(3):249–256. doi: 10.1097/ACI.0b013e32836093dd. [DOI] [PubMed] [Google Scholar]
- 10.Agache I, Sugita K, Morita H, Akdis M, Akdis CA. The complex type 2 endotype in allergy and asthma: from laboratory to bedside. Curr Allergy Asthma Rep. 2015;15(6):29. doi: 10.1007/s11882-015-0529-x. [DOI] [PubMed] [Google Scholar]
- 11.Agache I, Akdis CA. Endotypes of allergic diseases and asthma: an important step in building blocks for the future of precision medicine. Allergol Int. 2016;65(3):243–252. doi: 10.1016/j.alit.2016.04.011. [DOI] [PubMed] [Google Scholar]
- 12.Agache I, Strasser DS, Pierlot GM, Farine H, Izuhara K, Akdis CA. Monitoring inflammatory heterogeneity with multiple biomarkers for multidimensional endotyping of asthma. J Allergy Clin Immunol. 2018;141(1):442–445. doi: 10.1016/j.jaci.2017.08.027. [DOI] [PubMed] [Google Scholar]
- 13.Agache I, Rogozea L. Asthma biomarkers: do they bring precision medicine closer to the clinic? Allergy Asthma Immunol Res. 2017;9(6):466–476. doi: 10.4168/aair.2017.9.6.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agache I, et al. Serum IL-5 and IL-13 consistently serve as the best predictors for the blood eosinophilia phenotype in adult asthmatics. Allergy. 2016;71(8):1192–1202. doi: 10.1111/all.12906. [DOI] [PubMed] [Google Scholar]
- 15.Rodrigo-Munoz JM, et al. Asthma diagnosis using integrated analysis of eosinophil microRNAs. Allergy. doi: 10.1111/all.13570. doi: 10.1111/all.13570. [published online ahead of print July 24, 2018]. [DOI] [PubMed] [Google Scholar]
- 16.Zhang K, et al. Decreased epithelial and sputum miR-221-3p associates with airway eosinophilic inflammation and CXCL17 expression in asthma. Am J Physiol Lung Cell Mol Physiol. 2018;315(2):L253–L264. doi: 10.1152/ajplung.00567.2017. [DOI] [PubMed] [Google Scholar]
- 17.Kärner J, et al. Increased microRNA-323-3p in IL-22/IL-17-producing T cells and asthma: a role in the regulation of the TGF-β pathway and IL-22 production. Allergy. 2017;72(1):55–65. doi: 10.1111/all.12907. [DOI] [PubMed] [Google Scholar]
- 18.Dileepan M, Sarver AE, Rao SP, Panettieri RA, Subramanian S, Kannan MS. MicroRNA mediated chemokine responses in human airway smooth muscle cells. PLoS One. 2016;11(3):e0150842. doi: 10.1371/journal.pone.0150842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davis JS, et al. Circulating microRNAs and association with methacholine PC20 in the Childhood Asthma Management Program (CAMP) cohort. PLoS One. 2017;12(7):e0180329. doi: 10.1371/journal.pone.0180329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bartel S, Carraro G, Alessandrini F, Krauss-Etschmann S, Ricciardolo FLM, Bellusci S. miR-142-3p is associated with aberrant WNT signaling during airway remodeling in asthma. Am J Physiol Lung Cell Mol Physiol. 2018;315(2):L328–L333. doi: 10.1152/ajplung.00113.2018. [DOI] [PubMed] [Google Scholar]
- 21.Li JJ, et al. MicroRNA-9 regulates steroid-resistant airway hyperresponsiveness by reducing protein phosphatase 2A activity. J Allergy Clin Immunol. 2015;136(2):462–473. doi: 10.1016/j.jaci.2014.11.044. [DOI] [PubMed] [Google Scholar]
- 22.Kim RY, et al. MicroRNA-21 drives severe, steroid-insensitive experimental asthma by amplifying phosphoinositide 3-kinase-mediated suppression of histone deacetylase 2. J Allergy Clin Immunol. 2017;139(2):519–532. doi: 10.1016/j.jaci.2016.04.038. [DOI] [PubMed] [Google Scholar]
- 23.Martinez-Nunez RT, et al. Genome-wide posttranscriptional dysregulation by microRNAs in human asthma as revealed by frac-seq. J Immunol. 2018;201(1):251–263. doi: 10.4049/jimmunol.1701798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang XH, et al. Overexpression of miR-125b, a novel regulator of innate immunity, in eosinophilic chronic rhinosinusitis with nasal polyps. Am J Respir Crit Care Med. 2012;185(2):140–151. doi: 10.1164/rccm.201103-0456OC. [DOI] [PubMed] [Google Scholar]
- 25.Chen XF, et al. MiR-151a is involved in the pathogenesis of atopic dermatitis by regulating interleukin-12 receptor β2. Exp Dermatol. 2018;27(4):427–432. doi: 10.1111/exd.13276. [DOI] [PubMed] [Google Scholar]
- 26.Woodruff PG, et al. T-helper type 2-driven inflammation defines major subphenotypes of asthma. Am J Respir Crit Care Med. 2009;180(5):388–395. doi: 10.1164/rccm.200903-0392OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baines KJ, Simpson JL, Wood LG, Scott RJ, Gibson PG. Transcriptional phenotypes of asthma defined by gene expression profiling of induced sputum samples. J Allergy Clin Immunol. 2011;127(1):153–160.e1. doi: 10.1016/j.jaci.2010.10.024. [DOI] [PubMed] [Google Scholar]
- 28.Woodruff PG, et al. Genome-wide profiling identifies epithelial cell genes associated with asthma and with treatment response to corticosteroids. Proc Natl Acad Sci U S A. 2007;104(40):15858–15863. doi: 10.1073/pnas.0707413104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nicodemus-Johnson J, et al. Genome-wide methylation study identifies an IL-13-induced epigenetic signature in asthmatic airways. Am J Respir Crit Care Med. 2016;193(4):376–385. doi: 10.1164/rccm.201506-1243OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Modena BD, et al. Gene expression in relation to exhaled nitric oxide identifies novel asthma phenotypes with unique biomolecular pathways. Am J Respir Crit Care Med. 2014;190(12):1363–1372. doi: 10.1164/rccm.201406-1099OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Christenson SA, et al. Asthma-COPD overlap. Clinical relevance of genomic signatures of type 2 inflammation in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2015;191(7):758–766. doi: 10.1164/rccm.201408-1458OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hekking PP, et al. Pathway discovery using transcriptomic profiles in adult-onset severe asthma. J Allergy Clin Immunol. 2018;141(4):1280–1290. doi: 10.1016/j.jaci.2017.06.037. [DOI] [PubMed] [Google Scholar]
- 33.Cao Q, et al. STING positively regulates human ORMDL3 expression through TBK1-IRF3-STAT6 complex mediation. Exp Cell Res. 2018;370(2):498–505. doi: 10.1016/j.yexcr.2018.07.015. [DOI] [PubMed] [Google Scholar]
- 34.Okada N, et al. Distinct gene expression profiles and regulation networks of nasal polyps in eosinophilic and non-eosinophilic chronic rhinosinusitis. Int Forum Allergy Rhinol. 2018;8(5):592–604. doi: 10.1002/alr.22083. [DOI] [PubMed] [Google Scholar]
- 35.Kowalski ML, et al. Diagnosis and management of NSAID-Exacerbated Respiratory Disease (N-ERD)—a EAACI position paper. Allergy. 2019;74(1):28–39. doi: 10.1111/all.13599. [DOI] [PubMed] [Google Scholar]
- 36.Choi Y, et al. Surfactant protein D alleviates eosinophil-mediated airway inflammation remodeling in patients with aspirin-exacerbated respiratory disease. Allergy. 2019;74(1):78–88. doi: 10.1111/all.13458. [DOI] [PubMed] [Google Scholar]
- 37.Lee HY, et al. Identification of phenotypic clusters of nonsteroidal anti-inflammatory drugs exacerbated respiratory disease. Allergy. 2017;72(4):616–626. doi: 10.1111/all.13075. [DOI] [PubMed] [Google Scholar]
- 38.Rondon C, et al. Local allergic rhinitis is an independent rhinitis phenotype: The results of a 10-year follow-up study. Allergy. 2018;73(2):470–478. doi: 10.1111/all.13272. [DOI] [PubMed] [Google Scholar]
- 39.Wahn U, Bachert C, Heinrich J, Richter H, Zielen S. Real-world benefits of allergen immunotherapy for birch pollen-associated allergic rhinitis asthma. Allergy. 2019;74(3):594–604. doi: 10.1111/all.13598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karatzas K, et al. New European Academy of Allergy and Clinical Immunology definition on pollen season mirrors symptom load for grass and birch pollen-induced allergic rhinitis. Allergy. 2018;73(9):1851–1859. doi: 10.1111/all.13487. [DOI] [PubMed] [Google Scholar]
- 41.Yii ACA, Tay TR, Choo XN, Koh MSY, Tee AKH, Wang DY. Precision medicine in united airways disease: a “treatable traits” approach. Allergy. 2018;73(10):1964–1978. doi: 10.1111/all.13496. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Y, et al. Th2 cytokines orchestrate the secretion of MUC5AC and MUC5B in IL-5 positive chronic rhinosinusitis with nasal polyps. Allergy. 2019;74(1):131–140. doi: 10.1111/all.13489. [DOI] [PubMed] [Google Scholar]
- 43.Liao B, et al. Multidimensional endotypes of chronic rhinosinusitis and their association with treatment outcomes. Allergy. 2018;73(7):1459–1469. doi: 10.1111/all.13411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tan E, et al. Serum periostin levels in adults of Chinese descent: an observational study. Allergy Asthma Clin Immunol. 2018;14:87. doi: 10.1186/s13223-018-0312-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brunner PM, Guttman-Yassky E. Racial differences in atopic dermatitis. Ann Allergy Asthma Immunol. doi: 10.1016/j.anai.2018.11.015. doi: 10.1016/j.anai.2018.11.015. [published online ahead of print November 20, 2018]. [DOI] [PubMed] [Google Scholar]
- 46.Brown KR, et al. Endotypes of difficult-to-control asthma in inner-city African American children. PLoS One. 2017;12(7):e0180778. doi: 10.1371/journal.pone.0180778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Akdis CA, et al. Endotypes and phenotypes of chronic rhinosinusitis: a PRACTALL document of the European Academy of Allergy and Clinical Immunology and the American Academy of Allergy, Asthma & Immunology. J Allergy Clin Immunol. 2013;131(6):1479–1490. doi: 10.1016/j.jaci.2013.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Macauley MS, Crocker PR, Paulson JC. Siglec-mediated regulation of immune cell function in disease. Nat Rev Immunol. 2014;14(10):653–666. doi: 10.1038/nri3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen GY, et al. Broad and direct interaction between TLR and Siglec families of pattern recognition receptors and its regulation by Neu1. Elife. 2014;3:e04066. doi: 10.7554/eLife.04066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kiwamoto T, et al. Endogenous airway mucins carry glycans that bind Siglec-F and induce eosinophil apoptosis. J Allergy Clin Immunol. 2015;135(5):1329–1340.e9. doi: 10.1016/j.jaci.2014.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jia Y, et al. Expression of ligands for Siglec-8 and Siglec-9 in human airways and airway cells. J Allergy Clin Immunol. 2015;135(3):799–810.e7. doi: 10.1016/j.jaci.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mukherjee M, et al. Sputum autoantibodies in patients with severe eosinophilic asthma. J Allergy Clin Immunol. 2018;141(4):1269–1279. doi: 10.1016/j.jaci.2017.06.033. [DOI] [PubMed] [Google Scholar]
- 53.Kast JI, et al. Respiratory syncytial virus infection influences tight junction integrity. Clin Exp Immunol. 2017;190(3):351–359. doi: 10.1111/cei.13042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sugita K, et al. Type 2 innate lymphoid cells disrupt bronchial epithelial barrier integrity by targeting tight junctions through IL-13 in asthmatic patients. J Allergy Clin Immunol. 2018;141(1):300–310.e11. doi: 10.1016/j.jaci.2017.02.038. [DOI] [PubMed] [Google Scholar]
- 55.Yu QN, et al. ILC2 frequency and activity are inhibited by glucocorticoid treatment via STAT pathway in patients with asthma. Allergy. 2018;73(9):1860–1870. doi: 10.1111/all.13438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Steelant B, et al. Histamine and T helper cytokine-driven epithelial barrier dysfunction in allergic rhinitis. J Allergy Clin Immunol. 2018;141(3):951–963.e8. doi: 10.1016/j.jaci.2017.08.039. [DOI] [PubMed] [Google Scholar]
- 57.Wawrzyniak P, et al. Regulation of bronchial epithelial barrier integrity by type 2 cytokines and histone deacetylases in asthmatic patients. J Allergy Clin Immunol. 2017;139(1):93–103. doi: 10.1016/j.jaci.2016.03.050. [DOI] [PubMed] [Google Scholar]
- 58.Sweerus K, et al. Claudin-18 deficiency is associated with airway epithelial barrier dysfunction and asthma. J Allergy Clin Immunol. 2017;139(1):72–81.e1. doi: 10.1016/j.jaci.2016.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Arlian LG, Elder BL, Morgan MS. House dust mite extracts activate cultured human dermal endothelial cells to express adhesion molecules and secrete cytokines. J Med Entomol. 2009;46(3):595–604. doi: 10.1603/033.046.0326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kanemaru K, et al. Phospholipase Cδ1 regulates p38 MAPK activity and skin barrier integrity. Cell Death Differ. 2017;24(6):1079–1090. doi: 10.1038/cdd.2017.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pothoven KL, et al. Neutrophils are a major source of the epithelial barrier disrupting cytokine oncostatin M in patients with mucosal airways disease. J Allergy Clin Immunol. 2017;139(6):1966–1978.e9. doi: 10.1016/j.jaci.2016.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kwon BI, et al. Enhanced Th2 cell differentiation and function in the absence of Nox2. Allergy. 2017;72(2):252–265. doi: 10.1111/all.12944. [DOI] [PubMed] [Google Scholar]
- 63.Kortekaas Krohn I, et al. Emerging roles of innate lymphoid cells in inflammatory diseases: clinical implications. Allergy. 2018;73(4):837–850. doi: 10.1111/all.13340. [DOI] [PubMed] [Google Scholar]
- 64.Ozyigit LP, Morita H, Akdis M. Innate lymphocyte cells in asthma phenotypes. Clin Transl Allergy. 2015;5:23. doi: 10.1186/s13601-015-0068-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mjösberg JM, et al. Human IL-25- and IL-33-responsive type 2 innate lymphoid cells are defined by expression of CRTH2 and CD161. Nat Immunol. 2011;12(11):1055–1062. doi: 10.1038/ni.2104. [DOI] [PubMed] [Google Scholar]
- 66.Aron JL, Akbari O. Regulatory T cells and type 2 innate lymphoid cell-dependent asthma. Allergy. 2017;72(8):1148–1155. doi: 10.1111/all.13139. [DOI] [PubMed] [Google Scholar]
- 67.Uchida M, et al. Oxidative stress serves as a key checkpoint for IL-33 release by airway epithelium. Allergy. 2017;72(10):1521–1531. doi: 10.1111/all.13158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gorski SA, Hahn YS, Braciale TJ. Group 2 innate lymphoid cell production of IL-5 is regulated by NKT cells during influenza virus infection. PLoS Pathog. 2013;9(9):e1003615. doi: 10.1371/journal.ppat.1003615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Monticelli LA, et al. Innate lymphoid cells promote lung-tissue homeostasis after infection with influenza virus. Nat Immunol. 2011;12(11):1045–1054. doi: 10.1038/ni.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shen X, et al. Group 2 innate lymphoid cells promote airway hyperresponsiveness through production of VEGFA. J Allergy Clin Immunol. 2018;141(5):1929–1931.e4. doi: 10.1016/j.jaci.2018.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wallrapp A, et al. The neuropeptide NMU amplifies ILC2-driven allergic lung inflammation. Nature. 2017;549(7672):351–356. doi: 10.1038/nature24029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cephus JY, et al. Testosterone attenuates group 2 innate lymphoid cell-mediated airway inflammation. Cell Rep. 2017;21(9):2487–2499. doi: 10.1016/j.celrep.2017.10.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Laffont S, et al. Androgen signaling negatively controls group 2 innate lymphoid cells. J Exp Med. 2017;214(6):1581–1592. doi: 10.1084/jem.20161807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Banuelos J, et al. BCL-2 protects human and mouse Th17 cells from glucocorticoid-induced apoptosis. Allergy. 2016;71(5):640–650. doi: 10.1111/all.12840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gavino AC, Nahmod K, Bharadwaj U, Makedonas G, Tweardy DJ. STAT3 inhibition prevents lung inflammation, remodeling, and accumulation of Th2 and Th17 cells in a murine asthma model. Allergy. 2016;71(12):1684–1692. doi: 10.1111/all.12937. [DOI] [PubMed] [Google Scholar]
- 76.Komlósi ZI, et al. Human CD40 ligand-expressing type 3 innate lymphoid cells induce IL-10-producing immature transitional regulatory B cells. J Allergy Clin Immunol. 2018;142(1):178–194.e11. doi: 10.1016/j.jaci.2017.07.046. [DOI] [PubMed] [Google Scholar]
- 77.Mathews JA, et al. IL-33 drives augmented responses to ozone in obese mice. Environ Health Perspect. 2017;125(2):246–253. doi: 10.1289/EHP272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hong HY, et al. Local IL-25 contributes to Th2-biased inflammatory profiles in nasal polyps. Allergy. 2018;73(2):459–469. doi: 10.1111/all.13267. [DOI] [PubMed] [Google Scholar]
- 79.Rochman Y, et al. TSLP signaling in CD4+ T cells programs a pathogenic T helper 2 cell state. Sci Signal. 2018;11(521):eaam8858. doi: 10.1126/scisignal.aam8858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Weng CM, et al. Aryl hydrocarbon receptor activation by diesel exhaust particles mediates epithelium-derived cytokines expression in severe allergic asthma. Allergy. 2018;73(11):2192–2204. doi: 10.1111/all.13462. [DOI] [PubMed] [Google Scholar]
- 81.Zhu J, et al. Anti-allergic inflammatory activity of interleukin-37 is mediated by novel signaling cascades in human eosinophils. Front Immunol. 2018;9:1445. doi: 10.3389/fimmu.2018.01445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Deshpande DA, et al. Bitter taste receptors on airway smooth muscle bronchodilate by localized calcium signaling and reverse obstruction. Nat Med. 2010;16(11):1299–1304. doi: 10.1038/nm.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pan S, Sharma P, Shah SD, Deshpande DA. Bitter taste receptor agonists alter mitochondrial function and induce autophagy in airway smooth muscle cells. Am J Physiol Lung Cell Mol Physiol. 2017;313(1):L154–L165. doi: 10.1152/ajplung.00106.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lee RJ, et al. T2R38 taste receptor polymorphisms underlie susceptibility to upper respiratory infection. J Clin Invest. 2012;122(11):4145–4159. doi: 10.1172/JCI64240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hariri BM, et al. In vitro effects of anthocyanidins on sinonasal epithelial nitric oxide production and bacterial physiology. Am J Rhinol Allergy. 2016;30(4):261–268. doi: 10.2500/ajra.2016.30.4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Adappa ND, et al. Correlation of T2R38 taste phenotype and in vitro biofilm formation from nonpolypoid chronic rhinosinusitis patients. Int Forum Allergy Rhinol. 2016;6(8):783–791. doi: 10.1002/alr.21803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Adappa ND, et al. TAS2R38 genotype predicts surgical outcome in nonpolypoid chronic rhinosinusitis. Int Forum Allergy Rhinol. 2016;6(1):25–33. doi: 10.1002/alr.21666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wölfle U, Elsholz FA, Kersten A, Haarhaus B, Müller WE, Schempp CM. Expression and functional activity of the bitter taste receptors TAS2R1 and TAS2R38 in human keratinocytes. Skin Pharmacol Physiol. 2015;28(3):137–146. doi: 10.1159/000367631. [DOI] [PubMed] [Google Scholar]
- 89.Wölfle U, Haarhaus B, Schempp CM. Amarogentin displays immunomodulatory effects in human mast cells and keratinocytes. Mediators Inflamm. 2015;2015:630128. doi: 10.1155/2015/630128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wölfle U, et al. The herbal bitter drug gentiana lutea modulates lipid synthesis in human keratinocytes in vitro and in vivo. Int J Mol Sci. 2017;18(8):E1814. doi: 10.3390/ijms18081814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yoon SY, et al. Association between polymorphisms in bitter taste receptor genes and clinical features in Korean asthmatics. Respiration. 2016;91(2):141–150. doi: 10.1159/000443796. [DOI] [PubMed] [Google Scholar]
- 92.Mfuna Endam L, Filali-Mouhim A, Boisvert P, Boulet LP, Bossé Y, Desrosiers M. Genetic variations in taste receptors are associated with chronic rhinosinusitis: a replication study. Int Forum Allergy Rhinol. 2014;4(3):200–206. doi: 10.1002/alr.21275. [DOI] [PubMed] [Google Scholar]
- 93.Shaw L, et al. Personalized expression of bitter ‘taste’ receptors in human skin. PLoS One. 2018;13(10):e0205322. doi: 10.1371/journal.pone.0205322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ferrini ME, et al. CB2 receptors regulate natural killer cells that limit allergic airway inflammation in a murine model of asthma. Allergy. 2017;72(6):937–947. doi: 10.1111/all.13107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Frei RB, et al. Cannabinoid receptor 2 augments eosinophil responsiveness and aggravates allergen-induced pulmonary inflammation in mice. Allergy. 2016;71(7):944–956. doi: 10.1111/all.12858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Aisenberg WH, et al. Defining an olfactory receptor function in airway smooth muscle cells. Sci Rep. 2016;6:38231. doi: 10.1038/srep38231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Clemmer GL, et al. Measuring the corticosteroid responsiveness endophenotype in asthmatic patients. J Allergy Clin Immunol. 2015;136(2):274–281.e8. doi: 10.1016/j.jaci.2015.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tantisira KG, Drazen JM. Genetics and pharmacogenetics of the leukotriene pathway. J Allergy Clin Immunol. 2009;124(3):422–427. doi: 10.1016/j.jaci.2009.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tantisira KG, et al. TBX21: a functional variant predicts improvement in asthma with the use of inhaled corticosteroids. Proc Natl Acad Sci U S A. 2004;101(52):18099–18104. doi: 10.1073/pnas.0408532102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Levin AM, et al. Integrative approach identifies corticosteroid response variant in diverse populations with asthma. J Allergy Clin Immunol. doi: 10.1016/j.jaci.2018.09.034. doi: 10.1016/j.jaci.2018.09.034. [published online ahead of print October 24, 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Demenais F, et al. Multiancestry association study identifies new asthma risk loci that colocalize with immune-cell enhancer marks. Nat Genet. 2018;50(1):42–53. doi: 10.1038/s41588-017-0014-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Farzan N, et al. Rationale and design of the multiethnic Pharmacogenomics in Childhood Asthma consortium. Pharmacogenomics. 2017;18(10):931–943. doi: 10.2217/pgs-2017-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.McGeachie MJ, et al. Asthma remission: predicting future airways responsiveness using an miRNA network. J Allergy Clin Immunol. 2017;140(2):598–600.e8. doi: 10.1016/j.jaci.2017.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Keskin O, et al. Genetic associations of the response to inhaled corticosteroids in children during an asthma exacerbation. Pediatr Allergy Immunol. 2016;27(5):507–513. doi: 10.1111/pai.12566. [DOI] [PubMed] [Google Scholar]
- 105.Keskin O, et al. The efficacy of single-high dose inhaled corticosteroid versus oral prednisone treatment on exhaled leukotriene and 8-isoprostane levels in mild to moderate asthmatic children with asthma exacerbation. Allergol Immunopathol (Madr) 2016;44(2):138–148. doi: 10.1016/j.aller.2015.05.006. [DOI] [PubMed] [Google Scholar]
- 106.Hawcutt DB, et al. Susceptibility to corticosteroid-induced adrenal suppression: a genome-wide association study. Lancet Respir Med. 2018;6(6):442–450. doi: 10.1016/S2213-2600(18)30058-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hawcutt DB, Francis B, Pirmohamed M. Adrenal suppression with inhaled corticosteroids: the seed and the soil – Authors’ reply. Lancet Respir Med. 2018;6(6):e20. doi: 10.1016/S2213-2600(18)30149-8. [DOI] [PubMed] [Google Scholar]
- 108.Jayaram L, et al. Determining asthma treatment by monitoring sputum cell counts: effect on exacerbations. Eur Respir J. 2006;27(3):483–494. doi: 10.1183/09031936.06.00137704. [DOI] [PubMed] [Google Scholar]
- 109.James RG, et al. Deficient follistatin-like 3 secretion by asthmatic airway epithelium impairs fibroblast regulation and fibroblast-to-myofibroblast transition. Am J Respir Cell Mol Biol. 2018;59(1):104–113. doi: 10.1165/rcmb.2017-0025OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Birzele LT, et al. Environmental and mucosal microbiota and their role in childhood asthma. Allergy. 2017;72(1):109–119. doi: 10.1111/all.13002. [DOI] [PubMed] [Google Scholar]
- 111.Sampson HA, O’Mahony L, Burks AW, Plaut M, Lack G, Akdis CA. Mechanisms of food allergy. J Allergy Clin Immunol. 2018;141(1):11–19. doi: 10.1016/j.jaci.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 112.Rodriguez-Perez N, et al. Altered fatty acid metabolism and reduced stearoyl-coenzyme a desaturase activity in asthma. Allergy. 2017;72(11):1744–1752. doi: 10.1111/all.13180. [DOI] [PubMed] [Google Scholar]
- 113.Barcik W, Wawrzyniak M, Akdis CA, O’Mahony L. Immune regulation by histamine and histamine-secreting bacteria. Curr Opin Immunol. 2017;48:108–113. doi: 10.1016/j.coi.2017.08.011. [DOI] [PubMed] [Google Scholar]
- 114.Barcik W, et al. Histamine-secreting microbes are increased in the gut of adult asthma patients. J Allergy Clin Immunol. 2016;138(5):1491–1494.e7. doi: 10.1016/j.jaci.2016.05.049. [DOI] [PubMed] [Google Scholar]
- 115.Ferstl R, et al. Histamine receptor 2 modifies iNKT cell activity within the inflamed lung. Allergy. 2017;72(12):1925–1935. doi: 10.1111/all.13227. [DOI] [PubMed] [Google Scholar]
- 116.Schiavi E, et al. Exopolysaccharide from Bifidobacterium longum subsp. longum 35624™ modulates murine allergic airway responses. Benef Microbes. 2018;9(5):761–773. doi: 10.3920/BM2017.0180. [DOI] [PubMed] [Google Scholar]
- 117.Schiavi E, et al. The surface-associated exopolysaccharide of Bifidobacterium longum 35624 plays an essential role in dampening host proinflammatory responses and repressing local TH17 responses. Appl Environ Microbiol. 2016;82(24):7185–7196. doi: 10.1128/AEM.02238-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.McDonald VM, et al. Treatable traits can be identified in a severe asthma registry and predict future exacerbations. Respirology. 2019;24(1):37–47. doi: 10.1111/resp.13389. [DOI] [PubMed] [Google Scholar]
- 119.Zhang Y, et al. Chronic rhinosinusitis in Asia. J Allergy Clin Immunol. 2017;140(5):1230–1239. doi: 10.1016/j.jaci.2017.09.009. [DOI] [PubMed] [Google Scholar]
- 120.Dyjack N, et al. Minimally invasive skin tape strip RNA sequencing identifies novel characteristics of the type 2-high atopic dermatitis disease endotype. J Allergy Clin Immunol. 2018;141(4):1298–1309. doi: 10.1016/j.jaci.2017.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Huh D, Matthews BD, Mammoto A, Montoya-Zavala M, Hsin HY, Ingber DE. Reconstituting organ-level lung functions on a chip. Science. 2010;328(5986):1662–1668. doi: 10.1126/science.1188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Henry OYF, Villenave R, Cronce MJ, Leineweber WD, Benz MA, Ingber DE. Organs-on-chips with integrated electrodes for trans-epithelial electrical resistance (TEER) measurements of human epithelial barrier function. Lab Chip. 2017;17(13):2264–2271. doi: 10.1039/C7LC00155J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Benam KH, et al. Small airway-on-a-chip enables analysis of human lung inflammation and drug responses in vitro. Nat Methods. 2016;13(2):151–157. doi: 10.1038/nmeth.3697. [DOI] [PubMed] [Google Scholar]
- 124.Kreiner-Møller E, Strachan DP, Linneberg A, Husemoen LL, Bisgaard H, Bønnelykke K. 17q21 gene variation is not associated with asthma in adulthood. Allergy. 2015;70(1):107–114. doi: 10.1111/all.12537. [DOI] [PubMed] [Google Scholar]
- 125.Fuchs O, et al. The all age asthma cohort (ALLIANCE) - from early beginnings to chronic disease: a longitudinal cohort study. BMC Pulm Med. 2018;18(1):140. doi: 10.1186/s12890-018-0705-6. [DOI] [PMC free article] [PubMed] [Google Scholar]