Aspergillus fumigatus is the most important airborne human pathogenic fungus, causing thousands of deaths per year. Its lethality is due to late and often inaccurate diagnosis and the lack of efficient therapeutics. The failure of efficient prophylaxis and therapy is based on the ability of this pathogen to activate numerous salvage pathways that are capable of overcoming the different drug-derived stresses. A major role in the protection of A. fumigatus is played by melanins. Melanins are cell wall-associated macromolecules classified as virulence determinants. The understanding of the various signaling pathways acting in this organism can be used to elucidate the mechanism beyond melanin production and help to identify ideal drug targets.
KEYWORDS: Aspergillus fumigatus, GPCR, MAP kinases, melanin
ABSTRACT
The pathogenic fungus Aspergillus fumigatus is able to adapt to extremely variable environmental conditions. The A. fumigatus genome contains four genes coding for mitogen-activated protein kinases (MAPKs), which are important regulatory knots involved in diverse cellular responses. From a clinical perspective, MAPK activity has been connected to salvage pathways, which can determine the failure of effective treatment of invasive mycoses using antifungal drugs. Here, we report the characterization of the Saccharomyces cerevisiae Fus3 ortholog in A. fumigatus, designated MpkB. We demonstrate that MpkB is important for conidiation and that its deletion induces a copious increase of dihydroxynaphthalene (DHN)-melanin production. Simultaneous deletion of mpkB and mpkA, the latter related to maintenance of the cell wall integrity, normalized DHN-melanin production. Localization studies revealed that MpkB translocates into the nuclei when A. fumigatus germlings are exposed to caspofungin stress, and this is dependent on the cross-talk interaction with MpkA. Additionally, DHN-melanin formation was also increased after deletion of genes coding for the Gα protein GpaA and for the G protein-coupled receptor GprM. Yeast two-hybrid and coimmunoprecipitation assays confirmed that GpaA and GprM interact, suggesting their role in the MpkB signaling cascade.
INTRODUCTION
Aspergillus fumigatus is a saprophytic fungus mainly found in the soil and organic debris. This fungus is capable of producing myriads of airborne conidia that can survive in a wide range of environmental circumstances (1). The conidia are normally released into the air and, when inhaled by immunocompromised patients, can cause severe diseases, including invasive aspergillosis (IA). An increase in the incidence of IA has been observed in the last decades, and the mortality attributed to IA infections can reach 90%. IA is a multifactorial disease, and A. fumigatus has several phenotypic characteristics that make it an aggressive opportunistic pathogen (2). Several factors contribute to A. fumigatus virulence, such as production of dihydroxynaphthalene (DHN)-melanin, hypoxia resistance, ability to subtract environmental iron, toxin production, thermotolerance, and distinct surface molecules (3–7).
Mitogen-activated protein kinase (MAPK) pathways are important for the transmission, integration, and amplification of signals and are essential components involved in diverse cellular processes in eukaryotes (8). In fungi, MAPK pathways regulate cellular responses to different kinds of stresses (9–11). The central module of each MAPK signaling pathway consists of three protein kinases: a MAP kinase kinase kinase (MAPKKK), a MAP kinase kinase (MAPKK), and a MAPK. The MAPK cascades are normally triggered by upstream sensors (e.g., receptors) and end with the activation of downstream elements, such as transcriptional regulators (12).
MAPK signaling cascades have been well characterized in yeasts (13–16). In filamentous fungi, their function was mainly assigned to pheromone responses and filamentous growth, osmotic stress, and cell wall integrity. Additionally, it was demonstrated that MAPKs influence several phenotypes relevant for pathogenesis in both human and plant pathogens (9, 11).
A. fumigatus contains four MAPKs: MpkA, which mainly regulates cell wall integrity (17); MpkC and SakA, similar to the Saccharomyces cerevisiae Hog1, which are involved in the response to oxidative stress and in polyalcohol sugar metabolism (18–20); and MpkB, homologous to yeast Fus3, thus far uncharacterized (21). A cross talk interaction between SakA and MpkA has already been observed for the adaptation to the antimycotic drug caspofungin (22, 23). In addition, it was shown that MpkC and SakA cooperate in the responses to different types of stresses in A. fumigatus, including the response to osmotic and oxidative stress, high-temperature adaptation, cell wall damage, and virulence (23).
Previous investigations on Fus3/MpkB orthologs in several phytopathogenic fungi showed their conserved important role in plant infection (11). In parallel, in other important human pathogenic fungi, such as Candida albicans and Cryptococcus neoformans, the deletion of their FUS3 orthologs mainly caused the loss of mating efficiency and decrease of biofilm formation (24, 25). Of note, in the closely related species Aspergillus nidulans, the mpkB deletion not only inhibited sexual crossing, as expected, but also affected the production of relevant secondary metabolites, such as the mycotoxin sterigmatocystin, the antitumor agent terrequinone A, and penicillin (26, 27).
In the present study, we set out to analyze the function of the MpkB regulatory network in A. fumigatus. Deletion of mpkB increased sensitivity to the glucan synthase inhibitor caspofungin, suggesting a role in cell wall biosynthesis. Furthermore, we observed an increase of dark pigments during growth in liquid medium that was related to DHN-melanin production. Melanins are a class of dark-brown pigments often associated with the cell wall. Their main role is to protect the organisms from exogenous stressors, thereby contributing to the first line of defense against external hazards (7, 28). A. fumigatus produces two types of melanins: pyomelanin, which derives from the catabolism of tyrosine via the intermediate homogentisate, and DHN-melanin, which is produced as a polyketide derivative and is responsible for the gray-green color of the spores (7). Additionally, DHN-melanin plays a crucial role in conidial immune evasion and, consequently, fungal virulence (29–31). In the A. fumigatus genome, the genes coding for DHN-melanin biosynthetic enzymes, as well as those for pyomelanin, are clustered (32, 33).
Signaling pathways that regulate the expression of melanin biosynthesis genes are barely known. DHN-melanin was found to be regulated by cAMP signaling, and in A. fumigatus, two transcription factors responsible for its regulation were identified: DevR, a basic helix-loop-helix (bHLH) transcription factor, and RlmA, a MADS-box transcription factor actively involved in cell wall integrity (34, 35). The production of pyomelanin was also associated with cell wall stress, highlighting its function as a protective compound (36). Highly important here, we observed that the deletion of the gene coding for the Gα protein GpaA and the gene coding for the G protein-coupled receptor GprM resulted in phenotypes similar to that resulting from mpkB deletion, suggesting that they are involved in the same signaling pathway. Here, we present a detailed characterization of the MAPK MpkB in A. fumigatus that led to the identification of its physiological role and several components of its regulatory circuits.
RESULTS
MpkB represses DHN-melanin production and conidiation in liquid medium.
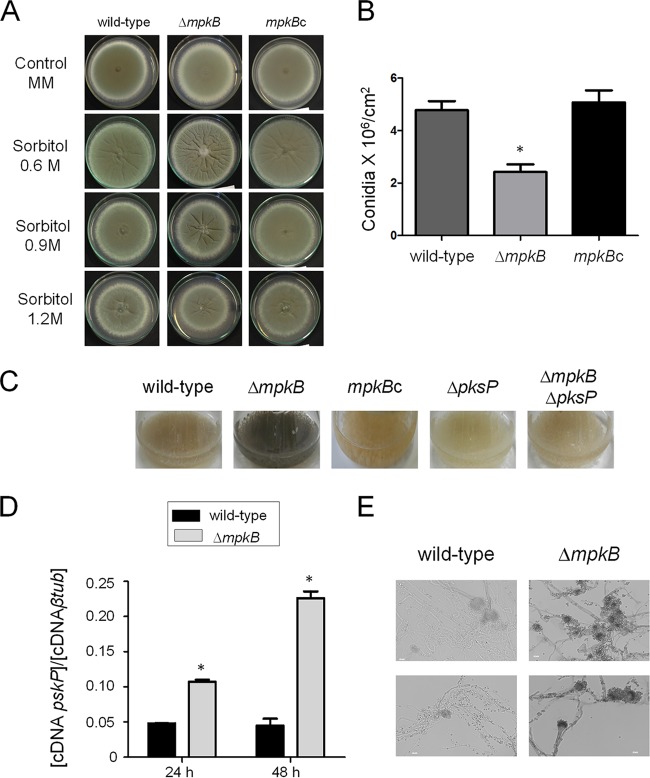
To investigate the function of mpkB in A. fumigatus, the complete gene was first deleted and then the mutant strain obtained was complemented by reinsertion of the wild-type gene. The deletion of mpkB in A. fumigatus did not affect the radial growth on solid minimal medium (MM) (Fig. 1A); however, a significant reduction in the number of conidia produced by the ΔmpkB strain was observed (Fig. 1B). Radial growth assays conducted by challenging the mutant with different exogenous stresses demonstrated that the growth of the ΔmpkB strain was not affected by high-temperature, cell wall, and oxidative stresses (Congo red, calcofluor white [CFW], SDS, menadione, paraquat, t-butyl hydroxyperoxide, and H2O2) (Fig. S1 and S2 in the supplemental material). Additionally, when the ΔmpkB strain was grown for 48 to 72 h in liquid MM, a dark pigment was produced and released (Fig. 1C). In order to test whether there was an increase of DHN-melanin in the ΔmpkB mutant, the pksP gene, encoding the polyketide synthase essential for conidial pigment formation and DHN-melanin biosynthesis, was deleted (7). Neither the ΔpksP mutant nor the ΔmpkB ΔpksP double mutant was able to produce the dark color (Fig. 1C). Furthermore, reverse transcription-quantitative PCR (qRT-PCR) analysis of the pksP transcription levels revealed increases of 2- to 5-fold in the ΔmpkB mutant compared to the levels in the wild type when both strains were grown in liquid MM for 24 and 48 h, respectively (Fig. 1D). Finally, based on the methods reported by Butler et al., we established a protocol for copper-silver staining to directly visualize the melanin produced by the mpkB mutant strain (37). As shown by the results in Fig. 1E, silver accumulation was higher in the hyphae, conidiophores, and conidia of the ΔmpkB mutant than in the wild-type strain, confirming that the accumulated black pigment was DHN-melanin.
FIG 1.
MpkB is important for repressing melanin production. (A) Phenotypic tests were performed by inoculating A. fumigatus conidia onto solid MM agar plates with or without sorbitol. Agar plates were incubated for 5 days at 37°C. (B) Numbers of conidia produced in the wild-type, ΔmpkB, and mpkBc (complemented) strains. The results are the averages of three repetitions ± standard deviations. One-way analysis of variance (ANOVA) with Tukey’s multiple comparison test was used for statistical analysis, and an asterisk represents a P value of <0.05. (C) A. fumigatus conidia were inoculated into liquid MM for 72 h at 37°C with 200 rpm shaking. (D) Results of qRT-PCRs showing the A. fumigatus pksP mRNA steady-state levels in the wild-type and ΔmpkB strains grown at 37°C for 24 and 48 h, respectively. The results represent the averages of three repetitions ± standard deviations and are expressed as the cDNA concentration of pksP divided by the cDNA concentration of the β-tubulin gene. The results are the averages of three repetitions ± standard deviation. One-way ANOVA with Bonferroni posttest was used for statistical analysis, and an asterisk represents a P value of <0.05. (E) Visualization of melanin by copper-silver staining of hyphae and conidiophores of wild-type and ΔmpkB strains grown for 48 h at 37°C. Bars, 10 µm.
Growth phenotypes of the wild type and MAPK and complemented mutants grown in the presence of Congo Red (CR) (A), calcofluor white (CFW) (B), and sodium dodecyl sulfate (SDS) (C). The strains were grown for 48 hours at 37°C on MM agar plates. The numbers of conidia are also reported. Download FIG S1, PDF file, 0.2 MB (222KB, pdf) .
Copyright © 2019 Manfiolli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Growth phenotypes of the wild type and ΔmpkA, ΔmpkB, and corresponding complemented mutants grown in liquid MM in the presence of t-butyl (A), menadione (B), paraquat (C), and hydrogen peroxide (D). The strains were grown for 48 hours at 37°C either on MM agar plates (A to C) or in liquid MM in 96-well plates (D). Numbers of conidia are also reported. Download FIG S2, PDF file, 0.3 MB (283.9KB, pdf) .
Copyright © 2019 Manfiolli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
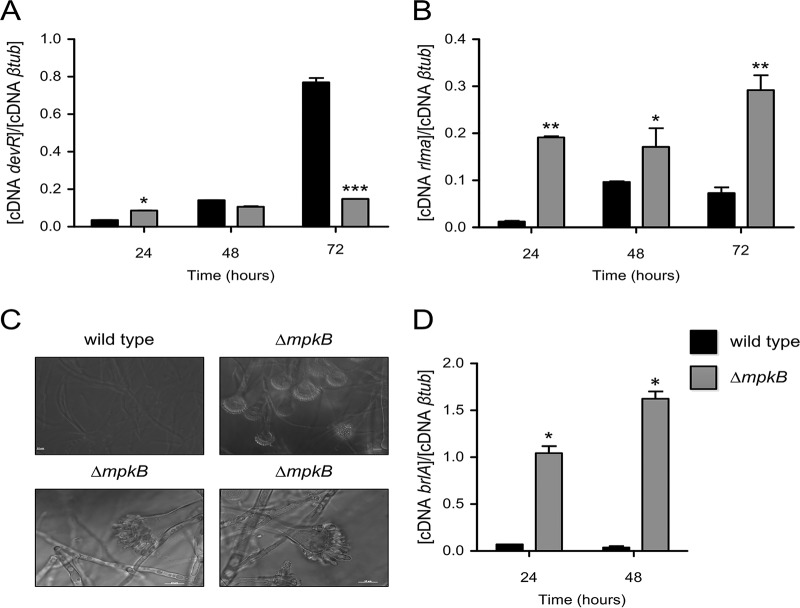
It was previously reported that two different transcription factors, named DevR and RlmA, regulate the DHN-melanin gene cluster (35). DevR belongs to the bHLH family, while RlmA is a classical MADS-box transcription factor. These two transcription factors regulate the DHN-melanin cluster both positively and negatively, depending on the recognized binding sites on the pksP promoter. Additionally, because the assigned binding sites lie close together on the pksP promoter region, a parallel binding of the two regulators was already excluded. qRT-PCR analysis showed that the devR mRNA steady-state level had doubled in the ΔmpkB strain compared to the level in the wild type in the first 24 h of incubation but decreased later on (Fig. 2A). In contrast, the rlmA mRNA level was about 10 to 15 times higher in the ΔmpkB strain than in the wild type (Fig. 2B). These results suggest that the lack of mpkB induced DHN-melanin production by affecting the expression of rlmA.
FIG 2.
mRNA steady-state levels of the transcription factors of devR, rlmA, and brlA genes. (A, B) Results of qRT-PCRs showing the A. fumigatus devR (A) and rlmA (B) mRNA accumulation in the wild-type and ΔmpkB strains after 24, 48, and 72 h of growth at 37°C. The results are the averages of three repetitions ± standard deviations. One-way ANOVA with Bonferroni posttest was used for statistical analysis. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (C) Hyphae and conidiophores of the wild-type and ΔmpkB strains grown for 48 h at 37°C. Bars, 10 µm. (D) brlA mRNA steady-state levels detected in the wild-type and ΔmpkB strains grown in MM at 37°C for 24 and 48 h, respectively. The results represent the averages of three repetitions ± standard deviations and are expressed as the cDNA concentration of brlA divided by the cDNA concentration of the β-tubulin gene; an asterisk represents a P value of <0.05.
DHN-melanin production is normally associated with conidiation. We also investigated whether the high melanin accumulation was accompanied by deregulated production of conidiophores. Indeed, in the ΔmpkB mutant, conidiophores and conidia were produced and released in liquid MM (Fig. 2C). This finding shows that the accumulation of the pigment in the medium was due to the secretion of melanin by the ΔmpkB mutant strain.
It was previously reported that the transcription factor BrlA is important in activating the development of conidiation in A. fumigatus (38). Expression studies showed that the brlA transcript is expressed 10 to 15 times more in liquid MM in the ΔmpkB mutant than in the wild-type strain (Fig. 2D). These results indicate that MpkB plays an important role in the prevention of conidium formation during active vegetative growth.
Gα protein GpaA and G protein-coupled receptor GprM are important for repressing DHN-melanin production.
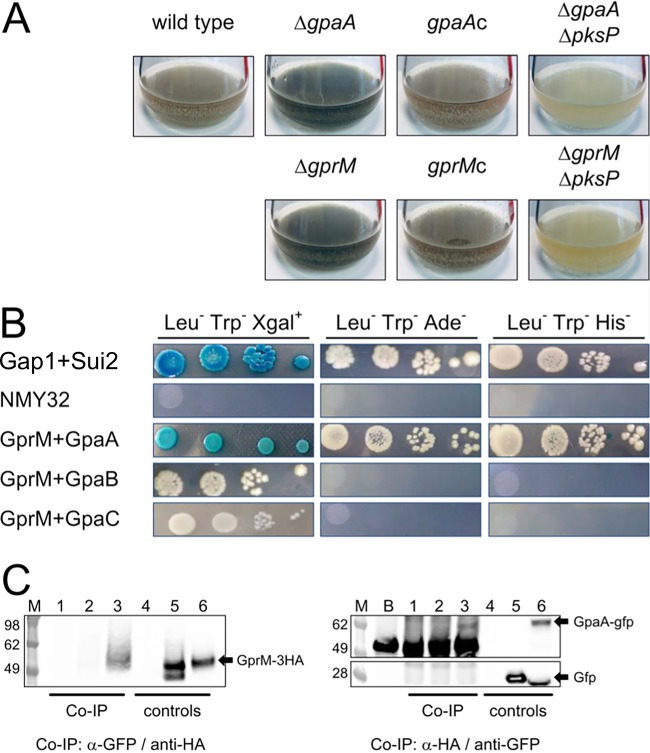
It has been previously reported that pksP transcription is connected to cAMP signaling via protein kinase A (PKA) (34). In order to identify further pathways involved in DHN-melanin regulation, we exploited the dark-color production as a readout system to identify additional signal proteins. Previous studies reported that the deletion of gpaB, coding for a Gα protein involved in the cAMP/PKA pathway, strongly decreased DHN-melanin production (39). Besides GpaB, A. fumigatus has two additional Gα proteins, GpaA and GpaC (40). Phenotypic analysis of mutant strains with deletions of the three Gα proteins revealed that only the ΔgpaA mutant showed increased pigment production in submerged culture (Fig. 3A). Parallel deletion of the pksP gene in the ΔgpaA mutant background confirmed that, in this strain also, the dark pigment was DHN-melanin (Fig. 3A).
FIG 3.
The Gα protein GpaA and the G protein-coupled receptor GprM are important for repressing melanin production. (A) A. fumigatus conidia from the reported strains were inoculated into liquid MM for 72 h at 37°C. (B) Split-ubiquitin yeast two-hybrid screen with A. fumigatus GprM as the bait and the three Gα proteins as preys. Two other yeast proteins, Gap1 and Sui2, previously shown as interacting by using a split-ubiquitin yeast two-hybrid screen, were used as positive controls (41). (C) Coimmunoprecipitation (co-IP) was performed with total protein extracted from the A. fumigatus wild-type strain (lanes 1 and 4), A. fumigatus expressing a dicistronic gene including the gfp- and the HA-tagged gprM (lanes 2 and 5), and A. fumigatus expressing a dicistronic gene including the gfp-tagged gpaA and the HA-tagged gprM (lanes 3 and 6). Co-IP performed with an anti-GFP antibody specifically isolates the GprM-3×HA protein only in the presence of the GFP-tagged GpaA (left, lane 3). Similarly, co-IP performed using an anti-HA antibody specifically isolates the GpaA-GFP protein, but not GFP alone (right, lane 3). The experiment also shows that the anti-GFP antibody used recognizes an unspecific band detectable also when the beads are not attached to any protein (right, lane B). However, because of the difference in size, a clear band showing the expressed GpaA-GFP protein was detected (right, lane 3). Lanes M, SeeBlue plus2 marker (ThermoFisher).
In eukaryotes, Gα proteins are normally associated with receptors, and in particular G protein-coupled receptors (GPCRs). We screened a comprehensive collection that included 12 A. fumigatus GPCR null mutants (Table 1). Spores for each mutant were inoculated into liquid medium at 37°C for 3 days with shaking (data not shown). Among the screened mutants, only the ΔgprM mutant produced more dark pigments than the wild type, and in this case also, ΔgprM complementation and the parallel deletion of the pksP gene confirmed that the phenotype was connected to DHN-melanin (Fig. 3A).
TABLE 1.
A. fumigatus strains used in this study
| Description | Target gene | Relevant genotypea | Reference or source |
|---|---|---|---|
| Wild type | CEA10 | 62 | |
| ΔakuB (recipient strain) | AFUB_019720 | CEA17 akuB::pyrG; PyrG+ | 63 |
| ΔmpkA | AFUB_070630 | ΔakuB mpkA::ptrA; PTR | 36 |
| mpkAc (complemented ΔmpkA strain) | ΔakuB ΔmpkA ΔmpkA::mpkA-hph; HygR PTR | 36 | |
| ΔmpkB | AFUB_078810 | ΔakuB mpkB::ptrA; PTR | This study |
| ΔmpkB::mpkB+ | ΔakuB ΔmpkB ΔmpkB::mpkB-hph; HygR | This study | |
| ΔpksP | AFUB_033290 | ΔakuB pksP::hph; HygR | 35 |
| ΔmpkB ΔpksP | ΔakuB ΔmpkB pksP::hph; HygR PTR | This study | |
| ΔmpkA ΔmpkB | ΔakuB ΔmpkB mpkA::ptrA; HygR PTR | This study | |
| ΔgpaA | AFUB_012620 | ΔakuB gpaA::ptrA; PTR | This study |
| gpaAc | ΔakuB ΔgpaA gpaA-hph; HygR PTR | This study | |
| ΔgpaA ΔpksP | ΔakuB ΔgpaA pksP::hph; HygR PTR | This study | |
| ΔgpaB | AFUB_012410 | ATCC 46645 gpaB::hph; HygR | 39 |
| ΔgpaC | AFUB_036760 | ΔakuB gpaC::ptrA; PTR | This study |
| ΔgprM | AFUB_090880 | ΔakuB gprM::ptrA; PTR | This study |
| gprMc | ΔakuB ΔgprM ΔgprM::gprM-hph; HygR | This study | |
| ΔgprM ΔpksP | ΔakuB ΔgprM pksP::hph; HygR PTR | This study | |
| mpkB-gfp | ΔakuB mpkB::mpkB-gfp-hph; HygR | This study | |
| ΔmpkA mpkB-gfp | ΔakuB mpkB-gfp mpkA::ptrA; HygR PTR | This study | |
| gpaA-gfp gprM-3×HA | CEA10 gpaA-gfp gprM-3×HA; PTR | This study | |
| gfp gprM-3×HA | CEA10 gfp gprM-3×HA; PTR | This study | |
| ΔhmgA | AFUB_021290 | ΔakuB hmgA::ptrA; PTR | 32 |
| ΔhppD | AFUB_021270 | ΔakuB hppD::ptrA; PTR | 32 |
| ΔgprA | AFUB_034900 | ΔakuB gprA::hph; HygR | This study |
| ΔgprB | AFUB_055410 | ΔakuB gprB::hph; HygR | This study |
| ΔgprC | AFUB_090350 | ΔakuB gprC::hph; HygR | This study |
| ΔgprD | AFUB_028290 | ΔakuB gprD::hph; HygR | This study |
| ΔgprH | AFUB_052640 | ΔakuB gprH::hph; HygR | This study |
| ΔgprI | AFUB_047630 | ΔakuB gprI::hph; HygR | This study |
| ΔgprJ | AFUB_007220 | ΔakuB gprJ::hph; HygR | This study |
| ΔgprK | AFUB_101830 | ΔakuB gprK::hph; HygR | This study |
| ΔgprO | AFUB_038590 | ΔakuB gprO::hph; HygR | This study |
| ΔgprP | AFUB_073100 | ΔakuB gprP::hph; HygR | This study |
| ΔnopA | AFUB_088000 | ΔakuB nopA::hph; HygR | This study |
HygR, hygromycin; PTR, pyrithiamine.
In order to check whether the GprM receptor and GpaA interact directly, we employed a split-ubiquitin yeast assay two-hybrid screen with A. fumigatus GprM as bait and the three Gα proteins as preys. This experiment clearly demonstrated that among the Gα proteins expressed by A. fumigatus, GprM interacts exclusively with GpaA (Fig. 3B). Two other yeast proteins, S. cerevisiae Gap1 and Sui2, previously shown to interact by using a split-ubiquitin yeast assay two-hybrid screen, were used as positive controls (41).
Finally, the interaction between the GprM receptor and GpaA was validated in A. fumigatus. We expressed a green fluorescent protein (GFP)-tagged gpaA gene with a hemagglutinin (HA)-tagged gprM as a dicistronic gene. The two fragments were spaced by the 2A peptide sequence and then placed downstream from a tetracycline-inducible promoter, TetON, optimized for fungal expression (42, 43). The plasmid obtained was used to transform A. fumigatus strain CEA10 by ectopic integration. By using anti-GFP antibody for the coimmunoprecipitation (co-IP) experiment, we could also isolate the expressed GprM-3×HA receptor (Fig. 3C, left). The opposite experiment, co-IP using the anti-HA antibody, gave the same results, with the GpaA-GFP protein identified by Western blotting (Fig. 3C, right). It is worth noticing that the anti-GFP antibody also recognized unspecific bands (∼50 kDa) from the beads; however, because of the difference in size compared to that of the GpaA-GFP protein, a clear band was still discernible.
Cross-talk interaction between the MpkB and MpkA pathways.
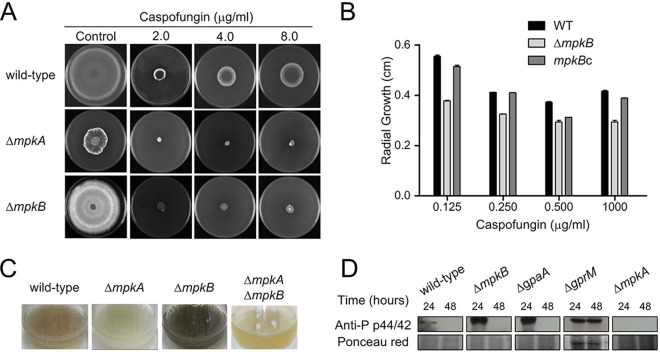
The screening conducted against the different cell wall-acting compounds revealed that the ΔmpkB mutant is more sensitive to the β-1,3-glucan synthase inhibitor caspofungin, with significant reduction of growth when compared to that of the recipient strain (Fig. 4A and B and Fig. S1). The ΔmpkB mutant had also lost the caspofungin paradoxical effect, a phenomenon where high caspofungin concentrations revert the anticipated inhibition of A. fumigatus growth (Fig. 4A and B) (44). This sensitivity was almost comparable with that observed for the ΔmpkA mutant; MpkA is the second p42/44 kinase encoded by A. fumigatus and mainly responsible for cell wall maintenance under stress conditions (45). Concerning MpkA, previously published transcriptomic studies reported that this kinase positively affects the expression of the DHN-melanin gene cluster (22, 46). These observations had not yet been proved by ad hoc experiments. We constructed an ΔmpkA ΔmpkB double deletion strain aiming to investigate possible interactions between the two kinases. As shown by the results in Fig. 4C, pigmentation of the medium was not observed for either the ΔmpkA or the ΔmpkA ΔmpkB mutant, suggesting that MpkA positively regulates DHN-melanin biosynthesis while MpkB negatively affects its production.
FIG 4.
Cross-talk interaction between MpkB and MpkA. (A) The wild-type, ΔmpkA, and ΔmpkB strains were grown for 5 days at 37°C in the presence of different caspofungin concentrations. (B) Growth defects in the ΔmpkB mutant were detected by measuring the radial diameter upon incubation with caspofungin and comparing with those of the wild type and the mpkBc complemented strain. Results are expressed as the average radial diameters of three repetitions ± standard deviations. Statistical analysis was performed by using two-way ANOVA with Bonferroni posttest. *, P < 0.001. (C) A. fumigatus conidia were inoculated into liquid MM for 72 h at 37°C with shaking at 200 rpm. (D) Immunoblot analyses for MpkA phosphorylation. The wild-type and the mutant strains were grown in MM for 24 and 48 h at 37°C. Anti-p44/42 MpkA antibody was used to detect the phosphorylation levels of MpkA. Ponceau red was used as a control for loading.
To investigate whether MpkB repression affects MpkA activation, we checked the MpkA phosphorylation levels in the mutants by immunoblot assay. We detected higher MpkA phosphorylation during the first 24 h of incubation for the ΔmpkB and ΔgpaA strains than for the wild type, and in the ΔgprM mutant, MpkA was shown to be highly activated even 48 h after incubation (Fig. 4D). These data confirmed that GpaA and GprM act upstream from MpkB and that the inhibition of this pathway increased MpkA phosphorylation, leading to higher DHN-melanin accumulation. The mechanism is still unknown; however, an indirect interaction is most likely, since binding between MpkA and MpkB was not detected by a coimmunoprecipitation assay performed with the two kinases (Fig. S3).
Coimmunoprecipitation assay performed with tagged MpkA and MpkB. (A) Diagnostic PCR to validate the in-locus homologous insertion of the mpkA-3×HA construct in the mpkB-gfp strain. (B) Phenotypic analysis of wild type, mpkB-gfp, and mpkB-gfp mpkA-3×HA strains grown on MM agar plates for 4 days at 37°C. (C, D) Affinity purification assays for 3×HA-tagged MpkA and GFP-tagged MpkB were performed with GFP-Trap beads (C) and anti-HA beads (D) to verify interactions. The coimmunoprecipitated proteins were analyzed using the indicated antibodies. The coimmunoprecipitation protocol used is reported in Text S1. Download FIG S3, PDF file, 0.1 MB (109.6KB, pdf) .
Copyright © 2019 Manfiolli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
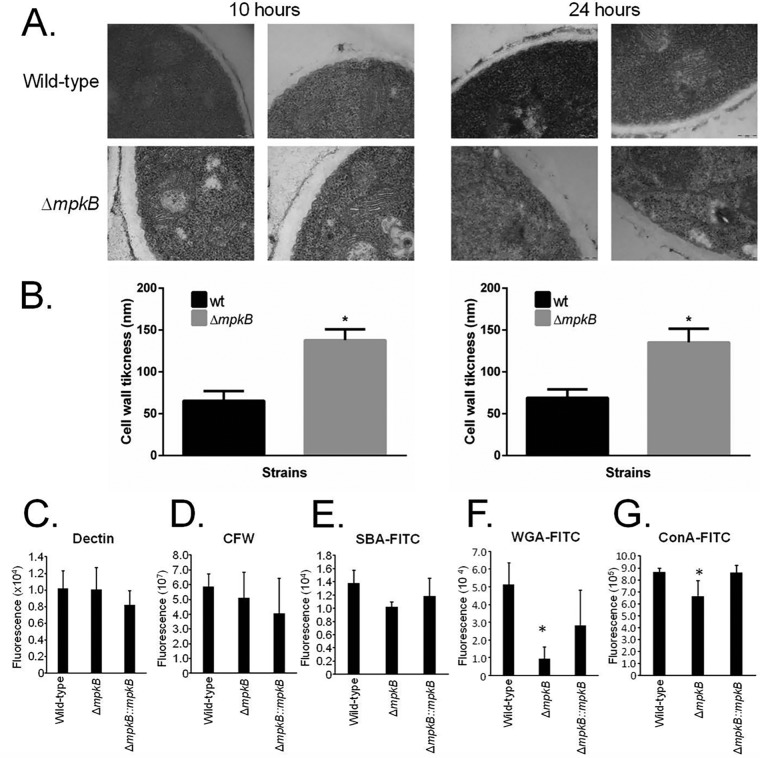
Transmission electron microscopy (TEM) analysis showed that untreated ΔmpkB germlings (10 and 24 h) grown in MM had approximately 2-fold-thicker cell walls than the wild type (Fig. 5A and B). Cell wall stains and lectins were used to identify differences in the content or exposure of different carbohydrates on the surface of the fungal cell wall in both the wild type and the ΔmpkB mutant. These included the following: (i) soybean agglutinin (SBA)-fluorescein isothiocyanate (FITC) (preferentially binds to oligosaccharide structures with terminal α- or β-linked N-acetylgalactosamine [GalNAc] and, to a lesser extent, galactose residues, which are important for recognizing galactosaminogalactan [GAG]); (ii) wheat germ agglutinin (WGA)-FITC (recognizing surface-exposed glucosamine [Glc]); (iii) ConA (concanavalin A)-FITC (recognizes α-linked mannose); (iv) soluble dectin-1 stain (recognizing β-glucans); and (v) CFW (recognizing chitin). There were no differences in the levels of β-1,3-glucan, chitin, or N-acetylgalactosamine in the wild-type, ΔmpkB, and complemented strains (Fig. 5C to E and G). In contrast, the ΔmpkB mutant had about 3-fold-less glucosamine and a lower level of α-linked mannose than the wild-type strain (Fig. 5F).
FIG 5.
The ΔmpkB mutant strain has an altered cell wall organization. (A) Transmission electron microscopy of mycelial sections of the A. fumigatus wild-type and ΔmpkB strains grown for 10 h or 24 h in MM at 37°C. (B) The cell wall thicknesses (nm) of 100 sections of different hyphal germlings (average of 4 sections per germling) were measured when grown under the same conditions as specified in the legend to panel A. The averages and standard deviations of the 50 measurements are presented. Statistical analysis was performed using the one-tailed, paired t test, comparing data to the results for the control condition (*, P < 0.00001). (C to G) Detection of different sugars exposed on the cell surface. Conidia were cultured in liquid MM to the hyphal stage, UV killed, and stained with CFW, soluble dectin-1, or specific probe to detect the content of exposed sugars. Experiments were performed in triplicate, and the results are displayed as mean values with standard errors (two-way ANOVA followed by Tukey’s; *, P < 0.05).
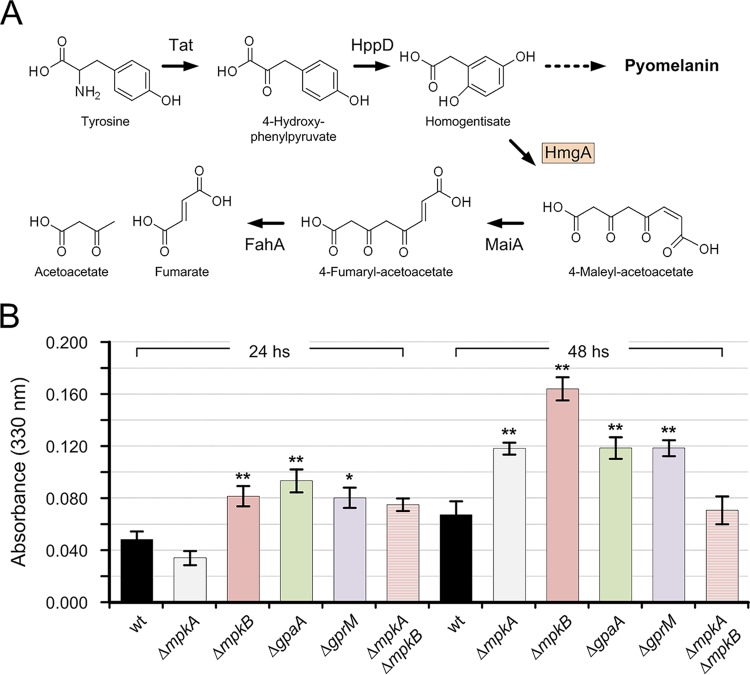
Finally, we investigated whether the misbalance in MpkA phosphorylation caused any effect on the production of pyomelanin. As previously reported, the cell wall stress induced by the deletion of mpkA promotes the formation of pyomelanin as a compensatory response (36). The increase in pyomelanin formation can be detected by testing the activity of the homogentisate 1,2-dioxygenase (HmgA) involved in tyrosine degradation (32). After adding l-tyrosine to the medium, a reduction in HmgA activity induced higher pyomelanin accumulation, and vice versa (Fig. 6A). By growing the mutants in liquid MM plus l-tyrosine for 24 h, we could not detect a significant change in HmgA activity for the mpkA deletion mutants. However, impairing MpkB signaling resulted in higher HmgA enzymatic activities for the ΔgprM, ΔgpaA, and ΔmpkB deletion mutants (Fig. 6B). This effect was conserved during the incubation time and was still detectable after 48 h. The mutant with the deletion of mpkA showed an increase of HmgA activity after 48 h of incubation, which is not in line with our previous results (36). For comparison to the results of the earlier activity test, we performed the HmgA activity test using smaller volumes (each sample had a total volume of 100 μl and was disposed in a 96-well plate) and reading the absorbance using a plate reader. However, aside from this incongruence, the double deletion of mpkA and mpkB restored the HmgA activity level, confirming the involvement of the cell wall integrity pathway in pyomelanin formation and highlighting once again the conflicting roles of the MpkA and MpkB pathways.
FIG 6.
Homogentisate dioxygenase (HmgA) activity assay. (A) Tyrosine degradation pathway showing pyomelanin production due to either an increase or decrease in HgmA (homogentisate dioxygenase) activity (Tat, tyrosine aminotransferase; HppD, 4-hydroxyphenylpyruvate dioxygenase; MaiA, 4-maleylacetoacetate; FahA, fumarylacetoacetate hydrolase). (B) Total proteins extracted from the wild type and ΔmpkA, ΔmpkB, ΔgpaA, ΔgprM, and ΔmpkA ΔmpkB mutant strains were assayed for HmgA activity. After 18 h of growth, 10 mM l-tyrosine was added to the culture. Samples were harvested, and the protein extracts were incubated with homogentisate for 35 min. Formation of maleylacetoacetate was monitored at 330 nm. Total protein extracted from the ΔhmgA and ΔhppD mutant strains was used as the negative and positive control, respectively (data not shown). HmgA activity was determined in protein extracts of mycelia harvested after 24 and 48 h postinoculation (average value ± standard deviation). Statistical significance was determined for the results of all of the experiments by Student’s t test. Significance of differences of data in comparison to the results for the wild-type strain is indicated as follows: *, P < 0.1; **, P < 0.01.
Cell wall stress enhances MpkB nuclear localization.
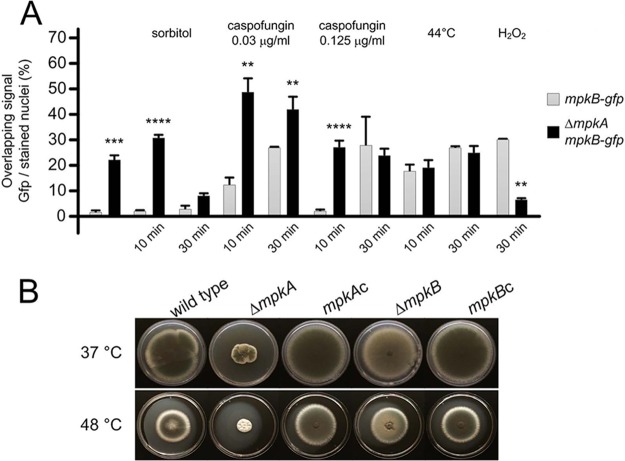
To assess the involvement and subcellular localization of MpkB during growth and under stress conditions, we generated a strain expressing MpkB-GFP under the control of its own endogenous promoter. This was achieved by replacing the wild-type allele with the mpkB-gfp cassette; the strain generated behaved identically to the wild-type strain (Fig. S3). When the MpkB-GFP strain was grown in MM for 16 h at 30°C, a very weak and diffuse fluorescent signal was observed in the cytosol and about 1% of the germlings’ stained nuclei overlapped with the GFP signal (Fig. 7A). The increase of MpkB nuclear localization was observed upon mpkA deletion, and this condition was similar when the growing mycelium was challenged with the osmotic stress inducer sorbitol. Moreover, the exposure to different caspofungin concentrations increased nuclear localization over time, with MpkB localized in almost one-third of the germlings 30 min after incubation. As for sorbitol, this phenomenon was strongly enhanced by mpkA deletion. These data confirmed that MpkB can activate compensatory pathways in the absence of MpkA during cell wall stress.
FIG 7.
The influence of MpkA on the MpkB-GFP nuclear translocation. (A) Conidia were grown for 16 h at 30°C before stress induction. The compounds used and the times of the induced stresses are also reported. The data represent the averages of three repetitions ± standard deviations. The nuclei of about 50 germlings were counted in each repetition (*, P < 0.05; **, P < 0.005; ***, P < 0.001; ****, P < 0.0001). Hoechst counterstaining was used to confirm the nuclear localization of the GFP signal. (B) The wild-type, ΔmpkA, and ΔmpkB strains were grown on MM for 5 days at 37 and 44°C, respectively.
Cell wall-impaired mutants are normally sensitive to heat shock (36). For the ΔmpkB strain, incubation at high temperatures affected the sporulation ability more than it did radial growth (Fig. 7B). MpkB nuclear localization was increased by incubating A. fumigatus at 44°C (Fig. 7A); however, the parallel deletion of mpkA did not affect MpkB activation, demonstrating that the interaction of the two pathways is stress related. This was even clearer when A. fumigatus was challenged with H2O2. As previously reported, hydrogen peroxide strongly induces MpkA activation, and the ΔmpkA mutant is very susceptible to oxidative stress (47). Here, we observed that H2O2 enhanced MpkB localization to the nucleus as well, but the lack of mpkA significantly decreased MpkB nuclear transport. It is possible that an additional salvage pathway that is not activated by MpkB operates during oxidative stress. This observation is supported by the fact that mpkB deletion did not affect H2O2 sensitivity (Fig. S2), suggesting that during oxidative stress, MpkB is transported into the nuclei but is likely still inactive.
A. fumigatus ΔmpkB is still virulent in a low-dose murine infection model.
In order to determine whether the lack of mpkB affects A. fumigatus virulence, the ΔmpkB strain, the mpkBc (complemented) strain, and the respective wild-type strain were tested in a murine infection model of IA (Fig. S4). The results of this experiment indicated that mpkB is not essential for virulence of A. fumigatus.
(A) Survival rates of mice infected with conidia from wild-type, ΔmpkB, and mpkBc strains. Mice in groups of 10 per strain were infected intranasally with a 20-μl suspension containing 2 × 105 conidia. (B) Histopathology of lung tissue from mice infected with the wild-type, ΔmpkB, and mpkBc strains plus the PBS control using periodic acid-Schiff (PAS) staining. All infected mice displayed normal fungal persistence compared to the uninfected mice. Download FIG S4, PDF file, 0.2 MB (158.9KB, pdf) .
Copyright © 2019 Manfiolli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Ethics statement.
The principles that guide our studies are based on the Declaration of Animal Rights ratified by the UNESCO in January 27, 1978, in its eighth and 14th articles. All protocols used in this study were approved by the local ethics committee for animal experiments from the Campus of Ribeirão Preto, Universidade de São Paulo (Permit Number: 08.1.1277.53.6; Studies on the interaction of A. fumigatus with animals). All animals were housed in groups of five within individually ventilated cages and were cared for in strict accordance with the principles outlined by the Brazilian College of Animal Experimentation (Princípios Eticos na Experimentacão Animal—Colégio Brasileiro de Experimentacão Animal, COBEA) and Guiding Principles for Research Involving Animals and Human Beings, American Physiological Society. All efforts were made to minimize suffering. Animals were clinically monitored at least twice daily and humanely sacrificed if moribund (defined by lethargy, dyspnea, hypothermia, and weight loss). All stressed animals were sacrificed by cervical dislocation.
DISCUSSION
A number of studies have characterized MAPKs with a focus on their role in pathogenesis and secondary metabolite production (9, 10, 48, 49). The pathogenic fungus A. fumigatus codes for four MAPKs, and until now, MpkB was the only one still uncharacterized. As shown by the experiments whose results are described here, the deletion of the A. fumigatus mpkB gene reduced conidium production on solid medium but induced the formation of conidiophores in submerged cultures. In A. nidulans, MpkB was found to be essential for sexual development, and the ΔmpkB mutant produced conidia in submerged cultures with constant brlA mRNA accumulation (50, 51). We also observed increased accumulation ofbrlA during growth of the A. fumigatus ΔmpkB mutant in submerged cultures, indicating that in both fungal systems, MpkB is important for temporal brlA expression. However, other than its role in spore development, MpkB had no impact on A. fumigatus virulence, since the ΔmpkB mutant showed wild-type-like virulence in a murine infection model. Previously, Atoui et al. (26) have shown that in the A. nidulans ΔmpkB mutant, the expression of genes involved in sterigmatocystin, penicillin, and terrequinone A biosynthesis, as well as the expression of laeA (loss of aflatoxin expression A, encodes global regulator of secondary metabolism), were significantly reduced (26). Similarly, MpkB also affects secondary metabolite production in A. fumigatus, in particular DHN-melanin, but surprisingly, does not confer resistance to oxidative stress agents. As shown herein, the MpkB-GFP translocation to the nucleus is affected by the presence of MpkA during cell wall stress. The negative interaction between MpkA and MpkB seems to be mutual in particular during cell wall stress; indeed, the deletion of mpkB increased MpkA phosphorylation, while the deletion of mpkA increased MpkB nuclear localization. Since MpkA phosphorylation was increased in the ΔmpkB mutant, the higher melanin production seems to be related to aberrant MpkA activation.
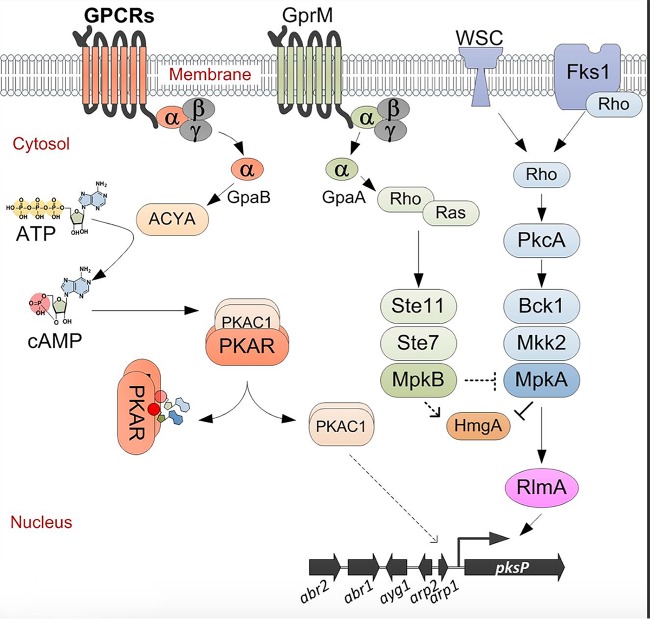
The involvement of the cell wall integrity signaling pathway in melanin production has already been hypothesized. The analysis of transcriptome data of ΔmpkA mutants and the involvement of the cell wall-related transcription factor RlmA in the DHN-melanin gene cluster activation already suggested that, besides the cAMP pathway, additional signal pathways are involved in melanin regulation (35, 46, 52). Taking into account that melanin production is lower in the ΔmpkA ΔmpkB double mutant than in the single mpkB deletion strain, this finding supports a model in which the expression of the DHN-melanin biosynthesis genes is controlled by the RlmA transcription factor (Fig. 8).
FIG 8.
Proposed model for the induction of DHN-melanin biosynthesis. The cAMP pathway activates the melanin cluster via protein kinase catalytic subunit 1 (PkaC1), as previously reported (39, 53). The GPCR GprM, which interacts with GpaA, activates the MpkB pathway. MpkB activation affects MpkA, which regulates the DHN-melanin gene cluster. Finally, the transcription factor RlmA directly regulates the expression of the melanin genes by directly binding the pksP promoter (35). Parallel to the expression of the DHN-melanin gene cluster, the activation of MpkB influences the HmgA activity, promoting the accumulation of pyomelanin. ACYA, adenylate cyclase; PKAR, PKA regulatory subunit.
Thus far, the cAMP/PKA pathway was the only regulatory signaling cascade associated with DHN-melanin production in A. fumigatus (34, 53). Null mutants with deletions of genes involved in this pathway, such as the G protein α subunit GpaB and adenylate cyclase AcyA, showed reductions in DHN-melanin production, while overexpression of the gene encoding the protein kinase A catalytic subunit 1 (PkaC1) induced the expression of the DHN-melanin cluster genes (Fig. 8) (27). This was also observed in other pathogenic fungi, such as C. neoformans, where mutations in the G protein Gpa1, adenylate cyclase Cac1, and/or the catalytic subunit of PKA Pka1 reduced the formation of melanins (54, 55).
Here, we report that an additional G protein α subunit (GpaA) and the GPCR GprM are also involved in the regulation of DHN-melanin production (Fig. 8). Accordingly, GprM physically interacts with GpaA, suggesting that melanin production in A. fumigatus is also mediated via the GprM/GpaA/MpkB pathway. Genome-wide analysis revealed that the A. fumigatus genome codes for three G protein α subunits, one G-β, and one G-γ, while it codes for at least 15 different GPCRs (53, 56). So far, gprD and gprC have been characterized in A. fumigatus, and their role was connected to hyphal morphogenesis and virulence (57). The high number of GPCRs encoded by fungal genomes makes their study difficult, because the majority of GPCR gene single deletion mutants do not show a discernible phenotype (data not shown). This was also experienced by studying a comprehensive Aspergillus flavus GPCR mutant library, where only a few mutants showed growth defects or impaired toxin production (58). Therefore, until now, there have only been a few examples of filamentous fungi for which the characterization of a signal pathway from the receptor to the activation of response genes has been reported.
In conclusion, here we report the first characterization of MpkB in A. fumigatus, showing its role in conidium formation and development and in the regulation of DHN-melanin production. Additionally, we demonstrate that MpkB interacts with the MpkA signaling pathway, particularly during the caspofungin stress response and melanin production. How this global cross talk affects transcriptional regulators needs to be elucidated, but in the future, the expression of DHN-melanin biosynthesis genes, with its known transcription factors, can be used to address these questions.
MATERIALS AND METHODS
Strains and media.
The A. fumigatus strains used in this study are described in Table 1. Detailed information about plasmids and the generation of mutant strains is reported in Text S1 and Fig. S5 in the supplemental material. Complete media were of the following two basic types: a complete yeast extract-glucose (YAG) medium (2% [wt/vol] glucose, 0.5% [wt/vol] yeast extract, 2% [wt/vol] agar, trace elements) with three variants (a YUU medium [YAG medium supplemented with 1.2 g/liter each of uracil and uridine] and liquid YG or YUU medium of the same composition but without agar) and a minimal medium (MM; 1% [wt/vol] glucose, original high nitrate salts, trace elements, 2% [wt/vol] agar, pH 6.5). The trace elements, vitamins, and nitrate salts used were described previously (59). S. cerevisiae strains were grown in synthetic dextrose (SD) medium (6.7 g Difco yeast nitrogen base [YNB] without amino acids, with distilled water to 950 ml, 20 g agar, and 50 ml sterile 40% [wt/vol] glucose).
Methods related to the murine infection model, plasmid construction, DNA manipulation, and Southern blot analysis are reported. Download Text S1, PDF file, 0.1 MB (148.2KB, pdf) .
Copyright © 2019 Manfiolli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Graphical representation of the mutated loci for the A. fumigatus transformant strains analyzed and the respective Southern blots. The genes of interest are shown in blue, their flanking genes are grey. The cassettes for resistance to pyrithiamine (ptrA) and hygromycin (hph) are labeled in green and pink, respectively. The probes used for Southern blot analyses are marked as red stripes. The DNA ladder used is indicated (Hyperladder 1 kb; Bioline). (A) The mpkB gene was deleted, and the mutant strain complemented with the native gene by ectopic integration. (B) The ΔmpkB strain was complemented by homologous recombination with the native mpkB gene fused to a gfp sequence (in yellow) and an nos terminator sequence (in black). (C) Deletion of the mpkA gene in the mpkB-gfp background and in the ΔmpkB strain. Southern blots were performed separately. The probe also anneals to an additional ∼2-kb band. (D) Deletion and complementation of the gprM locus; the complementation was obtained by homologous recombination. (E) Deletion and complementation of the gpaA locus. The complementation was performed by ectopic integration of the gpaA wild-type gene. (F) Disruption of the gpaC gene. (G) Disruption of the pksP gene in the wild-type, ΔmpkB, ΔgpaA, and ΔgprM strains. (H) The dicistronic genes including the gpaA-gfp gprM-3×HA and the gfp gprM-3×HA sequences were ectopically integrated into the A. fumigatus CEA10 genome. Positive transformants were validated by Western blot analysis using anti-GFP antibodies (left) and anti-HA antibodies (right). The predicted molecular weights of the gene products are also reported. Download FIG S5, PDF file, 0.4 MB (386.6KB, pdf) .
Copyright © 2019 Manfiolli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Conidium production.
Five microliters of 2 × 107 conidia of each strain was overlaid onto MM agar plates, followed by incubation at 37°C for 5 days. Fresh conidia were harvested in 10 ml of 0.01% (vol/vol) Tween 80, vortexed for 30 s, and filtered through miracloth. Conidial suspensions were spun for 5 min at 3,000 × g, and pellets were resuspended using 10 ml H2O. The numbers of conidia were counted using a hemacytometer. This experiment was performed in triplicate.
Phenotypic assays.
The phenotypes of the deletion mutants were evaluated either by measuring radial growth or by assessing the initial growth of a droplet of conidia from a serial dilution at different temperatures and in the presence or absence of oxidative and osmotic stressing agents. Dropout experiments were performed using 5-µl amounts of a 10-fold dilution series starting at a concentration of 2 × 107 for the wild-type and mutant strains spotted on different growth media and grown for 48 h at 37°C. The A. fumigatus crystal violet assay was performed as previously described (60, 61). All the experiments were performed at least three times. We also screened a collection of 12 GPCR mutants (Table 1) for the production of dark pigments by growing them in liquid MM for 48 h at 37°C.
Split-ubiquitin yeast two-hybrid assay.
The A. fumigatus gprM, gpaA, gpaB, and gpaC open reading frames were synthesized and cloned into the pTMBV4 and pDL2XN plasmids, respectively (41). The genes of interest were expressed in Saccharomyces cerevisiae strain NMY32. The NMY32 strain was initially transformed with the gprM-bearing plasmid and later with one of the gpaA-, gpaB-, or gpaC-bearing plasmids. Then, we used these transformants for the split-ubiquitin membrane-based yeast two-hybrid assay. First, these strains were inoculated onto SD-Leu-Trp selective culture medium and grown overnight. The absorbance of the inoculum was measured through spectrophotometry, and later, all samples were standardized to an optical density at 600 nm (OD600) of 1. Tenfold serial dilutions were made, and these dilutions were spotted on agar plates with different selective culture media, including SD-Leu-Trp plus X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside), SD-Leu-Trp-Ade, SD-Leu-Trp-His, SD-Leu-Trp-His plus 3-AT (3-amino-1,2,4-triazole), or SD-Leu-Trp-Ade-His medium. The agar plates were incubated for 3 days at 30°C.
Staining and microscopy.
The copper-silver staining experiments were performed by a modification of a previously described procedure (37). Briefly, the wild-type and ΔmpkB mutant strains were grown on solid MM on slides. After growth, the slide cultures were incubated in 10 mM copper sulfate solution at room temperature for 1 h and, after washing, treated with 1.0% (wt/vol) sodium sulfide solution for 1 h in the dark. Subsequently, the slide cultures were incubated for 4 min in a solution of 22 mg silver lactate and 170 mg hydroquinone (HQ) in a citrate buffer (0.1 M, pH 3.7) solution (Ag-HQ solution) at room temperature. The copper and sulfide treatments were followed by repeated washings (six times with 2 ml distilled water). Afterward, slides were visualized on a Zeiss Axio Observer Z1 fluorescence microscope. Bright-field images were captured with an AxioCam camera (Carl Zeiss) and processed using AxioVision software (version 4.8). For fluorescence experiments, MpkB-GFP strain conidia were cultivated on coverslips in 4 ml of YG medium for 16 h at 30°C. After incubation, subsets of coverslips with adherent germlings were left untreated or treated with 1.0 M sorbitol, different concentrations of H2O2, or 0.125 µg/ml caspofungin for different periods of time. Subsequently, the coverslips were rinsed with phosphate-buffered saline (PBS; 140 mM NaCl, 2 mM KCl, 10 mM NaHPO4, 1.8 mM KH2PO4, pH 7.4) and incubated for 3 min in a solution with Hoechst stain (Life Technologies) (12 μg/ml). After incubation with the dye, the coverslips were washed with PBS and mounted for examination. Slides were visualized on an Observer Z1 fluorescence microscope using a 100× objective oil immersion lens (for GFP, filter set 38 high efficiency [HE], excitation wavelengths of 450 to 490 nm, and emission wavelengths of 525/50 nm; for Hoechst stain, filter set 49, excitation wavelengths of 365 nm, and emission wavelengths of 420 to 470 nm). Differential interference contrast (DIC) images and fluorescence images were captured with an AxioCam camera and processed using AxioVision software (version 4.8).
Homogentisate dioxygenase assay.
A. fumigatus was grown in liquid MM at 37°C and 180 rpm shaking for 18 h. Afterwards, 10 mM l-tyrosine was added. Samples for the homogentisate dioxygenase assay were collected 6 and 30 h after induction (in total, 24 and 48 h of growth, respectively). The mycelia were harvested, washed with water, frozen in liquid nitrogen, and stored at −80°C overnight. For protein preparation, the mycelia were powdered in liquid nitrogen and resuspended in 50 mM KPO4, pH 7.0. After centrifugation at 16,000 × g, 4°C, for 15 min, the supernatants were collected and the protein concentrations were determined using the Coomassie plus protein assay (ThermoFisher Scientific) according to the manufacturer’s instructions. The homogentisate dioxygenase assay was performed as described previously (32), in a 9-well plate in a total volume of 100 µl with 2 µg of total protein for each sample. All measurements were done with three biological and three technical replicates. Proteins extracted from the A. fumigatus ΔhmgA and ΔhppD mutants were used as negative and positive controls, respectively (not shown) (32). All the measurements were performed using a ClarioSTAR plate reader (BMG Labtech).
Immunoblot analysis.
To assess the phosphorylation status of MpkA, freshly harvested conidia (1 × 107) of the wild-type and mutant strains were inoculated into 50 ml liquid MM at 37°C for 16 h with 180 rpm. Mycelia were ground in liquid nitrogen with pestle and mortar. Protein extraction was performed as previously described (36). Briefly, 0.5 ml lysis buffer containing 10% (vol/vol) glycerol, 50 mM Tris–HCl, pH 7.5, 1% (vol/vol) Triton X-100, 150 mM NaCl, 0.1% (wt/vol) SDS, 5 mM EDTA, 50 mM sodium fluoride, 5 mM sodium pyrophosphate, 50 mM β-glycerophosphate, 5 mM sodium orthovanadate, 1 mM phenylmethylsulfonyl fluoride (PMSF), and 1× cOmplete mini-protease inhibitor (Roche Applied Science) were added to the ground mycelium. Extracts were centrifuged at 20,000 × g for 40 min at 4°C. The supernatants were collected, and the protein concentrations were determined using the Bradford assay (Bio-Rad). Fifty micrograms of protein from each sample was resolved in a 12% (wt/vol) SDS–PAGE gel and transferred to polyvinylidene difluoride (PVDF) membranes (Merck Millipore). The phosphorylation state of MpkA was examined using anti-phospho-p44/42 MAPK antibody (Cell Signaling Technologies) following the manufacturer’s instructions using a 1:1,000 dilution in TBST buffer (137 mM NaCl, 20 mM Tris, 0.1% [vol/vol] Tween 20). Primary antibody was detected using a horseradish peroxidase (HRP)-conjugated secondary antibody raised in rabbit (Sigma). Chemoluminescent detection was achieved using the SuperSignal west pico chemiluminescent substrate (Thermo Scientific).
Coimmunoprecipitation of GpaA and GprM.
For co-IP studies of GpaA and GprM, C-terminally tagged GpaA-GFP and GprM-3×HA and GFP alone were constructed and gpaA-GFP::gprM-3×HA and GFP::gprM-3×HA strains generated. Amounts of 50 ml of MM were inoculated with 1 × 107 spores of the two mutant strains and the wild type and incubated at 37°C with shaking. After 16 h, 20 µg/ml doxycycline was added. Mycelia were harvested after an additional 4 h of incubation, flash-frozen, and stored at −80°C. For protein preparation, the mycelia were powdered in liquid nitrogen and resuspended in 500 µl Tris buffer (0.5 M NaCl, 0.1 M Tris, pH 8.0) including EDTA-free protease inhibitor cocktail (Roche), 1 mM AEBSF [4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride], and 1 mM EDTA. Samples were vortexed 3× for 10 s and afterwards centrifuged for 20 min at 4°C and 16,000 × g. The supernatant was removed, and protein concentration was determined using the Bradford assay (Pierce Coomassie-plus; Fisher Scientific). For reciprocal co-IP, protein A Dynabeads (Thermofisher Scientific) were incubated with anti-3×HA (Sigma) or anti-GFP B2 (SantaCruz Biotechnology) antibody for 30 min at room temperature. Before adding 100 µg of protein solution to 80 µl of beads for 3 h while shaking at 8°C, the beads were washed three times with resuspension buffer. After incubation, the beads were washed three times with resuspension buffer and resuspended in 20 µl buffer, and bound proteins were eluted by adding sample buffer and boiling at 95°C for 5 min. Afterwards, proteins were separated on X10 Bolt 4 to 12% Bis-Tris plus gels (Fisher Scientific) and transferred to a nitrocellulose membrane using an iBlot2 blotting system (ThermoFisher Scientific). GFP and GFP-tagged GpaA were detected using an HRP-linked anti-GFP B2 antibody (Santa Cruz Biotechnology) diluted 1:2,500 in blocking solution (5% [wt/vol] milk powder in TBST). 3×HA-tagged GprM was detected using the antibodies and dilutions described above. The antibodies were diluted in blocking solution. For controls for the anti-GFP antibody blotting, 20 µg of wild-type and GpaA-GFP::GprM-3×HA and 1 µg of GFP::GprM-3×HA protein preparations were used. For controls for the anti-3×HA antibody blotting, 2 µg of all protein preparations were used. Detection was performed using WesternSure premium chemiluminescent substrate (Li-Cor GmbH) and a Fusion system (Vilber Lourmat).
Primers used in this study. Download Table S1, PDF file, 0.1 MB (152.4KB, pdf) .
Copyright © 2019 Manfiolli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Plasmids used in this study. Download Table S2, PDF file, 0.1 MB (107.6KB, pdf) .
Copyright © 2019 Manfiolli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ACKNOWLEDGMENTS
We thank Daniela Hildebrandt and Carmen Schult for excellent technical assistance.
We thank São Paulo Research Foundation (FAPESP) for grants number 2016/07870-9 (G.H.G.), 2014/24951-7 (A.O.M.), and 2014/00789-6 (L.J.D.A.), as well as Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) (G.H.G.), both from Brazil. We thank the Leibniz Research Cluster (LRC), the Fund for Scientific Research Flanders (FWO), the Deutsche Forschungsgemeinschaft (DFG)-funded collaborative research center/Transregio (CRC/TR) 124 Human-pathogenic fungi and their human host–networks of interaction—FungiNet (project A1), the DFG-funded excellence graduate school Jena School for Communication, and the Federal Ministry of Research and Education (BMBF)-funded consortium InfectControl2020 for providing financial support.
None of the funding bodies had any role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
A.A.B., P.V.D., I.M., V.V., and G.H.G. designed the experiments and analyzed the data. A.O.M., F.S.S., T.F.D.R., S.S., S.H., M.F., M.S., L.J.D.A., T.H., M.C.R., and S.J. performed the experiments and analyzed the experimental data. G.H.G. and V.V. wrote the paper. All authors read and approved the final version of the paper.
The authors declare that they have no competing interests.
Footnotes
Citation Manfiolli AO, Siqueira FS, dos Reis TF, Van Dijck P, Schrevens S, Hoefgen S, Föge M, Straßburger M, de Assis LJ, Heinekamp T, Rocha MC, Janevska S, Brakhage AA, Malavazi I, Goldman GH, Valiante V. 2019. Mitogen-activated protein kinase cross-talk interaction modulates the production of melanins in Aspergillus fumigatus. mBio 10:e00215-19. https://doi.org/10.1128/mBio.00215-19.
REFERENCES
- 1.Brown GD, Denning DW, Gow NA, Levitz SM, Netea MG, White TC. 2012. Hidden killers: human fungal infections. Sci Transl Med 4:165rv113. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 2.Sugui JA, Kwon-Chung KJ, Juvvadi PR, Latgé J-P, Steinbach WJ. 2014. Aspergillus fumigatus and related species. Cold Spring Harb Perspect Med 5:a019786. doi: 10.1101/cshperspect.a019786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Voltersen V, Blango MG, Herrmann S, Schmidt F, Heinekamp T, Strassburger M, Krüger T, Bacher P, Lother J, Weiss E, Hünniger K, Liu H, Hortschansky P, Scheffold A, Löffler J, Krappmann S, Nietzsche S, Kurzai O, Einsele H, Kniemeyer O, Filler SG, Reichard U, Brakhage AA. 2018. Proteome analysis reveals the conidial surface protein CcpA essential for virulence of the pathogenic fungus Aspergillus fumigatus. mBio 9:e01557-18. doi: 10.1128/mBio.01557-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haas H. 2014. Fungal siderophore metabolism with a focus on Aspergillus fumigatus. Nat Prod Rep 31:1266–1276. doi: 10.1039/c4np00071d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hartmann T, Sasse C, Schedler A, Hasenberg M, Gunzer M, Krappmann S. 2011. Shaping the fungal adaptome–stress responses of Aspergillus fumigatus. Int J Med Microbiol 301:408–416. doi: 10.1016/j.ijmm.2011.04.008. [DOI] [PubMed] [Google Scholar]
- 6.Scharf DH, Heinekamp T, Remme N, Hortschansky P, Brakhage AA, Hertweck C. 2012. Biosynthesis and function of gliotoxin in Aspergillus fumigatus. Appl Microbiol Biotechnol 93:467–472. doi: 10.1007/s00253-011-3689-1. [DOI] [PubMed] [Google Scholar]
- 7.Heinekamp T, Thywißen A, Macheleidt J, Keller S, Valiante V, Brakhage AA. 2012. Aspergillus fumigatus melanins: interference with the host endocytosis pathway and impact on virulence. Front Microbiol 3:440. doi: 10.3389/fmicb.2012.00440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pearson G, Robinson F, Beers Gibson T, Xu BE, Karandikar M, Berman K, Cobb MH. 2001. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr Rev 22:153–183. doi: 10.1210/edrv.22.2.0428. [DOI] [PubMed] [Google Scholar]
- 9.Rispail N, Soanes DM, Ant C, Czajkowski R, Grunler A, Huguet R, Perez-Nadales E, Poli A, Sartorel E, Valiante V, Yang M, Beffa R, Brakhage AA, Gow NA, Kahmann R, Lebrun MH, Lenasi H, Perez-Martin J, Talbot NJ, Wendland J, Di Pietro A. 2009. Comparative genomics of MAP kinase and calcium-calcineurin signalling components in plant and human pathogenic fungi. Fungal Genet Biol 46:287–298. doi: 10.1016/j.fgb.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 10.Turra D, Segorbe D, Di Pietro A. 2014. Protein kinases in plant-pathogenic fungi: conserved regulators of infection. Annu Rev Phytopathol 52:267–288. doi: 10.1146/annurev-phyto-102313-050143. [DOI] [PubMed] [Google Scholar]
- 11.Hamel LP, Nicole MC, Duplessis S, Ellis BE. 2012. Mitogen-activated protein kinase signaling in plant-interacting fungi: distinct messages from conserved messengers. Plant Cell 24:1327–1351. doi: 10.1105/tpc.112.096156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levin DE. 2011. Regulation of cell wall biogenesis in Saccharomyces cerevisiae: the cell wall integrity signaling pathway. Genetics 189:1145–1175. doi: 10.1534/genetics.111.128264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brewster JL, Gustin MC. 2014. Hog1: 20 years of discovery and impact. Sci Signal 7:re7. doi: 10.1126/scisignal.2005458. [DOI] [PubMed] [Google Scholar]
- 14.Mok J, Zhu X, Snyder M. 2011. Dissecting phosphorylation networks: lessons learned from yeast. Expert Rev Proteomics 8:775–786. doi: 10.1586/epr.11.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saito H. 2010. Regulation of cross-talk in yeast MAPK signaling pathways. Curr Opin Microbiol 13:677–683. doi: 10.1016/j.mib.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 16.Correia I, Alonso-Monge R, Pla J. 2010. MAPK cell-cycle regulation in Saccharomyces cerevisiae and Candida albicans. Future Microbiol 5:1125–1141. doi: 10.2217/fmb.10.72. [DOI] [PubMed] [Google Scholar]
- 17.Valiante V, Macheleidt J, Foge M, Brakhage AA. 2015. The Aspergillus fumigatus cell wall integrity signaling pathway: drug target, compensatory pathways, and virulence. Front Microbiol 6:325. doi: 10.3389/fmicb.2015.00325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Posas F, Wurgler-Murphy SM, Maeda T, Witten EA, Thai TC, Saito H. 1996. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two-component” osmosensor. Cell 86:865–875. doi: 10.1016/S0092-8674(00)80162-2. [DOI] [PubMed] [Google Scholar]
- 19.Du C, Sarfati J, Latge JP, Calderone R. 2006. The role of the sakA (Hog1) and tcsB (sln1) genes in the oxidant adaptation of Aspergillus fumigatus. Med Mycol 44:211–218. doi: 10.1080/13693780500338886. [DOI] [PubMed] [Google Scholar]
- 20.Reyes G, Romans A, Nguyen CK, May GS. 2006. Novel mitogen-activated protein kinase MpkC of Aspergillus fumigatus is required for utilization of polyalcohol sugars. Eukaryot Cell 5:1934–1940. doi: 10.1128/EC.00178-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elion EA, Grisafi PL, Fink GR. 1990. FUS3 encodes a cdc2+/CDC28-related kinase required for the transition from mitosis into conjugation. Cell 60:649–664. doi: 10.1016/0092-8674(90)90668-5. [DOI] [PubMed] [Google Scholar]
- 22.Altwasser R, Baldin C, Weber J, Guthke R, Kniemeyer O, Brakhage AA, Linde J, Valiante V. 2015. Network modeling reveals cross talk of MAP kinases during adaptation to caspofungin stress in Aspergillus fumigatus. PLoS One 10:e0136932. doi: 10.1371/journal.pone.0136932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bruder Nascimento AC, Dos Reis TF, de Castro PA, Hori JI, Bom VL, de Assis LJ, Ramalho LN, Rocha MC, Malavazi I, Brown NA, Valiante V, Brakhage AA, Hagiwara D, Goldman GH. 2016. Mitogen activated protein kinases SakA(HOG1) and MpkC collaborate for Aspergillus fumigatus virulence. Mol Microbiol 100:841–859. doi: 10.1111/mmi.13354. [DOI] [PubMed] [Google Scholar]
- 24.Yi S, Sahni N, Daniels KJ, Pujol C, Srikantha T, Soll DR. 2008. The same receptor, G protein, and mitogen-activated protein kinase pathway activate different downstream regulators in the alternative white and opaque pheromone responses of Candida albicans. Mol Biol Cell 19:957–970. doi: 10.1091/mbc.e07-07-0688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang P, Perfect JR, Heitman J. 2000. The G-protein beta subunit GPB1 is required for mating and haploid fruiting in Cryptococcus neoformans. Mol Cell Biol 20:352–362. doi: 10.1128/MCB.20.1.352-362.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Atoui A, Bao D, Kaur N, Grayburn WS, Calvo AM. 2008. Aspergillus nidulans natural product biosynthesis is regulated by mpkB, a putative pheromone response mitogen-activated protein kinase. Appl Environ Microbiol 74:3596–3600. doi: 10.1128/AEM.02842-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bayram O, Bayram OS, Ahmed YL, Maruyama J, Valerius O, Rizzoli SO, Ficner R, Irniger S, Braus GH. 2012. The Aspergillus nidulans MAPK module AnSte11-Ste50-Ste7-Fus3 controls development and secondary metabolism. PLoS Genet 8:e1002816. doi: 10.1371/journal.pgen.1002816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nosanchuk JD, Stark RE, Casadevall A. 2015. Fungal melanin: what do we know about structure? Front Microbiol 6:1463. doi: 10.3389/fmicb.2015.01463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Akoumianaki T, Kyrmizi I, Valsecchi I, Gresnigt MS, Samonis G, Drakos E, Boumpas D, Muszkieta L, Prevost MC, Kontoyiannis DP, Chavakis T, Netea MG, van de Veerdonk FL, Brakhage AA, El-Benna J, Beauvais A, Latge JP, Chamilos G. 2016. Aspergillus cell wall melanin blocks LC3-associated phagocytosis to promote pathogenicity. Cell Host Microbe 19:79–90. doi: 10.1016/j.chom.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 30.Aimanianda V, Bayry J, Bozza S, Kniemeyer O, Perruccio K, Elluru SR, Clavaud C, Paris S, Brakhage AA, Kaveri SV, Romani L, Latge JP. 2009. Surface hydrophobin prevents immune recognition of airborne fungal spores. Nature 460:1117–1121. doi: 10.1038/nature08264. [DOI] [PubMed] [Google Scholar]
- 31.Thywißen A, Heinekamp T, Dahse H-M, Schmaler-Ripcke J, Nietzsche S, Zipfel PF, Brakhage AA. 2011. Conidial dihydroxynaphthalene melanin of the human pathogenic fungus Aspergillus fumigatus interferes with the host endocytosis pathway. Front Microbiol 2:96. doi: 10.3389/fmicb.2011.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmaler-Ripcke J, Sugareva V, Gebhardt P, Winkler R, Kniemeyer O, Heinekamp T, Brakhage AA. 2009. Production of pyomelanin, a second type of melanin, via the tyrosine degradation pathway in Aspergillus fumigatus. Appl Environ Microbiol 75:493–503. doi: 10.1128/AEM.02077-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langfelder K, Jahn B, Gehringer H, Schmidt A, Wanner G, Brakhage AA. 1998. Identification of a polyketide synthase gene (pksP) of Aspergillus fumigatus involved in conidial pigment biosynthesis and virulence. Med Microbiol Immunol 187:79–89. doi: 10.1007/s004300050077. [DOI] [PubMed] [Google Scholar]
- 34.Grosse C, Heinekamp T, Kniemeyer O, Gehrke A, Brakhage AA. 2008. Protein kinase A regulates growth, sporulation, and pigment formation in Aspergillus fumigatus. Appl Environ Microbiol 74:4923–4933. doi: 10.1128/AEM.00470-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Valiante V, Baldin C, Hortschansky P, Jain R, Thywißen A, Straßburger M, Shelest E, Heinekamp T, Brakhage AA. 2016. The Aspergillus fumigatus conidial melanin production is regulated by the bifunctional bHLH DevR and MADS-box RlmA transcription factors. Mol Microbiol 102:321–335. doi: 10.1111/mmi.13462. [DOI] [PubMed] [Google Scholar]
- 36.Valiante V, Jain R, Heinekamp T, Brakhage AA. 2009. The MpkA MAP kinase module regulates cell wall integrity signaling and pyomelanin formation in Aspergillus fumigatus. Fungal Genet Biol 46:909–918. doi: 10.1016/j.fgb.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 37.Butler MJ, Gardiner RB, Day AW. 2005. Fungal melanin detection by the use of copper sulfide-silver. Mycologia 97:312–319. doi: 10.1080/15572536.2006.11832806. [DOI] [PubMed] [Google Scholar]
- 38.Mah JH, Yu JH. 2006. Upstream and downstream regulation of asexual development in Aspergillus fumigatus. Eukaryot Cell 5:1585–1595. doi: 10.1128/EC.00192-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liebmann B, Muller M, Braun A, Brakhage AA. 2004. The cyclic AMP-dependent protein kinase A network regulates development and virulence in Aspergillus fumigatus. Infect Immun 72:5193–5203. doi: 10.1128/IAI.72.9.5193-5203.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lafon A, Han KH, Seo JA, Yu JH, d'Enfert C. 2006. G-protein and cAMP-mediated signaling in aspergilli: a genomic perspective. Fungal Genet Biol 43:490–502. doi: 10.1016/j.fgb.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 41.Van Zeebroeck G, Kimpe M, Vandormael P, Thevelein JM. 2011. A split-ubiquitin two-hybrid screen for proteins physically interacting with the yeast amino acid transceptor Gap1 and ammonium transceptor Mep2. PLoS One 6:e24275. doi: 10.1371/journal.pone.0024275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hoefgen S, Lin J, Fricke J, Stroe MC, Mattern DJ, Kufs JE, Hortschansky P, Brakhage AA, Hoffmeister D, Valiante V. 2018. Facile assembly and fluorescence-based screening method for heterologous expression of biosynthetic pathways in fungi. Metab Eng 48:44–51. doi: 10.1016/j.ymben.2018.05.014. [DOI] [PubMed] [Google Scholar]
- 43.Meyer V, Wanka F, van Gent J, Arentshorst M, van den Hondel CA, Ram AF. 2011. Fungal gene expression on demand: an inducible, tunable, and metabolism-independent expression system for Aspergillus niger. Appl Environ Microbiol 77:2975–2983. doi: 10.1128/AEM.02740-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steinbach WJ, Lamoth F, Juvvadi PR. 2015. Potential microbiological effects of higher dosing of echinocandins. Clin Infect Dis 61(Suppl 6):S669–S677. doi: 10.1093/cid/civ725. [DOI] [PubMed] [Google Scholar]
- 45.Ries LNA, Rocha MC, de Castro PA, Silva-Rocha R, Silva RN, Freitas FZ, de Assis LJ, Bertolini MC, Malavazi I, Goldman GH. 2017. The Aspergillus fumigatus CrzA transcription factor activates chitin synthase gene expression during the caspofungin paradoxical effect. mBio 8:e00705-17. doi: 10.1128/mBio.00705-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Müller S, Baldin C, Groth M, Guthke R, Kniemeyer O, Brakhage AA, Valiante V. 2012. Comparison of transcriptome technologies in the pathogenic fungus Aspergillus fumigatus reveals novel insights into the genome and MpkA dependent gene expression. BMC Genomics 13:519. doi: 10.1186/1471-2164-13-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jain R, Valiante V, Remme N, Docimo T, Heinekamp T, Hertweck C, Gershenzon J, Haas H, Brakhage AA. 2011. The MAP kinase MpkA controls cell wall integrity, oxidative stress response, gliotoxin production and iron adaptation in Aspergillus fumigatus. Mol Microbiol 82:39–53. doi: 10.1111/j.1365-2958.2011.07778.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Macheleidt J, Mattern DJ, Fischer J, Netzker T, Weber J, Schroeckh V, Valiante V, Brakhage AA. 2016. Regulation and role of fungal secondary metabolites. Annu Rev Genet 50:371–392. doi: 10.1146/annurev-genet-120215-035203. [DOI] [PubMed] [Google Scholar]
- 49.Valiante V. 2017. The cell wall integrity signaling pathway and its involvement in secondary metabolite production. J Fungi (Basel) 3:E68. doi: 10.3390/jof3040068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paoletti M, Seymour FA, Alcocer MJ, Kaur N, Calvo AM, Archer DB, Dyer PS. 2007. Mating type and the genetic basis of self-fertility in the model fungus Aspergillus nidulans. Curr Biol 17:1384–1389. doi: 10.1016/j.cub.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 51.Kang JY, Chun J, Jun SC, Han DM, Chae KS, Jahng KY. 2013. The MpkB MAP kinase plays a role in autolysis and conidiation of Aspergillus nidulans. Fungal Genet Biol 61:42–49. doi: 10.1016/j.fgb.2013.09.010. [DOI] [PubMed] [Google Scholar]
- 52.Rocha MC, Fabri JH, Franco de Godoy K, Alves de Castro P, Hori JI, Ferreira da Cunha A, Arentshorst M, Ram AF, van den Hondel CA, Goldman GH, Malavazi I. 2016. Aspergillus fumigatus MADS-box transcription factor rlmA is required for regulation of the cell wall integrity and virulence. G3 (Bethesda) 6:2983–3002. doi: 10.1534/g3.116.031112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liebmann B, Gattung S, Jahn B, Brakhage AA. 2003. cAMP signaling in Aspergillus fumigatus is involved in the regulation of the virulence gene pksP and in defense against killing by macrophages. Mol Genet Genomics 269:420–435. doi: 10.1007/s00438-003-0852-0. [DOI] [PubMed] [Google Scholar]
- 54.Alspaugh JA, Perfect JR, Heitman J. 1997. Cryptococcus neoformans mating and virulence are regulated by the G-protein alpha subunit GPA1 and cAMP. Genes Dev 11:3206–3217. doi: 10.1101/gad.11.23.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.D'Souza CA, Alspaugh JA, Yue C, Harashima T, Cox GM, Perfect JR, Heitman J. 2001. Cyclic AMP-dependent protein kinase controls virulence of the fungal pathogen Cryptococcus neoformans. Mol Cell Biol 21:3179–3191. doi: 10.1128/MCB.21.9.3179-3191.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grice CM, Bertuzzi M, Bignell EM. 2013. Receptor-mediated signaling in Aspergillus fumigatus. Front Microbiol 4:26. doi: 10.3389/fmicb.2013.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gehrke A, Heinekamp T, Jacobsen ID, Brakhage AA. 2010. Heptahelical receptors GprC and GprD of Aspergillus fumigatus are essential regulators of colony growth, hyphal morphogenesis, and virulence. Appl Environ Microbiol 76:3989–3998. doi: 10.1128/AEM.00052-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Affeldt KJ, Carrig J, Amare M, Keller NP. 2014. Global survey of canonical Aspergillus flavus G protein-coupled receptors. mBio 5:e01501. doi: 10.1128/mBio.01501-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Käfer E. 1977. The anthranilate synthetase enzyme complex and the trifunctional trpC gene of Aspergillus. Can J Genet Cytol 19:723–738. doi: 10.1139/g77-079. [DOI] [PubMed] [Google Scholar]
- 60.Mowat E, Butcher J, Lang S, Williams C, Ramage G. 2007. Development of a simple model for studying the effects of antifungal agents on multicellular communities of Aspergillus fumigatus. J Med Microbiol 56:1205–1212. doi: 10.1099/jmm.0.47247-0. [DOI] [PubMed] [Google Scholar]
- 61.Shopova I, Bruns S, Thywissen A, Kniemeyer O, Brakhage AA, Hillmann F. 2013. Extrinsic extracellular DNA leads to biofilm formation and colocalizes with matrix polysaccharides in the human pathogenic fungus Aspergillus fumigatus. Front Microbiol 4:141. doi: 10.3389/fmicb.2013.00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.d'Enfert C. 1996. Selection of multiple disruption events in Aspergillus fumigatus using the orotidine-5′-decarboxylase gene, pyrG, as a unique transformation marker. Curr Genet 30:76–82. doi: 10.1007/s002940050103. [DOI] [PubMed] [Google Scholar]
- 63.da Silva Ferreira ME, Kress MR, Savoldi M, Goldman MH, Hartl A, Heinekamp T, Brakhage AA, Goldman GH. 2006. The akuB(KU80) mutant deficient for nonhomologous end joining is a powerful tool for analyzing pathogenicity in Aspergillus fumigatus. Eukaryot Cell 5:207–211. doi: 10.1128/EC.5.1.207-211.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Growth phenotypes of the wild type and MAPK and complemented mutants grown in the presence of Congo Red (CR) (A), calcofluor white (CFW) (B), and sodium dodecyl sulfate (SDS) (C). The strains were grown for 48 hours at 37°C on MM agar plates. The numbers of conidia are also reported. Download FIG S1, PDF file, 0.2 MB (222KB, pdf) .
Copyright © 2019 Manfiolli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Growth phenotypes of the wild type and ΔmpkA, ΔmpkB, and corresponding complemented mutants grown in liquid MM in the presence of t-butyl (A), menadione (B), paraquat (C), and hydrogen peroxide (D). The strains were grown for 48 hours at 37°C either on MM agar plates (A to C) or in liquid MM in 96-well plates (D). Numbers of conidia are also reported. Download FIG S2, PDF file, 0.3 MB (283.9KB, pdf) .
Copyright © 2019 Manfiolli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Coimmunoprecipitation assay performed with tagged MpkA and MpkB. (A) Diagnostic PCR to validate the in-locus homologous insertion of the mpkA-3×HA construct in the mpkB-gfp strain. (B) Phenotypic analysis of wild type, mpkB-gfp, and mpkB-gfp mpkA-3×HA strains grown on MM agar plates for 4 days at 37°C. (C, D) Affinity purification assays for 3×HA-tagged MpkA and GFP-tagged MpkB were performed with GFP-Trap beads (C) and anti-HA beads (D) to verify interactions. The coimmunoprecipitated proteins were analyzed using the indicated antibodies. The coimmunoprecipitation protocol used is reported in Text S1. Download FIG S3, PDF file, 0.1 MB (109.6KB, pdf) .
Copyright © 2019 Manfiolli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A) Survival rates of mice infected with conidia from wild-type, ΔmpkB, and mpkBc strains. Mice in groups of 10 per strain were infected intranasally with a 20-μl suspension containing 2 × 105 conidia. (B) Histopathology of lung tissue from mice infected with the wild-type, ΔmpkB, and mpkBc strains plus the PBS control using periodic acid-Schiff (PAS) staining. All infected mice displayed normal fungal persistence compared to the uninfected mice. Download FIG S4, PDF file, 0.2 MB (158.9KB, pdf) .
Copyright © 2019 Manfiolli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Methods related to the murine infection model, plasmid construction, DNA manipulation, and Southern blot analysis are reported. Download Text S1, PDF file, 0.1 MB (148.2KB, pdf) .
Copyright © 2019 Manfiolli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Graphical representation of the mutated loci for the A. fumigatus transformant strains analyzed and the respective Southern blots. The genes of interest are shown in blue, their flanking genes are grey. The cassettes for resistance to pyrithiamine (ptrA) and hygromycin (hph) are labeled in green and pink, respectively. The probes used for Southern blot analyses are marked as red stripes. The DNA ladder used is indicated (Hyperladder 1 kb; Bioline). (A) The mpkB gene was deleted, and the mutant strain complemented with the native gene by ectopic integration. (B) The ΔmpkB strain was complemented by homologous recombination with the native mpkB gene fused to a gfp sequence (in yellow) and an nos terminator sequence (in black). (C) Deletion of the mpkA gene in the mpkB-gfp background and in the ΔmpkB strain. Southern blots were performed separately. The probe also anneals to an additional ∼2-kb band. (D) Deletion and complementation of the gprM locus; the complementation was obtained by homologous recombination. (E) Deletion and complementation of the gpaA locus. The complementation was performed by ectopic integration of the gpaA wild-type gene. (F) Disruption of the gpaC gene. (G) Disruption of the pksP gene in the wild-type, ΔmpkB, ΔgpaA, and ΔgprM strains. (H) The dicistronic genes including the gpaA-gfp gprM-3×HA and the gfp gprM-3×HA sequences were ectopically integrated into the A. fumigatus CEA10 genome. Positive transformants were validated by Western blot analysis using anti-GFP antibodies (left) and anti-HA antibodies (right). The predicted molecular weights of the gene products are also reported. Download FIG S5, PDF file, 0.4 MB (386.6KB, pdf) .
Copyright © 2019 Manfiolli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used in this study. Download Table S1, PDF file, 0.1 MB (152.4KB, pdf) .
Copyright © 2019 Manfiolli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Plasmids used in this study. Download Table S2, PDF file, 0.1 MB (107.6KB, pdf) .
Copyright © 2019 Manfiolli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.