Abstract
In Africa, cervical cancer and placental malaria (PM) are a major public health concern. There is currently no available PM vaccine and the marketed Human Papillomavirus (HPV) vaccines are prohibitively expensive. The idea of a combinatorial HPV and PM vaccine is attractive because the target population for vaccination against both diseases, adolescent girls, would be overlapping in Sub-Saharan Africa. Here we demonstrate proof-of-concept for a combinatorial vaccine utilizing the AP205 capsid-based virus-like particle (VLP) designed to simultaneously display two clinically relevant antigens (the HPV RG1 epitope and the VAR2CSA PM antigen). Three distinct combinatorial VLPs were produced displaying one, two or five concatenated RG1 epitopes without obstructing the VLP’s capacity to form. Co-display of VAR2CSA was achieved through a split-protein Tag/Catcher interaction without hampering the vaccine stability. Vaccination with the combinatorial vaccine(s) was able to reduce HPV infection in vivo and induce anti-VAR2CSA IgG antibodies, which inhibited binding between native VAR2CSA expressed on infected red blood cells and chondroitin sulfate A in an in vitro binding-inhibition assay. These results show that the Tag/Catcher AP205 VLP system can be exploited to make a combinatorial vaccine capable of eliciting antibodies with dual specificity.
Introduction
There is a striking overlap in the global prevalence of cervical cancer and malaria, particularly in Sub-Saharan Africa. Cervical cancer is caused by a sexually acquired infection with certain types of Human Papillomavirus (HPV)1, and causes more than 60,000 deaths annually in sub-Saharan Africa alone2. This region also accounts for 86% of global malaria cases (219 million), including 435,000 malaria related deaths per year3. Women are particularly vulnerable to malaria during pregnancy when P. falciparum infections cause placental malaria (PM) (reviewed by4 and5). Consequently, both cervical cancer and PM constitute major public health challenges in Sub-Saharan Africa, where people often have limited access to health care (e.g. for regular HPV screening or malaria testing), or the affected individuals may be incapable of paying for expensive vaccines or treatment2,6.
HPV vaccination programmes have been widely implemented in the developed world, which has caused a substantial decrease in the incidence of HPV infections2,7. The HPV vaccines based on the HPV L1 major capsid protein, which form Virus-Like Particles (VLPs), are extremely potent, possibly offering life-long protection after a single vaccine dose8,9. The high immunogenicity of the HPV vaccines is attributed to structural features of the L1 VLPs, which lead to the efficient generation of long-lived antigen-specific antibody-producing cells10–12. HPV L1 VLP vaccines strongly elicit neutralizing antibody responses against the included HPV genotypes represented in the vaccines, but offer little in the way of cross-protection against other HPV types (reviewed by13). The clinical data generated by the HPV L1 VLP vaccines has consequently stimulated a great interest in developing next-generation VLP-based vaccines with a similar capacity of inducing long-term antibody responses, but against a broader panel of HPV types.
Current efforts to develop a broadly protective prophylactic HPV vaccine focus on targeting the HPV minor capsid protein, L2, which contains multiple highly-conserved and broadly neutralizing epitopes (as reviewed by14). In particular, the N-terminus of the HPV L2 protein contains a major cross-neutralizing epitope, RG115. Several studies have shown anti-RG1 antibodies confer complete type-specific protection and moderate cross-protection against heterologous HPV types16,17. Recently, it was shown that displaying multiple conjoined L2 RG1 epitopes, deriving from different HPV types on the surface of a VLP backbone, broadened the protective immunity against diverse HPV types18. The RG1 epitope, specifically concatenated constructs comprised of the RG1 epitope from several HPV types, represents a good target antigen for the development of a broadly protective HPV vaccine. However, the RG1 epitope is only transiently exposed to the immune system during the attachment of HPV to the basement membrane19 and therefore, an RG1-based vaccine will likely need to elicit long-lived, high levels of circulating antibodies to be effective.
To date, there is no commercially available vaccine against PM, although two different vaccines, both including the minimal binding region of the P. falciparum adhesion protein, VAR2CSA, are being tested in clinical phase I trials (NCT02647489 and NCT02658253). VAR2CSA is responsible for the sequestration of P. falciparum parasites20 in the placenta of pregnant women by mediating binding to the placental chondroitin sulfate A (CSA)21. Accordingly, an effective VAR2CSA-based PM vaccine may need to elicit antibodies that inhibit the binding between placental CSA and VAR2CSA expressed on the surface of infected erythrocytes (iE)22–24.
Vaccines against PM and HPV have overlapping target populations and need to be inexpensive in order to be broadly employed in the sub-Saharan region25. Both vaccines should ideally be administered to pre-pubescent girls and elicit durable (decades-long) immune responses. Combining the two disease targets on a common, highly immunogenic VLP platform could be a strategy to induce long-term protection against both diseases, and will be beneficial in terms of production costs and deployment.
We have used the well-characterized AP205 capsid-based VLP platform26–30 to generate VLP based vaccines (coined 5x, 2x, and 1xL2-VLPs) that display both VAR2CSA and HPV L2 RG1 epitopes at high densities. Using mouse models, this study examined whether the VLP-based combinatorial vaccine design would allow for induction of high titered, dual-specificity antibody responses against the co-displayed antigens.
Results
Engineering 5x, 2x and 1xL2-VLPs
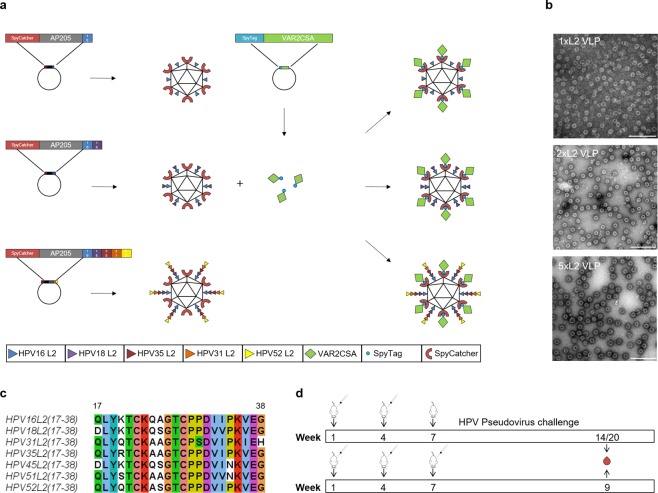
To produce VLPs which simultaneously display the L2-HPV and the VAR2CSA-P. falciparum protein, the SpyCatcher sequence and L2-HPV coding sequences were genetically fused to the 5′ and 3′ ends of the gene coding for AP205 capsid protein, respectively (Fig. 1a). The engineered genes were expressed in E. coli and the recombinant proteins spontaneously formed VLPs (Fig. 1b). Three different VLPs were produced. The SpyCatcher was fused at the N-terminus of the AP205 capsid protein, but the C-terminus was extended with either one (HPV16), two (HPV16 and 18) or five (HPV16, 18, 35, 31, 52) concatenated peptides derived from the highly conserved, cross-reactive RG1 epitope of the HPV L2 minor capsid protein (amino acid (aa) 17–38) (Fig. 1, panels a–c). The three SpyCatcher/L2-VLPs (termed 1xL2, 2x or 5xL2-VLP) were non-aggregated, had similar diameters (30–35 nm), and had low polydispersity (5xL2-VLP 47.4 nm with 10.1%Pd1, 2xL2-VLP, 45.1 nm with 21.9%Pd and 1xL2-VLP 40.4 nm with Pd% 6.9). The final yield of each VLPs was in the range of 25–100 mg/L culture and the VLPs were stable in 4 C for 14 days and in −80 °C for at least 12 months (Supplementary Fig. S1).
Figure 1.
(a) Schematic representation of the method used to create the combinatorial HPV and PM VLP vaccines. Three combinatorial HPV and PM VLP vaccines were created. Specifically, the AP205 capsid protein was genetically fused to SpyCatcher at the N-terminus whereas the C-terminus was genetically fused to either one (HPV16), two (HPV16 and 18) or five (HPV16, 18, 35, 31, 52) concatenated peptides derived from the highly conserved, cross-reactive epitope of the HPV L2 minor capsid protein (amino acid 17–38). Recombinant expression in E. coli resulted in formation of three distinct VLPs each displaying 180 L2 polypeptides and SpyCatcher proteins. Subsequent mixing of the PM antigen, VAR2CSA (genetically fused to SpyTag at the N-terminus) with VLPs resulted in covalent attachment of VAR2CSA to the surface of the VLPs. (b) Transmission electron microscopy images of the 1xL2-VLP, 2xL2-VLP and 5xL2-VLP. To verify the overall quality of the purified VLPs, the VLP samples were placed on carbon, adsorbed to a grid and negatively stained with 2% uranyl acetate. The three VLPs are non-aggregated and uniform in size (approximately 30–35 nm). Scale bar 200 nm. (c) Amino acid sequence alignment of the minor capsid protein L2 (aa 17–38) from HPV16, 18, 31, 35 and 52. The alignment was created using Jalview36 under a creative commons license (https://creativecommons.org/licenses/by-sa/3.0/legalcode). (d) Mouse immunization schedule. Top panel: Balb/C mice were immunized intramuscularly in a prime-boost regimen (week 1 and week 4) with an antigen dose of 5 µg (based on L2-VLP amount) consisting of either 5xL2-VLP (n = 25), 2xL2-VLP (n = 25), 5xL2-VLP-Var2 (n = 6) or 2xL2-VLP-Var2 (n = 6) VLPs. Three weeks following the last immunization, randomized groups of mice (n = 5) were infected vaginally with luciferase-expressing HPV pseudovirions. Blood samples collected at week 14 (for 5xL2-VLP-Var2 and 2xL2-VLP-Var2) or 20 (for 5 × -L2-VLP and 2xL2-VLP) were analyzed for anti-HPV L2 and anti-VAR2CSA IgG antibodies, both with regards to ELISA binding titers as well as in vitro parasite binding-inhibition. Lower panel: Balb/C mice were immunized intramuscularly in a prime-boost-boost regimen (weeks 1, 4 and 7) with a 3.14 µg antigen dose (based on L2-VLP amount) of 1xL2-VLP (n = 7). Blood samples were collected at week 9 to perform ELISA and in vitro PsV challenge.
In parallel, the 5′ end of VAR2CSA was genetically fused to SpyTag and expressed in E. coli. Mixing of recombinant SpyTag-VAR2CSA protein and the VLPs (i.e. 5xL2 or 2xL2), allowed for covalent attachment of VAR2CSA to the VLPs resulting in the formation of VLPs displaying both the HPV L2 and VAR2CSA antigens at high density (Fig. 1, and Supplementary Fig. S2). The combinatorial VLPs are stable and not aggregation prone (Supplementary Fig. S1).
Immunogenicity of the combinatorial HPV and PM VLP vaccines
The immunogenicity of the combinatorial VLP vaccines was assessed in mice, as described in Fig. 1d, and antibody responses were assessed by ELISA. All three vaccines elicited strong antibody responses against HPV L2. The 1xL2-VLP vaccine elicited antibodies with reactivity against the L2 peptides representing each of the HPV types we tested (16, 18 and 31) (Fig. 2b). As expected, antibody titers were high against the HPV16 L2 RG1 epitope included in the vaccine. The cross-reactivity against the HPV18 L2 was also high while cross-reactivity towards the HPV31 L2 RG1 epitope was lower. The 2xL2-VLP and 5xL2-VLP vaccines also induced antibodies with broad reactivity against the HPV L2 RG1 epitopes tested (i.e. 16, 18, 31, 35, 45 and 51) (Fig. 2a). The levels of anti-L2 IgG antibodies elicited by vaccines with and without co-display of VAR2CSA were comparable, indicating that co-display of VAR2CSA does not significantly reduce the anti-HPV L2 antibody response. There was no significant difference in the antibody levels against HPV16, 18, 31, 35, 45 L2 peptides elicited by the 5xL2 and 2xL2-VLP vaccines despite the fact that the 2xL2-VLP vaccine only contained the HPV16 and 18 L2 sequences. However, the 5xL2-VLP induced antibodies showing significantly higher reactivity to the HPV51 L2 RG1 epitope than the 2xL2-VLP (Fig. 2b). Finally, the combinatorial VLP vaccines (2xL2-VLP-VAR2CSA 5xL2-VLP-VAR2CSA) elicited high levels of anti-VAR2CSA IgG, confirming that the AP205 VLP platform can be utilized to generate immune responses against two distinct pathogenic targets simultaneously (Fig. 2c).
Figure 2.
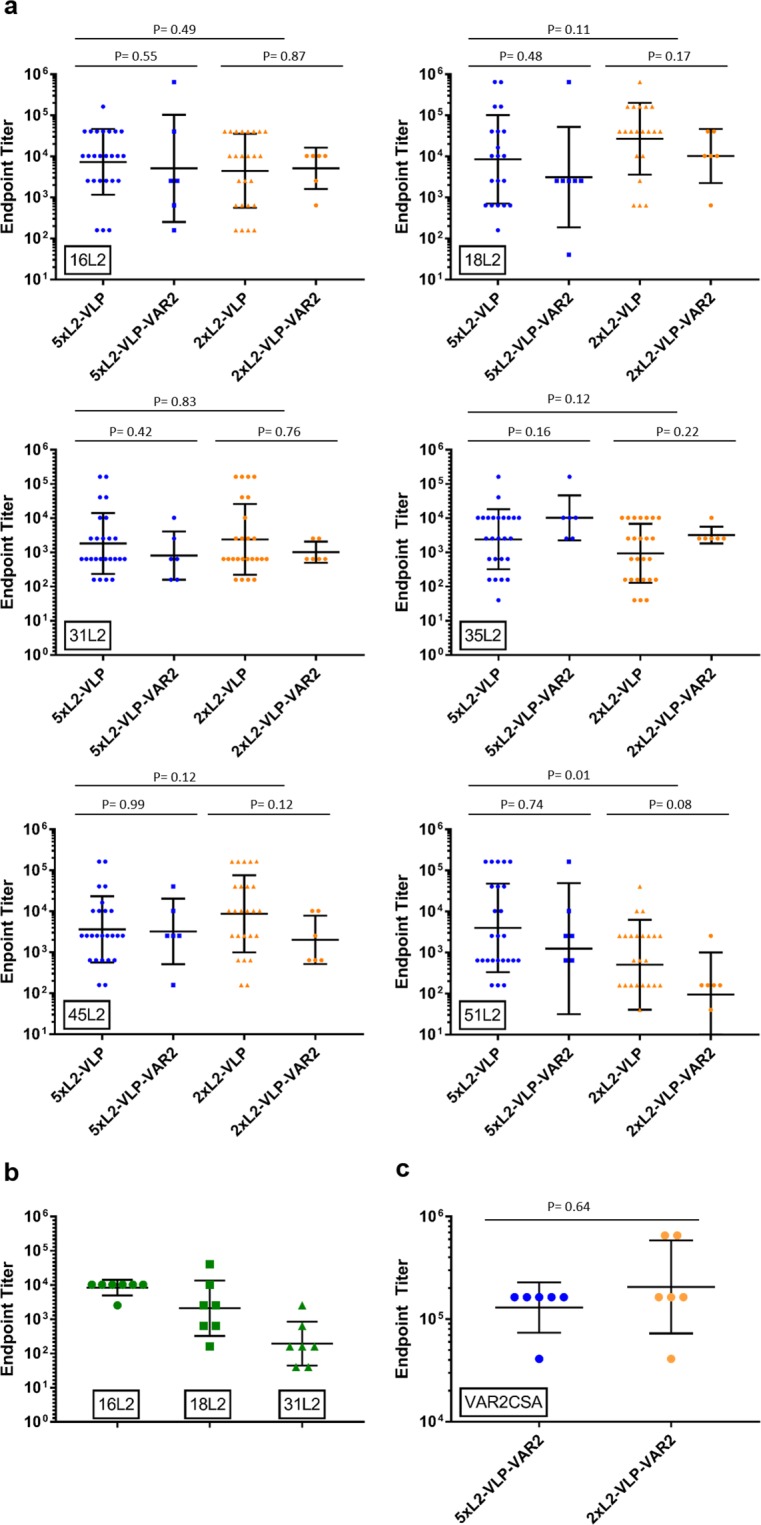
HPV and PM specific endpoint ELISA IgG binding titers. Endpoint antibody binding titers from individual mouse sera were calculated using a cut-off of O.D. = 0.1. Graphs show geometric mean and SD. Mann-Whitney Rank sum test was used for statistical analysis using significance cut-off value of 0.05. (a) Vaccine induced anti-HPV L2 binding titers measured in sera from mice immunized with 5xL2- and 2xL2 VLPs (with and without co-display of VAR2CSA). Peptide sequences derived from the HPV L2 RG1 epitope (aa 17–31) of a range of HPV types (i.e. 16, 18, 32, 35, 45, and 51) were genetically fused to a carrier protein (accession number WP_057363222, aa 44–338) and used as capture proteins in ELISA. (b) Vaccine induced anti-HPV16, 18 and 31 L2 IgG titers, measured by HPV specific ELISA. Mice immunized with 1xL2-VLPs. For measuring HPV type specific antibody responses ELISA plates were coated with synthetic peptides derived from the HPV L2 RG1 epitope (aa 17–31) of HPV types 16, 18 or 31. (c) Vaccine induced anti-VAR2CSA IgG titers measured in sera from mice immunized with 5xL2-VLP-VAR2CSA and 2xL2-VLP-VAR2CSA. VAR2CSA was used as capture protein for the ELISA.
Neutralization capacity of vaccine-induced anti-HPV L2
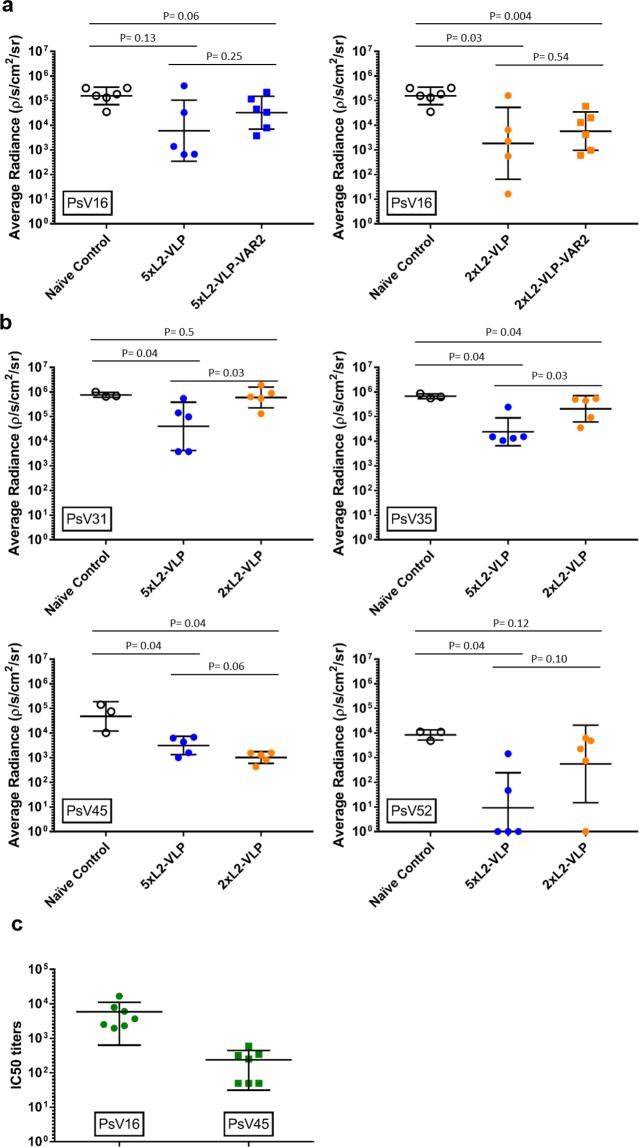
To test the capacity of vaccine-induced antibodies to neutralize HPV infection in vivo, mice immunized with 2xL2-VLP or 5xL2-VLP were challenged with a panel of luciferase expressing HPV pseudovirions (PsV) (i.e. PsV16, 31, 35, 45, 52) using a well-established PsV genital challenge mouse model17,18 (Fig. 3a,d). In this model the level of infection is indirectly determined by measuring the amount of light (radiance) emitted upon instilling luciferin. First, to test whether co-display of VAR2CSA affected the neutralization capacity of the vaccine-induced anti-HPV L2 antibody response, infection levels in mice immunized with L2-displaying VLPs with or without co-display of VAR2CSA were compared. There was no statistically significant difference in infection levels between mice immunized with L2-VLPs or with L2-VAR2CSA-VLPs (P = 0.25 and 0.54 for 5xL2-VLP and 2xL2-VLP, respectively) (Fig. 3a left and right panel). Further, mice vaccinated with the 2xL2-VLP vaccines showed a significant 84- and 27-fold reduction in infection levels compared to non-vaccinated control mice (P = 0.03 2xL2-VLP; P = 0.004 2xL2-VLP-VAR2CSA) (Fig. 3a, right panel). Although a similar trend could be seen in the 5xL2-VLP vaccinated mice, this difference was not significant (26-fold reduction P = 0.13 5xL2-VLP; 5-fold reduction P = 0.06 5xL2-VLP-VAR2CSA) (Fig. 3a, left panel).
Figure 3.
Neutralization capacity of vaccine-induced anti-HPV L2 antibody responses measured in vivo PsV genital challenge mouse model and in vitro L2-based PsV inhibition assay. Balb/C mice were first immunized and then challenged with HPV PsV, as described in Fig. 1d (top panel). Infection levels in vaccinated mice were measured against non-vaccinated i.e. naïve control mice. Graph shows average radiance for individual mice as well as geometric mean and SD for each group. (a) PsV infection in mice immunized with either the 5xL2-VLP (left panel) vaccine or the 2xL2-VLP vaccine (right panel) (with/without co-display of VAR2CSA) after challenge with HPV PsV16. (b) PsV infection in 5xL2-VLP or 2xL2-VLP immunized mice challenged with HPV PsV31, 35, 45, or 52. (c) Balb/C mice were immunized with 1xL2-VLP, as described in Fig. 1d (lower panel). Neutralization titers (IC50) against HPV PsV 16 and 45 were measured in mouse serum collected 9 weeks after prime vaccination using an in vitro L2-based PsV inhibition assay, as previously described35.
To investigate whether the breadth of protection against HPV infection was increased by adding L2 RG1 epitopes representing additional HPV types to the VLP vaccine design, infection levels in 5xL2-VLP and 2xL2-VLP vaccinated mice, were compared after challenging with PsV31, 35, 45, or 52. This comparison showed that vaccination with 5xL2-VLP vaccines resulted in a statistically significant, 15-to 916- fold reduction in infection levels of all included PsV types (P = 0.04 for PsV 31, 35, 45 and 52). By contrast, the 2xL2-VLP vaccine were generally less effective than the 5xL2-VLP vaccine at preventing infection with HPV PsV. Vaccination with 2xL2-VLP only caused a significant 3- and 47-fold reduction in PsV35 (P = 0.04) and 45 (P = 0.04) infection levels, respectively while no significant reduction was seen for PsV31 (P = 0.5) and 52 (P = 0.12) infection (Fig. 3b).
Sera from mice immunized with the 1xL2-VLP vaccine were tested in an in vitro HPV L2-based PsV inhibition assay. The tested sera showed both homologous PsV16 as well as heterologous neutralizing capacity. As expected, the neutralizing titers against the homologous PsV were higher compared to the heterologous PsV45 neutralization titers. (Fig. 3c).
Functional capacity of anti-VAR2CSA IgG responses
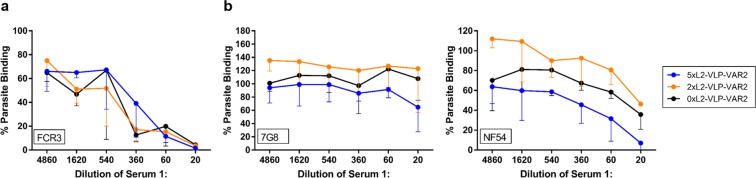
The biological activity of vaccine-induced anti-VAR2CSA IgG was tested using an assay measuring the inhibition of binding between P. falciparum infected erythrocytes expressing VAR2CSA and chondroitin sulfate A (CSA) (Fig. 4a). IgG induced by any of the two combinatorial vaccines (5xL2-VLP-Var2 or 2xL2-VLP-Var2) showed binding-inhibitory capacity. The binding inhibition induced by VLPs presenting both L2 and VARCSA, was comparable to the levels induced by VLPs presenting VAR2CSA only (serum from mice already published in27 was included in the assay as a reference and presented with the black line 0xL2-VLP-VAR2CSA in these graphs). Inhibition was most pronounced towards parasites of the FCR3 genotype on which the VAR2CSA sequence was based. Some heterologous inhibition was seen towards parasites of the NF54 genotype, but not towards parasites of the 7G8 genotype (Fig. 4b).
Figure 4.
Functional antibody response against VAR2CSA measured in vitro by a parasite binding-inhibition assay. A static parasite binding-inhibition assay was used to assess the capacity of vaccine-induced mouse antibodies to prevent binding between native VAR2CSA expressed by iE and the human receptor, chondroitin sulfate A (CSA)32,33. In brief, erythrocytes infected with VAR2CSA-expressing tritium-labelled P. falciparum were incubated either without serum (positive control) or with mouse sera in 3-fold dilutions starting at 1:20 in decorin-coated wells. Each assay measured mean iE binding and the percentage of binding iE, by dividing the test result with the mean value of iE in wells incubated without serum. Three repetitions of the assay were made for FCR3 and 7G8 and two for NF54. The graphs show the mean percentage of parasite binding across the technical repetitions of the assay. The error bars indicate the standard deviation of this mean. (a) Normalized iE binding of the homologous FCR3 strain after pre-incubation with different dilutions of pooled mouse sera of mice immunized with either 5xL2-VLP-VAR2CSA (n = 6, blue), 2xL2-VLP-VAR2CSA (n = 6, orange), or 0xL2-VLP-VAR2CSA (n = 5, black. previously published data27 here shown as reference). (b) Normalized iE binding of the heterologous 7G8 strain (left) and NF54 strain (right) after pre-incubation with the indicated dilutions of pooled serum samples as in (a).
Discussion
The aim of this study was to establish proof-of-concept for a VLP-based combinatorial vaccine eliciting functional antibody responses against both HPV and placental malaria. The unique attribute of the AP205 VLP, to accept genetic fusions at both the N- and C-terminus, was exploited to generate combinatorial VLP vaccines, which were designed to simultaneously display the placental malaria antigen (VAR2CSA) and between one and five concatenated HPV L2 sequences representing the cross-reactive RG1 epitope (aa 17–38). Specifically, L2 RG1 antigen sequences were based on the most prevalent oncogenic HPV types in the sub-Saharan African region.
The first question we sought to answer was whether the large genetic fusions to the AP205 capsid protein would hamper VLP assembly. To our knowledge the largest polypeptide that has previously been fused successfully to the C-terminus of AP205 is 55aa31 (NEF55) and co-display of antigenic polypeptides, accomplished by genetic fusion at both N- and C-terminus, has not previously been reported for any VLP. Remarkably, AP205 capsid proteins genetically fused to SpyCatcher (122 aa) at the N-terminus and to HPV L2 RG1 sequences (up to 118aa) at the C-terminus retained their ability to assemble into uniform, non-aggregated VLPs. These results confirm the exceptional capacity of the AP205 VLP to tolerate large genetic fusions at both the N-and C-terminus and underline the broad applicability of this VLP backbone for VLP-based vaccines.
Next, we investigated the ability of the combinatorial VLP based vaccines to generate functional antibody responses against the co-displayed antigen targets. To this end, a theoretical concern of the vaccine design was that one of the co-displayed antigens could dominate the immune response over the other and prevent induction of protective levels of antibodies against the less dominant. In particular, we speculated that covalent attachment of the large PM antigen (VAR2CSA) may sterically hinder access to the RG1 epitopes and/or dominate the immune response due to the high number of B and T-cell epitopes present in the antigen. However, the combinatorial vaccines all elicited high serum levels of anti-RG1 antibodies and co-display of VAR2CSA did not have any measurable effect on the induction of these antibodies. Finally, mice challenged with HPV16 PsV experienced a similar reduction in infection level after immunization with 2xL2-VLP with and without co-display of VAR2CSA. Concerning the PM antigen target, all combinatorial vaccines elicited high levels of anti-VAR2CSA antibodies, which were able to inhibit binding of both homologous and heterologous malaria parasite strains to their human receptor in vitro. Collectively, these results support the conceptual design of a VLP-based combinatorial HPV/PM vaccine.
Furthermore, using the PsV in vivo challenge model it was demonstrated that immunization with 5xL2-VLPs (comprising RG1 from HPV16, 18, 31, 35, 52) caused significant protection against a broader panel of HPV strains compared to immunization with 2xL2-VLPs (comprising RG1 from HPV16, 18). Whereas both vaccines caused a significant reduction in infection levels of PsV35 and PsV45 only the 5xL2-VLP vaccine induced significant protection against PsV31 and PsV52. Considering the RG1 sequence similarity across the five HPV genotypes included in the 5xL2-VLP vaccine design, HPV35 and HPV45 RG1 sequences show the highest similarity to the corresponding HPV16 and HPV18 sequences, respectively (Fig. 1c). The HPV45 and HPV18 RG1 sequences differ at only a single amino acid position (i.e. position 34) while the HPV16 and HPV35 sequences differ at two positions (i.e. positions 20/24). This could explain why antibody responses induced by the 5xL2-VLP and 2xL2-VLP vaccines are similar both in terms of anti-HPV35/45 antibody binding titers as well as their capacity to neutralize PsV35 and PsV45. In comparison, HPV31 and HPV52 RG1 sequence are less similar to the HPV16/18 RG1 sequences, which may explain why only the 5xL2-VLP vaccine could neutralize PsV of these genotypes. These results were not consistent with the vaccine-induced anti-HPV31 RG1 antibody titers measured by ELISA, which were similar in mice immunized with 5xL2-VLP and 2xL2-VLP vaccines. These data suggest that RG1 sequences may induce a polyclonal mixture of antibodies with different capacities for PsV neutralization.
A final consideration regarding the combinatorial vaccine design relates to the relative order of linked RG1 epitopes displayed on the VLP, which may have an effect on their immunogenicity. Our results showed that mice vaccinated with 5xL2-VLPs, where the HPV16 RG1 epitope was situated closest to the VLP backbone were not protected when challenged by PsV16. This could reflect that HPV16 RG1 was not accessible to B-cell receptors. On that basis, it may be possible to optimize the vaccine-induced anti-RG1 antibody response by changing the order of RG1 epitopes or introducing a longer linker between the VLP backbone and the first RG1 epitope.
In conclusion, our data supports the rational design of a VLP-based combinatorial HPV/PM vaccine with the potential to induce protection against HPV and PM in low-income settings.
Methods
Design, expression and purification of 5x, 2x and 1xL2-VLP
The RG1 epitope of HPV16 (QLYKTCKQAGTCPPDIIPKVEG), HPV16 and 18 (QLYKTCKQAGTCPPDIIPKVEGgsDLYKTCKQSGTCPPDVVPKVEG) or HPV16, 18, 35, 31 and 52 (QLYKTCKQAGTCPPDIIPKVEGgsDLYKTCKQSGTCPPDVVPKVEGgsQLYRTCKAAGTCPPDVIPKVEGgsQLYQTCKAAGTCPSDVIPKIEHgsQLYQTCKASGTCPPDVIPKVEG) was attached to the 3′ of the full-length SpyCatcher-AP205 gene27 by PCR. A small Glycine-Serine (GS) linker was inserted between L2 peptides followed by a Glycine-Glycine-Serine-Glycine (GGSG) linker between the SpyCatcher-VLPs and the L2 peptide(s). To create 1xL2-VLPs, the SpyCatcher-AP205 VLP served as template for the initial PCR, where GGSG-16L2 was added by including the nucleotide sequence for the L2 RG1 epitope of HPV16 in the reverse primer overhang.
The 5x and 2xL2-VLPs were created by PCR. The SpyCatcher-AP205 VLP sequence27 was amplified with a 5′ NcoI site and a 3′ overlap capable of annealing to the 5′ of a HPV16 RG1 sequence. In the second PCR reaction, a synthetic gene construct (GeneArt, Thermo Scientific) containing the concatenated L2 RG1 epitope of HPV16, 18, 35, 31, 52 was used to create an amplicon with a 5′ overlap, capable of annealing to the 3′ end of the SpyCatcher-AP205 sequence and containing a 3′ NotI cloning site. For 5xL2-VLP, the full sequence was amplified, whereas only the 16 and 18L2 sequences were amplified for creating the 2xL2-VLPs. In a third PCR reaction, the amplicons of the first and second PCR were mixed in order to amplify the 5x and 2xL2-VLP constructs.
The resulting 5xL2, 2xL2 and 1xL2 VLP genes were cloned into a pET-15b vector using NcoI and NotI restriction sites and the SpyCatcher-AP205-L2-pET-15b plasmid was transformed into E. coli One Shot® BL21 Star™ (DE3) cells (Thermo Scientific). The non-optimized small-scale upstream expression and downstream purification accomplished using Optiprep™ (Sigma) step gradients, as previously described27,28.
Design, expression and purification of SpyTag-VAR2CSA-HIS antigen
PCR added SpyTag (AHIVMVDAYKPTK) and a flexible GGS-linker 5′, as well as a V5 and hexa-histidine tag (HIS) 3′ to the gene coding for the DBL1-ID2A region (aa 9–1025) of VAR2CSA (FCR3 genotype)21 (herein termed VAR2CSA). The SpyTag-VAR2CSA-HIS gene sequence was subcloned into the pET-15b vector as described above, expressed in C3029H SHuffle T7 competent E.coli cells (New England Biolabs Inc.) and purified by IMAC and size exclusion chromatography (HiLoad Superdex 200 pg, GE Healthcare).
Quality assessment of 5x, 2x, and 1x-L2-VLPs
The 5x, 2x and 1x-L2-VLPs were assessed using negative stain transmission electron microscopy (TEM) as well as dynamic light scattering (DLS) as previously described27,28. Briefly, for TEM, the 5x, 2x and 1xL2-VLPs were adsorbed to 200-mesh carbon-coated grids. Grids were stained with 2% uranyl acetate (pH7.0) and analyzed with an accelerating voltage of 80 kV, using a CM 100 BioTWIN electron microscope (Phillips, Amsterdam).
For DLS measurements, the distribution of particle sizes was acquired (658 nm, at 25 °C, WYATT Technology, DynaPro NanoStar). The 5x and 2xL2-VLPs were analyzed with 20 runs and the 1xL2-VLP with 10 runs. The estimated diameter of the main particle population and the percent polydispersity (%Pd), was calculated for all three L2-VLPs.
Conjugation of VAR2CSA to 5x and 2xL2-VLPs
To generate the combinatorial HPV and PM VLP vaccines, purified SpyTag-VAR2CSA-His and 2x or 5xL2-VLPs were mixed in a 3:1 molar ratio (VAR2CSA:VLP) and incubated for 16 h at 4 °C in a phosphate-buffered saline (PBS), pH 8 and adjusted to 0.6 M NaCl.
Excess, unbound antigen was removed by dialysing (1000 kDa molecular weight cut-off, Spectrum Labs) against PBS pH8, 0.6 M NaCl.
Vaccine formulation
VLP vaccines (5xL2, 2xL2, 1xL2-VLP or the equivalent combinatorial vaccines) were formulated 1 hour prior to immunizations using 2%Alhydrogel (Brenntag) to a final concentration of 2 mg/ml aluminum hydroxide.
Mouse immunization Studies
8 week old, female BALB/c mice (Taconic, Denmark) were immunized intramuscularly (thigh muscle) with 3.14 µg antigen dose 1xL2 VLPs (based on the total L2-VLP amount in a prime-boost-boost regimen on week 1, 4 and 7 (Fig. 1d lower panel, n = 7). Blood samples were collected at week 9 and the in vitro HPV PsV challenge was performed on this serum. For the 5x and 2xL2-VLP vaccines (with and without VAR2CSA), 8 week old, female BALB/c mice (Jackson Laboratories, USA) were immunized with 5 µg (based on the total L2-VLP amount). Mice receiving the 5xL2 (n = 25) or 2xL2-VLP (n = 25) vaccines or the corresponding combinatorial vaccines (n = 6 in each group) were immunized in a prime-boost regimen on week 1 and 4 (Fig. 1d, upper panel). Blood samples were collected at week 5 and at week 14 (for 5xL2-VLP-Var2 and 2xL2-VLP-Var2) and week 20 (for 5 × -L2-VLP and 2xL2-VLP) to analyze HPV/VAR2CSA specific serum IgG titers and to perform the in vitro parasite binding-inhibition assay.
Serum immunoglobulin levels
Enzyme-linked immunosorbent assay (ELISA) was used to detect the level of anti-VAR2CSA and anti-L2 immunoglobulin (IgG) levels. For VAR2CSA specific IgG titers, 96-well microtiter plates (Nunc MaxiSorp) were coated with 0.1 µg/well recombinant VAR2CSA protein (no SpyTag). The L2 peptide (aa 17–31) of the 16, 18, 31, 35, 45 or 51 HPV types was genetically fused to the N-terminus of an unrelated mycobacterium tuberculosis antigen (accession number WP_057363222, aa 44–338). For 5xL2- and 2xL2-VLP studies, the recombinant fusion proteins were used as coat protein for all the ELISAs. For the 1xL2-VLP study, ELISA plates were coated with synthetic L2 peptides (aa 17–31) of the HPV16, 18 or 31, through conjugation of the biotinylated peptides to streptavidin as described elsewhere18.
Plates were developed with TMB and signal was measured at O.D.450. An O.D. cutoff 0.1 of was used to determine the endpoint titers of PM and HPV specific serum IgG.
Parasite binding-inhibition assay
An in vitro parasite binding-inhibition assay was employed to determine the capacity of antibodies induced by the 5x, and 2x-VLP-VAR2CSA vaccine to inhibit the binding between native VAR2CSA expressed on iE and CSA (decorin Sigma). For a detailed description of the assay see32,33. In short, panning of late-stage Plasmodium falciparum (homologous FCR3 or heterologous NF54 and 7G8 genotypes) parasites selected for a CSA-binding phenotype. CSA-binding parasites were labeled with Tritium and added to CSA-coated wells. Diluted, pooled VAR2CSA immune serum (5xL2-Var2 n = 6, 2xL2-Var2 n = 6 and 0xL2-Var2 n = 5) was added in triplicates. Unbound iE were washed off by a pipetting robot (Beckman-Coulter) and remaining iE were collected onto a filter plate (Perkin-Elmer). Liquid scintillation counting (Topcount NXT, Perkin-Elmer) determined the number of iE not inhibited by the VAR2CSA immune sera. Each assay measured mean iE binding and the percentage of binding iE was calculated by dividing the test result with the mean value of iE in wells incubated without serum.
The assay was repeated three times for FCR3 and 7G8 and twice for NF54. The data points represent the mean percentage parasite binding across the technical repetitions of the assay. Error bars represent the standard deviation of this mean. BSA coated wells and naïve sera were used as a negative control.
Pseudovirion Production
Type 16, 31, 35, 45 and 52 HPV PsV containing a reporter plasmid encoding for both GFP and luciferase were generated using HEK293TT cells as described elsewhere34.
The infectious unit (IU) count was determined by infecting HEK293TT cells for 48 h and measuring the percentage of infected (GFP-positive) cells by flow cytometry.
In vitro L2 based HPV pseudovirion neutralization assay
The capacity of anti-L2 antibodies to inhibit infection by the homologous HPV16 PsV as well as heterologous HPV45 PsV was determined for sera obtained from mice with 1xL2-VLP vaccine using a previously described L2-based HPV PsV neutralization assay35.
In vivo HPV Pseudovirion Neutralization Assay
Vaccine induced protection against HPV infection was evaluated in an in vivo PsV genital challenge model, as described previously17,18. Briefly, Balb/C mice previously immunized with 5xL2-, 2xL2-VLP, or the corresponding combinatorial vaccines, were randomized into challenge groups (n = 5 for 5xL2-VLP and 2xL2-VLP or n = 6 for 5xL2-VLP-Var2, 2xL2-VLP-Var2) starting approximately 3 weeks after the final boost. Five days prior to challenge, mice received 3 mg progesterone. Mice were briefly anesthetized using isoflurane and vaginally challenged with 6.3*106 infectious units (IU) PsV16, 3.15*107 IU PsV31, 8.84*106 IU PsV 35, 1.19*107 IU PsV45 or 6.2*105 IU PsV52. PsV infection was assessed under inhalation anesthesia (isoflurane) 48 hours after challenge by instilling 0.4 mg luciferin vaginally and measuring radiance (five minute exposure) using a Caliper IVIS Lumina II (Caliper Life Sciences). Background radiance was subtracted from this radiance value in order to determine average radiance (p/s/cm2/sr). Immunized mice were compared to groups of unvaccinated mice (n = 6 for PsV16 challenge and n = 3 for PsV31, 35, 45, and 52).
Ethical Statement
All animal studies were performed in accordance with Federation of European Laboratory Animal Science Associations (FELASA) and American Association for Laboratory Animal Science (AALAS) guidelines and regulations and were approved by the Danish Animal Experiments Inspectorate (Approval number: 2013-15-2934-00902/BES) and the University of New Mexico Institutional Animal Care and Use Committee (IACUC), Animal Welfare Assurance # D16–00228 (A3350-01) USDA Registration # 85-R-0014, Protocol No. 16-200498-HSC.
Statistical Analysis
GraphPadPrism7.0 was used for calculating geometric mean titers, geometric standard deviation, preparing all graphs and statistical calculations. The non-parametric, two tailed, Mann-Whitney Rank Sum Test was applied in order to determine statistical significance (P < 0.05).
Supplementary information
Acknowledgements
The authors would like to thank Anne Corfitz and Nahla Chehabi for their great technical assistance as well as Louise Goksøyr for constructive criticism of the manuscript. We would also like to thank John Schiller, NIH as well as Yuk-Ying (Susana) Pang, NIH, for assisting with the in vitro HPV pseudovirus assay. Furthermore, we would like to thank Emeline Ragonnaud, Centre for Medical Parasitology, and Christopher B. Buck, NIH, for technical advice on the production of HPV pseudovirus. We would like to acknowledge the Core Facility for Integrated Microscopy, Faculty of Health and Medical Sciences, University of Copenhagen for their assistance with the Transmission Electron Microscopy images and the Biophysics Facility—Protein Structure and Function Program from Center for Protein Research, Copenhagen, for assisting us with DLS measurements.
Author Contributions
C.M.J., J.P., S.T. and P.H.R.C. conducted all experiments. C.M.J., J.P., T.G.T., A.S., B.C., M.A.N. and A.F.S. designed the experiments. C.M.J., J.P. and A.F.S. wrote the paper. All authors contributed to analyzing the included data, reading as well as approving the final manuscript.
Data Availability
The datasets generated and analyzed during the current study are available from the corresponding author on reasonable request.
Competing Interests
C.M.J., S.T., T.G.T., A.S., M.A.N. and A.F.S. are inventors on a patent application concerning the VLP-display technology (WO2016112921 A1) licensed to AdaptVac. S.T. and A.F.S. are currently partially employed in AdaptVac. J.P., P.R.C. and B.C. declare no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Christoph M. Janitzek and Julianne Peabody contributed equally.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-41522-5.
References
- 1.Walboomers JM, et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999;189:12–19. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.WHO. World Health Organization; World Malaria Report 2018 Report No. ISBN 978-92-4-156565-3, (Geneva, 2018).
- 4.Desai M, et al. Epidemiology and burden of malaria in pregnancy. Lancet Infect Dis. 2007;7:93–104. doi: 10.1016/S1473-3099(07)70021-X. [DOI] [PubMed] [Google Scholar]
- 5.Moya-Alvarez V, Abellana R, Cot M. Pregnancy-associated malaria and malaria in infants: an old problem with present consequences. Malar J. 2014;13:271. doi: 10.1186/1475-2875-13-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anorlu RI. Cervical cancer: the sub-Saharan African perspective. Reprod Health Matters. 2008;16:41–49. doi: 10.1016/S0968-8080(08)32415-X. [DOI] [PubMed] [Google Scholar]
- 7.Bruni L, et al. Global estimates of human papillomavirus vaccination coverage by region and income level: a pooled analysis. Lancet Glob Health. 2016;4:e453–463. doi: 10.1016/S2214-109X(16)30099-7. [DOI] [PubMed] [Google Scholar]
- 8.Safaeian M, et al. Durable antibody responses following one dose of the bivalent human papillomavirus L1 virus-like particle vaccine in the Costa Rica Vaccine Trial. Cancer Prev Res (Phila) 2013;6:1242–1250. doi: 10.1158/1940-6207.CAPR-13-0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Safaeian, M. et al. Durability of Protection Afforded by Fewer Doses of the HPV16/18 Vaccine: The CVT Trial. J Natl Cancer Inst110, 10.1093/jnci/djx158 (2018). [DOI] [PMC free article] [PubMed]
- 10.Schiller J, Chackerian B. Why HIV virions have low numbers of envelope spikes: implications for vaccine development. PLoS Pathog. 2014;10:e1004254. doi: 10.1371/journal.ppat.1004254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amanna IJ, Slifka MK. Mechanisms that determine plasma cell lifespan and the duration of humoral immunity. Immunol Rev. 2010;236:125–138. doi: 10.1111/j.1600-065X.2010.00912.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schiller J, Lowy D. Explanations for the high potency of HPV prophylactic vaccines. Vaccine. 2018;36:4768–4773. doi: 10.1016/j.vaccine.2017.12.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schiller JT, Castellsague X, Garland SM. A review of clinical trials of human papillomavirus prophylactic vaccines. Vaccine. 2012;30(Suppl 5):F123–138. doi: 10.1016/j.vaccine.2012.04.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang RT, Schellenbacher C, Chackerian B, Roden RB. Progress and prospects for L2-based human papillomavirus vaccines. Expert Rev Vaccines. 2016;15:853–862. doi: 10.1586/14760584.2016.1157479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gambhira R, et al. A protective and broadly cross-neutralizing epitope of human papillomavirus L2. J Virol. 2007;81:13927–13931. doi: 10.1128/JVI.00936-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tumban E, et al. Preclinical refinements of a broadly protective VLP-based HPV vaccine targeting the minor capsid protein, L2. Vaccine. 2015;33:3346–3353. doi: 10.1016/j.vaccine.2015.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tumban E, Peabody J, Tyler M, Peabody DS, Chackerian B. VLPs displaying a single L2 epitope induce broadly cross-neutralizing antibodies against human papillomavirus. PLoS One. 2012;7:e49751. doi: 10.1371/journal.pone.0049751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhai L, et al. A novel candidate HPV vaccine: MS2 phage VLP displaying a tandem HPV L2 peptide offers similar protection in mice to Gardasil-9. Antiviral Res. 2017;147:116–123. doi: 10.1016/j.antiviral.2017.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kines RC, Thompson CD, Lowy DR, Schiller JT, Day PM. The initial steps leading to papillomavirus infection occur on the basement membrane prior to cell surface binding. Proc Natl Acad Sci USA. 2009;106:20458–20463. doi: 10.1073/pnas.0908502106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Viebig NK, et al. Disruption of var2csa gene impairs placental malaria associated adhesion phenotype. PLoS One. 2007;2:e910. doi: 10.1371/journal.pone.0000910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clausen TM, et al. Structural and functional insight into how the Plasmodium falciparum VAR2CSA protein mediates binding to chondroitin sulfate A in placental malaria. J Biol Chem. 2012;287:23332–23345. doi: 10.1074/jbc.M112.348839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duffy PE, Fried M. Antibodies that inhibit Plasmodium falciparum adhesion to chondroitin sulfate A are associated with increased birth weight and the gestational age of newborns. Infect Immun. 2003;71:6620–6623. doi: 10.1128/IAI.71.11.6620-6623.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Staalsoe T, et al. Variant surface antigen-specific IgG and protection against clinical consequences of pregnancy-associated Plasmodium falciparum malaria. Lancet. 2004;363:283–289. doi: 10.1016/S0140-6736(03)15386-X. [DOI] [PubMed] [Google Scholar]
- 24.Ndam NT, et al. Protective Antibodies against Placental Malaria and Poor Outcomes during Pregnancy, Benin. Emerg Infect Dis. 2015;21:813–823. doi: 10.3201/eid2105.141626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goldie SJ, et al. Health and economic outcomes of HPV 16,18 vaccination in 72 GAVI-eligible countries. Vaccine. 2008;26:4080–4093. doi: 10.1016/j.vaccine.2008.04.053. [DOI] [PubMed] [Google Scholar]
- 26.Brune KD, et al. Plug-and-Display: decoration of Virus-Like Particles via isopeptide bonds for modular immunization. Sci Rep. 2016;6:19234. doi: 10.1038/srep19234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thrane S, et al. Bacterial superglue enables easy development of efficient virus-like particle based vaccines. J Nanobiotechnology. 2016;14:30. doi: 10.1186/s12951-016-0181-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Janitzek CM, et al. Bacterial superglue generates a full-length circumsporozoite protein virus-like particle vaccine capable of inducing high and durable antibody responses. Malar J. 2016;15:545. doi: 10.1186/s12936-016-1574-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singh SK, et al. Improving the malaria transmission-blocking activity of a Plasmodium falciparum 48/45 based vaccine antigen by SpyTag/SpyCatcher mediated virus-like display. Vaccine. 2017;35:3726–3732. doi: 10.1016/j.vaccine.2017.05.054. [DOI] [PubMed] [Google Scholar]
- 30.Palladini A, et al. Virus-like particle display of HER2 induces potent anti-cancer responses. Oncoimmunology. 2018;7:e1408749. doi: 10.1080/2162402X.2017.1408749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tissot AC, et al. Versatile virus-like particle carrier for epitope based vaccines. PLoS One. 2010;5:e9809. doi: 10.1371/journal.pone.0009809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nielsen MA, et al. Induction of adhesion-inhibitory antibodies against placental Plasmodium falciparum parasites by using single domains of VAR2CSA. Infect Immun. 2009;77:2482–2487. doi: 10.1128/IAI.00159-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nielsen MA, Salanti A. High-Throughput Testing of Antibody-Dependent Binding Inhibition of Placental Malaria Parasites. Methods Mol Biol. 2015;1325:241–253. doi: 10.1007/978-1-4939-2815-6_20. [DOI] [PubMed] [Google Scholar]
- 34.Buck CB, Pastrana DV, Lowy DR, Schiller JT. Generation of HPV pseudovirions using transfection and their use in neutralization assays. Methods Mol Med. 2005;119:445–462. doi: 10.1385/1-59259-982-6:445. [DOI] [PubMed] [Google Scholar]
- 35.Day PM, et al. A human papillomavirus (HPV) in vitro neutralization assay that recapitulates the in vitro process of infection provides a sensitive measure of HPV L2 infection-inhibiting antibodies. Clin Vaccine Immunol. 2012;19:1075–1082. doi: 10.1128/CVI.00139-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analyzed during the current study are available from the corresponding author on reasonable request.