This study investigated the molecular mechanisms possibly associated with non-wild-type MICs for lefamulin among staphylococci and streptococci included in the lefamulin surveillance program from 2015 to 2016. A total of 2,919 Staphylococcus aureus, 276 coagulase-negative staphylococci (CoNS), 3,923 Streptococcus pneumoniae, 389 β-hemolytic, and 178 viridans group streptococci isolates were included in the surveillance studies.
KEYWORDS: BC-3781, pleuromutilins, vga(A), lsa(E)
ABSTRACT
This study investigated the molecular mechanisms possibly associated with non-wild-type MICs for lefamulin among staphylococci and streptococci included in the lefamulin surveillance program from 2015 to 2016. A total of 2,919 Staphylococcus aureus, 276 coagulase-negative staphylococci (CoNS), 3,923 Streptococcus pneumoniae, 389 β-hemolytic, and 178 viridans group streptococci isolates were included in the surveillance studies. Eleven (0.3% of all S. aureus) S. aureus isolates with lefamulin MICs above the staphylococcal epidemiological cutoff (ECOFF) value (>0.25 μg/ml) were selected for this study. Eight (72.7%) S. aureus (lefamulin MIC, 0.5 to 4 μg/ml) isolates carried vga(A or E), one isolate (MIC, 32 μg/ml) carried lsa(E), one isolate (MIC, 16 μg/ml) had an alteration in L4, and one strain (MIC, 0.5 μg/ml) did not carry any of the investigated resistance mechanisms. A total of 14 (5.1% of all CoNS) CoNS isolates had lefamulin MICs (0.5 to >32 μg/ml) above the ECOFF. Similar to S. aureus, 8 (57.1%) CoNS (lefamulin MIC, 1 to 8 μg/ml) isolates carried vga(A or B), while 2 isolates (MIC, 4 to 32 μg/ml) carried cfr. High genetic diversity was observed among staphylococci, although 3 S. aureus isolates belonged to sequence type 398 (ST398). Among the 3 Streptococcus agalactiae and 3 viridans group streptococci (0.1% of all streptococci surveyed) isolates selected for additional characterization, all but 1 isolate carried lsa(E). This study documents a low occurrence of surveillance isolates exhibiting a non-wild-type MIC for lefamulin, and among these isolates, vga and lsa(E) prevailed in staphylococci and streptococci, respectively.
INTRODUCTION
Lefamulin belongs to the pleuromutilin class of antibiotics, and its antibacterial profile covers the most relevant organisms causing community-acquired bacterial pneumonia (CABP), including Gram-positive, fastidious Gram-negative, and atypical respiratory pathogens (1–3). Lefamulin also shows in vitro activity against multidrug-resistant Neisseria gonorrhoeae and Mycoplasma genitalium (4, 5). Thus, in addition to the clinical utility for treating CABP, the characteristic lefamulin antibacterial profile fits treatment for acute bacterial skin and skin structure infections (ABSSSIs) and sexually transmitted diseases (6).
Lefamulin inhibits bacterial protein synthesis by binding the 23S ribosomal subunit at the A and P sites in the peptidyl transferase center (PTC) via 4 hydrogen bonds and other interactions. An “induced-fit” mechanism, which is characteristic for pleuromutilin antibiotics and causes the tight fit of these molecules to the target site, hinders the correct positioning of the tRNA and thereby prohibits peptide bond formation (7, 8).
Mechanisms mediating resistance to pleuromutilins include mutations within the domain V of the 23S rRNA, including methylation of the nucleotide A2503 by the methyl transferase Cfr (9). Mutations in the 23S rRNA at positions 2032, 2055, 2447, 2499, 2504, and 2572 were previously described to confer resistance to tiamulin in Brachyspira spp. (10), while alterations at positions 2055, 2447, 2504, and 2572 were associated with valnemulin resistance in Mycobacterium smegmatis (11). Ribosomal proteins L3 and L4 are not primarily pleuromutilin targets, but mutations within these molecules may alter the PTC structure and affect binding. L3 (rplC) at the amino acid positions 145, 148, 149, 152, 155, 157, 158, and 159 and L4 (rplD) at position 68 were associated with resistance (7, 10, 12–14). Moreover, ATP-binding cassette F (ABC-F) proteins, such as vga(A–E) and lsa(E), initially described as putative efflux pumps can cause pleuromutilin resistance by ribosomal protection (15, 16).
As part of the clinical development, the in vitro activity of lefamulin and comparator agents have been monitored against a global collection of Gram-positive and fastidious Gram-negative organisms causing CABP and ABSSSI through the SENTRY Antimicrobial Surveillance Program. This study evaluated the occurrence of staphylococci and streptococci displaying elevated lefamulin MICs or above the epidemiological cutoff (ECOFF) during the SENTRY Program from 2015 to 2016 and characterized the possible associated resistance mechanisms among non-wild-type surveillance isolates.
RESULTS
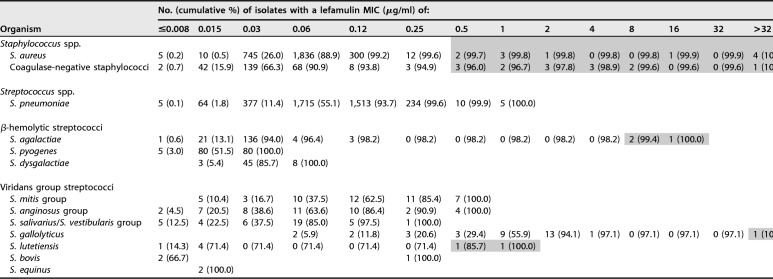
Lefamulin had MIC50 and MIC90 results of 0.06 and 0.12 μg/ml, respectively, with the majority (99.6%) of isolates displaying MICs of ≤0.008 to 0.25 μg/ml (Table 1). Eleven Staphylococcus aureus isolates showed lefamulin MICs above the ECOFF value (i.e., >0.25 μg/ml), and these isolates represented 0.3% of all S. aureus included in the 2015 and 2016 lefamulin surveillance programs (Table 1). When lefamulin was tested against coagulase-negative staphylococci (CoNS), a total of 14 isolates (4 species) had lefamulin MICs (0.5 to >32 μg/ml) above the ECOFF value (Tables 1, 2). The lefamulin MIC50 results obtained against streptococci varied depending on species or group of species (Table 1), and 3 Streptococcus agalactiae, 2 Streptococcus lutetiensis, and 1 Streptococcus gallolyticus isolates showed lefamulin MICs outside the wild-type distribution for the respective species and were further investigated.
TABLE 1.
Lefamulin MICs obtained during surveillance programs for 2015 and 2016a
Clinical isolates selected for further analysis with respective lefamulin MICs are highlighted.
bNC, not calculated.
TABLE 2.
MICs obtained for lefamulin and comparator agents tested against isolates included in the studya
| Collection no. | Species | MIC (µg/ml) by agent |
||||||
|---|---|---|---|---|---|---|---|---|
| Erythromycin | Clindamycin | Q-D | Linezolid | Lefamulin | Retapamulin | Chloramphenicol | ||
| 975498 | S. aureus | >8 | >64 | 0.5 | 1 | 0.5 (0.5) | ≤0.06 | 16 |
| 981256 | S. aureus | 0.12 | ≤0.5 | 0.5 | 1 | 0.5 (0.5) | 0.5 | 4 |
| 924825 | S. aureus | >8 | ≤0.5 | 0.5 | 1 | 1 (1) | 2 | 8 |
| 953474 | S. aureus | >8 | ≤0.5 | 0.5 | 0.5 | 1 (1) | 1 | 4 |
| 879822 | S. aureus | 0.12 | ≤0.5 | 0.5 | 1 | 2 (>1) | 4 | 8 |
| 913640 | S. aureus | >8 | >64 | 0.5 | 1 | 2 (>1) | 2 | 8 |
| 934242 | S. aureus | 0.12 | ≤0.5 | 0.5 | 0.5 | 2 (1) | 2 | 8 |
| 950457 | S. aureus | 0.12 | 8 | 1 | 1 | 4 (2) | >8 | 4 |
| 916083 | S. aureus | >8 | >64 | 1 | 0.25 | 16 (>1) | 8 | 8 |
| 976441 | S. aureus | >8 | >64 | 4 | 0.5 | 32 (16) | >8 | 64 |
| 972481 | S. aureus | 4 | 4 | 1 | 1 | >32 (>16) | >8 | 4 |
| 939671 | S. cohnii | >8 | ≤0.5 | 1 | 1 | 0.5 (2) | 1 | 4 |
| 939504 | S. epidermidis | >8 | ≤0.5 | ≤0.25 | 16 | 0.5 (1) | 0.25 | 16 |
| 947675 | S. epidermidis | >8 | 16 | ≤0.25 | 0.5 | 1 (0.5) | >8 | 4 |
| 951555 | S. epidermidis | >8 | >64 | 4 | 0.5 | 1 (0.5) | 1 | 4 |
| 955639 | S. epidermidis | 0.12 | ≤0.5 | 0.5 | 0.5 | 1 (1) | 1 | 4 |
| 956923 | S. epidermidis | >8 | 16 | ≤0.25 | 0.25 | 2 (0.5) | >8 | 2 |
| 949426 | S. epidermidis | ≤0.06 | 1 | ≤0.25 | 0.5 | 2 (2) | >8 | 4 |
| 938399 | S. epidermidis | >8 | ≤0.5 | ≤0.25 | 0.5 | 8 (4) | 8 | 2 |
| 952506 | S. epidermidis | >8 | 1 | ≤0.25 | 0.5 | 8 (4) | 8 | 4 |
| 958510 | S. epidermidis | ≤0.06 | 2 | ≤0.25 | 1 | 8 (2) | >8 | 4 |
| 934123 | S. epidermidis | 0.5 | >64 | 1 | 128 | 32 (8) | >8 | 64 |
| 939969 | S. haemolyticus | >8 | >64 | 4 | 2 | 4 (4) | 4 | 32 |
| 944662 | S. sciuri | >8 | >64 | 0.5 | 1 | 16 (8) | 8 | 32 |
| 941213 | S. sciuri | 0.25 | ≤0.5 | 1 | 1 | 32 (>16) | >8 | 4 |
| 960742 | S. lutetiensis | 0.03 | ≤0.5 | 1 | 1 | 0.5 (0.5) | 0.5 | 2 |
| 982012 | S. lutetiensis | ≤0.015 | 1 | 0.5 | 1 | 2 (1) | 2 | 2 |
| 971459 | S. agalactiae | >32 | >64 | 1 | 1 | 8 (8) | 8 | 2 |
| 935557 | S. agalactiae | 0.03 | 4 | 0.5 | 1 | 8 (8) | 4 | 2 |
| 935554 | S. agalactiae | 0.03 | 4 | 0.5 | 0.5 | 16 (16) | 4 | 2 |
| 965031 | S. gallolyticus | >32 | 4 | 1 | 2 | 32 (>16) | >8 | 4 |
Values within parentheses are the initial lefamulin MICs obtained during the surveillance studies. Q-D, quinupristin-dalfopristin.
Most S. aureus (7/11; 63.6%) isolates displaying lefamulin MICs of >0.25 μg/ml harbored vga(A) (lefamulin MIC, 0.5 to 4 μg/ml), while 2 strains carried either vga(E) (lefamulin MIC, >32 μg/ml) or the lsa(E) gene (lefamulin MIC, 32 μg/ml) (Table 3). Very little variability was observed in the S. aureus 23S rRNA nucleotide and ribosomal sequences. Overall, each isolate contained the same polymorphisms in the 23S rRNA (A21G, A1557T, and/or A2234G), while ribosomal proteins had wild-type sequences. The only exception was observed for isolate 916083, which had a V118A and an E147K in L4 (lefamulin MIC, 16 μg/ml). One S. aureus (975498) isolate with a lefamulin MIC of 0.5 μg/ml did not show any known resistance mechanisms associated with pleuromutilins. High genetic diversity was observed among staphylococci, although 3 S. aureus strains belonged to sequence type 398 (ST398).
TABLE 3.
Molecular epidemiology and resistance mechanism results for isolates included in this study
| Species | Isolate no. | Yr | MLSTa | Country | Lefamulin MIC (μg/ml) | Resistance determinantsb |
Ribosomal mutationsc |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Pleuromutilins | Other | 23S rRNA | L3 | L4 | L22 | ||||||
| S. aureus | 975498 | 2016 | 5 | United States | 0.5 | erm(A) | A21G, A1557T | WT | WT | WT | |
| S. aureus | 981256 | 2016 | 4335 | New Zealand | 0.5 | vga(A) | A21G, A1557T, A2234G | WT | WT | WT | |
| S. aureus | 924825 | 2015 | 88 | Australia | 1 | vga(A) | erm(C) | A21G, A1557T, A2234G | WT | WT | WT |
| S. aureus | 953474 | 2016 | 398 | France | 1 | vga(A) | erm(T) | A21G, A1557T, A2234G | WT | WT | WT |
| S. aureus | 879822 | 2015 | 1 | Slovenia | 2 | vga(A) | A21G, A2234G | WT | WT | WT | |
| S. aureus | 913640 | 2015 | 1148 | United States | 2 | vga(A) | erm(C) | A21G, A2234G | WT | WT | WT |
| S. aureus | 934242 | 2016 | 1148 | United States | 2 | vga(A) | A21G, A2234G | WT | WT | WT | |
| S. aureus | 950457 | 2016 | 97 | United States | 4 | vga(A) | A21G, A2234G | WT | WT | WT | |
| S. aureus | 916083 | 2015 | 5 | Korea | 16 | erm(A) | A21G, A1557T, A2234G | WT | V118A, E147K | WT | |
| S. aureus | 976441 | 2016 | 398 | Brazil | 32 | lsa(E) | erm(T), lnu(B) | A21G, A1557T, A2234G | WT | WT | WT |
| S. aureus | 972481 | 2016 | 398 | Germany | >32 | vga(E) | erm(A) | A21G, A1526G, A1557T | WT | WT | WT |
| S. cohnii | 939671 | 2016 | N/A | United States | 0.5 | msr(A) | A107G, A124G, T266G, C450T, T623C, A816G, T1261C, T1448A, T1549A | D108E, T190A, N193K, Y208F | N20S, A128T, A133T, V155I | WT | |
| S. epidermidis | 939504 | 2016 | 2 | Italy | 0.5 | msr(A) | G105A, G241T, T669C, T1236C, G2576T | WT | WT | WT | |
| S. epidermidis | 947675 | 2016 | 57 | United States | 1 | vga(A) | mph(C), msr(A) | G241T, T669C, T1236C | WT | WT | WT |
| S. epidermidis | 951555 | 2016 | 87 | Czech Republic | 1 | vga(A), vga(B) | erm(A) | C139T, G241T, T669C, T1236C | WT | WT | WT |
| S. epidermidis | 955639 | 2016 | 87 | Italy | 1 | vga(A), vga(B) | vat(B) | C139T, T669C, T1236C, C2809T | WT | WT | WT |
| S. epidermidis | 956923 | 2016 | 679 | Brazil | 2 | vga(A) | msr(A) | T669C, T1236C | V188I | WT | WT |
| S. epidermidis | 949426 | 2016 | 255 | United States | 2 | vga(A) | T669C, T1236C | WT | WT | WT | |
| S. epidermidis | 938399 | 2016 | 5 | United States | 8 | vga(A) | mph(C), msr(A) | C139T, T669C, T1236C | WT | WT | WT |
| S. epidermidis | 952506 | 2016 | 20 | Argentina | 8 | vga(A) | mph(C), msr(A) | C139T, T669C, T1236C, C1638T | WT | WT | WT |
| S. epidermidis | 958510 | 2016 | 487 | United States | 8 | vga(A) | G241T, T669C, T1236C, C1638T | WT | WT | WT | |
| S. epidermidis | 934123 | 2016 | 5 | United States | 32 | cfr | T669C, T1236C, C2534T | H146Q, V154L, A157R | G71_R72insG | WT | |
| S. haemolyticus | 939969 | 2016 | 3 | Mexico | 4 | cfr | mph(C), msr(A), erm(C) | C1486T, A2235G, T2882C | WT | WT | A29T |
| S. sciuri | 944662 | 2016 | N/A | Mexico | 16 | sal(A) | erm(C) | WT | WT | WT | A112D |
| S. sciuri | 941213 | 2016 | N/A | Australia | 32 | sal(A) | WT | WT | WT | WT | |
| S. lutetiensis | 960742 | 2016 | N/A | Belgium | 0.5 | lnu(C) | T225C, A2360G | WT | WT | WT | |
| S. lutetiensis | 982012 | 2016 | N/A | Argentina | 2 | lsa(E) | lnu(B), lnu(C) | WT | WT | WT | WT |
| S. agalactiae | 971459 | 2016 | 19 | Korea | 8 | lsa(E) | lnu(B), erm(B) | WT | WT | R1del, R2K | WT |
| S. agalactiae | 935557 | 2016 | 19 | Mexico | 8 | lsa(E) | lnu(B), erm(B) | WT | WT | R1del, R2K | WT |
| S. agalactiae | 935554 | 2016 | 19 | Mexico | 16 | lsa(E) | lnu(B), erm(B) | WT | WT | R1del, R2K | WT |
| S. gallolyticus | 965031 | 2016 | N/A | Spain | 32 | lsa(E) | lnu(B), erm(B) | C696T | WT | WT | WT |
MLST, multilocus sequence typing; N/A, not available.
MLSB (macrolide-lincosamide-streptogramin B) and pleuromutilin resistance genes screened as available at https://faculty.washington.edu/marilynr/ermwebA.pdf.
23S rRNA mutational analysis performed on nucleotide sequences (Escherichia coli numbering). Protein sequences analyzed for annotating L3, L4, and L22.
Both CoNS with a lefamulin MIC of 0.5 μg/ml, 1 Staphylococcus cohnii and 1 Staphylococcus epidermidis isolate, did not contain any known pleuromutilin resistance genes; however, both isolates had multiple alterations in the 23S rRNA or ribosomal proteins. Eight (57.1%) CoNS isolates contained acquired vga variants (lefamulin MIC, 1 to 8 μg/ml) (Table 3). The cfr gene was detected in 1 Staphylococcus haemolyticus (lefamulin MIC, 4 μg/ml) isolate and 1 S. epidermidis (lefamulin MIC, 32 μg/ml) isolate. The latter also showed multiple mutations in 23S rRNA, L3, and L4 (Table 3). Two Staphylococcus sciuri (lefamulin MIC, 16 to 32 μg/ml) isolates carried the intrinsic putative sal(A) gene (Table 3). In general, S. epidermidis isolates showed alterations in the 23S rRNA, such as G241T, T669C, and T1236C, that could be considered polymorphisms.
Among all streptococci surveyed, including 3,923 S. pneumoniae, 3 S. agalactiae, 2 S. lutetiensis, and 1 S. gallolyticus isolates, those with elevated lefamulin MICs (0.5 to 32 μg/ml) were selected for further evaluation (Tables 1 to 3). All but 1 of the selected streptococci carried lsa(E) (lefamulin MIC, 2 to 32 μg/ml) (Table 3). The S. lutetiensis isolate with a lefamulin MIC of 0.5 μg/ml carried lnu(C) and had a T225C and an A2360G in the 23S rRNA, while alterations within the ribosomal proteins evaluated were not detected (Table 3).
DISCUSSION
This 2-year (2015 to 2016) global surveillance program documents a small number of isolates showing a non-wild-type phenotype for lefamulin. Variants of the vga gene (8/11; 72.7%) were observed among most S. aureus isolates with lefamulin MICs above the ECOFF value (>0.25 μg/ml), while 2 isolates carried lsa(E) or L4 mutations (V118A and E147K). Two S. aureus (975498 and 981256) isolates displayed a lefamulin MIC of 0.5 μg/ml, but only the latter carried a vga(A) gene. Isolate 975498 only showed alterations in the 23S rRNA that was also observed in other S. aureus isolates included in the study, which are likely polymorphisms and not associated with pleuromutilin-resistance phenotypes. Furthermore, these locations are not associated with drug binding (7).
Staphylococci exhibiting elevated MICs to pleuromutilins, lincosamides, and streptogramin A (PLSA) usually carry the ATP-binding cassette F (ABC-F) proteins, such as those belonging to Vga, Lsa, or Sal families (15–17). In fact, similar to S. aureus, vga gene variants were also observed among most CoNS (8/14; 57.1%) or in 64.0% (16/25) of all staphylococci selected herein for further investigation. However, studies have demonstrated that alterations in the 23S rRNA and L3 ribosomal protein can also be responsible for decreased susceptibility to pleuromutilins (10–13, 18, 19), but in general, except for some polymorphisms observed in 23S rRNA, the S. aureus isolates included here showed 23S rRNA and ribosomal protein sequences equivalent to the respective reference strain.
Isolate 916083 displaying a lefamulin MIC of 16 μg/ml was the only S. aureus isolate with alterations in L4 (V118A and E147K). This isolate also exhibited elevated MICs for clindamycin, retapamulin, and erythromycin (Table 2). Previous studies linked L4 alterations with decreased susceptibility to tiamulin, chloramphenicol, and oxazolidinones (14, 20, 21). However, these previously reported alterations surrounded position K68, which is relatively close to the PTC and is responsible for stabilizing this region. V118 and E147 at L4 are located far from the PTC, but a hypothesis would be that L4 mutations may perturb the 3-dimensional structure of the 23S rRNA and minimize drug interaction (22). In fact, non-wild-type lefamulin MICs were obtained against 3 S. aureus surveillance isolates included in the SENTRY Program for 2010, and further investigations detected only the presence of L4 alterations in these older isolates. These isolates belonged to ST59 (lefamulin MIC, >16 μg/ml), carried A50G and V118A at L4 or ST398 (lefamulin MIC, 16 μg/ml), and had V118A and V142I at L4 (unpublished data). These data suggest that V118A, common to these S. aureus isolates from 2010, 2015, and 2016, may be associated with a decreased susceptibility to this agent. However, additional studies are needed to truly link this L4 alteration with the MICs presented here.
Two S. sciuri isolates showed lefamulin MICs of 16 to 32 μg/ml and did not carry any acquired resistance genes associated with the pleuromutilin phenotype. However, the sal(A) gene was detected in both isolates, and this gene was previously determined to be ubiquitous in this species and to cause decreased susceptibility to pleuromutilins and other agents (15, 23, 24). In addition, this gene has been detected in several staphylococcal species other than S. sciuri from animal and human origins, indicating that it has been mobilized to other bacterial species (15).
Both CoNS (939504 and 939671) isolates with a lefamulin MIC of 0.5 μg/ml had multiple alterations in the 23S rRNA. A G2576T was noted in isolate 939504, which is a well-known oxazolidinone resistance mechanism (14, 25) and known to affect tiamulin and valnemulin binding (11, 26). The binding effect for lefamulin appears to be less pronounced, likely because lefamulin appears to have more hydrogen bonds formed at the binding site than tiamulin and valnemulin. It also does not interact directly with G2576, although an alteration at this position causes a shift at the backbone of nucleotides from positions 2504 to 2507 (11), and these nucleotides interact with lefamulin (7). Importantly, isolate 939504 displayed a linezolid MIC of 16 μg/ml (Table 2), indicating the presence of G2576T in several 23S rRNA alleles (14); therefore, the lower lefamulin MIC was likely caused by a minimal effect of G2576T on drug binding rather than a low number of mutated ribosomes. As additional evidence, several staphylococci isolates included in past years of the SENTRY Program were characterized because of elevated linezolid MICs (≥8 μg/ml). Those isolates showing only G2576T had lefamulin MICs of 0.12 to 0.5 μg/ml (unpublished data), which are at the right side of the modal MIC (0.06 μg/ml) for S. aureus (Table 1). All 23S rRNA alterations observed in isolate 939671 are located outside the lefamulin binding site (13), and the L3 and L4 alterations detected have not been previously associated with resistance (19).
One S. epidermidis (lefamulin MIC, 32 μg/ml) and 1 S. haemolyticus (lefamulin MIC, 4 μg/ml) isolate carried cfr. This transferable gene confers a resistance phenotype to several classes of drugs (9), and its dissemination could jeopardize the clinical utility of several agents used in humans and animals. These study results corroborate those from large surveillance investigations that documented a low prevalence of cfr among Gram-positive isolates (25, 27, 28). Several studies reported sporadic outbreaks of cfr-carrying staphylococci; however, it was documented that the dissemination of such isolates are usually controlled by a combination of antibiotic stewardship and infection control measures (29–32).
All but 1 of the 6 streptococcal isolates selected for this study carried lsa(E). This gene has been reported among several Gram-positive isolates recovered from human and animal specimens (15, 24, 33). lsa(E) is usually part of a gene island that includes several resistance genes, including lnu(B) upstream (24, 34), which confers resistance to lincosamides. Among selected streptococcal isolates, pleuromutilin resistance mechanisms were not detected in S. lutetiensis 960742, except for 2 alterations in 23S rRNA (T225C and A2360G) located outside the lefamulin binding site (13). Isolate 960742 displayed a lefamulin MIC of 0.5 μg/ml, which is 32-fold higher than the modal MIC (0.015 μg/ml) shown for this species (Tables 1, 2).
In summary, this study showed a low prevalence of isolates exhibiting a non-wild-type MIC for lefamulin among Gram-positive isolates included in a 2-year global surveillance program. The non-wild-type phenotypes observed here could be generally explained by the presence of vga in staphylococci and lsa(E) in streptococci, which are more often detected among isolates collected from animals (15, 33–37). The association of acquired genes detected here with isolates from animal origins is further evidenced by the presence of 3 S. aureus isolates belonging to ST398, a lineage commonly responsible for infections in animals (16, 36–39). This study benchmarks the lefamulin activity against a global contemporary collection of Gram-positive surveillance isolates, as well as the rare instance of resistance genes associated with decreased susceptibility before clinical approval and use of this unique agent of the pleuromutilin class. Although the prevalence of surveillance isolates exhibiting non-wild-type MICs for lefamulin are rare, continued surveillance to monitor transferable genes and changes in MIC over time will provide valuable information for this new class of antibacterial agents for humans.
MATERIALS AND METHODS
Clinical isolates.
A total of 3,195 staphylococci and 4,489 streptococci isolates were included as part of the lefamulin surveillance program from 2015 to 2016. Based on the MIC distributions shown in Table 1, ECOFF values (≤0.25 μg/ml for S. aureus and CoNS) were calculated to define the lefamulin wild-type population of S. aureus and CoNS that included 99.9% of isolates within each group (40). Thus, staphylococcal isolates exhibiting lefamulin MICs of >0.25 μg/ml were selected for further molecular characterization (Table 1). Streptococci were selected based on species, and those isolates displaying elevated lefamulin MICs within a given species were selected for further molecular characterization (Table 1). Bacterial isolate identification was confirmed by matrix-assisted laser desorption ionization–time of flight mass spectrometry (Bruker Daltonics, Bremen, Germany) and genome sequencing.
Antimicrobial susceptibility testing.
Isolates were tested for susceptibility by broth microdilution methods, according to the recommendations of the Clinical and Laboratory Standards Institute (CLSI) (41). Frozen-form broth microdilution 96-well plates were manufactured by JMI Laboratories and contained cation-adjusted Mueller-Hinton broth (2.5% to 5% lysed horse blood added for testing streptococci). Isolates that met the inclusion criteria were retested for susceptibility in frozen-form panels containing extended ranges for lefamulin among other agents (Table 2). Bacterial inoculum density was monitored by colony counts to ensure an adequate number of cells for each testing event. MICs were validated by concurrently testing CLSI-recommended quality-control strains (42).
Characterization of resistance mechanisms by next-generation genome sequencing and analysis.
Selected isolates had total genomic DNA extracted with the fully automated Thermo Scientific KingFisher Flex magnetic particle processor (Cleveland, OH, USA), which was used as input material for library construction. DNA libraries were prepared using the Nextera library construction protocol (Illumina, San Diego, CA, USA) following the manufacturer’s instructions and were sequenced on a MiSeq sequencer (JMI Laboratories, North Liberty, IA, USA). FASTQ format sequencing files for each sample set were assembled independently using the de novo assembler SPAdes 3.9.0 (43), and an in-house-designed software program was applied to the assembled sequences to align against known macrolide-lincosamide-streptogramin B (MLSB) and pleuromutilin resistance genes, including tva(A) (44–46).
Sequences of 23S rRNA (PTC), rplC (L3), rplD (L4), and rplV (L22) were extracted from the assembled sequences and evaluated against corresponding sequences of susceptible wild-type reference strains. The analysis of 23S rRNA was performed based on nucleotide sequences (Escherichia coli numbering), while those from rRNA proteins were based on amino acid sequences. All intrinsic 23S rRNA target genes or ribosomal protein amino acid sequences were considered wild type if 100% identity was observed with the respective reference sequences. Nucleotide and amino acid differences were annotated when an identity of <100% was observed. Reference sequences were extracted from the following strains: S. aureus (NCTC 8325), S. epidermidis (ATCC 12228), S. cohnii (ATCC 29974), S. haemolyticus (JCSC1435), S. sciuri (ATCC 29062), S. lutetiensis (NCTC 13774), S. agalactiae (NEM316), and S. gallolyticus (ATCC 43143).
Multilocus sequence typing.
Multilocus sequence typing (MLST) was performed by extracting previously defined sets of 7 housekeeping gene fragments (approximately 500 bp) from each assembled sequence. Each fragment was compared with known allelic variants for each locus (housekeeping gene) on the MLST website (PubMLST, https://pubmlst.org). An allele sharing 100% genetic identity with a known variant received a numeric designation, and a 7-number sequence (1 for each housekeeping gene) formed an allelic profile, defined as STs.
Data availability.
This is an original work and the data set repository and published article in which the data set and/or code was originally described and have not been published previously. Upon request, and subject to certain criteria, conditions and exceptions, JMI Laboratories and Nabriva Therapeutics will provide access to the code and databases utilized here. This information may be requested 24 months after study completion and will be made available to researchers whose proposals meet the research criteria and other conditions and for which an exception does not apply, via a secure portal. To gain access, requestors must enter into an information access agreement with JMI Laboratories and/or Nabriva Therapeutics.
ACKNOWLEDGMENTS
The surveillance study and experiments performed in this study were supported by Nabriva Therapeutics.
The authors express appreciation to the following JMI employees for technical support or manuscript assistance: L. Flanigan, J. Oberholser, and C. Smith. Editorial support was provided by Lycely del C. Sepulveda-Torres at C4 MedSolutions, LLC (Yardley, PA), a CHC Group company, and funded by Nabriva Therapeutics.
JMI Laboratories was contracted to perform services in 2017 for Achaogen, Allecra Therapeutics, Allergan, Amplyx Pharmaceuticals, Antabio, API, Astellas Pharma, AstraZeneca, Athelas, Basilea Pharmaceutica, Bayer AG, Becton, Dickinson and Co., Boston Pharmaceuticals, CEM-102 Pharma, Cempra, Cidara Therapeutics, Inc., CorMedix, CSA Biotech, Cutanea Life Sciences, Inc., Entasis Therapeutics, Inc., Geom Therapeutics, Inc., GSK, Iterum Pharma, Medpace, Melinta Therapeutics, Inc., Merck & Co., Inc., MicuRx Pharmaceuticals, Inc., N8 Medical, Inc., Nabriva Therapeutics, Inc., NAEJA-RGM, Novartis, Paratek Pharmaceuticals, Inc., Pfizer, Polyphor, Ra Pharma, Rempex, Riptide Bioscience, Inc., Roche, Scynexis, Shionogi, Sinsa Labs, Inc., Skyline Antiinfectives, Sonoran Biosciences, Spero Therapeutics, Symbiotica, Synlogic, Synthes Biomaterials, TenNor Therapeutics, Tetraphase, The Medicines Company, Theravance Biopharma, VenatoRx Pharmaceuticals, Inc., Wockhardt, Yukon Pharma, Zai Laboratory, and Zavante Therapeutics, Inc. There are no speakers bureaus or stock options to declare for R.E.M., T.B.D., R.K.F. or H.S.S. S.P. and S.P.G. are employees of and hold stock in Nabriva Therapeutics plc.
Footnotes
For a companion article on this topic, see https://doi.org/10.1128/AAC.02161-18.
REFERENCES
- 1.Mendes RE, Farrell DJ, Flamm RK, Talbot GH, Ivezic-Schoenfeld Z, Paukner S, Sader HS. 2016. In vitro activity of lefamulin tested against Streptococcus pneumoniae with defined serotypes, including multidrug-resistant isolates causing lower respiratory tract infections in the United States. Antimicrob Agents Chemother 60:4407–4411. doi: 10.1128/AAC.00627-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Waites KB, Crabb DM, Duffy LB, Jensen JS, Liu Y, Paukner S. 2017. In vitro activities of lefamulin and other antimicrobial agents against macrolide-susceptible and macrolide-resistant Mycoplasma pneumoniae from the United States, Europe, and China. Antimicrob Agents Chemother 61:e2008-16. doi: 10.1128/AAC.02008-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sader HS, Paukner S, Ivezic-Schoenfeld Z, Biedenbach DJ, Schmitz FJ, Jones RN. 2012. Antimicrobial activity of the novel pleuromutilin antibiotic BC-3781 against organisms responsible for community-acquired respiratory tract infections (CARTIs). J Antimicrob Chemother 67:1170–1175. doi: 10.1093/jac/dks001. [DOI] [PubMed] [Google Scholar]
- 4.Paukner S, Gruss A, Jensen JS. 2018. In vitro activity of lefamulin against sexually transmitted bacterial pathogens. Antimicrob Agents Chemother 62:e02380-17. doi: 10.1128/AAC.02380-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacobsson S, Paukner S, Golparian D, Jensen JS, Unemo M. 2017. In vitro activity of the novel pleuromutilin lefamulin (BC-3781) and effect of efflux pump inactivation on multidrug-resistant and extensively drug-resistant Neisseria gonorrhoeae. Antimicrob Agents Chemother 61:e01497-17. doi: 10.1128/AAC.01497-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Veve MP, Wagner JL. 2018. Lefamulin: review of a promising novel pleuromutilin antibiotic. Pharmacotherapy 38:935–946. doi: 10.1002/phar.2166. [DOI] [PubMed] [Google Scholar]
- 7.Eyal Z, Matzov D, Krupkin M, Paukner S, Riedl R, Rozenberg H, Zimmerman E, Bashan A, Yonath A. 2016. A novel pleuromutilin antibacterial compound, its binding mode and selectivity mechanism. Sci Rep 6:39004. doi: 10.1038/srep39004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eyal Z, Matzov D, Krupkin M, Wekselman I, Paukner S, Zimmerman E, Rozenberg H, Bashan A, Yonath A. 2015. Structural insights into species-specific features of the ribosome from the pathogen Staphylococcus aureus. Proc Natl Acad Sci U S A 112:E5805–E5814. doi: 10.1073/pnas.1517952112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Long KS, Poehlsgaard J, Kehrenberg C, Schwarz S, Vester B. 2006. The cfr rRNA methyltransferase confers resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A antibiotics. Antimicrob Agents Chemother 50:2500–2505. doi: 10.1128/AAC.00131-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pringle M, Poehlsgaard J, Vester B, Long KS. 2004. Mutations in ribosomal protein L3 and 23S ribosomal RNA at the peptidyl transferase centre are associated with reduced susceptibility to tiamulin in Brachyspira spp. isolates. Mol Microbiol 54:1295–1306. doi: 10.1111/j.1365-2958.2004.04373.x. [DOI] [PubMed] [Google Scholar]
- 11.Long KS, Poehlsgaard J, Hansen LH, Hobbie SN, Bottger EC, Vester B. 2009. Single 23S rRNA mutations at the ribosomal peptidyl transferase centre confer resistance to valnemulin and other antibiotics in Mycobacterium smegmatis by perturbation of the drug binding pocket. Mol Microbiol 71:1218–1227. doi: 10.1111/j.1365-2958.2009.06596.x. [DOI] [PubMed] [Google Scholar]
- 12.Gentry DR, Rittenhouse SF, McCloskey L, Holmes DJ. 2007. Stepwise exposure of Staphylococcus aureus to pleuromutilins is associated with stepwise acquisition of mutations in rplC and minimally affects susceptibility to retapamulin. Antimicrob Agents Chemother 51:2048–2052. doi: 10.1128/AAC.01066-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Locke JB, Hilgers M, Shaw KJ. 2009. Mutations in ribosomal protein L3 are associated with oxazolidinone resistance in staphylococci of clinical origin. Antimicrob Agents Chemother 53:5275–5278. doi: 10.1128/AAC.01032-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Locke JB, Hilgers M, Shaw KJ. 2009. Novel ribosomal mutations in Staphylococcus aureus strains identified through selection with the oxazolidinones linezolid and torezolid (TR-700). Antimicrob Agents Chemother 53:5265–5274. doi: 10.1128/AAC.00871-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng F, Wang H, Liao Y, Li J, Feßler AT, Michael GB, Schwarz S, Wang Y. 2017. Detection and genetic environment of pleuromutilin-lincosamide-streptogramin A resistance genes in staphylococci isolated from pets. Front Microbiol 8:234. doi: 10.3389/fmicb.2017.00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mendes RE, Smith TC, Deshpande L, Diekema DJ, Sader HS, Jones RN. 2011. Plasmid-borne vga(A)-encoding gene in methicillin-resistant Staphylococcus aureus ST398 recovered from swine and a swine farmer in the United States. Diagn Microbiol Infect Dis 71:177–180. doi: 10.1016/j.diagmicrobio.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 17.Sharkey LKR, O'Neill AJ. 2018. Antibiotic resistance ABC-F proteins: bringing target protection into the limelight. ACS Infect Dis 4:239–246. doi: 10.1021/acsinfecdis.7b00251. [DOI] [PubMed] [Google Scholar]
- 18.Hillen S, Willems H, Herbst W, Rohde J, Reiner G. 2014. Mutations in the 50S ribosomal subunit of Brachyspira hyodysenteriae associated with altered minimum inhibitory concentrations of pleuromutilins. Vet Microbiol 172:223–229. doi: 10.1016/j.vetmic.2014.04.021. [DOI] [PubMed] [Google Scholar]
- 19.Klitgaard RN, Ntokou E, Nørgaard K, Biltoft D, Hansen LH, Trædholm NM, Kongsted J, Vester B. 2015. Mutations in the bacterial ribosomal protein l3 and their association with antibiotic resistance. Antimicrob Agents Chemother 59:3518–3528. doi: 10.1128/AAC.00179-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wolter N, Smith AM, Farrell DJ, Schaffner W, Moore M, Whitney CG, Jorgensen JH, Klugman KP. 2005. Novel mechanism of resistance to oxazolidinones, macrolides, and chloramphenicol in ribosomal protein L4 of the pneumococcus. Antimicrob Agents Chemother 49:3554–3557. doi: 10.1128/AAC.49.8.3554-3557.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Long KS, Vester B. 2012. Resistance to linezolid caused by modifications at its binding site on the ribosome. Antimicrob Agents Chemother 56:603–612. doi: 10.1128/AAC.05702-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gregory ST, Dahlberg AE. 1999. Erythromycin resistance mutations in ribosomal proteins L22 and L4 perturb the higher order structure of 23 S ribosomal RNA. J Mol Biol 289:827–834. doi: 10.1006/jmbi.1999.2839. [DOI] [PubMed] [Google Scholar]
- 23.Hot C, Berthet N, Chesneau O. 2014. Characterization of sal(A), a novel gene responsible for lincosamide and streptogramin A resistance in Staphylococcus sciuri. Antimicrob Agents Chemother 58:3335–3341. doi: 10.1128/AAC.02797-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feßler AT, Wang Y, Wu C, Schwarz S. 2018. Mobile lincosamide resistance genes in staphylococci. Plasmid 99:22–31. doi: 10.1016/j.plasmid.2018.06.002. [DOI] [PubMed] [Google Scholar]
- 25.Mendes RE, Deshpande LM, Jones RN. 2014. Linezolid update: stable in vitro activity following more than a decade of clinical use and summary of associated resistance mechanisms. Drug Resist Updat 17:1–12. doi: 10.1016/j.drup.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 26.Miller K, Dunsmore CJ, Fishwick CW, Chopra I. 2008. Linezolid and tiamulin cross-resistance in Staphylococcus aureus mediated by point mutations in the peptidyl transferase center. Antimicrob Agents Chemother 52:1737–1742. doi: 10.1128/AAC.01015-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mendes RE, Deshpande L, Streit JM, Sader HS, Castanheira M, Hogan PA, Flamm RK. 2018. ZAAPS programme results for 2016: an activity and spectrum analysis of linezolid using clinical isolates from medical centres in 42 countries. J Antimicrob Chemother 73:1880–1887. doi: 10.1093/jac/dky099. [DOI] [PubMed] [Google Scholar]
- 28.Pfaller MA, Mendes RE, Streit JM, Hogan PA, Flamm RK. 2017. Five-year summary of in vitro activity and resistance mechanisms of linezolid against clinically important Gram-positive cocci in the United States from the LEADER Surveillance Program (2011 to 2015). Antimicrob Agents Chemother 61:e00609-17. doi: 10.1128/AAC.00609-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanchez Garcia M, De la Torre MA, Morales G, Pelaez B, Tolon MJ, Domingo S, Candel FJ, Andrade R, Arribi A, Garcia N, Martinez Sagasti F, Fereres J, Picazo J. 2010. Clinical outbreak of linezolid-resistant Staphylococcus aureus in an intensive care unit. JAMA 303:2260–2264. doi: 10.1001/jama.2010.757. [DOI] [PubMed] [Google Scholar]
- 30.O'Connor C, Powell J, Finnegan C, O'Gorman A, Barrett S, Hopkins KL, Pichon B, Hill R, Power L, Woodford N, Coffey JC, Kearns A, O'Connell NH, Dunne CP. 2015. Incidence, management and outcomes of the first cfr-mediated linezolid-resistant Staphylococcus epidermidis outbreak in a tertiary referral centre in the Republic of Ireland. J Hosp Infect 90:316–321. doi: 10.1016/j.jhin.2014.12.013. [DOI] [PubMed] [Google Scholar]
- 31.Li X, Arias CA, Aitken SL, Galloway Pena J, Panesso D, Chang M, Diaz L, Rios R, Numan Y, Ghaoui S, DebRoy S, Bhatti MM, Simmons DE, Raad I, Hachem R, Folan SA, Sahasarabhojane P, Kalia A, Shelburne SA. 2018. Clonal emergence of invasive multidrug-resistant Staphylococcus epidermidis deconvoluted via a combination of whole-genome sequencing and microbiome analyses. Clin Infect Dis 67:398–406. doi: 10.1093/cid/ciy089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Inkster T, Coia J, Meunier D, Doumith M, Martin K, Pike R, Imrie L, Kane H, Hay M, Wiuff C, Wilson J, Deighan C, Hopkins KL, Woodford N, Hill R. 2017. First outbreak of colonization by linezolid- and glycopeptide-resistant Enterococcus faecium harbouring the cfr gene in a UK nephrology unit. J Hosp Infect 97:397–402. doi: 10.1016/j.jhin.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 33.Hawkins PA, Law CS, Metcalf BJ, Chochua S, Jackson DM, Westblade LF, Jerris R, Beall BW, McGee L. 2017. Cross-resistance to lincosamides, streptogramins A and pleuromutilins in Streptococcus agalactiae isolates from the USA. J Antimicrob Chemother 72:1886–1892. doi: 10.1093/jac/dkx077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wendlandt S, Kadlec K, Feßler AT, Schwarz S. 2015. Identification of ABC transporter genes conferring combined pleuromutilin-lincosamide-streptogramin A resistance in bovine methicillin-resistant Staphylococcus aureus and coagulase-negative staphylococci. Vet Microbiol 177:353–358. doi: 10.1016/j.vetmic.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 35.Li B, Wendlandt S, Yao J, Liu Y, Zhang Q, Shi Z, Wei J, Shao D, Schwarz S, Wang S, Ma Z. 2013. Detection and new genetic environment of the pleuromutilin-lincosamide-streptogramin A resistance gene lsa(E) in methicillin-resistant Staphylococcus aureus of swine origin. J Antimicrob Chemother 68:1251–1255. doi: 10.1093/jac/dkt015. [DOI] [PubMed] [Google Scholar]
- 36.Kadlec K, Pomba CF, Couto N, Schwarz S. 2010. Small plasmids carrying vga(A) or vga(C) genes mediate resistance to lincosamides, pleuromutilins and streptogramin A antibiotics in methicillin-resistant Staphylococcus aureus ST398 from swine. J Antimicrob Chemother 65:2692–2693. doi: 10.1093/jac/dkq365. [DOI] [PubMed] [Google Scholar]
- 37.Hauschild T, Fessler AT, Kadlec K, Billerbeck C, Schwarz S. 2012. Detection of the novel vga(E) gene in methicillin-resistant Staphylococcus aureus CC398 isolates from cattle and poultry. J Antimicrob Chemother 67:503–504. doi: 10.1093/jac/dkr446. [DOI] [PubMed] [Google Scholar]
- 38.Schwendener S, Perreten V. 2011. New transposon Tn6133 in methicillin-resistant Staphylococcus aureus ST398 contains vga(E), a novel streptogramin A, pleuromutilin, and lincosamide resistance gene. Antimicrob Agents Chemother 55:4900–4904. doi: 10.1128/AAC.00528-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith TC, Wardyn SE. 2015. Human infections with Staphylococcus aureus CC398. Curr Environ Health Rep 2:41–51. doi: 10.1007/s40572-014-0034-8. [DOI] [PubMed] [Google Scholar]
- 40.Turnidge J, Kahlmeter G, Kronvall G. 2006. Statistical characterisation of bacterial wild-type MIC value distributions and the determination of epidemiological cut-off values. Clin Microbiol Infect 12:418–425. doi: 10.1111/j.1469-0691.2006.01377.x. [DOI] [PubMed] [Google Scholar]
- 41.Clinical Laboratory Standards Institute. 2012. M07-A9. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 9th ed. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 42.Clinical Laboratory Standards Institute. 2018. M100Ed28E. Performance standards for antimicrobial susceptibility testing, 28th informational supplement. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 43.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roberts MC. 2017. Mechanisms of MLS resistance modified Aug. 1, 2018. https://faculty.washington.edu/marilynr/ermwebA.pdf.
- 45.Card RM, Stubberfield E, Rogers J, Nunez-Garcia J, Ellis RJ, AbuOun M, Strugnell B, Teale C, Williamson S, Anjum MF. 2018. Identification of a new antimicrobial resistance gene provides fresh insights Into pleuromutilin resistance in Brachyspira hyodysenteriae, aetiological agent of swine dysentery. Front Microbiol 9:1183. doi: 10.3389/fmicb.2018.01183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roberts MC. 2018. Efflux and inactivating genes modified Aug. 1, 2018. https://faculty.washington.edu/marilynr/ermweb2.pdf.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This is an original work and the data set repository and published article in which the data set and/or code was originally described and have not been published previously. Upon request, and subject to certain criteria, conditions and exceptions, JMI Laboratories and Nabriva Therapeutics will provide access to the code and databases utilized here. This information may be requested 24 months after study completion and will be made available to researchers whose proposals meet the research criteria and other conditions and for which an exception does not apply, via a secure portal. To gain access, requestors must enter into an information access agreement with JMI Laboratories and/or Nabriva Therapeutics.