AbaR-type genomic islands (AbaRs) are important elements responsible for antimicrobial resistance in Acinetobacter baumannii. This study performed a large-scale identification of AbaRs to understand their distribution and compositions of antimicrobial resistance genes.
KEYWORDS: antimicrobial resistance, genome, insertion site, mobile genetic element
ABSTRACT
AbaR-type genomic islands (AbaRs) are important elements responsible for antimicrobial resistance in Acinetobacter baumannii. This study performed a large-scale identification of AbaRs to understand their distribution and compositions of antimicrobial resistance genes. We identified 2.89-kb left-end and 1.87-kb right-end conserved sequences (CSs) and developed a bioinformatics approach to identify AbaRs, using the CSs as signatures, in 3,148 publicly available genomes. AbaRs were prevalent in A. baumannii, being found in 2,091 genomes. They were sparse in other Acinetobacter species and confined only to this genus. Results from 111 complete genomes showed that over 85% of AbaRs resided on chromosomes. The external flanks adjacent to the inverted repeats available in all identified CSs were mapped to an AbaR-free chromosome or searched in the NCBI database for empty loci to define insertion sites. Surprisingly, 84 insertion sites with diverse origins were revealed, including 51 scattered on the chromosome, 20 plasmid borne, 12 located on prophages, transposons, ISAba1, complex AbaRs, and genomic islands of other types, and one uncharacterized, and some were strongly associated with clonal lineages. Finally, we found 994 antimicrobial resistance genes covering 28 unique genes from 70.9% (299/422) of intact AbaRs currently available. The resistance gene profiles displayed an apparent clonal lineage-specific pattern, highlighting the distinct features of AbaRs in global clone 1 (GC1) and GC2. The tet(B) gene was highly specific to the AbaRs in GC2. In conclusion, AbaRs have diverse insertion sites on the chromosome and mobile genetic elements (MGEs) and display distinct antimicrobial resistance gene profiles in different clonal lineages.
INTRODUCTION
Acinetobacter baumannii is a nosocomial pathogen that infects and causes serious disease in immunocompromised people. A. baumannii is feared because an increasing number of clinical isolates are resistant against multiple antibiotics. AbaR-type genomic islands (AbaRs) are a class of important mobile genetic elements (MGEs) known to be involved in multiple antimicrobial resistance in A. baumannii (1–7). The noticeable plasticity of these elements facilitates the evolution of bespoke AbaRs that enable A. baumannii strains to thrive in the presence of multiple antibiotics and makes A. baumannii a formidable threat to the health of immunocompromised patients in clinical settings.
AbaRs are diverse, containing various resistance genes, and have distinct structural features in different epidemic clones (8–11). The AbaR3-type islands in global clone 1 (GC1) usually carry multiple resistance genes and have been mapped to a backbone transposon, Tn6019 (8, 9). AbaRs like AbaR4, which is mostly found in GC2 and carries the blaOXA-23 gene, differ from the AbaR3-type elements (4, 10, 12, 13) and have a Tn6022 backbone. A related element that carries no antibiotic resistance genes, Tn6021, has also been found (8, 12). More complex AbaR variants like Tn6166 (13) and Tn6167 (14) in GC2 that are associated with both Tn6022 and Tn6172 have been characterized and recently termed AbGRI1-type islands to distinguish them from the AbaR islands in GC1 (11, 15). Although these elements differ substantially in resistance gene content and organization and display distinct structural features, their backbones are closely related, and they share several open reading frames that constitute the so-called transposition module and 26-bp imperfect terminal inverted repeats (IRs) (9–11, 16). For simplicity, the abbreviation “AbaRs” will be used in this study to collectively refer to all of the AbaR-type genomic islands. Most AbaRs site-specifically disrupt the chromosomal comM gene, generating a target site duplication indicated by 5-bp perfect direct repeats (DRs). However, other chromosomal insertion sites, such as pho for some AbaR4-type islands in GC1 and the site identical to ACICU_02698 in an isolate belonging to clonal complex 10, also have been reported with distinct 5-bp DRs (10, 17). Plasmids are also reported to carry AbaRs. An AbaR4 carried by the conjugative plasmid pA85-3 has been found in a GC1 isolate (18). Moreover, the AbGIR-type elements are believed to have originated from a plasmid segment containing a Tn6022 and a Tn6172 that has transposed as a single unit into the comM locus of GC2 strains (11).
Compared to the exponential growth in sequenced A. baumannii genomes, only a limited number of AbaRs have been reported. This study has identified AbaRs from 3,148 publicly available genomes, allowing a large-scale analysis of AbaRs to understand their distribution and antimicrobial resistance features and to further correlate their clonal distribution with antimicrobial resistance signatures.
RESULTS
AbaRs contain conserved sequences at their terminal regions.
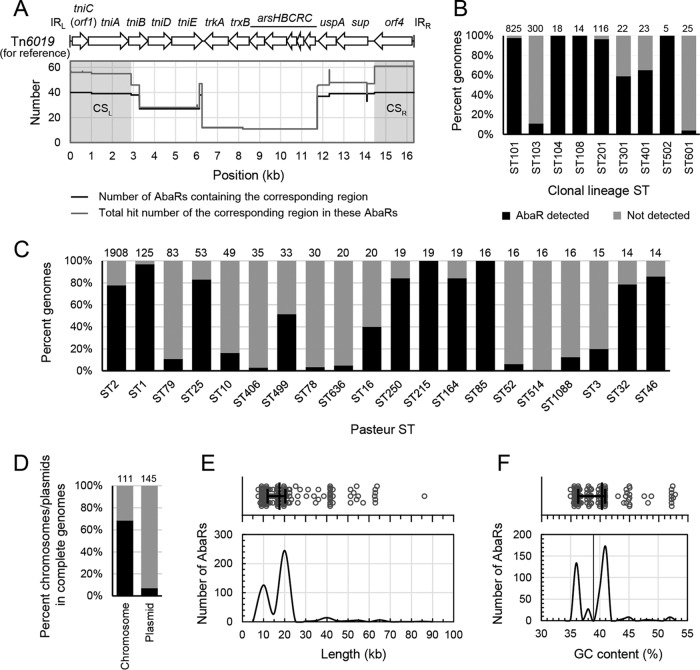
Comparison of 40 known AbaRs with the reference element Tn6019 that was known as the backbone of AbaR3-type elements in GC1 (8, 9), including the AbaR3, AbaR4 and AbGRI1 types (Table S1), revealed a 2.89-kb region at the left end and a 1.87-kb region at the right end that share conserved nucleotide sequences (Fig. 1A). The left-end conserved sequence (CSL) included the left-end inverted repeat (IRL), tniC (orf1), tniA, and partial tniB, while the right-end conserved sequence (CSR) contained orf4 and the right-end inverted repeat (IRR). The region containing uspA and sup adjacent to the CSR also appeared to be conserved. However, it mostly occurred along with an extra CSR within the internal region of a complex AbaR (e.g., AbGRI1-type elements), instead of located at the right terminal region of the entire element.
FIG 1.
Identification of AbaRs in Acinetobacter baumannii genomes. (A) Conserved sequences (CSs) of AbaRs obtained by aligning 40 known AbaRs against the reference element Tn6019. Left-end and right-end CSs (CSL and CSR, respectively) are indicated by gray shades. (B) Abundance of AbaRs in different clonal lineage sequence types (STs). (C) Abundance of AbaRs in different Pasteur STs. Only STs with top-20 genome numbers are shown. For panels B and C, the numbers above the histograms are the numbers of genomes belonging to the corresponding STs. Note that STs could not be determined in many unfinished genomes. For the results by Oxford ST, see Fig. S2. See Data Set S1 for results with detailed ST information. (D) Percentages of chromosomes and plasmids containing AbaRs based on the data from 111 complete genomes. The numbers above the histograms are the total numbers of chromosomes/plasmids. (E) Length distribution of AbaRs. (F) Guanine and cytosine content (GC %) distribution of AbaRs. For panels E and F, medians with interquartile ranges are given, and the data are based on 442 intact AbaRs.
AbaRs are prevalent in A. baumannii and are mostly chromosome borne.
The CSL and CSR were used for in silico AbaR identification (Fig. S1) in 3,148 unique A. baumannii genomes, including 111 complete and 3,037 draft genomes (Data Set S1). The CSs were identified in 2,091 (66.4%) genomes covering 100% (11/11) of clonal lineage sequence types (STs) (19), 45.7% (107/234) of Pasteur STs (20) and 59.0% (184/312) of Oxford STs (21) that could be detected (Data Sets S1 and S2). Note that STs could not be determined in many unfinished genomes. The Pasteur multilocus sequence typing (MLST) scheme was used as a reference method in this study (20). The abundances of AbaRs were different in different clones (Fig. 1B and C and S2) based on the current data. Of note, some multidrug-resistant epidemic clones showed extremely high abundance, butut it should be emphasized that the current genome database might be biased due to the overrepresentation of clinical isolates of certain STs. In other species, only a few AbaRs were found in a limited number of Acinetobacter species when searched against the NCBI nonredundant nucleotide database (Data Set S3).
Intact AbaRs (referring in this study to those with intact termini) were first identified in 111 complete A. baumannii genomes, and over 85% resided on chromosomes. In these genomes, we found 79 intact AbaRs on 76 chromosomes and 13 on 10 plasmids (Data Set S4). The AbaR-containing plasmids accounted for less than 7% (10/145) of all plasmids (Fig. 1D). Consistently, no AbaRs were detected on the additional 19 plasmids sequenced alone (Data Set S5). Most chromosomes or plasmids contained one intact AbaR, while a few harbored two. Among the AbaRs, 41 were identified as complex elements, as they contained extra CS copies in their internal regions and they were mostly found in the genomes of GC2 strains, which might be linked to the AbGRI1-type islands (Data Set S4). Interestingly, apart from comM, pho, ACICU_02698, and the three plasmid-borne insertion sites known previously, six novel insertion sites identical to ABR2091_2729, ABR2091_2130, AB57_05270, B7L39_19070, D721_p10064, and umuC were revealed (Data Set S4). Notably, AB57_05270 was a prophage-borne gene, whereas the last three listed were plasmid borne. The AbaRs previously reported in these genomes (Table S1) were all identified. Meanwhile, 34 AbaR remnants were found in 14 genomes, suggesting that they might have gone through complex genetic events. Similarly, nearly 80% (27/34) of remnants were located on chromosomes. We further managed to identify 331 intact AbaRs from 346 draft genomes (Data Set S4). Note that in draft genomes, an intact AbaR could be split on different contigs, which did not allow its identification as an intact element. Thus, intact AbaRs could only be defined in a limited portion of draft genomes (11.4%; 346/3037).
The 442 intact AbaRs were 7.66 to 86.24 kb (median, 17.63 kb) in length, and the distribution displayed two major peaks at the 7.5- to 12.5-kb and 17.5- to 22.5-kb intervals (Fig. 1E), which might correspond to certain types of AbaRs. Their GC contents varied from 34.94% to 52.80% (median, 40.32%), comparing to the ∼39% of A. baumannii genomes, the distribution likewise displayed three major peaks at 34.5% to 35.5%, 37.5% to 38.5% and 39.5% to 41.5% intervals (Fig. 1F).
AbaRs have diverse insertion sites.
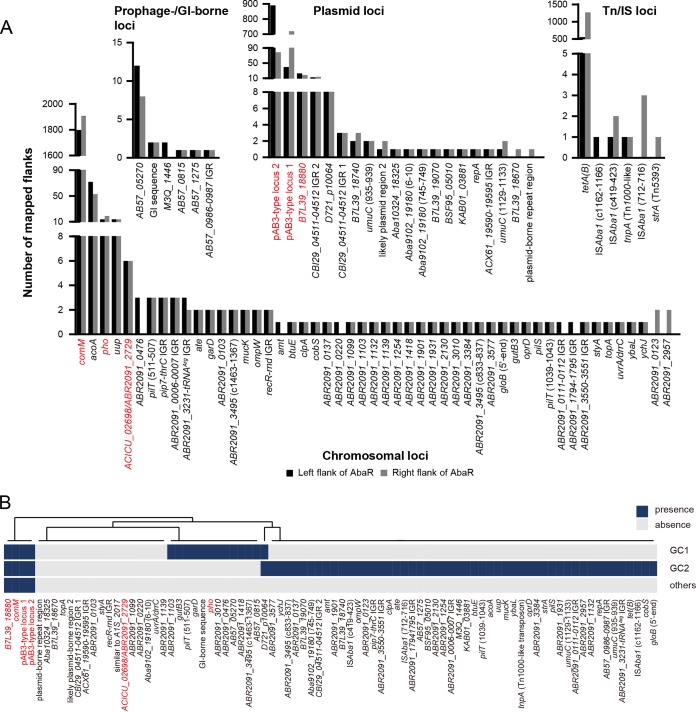
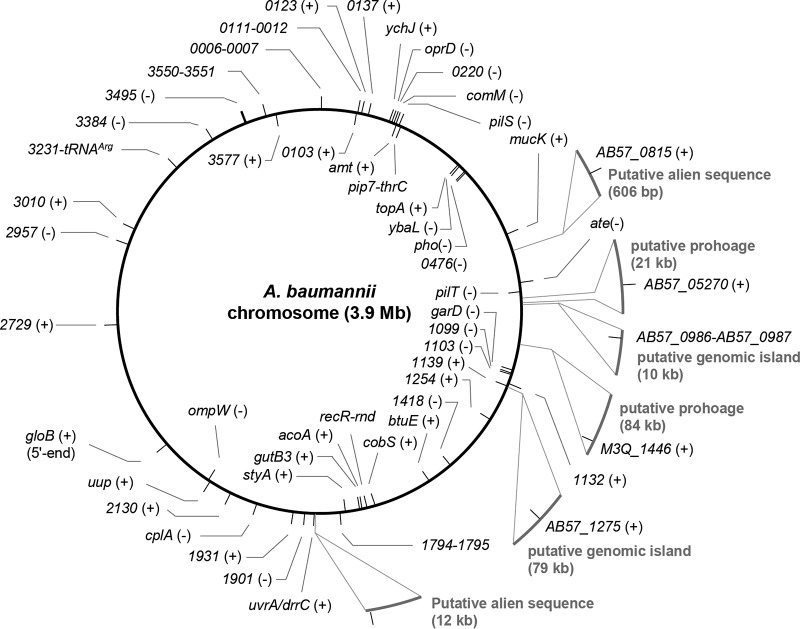
A large-scale search was performed on all of the retrieved A. baumannii genomes. The upstream and downstream sequences of AbaRs represented by the external flanks adjacent to the IRs available in 7,851 CS hits were mapped to an AbaR-free chromosome or otherwise searched in the NCBI database for empty loci to define insertion sites and DRs. A total of 7,749 (98.7%) flanks were mapped, and strikingly, 84 insertion sites were revealed (Fig. 2, Table S2, and Data Set S2). The insertion sites had diverse origins, as 51 were found to be chromosome borne, 20 plasmid borne, and 12 located on putative prophages, transposons, ISAba1, or genomic islands of other types, and they had distinct insertion frequencies (Fig. 2A). Most insertion sites were embedded in protein-coding genes. The transpositions generated 5- or 4-bp (only found in one locus) DRs (Fig. S3), except in one locus that was disturbed by Tn6021 on the pAB3 plasmid. The 5-bp left and right flanks of Tn6021 were 5′-ATTAT-3′ and 5′-ATAGT-3′, respectively. AbaRs had a strong preference for the comM gene. Insertions at acoA, pho, and uup were occasionally seen, but the frequencies had a sharp decrease. The other chromosome-borne insertion sites were rarely found to be occupied. AbaRs also occurred at high frequencies at the two loci seen on plasmid pAB3 and at the tet(B) gene that was mostly located within AbaRs (Data Set S4). We further mapped the insertion sites to different clonal lineages (Fig. 2A). Interestingly, of the 67 loci having clonal information, only five, including comM, the three known plasmid loci, and the locus identical to D721_p10064, were found in different clonal lineages, while the remaining loci were currently confined to either GC1 or GC2. However, for the majority of loci, the frequencies of occupancy were extremely low, which might not reflect actual clonal preference. However, several often-disrupted loci, such as tet(B), acoA, pho, and uup showed strong association with specific clonal lineages (Data Set S2). The disruption of tet(B), acoA, and pho was mostly associated with ST2 (GC2), while that of pho was only found in ST1 (GC1). Meanwhile, the disruption of tet(B) was seen in low frequencies in several other clones, most of which, like ST98, ST129, ST415, ST641, ST823, ST880, ST524, and ST922 could be mapped to GC2. The occupancy at acoA was also found in ST524. Notably, the chromosome-borne loci were diverse and scattered (Fig. 3). In addition, 1.3% of flanks could not be mapped to any known locus due to insufficient length or lack of hit.
FIG 2.
AbaRs have diverse insertion sites. (A) Numbers of CS flanks mapping to the insertion sites grouped by origins. For a name-unassigned gene, a representative locus tag identifier of an identical undisrupted gene is given. If there are two insertion sites in one gene or one insertion site, the positions of the corresponding DR sequences are given in brackets for distinction; the letter “c” means complement where applicable. Loci that have been reported are labeled in red. The locus that used to be represented by ACICU_02698 is tagged here with ABR2091_2729 to be consistent with other chromosomal loci. B7L39_18880 corresponds to the locus reported on pA85-3. The pAB3-type locus 1 and pAB3-type locus 2 correspond to the ones on plasmid pAB3 disrupted by Tn6021 and Tn6174, respectively. Data for the locus with a similar sequence to A1S_2017 are not shown, as the origin of the locus could not be precisely determined at present. (B) Occupancy distribution of the insertion sites in different clonal lineages. GC, global clone. See Data Set S2 for detailed ST information. GI, genomic island; Tn, transposon; IS, insertion sequence. IGR, intergenic region.
FIG 3.
Chromosomal locations of AbaR insertion sites. The schematic is drawn based on the chromosome of an AbaR-free strain, R2091 (GenBank accession number LN997846). Short lines indicate the insertion of AbaR with positive (outside) or negative (inside) orientation. Numbers are ABR2091 locus tag identifiers representing the insertion sites that mapped to name-unassigned genes in R2091. Note that in R2091, those loci are actually empty. A + or − indicates the orientation of each gene. AbaR orientation is defined in accord with that of the transposase genes. Gray arcs indicate alien DNA integrated into some of the A. baumannii chromosomes.
The antimicrobial resistance gene profiles of AbaRs are associated with host clonal lineages.
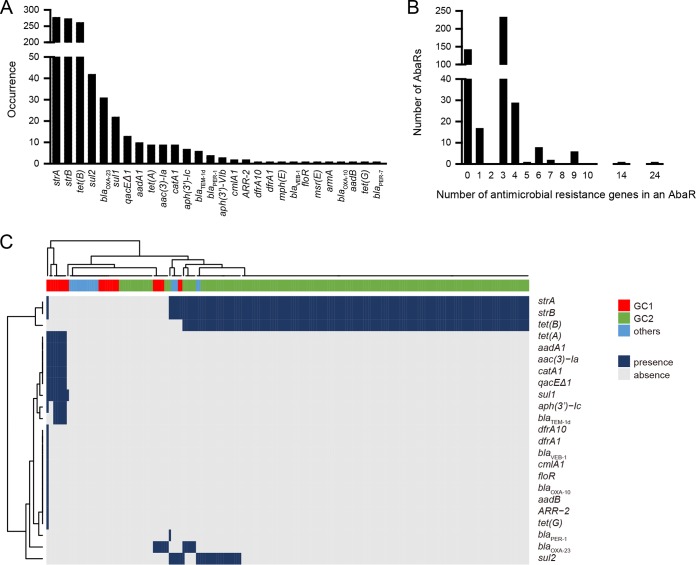
A total of 994 antimicrobial resistance genes covering 28 unique ones was identified from 70.9% (299/422) of the intact AbaRs (Fig. 4A). The strA, strB, and tet(B) genes were predominant in these AbaRs. Notably, as an insertion site within AbaRs, tet(B) often appeared in an occupied form and was truncated by 6 bp at the 3′ end, but it recruited a new stop codon from the CSR. Most resistant AbaRs contained three or four resistance genes, but they were capable of hosting up to 24 resistance genes (Fig. 4C). We further correlated their resistance gene profiles with host clones, where applicable. Interestingly, an apparent clonal lineage-specific pattern was found (Fig. 4C). GC2 was associated with an strA+ strB+ tet(B)+ signature, while GC1 was associated with a different and wide spectrum of resistance genes. The tet(B) gene seemed to be specific to AbaRs in GC2. The blaOXA-23 gene was found on AbaRs in both GC1 and GC2. Nonresistant AbaRs were found in nearly all available clonal lineages.
FIG 4.
Antimicrobial resistance genes identified on AbaRs. (A) Occurrence of the antimicrobial resistance genes in the 442 intact AbaRs. The tet(B) gene includes its intact form and its variant after truncation. (B) Distribution of resistance gene numbers. (C) The resistance gene profiles of AbaRs in different clonal lineages. See Data Set S4 for detailed ST information. Profiles of the AbaRs without host clonal information are not shown. GC, global clone.
DISCUSSION
This study developed a bioinformatics method to identify AbaRs in 3,148 publicly available A. baumannii genomes and analyzed their distribution, insertion sites, and features of antimicrobial resistance gene profiles.
AbaRs are known to be diverse but have been mapped to several closely related transposon backbones (8–11). This study has identified conserved sequences at the ends of AbaRs that actually reflect the relatedness of their backbones. Thus, the term AbaR (AbaR-type genomic island) used in this study broadly refers to a class of mobile genetic elements whose structures are based on these closely related backbones. AbaRs are prevalent in A. baumannii and are likely confined to the genus Acinetobacter. AbaRs seems to have distinct distributions in different clones, but further epidemiological studies are required, as the current A. baumannii genome database may be biased toward clinical isolates of certain clones. AbaRs occurred in non-baumannii Acinetobacter strains at low frequencies. Only a few AbaRs have been reported in A. nosocomialis and A. seifertii clinical isolates (22). These results may suggest that the interspecies transfer of AbaRs is infrequent. It has also been suggested that AbaRs evolve independently in different epidemic clones. Most AbaRs are located on chromosomes and fewer were reported on plasmid (11, 18). AbaRs are not self-transmissible elements like conjugative plasmids or integrative and conjugative elements. The AbaR4 carried by a conjugative plasmid is also found in a GC1 isolate, suggesting plasmid-mediated dissemination (18). However, we found that AbaRs do not commonly occur on plasmids, which may reduce their chance to be mobilized, partially contributing to infrequent interspecies transfer.
AbaRs have unexpectedly diverse insertion sites. There were six insertion sites reported in previous studies (1, 11, 17, 18, 23). However, the large-scale analysis in this study allows the identification of uncommon and cryptic sites, and thus 84 insertion sites in total were revealed. Consistent with previous reports, we found that most of the elements disrupted the comM gene. Other insertion sites displayed varied frequencies of occupancy. Interestingly, apart from chromosome- and plasmid-borne insertion sites, we also found several loci related to mobile genetic elements (MGEs), including genomic islands, transposons, and an insertion sequence, which may suggest that the incoming MGEs could expand the hosts’ capacity to accommodate AbaRs and/or mobilize AbaRs. In addition, our analysis utilized the flanking sequences of all identified CSs, which included the CSs within the internal regions of complex AbaRs. Therefore, we found several insertion sites located within complex AbaRs. Note that the split parts of the two loci (here designated pAB3-type locus 1 and locus 2) reported on pAB3 could belong to actual plasmid-borne sites or exist in the internal parts of the AbGRI1-type elements (11). Consistently, the occurrence of the matching split parts of each of the two loci were largely uneven. The tet(B) locus is likely an AbaR-borne insertion hot spot. In many reported AbaRs, such as Tn6166, the tet(B) locus has been seen to be truncated and immediately adjacent to an internal IRR, with the last 6 bp of the gene missing (14). Here, however, we found a full set of split parts for tet(B) in some complex AbaRs, which may provide insight into the formation of those complex AbaRs.
It is known that AbaRs carry multiple antimicrobial resistance genes. Indeed, we have identified diverse resistance genes on over 70% of available AbaRs. Moreover, the resistance gene profiles of these AbaRs follow specific patterns, allowing the grouping of the elements into distinct clades and correlation of the resistance gene profiles with clonal lineages. Our findings are highly consistent with those of previous reports. A highly mosaic multiple antimicrobial resistance region located between two copies of Tn6018 has been widely seen in AbaR3-type elements in GC1 (24), whereas in GC2, the AbaRs usually contain sul2, tet(B), strA, strB, and/or blaOXA-23 (10), and the multiple antimicrobial resistance is also attributed to AbGRI2 and/or AbGRI3 elements (15, 25). The patterns in the minor clones seem to be similar to that in GC2. These results again reiterate the distinct features of AbaRs in different clones and provide strong support to help correlate the resistance phenotypes of A. baumannii infections with genetic determinants, but future studies focusing on comprehensive analyses of AbaR structures are required.
Overall, the findings of our study will advance understanding of the distribution, insertion sites, and features of antimicrobial resistance gene profiles of AbaRs.
MATERIALS AND METHODS
Genomic data.
A total of 3,258 A. baumannii genomes were retrieved from the NCBI database (ftp://ftp.ncbi.nih.gov/genomes/). After removing duplicated genomes and those that did not pass the RefSeq criteria, 3,148 unique genomes were included for analysis (Data Set S1). Meanwhile, 19 unique plasmids sequenced alone were also retrieved from the NCBI database (Data Set S5).
In silico multilocus sequence typing (MLST) and clonal lineage sequence typing of A. baumannii genomes.
A. baumannii MLST was performed with both Pasteur (20) and Oxford (21) schemes. Allele sequences and MLST profile definitions were downloaded from the Bacterial Isolate Genome Sequence Database (BIGSdb; http://bigsdb.pasteur.fr/) (26). A clonal lineage sequence typing scheme utilizing ompA, csuE, and blaOXA-51-like genes was performed (19). A feature of this scheme was that it could distinguish global clones. For all schemes, alleles in the A. baumannii genomes were identified with BLASTn using criteria of 100% identity and 100% coverage. STs were then defined according to the allele profiles using an in-house Perl script.
Identification of conserved sequences in known AbaRs.
A total of 59 known AbaRs collected from published literatures or by text searching in the GenBank database (Table S1) were subjected to manual review to refine precise boundaries. Forty completely sequenced intact AbaRs were selected. The Tn6019 (sequence extracted from AbaR5) (8, 27) was used as the reference element. The 40 elements were aligned against Tn6019 with Mega BLAST (version 2.3.0+) (28) (E value, ≤0.001) to obtain conserved sequences. In this study, orientation of AbaRs was defined in accord with that of the tniA through tniE genes, and IRL referred to the IR upstream of those transposase genes.
Identification of AbaRs in genomic sequences.
We developed an AbaR identification pipeline with Perl (Fig. S1). The conserved sequences (CSs) locating at both ends of AbaRs were used as a query data set for AbaR identification. CSs were searched in genomic sequences with Mega BLAST (E value, ≤0.001). Fragmental hits adjacent or close (with a distance of ≤2 kb) to each other that could make up one CS and matching an insertion, deletion, or indel scenario were assembled as one CS hit (with the fragments in between if applicable). The occupancy at the known insertion sites comM and pho were examined. Cooccurrence of a left-end CS (CSL) and a right-end CS (CSR) with the same orientation would allow the identification of a complete element and recognition of its precise boundaries. Thus, the identified CSL and CSR with the same orientation were paired within a 150-kb window. However, a complex AbaR containing extra copies of CSs or two individual AbaRs close to each other could generate more than one CSL and/or CSR hits within the defined window, resulting in many combination possibilities. To obtain reliable pairing in such cases, hits with a matching insertion site were paired first, followed by hits with matching DRs, and last by the remaining hits. The hits located between two paired hits were excluded for subsequent pairing. The identification results were manually checked.
Identification of insertion sites and direct repeats.
Where applicable, the up-to-100-bp external sequences flanking the invert repeats (IRs) of CS hits were extracted to identify insertion sites. Those flanks were mapped to the chromosome of the AbaR-free A. baumannii strain R2091 (GenBank accession number LN997846) or searched in the NCBI Nucleotide online database to identify empty sites using BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The overlapping sequences of a left flank and a right flank matching to the same loci were defined as DRs. Results were manually checked.
Profiling of antimicrobial resistance genes.
Antimicrobial resistance genes were identified with the ResFinder (29) database, supplemented with the qacEΔ1 and qacE gene sequences, by using an in-house Perl script. Results were manually checked.
Dendrograms.
Binary heat maps indicating presence or absence were generated with the ComplexHeatmap package (30) in R. The accompanied dendrograms were drawn with default settings.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (grants 81702037 to D.B. and 81472501 and 81772849 to Q.W.), the Pandeng Research Foundation of Shanghai Tenth People’s Hospital (grant 2018SYPDRC015 to D.B.) and the 973 program, Ministry of Science and Technology, China (grant 2015CB554202 to H.Y.O.).
We thank Tobias Kieser and two anonymous reviewers for constructive suggestions.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.02526-18.
REFERENCES
- 1.Fournier PE, Vallenet D, Barbe V, Audic S, Ogata H, Poirel L, Richet H, Robert C, Mangenot S, Abergel C, Nordmann P, Weissenbach J, Raoult D, Claverie JM. 2006. Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet 2:e7. doi: 10.1371/journal.pgen.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kochar M, Crosatti M, Harrison EM, Rieck B, Chan J, Constantinidou C, Pallen M, Ou HY, Rajakumar K. 2012. Deletion of TnAbaR23 results in both expected and unexpected antibiogram changes in a multidrug-resistant Acinetobacter baumannii strain. Antimicrob Agents Chemother 56:1845–1853. doi: 10.1128/AAC.05334-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim DH, Choi JY, Kim HW, Kim SH, Chung DR, Peck KR, Thamlikitkul V, So TM, Yasin RM, Hsueh PR, Carlos CC, Hsu LY, Buntaran L, Lalitha MK, Song JH, Ko KS. 2013. Spread of carbapenem-resistant Acinetobacter baumannii global clone 2 in Asia and AbaR-type resistance islands. Antimicrob Agents Chemother 57:5239–5246. doi: 10.1128/AAC.00633-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saule M, Samuelsen O, Dumpis U, Sundsfjord A, Karlsone A, Balode A, Miklasevics E, Karah N. 2013. Dissemination of a carbapenem-resistant Acinetobacter baumannii strain belonging to international clone II/sequence type 2 and harboring a novel AbaR4-like resistance island in Latvia. Antimicrob Agents Chemother 57:1069–1072. doi: 10.1128/AAC.01783-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wright MS, Haft DH, Harkins DM, Perez F, Hujer KM, Bajaksouzian S, Benard MF, Jacobs MR, Bonomo RA, Adams MD. 2014. New insights into dissemination and variation of the health care-associated pathogen Acinetobacter baumannii from genomic analysis. mBio 5:e00963–e00913. doi: 10.1128/mBio.00963-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan AP, Sutton G, DePew J, Krishnakumar R, Choi Y, Huang XZ, Beck E, Harkins DM, Kim M, Lesho EP, Nikolich MP, Fouts DE. 2015. A novel method of consensus pan-chromosome assembly and large-scale comparative analysis reveal the highly flexible pan-genome of Acinetobacter baumannii. Genome Biol 16:143. doi: 10.1186/s13059-015-0701-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li H, Liu F, Zhang Y, Wang X, Zhao C, Chen H, Zhang F, Zhu B, Hu Y, Wang H. 2015. Evolution of carbapenem-resistant Acinetobacter baumannii revealed through whole-genome sequencing and comparative genomic analysis. Antimicrob Agents Chemother 59:1168–1176. doi: 10.1128/AAC.04609-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Post V, White PA, Hall RM. 2010. Evolution of AbaR-type genomic resistance islands in multiply antibiotic-resistant Acinetobacter baumannii. J Antimicrob Chemother 65:1162–1170. doi: 10.1093/jac/dkq095. [DOI] [PubMed] [Google Scholar]
- 9.Krizova L, Dijkshoorn L, Nemec A. 2011. Diversity and evolution of AbaR genomic resistance islands in Acinetobacter baumannii strains of European clone I. Antimicrob Agents Chemother 55:3201–3206. doi: 10.1128/AAC.00221-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seputiene V, Povilonis J, Suziedeliene E. 2012. Novel variants of AbaR resistance islands with a common backbone in Acinetobacter baumannii isolates of European clone II. Antimicrob Agents Chemother 56:1969–1973. doi: 10.1128/AAC.05678-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamidian M, Hall RM. 2017. Origin of the AbGRI1 antibiotic resistance island found in the comM gene of Acinetobacter baumannii GC2 isolates. J Antimicrob Chemother 72:2944–2947. doi: 10.1093/jac/dkx206. [DOI] [PubMed] [Google Scholar]
- 12.Hamidian M, Hall RM. 2011. AbaR4 replaces AbaR3 in a carbapenem-resistant Acinetobacter baumannii isolate belonging to global clone 1 from an Australian hospital. J Antimicrob Chemother 66:2484–2491. doi: 10.1093/jac/dkr356. [DOI] [PubMed] [Google Scholar]
- 13.Nigro SJ, Hall RM. 2012. Antibiotic resistance islands in A320 (RUH134), the reference strain for Acinetobacter baumannii global clone 2. J Antimicrob Chemother 67:335–338. doi: 10.1093/jac/dkr447. [DOI] [PubMed] [Google Scholar]
- 14.Nigro SJ, Hall RM. 2012. Tn6167, an antibiotic resistance island in an Australian carbapenem-resistant Acinetobacter baumannii GC2, ST92 isolate. J Antimicrob Chemother 67:1342–1346. doi: 10.1093/jac/dks037. [DOI] [PubMed] [Google Scholar]
- 15.Blackwell GA, Nigro SJ, Hall RM. 2015. Evolution of AbGRI2-0, the progenitor of the AbGRI2 resistance island in global clone 2 of Acinetobacter baumannii. Antimicrob Agents Chemother 60:1421–1429. doi: 10.1128/AAC.02662-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bonnin RA, Poirel L, Nordmann P. 2012. AbaR-type transposon structures in Acinetobacter baumannii. J Antimicrob Chemother 67:234–236. doi: 10.1093/jac/dkr413. [DOI] [PubMed] [Google Scholar]
- 17.Zhu L, Yan Z, Zhang Z, Zhou Q, Zhou J, Wakeland EK, Fang X, Xuan Z, Shen D, Li QZ. 2013. Complete genome analysis of three Acinetobacter baumannii clinical isolates in China for insight into the diversification of drug resistance elements. PLoS One 8:e66584. doi: 10.1371/journal.pone.0066584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamidian M, Kenyon JJ, Holt KE, Pickard D, Hall RM. 2014. A conjugative plasmid carrying the carbapenem resistance gene blaOXA-23 in AbaR4 in an extensively resistant GC1 Acinetobacter baumannii isolate. J Antimicrob Chemother 69:2625–2628. doi: 10.1093/jac/dku188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turton JF, Gabriel SN, Valderrey C, Kaufmann ME, Pitt TL. 2007. Use of sequence-based typing and multiplex PCR to identify clonal lineages of outbreak strains of Acinetobacter baumannii. Clin Microbiol Infect 13:807–815. doi: 10.1111/j.1469-0691.2007.01759.x. [DOI] [PubMed] [Google Scholar]
- 20.Diancourt L, Passet V, Nemec A, Dijkshoorn L, Brisse S. 2010. The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS One 5:e10034. doi: 10.1371/journal.pone.0010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartual SG, Seifert H, Hippler C, Luzon MA, Wisplinghoff H, Rodriguez-Valera F. 2005. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J Clin Microbiol 43:4382–4390. doi: 10.1128/JCM.43.9.4382-4390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim DH, Ko KS. 2015. AbaR-type genomic islands in non-baumannii Acinetobacter species isolates from South Korea. Antimicrob Agents Chemother 59:5824–5826. doi: 10.1128/AAC.01175-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adams MD, Goglin K, Molyneaux N, Hujer KM, Lavender H, Jamison JJ, MacDonald IJ, Martin KM, Russo T, Campagnari AA, Hujer AM, Bonomo RA, Gill SR. 2008. Comparative genome sequence analysis of multidrug-resistant Acinetobacter baumannii. J Bacteriol 190:8053–8064. doi: 10.1128/JB.00834-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamidian M, Hall RM. 2018. The AbaR antibiotic resistance islands found in Acinetobacter baumannii global clone 1—structure, origin and evolution. Drug Resist Updat 41:26–39. doi: 10.1016/j.drup.2018.10.003. [DOI] [PubMed] [Google Scholar]
- 25.Blackwell GA, Holt KE, Bentley SD, Hsu LY, Hall RM. 2017. Variants of AbGRI3 carrying the armA gene in extensively antibiotic-resistant Acinetobacter baumannii from Singapore. J Antimicrob Chemother 72:1031–1039. doi: 10.1093/jac/dkw542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jolley KA, Maiden MC. 2010. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Post V, Hall RM. 2009. AbaR5, a large multiple-antibiotic resistance region found in Acinetobacter baumannii. Antimicrob Agents Chemother 53:2667–2671. doi: 10.1128/AAC.01407-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Z, Schwartz S, Wagner L, Miller W. 2000. A greedy algorithm for aligning DNA sequences. J Comput Biol 7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- 29.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gu Z, Eils R, Schlesner M. 2016. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.