Abstract
Objective
The gut microbiota is an important influencing factor of metabolic health. Although dietary interventions with probiotics, prebiotics, and synbiotics can be effective means to regulate obesity and associated comorbidities, the underlying shifts in gut microbial communities, especially at the functional level, have not been characterized in great details. In this study, we sought to investigate the effects of synbiotics on the regulation of gut microbiota and the alleviation of high-fat diet (HFD)-induced metabolic disorders in mice.
Methods
Specific pathogen-free (SPF) male C57BL/6J mice were fed diets with either 10% (normal diet, ND) or 60% (high-fat diet, HFD) of total calories from fat (lard). Dietary interventions in the HFD-fed mice included (i) probiotic (Bifidobacterium animalis subsp. lactis and Lactobacillus paracasei subsp. paracasei DSM 46331), (ii) prebiotic (oat β-glucan), and (iii) synbiotic (a mixture of i and ii) treatments for 12 weeks. Besides detailed characterization of host metabolic parameters, a multi-omics approach was used to systematically profile the microbial signatures at both the phylogenetic and functional levels using 16S rRNA gene sequencing, metaproteomics and targeted metabolomics analysis.
Results
The synbiotic intervention significantly reduced body weight gain and alleviated features of metabolic complications. At the phylogenetic level, the synbiotic treatment significantly reversed HFD-induced changes in microbial populations, both in terms of richness and the relative abundance of specific taxa. Potentially important species such as Faecalibaculum rodentium and Alistipes putredinis that might mediate the beneficial effects of the synbiotic were identified. At the functional level, short-chain fatty acid and bile acid profiles revealed that all dietary interventions significantly restored cecal levels of acetate, propionate, and butyrate, while the synbiotic treatment reduced the bile acid pools most efficiently. Metaproteomics revealed that the effects of the synbiotic intervention might be mediated through metabolic pathways involved in carbohydrate, amino acid, and energy metabolisms.
Conclusions
Our results suggested that dietary intervention using the novel synbiotic can alleviate HFD-induced weight gain and restore gut microbial ecosystem homeostasis phylogenetically and functionally.
Keywords: Dietary intervention, Gut microbiota, High-fat diet, Obesity, Synbiotics
Abbreviations: 3-dehydroCDCA, 3-dehydrochenodeoxycholic acid; 7-oxoDCA, 7-oxodeoxycholic acid; alloCA, Allocholic acid; BAs, bile acids; CA, Cholic acid; CA-7S, Cholic acid 7-sulfate; DCA, Deoxycholic acid; eAT, epididymal adipose tissue; HDCA, Hyodeoxycholic acid; LBP, lipopolysaccharide-binding protein; LCA, Lithocholic acid; MCP-1, monocyte chemotactic protein 1; MDCA, Murideoxycholic acid; SCFAs, short-chain fatty acids; TCA, Taurocholic acid; TCDCA, Taurochenodeoxycholic acid; TDCA, Taurodeoxycholic acid; THDCA, Taurohyodeoxycholic acid; TNF-α, tumor necrosis factor-α; TUDCA, Tauroursodeoxycholic acid; UDCA, Ursodeoxycholic acid; αMCA, α-Muricholic acid; αTMCA, α-Tauromuricholic acid; βMCA, β-Muricholic acid; βTMCA, β-Tauromuricholic acid; ωMCA, ω-Muricholic acid; ωTMCA, ω-Tauromuricholic acid
Highlights
-
•
Synbiotic (SYN) intervention alleviated HFD-induced metabolic disorders.
-
•
SYN regulated the microbial population at the diversity and taxonomic levels.
-
•
HFD reduces SCFAs production which is restored by dietary interventions.
-
•
HFD promotes bile acids formation which is ameliorated by SYN.
-
•
SYN regulated microbial functional activities and metabolic pathways.
1. Introduction
The obesity epidemic has become a major global health concern with substantial increases in its prevalence and severity during the past decades [1], [2]. It is a multi-factorial condition, which is linked with surplus of energy uptake and influenced by a complex interplay of genetic, epigenetic, dietary, lifestyle and environmental factors. It is associated not only with a variety of important chronic metabolic disorders, including type-2 diabetes, non-alcoholic fatty liver diseases, and cardiovascular diseases, but also with an increased incidence of a variety of cancers [3]. So far no gold standard treatment beside bariatric surgery for morbidly obese patients has been found but this can have strong side effects. Therefore, finding effective non-surgical therapies, such as dietary intervention [4], is in urgent demand. Accumulating insights have shed light on an entirely new perspective suggesting that the gut microbiota plays a key role in the regulation of host metabolism [5], [6], [7]. Microbiota can influence the whole-body metabolism by affecting energy balance and immune system [8], [9]. Understanding the effect of gut microbiota on the pathogenesis of obesity is of utmost importance for future development of microbiota-targeted approaches towards its therapy and prevention.
Dietary interventions in obesity using probiotics or prebiotics to specifically target the gut microbiota have gained wide attentions because of their potential ability to re-establish gut homeostasis [10], [11]. Probiotics are defined as living microorganisms that when administered in sufficient amounts conferring a health benefit to the host [12]. Probiotics have been shown to interact with endogenous gut bacteria, which may affect metabolic pathways involved in the regulation of fat metabolism and obesity development [13]. Early studies, in both animals and humans, have revealed the potential effectiveness of probiotic administration on high-fat diet (HFD)-induced obesity and metabolic complications [14], [15], [16], [17]. Bifidobacterium lactis and Lactobacillus paracasei were previously shown to affect gut microbiota in mice and concomitantly attenuate obesity comorbidities [18]. However, the relationships between these probiotic interventions and the gut microbiota in the context of obesity have not yet to be investigated at the functional level. Prebiotics are non-digestible food ingredients or substances that can selectively stimulate the growth and/or activity of beneficial bacteria in the intestinal tract [19]. By modulating the gut microbiota, prebiotics usually influence the production of short-chain fatty acids (SCFAs) with consequences on gut barrier functions and immune responses [20]. Typical prebiotics such as oligofructose have been found to modulate the gut microbiota to counteract HFD-induced inflammation and related metabolic disturbances in C57BL/6J mice [21] and potentially in obese human adults [22]. Oat β-glucan has gained interest recently due to its beneficial role in insulin resistance, dyslipidemia, hypertension, and obesity-associated metabolic disorders [23], [24]. Recently, it has been reported to significantly decrease body weight and alter blood lipids profiles in HFD-induced obese mice, accompanied by increased colonic SCFA concentrations and the occurrence of Lactobacillus, indicating its prebiotic property and anti-obesity potential [25]. However, these investigations focused primarily on the host without a comprehensive analysis of the prebiotic-induced changes in the gut microbiota. Synbiotics are dietary supplements combining probiotics and prebiotics in a form of synergism. A synbiotic approach based on the effects of oligofructose and Bifidobacterium animalis has been applied to modify gut microbiota and attenuate glycemia in obese rats [26]. A recent clinical trial demonstrated that probiotic and synbiotic supplementations controlled body fat mass, reduced waist circumference and food intake in overweight and obese adults [27]. Hence, synbiotic intervention in obesity-related comorbidities is a potential promising strategy. However, this field is still in its infancy and the detailed characterization of host- and microbiota-related molecular mechanisms remains to be investigated.
In this study, we investigated how the development of HFD-induced obesity and associated metabolic disturbances can be improved by dietary intervention with a novel synbiotic. We performed controlled dietary interventions in mice with either two probiotic strains (B. animalis subsp. lactis DSM 10140 and L. paracasei subsp. paracasei DSM 46331), or a prebiotic (oat β-glucan), or a mixture thereof (synbiotic). Besides detailed characterization of host metabolic parameters, the gut microbial communities were comprehensively analyzed at both the phylogenetic and functional levels to decipher gut microbiota profiles associated with the dietary interventions in the context of obesity.
2. Materials and methods
2.1. Animal experiments
The experimental procedures were approved by the Animal Experimentation Ethics Committee of The Chinese University of Hong Kong (Ref NO. 15-023-MIS). All animals were housed in the facilities of the Laboratory Animal Services Centre at the Chinese University of Hong Kong.
After one week of acclimatization, sixty 8-week-old, male, specific pathogen-free (SPF) C57BL/6J mice were randomly divided into five groups (n = 12 mice per group). One control group and the other 4 groups were fed diets with 10% (normal diet, ND) and 60% (high-fat diet, HFD) of total calories from fat (lard) purchased from Harlan (TD.08806 and TD.06414), respectively. The composition of experimental diet (Table S1) is provided in the Supplementary Materials and Methods. After another week of acclimatization with the diets, three groups of HFD-fed mice were subjected to a 12-week dietary intervention with daily oral administration (by gavage) of pro-, pre-, and synbiotics, respectively. B. animalis subsp. lactis DSM 10140 and L. paracasei subsp. paracasei DSM 46331 (DSMZ, Braunschweig, Germany) were used for the probiotic group (PRO) at a dose of each 108 cells per day. Oat β-glucan (80% purity, Green Stone Swiss Co., Ltd, Shanghai, China) was used for the prebiotic group (PRE) at a dose of 1 g/kg body weight per day. The synbiotic group (SYN) received a combined dose of the pro- and prebiotics. The ND and HFD control groups received the same amount of placebo by gavage. Body weight and caloric intake of the animals were measured weekly. Fecal and cecal samples were collected for microbial analysis at the end of the 12th week. Details on the bacterial suspensions preparation and sample collection procedures are provided in the Supplementary Materials and Methods.
2.2. Assessment of host metabolic parameters
Fasting blood glucose levels were measured with a blood glucose meter (Accu-Check; Roche Diagnostics, Mannheim, Germany). Oral glucose tolerance tests (OGTT) were conducted as described in the Supplementary Materials and Methods. Fasting insulin (Mercodia, Uppsala, Sweden) and lipopolysaccharide (LPS)-binding protein (LBP; Cell Sciences, Canton, MA, USA) in serum were determined using ELISA kits, following the manufacturer's instructions. Serum cholesterol and triglycerides were enzymatically determined using commercial kits (Stanbio Laboratory, Boerne, USA). Host mRNA gene expression of inflammatory markers, including tumor necrosis factor-alpha (TNF-α), CD11c, monocyte chemotactic protein-1 (MCP-1), and LPS-binding protein (LBP), in epididymal adipose tissue (eAT), liver, and jejunum were measured by RT-qPCR as described in the Supplementary Materials and Methods. Histological images of hematoxylin and eosin (H & E)-stained liver and eAT sections were acquired with a light microscope (Nikon ECLIPSE 80i, Nikon Instruments Inc., Melville, NY, USA). Adipocyte size was obtained from perimeter tracings using the Image J software (NIH, Bethesda, MD, USA).
2.3. High-throughput 16S ribosomal RNA amplicon sequencing
The feces collected at baseline and the 12th week from each mouse were sequenced. Isolation of metagenomic DNA and Illumina sequencing of the V3-V4 regions of 16S rRNA genes were performed as described previously [28]. Data were analyzed using IMNGS and Rhea [29], [30]. Details on wet lab procedures and bioinformatic analysis are provided in the Supplementary Materials and Methods.
2.4. Mass spectrometry for targeted metabolites analysis
The concentrations of short-chain fatty acids and bile acids in the caecum were measured using ultra-high performance liquid chromatography (UHPLC; Acquity UPLC, Waters, Milford, MA, USA) coupled with mass spectrometry (MS) as described previously [31], [32]. The cecal samples used were collected at the 12th week from each mouse, with n = 12 mice per group. Detailed procedures are provided in the Supplementary Materials and Methods.
2.5. Mass spectrometry for metaproteome analysis
Cecal metaproteomes were assessed using nano-HPLC system (UltiMate 3000 RSLCnano, Dionex/Thermo Fisher Scientific, Idstein, Germany) coupled with a QExactive HF (Thermo Fisher Scientific, San Jose, CA, USA) mass spectrometer. Protein extraction, LC-MS/MS measurements and bioinformatic analysis were performed as described earlier [33] and explained in more detail in the Supplementary Materials and Methods. The cecal samples used were collected at the 12th week after dietary intervention from each mouse, with n = 6 mice per group.
2.6. Statistical analysis of data
For host parameters and metabolites data, statistical significances were assessed by one-way ANOVA followed by Tukey post-hoc test (SPSS Inc., Chicago, IL, USA). Differences were considered significant when the p-values are below 0.05. For sequencing and metaproteomic data, statistical tests were done in the R programming environment. P-values were calculated using non-parametric Kruskal–Wallis Rank Sum Test. Wilcoxon Rank Sum Test or independent two-sided Student's t-test was used for pairwise comparison. P-values were corrected for multiple testing using the Benjamini-Hochberg method. P-values below 0.05 were considered to be statistically significant. Non-parametrical multiple dimensional scaling plots were computed using the package vegan and ade4. Details of the bioinformatic analysis are provided in the Supplementary Materials and Methods.
2.7. Accession number
The sequencing data generated in the present study are available at the European Nucleotide Archive (ENA) database under accession number PRJEB26534 (ERP108526). The mass spectrometry metaproteomics data were deposited to the ProteomeXchange Consortium via the PRIDE [34] partner repository and are available under the dataset identifier PXD009564.
3. Results
3.1. Synbiotic intervention significantly reversed HFD-induced weight gain and associated metabolic disorders
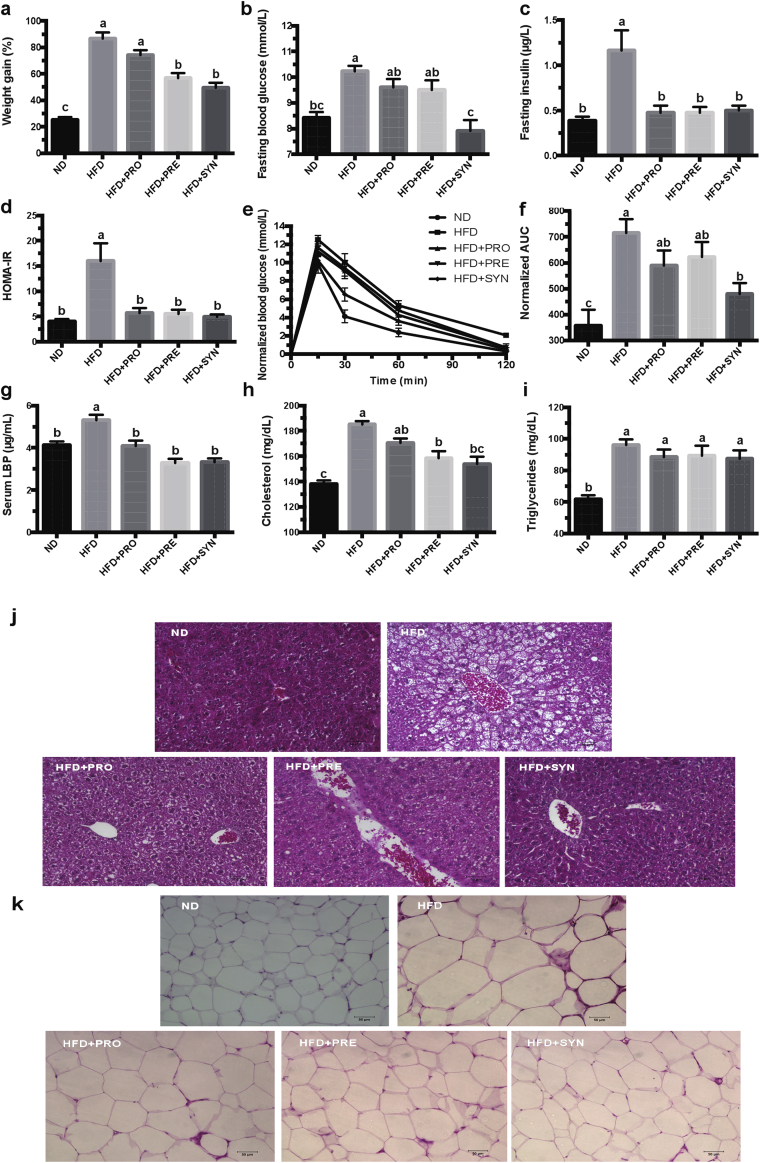
To validate the effects of the pro-, pre- and synbiotics on HFD-induced weight gain and metabolic disorders, host metabolic parameters were measured during and at the end of mice experiments. After 12 weeks of intervention, HFD-fed mice significantly gained more body weight and developed features of metabolic complications when compared with ND-fed mice (Figure 1 and Figure S1). Dietary interventions with pre- and synbiotic significantly attenuated most of those HFD-induced features, resulting in decreased body weight gain (Figure 1A) despite similar energy intake (Figure S2), decreased liver and eAT weights (Figure S3), improved HOMA-IR score (Figure 1D), and decreased fasting insulin (Figure 1C), LBP (Figure 1G) and cholesterol (Figure 1H) levels in serum. Compared with the pro- and prebiotic treatments, the synbiotic showed the highest potency in decreasing fasting blood glucose (Figure 1B) and improving glucose tolerance in OGTT (Figure 1E and F). Although the probiotic attenuated the body weight gain only slightly, it was associated with reduced fasting insulin (Figure 1C) and serum LBP levels (Figure 1G). Dietary interventions also protected against HFD-induced hepatic steatosis (Figure 1J) and adipocyte size increase in eAT (Figure 1K and Figure S4). As determined by quantitative (q) PCR, the fecal level of each probiotic strain increased significantly during the pro- and synbiotic interventions (Figure S5), indicating their presence in the mouse intestine throughout the intervention period. Taken together, these results indicate that dietary interventions, especially the synbiotic, alleviated HFD-induced weight gain accompanied by the enhancement of glucose-insulin homeostasis and protection against hepatic steatosis.
Figure 1.
Synbiotic intervention significantly reversed HFD-induced weight gain and associated metabolic syndrome parameters after 12 weeks of intervention. (a) Body weight gain as the percentage of baseline weight for each mouse. (b) Fasting blood glucose. (c) Fasting insulin. (d) HOMA-IR, calculated by fasting blood glucose (mmol/L) x fasting insulin (mU/L)/22.5. (e) Curve of normalized OGTT (Oral glucose tolerance tests). Data are normalized by subtracting the baseline value (fasting blood glucose) from the measured value in each mouse. (f) Areas under the curve (AUC) of normalized OGTT. The original Curve of OGTT and AUC of OGTT are available in the Supplementary Figures S12 and S13. (g-i) Levels of LBP (g), cholesterol (h) and triglycerides (i) in serum. Hematoxylin and eosin-stained sections of the liver (j) and epididymal adipose tissue (eAT) (k). Data are shown as means ± SEM. Groups with different letters are significantly different from each other as assessed by one-way ANOVA followed by Tukey post hoc test (P < 0.05). n = 12 or n = 8 (for g-k) mice per group.
3.2. Pro-, pre- and synbiotic interventions alleviated HFD-induced inflammation in eAT, liver and jejunum
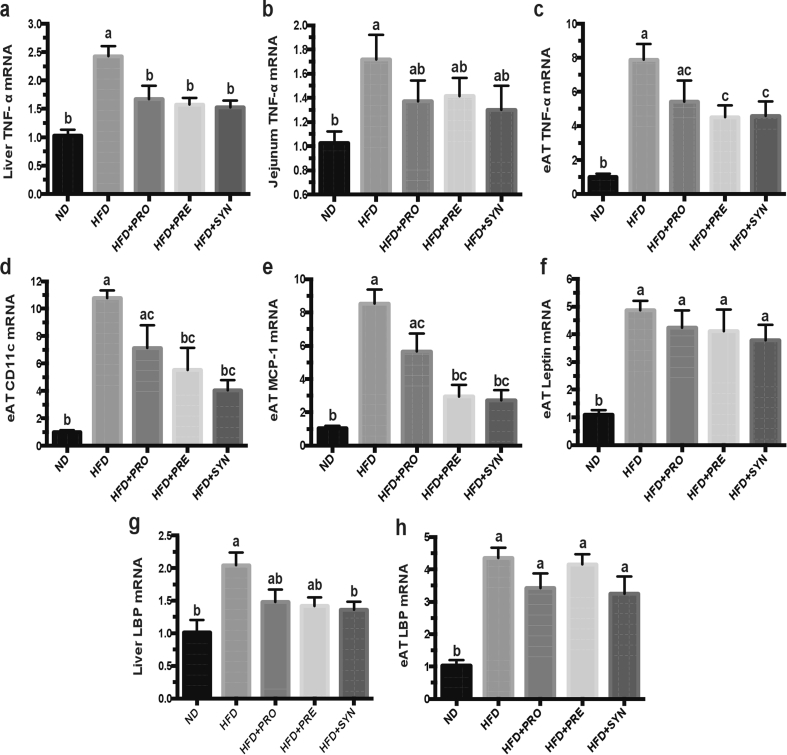
Obesity and its associated metabolic disorders are usually characterized by low-grade, chronic systemic inflammation. To assess effects of the dietary interventions on HFD-induced inflammation, the inflammation-related makers were quantified by qPCR in tissues of eAT, liver and jejunum. Compared with ND-fed mice, HFD-fed mice were characterized by significantly increased gene expressions of all traced makers. All dietary interventions markedly reduced TNF-a gene expression level in the liver (Figure 2A). Only the synbiotic intervention significantly attenuated LBP gene expression level in the liver (Figure 2G). In eAT, except for the genes of leptin and LBP, both the pre- and synbiotic treatments significantly decreased the gene expressions of TNF-a (Figure 2C), CD11c (Figure 2D), and MCP-1 (Figure 2E), while the probiotic treatment did not reach statistical significance. These results suggest the anti-inflammatory capabilities of pro-, pre- and synbiotic interventions.
Figure 2.
Probiotic, prebiotic and synbiotic interventions alleviated HFD-induced inflammation in eAT, liver and jejunum. (a–c) Gene expression levels of TNF-α in liver (a), jejunum (b) and eAT (c); (d-f) Gene expression levels of CD11c (d), MCP-1 (e) and leptin (f) in eAT; (g-h) Gene expression levels of LBP in liver (g) and eAT (h) by RT-qPCR. All mRNA quantification data were normalized to the housekeeping gene GAPDH. Gene expression levels were expressed as values relative to the ND group. Data are shown as means ± SEM. With n = 8 mice per group. Groups with different letters are significantly different from each other as assessed by one-way ANOVA (Tukey post hoc test, P < 0.05). Abbreviations: TNF- α: tumor necrosis factor-α; eAT: epididymal adipose tissue; MCP-1: monocyte chemotactic protein 1; LBP: lipopolysaccharide-binding protein.
3.3. Pro-, pre- and synbiotic interventions counteracted HFD-induced shifts in the gut microbiota
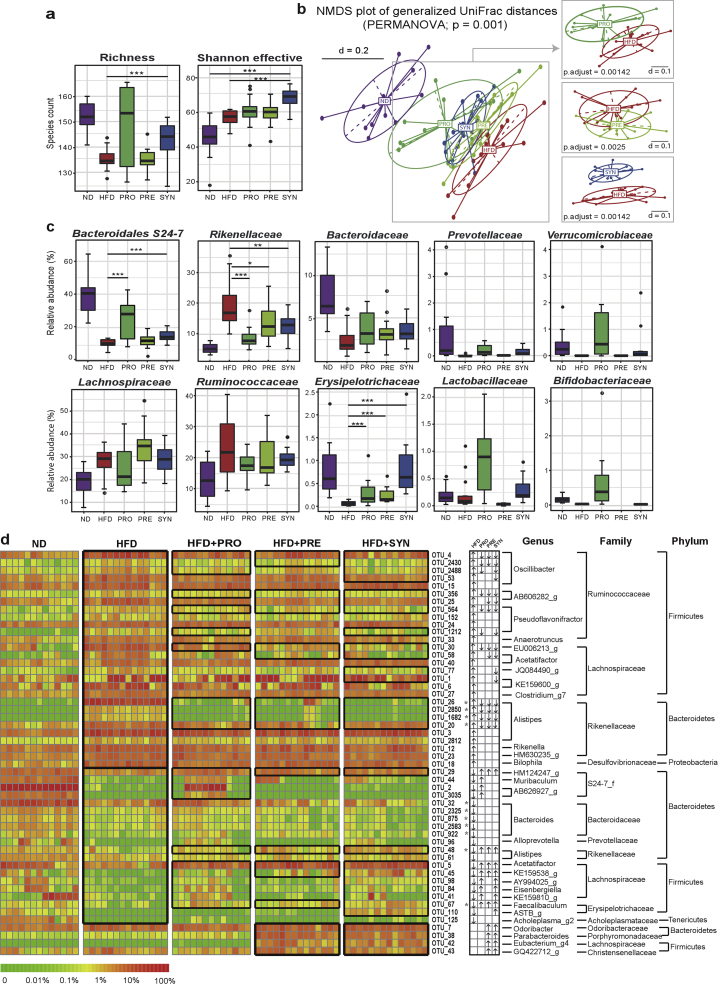
To assess the impact of the pro-, pre- and synbiotic treatments on gut microbiota structure, we sequenced the V3-V4 region of 16S rRNA genes amplified from feces collected before and after dietary intervention. After quality- and chimera-check as well as OTU filtering, we obtained a total of 2,703,373 amplicon sequences (19,590 ± 4,985 per sample) clustering in a total of 224 OTUs at 97% sequence identity. HFD significantly affected alpha-diversity, leading to a decrease in richness, yet an increase in the Shannon effective diversity (Figure 3A). The synbiotic intervention significantly attenuated the reduction in richness and showed the highest Shannon effective diversity. Beta-diversity analysis based on phylogenetic distances revealed that samples clustered according to diets and interventions (Figure 3B). The most distinct clustering was observed between the ND and HFD. Dietary interventions significantly shifted the overall microbial profiles from HFD to ND, with SYN being the most separated from HFD. Of note, all mice fed the HFD had overlapping phylogenetic makeup of the gut microbiota before intervention (Figure S6), indicating that changes observed after 12 weeks of intervention were indeed due to the dietary interventions. To see whether the differences observed at the level of diversity were associated with changes in specific microbes, we tested differences in taxonomic groups and single OTUs. The relative abundance of sequences classified within the phylum Bacteroidetes was significantly decreased and that of Firmicutes increased by HFD (Figure S7). At the family level, the decrease in Bacteroidetes was reflected by a significant decrease in the relative abundances of Bacteroidaceae, Bacteroidales S24-7 group, and Prevotellaceae (Figure 3C). The pro- and synbiotics significantly restored the level of S24-7. In contrast, Rikenellaceae was elevated upon HFD feeding, which was significantly reversed by all dietary interventions. The HFD-induced increase in Firmicutes was characterized at the family level by an increase in the relative abundances of Lachnospiraceae and Ruminococcaceae (Figure 3C). In contrast, HFD reduced the relative abundance of Lactobacillaceae, Bifidobacteriaceae, Erysipelotrichaceae and Verrucomicrobiaceae. All interventions, with the synbiotic one in particular, significantly restored the level of Erysipelotrichaceae (Figure 3C). Sequences within the families of Lactobacillaceae and Bifidobacteriaceae showed higher relative abundances in the probiotic group, most likely due to the detection of probiotic strains. At the level of phylotypes, a total of 52 species-level OTUs were significantly altered by the HFD and dietary interventions (Figure 3D and Table S4). Compared with ND, HFD was associated with a significant increase in the relative abundance of 28 OTUs (representing 13 genera, 4 families and 3 phyla), 16 of which were reversed by at least one intervention, and with a significant decrease in 20 OTUs (representing 14 genera, 7 families and 3 phyla), of which 12 were reversed after dietary intervention. Among these, 22, 16 and 25 OTUs were significantly reversed by the pro-, pre- and synbiotic interventions, respectively (Figure 3D). Besides, 4 OTUs (representing 4 genera, 4 families and 2 phyla) were particularly enriched in PRE and SYN. Taken together, the dietary interventions significantly altered gut microbiota profiles in response to HFD at the levels of microbial richness, taxonomic composition and OTUs, indicating the effectiveness of the pro-, pre- and synbiotic interventions in regulating gut microbial populations.
Figure 3.
Probiotic, prebiotic and synbiotic interventions counteracted HFD-induced shifts in the gut microbiota. (a) Richness and Shannon effective diversity. (b) Non-metric multidimensional scaling (NMDS) plots based on generalized UniFrac distances. (c) Relative abundances of dominant bacterial families with significant differences between groups. First four taxa in each row belong to the phyla of Bacteroidetes and Firmicutes, respectively. (d) Relative abundances of the 52 OTUs that were significantly altered by HFD and reversed by interventions. Their top-hit taxa according to EzTaxon (at genus, family and phylum levels) analysis are depicted on the right. Detailed taxonomic annotation information is available in the Supplementary Table S4. In the heatmap, the black boxes highlight the OTUs that were significantly altered by HFD, and those were significantly reversed by interventions, also that were particularly enriched in PRE and SYN. Arrows (“↑” and “↓”) in the boxes black boxes highlight OTUs significantly altered by the HFD and reversed by the funational food interventions. P-values were calculated using ANOVA on Ranks followed by Wilcoxon Rank–Sum Test and corrected using Benjamini-Hochberg method. With n = 12 mice per group. *<0.05, **<0.01, ***<0.001.
3.4. The synbiotic intervention significantly affected gut microbial ecosystem at the metabolic level
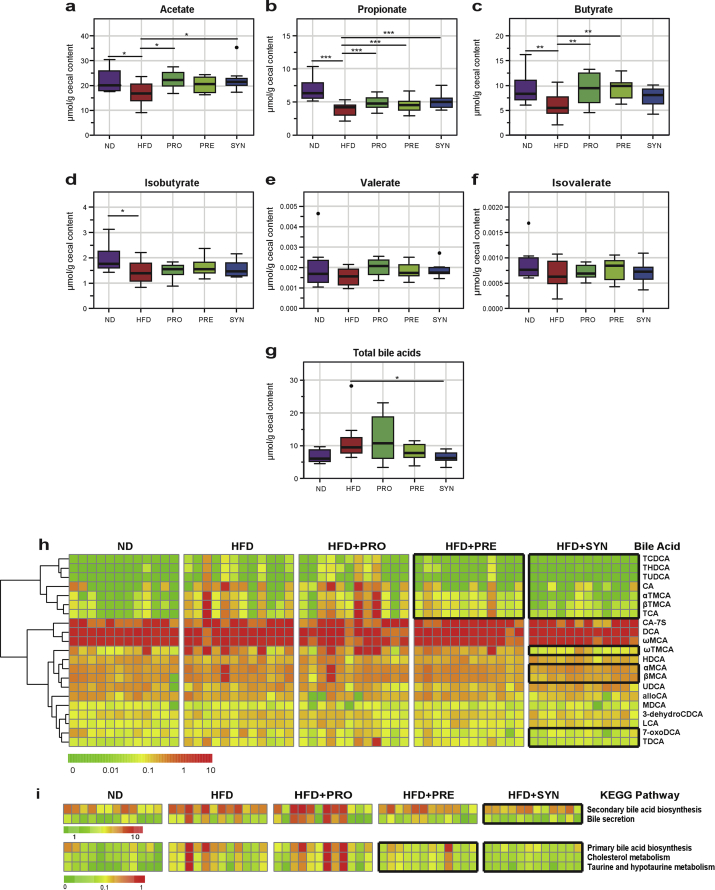
To assess the impact of dietary interventions on gut microbial ecosystem at the metabolic level, two major groups of gut microbiota-dependent metabolites in mouse cecal contents were quantified: the short-chain fatty acids (SCFAs) and bile acids (BAs). HFD feeding was associated with a significant reduction in the concentrations of acetate, propionate, butyrate, and iso-butyrate. Dietary interventions significantly restored the levels of acetate (Figure 4A), propionate (Figure 4B), and butyrate (Figure 4C). HFD also increased the level of total BA, which was significantly reduced by the synbiotic (Figure 4G). HFD increased most of the identified 21 BAs (Figure 3H). The ability of synbiotic to reverse total BA level could be reflected by controlling the levels of βMCA, 7-oxoDCA and other BAs (Figure 4H). BA-related pathways analysis based on the BA concentrations showed the same trend with a pronounced effect in SYN (Figure 4I). Taken together, these results revealed that dietary interventions, especially the synbiotic one, affected key metabolic activities of the gut microbial ecosystem beyond the changing at the phylogenetic level.
Figure 4.
Synbiotic intervention significantly affected the SCFAs and bile acids profiles in the cecum in response to HFD. (a–f) Levels of acetate (a), propionate (b), butyrate (c), isobutyrate (d), valerate (e), isovalerate (f). (g) Levels of total bile acids. (h) Overall patterns of the 21 detected bile acids. (i) Bile acids-related KEGG pathways that were calculated based on bile acids concentrations. In the heatmap, the black boxes highlight the bile acids or pathways that were particularly reversed by the prebiotic and synbiotic interventions. Data are shown as μmol/g wet weight of cecal content. With n = 12 mice per group. *<0.05, **<0.01, ***<0.001 by one-way ANOVA (Tukey post hoc test). Abbreviations: THDCA: Taurohyodeoxycholic acid; TCDCA: Taurochenodeoxycholic acid; TUDCA: Tauroursodeoxycholic acid; CA: Cholic acid; αTMCA: α-Tauromuricholic acid; βTMCA: β-Tauromuricholic acid; TCA: Taurocholic acid; CA-7S: Cholic acid 7-sulfate; DCA: Deoxycholic acid; ωMCA: ω-Muricholic acid; MDCA: Murideoxycholic acid; 3-dehydroCDCA: 3-dehydrochenodeoxycholic acid; LCA: Lithocholic acid; 7-oxoDCA: 7-oxodeoxycholic acid; TDCA: Taurodeoxycholic acid; alloCA: Allocholic acid; UDCA: Ursodeoxycholic acid; ωTMCA: ω-Tauromuricholic acid; HDCA: Hyodeoxycholic acid; βMCA: β-Muricholic acid; αMCA: α-Muricholic acid.
3.5. The synbiotic intervention significantly affected gut metaproteomes
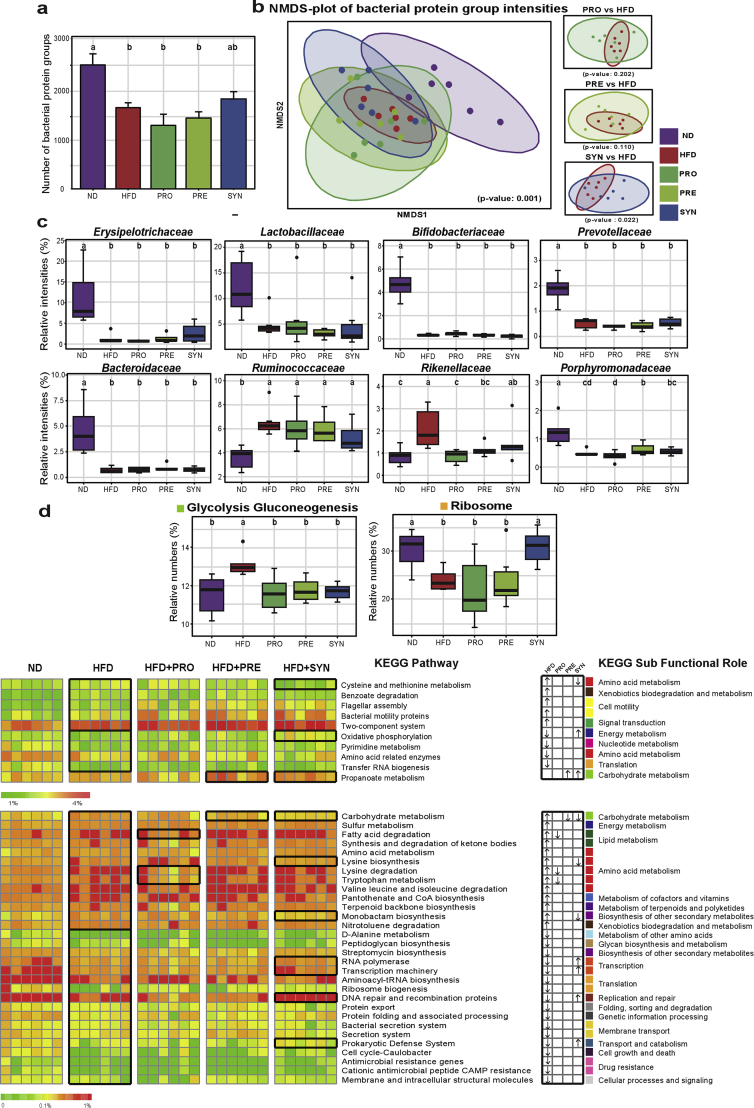
To further assess the impact of dietary interventions on gut microbial ecosystem at the functional level, the cecal metaproteomes were profiled. A total of 11944 protein groups and 167 KEGG pathways were identified in all samples. The numbers of protein groups assigned to different taxa levels are shown in Figure S8. Compared with ND, HFD significantly decreased the number of identified bacterial protein groups (Figure 5A). Beta-diversity analysis based on protein group intensities also revealed a clear effect of the HFD. Consistent with the microbial phylogenetic profiles, the most distinct clustering linked to intervention was observed for SYN compared with HFD (Figure 5B). We further explored the changes of functional activities within specific bacterial groups. Some important families, such as Erysipelotrichaceae, Lactobacillaceae, Bifidobacteriaceae, and Prevotellaceae (Figure 5C), had higher functional activities although they were observed at relative low abundances (Figure 3C). In contrast, other families had lower functional activities relative to their total abundances (Figure 5C). Furthermore, at the level of KEGG pathway, 42 pathways were significantly altered by the HFD and dietary interventions (Figure 5D and Table S5). Compared with ND, HFD significantly increased 19 pathways and decreased the other 22 pathways. Among these pathways, there were four, two and eleven pathways significantly reversed by the pro-, pre- and synbiotic treatments, respectively (Figure 5D). In the dominant pathways, HFD significantly elevated the Glycolysis/Gluconeogenesis pathway but significantly decreased the Ribosome. All dietary interventions significantly reversed the impact of HFD on Glycolysis/Gluconeogenesis, while the synbiotic one significantly restored the Ribosome (Figure 5D). In conclusion, metaproteome analyses further revealed the capabilities of dietary treatments, especially the synbiotic one, in modulating metabolic pathways and functional activities of gut microbial ecosystem.
Figure 5.
Synbiotic intervention significantly altered gut metaproteomes in response to HFD. (a) Mean number of identified bacterial protein groups in five groups of mice. (b) Non-metric multidimensional scaling (NMDS) plots based on bacterial protein group intensities. (c) Relative intensities of dominated bacterial taxa (at the family level) with significant changes based on relative protein group intensities. Taxa in the first and second rows were up- and down-regulated, respectively, compared with the results at phylogenetic level. (d) The relative numbers of 42 important KEGG pathways that were significantly altered by HFD and reversed by interventions, their KEGG sub-functional role are depicted and color-coded on the right. The detailed information of these 42 significant pathways is available in the Supplementary Table S5. Pathways are displayed in high (>5%), medium and low (<1%) portions using boxplots (for high) and heatmaps (for medium and low). In the heatmaps, the black boxes highlight the pathways that were significantly altered by HFD, and those were significantly reversed by interventions, also that was particularly enriched in PRE and SYN. Arrows (“↑” and “↓”) in the boxes indicate the changing directions (increased and decreased) of HFD vs ND, PRO vs HFD, PRE vs HFD, SYN vs HFD, respectively. Data are shown as means ± SEM in (a) and as relative numbers (%) based on relative protein group numbers in (d). With n = 6 mice per group. Groups with different letters are significantly different from each other assessed by Kruskal–Wallis test followed by post-hoc independent two-sided Students test (for a) or Fisher LSD (P < 0.05).
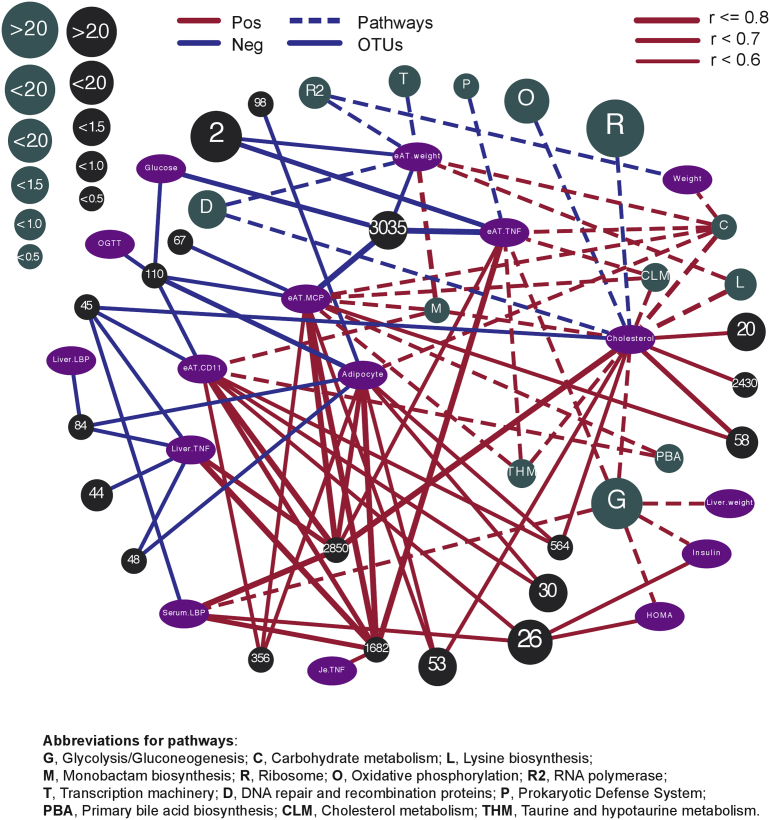
3.6. OTUs and KEGG pathways rescued by the dietary interventions are significantly associated with host metabolic disorder parameters
The microbial results were further integrated with host parameters using Pearson's correlation analysis. A total of 19 OTUs and 13 pathways that were significantly reversed by at least one intervention had significant (P < 0.05) and strong (r > 0.5) correlations with host metabolic parameters (Figures S10 and S11). These two correlation results were integrated and visualized in Cytoscape 3.6.0, the upper left and lower right areas are those negatively and positively correlated with obesity (Figure 6). From the network, it was found that the relative abundances of OTU2850/1682/26 (Alistipes timonensis) were positively correlated with most of the host parameters and the correlations were very strong, indicating an important link of this microbe to obesity. Other OTUs, including 3035, 110, 67, and 48, were negatively correlated with obesity, suggesting their potential role as probiotics for future preventive and therapeutic applications. In pathways, the pathways of Glycolysis/Gluconeogenesis, Lysine biosynthesis and Primary bile acid biosynthesis were positively correlated with obesity, while Ribosome, Oxidative phosphorylation, RNA polymerase and Transcription machinery were negatively correlated with obesity (Figure 6). The network highlighted that the impact of dietary interventions used in this study are mediated through the regulation of obesity-associated bacteria, which in turn, modulates the functional activities of gut microbial ecosystem in terms of carbohydrate, amino acid, lipid and energy metabolisms.
Figure 6.
Correlation network: specific OTUs and KEGG pathways reversed by the dietary interventions are significantly correlated with host metabolic syndrome parameters. Variables are color-coded as follows: OTUs, black; pathways, dark green; host parameters, purple. Solid lines indicate the correlations between OTUs and host metabolic parameters; dash lines indicate the correlations between pathways and host metabolic parameters. Only OTUs and pathways that have significant (p < 0.05) and strong (r > 0.5) correlations and that were significantly reversed by at least one intervention compared with HFD are displayed. Red and blue edges indicate positive and negative correlations, respectively. The size of the OTUs and pathways are proportional to their relative abundances (%) and relative numbers (%), respectively. The width of edges indicates the intensities of correlation with the wider the edges, the stronger the correlation. Details on statistics including P value and corrected P value are provided in the Supplementary Table S6. OTU numbers are used to indicate OTUs. Abbreviations for pathways: G, Glycolysis/Gluconeogenesis; C, Carbohydrate metabolism; L, Lysine biosynthesis; M, Monobactam biosynthesis; R, Ribosome; O, Oxidative phosphorylation; R2, RNA polymerase; T, Transcription machinery; D, DNA repair and recombination proteins; P, Prokaryotic Defense System; PBA, Primary bile acid biosynthesis; CLM, Cholesterol metabolism; THM, Taurine and hypotaurine metabolism; Abbreviations for host parameters: Weight, Weight gain; Glucose, Fasting blood glucose; Insulin, Fasting insulin; HOMA, HOMA-IR; OGTT, AUC of OGTT; Adipocyte, eAT adipocyte size; Liver.TNF, Liver TNF-α mRNA; Je.TNF, Jejunum TNF-α mRNA; eAT.TNF, eAT TNF-α mRNA; eAT.CD11, eAT CD11c mRNA; eAT.MCP, eAT MCP-1 mRNA.
4. Discussion
The primary aim of this study was to assess the impact of dietary interventions on diet-induced obesity and comorbidities in mice, along with detailed characterization of the gut microbiota. Whereas the effects of the selected probiotic strains and oat β-glucans were separately reported elsewhere in the context of obesity and metabolic syndrome [18], [25], [35], the novelty of the present study is threefold: (i) a novel synbiotic candidate was used and comprehensive mouse experiments were conducted to investigate its impact on obesity in comparison with individual pro- and prebiotic components; (ii) the gut microbial ecosystem was deeply characterized at both the phylogenetic and functional (metabolic and metaproteomic) levels in the context of obesity and dietary interventions; (iii) multi-omics data were further integrated and the important taxa and metabolic pathways that mediated the beneficial effects of the novel synbiotic were identified.
Adiposity and metabolic complication have been associated with low bacterial richness in the gut [36], [37], [38]. A similar result was observed in the present study, which was compensated by dietary intervention with the synbiotic. At the taxonomic level, relative abundance of the family Rikenellaceae was previously found to be elevated in the gut microbiota of HFD-induced obese mice [39], [40], which is consistent with our findings. Bacteria within family S24-7 (order Bacteroidales) are abundant members in the mouse gut that are efficient primary fermenter capable of producing acetate, propionate, and succinate [41]. Increased relative abundances of S24-7 have been described in HFD-fed diabetes-sensitive mice supplemented with gluco-oligosaccharide [42], in mice fed a low-fat diet associated with increased exercise [43], and in mice following treatment-induced remission of colitis [44]. Taken together, the effect of interventions on microbiota was associated with significantly reducing the obesity-associated family of Rikenellaceae and restoring the levels of SCFA producer S24-7. At the level of OTUs, the relative abundance of A. timonensis (OTU26/2850/1682) [45] and Alistipes inops (OTU20) [46] were markedly elevated by the HFD, which were reversed by the interventions and positively correlated with many host metabolic syndrome parameters, suggesting their potential role as biomarkers for early screening of metabolic disorders. In contrast, the relative abundance of Bacteroides vulgatus (OTU32/2325/875) [47], Bacteroides dorei (OTU2583) [48], and Bacteroides oleiciplenus (OTU922) [49] were significantly decreased by the HFD. B. vulgatus is generally considered to be a beneficial gut commensal, with specific strains of B. vulgatus shown to be reduced in gut microbiome of Crohn's disease patients [44], be capable of protecting against colitis [50] and be identified as potential new probiotic for the treatment of inflammatory and infectious gastrointestinal disorders [51]. Furthermore, we observed that the interventions significantly restored the HFD-reduced species of Faecalibaculum rodentium (OTU67) [52] and Alistipes putredinis (OTU48) [53]. F. rodentium is a gram-positive obligate anaerobe within the genus of Faecalibaculum. It has higher fermentation ability, especially butyrate production, than other related organisms and has been hypothesized as the main replacer of Lactobacillus and Bifidobacterium between the early and late stages of life, along with a shift from lactate metabolism to increased SCFAs production and carbohydrate metabolism [54]. A. putredinis has been demonstrated to be the major bacterium that governs the changes in bacterial community composition by cruciferous vegetable consumption [55]. In the present study, the relative abundances of A. putredinis and F. rodentium were negatively correlated with host metabolic syndrome parameters, possibly suggesting the important role of these two species in alleviating HFD-driven metabolic disturbances and indicating their potential role as probiotics. Taken together, several potentially important OTUs were identified for future investigations as either biomarker of metabolic disorders or as probiotics for preventive and therapeutic applications.
One potential mechanism underlying the regulation of host metabolism by the gut microbiota is through the production of bioactive metabolites and their regulatory effects. The SCFAs and BAs are two important groups of microbiome-derived metabolites with profound impacts on microbial homeostasis and host metabolism [56], [57]. The major microbial fermentation products of dietary fibers in the gut are SCFAs, in particular, acetate, propionate, and butyrate [58]. However, when fermentable dietary fibers are in short supply, microbes can switch to consuming energetically less favorable sources, such as amino acids or dietary fats [59]. This might explain several associations we observed in HFD-fed mice: the association between the decrease in SCFAs concentrations and the increase in metabolic pathways involved in amino acid and lipid metabolisms. Dietary supplementations with acetate [60], butyrate and propionate [61] have been demonstrated to protect against diet-induced obesity and regulate gut hormones in rodents. In humans, a significant reduction in weight gain was observed after long-term supplementation with inulin-propionate ester, which can be metabolized by the microbiota to propionate in the colon [62]. In the present study, all dietary interventions significantly restored the cecal levels of acetate, propionate, and butyrate, indicating that our interventions might regulate the gut microbiota by favoring the growth and/or activity of SCFAs producers, such as F. rodentium, A. putredinis and bacteria within the family of S24-7.
During the last decades, it has become clear that BAs not only serve as detergent to facilitate the absorption of lipids, but also act as signaling molecules. Recent findings supported the important role of the cross-talk between BAs and gut microbiota in regulating host metabolism [57], [63], [64]. Changes in BAs and their metabolism have been implicated in irritable bowel syndrome [65], Crohn's disease [66], and diabetes [67], [68]. Furthermore, the activation of FXR, the cognate receptor of BAs, has been reported to improve metabolic syndrome in mouse models [69], [70], providing additional insight into the effect of the gut microbiota on metabolic syndrome. By investigating alterations of the overall bile acid pools in the context of obesity and with dietary interventions, we observed that the synbiotic significantly reduced the concentrations of total BAs and a number of BAs, including TCDCA, TCA, TUDCA and TDCA. BAs regulate important metabolic pathways, including those involved in drug, lipid, carbohydrate, and energy metabolism and transport [71]. BAs can also influence the microbiota composition. It has been reported that increased BA concentrations resulted in an increase in Firmicutes and a decrease in Bacteroidetes [72], which was also observed in the microbiota of our HFD mice. Gram-negative bacteria are currently thought to be more resistant to BAs than Gram-positive microorganisms [73]. This indicates that the synbiotic might modulate microbiota via the down-regulation of BAs, which in turn creates a microbial environment that favors the growth of some Gram-positive microbes.
Compared with metaproteomics, which measures proteins - the direct effectors of biological processes, provides an opportunity to characterize gut microbial functions and activities at a deeper level. The clinical importance of interrogating gut microbiota using metaproteomics has been highlighted recently [74]. To our knowledge, this is the first study that illustrates the effects of synbiotics on the gut microbiota in the context of metabolic disorders using metaproteomics. Consistent with a metaproteomic analysis by Kolmeder and colleagues [75], we found that Firmicutes (60–70% of assigned protein groups) as the most prominent phylum as compared with Bacteroidetes (2–15%). In present study, HFD affected both total and active bacteria with around 60% Firmicutes and 40% Bacteroidetes in HFD compared with 40% Firmicutes and 60% Bacteroidetes in ND. At the functional level, Firmicutes showed more activity in HFD (60–70% Firmicutes and 2–4% Bacteroidetes) compared with ND (50–60% Firmicutes and 5–15% Bacteroidetes). These indicated that the abundances and activities of many bacteria within the phylum of Bacteroidetes were decreased or even depleted due to HFD. These differences between the phylogenetic and functional levels highlight the importance of comparative analysis of gut microbiota at both levels, as whether a community member is active or not is not evident in genomics data alone. When assessing the metaproteome of the important taxa identified, the protein groups originating from F. rodentium were found in many important pathways, including the Butanoate and Propanoate metabolisms, Ribosome, Amino acid related enzymes, and DNA repair and recombination proteins. Most of F. rodentium-originated proteins involved in those pathways were only detected in ND but were depleted in HFD while a few were detected again in SYN. This pattern was similar to that of proteins derived from Bifidobacterium spp. and Lactobacillus spp. in those pathways. These results further support the important role of F. rodentium (OUT 67) in the regulation of metabolic pathways. However, up to now large parts of the microbiome is still not deciphered which could mean that changes in the metaproteome can be mediated by other still unknown bacteria.
At the pathway level, in consistence with a previous functional study of gut microbial communities in lean and obese guts using metaproteomics [76], we found the cell mobility, and carbohydrate and lipid metabolisms were significantly increased by HFD, while the nucleotide metabolism was decreased. In agreement with another metaproteomic analysis of gut microbiota in mice fed with HFD [39], a significant increase in functions involved in amino acid metabolism and cell motility by HFD, while a decrease in energy and nucleotide metabolisms were observed. It has been proposed that when compared with African rural diets that are high in complex carbohydrates, a westernized diet is associated with metagenomes enriched in amino acid- and simple sugar-degrading enzymes [77]. Hence, our data are consistent with the literature and we proposed that HFD could induce a striking change in the functional activities of the gut ecosystem, which is characterized mainly by an increase in cell motility, and amino acid, carbohydrate and lipid metabolisms as well as a decrease in energy and nucleotide metabolisms. In the present study, the synbiotic significantly reversed 11 KEGG pathways that are involved in carbohydrate, amino acid, and energy metabolisms, biosynthesis of other secondary metabolites, transcription, translation, replication and repair, as well as transport and catabolism.
Altogether, the dietary interventions with the pro-, pre- and synbiotics used in the present study alleviated HFD-induced metabolic disorders and concomitantly modulated the gut microbiota at different levels: firstly, regulating the microbial population at the diversity and taxonomic levels, including a significant reduction in the obesity-associated taxa (such as A. timonensis, A. inops and Rikenellaceae), and a restoration of carbohydrate-degrading and SCFA-producing taxa (such as F. rodentium, A. putredinis and Bacteroidales S24-7); secondly, regulating important microbial metabolites including (i) SCFAs by restoring the concentrations of acetate, propionate, and butyrate and (ii) total BA by controlling the levels of primary and secondary BAs; thirdly, regulating the functional activity of gut ecosystem, mostly via the reorganization of important metabolic pathways including carbohydrate, amino acid, and energy metabolisms; fourthly, diet-mediated modulation in the cross-talk between microbial communities and their metabolites leading to the functional activity alterations of gut ecosystem, which in turn, forms an environment that favors the growth of particular microbes and the formation of certain metabolites.
Relative to pro- and prebiotics, the synbiotic treatment seemed to be more potent in attenuating weight gain, which was linked to a more efficient regulation of gut microbiota with regard to (i) regulating microbial populations by significantly increasing microbial richness and reversing more OTUs; (ii) controlling the levels of total BA and several BAs; (iii) regulating microbial functions by significantly shifting the overall metaproteome profiles away from HFD and reversing more metabolic pathways.
In summary, we propose a novel dietary strategy with synbiotic intervention for the improvement of HFD-induced weight gain, and thoroughly elucidated its effects on the regulation of the gut microbial ecosystem at multiple aspects (including genes, proteins, and metabolites) in a mouse model. This study provides a resourceful, multi-dimensional dataset that will facilitate future investigations of the relationship among diets, gut microbiota, and obesity, as well as future applications of dietary intervention in clinical prevention or therapy of obesity and its associated metabolic disorders.
Author contributions
Xinxin Ke designed the project, conducted the animal experiments, analyzed and integrated all the datasets, finalized the figures, and wrote the original draft of the manuscript; Alesia Walker and Philippe Schmitt-Kopplin helped with targeted metabolites data acquisition and analysis; lias Lagkouvardos and Thomas Clavel contributed to the 16S rRNA gene sequence data acquisition and analysis; Sven-Bastiaan Haange, Martin von Bergen, and Nico Jehmlich supported for metaproteomics data acquisition and analysis; Yuwen Liu and Xin He helped with the bioinformatic analysis and the interpretation of data; Peter C.K. Cheung directed and supervised the entire project. All authors reviewed and edited the manuscript.
Acknowledgements
We thank Mr. John Tse and Mr. L.W. Lam from the Laboratory Animal Services Centre in the Chinese University of Hong Kong for their help on the animal experiments and Klaus Neuhaus at the ZIEL Institute for Food and Health of the Technical University of Munich for help with 16S rRNA gene sequencing.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2019.01.012.
Conflict of interest
The authors declare no conflict of interest.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Ng M., Fleming T., Robinson M., Thomson B., Graetz N., Margono C. Global, regional, and national prevalence of overweight and obesity in children and adults during 1980-2013: a systematic analysis for the Global Burden of Disease Study 2013. The Lancet. 2014;384(9945):766–781. doi: 10.1016/S0140-6736(14)60460-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Skinner A.C., Perrin E.M., Skelton J.A. Prevalence of obesity and severe obesity in US children, 1999-2014. Obesity. 2016;24(5):1116–1123. doi: 10.1002/oby.21497. [DOI] [PubMed] [Google Scholar]
- 3.Kotzampassi K., Giamarellos-Bourboulis E.J., Stavrou G. Obesity as a consequence of gut bacteria and diet interactions. ISRN Obesity. 2014 doi: 10.1155/2014/651895. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4.Mardinoglu A., Wu H., Bjornson E., Zhang C., Hakkarainen A., Räsänen S.M. An integrated understanding of the rapid metabolic benefits of a carbohydrate-restricted diet on hepatic steatosis in humans. Cell Metabolism. 2018;27(3):559–571. doi: 10.1016/j.cmet.2018.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cani P.D., Bibiloni R., Knauf C., Waget A., Neyrinck A.M., Delzenne N.M. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes. 2008;57(6):1470–1481. doi: 10.2337/db07-1403. [DOI] [PubMed] [Google Scholar]
- 6.Ridaura V.K., Faith J.J., Rey F.E., Cheng J., Duncan A.E., Kau A.L. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341(6150):1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tilg H., Kaser A. Gut microbiome, obesity, and metabolic dysfunction. The Journal of Clinical Investigation. 2011;121(6):2126–2132. doi: 10.1172/JCI58109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Everard A., Cani P.D. Diabetes, obesity and gut microbiota. Best Practice & Research Clinical Gastroenterology. 2013;27(1):73–83. doi: 10.1016/j.bpg.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 9.Parekh P.J., Arusi E., Vinik A.I., Johnson D.A. The role and influence of gut microbiota in pathogenesis and management of obesity and metabolic syndrome. Frontiers in Endocrinology. 2014;5:47. doi: 10.3389/fendo.2014.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Molinaro F., Paschetta E., Cassader M., Gambino R., Musso G. Probiotics, prebiotics, energy balance, and obesity: mechanistic insights and therapeutic implications. Gastroenterology Clinics. 2012;41(4):843–854. doi: 10.1016/j.gtc.2012.08.009. [DOI] [PubMed] [Google Scholar]
- 11.Delzenne N.M., Neyrinck A.M., Bäckhed F., Cani P.D. Targeting gut microbiota in obesity: effects of prebiotics and probiotics. Nature Reviews Endocrinology. 2011;7(11):639–646. doi: 10.1038/nrendo.2011.126. [DOI] [PubMed] [Google Scholar]
- 12.Bienenstock J., Gibson G., Klaenhammer T.R., Walker W.A., Neish A.S. New insights into probiotic mechanisms: a harvest from functional and metagenomic studies. Gut Microbes. 2013;4(2):94–100. doi: 10.4161/gmic.23283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arora T., Singh S., Sharma R.K. Probiotics: interaction with gut microbiome and antiobesity potential. Nutrition. 2013;29(4):591–596. doi: 10.1016/j.nut.2012.07.017. [DOI] [PubMed] [Google Scholar]
- 14.An H.M., Park S.Y., Lee D.K., Kim J.R., Cha M.K., Lee S.W. Antiobesity and lipid-lowering effects of Bifidobacterium spp. in high fat diet-induced obese rats. Lipids in Health and Disease. 2011;10:1–8. doi: 10.1186/1476-511X-10-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoo S.R., Kim Y.J., Park D.Y., Jung U.J., Jeon S.M., Ahn Y.T. Probiotics L. plantarum and L. curvatus in combination alter hepatic lipid metabolism and suppress diet-induced obesity. Obesity. 2013;21(12):2571–2578. doi: 10.1002/oby.20428. [DOI] [PubMed] [Google Scholar]
- 16.Kadooka Y., Sato M., Imaizumi K., Ogawa A., Ikuyama K., Akai Y. Regulation of abdominal adiposity by probiotics (Lactobacillus gasseri SBT2055) in adults with obese tendencies in a randomized controlled trial. European Journal of Clinical Nutrition. 2010;64(6):636–643. doi: 10.1038/ejcn.2010.19. [DOI] [PubMed] [Google Scholar]
- 17.Cani P.D., Neyrinck A.M., Fava F., Knauf C., Burcelin R.G., Tuohy K.M. Selective increases of bifidobacteria in gut microflora improve high-fat-diet-induced diabetes in mice through a mechanism associated with endotoxaemia. Diabetologia. 2007;50(11):2374–2383. doi: 10.1007/s00125-007-0791-0. [DOI] [PubMed] [Google Scholar]
- 18.Wang J., Tang H., Zhang C., Zhao Y., Derrien M., Rocher E. Modulation of gut microbiota during probiotic-mediated attenuation of metabolic syndrome in high fat diet-fed mice. The ISME Journal. 2015;9:1–15. doi: 10.1038/ismej.2014.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bindels L.B., Delzenne N.M., Cani P.D., Walter J. Towards a more comprehensive concept for prebiotics. Nature Reviews Gastroenterology and Hepatology. 2015;12(5):303–310. doi: 10.1038/nrgastro.2015.47. [DOI] [PubMed] [Google Scholar]
- 20.Geurts L., Neyrinck A.M., Delzenne N.M., Knauf C., Cani P.D. Gut microbiota controls adipose tissue expansion, gut barrier and glucose metabolism: novel insights into molecular targets and interventions using prebiotics. Beneficial Microbes. 2013;5(1):3–17. doi: 10.3920/BM2012.0065. [DOI] [PubMed] [Google Scholar]
- 21.Everard A., Lazarevic V., Gaïa N., Johansson M., Ståhlman M., Backhed F. Microbiome of prebiotic-treated mice reveals novel targets involved in host response during obesity. The ISME Journal. 2014;8(10):2116–2130. doi: 10.1038/ismej.2014.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parnell J.A., Reimer R.A. Weight loss during oligofructose supplementation is associated with decreased ghrelin and increased peptide YY in overweight and obese adults. The American Journal of Clinical Nutrition. 2009;89(6):1751–1759. doi: 10.3945/ajcn.2009.27465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khoury D.E., Cuda C., Luhovyy B., Anderson G. Beta glucan: health benefits in obesity and metabolic syndrome. Journal of Nutrition and Metabolism. 2011;2012:851362. doi: 10.1155/2012/851362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daou C., Zhang H. Oat beta-glucan: its role in health promotion and prevention of diseases. Comprehensive Reviews in Food Science and Food Safety. 2012;11(4):355–365. [Google Scholar]
- 25.Ji-Lin D., Ying-ying Z., Lin L., Rui-ling S., Hong L. Effect of oat soluble and insoluble β-glucan on lipid metabolism and intestinal lactobacillus in high-fat diet-induced obese mice. Journal of Food and Nutrition Research. 2014;2(8):510–516. [Google Scholar]
- 26.Bomhof M.R., Saha D.C., Reid D.T., Paul H.A., Reimer R.A. Combined effects of oligofructose and Bifidobacterium animalis on gut microbiota and glycemia in obese rats. Obesity. 2014;22(3):763–771. doi: 10.1002/oby.20632. [DOI] [PubMed] [Google Scholar]
- 27.Stenman L.K., Lehtinen M.J., Meland N., Christensen J.E., Yeung N., Saarinen M.T. Probiotic with or without fiber controls body fat mass, associated with serum zonulin, in overweight and obese adults-randomized controlled trial. EBioMedicine. 2016;13:190–200. doi: 10.1016/j.ebiom.2016.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lagkouvardos I., Kläring K., Heinzmann S.S., Platz S., Scholz B., Engel K.H. Gut metabolites and bacterial community networks during a pilot intervention study with flaxseeds in healthy adult men. Molecular Nutrition & Food Research. 2015;59(8):1614–1628. doi: 10.1002/mnfr.201500125. [DOI] [PubMed] [Google Scholar]
- 29.Lagkouvardos I., Joseph D., Kapfhammer M., Giritli S., Horn M., Haller D. IMNGS: a comprehensive open resource of processed 16S rRNA microbial profiles for ecology and diversity studies. Scientific Reports. 2016;6:33721. doi: 10.1038/srep33721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lagkouvardos I., Fisher S., Kumar N., Clavel T. Rhea: a transcript and modular R pipeline for microbial profiling based on 16S rRNA gene amplicons. Peer J. 2017;5:e2836. doi: 10.7717/peerj.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kespohl M., Vachharajani N., Luu M., Harb H., Pautz S., Wolff S. The microbial metabolite butyrate induces expression of Th1-associated factors in CD4+ T cells. Frontiers in Immunology. 2017;8:1036. doi: 10.3389/fimmu.2017.01036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sillner N., Walker A., Koch W., Witting M., Schmitt-Kopplin P. Metformin impacts cecal bile acid profiles in mice. Journal of Chromatography B. 2018;1083:35–43. doi: 10.1016/j.jchromb.2018.02.029. [DOI] [PubMed] [Google Scholar]
- 33.Schaubeck M., Clavel T., Calasan J., Lagkouvardos I., Haange S.B., Jehmlich N. Dysbiotic gut microbiota causes transmissible Crohn's disease-like ileitis independent of failure in antimicrobial defence. Gut. 2015 doi: 10.1136/gutjnl-2015-309333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.VizcaÃno J.A., Csordas A., Del-Toro N., Dianes J.A., Griss J., Lavidas I. 2016 update of the PRIDE database and its related tools. Nucleic Acids Research. 2016;44(D1):D447–D456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aoe S., Ichinose Y., Kohyama N., Komae K., Takahashi A., Yoshioka T. Effects of β-glucan content and pearling of barley in diet-induced obese mice. Cereal Chemistry. 2017;94(6):956–962. [Google Scholar]
- 36.Le Chatelier E., Nielsen T., Qin J., Prifti E., Hildebrand F., Falony G. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500(7464):541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- 37.Cotillard A., Kennedy S.P., Kong L.C., Prifti E., Pons N., Le Chatelier E. Dietary intervention impact on gut microbial gene richness. Nature. 2013;500(7464):585–588. doi: 10.1038/nature12480. [DOI] [PubMed] [Google Scholar]
- 38.Turnbaugh P.J., Bäckhed F., Fulton L., Gordon J.I. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host & Microbe. 2008;3(4):213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim K.A., Gu W., Lee I.A., Joh E.H., Kim D.H. High fat diet-induced gut microbiota exacerbates inflammation and obesity in mice via the TLR4 signaling pathway. PLoS One. 2012;7(10):e47713. doi: 10.1371/journal.pone.0047713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Daniel H., Gholami A.M., Berry D., Desmarchelier C., Hahne H., Loh G. High-fat diet alters gut microbiota physiology in mice. The ISME Journal. 2013;8(2):295–308. doi: 10.1038/ismej.2013.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ormerod K.L., Wood D.L., Lachner N., Gellatly S.L., Daly J.N., Parsons J.D. Genomic characterization of the uncultured Bacteroidales family S24-7 inhabiting the guts of homeothermic animals. Microbiome. 2016;4(1):36. doi: 10.1186/s40168-016-0181-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Serino M., Luche E., Gres S., Baylac A., Bergé M., Cenac C. Metabolic adaptation to a high-fat diet is associated with a change in the gut microbiota. Gut. 2012;61(4):543–553. doi: 10.1136/gutjnl-2011-301012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Evans C.C., LePard K.J., Kwak J.W., Stancukas M.C., Laskowski S., Dougherty J. Exercise prevents weight gain and alters the gut microbiota in a mouse model of high fat diet-induced obesity. PLoS One. 2014;9(3):e92193. doi: 10.1371/journal.pone.0092193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rooks M.G., Veiga P., Wardwell-Scott L.H., Tickle T., Segata N., Michaud M. Gut microbiome composition and function in experimental colitis during active disease and treatment-induced remission. The ISME Journal. 2014;8(7):1403–1417. doi: 10.1038/ismej.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lagier J.C., Armougom F., Mishra A.K., Nguyen T.T., Raoult D., Fournier P.E. Non-contiguous finished genome sequence and description of Alistipes timonensis sp. nov. Standards in Genomic Science. 2012;6(3):315–324. doi: 10.4056/sigs.2685971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shkoporov A.N., Chaplin A.V., Khokhlova E.V., Shcherbakova V.A., Motuzova O.V., Bozhenko V.K. Alistipes inops sp. nov. and Coprobacter secundus sp. nov., isolated from human faeces. International Journal of Systematic and Evolutionary Microbiology. 2015;65(12):4580–4588. doi: 10.1099/ijsem.0.000617. [DOI] [PubMed] [Google Scholar]
- 47.Cuív P.Ó., Klaassens E.S., Durkin A.S., Harkins D.M., Foster L., McCorrison J. Draft genome sequence of Bacteroides vulgatus PC510, a strain isolated from human feces. Journal of Bacteriology. 2011;193(15):4025–4026. doi: 10.1128/JB.05256-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bakir M.A., Sakamoto M., Kitahara M., Matsumoto M., Benno Y. Bacteroides dorei sp. nov., isolated from human faeces. International Journal of Systematic and Evolutionary Microbiology. 2006;56(7):1639–1643. doi: 10.1099/ijs.0.64257-0. [DOI] [PubMed] [Google Scholar]
- 49.Watanabe Y., Nagai F., Morotomi M., Sakon H., Tanaka R. Bacteroides clarus sp. nov., Bacteroides fluxus sp. nov. and Bacteroides oleiciplenus sp. nov., isolated from human faeces. International Journal of Systematic and Evolutionary Microbiology. 2010;60(8):1864–1869. doi: 10.1099/ijs.0.015107-0. [DOI] [PubMed] [Google Scholar]
- 50.Waidmann M., Bechtold O., Frick Js., Lehr Ha., Schubert S., Dobrindt U. Bacteroides vulgatus protects against Escherichia coli-induced colitis in gnotobiotic interleukin-2-deficient mice. Gastroenterology. 2003;125(1):162–177. doi: 10.1016/s0016-5085(03)00672-3. [DOI] [PubMed] [Google Scholar]
- 51.Frick J.S., Fink K., Kahl F., Niemiec M.J., Quitadamo M., Schenk K. Identification of commensal bacterial strains that modulate Yersinia enterocolitica and dextran sodium sulfate-induced inflammatory responses: implications for the development of probiotics. Infection Immunology. 2007;75(7):3490–3497. doi: 10.1128/IAI.00119-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chang D.H., Rhee M.S., Ahn S., Bang B.H., Oh J.E., Lee H.K. Faecalibaculum rodentium gen. nov., sp. nov., isolated from the faeces of a laboratory mouse. Antonie van Leeuwenhoek. 2015;108(6):1309–1318. doi: 10.1007/s10482-015-0583-3. [DOI] [PubMed] [Google Scholar]
- 53.Rautio M., Eerola E., Väisänen-Tunkelrott M.L., Molitoris D., Lawson P., Collins M.D. Reclassification of Bacteroides putredinis (Weinberg et al., 1937) in a new genus Alistipes gen. nov., as Alistipes putredinis comb. nov., and description of Alistipes finegoldii sp. nov., from human sources. Systematic and Applied Microbiology. 2003;26(2):182–188. doi: 10.1078/072320203322346029. [DOI] [PubMed] [Google Scholar]
- 54.Lim S., Chang D.H., Ahn S., Kim B.C. Whole genome sequencing of “Faecalibaculum rodentium” ALO17, isolated from C57BL/6J laboratory mouse feces. Gut Pathogens. 2016;8(1):3. doi: 10.1186/s13099-016-0087-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li F., Hullar M.A., Schwarz Y., Lampe J.W. Human gut bacterial communities are altered by addition of cruciferous vegetables to a controlled fruit-and vegetable-free diet. The Journal of Nutrition. 2009;139(9):1685–1691. doi: 10.3945/jn.109.108191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koh A., Vadder F.D., Kovatcheva-Datchary P., Bäckhed F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell. 2016;165(6):1332–1345. doi: 10.1016/j.cell.2016.05.041. [DOI] [PubMed] [Google Scholar]
- 57.Staley C., Weingarden A.R., Khoruts A., Sadowsky M.J. Interaction of gut microbiota with bile acid metabolism and its influence on disease states. Applied Microbiology and Biotechnology. 2017;101(1):47–64. doi: 10.1007/s00253-016-8006-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cummings J., Pomare E., Branch W., Naylor C., Macfarlane G. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut. 1987;28(10):1221–1227. doi: 10.1136/gut.28.10.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cummings J., Macfarlane G. The control and consequences of bacterial fermentation in the human colon. Journal of Applied Microbiology. 1991;70(6):443–459. doi: 10.1111/j.1365-2672.1991.tb02739.x. [DOI] [PubMed] [Google Scholar]
- 60.Yamashita H., Fujisawa K., Ito E., Idei S., Kawaguchi N., Kimoto M. Improvement of obesity and glucose tolerance by acetate in Type 2 diabetic Otsuka Long-Evans Tokushima Fatty (OLETF) rats. Bioscience, Biotechnology, and Biochemistry. 2007;71(5):1236–1243. doi: 10.1271/bbb.60668. [DOI] [PubMed] [Google Scholar]
- 61.Lin H.V., Frassetto A., Kowalik E.J., Jr., Nawrocki A.R., Lu M.M., Kosinski J.R. Butyrate and propionate protect against diet-induced obesity and regulate gut hormones via free fatty acid receptor 3-independent mechanisms. PLoS One. 2012;7(4):e35240. doi: 10.1371/journal.pone.0035240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chambers E.S., Viardot A., Psichas A., Morrison D.J., Murphy K.G., Zac-Varghese S.E. Effects of targeted delivery of propionate to the human colon on appetite regulation, body weight maintenance and adiposity in overweight adults. Gut. 2015;64(11):1744–1754. doi: 10.1136/gutjnl-2014-307913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thomas C., Pellicciari R., Pruzanski M., Auwerx J., Schoonjans K. Targeting bile-acid signalling for metabolic diseases. Nature Reviews Drug Discovery. 2008;7(8):678. doi: 10.1038/nrd2619. [DOI] [PubMed] [Google Scholar]
- 64.Nie Y-f., Hu J., Yan X-h. Cross-talk between bile acids and intestinal microbiota in host metabolism and health. Journal of Zhejiang University-Science B. 2015;16(6):436–446. doi: 10.1631/jzus.B1400327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Duboc H., Rainteau D., Rajca S., Humbert L., Farabos D., Maubert M. Increase in fecal primary bile acids and dysbiosis in patients with diarrhea-predominant irritable bowel syndrome. Neuro-Gastroenterology and Motility. 2012;24(6):513–520. doi: 10.1111/j.1365-2982.2012.01893.x. [DOI] [PubMed] [Google Scholar]
- 66.Labbé A., Ganopolsky J.G., Martoni C.J., Prakash S., Jones M.L. Bacterial bile metabolising gene abundance in Crohn's, ulcerative colitis and type 2 diabetes metagenomes. PLoS One. 2014;9(12):e115175. doi: 10.1371/journal.pone.0115175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Knop F.K. Bile-induced secretion of glucagon-like peptide-1: pathophysiological implications in type 2 diabetes? American Journal of Physiology-Endocrinology and Metabolism. 2010;299(1):E10–E13. doi: 10.1152/ajpendo.00137.2010. [DOI] [PubMed] [Google Scholar]
- 68.Prawitt J., Caron S., Staels B. Bile acid metabolism and the pathogenesis of type 2 diabetes. Current Diabetes Reports. 2011;11(3):160–166. doi: 10.1007/s11892-011-0187-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Porez G., Prawitt J., Gross B., Staels B. Bile acid receptors as targets for the treatment of dyslipidemia and cardiovascular disease: thematic review series: new lipid and lipoprotein targets for the treatment of cardiometabolic diseases. Journal of Lipid Research. 2012;53(9):1723–1737. doi: 10.1194/jlr.R024794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Del Bas J.M., Ricketts M.L., Vaqué M., Sala E., Quesada H., Ardevol A. Dietary procyanidins enhance transcriptional activity of bile acid-activated FXR in vitro and reduce triglyceridemia in vivo in a FXR-dependent manner. Molecular Nutrition & Food Research. 2009;53(7):805–814. doi: 10.1002/mnfr.200800364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hylemon P.B., Zhou H., Pandak W.M., Ren S., Gil G., Dent P. Bile acids as regulatory molecules. Journal of Lipid Research. 2009;50(8):1509–1520. doi: 10.1194/jlr.R900007-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ridlon J.M., Alves J.M., Hylemon P.B., Bajaj J.S. Cirrhosis, bile acids and gut microbiota: unraveling a complex relationship. Gut Microbes. 2013;4(5):382–387. doi: 10.4161/gmic.25723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Begley M., Gahan C.G., Hill C. The interaction between bacteria and bile. FEMS Microbiology Reviews. 2005;29(4):625–651. doi: 10.1016/j.femsre.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 74.Haange S.B., Jehmlich N. Proteomic interrogation of the gut microbiota: potential clinical impact. Expert Review of Proteomics. 2016;13:535–537. doi: 10.1080/14789450.2016.1190652. [DOI] [PubMed] [Google Scholar]
- 75.Kolmeder C.A., De Been M., Nikkilä J., Ritamo I., Mättö J., Valmu L. Comparative metaproteomics and diversity analysis of human intestinal microbiota testifies for its temporal stability and expression of core functions. PLoS One. 2012;7(1):e29913. doi: 10.1371/journal.pone.0029913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ferrer M., Ruiz A., Lanza F., Haange S.B., Oberbach A., Till H. Microbiota from the distal guts of lean and obese adolescents exhibit partial functional redundancy besides clear differences in community structure. Environmental Microbiology. 2013;15(1):211–226. doi: 10.1111/j.1462-2920.2012.02845.x. [DOI] [PubMed] [Google Scholar]
- 77.Yatsunenko T., Rey F.E., Manary M.J., Trehan I., Dominguez-Bello M.G., Contreras M. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.