Summary
DNA rearrangements resulting in human genome structural variants (SVs) are caused by diverse mutational mechanisms. We used long- and short-read sequencing technologies to investigate end products of de novo chromosome 17p11.2 rearrangements and query the molecular mechanisms underlying both recurrent and non-recurrent events. Evidence for an increased rate of clustered single nucleotide variant (SNV) mutation in cis with non-recurrent rearrangements was found. Indel and SNV formation are associated with both copy number gains and losses of 17p11.2, occur up to ~1 Mb away from the breakpoint junctions, and favor C>G transversion substitutions; results suggesting that single stranded DNA is formed during the genesis of the SV and providing compelling support for a microhomology-mediated break-induced replication (MMBIR) mechanism for SV formation. Our data show an additional mutational burden of MMBIR consisting of hypermutation confined to the locus and manifesting as SNVs and indels predominantly within genes.
Graphical Abstract

Introduction
The systematic analysis of DNA breakpoint junctions has delineated several mutagenic mechanisms whereby Structural Variants (SVs) may arise in the human genome (Collins et al., 2017; Conrad et al., 2010; Kidd et al., 2010; Lupski, 1998, 2015). Two general classifications for SVs derived from breakpoint junction mapping, i.e. recurrent and non-recurrent events, have been used to define genomic rearrangements. Recurrent events manifest with the same size and genomic content in unrelated individuals; this is in contrast with non-recurrent rearrangements with distinct sizes and genomic content for which a smallest region of overlap is shared among subjects with the same clinical phenotypic trait.
The underlying molecular mechanisms and resulting features associated with the formation of non-recurrent rearrangements are still being defined (Conrad et al., 2010; Kloosterman et al., 2015). Mechanistically, Non-Homologous End Joining (NHEJ), Microhomology-Mediated End Joining (MMEJ) (Lieber, 2010), and replicative repair/recombination processes such as Break Induced Replication (BIR), Fork Stalling and Template Switching (FoSTeS), and Microhomology-Mediated Break Induced Replication (MMBIR) (Hastings et al., 2009; Lee et al., 2007; Sakofsky et al., 2015) have been implicated. A prominent role for aberrant repair during replication is provided by SVs that contain more than two breakpoint junctions formed in the same mutational event termed Complex Genomic Rearrangements (CGRs) (Zhang et al., 2009b). CGRs exhibit further amplification of a given genomic locus associated with additional complexities, including inversions and insertions of templated segments at the junctions (reviewed in Carvalho and Lupski, 2016). These SV features likely reflect replicative processes and iterative short- and long-range template switching (TS) characterized by the presence of microhomology at the recombinant join-points (Kloosterman et al., 2011; Liu et al., 2011a; Zhang et al., 2009a; Zhang et al., 2009b).
The consequences of replication-based repair underlying genomic SV may extend beyond DNA rearrangement within a given locus. For instance, yeast studies identified a large increase of one base pair frameshifts and base substitutions attributed to accumulation of single-stranded DNA intermediates and decreased mismatch repair during BIR (Deem et al., 2011; Saini et al., 2013; Sakofsky et al., 2014). Similarly, in humans, non-recurrent SVs resulting in copy number gain display increased rates of single-nucleotide variants (SNVs) and small insertion/deletions (indels) flanking breakpoint junctions (Beck et al., 2015; Brandler et al., 2016; Carvalho et al., 2013; Carvalho et al., 2011; Dhokarh and Abyzov, 2016; Liu et al., 2017; Wang et al., 2015b). The ability for findings in human and yeast to bridge organisms points to the conservation of mutagenic BIR at the level of mechanism and enzymatic requirements (Costantino et al., 2014; Roumelioti et al., 2016), although the length and spectrum of the hypermutation tract accompanying SVs requires further characterization. Moreover, it remains unknown whether MMBIR in humans undergoes extensive synthesis subject to mutagenic processes which may have relevant biological consequences for both mutagenesis and disease studies. Such questions required technological innovations to enable robust resolution of various sized SVs in a genome laden with low-copy repeats (LCRs) and repetitive sequences, while also enabling detection of de novo indel and SNV mutations within megabase tracts of DNA (English et al., 2015; Wang et al., 2015a).
We hypothesize that synthesis of DNA during MMBIR and BIR could extend over substantial genomic distances. We also postulate that although SNVs and indels proximal to the breakpoint could be explained via a variety of mechanisms of double strand break repair (Chan and Gordenin, 2015; Conrad et al., 2010; Deem et al., 2011; Ponder et al., 2005; Sinha et al., 2017; Strathern et al., 1995), long tracts of mutation would strongly support BIR or MMBIR in the formation of genomic rearrangements (Deem et al., 2011; Mayle et al., 2015). We therefore tested the hypothesis that mutation rate increases might extend for hundreds of kilobase pairs on the same allele as the SV, consistent with replication-based mechanisms of DNA repair.
To investigate the mutational spectrum accompanying SV formation, we interrogated the genomes of individuals who present with Potocki-Lupski syndrome (PTLS, MIM#610883; https://www.omim.org) or Smith-Magenis syndrome (SMS, MIM#182290) due to the de novo duplication or deletion at 17p11.2 using molecular cytogenetics and both short- and long-read DNA sequencing technologies. This experimental approach allowed the genomic focus to be relatively small (megabases Mb vs Gigabases Gb), and therefore could be applied at scale for our analyses (SV characterization and junction sequencing, SNV, indels, and phasing). We identified breakpoint junctions within previously intractable loci and re-affirm the predominance of microhomology and microhomeology (perfectly matching sequences or up to a 40% mismatch in nucleotide sequence), underscoring the role of TS in generating these disease-causing CNVs. As predicted, a high SNV rate was observed in cis with non-recurrent SVs. Moreover, the length of the affected DNA strand was shown to extend for up to 1 Mb from the SV breakpoint junctions, and the mutations tend to occur within genes. In summary, our findings support the contention that replication-based repair underlying human genome SV formation can lead to long tracts of mutagenic synthesis. These data show how de novo SNV mutation rates can vary in the genome, how hypermutation can be regionally confined, and add further dimensions to understanding the mutational spectra due to de novo non-recurrent SVs.
Results
Cohort of individuals harboring 17p11.2 rearrangements
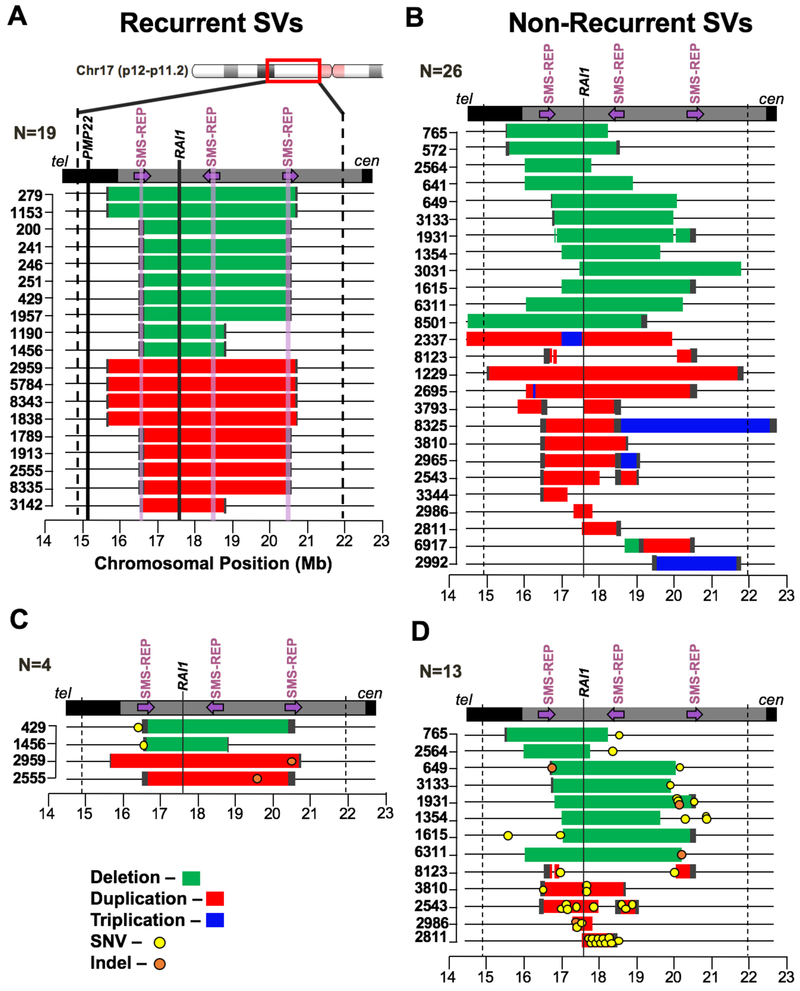
We examined 45 trios (DNA available from mother, father, and proband) with rearrangements of 17p11.2 for genomic analyses of de novo SV mutagenesis. The 45 de novo CNV trios can be divided into their types of CNV rearrangements: 19 trios with probands harboring common or uncommon recurrent rearrangements (Figure 1A), and 26 trios with probands harboring non-recurrent rearrangements (Figure 1B). All CNVs map to 17p11.2, and ~90% (41/45) encompass the main dosage sensitive gene mapping to the region, RAI1 (Figure 1). The control cohort of 10 trios we assembled consists of probands that lack de novo CNV within the 17p region.
Figure 1-. Non-recurrent rearrangements display increased SNV and indel mutations.
A) Array comparative genomic hybridization (aCGH) data for 19 de novo recurrent rearrangements (10 SMS deletions in green and 9 PTLS duplications in red) are depicted. The bounds of the capture region are shown with dashed black lines. The three SMS repeats are denoted with arrows indicating their orientation on chromosome 17p11.2 (purple translucent vertical lines depict these regions in panel A only). The established dosage-sensitive gene underlying SMS and PTLS, RAI1 is denoted with a black vertical line (another gene in the region, PMP22, is also indicated in panel A). B) 26 de novo non-recurrent rearrangements (14 PTLS and 12 SMS) are depicted. Deleted regions are shown in green, duplicated regions in red, and triplicated regions in blue. C) Local, de novo Single Nucleotide Variant (SNV) and indel mutations are shown in the context of the 4/19 recurrent structural variants (SVs) they occurred with. D) Local, de novo SNV and indel mutations are shown in the context of the 13/26 non-recurrent SV events that harbored concurrent SNV mutational events. All mutations in SMS cases occur within the non-deleted regions; all mutations in PTLS cases occur within duplicated regions except one mutation in BAB2811. See also Figure S1 and Table S3).
SV and SNV sequencing strategy to uncover de novo events at 17p11.2
The genomic DNA from 55 trios (45 CNV carriers and 10 non-CNV controls; Table S1) were subjected to high-density oligonucleotide array comparative genomic hybridization (aCGH). This analysis verified that SMS and PTLS associated SV were de novo and determined the approximate genomic coordinates of CNV breakpoint junctions. All 45 individuals carrying CNVs contained at least one breakpoint within a 7 Mb interval from Ch17:14,890,000–21,890,000 in hg19 (GRCh37). These coordinates bound the capture design and reagents used for our combined short/long-read genomic sequencing approach, and define the regional analyses. The regional interval is centered on the ~3.6 Mb deleted or duplicated in common recurrent 17p11.2 rearrangements, with an additional ~1.75 Mb flanking either side (Figure 1A); the callable portion of this region is 6.074 Mb (Table S2) (Wang et al., 2015a).
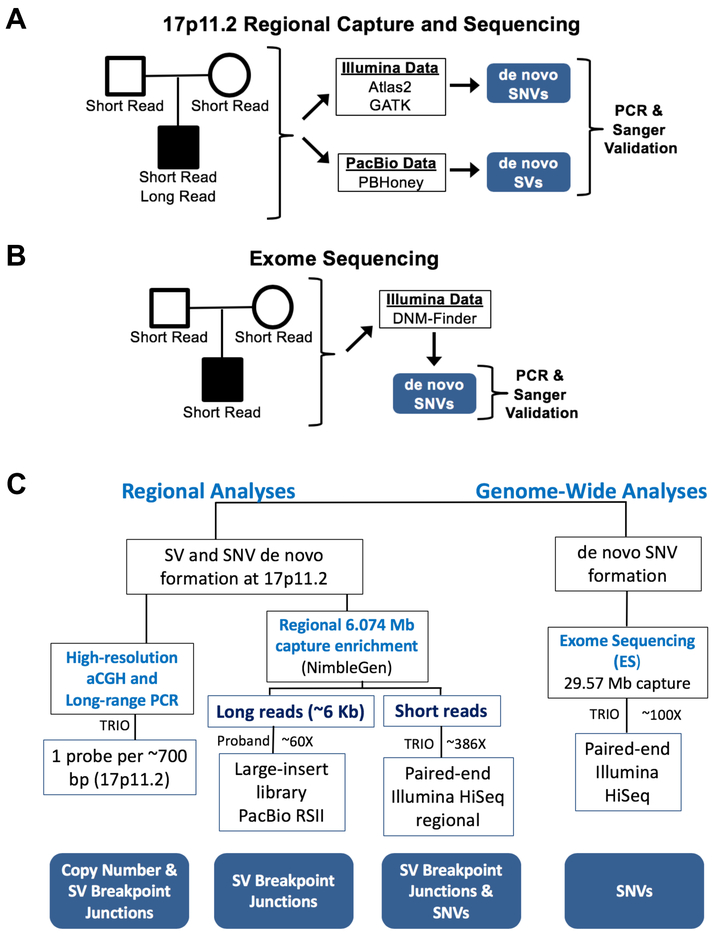
After aCGH analysis, the 55 trios chosen for the study were then subjected to multiple sequencing approaches to determine additional CNV/SV breakpoints as well as de novo SNV in their genomes (Figure 1C and 1D and Figure S1). Each individual within the 55 trios had Exome Sequencing (ES) and our targeted regional capture and Illumina sequencing performed (Figure 2A and 2B and Table S1). Additionally, genomic DNA purified from blood obtained from the 26 probands with a non-recurrent CNV was subjected to regional capture and PacBio sequencing (Wang et al., 2015a) (Figure 2A and 2C). The PacBio approach results in relatively long DNA sequencing reads (~5.5 kb average library insert size) that were analyzed by PBHoney to identify structural variation breakpoints (English et al., 2014).
Figure 2-. Sequencing and analysis strategy.
A) Schematic of regional capture and sequencing of trios. Regional capture and Illumina sequencing was performed on each individual in the study. From these data, regional de novo SNVs and indel mutations were ascertained. Additionally, 7 Mb capture and PacBio sequencing of the probands was performed to identify SV junctions. SNVs, indels, and SV junctions were experimentally verified to be de novo with PCR and Sanger sequencing, and SNV and indel mutations were visualized in the PacBio data from the same individuals. B) Exome sequencing of the trios was conducted to identify genome-wide de novo mutational burden in each trio in the study; all mutations were confirmed with PCR and Sanger sequencing. C) A flow diagram shows the regional genomic sequencing strategy employed for the 26 individuals studied with non-recurrent mutations. See also Table S1.
Resolving complex genomic rearrangements in repeat sequences
The 26 SMS and PTLS individuals harboring non-recurrent rearrangements of 17p11.2 contained a constellation of diverse rearrangements, including CGRs. Our targeted aCGH approach discerned the configuration of the rearrangements in the 26 individuals, including apparently simple deletions (DEL) and tandem duplications (DUP) (14; 10 DEL and 4 DUP), DUP-normal (NML)-DUP/inverted (INV) (2), DUP-triplication (TRP) (2), DEL-DUP (2), and one each of the following; DEL-NML-DEL (contains an inversion), DEL-NML-DELNML-DEL, DUP-TRP-DUP, TRP, DUP-NML-DUP-NML-DUP, and a very complex rearrangement consisting of DEL-NML-DUP-NML-DEL-NML-DUP-TRP-DUP-TRP-DUP, reminiscent of a chromoanasynthesis/chromothripsis-like event, and an unbalanced translocation (Data S1 and Table S3) (Yuan et al., 2015).
In summary, of the 42 apparent junctions seen by aCGH, as evidenced by copy number transition states in these 26 individuals, 30 were identified by our capture and PacBio sequencing approach. Of these 30 junctions, 14 were previously discerned by aCGH and PCR, and 16 were newly delineated using our long-read strategy (Table 1, Table S3, and Data S1). Importantly, of the 25 junctions contained in LCRs apparent via aCGH in the current cohort, only 2 were previously identified at nucleotide resolution using long-range PCR and sequencing for breakpoint analyses (Liu et al., 2011b; Shaw and Lupski, 2005); here, 12 additional LCR-mediated junctions (14/25 LCR-containing junctions in total) were resolved using capture and long-read sequencing approaches (Table 1). One of the junctions within an LCR was previously discerned using PacBio-LITS (BAB2695_DUP) (Wang et al., 2015a). Six of the 30 junctions were Alu-Alu-mediated (Song et al., 2018), and 17 others contained microhomologies or microhomeologies of 1 or more base pairs at their breakpoints (Table 1, Table S4); microhomeologies defined as in (Liu et al., 2017).
Table 1-.
Breakpoint characteristics of non-recurrent 17p11.2 SVs
| SMS patient (BAB) | CNV Coordinates Start | CNV Coordinates End | Previously published jet | How First Discovered | Microhomology/Microhomeology | LCR involvement | Parent of origin |
| 572 | 15442169–15538711 | 18285665–18529905 | No | ND | - | Prox-CMT1A REP and Middle_REP | Paternal |
| 16066603 | 18517335 | No | PacBio-Honey | GG-ACaAATGT | One side LCR (Middle_REP) | Paternal | |
| 16808862 | 16845031 | Yesa | aCGH/PCR | CTCCaATT | One side LCR (16800001–16810027) | Paternal | |
| 15441705 | 18005708 | Yesb | aCGH/PCR | AluY (LCR)-AluSg | One side LCR (15422954–15470900) | Paternal | |
| 17141946 | 19676163 | Yesb | aCGH/PCR | AluSc-AluY | No | Paternal | |
| 16035839 | 17868741 | Yesa | aCGH/PCR | AGAgAACCAc- | No | Paternal | |
| 16936331 | 16957539 | Yesa | aCGH/PCR | T | No | Paternal | |
| 17528139 | 21442150 | Yesa | aCGH/PCR | GAG | No | Paternal | |
| 16916256 | 19710487 | No | PacBio-Honey | TCA | One side LCR (chr17:16905297–16921134) | Paternal | |
| 17179369 | 20415088 | No | PacBio-Honey | CCccAG | One side LCR (Prox_REP) | Paternal | |
| 16008637 | 20245447 | No | PacBio-Honey | ATGA-GcTG | One side LCR (Prox_REP) | Paternal | |
| 13845144 | 19105614 | No | PacBio | CAGT | One side LCR (REPA/B) | Maternal | |
| CNV Coordinates Start | CNV Coordinates End | Previously published jet | How First Discovered | Microhomology/Microhomeology | LCR involvement | Parent of origin | |
| 16196568 | 16237498 | Yesc | aCGH/PCR | Jct1– 31 bp AluY-AluY | No | Paternal, interg | |
| 16596682 – 16700366 | 18322741 – 18672674 | No | ND | - | Distal REP to Middle_REP | Paternal, interg | |
| 18922955–19139789 | 21508001 – 21692001 | No | ND | - | REPA/B and gap containing LCR | ND- | |
| 16654065 | 17326680 | No | PacBio-Honey | TCT | One side LCR (Distal_REP) | ND | |
| 16203013 | 16596682–16757352 | Yesd | PacBio-Honey | Jct1- TT…(74 bp)…ATC | No | Estimated Paternal | |
| 0 | 565389 – 566912 | Yese | aCGH/PCR | Jct- AluSp T-rich repeats:18 bp insertion | No | Maternal, intrag | |
| 18517199–18530187 | 18921809–19140804 | No | ND | - | Middle REP to REPA/B | Estimated Maternal | |
| 16596682–16757352 | 18322741 – 18672674 | No | ND | - | Distal to Middle REP | Estimated Maternal | |
| 15075315 | 15113726 | Yesc | aCGH/PCR | Jct1-ACCTTC | No | Estimated Maternalf | |
| 16596682–16757352 | 18037186 | No | PacBio-Honey | Jct1–29bp, AluSz-AluSx | One side LCR (REPA/B) | Maternal, interg | |
| 18285665–18529905 | 19015148 | No | ND | - | Distal and Middle REPs | ||
| 17568703 | 18320581 | No | PacBio-Honey | CA-CA-CA | Middle_REP | Estimated Maternal | |
| 17421889 | 17833037 | No | PacBio-Honey | A | No | Estimated Maternal | |
| 16688590 | 18555065 | No | PacBio-Honey | AtacATgAT | Distal _REP and REPA/B | Estimated Maternal | |
| 16596682–16757352 | 16881155 | No | PacBio-Honey | Jct1-CAGG insertion and 25 bp, AluY-AluY | No | Estimated Paternal | |
found previously (Liu et al., 2011b)
found previously (Shaw and Lupski, 2005)
found previously (Zhang et al., 2009b)
found previously (Wang et al., 2015a)
found previously (Yuan et al., 2015)
found previously (Potocki et al., 2007) to be paternal; BAB numbers of parents were switched in the 2007 study; therefore, the previous and current data are congruent.
found previously (Sun et al., 2013)
coordinates in hg38, jct in gap region in hg19
All coordinates are in hg19 unless indicated. Abbreviations in the table: DUP, duplication; TRP, triplication; DEL, deletion; ND, not-determined; Jct, junction; aCGH, array comparative genomic hybridization; intra, intrachromosomal; inter, interchromosomal.
BAB1229 contained a breakpoint that occurred within a gap sequence in hg19 which required re-mapping the reads to hg38. PBHoney called the duplication junction, subsequently validated by PCR and Sanger sequencing, that mapped within a simple repeat present within the hg19 gap segment (Data S1). Many of the SV breakpoints described in this study displayed aCGH patterns and breakpoint junctions that appear to describe CGRs; however, apparent complexity may be a product of the structural haplotype that a given SV has arisen upon (Zhang et al., 2009b). For example, BAB641 was found to contain a deletion rearrangement on a putative inversion haplotype that is supported by fosmid mapping data (Kidd et al., 2010), and leads to an inversion junction (Data S1).
Parent of origin of 17p11.2 deletions and duplications
Determination of the parent of origin revealed a striking contrast between non-recurrent deletions and duplications (Table 1, Table S3), that is not observed in recurrent events. SMS cases were derived from the paternal chromosome in >90% of the deletions (11/12), whereas the phased duplication cases (12/14; see STAR methods) were inherited from either parent (5 paternal, 7 maternal, Figure 3 and Table S3). These data are concordant with previous observations of rearrangements at 17p11.2 (Stankiewicz et al., 2003; Sun et al., 2013), and underscore potential mechanistic differences between de novo non-recurrent deletion and duplication CNV formation. Recurrent PTLS associated rearrangements were distributed between paternal (6/9) and maternal (3/9) haplotypes (Table S3). Similarly, recurrent SMS rearrangements occur on either paternal (7/10) or maternal (3/10) haplotypes (Table S3) (Shaw et al., 2002). Previous studies have noted both SV and SNV bias towards mutation on paternal haplotypes (Kloosterman et al., 2015; Stankiewicz et al., 2003). In both non-recurrent and recurrent rearrangements, we did not observe correlation between paternal age and de novo SVs. The average age of fathers of non-recurrent deletion carriers was 29y, control individuals was 31y and all other individuals was 30y; these groups are not statistically significantly different.
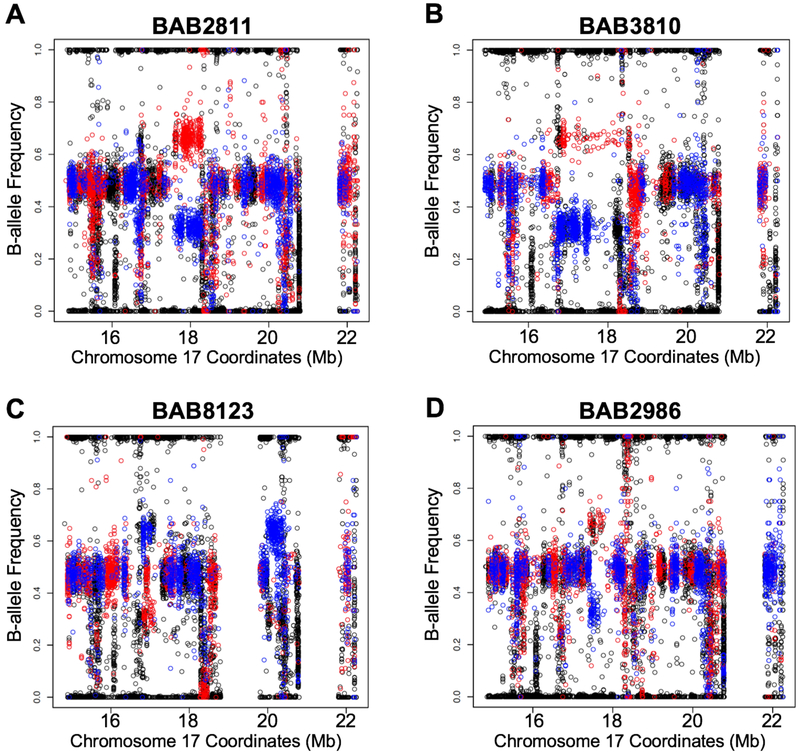
Figure 3-. Regional B-allele frequency and genotype information allows SV phasing.
The phasing data for selected individuals carrying duplications is shown; red dots represent maternal and blue dots paternal informative SNPs (black dots are non-informative). The x-axis represents the coordinates (hg19 genomic position) along the 17p11.2 capture region, and the y-axis is the B-allele frequency. A) BAB2811 carries a duplication on the maternal haplotype; we phased 11 SNVs in cis with this SV, including one outside of the duplicated region. B) BAB3810 carries a duplication on the maternal haplotype; one SNV was in cis with this SV junction. C) BAB8123 carries a duplication on the paternal haplotype; two SNVs were in cis with this SV. D) BAB2986 carries a duplication on the maternal haplotype; two SNVs were in cis with this SV. See Table S3 for SNV phasing data for these probands indicating that SV and SNV occurred de novo in cis with the SV (Table S3).
SNV and indel mutations delineate an extended tract of increased mutation rate
In the ~7 Mb capture sequence interval of 17p11.2, 46 putative de novo SNVs and indels were identified computationally in 17 of the 45 individuals with de novo SV; all were verified experimentally by PCR and Sanger sequencing in trios (Figure 1). These data suggested a high rate of de novo SNV accompanying de novo SV. Genome-wide de novo mutations in the 55 individuals were measured by trio ES, and ranged from zero to five mutations per individual. Given the 29.57 Mb callable capture region targeted by VCRome 2.1 for ES (Bainbridge et al., 2011; Yang et al., 2014), the empirically determined background mutation rates (including both SNVs and indels) were 2.21×10−8 for individuals carrying non-recurrent rearrangements, 2.58×10−8 for individuals carrying recurrent SVs, and 3.21×10−8 for control individuals. These values are similar to the average rate of ~1–3×10−8 SNV mutations per base pair per generation (Shendure and Akey, 2015).
We next compared regional mutation occurrence for the three groups of individuals to each other and to their respective ES rates to examine whether SVs were associated with increased local mutation rates (Table 2). Non-recurrent rearrangement mutation rates differed widely from all others (Table 2, Table S3). Within the callable region (6.074 Mb) of our 7 Mb capture design on 17p11.2, control individuals and individuals with recurrent rearrangements largely lacked de novo SNVs or indels (zero were detected in 15/19 individuals carrying recurrent rearrangements and 8/10 control individuals). The observed mutation rate across all 19 recurrent (4 within 231 Mb regional sequence, or 1.73×10−8, Figure 1C) and 10 control individuals (2 within 121 Mb of regional sequence, or 1.65×10−8) are close to the published rates of ~1–3×10−8.
Table 2-.
Significance of observed mutations
| Data Type | Type of CNV (# of subjects) | Mutation # (TS:TV:indel) | What is being queried | Test | P-value |
|---|---|---|---|---|---|
| ES | Non-Recurrent (26) | 34 (27:3:4) | Total Reg NR (n=26) > CTRL | Poisson | 5.3 × 10−24 |
| Recurrent (19) | 29 (23:5:1) | Total Reg NR (n=26) > R | Poisson | 3.5 × 10−23 | |
| Control (10) | 19 (9:6:4) | Reg NR (n=24) > R | Poisson | 4.7 × 10−9 | |
| Reg NR (n=2) > R | Poisson | 4.0 × 10−25 | |||
| Regional Capture Seq | Non-Recurrent (24) | 23 (6:13:4) | Total Reg NR (n=26) > exome | Poisson | 2.2 × 10−19 |
| Non-Recurrent (2) | 19 (6:13:0) | Reg NR (n=24) > exome | Poisson | 4.8 × 10−7 | |
| Recurrent (19) | 4 (2:0:2) | Reg NR (n=2) > exome | Poisson | 2.5 × 10−25 | |
| Control (10) | 2 (0:1:1) | Reg R (n=19) = exome | Poisson | 0.541 | |
| Reg CTRL (n=10) = exome | Poisson | 0.605 | |||
| TV/TS Reg NR (26) > NR exome | Binomial | 8.0 × 10−18 | |||
| TV/TS Reg NR (24) > NR exome | Binomial | 1.5 × 10−9 | |||
| TV/TS Reg NR (2) > NR exome | Binomial | 1.5 × 10−9 |
Abbreviations: ES = Exome Sequencing; Reg = Regional; NR = Non-Recurrent; R = Recurrent; CTRL = Control; TS = Transition; TV = Transversion; Seq = Sequencing
Conversely, the 26 individuals with de novo non-recurrent rearrangements contained 38 de novo SNVs and 4 indel mutations (present in 13 of the 26 individuals) within the callable capture region (Table 3, Figure 1D). This generates a mutation rate within the locus of 42/316 Mb or 1.33×10−7 which is ~ten-fold higher than recurrent or control individuals. These mutations were distributed unevenly in the 26 individuals, with two outliers (BAB2543 and BAB2811) containing 19 of the 42 mutations. These two outliers present a locus-specific mutation rate of 7.8×10−7, while the 24 remaining individuals have an average locus-specific mutation rate of 7.89×10−8;; a comparison of mutation rates under a Poisson model suggests the two groups are different (Poisson-based P-value of 3.12×10−13; Table 3). Importantly, SNV and indel phasing in 28 out of 42 mutations in 10 individuals with non-recurrent rearrangements found that they are in cis with the breakpoint junctions (Table S3 and Figure S3). For de novo SNV located distant from breakpoint junctions, phasing was often complicated by: i) the TS inherent to the SV mutagenesis mechanism, ii) potential recombination occurring in the genomic interval between the junction and the SNV, and iii) the LCR-rich architecture of proximal 17p. To overcome these challenges, which are particularly relevant to duplications and triplications (Carvalho et al., 2015), we developed phasing methods using both short- and long-read sequencing data (see STAR Methods and Figure 3).
Table 3-.
Regional de novo mutations (red) occurring with de novo CNV of 17p11.2
| NR SMS Patient | Coordinate (hg19) | REF | ALT | Context | Type of Mutation | Distance to jct (bp) |
| BAB649 | 16808831 | TTGTGTCTCTGT GTCTCTAT | T--GTGTCTCTA | TTGTGTCTCTGTGTCTCTAT | indel | 31 |
| BAB765 | 18597960 | A | C | GGAAAAGGGT | TV | 592,252 |
| BAB1354 | 20752404 | G | A | TGAAAGGAAA | TS | 1,076,241 |
| BAB2564 | 18391015 | C | T | TCCACCGTGT | TS | 522,274 |
| BABA1931 | 20028497 | A | C | CAAAAAACAA | TV | 24,895 |
| BAB3133 | 19785197 | G | T | AGATCGAGACa | TV | 74,710 |
| BAB1615 | 15548131 | T | C | TGTTGTTCTT | TS | 1,631,238 |
| BAB6311 | 20245463 | TACAAAAATG | TAC-AAAATG | TACAAAAATG | indel | 16 |
| NR PTLS Patient | Coordinate (hg19) | REF | ALT | Context | Type of Mutation | Distance to jct (bp) |
| BAB2543 | 17095142 | G | C | CACAAGCTTCA | TV | 337,790 |
| BAB2811 | 17791392 | G | C | ACTGAGGGTGG | TV | 222,689 |
| BAB2986 | 17433807 | C | CA | GTCTCCAAAAA | indel | 11,918 |
| BAB3810 | 16688623 | A | C | TTCTAAGTGGA | TV | 33 |
| BAB8123 | 17013700 | A | G | ATAAAATCCAC | TS | 60,084 |
| Recurrent Patient | Coordinate (hg19) | REF | ALT | Context | Type of Mutation | Distance to jct (bp) |
| BAB429 | 16427186 | C | T | TAATCCCAG | TS | 159495 |
| BAB1456 | 16772881 | G | A | TGGAGGTGC | TS | 191633 |
| BAB2555 | 19661617 | CT | C | ACCCCTGGTGA | indel | 780687 |
| BAB2959 | 20554243 | CA | C | CAGGCAAGCC | indel | 27206 |
denotes APOBEC-like signature
Abbreviations: NR = Non-Recurrent; TS = Transition; TV = Transversion; REF = Reference; ALT = Alternate; jct = junction
The density of regional SNVs and indels for both recurrent and control individuals in this study showed no significant difference when compared with their genome-wide exome rates of mutation (Poisson two-tailed P-values of 0.541 and 0.605, respectively). In the absence of the two hypermutation outliers, the 24 individuals with non-recurrent mutations still had a statistically significant increase (Poisson one-tailed P-value of 4.76×10−7) in their point mutation rate at 17p11.2 (7.89×10−8) with respect to their ES rate (32 mutations in the callable exome region; 32/1419 Mb or 2.25×10−8). This comparison is a conservative estimate, given that ES rates are likely higher than regional genomic mutation rates (Shendure and Akey, 2015); however, this controlled for genome-wide mutator phenotypes in the individuals in the study. Importantly, when we calculated the difference between the regional non-recurrent mutation rate and that of the regional recurrent or control individuals, the values are also statistically significant (Table 2), consistent with different mechanisms for generating non-recurrent and recurrent rearrangements. The mutations in the 26 non-recurrent individuals are largely within 1 Mb of the junctions (Table 3); therefore, these are conservative calculations of the local mutation rates and significance. In aggregate, these data indicate that non-recurrent rearrangements are accompanied by significantly elevated mutation rates that are constrained to the locus of the CNV, i.e. genomic region-specific SNV hypermutation.
The de novo regional mutations associated with non-recurrent rearrangements were often (33/42) located further than 20 kb away from breakpoint junctions (Figure 4A and 4B). Only the 9 mutations within ~20 kb of a breakpoint junction would likely be observed using PCR amplification and sequencing of SV breakpoints. Therefore, most of the SNV variation accompanying de novo CNV/SV formation would not be captured by long-range PCR. Interestingly, all four indels were located within these breakpoint-proximal regions (Figure 4, Table 3). Two of these four indel mutations were within polyA tracts consistent with polymerase slippage events, as previously postulated for replicative repair (Carvalho et al. 2013) (Table 1 and Table S3). The other two indels are a deletion of an 8 bp tandem duplication and a 10 bp tandem duplication, also suggesting a slippage mechanism.
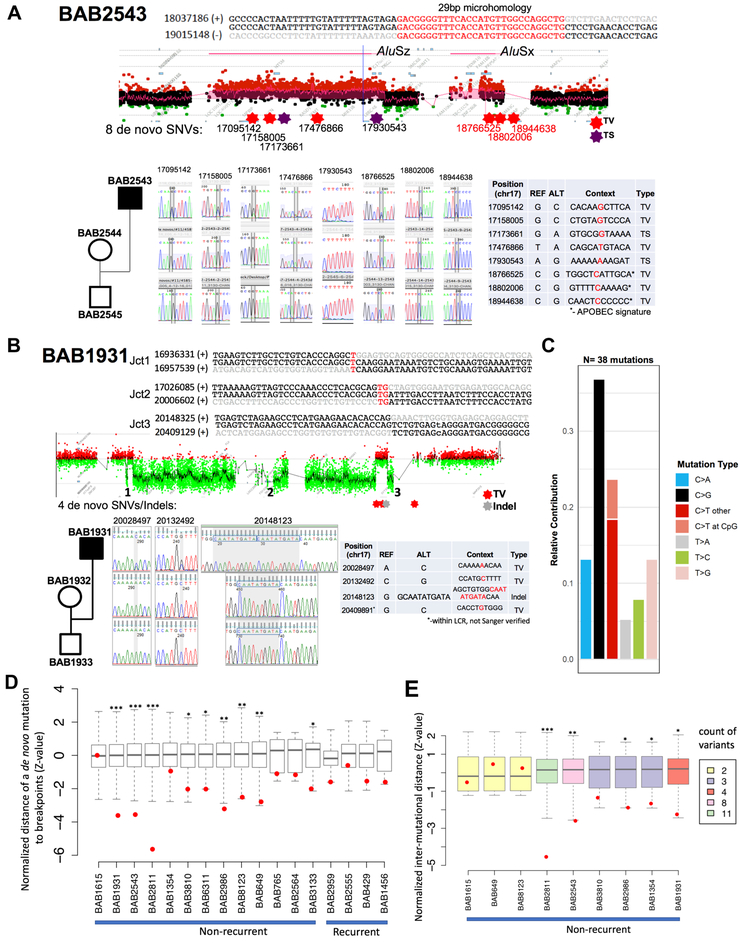
Figure 4-. SNVs and indels accompany SV formation.
A) BAB2543 carries two duplications in an inverted orientation separated with a copy-neutral segment (DUP-NML-DUP/INV). Breakpoint junction (jct) 1 maps to inverted SMSREP LCRs, and evaded sequencing attempts. Breakpoint jct 2 was mediated by inverted Alu repeats and forms an Alu-Alu chimera; junction sequence is characterized by 29 bp of microhomology. Eight de novo mutations have also been characterized within 17p11.2; Sanger sequencing electropherogram confirming each SNV is shown along with the location, genomic context and type. B) BAB1931 carries three deletions interspersed with copy-neutral segments (DEL-NML-DEL-NML-DEL). SV breakpoints display one, two and zero bp of microhomology at the junctions, and jct3 was previously uncharacterized. The four de novo SNVs and indels present in the proband and Sanger sequencing electropherogram confirmation are depicted below. The SNV at 20409881 was not independently confirmed by using a PCR/Sanger sequencing strategy due to its presence within an LCR; however, it was observed in both Illumina and PacBio sequencing data and shown to be de novo in the trio Illumina sequencing data. C) Plot shows the relative contribution of each SNV transition and transversion observed de novo in the non-recurrent individuals. Overall abundance of C>G mutations can be readily observed. D) Enrichment of de novo SNVs in proximity to SV breakpoints was observed in the genomes of 9 out of 13 subjects with non-recurrent SV. This enrichment was not observed for de novo SNVs (N=4) detected in the subjects carrying recurrent SVs. The normalized statistics (Z-value) for each simulation and observation (red dot) is displayed with the box plots. E) Mutational clustering was examined in individuals with more than one de novo SNV. SNV mutations show statically significant clustering in 5 out of 9 NR rearrangements. The normalized statistics (Z-value) for each simulation and the observation (red dot) are plotted. The box plots were colored according to the number of de novo mutations detected in each subject. (*) P ≤ 0.05; (**) P ≤ 0.01; (***) P ≤ 0.001. See also STAR Methods, Figure S2 and Data S1.
Spectrum of hypermutation in regional de novo SNV mutations
In addition to the increased regional rate of SNV mutation, the type of variant alterations we observe differ from the de novo mutational spectrum found in intergenerational human genomes (Campbell and Eichler, 2013; Goldmann et al., 2016). In general, transitions dominate the landscape of de novo SNVs in human genomes, with a noted increase in mutations at CpG to TpG dinucleotides resulting from cytosine deamination (Shendure and Akey, 2015). ES of non-recurrent (27 transitions/30 total SNV mutations; 90%), recurrent (23/28; 82%), and control (9/15; 60%) individuals all display a high proportion of de novo transition mutations. Remarkably, in non-recurrent rearrangements the de novo SNV mutations present within the regional capture exhibit a higher transversion rate than controls as evidenced by an approximate 2/3 ratio of transversions (26/38 total SNV mutations) (Figure 4C). The transversion rate in the 17p11.2 region is significantly greater in individuals with non-recurrent rearrangements at this locus than their mutation rate from ES (binomial one-tailed P-value of 8×10−18 See Table 2 and Figure 4C). Conversely, the rate of transversion mutations observed in the regional data of recurrent and control individuals do not significantly differ from the exomes of these individuals (binomial two-tailed P-values of 1 and 0.4 respectively).
When we examined the 42 de novo SNV and indel mutations present within the callable capture region in individuals harboring non-recurrent rearrangements, we found that more than half of the transversions (14/26) display a C to G or G to C signature; 11/18 of these were present in PTLS genomes (3/8 are in SMS genomes). The expected ratio of this signature would be ¼ or 6 – 7 of the 26 validated transversions. An excess of C to G or G to C transversion mutations has been observed in the context of kataegis (Nik-Zainal et al., 2012; Sakofsky et al., 2014) mediated by deamination in the genome. Furthering this observation, 6 mutations overall (including three sequential mutations in BAB2543) are in the context of an APOBEC signature (TCA, TCC, TCT, or TCG) and five exhibit a C to G transversion signature (Figure 4C, Table 3). The actions of APOBEC proteins have been associated with cytosine deamination in kataegis (Nik-Zainal et al., 2012), however most of the transversions observed in our cohort did not fit the recognition signature for APOBEC proteins.
We also addressed the proximity to breakpoints and the clustering (i.e. the distance between the mutations) of the de novo SNV and indel mutations in the callable capture and sequencing region. We found that mutations were significantly enriched near breakpoints in 9/13 of the non-recurrent SV cases, but none (n=4) of the recurrent cases that contained de novo SNVs and indels in the 17p11.2 capture region (Figure 4D and Figure S2). Furthermore, we also found significant clustering of mutations in 5 of the 9 non-recurrent SVs with 2 or more SNV (Figure 4E).
Finally, the majority of regional mutations in the non-recurrent cohort are contained within RefSeq genes (34/42 Supplemental Table S3), with 30/34 occurring within introns, and three within 3’UTRs. This finding is intriguing, as only 53.3% of the callable capture region is spanned by RefSeq genes, yet 81% of events occur within the genic regions within the callable capture region (binomial one-tailed P-value of 0.00018). One of these 34 genic events resulted in a de novo regional missense mutation (g. 18391015LGALS9C: NM_001040078.2:exon4:c.C388T:p.R130C) in SMS patient BAB2564. This nucleotide change was observed in both exome and capture sequencing data, was confirmed by Sanger sequencing like all de novo SNVs identified from our trio analyses, and is predicted by conceptual translation to lead to an arginine to cysteine amino acid substitution in LGALS9C.
Discussion
We have used orthogonal DNA sequencing technologies to study rearrangement structures and resultant mutagenesis at 17p11.2. These data, in conjunction with the presence of microhomology or microhomeology at 23/30 of the discerned non-recurrent rearrangement junctions (Table 1 and Table S4), suggests that base-pairing facilitates primer annealing enabling template switching during replicative repair (Lee et al., 2007; Liu et al., 2017; Slack et al., 2006). Long read sequence data were critical when investigating breakpoints with one or both ends within LCRs (Wang et al., 2015a), and improved our calling of junctions by more than two-fold. These analyses allowed us both to infer mechanisms of breakpoint formation and potential genomic structures generated in CGR formation.
Intriguingly, the four indel mutations observed in non-recurrent SVs mapped <12 Kb from breakpoint junctions, and tended to occur within homopolymeric tracts; this likely reflects replication slippage events (Carvalho et al., 2013). The mutations proximal to the breakpoint junctions of non-recurrent rearrangements could be due to a reduced processivity polymerase that is more prone to detaching from the template and that switches to a more processive polymerases. Indeed, more frequent template switches are observed within the first 10 kb of BIR (Carvalho and Lupski, 2016; Carvalho et al., 2013; Sakofsky et al., 2015; Smith et al., 2007). Mutations near breakpoints differ from the SNV mutational spectrum distal to breakpoint junctions, where transversion point mutations dominate. We found that the SNV tracts associated with replication-based repair are in cis with the SV (Figure 3 and Table S3), and may extend for hundreds of kilobase pairs after a template switch. The 17p11.2 genomic region has a replication length of ~1 Mb as suggested by the strandedness of SNVs with exonuclease mutations in Pol ε (Shinbrot et al., 2014); this is consistent with the length of the mutation tracts in these individuals.
Here, we suggest that we are encountering a new signature of MMBIR mutagenesis. More processive synthesis of BIR likely begins further from the strand invasion site (Lydeard et al., 2007; Lydeard et al., 2010; Smith et al., 2007). BIR and MMBIR-associated mutagenesis tested with mutational reporters has been attributed to the migrating bubble that is formed by homologous recombination (Saini et al., 2013; Sakofsky et al., 2015; Wilson et al., 2013). In this context, polymerase errors made on the leading strand may not be repaired effectively, because they would occur on conservatively segregating DNA, which would not allow mismatch correction to be targeted to the correct strand for repair (Kuzminov, 1995). Alternatively, the bubble could migrate prior to lagging-strand synthesis (Saini et al., 2013; Wilson et al., 2013), and then the nascent leading strand becomes the template, and errors will not be corrected (Malkova and Ira, 2013). The observations of SV mutagenesis accompanying non-recurrent deletions or duplications of the human genome presented here in a non-selected experimental manner describe a novel SNV mutational mechanism and mutational signature.
During BIR and MMBIR, an extensive length of the genome may experience single-strandedness because the displacement loop (D-loop) can proceed for long distances. Single-stranded DNA is associated with the D-loop because lagging-strand synthesis at this structure is delayed (Malkova and Ira, 2013). BIR and potentially MMBIR generate regions of conservative segregation of old and new DNA strands (Saini et al., 2013; Smith et al., 2007; Wilson et al., 2013) as opposed to the semi-conservative mode of segregation seen in conventional DNA replication (Meselson and Stahl, 1958). The ratio of transversions to transition mutations (26:12) and the predominance of C>G and C>T mutations (22/42) within the 26 SMS and PTLS patients with non-recurrent rearrangements is reminiscent of kataegis, or clustered mutational showers that can stretch to greater than 100 Kb from rearrangement breakpoint junctions and consist of more than two such mutations (Chan and Gordenin, 2015; Nik-Zainal et al., 2012; Sakofsky et al., 2014) (see Figure 4). These processes could explain 6 of the 22 C>G or C>T mutations (Table 3), suggesting a potential role for spontaneous cytosine deamination in the genome during MMBIR or BIR.
Of the 26 individuals harboring non-recurrent rearrangements in this study, 13 lacked detectable de novo mutations that occurred concomitant with SV formation. These may represent random variation, mutations outside the callable regional data, or variation in repair mechanism. During replication, an oncoming fork or cleavage of a re-initiated fork by a structure-specific endonuclease (Mayle et al., 2015; Roseaulin et al., 2008) could lead to a restricted length or even a lack of mutagenic synthesis, and could occur after the initial template switch to limit SNV mutation. Microhomology-mediated end-joining (MMEJ) might be considered as causative for cases lacking detectable mutations in the 7 Mb capture region. MMEJ can readily explain deletions, but involves limited DNA synthesis and therefore is difficult to associate with the formation of duplications and triplications. Eight of the 13 cases lacking SNV mutations have triplicated (BAB2965, BAB2992, BAB2695 and BAB8325) or duplicated (BAB3344, BAB3793, BAB6917, and BAB1229) regions. MMEJ can lead to chromosomal rearrangements with microhomology, deletions, and templated insertions at the junctions (Sfeir and Symington, 2015). Although the spectrum of junction sequences is similar between MMEJ and MMBIR, SNV is only reported up to 14 kb from microhomology in yeast MMEJ events (Sinha et al., 2017). Therefore, we propose that the events lacking SNV and indel mutations may also occur by MMBIR or BIR, but that a converging replication fork or resolution of the recombination structure could limit the mutation tract in these cases (Correa et al., 2018; Mayle et al., 2015). The absence of long-distance SNV associated with recurrent events suggests that most do not occur by BIR, but rather by homologous reciprocal exchange (crossing over) (Shaw et al., 2002).
We observed an order of magnitude difference in the increase in mutation rate among the non-recurrent SV cohort. There are potentially different subsets of SNV formation driven by non-recurrent rearrangement, ranging from ~5 to 50-fold higher rates of mutation (Table 2). The correlates of these differing rates – whether genomic context related or otherwise – remain to be pursued. The sequence changes accompanying SVs extend the clinical genomics implications accompanying non-recurrent CNV formation because SNV could potentially disrupt function of genes mapping 1 Mb from an SV junction. These mutational processes could also be important in somatic mutagenesis in cancer. Most of the 42 de novo SNVs and indels were present within genes (33/42 or ~80%), and therefore could potentially effect splicing or coding regions within a megabase of the breakpoint junctions; in fact, one of the observed mutations results in a missense amino acid substitution in LGALS9C. Finding that a preponderance of SNVs associated with SV are within genes suggests that their occurrence could be related to transcription, and perhaps to transcription/BIR collision.
Future studies of the mechanisms of SV formation would benefit from using technology that encompasses the advantages of the combination of methods described here. Moreover, our study indicates that current genome-wide assays being implemented for research studies of genomic disorders, Mendelian disease, and cancer, and applied translationally in clinical genomics assays are missing genetic variation that could have potential pathogenic consequences. Further development of human genome sequencing technologies that can capture the full range of variant types is warranted.
In conclusion, we provide evidence suggesting mutation associated with non-recurrent SV formation at 17p11.2 results in a 5–50 fold higher SNV mutation rate than that observed with recurrent SV, consistent with error-prone MMBIR and contrasting with regular replication through the region. Non-recurrent rearrangements occur throughout the genome and several have been shown to have concomitant de novo SNV in proximity to breakpoint junctions (Abyzov et al., 2015; Beck et al., 2015; Brandler et al., 2016; Carvalho et al., 2013; Yuen et al., 2016). For such events, we anticipate instances wherein a similar mutational spectrum, clustering of SNV, and extension over megabase distances may be found as observed here. Within a few kilobase-pairs of a junction, we see indels attributable to polymerase slippage, as described before (Carvalho et al., 2013). Within about one megabase-pair of the junction we find loose clusters of kataegis-like mutations possibly due to deamination of single-stranded DNA (Sakofsky et al., 2014), and we see widely distributed base substitution mutations of all types that might occur because of the unusual mechanism of BIR and MMBIR as described above. These data would be difficult to explain with any currently available model other than MMBIR. Overall, the data confirm a major prediction of the MMBIR model, that SVs formed by MMBIR will show SNV mutation over replicore-sized genomic distances. The conclusion that CNVs arise by MMBIR applies to half and possibly all non-recurrent events at 17p11.2. Finally, these mutational processes have potential phenotypic consequences for patients with genomic disorders, due to gene mutations mapping large distances from SV breakpoint junctions.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Requests for further information on analyses, DNA samples, and requests for raw sequencing data may be directed to, and will be fulfilled by Lead Contact James R. Lupski (jlupski@bcm.edu). Regional PacBio and Illumina sequencing as well as Exome Sequencing data for the individuals in this study are not deposited as the Institutional Review Board at Baylor College of Medicine has restricted the deposition of these data.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Study subjects provided informed consent for molecular and genomic analyses under the Baylor College of Medicine Institutional Review Board-approved protocols H-9170 or H-29697. The individuals in the study were selected from an overall cohort of 266 individuals (134 and 132 clinically identified Potocki Lupski Syndrome (PTLS) and Smith Magenis Syndrome (SMS) patients, respectively) who presented with PTLS due to 17p11.2 duplication, SMS due to deletion of this region, those who have pathogenic point mutations in RAI1 leading to SMS (Slager et al., 2003), or individuals with overlapping clinical phenotypes (Loviglio et al., 2016). The individuals in this study are generally diagnosed and are enrolled in research participation as pediatric cases, and were acquired and interrogated agnostic of sex. The study consists of 25 female probands (11 with non-recurrent rearrangements, 8 with recurrent rearrangements and 6 control individuals) and 30 male probands (15 with non-recurrent rearrangements, 11 with recurrent rearrangements, and 4 control individuals); SMS and PTLS have similar penetrance regardless of sex. The parents of the probands consist of both mothers and fathers and are therefore equally distributed by sex. Results for exome sequencing in nine of the control individuals were detailed previously (Loviglio et al., 2016; White et al., 2018).
METHOD DETAILS
Array comparative genomic hybridization to determine extent of genomic rearrangements
Arrays were designed using the Agilent eArray website (https://earray.chem.agilent.com/earray/); we used a custom tiling-path 4×180K oligonucleotide format (AMADID# 032121), a 4×44K format (AMADID# 017821), and a 8×60K format (AMADID# 043020) to investigate copy number alterations at 17p11.2 (Yuan et al., 2015). These aCGH platforms were used to interrogate patients and their parents at a resolution of one probe every ~200–700 bp. Arrays were performed using genomic DNAs from each trio of individuals paired with sex-matched control DNAs from Coriell, NA15510 and NA10851. These analyses both identified the extent of the rearranged segments of DNA, allowing for selection for our study based upon the location of breakpoint junctions, and also determined that the rearrangements were de novo in the proband. To perform the targeted array interrogation, 1.2 μg of high quality patient or parental and sex-matched control DNA was fragmented using digestion with AluI (5U) and RsaI (5U) (Promega) at 37°C for 2 hours. Digestion was validated with agar ose gel electrophoresis. Fragmented DNAs were then labeled with Cy5 (interrogated DNA) and Cy3 (control DNA) using the BioPrime Array CGH Genomic Labeling Module (Life Technologies) and Cyanine Smart Pack dCTP (PerkinElmer NEL620001KT) according to the manufacturer’s instructions (Agilent Technologies Oligonucleotide Array-Based CGH for Genomic DNA Analysis, Version 7.2). Labeling was conducted at 37°C for 2 hours, and was followed by purification using a 30 kDa cut off filter (Amicon Ultra 0.5 mL from Millipore). DNA quantities and labeling efficiencies were then determined using a NanoDrop ND-1000 spectrophotometer. Fluorescent dye labeled patient or parent and sex-matched reference DNAs were combined with 5 μg of human Cot-1 DNA (Life Technologies) to block repetitive sequences. The resultant DNA mixture was then boiled for 5 minutes at 95°C with Agilent 10X blocking agent and 2X Agilent Hi-RPM hybridization buffer, and then incubated for 30 minutes at 37°C. This mixture was then applied to the relevant array format, placed in a hybridization chamber, and incubated while rotating for 40 hours at 65°C in an Agilent hybridization oven (Agilent G2545). After the hybridization, array slides were then washed according to manufacturer’s instructions with Agilent OligoCGH Wash buffers 1 and 2, as well as acetonitrile (Sigma). Slides with comparatively hybridized DNA samples were then scanned using an Agilent SureScan Microarray Scanner (Agilent model G2565CA), and image extraction was performed using the accompanying feature extraction software (Version 11.5, Agilent) to produce array files for analysis. Copy number alterations and approximate breakpoint junction locations were visualized using Agilent Genomic Workbench, and all files were hand curated for the extent of the rearrangement imaged as indicated by significant deviations in the normalized log2 ratio of query/control fluorescence in a 5 kb sliding window. The results from the aCGH experiments helped to guide analysis of the PacBio and short-read Illumina sequencing data to identify breakpoint junctions. Breakpoint junction analysis for some subjects, resolved by targeted long-range PCR and Sanger sequencing guided by aCGH, has been reported previously (Liu et al., 2011b; Shaw and Lupski, 2005; Yuan et al., 2015).
Genome-wide and targeted sequencing methodologies
Capture was performed using a previously designed NimbleGen SeqCap EZ library targeted to Chromosome 17:14,890,000–21,890,000 in hg19 (Wang et al., 2015a). This capture reagent, when coupled with PCR amplification of 6 kb libraries from an individual and a single molecule real time (SMRT) sequencing approach yields reads that are 85% on-target for the capture locus. Sequencing was performed using the PacBio RSII machine. The capture reagent and regionally captured genomic sequence from each subject was also utilized for Illumina HiSeq short-read sequencing.
For the regional 7 Mb capture as well as for the ES capture design, the overall callable bases were determined using ExCID (https://github.com/cbuhay/ExCID) (Sanghvi et al., 2018). These values were determined by intersection of all batched data, and elimination of captured regions with less than 20x coverage with mapping quality ≥1 in 10% of the samples. This analysis was first completed for non-recurrent, recurrent, and control individuals separately. We then merged the non-callable regions from these three groups of individuals, and removed them from the targeted capture and exome capture regions. This analysis led to a callable regional capture (6.074 Mb; see Table S2) and a callable ES (29.57 Mb) region per individual that were then used for mutation rate calculations and statistical analyses.
To identify de novo SNV and indel changes in the genomes of the 55 trios, we used Burrows-Wheeler Alignment (BWA) to map reads to GRCh37. We called variants with both Atlas2 (Challis et al., 2012; Reid et al., 2014) and GATK (McKenna et al., 2010) in parallel, and then computationally derived potential de novo variants present in the proband using the in-house developed de novo mutation-finder (DNM-Finder; https://github.com/BCMLupskilab/DNM-Finder) (Eldomery et al., 2017). For variants in either the ES data or in the regional HiSeq short-read data, we employed both variant calling algorithms (Table S3) to identify de novo mutations in silico and validated by orthogonal experiments using Sanger sequencing of trios (Figure 2). CNV identification was also validated using ES data and using the XHMM algorithm (Fromer et al., 2012). XHMM confirmed the size and genomic region of each CNV in the study.
Phasing structural variants
Structural variants in individuals with Smith Magenis Syndrome (SMS) and Potocki Lupski Syndrome (PTLS) were phased using informative genotype data provided by GATK trio joint-calling of the deleted and duplicated segments within the regional 7 Mb capture. For SMS individuals, the remaining allele called by GATK represented the non-deleted segment, and therefore the deleted haplotype came from the alternate parental genome. For some individuals, phasing was previously determined using microsatellite and restriction fragment length polymorphism data in conjunction with family analyses (Shaw et al., 2002; Sun et al., 2013), which also provided more detailed information such as intrachromosomal and interchromosomal origin of duplications. For the remaining individuals with duplications, we used GATK trio joint-calling to phase informative SNVs within the 7 Mb (Figure 3, Table S3). The duplications were then phased using variant/reference expected ratios (B-allele frequencies) within the SVs for intrachromosomal SVs. For instance, a copy number neutral region will present with three B-allele frequencies: for homozygous calls, 0 or 1, and heterozygous calls 0.5. Duplications present with B-allele frequencies consistent with homozygous calls (0 and 1) and heterozygous calls (0.33 and 0.66, no 0.5). Intrachromosomal duplications will show a skewed B-allele frequency pattern of 0.66 originating from the parent of origin of the duplication and will lack frequencies of 0.33 which represent the non-duplicated allele (Figure 3). For the purpose of phasing SVs, we analyzed SNPs within the duplication with allelic ratios larger than 0.65 and summarized the number of maternal vs. paternal alleles based on GATK for each proband (Table 1 and Table S3 estimated parent of origin values). Interchromosomal duplications are challenging to phase using our GATK genotyping and B-allele frequency approach due to the great number of non-informative SNVs; however, we have estimated interchromosomal duplications (Table 1 and Table S3) and have found them to be consistent with previous data.
Phasing single nucleotide variants and indels
De novo SNVs and indels were phased using one (indels) or two independent approaches. For variants near the breakpoint junctions, within the overlapping range of a single PacBio read containing the junction (3/4 indels and two SNVs), we could promptly phase them to the SVs and confirm these data in the junction PCR and Sanger sequencing data. For majority of the de novo SNVs, which generally map >10 Kb away of the breakpoint junctions, we used information from inherited nearby SNVs either by PCR amplification and sequencing of the inherited and de novo SNV, or by investigating the presence of these inherited SNVs in the Illumina short read-data and in the PacBio long-reads. In addition, we also phased 22 de novo SNVs (Estimated Maternal or Paternal, Table S3, column P) as follows. The raw PacBio reads were mapped using NGMLR (Sedlazeck et al., 2018) to hg19. Subsequently, the Illumina based SNPs called by Atlas2 were phased using HapCUT2 (Edge et al., 2017) based on the mapped PacBio reads per proband. HapCUT2 assigns phase blocks (i.e. regions where SNPs could be phased together), but is not able to indicate haplotype combinations between blocks. Phased SNPs were then compared to the parental informative SNPs based on xAtlas (Farek et al., 2018) and GATK. This step was done with the SURVIVOR module parent_phasing (Jeffares et al., 2017). For each SNV reported by HapCUT2 we annotate the allele ratio and quality information in the proband based on the Atlas2 information. Furthermore, we annotate each SNV if it is an informative parental SNP from the mother or father. This file was generated for each proband. In summary, we looked at the HapCUT2 phase block assignment of the de novo SNV and summarized the overlap of the assigned phase block with the parental informative SNV using GATK.
Breakpoint PCR and confirmation of single nucleotide variants
All putative de novo variants called by Atlas2 and GATK identified by DNM-Finder (Eldomery et al., 2017) were examined in the Integrative Genomics Viewer (IGV), and were determined to be present in the PacBio data of the proband. If the variant was computationally determined to be de novo, forward and reverse primers were designed for SNVs. CNV breakpoint junctions were similarly verified using coordinates discerned from aCGH data, and PBhoney (English et al., 2014). Primer pairs for deletions and duplications were designed with respect to the hg19 reference, and were checked for uniqueness using the UCSC genome browser. PCR for breakpoint junctions was performed using TaKaRa LA Taq (Clontech). PCR for Sanger confirmation of SNVs was performed using HotStarTaq (Qiagen). All breakpoint PCRs utilized parental samples for negative controls, and SNV validations were performed for the trio of individuals in the study. In conjunction with breakpoint junction determination, PacBio data were used to phase local SNV data using IGV and long distance PCR. When the haplotype on which the SV was derived was identified, local phasing could be conducted with informative SNVs in cis with the de novo mutations present in the patient.
Statistical analyses for mutation rate estimates
In order to determine the mutation rate per base pair in each group of patients, we first calculated the diploid callable genomic regional size for both exome and targeted regions. These individual callable regions (59.14 Mb for diploid ES and 12.148 Mb for diploid regional data) were then used to determine the rate by dividing the total number of de novo mutations (SNV and indels) in each group of patients by the callable diploid capture region multiplied by the number of individuals in the group.
For the Poisson test p-value calculations, we compared the average mutation rate per base pair between two groups of individuals. First, the expected number of mutations (expected) was calculated as the number of mutations per base pair (second group) multiplied by the total size of diploid genome callable region size in the second group. Then the deviance between the total number of mutations observed in the first group (observed) and the expected number of mutations (expected) was calculated by this formula: (observed-expected)/(expected1/2).
Statistical analyses for clustering of mutations and proximity to breakpoints
To test whether the proximity to the nearest junction (Figure 4D) and the clustering present in individuals with more than one SNV or indel mutation accompanying SV formation (Figure 4E) were significant, we generated non-overlapping 1 Kb-sized bins across the callable 7 Mb capture/sequencing region. Each window is weighted by both sequencing coverage and the mutation rate, which was estimated by the density of high-quality SNVs in that window in 103 unaffected parents and control patients (N=10) without any CNV. For those windows with no SNVs detected in control patients, we set the mutation rate as 10−5.
10,000 simulations for each subject were generated by performing weighted random sampling for the same number of de novo mutations using 1 Kb-sized moving average windows. We then computed three statistics: (1) the mean value of the distances between each mutation to the closest SV breakpoint, (2) the mean value of the inter-mutational distances for individuals with more than one de novo mutation detected, (3) the Mahalanobis distance between each mutation and the bi-variate distribution of the 10,000 simulations for both types of distances discussed above.
For the first two statistics, we first normalized the distance values among 10,000 simulations by calculating Z-values for the measurements from each subject and tested the significance using the function pnorm(x) in R, where x is the calculated Z-value for the observation. For the bi-variant analysis, we tested the significance of the mutation’s being an outlier compared to the distribution of 10,000 simulations using a chi-square test.
DATA AND SOFTWARE AVAILABILITY
Microarray data generated in this study are available through GEO under the accession number GSE125120.
Supplementary Material
Data S1- Junctions and array comparative genomic hybridization data for each individual in the study; related to Table 1 and Figure 4
Figure S1. Breakpoints and relative positions of SNVs and Indels; related to Figure 1. The precisely mapped breakpoints and de novo SNVs and indels that accompany the genomic rearrangements are depicted. Breakpoints are depicted with blue vertical lines, and de novo mutations with red stars. For recurrent SVs, the breakpoint is drawn at the mid-point of the LCR involved in the rearrangement.
Figure S2. Distribution of regional de novo mutations within the capture region; related to Figure 4. A) The proximity of each de novo SNV and indel with respect to the nearest junction is plotted in the histogram. B) The analysis shows that 5/9 of the regional de novo patient mutations are significantly enriched near the breakpoints of junctions and separated from the distribution of 10,000 simulations. The figures plot each subject (red dot) vs. the 10,000 simulations (grey dots) in the bi-dimensional space of the Mahalanobis distance between de novo mutations and SV breakpoints (x-axis) and inter-mutational distance (y-axis). The p-value from a chi-square test is shown on top of each panel.
Table S2- Callable regions within the regional Illumina sequencing data; related to STAR Methods - Sequencing methodologies
Table S3- Junction and point mutation data for each individual in the study; related to Table3 and Figure 3
Orthogonal DNA sequencing approaches are required to observe all variant types
De novo SNVs and indels accompany non-recurrent Structural Variants (SVs) in cis
SV-associated SNVs are primarily within genes and occur over Mb sized distances
MMBIR involves extensive DNA replication resulting in regional hypermutation
Newly occurring structural variants with human genomes spawn extensive, local single nucleotide changes leading to an enhanced mutational burden within proximal genes.
Acknowledgements
We thank the patients and family members who participated in this study. This work was supported in part by National Institute of General Medical Sciences grant R01GM106373 to P.J.H. and J.R.L., R01GM80600 to G.I., and R00GM120453 to C.R.B., grants from the National Institute of Neurological Disorders and Stroke to J.R.L. (R01NS058529 and R35NS105078), a National Human Genome Research grant (U54 HG003273) to R.A.G., and a joint National Human Genome Research Institute/National Heart Blood and Lung Institute (https://www.nhlbi.nih.gov/) grant (UM1HG006542) to the Baylor Hopkins Center for Mendelian Genomics. C.R.B. was previously supported by both the Brain Disorders and Development training grant at BCM (Grant number 2T32NS043124–11), and an HHMI fellowship from the Damon Runyon Cancer Research Foundation (DRG 2155–13).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
Baylor College of Medicine (BCM) and Miraca Holdings have formed a joint venture with shared ownership and governance of the Baylor Genetics (BG), which performs clinical microarray analysis and clinical exome sequencing. C.A.S. is an employee of BCM and derives support through a professional services agreement with the BG. J.R.L. serves on the Scientific Advisory Board of the BG. J.R.L. has stock ownership in 23andMe, is a paid consultant for Regeneron Pharmaceuticals, has stock options in Lasergen, and is a co-inventor on multiple United States and European patents related to molecular diagnostics for inherited neuropathies, eye diseases, and bacterial genomic fingerprinting.
References
- Abyzov A, Li S, Kim DR, Mohiyuddin M, Stutz AM, Parrish NF, Mu XJ, Clark W, Chen K, Hurles M, et al. (2015). Analysis of deletion breakpoints from 1,092 humans reveals details of mutation mechanisms. Nat Commun 6, 7256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bainbridge MN, Wang M, Wu Y, Newsham I, Muzny DM, Jefferies JL, Albert TJ, Burgess DL, and Gibbs RA (2011). Targeted enrichment beyond the consensus coding DNA sequence exome reveals exons with higher variant densities. Genome Biol 12, R68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck CR, Carvalho CM, Banser L, Gambin T, Stubbolo D, Yuan B, Sperle K, McCahan SM, Henneke M, Seeman P, et al. (2015). Complex genomic rearrangements at the PLP1 locus include triplication and quadruplication. PLoS Genet 11, e1005050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandler WM, Antaki D, Gujral M, Noor A, Rosanio G, Chapman TR, Barrera DJ, Lin GN, Malhotra D, Watts AC, et al. (2016). Frequency and Complexity of De Novo Structural Mutation in Autism. Am J Hum Genet 98, 667–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell CD, and Eichler EE (2013). Properties and rates of germline mutations in humans. Trends Genet 29, 575–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho CM, and Lupski JR (2016). Mechanisms underlying structural variant formation in genomic disorders. Nat Rev Genet 17, 224–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho CM, Pehlivan D, Ramocki MB, Fang P, Alleva B, Franco LM, Belmont JW, Hastings PJ, and Lupski JR (2013). Replicative mechanisms for CNV formation are error prone. Nat Genet 45, 1319–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho CM, Pfundt R, King DA, Lindsay SJ, Zuccherato LW, Macville MV, Liu P, Johnson D, Stankiewicz P, Brown CW, et al. (2015). Absence of heterozygosity due to template switching during replicative rearrangements. Am J Hum Genet 96, 555–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho CM, Ramocki MB, Pehlivan D, Franco LM, Gonzaga-Jauregui C, Fang P, McCall A, Pivnick EK, Hines-Dowell S, Seaver LH, et al. (2011). Inverted genomic segments and complex triplication rearrangements are mediated by inverted repeats in the human genome. Nat Genet 43, 1074–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challis D, Yu J, Evani US, Jackson AR, Paithankar S, Coarfa C, Milosavljevic A, Gibbs RA, and Yu F (2012). An integrative variant analysis suite for whole exome next-generation sequencing data. BMC bioinformatics 13, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K, and Gordenin DA (2015). Clusters of Multiple Mutations: Incidence and Molecular Mechanisms. Annual review of genetics 49, 243–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins RL, Brand H, Redin CE, Hanscom C, Antolik C, Stone MR, Glessner JT, Mason T, Pregno G, Dorrani N, et al. (2017). Defining the diverse spectrum of inversions, complex structural variation, and chromothripsis in the morbid human genome. Genome Biol 18, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad DF, Bird C, Blackburne B, Lindsay S, Mamanova L, Lee C, Turner DJ, and Hurles ME (2010). Mutation spectrum revealed by breakpoint sequencing of human germline CNVs. Nat Genet 42, 385–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa R, Thornton PC, Rosenberg SM, and Hastings PJ (2018). Oxygen and RNA in stress-induced mutation. Curr Genet 64, 769–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantino L, Sotiriou SK, Rantala JK, Magin S, Mladenov E, Helleday T, Haber JE, Iliakis G, Kallioniemi OP, and Halazonetis TD (2014). Break-induced replication repair of damaged forks induces genomic duplications in human cells. Science 343, 88–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deem A, Keszthelyi A, Blackgrove T, Vayl A, Coffey B, Mathur R, Chabes A, and Malkova A (2011). Break-induced replication is highly inaccurate. PLoS Biol 9, e1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhokarh D, and Abyzov A (2016). Elevated variant density around SV breakpoints in germline lineage lends support to error-prone replication hypothesis. Genome Res 26, 874–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edge P, Bafna V, and Bansal V (2017). HapCUT2: robust and accurate haplotype assembly for diverse sequencing technologies. Genome Res 27, 801–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldomery MK, Coban-Akdemir Z, Harel T, Rosenfeld JA, Gambin T, Stray-Pedersen A, Kury S, Mercier S, Lessel D, Denecke J, et al. (2017). Lessons learned from additional research analyses of unsolved clinical exome cases. Genome Med 9, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English AC, Salerno WJ, Hampton OA, Gonzaga-Jauregui C, Ambreth S, Ritter DI, Beck CR, Davis CF, Dahdouli M, Ma S, et al. (2015). Assessing structural variation in a personal genome-towards a human reference diploid genome. BMC genomics 16, 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English AC, Salerno WJ, and Reid JG (2014). PBHoney: identifying genomic variants via long-read discordance and interrupted mapping. BMC bioinformatics 15, 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farek J, Hughes D, Mansfield A, Krasheninina O, Nasser W, Sedlazeck FJ, Khan Z, Venner E, Metcalf G, Boerwinkle E, et al. (2018). xAtlas: Scalable small variant calling across heterogeneous next-generation sequencing experiments. BioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromer M, Moran JL, Chambert K, Banks E, Bergen SE, Ruderfer DM, Handsaker RE, McCarroll SA, O’Donovan MC, Owen MJ, et al. (2012). Discovery and statistical genotyping of copy-number variation from whole-exome sequencing depth. Am J Hum Genet 91, 597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmann JM, Wong WS, Pinelli M, Farrah T, Bodian D, Stittrich AB, Glusman G, Vissers LE, Hoischen A, Roach JC, et al. (2016). Parent-of-origin-specific signatures of de novo mutations. Nat Genet 48, 935–939. [DOI] [PubMed] [Google Scholar]
- Hastings PJ, Ira G, and Lupski JR (2009). A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet 5, e1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffares DC, Jolly C, Hoti M, Speed D, Shaw L, Rallis C, Balloux F, Dessimoz C, Bahler J, and Sedlazeck FJ (2017). Transient structural variations have strong effects on quantitative traits and reproductive isolation in fission yeast. Nat Commun 8, 14061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd JM, Graves T, Newman TL, Fulton R, Hayden HS, Malig M, Kallicki J, Kaul R, Wilson RK, and Eichler EE (2010). A human genome structural variation sequencing resource reveals insights into mutational mechanisms. Cell 143, 837–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman WP, Francioli LC, Hormozdiari F, Marschall T, Hehir-Kwa JY, Abdellaoui A, Lameijer EW, Moed MH, Koval V, Renkens I, et al. (2015). Characteristics of de novo structural changes in the human genome. Genome Res 25, 792–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman WP, Guryev V, van Roosmalen M, Duran KJ, de Bruijn E, Bakker SC, Letteboer T, van Nesselrooij B, Hochstenbach R, Poot M, et al. (2011). Chromothripsis as a mechanism driving complex de novo structural rearrangements in the germline. Hum Mol Genet 20, 1916–1924. [DOI] [PubMed] [Google Scholar]
- Kuzminov A (1995). Collapse and repair of replication forks in Escherichia coli. Mol Microbiol 16, 373–384. [DOI] [PubMed] [Google Scholar]
- Lee JA, Carvalho CM, and Lupski JR (2007). A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell 131, 1235–1247. [DOI] [PubMed] [Google Scholar]
- Lieber MR (2010). The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem 79, 181–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Erez A, Nagamani SC, Dhar SU, Kolodziejska KE, Dharmadhikari AV, Cooper ML, Wiszniewska J, Zhang F, Withers MA, et al. (2011a). Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell 146, 889–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Lacaria M, Zhang F, Withers M, Hastings PJ, and Lupski JR (2011b). Frequency of nonallelic homologous recombination is correlated with length of homology: evidence that ectopic synapsis precedes ectopic crossing-over. Am J Hum Genet 89, 580–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Yuan B, Carvalho CM, Wuster A, Walter K, Zhang L, Gambin T, Chong Z, Campbell IM, Coban Akdemir Z, et al. (2017). An Organismal CNV Mutator Phenotype Restricted to Early Human Development. Cell 168, 830–842 e837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loviglio MN, Beck CR, White JJ, Leleu M, Harel T, Guex N, Niknejad A, Bi W, Chen ES, Crespo I, et al. (2016). Identification of a RAI1-associated disease network through integration of exome sequencing, transcriptomics, and 3D genomics. Genome Med 8, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski JR (1998). Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet 14, 417–422. [DOI] [PubMed] [Google Scholar]
- Lupski JR (2015). Structural variation mutagenesis of the human genome: Impact on disease and evolution. Environmental and molecular mutagenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydeard JR, Jain S, Yamaguchi M, and Haber JE (2007). Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature 448, 820–823. [DOI] [PubMed] [Google Scholar]
- Lydeard JR, Lipkin-Moore Z, Sheu YJ, Stillman B, Burgers PM, and Haber JE (2010). Break-induced replication requires all essential DNA replication factors except those specific for pre-RC assembly. Genes & development 24, 1133–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkova A, and Ira G (2013). Break-induced replication: functions and molecular mechanism. Curr Opin Genet Dev 23, 271–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayle R, Campbell IM, Beck CR, Yu Y, Wilson M, Shaw CA, Bjergbaek L, Lupski JR, and Ira G (2015). DNA REPAIR. Mus81 and converging forks limit the mutagenicity of replication fork breakage. Science 349, 742–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. (2010). The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20, 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meselson M, and Stahl FW (1958). The Replication of DNA in Escherichia Coli. Proc Natl Acad Sci U S A 44, 671–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, Jones D, Hinton J, Marshall J, Stebbings LA, et al. (2012). Mutational processes molding the genomes of 21 breast cancers. Cell 149, 979–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponder RG, Fonville NC, and Rosenberg SM (2005). A switch from high-fidelity to error-prone DNA double-strand break repair underlies stress-induced mutation. Mol Cell 19, 791–804. [DOI] [PubMed] [Google Scholar]
- Potocki L, Bi W, Treadwell-Deering D, Carvalho CM, Eifert A, Friedman EM, Glaze D, Krull K, Lee JA, Lewis RA, et al. (2007). Characterization of Potocki-Lupski syndrome (dup(17)(p11.2p11.2)) and delineation of a dosage-sensitive critical interval that can convey an autism phenotype. Am J Hum Genet 80, 633–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid JG, Carroll A, Veeraraghavan N, Dahdouli M, Sundquist A, English A, Bainbridge M, White S, Salerno W, Buhay C, et al. (2014). Launching genomics into the cloud: deployment of Mercury, a next generation sequence analysis pipeline. BMC bioinformatics 15, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseaulin L, Yamada Y, Tsutsui Y, Russell P, Iwasaki H, and Arcangioli B (2008). Mus81 is essential for sister chromatid recombination at broken replication forks. The EMBO journal 27, 1378–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roumelioti FM, Sotiriou SK, Katsini V, Chiourea M, Halazonetis TD, and Gagos S (2016). Alternative lengthening of human telomeres is a conservative DNA replication process with features of break-induced replication. EMBO Rep 17, 1731–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini N, Ramakrishnan S, Elango R, Ayyar S, Zhang Y, Deem A, Ira G, Haber JE, Lobachev KS, and Malkova A (2013). Migrating bubble during break-induced replication drives conservative DNA synthesis. Nature 502, 389–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakofsky CJ, Ayyar S, Deem AK, Chung WH, Ira G, and Malkova A (2015). Translesion Polymerases Drive Microhomology-Mediated Break-Induced Replication Leading to Complex Chromosomal Rearrangements. Mol Cell 60, 860–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakofsky CJ, Roberts SA, Malc E, Mieczkowski PA, Resnick MA, Gordenin DA, and Malkova A (2014). Break-induced replication is a source of mutation clusters underlying kataegis. Cell Rep 7, 1640–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanghvi RV, Buhay CJ, Powell BC, Tsai EA, Dorschner MO, Hong CS, Lebo MS, Sasson A, Hanna DS, McGee S, et al. (2018). Characterizing reduced coverage regions through comparison of exome and genome sequencing data across 10 centers. Genet Med 20, 855–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedlazeck FJ, Rescheneder P, Smolka M, Fang H, Nattestad M, von Haeseler A, and Schatz MC (2018). Accurate detection of complex structural variations using single-molecule sequencing. Nat Methods 15, 461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A, and Symington LS (2015). Microhomology-Mediated End Joining: A Back-up Survival Mechanism or Dedicated Pathway? Trends Biochem Sci 40, 701–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw CJ, Bi W, and Lupski JR (2002). Genetic proof of unequal meiotic crossovers in reciprocal deletion and duplication of 17p11.2. Am J Hum Genet 71, 1072–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw CJ, and Lupski JR (2005). Non-recurrent 17p11.2 deletions are generated by homologous and non-homologous mechanisms. Hum Genet 116, 1–7. [DOI] [PubMed] [Google Scholar]
- Shendure J, and Akey JM (2015). The origins, determinants, and consequences of human mutations. Science 349, 1478–1483. [DOI] [PubMed] [Google Scholar]
- Shinbrot E, Henninger EE, Weinhold N, Covington KR, Goksenin AY, Schultz N, Chao H, Doddapaneni H, Muzny DM, Gibbs RA, et al. (2014). Exonuclease mutations in DNA polymerase epsilon reveal replication strand specific mutation patterns and human origins of replication. Genome Res 24, 1740–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S, Li F, Villarreal D, Shim JH, Yoon S, Myung K, Shim EY, and Lee SE (2017). Microhomology-mediated end joining induces hypermutagenesis at breakpoint junctions. PLoS Genet 13, e1006714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack A, Thornton PC, Magner DB, Rosenberg SM, and Hastings PJ (2006). On the mechanism of gene amplification induced under stress in Escherichia coli. PLoS Genet 2, e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slager RE, Newton TL, Vlangos CN, Finucane B, and Elsea SH (2003). Mutations in RAI1 associated with Smith-Magenis syndrome. Nat Genet 33, 466–468. [DOI] [PubMed] [Google Scholar]
- Smith CE, Llorente B, and Symington LS (2007). Template switching during reak-induced replication. Nature 447, 102–105. [DOI] [PubMed] [Google Scholar]
- Song X, Beck CR, Du R, Campbell IM, Coban-Akdemir Z, Gu S, Breman AM, Stankiewicz P, Ira G, Shaw CA, et al. (2018). Predicting human genes susceptible to genomic instability associated with Alu/Alu-mediated rearrangements. Genome Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz P, Shaw CJ, Dapper JD, Wakui K, Shaffer LG, Withers M, Elizondo L, Park SS, and Lupski JR (2003). Genome architecture catalyzes nonrecurrent chromosomal rearrangements. Am J Hum Genet 72, 1101–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathern JN, Shafer BK, and McGill CB (1995). DNA synthesis errors associated with double-strand-break repair. Genetics 140, 965–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Liu P, Jia X, Withers MA, Jin L, Lupski JR, and Zhang F (2013). Replicative mechanisms of CNV formation preferentially occur as intrachromosomal events: evidence from Potocki-Lupski duplication syndrome. Hum Mol Genet 22, 749–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Beck CR, English AC, Meng Q, Buhay C, Han Y, Doddapaneni HV, Yu F, Boerwinkle E, Lupski JR, et al. (2015a). PacBio-LITS: a large-insert targeted sequencing method for characterization of human disease-associated chromosomal structural variations. BMC genomics 16, 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Su P, Hu B, Zhu W, Li Q, Yuan P, Li J, Guan X, Li F, Jing X, et al. (2015b). Characterization of 26 deletion CNVs reveals the frequent occurrence of micro-mutations within the breakpoint-flanking regions and frequent repair of double-strand breaks by templated insertions derived from remote genomic regions. Hum Genet 134, 589–603. [DOI] [PubMed] [Google Scholar]
- White JJ, Mazzeu JF, Coban-Akdemir Z, Bayram Y, Bahrambeigi V, Hoischen A, van Bon BWM, Gezdirici A, Gulec EY, Ramond F, et al. (2018). WNT Signaling Perturbations Underlie the Genetic Heterogeneity of Robinow Syndrome. Am J Hum Genet 102, 27–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MA, Kwon Y, Xu Y, Chung WH, Chi P, Niu H, Mayle R, Chen X, Malkova A, Sung P, et al. (2013). Pif1 helicase and Poldelta promote recombination-coupled DNA synthesis via bubble migration. Nature 502, 393–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Muzny DM, Xia F, Niu Z, Person R, Ding Y, Ward P, Braxton A, Wang M, Buhay C, et al. (2014). Molecular findings among patients referred for clinical whole-exome sequencing. Jama 312, 1870–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan B, Harel T, Gu S, Liu P, Burglen L, Chantot-Bastaraud S, Gelowani V, Beck CR, Carvalho CM, Cheung SW, et al. (2015). Nonrecurrent 17p11.2p12 Rearrangement Events that Result in Two Concomitant Genomic Disorders: The PMP22-RAI1 Contiguous Gene Duplication Syndrome. Am J Hum Genet 97, 691–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen RK, Merico D, Cao H, Pellecchia G, Alipanahi B, Thiruvahindrapuram B, Tong X, Sun Y, Cao D, Zhang T, et al. (2016). Genome-wide characteristics of de novo mutations in autism. NPJ Genom Med 1, 160271–1602710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Carvalho CM, and Lupski JR (2009a). Complex human chromosomal and genomic rearrangements. Trends Genet 25, 298–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Khajavi M, Connolly AM, Towne CF, Batish SD, and Lupski JR (2009b). The DNA replication FoSTeS/MMBIR mechanism can generate genomic, genic and exonic complex rearrangements in humans. Nat Genet 41, 849–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1- Junctions and array comparative genomic hybridization data for each individual in the study; related to Table 1 and Figure 4
Figure S1. Breakpoints and relative positions of SNVs and Indels; related to Figure 1. The precisely mapped breakpoints and de novo SNVs and indels that accompany the genomic rearrangements are depicted. Breakpoints are depicted with blue vertical lines, and de novo mutations with red stars. For recurrent SVs, the breakpoint is drawn at the mid-point of the LCR involved in the rearrangement.
Figure S2. Distribution of regional de novo mutations within the capture region; related to Figure 4. A) The proximity of each de novo SNV and indel with respect to the nearest junction is plotted in the histogram. B) The analysis shows that 5/9 of the regional de novo patient mutations are significantly enriched near the breakpoints of junctions and separated from the distribution of 10,000 simulations. The figures plot each subject (red dot) vs. the 10,000 simulations (grey dots) in the bi-dimensional space of the Mahalanobis distance between de novo mutations and SV breakpoints (x-axis) and inter-mutational distance (y-axis). The p-value from a chi-square test is shown on top of each panel.
Table S2- Callable regions within the regional Illumina sequencing data; related to STAR Methods - Sequencing methodologies
Table S3- Junction and point mutation data for each individual in the study; related to Table3 and Figure 3
Data Availability Statement
Microarray data generated in this study are available through GEO under the accession number GSE125120.