Abstract
Starvation is a common stress in fish. The underlying molecular mechanisms associated with growth depression caused by feeding restriction and compensatory growth are not well understood. We investigated the effect of fasting and refeeding on the transcriptome profiles of brain in juvenile S. hollandi using RNA-seq. A total of 4.73 × 108 raw reads were obtained from nine brain samples. De novo transcriptome assembly identified 387,085 unigenes with 2.1×109 nucleotides. A total of 936 annotated unigenes showed significantly differential expression among the control, fasting, and fasting-refeeding groups. The down-regulated differentially expressed genes (DEGs) during fasting were mainly associated with cell cycle, DNA replication, and mitosis. The up-regulated DEGs were mainly related to glucose and lipid metabolism, material transportation, and transcription factors. Most decreased DEGs during fasting were restored to normal levels after refeeding. Comparing with the control group, genes associated with protein synthesis, stimulus response, and carbohydrate metabolism were significantly over-expressed and pro-opio melanocortin (POMC) was down-regulated during the refeeding period. In conclusion, fish mobilized stored energetic materials and reduced energy consumption to prolong survival during fasting. After refeeding, the down-regulation of DEGs, e.g., POMC may be associated with compensatory growth. Up-regulation of DEGs related to ribosomal protein, stimulus response, and carbohydrate metabolism may contribute to eliminate negative effect of starvation on brain. This study provided the first transcriptome data related with impact of short-time starvation and refeeding in S. hollandi brains.
Introduction
Starvation is a common stress in fish. Extreme aquatic conditions, such as unsuitable temperature, hypoxia, and high population density, can affect the feeding behavior of aquatic animals. The starvation condition is strongly associated with the mobilization of the body’s energy reserves, which are mainly carbohydrates, fats, and proteins[1]. During starvation, organisms usually maintain metabolic homeostasis by changing their enzymatic activities and hormone levels, and activating various physiological and biochemical adaptive mechanisms[2]. In previous studies, the impacts of starvation on transcriptome profiles have been investigated in several fish species, such as the yellow croaker (Larimichthys crocea)[3], rainbow trout (Oncorhynchus mykiss)[4], and channel catfish (Ictalurus punctatus)[5]; however, these reports mainly focused on liver and muscle tissues[3,4].
Compensatory growth is defined as the physiological process of accelerated growth when food intake is restored following a period of food deprivation[6]. The phenomenon is ubiquitous in teleosts such as European minnow (Phoxinus phoxinus) [7], gibel carp (Carassius auratus gibelio)[8], barramundi (Lates calcarifer)[9], channel catfish (Ictalurus punctatus)[10], tongue sole (Cynoglossus semilaevis)[11], and Atlantic salmon (Salmo salar)[12]. Information regarding the biological and molecular mechanisms of modulating the exaggerated growth phenotype remains limited. Recently, studies on fine flounder (Paralichthys adspersus)[13], grass carp (Ctenopharyngodon idella)[14], and rainbow trout[4], have revealed changes in the transcriptome profile of muscle during the fasting and refeeding period. He et al. described the global gene expression patterns of liver following compensatory growth in grass carp and identified several genes potentially associated with compensatory growth [14].
Spinibarbus hollandi, an endemic Cyprinidae species in southeastern China, is mainly distributed in the provinces of Anhui, Hubei, Hunan, Fujian, Guangdong, and Guangxi. S. hollandi is an easy-to-raise omnivore, and has already attracted increasing attention owing to its high nutritional and medicinal value. However, the low growth rate has severely limited its farming[15]. In recent years, researchers attempted to enhance the growth rate of S. hollandi by improving the rearing condition and via cross-breeding. We have developed molecular markers associated with growth traits in S. hollandi[15] and recently observed the compensatory growth process in this species. Currently, studies on the effects of fasting on the growth rate and the compensatory growth of S. hollandi are rare. The underlying molecular mechanisms associated with growth depression caused by feeding restriction and compensatory growth are not well understood.
The next-generation sequencing technology has revolutionized the field of genomics. RNA-Seq can be used to analyze the transcriptome of non-model species without requiring their genomic information [16–20]. Brain plays an important role in maintaining energy balance during the fasting and refeeding period. The aim of this study was to investigate the changes of transcriptome profile of S. hollandi brain in response to fasting and refeeding. Our study provides an important dataset for understanding the molecular mechanism of starvation response and compensatory growth in fish.
Materials and methods
Fasting treatment and sampling
S. hollandi fish at about one-month-old were obtained from Shaoguan Fisheries Research Institute, Guangdong Province, China. The fish were sexually immature and had an approximate weight of 3–5 cm. 210 fish were equally distributed to seven aquariums containing 100 L of continuous flow, filtered water with aeration. The fish were housed at 26°C with a 14:10 h light/dark photoperiod. The fish were fed with a commercial pellet containing 35% crude protein, 4% crude fat, 14% crude fiber, 14% crude ashes, and 12% humidity (Qicai Pet Products Co., Ltd., Guangzhou, China), twice daily to apparent satiation at 7:00 am and 5:00 pm, for a 2-week acclimation. Of the seven aquariums, four were served as control groups, one was used as the fasting group and the remaining two aquariums were used as the refeeding groups.
Fish in the control groups were fed to apparent satiation everyday as mentioned above. The fish were sequentially sampled on day 0 (C0), day 7 (C7), day 14 (C14), and day 40 (C40). Fish in the fasting group were deprived of feed for 7 days, and then sampled (F7). The fish in the refeeding groups were fed twice per day to satiation after 7 days of fasting. The fish were sampled after 7 (R7) and 33 days refeeding (R33). All seven groups were then anesthetized by an overdose (100 ppm) of eugenol (Sangon Biotech (Shanghai) Co., Ltd., Shanghai, China). The body weights of each group were measured prior to sampling (n = 8). Brain tissues were rapidly isolated, immediately frozen in liquid nitrogen, and stored at -80°C.
RNA isolation
Total RNA of brain tissues was extracted using RNAiso reagents (Takara Biomedical Technology Co., Ltd., Dalian, China), following the manufacturer’s instructions. The quantity and quality of RNA samples were determined using Epoch Microplate Spectrophotometer (BioTek Instruments, Inc., USA), Agilent 2100 instruments and electrophoresis using 1% agarose gel. RNA samples with 28S/18S ratio >1.0, optical density (OD) 260/280 values >1.8 and <2.0, and RNA yield > 5 μg were used for subsequent transcriptome analysis.
cDNA library preparation and sequencing
RNA samples of the C7, F7, and R7 groups were used for RNA-seq analysis. To minimize the variation among individuals, equal quantities of RNA from 5 samples belonging to the same group were pooled, and 3 replicates of mixed RNA pools were prepared for each group. A total of 9 RNA pools were used for cDNA library preparation. mRNA was purified from total RNA using oligo (dT) magnetic beads and fragmented using fragmentation buffer. First-strand cDNA was synthesized using SuperScript II Reverse Transcriptase (Applied Biosystems Ltd., USA) followed by second strand generation using the components dNTPs, buffer, DNA polymerase I, and RNase H. After purification, adapters were ligated to the cDNA, and then enriched using PCR. The nine cDNA libraries were sequenced on an Illumina-Hiseq 2000 platform using the 150 bp paired-end approach.
Data processing and analysis
Clean reads were generated by removing low-quality reads reads containing adapter and ploy-N from raw data. Clean reads were de novo assembled using Trinity software version 2.5.1 [21]. The assembled transcripts were identified using Blast+ (version: ncbi-blast-2.2.28+) against NCBI non-redundant (Nr, e-value <1e-5), Clusters of Orthologous Groups of proteins (COG, e-value <1e-3), Kyoto Encyclopedia of Genes and Genomes (KEGG, e-value <1e-5), and Swiss-Prot databases (e-value <1e-5). Gene ontology (GO) annotation was performed using Blast2GO with an e-value threshold of 1e-6.
Total number of mapped reads for each transcript was determined and normalized, to calculate the expected number of Fragment Per Kilobase of transcript sequence per million base pairs sequenced (FPKM) by using RSEM V1.2.15 [22]. Differential expression analyses of three groups (C7, F7, and R7) were performed using DESeq R [23]. The adjusted p-value (p-adj) with a cut-off of 0.05 was used to identify significant differentially expressed genes (DEGs). Pathway enrichments of DEGs were analyzed using the KEGG database.
RNA-seq validation by quantitative real-time PCR analysis
Six DEGs (CDC6, CDC20, MCM2, DHCR7, FABP7, HSP70) belonging to different functional classification were selected and analyzed using the software FasParser [24] to identify gene expression changes in seven groups (C0, C7, C14, C40, F7, R7, and R33) through quantitative real-time PCR (qRT-PCR). A total of 10 DEGs including the above six genes, and TFRC1, TUBB5, MCH1, and ACSBG2, were used to verify RNA-seq results. It was performed by detecting expression changes of these DEGs during fasting (C7 vs. F7) and refeeding (C7 vs. R7) using qRT-PCR. Three biological replicates were used for each RNA sample and the beta-actin gene was used as an internal control. Primers used for qRT-PCR analysis are shown in S1 Table. The qRT-PCR analysis was performed using ABI 7000 platform with 20 μL reactions containing the following components: 100 ng of cDNA, 10 μL Power SYBR Green PCR Master Mix (Vazyme Biotech, Nanjing, China), 0.3 μL of each primer (10 μmol/L), and 7.4 μL double-distilled water. The reaction procedure followed by 95°C for 30 s, 40 cycles at 95°C for 10s and 60°C for 30s. After amplification, a melting curve was obtained by default processes in ABI7000. All samples were analyzed in triplicate, and fold changes were calculated using the comparative Ct method (2−ΔΔCt method) [25]. All data are provided in terms of relative mRNA expression as mean ± S.E. (n = 3).
Results
Changes of body weights during fasting and refeeding periods
The body weights of fish in the control groups (C7, C14, and C40) and the test groups (F7, R7, and R33) were measured. The growth curves for these groups were shown in Fig 1. At the end of 7-day fasting period, the body weight of C7 and F7 were 1.57 g and 1.16 g, respectively. There was a significantly difference in body weights between groups (p < 0.05). After 7-day refeeding, the body weight of C14 and R7 were 1.94 g and 1.63 g. After 40 days, the body weights for C40 and R33 was 3.34g and 3.28 g, respectively. However, no significant difference between groups was detected at the same time points.
Fig 1. Growth curves of Spinibarbus hollandi under continuous feeding and fasting-refeeding.
Asterisk indicated significant difference between two groups (p < 0.05); blue line represented the growth curve for the continuous feeding groups; red line indicated the growth curve for the fasting-refeeding groups.
RNA-seq data analysis
Through high-throughput paired-end sequencing, 4.93 × 108 raw reads (150 bp) were yielded from nine brain samples. After removing low-quality sequences, 4.73 × 108 clean reads with 71.01 Gb were generated. The Q20 and Q30 of all samples were above 96.27% and 90.97%, respectively. The sequence data were deposited in the NCBI SRA database, under the accession number, SRP156354.
After de novo assembly, 864,488 transcripts (ranging from 201 to 23,693 bp) were generated. The longest transcript of a gene was defined as a unigene for further analysis. A total of 387,085 unigenes with 2.1×109 nucleotides were obtained, with an average length of 568 bp. The N50 and N90 were 752 and 254 bp, respectively.
A total of 349,336 (90.24%) unigenes could be annotated in at least one of the following database: Nr, Nt, Pfam, KOG, Swiss-Prot, KEGG, and GO. A total of 10,438 (6.65%) unigenes were annotated in all these databases. Among of all unigenes, 76,988 (19.88%) unigenes were matched in the Nr database. Of which, nearly 58.6% could be annotated with sequences from Danio rerio, followed by those from Astyanax mexicanus (5.5%), Clupea harengus (2.8%), Mus musculus (2.4%), and Oncorhynchus mykiss (2.3%).
Functional prediction and classification of all unigenes were performed using the GO database. A total of 71,164 unigenes were classified into 56 GO terms, including 10 molecular function terms, 20 cellular component terms, and 26 biological process terms. Cellular process (39,913), binding (37,484), single-organism process (32,048), metabolic process (31,836), and cell (24,674), were the five most abundant functional terms at the second GO level (S1 Fig).
Furthermore, all unigenes were aligned to COG database for phylogenetic classification. In total, 22,886 unigenes were divided into 26 functional categories (S2 Fig). The largest category was signal transduction mechanisms with 4,653 unigenes followed by general function prediction (3,511), posttranslational modification, protein turnover, chaperones (2,283), intracellular trafficking, secretion, and vesicular transport (1,725).
To interpret the biological pathways, we annotated all assembled unigenes in the KEGG database, and 39,265 unigenes were matched in level 2 of the KEGG pathway. Signal transduction (6744), endocrine system (3302), cellular community (2976), immune system (2643), and nervous system (2432) were the five most significant matched catalogs in the KEGG pathway (S2 Table).
Identification of DEGs between control and fasting group
A total of 936 annotated unigenes showed significantly differential expression among the C7, F7, and R7 groups. Of the DEGs, 464 were significantly expressed between C7 and F7 (fasting period), 491 were significantly expressed between F7 and R7 (refeeding period), and 230 were significantly expressed between C7 and R7.
After fasting, of the 464 DEGs, 169 and 295 genes were up-regulated and down-regulated, respectively. Most of the genes associated with cell division were significantly down-regulated (Table 1). These genes were related to various key proteins involved in the cell cycle (CCNA2, CDC20, CDC25A, CDC6, CDK1, CDK2, CHEK2, E2F3, PLK1, and PTTG1), DNA replication (MCM2, MCM3, MCM4, MCM5-B, MCM7-A, PCNA) and mitosis (ESPL1, MAD2L1, NCAPD2, NCAPG, MAD2L1, SMC2, and SMC4).
Table 1. Down-regulated DEGs involved in cell division after 7-day fasting.
| Gene name | Gene ID | Gene description | log2FC | padj |
|---|---|---|---|---|
| CCNA2 | TRINITY_DN98393_c2_g1 | Cyclin-A2 | 1.235 | 1.7884E-02 |
| CDC20 | TRINITY_DN101219_c0_g1 | Cell division cycle protein 20 homolog | 1.2824 | 2.6243E-06 |
| CDC25A | TRINITY_DN97504_c0_g1 | M-phase inducer phosphatase 1 | 1.3734 | 1.1450E-05 |
| CDC6 | TRINITY_DN100724_c0_g3 | Cell division control protein 6 homolog | 1.3301 | 1.4772E-02 |
| CDK1 | TRINITY_DN90363_c1_g2 | Cyclin-dependent kinase 1 | 1.2218 | 7.9794E-05 |
| CDK2 | TRINITY_DN73824_c1_g1 | Cyclin-dependent kinase 2 | 1.1128 | 1.4598E-02 |
| CHEK2 | TRINITY_DN99705_c1_g2 | Serine/threonine-protein kinase Chk2 | 1.4801 | 2.7848E-02 |
| E2F3 | TRINITY_DN109456_c1_g1 | Transcription factor E2F3 | 1.3032 | 3.6578E-03 |
| ESPL1 | TRINITY_DN110892_c9_g2 | Separin | 1.5113 | 1.0703E-02 |
| MAD2L1 | TRINITY_DN84178_c0_g1 | Mitotic spindle assembly checkpoint protein MAD2A | 1.0504 | 4.9257E-02 |
| MCM2 | TRINITY_DN107178_c1_g1 | DNA replication licensing factor mcm2 | 1.6036 | 5.2371E-15 |
| MCM2 | TRINITY_DN107178_c1_g4 | DNA replication licensing factor mcm2 | 1.7659 | 7.7048E-03 |
| MCM3 | TRINITY_DN86555_c1_g1 | DNA replication licensing factor MCM3 | 1.3235 | 6.8196E-08 |
| MCM4 | TRINITY_DN108416_c1_g1 | DNA replication licensing factor mcm4 | 1.4637 | 5.6243E-09 |
| MCM4 | TRINITY_DN108416_c1_g2 | DNA replication licensing factor mcm4 | 1.5125 | 3.2801E-06 |
| MCM5-B | TRINITY_DN108607_c2_g1 | DNA replication licensing factor mcm5-B | 1.6463 | 6.7690E-08 |
| MCM7-A | TRINITY_DN94788_c0_g1 | DNA replication licensing factor mcm7-A | 0.86554 | 2.6866E-05 |
| NCAPD2 | TRINITY_DN109869_c3_g2 | Condensin complex subunit 1 | 0.9826 | 1.5797E-03 |
| NCAPG | TRINITY_DN107734_c1_g2 | Condensin complex subunit 3 | 1.6892 | 5.3557E-09 |
| NCAPG | TRINITY_DN107734_c1_g3 | Condensin complex subunit 3 | 1.368 | 9.6760E-03 |
| PCNA | TRINITY_DN97153_c1_g2 | Proliferating cell nuclear antigen | 0.85759 | 4.0344E-05 |
| PLK1 | TRINITY_DN101420_c0_g1 | Serine/threonine-protein kinase PLK1 | 1.5956 | 1.9129E-03 |
| PLK1 | TRINITY_DN101420_c0_g3 | Serine/threonine-protein kinase PLK1 | 1.3167 | 5.6127E-05 |
| PTTG1 | TRINITY_DN85037_c0_g1 | Securin | 1.1932 | 1.7091E-02 |
| SMC2 | TRINITY_DN107599_c0_g1 | Structural maintenance of chromosomes protein 2 | 1.4581 | 5.9764E-11 |
| SMC4 | TRINITY_DN108206_c1_g1 | Structural maintenance of chromosomes protein 4 | 0.90865 | 8.2643E-05 |
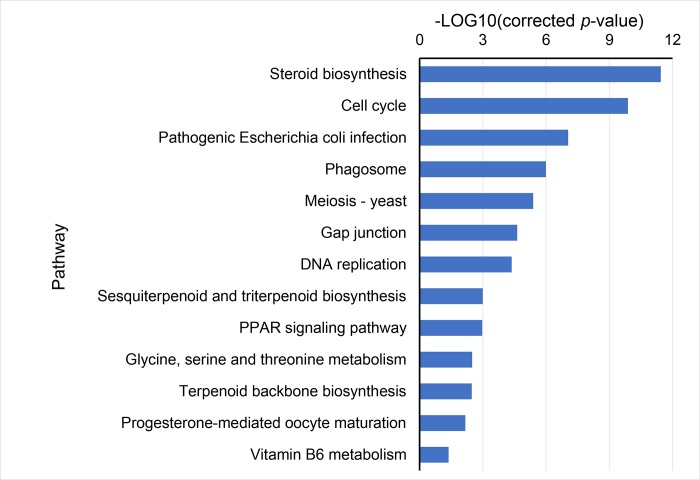
Except the above catalogs, KEGG pathway analysis identified that the down-regulated genes were also enriched in other pathways, such as steroid biosynthesis, pathogenic Escherichia coli infection, phagosome, gap junction, and sesquiterpenoid and triterpenoid backbone biosynthesis (Fig 2).
Fig 2. Classification of down-regulated DEGs during fasting according to KEGG database.
The up-regulated DEGs were annotated and analyzed (S3 Table). We observed several up-regulated DEGs involved in regulating glucose and lipid metabolism (PDK4, ADIPOR1, ARRDC3, PMM1, ZBTB16, and BBOX 1), material transportation (SLC43A2, SLC16A3, SLC19A3), atrophy (FBXO32), protein synthesis (EEF2K, PRKCA), and autophagy (GABARAPL1, TP53INP2) (Table 2).
Table 2. Up-regulated DEGs after 7-day fasting in brain.
| Gene name | Gene ID | Gene description | log2FC |
|---|---|---|---|
| PDK4 | TRINITY_DN95752_c2_g2 | pyruvate dehydrogenase kinase isoenzyme 4 | 1.8265 |
| ADIPOR1 | TRINITY_DN92796_c0_g1 | Adiponectin receptor protein 1 | 0.7845 |
| ARRDC3 | TRINITY_DN107064_c1_g2 | Arrestin domain-containing protein 3 | 0.84685 |
| PMM1 | TRINITY_DN101868_c1_g2 | Phosphomannomutase 1 | 1.0098 |
| ZBTB16 | TRINITY_DN106696_c2_g1 | Zinc finger and BTB domain-containing protein 16 | 0.76865 |
| SLC16A3 | TRINITY_DN107827_c3_g1 | Monocarboxylate transporter 4 | 0.79206 |
| SLC19A3 | TRINITY_DN89387_c3_g3 | Thiamine transporter 2 | 0.94927 |
| PDE5A | TRINITY_DN95368_c1_g1 | cGMP-specific 3',5'-cyclic phosphodiesterase | 0.90502 |
| FBXO32 | TRINITY_DN93305_c1_g1 | F-box only protein 32 | 1.2457 |
| EEF2K | TRINITY_DN106107_c1_g1 | Eukaryotic elongation factor 2 kinase | 1.1637 |
| PRKCA | TRINITY_DN107660_c5_g2 | Protein kinase C alpha type | 0.57357 |
| GABARAPL1 | TRINITY_DN75369_c2_g1 | Gamma-aminobutyric acid receptor-associated protein-like 1 | 0.86148 |
Identification of DEGs between fasting and refeeding group
After 7-day refeeding, of the 491 DEGs, 374 and 117 genes were up-regulated and down-regulated, respectively. The up-regulated DEGs were remarkably enriched in antigen processing and presentation, cell cycle, steroid biosynthesis, protein processing in endoplasmic reticulum, DNA replication, mitosis, pathogenic E. coli infection, and terpenoid backbone biosynthesis (Fig 3).
Fig 3. Classification of up-regulated DEGs after 7-day refeeding according to KEGG database.
The down-regulated DEGs were identified and annotated (S4 Table). These genes included some of the genes up-regulated during the fasting period, such as FBXO32, EEF2K, PDK4, ZNF778, ZBTB16, SLC43A2, SLC19A3, SLC16A3, PMM1, and GABARAPL1. The ATP-binding cassette transporters (ABCA1, ABCC5), IGFBP1, IRS2, JARID2, ACACB, CHKA, PDK2, PPARA were also down-regulated.
Identification of DEGs between control and refeeding group
DEGs between the control and refeeding group were identified and annotated (S5 Table). DEGs encoding ribosomal protein (RPS11, RPS12, RPS13, RPS15, RPLP0, RPLP2, RPL10A, RPL12, RPL13, and RPL13A), the heat shock protein (HSP) family (HSP70, HSC71, HSC71, HSPA8, HSP90A1, and HSP90AA1) and enzymes associated with glycolysis and gluconeogenesis (LDHB, TPI1, GAPDH, PGAM1, LDHA, PGK1, ALDOA, GPI) were all up-regulated (Table 3). The expression of MT-ND5, MT-CO1, ITM2A, ACTG1, and IL32 were also significantly increased. Several DEGs associated with the electron transport chain (MT-CO3, MT-ND3, MT-ND6) were down-regulated, and the expression of GTF2IRD2, JARID2, JARID2B, POMCA, STMN2, and UCHL1 were also significantly decreased.
Table 3. Up-regulated genes in the refeeding group comparing with the control group.
| Gene name | Gene ID | Gene description | log2FC |
|---|---|---|---|
| Ribosomal protein | |||
| RPSA | TRINITY_DN88004_c0_g3 | 40S ribosomal protein SA | 4.535942951 |
| RPS9 | TRINITY_DN94296_c1_g2 | 40S ribosomal protein S9 | 4.18762245 |
| RPS8 | TRINITY_DN69062_c0_g1 | 40S ribosomal protein S8 | 2.33022993 |
| RPS7 | TRINITY_DN87535_c1_g2 | 40S ribosomal protein S7 | 5.265127258 |
| RPS5 | TRINITY_DN80306_c3_g4 | 40S ribosomal protein S5 | 3.929266576 |
| RPS3A | TRINITY_DN77584_c1_g3 | 40S ribosomal protein S3a | 4.032269037 |
| RPS3 | TRINITY_DN94897_c1_g6 | 40S ribosomal protein S3 | 4.308172588 |
| RPS3 | TRINITY_DN94897_c1_g5 | 40S ribosomal protein S3 | 3.255821893 |
| RPS25 | TRINITY_DN98335_c1_g1 | 40S ribosomal protein S25 | 4.757039305 |
| RPS24 | TRINITY_DN75798_c3_g1 | 40S ribosomal protein S24 | 2.163325301 |
| RPS21 | TRINITY_DN69959_c0_g2 | 40S ribosomal protein S21 | 3.569696459 |
| RPS20 | TRINITY_DN96702_c4_g9 | 40S ribosomal protein S20 | 3.858889849 |
| RPS2 | TRINITY_DN82419_c5_g1 | 40S ribosomal protein S2 | 6.7053945 |
| RPS2 | TRINITY_DN82419_c5_g2 | 40S ribosomal protein S2 | 3.589301509 |
| RPS18 | TRINITY_DN90356_c0_g5 | 40S ribosomal protein S18 | 5.417252941 |
| RPS16 | TRINITY_DN49145_c0_g2 | 40S ribosomal protein S16 | 4.748258981 |
| RPS15A | TRINITY_DN102334_c0_g1 | 40S ribosomal protein S15a | 3.784354695 |
| RPS15 | TRINITY_DN80664_c0_g4 | 40S ribosomal protein S15 | 4.829265745 |
| RPS13 | TRINITY_DN72902_c0_g2 | 40S ribosomal protein S13 | 3.677167149 |
| RPS12 | TRINITY_DN79746_c3_g1 | 40S ribosomal protein S12 | 4.387961359 |
| RPS11 | TRINITY_DN99507_c12_g4 | 40S ribosomal protein S11 | 4.506353942 |
| RPLP2 | TRINITY_DN102307_c1_g5 | 60S acidic ribosomal protein P2 | 3.712851556 |
| RPLP0 | TRINITY_DN81984_c0_g1 | 60S acidic ribosomal protein P0 | 1.534808896 |
| RPL9 | TRINITY_DN71792_c0_g2 | 60S ribosomal protein L9 | 3.867069267 |
| RPL8 | TRINITY_DN82367_c6_g1 | 60S ribosomal protein L8 | 6.943147623 |
| RPL7 | TRINITY_DN85697_c1_g1 | 60S ribosomal protein L7 | 4.733797505 |
| RPL6 | TRINITY_DN50400_c0_g1 | 60S ribosomal protein L6 | 4.861227838 |
| RPL4 | TRINITY_DN68389_c0_g1 | 60S ribosomal protein L4 | 5.294078581 |
| RPL39 | TRINITY_DN59795_c0_g1 | 60S ribosomal protein L39 | 4.060313367 |
| RPL38 | TRINITY_DN87593_c1_g5 | 60S ribosomal protein L38 | 3.262147471 |
| RPL36 | TRINITY_DN54848_c0_g2 | 60S ribosomal protein L36 | 3.633289825 |
| RPL35 | TRINITY_DN85158_c0_g1 | 60S ribosomal protein L35 | 4.190676949 |
| RPL32-PS | TRINITY_DN89995_c3_g4 | 60S ribosomal protein L32 | 3.419806477 |
| RPL31 | TRINITY_DN65558_c0_g1 | 60S ribosomal protein L31 | 3.867236433 |
| RPL3 | TRINITY_DN93473_c1_g3 | 60S ribosomal protein L3 | 5.685872966 |
| RPL28 | TRINITY_DN8069_c0_g1 | 60S ribosomal protein L28 | 4.38506942 |
| RPL27A | TRINITY_DN68249_c0_g1 | 60S ribosomal protein L27a | 4.642412735 |
| RPL26 | TRINITY_DN92477_c0_g2 | 60S ribosomal protein L26 | 4.480991296 |
| RPL18A | TRINITY_DN75517_c3_g5 | 60S ribosomal protein L18a | 6.229154361 |
| RPL18 | TRINITY_DN65120_c0_g1 | 60S ribosomal protein L18 | 4.311837585 |
| RPL17 | TRINITY_DN64100_c0_g1 | 60S ribosomal protein L17 | 3.933855363 |
| RPL15 | TRINITY_DN76924_c1_g5 | 60S ribosomal protein L15 | 2.389800649 |
| RPL13A | TRINITY_DN72454_c1_g2 | 60S ribosomal protein L13a | 5.651412321 |
| RPL13 | TRINITY_DN97764_c3_g1 | 60S ribosomal protein L13 | 3.503720206 |
| RPL12 | TRINITY_DN83096_c3_g3 | 60S ribosomal protein L12 | 5.010585117 |
| RPL10A | TRINITY_DN85223_c3_g1 | 60S ribosomal protein L10a | 3.896006318 |
| Heat shock protein family | |||
| HSPA8 | TRINITY_DN100126_c1_g2 | Heat shock cognate 71 kDa protein | 0.974477412 |
| HSP90AA1 | TRINITY_DN89892_c3_g3 | Heat shock protein HSP 90-alpha | 0.971776669 |
| HSP90A1 | TRINITY_DN109438_c3_g6 | Heat shock protein HSP 90-alpha 1 | 0.842457828 |
| HSP90A1 | TRINITY_DN89892_c3_g1 | Heat shock protein HSP 90-alpha 1 | 0.799479292 |
| HSP70 | TRINITY_DN111407_c8_g7 | Heat shock 70 kDa protein | 1.041022237 |
| HSP70 | TRINITY_DN111407_c8_g5 | Heat shock 70 kDa protein 1 | 1.012967973 |
| HSC71 | TRINITY_DN84773_c1_g3 | Heat shock cognate 70 kDa protein | 0.816312679 |
| HSC71 | TRINITY_DN90379_c0_g3 | Heat shock cognate 70 kDa protein | 0.742926524 |
| DNAJA4 | TRINITY_DN98506_c3_g1 | DnaJ homolog subfamily A member 4 | 1.352175205 |
| glycolysis and gluconeogenesis | |||
| ALDOA | TRINITY_DN93020_c1_g10 | Fructose-bisphosphate aldolase A | 3.825625389 |
| GAPDH | TRINITY_DN73283_c0_g1 | Glyceraldehyde-3-phosphate dehydrogenase | 2.767387832 |
| GPI | TRINITY_DN96702_c4_g7 | Glucose-6-phosphate isomerase | 4.875604882 |
| LDHA | TRINITY_DN84691_c6_g1 | L-lactate dehydrogenase A chain | 7.697424007 |
| LDHB | TRINITY_DN21161_c0_g1 | L-lactate dehydrogenase B chain | 3.711726558 |
| PGAM1 | TRINITY_DN79644_c1_g1 | Phosphoglycerate mutase 1 | 6.856235293 |
| PGK1 | TRINITY_DN85707_c4_g2 | Phosphoglycerate kinase 1 | 4.479693518 |
| ENO1 | TRINITY_DN76924_c1_g6 | Alpha-enolase | 7.645409467 |
| TPI1 | TRINITY_DN70056_c0_g3 | Triosephosphate isomerase | 3.876363706 |
qRT-PCR analysis of key genes
To investigate the expression profiles of key DEGs under the continuous feeding condition and fasting-refeeding condition, genes associated with cell cycle (CDC6 and CDC20), DNA replication (MCM2), steroid biosynthesis (DHCR7), fat metabolism (FABP7), and stimulus response (HSP70) were analyzed using qRT-PCR. All of these genes (CDC6, CDC20, MCM2, DHCR7, FABP7, and HSP70) were significantly down-regulated after 7-day fasting. CDC20 and HSP70 were sharply increased and higher than the control group after 7-day refeeding. On day 33 of refeeding, HSP70 remained significantly higher in the refeeding group than the control group. CDC20, however, was restored to normal level, and other genes were also restored and stable after refeeding. All genes had stable expressions under the continuous feeding condition (Fig 4).
Fig 4. Analysis of the expression profiles of key genes using qRT-PCR.
Asterisk indicates significant difference (p < 0.05), a~f represent the expression profiles of CDC6, CDC20C, MCM2, DHCR7, FABP7, and HSP70, respectively.
Confirmation of DEGs by qRT-PCR
To confirm the RNA-Seq results, 10 DEGs in different pathways were selected for qRT-PCR verification. Expression changes of these genes at fasting and refeeding were analyzed. The changes in gene expression obtained from qRT-PCR were consistent with the RNA-Seq under fasting and refeeding conditions, with R2 values of 0.7315 and 0.8251 (Fig 5), respectively. The results of the qRT-PCR analyses confirmed the reliability and accuracy of the data obtained by RNA-seq.
Fig 5. Correlation between results of RNA-seq and qRT-PCR.
a and b represent correlations between control and fasting groups, and between control and refeeding groups, respectively. X-axis numbers represent the log2 (fold change) values from RNA-seq results. Y-axis numbers represent the log2 (fold change) values from qRT-PCR results.
Discussion
1. Changes in transcriptome profile of fish brain after 7-day fasting
The present study used RNA-Seq to investigate the impact of fasting and refeeding on transcriptome profiles of brain in S. hollandi. The global gene expression pattern of fish brain under continuous feeding condition, fasting condition, and refeeding condition were displayed. After 7-day fasting, 295 down-regulated and 169 up-regulated genes were identified in the brain.
The down-regulated DEGs were mainly enriched in the pathway of cell cycle, DNA replication, mitosis, steroid biosynthesis, immunity, gap junction, and sesquiterpenoid and triterpenoid backbone biosynthesis. Cyclins are important factors in the control of cell cycle, and can be activated by cyclin-dependent kinase enzymes [26]. Cyclin A2 (CCNA2), Cyclin-dependent kinase 1 (CDK1), and Cyclin-dependent kinase 2 (CDK2) were down-regulated during fasting. CHEK2, E2F3, and ESPL1 play crucial roles in the control of cell cycle[27–29]. CDC20 is essential for the regulation of cell division; however, its most important function is to promote chromatid separation [30]. CDC6 is crucial to the maintenance of checkpoint mechanisms in the cell cycle and the loading of minichromosome maintenance (MCM) proteins onto DNA [31]. MCM, a DNA helicase composed of six subunits, is essential for genomic DNA replication. During fasting, MCM2, MCM3, MCM4, MCM5-B, and MCM7-A were all down-regulated. Mitosis is a part of the cell cycle wherein replicated sister chromatids are separated into two chromosomes. Several genes related to mitosis (NCAPD2, NCAPG, PTTG1, SMC2, and SMC4) were down-regulated. NCAPD2, NCAPG, SMC2, and SMC4 were involved in chromosome condensation [32,33]. Securin, encoded by PTTG1, is a protein involved in the control of the metaphase-anaphase transition[34]. It has been widely reported that cell proliferation can be inhibited by starvation in mammal and fish [2,4]. The down-regulation of DEGs associated with steroid biosynthesis, immunity, and other functions may reduce energy expenditure and prolong the survival of S. hollandi.
Several genes associated with regulating glucose and lipid metabolism (PDK4, ADIPOR1, ARRDC3, PMM1, and BBOX1) were markedly up-regulated. PDK4 decreases glucose utilization and increases fat metabolism through inhibiting pyruvate dehydrogenase. This plays an important role in regulating the shift in fuel economy to prolonged fasting and starvation [35]. ADIPOR1 is essential in the regulation of normal glucose and fat homeostasis, and for maintaining normal body weight during the fasting period[36]. ARRDC3 can decrease energy expenditure through decrease in the thermogenesis of adipose tissues [37] and PMM1 has a phosphoglucomutase activity, which converts glucose-1-P into glucose-6-P[38]. BBOX1 is an enzyme involved in the biosynthesis of L-carnitine, a key molecule in fatty acid metabolism [39]. ZBTB16, a transcriptional repressor, plays a key role in various biological processes such as adipogenesis, regulation of lipid levels, and insulin sensitivity [40]. Maintaining metabolic homeostasis is essential for the survival of fish during fasting, and similar results have been reported in the rainbow trout, yellow croaker, and grass crap [3,14,41].
Three nutrition transport genes (SLC43A2, SLC16A3, and SLC19A3) were up-regulated. These trans-membrane proteins are substrate-specific and their increased expression may aid in the mobilization of nutrients. Several genes associated with ketone body metabolism (SLC16A3 and PDK4) were up-regulated, and brain energy metabolism usually changes toward oxidation of ketone bodies during starvation [42]. Atrophy and autophagy can be induced by starvation, and protein synthesis can be inhibited. EEF2K can inhibit the eukaryotic elongation factor 2 (EEF2), an essential factor for protein synthesis. FBXO32 is associated with muscle atrophy during the fasting period [43] and plays a similar function in brain. GABARAPL1 and TP53INP2 are essential genes for autophagosome maturation [44,45]. These DGEs can assist in alleviating starvation stress in fish. The change of transcriptome profiles associated metabolism and mobilization of stored energy were similar to the liver tissues [3].
2. Changes in transcriptome profiles of fish brain after refeeding
After refeeding, most DEGs down-regulated after fasting were restored to normal levels. Many of the genes associated with immunity and protein synthesis were up-regulated, and are known to play important roles in antigen processing and presentation, protein processing in endoplasmic reticulum and pathogenic E. coli infection. An improvement in disease resistance capability and immune status was observed in the refeeding fish. These fish were also seen to outperform the fish that were continuously fed.
Genes such as ABCA1, ABCC5, IGFBP1, IRS2, JARID2, ACACB, CHKA, PDK2, and PPARA, were down-regulated, implying these genes may not be key in fish recovery.
DEGs between control group and refeeding group were also analyzed. Many ribosomal proteins were over-expressed in the refeeding group. These proteins consisted of the ribonucleoprotein complexes and involved in the cellular process of translation. Stress-specific alterations in ribosomal proteins were reported in many species, the over-expressions of ribosomal proteins contributed to the starvation response. Similar result has been observed in the muscle of rainbow trout [46].
Several members of the heat shock proteins were over-expressed after refeeding. HSP is an important protein in response to the exposure to stressful conditions. Many members of HSP act as chaperones to assist new proteins, ensuring them correct folding and preventing protein damage by cell stress [47]. The over-expression of HSP can enhance translational efficiency and increase the ability of stimulus response.
Glycolysis and oxidative phosphorylation are the main metabolic pathways for releasing energy in organisms and provide energy for various physiological activities. The up-regulation of LDHB, TPI1, GAPDH, PGAM1, LDHA, PGK1, ALDOA, GPI, MT-ND5, and MT-CO1 was beneficial for the production and restoration of energy.
In down-regulated DEGs, proopiomelanocortin (POMC) is an important precursor polypeptide, which can be enzymatically cleaved into many peptides, including melanotropins (α-MSH, β-MSH, and γ-MSH), adrenocorticotropin (ACTH), lipotropins, and endorphins [48]. These ligands closely associated with energy balance, glucose homeostasis and feeding behavior by negative regulation of energy metabolism [15,49]. The down-regulation of POMC is beneficial to diet intake.
3. Analyses of expression profiles of key DEGs using qRT-PCR
Expression profiles of six key DEGs were investigated using qRT-PCR. CDC6, CDC20, MCM2, DHCR7, and FABP7 were down-regulated during fasting and restored after refeeding. DHCR7 plays a vital role in the cholesterol biosynthesis pathway, which is part of the steroid biosynthesis pathway [50]. Starvation inhibited steroid biosynthesis; thus, DHCR7 was down-regulated during fasting and restored after refeeding. FABP7 was down-regulated during fasting, and is most likely because the ketone body was the main energetic materials of brain during starvation. FABP3 was also down-regulated during fasting. CDC20, CDC6, and MCM2 were all down-regulated because of the blockage of cell division by starvation. After refeeding, the expression of CDC20 was higher than in the control group, despite there was no significant difference. HSP70 is important for protein folding, and protects cells from stress. Thus, the over-expression of HSP70, after refeeding, can protect organisms from undergoing stress.
Conclusion
A transcriptome analysis was carried out to reveal the effects of fasting and refeeding on brains of juvenile S. hollandi. During fasting, the fish significantly reduced gene expression in the cell cycle, DNA replication, mitosis, steroid synthesis, and other physiological activities. After 7-day refeeding, the up-regulated DEGs were remarkably enriched in antigen processing and presentation, cell cycle, steroid biosynthesis, protein processing in endoplasmic reticulum, DNA replication, mitosis, pathogenic infection, and terpenoid backbone biosynthesis.
The study indicated that growth was inhibited during starvation since the fish mobilized the stored energy to prolong survival. After refeeding, the energy was stored again, and compensatory growth occurred, the body weight was recovered. In response to the starvation and feeding, a great deal of related genes and pathways were changed. This study provided the first transcriptome data on impacts of short-time starvation and refeeding to fish brains.
Supporting information
(TIF)
(TIF)
(DOCX)
(DOCX)
(DOCX)
(XLSX)
(XLSX)
(DOCX)
Acknowledgments
We thank the anonymous reviewers for their helpful comments on this work. This work is supported by the National Public Industry Major Projects of China (No.201303048) and the Guangdong Provincial Science and Technology Program (Nos. 2014A020208145).
Data Availability
All the read data were available at NCBI SRA database under the SRA accession number: SRP156354.
Funding Statement
This work is supported by the National Public Industry Major Projects of China (No.201303048;received by HS) and the Guangdong Provincial Science and Technology Program (Nos. 2014A020208145;received by HS).The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Takahashi LS, Biller JD, Criscuolo-Urbinati E, Urbinati EC (2011) Feeding strategy with alternate fasting and refeeding: Effects on farmed pacu production. Journal of Animal Physiology and Animal Nutrition 95: 259–266. 10.1111/j.1439-0396.2010.01050.x [DOI] [PubMed] [Google Scholar]
- 2.Keogh K, Kenny DA, Cormican P, Kelly AK, Waters SM (2016) Effect of dietary restriction and subsequent re-alimentation on the transcriptional profile of hepatic tissue in cattle. BMC Genomics 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qian B, Xue L, Huang H (2016) Liver transcriptome analysis of the large yellow croaker (Larimichthys crocea) during fasting by using RNA-seq. PLoS ONE 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rescan PY, Montfort J, Rallière C, Le Cam A, Esquerré D, Hugot K (2007) Dynamic gene expression in fish muscle during recovery growth induced by a fasting-refeeding schedule. BMC Genomics 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu L, Li C, Su B, Beck BH, Peatman E (2013) Short-term feed deprivation alters immune status of surface mucosa in channel catfish (Ictalurus punctatus). PLoS ONE 8: e74581 10.1371/journal.pone.0074581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Won ET, Borski RJ (2013) Endocrine regulation of compensatory growth in fis. Frontiers in Endocrinology 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Russell NR, Wootton RJ (1992) Appetite and growth compensation in the European minnow, Phoxinus phoxinus (Cyprinidae), following short periods of food restriction. Environmental Biology of Fishes 34: 277–285. [Google Scholar]
- 8.Xie S, Zhu X, Cui Y, Wootton RJ, Lei W, Yang Y (2001) Compensatory growth in the gibel carp following feed deprivation: Temporal patterns in growth, nutrient deposition, feed intake and body composition. Journal of Fish Biology 58: 999–1009. [Google Scholar]
- 9.Tian X, Qin JG (2003) A single phase of food deprivation provoked compensatory growth in barramundi Lates calcarifer. Aquaculture 224: 169–179. [Google Scholar]
- 10.Reigh RC, Williams MB, Jacob BJ (2006) Influence of repetitive periods of fasting and satiation feeding on growth and production characteristics of channel catfish, Ictalurus punctatus. Aquaculture 254: 506–516. [Google Scholar]
- 11.Tian X, Fang J, Dong S (2010) Effects of starvation and recovery on the growth, metabolism and energy budget of juvenile tongue sole (Cynoglossus semilaevis). Aquaculture 310: 122–129. [Google Scholar]
- 12.Johansen SJS, Ekli M, Stangnes B, Jobling M (2001) Weight gain and lipid deposition in Atlantic salmon, Salmo salar, during compensatory growth: Evidence for lipostatic regulation? Aquaculture Research 32: 963–974. [Google Scholar]
- 13.Mendez KN, Zuloaga R, Valenzuela CA, Bastias-Molina M, Meneses C, Vizoso P, et al. (2018) RNA-seq analysis of compensatory growth in the skeletal muscle of fine flounder (Paralichthys adspersus). Aquaculture 490: 270–280. [Google Scholar]
- 14.He L, Pei Y, Jiang Y, Li Y, Liao L, Zhu Z, et al. (2015) Global gene expression patterns of grass carp following compensatory growth. BMC Genomics 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Y, Li Q, Shu H, Zhou H, Li X, Hou L (2017) Characterization of the melanocortin-4 receptor gene from Spinibarbus hollandi and the association between its polymorphisms and S. hollandi growth traits. Fisheries Science 83: 967–976. [Google Scholar]
- 16.Qian X, Ba Y, Zhuang Q, Zhong G (2014) RNA-seq technology and its application in fish transcriptomics. OMICS A Journal of Integrative Biology 18: 98–110. 10.1089/omi.2013.0110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang XL, Ikhwanuddin M, Li XC, Lin F, Wu QY, Zhang YL, et al. (2018) Comparative transcriptome analysis provides insights into differentially expressed genes and long non-coding rnas between ovary and testis of the mud crab (scylla paramamosain). Marine Biotechnology 20: 20–34. 10.1007/s10126-017-9784-2 [DOI] [PubMed] [Google Scholar]
- 18.Ye H, Xiao SJ, Wang XQ, Wang ZY, Zhang ZS, Zhu CK, et al. (2018) Characterization of spleen transcriptome of schizothorax prenanti during aeromonas hydrophila infection. Marine Biotechnology 20: 246–256. 10.1007/s10126-018-9801-0 [DOI] [PubMed] [Google Scholar]
- 19.Yu LY, Xu DD, Ye H, Yue HM, Ooka S, Kondo H, et al. (2018) Gonadal transcriptome analysis of pacific abalone haliotis discus discus: Identification of genes involved in germ cell development. Marine Biotechnology 20: 467–480. 10.1007/s10126-018-9809-5 [DOI] [PubMed] [Google Scholar]
- 20.Yue CY, Li Q, Yu H (2018) Gonad transcriptome analysis of the pacific oyster Crassostrea gigas identifies potential genes regulating the sex determination and differentiation process. Marine Biotechnology 20: 206–219. 10.1007/s10126-018-9798-4 [DOI] [PubMed] [Google Scholar]
- 21.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature Biotechnology 29: 644–652. 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li B, Dewey CN (2011) RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12: 323 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biology 11: R106 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun YB (2017) FasParser: a package for manipulating sequence data. Zool Res 38: 110–112. 10.24272/j.issn.2095-8137.2017.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramakers C, Ruijter JM, Lekanne Deprez RH, Moorman AFM (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neuroscience Letters 339: 62–66. [DOI] [PubMed] [Google Scholar]
- 26.Nigg EA (1995) Cyclin‐dependent protein kinases: Key regulators of the eukaryotic cell cycle. BioEssays 17: 471–480. 10.1002/bies.950170603 [DOI] [PubMed] [Google Scholar]
- 27.Leone G, DeGregori J, Yan Z, Jakoi L, Ishida S, Williams RS, et al. (1998) E2F3 activity is regulated during the cell cycle and is required for the induction of S phase. Genes and Development 12: 2120–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dai XX, Duan X, Liu HL, Cui XS, Kim NH, Sun SC (2014) Chk2 regulates cell cycle progression during mouse oocyte maturation and early embryo development. Molecules and Cells 37: 126–132. 10.14348/molcells.2014.2259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zou H, McGarry TJ, Bernal T, Kirschner MW (1999) Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science 285: 418–421. [DOI] [PubMed] [Google Scholar]
- 30.Weinstein J, Jacobsen FW, Hsu-Chen J, Wu T, Baum LG (1994) A novel mammalian protein, p55CDC, present in dividing cells is associated with protein kinase activity and has homology to the Saccharomyces cerevisiae cell division cycle proteins Cdc20 and Cdc4. Molecular and Cellular Biology 14: 3350–3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oehlmann M, Score AJ, Blow JJ (2004) The role of Cdc6 in ensuring complete genome licensing and S phase checkpoint activation. Journal of Cell Biology 165: 181–190. 10.1083/jcb.200311044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmiesing JA, Ball AR Jr, Gregson HC, Alderton JM, Zhou S, Yokomori K (1998) Identification of two distinct human SMC protein complexes involved in mitotic chromosome dynamics. Proceedings of the National Academy of Sciences of the United States of America 95: 12906–12911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmiesing JA, Gregson HC, Zhou S, Yokomori K (2000) A human condensin complex containing hCAP-C-hCAP-E and CNAP1, a homolog of Xenopus XCAP-D2, colocalizes with phosphorylated histone H3 during the early stage of mitotic chromosome condensation. Molecular and Cellular Biology 20: 6996–7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu D, Hsiao JY, Davey NE, van Voorhis VA, Foster SA, Tang C, et al. (2014) Multiple mechanisms determine the order of APC/C substrate degradation in mitosis. Journal of Cell Biology 207: 23–39. 10.1083/jcb.201402041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Faherty SL, Villanueva-Cañas JL, Blanco MB, Albà MM, Yoder AD (2018) Transcriptomics in the wild: Hibernation physiology in free-ranging dwarf lemurs. Molecular Ecology 27: 709–722. 10.1111/mec.14483 [DOI] [PubMed] [Google Scholar]
- 36.Bjursell M, Ahnmark A, Bohlooly-Y M, William-Olsson L, Rhedin M, Peng XR, et al. (2007) Opposing effects of adiponectin receptors 1 and 2 on energy metabolism. Diabetes 56: 583–593. 10.2337/db06-1432 [DOI] [PubMed] [Google Scholar]
- 37.Patwari P, Emilsson V, Schadt EE, Chutkow WA, Lee S, Marsili A, et al. (2011) The arrestin domain-containing 3 protein regulates body mass and energy expenditure. Cell Metabolism 14: 671–683. 10.1016/j.cmet.2011.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cromphout K, Vleugels W, Heykants L, Schollen E, Keldermans L, Sciot R, et al. (2006) The normal phenotype of Pmm1-deficient mice suggests that Pmm1 is not essential for normal mouse development. Molecular and Cellular Biology 26: 5621–5635. 10.1128/MCB.02357-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee H, Kim HK, Kwon JT, Park S, Park HJ, Kim SK, et al. (2018) BBOX1 is down-regulated in maternal immune-activated mice and implicated in genetic susceptibility to human schizophrenia. Psychiatry Research 259: 197–202. 10.1016/j.psychres.2017.10.018 [DOI] [PubMed] [Google Scholar]
- 40.Šeda O, Šedová L, Vcelák J, Vanková M, Liška F, Bendlová B (2017) ZBTB16 and metabolic syndrome: A network perspective. Physiological Research 66: S357–S365. [DOI] [PubMed] [Google Scholar]
- 41.Johansen KA, Overturf K (2006) Alterations in expression of genes associated with muscle metabolism and growth during nutritional restriction and refeeding in rainbow trout. Comparative Biochemistry and Physiology—B Biochemistry and Molecular Biology 144: 119–127. [DOI] [PubMed] [Google Scholar]
- 42.Hasselbalch SG, Madsen PL, Hageman LP, Olsen RS, Justesen N, Holm S, et al. (1996) Changes in cerebral blood flow and carbohydrate metabolism during acute hyperketonemia. American Journal of Physiology—Endocrinology and Metabolism 270: E746–E751. [DOI] [PubMed] [Google Scholar]
- 43.Cleveland BM, Evenhuis JP (2010) Molecular characterization of atrogin-1/F-box protein-32 (FBXO32) and F-box protein-25 (FBXO25) in rainbow trout (Oncorhynchus mykiss): Expression across tissues in response to feed deprivation. Comparative Biochemistry and Physiology—B Biochemistry and Molecular Biology 157: 248–257. [DOI] [PubMed] [Google Scholar]
- 44.Chakrama FZ, Seguin-Py S, Le Grand JN, Fraichard A, Delage-Mourroux R, Despouy G, et al. (2010) GABARAPL1 (GEC1) associates with autophagic vesicles. Autophagy 6: 495–505. 10.4161/auto.6.4.11819 [DOI] [PubMed] [Google Scholar]
- 45.Sancho A, Duran J, García-España A, Mauvezin C, Alemu EA, Lamark T, et al. (2012) Dor/tp53inp2 and tp53inp1 constitute a metazoan gene family encoding dual regulators of autophagy and transcription. PLoS ONE 7: e34034 10.1371/journal.pone.0034034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rescan PY, Cam A, Rallière C, Montfort J (2017) Global gene expression in muscle from fasted/refed trout reveals up-regulation of genes promoting myofibre hypertrophy but not myofibre production. BMC Genomics 18: 447 10.1186/s12864-017-3837-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu C (1995) Heat shock transcription factors: Structure and regulation. Annual Review of Cell and Developmental Biology. pp. 441–469. 10.1146/annurev.cb.11.110195.002301 [DOI] [PubMed] [Google Scholar]
- 48.Chakraborty AK, Funasaka Y, Slominski A, Ermak G, Hwang J, Pawelek JM, et al. (1996) Production and release of proopiomelanocortin (POMC) derived peptides by human melanocytes and keratinocytes in culture: Regulation by ultraviolet B. Biochimica et Biophysica Acta—Molecular Cell Research 1313: 130–138. [DOI] [PubMed] [Google Scholar]
- 49.Varela L, Horvath TL (2012) Leptin and insulin pathways in POMC and AgRP neurons that modulate energy balance and glucose homeostasis. EMBO Reports 13: 1079–1086. 10.1038/embor.2012.174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prabhu AV, Sharpe LJ, Brown AJ (2014) The sterol-based transcriptional control of human 7-dehydrocholesterol reductase (DHCR7): Evidence of a cooperative regulatory program in cholesterol synthesis. Biochimica et Biophysica Acta—Molecular and Cell Biology of Lipids 1841: 1431–1439. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
(DOCX)
(DOCX)
(DOCX)
(XLSX)
(XLSX)
(DOCX)
Data Availability Statement
All the read data were available at NCBI SRA database under the SRA accession number: SRP156354.