Abstract
Despite considerable recent advancements in colorectal cancer (CRC) therapy, the prognosis of patients with advanced disease remains poor. Further understanding of the molecular mechanisms and treatment strategies of this disease is required. Zinc finger protein 692 (ZNF692), also known as AREBP and Zfp692, was first reported to have an important role in gluconeogenesis. A recent study demonstrated that ZNF692 is overexpressed in lung adenocarcinoma (LUAD) tissues and that ZNF692 knockdown inhibited LUAD cell proliferation, migration, and invasion both in vitro and in vivo. However, the role of ZNF692 in colon adenocarcinoma (COAD) remains unclear. The present study revealed that ZNF692 was upregulated in COAD tissues and cells and that high ZNF692 expression was significantly correlated with lymph node metastasis, distant metastasis and tumor stage in COAD patients. Gain- and loss-of-function experiments were employed to identify the function of ZNF692 in COAD progression. In vitro and in vivo assays revealed that ZNF692 promoted COAD cell proliferation, migration and invasion. Furthermore, western blot analysis demonstrated that the effects of ZNF692 were mediated by upregulating cyclin D1, cyclin-dependent kinase 2 (CDK2) and matrix metalloproteinase-9 (MMP-9) and by downregulating p27Kip1 through the phosphoinositide 3-kinase/AKT signaling pathway. Collectively, these data indicated that ZNF692 may serve as a novel oncogene and a potential treatment target in COAD patients.
Keywords: zinc finger protein 692, colon adenocarcinoma, cell cycle, phosphoinositide 3-kinase, AKT serine/threonine kinase, tumorigenesis
Introduction
Colorectal cancer (CRC) is one of the most common gastrointestinal malignancies worldwide, ranking as the third most common cancer in males and the second most common in females, with 60% of cases occurring in developed nations (1). Previous studies have demonstrated that the incidence of CRC has increased considerably over the past few decades (2,3). Histopathologically, adenocarcinoma is the most common diagnosis (4). Despite considerable recent advancements in CRC therapy, the prognosis of patients with advanced disease remains poor (5). Although many altered pathways (6,7) and aberrantly expressed genes (8,9) involved in CRC have been identified, determining which factors are the most important in CRC development is difficult, due to its complex genetic background. Thus, the identification of new biomarkers for the diagnosis, prognostic prediction, and therapeutic targeting of CRC would have great clinical relevance.
The present study queried the UALCAN portal (10) (http://ualcan.path.uab.edu/index.html) with the aim to obtain such a target gene. Through analysis of publicly available data from the UALCAN database, zinc finger protein 692 (ZNF692) was identified to be overexpressed in the following fourteen malignancies: bladder urothelial carcinoma, colon adenocarcinoma (COAD), lung adenocarcinoma (LUAD), renal clear cell carcinoma, prostate adenocarcinoma, lung squamous cell carcinoma, head and neck squamous cell carcinoma, renal papillary cell carcinoma, esophageal carcinoma, hepatocellular carcinoma, rectal adenocarcinoma, uterine corpus endometrial carcinoma, cholangiocarcinoma, and gastric adenocarcinoma. Furthermore, ZNF692 was associated with an unfavorable prognosis in four malignancies: COAD, renal clear cell carcinoma, hepatocellular carcinoma and adrenocortical carcinoma. A previous comprehensive analysis from our group of RNA sequencing (RNA-seq) data from the Cancer Genome Atlas (TCGA) also confirmed that ZNF692 is overexpressed in COAD (11). The gene encoding ZNF692, also known as AREBP and Zfp692, is located on chromosomal region 1q44 and produces a 2,208 bp mRNA expressing a 524 amino acid protein. ZNF692 belongs to the Krüppel C2H2-type zinc finger protein family, it contains five C2H2-type zinc finger repeats and is predicted to be involved in transcriptional regulation. Accumulating evidence has suggested that this type of zinc finger protein has important biological functions in tumor formation and progression (12,13). For example, zinc finger protein 639 (also known as ZASC1), a novel member of the Krüppel C2H2-type zinc finger protein family, was reported to be an independent prognostic indicator in esophageal squamous cell (ESC) carcinoma; furthermore, ZASC1 upregulation promoted the growth of ESC cells, while ZASC1 downregulation produced the opposite effect (14). In addition, zinc finger X-chromosomal protein (ZFX), a novel member of this family, was demonstrated to be highly expressed in osteosarcoma (15), hepatocellular carcinoma (16), glioblastoma (17), and CRC (18). Finally, these studies indicated that ZFX expression was positively correlated with advanced stage and poor survival and had important roles in tumor progression. Considering that ZNF692 is overexpressed in many tumors and the important roles of this family of proteins in cancer, it can be hypothesized that ZNF692 may be involved in the tumorigenesis of COAD.
The present study demonstrated that ZNF692 expression was upregulated in COAD tissues and cells. High ZNF692 expression was correlated with lymph node metastasis, distant metastasis and tumor stage in patients with COAD. In addition, ZNF692 knockdown attenuated cell proliferation, migration and invasion in vitro and in vivo; conversely, ZNF692 upregulation produced the opposite effects. Flow cytometric analysis revealed that ZNF692 knockdown induced G1-phase cell cycle arrest in COAD cells, whereas ZNF692 overexpression promoted G1/S phase transition. Furthermore, the present results demonstrated that ZNF692 acts via regulating cyclin D1, cyclin-dependent kinase 2 (CDK2), p27Kip1 and matrix metalloproteinase-9 (MMP-9) expression, through activation of the phosphoinositide 3-kinase (PI3K)/AKT serine/threonine kinase (AKT) signaling pathway. The PI3K/AKT inhibitor LY294002 markedly reversed ZNF692-induced cyclin D1 and MMP-9 expression. These results suggested that ZNF692 may have an important role in the progression and metastasis of COAD and might be a new therapeutic target in patients with this disease.
Materials and methods
Bioinformatics analysis
The target gene was obtained by querying the UALCAN portal (http://ualcan.path.uab.edu.). The detailed procedures for querying the UALCAN portal have been previously described (10). TCGA RNA-seq V2 exon data (level 3, raw count) and clinical information of COAD patients were downloaded from the TCGA data portal (https://tcga-data.nci.nih.gov/). The data were compared with the annotation information of the mRNA chromosomal location of the Gencode (v25) database (https://www.gencodegenes.org/). The raw count was normalized to log-counts per million (CPM) values using the R package edgeR (19) (http://www.bioconductor.org/packages/release/bioc/html/edgeR.html). The detailed procedures for bioinformatics analyses have been described in a previous study (11). Tumor staging was based on the guidelines of the American Joint Committee on Cancer (AJCC) staging system, 7th edition (20). Data for 326 samples (285 tumor samples and 41 normal tissue samples) were downloaded. Of these, 194 patients who did not undergo radiotherapy prior to surgery with complete information (age, sex, pathologic stage, pathology T/N/M stage) were included in the present study. The clinicopathological features of these patients are presented in Table I.
Table I.
Clinicopathological features of 194 patients with colon adenocarcinoma.
| Characteristics | Value |
|---|---|
| Age, years (mean ± standard deviation) | 63.94±13.29 |
| Sex (male/female) | 105/89 |
| Pathologic_T (T1/T2/T3/T4) | 5/34/128/27 |
| Pathologic_N (N0/N1/N2) | 118/51/25 |
| Pathologic_M (M0/M1) | 158/36 |
| Stage (I/II/III/IV) | 37/76/45/36 |
Patients
The study was approved by the Ethics Committee of the First Affiliated Hospital of Harbin Medical University (Harbin, China). Written informed consent was obtained from all subjects. A total of eight fresh COAD tissues and paired adjacent normal tissues were collected from patients who underwent colon resection from March 2017 to June 2017 at The First Affiliated Hospital of Harbin Medical University. The mean age of the patients was 63.5 years (range, 42–78 years), and the male-to-female ratio was 3:5. All tissues were snap-frozen and stored at −80°C until use. None of the participants received chemotherapy or radiotherapy prior to surgery.
Cell culture and reagents
The human intestinal epithelial cell line NCM460 and COAD cell lines HCT116, LoVo and DLD-1 were obtained from Shanghai Fuheng Biological Technology Co., Ltd. (Shanghai, China). NCM460 and DLD-1 cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Thermo Fisher Scientific, Inc., Waltham, MA, USA). LoVo cells were cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.), and HCT116 cells were cultured in McCoy's 5A medium (Thermo Fisher Scientific, Inc.). All culture media were supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) and antibiotics (100 U/ml penicillin and 100 mg/ml streptomycin). The cells were incubated at 37°C in a humidified chamber containing 5% CO2. The PI3K inhibitor LY294002 was obtained from Abcam (cat. no. ab146593; Abcam, Cambridge, UK).
Lentivirus-mediated RNA interference and transfection of overexpression plasmids
The lentiviral vectors were designed and produced by GeneChem Co., Ltd. (Shanghai, China), and transfections were performed following the manufacturer's protocol. The short hairpin RNA (shRNA) sequences were as follows: Lv-sh-ZNF692 #1, TTGTCATCCACAGACGTAT; Lv-sh-ZNF692 #2, AGTATTTGAATCACCACAA; and Lv-sh-ZNF692 #3, AGATTTGGAATCTGAGCAT. The sequence of the negative control shRNA (Lv-NC) was TTC TCCGAACGTGTCACGT. In addition, the full-length human ZNF692 cDNA was inserted into the pcDNA3.1 vector to generate the overexpressing pcDNA ZNF692 construct. A total of 1×106 HCT116 cells were seeded in 6-well plates and cultured for 24 h to 80–90% confluence. Then, the cells were transfected with negative control pcDNA (4 µg/well) or pcDNA ZNF692 (4 µg/well) by using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). At 48 h post-transfection, the cells were harvested and the transfection efficiency was evaluated by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and western blotting.
Immunohistochemistry (IHC)
Briefly, formalin-fixed and paraffin-embedded tumor tissues were cut into 4 µm-thick sections. The sections were deparaffinized by xylene, rehydrated in a graded series of ethanol, incubated with 3% H2O2 for 30 min, blocked with 3% bovine serum albumin for 1 h, and incubated with an anti-Ki-67 antibody (polyclonal, rabbit anti-human IgG; 1:50; cat. no. WL0280a; Wanleibio Co., Ltd., Shenyang, China) at 4°C overnight. The next day, the tissue sections were incubated with a secondary antibody (biotin-labeled goat anti-rabbit IgG; 1:200; cat. no. A0277; Beyotime Institute of Biotechnology, Shanghai, China) and streptavidin-HRP (HRP-labeled streptavidin; 1:200; cat. no. A0303; Beyotime Institute of Biotechnology) at room temperature for 30 min. Color was developed using a 3,3′-diaminobenzidine (DAB) Substrate kit (cat. no. DA1010; Beijing Solarbio Science and Technology Co., Ltd., Beijing, China). The tissue sections were counterstained with hematoxylin. IHC analysis was performed as previously described (21).
RNA extraction and RT-qPCR
Total RNA was extracted from tissues or cells using a total RNA extraction kit (cat. no. AP-MNMS-RNA-250; Axygen BioScience Inc., Union City, CA, USA) following the manufacturer's instructions. RNA was reverse transcribed to first-strand cDNA using ReverTra Ace qPCR RT Master Mix with gDNA Remover (cat. no. FSQ-301; Toyobo Co., Ltd., Osaka, Japan). Quantitative detection of ZNF692 was performed using cDNA with the THUNDERBIRD SYBR qPCR Mix (cat. no. QPS-201; Toyobo Co., Ltd.) with an ABI 7500 Fast system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The following thermal cycling protocol was used: heating at 95°C for 60 sec, followed by denaturation at 95°C for 15 sec, annealing at 60°C for 30 sec and elongation at 72°C for 60 sec. The sequences of the forward and reverse primers used for reactions were as follows: ZNF692, forward, 5′-CTCCAACCGGCA GTAT TTG-3′ and reverse, 5′-GTGCTCCTTCAGGTG TTTCTT-3′; and GAPDH, forward, 5′-GCTCTCTGCTCCT CCTGTTC-3′ and reverse, 5′-ACGACCAAATCCGTTGA CTC-3′. The primers were provided by Sangon Biotech (Shanghai, China). The relative expression level of ZNF692 was calculated using the comparative cycle threshold (2−ΔΔCq) method (22). GAPDH was used as the internal control.
Cell proliferation analysis
Cell proliferation was monitored using Cell Counting Kit-8 (CCK-8; cat. no. HY-K0301; MedChemExpress Monmouth Junction, NJ, USA) assays. DLD-1 and LoVo cells transfected with Lv-sh-ZNF692 #1 (3,000 cells/well) and HCT116 cells transfected with pcDNA ZNF692 were seeded in a 96-well plate for 24 h and cultured in normal medium. Next, 10 µl CCK-8 assay solution was added to each well once daily for 4 days, and the cells were incubated for 2 h after addition of CCK-8 solution, according to the manufacturer's instructions. The relative number of surviving cells was assessed by measuring the absorbance at a wavelength of 450 nm.
Colony formation assays
For colony formation assays, cells were seeded in 6-well plates at a density of 500 cells per well and cultured at 37°C for 14 days. The colonies formed were fixed with methyl alcohol and stained with 0.1% crystal violet for 20 min. The number of visible colonies was counted manually. The experiment was performed in triplicate.
Flow cytometric analysis of the cell cycle
This analysis was conducted with a cell cycle detection reagent kit (cat. no. FXP0211; 4A Biotech, Beijing, China). Transfected cells and corresponding control cells were seeded in 6-well plates and cultured for 24 h to 100% confluence. After washing with PBS, the cells were digested using 0.25% trypsin and then collected and washed 2 times with 1 ml precooled PBS. The cells were fixed with 0.5% paraformaldehyde for 15 min and permeabilized with precooled 70% ethanol at 4°C overnight. The cells were centrifuged at 60 × g for 10 min, and the ethanol was removed. Finally, 40 µg/ml propidium iodide (PI) solution and 2.5 mg/ml RNase A were added to the cells, mixed in the dark and left at 37°C for 30 min. The DNA content of the cells was quantified by flow cytometry (FACSCalibur; BD Biosciences, Franklin Lakes, NJ, USA), and the data were processed using ModFit LT software (Verity Software House Inc., Topsham, ME, USA).
Wound healing assay
Transfected cells and corresponding control cells (1×106 cells) were seeded in 6-well plates and cultured for 24 h to 90% confluence. Then, wounds were created using a 200 µl plastic pipette tip, and images were acquired at 0 and 36 h after wounding using a microscope. The speed of wound closure in multiple fields reflected the migratory ability of the tumor cells. The assay was repeated three times.
Migration and invasion assays
Transfected cells and the corresponding control cells (5×104 cells) suspended in 200 µl of serum-free medium were cultured in the top chamber with or without BD Matrigel (BD Biosciences) coating, while 700 µl of base medium supplemented with 10% FBS was added to the lower chamber. After 24 h, the cells were fixed with methanol, and the cells in the upper chamber were swabbed. The cells in the lower chamber were subsequently stained with a 0.1% crystal violet solution and imaged. Five random fields (magnification, ×100) were observed by microscopy, and the average number of cells in the five fields was used for quantitative analysis. The experiment was repeated three times.
Immunofluorescence (IF)
For IF, formalin-fixed and paraffin-embedded tumor tissues were cut into 5 µm thick sections. The sections were deparaffinized by xylene for 15 min, rehydrated in 95, 85 and 75% ethanol for 1 min, respectively. Then the slices were washed with PBS and blocked with 10% normal goat serum. After 15 min, the slices were incubated with an anti-ZNF692 antibody (polyclonal, rabbit anti-human IgG; 1:100; cat. no. NBP2-56800; Novus Biologicals, Littleton, CO, USA) overnight at 4°C. The slices were subsequently incubated with Cy3-tagged fluorescent secondary antibodies (1:200; cat. no. A0516; Beyotime Institute of Biotechnology) for 1 h in the dark. Then, the slices were washed with PBS for 15 min and counterstained with DAPI, and the IF signals were visualized with a fluorescence microscope (DP73; Olympus Corporation, Tokyo, Japan).
Western blotting
Total protein was extracted from the samples using radioimmunoprecipitation assay (RIPA) lysis buffer (cat. no. P0013B; Beyotime Institute of Biotechnology) with protease and phosphatase inhibitors. Protein concentration was measured with a Bradford protein assay kit (cat. no. P0012S; Beyotime Institute of Biotechnology). Equal amounts of protein (40 µg/lane) were separated by 10% SDS-PAGE (cat. no. P0012A; Beyotime Institute of Biotechnology) and transferred onto polyvinylidene difluoride membranes (Millipore, Billerica, MA, USA). The membranes were blocked with 5% non-fat milk for 1 h at room temperature and then incubated with the following antibodies overnight at 4°C: anti-ZNF692 (polyclonal, rabbit anti-human IgG; 1:1,000; cat. no. NBP2-56800; Novus Biologicals), anti-p27 (polyclonal, rabbit anti-human IgG; 1:500; cat. no. WL01769; Wanleibio Co., Ltd.), anti-cyclin D1 (polyclonal, rabbit anti-human IgG; 1:800; cat. no. WL01580; Wanleibio Co., Ltd.), anti-CDK2 (polyclonal, rabbit anti-human IgG; 1:1000; cat. no. WL02028; Wanleibio Co., Ltd.), anti-AKT (polyclonal, rabbit anti-human IgG; 1:500; cat. no. WL01619; Wanleibio Co., Ltd.), anti-phsphorylated (p-) AKT(Ser473) (polyclonal, rabbit anti-human IgG; 1:500; cat. no. WLP001a; Wanleibio Co., Ltd.), anti-MMP-9 (polyclonal, rabbit anti-human IgG; 1:800; cat. no. WL01580; Wanleibio Co., Ltd.), anti-extracellular signal-regulated kinase (ERK) 1/2 (polyclonal, rabbit anti-human IgG; 1:500; cat. no. WL02195; Wanleibio Co., Ltd.), anti-p-ERK1/2 (polyclonal, rabbit anti-human IgG; 1:500; cat. no. WL02368; Wanleibio Co., Ltd.), anti-c-Jun N-terminal kinase (JNK; polyclonal, rabbit anti-human IgG; 1:500; cat. no. WL01295; Wanleibio Co., Ltd.), anti-p-JNK (polyclonal, rabbit anti-human IgG; 1:500; cat. no. WL01813; Wanleibio Co., Ltd.), anti-GAPDH (monoclonal, mouse anti-human IgG; 1:1,000; cat. no. KC-5G4; KangChen Biotech, Shanghai, China) and anti-β-actin (polyclonal, mouse anti-human IgG; 1:1,000; cat. no. TA-09; ZSGB-BIO, Beijing, China). The membranes were subsequently incubated with anti-rabbit secondary antibody (peroxidase-conjugated goat anti-rabbit IgG; 1:10,000; cat. no. ZB-2301; ZSGB-BIO) or anti-mouse secondary antibody (peroxidase-conjugated goat anti-mouse IgG; 1:10,000; cat. no. ZB-2305; ZSGB-BIO), and protein signals were visualized by enhanced chemiluminescence detection with a Bio-Rad gel imaging system (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Experimental mouse model
A total of 10 six-week-old female BALB/c nude mice (20 g) were obtained from Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China) and maintained under specific pathogen-free conditions. The experimental protocol was reviewed and approved by the Institutional Animal Care and Use Committee of Harbin Medical University (Harbin, China). Briefly, 1×106 cells stably transfected with DLD-1-Lv-NC or DLD-1-Lv-sh-ZNF692 #1 in 0.1 ml PBS were subcutaneously injected into the back of nude mice. Tumor length (L) and width (W) were measured every 5 days, and the tumor volume was calculated using the following formula: volume = L×W2×0.5. After 4 weeks, the mice were sacrificed, and tumor volume and weight were measured.
Statistical analysis
Data are presented as the mean ± standard deviation and were analyzed using IBM SPSS Statistics, version 23.0 (IBM Corp., Armonk, NY, USA). GraphPad Prism 6 (GraphPad Software, La Jolla, CA, USA) was applied to generate graphs. Statistical significance was determined using the Chi-square test and two-tailed Student's t-test. Multiple groups were analyzed using one-way analysis of variance followed by the Least Significance Difference (LSD) post hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
ZNF692 expression is upregulated in COAD cell lines and tissues
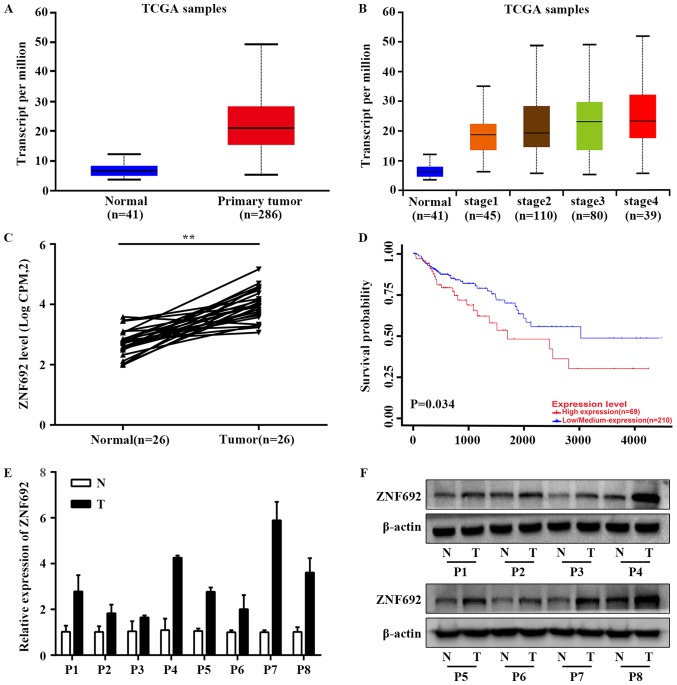
Based on TCGA RNA-seq data available from UALCAN, expression analysis demonstrated that the mean expression values of ZNF692 were significantly higher in COAD tissues compared with adjacent normal tissues (P<0.001; Fig. 1A and B), consistent with previous research results from our group (11) (P<0.001; Fig. 1C). The Kaplan-Meier plot of overall survival (OS) in patients with COAD provided by UALCAN revealed that COAD patients with higher ZNF692 expression had poorer prognoses compared with patients with lower ZNF692 expression (P<0.05; Fig. 1D). This association was also evident in three other types of cancer, namely, adrenocortical carcinoma, renal clear cell carcinoma and hepatocellular carcinoma (Fig. S1). Furthermore, ZNF692 was highly expressed in thirteen other types of cancer, including bladder urothelial carcinoma, LUAD, renal clear cell carcinoma, prostate adenocarcinoma, lung squamous cell carcinoma, head and neck squamous cell carcinoma, renal papillary cell carcinoma, esophageal carcinoma, hepatocellular carcinoma, rectal adenocarcinoma, uterine corpus endometrial carcinoma, cholangiocarcinoma, and gastric adenocarcinoma (Fig. S2). We thus speculated that ZNF692 is highly expressed in COAD and has an important role in COAD development. Therefore, ZNF692 expression was further validated in 8 paired COAD and adjacent normal tissues and the results demonstrated that ZNF692 mRNA and protein expression was markedly upregulated in COAD tissue samples compared with the matched adjacent normal tissue samples (Fig. 1E and F).
Figure 1.
ZNF692 is upregulated in COAD patient tissues. (A) ZNF692 is overexpressed in COAD tissues in a TCGA COAD dataset available from UALCAN. (B) ZNF692 expression in COAD cases based on individual cancer stage in a TCGA COAD dataset available from UALCAN. (C) Correlation of ZNF692 expression in COAD tissue specimens (n=26) and matched normal tissues (n=26) in the downloaded TCGA dataset. (D) Kaplan–Meier plot of OS of patients with COAD stratified by ZNF692 expression from the data available from UALCAN. (E) Reverse transcription-quantitative polymerase chain reaction and (F) western blotting assays confirmed the ZNF692 overexpression in patient COAD tissues compared with adjacent normal tissues. GAPDH and β-actin were used as endogenous controls. The experiments were performed three times, and the data are presented as the mean ± standard deviation. **P<0.01. ZNF692, zinc finger protein 692; COAD, colon adenocarcinoma; TCGA, The Cancer Genome Atlas; OS, overall survival; T, tumor tissue; N, normal tissue.
Higher ZNF692 expression is correlated with advanced clinicopathological features in COAD patients
To explore the role of ZNF692 in COAD, ZNF692 mRNA expression was examined in human COAD tissues (194 samples) downloaded from TCGA. The clinicopathological data of all these patients are listed in Table I. The relative expression levels of ZNF692 in tumor tissues were categorized as low or high relative to the median value of ZNF692 expression. As presented in Table II, a Chi-square test indicated that increased ZNF692 levels correlated significantly with lymph node metastasis (N0 vs N1-N3, P<0.05), distant metastasis (M0 vs M1, P<0.01) and tumor stage (I-II vs III-IV, P<0.01). Other clinicopathological characteristics, such as age, sex and tumor invasion depth, were not significant.
Table II.
Association between the expression of ZNF692 and clinicopathological features in 194 patients with colon adenocarcinoma.
| Characteristics | ZNF692 expression
|
χ2 | P-value | |
|---|---|---|---|---|
| High | Low | |||
| Sex | 1.682 | 0.195 | ||
| Male | 57 | 48 | ||
| Female | 40 | 49 | ||
| Age, years | 1.065 | 0.302 | ||
| ≤60 | 34 | 41 | ||
| >60 | 63 | 56 | ||
| Tumor invasion depth | 0.289 | 0.591 | ||
| T1–T2 | 18 | 21 | ||
| T3–T4 | 79 | 76 | ||
| Lymph node metastasis | 5.538 | 0.019a | ||
| N0 | 51 | 67 | ||
| N1–N3 | 46 | 30 | ||
| Distant metastasis | 8.731 | 0.003b | ||
| M0 | 71 | 87 | ||
| M1 | 26 | 10 | ||
| TNM stages | 7.651 | 0.006b | ||
| I–II | 47 | 66 | ||
| III–IV | 50 | 31 | ||
P<0.05
P<0.01 by Chi-square test. ZNF692, zinc finger protein 692.
Silencing ZNF692 suppresses the proliferative ability of COAD cells in vitro
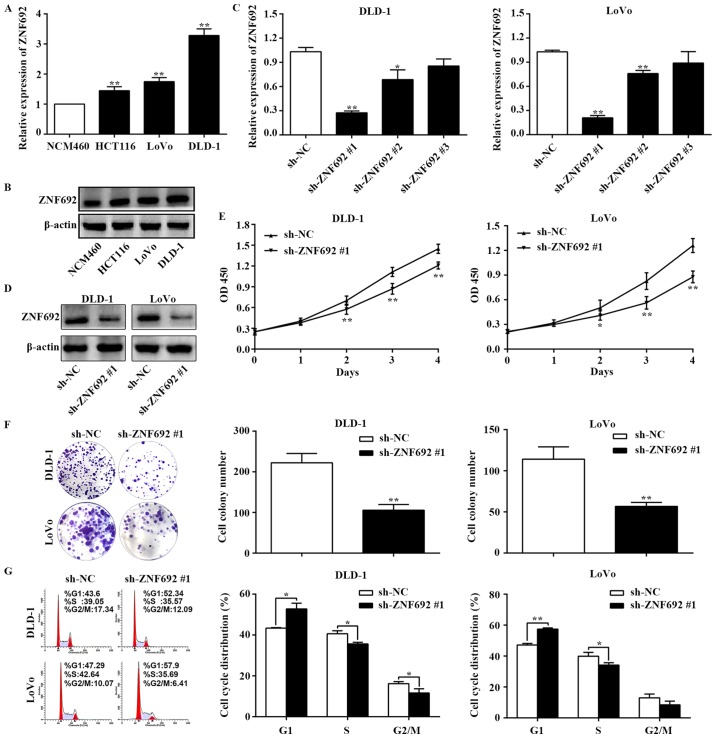
As expected, ZNF692 expression was upregulated in COAD cell lines compared with NCM460 normal colon epithelial cells (Fig. 2A and B). The DLD-1 and LoVo cell lines were selected for further experiments, since they exhibited the highest expressions levels of ZNF692. To gain insights into the biological functions of ZNF692 in COAD cells, Lv-sh-NC and Lv-sh-ZNF692 #1, #2, and #3 lentiviral constructs were introduced into DLD-1 and LoVo cells. Knockdown of ZNF692 was confirmed by RT-qPCR and western blotting (Fig. 2C and D). Lv-sh-ZNF692 #1 was selected for further study because it resulted in the highest knockdown efficiency (P<0.01). To assess the effect of ZNF692 on the proliferative ability of COAD cells, CCK-8 and colony formation assays were performed. The results of these assays demonstrated that ZNF692 knockdown cells displayed reduced viability and colony formation efficiency (P<0.01; Fig. 2E and F). To determine the pro-proliferative effect of ZNF692, flow cytometric analysis of the cell cycle was performed. As illustrated in Fig. 2G, ZNF692 silencing resulted in G1 arrest in COAD cells (P<0.05).
Figure 2.
ZNF692 knockdown inhibits the proliferation and colony formation of COAD cells. (A) mRNA and (B) protein expression levels of ZNF692 were evaluated by RT-qPCR and western blotting, respectively, in NCM460 cells and COAD cell lines. (C) RT-qPCR and (D) western blot analysis of the transfection efficiency of Lv-shRNA-ZNF692 in DLD-1 and LoVo cells. (E) CCK-8 assays of sh-NC- and sh-ZNF692 #1-transfected DLD-1 and LoVo cells. (F) Analysis of the colony formation capacity of sh-NC- and sh-ZNF692 #1-transfected DLD-1 and LoVo cells. (G) Flow cytometry analysis of the cell cycle phase distribution of DLD-1 and LoVo cells following the indicated transfections. The experiments were performed three times, and the data are presented as the mean ± standard deviation. *P<0.05 and **P<0.01 vs sh-NC. ZNF692, zinc finger protein 692; COAD, colon adenocarcinoma; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; CCK-8, Cell Counting Kit-8; shRNA, short hairpin RNA; NC, negative control; OD, optical density.
ZNF692 knockdown inhibits the migration and invasion of COAD cells in vitro
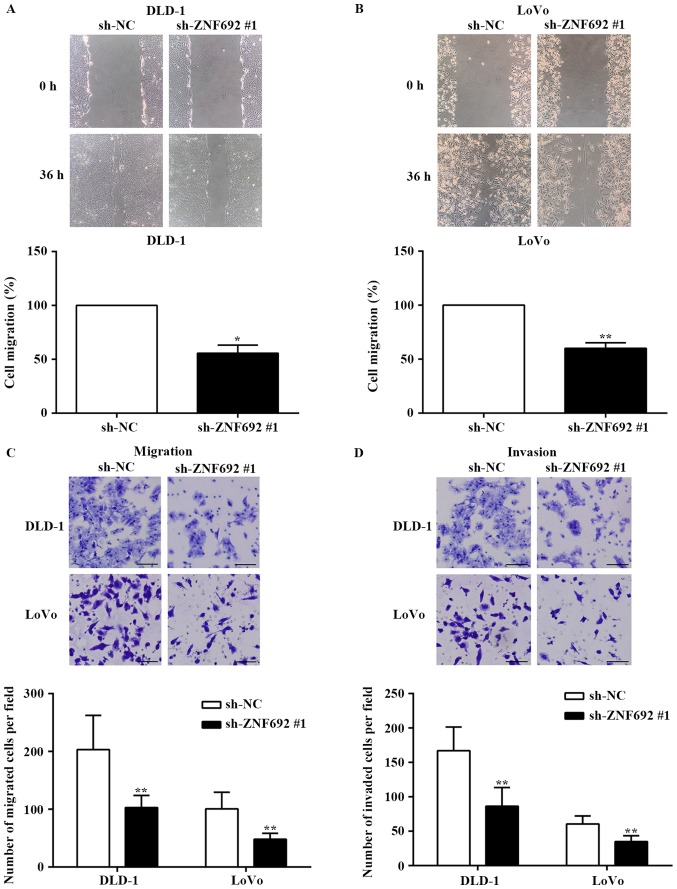
To examine the effect of ZNF692 on cell migration, wound healing assays and Transwell migration assays were employed. The results demonstrated that ZNF692 knockdown decreased the migration of DLD-1 (P<0.05) and LoVo cells (P<0.01) compared with the control group (Fig. 3A and C). Furthermore, Transwell invasion assays revealed that the invasive potential was also significantly decreased in DLD-1 and LoVo cells transfected with sh-ZNF692 #1, compared with cells transfected with the sh-NC control (P<0.01; Fig. 3D).
Figure 3.
ZNF692 knockdown inhibits the migration and invasion of COAD cells. (A) Wound healing assays revealed that ZNF692 knockdown reduced the migration of DLD-1 and (B) LoVo cells. (C) Transwell migration and (D) invasion assays revealed that ZNF692 knockdown reduced the migration and invasion of DLD-1 and LoVo cells. Magnification, ×100. Scale bar, 100 µm. The experiments were performed three times, and the data are presented as the mean ± standard deviation. *P<0.05 and **P<0.01 vs sh-NC. ZNF692, zinc finger protein 692; COAD, colon adenocarcinoma; sh, short hairpin RNA; NC, negative control.
ZNF692 promotes COAD cell proliferation, migration and invasion in vitro
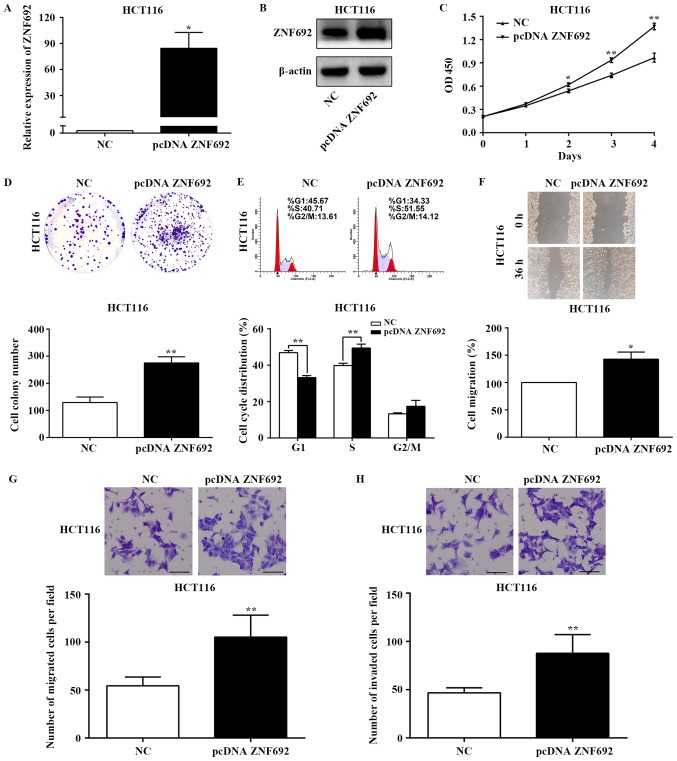
Having confirmed the inhibitory effects of ZNF692 knockdown in COAD cells, HCT116 cells, which exhibit low levels of ZNF692 (Fig. 1), were employed next in order to overexpress ZNF692 and to evaluate the effects on cell proliferation, migration, and invasion. The transfection efficiency of the ZNF692 overexpression constructs was confirmed by RT-qPCR and western blot analysis (P<0.05; Fig. 4A and B). Functional analyses demonstrated that ZNF692 overexpression significantly increased proliferation (P<0.01; Fig. 4C), colony formation (P<0.01; Fig. 4D), cell cycle progression to the S phase (P<0.01; Fig. 4E), migration (P<0.05; Fig. 4F and G) and invasion (P<0.01; Fig. 4H) in HCT116 cells. These results demonstrate that ZNF692 promotes COAD cell proliferation, migration and invasion in vitro.
Figure 4.
ZNF692 overexpression promotes the growth, migration and invasion of colon adenocarcinoma cells. HCT116 cells were transfected with either the pcDNA-ZNF692 overexpressing vector or an empty vector control. (A) RT-qPCR and (B) western blot analysis of the transfection efficiency of pcDNA-ZNF692 in HCT116 cells. (C) Proliferation and (D) colony formation capability were estimated by CCK-8 and colony formation assays, respectively. (E) Flow cytometry showed the distribution of pcDNA-ZNF692-transfected HCT116 cells in the G1, S, and G2/M phases of the cell cycle. (F) Cell migration ability was assessed by wound healing and (G) Transwell migration assays. (H) Invasion was assessed by the Transwell invasion assay in pcDNA-ZNF692-transfected HCT116 cells and control cells. Magnification, ×100. Scale bar, 100 µm. The experiments were performed three times, and the data are presented as the mean ± standard deviation. *P<0.05 and **P<0.01 vs NC. ZNF692, zinc finger protein 692; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; CCK-8, Cell Counting Kit-8; NC, negative control; OD, optical density.
Effect of ZNF692 knockdown on tumor growth in vivo
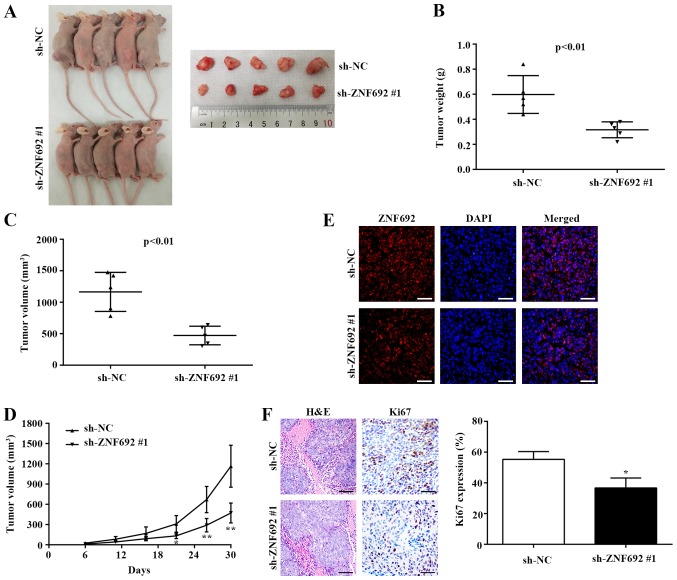
The role of ZNF692 in tumorigenicity was further explored in vivo. To this end, DLD-1 cells were stably transfected with Lv-NC or Lv-sh-ZNF692 #1 and subcutaneously injected into the back of nude mice. As presented in Fig. 5A, the tumors arising from cells transfected with sh-ZNF692 #1 were markedly smaller compared with those arising from cells transfected with sh-NC. Quantification of the tumor weights and volumes also demonstrated that the Lv-NC group tumors were significantly larger compared with the sh-ZNF692 #1 group (P<0.01; Fig. 5B and C). In addition, the xenograft tumor growth curve indicated that the tumors from the sh-ZNF692 #1-transfected group grew substantially more slowly, in a time-dependent manner, compared with the tumors from the sh-NC group (Fig. 5D). IF analysis results revealed that ZNF692 localized to the nucleus and that ZNF692 expression was markedly decreased in tumors arising from cells transfected with sh-ZNF692 #1 (Fig. 5E). According to IHC analysis, ZNF692 knockdown significantly decreased the expression of the cell proliferation marker Ki-67 (Fig. 5F).
Figure 5.
ZNF692 knockdown inhibits xenograft tumor growth in nude mice. (A) ZNF692 knockdown reduced DLD-1 cell-derived xenograft tumor growth in nude mice (n=5). (B) Statistical comparison of differences in tumor weights and (C) volumes between the sh-NC and sh-ZNF692 #1 groups. (D) Growth curve of DLD-1 xenograft tumors monitored in the sh-NC and sh-ZNF692 #1 groups. (E) Expression levels and nuclear localization of ZNF692 protein were determined using immunofluorescence in xenograft tumor tissues in the sh-NC and sh-ZNF692 #1 groups. Magnification, ×400. Scale bar, 50 µm. (F) HE staining (magnification, ×200; scale bar, 100 µm) and immunohistochemistry analysis of Ki-67 expression (magnification, ×400; scale bar, 50 µm) in xenograft tumor tissues in the sh-NC and sh-ZNF692 #1 groups. The data are presented as the mean ± standard deviation. *P<0.05 and **P<0.01 vs sh-NC. ZNF692, zinc finger protein 692; sh, short hairpin; NC, negative control; HE, hematoxylin and eosin.
ZNF692 promotes COAD cell proliferation, migration and invasion by regulating cyclin D1, p27 and MMP-9 expression through the PI3K/AKT pathway
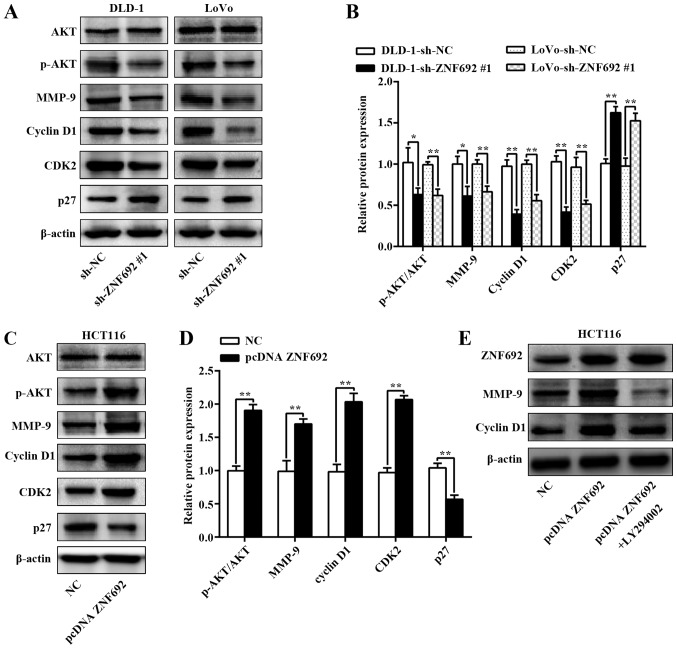
To further explore the molecular mechanisms by which ZNF692 mediates biological behavior, the PI3K/AKT and mitogen-activated protein kinase pathways were assessed using western blot analysis in COAD cells with ZNF692 knockdown or overexpression. As illustrated in Fig. 6A-D, ZNF692 knockdown decreased AKT phosphorylation in DLD-1 (P<0.05) and LoVo (P<0.01) cells, but the opposite results were obtained when ZNF692 was upregulated in HCT116 cells (P<0.01). However, total AKT expression was not affected. No changes were observed in p-ERK1/2 or p-JNK expression, indicating that ZNF692 promoted the PI3K/AKT pathway independently (Fig. S3). Then, several key PI3K/AKT downstream molecules were explored, namely cyclin D1, CDK2, p27Kip1 and MMP-9. As illustrated in Fig. 6A-D, cyclin D1, CDK2 and MMP-9 protein expression was significantly inhibited and p27Kip1 protein expression was upregulated in ZNF692-silenced COAD cells; ZNF692 overexpression produced the opposite effect in HCT116 cells. Additionally, LY294002 was used to investigate whether ZNF692 regulating COAD cells depends on PI3K/AKT signaling. As presented in Fig. 6E, cyclin D1 and MMP-9 expression induced by ectopic ZNF692 expression was significantly reversed following treatment of HCT116 cells with LY294002 (20 µM).
Figure 6.
ZNF692 is involved in regulating the PI3K/AKT signaling pathway. (A) Representative images and (B) quantification from western blot analysis of the effects of ZNF692 knockdown on the PI3K/AKT pathway and the downstream factors cyclin D1, CDK2, p27 and MMP-9 in DLD-1 and LoVo cells. (C) Representative images and (D) quantification from western blot analysis of the effects of ZNF692 overexpression on the PI3K/AKT pathway and the downstream transcription factors cyclin D1, CDK2, p27 and MMP-9 in HCT116 cells. (E) Western blot analysis demonstrated that LY294002 effectively decreased the expression levels of cyclin D1 and MMP-9 induced by ZNF692 overexpression. The experiments were performed three times, and the data are presented as the mean ± standard deviation. *P<0.05 and **P<0.01 with comparisons indicated by brackets. ZNF692, zinc finger protein 692; PI3K; phosphoinositide 3-kinase, AKT, AKT serine/threonine kinase; CDK, cyclin-dependent kinase; MMP, matrix metalloproteinase; p-, phosphorylated; sh, short hairpin; NC, negative control.
Discussion
ZNF692 was first reported to have an important role in gluconeogenesis by Inoue and Yamauchi (23), who demonstrated that the transcription factor ZNF692 is phosphorylated at Ser470 by AMP-activated protein kinase (AMPK) and then represses expression of phosphoenolpyruvate carboxykinase (PEPCK), a key enzyme in gluconeogenesis (23,24). PEPCK is reportedly downregulated in human colon cancer cell lines and is associated with 5-fluorouracil (5-FU) resistance and cell proliferation (25,26). Huang et al (27) recently performed gene expression analysis and reported that ZNF692 is involved in the relapse of Wilms tumors. Zhang et al (28) demonstrated that ZNF692 expression is elevated in LUAD tissues, and ZNF692 downregulation suppresses LUAD cell proliferation, migration and invasion in vitro and inhibits the tumorigenicity of LUAD cells in vivo. However, the function of ZNF692 in carcinogenesis and tumor progression in COAD has remained unclear.
In the current study, ZNF692 expression was investigated in human COAD tissues and cell lines and compared with paired normal tissues from the patients and a human intestinal epithelial cell line, respectively. The results revealed that all COAD tissues and cell lines exhibited high ZNF692 expression compared with the respective controls. To the best of our knowledge, the present study is the first to demonstrate that ZNF692 is upregulated in COAD tissues and cell lines, which is consistent with the results of the TCGA data analysis. In addition, ZNF692 expression was significantly correlated with lymph node metastasis, distant metastasis and tumor stage in COAD patients. As high ZNF692 expression was associated with an aggressive tumor phenotype in COAD, it was speculated that ZNF692 may have an important role in tumor biology. Therefore, functional in vitro and in vivo experiments were conducted to investigate the role of ZNF692 in COAD cell growth, migration and invasion. As expected, the results revealed that ZNF692 knockdown suppressed COAD cell proliferation, migration and invasion and reduced xenograft tumor growth, whereas ZNF692 overexpression enhanced cell proliferation, migration and invasion. Furthermore, ZNF692 inhibited COAD cell growth by inducing G1 phase arrest. Therefore, the present observations strongly suggest that ZNF692 functions as an oncogene in COAD and may be a novel prognostic indicator for this disease.
To explore the molecular mechanism through which ZNF692 contributes to cell proliferation in COAD, potential target proteins in cell cycle regulation were investigated. The cell cycle is divided into four phases and is regulated by a series of checkpoints involving cyclins and CDKs (29,30). Entry into the G1 phase from the G0 phase is dependent on the cyclin D1-CDK4/CDK6 complex (30,31), whereas the cyclin E/CDK2 complex serves an important role in the transition from the G1 phase to the S phase (32). In the present study, ZNF692 expression was up- or downregulated and then cell cycle-related protein expression was probed. Western blot analysis revealed that cyclin D1 and CDK2 expression levels were reduced or elevated following the downregulation or upregulation of ZNF692, respectively. The present results demonstrated that ZNF692 blocked cell cycle progression in the G1 phase by altering the expression levels of cyclin D1 and CDK2 in COAD cells. p27Kip1 is a member of the kinase inhibitor protein (KIP) family, and many studies have reported that p27Kip1 blocks cell cycle progression by inhibiting the activity of cyclin-CDK complexes (33,34). The current western blot results indicated that ZNF692 silencing significantly increased the expression of p27Kip1. Furthermore, ZNF692 overexpression decreased p27Kip1 levels. These data suggest that p27Kip1 may be a major downstream effector of ZNF692.
The PI3K/AKT pathway is one of the most frequently deregulated pathways in cancer (35-37). PI3K transduces various signals, such as growth factors and cytokines, from the extracellular matrix (ECM) into the intracellular environment, which in turn results in the phosphorylation of AKT (38,39). Multiple studies have reported that the PI3K/AKT pathway can enhance cancer cell proliferation via the induction of cyclin D1 and CDK2 expression and repression of p27Kip1 (40-42). Thus, the present study examined the effects of ZNF692 on the PI3K/AKT pathway. The results demonstrated that sh-ZNF692 #1 significantly decreased p-AKT levels in DLD-1 and LoVo cells, but did not affect total AKT protein expression. However, ectopic overexpression of ZNF692 increased p-AKT protein expression. Therefore, these findings indicated that ZNF692 may have an oncogenic role in COAD by promoting the upregulation of cyclin D1 and CDK2 and the downregulation of p27Kip1 through the PI3K/AKT pathway. This hypothesis was also supported by the addition of LY294002, which dramatically reversed the ZNF692-induced cyclin D1 expression.
Invasion and metastasis are predominant characteristics of cancer and the greatest challenge in its clinical management (43,44). In the present study, the in vitro functional experiments wound healing assays and Transwell assays were employed, and the results demonstrated that the migration and invasion capabilities of COAD cells were closely dependent to the ZNF692 expression levels. These results are in line with the clinical findings that ZNF692 correlates significantly with lymph node metastasis and distant metastasis. It was thus speculated that ZNF692 may have an important role in the invasion and metastasis of COAD. MMPs are key enzymes that degrade the ECM barrier, enabling cancer cells to invade and metastasize (45). MMP-9, also known as gelatinase-B, is well known for its role in basement membrane degradation (46). Multiple studies have reported that MMP-9 is associated with tumor invasion and metastasis (47–49). Accumulating data have demonstrated that MMP-9 functions downstream of the PI3K/AKT pathway to regulate tumor cell migration and invasion (50,51). To decipher the molecular mechanism through which ZNF692 contributes to cell migration and invasion, the MMP-9 protein expression levels were analyzed by western blotting, in the presence or absence of LY294002. The results suggested that ZNF692 increased the migration and invasion of COAD cells potentially by increasing MMP-9 expression via the PI3K/AKT pathway.
Despite the noteworthy findings, the present study has several limitations to be addressed in future research. First, the TCGA RNA-seq data on ZNF692 prognosis analysis should be further confirmed by RT-qPCR at the level of transcription in another set of COAD patients with complete follow-up information. Second, the protein expression levels of ZNF692 in COAD tissues should be detected by IHC to confirm the association between clinicopathological features and ZNF692 expression.
In summary, the present findings revealed that ZNF692 expression is significantly upregulated in COAD tissues and cell lines and that high ZNF692 expression is significantly associated with lymph node metastasis, distant metastasis and tumor stage. Furthermore, ZNF692 promoted COAD cell proliferation, migration and invasion via the PI3K/AKT pathway. Thus, ZNF692 may serve as a novel oncogene and a potential treatment target in patients with COAD.
Supplementary Materials
Acknowledgments
Not applicable.
Funding
This work was supported by the Scientific Planning Project of Heilongjiang Province (grant no. 201713) and the Medical Science and Technology Project of Heilongjiang Province (grant no. 2004-093)
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
DP, YX and YZ conceived and designed the research, YX, SR, ZZ and LA conducted the experiments, FJ and WS were involved in the analysis and interpretation of data, YX was involved in drafting the manuscript, ZZ and WS revised the manuscript for important intellectual content. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The study involving the use of human tissue was approved by the Ethics Committee of the First Affiliated Hospital of Harbin Medical University (Harbin, China). Written informed consent was obtained from all subjects. Protocols involving the use of animals were reviewed and approved by the Institutional Animal Care and Use Committee of Harbin Medical University (Harbin, China). All applicable international, national, and/or institutional guidelines for the care and use of animals were followed. All procedures performed in the studies were in accordance with the 1964 Declaration of Helsinki and its later amendments.
Patient consent for publication
Informed consent was signed by the patients and/or guardians.
Competing interests
The authors declare that they have no conflicts of interest.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 3.Ahnen DJ, Wade SW, Jones WF, Sifri R, Mendoza Silveiras J, Greenamyer J, Guiffre S, Axilbund J, Spiegel A, You YN. The increasing incidence of young-onset colorectal cancer: A call to action. Mayo Clin Proc. 2014;89:216–224. doi: 10.1016/j.mayocp.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 4.Rasool S, Kadla SA, Rasool V, Ganai BA. A comparative overview of general risk factors associated with the incidence of colorectal cancer. Tumour Biol. 2013;34:2469–2476. doi: 10.1007/s13277-013-0876-y. [DOI] [PubMed] [Google Scholar]
- 5.Ross JS, Torres-Mora J, Wagle N, Jennings TA, Jones DM. Biomarker-based prediction of response to therapy for colorectal cancer: Current perspective. Am J Clin Pathol. 2010;134:478–490. doi: 10.1309/AJCP2Y8KTDPOAORH. [DOI] [PubMed] [Google Scholar]
- 6.Rahmani F, Avan A, Hashemy SI, Hassanian SM. Role of Wnt/β-catenin signaling regulatory microRNAs in the pathogenesis of colorectal cancer. J Cell Physiol. 2018;233:811–817. doi: 10.1002/jcp.25897. [DOI] [PubMed] [Google Scholar]
- 7.Mei ZB, Duan CY, Li CB, Cui L, Ogino S. Prognostic role of tumor PIK3CA mutation in colorectal cancer: A systematic review and meta-analysis. Ann Oncol. 2016;27:1836–1848. doi: 10.1093/annonc/mdw264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen H, Xu J, Hong J, Tang R, Zhang X, Fang JY. Long noncoding RNA profiles identify five distinct molecular subtypes of colorectal cancer with clinical relevance. Mol Oncol. 2014;8:1393–1403. doi: 10.1016/j.molonc.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Staub E, Gröne J, Mennerich D, Röpcke S, Klamann I, Hinzmann B, Castanos-Velez E, Mann B, Pilarsky C, Brümmendorf T, et al. A genome-wide map of aberrantly expressed chromosomal islands in colorectal cancer. Mol Cancer. 2006;5:37. doi: 10.1186/1476-4598-5-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK, Varambally S. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xing Y, Zhao Z, Zhu Y, Zhao L, Zhu A, Piao D. Comprehensive analysis of differential expression profiles of mRNAs and lncRNAs and identification of a 14-lncRNA prognostic signature for patients with colon adenocarcinoma. Oncol Rep. 2018;39:2365–2375. doi: 10.3892/or.2018.6324. [DOI] [PubMed] [Google Scholar]
- 12.Dahmane N, Lee J, Robins P, Heller P, Ruiz i Altaba A. Activation of the transcription factor Gli1 and the Sonic hedgehog signalling pathway in skin tumours. Nature. 1997;389:876–881. doi: 10.1038/39918. [DOI] [PubMed] [Google Scholar]
- 13.Nonet GH, Stampfer MR, Chin K, Gray JW, Collins CC, Yaswen P. The ZNF217 gene amplified in breast cancers promotes immortalization of human mammary epithelial cells. Cancer Res. 2001;61:1250–1254. [PubMed] [Google Scholar]
- 14.Imoto I, Yuki Y, Sonoda I, Ito T, Shimada Y, Imamura M, Inazawa J. Identification of ZASC1 encoding a Krüppel-like zinc finger protein as a novel target for 3q26 amplification in esophageal squamous cell carcinomas. Cancer Res. 2003;63:5691–5696. [PubMed] [Google Scholar]
- 15.Jiang R, Wang JC, Sun M, Zhang XY, Wu H. Zinc finger X-chromosomal protein (ZFX) promotes solid agar colony growth of osteosarcoma cells. Oncol Res. 2012;20:565–570. doi: 10.3727/096504013X13775486749290. [DOI] [PubMed] [Google Scholar]
- 16.Lai KP, Chen J, He M, Ching AK, Lau C, Lai PB, To KF, Wong N. Overexpression of ZFX confers self-renewal and chemoresistance properties in hepatocellular carcinoma. Int J Cancer. 2014;135:1790–1799. doi: 10.1002/ijc.28819. [DOI] [PubMed] [Google Scholar]
- 17.Fang X, Huang Z, Zhou W, Wu Q, Sloan AE, Ouyang G, McLendon RE, Yu JS, Rich JN, Bao S. The zinc finger transcription factor ZFX is required for maintaining the tumorigenic potential of glioblastoma stem cells. Stem Cells. 2014;32:2033–2047. doi: 10.1002/stem.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan X, Shan Z, Yan L, Zhu Q, Liu L, Xu B, Liu S, Jin Z, Gao Y. High expression of Zinc-finger protein X-linked promotes tumor growth and predicts a poor outcome for stage II/III colorectal cancer patients. Oncotarget. 2016;7:19680–19692. doi: 10.18632/oncotarget.7547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th Edition of the AJCC Cancer Staging Manual and the Future of TNM. Ann Surg Oncol. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 21.Wang X, Sun D, Tai J, Chen S, Yu M, Ren D, Wang L. TFAP2C promotes stemness and chemotherapeutic resistance in colorectal cancer via inactivating hippo signaling pathway. J Exp Clin Cancer Res. 2018;37:27. doi: 10.1186/s13046-018-0683-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Inoue E, Yamauchi J. AMP-activated protein kinase regulates PEPCK gene expression by direct phosphorylation of a novel zinc finger transcription factor. Biochem Biophys Res Commun. 2006;351:793–799. doi: 10.1016/j.bbrc.2006.10.124. [DOI] [PubMed] [Google Scholar]
- 24.Shirai T, Inoue E, Ishimi Y, Yamauchi J. AICAR response element binding protein (AREBP), a key modulator of hepatic glucose production regulated by AMPK in vivo. Biochem Biophys Res Commun. 2011;414:287–291. doi: 10.1016/j.bbrc.2011.08.120. [DOI] [PubMed] [Google Scholar]
- 25.Blouin JM, Bortoli S, Nacfer M, Collinet M, Penot G, Laurent-Puig P, Forest C. Down-regulation of the phosphoenolpyruvate carboxykinase gene in human colon tumors and induction by omega-3 fatty acids. Biochimie. 2010;92:1772–1777. doi: 10.1016/j.biochi.2010.07.011. [DOI] [PubMed] [Google Scholar]
- 26.Park JW, Kim SC, Kim WK, Hong JP, Kim KH, Yeo HY, Lee JY, Kim MS, Kim JH, Yang SY, et al. Expression of phosphoenol-pyruvate carboxykinase linked to chemoradiation susceptibility of human colon cancer cells. BMC Cancer. 2014;14:160. doi: 10.1186/1471-2407-14-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang CC, Gadd S, Breslow N, Cutcliffe C, Sredni ST, Helenowski IB, Dome JS, Grundy PE, Green DM, Fritsch MK, et al. Predicting relapse in favorable histology Wilms tumor using gene expression analysis: A report from the Renal Tumor Committee of the Children's Oncology Group. Clin Cancer Res. 2009;15:1770–1778. doi: 10.1158/1078-0432.CCR-08-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Q, Zheng X, Sun Q, Shi R, Wang J, Zhu B, Xu L, Zhang G, Ren B. ZNF692 promotes proliferation and cell mobility in lung adenocarcinoma. Biochem Biophys Res Commun. 2017;490:1189–1196. doi: 10.1016/j.bbrc.2017.06.180. [DOI] [PubMed] [Google Scholar]
- 29.Kaproth-Joslin KA, Li X, Reks SE, Kelley GG. Phospholipase C delta 1 regulates cell proliferation and cell-cycle progression from G1- to S-phase by control of cyclin E-CDK2 activity. Biochem J. 2008;415:439–448. doi: 10.1042/BJ20080233. [DOI] [PubMed] [Google Scholar]
- 30.Sherr CJ. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 31.Bansal R, Marin-Husstege M, Bryant M, Casaccia-Bonnefil P. S-phase entry of oligodendrocyte lineage cells is associated with increased levels of p21Cip1. J Neurosci Res. 2005;80:360–368. doi: 10.1002/jnr.20454. [DOI] [PubMed] [Google Scholar]
- 32.Graña X, Reddy EP. Cell cycle control in mammalian cells: Role of cyclins, cyclin dependent kinases (CDKs), growth suppressor genes and cyclin-dependent kinase inhibitors (CKIs) Oncogene. 1995;11:211–219. [PubMed] [Google Scholar]
- 33.Bagui TK, Mohapatra S, Haura E, Pledger WJ. P27Kip1 and p21Cip1 are not required for the formation of active D cyclin-cdk4 complexes. Mol Cell Biol. 2003;23:7285–7290. doi: 10.1128/MCB.23.20.7285-7290.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Slingerland JM, Hengst L, Pan CH, Alexander D, Stampfer MR, Reed SI. A novel inhibitor of cyclin-Cdk activity detected in transforming growth factor beta-arrested epithelial cells. Mol Cell Biol. 1994;14:3683–3694. doi: 10.1128/MCB.14.6.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whitman M, Kaplan DR, Schaffhausen B, Cantley L, Roberts TM. Association of phosphatidylinositol kinase activity with polyoma middle-T competent for transformation. Nature. 1985;315:239–242. doi: 10.1038/315239a0. [DOI] [PubMed] [Google Scholar]
- 36.Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7:606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- 37.Wood LD, Parsons DW, Jones S, Lin J, Sjöblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 38.Simpson DR, Mell LK, Cohen EE. Targeting the PI3K/AKT/mTOR pathway in squamous cell carcinoma of the head and neck. Oral Oncol. 2015;51:291–298. doi: 10.1016/j.oraloncology.2014.11.012. [DOI] [PubMed] [Google Scholar]
- 39.Luo J, Manning BD, Cantley LC. Targeting the PI3K-Akt pathway in human cancer: Rationale and promise. Cancer Cell. 2003;4:257–262. doi: 10.1016/S1535-6108(03)00248-4. [DOI] [PubMed] [Google Scholar]
- 40.Lenferink AE, Busse D, Flanagan WM, Yakes FM, Arteaga CL. ErbB2/neu kinase modulates cellular p27(Kip1) and cyclin D1 through multiple signaling pathways. Cancer Res. 2001;61:6583–6591. [PubMed] [Google Scholar]
- 41.Wierød L, Rosseland CM, Lindeman B, Oksvold MP, Grøsvik H, Skarpen E, Huitfeldt HS. CDK2 regulation through PI3K and CDK4 is necessary for cell cycle progression of primary rat hepatocytes. Cell Prolif. 2007;40:475–487. doi: 10.1111/j.1365-2184.2007.00451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang L, Cao XX, Chen Q, Zhu TF, Zhu HG, Zheng L. DIXDC1 targets p21 and cyclin D1 via PI3K pathway activation to promote colon cancer cell proliferation. Cancer Sci. 2009;100:1801–1808. doi: 10.1111/j.1349-7006.2009.01246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gupta GP, Massagué J. Cancer metastasis: Building a framework. Cell. 2006;127:679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 44.Stacker SA, Achen MG, Jussila L, Baldwin ME, Alitalo K. Metastasis: Lymphangiogenesis and cancer metastasis. Nature Reviews Cancer. 2002;2:573. doi: 10.1038/nrc863. [DOI] [PubMed] [Google Scholar]
- 45.Deryugina EI, Quigley JP. Matrix metalloproteinases and tumor metastasis. Cancer Metastasis Rev. 2006;25:9–34. doi: 10.1007/s10555-006-7886-9. [DOI] [PubMed] [Google Scholar]
- 46.Rundhaug JE. Matrix metalloproteinases, angiogenesis, and cancer: commentary re: A. C. Lockhart et al., Reduction of wound angiogenesis in patients treated with BMS-275291, a broad spectrum matrix metalloproteinase inhibitor. Clin. Cancer Res. 2003;9:00–00. [PubMed] [Google Scholar]; Clin Cancer Res. 2003;9:551–554. [PubMed] [Google Scholar]
- 47.Aparna M, Rao L, Kunhikatta V, Radhakrishnan R. The role of MMP-2 and MMP-9 as prognostic markers in the early stages of tongue squamous cell carcinoma. J Oral Pathol Med. 2015;44:345–352. doi: 10.1111/jop.12245. [DOI] [PubMed] [Google Scholar]
- 48.Morgia G, Falsaperla M, Malaponte G, Madonia M, Indelicato M, Travali S, Mazzarino MC. Matrix metalloproteinases as diagnostic (MMP-13) and prognostic (MMP-2, MMP-9) markers of prostate cancer. Urol Res. 2005;33:44–50. doi: 10.1007/s00240-004-0440-8. [DOI] [PubMed] [Google Scholar]
- 49.Herszényi L, Hritz I, Lakatos G, Varga MZ, Tulassay Z. The behavior of matrix metalloproteinases and their inhibitors in colorectal cancer. Int J Mol Sci. 2012;13:13240–13263. doi: 10.3390/ijms131013240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee CW, Lin CC, Lin WN, Liang KC, Luo SF, Wu CB, Wang SW, Yang CM. TNF-α induces MMP-9 expression via activation of Src/EGFR, PDGFR/PI3K/Akt cascade and promotion of NF-kappaB/p300 binding in human tracheal smooth muscle cells. Am J Physiol Lung Cell Mol Physiol. 2007;292:L799–L812. doi: 10.1152/ajplung.00311.2006. [DOI] [PubMed] [Google Scholar]
- 51.Yoo YA, Kang MH, Lee HJ, Kim BH, Park JK, Kim HK, Kim JS, Oh SC. Sonic hedgehog pathway promotes metastasis and lymphangiogenesis via activation of Akt, EMT, and MMP-9 pathway in gastric cancer. Cancer Res. 2011;71:7061–7070. doi: 10.1158/0008-5472.CAN-11-1338. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.