Abstract
The cancer susceptibility candidate 9 (CASC9) gene has been reported to exert an oncogenic effect in several types of cancer. However, its role in lung squamous cell carcinoma (LUSC) is unknown. Therefore, the present study examined the expression of CASC9 in LUSC and non-cancer tissues by reverse transcription-quantitative polymerase chain reaction assays and by data mining of high-throughput public databases, including The Cancer Genome Atlas, the Gene Expression Omnibus, ArrayExpress and the Cancer Cell Line Encyclopedia. In vitro experiments were conducted to investigate the effects of CASC9 on the viability and the proliferation of LUSC cells. Furthermore, consulting the alteration status of CASC9 in LUSC from cBioPortal, functional enrichment analysis of co-expressed genes, prediction of potential transcription factors, and inspection of adjacent protein-coding genes were conducted to explore the potential molecular mechanism of CASC9 in LUSC. The results revealed that CASC9 was overexpressed in LUSC tissue, and significantly associated with the malignant progression of LUSC. In vitro experiments demonstrated that CASC9 knockdown by RNA interference attenuated the viability and proliferation of LUSC cells. Multiple copies of CASC9 gene were detected in 4 of 179 available sequenced LUSC cases. A functional enrichment analysis of 200 co-expressed genes indicated that these genes were significantly associated with terms, including 'cell-cell junction organization', 'desmosome organization', 'epidermis development', 'Hippo signaling pathway', 'pathogenic Escherichia coli infection' and 'PID HIF1 TF pathway'. Three genes, Fos-related antigen 2 (FOSL2), SWI/SNF complex subunit SMARCC2, and transcription factor COE1 (EBF1), were predicted by lncRNAMap to be associated with CASC9. Among these, the expression of FOSL2 and EBF1 was positively and negatively correlated with the expression of CASC9, respectively. Two adjacent protein-coding genes, cysteine-rich secretory protein LCCL domain-containing 1 and hepatocyte nuclear factor 4-γ, were also positively correlated with CASC9 expression. In conclusion, the present data suggest that CASC9 serves as an oncogene in LUSC and may be a promising target for alternative therapeutic options for patients with this condition.
Keywords: cancer susceptibility candidate 9, lung squamous cell carcinoma, reverse transcription-quantitative polymerase chain reaction, integrated meta-analysis, in vitro experiments
Introduction
Lung cancer (LC) is one of the most common malignant tumors in the world (1,2). According to data compiled by the American Cancer Society, there were 234,030 estimated new LC cases and 154,050 estimated LC-associated mortalities in the USA in 2018 (3). In terms of histology, there are two types of LC: Non-small cell LC (NSCLC) and small cell LC, of which the former makes up 85% of all cases (4). NSCLC is comprised of two subtypes: Lung squamous cell carcinoma (LUSC) and lung adenocarcinoma (LUAD) (3,5). Drugs targeting mutated versions of the epidermal growth factor receptor (EGFR), GTPase KRas and ALK tyrosine kinase receptor proteins mutations have already demonstrated beneficial effects in patients with LUAD (6). However, these drugs are not applicable to LUSC, which is associated with poor survival rates (7), highlighting the urgent requirement for effective diagnostic and therapeutic targets for LUSC.
Long non-coding RNAs (lncRNAs) are a class of non-protein-coding RNAs that control gene expression in complex ways (8,9). Numerous studies have revealed that the ectopic expression of lncRNAs is implicated in human cancer (10-13). As a member of the lncRNA family, the cancer susceptibility candidate 9 (CASC9) gene, located on chromosome 8q21.11, was originally observed to be upregulated in esophageal squamous cell carcinoma (ESCC) (14). In addition, CASC9 was reported to constitute a crucial component in the tumorigenesis of other types of cancer, including gastric and nasopharyngeal cancer (15,16). With regard to NSCLC, recent studies have discovered that CASC9 was associated with the resistance of NSCLC cells to EGFR tyrosine kinase inhibitors, and that it served important roles in the proliferation and metastasis of LUAD cells (17,18). Although studies on CASC9 in NSCLC have reported promising results, they have mainly focused on LUAD, and studies on the role of CASC9 in LUSC are lacking.
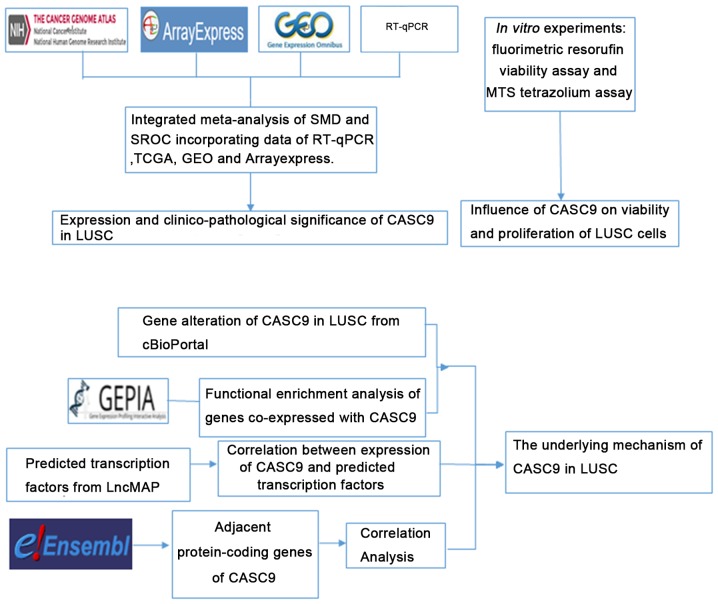
In the present study, the clinicopathological effect of CASC9 in LUSC was investigated using reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and data mining of public databases, including The Cancer Genome Atlas (TCGA), the Gene Expression Omnibus (GEO), ArrayExpress, and the Cancer Cell Line Encyclopedia (CCLE). Furthermore, the functional influence of CASC9 on LUSC, and the underlying mechanism, were explored through in vitro experiments, investigation of the mutation status of CASC9 in LUSC from cBioPortal, functional enrichment analysis of co-expressed genes using Gene Expression Profiling Interactive Analysis (GEPIA), prediction of potential transcription factors, and inspection of adjacent protein-coding genes. The design of the current study is illustrated in Fig. 1.
Figure 1.
Flowchart of the design of the current study. SMD, standardized mean difference; SROC, summarized receiver operating characteristic; TCGA, The Cancer Genome Atlas; GEO, Gene Expression Omnibus; LUSC, refers to lung squamous cell carcinoma; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; GEPIA, Gene Expression Profiling Interactive Analysis; CASC9, cancer susceptibility candidate 9.
Materials and methods
Tissue samples
A total of 20 patients first diagnosed with LUSC (age range, 35-68 years; mean, 51 years; 12 male and 8 female) attending the First Affiliated Hospital of Guangxi Medical University (Nanning, China) were included in the present study. The patients were receiving no medication and underwent radical resection of lung cancer between August 2017 and March 2018. Tumor and matched non-cancer lung tissues were obtained from these patients during the radical resection surgery. The samples were fixed in 10% buffered formalin under ambient temperature for 16 h and paraffin-embedded. All patients provided signed informed consent and approval of this study was granted by the Ethics Committee of the First Affiliated Hospital of Guangxi Medical University.
RT-qPCR
The isolation and relative quantification of RNA was conducted based on methods described previously (19). RT and qPCR kits were used to examine the CASC9 expression levels in LUSC and adjacent non-cancer tissues, following the manufacturers' protocols. qPCR was performed on an ABI 7900 Real-time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA) using the LightCycler® FastStart DNA Master plus SYBR Green kit (Roche Diagnostics, Basel, Switzerland). The denaturation, annealing and extension steps of the 40 PCR cycles were set at 95°C for 10 sec, 60°C for 5 sec and 72°C for 5 sec, following a 10-min hot start at 95°C. The primers for CASC9 and the reference gene GAPDH were as follows: CASC9 forward, 5′-AAAACCAGGTGGGACCCAGA-3′; reverse, 5′-TGATCAGAAGAAGAGGGGCA-3′; GAPDH forward, 5′-ACCCACTCCTCCACCTTTG-3′; and reverse, 5′-CTCTTGTGCTCTTGCTGGG-3′. CASC9 expression was calculated according to the formula: 2−ΔCq= 2−(CqCASC9-CqGAPDH), where the Cq value is the quantification cycle number (20).
Evaluation of the clinicopathological associations of CASC9 in LUSC, using TCGA data
TCGA (http://cancergenome.nih.gov/), an extensive reservoir of DNA methylation, exome sequencing, single nucleotide polymorphism array, RNA-seq and microRNA-seq data (21,22), assists researchers in conducting studies using complicated cancer genomics profiles. In the present study, log2(x+1)-transformed level 3 transcripts per million reads (TPM) RNA-seq data of CASC9 expression in 501 LUSC and 49 adjacent normal tissues as well as the clinicopathological data of the 501 patients from whom the samples were obtained (TCGA-LUSC), were collected from TCGA data portal (https://portal.gdc.cancer.gov/) and analyzed using SPSS version 22.0 (IBM Corp., Armonk, NY, USA).
Integrated meta-analysis of CASC9 expression in LUSC and non-cancer tissues
To comprehensively appraise the differential expression of CASC9 in LUSC and non-cancer tissues, expression data of CASC9 in LUSC and non-cancer tissues published before September 19, 2018 were searched in the GEO (https://www.ncbi.nlm.nih.gov/gds/), ArrayExpress (https://www.ebi.ac.uk/arrayexpress/) and literature databases, including Pubmed (https://www.ncbi.nlm.nih.gov/pubmed), Google Scholar (https://scholar.google.com/), Wiley Online Library (https://onlinelibrary.wiley.com/), Cochrane Library (https://www.cochranelibrary.com/library), Web of Science (http://apps.webofknowledge.com), Embase (https://www.embase.com), Ebsco (https://www.ebsco.com/), Chinese VIP (http://www.cqvip.com/), China National Knowledge Infrastructure (http://www.cnki.net/), Sinomed (http://www.sinomed.ac.cn/) and Wang Fang (http://www.wanfangdata.com.cn/index.html). The search terms used in the GEO were: ('Lung neoplasms'[Mesh]) AND ('microarray analysis'[Mesh] OR 'tissue array analysis'[Mesh] OR 'transcriptome'[Mesh] OR 'sequence analysis, RNA'[Mesh] OR 'high-throughput nucleotide sequencing'[Mesh]). The key words for searching in ArrayExpress were: ('Cancer' OR 'carcinoma' OR 'adenocarcinoma' OR 'tumour' OR 'tumor' OR 'malignanc*' OR 'neoplas*') AND ('lung' OR 'pulmonary' OR 'respiratory' OR 'respiration' OR 'aspiration' OR 'bronchi' OR 'bronchioles' OR 'alveoli' OR 'pneumocytes' OR 'air way'). Regarding the literature survey, the search terms used were as follows: ('CASC9' OR 'cancer susceptibility 9' OR 'ESSCAL1' OR 'ESCCAL-1' OR 'LINC0098') AND ('lung' OR 'pulmonary' OR 'respiratory' OR 'respiration' OR 'aspiration' OR 'bronchi' OR 'bronchioles' OR 'alveoli' OR 'pneumocytes' OR 'air way') AND ('cancer' OR 'carcinoma' OR 'adenocarcinoma' OR 'tumour' OR 'tumor' OR 'malignanc*' OR 'neoplas*'). Studies using human-derived samples offering sufficient CASC9 expression data (where the numbers of LUSC and non-cancer cases exceeded 3) in LUSC and non-cancer samples for the calculation of a standardized mean difference (SMD) were included. Basic information, as well as expression and diagnostic data, including accession ID, first author, publication year, country, experiment type, sample type, platform, number of cases in the cancer or non-cancer groups, mean ± SD of CASC9 expression in the cancer or non-cancer groups, true positives, false positives, false negatives and true negatives, were extracted from the included studies according to methods described previously (23). An SMD with the 95% confidence interval (CI) and summarized receiver operating characteristic (SROC) curves were produced for the integrated meta-analysis, consisting of the in-house RT-qPCR data, TCGA data and microarray chip data from ArrayExpress, as described previously (23).
Cell line data of CASC9 expression from CCLE
The expression data of CASC9 in all available LC cell lines was downloaded from CCLE (https://portals.broadinstitute.org/ccle/about) on October 5, 2018. The original data included 192 LC cell lines. Due to contamination or misidentification of the PC-14 cell line being reported by the International Cell Line Authentication Committee, Database of Cross-Contaminated or Misidentified Cell Lines (http://iclac.org/databases/cross-contaminations/), it was excluded from the present expression analysis. Finally, the expression data of CASC9 from a total of 191 cell lines were merged into a heat map using GraphPad Prism 7 (GraphPad Software, Inc., La Jolla, CA, USA).
Cell transfection and in vitro experiments
The human LUSC H226 cells were purchased from the American Type Culture Collection (Manassas, VA, USA), cultured in Dulbecco's modified Eagle's medium (Gibco; Thermo Fisher Scientific, Inc.) and supplemented with 10% fetal bovine serum (Biological Industries, Kibbutz Beit Haemek, Israel) and penicillin-streptomycin at 37°C under a humidified atmosphere of 5% CO2. Each in vitro experiment was performed in triplicate. The H226 cells were seeded into 96-well plates, at 2.5×103 cells per well, and incubated at 37°C for 24 h prior to transfection. Transfections of mock control, scrambled small interfering (si)RNAs, and CASC9 siRNAs (Ambion; Thermo Fisher Scientific, Inc.) were performed in H226 cells at a concentration of 200 nM for 96 h using the CombiMag Magnetofection™ transfection kit (OZBiosciences SAS, Marseille, France), according to the manufacturer's protocol. The siRNAs were designed for the best efficiency to knockdown CASAC9 using the online tool InvivoGen siRNA Wizard Software version 3.1 (https://www.invivogen.com/sirnawizard/index.php), and 4 siRNAs were finally selected. Scrambled siRNAs were included to serve as negative controls for each of these. The siRNAs sequences are listed in Table SI. The 4 selected CASC9 siRNAs were pooled for use in subsequent experiments, as were the scrambled siRNAs. To investigate the influence of CASC9 on the viability and proliferation of H226 cells, fluorimetric resorufin viability and MTS tetrazolium assays were conducted as described previously (24-27).
cBioPortal gene alteration of CASC9 in LUSC tissue
The gene alteration status of CASC9 in LUSC (dataset TCGA, provisional) was acquired from cBioPortal version 2.0.1 (http://www.cbioportal.org) (28). The distribution of the CASC9 alteration in 179 sequenced patients with LUSC was visualized using the OncoPrint module of cBioPortal.
Functional enrichment analysis of co-expressed genes
Genes co-expressed with CASC9 were identified from the GEPIA. The biological functions of the co-expressed genes were investigated via the enrichment annotation modules in Metascape (http://metascape.org) (Kyoto Encyclopedia of Genes and Genomes Pathway, GO Biological Processes, Reactome Pathway Database, Canonical Pathways, and CORUM). Terms with P<0.01 and the number of enriched genes >3 were considered to be significantly associated with the genes.
Prediction of potential transcription factors for CASC9
Potential transcription factors associated with CASC9 were identified by referring to a series of databases, including starBase (http://starbase.sysu.edu.cn/), TransmiR (http://www.cuilab.cn/transmir), TRED (http://rulai.cshl.edu/TRED), ITFP (https://omictools.com/itfp-tool), TFe (http://www.cisreg.ca/tfe), AnimalTFDB (http://bioinfo.life.hust.edu.cn/AnimalTFDB/) and LncMAP (http://bio-bigdata.hrbmu.edu.cn/LncMAP/). HT-Seq TPM RNA-seq data of predicted transcription factors in LUSC were downloaded from TCGA, and Pearson's correlation analysis was performed in GraphPad Prism 7 to evaluate the correlation between them and CASC9 expression.
Correlations between CASC9 and adjacent protein-coding genes
Protein-coding genes adjacent to CASC9 were searched using the Ensemble Genome Browser (http://grch37.ensembl.org/index.html) and the HT-Seq TPM RNA-seq data of these genes in LUSC were downloaded from TCGA. The association between CASC9 expression and that of the identified adjacent protein-coding genes was analyzed by Pearson's correlation tests in GraphPad Prism 7.
Statistical analysis
The statistical analyses for the RT-qPCR and TCGA data were performed in SPSS version 22.0. The expression values of CASC9 in LUSC and non-cancer tissues are presented as the mean ± SD. Paired t-tests were conducted to compare the expression of CASC9 between LUSC and non-cancer tissues, as derived from the RT-qPCR assays. Regarding the clinicopathological effect of CASC9 expression in LUSC from TCGA data, the Mann-Whitney test was employed to evaluate the differential expression of CASC9 in two subgroups of clinicopathological variables. When a clinicopathological variable contained >3 subgroups, the Kruskal-Wallis test was performed. To assess the ability of CASC9 to distinguish LUSC from non-cancer tissue, ROC curves were created. The discernment capacity of CASC9 for LUSC increased with the area under the curves (AUC) varying from 0.5-1.0. The impact of high versus low CASC9 expression (divided by the average CASC9 expression value) on the survival rate of patients with LUSC was evaluated using the Kaplan-Meier survival curves. Multivariate Cox regression analysis was performed to judge whether CASC9 expression could serve as a prognostic indicator for LUSC independent of clinical variables. Two-way analysis of variance and Bonferroni post-tests were performed for the comparison of the groups in the fluorimetric resorufin viability and MTS tetrazolium assays. P<0.05 was considered to indicate statistically significant differences.
Results
RT-qPCR
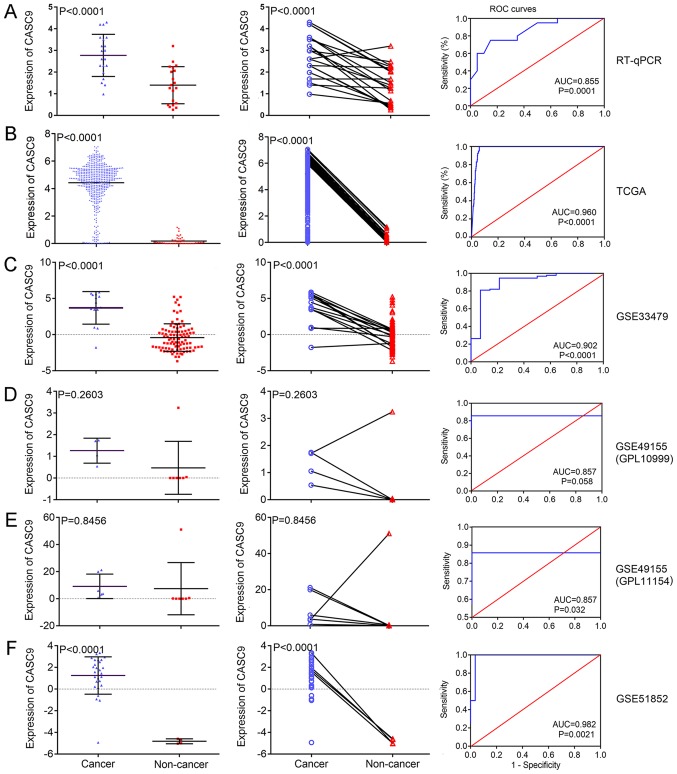
Fig. 2 illustrates the finding that the expression level of CASC9 in LUSC tissues (relative expression, 2.771±0.974) was significantly higher than that in paired, non-cancer tissues (relative expression, 1.397±0.857) (P<0.001; Fig. 2A). ROC curves with an AUC value of 0.855 indicated the diagnostic ability of CASC9 expression levels to distinguish LUSC tissues from non-cancer tissues (P<0.001; Fig. 2A, right panel).
Figure 2.
CASC9 expression and its diagnostic ability in LUSC for all the investigated datasets. The scatter plots (left panels) and wiring diagrams (middle panels) present the differential expression levels of CASC9 in LUSC samples and non-cancer samples for (A) RT-qPCR, (B) TCGA and GEO microarray (C) GSE33479, (D) GSE49155 (GPL10999), (E) GSE49155 (GPL11154) and (F) GSE51852 data. The ROC curves (right panels) demonstrate the capacity of CASC9 to identify LUSC from normal samples. CASC9, cancer susceptibility candidate 9; LUSC, lung squamous cell carcinoma; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; TCGA, The Cancer Genome Atlas; GEO, Gene Expression Omnibus; ROC, receiver operating characteristic; AUC, area under the curve.
Evaluation of the clinicopathological influence of CASC9 in LUSC from TCGA data
Compared with adjacent normal lung tissue, CASC9 was significantly upregulated in LUSC tissue (relative expression, 0.190±0.282 vs. 4.420±1.528; P<0.001; Fig. 2B). Apart from the differential expression between LUSC and normal tissue, overexpression of CASC9 in LUSC was observed to be significantly associated with several clinicopathological parameters of LUSC, including sex (P=0.029), clinical stage (P<0.001), distant metastasis (P=0.004) and lymph node metastasis (P=0.039) (Table I). Male patients and patients with advanced clinical stage (III-IV), distant metastasis and lymph node metastasis exhibited notably higher expression of CASC9. No significant associations were observed between the expression levels of CASC9 and other clinicopathological variables. The ROC curves in Fig. 2 suggest that the measurement of CASC9 performed well in discriminating LUSC from normal tissue (AUC=0.960; P<0.001). There was no significant difference between the survival rates of patients divided into high- and low-expression groups according to the average CASC9 expression value (P=0.189; Fig. S1) and the multivariate Cox regression analysis revealed that age (HR, 0.480; 95% CI, 0.269-0.855; P=0.013) and tumor location (HR, 1.850; 95% CI, 1.080-3.169; P=0.025) were independent prognostic factors for LUSC (Table II).
Table I.
Clinicopathological variables and CASC9 expression in lung squamous cell carcinoma data from The Cancer Genome Atlas.
| Clinicopathological feature | n | CASC9 expression
|
P-value | |
|---|---|---|---|---|
| Mean ± SD | z-score | |||
| Tissue type | 10.921 | <0.001 | ||
| Cancer | 501 | 4.420±1.528 | ||
| Normal | 49 | 0.190±0.282 | ||
| Sex | −2.181 | 0.029 | ||
| Male | 370 | 4.503±1.472 | ||
| Female | 131 | 4.187±1.627 | ||
| Age, yearsa | −0.187 | 0.852 | ||
| >60 | 405 | 4.072±1.864 | ||
| <60 | 87 | 4.045±1.923 | ||
| Tumor location | −0.929 | 0.353 | ||
| Central | 145 | 4.179±1.845 | ||
| Peripheral | 94 | 3.911±1.964 | ||
| Stage | −4.040 | <0.001 | ||
| I-II | 412 | 3.987±1.885 | ||
| III-IV | 85 | 4.795±1.396 | ||
| Distant metastasis | −2.843 | 0.004 | ||
| M0-MX | 489 | 4.043±1.889 | ||
| M1-M1b | 7 | 5.691±0.770 | ||
| Lymph node metastasis | −2.064 | 0.039 | ||
| N0-NX | 325 | 4.328±1.546 | ||
| N1-N3 | 176 | 4.592±1.457 | ||
| Primary tumor stage | −1.077 | 0.281 | ||
| T1-T2 | 407 | 4.397±1.521 | ||
| T3-T4 | 94 | 4.524±1.517 | ||
| Anatomical classification | 3.253 | 0.661 | ||
| Bronchial | 10 | 4.387±1.586 | ||
| L-lower | 77 | 4.356±1.772 | ||
| L-upper | 134 | 4.292±1.628 | ||
| R-lower | 107 | 4.118±1.834 | ||
| R-middle | 16 | 3.636±2.277 | ||
| R-upper | 139 | 3.834±2.048 | ||
Data available for 492 cases. Comparisons were performed using a Mann-Whitney test, with the exception of the anatomical classification, which was analyzed using a Kruskal-Wallis test. CASC9, cancel susceptibility candidate 9; SD, standard deviation; L, left; R, right.
Table II.
Multivariate Cox regression analysis of clinical parameters and CASC9 expression in TCGA cohort of patients with lung squamous cell carcinoma.
| Variablea | HR (95% CI) | P-value |
|---|---|---|
| CASC9 expression (high vs. low) | 1.308 (0.790-2.168) | 0.297 |
| Sex | 1.177 (0.681-2.034) | 0.560 |
| Age, years | 0.480 (0.269-0.855) | 0.013 |
| Tumor location | 1.850 (1.080-3.169) | 0.025 |
| Tumor stage | 1.886 (0.991-3.590) | 0.053 |
| Distant metastasis | 2.382 (0.321-17.689) | 0.396 |
| Lymph node metastasis | 0.842 (0.492-1.441) | 0.529 |
| Primary tumor stage | 1.582 (0.843-2.971) | 0.153 |
| Anatomical subdivision | 1.024 (0.874-1.201) | 0.768 |
The subcategories of the variables being compared are as listed in Table I. CASC9, cancer susceptibility candidate 9; HR, hazard ratio; CI, confidence interval.
Integrated meta-analysis for the expression difference of CASC9 between LUSC and non-cancer tissue
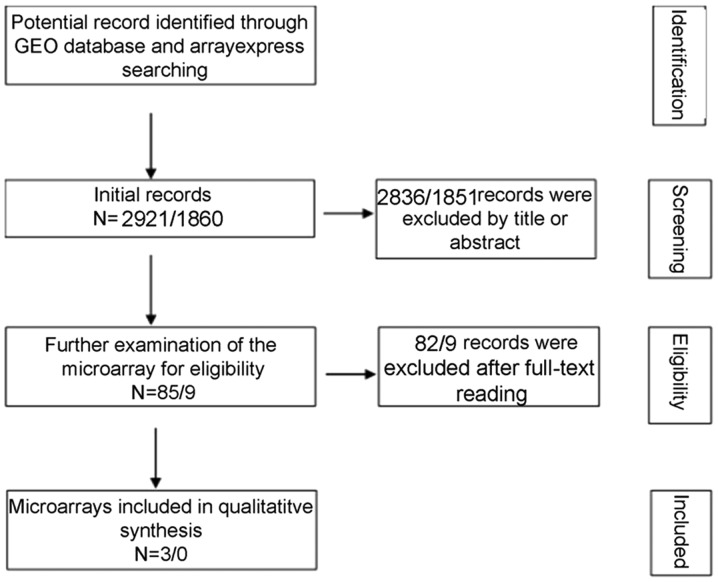
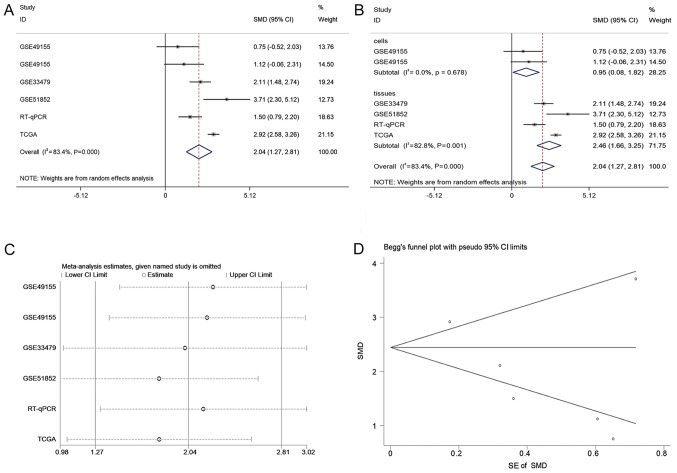
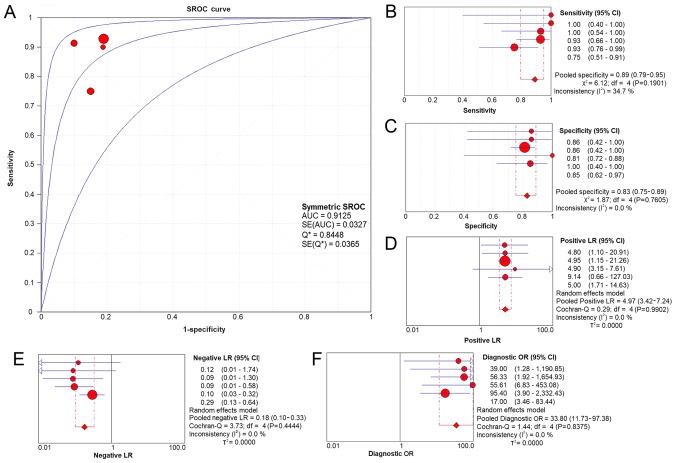
A total of 2,921 and 1,860 studies were identified in the initial searches in GEO and ArrayExpress, respectively. Of these, 85 GEO microarray chips and 9 ArrayExpress chips were included following the scanning of titles and abstracts. Finally, 3 GSE datasets, GSE49155, GSE33479 and GSE51852, were included (29,30). The distribution of CASC9 expression and its ability to discriminate between LUSC and non-cancer tissue in each GSE dataset are displayed in Fig. 2. A flowchart of the selection process for appropriate microarray chips is illustrated in Fig. 3. No study from the literature survey was included in the final selection. The in-house RT-qPCR, TCGA and the included GEO microarray data were merged into a large pool containing 574 LUSC cases and 182 non-cancer cases for the integrated meta-analysis. Information on the three cohorts is listed in Table III. As demonstrated in Fig. 4A, the SMD generated from all the cohorts verified the overexpression of CASC9 in LUSC tissues, albeit with significant heterogeneity (SMD, 2.04; 95% CI, 1.27-2.81; I2=83.4%; P<0.001). A subgroup analysis based on sample type and a sensitivity analysis failed to trace the source of the heterogeneity (Fig. 4B and C). No heterogeneity was observed in two studies on patient LUSC cells (I2=0.0%; P=0.678), in contrast with the large heterogeneity from the tissue data (I2=82.8%; P=0.001). The reason for this is that the two studies with different platforms in the cells subgroup were from the same GSE dataset (GSE49155). Begg's funnel plot indicated no publication bias (P=0.348). According to the evaluation with the SROC curves in Fig. 5, a high AUC (0.9125) revealed the marked ability of CASC9 in distinguishing LUSC from non-cancer tissues. The aggregated sensitivity, specificity, positive likelihood ratio, negative likelihood ratio and diagnostic odds ratio were 0.89, 0.83, 4.97, 0.18 and 33.80, respectively (Fig. 5B-F).
Figure 3.
Flowchart of the integrated meta-analysis data selection. A total of 2,921 and 1,860 studies appeared as the initial records from the GEO and ArrayExpress, respectively. Following the screening of titles and abstracts, 2,836 and 1,851 studies from GEO and ArrayExpress, respectively, were excluded. Finally, 3 GSE datasets were enrolled for the meta-analysis following the process of full-text reviewing. GEO, Gene Expression Omnibus.
Table III.
Basic, expression and diagnostic information of all datasets.
| Cancer
|
Normal
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dataset | Sample type | Study | Sequencing or microarray platform | n | Mean ± SD CASC9 expression | n | Mean ± SD CASC9 expression | TP | FP | FN | TN | Refs. |
| GSE49155 | Cells | Ooi et al, 2014 | GPL10999 | 4 | 1.263±0.578 | 7 | 0.470±1.223 | 4 | 1 | 0 | 6 | (31) |
| GSE49155 | Cells | Ooi et al, 2014 | GPL11154 | 6 | 5.976±7.863 | 7 | 0.028±0.070 | 6 | 1 | 0 | 6 | (31) |
| GSE33479 | Tissues | N/A | GPL6480 | 14 | 3.706±2.256 | 95 | −0.415±1.908 | 13 | 18 | 1 | 77 | |
| GSE51852 | Tissues | Arima et al, 2014 | GPL6480 | 28 | 1.255±1.725 | 4 | −4.824±0.231 | 26 | 0 | 2 | 4 | (32) |
| RT-qPCR | Tissues | Present study | N/A | 20 | 2.771±0.974 | 20 | 1.397±0.857 | 15 | 3 | 5 | 17 | - |
| TCGA | Tissues | N/A | Seq-TPM | 502 | 4.422±1.518 | 49 | 0.185±0.282 | 465 | 0 | 37 | 49 | - |
N/A, not applicable; SD, standard deviation; TP, true positive; FP, false positive; FN, false negative; TN, true negative; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; TPM, transcripts per million reads.
Figure 4.
Integrated meta-analysis for CASC9 expression in lung squamous cell carcinoma. (A) The forest plot for the integrated meta-analysis. (B) Forest plot of subgroup analysis based on sample type. (C) Sensitivity analysis. (D) Begg's funnel plot. CASC9, cancer susceptibility candidate 9; SMD, standardized mean difference; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; TCGA, The Cancer Genome Atlas; CI, confidence interval; SE, standard error.
Figure 5.
SROC curves and pooled diagnostic indices for the integrated meta-analysis. (A) The AUC of the SROC curves was 0.9520, indicating a significant differentiating capacity of CASC9 in LUSC. (B) The pooled sensitivity for the included studies was 0.93 (0.91-0.95). (C) The pooled specificity was 0.87 (0.81-0.91). (D) The pooled positive LR was 6.78 (2.37-19.36). (E) The pooled negative LR was 0.11 (0.05-0.25). (F) The pooled diagnostic OR was 66.23 (17.54-250.11). SROC, summarized receiver operating characteristic; AUC, area under the curve; SE, standard error; Q, quartile; CI, confidence interval; df, degrees of freedom; LR, likelihood ratio; OR, odds ratio.
Cell line data of CASC9 expression from the CCLE
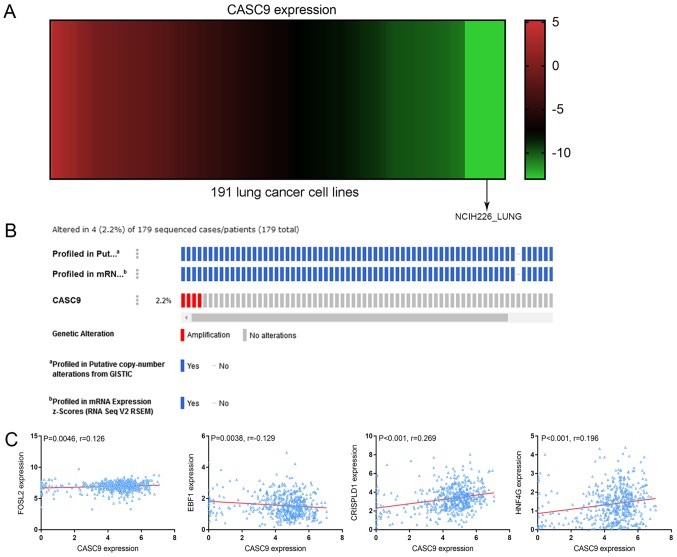
The expression of CASC9 in each of the 191 LC cell lines was obtained and plotted in a heat map. The spectrum of colors ranged from red to green, reflecting a wide range of CASC9 expression from high to low (Fig. 6A).
Figure 6.
CASC9 expression heat-map, gene alteration prediction and correlation analysis. (A) Heat-map of CASC9 expression in 191 lung cancer cell lines arranged from high to low, corresponding to the spectrum of colors from red to green. (B) A total of 4 (2.2%) incidences of CASC9 amplification were revealed in 179 sequenced cases. (C) Correlation between the expression of CASC9 and predicted transcription factors or adjacent protein-coding genes. CASC9 was positively correlated with FOSL2 (r=0.126; P=0.0046), CRISPLD1 (r=0.269; P<0.001) and HNF4G (r=0.196; P<0.001), and negatively correlated with EBF1 (r=−0.129; P=0.0038). CASC9, cancer susceptibility candidate 9; GISTIC, genomic identification of significant targets in cancer; FOSL2, Fos-related antigen 2; EBF1, transcription factor COE1; CRISPLD1, cysteine-rich secretory protein LCCL domain-containing 1; HNF4G, hepatocyte nuclear factor 4-γ.
Gene alteration of CASC9 in LUSC tissue from cBioPortal
The alteration profiles of CASC9 from OncoPrint revealed that the CASC9 sequence was altered in 4 (2.2%) out of the 179 sequenced cases. The 4 cases of alteration all belonged to the category of amplification, i.e. multiple copies of the complete CASC9 gene were naturally occurring in 4 of the 179 sequenced cases (Fig. 6B).
Prediction of potential transcription factors for CASC9
Based on the prediction results from LncMAP, 3 transcription factors, including Fos-related antigen 2 (FOSL2), SWI/SNF complex subunit SMARCC2, and transcription factor COE1 (EBF1), are likely to be involved in the regulatory effect of CASC9 on downstream molecules, including diacylglycerol kinase a, transient receptor potential cation channel subfamily V member 4, ankyrin repeat domain-containing protein SOWAHC, semaphorin-3G and DNA-binding protein Ikaros (Table IV). No prediction results of transcription factors associated with CASC9 were produced by other online programs. The correlation analyses demonstrated a positive correlation between FOSL2 and CASC9 expression (r=0.126; P=0.0046) and a negative correlation with EBF1 (r=−0.129; P=0.0038) (Fig. 6C). No significant correlations between SMARCC2 and CASC9 were observed (data not shown).
Table IV.
Potential transcription factors and genes associated with CASC9 (lncRNA ID, ENSG00000249395) from LncMAP.
| TF ID | TF name | Gene ID | Gene name | Correlation coefficient (r) according to CASC9 expression
|
Score | P-value | FDR | |
|---|---|---|---|---|---|---|---|---|
| Low expression | High expression | |||||||
| EN S G00000075426 | FOSL2 | ENSG00000065357 | DGKA | 0.513 | −0.002 | 0.995 | <0.001 | <0.001 |
| ENSG00000075426 | FOSL2 | ENSG00000111199 | TRPV4 | 0.600 | −0.098 | 1.000 | <0.001 | <0.001 |
| ENSG00000075426 | FOSL2 | ENSG00000198142 | SOWAHC | 0.607 | −0.082 | 1.000 | <0.001 | <0.001 |
| ENSG00000139613 | SMARCC2 | ENSG00000010319 | SEMA3G | 0.079 | 0.588 | 0.997 | <0.001 | <0.001 |
| ENSG00000164330 | EBF1 | ENSG00000185811 | IKZF1 | 0.608 | 0.135 | 0.995 | <0.001 | <0.001 |
CASC9, cancer susceptibility candidate 9; TF, transcription factor; FDR, false discovery rate; FOSL2, Fos-related antigen 2; SMARCC2, SWI/SNF complex subunit SMARCC2; EBF1, transcription factor COE1; DGKA, diacylglycerol kinase a; TRPV4, transient receptor potential cation channel subfamily V member 4; SOWAHC, ankyrin repeat domain-containing protein SOWAHC; SEMA3G, semaphorin-3G; IKZF1, DNA-binding protein Ikaros.
In vitro experiments
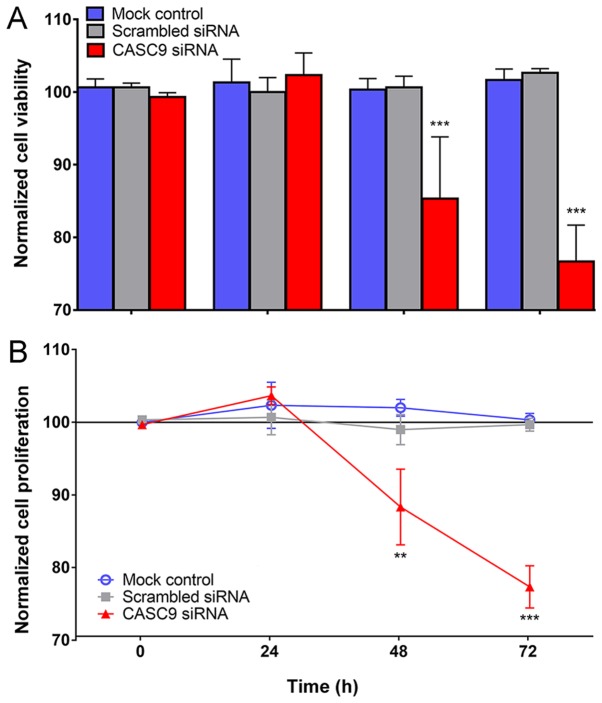
The transfection efficiency of the 4 selected CASC9 siRNAs was >80% at 96 h, as determined by RT-qPCR. It can be observed that CASC9 mRNA expression was obviously lower in CASC9 siRNA group than in scrambled siRNA group and blank control at 48, 72 and 96 h (Fig. S2). Fig. 7 illustrates the behavioral changes in LUSC cells caused by the siRNA knockdown of CASC9 in terms of cell viability and proliferation. The viability of the H226 cells in the CASC9 siRNA group decreased significantly after 48 and 72 h (both P<0.001), while no significant change of cell viability occurred in H226 cells among the mock control (P=0.9821) and scrambled siRNA group (P=0.8790) at 72 h (Fig. 7A). Similarly, compared with the mock and scrambled siRNA controls, a substantial decrease in the cell proliferation of H226 cells of CASC9 siRNAs group was recorded using the MTS tetrazolium assay at 48 (P<0.01) and 72 h (P<0.001) (Fig. 7B).
Figure 7.
Cell viability and proliferation measurements following CASC9 knockdown. The viability and proliferation of H226 cells with siRNA down- regulation of CASC9 were measured using fluorimetric resorufin viability and MTS tetrazolium assays. (A) The influence of CASC9 silencing on (A) the viability and (B) the proliferation of H226 cells. The data are normalized to the blank control at the corresponding time point, and are presented as the average of three experiments and the standard deviation (error bars). **P<0.01 and ***P<0.001, compared with the mock control or scrambled siRNAs groups at the same time point. CASC9, cancer susceptibility candidate 9; siRNA, small interfering RNA.
Functional enrichment analysis of genes co-expressed with CASC9
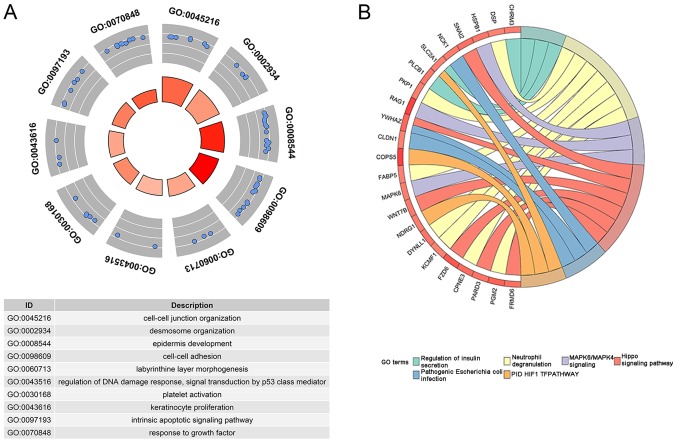
With the aid of GEPIA, 200 genes were identified to be co-expressed with CASC9. An enrichment analysis of the 200 genes indicated 16 terms of biological processes and pathways significantly associated with them (Table V; Fig. 8). Among the 16 significant terms, the top 3 biological processes were 'cell-cell junction organization', 'desmosome organization' and 'epidermis development', and the top 3 pathways were 'Hippo signaling pathway', 'pathogenic Escherichia coli infection', and 'PID HIF1 TF pathway' (all P<0.01).
Table V.
Metascape enrichment analysis for the co-expressed genes of cancer susceptibility candidate 9.
| GO ID | Category | Term | Count of enriched genes | Log10(P) |
|---|---|---|---|---|
| GO:0045216 | GO Biological Processes | 'Cell-cell junction organization' | 9 | −7.340 |
| GO:0002934 | GO Biological Processes | 'Desmosome organization' | 4 | −7.010 |
| GO:0008544 | GO Biological Processes | 'Epidermis development' | 13 | −6.630 |
| GO:0098609 | GO Biological Processes | 'Cell-cell adhesion' | 15 | −5.420 |
| hsa04390 | KEGG Pathway | 'Hippo signaling pathway' | 6 | −4.030 |
| GO:0060713 | GO Biological Processes | 'Labyrinthine layer morphogenesis' | 3 | −3.960 |
| GO:0043516 | GO Biological Processes | 'Regulation of DNA damage response, signal transduction by p53 class mediator' | 3 | −3.420 |
| GO:0030168 | GO Biological Processes | 'Platelet activation' | 5 | −3.100 |
| GO:0043616 | GO Biological Processes | 'Keratinocyte proliferation' | 3 | −3.020 |
| hsa05130 | KEGG Pathway | 'Pathogenic Escherichia coli infection' | 3 | −2.650 |
| GO:0097193 | GO Biological Processes | 'Intrinsic apoptotic signaling pathway' | 6 | −2.610 |
| M255 | Canonical Pathways | 'PID HIF1 TF pathway' | 3 | −2.420 |
| R-HSA-422356 | Reactome Gene Sets | Regulation of insulin secretion | 3 | −2.190 |
| GO:0070848 | GO Biological Processes | Response to growth factor | 9 | −2.130 |
| R-HSA-6798695 | Reactome Gene Sets | Neutrophil degranulation | 7 | −2.110 |
| R-HSA-5687128 | Reactome Gene Sets | MAPK6/MAPK4 signaling | 3 | −2.060 |
GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; MAPK, mitogen-activated protein kinase.
Figure 8.
GO analysis and pathway enrichment. (A) Nodes in the concentric circle graph represent co-expressed genes clustered in specific biological process terms. The inner sectors with larger size and darker color represented more significant enrichment. (B) Ribbons with different colors corresponded to different enriched pathways terms from Metascape. GO, gene ontology.
Associations between CASC9 and adjacentprotein-coding genes
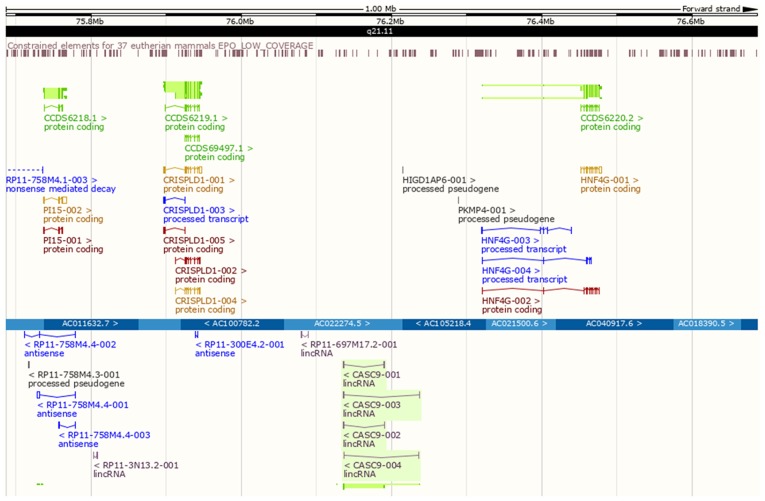
Location-based displays (chromosome 8, 75,686,308-76,686,308) in the Ensemble Genome Browser unfolded the positional associations between CASC9 and adjacent protein-coding genes. Two such genes, cysteine-rich secretory protein LCCL domain-containing 1 (CRISPLD1; chromosome 8, 75,896,750-75,946,793) and hepatocyte nuclear factor 4-γ (HNF4G; chromosome 8, 76,320,271-76,476,562), were located 188,846 bp downstream and 129,575 bp upstream of CASC9 (chromosome 8, 76,135,639-76,190,696), respectively (Fig. 9). Correlation analyses indicated positive relationships between CASC9 and CRISPLD1 (r=0.269, P<0.001) or HNF4G expression (r=0.196; P<0.001) (Fig. 6C).
Figure 9.
Genomic locations of CASC9 and adjacent protein-coding genes. CRISPLD1 (chromosome 8, 75,896,750-75,946,793) and HNF4G (chromosome 8, 76,320,271-76,476,562) were 188,846 bp downstream and 129,575 bp upstream from CASC9 (chromosome 8, 76,135,639-76,190,696), respectively. CASC9, cancer susceptibility candidate 9; CRISPLD1, cysteine-rich secretory protein LCCL domain-containing 1; HNF4G, hepatocyte nuclear factor 4-γ.
Discussion
There is a large amount of evidence that lncRNAs are involved in the development of human cancer, with their regulatory effect on gene expression. Therefore, identifying tumor-associated lncRNAs and investigating the role of these molecules in the onset and progression of human cancer may facilitate the discovery of novel diagnostic and therapeutic biomarkers for LUSC.
The present study concentrated on CASC9, a cancer-associated lncRNA with carcinogenic function in several types of human cancer that has not been studied in LUSC. The expression level of CASC9 between LUSC and non-cancer tissue was first examined through RT-qPCR. The results demonstrated that CASC9 is overexpressed in LUSC tissue compared with non-cancer tissues. The statistical analysis of TCGA data and the integrated meta-analysis also reported significantly elevated CASC9 expression in LUSC tissues, supporting the RT-qPCR data. One of the highlights of the present study lies in the integrated meta-analysis incorporating in-house RT-qPCR, TCGA and GEO data. Evaluating the expression of CASC9 between LUSC and non-cancer tissues with the combined methods of data excavation from various public databases balanced the limited sample size of the clinical specimens obtained for the present study to a certain extent. The results of the analysis of the association between CASC9 expression and the clinicopathological parameters of LUSC imply that upregulated CASC9 may promote the malignant development of LUSC. The ROC and SROC curves for TCGA data reflected the capacity of CASC9 to distinguish LUSC from non-cancer tissues. These findings suggest that CASC9 may be applied as a therapeutic target for LUSC patients.
To gain deeper insights into the oncogenic influence of CASC9 on LUSC, in vitro experiments were performed investigating its functional role of CASC9 in this disease. Knockdown of CASC9 significantly diminished the viability and proliferation of H226 cells. This provided indirect evidence reinforcing the effect of CASC9 on the viability and growth of LUSC cells. Previous studies have probed into the influence of CASC9 on the cell growth in other cancer types, including ESCC and LUAD (19,31). The study conducted by Wu et al (31) demonstrated the negative correlation between CASC9 and programmed cell death protein 4 (PDCD4) in ESCC. PDCD4 is a tumor suppressor gene that participates in the regulation of apoptosis, proliferation and the cell cycle (32-34). Wu et al (31) further proposed that CASC9 may downregulate PDCD4 expression by recruiting histone-lysine N-methyltransferase EZH2 to augment the proliferative ability of ESCC. It was hypothesized that the regulatory association between CASC9 and PDCD4 is be a possible explanation for the effect of CASC9 on the viability and proliferation of LUSC cells.
Following the in vitro experiments, the molecular mechanism of CASC9 in LUSC was investigated. The 2.2% incidence of naturally occurring gene duplication of CASC9 in LUSC samples was consistent with the upregulation of this lncRNA, providing a possible explanation for its overexpression in this cancer type. Since the functions of lncRNAs are dependent on binding to proteins (35,36), a functional enrichment analysis of genes co-expressed with CASC9 is conducive to comprehending the molecular basis of CASC9-associated carcinogenesis of LUSC. From the annotation results, the top biological processes significantly clustered with the genes co-expressed with CASC9 were mainly associated with the epithelial-mesenchymal transition (EMT). Gao et al (37) reported that CASC9 promotes ESCC proliferation and metastasis by modulating the EMT signaling pathway. The combination of the functional annotation results from the present study and the literature search suggest that CASC9 is involved in the dysregulation of the EMT process in LUSC. Apart from the aforementioned biological processes, the functional enrichment analysis revealed significantly assembled pathways, including the Hippo signaling and mitogen-activated protein kinase (MAPK)6/MAPK4 signaling pathways, which serve roles in the promotion or suppression of tumors (38,39). It is speculated that CASC9 contributes to the initiation and progression of LUSC by participating in these biological processes and pathways. Other noteworthy aspects of the CASC9-centered tumorigenesis of LUSC are its upstream mediators and adjacent protein-coding genes. Potential factors that regulated the transcriptional activity of CASC9 were searched in a number of online programs. However, only lncRNAMap provided prediction results of CASC9-associated downstream transcription factors and mRNAs. Two of the predicted transcription factors were linked to NSCLC. The abnormal expression of SMARCC2 was discovered in squamous NSCLC tissue, and FOSL2 is required for transforming growth factor p1-induced migration in NSCLC (40,41). The other transcription factor, EBF1, is a B-lineage transcriptional regulator involved in B-cell acute lymphoblastic leukemia (42). Although only EBF1 expression was negatively correlated with CASC9 expression in LUSC, and none of the predicted transcription factors or mRNAs were confirmed in the literature search to be targeted by CASC9, these predicted downstream molecules hold important implications for the pathogenesis of LUSC. The association between CASC9 and adjacent protein-coding genes was also analyzed and CASC9 expression was positively correlated with two such genes, CRISPLD1 and HNF4G. Notably, HNF4G was reported in a previous study to serve an oncogenic role in LC by promoting cell proliferation (43). Therefore, it was hypothesized that CASC9 modulates the expression of HNF4G or interacts with it to affect the occurrence and progression of LUSC. Additionally, CASC9 has been reported to enhance the malignant potential of human cancer types, including breast cancer, hepatocellular carcinoma and ESCC, by interacting with numerous target genes, such as EZH2, heterogeneous nuclear ribonucleoprotein L and laminin subunit γ-2 (44-46). Although these target genes were not revealed in the bioinformatics analysis of the present study, the association between them and CASC9 in LUSC is also worth exploring in future studies.
Although certain notable findings have been revealed, the limitations of the present study lie in the following aspects: Only the discernment capacity of CASC9 to distinguish LUSC from non-cancer tissues was assessed, and there was not sufficient evidence to support its diagnostic value in this disease. The serum CASC9 levels in patients should be investigated in future experiments for the evaluation of the diagnostic significance. Additionally, in the in vitro experiments, only CASC9 knockout was conducted. To comprehensively investigate the influence of CASC9 on the biological function of LUSC cells, overexpression experiment should also be performed. Furthermore, using >3 types of cells lines in the in vitro experiment would be more conclusive than a single cell line. Further in vitro and in vivo experiments are required to verify the influence of CASC9 on the aforementioned biological processes and pathways, as well as the targeting regulatory association between CASC9 and the predicted transcription factors or adjacent protein-coding genes.
In summary, the present study revealed the overexpression and clinicopathological significance of CASC9 in LUSC for the first time. The role of CASC9 as a cancer-promoting factor in LUSC may be accomplished by strengthening the viability and proliferation capacity of the tumor cells. A functional enrichment analysis of co-expressed genes and the correlation between the expression of CASC9 and predicted transcription factors or adjacent protein-coding genes inferred a potential molecular mechanism of CASC9-associated LUSC tumorigenesis.
Supplementary Materials
Acknowledgments
Not applicable.
Funding
The present study was supported by the Natural Science Foundation of Guangxi, China (grant nos. 2017GXNSFAA 198016, 2015GXNSFCA139009 and 2017GXNSFAA198067), the Fund of National Natural Science Foundation of China (grant no. NSFC 81560469), the Guangxi Zhuang Autonomous Region Health and Family Planning Commission Self-financed Scientific Research Project (grant no. Z20180979), a Guangxi Medical University Training Program for Distinguished Young Scholars, a Medical Excellence Award Funded by the Creative Research Development Grant from the First Affiliated Hospital of Guangxi Medical University, and a Future Academic Stars Project from Guangxi Medical University (grant no. WLXSZX18068).
Availability of data and materials
The datasets generated and/or analyzed during the current study were TCGA-LUSC (https://portal.gdc.cancer.gov/) GSE49155, GSE33479 and GSE51852 (https://www.ncbi.nlm.nih.gov/gds/). The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
GC, KLW and LG designed the study. LG, JHZ, FCM, JL, HWZ and SX performed the analyses and calculations. LG and YNG reviewed the microarray and RNA-sequencing sources in the meta-analysis, and contributed to the writing of the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The patients involved in the present study provided signed informed consent and approval was granted by the Ethics Committee of the First Affiliated Hospital of Guangxi Medical University, Nanning, China (approval no. 2015-KY-NSFC-059).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Perez-Moreno P, Brambilla E, Thomas R, Soria JC. Squamous cell carcinoma of the lung: molecular subtypes and therapeutic opportunities. Clin Cancer Res. 2012;18:2443–2451. doi: 10.1158/1078-0432.CCR-11-2370. [DOI] [PubMed] [Google Scholar]
- 2.Guo NL, Dowlati A, Raese RA, Dong C, Chen G, Beer DG, Shaffer J, Singh S, Bokhary U, Liu L, et al. A predictive 7-gene assay and prognostic protein biomarkers for non-small cell lung cancer. EBioMedicine. 2018;32:102–110. doi: 10.1016/j.ebiom.2018.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 4.Yan X, Shen H, Jiang H, Hu D, Wang J, Wu X. YXQ-EQ induces apoptosis and inhibits signaling pathways important for metastasis in non-small cell lung carcinoma cells. Cell Physiol Biochem. 2018;49:911–919. doi: 10.1159/000493223. [DOI] [PubMed] [Google Scholar]
- 5.Wu C, Li X, Zhang D, Xu B, Hu W, Zheng X, Zhu D, Zhou Q, Jiang J, Wu C. IL-1beta-mediated up-regulation of WT1D via miR-144-3p and their synergistic effect with NF-kappaB/COX-2/HIF-1alpha pathway on cell proliferation in LUAD. Cell Physiol Biochem. 2018;48:2493–2502. doi: 10.1159/000492687. [DOI] [PubMed] [Google Scholar]
- 6.Lee B, Lee T, Lee SH, Choi YL, Han J. Clinicopathologic characteristics of EGFR, KRAS, and ALK alterations in 6,595 lung cancers. Oncotarget. 2016;7:23874–23884. doi: 10.18632/oncotarget.8074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hou Z, Xu C, Xie H, Xu H, Zhan P, Yu L, Fang X. Long noncoding RNAs expression patterns associated with chemo response to cisplatin based chemotherapy in lung squamous cell carcinoma patients. PLoS One. 2014;9:e108133. doi: 10.1371/journal.pone.0108133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao J, Liu Y, Zhang W, Zhou Z, Wu J, Cui P, Zhang Y, Huang G. Long non-coding RNA Linc00152 is involved in cell cycle arrest, apoptosis, epithelial to mesenchymal transition, cell migration and invasion in gastric cancer. Cell Cycle. 2015;14:3112–3123. doi: 10.1080/15384101.2015.1078034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan CN, Ma L, Liu N. Systematic analysis of lncRNA-miRNA-mRNA competing endogenous RNA network identifies four-lncRNA signature as a prognostic biomarker for breast cancer. J Transl Med. 2018;16:264. doi: 10.1186/s12967-018-1640-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang YK, Yu JC. Circulating microRNAs and long non-coding RNAs in gastric cancer diagnosis: An update and review. World J Gastroenterol. 2015;21:9863–9886. doi: 10.3748/wjg.v21.i34.9863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, Sehgal L, Jain N, Khashab T, Mathur R, Samaniego F. LncRNA MALAT1 promotes development of mantle cell lymphoma by associating with EZH2. J Transl Med. 2016;14:346. doi: 10.1186/s12967-016-1100-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong L, Ding H, Li Y, Xue D, Liu Y. LncRNA TINCR is associated with clinical progression and serves as tumor suppressive role in prostate cancer. Cancer Manag Res. 2018;10:2799–2807. doi: 10.2147/CMAR.S170526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang X, Qi M, Wu R, Liu A, Chen D, Tang L, Chen J, Hu X, Li W, Zhan L, et al. Long non-coding RNA CUDR promotes malignant phenotypes in pancreatic ductal adenocarcinoma via activating AKT and ERK signaling pathways. Int J Oncol. 2018;53:2671–2682. doi: 10.3892/ijo.2018.4574. [DOI] [PubMed] [Google Scholar]
- 14.Cao W, Wu W, Shi F, Chen X, Wu L, Yang K, Tian F, Zhu M, Chen G, Wang W, et al. Integrated analysis of long noncoding RNA and coding RNA expression in esophageal squamous cell carcinoma. Int J Genomics. 2013;2013:480534. doi: 10.1155/2013/480534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Su X, Li G, Liu W. The long noncoding RNA cancer susceptibility candidate 9 promotes nasopharyngeal carcinogenesis via stabilizing HIF1a. DNA Cell Biol. 2017;36:394–400. doi: 10.1089/dna.2016.3615. [DOI] [PubMed] [Google Scholar]
- 16.Shang C, Sun L, Zhang J, Zhao B, Chen X, Xu H, Huang B. Silence of cancer susceptibility candidate 9 inhibits gastric cancer and reverses chemoresistance. Oncotarget. 2017;8:15393–15398. doi: 10.18632/oncotarget.14871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma P, Zhang M, Nie F, Huang Z, He J, Li W, Han L. Transcriptome analysis of EGFR tyrosine kinase inhibitors resistance associated long noncoding RNA in non-small cell lung cancer. Biomed Pharmacother. 2017;87:20–26. doi: 10.1016/j.biopha.2016.12.079. [DOI] [PubMed] [Google Scholar]
- 18.Zhou J, Xiao H, Yang X, Tian H, Xu Z, Zhong Y, Ma L, Zhang W, Qiao G, Liang J. Long noncoding RNA CASC9.5 promotes the proliferation and metastasis of lung adenocarcinoma. Sci Rep. 2018;8:37. doi: 10.1038/s41598-017-18280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiong DD, Li ZY, Liang L, He RQ, Ma FC, Luo DZ, Hu XH, Chen G. The lncRNA NEAT1 accelerates lung adenocarcinoma deterioration and binds to mir-193a-3pas a competitive endogenous RNA. Cell Physiol Biochem. 2018;48:905–918. doi: 10.1159/000491958. [DOI] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Tomczak K, Czerwinska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): An immeasurable source of knowledge. Contemp Oncol (Pozn) 2015;19A:A68–A77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hutter C, Zenklusen JC. The Cancer Genome Atlas: Creating lasting value beyond its data. Cell. 2018;173:283–285. doi: 10.1016/j.cell.2018.03.042. [DOI] [PubMed] [Google Scholar]
- 23.He RQ, Gao L, Ma J, Li ZY, Hu XH, Chen G. Oncogenic role of miR-183-5p in lung adenocarcinoma: A comprehensive study of qPCR, in vitro experiments and bioinformatic analysis. Oncol Rep. 2018;40:83–100. doi: 10.3892/or.2018.6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dang Y, Luo D, Rong M, Chen G. Underexpression of miR-34a in hepatocellular carcinoma and its contribution towards enhancement of proliferating inhibitory effects of agents targeting c-MET. PLoS One. 2013;8:e61054. doi: 10.1371/journal.pone.0061054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rong M, Chen G, Dang Y. Increased miR-221 expression in hepatocellular carcinoma tissues and its role in enhancing cell growth and inhibiting apoptosis in vitro. BMC Cancer. 2013;13:21. doi: 10.1186/1471-2407-13-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang S, He R. Synergistic effect of MiR-146a mimic and cetuximab on hepatocellular carcinoma cells. BioMed Research International. 2014;2014:384121. doi: 10.1155/2014/384121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dang YW, Zeng J, He RQ, Rong MH, Luo DZ, Chen G. Effects of miR-152 on cell growth inhibition, motility suppression and apoptosis induction in hepatocellular carcinoma cells. Asian Pac J Cancer Prev. 2014;15:4969–4976. doi: 10.7314/APJCP.2014.15.12.4969. [DOI] [PubMed] [Google Scholar]
- 28.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBio- Portal. Sci Signal. 2013;6:pl 1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ooi AT, Gower AC, Zhang KX, Vick JL, Hong L, Nagao B, Wallace WD, Elashoff DA, Walser TC, Dubinett SM, et al. Molecular profiling of premalignant lesions in lung squamous cell carcinomas identifies mechanisms involved in stepwise carcinogenesis. Cancer Prev Res (Phila) 2014;7:487–495. doi: 10.1158/1940-6207.CAPR-13-0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arima C, Kajino T, Tamada Y, Imoto S, Shimada Y, Nakatochi M, Suzuki M, Isomura H, Yatabe Y, Yamaguchi T, et al. Lung adenocarcinoma subtypes definable by lung development-related miRNA expression profiles in association with clinicopathologic features. Carcinogenesis. 2014;35:2224–2231. doi: 10.1093/carcin/bgu127. [DOI] [PubMed] [Google Scholar]
- 31.Wu Y, Hu L, Liang Y, Li J, Wang K, Chen X, Meng H, Guan X, Yang K, Bai Y. Up-regulation of lncRNA CASC9 promotes esophageal squamous cell carcinoma growth by negatively regulating PDCD4 expression through EZH2. Mol Cancer. 2017;16:150. doi: 10.1186/s12943-017-0715-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lankat-Buttgereit B, Göke R. The tumour suppressor Pdcd4: Recent advances in the elucidation of function and regulation. Biol Cell. 2009;101:309–317. doi: 10.1042/BC20080191. [DOI] [PubMed] [Google Scholar]
- 33.Allgayer H. Pdcd4, a colon cancer prognostic that is regulated by a microRNA. Crit Rev Oncol Hematol. 2010;73:185–191. doi: 10.1016/j.critrevonc.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 34.Hwang SK, Minai-Tehrani A, Lim HT, Shin JY, An GH, Lee KH, Park KR, Kim YS, Beck GR, Jr, Yang HS, et al. Decreased level of PDCD4 (programmed cell death 4) protein activated cell proliferation in the lung of A/J mouse. J Aerosol Med Pulm Drug Deliv. 2010;23:285–293. doi: 10.1089/jamp.2009.0778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaneko S, Li G, Son J, Xu CF, Margueron R, Neubert TA, Reinberg D. Phosphorylation of the PRC2 component Ezh2 is cell cycle-regulated and up-regulates its binding to ncRNA. Genes Dev. 2010;24:2615–2620. doi: 10.1101/gad.1983810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao GD, Liu XY, Lin Y, Liu HF, Zhang GJ. LncRNA CASC9 promotes tumorigenesis by affecting EMT and predicts poor prognosis in esophageal squamous cell cancer. Eur Rev Med Pharmacol Sci. 2018;22:422–429. doi: 10.26355/eurrev_201801_14191. [DOI] [PubMed] [Google Scholar]
- 38.Kostenko S, Dumitriu G, Moens U. Tumour promoting and suppressing roles of the atypical MAP kinase signalling pathway ERK3/4-MK5. J Mol Signal. 2012;7:9. doi: 10.1186/1750-2187-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu Y, Yang C, Yang S, Cheng F, Rao J, Wang X. miR-665 promotes hepatocellular carcinoma cell migration, invasion, and proliferation by decreasing Hippo signaling through targeting PTPRB. Cell Death Dis. 2018;9:954. doi: 10.1038/s41419-018-0978-y. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Wang J, Sun D, Wang Y, Ren F, Pang S, Wang D, Xu S. FOSL2 positively regulates TGF-ß1 signalling in non-small cell lung cancer. PLoS One. 2014;9:e112150. doi: 10.1371/journal.pone.0112150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Remmelink M, Mijatovic T, Gustin A, Mathieu A, Rombaut K, Kiss R, Salmon I, Decaestecker C. Identification by means of cDNA microarray analyses of gene expression modifications in squamous non-small cell lung cancers as compared to normal bronchial epithelial tissue. Int J Oncol. 2005;26:247–258. [PubMed] [Google Scholar]
- 42.Georgopoulos K. Ebf1 in DNA repair and leukemogenesis. Blood. 2015;125:3969–3971. doi: 10.1182/blood-2015-05-639427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang J, Zhang J, Xu L, Zheng Y, Ling D, Yang Z. Expression of HNF4G and its potential functions in lung cancer. Oncotarget. 2017;9:18018–18028. doi: 10.18632/oncotarget.22933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang B, Li Y, Qu X, Zhu H, Tan Y, Fan Q, Jiang Y, Liao M, Wu X. Long noncoding RNA cancer susceptibility candidate 9 promotes doxorubicin-resistant breast cancer by binding to enhancer of zeste homolog 2. Int J Mol Med. 2018;42:2801–2810. doi: 10.3892/ijmm.2018.3812. [DOI] [PubMed] [Google Scholar]
- 45.Klingenberg M, Groß M, Goyal A, Polycarpou-Schwarz M, Miersch T, Ernst AS, Leupold J, Patil N, Warnken U, Allgayer H, et al. The long noncoding RNA cancer susceptibility 9 and RNA binding protein heterogeneous nuclear ribonucleoprotein L form a complex and coregulate genes linked to AKT signaling. Hepatology. 2018;68:1817–1832. doi: 10.1002/hep.30102. [DOI] [PubMed] [Google Scholar]
- 46.Liang Y, Chen X, Wu Y, Li J, Zhang S, Wang K, Guan X, Yang K, Bai Y. LncRNA CASC9 promotes esophageal squamous cell carcinoma metastasis through upregulating LAMC2 expression by interacting with the CREB-binding protein. Cell Death Differ. 2018;25:1980–1995. doi: 10.1038/s41418-018-0084-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study were TCGA-LUSC (https://portal.gdc.cancer.gov/) GSE49155, GSE33479 and GSE51852 (https://www.ncbi.nlm.nih.gov/gds/). The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.