Abstract
The vitamin A derivative 11-cis-retinaldehyde plays a pivotal role in vertebrate vision by serving as the chromophore of rod and cone visual pigments. In the initial step of vision, a photon is absorbed by this chromophore resulting in its isomerization to an all-trans state and consequent activation of the visual pigment and phototransduction cascade. Spent chromophore is released from the pigments through hydrolysis. Subsequent photon detection requires the delivery of regenerated 11-cis-retinaldehyde to the visual pigment. This trans-cis conversion is achieved through a process known as the visual cycle. In this review, we will discuss the enzymes, binding proteins and transporters that enable the visual pigment renewal process with a focus on advances made during the past decade in our understanding of their structural biology.
OVERVIEW
Vision is one of the fundamental senses that enables the perception of the surrounding environment. The human retina is lined with millions of photoreceptors expressing opsins bound to the visual chromophore, 11-cis-retinaldehyde [1]. Absorption of a photon by opsin in the photoreceptors causes isomerization of the visual chromophore to an all-trans configuration [2]. This initial photochemical reaction triggers the activation of the signal transduction cascade that eventually leads to transmission of a visual signal to the brain but leaves the opsin insensitive to further light stimulation [3]. Sustained vision thus requires continuous renewal of the visual chromophore upon light exposure [4]. The classical visual cycle is a long-known enzymatic pathway that serves to produce 11-cis-retinaldehyde for both rod and cone visual pigments [5–9]. Photoreceptors and the retinal pigment epithelium (RPE) harbor the molecular components that achieve this chemical conversion [10]. The reactions of the visual cycle include Schiff base formation and hydrolysis, alcohol/aldehyde redox chemistry, esterification, and isomerization coupled to ester cleavage. Besides chemical transformations, retinoid trafficking within and between cells is also critically important for efficient visual cycle function.
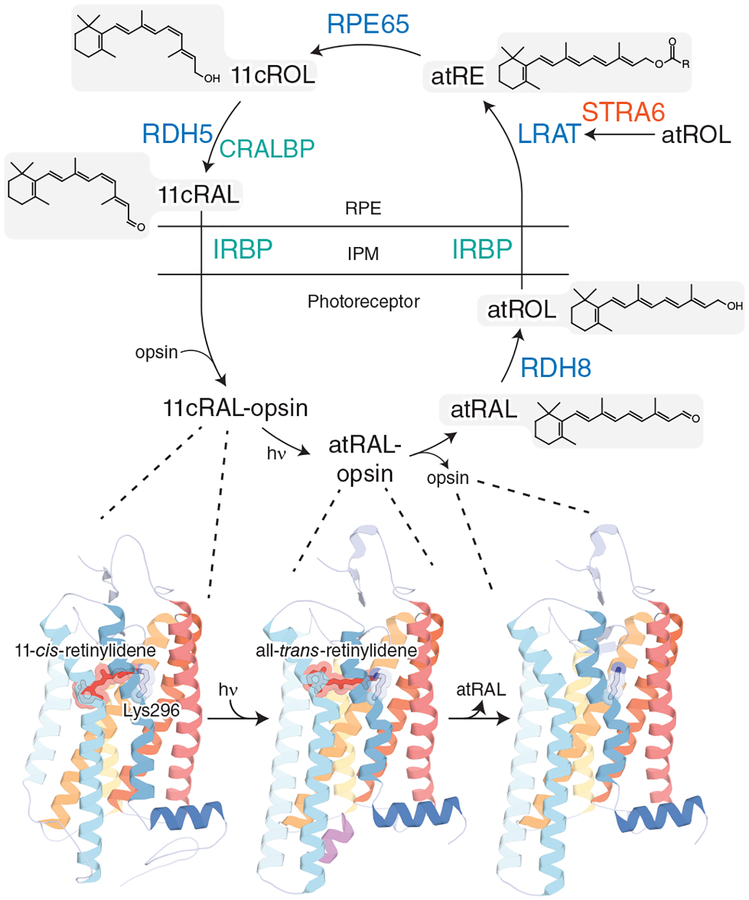
For the visual cycle to ensue, rod and cone opsins in photoreceptor outer segment disk membranes must first undergo structural changes to liberate the chromophore. The crystal structure of bovine rhodopsin reported in 2000 [11], (Fig. 1, leftmost structure), provided the first high-resolution picture of visual pigment structure and enabled an appreciation of the molecular interactions involved in the perception of light. Rhodopsin, a G protein-coupled receptor, contains an 11-cis-retinaldehyde prosthetic group that is bound to the opsin protein through residue Lys296 as an 11-cis-retinylidene Schiff base [12, 13]. Following light activation which produces alterations in the opsin protein structure [14, 15] (Fig. 1, middle structure), rhodopsin decomposes to form opsin protein [16] (Fig. 1, rightmost structure) and all-trans-retinaldehyde through a series of intermediates [17]. Cone photoreceptors have opsins homologous to rhodopsin in rods [18] and the process of photoactivation and phototransduction is thought to be basically conserved between the two types of photoreceptors [19, 20]. Once released into the photoreceptor cytoplasm, all-trans-retinaldehyde is reduced to all-trans-retinol by retinol dehydrogenase (RDH) 8 aided by the ATP-binding cassette transporter A4 (ABCA4), in the first step of the retinoid cycle.
Figure 1. Steps in the classical visual cycle.
The top schematic outlines the individual chemical steps and proteins mediating the reaction and transport pathways of the classical visual cycle. Abbreviations are as follows: RAL, retinaldehyde; ROL, retinol; RE, retinyl ester; 11c, 11-cis; at, all-trans. The lower part of the figure shows key states of the rod visual pigment as it progresses from the ground state (PDB accession code: 1U19), through a photoactivated intermediate (PDB accession code: 3PQR), to the free opsin molecule (PDB accession code: 3CAP), as determined by X-ray crystallography.
Retinoids, being hydrophobic in nature, do not readily diffuse across aqueous environments between cellular compartments and instead rely on binding proteins to facilitate the process [21]. Interphotoreceptor retinoid-binding protein (IRBP) facilitates retinoid transport between photoreceptors and RPE by sequestering them in the interphotoreceptor matrix (IPM) [22, 23]. IRBP transports all-trans-retinol to the RPE where it enters the cell forming a complex with cellular retinol-binding protein (CRBP) 1 and is metabolized by lecithin:retinol acyltransferase (LRAT), which catalyzes the esterification of retinols using a phospholipid fatty acid donor. The resulting retinyl esters then act as substrates for retinal pigment epithelium-specific 65 kDa protein (RPE65), which catalyzes the formation of 11-cis-retinol. This reaction involves ester cleavage coupled to trans-cis double bond isomerization. After this isomerization reaction, 11-cis-retinol is oxidized back to 11-cis-retinaldehyde by RDH5 and transported out of the RPE. Cellular retinaldehyde-binding protein (CRALBP) functions as a retinoid carrier in the latter two reactions, assisting the oxidation of 11-cis-retinol to 11-cis-retinaldehyde and preventing uncontrolled storage of 11-cis-retinol by esterification. CRALBP binds nascent 11-cis-retinaldehyde protecting it from unwanted isomerization and facilitating its transport from RPE back to photoreceptors. IRBP transports 11-cis-retinaldehyde back to the photoreceptors where it combines with opsin to form a ground-state rhodopsin molecule ready to be activated by another photon. In addition, the transporter protein stimulated by retinoic acid 6 (STRA6), localized on the basolateral membrane of the RPE, works in concert with retinol-binding protein 4 (RBP4) and CRBP1 to facilitate uptake of retinol from the choriocapillaris which is critical for replacement of ocular retinoids lost to off-pathway side reactions. Mutations leading to various retinal pathologies have been found for many visual cycle components [24, 25]. For recent reviews of the classical visual cycle, see [10, 26, 27].
In the present review, we discuss classical visual cycle components for which recent progress (i.e. over the past decade) has been made in understanding the structural basis of their retinoid-processing activity. We do not attempt to cover other potential sources of 11-cis-retinaldehyde including cone-specific [28] and light-dependent pathways [29, 30], as the structural biology of these visual chromophore synthetic processes is still in its infancy.
RETINOL DEHYDROGENASES
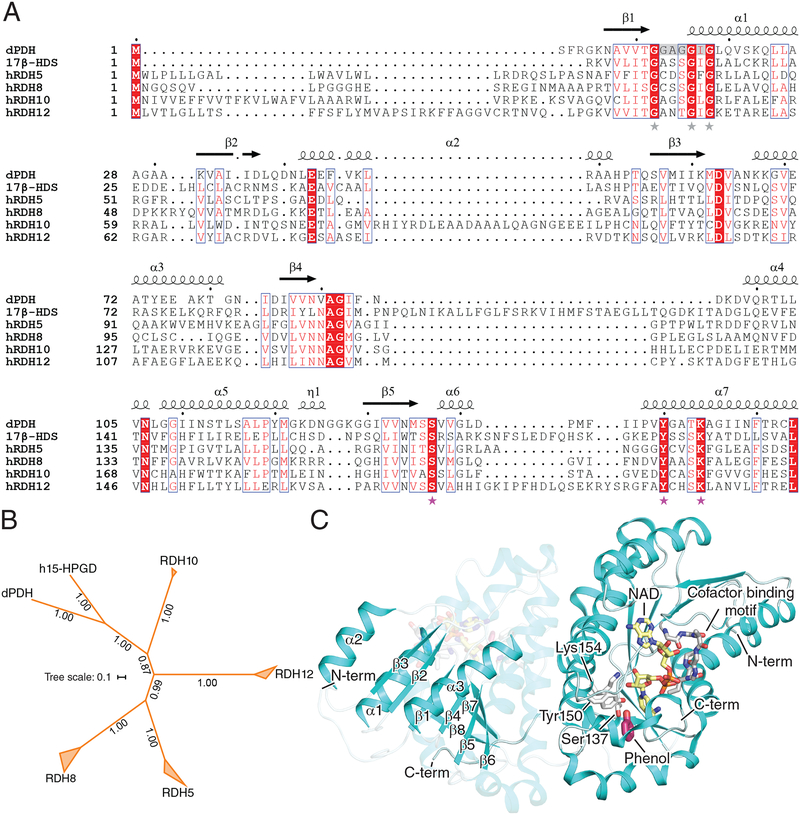
RDHs perform key redox reactions essential for visual pigment regeneration. They belong to the short-chain dehydrogenase/reductase (SDR) superfamily of enzymes and sequence alignment shows the presence of highly conserved motifs between 17β-hydroxysteroid dehydrogenases (17β-HSDs) and RDHs (Fig. 2A) [31]. These include the catalytic Ser, Tyr and Lys motif as well as the nucleotide-binding motif G-X-X-X-G-X-G. Genes encoding SDR enzymes known to be major players in classical visual cycle function, namely RDH5, RDH8, RDH10, and RDH12, are believed to have their origin in the last common ancestor of vertebrates, as orthologs of these genes appear to be absent in cephalochordates and cnidaria [32]. Nevertheless, SDR enzymes with retinol dehydrogenase activity are present in invertebrates such as Drosophila melanogaster indicating that retinol substrate specificity in this enzyme superfamily has been acquired more than once by convergent evolution (Fig. 2B). Besides the major visual cycle-associated RDHs listed above, several other RDHs are expressed in the vertebrate retina. In fact, knockout mice studies show that these could be part of compensatory pathways in the absence of one or more RDH. RDH8 in the photoreceptor outer segments and RDH5 in the RPE were initially identified to play roles in visual chromophore production, although RDH10, RDH11 and RDH12 may also be important in assisting retinoid metabolism within the classical visual cycle ([33] and references cited therein).
Figure 2. Structure and enzymology of RDH enzymes.
A) Alignment of RDH and RDH-related protein sequences. The catalytic motif Ser-Tyr-Lys is marked by pink stars whereas the cofactor-binding motif G-X-X-X-G-X-G is marked by grey stars. The alignment was generated by Clustal Omega and annotated in Espript. Secondary structure elements marked on the top of the alignment are derived from the crystal structure of dPDH (PDB accession code: 5ILG). Portions of the C-termini of the sequences are omitted for clarity. Abbreviations are as follows: dPDH, Drosophila melanogaster pigment cell-enriched dehydrogenase isoform c (NCBI RefSeq: NP_001137959.1); 17β-HSD, human 17β-hydroxysteroid dehydrogenase (Genebank ID: CAC88111.1) B) Phylogeny of RDH and RDH-related proteins estimated by the program MrBayes. The RDH clades include orthologs from Homo sapiens, Bos taurus, Danio rerio, Xenopus tropicalis, and Gallus gallus, except for the RDH12 clade for which a X. tropicalis ortholog was not identified. The protein sequences were aligned with Clustal omega using default parameters and the phylogeny was computed assuming a uniform prior, equal rate of substitution across sites and WAG substitution matrix (chosen by Markov chain sampling). The posterior probability distribution was estimated by Markov Chain Monte Carlo sampling for 839,000 generations with samples taken every 100 generations after a burn-in fraction of 25%, at which point an average standard deviation of split frequencies of 0.004 was reached indicating convergence. The majority rule consensus tree is shown in the figure with posterior probabilities for each split displayed along the branches. The two side lengths of the triangles representing collapsed nodes are proportional to the distances of the closest and furthest terminal nodes. The scale bar represents the average number of changes per site. Abbreviations are as follows: h15-HPGD, human 15-hydroxyprostaglandin dehydrogenase (NCBI RefSeq: NP_000851.2). C) Crystal structure of dPDH (PDB accession code: 5ILG) displayed in Pymol as a dimer. (Left monomer) The Rossmann fold (β1-α1-β2-α2-β3-α3-β4) is highlighted as well as the substrate-binding site comprised in part by β strands 5, 6 and 7. (Right monomer) Displayed as sticks is the catalytic triad of Ser137, Tyr150 and Lys154 in white, the substrate-mimetic phenol in pink, the NAD cofactor in yellow, and its binding motif (G-X-X-X-G-X-G) in grey highlighting a funnel-shaped binding cavity.
RDH8 (also known as photoreceptor RDH or prRDH) catalyzes the reduction of all-trans-retinaldehyde to all-trans-retinol in the first enzymatic step in the visual cycle. It was cloned from a bovine retinal cDNA library [34]. RDH8 is selective for all-trans-retinol and spares 11-cis-retinaldehyde that is transported back to the photoreceptors, ensuring it is not reduced before it conjugates with opsin. The presence of a Ser residue in the catalytic binding pocket suggested NADP/H cofactor selectivity for this enzyme [34], which was confirmed by kinetic studies [35]. Accumulation of all-trans-retinaldehyde during high levels of illumination suggests that this reaction rate limits the visual cycle under such conditions, whereas other steps may be rate limiting under different conditions [35–37]. Although, RDH8 is the main contributor to all-trans-retinaldehyde reduction in the photoreceptors, RDH12 also contributes up to 30% of RDH activity [33]. No retinal diseases have been linked to mutations in RDH8; however, mutations in RDH12 have been associated with Leber congenital amaurosis (LCA), characterized by progressive photoreceptor degeneration and severe macular dystrophy [38].
Electron microscopy revealed no differences in the retinal structure of Rdh8−/−, Rdh8+/− and Rdh+/+ mice [39]. Although, RDH activity is reduced in Rdh8−/− cells compared to Rdh8+/+, specificity for NADPH is maintained, and the lack of retinal degeneration in Rdh8 knockout mice is probably due to sustained 11-cis-retinaldehyde recovery. Similarly, Rdh12−/− mice do not exhibit a phenotype resembling LCA in humans. These observations prompted researchers to study the phenotype of Rdh8−/− Rdh12−/− mice. Although only mild light-induced degeneration was noticed, accumulation of N-retinylidene-N-retinylethanolamine (A2E) was observed in these double knockout mice. A2E, made from two molecules of all-trans-retinaldehyde and one molecule of phosphatidylethanolamine (PE), tends to build up in photoreceptor outer segments due to inefficient all-trans-retinaldehyde clearance resulting in retinaldehyde toxicity. The ABCA4 transporter located in photoreceptor disk rims functions to transport all-trans-retinal-PE adducts from the photoreceptor intradiscal space to the cytoplasm where they dissociate allowing retinaldehyde to be reduced by RDHs. The Abca4−/− mouse model of Stargardt disease and Rdh8−/− Rdh12−/− mice exhibit similar phenotypes as expected from the sequential actions of these proteins in retinaldehyde clearance. Retinaldehyde generated in the photoreceptor outer segments was found to seep into the inner segments [40]. RDH12 localizes to rod inner segments and likely plays a key role in reducing retinaldehyde that diffuses to the inner segment, which could explain the severe retinal phenotype observed in patients with loss of function RDH12 mutations.
RDH5 (also referred to as 11-cis-RDH) catalyzes the final enzymatic step of the visual cycle, oxidation of 11-cis-retinol to 11-cis-retinaldehyde. The activity of 11-cis-RDH was localized to the microsomes of RPE [41]. RDH5 is a 32 kDa membrane-bound protein and was the first retinol-metabolizing enzyme identified in the SDR family [42]. It was isolated in a complex with p63 (later renamed RPE65). RDH5 prefers NAD/H as a cofactor, which can be explained by the presence of an aspartate at position 37 in place of a serine (as in RDH8) in the cofactor-binding pocket preventing accommodation of the 2’ phosphate group from NADP/H [34]. Although retinol/retinaldehyde interconversion is thermodynamically almost neutral, the high ratio of NAD+ to NADH in the cytosol [43] explains the tendency of RDH5 to operate in the oxidative direction in vivo opposite to that of the NADP/H-utilizing RDH8. RDH10 and RDH11 are also expressed in the RPE and are known to have 11-cis-retinol dehydrogenase activity [44, 45].
Mutations in RDH5 are associated with fundus albipunctatus, a rare form of congenital stationary night blindness characterized by the presence of white spots or flecks on fundus examination and slowly progressing retinal degeneration [46]. Studies of patients suffering from this condition showed delayed rod and cone dark adaptation after exposure to bright light resulting from slowed production of 11-cis-retinaldehyde [47]. Homology modeling of RDH5 traces some of these mutations to residues likely involved in dimer formation. A more recent study of a 16-year-old Polish female patient suffering from night blindness, reported a Tyr175Phe mutation which affects the invariant Tyr residue present in the catalytic triad [48].
Electron microscopy reveals no differences in retinal structure between Rdh5−/−, Rdh5+/− and Rdh5+/+ mice [49]. In addition, the rate of rhodopsin recovery in the dark after intense bright light exposure is similar in Rdh5−/−, Rdh5+/− and Rdh5+/+ mice. Rdh5−/− mice do not display phenotypic changes associated with fundus albipunctatus. Deficient 11-cis-RDH activity can result in increased levels of free 11/13-cis-retinol which LRAT subsequently converts to 11/13-cis retinyl esters. Although, 11-cis-retinol may be oxidized to 11-cis-retinaldehyde by an alternative enzyme, 13-cis-retinyl esters accumulate in RDH5−/− mice [50]. Rdh5−/− Rdh11−/− mice exhibit delayed dark adaptation after prolonged bleaching as well as an elevation in 11-cis-retinol and 11-cis-retinyl esters [45]. However, dark adapted levels of 11-cis-retinaldehyde remained normal. This could be attributed to the fact that the oxidation reaction is still able to catch up, given the slower, rate limiting reduction of all-trans-retinaldehyde to retinol. This is also indicative of the presence of an alternate enzyme capable of carrying out this reaction in the absence of RDH5 and RDH11. Global and RPE-conditional Rdh10−/− mice do not exhibit retinal degeneration, nor do Rdh10−/− Rdh5−/− mice recapitulate the RDH5 mutation phenotype in humans [44].
No three dimensional structure of a vertebrate RDH has been reported to date due in part to the difficulty in expressing and purifying of these membrane-associated enzymes. Recently, an invertebrate homolog from Drosophila melanogaster with RDH activity, the pigment cell-enriched dehydrogenase isoform c (dPDH), was crystallized [51]. Arthropods like D. melanogaster have rhabdomeric photoreceptors as opposed to the ciliary variety in vertebrates. These rhabdomeres consist of microvillar structures containing bistable rhodopsins that activate phospholipase C driving a phosphatidylinositol-related cascade instead of the cyclic nucleotide/Ca2+ signaling employed in vertebrates. Also, unlike the vertebrate visual cycle, all-trans-retinaldehyde (3-OH) is reduced to all-trans-retinol by PDH in retinal pigment cells [52]. Although this protein has retinol dehydrogenase activity, it is evolutionarily more closely related to hydroxyprostaglandin dehydrogenases based on reciprocal BLAST searches and phylogenetic analysis (Fig. 2B). dPDH was crystallized in the presence of NADH and phenol (Fig. 2C). The N-terminus of PDH forms a Rossmann fold consisting of a βαβ pattern (β1-α1-β2-α2-β3-α3-β4) like other SDR enzymes. Heterologous expression of PDH resulted in an apparent monomeric species that nevertheless was found as a dimeric assembly in crystallo. Dimerization is known to be crucial for SDR activity and accordingly retinaldehyde reductase activity was only observed from crystalline enzyme samples. dPDH binds to NAD in an energetically favorable conformation with the active site Ser137 forming a hydrogen bond with the amide group of the cofactor in a binary state. Upon binding of the substrate-mimetic compound phenol, NAD flips so that its reactive hydrogen is accessible to substrate. Tyr150 acts as an acid in the catalytic cycle by donating its hydroxyl proton to the substrate while Lys154 lowers the pKa of the Tyr150 hydroxyl group setting up a proton relay mechanism. Ser137 stabilizes the carbonyl group of the substrate [53]. The β sheet in the Rossmann fold region is stabilized by cofactor binding as observed by hydrogen-deuterium exchange experiments. The substrate binds in the loop region between β strands 5, 6 and 7, which was observed to be flexible. The NAD/H-binding site in dPDH is funnel shaped and lined by hydrophilic residues, whereas the substrate entry site is mostly hydrophobic enabling access to membrane dissolved retinoids. Positively charged residues in the cofactor-binding site are decisive in cofactor specificity [54]. However, the presence of uncharged residues in the cofactor-binding site of dPDH provide no indication of a specific preference for either NAD/H or NADP/H. Given the dual substrate and cofactor binding specificities of the RDHs, it is of interest to study the active site structure of other members of this group.
LECITHIN:RETINOL ACYLTRANSFERASE
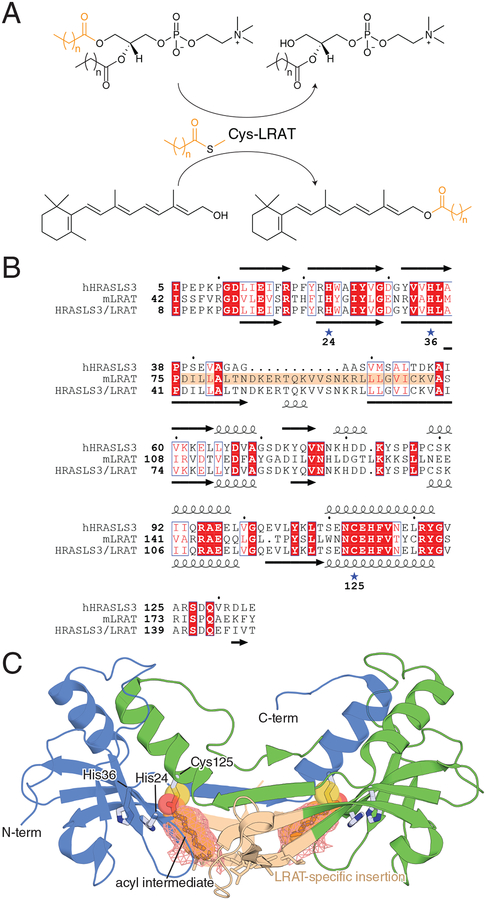
LRAT is one of the two indispensable enzymes of the classical visual cycle [24, 55, 56]. The activity of this protein was characterized first in rat small intestine [57] and was later found in the RPE [58], from which the gene was cloned [59]. This vertebrate-specific enzyme [32] of the N1pC/P60 papain-like thiol protease protein superfamily [60] catalyzes the formation of retinyl esters from retinol and phosphatidylcholine substrate with sn-1 specificity. The enzyme employs a His/His/Cys nucleophile [61] catalytic triad to form a thioacyl intermediate [62] that is transferred to retinol via a bi-bi ping-pong-type mechanism [63] (Fig. 3A). LRAT is capable of esterifying other geometric isomers of retinol as well as amidating retinol derivatives [64]. This activity differs from other members of the HRAS-like suppressor (HRASLS) family, which act as either phospholipases or phospholipid acyltransferases.
Figure 3. Structure and enzymology of LRAT.
A) Physiological reaction catalyzed by LRAT. B) Sequence alignment of human HRASLS3 (hHRASLS3), mouse LRAT (mLRAT), and the HRASLS3/LRAT chimera. The LRAT-specific sequence mentioned in the main text is marked with a brown background. His and Cys residues constituting the catalytic triad of LRAT-family proteins are marked by stars. Numbering under the stars refers to the sequence of HRASLS3/LRAT. Secondary structure elements for HRASLS3 and HRASLS3/LRAT are shown above and below the respective sequences. C) Crystal structure of the HRASLS3-LRAT chimera protein. The two chains of the dimer are colored blue and green. The Cys146 acyl intermediate is shown in stick and transparent van der Waals sphere representation within the hydrophobic pocket, which is outlined by salmon-colored mesh.
LRAT retinyl ester synthase activity serves two crucial functions in the eye: 1) the esterification of all-trans-retinol by LRAT provides a key thermodynamic driving force for retinol uptake through the RBP4-STRA6 transport system [65] and 2) all-trans-retinyl esters, which mainly consist of palmitate and to a lesser extent myristate and sterate esters, serve as substrates for the visual cycle isomerase RPE65 [66, 67]. The absence of these functions is conspicuous in the eyes of humans and animal models with loss of function LRAT mutations, which exhibit a paucity of both all-trans-retinyl esters and 11-cis-retinylaldehyde resulting in visual dysfunction and retinal degeneration. In humans, LRAT mutations cause recessive forms of retinitis pigmentosa (RP) or LCA depending upon the severity of functional disruption [68]. Lrat-knockout mice phenocopy the patient disease state with an essential absence of ocular retinoids leading to both rod and cone photoreceptor degeneration [69].
LRAT is an integral membrane protein containing a C-terminal membrane-spanning helix as well as monotopic membrane-binding elements at its N-terminus [70]. The protein resides in the endoplasmic reticulum of the RPE [58] forming a functional dimer [71]. A three-dimensional structure of native LRAT has not been reported to date. However, key structural insights into LRAT function were obtained through determination of the crystal structure of an HRASLS3/LRAT chimera protein [72] in which an LRAT-specific sequence (residues 76–105 of murine LRAT) was substituted for residues (39–57) of human HRASLS3, whose structure has previously been determined by X-ray crystallography [73, 74] (Fig. 3B). This sequence substitution conferred LRAT activity to the chimeric protein, which unlike native LRAT, was amenable to crystallization [72]. The X-ray crystal structure of the chimera revealed dramatic structural changes relative to native HRASLS3 whereby the LRAT-specific sequence mediated the formation of a domain-swapped parallel dimeric assembly not seen in HRASLS structures (Fig. 3C). This arrangement resulted in a mixed catalytic triad whereby the two His residues were contributed from one subunit and the catalytic Cys nucleophile from the other. The thiol nucleophile resides at the end of a hydrophobic pocket that is partially formed by side chains originating from the LRAT sequence insertion with the pocket entry of both members of the dimer pair in proximity to the lipid membrane. This structural feature probably helps ensure that lecithin:retinol acyl transfer predominates over hydrolysis by shielding the thioester intermediate from water. The thioester intermediate was visualized in the structure through crystallizing the chimera in the presence of a synthetic phospholipid substrate, diheptanoyl phosphatidylcholine (DHPC). The structure revealed a heptanoyl chain in a fully extended conformation occupying the abovementioned hydrophobic pocket. His36 and Cys125 were found in conformations similar to the corresponding residues in HRASLS3, whereas His24 was rotated by ~55° away from the thiol nucleophile to accommodate the carbonyl oxygen of the acyl moiety. Notably, the acyl group occupies nearly the full volume of the pocket, particularly near its base. Hence, the ability of the retinol nucleophile to access the acyl intermediate appears to require conformational changes not observed in this specific crystal structure. Although it is possible that retinol gains access to the acyl group via direct partitioning from the lipid bilayer, kinetic data have also suggested that retinol could be delivered to LRAT directly by CRBP [75], which would presumably require conformational changes to the cytosol-facing side of the protein. Resolution of this issue will require additional kinetic and structural analyses including potential complex structures with retinoid-binding proteins.
Retinal pigment epithelium-specific 65 kda protein
RPE65 carries out the hallmark trans-cis isomerization step of the visual cycle and is the second indispensable enzyme of this pathway [76]. RPE65 was cloned from bovine RPE in the early 1990’s [77], but it wasn’t until 2015 that its identity as the visual cycle isomerase was generally accepted despite substantial research on this protein in the interim [78–80]. The association of RPE65 mutations with RP and LCA2 was a major motive for investigations of RPE65 structure and function [81, 82]. In 2001, an RPE65 knockout mouse was generated by the Redmond laboratory, which recapitulated the phenotypes observed in humans with homozygous RPE65 loss-of-function mutations: severe visual chromophore deficiency, accumulation of retinyl esters in the RPE, attenuated ERG responses and loss of visual function [83]. In both mice and humans, prolonged visual chromophore starvation ultimately leads to photoreceptor cell death. Extensive research on RPE65-associated RP/LCA has led to an AAV-based gene therapy for this disease, which is the first FDA-approved gene therapy for an inherited condition [84].
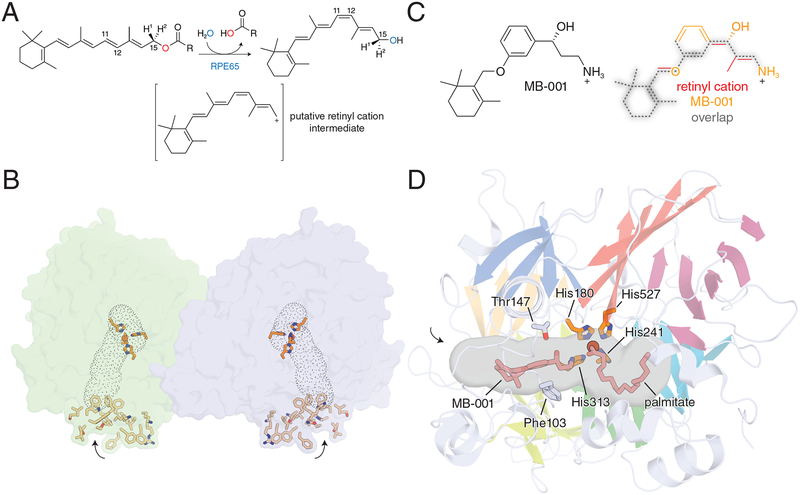
RPE65 is a vertebrate-specific member of the carotenoid cleavage oxygenase (CCO) superfamily [32, 85], although its catalytic activity is different from other CCO enzymes, and it does not use dioxygen as a co-substrate [86]. Rather than cleaving carotenoids, RPE65 evolved to catalyze the hydrolytic cleavage and isomerization of all-trans-retinyl esters [67, 87] to form 11-cis-retinol and a fatty acid secondary product in a reaction that is thought to occur through a retinyl cation intermediate [88, 89] (Fig. 4A). Isotope labeling studies have demonstrated that this reaction involves an atypical ester hydrolysis reaction involving O-alkyl cleavage [88], which is essential for generation of the putative cationic intermediate [89]. This dramatic change in catalytic activity is consistent with the rapid evolution of RPE65 following its split from a beta carotene oxygenase-like ancestor [90]. Like its carotenoid-cleaving relatives, RPE65 contains a 4-His coordinated Fe(II) center [91, 92]. The iron cofactor is accessible through a hydrophobic cavity that enters the protein from a surface comprised of several hydrophobic and positively charged residues [93] (Fig. 4B). The intrinsic physicochemical properties of this surface potentially together with contributions made from an S-palmitoyl group located in the same region [94] are likely responsible for the affinity of RPE65 for lipid membranes [95]. The activity of RPE65 critically depends on the presence of phospholipid membranes [96], which can modulate the structure of the RPE65 molecule [91, 97]. Crystal structures of RPE65 indicate that the protein is functionally dimeric [91], although in vivo evidence for this proposal is lacking. The mechanistic details of retinoid isomerization by RPE65 have been investigated extensively over the past few decades. Recent crystallographic work using retinoid-mimetic compounds including MB-001 (Fig. 4C) have helped provide a structural biology framework in which to interpret the large amount of biochemical and kinetic data that have been gathered for RPE65 [98].
Figure 4. Structure and enzymology of RPE65.
A) Scheme of the RPE65-catalyzed retinoid isomerization reaction. B) Crystallographic structure of RPE65 showing the dimeric assembly and the parallel-oriented membrane binding surfaces that surround the opening to the active site cavity (outlined by grey dots). Residues thought to mediate RPE65 membrane binding are shown as sticks. The entrances to the active site pockets are marked by arrows. C) Comparison of MB-001 with the putative retinyl cation intermediate from panel A showing similarity in the core structure of the two molecules. D) Details of the RPE65 active site cavity showing the binding sites for MB-001/emixustat and palmitate. Residues Phe103 and Thr147, which are thought to be important contributors to the 11-cis isomerization stereoselectivity of RPE65 are shown as grey sticks, while the critical iron-binding His residues are shown as orange sticks. The entrance to the active site pocket is marked by an arrow. Images of the RPE65 crystal structure were generated in PyMOL using coordinates deposited under PDB accession code 4RSE.
MB-001 is a potent competitive RPE65 inhibitor that belongs to a class of emixustat and retinylamine-related visual cycle modulators [98]. These molecules share a core structural similarity including the presence of a terminal primary amine functionality that is partially charged at physiological pH and was designed into these molecules to mimic the putative retinyl cation transition state of the RPE65-catalyzed reaction (Fig. 4C). MB-001 as well as emixustat were successfully co-crystallized with bovine RPE65, which allowed delineation of the retinoid-binding region of the active site pocket (Fig. 4D). The presence of this inhibitor resulted in the serendipitous trapping of a palmitate molecule in the adjacent region of the active site pocket where its carboxylate group was found in complex with the iron center. The simultaneous presence of these two ligands clarified the orientation of retinyl ester binding to the RPE65 active site during catalysis, which had been ambiguous. Additionally, the structures provided strong evidence for a role of the iron cofactor in facilitating ester cleavage through its Lewis acid property as hypothesized based on data from prior structural studies [93]. These structures also allowed interpretation ofmutagenesis studies examining residues important for the 11-cis stereoselectivity of RPE65 [89, 99, 100]. Many of the critical residues found in these studies were located within the MB-001/emixustat-binding pocket. Phe103 and Thr147 appear to play a particularly important role in providing charge stabilization at the retinoid C11 position to allow 11-cis-selective isomerization. Mutation of these and other residues change the retinol isomer product distribution of RPE65, which was known not to be 11-cis specific even in wild-type form [88, 89].
CELLULAR RETINALDEHYDE-BINDING PROTEIN
CRALBP (encoded by the RLBP1 gene) is a soluble cis-retinoid-binding protein composed of 316 amino acid residues, excluding the initiating Met residue which is absent in the mature protein [101, 102]. The discovery and properties of CRALBP have been reviewed elsewhere [103, 104]. The protein was first purified from bovine retina and RPE [105] where it plays an integral role in rod and cone visual pigment renewal. Immunohistochemical studies localized CRALBP to RPE and Müller glia [106]. The CRALBP protein sequence was determined by direct peptide analysis as well as DNA sequencing from human and bovine retinal cDNAs revealing a conserved primary sequence without sequence similarity to previously characterized proteins [101, 102]. CRALBP belongs to a family of transfer proteins containing the CRAL-TRIO domain (named after CRALBP and the TRIO guanine exchange factor) that typically bind small lipophilic molecules. When isolated from RPE homogenates CRALBP contains only 11-cis-retinaldehyde whereas ~25% of the protein isolated from neural retinal contains 11-cis-retinol with essentially the remainder being occupied with 11-cis-retinaldehyde [107]. CRALBP binds 11-cis-retinaldehyde with highest affinity (Kd < 20 nM) and 11-cis-retinol and 9-cis-retinaldehyde with Kd values of ~50 nM [103]. The tight binding of 11-cis-retinoids to CRALBP necessitates a mechanism to allow their release from the protein during visual cycle flux. Retinoid release was found to be enhanced by acidic phospholipids with phosphatidic acid and phosphatidylserine being most effective [108]. Thus, it is likely that cellular membranes play an important role in triggering retinoid dissociation from CRALBP allowing transmembrane diffusion.
Mutations in the RLBP1 gene, some which can either enhance or abolish retinoid-binding affinity [109], result a number of autosomal recessive retinopathies including RP, retinitis punctata albescens, Bothnia dystrophy, fundus albipunctatus, and Newfoundland rod/cone dystrophy[24]. This variability in disease severity and manifestations may be attributable to different functional defects induced by particular mutations as well as differing patient genetic backgrounds [110]. Molecular genetic analysis of a family with non-syndromic autosomal recessive RP identified a G4763A nucleotide substitution in RLBP1, leading to the replacement of an Arg with Gln at residue 150 [111]. These patients have classical retinitis punctata albescens with white dot-like deposits observed in their fundus, early childhood night blindness, and macular degeneration. Subsequent studies found several novel mutations associated with recessively inherited retinitis punctata albescens [112, 113]. Prior to retinal degeneration, RP patients with a pathological mutation in RLBP1 display abnormally slow dark adaptation, requiring 12–24 h for dark adaptation whereas normal individuals need less than 1 h. The Arg233Trp substitution is associated with a phenotypically distinct variant of retinitis punctata albescens known as Bothnia dystrophy found among a population residing in Sweden [114]. In pre-clinical studies, treatment of Rlbp1−/− mice with an AAV8-hRLBP1 viral vector delivered subretinally improved the rate of both cone and rod dark adaptation [113].
Rlbp1−/− mice have provided important insights into the role of CRALBP in the visual cycle [115]. Although dark-adapted photosensitivity in Rlbp1−/− mice was normal, 11-cis-retinaldehyde production after illumination, which reflects the rate visual pigment renewal, was delayed by more than 10-fold leading to very slow dark adaptation. These results are consistent with biochemical studies that showed effective isomerization by the retinoid isomerase (RPE65) requires this binding protein [116]. In vitro retinoid isomerization is strongly facilitated by CRALBP likely due to relief of RPE65 product inhibition by sequestering 11-cis-retinol product. When raised in cyclic light/dark conditions Rlbp1−/− mice showed no evidence of photoreceptor degeneration, which contrasts with the CRALBP-associated disease phenotype observed in humans [25]. Subsequent characterization of Rlbp1−/− mice revealed a greater than initially appreciated effect on cone photoreceptor structure, function, and viability. The knockout mice were found to display aberrant localization of M-opsin, M-cone cell death, changes in M-cone cell dark adaptation rate, and impaired cone-driven visual behavior and light responses. Viral vector restoration of Cralbp expression specifically in Müller glia, but not in RPE cells, rescued intraretinal visual cycle function and M-cone sensitivity in Rlbp1−/− mice [117].
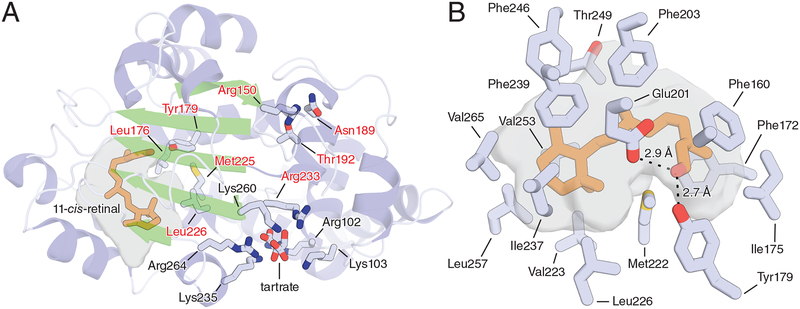
The tertiary structure of CRALBP and its interaction with retinaldehyde were characterized by NMR spectroscopy [118] and later by X-ray crystallography [119]. Crystal structures were solved for both wild-type human CRALBP and the R233W pathogenic mutant as binary complexes with the endogenous ligand 11-cis-retinaldehyde [119] (Fig. 5A). Arg233 is located 15 Å from the retinoid-binding pocket. The Arg233Trp substitution causes a domino-like reorganization of the retinoid-binding site and impairs ligand release [109]. Arg233 is also a component of the cationic residue cluster on CRALBP that is thought to bind acidic phospholipids triggering retinoid release [108]. The Bothnia retinal dystrophy-associated Arg233Trp mutation perturbed the structure of the cationic cluster, which may impair membrane-induced retinoid dissociation [119]. This demonstated communication between the cationic residue cluster and the retinoid-binding pocket suggest an allosteric mechanism of retinoid release upon CRALBP interaction with negatively charged phospholipids, although competitive displacement remains a possibility [108]. Due to its hydrophobic nature, 11-cis-retinaldehyde is sequestered from the solvent within the CRALBP retinoid-binding pocket, which has a volume of ~650 Å3. In wild-type CRALBP, the electron density map for the C11–C15 terminal tail of the chromophore is undefined, suggesting flexible binding. By contrast, clear density was observed for the entire 11-cis-retinaldehyde molecule in Arg233Trp CRALBP suggesting more stable binding, which is consistent with biochemical data [109]. 11-cis-retinaldehyde is found in a 6-s-trans, 11-cis, and twisted 12-s-cis configuration, and the polyene chain is planar up to carbon 12. The β-ionone ring is surrounded and immobilized by multiple van der Waals interactions that twist the ring by about 7° around the C6–C7 bond relative to the plane of the polyene chain (Fig. 5B). The carbonyl oxygen of the aldehyde serves as the hydrogen bond acceptor for the phenol group of Tyr179 with the acidic oxygen of Glu201 also within hydrogen bonding distance of the aldehyde group. The higher affinity of CRALBP for 11-cis-retinaldehyde compared to 11-cis-retinol may be explained by differences in hydrogen bonding and pi stacking interactions between the alcohol and aldehyde functional groups. The 11-cis-retinaldehyde configuration is preferable as compared to 9-cis-retinaldehyde owing to shape complementarity between the binding pocket and the 11-cis-retinaldehyde with a 45° out-of-plane twist between C12 and C13 being sterically unfavorable for the 9-cis isomer to assume. This is also the reason that 13-cis-retinaldehyde or all-trans-retinaldehyde do not bind tightly to CRALBP.
Figure 5. Structural basis of 11-cis-retinoid binding by CRALBP.
A) Overall structure of wild-type human CRALBP. The cavities housing the bound 11-cis-retinaldehyde (orange sticks) are shown as a grey surface. Residues known to be mutated in retinal diseases are labeled with red text. Residues labeled in black (as well as Arg233) make up a basic patch on the protein surface that is thought to bind acidic phospholipids to facilitate retinoid release from the protein. The structure features a bound tartrate molecule at the center of the basic patch which possibly mimics the structure of an acidic phospholipid head group. B) Close-up view of the retinoid-binding cavity of CRALBP showing residues responsible for forming the bend geometry of the cavity as well as those involved in dipolar interactions with the terminal aldehyde moiety. Residue numbering refers to the mature protein lacking the initiating Met residue.
STIMULATED BY RETINOIC ACID 6
Retinol mainly circulates in the blood bound to RBP4 [120–122]. This complex formation effectively solubilizes retinol in plasma and protects it from enzymatic and oxidative reactions. The passage of retinol through the cellular membrane can occur by a simple diffusion due to its hydrophobicity, but there is also evidence to support a receptor-mediated uptake of retinol [123, 124]. The search for the receptor for RBP4 took place over many years and its existence was controversial. However, in 2007 the holo-RBP4 receptor was identified using a photo-crosslinking approach [120]. This protein, known as STRA6 for stimulated by retinoic acid 6, was originally identified in embryonic carcinoma cells but remained functionally uncharacterized [125]. STRA6 is expressed in a variety of organs including the eye, brain, adipose tissue, spleen, kidney, testis, and adult female genital tract [120, 125]. STRA6 was shown to catalyze the release of retinol from RBP4 and to facilitate retinol translocation across the cell membrane to the cytosol where it associates with CRBPs [126], reviewed in [127]. Esterification of retinol by LRAT improves the efficiency of STRA6-mediated transport by increasing the amount of available apo-CRBP [65, 120].
Mutations in STRA6 are associated with defects in multiple organs as demonstrated by Matthew-Wood syndrome [128–130]. This disease is characterized by anophthalmia, mental disability, congenital heart defects, lung hypoplasia, short stature, and even embryonic lethality. Interestingly, Stra6 loss of function causes less severe phenotypes in mice, primarily affecting the eye. Stra6 knockout mice exhibited thinning of the rod outer and inner segments, reduction of retinoid content, a hypertrophic vitreous, and formation of dense vasculature in the vitreous humor [131, 132]. These observations suggest that STRA6 is not the only transporter facilitating the uptake of retinol in mice.
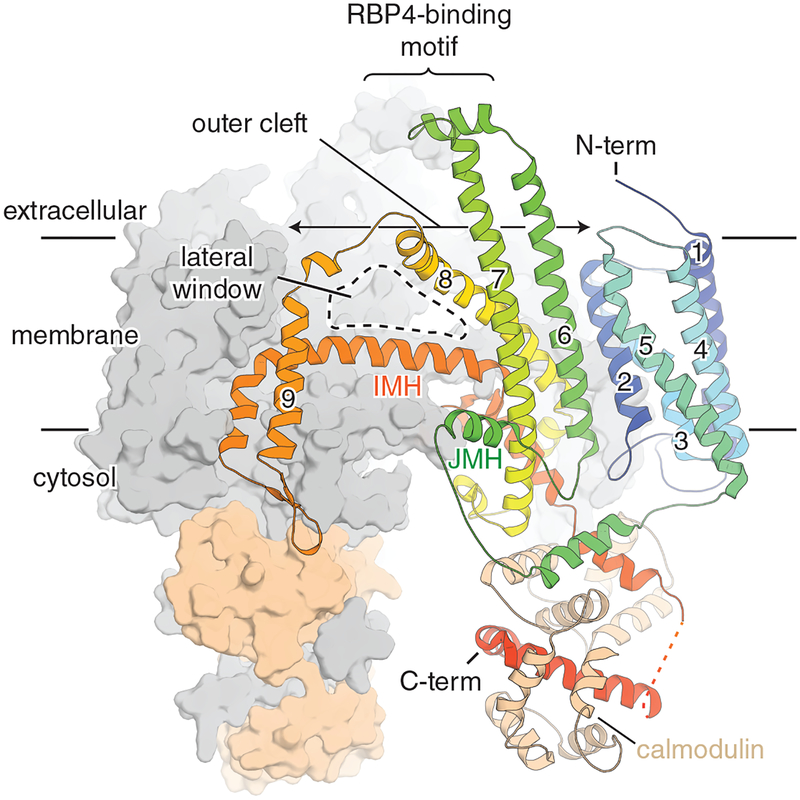
Biochemical and transgenic animal studies contributed to our understanding of STRA6 function, but the absence of structural information left many unresolved issues regarding its mechanism of retinol transport. A 3.9 Å cryo-EM structure of zebrafish STRA6 was recently solved [133] (Fig. 6). The structure shows that STRA6 forms an intricately associated dimer comprised of 18 transmembrane helices (9 per protomer) and 2 intramembrane helices (1 per protomer) that interact at the central dimer interface [133]. The structure showed a large hydrophobic cleft on the extracellular side, which could be a site for release of retinol from RBP4 [133]. This cleft extends down about halfway through the transmembrane region of the protein and is exposed to the membrane through a lateral window [133]. This observation suggests a pathway for retinol to diffuse into the transmembrane after delivery by RBP4. The cytoplasmic side of STRA6 was serendipitously found to be associated with calmodulin (CaM) in an unconventional arrangement [133]. The role of CaM in this complex remains elusive as the association between Ca2+ and retinol uptake through STRA6 has not been rigorously investigated. Overall, the structure of STRA6 suggests a mechanism for retinol release from holo-RBP and diffusion of retinol into the membrane [133]. Several lines of evidence suggested STRA6 is a ligand-activated signaling receptor which leads to activation of the JAK/STAT signaling pathway and regulation of transcription [134, 135]. Thus, it was suggested that STRA6 acts as a cytokine receptor that links retinol transport with cellular signaling [136]. Further studies will reveal the role of STRA6 in signaling, the functional significance of its association with CaM and its regulation.
Figure 6. Cryo-electron microscopy structure of STRA6.
One STRA6-calmodulin complex is shown in surface representation (colored in grey and wheat, respectively). The second in shown in cartoon representation. Transmembrane helices are numbered. Abbreviations are as follows: IMH, intramembrane helix; JMH, juxtamembrane helix.
CONCLUSIONS AND PROSPECTS
The past decade has witnessed several major advances in our understanding of the molecular basis of visual cycle activity. High resolution structures are now available for most of the major visual cycle enzymes and binding proteins. Combined with extensive biochemical information about the visual cycle, these structures have provided a new level of sophistication in our understanding of the chemistry of the first steps of vision. Despite this remarkable progress, there are still many fundamental aspects of visual cycle function that require further investigation. Specifically, the intra- and inter-cellular transport of retinoids within the visual cycle remain an area of significant uncertainty.
Retinoid transport between photoreceptor outer segments and the RPE is a critical aspect of visual cycle function that remains poorly understood. IRBP (encoded by the RBP3 gene) is thought to play a role in the transfer of retinoids between photoreceptors and RPE [137, 138]. The structure of IRBP modules from X. laevis [139] and D. rerio [140] have been determined by X-ray crystallography. However, the structure of the full-length protein has not been determined at high resolution and the retinoid-binding sites within the protein remain unclear. Given the large size and apparently flexible nature of IRBP, it could be a good candidate for structural analysis by cryo-EM. Although there is ample evidence supporting a role for IRBP in retinoid trafficking, it is important to note that this protein is dispensable for both classical [141] and cone-specific [142] visual cycle function.
Structural analysis of STRA6 using molecular biology [143, 144] and cryo-EM [133] techniques has profoundly improved our understanding of retinoid transport between the RPE and the circulation. Despite this progress, the details of retinoid transfer from RBP4 through STRA6 to CRBP remain to be elucidated. The exact retinoid passageway through STRA6 requires further clarification as does the role of calmodulin in STRA6 structure, function and regulation. Additionally, it remains unclear whether retinol is accepted by CRBP1 directly from STRA6 or alternatively is obtained from the membrane. Likewise, the mechanism of retinol delivery by CRBP1 to LRAT remains unclear as mentioned earlier [145]. Recent progress in the structural biology of retinol binding and release by CRBP1 [146] is likely to facilitate advances in understanding these processes as they relate to in vivo visual cycle activity. A related issue concerns the mechanism by which CRALBP accepts cis-retinoids generated by RPE65 and 11-cis-RDHs. While there is evidence suggesting that CRALBP may directly accept retinoids from these proteins [88, 115], the structure of RPE65 and the predicted structure of RDH5 would indicate that product uptake and release would occur instead via interaction with the lipid bilayer rather than a binding protein. The basic patch on CRALBP would provide a mechanism for placing the protein in proximity to membranes to accept these ligands. The dynamic changes in CRALBP structure that occur in response to acidic phospholipid binding enabling cis-retinoid release remain to be determined [26]. Advances in expression and purification of visual cycle enzymes and binding proteins may enable the issue of complex formation between visual cycle components to be revisited using modern high resolution structural biology techniques [147].
Retinoid trafficking is modulated by the visual pigment homologs, retinal G protein-coupled receptor (RGR) [148, 149] and peropsin [150], although the molecular basis of these effects are not understood. Besides, this modulator effect, RGR has previously been characterized as a retinaldehyde photoisomerase similar in function to squid retinochrome [29]. Although the physiological relevance of RGR photoisomerase activity in direct visual pigment renewal has been debated understanding the structure of this protein may provide important insights into an RPE65-independent pathway of 11-cis-retinaldehyde formation and the mechanism by which this conversion could modulate G protein signaling in the RPE and Müller glia.
ACKNOWLEDGEMENTS
We thank members of the Kiser and Palczewski laboratories for helpful comments on this manuscript.
FUNDING INFORMATION
This work was supported by funding from the National Institutes of Health EY009339 (KP and PDK), EY025451 (KP), and the Department of Veterans Affairs BX002683 (PDK). K.P. is John H. Hord Professor of Pharmacology.
ABBREVIATIONS LIST
- 17β-HSD
17-β-hydroxysteroid dehydrogenase
- A2E
N-retinylidene-N-retinylethanolamine
- ABCA4
ATP-binding cassette transporter A4
- CRALBP
cellular retinaldehyde-binding protein
- CRBP
cellular retinol-binding protein
- dPDH
Drosophila melanogaster pigment cell-enriched dehydrogenase, isoform c
- HRASLS
HRAS-like suppressor
- IPM
interphotoreceptor matrix
- IRBP
interphotoreceptor retinoid-binding protein
- LCA
Leber congenital amaurosis
- LRAT
lecithin:retinol acyltransferase
- RBP4
retinol-binding protein 4
- RDH
retinol dehydrogenase
- RGR
retinal G protein-coupled receptor
- RPE
retinal pigment epithelium
- RPE65
retinal pigment epithelium-specific 65 kDa protein
- SDR
short-chain dehydrogenase/reductase
- STRA6
stimulated by retinoic acid 6
Footnotes
DECLARATIONS OF INTEREST
K.P. is an inventor of US Patent No. 8722669 - “Compounds and methods of treating ocular disorders” and US Patent No. 20080275134 - “Methods for treatment of retinal degenerative disease” issued to Case Western Reserve University (CWRU) whose values may be affected by this publication. CWRU may license this technology for commercial development. K.P. is an inventor of the U.S. Patent No. 7706863 – “Methods for assessing a physiological state of a mammalian retina”, and U.S. Patent No. 8346345 B2 – “Methods for assessing a physiological state of a mammalian retina” whose values may be affected by this publication.
REFERENCES
- 1.Rodieck RW (1998) The first steps in seeing. Sinauer Associates, Sunderland, Mass. [Google Scholar]
- 2.Hubbard R and Wald G (1952) Cis-trans isomers of vitamin A and retinene in the rhodopsin system. J. Gen. Physiol 36, 269–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yau KW (1994) Phototransduction mechanism in retinal rods and cones. The Friedenwald Lecture. Invest. Ophthalmol. Vis. Sci 35, 9–32 [PubMed] [Google Scholar]
- 4.(!!! INVALID CITATION !!! [4])
- 5.Kiser PD, Zhang J, Sharma A, Angueyra JM, Kolesnikov AV, Badiee M, Tochtrop GP, Kinoshita J, Peachey NS, Li W, Kefalov VJ and Palczewski K (2018) Retinoid isomerase inhibitors impair but do not block mammalian cone photoreceptor function. J. Gen. Physiol 150, 571–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jacobson SG, Aleman TS, Cideciyan AV, Heon E, Golczak M, Beltran WA, Sumaroka A, Schwartz SB, Roman AJ, Windsor EA, Wilson JM, Aguirre GD, Stone EM and Palczewski K (2007) Human cone photoreceptor dependence on RPE65 isomerase. Proc. Natl. Acad. Sci. U.S.A 104, 15123–15128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wenzel A, von Lintig J, Oberhauser V, Tanimoto N, Grimm C and Seeliger MW (2007) RPE65 is essential for the function of cone photoreceptors in NRL-deficient mice. Invest Ophthalmol Vis Sci 48, 534–542 [DOI] [PubMed] [Google Scholar]
- 8.Seeliger MW, Grimm C, Stahlberg F, Friedburg C, Jaissle G, Zrenner E, Guo H, Reme CE, Humphries P, Hofmann F, Biel M, Fariss RN, Redmond TM and Wenzel A (2001) New views on RPE65 deficiency: the rod system is the source of vision in a mouse model of Leber congenital amaurosis. Nat. Genet 29, 70–74 [DOI] [PubMed] [Google Scholar]
- 9.Kuhne W (1977) Chemical processes in the retina. Vision Res. 17, 1269–1316 [DOI] [PubMed] [Google Scholar]
- 10.Kiser PD, Golczak M and Palczewski K (2014) Chemistry of the retinoid (visual) cycle. Chem. Rev 114, 194–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M and Miyano M (2000) Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 289, 739–745 [DOI] [PubMed] [Google Scholar]
- 12.Wang JK, McDowell JH and Hargrave PA (1980) Site of attachment of 11-cis-retinal in bovine rhodopsin. Biochemistry. 19, 5111–5117 [DOI] [PubMed] [Google Scholar]
- 13.Bownds D (1967) Site of attachment of retinal in rhodopsin. Nature. 216, 1178–1181 [DOI] [PubMed] [Google Scholar]
- 14.Choe HW, Kim YJ, Park JH, Morizumi T, Pai EF, Krauss N, Hofmann KP, Scheerer P and Ernst OP (2011) Crystal structure of metarhodopsin II. Nature. 471, 651–655 [DOI] [PubMed] [Google Scholar]
- 15.Salom D, Lodowski DT, Stenkamp RE, Le Trong I, Golczak M, Jastrzebska B, Harris T, Ballesteros JA and Palczewski K (2006) Crystal structure of a photoactivated deprotonated intermediate of rhodopsin. Proc. Natl. Acad. Sci. U.S.A 103, 16123–16128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scheerer P, Park JH, Hildebrand PW, Kim YJ, Krauss N, Choe HW, Hofmann KP and Ernst OP (2008) Crystal structure of opsin in its G-protein-interacting conformation. Nature. 455, 497–502 [DOI] [PubMed] [Google Scholar]
- 17.Ernst OP, Lodowski DT, Elstner M, Hegemann P, Brown LS and Kandori H (2014) Microbial and animal rhodopsins: structures, functions, and molecular mechanisms. Chem. Rev 114, 126–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nathans J, Thomas D and Hogness DS (1986) Molecular genetics of human color vision: the genes encoding blue, green, and red pigments. Science. 232, 193–202 [DOI] [PubMed] [Google Scholar]
- 19.Hofmann L and Palczewski K (2015) Advances in understanding the molecular basis of the first steps in color vision. Prog. Retin. Eye Res 49, 46–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ingram NT, Sampath AP and Fain GL (2016) Why are rods more sensitive than cones? J. Physiol 594, 5415–5426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noy N (2000) Retinoid-binding proteins: mediators of retinoid action. Biochem. J 348 Pt 3, 481–495 [PMC free article] [PubMed] [Google Scholar]
- 22.Gonzalez-Fernandez F and Ghosh D (2008) Focus on Molecules: interphotoreceptor retinoid-binding protein (IRBP). Exp. Eye Res 86, 169–170 [DOI] [PubMed] [Google Scholar]
- 23.Gonzalez-Fernandez F, Kittredge KL, Rayborn ME, Hollyfield JG, Landers RA, Saha M and Grainger RM (1993) Interphotoreceptor retinoid-binding protein (IRBP), a major 124 kDa glycoprotein in the interphotoreceptor matrix of Xenopus laevis. Characterization, molecular cloning and biosynthesis. J. Cell Sci 105 (Pt 1), 7–21 [DOI] [PubMed] [Google Scholar]
- 24.Travis GH, Golczak M, Moise AR and Palczewski K (2007) Diseases caused by defects in the visual cycle: retinoids as potential therapeutic agents. Annu. Rev. Pharmacol. Toxicol 47, 469–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kiser PD and Palczewski K (2016) Retinoids and Retinal Diseases. Annu. Rev. Vis. Sci 2, 197–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saari JC (2012) Vitamin A metabolism in rod and cone visual cycles. Annu. Rev. Nutr 32, 125–145 [DOI] [PubMed] [Google Scholar]
- 27.Wright CB, Redmond TM and Nickerson JM (2015) A History of the Classical Visual Cycle. Prog. Mol. Biol. Transl. Sci 134, 433–448 [DOI] [PubMed] [Google Scholar]
- 28.Wang JS and Kefalov VJ (2011) The cone-specific visual cycle. Prog. Retin. Eye Res 30, 115–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen P, Hao W, Rife L, Wang XP, Shen D, Chen J, Ogden T, Van Boemel GB, Wu L, Yang M and Fong HK (2001) A photic visual cycle of rhodopsin regeneration is dependent on Rgr. Nat. Genet 28, 256–260 [DOI] [PubMed] [Google Scholar]
- 30.Kaylor JJ, Xu T, Ingram NT, Tsan A, Hakobyan H, Fain GL and Travis GH (2017) Blue light regenerates functional visual pigments in mammals through a retinyl-phospholipid intermediate. Nat. Commun 8, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haller F, Moman E, Hartmann RW, Adamski J and Mindnich R (2010) Molecular framework of steroid/retinoid discrimination in 17beta-hydroxysteroid dehydrogenase type 1 and photoreceptor-associated retinol dehydrogenase. J. Mol. Biol 399, 255–267 [DOI] [PubMed] [Google Scholar]
- 32.Albalat R (2012) Evolution of the genetic machinery of the visual cycle: a novelty of the vertebrate eye? Mol. Biol. Evol 29, 1461–1469 [DOI] [PubMed] [Google Scholar]
- 33.Maeda A, Maeda T, Sun W, Zhang H, Baehr W and Palczewski K (2007) Redundant and unique roles of retinol dehydrogenases in the mouse retina. Proc. Natl. Acad. Sci. U.S.A 104, 19565–19570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rattner A, Smallwood PM and Nathans J (2000) Identification and characterization of all-trans-retinol dehydrogenase from photoreceptor outer segments, the visual cycle enzyme that reduces all-trans-retinal to all-trans-retinol. J. Biol. Chem 275, 11034–11043 [DOI] [PubMed] [Google Scholar]
- 35.Palczewski K, Jager S, Buczylko J, Crouch RK, Bredberg DL, Hofmann KP, Asson-Batres MA and Saari JC (1994) Rod outer segment retinol dehydrogenase: substrate specificity and role in phototransduction. Biochemistry. 33, 13741–13750 [DOI] [PubMed] [Google Scholar]
- 36.Saari JC, Garwin GG, Van Hooser JP and Palczewski K (1998) Reduction of all-trans-retinal limits regeneration of visual pigment in mice. Vision Res. 38, 1325–1333 [DOI] [PubMed] [Google Scholar]
- 37.Chen C, Blakeley LR and Koutalos Y (2009) Formation of all-trans retinol after visual pigment bleaching in mouse photoreceptors. Invest Ophthalmol Vis Sci. 50, 3589–3595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Janecke AR, Thompson DA, Utermann G, Becker C, Hubner CA, Schmid E, McHenry CL, Nair AR, Ruschendorf F, Heckenlively J, Wissinger B, Nurnberg P and Gal A (2004) Mutations in RDH12 encoding a photoreceptor cell retinol dehydrogenase cause childhood-onset severe retinal dystrophy. Nat. Genet 36, 850–854 [DOI] [PubMed] [Google Scholar]
- 39.Maeda A, Maeda T, Imanishi Y, Kuksa V, Alekseev A, Bronson JD, Zhang H, Zhu L, Sun W, Saperstein DA, Rieke F, Baehr W and Palczewski K (2005) Role of photoreceptor-specific retinol dehydrogenase in the retinoid cycle in vivo. J. Biol. Chem 280, 18822–18832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen C, Thompson DA and Koutalos Y (2012) Reduction of all-trans-retinal in vertebrate rod photoreceptors requires the combined action of RDH8 and RDH12. J. Biol. Chem 287, 24662–24670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zimmerman WF, Lion F, Daemen FJ and Bonting SL (1975) Biochemical aspects of the visual process. XXX. Distribution of stereospecific retinol dehydrogenase activities in subcellular fractions of bovine retina and pigment epithelium. Exp. Eye Res 21, 325–332 [DOI] [PubMed] [Google Scholar]
- 42.Simon A, Hellman U, Wernstedt C and Eriksson U (1995) The Retinal-Pigment Epithelial-Specific 11-Cis Retinol Dehydrogenase Belongs to the Family of Short-Chain Alcohol Dehydrogenases. J. Biol. Chem 270, 1107–1112 [PubMed] [Google Scholar]
- 43.Williamson DH, Lund P and Krebs HA (1967) The redox state of free nicotinamide-adenine dinucleotide in the cytoplasm and mitochondria of rat liver. Biochem. J 103, 514–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sahu B, Sun W, Perusek L, Parmar V, Le YZ, Griswold MD, Palczewski K and Maeda A (2015) Conditional Ablation of Retinol Dehydrogenase 10 in the Retinal Pigmented Epithelium Causes Delayed Dark Adaption in Mice. J. Biol. Chem 290, 27239–27247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim TS, Maeda A, Maeda T, Heinlein C, Kedishvili N, Palczewski K and Nelson PS (2005) Delayed dark adaptation in 11-cis-retinol dehydrogenase-deficient mice: a role of RDH11 in visual processes in vivo. J. Biol. Chem 280, 8694–8704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gonzalez-Fernandez F, Kurz D, Bao Y, Newman S, Conway BP, Young JE, Han DP and Khani SC (1999) 11-cis retinol dehydrogenase mutations as a major cause of the congenital night-blindness disorder known as fundus albipunctatus. Mol. Vis 5, 41. [PubMed] [Google Scholar]
- 47.Yamamoto H, Simon A, Eriksson U, Harris E, Berson EL and Dryja TP (1999) Mutations in the gene encoding 11-cis retinol dehydrogenase cause delayed dark adaptation and fundus albipunctatus. Nat. Genet 22, 188–191 [DOI] [PubMed] [Google Scholar]
- 48.Skorczyk-Werner A, Pawlowski P, Michalczuk M, Warowicka A, Wawrocka A, Wicher K, Bakunowicz-Lazarczyk A and Krawczynski MR (2015) Fundus albipunctatus: review of the literature and report of a novel RDH5 gene mutation affecting the invariant tyrosine (p.Tyr175Phe). J. Appl. Genet 56, 317–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Driessen CA, Winkens HJ, Hoffmann K, Kuhlmann LD, Janssen BP, Van Vugt AH, Van Hooser JP, Wieringa BE, Deutman AF, Palczewski K, Ruether K and Janssen JJ (2000) Disruption of the 11-cis-retinol dehydrogenase gene leads to accumulation of cis-retinols and cis-retinyl esters. Mol. Cell. Biol 20, 4275–4287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jang GF, Van Hooser JP, Kuksa V, McBee JK, He YG, Janssen JJ, Driessen CA and Palczewski K (2001) Characterization of a dehydrogenase activity responsible for oxidation of 11-cis-retinol in the retinal pigment epithelium of mice with a disrupted RDH5 gene. A model for the human hereditary disease fundus albipunctatus. J. Biol. Chem 276, 32456–32465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hofmann L, Tsybovsky Y, Alexander NS, Babino D, Leung NY, Montell C, Banerjee S, von Lintig J and Palczewski K (2016) Structural Insights into the Drosophila melanogaster Retinol Dehydrogenase, a Member of the Short-Chain Dehydrogenase/Reductase Family. Biochemistry. 55, 6545–6557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang T and Montell C (2007) Phototransduction and retinal degeneration in Drosophila. Pflug. Arch. Eur. J. Phy 454, 821–847 [DOI] [PubMed] [Google Scholar]
- 53.Schafer M, Stevenson CEM, Wilkinson B, Lawson DM and Buttner MJ (2016) Substrate-Assisted Catalysis in Polyketide Reduction Proceeds via a Phenolate Intermediate. Cell Chem. Biol 23, 1091–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haeseleer F, Jang GF, Imanishi Y, Driessen C, Matsumura M, Nelson PS and Palczewski K (2002) Dual-substrate specificity short chain retinol dehydrogenases from the vertebrate retina. J. Biol. Chem 277, 45537–45546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ruiz A and Bok D (2010) Focus on molecules: lecithin retinol acyltransferase. Exp. Eye Res 90, 186–187 [DOI] [PubMed] [Google Scholar]
- 56.Sears AE and Palczewski K (2016) Lecithin:Retinol Acyltransferase: A Key Enzyme Involved in the Retinoid (visual) Cycle. Biochemistry. 55, 3082–3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.MacDonald PN and Ong DE (1988) A lecithin:retinol acyltransferase activity in human and rat liver. Biochem. Biophys. Res. Commun 156, 157–163 [DOI] [PubMed] [Google Scholar]
- 58.Saari JC and Bredberg DL (1989) Lecithin:retinol acyltransferase in retinal pigment epithelial microsomes. J. Biol. Chem 264, 8636–8640 [PubMed] [Google Scholar]
- 59.Ruiz A, Winston A, Lim YH, Gilbert BA, Rando RR and Bok D (1999) Molecular and biochemical characterization of lecithin retinol acyltransferase. J. Biol. Chem 274, 3834–3841 [DOI] [PubMed] [Google Scholar]
- 60.Anantharaman V and Aravind L (2003) Evolutionary history, structural features and biochemical diversity of the NlpC/P60 superfamily of enzymes. Genome Biol. 4, R11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mondal MS, Ruiz A, Bok D and Rando RR (2000) Lecithin retinol acyltransferase contains cysteine residues essential for catalysis. Biochemistry. 39, 5215–5220 [DOI] [PubMed] [Google Scholar]
- 62.Golczak M and Palczewski K (2010) An acyl-covalent enzyme intermediate of lecithin:retinol acyltransferase. J. Biol. Chem 285, 29217–29222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shi YQ, Hubacek I and Rando RR (1993) Kinetic mechanism of lecithin retinol acyl transferase. Biochemistry. 32, 1257–1263 [DOI] [PubMed] [Google Scholar]
- 64.Golczak M, Imanishi Y, Kuksa V, Maeda T, Kubota R and Palczewski K (2005) Lecithin:retinol acyltransferase is responsible for amidation of retinylamine, a potent inhibitor of the retinoid cycle. J. Biol. Chem 280, 42263–42273 [DOI] [PubMed] [Google Scholar]
- 65.Amengual J, Golczak M, Palczewski K and von Lintig J (2012) Lecithin: Retinol Acyltransferase Is Critical for Cellular Uptake of Vitamin A from Serum Retinol-binding Protein. J. Biol. Chem 287, 24216–24227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Trehan A, Canada FJ and Rando RR (1990) Inhibitors of retinyl ester formation also prevent the biosynthesis of 11-cis-retinol. Biochemistry. 29, 309–312 [DOI] [PubMed] [Google Scholar]
- 67.Moiseyev G, Crouch RK, Goletz P, Oatis J Jr., Redmond TM and Ma JX (2003) Retinyl esters are the substrate for isomerohydrolase. Biochemistry. 42, 2229–2238 [DOI] [PubMed] [Google Scholar]
- 68.Thompson DA, Li Y, McHenry CL, Carlson TJ, Ding X, Sieving PA, Apfelstedt-Sylla E and Gal A (2001) Mutations in the gene encoding lecithin retinol acyltransferase are associated with early-onset severe retinal dystrophy. Nat. Genet 28, 123–124 [DOI] [PubMed] [Google Scholar]
- 69.Batten ML, Imanishi Y, Maeda T, Tu DC, Moise AR, Bronson D, Possin D, Van Gelder RN, Baehr W and Palczewski K (2004) Lecithin-retinol acyltransferase is essential for accumulation of all-trans-retinyl esters in the eye and in the liver. J. Biol. Chem 279, 10422–10432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Moise AR, Golczak M, Imanishi Y and Palczewski K (2007) Topology and membrane association of lecithin: retinol acyltransferase. J. Biol. Chem 282, 2081–2090 [DOI] [PubMed] [Google Scholar]
- 71.Jahng WJ, Cheung E and Rando RR (2002) Lecithin retinol acyltransferase forms functional homodimers. Biochemistry. 41, 6311–6319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Golczak M, Sears AE, Kiser PD and Palczewski K (2015) LRAT-specific domain facilitates vitamin A metabolism by domain swapping in HRASLS3. Nat. Chem. Biol 11, 26–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Golczak M, Kiser PD, Sears AE, Lodowski DT, Blaner WS and Palczewski K (2012) Structural Basis for the Acyltransferase Activity of Lecithin:Retinol Acyltransferase-like Proteins. J. Biol. Chem 287, 23790–23807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pang XY, Cao J, Addington L, Lovell S, Battaile KP, Zhang N, Rao JL, Dennis EA and Moise AR (2012) Structure/function relationships of adipose phospholipase A2 containing a cys-his-his catalytic triad. J. Biol. Chem 287, 35260–35274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Napoli JL (2016) Functions of Intracellular Retinoid Binding-Proteins. Subcell. Biochem 81, 21–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Redmond TM (2009) Focus on Molecules: RPE65, the visual cycle retinol isomerase. Exp. Eye Res 88, 846–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hamel CP, Tsilou E, Pfeffer BA, Hooks JJ, Detrick B and Redmond TM (1993) Molecular cloning and expression of RPE65, a novel retinal pigment epithelium-specific microsomal protein that is post-transcriptionally regulated in vitro. J. Biol. Chem 268, 15751–15757 [PubMed] [Google Scholar]
- 78.Redmond TM, Poliakov E, Yu S, Tsai JY, Lu Z and Gentleman S (2005) Mutation of key residues of RPE65 abolishes its enzymatic role as isomerohydrolase in the visual cycle. Proc. Natl. Acad. Sci. U.S.A 102, 13658–13663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Moiseyev G, Chen Y, Takahashi Y, Wu BX and Ma JX (2005) RPE65 is the isomerohydrolase in the retinoid visual cycle. Proc. Natl. Acad. Sci. U.S.A 102, 12413–12418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jin M, Li S, Moghrabi WN, Sun H and Travis GH (2005) Rpe65 is the retinoid isomerase in bovine retinal pigment epithelium. Cell. 122, 449–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Marlhens F, Bareil C, Griffoin J-M, Zrenner E, Amalric P, Eliaou C, Liu S-Y, Harris E, Redmond TM, Arnaud B, Claustres M and Hamel CP (1997) Mutations in RPE65 cause Leber’s congenital amaurosis. Nat. Genet 17, 139–141 [DOI] [PubMed] [Google Scholar]
- 82.Gu SM, Thompson DA, Srikumari CR, Lorenz B, Finckh U, Nicoletti A, Murthy KR, Rathmann M, Kumaramanickavel G, Denton MJ and Gal A (1997) Mutations in RPE65 cause autosomal recessive childhood-onset severe retinal dystrophy. Nat. Genet 17, 194–197 [DOI] [PubMed] [Google Scholar]
- 83.Redmond TM, Yu S, Lee E, Bok D, Hamasaki D, Chen N, Goletz P, Ma JX, Crouch RK and Pfeifer K (1998) Rpe65 is necessary for production of 11-cis-vitamin A in the retinal visual cycle. Nat. Genet 20, 344–351 [DOI] [PubMed] [Google Scholar]
- 84.Russell S, Bennett J, Wellman JA, Chung DC, Yu ZF, Tillman A, Wittes J, Pappas J, Elci O, McCague S, Cross D, Marshall KA, Walshire J, Kehoe TL, Reichert H, Davis M, Raffini L, George LA, Hudson FP, Dingfield L, Zhu X, Haller JA, Sohn EH, Mahajan VB, Pfeifer W, Weckmann M, Johnson C, Gewaily D, Drack A, Stone E, Wachtel K, Simonelli F, Leroy BP, Wright JF, High KA and Maguire AM (2017) Efficacy and safety of voretigene neparvovec (AAV2-hRPE65v2) in patients with RPE65-mediated inherited retinal dystrophy: a randomised, controlled, open-label, phase 3 trial. Lancet. 390, 849–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Poliakov E, Gubin AN, Stearn O, Li Y, Campos MM, Gentleman S, Rogozin IB and Redmond TM (2012) Origin and evolution of retinoid isomerization machinery in vertebrate visual cycle: hint from jawless vertebrates. PLoS ONE. 7, e49975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sui X, Golczak M, Zhang J, Kleinberg KA, von Lintig J, Palczewski K and Kiser PD (2015) Utilization of Dioxygen by Carotenoid Cleavage Oxygenases. J. Biol. Chem 290, 30212–30223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Deigner PS, Law WC, Canada FJ and Rando RR (1989) Membranes as the energy source in the endergonic transformation of vitamin A to 11-cis-retinol. Science. 244, 968–971 [DOI] [PubMed] [Google Scholar]
- 88.McBee JK, Kuksa V, Alvarez R, de Lera AR, Prezhdo O, Haeseleer F, Sokal I and Palczewski K (2000) Isomerization of all-trans-retinol to cis-retinols in bovine retinal pigment epithelial cells: dependence on the specificity of retinoid-binding proteins. Biochemistry. 39, 11370–11380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Redmond TM, Poliakov E, Kuo S, Chander P and Gentleman S (2010) RPE65, visual cycle retinol isomerase, is not inherently 11-cis-specific: support for a carbocation mechanism of retinol isomerization. J. Biol. Chem 285, 1919–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Poliakov E, Soucy J, Gentleman S, Rogozin IB and Redmond TM (2017) Phylogenetic analysis of the metazoan carotenoid oxygenase superfamily: a new ancestral gene assemblage of BCO-like (BCOL) proteins. Sci. Rep 7, 13192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kiser PD, Farquhar ER, Shi W, Sui X, Chance MR and Palczewski K (2012) Structure of RPE65 isomerase in a lipidic matrix reveals roles for phospholipids and iron in catalysis. Proc. Natl. Acad. Sci. U.S.A 109, E2747–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Moiseyev G, Takahashi Y, Chen Y, Gentleman S, Redmond TM, Crouch RK and Ma JX (2006) RPE65 is an iron(II)-dependent isomerohydrolase in the retinoid visual cycle. J. Biol. Chem 281, 2835–2840 [DOI] [PubMed] [Google Scholar]
- 93.Kiser PD, Golczak M, Lodowski DT, Chance MR and Palczewski K (2009) Crystal structure of native RPE65, the retinoid isomerase of the visual cycle. Proc. Natl. Acad. Sci. U.S.A 106, 17325–17330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Takahashi Y, Moiseyev G, Ablonczy Z, Chen Y, Crouch RK and Ma JX (2009) Identification of a novel palmitylation site essential for membrane association and isomerohydrolase activity of RPE65. J. Biol. Chem 284, 3211–3218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kiser PD and Palczewski K (2010) Membrane-binding and enzymatic properties of RPE65. Prog. Retin. Eye Res 29, 428–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Golczak M, Kiser PD, Lodowski DT, Maeda A and Palczewski K (2010) Importance of membrane structural integrity for RPE65 retinoid isomerization activity. J. Biol. Chem 285, 9667–9682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nikolaeva O, Moiseyev G, Rodgers KK and Ma JX (2011) Binding to lipid membrane induces conformational changes in RPE65: implications for its isomerohydrolase activity. Biochem. J 436, 591–597 [DOI] [PubMed] [Google Scholar]
- 98.Kiser PD, Zhang J, Badiee M, Li Q, Shi W, Sui X, Golczak M, Tochtrop GP and Palczewski K (2015) Catalytic mechanism of a retinoid isomerase essential for vertebrate vision. Nat. Chem. Biol 11, 409–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chander P, Gentleman S, Poliakov E and Redmond TM (2012) Aromatic residues in the substrate cleft of RPE65 protein govern retinol isomerization and modulate its progression. J. Biol. Chem 287, 30552–30559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Takahashi Y, Moiseyev G, Nikolaeva O and Ma JX (2012) Identification of the key residues determining the product specificity of isomerohydrolase. Biochemistry. 51, 4217–4225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Crabb JW, Goldflam S, Harris SE and Saari JC (1988) Cloning of the cDNAs encoding the cellular retinaldehyde-binding protein from bovine and human retina and comparison of the protein structures. J. Biol. Chem 263, 18688–18692 [PubMed] [Google Scholar]
- 102.Crabb JW, Johnson CM, Carr SA, Armes LG and Saari JC (1988) The complete primary structure of the cellular retinaldehyde-binding protein from bovine retina. J. Biol. Chem 263, 18678–18687 [PubMed] [Google Scholar]
- 103.Saari JC and Crabb JW (2005) Focus on molecules: cellular retinaldehyde-binding protein (CRALBP). Exp. Eye Res 81, 245–246 [DOI] [PubMed] [Google Scholar]
- 104.Saari JC (2016) Vitamin A and Vision. Subcell. Biochem 81, 231–259 [DOI] [PubMed] [Google Scholar]
- 105.Saari JC and Bredberg DL (1988) Purification of cellular retinaldehyde-binding protein from bovine retina and retinal pigment epithelium. Exp. Eye Res 46, 569–578 [DOI] [PubMed] [Google Scholar]
- 106.Bunt-Milam AH and Saari JC (1983) Immunocytochemical localization of two retinoid-binding proteins in vertebrate retina. J. Cell Biol 97, 703–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Saari JC, Bredberg L and Garwin GG (1982) Identification of the endogenous retinoids associated with three cellular retinoid-binding proteins from bovine retina and retinal pigment epithelium. J. Biol. Chem 257, 13329–13333 [PubMed] [Google Scholar]
- 108.Saari JC, Nawrot M, Stenkamp RE, Teller DC and Garwin GG (2009) Release of 11-cis-retinal from cellular retinaldehyde-binding protein by acidic lipids. Mol. Vis 15, 844–854 [PMC free article] [PubMed] [Google Scholar]
- 109.Golovleva I, Bhattacharya S, Wu Z, Shaw N, Yang Y, Andrabi K, West KA, Burstedt MS, Forsman K, Holmgren G, Sandgren O, Noy N, Qin J and Crabb JW (2003) Disease-causing mutations in the cellular retinaldehyde binding protein tighten and abolish ligand interactions. J. Biol. Chem 278, 12397–12402 [DOI] [PubMed] [Google Scholar]
- 110.Thompson DA and Gal A (2003) Vitamin A metabolism in the retinal pigment epithelium: genes, mutations, and diseases. Prog. Retin. Eye Res 22, 683–703 [DOI] [PubMed] [Google Scholar]
- 111.Maw MA, Kennedy B, Knight A, Bridges R, Roth KE, Mani EJ, Mukkadan JK, Nancarrow D, Crabb JW and Denton MJ (1997) Mutation of the gene encoding cellular retinaldehyde-binding protein in autosomal recessive retinitis pigmentosa. Nat. Genet 17, 198–200 [DOI] [PubMed] [Google Scholar]
- 112.Morimura H, Berson EL and Dryja TP (1999) Recessive mutations in the RLBP1 gene encoding cellular retinaldehyde-binding protein in a form of retinitis punctata albescens. Invest. Ophthalmol. Vis. Sci 40, 1000–1004 [PubMed] [Google Scholar]
- 113.Choi VW, Bigelow CE, McGee TL, Gujar AN, Li H, Hanks SM, Vrouvlianis J, Maker M, Leehy B, Zhang Y, Aranda J, Bounoutas G, Demirs JT, Yang J, Ornberg R, Wang Y, Martin W, Stout KR, Argentieri G, Grosenstein P, Diaz D, Turner O, Jaffee BD, Police SR and Dryja TP (2015) AAV-mediated RLBP1 gene therapy improves the rate of dark adaptation in Rlbp1 knockout mice. Mol. Ther. Methods Clin. Dev 2, 15022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Burstedt MS, Sandgren O, Holmgren G and Forsman-Semb K (1999) Bothnia dystrophy caused by mutations in the cellular retinaldehyde-binding protein gene (RLBP1) on chromosome 15q26. Invest. Ophthalmol. Vis. Sci 40, 995–1000 [PubMed] [Google Scholar]
- 115.Saari JC, Nawrot M, Kennedy BN, Garwin GG, Hurley JB, Huang J, Possin DE and Crabb JW (2001) Visual cycle impairment in cellular retinaldehyde binding protein (CRALBP) knockout mice results in delayed dark adaptation. Neuron. 29, 739–748 [DOI] [PubMed] [Google Scholar]
- 116.Stecher H, Gelb MH, Saari JC and Palczewski K (1999) Preferential release of 11-cis-retinol from retinal pigment epithelial cells in the presence of cellular retinaldehyde-binding protein. J. Biol. Chem 274, 8577–8585 [DOI] [PubMed] [Google Scholar]
- 117.Xue Y, Shen SQ, Jui J, Rupp AC, Byrne LC, Hattar S, Flannery JG, Corbo JC and Kefalov VJ (2015) CRALBP supports the mammalian retinal visual cycle and cone vision. J. Clin. Invest 125, 727–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wu Z, Yang Y, Shaw N, Bhattacharya S, Yan L, West K, Roth K, Noy N, Qin J and Crabb JW (2003) Mapping the ligand binding pocket in the cellular retinaldehyde binding protein. J. Biol. Chem 278, 12390–12396 [DOI] [PubMed] [Google Scholar]
- 119.He XQ, Lobsiger J and Stocker A (2009) Bothnia dystrophy is caused by domino-like rearrangements in cellular retinaldehyde-binding protein mutant R234W. Proc. Natl. Acad. Sci. U.S.A 106, 18545–18550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kawaguchi R, Yu J, Honda J, Hu J, Whitelegge J, Ping P, Wiita P, Bok D and Sun H (2007) A membrane receptor for retinol binding protein mediates cellular uptake of vitamin A. Science. 315, 820–825 [DOI] [PubMed] [Google Scholar]
- 121.Kanai M, Raz A and Goodman DS (1968) Retinol-binding protein: the transport protein for vitamin A in human plasma. J. Clin. Invest 47, 2025–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Blomhoff R, Green MH, Berg T and Norum KR (1990) Transport and storage of vitamin A. Science. 250, 399–404 [DOI] [PubMed] [Google Scholar]
- 123.Heller J (1975) Interactions of plasma retinol-binding protein with its receptor. Specific binding of bovine and human retinol-binding protein to pigment epithelium cells from bovine eyes. J. Biol. Chem 250, 3613–3619 [PubMed] [Google Scholar]
- 124.Heller M and Bok D (1976) A specific receptor for retinol binding protein as detected by the binding of human and bovine retinol binding protein to pigment epithelial cells. Am. J. Ophthalmol 81, 93–97 [DOI] [PubMed] [Google Scholar]
- 125.Bouillet P, Sapin V, Chazaud C, Messaddeq N, Decimo D, Dolle P and Chambon P (1997) Developmental expression pattern of Stra6, a retinoic acid-responsive gene encoding a new type of membrane protein. Mech. Dev 63, 173–186 [DOI] [PubMed] [Google Scholar]
- 126.Kawaguchi R, Zhong M, Kassai M, Ter-Stepanian M and Sun H (2012) STRA6-catalyzed vitamin A influx, efflux, and exchange. J. Membr. Biol 245, 731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chelstowska S, Widjaja-Adhi MA, Silvaroli JA and Golczak M (2016) Molecular Basis for Vitamin A Uptake and Storage in Vertebrates. Nutrients. 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Golzio C, Martinovic-Bouriel J, Thomas S, Mougou-Zrelli S, Grattagliano-Bessieres B, Bonniere M, Delahaye S, Munnich A, Encha-Razavi F, Lyonnet S, Vekemans M, Attie-Bitach T and Etchevers HC (2007) Matthew-Wood syndrome is caused by truncating mutations in the retinol-binding protein receptor gene STRA6. Am. J. Hum. Genet 80, 1179–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Pasutto F, Sticht H, Hammersen G, Gillessen-Kaesbach G, Fitzpatrick DR, Nurnberg G, Brasch F, Schirmer-Zimmermann H, Tolmie JL, Chitayat D, Houge G, Fernandez-Martinez L, Keating S, Mortier G, Hennekam RC, von der Wense A, Slavotinek A, Meinecke P, Bitoun P, Becker C, Nurnberg P, Reis A and Rauch A (2007) Mutations in STRA6 cause a broad spectrum of malformations including anophthalmia, congenital heart defects, diaphragmatic hernia, alveolar capillary dysplasia, lung hypoplasia, and mental retardation. Am. J. Hum. Genet 80, 550–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chassaing N, Golzio C, Odent S, Lequeux L, Vigouroux A, Martinovic-Bouriel J, Tiziano FD, Masini L, Piro F, Maragliano G, Delezoide AL, Attie-Bitach T, Manouvrier-Hanu S, Etchevers HC and Calvas P (2009) Phenotypic spectrum of STRA6 mutations: from Matthew-Wood syndrome to non-lethal anophthalmia. Hum. Mutat 30, E673–681 [DOI] [PubMed] [Google Scholar]
- 131.Amengual J, Zhang N, Kemerer M, Maeda T, Palczewski K and Von Lintig J (2014) STRA6 is critical for cellular vitamin A uptake and homeostasis. Hum. Mol. Genet 23, 5402–5417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ruiz A, Mark M, Jacobs H, Klopfenstein M, Hu J, Lloyd M, Habib S, Tosha C, Radu RA, Ghyselinck NB, Nusinowitz S and Bok D (2012) Retinoid content, visual responses, and ocular morphology are compromised in the retinas of mice lacking the retinol-binding protein receptor, STRA6. Invest. Ophthalmol. Vis. Sci 53, 3027–3039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chen Y, Clarke OB, Kim J, Stowe S, Kim YK, Assur Z, Cavalier M, Godoy-Ruiz R, von Alpen DC, Manzini C, Blaner WS, Frank J, Quadro L, Weber DJ, Shapiro L, Hendrickson WA and Mancia F (2016) Structure of the STRA6 receptor for retinol uptake. Science. 353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Noy N (2015) Signaling by retinol and its serum binding protein. Prostaglandins Leukot. Essent. Fatty Acids 93, 3–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Berry DC, Jin H, Majumdar A and Noy N (2011) Signaling by vitamin A and retinol-binding protein regulates gene expression to inhibit insulin responses. Proc. Natl. Acad. Sci. U.S.A 108, 4340–4345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Berry DC, O’Byrne SM, Vreeland AC, Blaner WS and Noy N (2012) Cross talk between signaling and vitamin A transport by the retinol-binding protein receptor STRA6. Mol. Cell. Biol 32, 3164–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Shaw NS and Noy N (2001) Interphotoreceptor retinoid-binding protein contains three retinoid binding sites. Exp. Eye Res 72, 183–190 [DOI] [PubMed] [Google Scholar]
- 138.Chen C, Adler L. t., Goletz P, Gonzalez-Fernandez F, Thompson DA and Koutalos Y (2017) Interphotoreceptor retinoid-binding protein removes all-trans-retinol and retinal from rod outer segments, preventing lipofuscin precursor formation. J. Biol. Chem 292, 19356–19365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Loew A and Gonzalez-Fernandez F (2002) Crystal structure of the functional unit of interphotoreceptor retinoid binding protein. Structure. 10, 43–49 [DOI] [PubMed] [Google Scholar]
- 140.Ghosh D, Haswell KM, Sprada M and Gonzalez-Fernandez F (2015) Structure of zebrafish IRBP reveals fatty acid binding. Exp. Eye Res 140, 149–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ripps H, Peachey NS, Xu X, Nozell SE, Smith SB and Liou GI (2000) The rhodopsin cycle is preserved in IRBP “knockout” mice despite abnormalities in retinal structure and function. Vis. Neurosci 17, 97–105 [DOI] [PubMed] [Google Scholar]
- 142.Kolesnikov AV, Tang PH, Parker RO, Crouch RK and Kefalov VJ (2011) The mammalian cone visual cycle promotes rapid M/L-cone pigment regeneration independently of the interphotoreceptor retinoid-binding protein. J. Neurosci 31, 7900–7909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kawaguchi R, Yu J, Wiita P, Honda J and Sun H (2008) An essential ligand-binding domain in the membrane receptor for retinol-binding protein revealed by large-scale mutagenesis and a human polymorphism. J. Biol. Chem 283, 15160–15168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kawaguchi R, Yu J, Wiita P, Ter-Stepanian M and Sun H (2008) Mapping the membrane topology and extracellular ligand binding domains of the retinol binding protein receptor. Biochemistry. 47, 5387–5395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Napoli JL (2017) Cellular retinoid binding-proteins, CRBP, CRABP, FABP5: Effects on retinoid metabolism, function and related diseases. Pharmacol. Ther 173, 19–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Silvaroli JA, Arne JM, Chelstowska S, Kiser PD, Banerjee S and Golczak M (2016) Ligand Binding Induces Conformational Changes in Human Cellular Retinol-binding Protein 1 (CRBP1) Revealed by Atomic Resolution Crystal Structures. J. Biol. Chem 291, 8528–8540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Cascella M, Barfuss S and Stocker A (2013) Cis-retinoids and the chemistry of vision. Arch. Biochem. Biophys 539, 187–195 [DOI] [PubMed] [Google Scholar]
- 148.Wenzel A, Oberhauser V, Pugh EN Jr., Lamb TD, Grimm C, Samardzija M, Fahl E, Seeliger MW, Reme CE and von Lintig J (2005) The retinal G protein-coupled receptor (RGR) enhances isomerohydrolase activity independent of light. J. Biol. Chem 280, 29874–29884 [DOI] [PubMed] [Google Scholar]
- 149.Radu RA, Hu J, Peng J, Bok D, Mata NL and Travis GH (2008) Retinal pigment epithelium-retinal G protein receptor-opsin mediates light-dependent translocation of all-trans-retinyl esters for synthesis of visual chromophore in retinal pigment epithelial cells. J. Biol. Chem 283, 19730–19738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Cook JD, Ng SY, Lloyd M, Eddington S, Sun H, Nathans J, Bok D, Radu RA and Travis GH (2017) Peropsin modulates transit of vitamin A from retina to retinal pigment epithelium. J. Biol. Chem 292, 21407–21416 [DOI] [PMC free article] [PubMed] [Google Scholar]