Summary:
The carboxy-terminal domain (CTD) of RNA polymerase II (Pol II) is composed of repeats of the consensus YSPTSPS, and is an essential binding scaffold for transcription-associated factors. Metazoan CTDs have well-conserved lengths and sequence compositions arising from the evolution of divergent motifs, features thought to be essential for development. To the contrary, we show that a truncated CTD composed solely of YSPTSPS repeats supports Drosophila viability, but a CTD with enough YSPTSPS repeats to match the length of the wild-type Drosophila CTD is defective. Furthermore, a fluorescently-tagged CTD lacking the rest of Pol II dynamically enters transcription compartments, indicating that the CTD functions as a signal sequence. However, CTDs with too many YSPTSPS repeats are more prone to localize to static nuclear foci separate from the chromosomes. We propose that the sequence complexity of the CTD offsets aberrant behavior caused by excessive repetitive sequences without compromising its targeting function.
Keywords: Pol II CTD, consensus heptads, divergent motifs, signal sequence, transcription compartment, Drosophila genetics
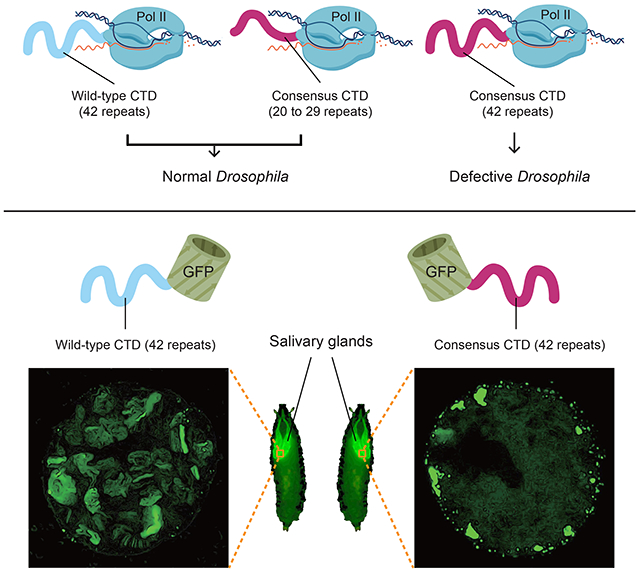
Graphical Abstract
eTOC blurb:
Using Drosophila, Lu et al. show that RNA Polymerase II lacking conserved divergent motifs supports development, that the C-terminal domain alone is localized to sites of transcription and illuminate the roles of divergent C-terminal domain motifs in supporting proper Pol II function and prevention of aberrant self-assembly.
Introduction:
Transcription in eukaryotes involves coordination of RNA synthesis, RNA processing and modulation of chromatin structure. The CTD of the largest Pol II subunit, Rpb1, plays a central role by serving as a landing pad for many of the proteins involved in these processes (Corden, 2013; Egloff et al., 2012; Eick and Geyer, 2013; Harlen and Churchman, 2017; Zaborowska et al., 2016). There is considerable variation in the amino acid sequence composition of CTDs across different evolutionary lineages (Yang and Stiller, 2014). For example, the yeast CTD and the proximal half of the mammalian CTD are composed primarily of the repeating consensus heptad, YSPTSPS, whereas the distal half of the mammalian CTD consists primarily of divergent motifs, which differ from the consensus at one or more positions (Figure 1A). In addition, the length of the CTD roughly scales with developmental complexity (Yang and Stiller, 2014). The divergent motifs and CTD length are postulated to have evolved to provide additional layers of control for gene expression programs essential for development of multicellular organisms (Chapman et al., 2008; Corden, 2013; Dias et al., 2015; Egloff et al., 2012; Eick and Geyer, 2013; Schröder et al., 2013; Simonti et al., 2015; Sims et al., 2011; Zaborowska et al., 2016; Zhao et al., 2016), although this remains largely untested.
Figure 1.
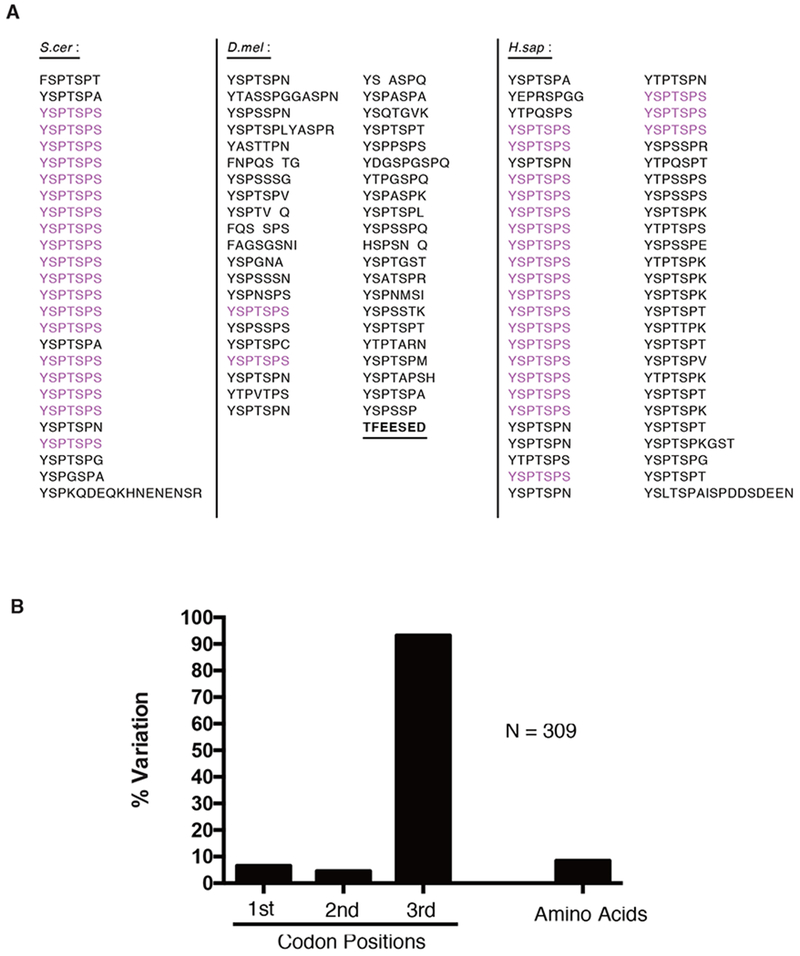
The CTD of Drosophila is composed primarily of divergent motifs that are under selective pressure
(A) Display of the CTD of S. cerevisiae (26 repeats), D. melanogaster (42 repeats) and H sapien (52 repeats) with consensus heptads highlighted in magenta. The acidic tip (underlined) was retained in all consensus CTD mutants described throughout this study.
(B) Variation in the codon positions and amino acids of the Drosophila CTD. Variation was defined as having at least one sequence variant among 12 species of Drosophila.
See also Figure S1.
Here, we assess the significance of sequence complexity and length of the CTD in animal development and viability by using Drosophila as a model. The Drosophila CTD contains 42 repeats, only 2 of which match the consensus heptad (Figure 1A). Surprisingly, CTDs composed solely of YSPTSPS repeats suffice to support fly development. In addition, although consensus CTDs at approximately half wild-type lengths fully support development, consensus CTDs at wild-type lengths do not. Thus, too many consensus heptads is deleterious, and this can be counteracted by substituting consensus heptads with divergent motifs. These results argue against the notion that highly-conserved, lineage-specific divergent motifs and lengths of the CTD are essential for metazoans. To gain insight into the functional differences between various CTDs, we analyzed their behaviors when expressed separately from the rest of Pol II. We observe that a GFP-tagged wild-type Drosophila CTD dynamically enters transcription compartments. This defines the CTD as a signal sequence that could serve to target Pol II to sites of active transcription. Furthermore, CTDs harboring too many consensus heptads form static nuclear foci when fused to GFP, and fail to support viability in the context of Pol II. Our results suggest the sequence of the CTD has evolved to balance length and composition in order to dynamically target Pol II to transcription compartments without resulting in aggregation.
Results:
A CTD Solely Composed of 29 Repeats of Consensus Heptads Is Sufficient for Viability of Drosophila
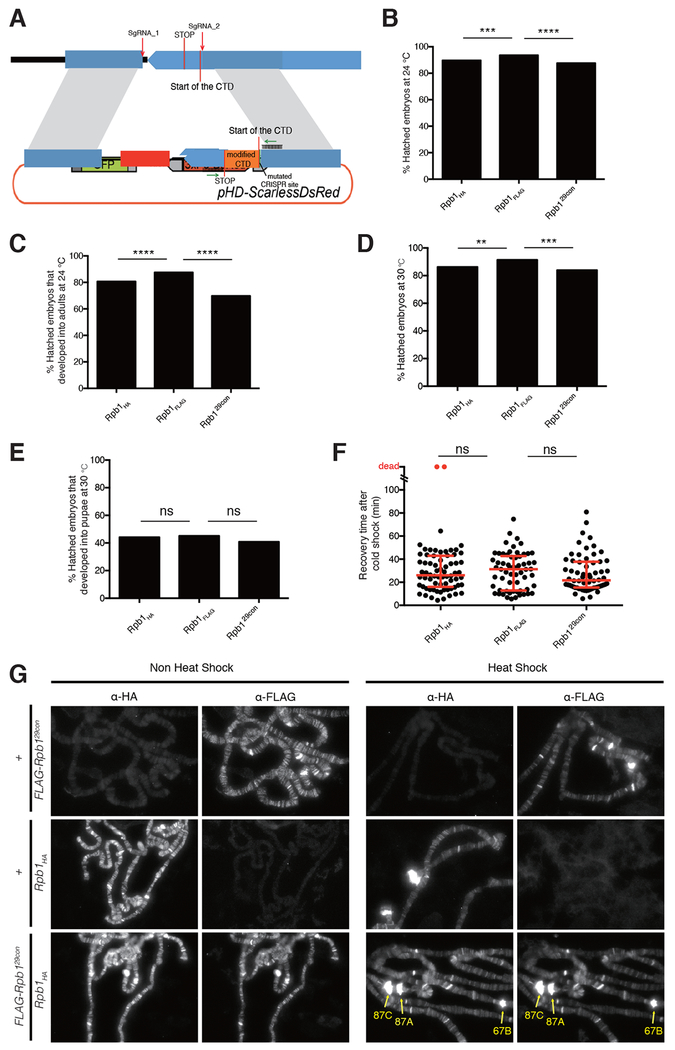
Differences between the levels of amino acid sequence conservation and the underlying nucleic acid conservation for Drosophila species indicates that the amino acid sequence of the CTD is subject to strong purifying selection (Figure 1B and Figure S1). This observation, and prior analysis revealing lineage specific conservation of divergent motifs in metazoans (Yang and Stiller, 2014), suggests the diversity of motifs is adaptive. Leveraging the sequence complexity of the Drosophila CTD, we set out to test the long-standing hypothesis that the divergent motifs enriched among metazoan CTDs and conserved within lineages serve crucial roles in development. Previously, we found that ectopic expression of an RNAi-resistant Rpb1 mutant with as few as 29 Drosophila motifs, including the two consensus heptads, rescued lethality caused by depleting endogenous Rpb1 with RNAi (Gibbs et al., 2017). To assess whether viability was conferred by the remaining divergent motifs, we matched the length of this mutant by replacing the Drosophila CTD with a FLAG-tagged, all-consensus CTD (Rpb129con hereafter) composed of 29 YSPTSPS heptads. Following CRISPR-Cas9-mediated replacement of sequences encoding the wild-type CTD, we obtained homozygous viable flies whose only Rpb1 contained 29 consensus heptads (Figure 2A). For comparison, two fly lines with either an HA tag or a FLAG tag appended to the C-terminus of a wild-type Drosophila CTD were generated (Rpb1HA and Rpb1FLAG respectively hereafter). Rpb129con flies were indistinguishable in morphology from these wild-type counterparts. Hatch rates and percentages of Rpb129con individuals that completed development when grown at 24°C were only slightly less than wild-type (Figure 2B and 2C). When raised at 30°C, these three fly lines were also morphologically indistinguishable, showed similar hatch rates and similar percentages of individuals developing into pupae (Figure 2D and 2E). In addition, Rpb129con flies recovered normally after cold shock (Figure 2F). Rpb129con flies were mated to Rpb1HA flies to generate larvae expressing both forms of Pol II. Immunostaining of polytene chromosomes for the two forms of Rpb1 with anti-FLAG and anti-HA antibodies revealed nearly identical staining patterns both before and after heat shock (Figure 2G). In contrast to the hypothesized role of divergent motifs in metazoa, these observations show that Pol II harboring a fully-consensus CTD is sufficient to execute the complex gene regulatory programs necessary for development, reproduction and thermotolerance.
Figure 2.
Replacing endogenous Rpb1 with one that has 29 consensus heptads produces healthy homozygous flies
(A) Schematic for CRISPR-mediated replacement of the endogenous Drosophila CTD. Rpb1HA and Rpb1FLAG have the wild-type Drosophila CTD sequence with a HA or FLAG-tag attached to the end of the C-terminus. Rpb129con has 29 YSPTSPS motifs substituted for the wild-type CTD and a FLAG-tag attached to the end of the C-terminus. The sequence of the CTD (orange) in each fly line was verified by sequencing the PCR product amplified from genomic DNA with primers flanking the CTD (green arrows).
(B) Hatch rates of embryos at 24°C, measured 36h after egg deposition, n>300 for each genotype.
(C) Percentages of hatched embryos that developed into adults when raised at 24°C, n>200 for each genotype.
(D) Hatch rates of embryos at 30°C, measured 36h after egg deposition, n>300 for each genotype.
(E) Percentages of hatched embryos that developed into pupae when raised at 30°C, n>200 for each genotype.
(F) Quantification of recovery time after 14h of cold shock on ice, n=70 for Rpb1HA, n=64 for Rpb1FLAG; n=64 for Rpb129con (equal numbers of male and female adults were assayed). Dots represent individual adults. Red bars show the medians and interquartile ranges.
(G) Immunofluorescence with anti-HA and anti-FLAG antibodies on polytene chromosomes derived from salivary glands of third-instar larvae (FLAG-Rpb129con/Rpb1HA) under non heat shock and heat shock conditions. Chromosomes derived from salivary glands lacking either tagged form of Rpb1 (FLAG-Rpb129con/+ or Rpb1HA/+) served as negative controls. The heat shock puffs are readily visible as pointed by yellow arrows.
Two-sided chi-squared tests were used for (B) to (E). Mann-Whitney U tests were used for (F). ns: not significant (p>0.05), *p < 0.05, **p < 0.01, ***p < 0.001 or ****p < 0.0001 values were considered statistically significant from Rpb1FLAG.
A Consensus CTD Matching the Wild-type Length of the Drosophila CTD Is Defective
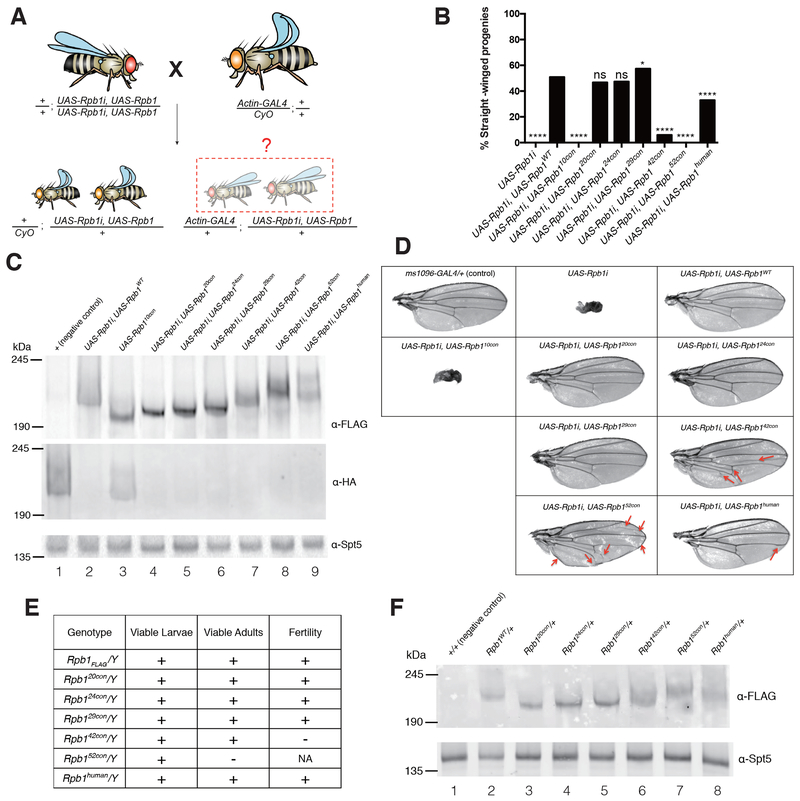
Like sequence complexity, the length of the CTD is highly conserved within taxonomic lineages, and is also thought to be essential for viability (Chapman et al., 2008; Corden, 2013; Eick and Geyer, 2013; Liu et al., 2010; Yang and Stiller, 2014; Zaborowska et al., 2016). However, the role of conserved CTD length has been difficult to study in developmentally complex organisms, where changing CTD length also alters motif composition. Our finding that Pol II with a fully-consensus CTD functions in the fly enabled us to directly test the role of altered CTD length on fly viability, divorced from the confounding influence of altered numbers of divergent motifs. To this end, we used our previously described RNAi rescue assay (Gibbs et al., 2017; Portz et al., 2017) to test if CTDs with 10, 20, 24, 42 or 52 consensus heptads (Rpb110con, Rpb120con, Rpb124con, Rpb142con and Rpb152con) could rescue flies from the lethality caused by depleting endogenous Rpb1 with RNAi (Figure 3A). Act-GAL4-mediated expression of the RNAi against Rpb1 in all tissues resulted in complete lethality (Figure 3B, UAS-Rpb1i). Co-expression of RNAi-resistant forms of Rpb120con or Rpb124con restored viability, whereas co-expression of Rpb110con or Rpb152con did not (Figure 3B). In contrast to Rpb152con, the human CTD that also has 52 repeats (Rpb1human) was able to restore viability (Figure 3B), raising the possibility that the divergent motifs counteract defects caused by having too many consensus heptads. Likewise, the Rpb142con, a mutant closely matching the length of the Drosophila CTD, restored viability into adulthood but was markedly less efficient than Rpb1WT (Figure 3B and Figure S2A). Importantly, Rpb110con, Rpb142con and Rpb152con were expressed at levels similar to or greater than Rpb1WT, suggesting that the failure to rescue was not due to the instability of these mutants (Figure 3C, cf lanes 4, 7 and 8 to lane 2). In addition, endogenous Rpb1 was efficiently depleted (Figure 3C, cf lane 1 to lanes 2 and 4 to 9). Therefore, the differences in rescue efficiency was not due to variation in the levels of Rpb1 knock down.
Figure 3.
CTDs with too few or too many consensus heptads are dysfunctional
(A) Schematic of the ubiquitous Rpb1i rescue assay. The Act-GAL4 transgene expresses GAL4 throughout development and drives expression of UAS-associated transgenes. UAS-Rpb1i is a GAL4-activated transgene encoding RNAi that knocks down expression of endogenous Rpb1. Rpb1i expression in the absence of ectopically expressed Rpb1 eliminates all straight-winged adult progeny (Act-GAL4/+; UAS-Rpb1i/+). Co-expression of a functional derivative of Rpb1 that has been rendered resistant to the RNAi by synonymous mutations rescues the lethality and yields straight-winged progeny.
(B) Quantification of the results from the Rpb1i rescue assay, n>150 for each genotype. 10con, 20con, 24con, 29con, 42con, and 52con designate Rpb1 derivatives with 10 to 52 consensus heptads. WT designates the wild-type Drosophila CTD and human designates the human CTD. Two-sided chi-squared tests, ns: not significant (p>0.05), *p < 0.05, **p < 0.01, ***p < 0.001 or ****p < 0.0001 values were considered statistically significant from UAS-Rpb1i, UAS-Rpb1WT.
(C) Western blots of late pupae showing the expression of FLAG-tagged, RNAi-resistant forms of Rpb1 derivatives, and the knockdown of the RNAi-sensitive Rpb1HA. Spt5 serves as a loading control. UAS-Rpb1 and Act-GAL4 were each marked with w+, thus allowing late pupae with Act-GAL4 to be distinguished from their CyO counterparts by an additional copy of w+.
(D) Testing the function of various Rpb1 derivatives in Drosophila wings. Wing-specific expression was driven by ms1096-GAL4. Red arrows indicate creases in the wing or changes in the vein path.
(E) Functionality of CRISPR derivatives of Rpb1 in hemizygous males. The Rpb1 gene resides on the X chromosome.
(F) Western blot showing the expression of FLAG-tagged, CRISPR derivatives of Rpb1 in late pupae. Spt5 serves as a loading control.
See also Figure S2.
We also performed a wing-specific knockdown of the endogenous Rpb1 and tested the effects of co-expressing various forms of mutant Rpb1. The wing-specific knockdown of Rpb1 gave rise to severely distorted, miniature-sized wings that were rescued by the expression of Rpb1WT, Rpb120con, Rpb124con and Rpb129con. As was observed in the whole-animal experiments, expression of Rpb110con failed to rescue wing defects. Conversely, expression of either Rpb142con or Rpb152con in the wing partially rescued the RNAi phenotype (Figure 3D). This suggests that the basis for the defects caused by reducing the number of consensus heptads may be different from defects caused by having too many consensus heptads.
To definitively test if the simplified CTDs can replace the functions of the wild-type CTD, we used CRISPR-Cas9 to introduce various mutations into the endogenous Rpb1 locus in flies. Rpb120con, Rpb124con, Rpb142con and Rpb152con were introduced whereas two attempts to introduce Rpb110con were unsuccessful. As was the case for Rpb129con, Rpb120con and Rpb124con supported development and proliferation under normal growth conditions (Figure 3E, Figure S2B, S2C and S2G). They also functioned similarly to Rpb1FLAG in response to stress (Figure S2D, S2E and S2F). Hemizygous males carrying Rpb142con were sterile whereas Rpb152con was unable to support development into adulthood. In contrast to Rpb152con, Rpb1human substituted for the endogenous Rpb1 to produce homozygous viable adults that were fertile (Figure 3E, Figure S2G). The dysfunction of Rpb142con and Rpb152con was not simply due to under-expression, as all CRISPR derivatives of Rpb1 were expressed at comparable levels (Figure 3F). Collectively, these results suggest that while a fly can survive without divergent motifs, too many consensus heptads can be deleterious, even at CTD lengths comparable to the wild-type length.
The Drosophila CTD Dynamically Enters Sites of Active Transcription Independent of the Body of Pol II
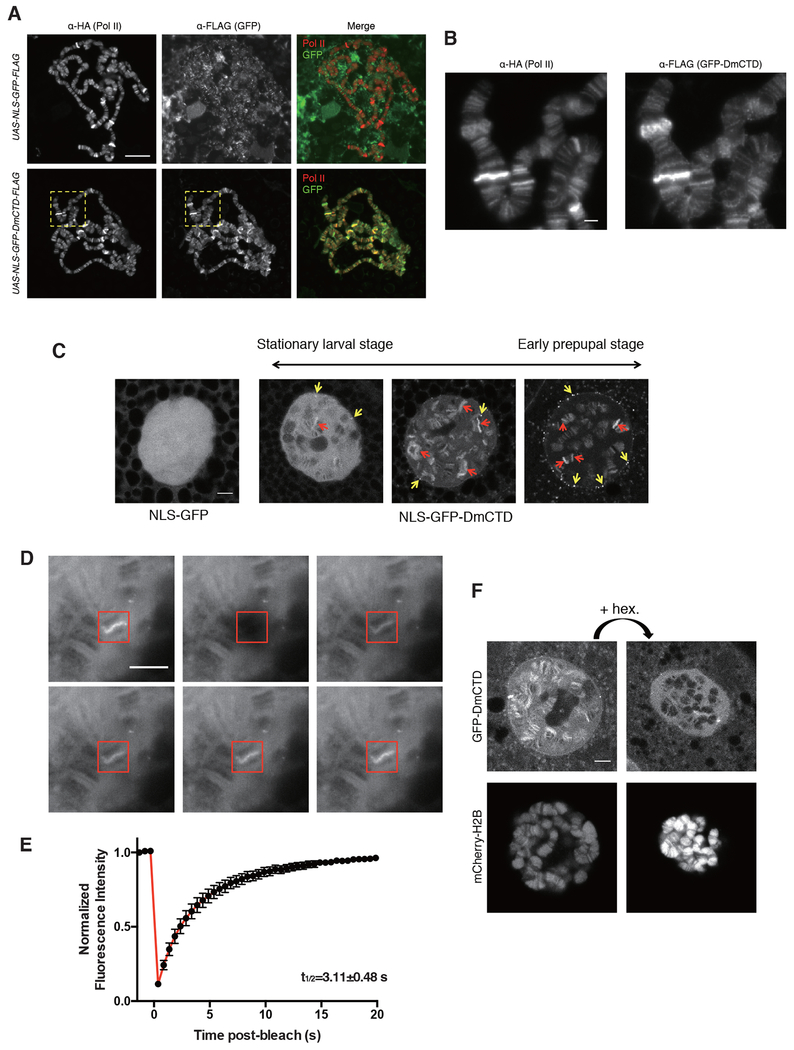
Results from previous studies have lead to conjecture that transcription occurs in “transcription compartments” or “transcription factories” (Papantonis and Cook, 2013; Yao et al., 2007). More recent findings suggest that these could be phase-separated liquids formed by networks of weak interactions between RNA and proteins with multivalent, low complexity regions (Burke et al., 2015; Cho et al., 2018; Chong et al., 2018; Kwon et al., 2013; Lu et al., 2018; Sabari et al., 2018). In vitro, the CTD has been shown to partition into liquid and gel phases comprised of other low complexity domains and this partitioning is influenced by both the length and sequence composition of the CTD (Boehning et al., 2018; Kwon et al., 2013). Since transcriptionally active loci are easily observed as puffs on polytene chromosomes in Drosophila salivary glands, we explored whether the CTD could serve to partition Pol II into sites of active transcription and, if so, whether this might provide insight into the functionality of our various CTD derivatives of Rpb1. To this end, we tagged the Drosophila CTD with green fluorescent protein and expressed this GFP-DmCTD protein or a GFP control in salivary glands. Each protein had a nuclear localization signal (NLS) and a FLAG-tag, and both were expressed at comparable levels and localized to nuclei (Figure S3). Indirect immunofluorescence microscopy of spread chromosomes revealed that the GFP-DmCTD co-localized with Pol II at puffs whereas GFP alone did not (Figure 4A and 4B). Live cell imaging using confocal microscopy showed that the GFP-DmCTD associated with puffs while GFP alone was dispersed throughout the nucleus (Figure 4C). Thus, the CTD alone can function as a signal sequence that facilitates the interaction of Pol II with transcriptionally active loci.
Figure 4.
GFP-DmCTD is dynamically associated with sites of active transcription and sensitive to 1,6-hexanediol
(A) Indirect immunofluorescence detection of HA-tagged Pol II and FLAG-tagged NLS-GFP derivatives on fixed and spread polytene chromosomes. Salivary glands containing the polytene chromosomes were dissected from progeny derived from matings of Rpb1HA, da-GAL4 to UAS-NLS-GFP-DmCTD-FLAG or UAS-NLS-GFP-FLAG. See STAR methods for details.
(B) Magnified view of yellow boxes in (B).
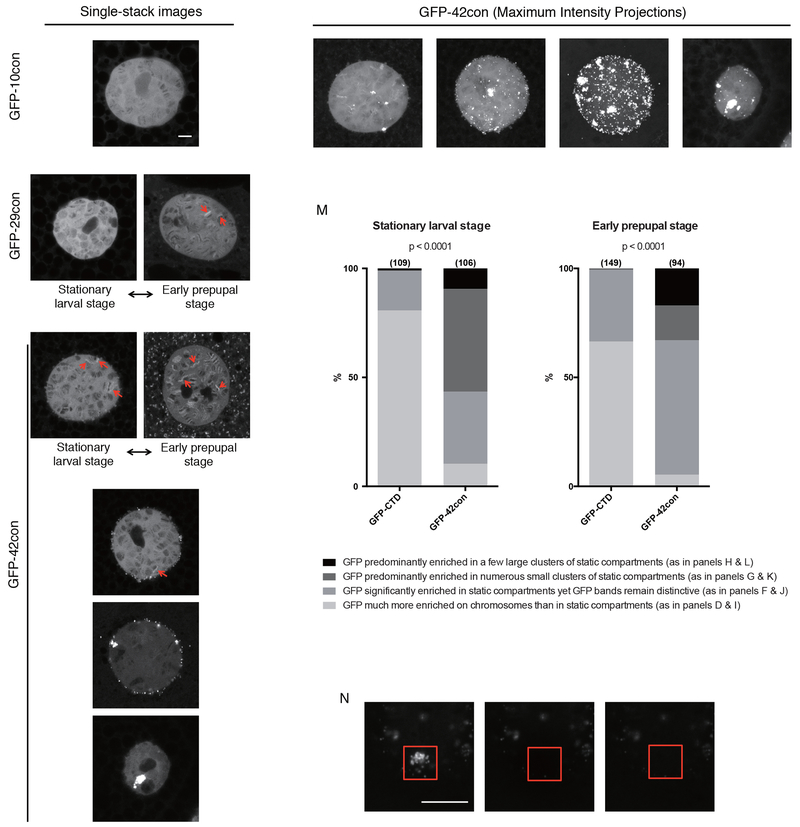
(C) Single-stack confocal images of nuclei in live salivary glands expressing either NLS-GFP or NLS-GFP-DmCTD. >99% of cells expressing the NLS-GFP-DmCTD from the stationary larval stage have a range of appearances that resemble the left and middle panels whereas >99% cells from early prepupal stage have a range of appearances that resemble the middle and right panels. Examples represent the range of appearances observed. Red and yellow arrows indicate examples of chromosomal puffs and extrachromosomal foci that show enrichment of GFP-tagged CTD respectively.
(D and E) FRAP images (D) and average GFP intensity at GFP-DmCTD puffs following photobleaching (E). The red box indicates a bleached region. Data are from 15 puffs in 15 different salivary glands. Error bars represents SEM.
(F) Treatment with 1% 1,6-hexanediol disrupts the association of GFP-DmCTD with chromosomes. Fluorescence from mCherry-labeled H2B indicates that the chromosomes remain intact following 1,6-hexanediol treatment. Each image is a single Z stack.
Scale bar in (A) represents 50 μm, all other scale bars represent 5 μm.
See also Figure S3 and S4 and Movie S1, S2 and S3.
Previous analyses of Pol II interactions with puffs in live cells showed dynamic exchange of Pol II between the puff and the surroundings; fluorescence recovery after photobleaching (FRAP) showed fluorescence recovery times for Pol II of approximately 1 to 2 minutes (Yao et al., 2007). In contrast, our GFP-DmCTD displayed recovery times at puffs of a few seconds (Figures 4D and 4E). The difference in recovery times is not unexpected given that Pol II in the puffs is engaged in transcription.
In addition to its association with puffs, the GFP-DmCTD also formed foci that appeared separate from the chromosomes (Figure 4C, yellow arrows). FRAP analysis showed that some GFP-DmCTD-containing foci are dynamic while others are static (Figure S4A, Movie S1 and S2). Dynamic foci that recovered fluorescence did so with times of several seconds, similar to the recovery time for fluorescence at puffs. In contrast, static foci did not recover fluorescence. Dynamic foci tended to be in the space between the chromosomes and nuclear periphery whereas static foci tended to be at the nuclear periphery.
Several recent studies provide evidence that the CTD can partition into phase separated liquid droplets (Boehning et al., 2018; Burke et al., 2015; Lu et al., 2018). The dynamic nature of the interaction of GFP-DmCTD with chromosomes and a subset of foci suggests that the CTD could be partitioning into phase-separated liquid compartments. To test this, we examined the effect of treating glands with 1,6-hexanediol, a chemical that has been shown to disperse proteins from phase-separated compartments (Boehning et al., 2018; Cho et al., 2018; Kroschwald et al., 2017; Lu et al., 2018; Sabari et al., 2018; Strom et al., 2018). Treatment of intact glands with 1,6-hexanediol resulted in significant loss of GFP-DmCTD from chromosomes (Figure 4F, upper panels) and the disappearance of a subset of GFP-DmCTD foci (Figure S4B). Although the nucleus shrunk in size upon treatment with 1,6-hexanediol, the histone banding pattern was not disrupted (Figure 4F, lower panels), indicating that the dissociation of the GFP-DmCTD from chromosomes was not due to a global disruption of chromosome structure.
Consensus CTDs at Wild-type Lengths Tend to Localize to Static Nuclear Foci Separate from Chromosomes
We next investigated the behavior of the consensus CTDs by fusing them to GFP and expressing them in salivary glands. Unlike GFP-DmCTD, which was enriched at a few puffs during stationary larval stage and tended to be enriched at more puffs during early prepupal stage (Figure 4C), GFP-10con was not enriched on puffs and showed no increase in concentration on chromosomes over what was observed in the nucleoplasm (Figure 5A). Similar to GFP-DmCTD, GFP-20con, GFP-24con and GFP-29con were also enriched on puffs, although the enrichment was not observed until early prepupal stage (Figure 5B and 5C, Figure S5A to S5D). This argues that the increase in CTD length facilitates targeting to sites of active transcription.
Figure 5.
Varying the CTD sequence composition alters its distribution in the nucleus(A to H) Single-stack confocal images of nuclei in live salivary glands expressing NLS-GFP-10con (A), NLS-GFP-29con (B and C) and NLS-GFP-42con (D to H). B and D were collected from the stationary larval stage whereas C and E were collected from the early prepupal stage. Examples depicted in A, F, G and H can be observed in both stages. Red arrows indicate bright puffs that show GFP enrichment over the surroundings.
(I to L) Maximum intensity projections of the same nuclei shown in D, F, G and H, respectively.
(M) GFP-42con cells are more prone to partition in extrachromosomal foci, many of which do not recover after photobleaching as shown in (N). Sample sizes are indicated in parentheses. Darker shades of gray indicate that more CTD associates with static compartments than with chromosomes. p<0.0001, two-sided chi-squared tests.
(N) FRAP images on a cluster of GFP-42con foci. Red box indicates bleached region.
All scale bars represent 5 μm.
See also Figure S3 and S5 and Movie S4.
The distributions of the GFP-42con and GFP-52con derivatives in salivary glands were very different from the GFP-DmCTD. Whereas GFP-DmCTD was enriched in puffs in most cells, GFP-42con or GFP-52con was more frequently localized in extrachromosomal foci than on chromosomes (Figure 5D to 5M, Figure S5E to S5N). Much of the foci containing GFP-42con did not recover after photobleaching (Figure 5N, Movie S4), suggesting that consensus CTDs at wild-type lengths are more prone to localize in static foci. Together, our results with GFP-Ctd fusions supports the hypothesis that additional CTD length improves targeting to transcription compartments, but CTDs above a length threshold form less dynamic, possibly aggregated structures. This propensity for aggregation is counteracted by the divergent motifs that predominate in metazoa.
Discussion:
Consensus Heptad Repeats Are Sufficient to Satisfy Requirements for Multicellularity
Our functional analysis of the CTD in Drosophila challenges the long-standing proposition that features of the CTD unique to multicellular organisms are essential for development. We show that CTDs of 20 to 29 consensus heptads support Drosophila viability, development and thermotolerance. In contrast, CTDs of 10, 42 or 52 consensus heptads do not. It is unlikely that the viability of our fully-consensus CTD flies is due to compensatory mutations that have occurred elsewhere in the genome because the generation of healthy fly lines containing the 20, 24, and 29 consensus heptads using CRISPR did not require screening an inordinate number of candidates. Moreover, these results are in accordance with the RNAi rescue results in which the selective pressure is only applied during one generation when flies are mated to a GAL4 driver line.
Remarkably, the optimal range in the number of consecutive consensus heptads that functions in Drosophila is similar to the number of consecutive consensus and near-consensus heptads found in a wide spectrum of eukaryotes spanning from yeast to plants (Yang and Stiller, 2014). We propose that this consecutive array of consensus heptads functions as a single unit. Otherwise, it is difficult to account for why the number of consecutive heptads that is conserved within a given taxon far exceeds the sizes of the binding sites for known proteins. For example, a stretch of 15 consensus heptads is conserved between human and mouse yet the known binding sites for proteins that associate with the CTD only span three heptads or less (Jasnovidova and Stefl, 2013). In addition, CTD mutant strains of yeast with varied numbers of consensus heptads spontaneously acquire additional mutations that result in CTDs approximating the natural number of 26 (Morrill et al., 2016; Nonet and Young, 1989). How this consecutive array of repeats functions as a single unit is not known. One possibility is that it provides for phosphorylation-dependent, highly cooperative regulation of factor binding (Lenz and Swain, 2006). This entropically driven mechanism of regulation could provide switch-like binding and release of factors dictated by threshold levels of phosphorylation.
Functionality of Divergent Motifs Is Dictated by Their Sequence Context
The divergent motifs of Drosophila likely function within the specific context established by the overall amino acid sequence of the Drosophila CTD. Otherwise, it is difficult to reconcile how all of the divergent motifs can be replaced with consensus heptads yet the natural sequence of the CTD is highly conserved between distantly related species of Drosophila. Likewise, in spite of the high sequence conservation within the chordate lineage, the CTD in human cells grown in culture can be replaced with an all consensus CTD without losing viability (Chapman et al., 2005). We envision several ways in which the functions of the divergent motifs could be context dependent. Our SAXS analysis of the Drosophila CTD reveals that it adopts a compact random coil structure. This implies the occurrence of transient intramolecular interactions (Portz et al., 2017). In addition, the CTD self-associates and this self-association could contribute to the clustering of Pol II molecules that has been linked to transcription (Boehning et al., 2018). These intra- and intermolecular interactions could extend over the entirety of CTD thus placing constraints on the sequence composition throughout the CTD. Another possibility is that divergent motifs themselves or through interaction with other proteins serve as binding sites for the same cadre of proteins that otherwise directly bind consensus heptads. This would explain why a fully consensus CTD functions in place of the natural Drosophila counterpart. A corollary to this hypothesis is that the divergent motifs do not provide unique regulatory functions; they simply provide alternative pathways for interactions normally provided by consensus heptads. Divergent motifs could have emerged as a consequence of constructive neutral evolution (Gray et al., 2010) in which a chance mutation resulted in loss of consensus heptads but fortuitously produced motifs that associated with a protein providing an alternate to the lost consensus heptads. Future investigations should seek to determine if known proteins that recognize consensus heptads also associate with divergent motifs and whether there are adaptor proteins selectively binding divergent motifs in the Drosophila CTD that in turn serve to recruit proteins which in other species bind directly to consensus heptads.
Targeting of the CTD to Transcription Compartments
Our finding that the functionality of the consensus CTD is dependent on the number of consensus heptads prompted us to explore the behavior of the various CTDs when they were expressed separately from the rest of Pol II. We were motivated by previous reports that the CTD partitions into liquid phase separated droplets and hydrogels (Burke et al., 2015; Kwon et al., 2013), and the proposition that the highly transcribed regions of Drosophila polytene chromosomes known as puffs could be liquid phase separated compartments (Harlen and Churchman, 2017).
We find that GFP-tagged versions of the CTD associate with puffs and the characteristics of these interactions are consistent with puffs being phase separated compartments. First, treatment of the salivary glands with 1,6-hexanediol caused the CTD to disperse from puffs, a behavior consistent with that of proteins partitioning into liquid phase separated domains (Boehning et al., 2018; Chong et al., 2018; Lu et al., 2018; Sabari et al., 2018). Second, FRAP analysis showed that the CTD’s association with puffs is dynamic and has a recovery time after photobleaching of several seconds. This is similar to the recovery times reported for the CTD when it partitions into phase separated droplets formed by the low complexity domain of the FUS protein (Burke et al., 2015). Finally, while CTDs composed of 20, 24, or 29 consensus heptads become enriched in puffs, a CTD composed of 10 consensus heptads did not. This dependence on length is consistent with the puff interaction being dependent on the valency of the CTD. Liquid phase separation in biological systems is driven by networks of weak interactions between multivalent molecules (Banani et al., 2017; Li et al., 2012). The capacity of a protein to become enriched in a phase separated domain depends on its capacity to interact favorably with the network. We speculate that the CTD with 10 consensus heptads lacks sufficient valency to interact favorably with other components in the puff.
We propose that the CTD is functioning as a signal sequence and is serving to target Pol II to highly transcribed regions of the genome. Several molecules are known to reside at puffs that could be involved in establishing a compartment that is targeted by the CTD. One is the CTD itself since it was recently shown that the CTD self-associates into phase separated droplets in the presence of crowding agents and these droplets recruit Pol II (Boehning et al., 2018). Another is poly (ADP-ribose) whose chains are known to contribute to puff formation (Tulin and Spradling, 2003) and have been shown to nucleate phase separation (Altmeyer et al., 2015; McGurk et al., 2018). Yet another could be FUS which has been shown to be present at puffs and which forms phase separated droplets that can enrich the CTD (Burke et al., 2015; Immanuel et al., 1995). Notably, all three of these components of puffs are also found at transcribed loci in diploid cells indicating transcription compartments analogous to puffs could form in these cells (Petesch and Lis, 2012; Schwartz et al., 2012).
Divergent Motifs Counteract Aggregation of the CTD
In addition to its enrichment at puffs, we observed that GFP-tagged CTD formed foci that were clearly separate from the chromosomes. FRAP analysis showed that some of these foci were dynamic while others were not. The recovery time for the dynamic foci was comparable to the recovery times detected at puffs. Future experiments with fluorescently labeled Pol II should reveal whether the foci formed by the CTD also include Pol II, or other transcription machinery harboring the multivalent, low complexity domains so often associated with phase separation.
The abundance of CTD foci increases dramatically when the number of consensus heptads in the CTD increases from 29 to 42 or 52. In some cases, a small number of large clumps are observed instead of multiple foci as if the foci have coalesced. The increase in the number of foci and formation of large clumps is consistent with the known behaviour of multivalent polymers where expansion of repeat numbers can cause molecules to transition from being dynamic to forming static, dysfunctional aggregates (Weber and Brangwynne, 2012). In contrast to the 42 consensus CTD, the Drosophila CTD, which is comparable in length, does not form large numbers of foci or clumps. This raises the possibility that divergent motifs function to counteract the tendency of the multivalent CTD to form dysfunctional aggregates. We speculate that the formation of such aggregates could be the reason that Pol II with the 42 or 52 consensus heptads result respectively in defects in male fertility and pupal development.
Drosophila Provides a Tractable Model for Investigating CTD Function
It has been almost 4 decades since the CTD was first described and its function remains enigmatic. Virtually everything we know about the CTD’s function has focused on the consensus heptads in yeast and mammalian cells grown in culture. Our results now establish Drosophila as a powerful model system for investigating CTD function. Specifically, imaging Drosophila polytene chromosomes in intact nuclei represents a democtratizible system for the simultaneous visualization of the nucleoplasm and complete genome with common and accessible microscopy. The platform introduced here is amenable to exploring the partitioning, miscibility and dynamics of any number of low complexity domains associated with the transcription machinery, with the ability to map localization to precise genomic loci, discern between heterochromatic, euchromatic, and nucleoplasmic compartments, and to tune expression levels and timing with the available suite of Drosophila genetic tools.
STAR Methods:
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to the Lead Contact, David S. Gilmour (dsg11@psu.edu).
Experimental Model and Subject Details
Drosophila strains
UAS-Rpb1i, Act-GAL4 [w; Act-GAL4 (w+)/CyO], ms1096-GAL4 and da-Gal4 fly lines were obtained from the Bloomington Stock Center (Bloomington 36830, 4414, 8860 and 55850 respectively). UAS-Rpb1i, UAS-Rpb1WT and UAS-Rpb1i, UAS-Rpb1human lines were previously described (Portz et al., 2017). DNA encoding either mutant Rpb1 with double FLAG-tags at the C-terminus, or various NLS-GFP derivatives were subcloned into the pUAST-attB vector, followed by transformation into the PhiC31 attP 86Fb site (Bischof et al., 2007). Rpb1i-resistance of transgenic Rpb1 was achieved by changing the part of the coding sequence of Rpb1 that is targeted by the 21 nucleotide shRNA (sense strand: AACGGTGAAACTGTCGAACAA) to AACCGTCAAGTTGAGCAACAA. The UAS-Rpb1i, UAS-Rpb1 line, as well as the w, Rpb1HA; Act-GAL4 (w+)/CyO and Rpb1HA; da-GAL4 lines were generated by routine matings and meiotic recombination. All NLS-GFP derivatives have a Strep-tag appended to their C-terminus in addition to the FLAG tag.
CRISPRed Rpb1 fly lines were generated through injection of DNA mixtures into a Cas9 expressing fly line (Bloomington 51324). As previously described (Gratz et al., 2014), each DNA mixture contains two CRISPR targeting vectors that express sgRNAs and a homology donor vector. The two CRISPR targets were selected from a list of targets with zero predicted potential off-target sites using flyCRISPR Optimal Target Finder (http://tools.flycrispr.molbio.wisc.edu/targetFinder/, Maximum Stringency, NGG PAM sequence only). Complementary strands encoding the sgRNAs were annealed, phosphorylated and cloned into the BbsI site of pU6-BbsI-chiRNA (Addgene plasmid 45946). The combination of Rpb1_fw1: 5’-CTTCGTAGGGATTTGAGAGCCAGTG-3’ and Rpb1_rev1: 5’-AAACCACTGGCTCTCAAATCCCTAC-3’ directed cleavage downstream of the 3’UTR of Rpb1. The combination of Rpb1_fw2: 5’-CTTCGAGAAACACTCGGCGAGGCT-3’and Rpb1_rev2: 5’-AAACAGCCTCGCCGAGTGTTTCTC-3’ directed cleavage adjacent to the start of the Drosophila CTD. Two homology arms, each about 1kb in length were PCR amplified from strain w1118. The homology arm adjacent to the 3’UTR of Rpb1 was cloned into the Aarl site of pHD-ScarlessDsRed (DGRC plasmid 1364). The other homology arm was cloned into the Sapl site alongside desired mutations in the CTD and the CRISPR targeting site (Figure 2A). Candidate recombinant flies were identified by expression of the DsRed marker in eyes. Individual DsRed candidates were out-crossed to strain w1118. Desired candidates were verified by sequencing and western blot. The insertion of the DsRed cassette disrupts the gene adjacent to the 3’UTR of Rpb1, which to our knowledge does not affect the interpretation of our data. The Cas9 transgene on the third chromosome was subsequently removed by selection against the 3xP3-GFP marker. Rpb142con and Rpb152con alleles were maintained over FM7(Tb) balancer (BDSC 36337).
Illustration of the genetic mating scheme in Figure 3A was generated using Genotype Builder (Roote and Prokop, 2013).
Method Details
Sequence alignment
Previously published amino acid and nucleotide sequences of the CTDs of 12 species of Drosophila were downloaded from FLYBASE and aligned using Clustal Omega.
PCR genotyping
DNA was extracted from male individuals of desired genotypes as previously described (Gloor et al., 1993), and analyzed with the following primers: fw: 5’-CGCCTTCGGCTGCATCGG-3’ and rev: 5’-ACAAAGATCCTCTAGAGGTACCCTCGAGC-3’ (Figure S2A); fw: 5’-GGCGATCGAGCGTAGTCGGTACTT-3’ and rev: 5’-CCAGGACCTTCGATGTCGCCGTATTT-3’ (Figure S2G). The resulting PCR products were isolated and sequenced to verify the presence of the desired mutations.
Western blotting
Lysate was prepared from late pupae (Figure 3, C and F) as previously described (Gibbs et al., 2017). Samples were subsequently loaded onto a 7% Tris-Acetate gel (Figure 3, C and F). Rpb1HA was detected with mouse anti-HA antibody monoclonal antibody (1:2500, Pierce, 26183, Thermo Scientific). Spt5 and Rpb3 were detected with rabbit anti-Spt5 antibody (1:3000) and rabbit anti-Rpb3 antibody (1:3000) respectively (Qiu and Gilmour, 2017). Expressions of FLAG Rpb1 were detected with mouse anti-FLAG M2 antibody (1:2500; Sigma). Blots were subsequently probed with goat anti-rabbit IgG (1:3000; Alexa Fluor 488) and goat anti-mouse IgG (1:3000; Alexa Fluor 647) and visualized with a Typhoon (GE Healthcare).
Immunofluorescence
For polytene chromosome staining, salivary glands from wandering third-instar larvae were dissected and squashed as previously described (Schwartz et al., 2004). Slides were then incubated with rabbit anti-HA antibody (1:100; ThermoFisher 71-5500) and mouse anti-FLAG M2 antibody (1:200; Sigma) overnight at 4°C. For Figure 2G, the slides were subsequently probed with goat anti-rabbit IgG (1:200; Alexa Fluor 488) and goat anti-mouse IgG (1:200; Alexa Fluor 568) for 4 hours at room temperature. For Figure 4A and 4B, the slides were subsequently probed with goat anti-mouse IgG (1:200; Alexa Fluor 488) and goat anti-rabbit IgG (1:200; Alexa Fluor 568) for 4 hours at room temperature. Samples were imaged on a CARV II spinning disc confocal (BD Biosystems) and adjusted for brightness and contrast using ImageJ.
Animal assays
For Figure 2D and E, animals were cultured at 30°C and 33% humidity. For Figure 3D, animals were cultured at 18°C and 33% humidity. Wings were dissected from 1 to 3 day old females. For cold shock analysis in Figure 2F, 0-6h old _adults were aged for 96h, placed on ice for 14h and recover at room temperature (21°C). Recovery of movement was monitored over the course of at least 4 hours with a video camera. Recovery time was defined as the time point at which an individual started to move. For the rest of the figures, animals were maintained at 24°C and 65% humidity.
Drosophila live imaging
Larvae were raised in a non-crowded vial with standard cornmeal culture media. Prior to dissection, wandering third instar larvae expressing a GFP-tagged CTD derivative fusion and mCherry tagged H2B-CTD (Sgs3-GAL4, UAS-mCh-H2B/UAS-NLS-GFP-DmCTD) were transferred to a fresh vial. The selected larvae tended to migrate away from the food and always developed into early prepupae within 1 to 6 hours if left undissected, which corresponded to the “stationary larvae stage” (Fletcher and Thummel, 1995). Newly formed prepupae (<1 hour post puparium formation) were selected as the “early prepupae stage”. Individuals at this stage have transparent glands that can be clearly distinguished from prepupae of later stages, which have white non-transparent glands. Glands were isolated and imaged in M3 medium (Sigma Aldrich S8398) supplemented with 2.5 g of bactopeptone and 1 g of yeast extract per liter medium (M3+BPYE). Confocal images were acquired using a Zeiss LSM800 with a 63X oil objective (NA1.4) and adjusted for brightness and contrast using ImageJ. For consistency in comparison, all images of single nuclei were collected from the distal ⅓ of the salivary glands. Polytene nuclei were imaged at a depth no greater than 60 μm into the salivary gland tissue. For Figure 4C, Figure 5 and Figure S5, the intensity of GFP was adjusted based on the intensity of mCherry-H2B to correct for minor intensity differences produced by variation in imaging depth. Non-confocal images in Figure S3 were collected at the same exposure times on a Carl Zeiss Axioskop 40 microscope and adjusted for brightness and contrast using ImageJ.
Fluorescence recovery after photobleaching (FRAP)
Salivary glands were prepared as above. Three images were taken before bleaching. On the fourth frame, a square region of approximately 4 μm X 4 μm was bleached with full laser power and recovery was recorded at 0.2% power, at 0.5 second intervals for 90 seconds. Normalized fluorescence intensity was calculated with a two-step normalization: first by normalizing the fluorescence intensity of GFP-DmCTD puffs to that of pre-bleach, second by normalizing this ratio to the ratio obtained from an unbleached region to correct for the subtle loss in fluorescence intensity due to bleaching. For calculating half time recoveries, normalized values from each recording were separately fit to a single exponential model, and half time recovery was presented as mean ± standard error.
1,6-Hexanediol treatments
Salivary glands were prepared as above and immediately imaged in M3+BPYE (before treatment). Salivary glands were then transferred to the same medium containing 1% 1,6-hexanediol (Alfa Aesar A12439), incubated for approximately 20 min and imaged again (after treatment).
Quantification and Statistical Analysis
Statistical analyses were performed using the Prism 6 software. Two-sided chi-squared tests were used for the analyses in Figure 2B to E, Figure 3B, Figure 5M, Figure S2B to S2E and Figure S5N. Mann-Whitney U tests were used for the analyses in Figure 2F and Figure S2F. The *p < 0.05, **p < 0.01, ***p < 0.001 or ****p < 0.0001 values were considered statistically significant from Rpb1FLAG or UAS-Rpb1i, UAS-Rpb1WT.
Supplementary Material
Movie. S1. FRAP movie on static GFP-DmCTD foci, related to Figure 4 and Figure S4A (upper).
Movie. S2. FRAP movie on dynamic GFP-DmCTD foci, related to Figure 4 and Figure S4A (lower).
Movie. S3. Representative FRAP movie on a GFP-DmCTD puff, related to Figure 4D.
Movie. S4. Example of foci containing GFP-42con that did not recover after photobleaching, related to Figure 5N.
Highlights:
Consensus heptads are sufficient to support Drosophila development
Too many consensus heptads are deleterious
The wild-type Drosophila CTD alone localizes to sites of active transcription
CTDs with too many consensus heptads tend to localize to extrachromosomal foci
Acknowledgments:
We thank M. M. Rolls, C. M. Thomas, and Z. C. Lai for imaging support; C. Feng for CRISPR and live imaging suggestions; D. G. Baumann, Y. Qiu, R. Dollinger and J. C. Reese for comments on the manuscript. The Sgs3-GAL4, UAS-mCh-H2B/TM6 fly line is a generous gift from C. Peter Verrijzer. We thank the TRiP at Harvard Medical School (NIH/NIGMS R01-GM084947) for providing the transgenic Rpb1 RNAi fly stock used in this study. This work was funded by NIH GM047477 (to D.S.G.).
Footnotes
Declaration of Interests:
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and Notes:
- Altmeyer M, Neelsen KJ, Teloni F, Pozdnyakova I, Pellegrino S, Grøfte M, Rask M-BD, Streicher W, Jungmichel S, Nielsen ML, et al. (2015). Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose). Nat. Commun. 6, 8088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani SF, Lee HO, Hyman AA, and Rosen MK (2017). Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, and Basler K (2007). An optimized transgenesis system for Drosophila using germ-line-specific φC31 integrases. Proc. Natl. Acad. Sci. U. S. A. 104, 3312–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehning M, Dugast-Darzacq C, Rankovic M, Hansen AS, Yu T, Marie-Nelly H, McSwiggen DT, Kokic G, Dailey GM, Cramer P, et al. (2018). RNA polymerase II clustering through carboxy-terminal domain phase separation. Nat. Struct. Mol. Biol. 25, 833–840. [DOI] [PubMed] [Google Scholar]
- Burke KA, Janke AM, Rhine CL, and Fawzi NL (2015). Residue-by-Residue View of In Vitro FUS Granules that Bind the C-Terminal Domain of RNA Polymerase II. Mol. Cell 60, 231–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman RD, Conrad M, and Eick D (2005). Role of the mammalian RNA polymerase II C-terminal domain (CTD) nonconsensus repeats in CTD stability and cell proliferation. Mol. Cell. Biol. 25, 7665–7674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman RD, Heidemann M, Hintermair C, and Eick D (2008). Molecular evolution of the RNA polymerase II CTD. Trends Genet. 24, 289–296. [DOI] [PubMed] [Google Scholar]
- Cho W-K, Spille J-H, Hecht M, Lee C, Li C, Grube V, and Cisse II (2018). Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science. 361, 412–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong S, Dugast-Darzacq C, Liu Z, Dong P, Dailey GM, Cattoglio C, Heckert A, Banala S, Lavis L, Darzacq X, et al. (2018). Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science eaar2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corden JL (2013). RNA polymerase II C-terminal domain: Tethering transcription to transcript and template. Chem. Rev. 113, 8423–8455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias JD, Rito T, Torlai Triglia E, Kukalev A, Ferrai C, Chotalia M, Brookes E, Kimura H, and Pombo A (2015). Methylation of RNA polymerase II non-consensus Lysine residues marks early transcription in mammalian cells. Elife 4, e11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff S, Dienstbier M, and Murphy S (2012). Updating the RNA polymerase CTD code: adding gene-specific layers. Trends Genet. 28, 333–341. [DOI] [PubMed] [Google Scholar]
- Eick D, and Geyer M (2013). The RNA polymerase II carboxy-terminal domain (CTD) code. Chem. Rev. 113, 8456–8490. [DOI] [PubMed] [Google Scholar]
- Fletcher JC, and Thummel CS (1995). The Drosophila E74 gene is required for the proper stage- and tissue-specific transcription of ecdysone-regulated genes at the onset of metamorphosis. Development 121, 1411–1421. [DOI] [PubMed] [Google Scholar]
- Gibbs EB, Lu F, Portz B, Fisher MJ, Medellin BP, Laremore TN, Zhang YJ, Gilmour DS, and Showalter SA (2017). Phosphorylation induces sequence-specific conformational switches in the RNA polymerase II C-terminal domain. Nat. Commun. 8, 15233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloor GB, Preston CR, Johnson-Schlitz DM, Nassif NA, Phillis RW, Benz WK, Robertson HM, and Engels WR (1993). Type I repressors of P element mobility. Genetics 135, 81–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz SJ, Ukken FP, Rubinstein CD, Thiede G, Donohue LK, Cummings AM, and O’Connor-Giles KM (2014). Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics 196, 961–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MW, Lukes J, Archibald JM, Keeling PJ, and Doolittle WF (2010). Cell biology. Irremediable complexity? Science 330, 920–921. [DOI] [PubMed] [Google Scholar]
- Harlen KM, and Churchman LS (2017). The code and beyond: transcription regulation by the RNA polymerase II carboxy-terminal domain. Nat. Rev. Mol. Cell Biol. 18, 263–273. [DOI] [PubMed] [Google Scholar]
- Immanuel D, Zinszner H, and Ron D (1995). Association of SARFH (sarcomaassociated RNA-binding fly homolog) with regions of chromatin transcribed by RNA polymerase II. Mol. Cell. Biol. 15, 4562–4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasnovidova O, and Stefl R (2013). The CTD code of RNA polymerase II: a structural view. Wiley Interdiscip. Rev. RNA 4, 1–16. [DOI] [PubMed] [Google Scholar]
- Kroschwald S, Maharana S, and Simon A (2017). Hexanediol: a chemical probe to investigate the material properties of membrane-less compartments. Matters 3, e201702000010. [Google Scholar]
- Kwon I, Kato M, Xiang S, Wu L, Theodoropoulos P, Mirzaei H, Han T, Xie S, Corden JL, and McKnight SL (2013). Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell 155, 1049–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz P, and Swain PS (2006). An entropic mechanism to generate highly cooperative and specific binding from protein phosphorylations. Curr. Biol. 16, 2150–2155. [DOI] [PubMed] [Google Scholar]
- Li P, Banjade S, Cheng H-C, Kim S, Chen B, Guo L, Llaguno M, Hollingsworth JV, King DS, Banani SF, et al. (2012). Phase transitions in the assembly of multivalent signalling proteins. Nature 483, 336–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Kenney JM, Stiller JW, and Greenleaf AL (2010). Genetic organization, length conservation, and evolution of RNA polymerase II carboxyl-terminal domain. Mol. Biol. Evol. 27, 2628–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Yu D, Hansen AS, Ganguly S, Liu R, Heckert A, Darzacq X, and Zhou Q (2018). Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II. Nature 558, 318–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGurk L, Gomes E, Guo L, Mojsilovic-Petrovic J, Tran V, Kalb RG, Shorter J, and Bonini NM (2018). Poly(ADP-Ribose) Prevents Pathological Phase Separation of TDP-43 by Promoting Liquid Demixing and Stress Granule Localization. Mol. Cell 71, 703–717. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrill SA, Exner AE, Babokhov M, Reinfeld BI, and Fuchs SM (2016). DNA Instability Maintains the Repeat Length of the Yeast RNA Polymerase II C-terminal Domain. J. Biol. Chem. 291, 11540–11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet ML, and Young RA (1989). Intragenic and extragenic suppressors of mutations in the heptapeptide repeat domain of Saccharomyces cerevisiae RNA polymerase II. Genetics 123, 715–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papantonis A, and Cook PR (2013). Transcription factories: genome organization and gene regulation. Chem. Rev. 113, 8683–8705. [DOI] [PubMed] [Google Scholar]
- Petesch SJ, and Lis JT (2012). Activator-induced spread of poly(ADP-ribose) polymerase promotes nucleosome loss at Hsp70. Mol. Cell 45, 64–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portz B, Lu F, Gibbs EB, Mayfield JE, Rachel Mehaffey M, Zhang YJ, Brodbelt JS, Showalter SA, and Gilmour DS (2017). Structural heterogeneity in the intrinsically disordered RNA polymerase II C-terminal domain. Nat. Commun. 8, 15231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, and Gilmour DS (2017). Identification of regions in the Spt5 subunit of DSIF that are involved in promoter proximal pausing. J. Biol. Chem. 292, 5555–5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roote J, and Prokop A (2013). How to design a genetic mating scheme: a basic training package for Drosophila genetics. G3 3, 353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabari BR, Dall’Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K, Abraham BJ, Hannett NM, Zamudio AV, Manteiga JC, et al. (2018). Coactivator condensation at super-enhancers links phase separation and gene control. Science eaar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder S, Herker E, Itzen F, He D, Thomas S, Gilchrist DA, Kaehlcke K, Cho S, Pollard KS, Capra JA, et al. (2013). Acetylation of RNA polymerase II regulates growth-factor-induced gene transcription in mammalian cells. Mol. Cell 52, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz BE, Werner JK, and Lis JT (2004). Indirect immunofluorescent labeling of Drosophila polytene chromosomes: visualizing protein interactions with chromatin in vivo. Methods Enzymol. 376, 393–404. [DOI] [PubMed] [Google Scholar]
- Schwartz JC, Ebmeier CC, Podell ER, Heimiller J, Taatjes DJ, and Cech TR (2012). FUS binds the CTD of RNA polymerase II and regulates its phosphorylation at Ser2. Genes Dev. 26, 2690–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonti CN, Pollard KS, Schröder S, He D, Bruneau BG, Ott M, and Capra JA (2015). Evolution of lysine acetylation in the RNA polymerase II C-terminal domain. BMC Evol. Biol. 15, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ, Rojas LA, Beck DB, Bonasio R, Schüller R, Drury WJ 3rd, Eick D, and Reinberg D (2011). The C-terminal domain of RNA polymerase II is modified by site-specific methylation. Science 332, 99–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom AR, Emelyanov AV, Mir MR, Fyodorov DV, Darzacq XR, and Karpen GH (2017). Phase Separation Drives Heterochromatin Domain Formation. Nature 547, 241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulin A, and Spradling A (2003). Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science 299, 560–562. [DOI] [PubMed] [Google Scholar]
- Weber SC, and Brangwynne CP (2012). Getting RNA and protein in phase. Cell 149, 1188–1191. [DOI] [PubMed] [Google Scholar]
- Yang C, and Stiller JW (2014). Evolutionary diversity and taxon-specific modifications of the RNA polymerase II C-terminal domain. Proc. Natl. Acad. Sci. U. S. A. 111, 5920–5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Ardehali MB, Fecko CJ, Webb WW, and Lis JT (2007). Intranuclear distribution and local dynamics of RNA polymerase II during transcription activation. Mol. Cell 28, 978–990. [DOI] [PubMed] [Google Scholar]
- Zaborowska J, Egloff S, and Murphy S (2016). The pol II CTD: new twists in the tail. Nat. Struct. Mol. Biol. 23, 771–777. [DOI] [PubMed] [Google Scholar]
- Zhao DY, Gish G, Braunschweig U, Li Y, Ni Z, Schmitges FW, Zhong G, Liu K, Li W, Moffat J, et al. (2016). SMN and symmetric arginine dimethylation of RNA polymerase II C-terminal domain control termination. Nature 529, 48–53. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie. S1. FRAP movie on static GFP-DmCTD foci, related to Figure 4 and Figure S4A (upper).
Movie. S2. FRAP movie on dynamic GFP-DmCTD foci, related to Figure 4 and Figure S4A (lower).
Movie. S3. Representative FRAP movie on a GFP-DmCTD puff, related to Figure 4D.
Movie. S4. Example of foci containing GFP-42con that did not recover after photobleaching, related to Figure 5N.