Enteropathogenic Escherichia coli (EPEC) is a major cause of diarrhea in children from developing countries and presents high genetic variability. We aimed to characterize the EPEC virulence-related gene (VRG) distribution and copathogens associated with diarrhea and nutrition-related outcomes in children from the low-income Brazilian semiarid region.
KEYWORDS: enteropathogenic Escherichia coli, diarrhea, virulence genes
ABSTRACT
Enteropathogenic Escherichia coli (EPEC) is a major cause of diarrhea in children from developing countries and presents high genetic variability. We aimed to characterize the EPEC virulence-related gene (VRG) distribution and copathogens associated with diarrhea and nutrition-related outcomes in children from the low-income Brazilian semiarid region. A cross-sectional case-control study of diarrhea was conducted in 1,191 children aged 2 to 36 months from the northeast region of Brazil. Stool samples were collected and clinical, epidemiological, and anthropometric data were identified from each child. A broad molecular evaluation of enteropathogens was performed, and EPEC-positive samples were further investigated for 18 VRGs using five multiplex PCRs. EPEC was detected in 28.2% of the study population, with similar proportions among cases and controls. Typical EPEC (tEPEC) infections were more often associated with diarrhea than atypical EPEC (aEPEC) infections, while aEPEC infections presented a higher prevalence. The VRG ler, a negative regulator of the locus of enterocyte effacement, was associated with the absence of diarrhea in aEPEC-positive children; espB, a major component of the type 3 secretion system, was associated with diarrhea in tEPEC-positive children; the presence of procolonization VRGs—the combination of cesT positivity, espP negativity, and the presence of the map gene—was associated with undernutrition; and Campylobacter spp., norovirus, and enteroaggregative E. coli (EAEC) coinfections were associated with increased clinical severity in EPEC-infected children. These data identified tEPEC strains associated with diarrhea and specific VRGs of EPEC (ler, espB, cesT, and map genes) and Campylobacter spp., norovirus, and EAEC to be major contributors to diarrhea and undernutrition in children from a low-income Brazilian region.
INTRODUCTION
Diarrhea is a major cause of morbidity and mortality in children worldwide, particularly in low- and middle-income countries (1). The Global Enteric Multicenter Study reported that enteropathogenic Escherichia coli (EPEC) is the leading cause of mortality associated with diarrhea in children under 12 months of age from developing countries in Africa and Asia (2).
EPEC bacteria are characterized by a lesion named the attaching-and-effacing lesion on the intestinal epithelium and are negative for heat-labile or heat-stable enterotoxin production (3). EPEC can be divided into two subgroups, atypical EPEC (aEPEC) and typical EPEC (tEPEC), depending on the presence of the EPEC adherence factor virulence plasmid encoding bundle-forming pili (4). While tEPEC is generally associated with severe diarrhea, aEPEC presents a large diversity of clinical outcomes, ranging from prolonged diarrhea to asymptomatic colonization in different settings (4, 5). A clear understanding of EPEC subtypes and epidemiology is missing due to the lack of discrimination in some studies (6).
Virulence-related genes (VRGs) that contribute to pathobiological mechanisms of EPEC disease are mainly located on the pathogenicity island named the locus of enterocyte effacement (LEE). Genes located on the LEE encode structural components of a type III secretion-translocation apparatus, factors enabling the bacterium to adhere intimately to intestinal epithelial cells (eae and tir genes), secreted and effector proteins (espF, espG, espH, map, and espZ genes), and translocator proteins (espA, espB, and espD genes) (7). Additionally, other genes not located on the LEE are also involved in the modulation of major cellular processes, including inhibition of endoplasmic proteins (espI and nleA), proinflammatory reticulum signaling (nleB, nleC, nleD, and nleE), and phagocytosis (espJ and nleH), which favor EPEC colonization and disease (8, 9).
The genetic diversity of EPEC strains has been highlighted (10–13), and further studies showed that specific EPEC VRGs may be associated with different clinical outcomes (4, 12, 14, 15). In this study, we aimed to characterize the EPEC virulence-related gene (VRG) distribution and the copathogens associated with diarrhea and nutrition-related outcomes in children aged 2 to 36 months. We conducted a case-control study of diarrhea in the low-income Brazilian semiarid region and used unprecedented multiplex PCR (mPCR) panels covering 18 different EPEC VRGs.
MATERIALS AND METHODS
Geographic location, study design, and ethical approval.
The present study originated from a previous cross-sectional case-control study of diarrhea (16), which was conducted in cities from the four states of the northeast region of Brazil: Cajazeiras (Paraiba), Crato (Ceará), Ouricuri (Pernambuco), Patos (Paraiba), Picos (Piauí), and Sousa (Paraiba). These cities were included in this study since they were representatives of the semiarid region of Brazil and had more than 50,000 inhabitants.
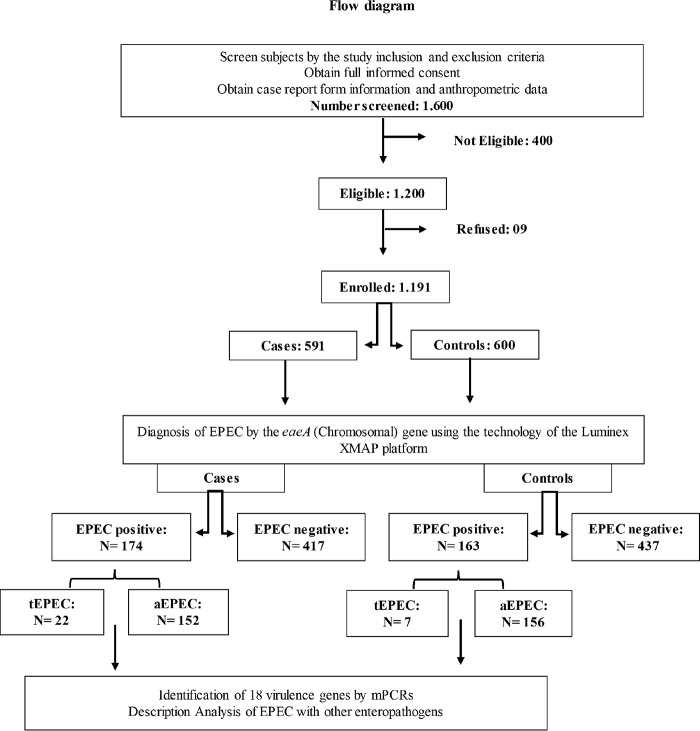
Health care workers collected fecal samples from children between 2 and 36 months old in health care units or during active surveillance from November 2009 to February 2012. An interview was conducted during subject enrollment, and detailed health information, such as demographic information, environmental information, socioeconomic status, breast-feeding status, clinical conditions, occurrence of diarrhea episodes, and anthropometric measurements, were collected via a standardized questionnaire form. The study had the following inclusion criteria for diarrhea cases: the child had to have had three or more liquid stools in the last 24 h, and written consent had to be provided. The inclusion criteria for the controls were as follows: the child did not present with diarrhea in the previous 2 weeks, and written consent was provided. Prior to enrollment, the children had no chronic illness and had not been admitted to health care facilities for more than 12 h. Children from the study stayed in the health care units during the whole duration of the consultation. A total of 1,191 age- and gender-matched participants (591 cases and 600 controls) were included in the study, and their fecal samples were collected (Fig. 1).
FIG 1.
Study protocol. Data on demographic, socioeconomic, and child health characteristics were collected.
This research was approved by the National Commission on Ethics in Research of Brazil and the Research Ethics Committee of the Federal University of Ceará (craft no. 550/2006, protocol no. 238/05). The parents or guardians of the children provided written informed consent during fecal specimen collection.
Clinical and nutritional evaluation.
The children’s clinical information was collected to evaluate diarrhea severity and other associated symptoms (dehydration, fever, vomiting, abdominal pain, restlessness, weakness, respiratory symptoms, mucous and bloody stools). Fever was confirmed by a body temperature measurement of greater than 37.5°C (16). This careful evaluation was based on the procedures done by a previous large multicenter study of diarrhea etiology in children from developing countries (17). A standardized questionnaire form was administered by previously trained clinical staff when interviewing the parents or guardians of the children. The nutritional status was assessed by calculating the anthropometric weight-for-age Z-score (WAZ), height-for-age Z-score (HAZ), and weight-for-height Z-score (WHZ) (18).
Diagnosis of EPEC infection.
Fecal DNA was extracted from stool samples using a QIAamp DNA stool minikit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. DNA quality and quantity were checked using the spectrophotometric method (NanoDrop 2000 spectrophotometer; Thermo Fisher Scientific, MA, USA). Extracted DNA was stored at −20°C until further analysis.
The diagnosis of enteropathogenic Escherichia coli (EPEC) infection was established using the Luminex Bio-Plex system technology (Bio-Rad, CA, USA) on the DNA samples. Both forward and reverse primers were biotinylated on the 5′ end, and probes were amine modified at the 5′ end and included 12 carbon spacers to enable coupling to the carboxylated fluorescent microspheres (Bio-Rad). mPCRs and hybridization procedures were then performed using a Bio-Plex 200 system (Bio-Rad). The results were reported as the microsphere-specific median fluorescence intensity (MFI) and corrected using the background bead fluorescence. The corrected MFI (cMFI) was calculated as follows: cMFI = (MFIanalyte – MFInegative control)/MFInegative control. Positive samples had cMFI values greater than 3. Positive controls (a DNA template from EPEC) and negative controls (nuclease-free water) were included in every run.
The identification of EPEC was made concomitantly with a screening for other 16 enteropathogens, divided into different panels: the virus panel (rotavirus, adenovirus, norovirus, astrovirus, and sapovirus) (19), bacterial panel 1 (pathogenic Escherichia coli) (20), bacterial panel 2 (Shigella spp., Salmonella spp., Campylobacter spp., Aeromonas spp., and Vibrio spp.) (21), and the protozoal panel (Giardia spp., Cryptosporidium spp., and Entamoeba histolytica) (22).
Detection of EPEC virulence-related genes.
The DNA fecal samples positive for EPEC were tested, using mPCRs, for the presence of 18 VRGs. Five mPCRs were used, where specific markers were sought among the EPEC nucleotide sequences available in GenBank (NCBI, Bethesda, MD, USA) (see Table S1 in the supplemental material). New primers were designed using OligoPerfect Designer software (Thermo Fisher Scientific, MA, USA) and were synthesized by Invitrogen (São Paulo, Brazil) by consideration of similar melting temperatures. A Basic Local Alignment Search Tool (NCBI, Bethesda, MD, USA) search against the GenBank nucleotide database was used to confirm target specificity. Prior to testing and optimization, all primer pairs were evaluated using uniplex PCRs with their respective positive controls.
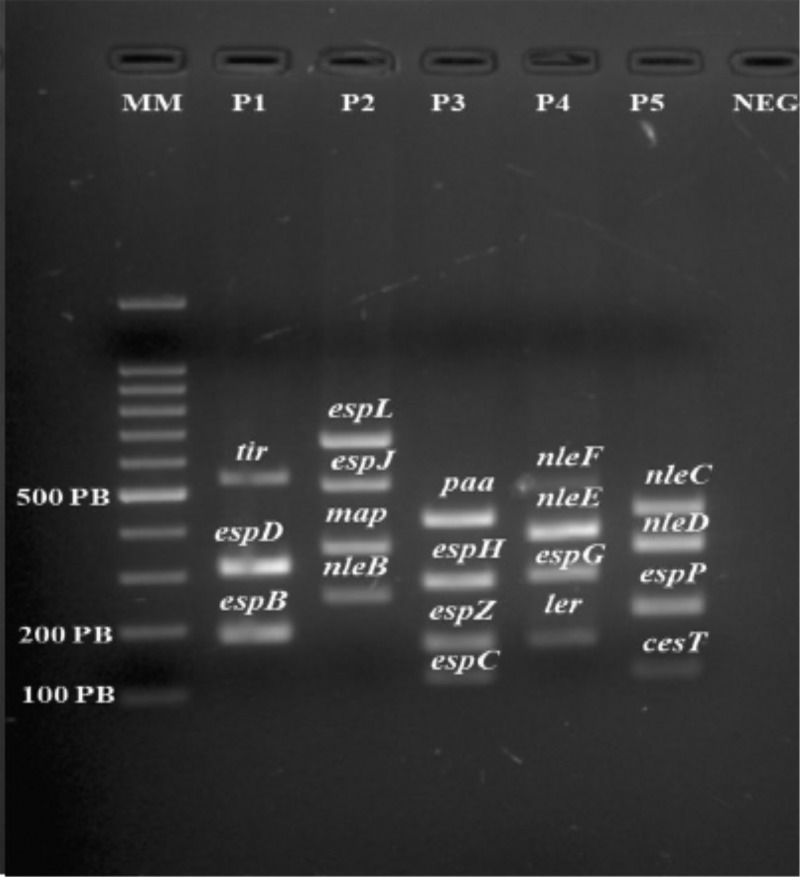
After optimization, each mPCR tube contained a 25-μl reaction mixture comprising 12.5 μl of master mix (Qiagen multiplex PCR kit; Qiagen, CA, USA), 2.5 μl of Q solution (Qiagen, CA, USA), and 2.0 μl of DNA, in addition to primers (2 mM) and water. The PCR products were visualized and photographed (ChemiDoc XRS; Bio-Rad Laboratories, CA, USA) after electrophoresis using an ethidium bromide-stained 2% agarose gel in 1× Tris acetate-EDTA buffer (Fig. 2). DNA from EPEC strain 2348/69 was used as a positive control, and autoclaved Milli-Q water was used as a negative control.
FIG 2.
Visualization of the bands obtained after PCR amplification of genes encoding virulence factors. Lanes: MM, molecular markers; P1 to P5, patients 1 to 5, respectively; NEG, negative control. PB, number of base pairs.
Data analysis.
All the data generated were coded in an Excel spreadsheet (Microsoft, New York, NY, USA) and added by two independent typists and then verified through the intersection of these spreadsheets. Fisher’s exact test, the odds ratio (OR), and the Mann-Whitney U test were used to compare the data derived from case and control children regarding the presence of VRGs. Statistical analyses were performed using Statistical Package for the Social Sciences software, version 20.0 (SPSS Inc., Chicago, IL), and GraphPad Prism software (version 6.00 for Windows; GraphPad Software, San Diego, CA, USA) was used for complementary statistical analysis, table formatting, and figure preparation. Anthropometric Z-scores (HAZ, WAZ, and WHZ) were calculated using Epi Info software (version 6.0; Centers for Disease Control and Prevention, Atlanta, GA) and data from the World Health Organization Multi-Country Growth Reference Study (18). For categorical data analysis, Z-scores less than −2 were considered low, reflecting undernutrition (1), while Z-scores above −1 were interpreted to be from nourished subjects.
In order to evaluate the correlations of VRG combinations with clinical parameters, the Classification and Regression Tree (CART software, pro version 6.0; Salford Systems, San Diego, CA) was used (23, 24). ORs with 95% confidence intervals (CI) are shown to assess for the risk found between a variable and the outcome analyzed. A P value of <0.05 was considered statistically significant and was used in the statistical analysis.
RESULTS
EPEC prevalence and characterization of EPEC-positive population.
EPEC was detected in 28.2% (337/1,191) of all samples, 29.4% (174/591) of samples from cases, and 27.1% (163/600) of samples from controls. Only 8.6% (29/337) of the samples were classified as having tEPEC, while 91.4% (308/337) were classified as having aEPEC. Among the children, 50.4% were male and the age range was 2 to 36 months (mean, 16.93 months; median, 15.84 months), with 37.3% (126/337) of the children being between 12 and 24 months old. Children aged 2 to 12 months were significantly associated with diarrhea cases (P = 0.0055; OR = 2.02; 95% CI = 1.24 to 3.29). The monthly family income of the vast majority (85%, 287/337) was below US$303, which corresponds to the minimum wage in Brazil at the time of the study (Table 1).
TABLE 1.
Characterization of the EPEC-positive population of children with and without diarrhea (cases and controls) from the Brazilian semiarid region regarding sex, age, and family monthly income
| Characteristic | No. (%) of subjects |
P valuea | OR (95% CI)b | ||
|---|---|---|---|---|---|
| Total (n = 337) | Case (n = 174) |
Control (n= 163) |
|||
| Sex | |||||
| Male | 170 (50.4) | 94 (54.1) | 76 (46.6) | 0.1916 | 1.34 (0.87–2.06) |
| Female | 167 (49.5) | 80 (45.9) | 87 (53.3) | ||
| Age (mo) | |||||
| 2–12 | 97 (28.7) | 62 (35.6) | 35 (21.4) | 0.0055 | 2.02 (1.24–3.29) |
| 12.1–24 | 126 (37.3) | 63 (36.2) | 63 (38.6) | 0.6540 | 0.90 (0.57–1.40) |
| 24.1–36 | 114 (33.8) | 49 (28.1) | 65 (39.8) | 0.0285 | 0.59 (0.37–0.93) |
| Family monthly income (times minimum wagec ) | |||||
| >0.5 and ≤2 | 287 (85.1) | 148 (85.1) | 139 (85.2) | 1.0000 | 0.98 (0.53–1.79) |
| >2 and ≤5 | 48 (14.2) | 25 (14.3) | 23 (14.1) | 1.0000 | 1.02 (0.55–1.88) |
| No answer | 2 (0.59) | 1 (0.57) | 1 (0.61) | ||
P values were obtained from Fisher’s exact tests (a P value of <0.05 was significant). Values with statistical significance are highlighted in bold.
CI, confidence interval; OR, odds ratio.
Minimum wage corresponds to the minimum wage in Brazil.
Clinical evaluation of typical and atypical EPEC infections.
We evaluated the clinical outcomes associated with EPEC subtype infections. In the EPEC-positive population, tEPEC was more frequently associated with the cases (12.0% [22/174] for cases versus 4.09% [7/163] for controls; P = 0.0065; OR = 3.22; 95% CI = 1.33 to 7.75), while aEPEC was more frequently associated with the controls (91.2% [156/163] for controls versus 83.5% [152/174] for cases; P = 0.0374; OR = 2.05; 95% CI = 0.96 to 4.37). Among the children with diarrhea, children infected with aEPEC showed a significantly higher number of days of duration of diarrhea than children with tEPEC infections (4.28 versus 3.77 days; P = 0.0102). There were no significant differences in the other symptoms between children with aEPEC infections and children with tEPEC infections (Table 2).
TABLE 2.
Clinical characterization of EPEC-positive children from Brazilian semiarid region infected with typical versus atypical EPEC isolatesa
| Characteristic | Values for children infected with: |
P valueb | ||
|---|---|---|---|---|
| Total (n = 174) | Typical EPEC (n = 22) | Atypical EPEC (n = 152) | ||
| Mean ± SD Z-score for the following anthropometric measures: | ||||
| HAZ | −0.5277 ± 1.599 | −0.1786 ± 1.526 | −0.5786 ± 1.608 | 0.1546 |
| WAZ | 0.2498 ± 1.194 | 0.2686 ± 1.158 | 01205 ± 1.436 | 0.5843 |
| WHZ | 0.7541 ± 1.550 | 0.3491 ± 1.453 | 0.8135 ± 1.559 | 0.0094 |
| Diarrhea | ||||
| Mean ± SD no. of days of diarrhea duration | 4.21 ± 1.376 | 3.77 ± 1.445 | 4.28 ± 1.359 | 0.0102 |
| Mean ± SD no. of episodes/day | 4.49 ± 0.831 | 4.31 ± 0.646 | 4.52 ± 0.853 | 0.1460 |
| No. (%) of children with mucus in stool | 12 (6.8) | 2 (9.09) | 10 (6.5) | 0.6510 |
| No. (%) of children with the following symptoms; | ||||
| Fever (≥37.3°C) | 56 (32.1) | 6 (27.2) | 50 (32.9) | 0.8076 |
| Abdominal pain | 19 (10.9) | 3 (13.6) | 16 (10.5) | 0.7130 |
| Vomiting | 66 (37.9) | 7 (31.8) | 59 (38.8) | 0.6410 |
| Dehydration | 71 (40.8) | 6 (27.2) | 65 (42.7) | 0.2455 |
| Respiratory symptoms | 35 (20.1) | 7 (31.8) | 28 (18.4) | 0.1582 |
| No. (%) of children with the following behavioral signs: | ||||
| Inquietude | 63 (36.2) | 4 (18.1) | 59 (38.8) | 0.0945 |
| Weakness | 38 (21.8) | 5 (22.7) | 33 (21.7) | 1.0000 |
HAZ, height-for-age Z-score; WAZ, weight-for-age Z-score; WHZ, weight-for-height Z-score; EPEC, enteropathogenic Escherichia coli.
P values were obtained from Fisher’s exact tests (a P value of <0.05 was considered significant). Values with statistical significance are highlighted in bold.
We evaluated the impact of EPEC infections on the nutritional status of the study population. Children with an EPEC-positive diagnosis showed lower HAZ values than children without EPEC infection (P = 0.0200). When comparing anthropometric Z-scores between the two types of EPEC infection, aEPEC-infected children showed significantly lower WHZ values than tEPEC-infected children (0.12 ± 1.42 versus 0.71 ± 1.57; P = 0.0200) (Table 2). It is important to mention that the children evaluated here could not be identified as undernourished based on the anthropometric cutoff Z-scores (1).
EPEC VRG distribution.
All EPEC-positive samples were positive for at least one VRG. A total of 10.3% of the samples (35/337) were positive for up to 11 different VRGs, while 8.3% (28/337) were positive for 10 VRGs. The most prevalent VRG was ler (72.1%), followed by espG (61.1%), cesT (59.6%), and espB (55.1%). The least prevalent VRG was nleD (13%). A complete list of the EPEC VRGs and their prevalence can be seen in Table 3.
TABLE 3.
Prevalence of VRGs of EPEC among children with and without diarrhea (cases and controls) from Brazilian semiarid regiona
| VRG | Encoded factor | No. (%) of children |
Univariable analysis |
Multivariable analysis |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cases (n = 174) |
Controls (n = 163) |
Total (n = 337) |
P value | OR | 95% CI | P value | OR | 95% CI | ||
| espC | Enterotoxin | 76 (43.6) | 66 (40.4) | 142 (42.1) | 0.5820 | 1.14 | 0.73–1.75 | 0.212 | 1.35 | 0.84–2.18 |
| espP | Protease | 67 (38.5) | 72 (44.1) | 139 (41.2) | 0.3197 | 0.79 | 0.51–1.22 | 0.091 | 0.66 | 0.41–1.06 |
| tir | Intimin | 27 (15.5) | 22 (13.4) | 49 (14.5) | 0.6446 | 1.17 | 0.64–2.16 | 0.514 | 1.23 | 0.65–2.3 |
| map | LEE-encoded effector | 99 (56.8) | 90 (55.2) | 189 (56.0) | 0.8262 | 1.07 | 0.69–1.64 | 0.73 | 1.08 | 0.69–1.67 |
| espH | LEE-encoded effector | 80 (46.0) | 76 (46.6) | 156 (46.2) | 0.9134 | 0.97 | 0.63–1.49 | 0.706 | 0.92 | 0.59–1.42 |
| espB | T3SS effector | 100 (57.4) | 86 (52.7) | 186 (55.1) | 0.4430 | 1.21 | 0.78–1.86 | 0.56 | 1.14 | 0.72–1.82 |
| espD | T3SS effector | 69 (39.6) | 56 (34.3) | 125 (37.0) | 0.3668 | 1.25 | 0.80–1.05 | 0.246 | 1.31 | 0.82–2.08 |
| espZ | T3SS effector | 74 (42.5) | 61 (37.4) | 135 (40.0) | 0.3741 | 1.23 | 0.79–1.91 | 0.376 | 1.23 | 0.77–1.96 |
| nleB | T3SS effector | 66 (37.9) | 53 (32.5) | 119 (35.3) | 0.3072 | 1.26 | 0.80–1.98 | 0.496 | 1.17 | 0.73–1.87 |
| cesT | T3SS effector | 103 (59.1) | 98 (60.1) | 201 (59.6) | 0.9117 | 0.96 | 0.62–1.48 | 0.511 | 0.85 | 0.54–1.35 |
| ler | LEE-encoded regulators | 119 (68.3) | 124 (76.0) | 243 (72.1) | 0.1445 | 0.68 | 0.42–1.10 | 0.057 | 0.61 | 0.37–1.01 |
| espJ | No LEE | 49 (28.1) | 52 (31.9) | 101 (29.9) | 0.4769 | 0.83 | 0.52–1.33 | 0.498 | 0.83 | 0.48–1.42 |
| espL | No LEE | 70 (40.2) | 61 (37.4) | 131 (38.8) | 0.6549 | 1.12 | 0.72–1.74 | 0.293 | 1.31 | 0.78–2.19 |
| espG | No LEE | 107 (61.4) | 99 (60.7) | 206 (61.1) | 0.9113 | 1.03 | 0.66–1.60 | 0.775 | 0.93 | 0.58–1.49 |
| nleE | No LEE | 52 (29.8) | 51 (31.2) | 103 (30.5) | 0.8135 | 0.93 | 0.58–1.48 | 0.891 | 0.96 | 0.58–1.59 |
| nleF | No LEE | 65 (37.3) | 63 (38.6) | 130 (38.5) | 0.8231 | 0.94 | 0.60–1.47 | 0.9 | 0.97 | 0.60–1.54 |
| nleD | No LEE | 28 (16.0) | 16 (9.8) | 44 (13.0) | 0.1058 | 1.76 | 0.91–3.39 | 0.052 | 2.02 | 0.99–4.11 |
| nleC | No LEE | 53 (30.4) | 55 (33.7) | 108 (32.0) | 0.5599 | 0.86 | 0.54–1.36 | 0.173 | 0.69 | 0.41–1.16 |
LEE, locus of enterocyte effacement; T3SS, type III secretion system; CI, confidence interval; OR, odds ratio.
When comparing the VRG distribution between tEPEC- and aEPEC-positive samples, we observed that aEPEC-positive samples harbored more genes (P = 0.0304). Specifically, aEPEC-positive samples were significantly associated with the presence of map (57.7% [178/308] versus 37.9% [11/29] for tEPEC-positive samples; P = 0.0498; OR = 2.24; 95% CI = 1.02 to 4.90) (see Table S2 in the supplemental material). Albeit no single VRG was associated with either the cases or the controls in the total population, when analyzing the subgroup of children infected with aEPEC, the ler gene presence was significantly associated with control children (75.6% [118/156] versus 70.3% [107/152] for tEPEC-positive children; P = 0.0376; OR = 1.64; 95% CI = 1.03 to 2.60) (Table S3). In tEPEC-infected children, samples positive for the espB gene were significantly associated with case children (diarrhea occurrence) (63.6% [14/22] versus 57.1% [4/7] for control children; P = 0.0281; OR = 3.47; 95% CI = 1.12 to 10.80) (Table S4). Further investigation using CART analysis of VRG combinations that could be associated with clinical symptoms did not show any significant findings.
We also investigated whether a specific VRG could play a role in the decrements of the anthropometric Z-scores in the total EPEC-positive population. Children infected with bacteria harboring the cesT gene but lacking the espP gene were associated with a WAZ of less than −2 (69.2% [9/13] versus 26.3% [74/281]; P = 0.0020; OR = 6.29; 95% CI = 1.88 to 21.06). In addition, children infected with bacteria harboring map were associated with a HAZ of less than −2 (76.4% [26/34] versus 42.3% [36/85]; P = 0.0010; OR = 4.42; 95% CI = 1.79 to 10.90). The analysis of VRG combinations associated with undernutrition within the control children did not show any significant results.
Coenteropathogen distribution.
We observed a high prevalence of coenteropathogens in children with EPEC infections: 92.9% (313/337) and 57.2% (193/337) of the EPEC-infected children showed at least one and two other copathogens, respectively. All EPEC-positive samples were positive for at least another pathogen. Enteroaggregative E. coli (EAEC) was the major copathogen detected (74.7%, 252/337), followed by Salmonella spp. (20.8%, 70/337). Vibrio cholerae was the least common copathogen found (0.6%, 2/337) (Table 4). In order to evaluate which copathogens could be associated with EPEC infection and its outcomes, we analyzed the findings for the enteric copathogens detected by the Bio-Plex panel. We evaluated whether the detection of any given copathogen correlated with EPEC-positive samples and found that EAEC was significantly associated with EPEC (70.9% [252/355] of EPEC-positive samples contained EAEC, whereas 47.6% [398/836] of EPEC-negative samples contained EAEC; P < 0.0001; OR = 2.69; 95% CI = 2.06 to 3.51). The presence of copathogens was also evaluated regarding clinical symptoms. Interestingly, Campylobacter spp. and norovirus coinfection was associated with cases (diarrhea occurrence) in EPEC-infected children by a univariate analysis (P = 0.0309, OR = 2.51, and 95% CI = 1.07 to 5.88 for Campylobacter spp.; P = 0.0209, OR = 5.43, and CI =1.18 to 24.91 for norovirus) (Table 4). Additionally, EAEC coinfection was associated with abdominal pain in EPEC-positive children (P = 0.0056; OR = 2.48; 95% CI = 1.300 to 4.749). No other copathogen was associated with any other clinical symptoms.
TABLE 4.
Prevalence of enterocopathogens of EPEC infections in children with and without diarrhea (cases and controls) from Brazilian semiarid regiona
| Copathogen | No. (%) of children |
Univariable analysis |
Multivariable analysis |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Cases (n = 174) | Controls (n = 163) | Total (n = 337) |
P valueb | OR | 95% CI | P valueb | OR | 95% CI | |
| Bacteria | |||||||||
| EAEC | 130 (74.7) | 112 (68.7) | 252 (74.7) | 0.2284 | 1.34 | 0.83–2.16 | 0.575 | 1.15 | 0.69–1.90 |
| EIEC/Shigella | 20 (11.5) | 25 (15.3) | 45 (13.3) | 0.3381 | 0.71 | 0.38–1.34 | 0.755 | 0.89 | 0.42–1.85 |
| ETEC | 25 (14.3) | 27 (16.5) | 52 (15.4) | 0.6514 | 0.84 | 0.46–1.52 | 0.516 | 0.81 | 0.43–1.51 |
| Salmonella spp. | 32 (18.3) | 38 (23.3) | 70 (20.8) | 0.2846 | 0.74 | 0.43–1.25 | 0.517 | 0.81 | 0.44–1.50 |
| Campylobacter spp. | 20 (11.5) | 8 (5.0) | 28 (8.3) | 0.0309 | 2.51 | 1.07–5.88 | 0.027 | 2.72 | 1.12–6.61 |
| Aeromonas spp. | 7 (4.1) | 7 (4.3) | 14 (4.1) | 1.0000 | 0.93 | 0.32–2.72 | 0.765 | 0.83 | 0.25–2.72 |
| Vibrio cholerae | 1 (0.6) | 1 (0.6) | 2 (0.6) | 1.0000 | 0.93 | 0.05–15.11 | 1.0000 | 1.00 | 0.52–19.2 |
| Viruses | |||||||||
| Adenovirus | 6 (3.4) | 1 (0.6) | 7 (2.1) | 0.1225 | 5.78 | 0.68–48.61 | 0.165 | 4.69 | 0.53–41.5 |
| Astrovirus | 4 (2.3) | 3 (1.8) | 7 (2.1) | 1.0000 | 1.25 | 0.27–5.69 | 0.710 | 1.33 | 0.29–6.12 |
| Norovirus | 11 (6.3) | 2 (1.2) | 13 (3.9) | 0.0209 | 5.43 | 1.1824.91 | 0.032 | 5.33 | 1.15–24.69 |
| Rotavirus | 21 (12.1) | 10 (6.1) | 31 (9.2) | 0.0883 | 2.10 | 0.95–4.60 | 0.111 | 1.92 | 0.86–4.30 |
| Sapovirus | 6 (3.4) | 4 (2.4) | 10 (2.3) | 0.7515 | 1.42 | 0.39–5.12 | 0.090 | 6.23 | 0.75–51.7 |
| Protozoa | |||||||||
| Entamoeba spp. | 3 (1.7) | 1 (0.6) | 4 (1.2) | 0.6236 | 2.84 | 0.29–27.62 | 0.333 | 3.09 | 0.31–30.3 |
| Cryptosporidium spp. | 10 (5.8) | 10 (6.1) | 20 (6.0) | 1.0000 | 0.93 | 0.37–2.30 | 0.750 | 0.86 | 0.34–2.15 |
| Giardia lamblia | 33 (19.0) | 25 (15.3) | 58 (17.2) | 0.3905 | 1.29 | 0.73–2.28 | 0.228 | 1.42 | 0.80–2.53 |
EAEC, enteroaggregative Escherichia coli; EIEC, enteroinvasive Escherichia coli; ETEC, enterotoxigenic Escherichia coli; CI, confidence interval; OR, odds ratio.
P values were obtained from Fisher’s exact tests (a P value of <0.05 was considered significant). Values with statistical significance are highlighted in bold.
DISCUSSION
EPEC represents a highly important group of enteropathogens that present a high degree of genetic variability and that are associated with a broad range of clinical outcomes (12). We investigated EPEC genetics and suggested specific VRGs and coenteropathogens that may be responsible for clinical outcomes in both tEPEC and aEPEC infections in children from the Brazilian semiarid region, which is one of the poorest areas of Brazil, showing precarious sanitation and a population with a low socioeconomic status (25). In parallel, we described the most prevalent enteropathogens in this population and found tEPEC to be one of the major causes of diarrhea (16).
The high prevalence of EPEC in this study was mostly characterized by the high frequency of aEPEC rather than tEPEC. Several studies have reported the same observation (26–28). The overall EPEC burden was not associated with diarrhea. However, in the EPEC-positive population, children under 12 months of age were positively correlated with the occurrence of diarrhea. This is also in agreement with studies that support the suggestion that EPEC diarrhea is more frequent during the first year of life (2, 4, 26, 29–31).
Although total EPEC isolate detection was not associated with diarrhea in this study, infection with tEPEC showed a significant association with diarrhea in the EPEC-positive population, while infection with aEPEC was associated with the controls. The stronger association with diarrhea is commonly observed more often with tEPEC than with aEPEC (2), while the former is associated with nonsevere diarrhea and asymptomatic infections (32). Interestingly, when considering only the population with diarrhea, aEPEC was associated with a slightly longer duration of diarrhea than tEPEC, which further corroborates the findings of previous studies (26, 33). Our data support the importance of discriminating tEPEC and aEPEC infections, as the related outcomes can be highly different (6, 34).
Attempts to identify specific EPEC genetic markers that could predict severe outcomes of infection were described in the past (4, 12, 14, 35). Here, we employed an unprecedented mPCR panel covering 18 different EPEC virulence strategies, including LEE- and non-LEE-encoded effectors: proteases, chaperones, regulators, and other type 3 secretion system (T3SS) components. The VRG distribution in the samples reflected the high degree of heterogeneity of these bacteria. Importantly, the presence of the ler gene, a negative autoregulator of the LEE1 operon (36), was associated with aEPEC-infected children without diarrhea. Additionally, the espB gene was associated with diarrhea in tEPEC-positive children. These results are strongly supported by their biological plausibility. The Aar-encoding gene, a correlate of Ler (a H-NS homolog) in enteroaggregative E. coli (EAEC) that negatively regulates EAEC virulence, has been consistently associated with less severe outcomes in children from Brazil and Mali (23, 37). espB is a major part of the T3SS machinery, contributing to the injection of several effector proteins into the host, and also induces cell death in immune cells (38). Albeit the power of the association was not substantially strong, these findings suggest that these genes are the major components in the EPEC virulence arsenal associated with diarrhea and reinforce the differences in the pathogenesis between tEPEC and aEPEC. Interestingly, when trying to investigate whether some VRGs would correlate with tEPEC- or aEPEC-positive samples, we found that map, a gene located on the LEE, was the only one differently distributed between the two groups, being more prevalent in aEPEC isolates. Other studies have identified specific genetic markers associated with EPEC subtypes (39, 40), showing that genes both located on the LEE and not located on the LEE could be associated with either tEPEC or aEPEC. In general, aEPEC bacteria show more variability than tEPEC bacteria (39–41).
Another focus of this study was evaluation of whether EPEC infections could be associated with anthropometric Z-score decrements. We observed a decrease in HAZ levels in EPEC-infected children compared to those in non-EPEC-infected children, and this was mainly attributed to aEPEC, likely due to its higher prevalence than tEPEC in the study. Furthermore, aEPEC infection was associated with decreases in WHZ. Indeed, this is the first study to show the burdens of EPEC on undernutrition. Other studies have tried to associate enteropathogens, including diarrheagenic E. coli, such as enteroaggregative E. coli (EAEC), with undernutrition in child populations (42, 43). Recent data emerging from MAL-ED studies have reinforced the importance of even asymptomatic enteric infections as a cause of growth impairment in children from developing countries (44, 45), which was observed due to the use of molecular diagnostic techniques instead of conventional techniques (46, 47). Indeed, the higher prevalence of aEPEC than tEPEC in asymptomatic children was recently supported by Rogawski and colleagues (47) and requires further investigation. aEPEC is a very heterogeneous group of bacteria, and the pathogenic ability of some aEPEC isolates may be related to the specific genetic sequences of the various serotypes (15). The zoonotic potential of aEPEC has also been described and may help to explain its high prevalence in many settings (13, 15).
The influence of VRGs on the parameters of undernutrition was further assessed. We found two different VRG combinations associated with anthropometric score decrements. EPEC infections positive for cesT but negative for espP were associated with undernutrition in the study population. CesT is a major chaperone for T3SS effectors, including Tir, which is essential for intestinal colonization (48), and thus may be associated with persistent infection that could lead to chronic malabsorption and undernutrition. On the other hand, espP, which encodes the autotransporter serine protease EspP, is found more commonly in aEPEC and rarely in tEPEC (49), and this may suggest that its absence could be correlated with more severe outcomes. Additionally, map was associated with undernutrition. Map is a LEE-encoded effector protein which interacts with mitochondria and changes their morphology (50), but it is also reported to maintain colonization in vivo (51). These findings suggest the role of specific procolonization virulence genes, cesT and map, in the induction of potentially persistent EPEC infections, which may be linked with undernutrition. Indeed, it also reflects the highly complex interactions between EPEC virulence strategies and host inflammatory responses that lead to different clinical outcomes. Of note, this is the first study to correlate EPEC VRGs with undernutrition in children.
The diverse clinical outcomes of EPEC infections may also be influenced by infection with other copathogens (52). Therefore, we aimed to investigate whether specific coenteropathogens could correlate with EPEC infection and its outcomes. Campylobacter spp. and norovirus were associated with diarrhea, and EAEC was associated with increased abdominal pain in EPEC-infected children. These observations corroborate recent data that highlighted the importance of coinfections in EPEC-associated diarrhea in children from developing countries (53). Further, it supports EAEC as a major copathogen that may exacerbate the deleterious effects of other enteric infections in children (54). Importantly, Campylobacter spp. and EAEC were the two major enteropathogens associated with growth decrements in a large multicenter study of the etiology of environmental enteropathy in children from developing countries (44), and this study reinforces the major burden of these pathogens.
This study has the potential limitation of using DNA extracted directly from stool samples instead of the EPEC isolates, which would yield more precise findings about a single EPEC bacterium. Moreover, other Enterobacteriaceae, especially other diarrheagenic E. coli strains, such as enterohemorrhagic E. coli, EAEC, and enterotoxigenic E. coli, and Shigella, might share similar virulence genetic sequences with EPEC (55–58). However, it is important to highlight that polymicrobial infections may occur in settings with high levels of environmental contamination, as in this study, which supports the approach used here. Moreover, the high rates of coinfections suggest that other pathogens and markers play a role in infection outcomes and indicate the likely multifactorial etiology of diarrhea and enteropathy in EPEC-infected children. Finally, even though a wide evaluation of other enteric pathogens was employed, tests for other pathogens, such as Yersinia spp. and Plesiomonas shigelloides, were not performed.
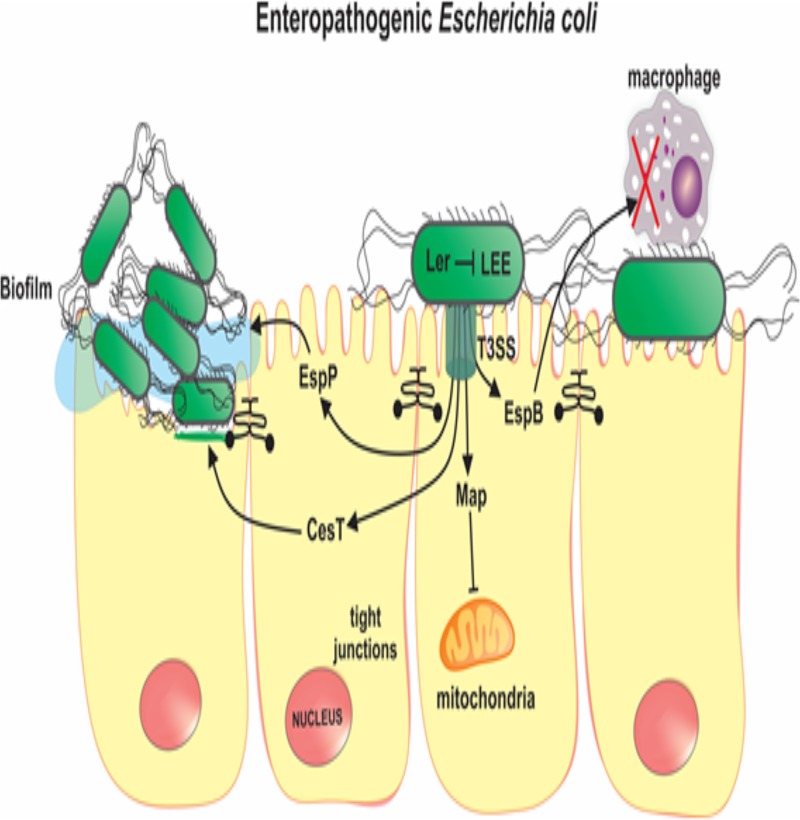
In conclusion, this study characterized EPEC infections in a large number of children with and without diarrhea from the low-income Brazilian semiarid region. The findings reveal that tEPEC is strongly associated with diarrhea, while aEPEC presents a high prevalence. Importantly, infection with any type of EPEC isolate was associated with decreases in HAZ compared with that in children without EPEC infections. However, aEPEC was associated with a lower WHZ than tEPEC. Furthermore, by using a panel for the detection of a broad range of EPEC VRGs, we found that ler, a negative regulator of LEE, was associated with the absence of diarrhea in aEPEC-positive children; espB, a major component of T3SS, was associated with diarrhea in tEPEC-positive children; the presence of the procolonization virulence genes cesT and map was associated with undernutrition; and Campylobacter spp., norovirus, and EAEC coinfections were associated with increased clinical severity in EPEC-infected children. The data presented in this study increase the understanding of the burden and genetic determinants of EPEC infections in children from developing countries. Furthermore, these data identified specific virulence genes of EPEC (ler, espB, cesT, and map) (Fig. 3) and coenteropathogens to be major contributors to diarrhea and undernutrition in children from the low-income Brazilian semiarid region.
FIG 3.
Model of the pathobiology of enteropathogenic E. coli (EPEC) infection, highlighting major virulence genes that were associated with the clinical outcomes in the present study. Ler is a negative autoregulator of the locus of enterocyte effacement (LEE), on which several EPEC virulence genes are located, including components of the type III secretion system (T3SS); thus, Ler can attenuate the virulence of EPEC strains; Map is a LEE-encoded effector protein that changes mitochondrial functions in the host; CesT is a major chaperone for T3SS effectors which facilitates EPEC intestinal colonization; EspB is a component of the T3SS apparatus and induces immune cell death; EspP is an autotransporter serine-protease which contributes to biofilm formation and host defense modulation.
Supplementary Material
ACKNOWLEDGMENTS
We thank the staff and field team for their important contributions to this study.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.01777-18.
REFERENCES
- 1.Lanata CF, Fischer-Walker CL, Olascoaga AC, Torres CX, Aryee MJ, Black RE, Child Health Epidemiology Reference Group of the World Health Organization and UNICEF. 2017. Global causes of diarrheal disease mortality in children <5 years of age: a systematic review. PLoS One 8:e72788. doi: 10.1371/journal.pone.0072788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, Wu Y, Sow SO, Sur D, Breiman RF, Faruque ASG, Zaidi AKM, Saha D, Alonso PL, Tamboura B, Sanogo D, Onwuchekwa U, Manna B, Ramamurthy T, Kanungo S, Ochieng JB, Omore R, Oundo JO, Hossain A, Das SK, Ahmed S, Qureshi S, Quadri F, Adegbola RA, Antonio M, Hossain MJ, Akinsola A, Mandomando I, Nhampossa T, Acácio S, Biswas K, O'Reilly CE, Mintz ED, Berkeley LY, Muhsen K, Sommerfelt H, Robins-Browne RM, Levine MM. 2013. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 382:209–222. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- 3.Chen HD, Frankel G. 2005. Enteropathogenic Escherichia coli: unravelling pathogenesis. FEMS Microbiol Rev 29:83–98. doi: 10.1016/j.femsre.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 4.Donnenberg MS, Hazen TH, Farag TH, Panchalingam S, Antonio M, Hossain A, Mandomando I, Ochieng JB, Ramamurthy T, Tamboura B, Zaidi A, Levine MM, Kotloff K, Rasko DA, Nataro JP. 2015. Bacterial factors associated with lethal outcome of enteropathogenic Escherichia coli infection: genomic case-control studies. PLoS Negl Trop Dis 9:e0003791. doi: 10.1371/journal.pntd.0003791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hernandes RT, Elias WP, Vieira MA, Gomes TA. 2009. An overview of atypical enteropathogenic Escherichia coli. FEMS Microbiol Lett 297:137–149. doi: 10.1111/j.1574-6968.2009.01664.x. [DOI] [PubMed] [Google Scholar]
- 6.Gomes TA, Elias WP, Scaletsky IC, Guth BE, Rodrigues JF, Piazza RM, Ferreira LC, Martinez MB. 2016. Diarrheagenic Escherichia coli. Braz J Microbiol 47:3–30. doi: 10.1016/j.bjm.2016.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ochoa TJ, Contreras CA. 2011. Enteropathogenic E. coli (EPEC) infection in children. Curr Opin Infect Dis 24:478–483. doi: 10.1097/QCO.0b013e32834a8b8b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dean P, Kenny B. 2009. The effector repertoire of enteropathogenic E. coli: ganging up on the host cell. Curr Opin Microbiol 12:101–109. doi: 10.1016/j.mib.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wong AR, Pearson JS, Bright MD, Munera D, Robinson KS, Lee SF, Frankel G, Hartland EL. 2011. Enteropathogenic and enterohaemorrhagic Escherichia coli: even more subversive elements. Mol Microbiol 80:1420–1438. doi: 10.1111/j.1365-2958.2011.07661.x. [DOI] [PubMed] [Google Scholar]
- 10.Dutta S, Pazhani GP, Nataro JP, Ramamurthy T. 2015. Heterogenic virulence in a diarrheagenic Escherichia coli: evidence for an EPEC expressing heat-labile toxin of ETEC. Int J Med Microbiol 305:47–54. doi: 10.1016/j.ijmm.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 11.Hazen TH, Kaper JB, Nataro JP, Rasko DA. 2015. Comparative genomics provides insight into the diversity of the attaching and effacing Escherichia coli virulence plasmids. Infect Immun 83:4103–4117. doi: 10.1128/IAI.00769-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hazen TH, Donnenberg MS, Panchalingam S, Antonio M, Hossain A, Mandomando I, Ochieng JB, Ramamurthy T, Tamboura B, Qureshi S, Quadri F, Zaidi A, Kotloff KL, Levine MM, Barry EM, Kaper JB, Rasko DA, Nataro JP. 2016. Genomic diversity of EPEC associated with clinical presentations of differing severity. Nat Microbiol 1:15014. doi: 10.1038/nmicrobiol.2015.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watson VE, Jacob ME, Flowers JR, Strong SJ, DebRoy C, Gookin JL. 2017. Association of atypical enteropathogenic Escherichia coli with diarrhea and related mortality in kittens. J Clin Microbiol 55:2719–2735. doi: 10.1128/JCM.00403-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cavalcante PA, Prata MMG, Medeiros PHQS, Alves da Silva AV, Quetz JS, Reyes MAV, Rodrigues TS, Santos AKS, Ribeiro SA, Veras HN, Bona MD, Amaral MSMG, Rodrigues FAP, Lima IFN, Havt A, Lima AAM. 2018. Intestinal cell migration damage induced by enteropathogenic Escherichia coli strains. Braz J Med Biol Res 51:e7423. doi: 10.1590/1414-431X20187423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu Y, Bai X, Jin Y, Hu B, Wang H, Sun H, Fan R, Fu S, Xiong Y. 2017. High prevalence of virulence genes in specific genotypes of atypical enteropathogenic Escherichia coli. Front Cell Infect Microbiol 7:109. doi: 10.3389/fcimb.2017.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lima AAM, Oliveira DB, Quetz JS, Havt A, Prata MMG, Lima IFN, Soares AM, Filho JQ, Lima NL, Medeiros PHQS, Santos AKS, Veras HN, Gondim RNDG, Pankov RC, Bona MD, Rodrigues FAP, Moreira RA, Moreira AC, Bertolini M, Bertolini LR, Freitas VJF, Houpt ER, Guerrant RL. 2019. Etiology and severity of diarrheal diseases in infants at the semiarid region of Brazil: a case-control study. PLoS Negl Dis 13:e0007154. doi: 10.1371/journal.pntd.0007154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.MAL-ED Network Investigators. 2014. The MAL-ED study: a multinational and multidisciplinary approach to understand the relationship between enteric pathogens, malnutrition, gut physiology, physical growth, cognitive development, and immune responses in infants and children up to 2 years of age in resource-poor environments. Clin Infect Dis 9:193–206. doi: 10.1093/cid/ciu653. [DOI] [PubMed] [Google Scholar]
- 18.World Health Organization. 2006. WHO child growth standards: methods and development: length/height-for-age, weight-for-age, weight-for-length, weight-for-height and body mass index-for-age. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 19.Liu J, Kibiki G, Maro V, Maro A, Kumburu H, Swai N, Taniuchi M, Gratz J, Toney D, Kang G, Houpt E. 2011. Multiplex reverse transcription PCR Luminex assay for detection and quantitation of viral agents of gastroenteritis. J Clin Virol 50:308–313. doi: 10.1016/j.jcv.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taniuchi M, Sobuz SU, Begum S, Platts-Mills JA, Liu J, Yang Z, Wang XQ, Petri WA, Haque R, Houpt ER. 2013. Etiology of diarrhea in Bangladeshi infants in the first year of life analyzed using molecular methods. J Infect Dis 208:1794–1802. doi: 10.1093/infdis/jit507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu J, Gratz J, Maro A, Kumburu H, Kibiki G, Taniuchi M, Howlader AM, Sobuz SU, Haque R, Talukder KA, Qureshi S, Zaidi A, Haverstick DM, Houpt ER. 2012. Simultaneous detection of six diarrhea-causing bacterial pathogens with an in-house PCR-Luminex assay. J Clin Microbiol 50:98–103. doi: 10.1128/JCM.05416-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taniuchi M, Verweij JJ, Noor Z, Sobuz SU, Lieshout L, Petri WA, Haque R, Houpt ER. 2011. High throughput multiplex PCR and probe-based detection with Luminex beads for seven intestinal parasites. Am J Trop Med Hyg 84:332–337. doi: 10.4269/ajtmh.2011.10-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Havt A, Lima IFN, Medeiros PHQS, Clementino MA, Santos AK, Amaral MS, Veras HN, Prata MM, Lima NL, Di Moura A, Leite AM, Soares AM, Filho JQ, Houpt ER, Nataro JP, Guerrant RL, Lima AA. 2017. Prevalence and virulence gene profiling of enteroaggregative Escherichia coli in malnourished and nourished Brazilian children. Diagn Microbiol Infect Dis 89:98–105. doi: 10.1016/j.diagmicrobio.2017.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lima IFN, Boisen N, Silva JDQ, Havt A, de Carvalho EB, Soares AM, Lima NL, Mota RMS, Nataro JP, Guerrant RL, Lima AAM. 2013. Prevalence of enteroaggregative Escherichia coli and its virulence-related genes in a case-control study among children from north-eastern Brazil. J Med Microbiol 62:683–693. doi: 10.1099/jmm.0.054262-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mendes PS, Ribeiro HDAC, Mendes CM. 2013. Temporal trends of overall mortality and hospital morbidity due to diarrheal disease in Brazilian children younger than 5 years from 2000 to 2010. J Pediatr 89:315–325. doi: 10.1016/j.jped.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 26.Nguyen RN, Taylor LS, Tauschek M, Robins-Browne RM. 2006. Atypical enteropathogenic Escherichia coli. Infection and prolonged diarrhea in children. Emerg Infect Dis 12:597–603. doi: 10.3201/eid1204.051112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu J, Torres AG. 2015. Enteropathogenic Escherichia coli: foe or innocent bystander. Clin Microbiol Infect 21:729–734. doi: 10.1016/j.cmi.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Platts-Mills JA, Babji S, Bodhidatta L, Gratz J, Haque R, Havt A, McCormick BJ, McGrath M, Olortegui MP, Samie A, Shakoor S, Mondal D, Lima IF, Hariraju D, Rayamajhi BB, Qureshi S, Kabir F, Yori PP, Mufamadi B, Amour C, Carreon JD, Richard SA, Lang D, Bessong P, Mduma E, Ahmed T, Lima AA, Mason CJ, Zaidi AK, Bhutta ZA, Kosek M, Guerrant RL, Gottlieb M, Miller M, Kang G, Houpt ER, MAL-ED Network Investigators. 2015. Pathogen-specific burdens of community diarrhea in developing countries: a multisite birth cohort study (MAL-ED). Lancet Glob Health 3:e564–e575. doi: 10.1016/S2214-109X(15)00151-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ochoa TJ, Ecker L, Barletta F, Mispireta ML, Gil AI, Contreras C, Molina M, Amemiya I, Verastegui H, Hall ER, Cleary TG, Lanata CF. 2009. Age-related susceptibility to infection with diarrheagenic E. coli infants from peri-urban areas of Lima, Peru. Clin Infect Dis 49:1694–1702. doi: 10.1086/648069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barletta F, Ochoa TJ, Mercado E, Ruiz J, Ecker L, Lopez G, Mispireta M, Gil AI, Lanata CG, Cleary TG. 2011. Quantitative real-time polymerase chain reaction for enteropathogenic Escherichia coli: a tool for investigation of asymptomatic versus symptomatic infections. Clin Infect Dis 53:1223–1229. doi: 10.1093/cid/cir730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu J, Platts-Mills JA, Juma J, Kabir F, Nkeze J, Okoi C, Operario DJ, Uddin J, Ahmed S, Alonso PL, Antonio M, Becker SM, Blackwelder WC, Breiman RF, Faruque ASG, Fields B, Gratz J, Haque R, Hossain A, Hossain MJ, Jarju S, Qamar F, Iqbal NT, Kwambana B, Mandomando I, McMurry TL, Ochieng C, Ochieng JB, Ochieng M, Onyango C, Panchalingam S, Kalam A, Aziz F, Qureshi S, Ramamurthy T, Roberts JH, Saha D, Sow SO, Stroup SE, Sur D, Tamboura B, Taniuchi M, Tennant SM, Toema D, Wu Y, Zaidi A, Nataro JP, Kotloff KL, Levine MM, Houpt ER. 2016. Use of quantitative molecular diagnostic methods to identify causes of diarrhea in children: a reanalysis of the GEMS case-control study. Lancet 388:1291–1301. doi: 10.1016/S0140-6736(16)31529-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gosh P, Ali A. 2010. Isolation of atypical enteropathogenic Escherichia coli from children with and without diarrhea in Delhi and the National Capital Region, India. J Med Microbiol 59:1156–1162. doi: 10.1099/jmm.0.014530-0. [DOI] [PubMed] [Google Scholar]
- 33.Afset JE, Bevanger L, Romundstad P, Bergh K. 2004. Association of atypical enteropathogenic Escherichia coli (EPEC) with prolonged diarrhoea. J Med Microbiol 53:1137–1144. doi: 10.1099/jmm.0.45719-0. [DOI] [PubMed] [Google Scholar]
- 34.Donnenberg MS, Finlay BB. 2013. Combating enteropathogenic Escherichia coli (EPEC) infections: the way forward. Trends Microbiol 21:317–319. doi: 10.1016/j.tim.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Afset JE, Bruant G, Brousseau R, Harel J, Anderssen E, Bevanger L, Bergh K. 2006. Identification of virulence genes linked with diarrhea due to atypical enteropathogenic Escherichia coli by DNA microarray analysis and PCR. J Clin Microbiol 44:3703–3711. doi: 10.1128/JCM.00429-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berdichevsky T, Friedberg D, Nadler C, Rokney A, Oppenheim A, Rosenshine I. 2005. Ler is a negative autoregulator of the LEE1 operon in enteropathogenic Escherichia coli. J Bacteriol 187:349–357. doi: 10.1128/JB.187.1.349-357.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boisen N, Scheutz F, Rasko DA, Redman JC, Persson S, Simon J, Kotloff KL, Levine MM, Sow S, Tamboura B, Toure A, Malle D, Panchalingam S, Krogfelt KA, Nataro JP. 2012. Genomic characterization of enteroaggregative Escherichia coli from children in Mali. J Infect Dis 205:431–444. doi: 10.1093/infdis/jir757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baumann D, Salia H, Greune L, Norkowski S, Korner B, Uckeley ZM, Frankel G, Guenot M, Ruter C, Schmidt MA. 2018. Multitalented EspB of enteropathogenic Escherichia coli (EPEC) enters cells autonomously and induces programmed cell death in human monocytic THP-1 cells. Int J Med Microbiol 308:387–404. doi: 10.1016/j.ijmm.2018.03.005. [DOI] [PubMed] [Google Scholar]
- 39.Salvador FA, Hernandes RT, Vieira MA, Rockstroh AC, Gomes TA. 2014. Distribution of non-LEE-encoded type 3 secretion system dependent effectors in enteropathogenic Escherichia coli. Braz J Microbiol 45:851–855. doi: 10.1590/S1517-83822014000300014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vieira MA, Salvador FA, Silva RM, Irino K, Vaz TM, Rockstroh AC, Guth BE, Gomes TA. 2010. Prevalence and characteristics of the O122 pathogenicity island in typical and atypical enteropathogenic Escherichia coli strains. J Clin Microbiol 48:1452–1455. doi: 10.1128/JCM.01944-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abreu AG, Bueris V, Porangaba TM, Sircili MP, Navarro-Garcia F, Elias WP. 2013. Autotransporter protein-encoding genes of diarrheagenic Escherichia coli are found in both typical and atypical enteropathogenic E. coli strains. Appl Environ Microbiol 79:411–414. doi: 10.1128/AEM.02635-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Platts-Mills JA, Taniuchi M, Uddin J, Sobuz SU, Mahfuz M, Gaffar SA, Mondal D, Hossain MI, Islam MM, Ahmed AS, Petri WA, Haque R, Houpt ER, Ahmed T. 2017. Association between enteropathogens and malnutrition in children aged 6-23 mo in Bangladesh: a case-control study. Am J Clin Nutr 105:1132–1138. doi: 10.3945/ajcn.116.138800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lima AAM, Leite ÁM, Di Moura A, Lima NL, Soares AM, Abreu CB, Filho JQ, Mota RMS, Lima IFN, Havt A, Medeiros PHQS, Prata MMG, Guedes MM, Cavalcante PA, Veras HN, Santos AKS, Moore SR, Pinkerton RC, Houpt ER, Guerrant RL. 2017. Determinants variables, enteric pathogen burden, gut function, and immune-related inflammatory biomarkers associated with childhood malnutrition: a prospective case-control study in northeastern Brazil. Pediatr Infect Dis J 36:1177–1185. doi: 10.1097/INF.0000000000001569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.MAL-ED Network Investigators. 2017. Relationship between growth and illness, enteropathogens and dietary intakes in the first 2 years of life: findings from the MAL-ED birth cohort study. BMJ Glob Health 2:e000370. doi: 10.1136/bmjgh-2017-000370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kosek MN, MAL-ED Network Investigators. 2017. Causal pathways from enteropathogens to environmental enteropathy: findings from the MAL-ED birth cohort study. EBioMedicine 18:109–117. doi: 10.1016/j.ebiom.2017.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li LL, Liu N, Humphries EM, Yu JM, Li S, Lindsay BR, Stine OC, Duan ZJ. 2016. Aetiology of diarrhoeal disease and evaluation of viral-bacterial coinfection in children under 5 years old in China: a matched case-control study. Clin Microbiol Infect 22:381.e9–381.e16. doi: 10.1016/j.cmi.2015.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rogawski ET, Liu J, Platts-Mills JA, Kabir F, Lertsethtakarn P, Siguas M, Khan SS, Praharaj I, Murei A, Nshama R, Mujaga B, Havt A, Maciel IA, Operario DJ, Taniuchi M, Gratz J, Stroup SE, Roberts JH, Kalam A, Aziz F, Qureshi S, Islam MO, Sakpaisal P, Silapong S, Yori PP, Rajendiran R, Benny B, McGrath M, Seidman JC, Lang D, Gottlieb M, Guerrant RL, Lima AAM, Leite JP, Samie A, Bessong PO, Page N, Bodhidatta L, Mason C, Shrestha S, Kiwelu I, Moduma ER, Igbal NT, Bhutta ZA, Ahmed T, Haque R, Kang G, Kosek MN, Houpt ER, MAL-ED Network Investigators. 2018. Use of quantitative molecular diagnostic methods to investigate the effect of enteropathogen infections on linear growth in children in low-resource settings: longitudinal analysis of results from the MAL-ED cohort study. Lancet Glob Health 6:1319–1328. doi: 10.1016/S2214-109X(18)30351-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thomas NA, Deng W, Puente JL, Frey EA, Yip CK, Strynadka NC, Finlay BB. 2005. CesT is a multi-effector chaperone and recruitment factor required for the efficient type III secretion of both LEE- and non-LEE-encoded effectors of enteropathogenic Escherichia coli. Mol Microbiol 57:1762–1779. doi: 10.1111/j.1365-2958.2005.04802.x. [DOI] [PubMed] [Google Scholar]
- 49.Weiss A, Brockmeyer J. 2012. Prevalence, biogenesis, and functionality of the serine protease autotransporter EspP. Toxins 5:25–48. doi: 10.3390/toxins5010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Papatheodorou P, Domańska G, Oxle M, Mathieu J, Selchow O, Kenny B, Rassow J. 2006. The enteropathogenic Escherichia coli (EPEC) Map effector is imported into the mitochondrial matrix by the TOM/Hsp70 system and alters organelle morphology. Cell Microbiol 8:677–689. doi: 10.1111/j.1462-5822.2005.00660.x. [DOI] [PubMed] [Google Scholar]
- 51.Nguyen M, Rizvi J, Hecht G. 2015. Expression of enteropathogenic Escherichia coli Map is significantly different than that of other type III secreted effectors in vivo. Infect Immun 83:130–137. doi: 10.1128/IAI.02467-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lääveri T, Pakkanen SH, Antikainen J, Riutta J, Mero S, Kirveskari J, Kantele A. 2014. High number of diarrhoeal co-infections in travelers to Benin, West Africa. BMC Infect Dis 14:81. doi: 10.1186/1471-2334-14-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Platts-Mills JA, Liu J, Rogawski ET, Kabir F, Lertsethtakarn P, Siguas M, Khan SS, Praharaj I, Murei A, Nshama R, Mujaga B, Havt A, Maciel IA, McMurry TL, Operario DJ, Taniuchi M, Gratz J, Stroup SE, Roberts JH, Kalam A, Aziz F, Qureshi S, Islam MO, Sakpaisal P, Silapong S, Yori PP, Rajendiran R, Benny B, McGrath M, McCormick BJJ, Seidman JC, Lang D, Gottlieb M, Guerrant RL, Lima AAM, Leite JP, Samie A, Bessong PO, Page N, Bodhidatta L, Mason C, Shrestha S, Kiwelu I, Mduma ER, Igbal NT, Bhutta ZA, Ahmed T, Haque R, Kang G, Kosek MN, Houpt ER, MAL-ED Network Investigators. 2018. Use of quantitative molecular diagnostic methods to assess the aetiology, burden, and clinical characteristics of diarrhoea in children in low-resource settings: a reanalysis of the MAL-ED cohort study. Lancet Glob Health 6:e1309–e1318. doi: 10.1016/S2214-109X(18)30349-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lima AAM, Soares AM, Filho JQS, Havt A, Lima IFN, Lima NL, Abreu CB, Junior FS, Mota RMS, Pan WK-Y, Troeger C, Medeiros PHQS, Veras HN, Prata MA, McCormick BJJ, McGrath M, Rogawski ET, Houpt ER, Platts-Mills JA, Gratz J, Samie A, Bessong P, Babji S, Kang G, Qureshi S, Shakoor S, Bhutta ZA, Haque R, Ahmed T, Mduma ER, Svensen E, Kosek M, Yori PP, Bodhidatta L, Jasmin S, Mason CJ, Lang D, Gottlieb M, Guerrant RL. 2018. Enteroaggregative Escherichia coli subclinical infection and coinfections and impaired child growth in the MAL-ED Cohort Study. J Pediatr Gastroenterol Nutr 66:325–333. doi: 10.1097/MPG.0000000000001717. [DOI] [PubMed] [Google Scholar]
- 55.Andrade FB, Abreu AG, Nunes KO, Gomes TAT, Piazza RMF, Elias WP. 2017. Distribution of serine protease autotransporters of Enterobacteriaceae in typical and atypical enteroaggregative Escherichia coli. Infect Genet Evol 50:83–86. doi: 10.1016/j.meegid.2017.02.018. [DOI] [PubMed] [Google Scholar]
- 56.Bugarel M, Beutin L, Scheutz F, Loukiadis E, Fach P. 2011. Identification of genetic markers for differentiation of Shiga toxin-producing, enteropathogenic, and avirulent strains of Escherichia coli O26. Appl Environ Microbiol 77:2275–2281. doi: 10.1128/AEM.02832-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hazen TH, Daugherty SC, Shetty AC, Nataro JP, Rasko DA. 2017. Transcriptional variation of diverse enteropathogenic Escherichia coli isolates under virulence-inducing conditions. Systems 2:e00024-17. doi: 10.1128/mSystems.00024-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mattock E, Blocker AJ. 2017. How do the virulence factors of Shigella work together to cause disease? Front Cell Infect Microbiol 7:63. doi: 10.3389/fcimb.2017.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.