Nosocomial infections of Elizabethkingia species can have fatal outcomes if not identified and treated properly. The current diagnostic tools available require culture and isolation, which can extend the reporting time and delay treatment.
KEYWORDS: Elizabethkingia, differential assay, molecular diagnostics, real-time PCR
ABSTRACT
Nosocomial infections of Elizabethkingia species can have fatal outcomes if not identified and treated properly. The current diagnostic tools available require culture and isolation, which can extend the reporting time and delay treatment. Using comparative genomics, we developed an efficient multiplex real-time PCR for the simultaneous detection of all known species of Elizabethkingia, as well as differentiating the two most commonly reported species, Elizabethkingia anophelis and Elizabethkingia meningoseptica.
INTRODUCTION
Elizabethkingia is a genus in the Flavobacteriaceae family of Bacteroidetes and represents a separate lineage from the Chryseobacterium-Bergeyella-Riemerella branch (1). Named after Elizabeth King, the first chief of the Special Bacteriology Reference Laboratory (SBRL) of the Centers for Disease Control and Prevention (CDC), these Gram-negative bacteria are widely distributed in the environment. The type species for the genus, Elizabethkingia meningoseptica, was first identified by its namesake in 1959 as a cause of meningitis outbreaks among hospitalized newborns, and at the time placed in the genus Flavobacterium (2). The genus Elizabethkingia is comprised of three medically important species, E. anophelis, E. meningoseptica, and E. miricola. Outbreaks caused by this genus are generally associated with hospital water sources in adult critical care units which are attributable to the tolerance demonstrated by these species to such oligotrophic conditions (3–5). As a result of the bacteria’s multidrug resistance, these infections can have high mortality rates if not treated properly (6).
Isolates from this genus are phenotypically very similar and historically were separated into five groups (E. meningoseptica and genomospecies 1 to 4) distinguished by DNA-DNA hybridization. Recently, through the use of whole-genome sequencing and optical mapping, the genus was reassessed taxonomically (7–9). Initially, genomospecies 1 consisted of E. anophelis and E. endophytica; however, Doijad et al. determined E. endophytica was a synonym of E. anophelis (7). Nicholson et al. further assessed the genus and among other organizational changes proposed the addition of three new species “Elizabethkingia bruuniana” (previously genomospecies 3), “Elizabethkingia ursingii” (genomospecies 4), and “Elizabethkingia occulta” (an atypical isolate originally grouped within genomospecies 4) (9). As of publication, the authors are in the process of requesting the addition of these names to the list of validly described species. Due to phenotypic similarities, Elizabethkingia anophelis has been frequently misidentified as E. meningoseptica by traditional identification techniques and automated systems; thus, E. anophelis is thought to be underreported as a cause of disease. Recent investigations into the prevalence and clinical significance of this species have determined E. anophelis accounts for a significant proportion of Elizabethkingia infections, and a more severe clinical presentation compared to infections from other Elizabethkingia species (10). Conversely, isolates from the “E. miricola cluster” (E. miricola, “E. bruuniana,” “E. occulta,” and “E. ursingii”) have been found to commonly colonize the lungs of cystic fibrosis patients, in many cases without causing disease (11, 12). This variability in clinical presentation makes it important to rapidly and accurately identify the species of an Elizabethkingia isolate. Modern and specific identification methods for identifying Elizabethkingia species, such as conventional PCR assays (10), 16S rRNA gene sequence analysis, and matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry, all require isolation and cultivation of the organism. Consequently, correct identification of an Elizabethkingia infection may take a week or more and is dependent on the availability of an up-to-date and accurately curated database of sequences and MALDI-TOF spectra.
Between 2015 and 2016, the largest documented outbreak of E. anophelis was confirmed in 65 patients from Wisconsin, Illinois, and Michigan in the United States (13) and was associated with 20 fatalities (https://www.cdc.gov/elizabethkingia/outbreaks/). A joint investigation conducted by the local and state public health departments and CDC assessed many potential sources of the outbreak, including health care products, personal care products, food, tap water, and person-to-person transmission (14). Ultimately the source was not determined, further demonstrating the need for a fast, sensitive, and specific method of identifying the presence of this organism without the need for culture and isolation.
Our aim was to develop a method for rapid detection of the presence of Elizabethkingia bacteria directly from primary specimens. Here we present a multiplex real-time PCR assay that detects all Elizabethkingia species while simultaneously differentiating E. anophelis and E. meningoseptica. By eliminating the need for culture and isolation, this assay would enable clinicians to prescribe appropriate treatment to Elizabethkingia infections at least 1 day sooner.
MATERIALS AND METHODS
Comparative genomics and quantitative PCR (qPCR) assay design.
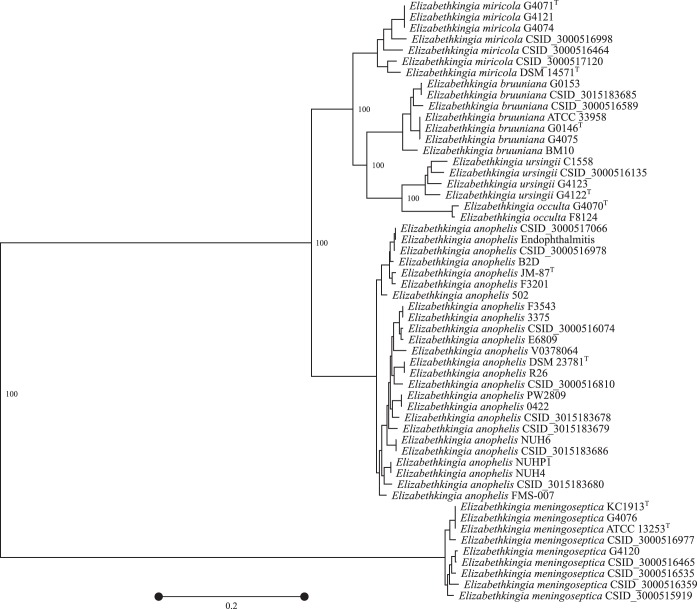
Elizabethkingia whole-genome sequences and alignments generated by SBRL (9) and publicly available from NCBI’s GenBank (15) were included in our analysis (see Table S1 in the supplemental material). Single-copy essential proteins (n = 107) from each isolate’s proteome (n = 78) were extracted with HMMER3 using HMMs trained from 5,372 bacterial genomes in NCBI’s GenBank (16). The Biopython module was used to fetch the corresponding gene sequence for each protein identified in order to identify genus and species targets (17). Each gene for all isolates was individually aligned with MUSCLE (18) prior to concatenating all sequences together. The core alignment was then used to construct a phylogenetic tree (Fig. 1) with PhyML 3 (19) and RAxML 8 (20) with the JC69 substitution model. PhyML was first used to build a starting tree and to calculate sequence statistics such as the transition-to-transversion ratio (kappa), the gamma shape (alpha), and nucleotide frequencies. Topology searching (1,000 iterations) was performed by RAxML. The best tree, based on the log likelihood metric, was given to PhyML, along with the calculated sequence statistics to optimize branch lengths. Finally, 250 bootstrap replicates were performed and projected onto the tree.
FIG 1.
Phylogenetic tree of 81 core singleton genes (6,555 nucleotide variant sites) in Elizabethkingia isolates. Bootstrap values, expressed as a percentage, are only shown for taxonomic partitions. A superscript “T” denotes a type strain. The distance scale bar represents the number of substitutions per variable site.
For each of the essential genes extracted, primers and probes were designed using PrimerSelect version 12.3.1 (DNAStar, Madison, WI). Products were then queried with MoleBlast (21) to assess the potential for amplifying near neighbors. Initial computational PCR simulations were facilitated by Visual OMP (DNA Software, Inc., Ann Arbor, MI) and De-MetaST-BLAST (22). The secY gene is conserved in Elizabethkingia and was chosen as a representative target of all members of the genus. The genes elongation factor 4 (lepA) and phenylalanine-tRNA ligase, beta subunit (pheT) were selected to differentiate E. anophelis from E. meningoseptica (Table 1 ). The primers and probes were synthesized by the Biotechnology Core Facility Branch at CDC.
TABLE 1.
Oligonucleotide sequences
| Gene target | Target organism | Primer/probe ID | Fluorophore/quencher | Final reaction concn (μM) | Product size (bp) | Primer sequence (5′-3′) |
|---|---|---|---|---|---|---|
| secY | Elizabethkingia sp. | SECYF1_4 | 0.01 | GTTTTTACGTTCACGCTCATCTTGGT | ||
| SECY R2 | 0.07 | 146 | AGTAAGCCTAAAAGCCCAGAAG | |||
| SECYP2_5 | FAM/BHQ1 | 0.05 | TTGCAAGTATACAGAACCAAGGAGGAAGCAAG | |||
| pheT | E. meningoseptica | TIGR472_F7 | 0.10 | TTTAAACTGGATGTGGAAGATGCTGAT | ||
| TIGR472_R1_2 | 0.05 | 90 | CCACTCTGGGGACTCTTCTACCTGT | |||
| TIGR472_P3 | Quasar 670/BHQ3 | 0.05 | GCGTTATCTGGGAGCTGTAATTGAAGG | |||
| lepA | E. anophelis | TIGR1393F22 | 0.07 | CATGTGAAGGGGCGCTACTTATTGT | ||
| T1393R3WT | 0.10 | 142 | TCAGGGTTTGCAGAAGGAAGGTC | |||
| TIGR1393P1 | Cal Red 610/BHQ1 | 0.02 | ACCTGGCTTTGGAAAATGACCTTACC |
Assay optimization and validation.
All bacterial strains were grown on heart infusion agar (HIA) with 5% rabbit blood in a candle jar for 24 h at 35°C. Total nucleic acid from all bacterial strains, including the positive controls E. anophelis DSM 23781T and E. meningoseptica ATCC 13253T, was extracted using QIAamp DNA Mini kit (Qiagen, Valencia, CA) according to the manufacturer’s protocols. The DNA concentration was determined using a Qubit 2.0 fluorometer (Life Technologies, Grand Island, NY).
The PCR assay was performed in a Bio-Rad CFX 96 Touch thermal cycler (Bio-Rad, Hercules, CA) using the PerfeCTa MultiPlex qPCR SuperMix (Quanta Biosciences, Gaithersburg, MD). Amplification was done in 25-µl reaction volumes consisting of 10 µl of the PerfeCTa SuperMix, 5 µl of PCR-grade water, 5 µl of the primer-probe mixture (equal volumes of primer and probe; the concentrations are presented in Table 1), and 5 µl of template DNA within the concentration range of 50 to 0.0005 pg/µl. The cycling conditions were as follows: initial denaturation at 95°C for 3 min, followed by 40 cycles of 95°C for 10 s and 64°C for 30 s. Fluorescence data were acquired at the end of the annealing step of each cycle.
The qPCR amplicons were analyzed by agarose gel electrophoresis, and the remaining 20-µl reaction mixtures were purified and eluted into 100 µl of elution buffer using a Macherey-Nagel NucleoVac 96 vacuum manifold and NucleoSpin 8 extract II (Macherey-Nagel, Inc., Bethlehem, PA) according to the manufacturer’s protocols. The sequencing reactions were set up using 2 μl of purified PCR products, 2 μl of BigDye Terminator v1.1 and v3.1 5× sequencing buffer, 2 µl of BigDye Terminator v3.1 cycle sequencing ready reaction mix, 1 μl of sequencing primer at an initial concentration of 3.2 pmol, and 13 μl of PCR-grade water (Life Technologie); thermal cycler conditions followed the manufacturer’s instructions. The reaction mixtures were subsequently purified using Sephadex G-50 (Sigma-Aldrich, St. Louis, MO), analyzed using ABI 3730 DNA analyzer (Applied Biosystems, Life Technologies), and aligned using Geneious version 8.1.6 (Biomatters, Inc., Auckland, NZ).
Cloning and sequencing.
Control plasmids were constructed by ligating the secY, lepA, and pheT amplicons into pCR2.1 using the TOPO TA cloning kit (Life Technologies). The integrity of the insertion site and the sequences of the fragments were verified via PCR and DNA sequencing using primers M13 reverse and T7 promoter, which flank the gene insert. The plasmids were subsequently used to determine the limit of detection of the assay and served as positive controls for each real-time assay. Since the purified plasmid may contain a small amount of Escherichia coli DNA carried over from the competent cells used to replicate the plasmid, the assay was tested to ensure that it would not amplify E. coli DNA, using ATCC 25922.
Standard curves and amplification efficiency.
For quantification, 10-fold serial dilutions (101 to 105 genomic equivalents [GE]/μl) of known E. anophelis and E. meningoseptica purified DNA and control plasmids were tested in triplicate using the singleplex and multiplex formats. PCR efficiency was estimated by testing at least ten replicates of the 10-fold dilution series in different runs. The quantitative range was determined from the range of 10-fold dilutions that gave optimal linearity, efficiency, standard deviation, and correlation coefficient (R2) between replicates. The dilution series was used to create a standard curve for determining the analytical limit of detection.
Analytical sensitivity of bacterial culture suspensions was estimated using artificial specimens of Hanks' balanced salt solution spiked and vortexed, before extraction, with target whole-cell bacterial species. Colony counts were performed using the 6 × 6 drop plate procedure (23) starting from a 0.5 McFarland standard. These were tested at six different bacterial concentrations in triplicate per run, for three independent runs. The cycle quantification (Cq) values for the bacterial concentrations (total, n = 72) were compared to the Cq values of comparable target plasmid concentrations. Viable cell concentrations (CFU/µl) were assumed to be equivalent to the GE/µl, because all of the bacterial target genes occur as a single copy per genome.
Analytical specificity was measured by testing DNA extracted from the panel of positive- and negative-control isolates detailed in Table S2. This panel consisted of 91 control isolates that were either closely related to the target species or represented a wide range of pathogenic isolates submitted to the CDC SBRL and identified using 16S rRNA gene sequence analysis.
DNA extracts of E. anophelis, E. meningoseptica, and one strain from three of the nearest genera within the family Flavobacteriaceae (Riemerella anatipestifer, Chryseobacterium gleum, and Empedobacter brevis) were used to assess the performance of the assay in mixed infections. DNA extracts of the three target strains were mixed at equal concentrations of 101, 102, and 103 GE/µl. Unequal mixtures were evaluated by comparing E. anophelis, E. meningoseptica, and the most closely related species outside the genus, Riemerella anatipestifer, with the first, second, and third strains being mixed at concentrations of 1.5 × 101, 1.5 × 102, and 1.5 × 103 GE/µl, respectively. Unequal mixtures were then repeated at these concentrations for all potential combinations of the three strains. To evaluate between-run and within-run variations in the PCR process, extracts were tested in duplicate or triplicate per run for four runs on different days with the same operator and equipment. The resulting data were reported as arithmetic means.
RESULTS
Amplification efficiency.
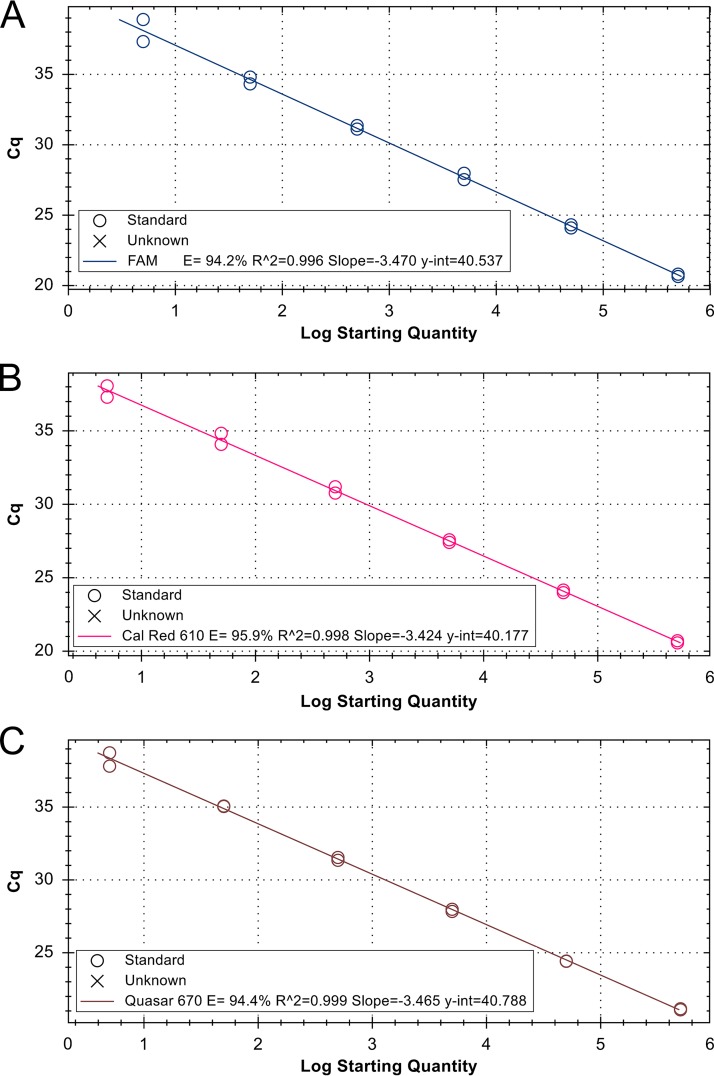
Standard curves were generated to evaluate and compare the efficiency of the real-time assay in singleplex and multiplex formats. The slopes of the linear regression curves were −3.47 for the Elizabethkingia genus-wide target, −3.42 for the E. anophelis-specific target, and −3.46 for the E. meningoseptica target. Amplification efficiencies were 94.2, 95.9, and 94.4% for Elizabethkingia spp., E. anophelis, and E. meningoseptica, with R2 values of 0.995, 0.998, and 0.999, respectively (Fig. 2).
FIG 2.
Standard curves for multiplex real-time PCR in pure culture for efficiency evaluation. (A) Standard curve for target secY, Elizabethkingia genus-wide assay. (B) Standard curve for target lepA to detect only E. anophelis. (C) Standard curve for target pheT to detect only E. meningoseptica.
Analytical sensitivity.
The analytical limit of detection for the multiplex assay with 95% confidence was above 5 GE/µl for all targets. Within the quantitative range of 5 to 500,000 GE/µl, all replicates tested were positive for their intended gene target. Outside of the quantitative range, the assays were less specific and less efficient. Below the 5 GE/µl range, false negatives occasionally occurred (i.e., replicates were not consistently detected), and above 500,000 GE/µl there were some false-positive results.
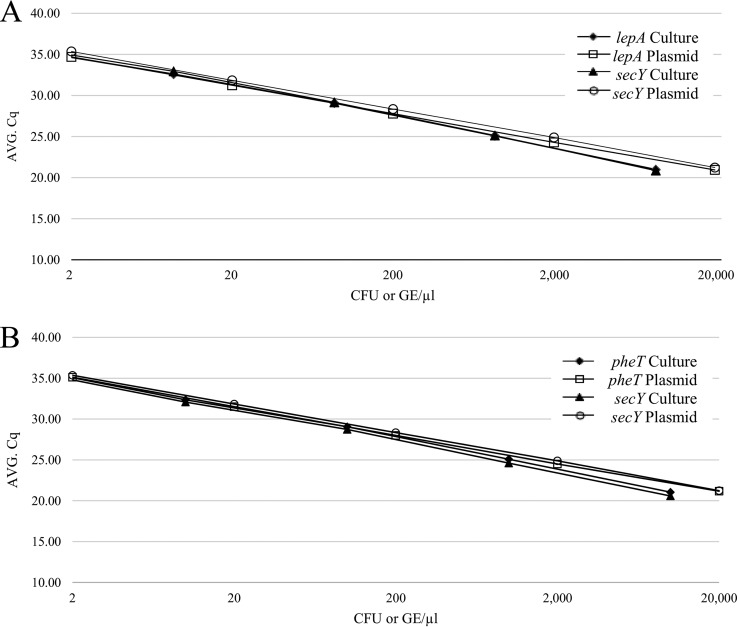
Analytical sensitivity of bacterial culture suspensions for the multiplex assay with 95% confidence averaged 8.6 CFU/µl for E. anophelis (both lepA and secY targets) and 10 CFU/µl for E. meningoseptica (both pheT and secY targets). The standard curves of all bacterial suspensions were comparable to the plasmid results at the corresponding log (Fig. 3).
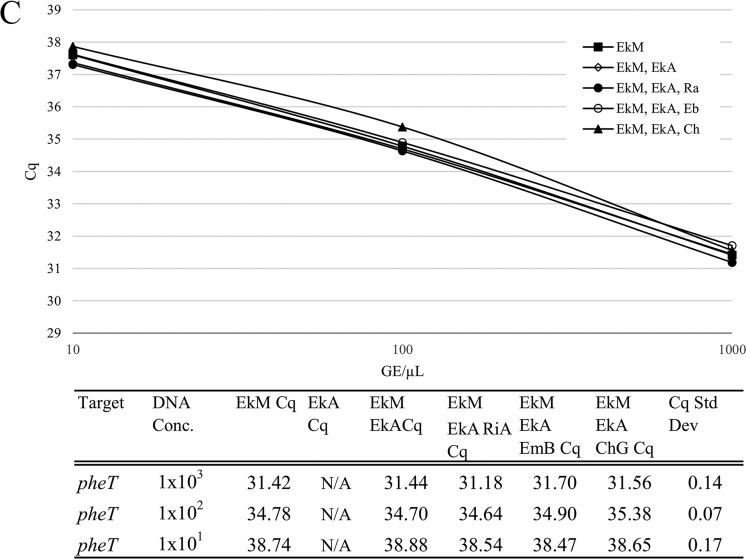
FIG 3.
Comparison of analytical sensitivity using bacterial culture suspensions versus plasmid DNA. (A) Standard curves for detection of E. anophelis (lepA target detects only E. anophelis, secY detects all Elizabethkingia species). (B) Standard curves for detection of E. meningoseptica (pheT target detects only E. meningoseptica, secY detects all Elizabethkingia species).
Analytical specificity.
The assay correctly detected all of the Elizabethkingia strains, and it differentiated E. anophelis and E. meningoseptica from the rest of the genus. For all of the controls listed in Table 2, including the type strain of each Elizabethkingia species, DNA amplification was observed only for intended targets within the quantitative range of 5 to 500,000 GE/µl. The secY target amplified all validated species of Elizabethkingia, while the pheT and lepA targets only amplified their intended species of E. meningoseptica and E. anophelis, respectively. There were no cross-reactions between targets.
TABLE 2.
Assessment of assay performance in unequal triple mixtures: Elizabethkingia species target secYa
| Target sample (EkA) concn | Cq |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Target | EkM, 1.5 × 103 | EkA, 1.5 × 103; EkM, 1.5 × 102; RiA, 1.5 × 101 | EkA, 1.5 × 102; EkM, 1.5 × 103; RiA, 1.5 × 101 | EkA, 1.5 × 102; EkM, 1.5 × 101; RiA, 1.5 × 103 | EkA, 1.5 × 101; EkM, 1.5 × 103; RiA, 1.5 × 102 | EkM, 1.5 × 102 | EkA, 1.5 × 101; EkM, 1.5 × 102; RiA, 1.5 × 103 | EkA, 1.5 × 102; EkM, 1.5 × 101; RiA, 1.5 × 103 | EkM, 1.5 × 101 | SD | |
| 1.5 × 103 | 29.26 | 29.74 | 29.39 | 29.55 | 29.72 | 29.23 | 0.21 | ||||
| 1.5 × 102 | 32.83 | 33.39 | 33.05 | 33.69 | 0.28 | ||||||
| 1.5 × 101 | 36.09 | 37.26 | 0.82 | ||||||||
DNA extracts of E. anophelis (EkA), E. meningoseptica (EkM), and the most closely related species from the nearest genus within the family Flavobacteriaceae, Riemerella anatipestifer (RiA), in unequal mixtures of 1.5 × 101, 1.5 × 102, and 1.5 × 103 GE/µl.
Detection of targets in equal and unequal mixtures.
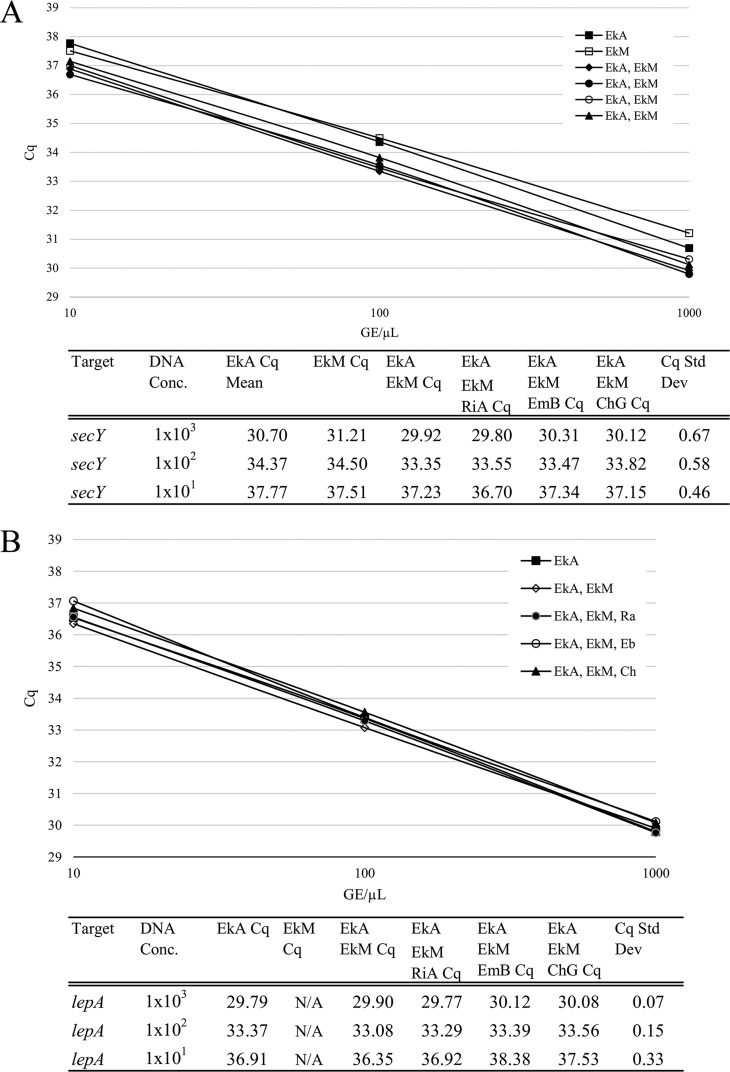
As shown in Fig. 4, the Cq value for each target in an equal mixture was comparable to the Cq value for that target alone. The Cq comparison of DNA extracts of either E. anophelis, E. meningoseptica, or the two Elizabethkingia species and Riemerella anatipestifer, Chryseobacterium gleum, or Empedobacter brevis in equal mixtures of 101, 102, and 103 GE/µl concentrations were within one cycle (expressed as standard deviation) of the pure desired target.
FIG 4.
Assessment of assay performance in equally mixed suspensions. DNA extracts of E. anophelis (EkA), E. meningoseptica (EkM), and strains from three of the nearest genera within the family Flavobacteriaceae—Riemerella anatipestifer (RiA), Chryseobacterium gleum (ChG), and Empedobacter brevis (EmB)—in equal mixtures of 1 × 101, 1 × 102, and 1 × 103 GE/μl concentrations were examined. (A) Elizabethkingia species target secY. (B) E. anophelis target lepA. (C) E. meningoseptica target pheT.
This was also observed in Cq comparisons between the pure target sample and competing strains in different concentrations. Tables 3 and 4 show the mixtures in which E. anophelis and E. meningoseptica specific gene targets were detected within one cycle of the corresponding concentration of the pure target sample regardless of the concentration of the intended or competing species. Similarly, in Table 2, when the two species of Elizabethkingia were both mixed with a competing Riemerella anatipestifer in different concentrations, the Elizabethkingia species with the higher concentration of DNA was detected within one cycle of the pure target sample concentration. Therefore, all Elizabethkingia DNA within a triple mixture could be accurately detected despite the presence of DNA from other species, even when at lower concentrations.
TABLE 3.
Assessment of assay performance in unequal triple mixtures: E. anophelis target lepAa
| Target sample (EkA) concn | Cq |
|||||||
|---|---|---|---|---|---|---|---|---|
| Target | EkA, 1.5 × 103; EkM, 1.5 × 101; RiA, 1.5 × 102 | EkA, 1.5 × 103; EkM, 1.5 × 102; RiA, 1.5 × 101 | EkA, 1.5 × 102; EkM, 1.5 × 103; RiA, 1.5 × 101 | EkA, 1.5 × 102; EkM, 1.5 × 101; RiA, 1.5 × 103 | EkA, 1.5 × 101; EkM, 1.5 × 103; RiA, 1.5 × 102 | EkA, 1.5 × 101; EkM, 1.5 × 102; RiA, 1.5 × 103 | SD | |
| 1.5 × 103 | 28.14 | 28.29 | 28.20 | 0.07 | ||||
| 1.5 × 102 | 31.67 | 31.49 | 32.01 | 0.26 | ||||
| 1.5 × 101 | 34.72 | 34.53 | 34.84 | 0.16 | ||||
DNA extracts of E. anophelis (EkA), E. meningoseptica (EkM), and the most closely related species from the nearest genus within the family Flavobacteriaceae, Riemerella anatipestifer (RiA), in unequal mixtures of 1.5 × 101, 1.5 × 102, and 1.5 × 103 GE/µl.
TABLE 4.
Assessment of assay performance in unequal triple mixtures: E. meningoseptica target pheTa
| Target sample (EkM) concn | Cq |
|||||||
|---|---|---|---|---|---|---|---|---|
| Target | EkM, 1.5 × 103; EkA, 1.5 × 101; RiA, 1.5 × 102 | EkM, 1.5 × 103; EkA, 1.5 × 102; RiA, 1.5 × 101 | EkM, 1.5 × 102; EkA, 1.5 × 101; RiA, 1.5 × 103 | EkM, 1.5 × 102; EkA, 1.5 × 103; RiA, 1.5 × 101 | EkM, 1.5 × 101; EkA, 1.5 × 102; RiA, 1.5 × 103 | EkM, 1.5 × 101; EkA, 1.5 × 103; RiA, 1.5 × 102 | SD | |
| 1.5 × 103 | 30.10 | 30.25 | 30.17 | 0.08 | ||||
| 1.5 × 102 | 33.55 | 33.73 | 33.79 | 0.12 | ||||
| 1.5 × 101 | 36.27 | 37.08 | 37.49 | 0.62 | ||||
DNA extracts of E. anophelis (EkA), E. meningoseptica (EkM), and the most closely related species from the nearest genus within the family Flavobacteriaceae, Riemerella anatipestifer (RiA), in unequal mixtures of 1.5 × 101, 1.5 × 102, and 1.5 × 103 GE/µl.
DISCUSSION
Members of the genus Elizabethkingia are frequently found in aquatic environments, having been isolated from various animals associated with water, including fish, turtles, frogs, and hematophagous flies (24–26), as well as from hospital water systems in Greece (27). Two of the medically relevant species, E. anophelis and E. meningoseptica, have commonly been linked with health care associated infections, and it has been proposed that hospital water supply systems may act as a reservoir for these pathogens (3, 27, 28). The association with hospitals and potentially immunosuppressed patients coupled with the resistance to most antibiotics can result in poor infection outcomes if not identified and treated quickly (6). Currently, few molecular methods are available for typing Elizabethkingia strains without the need to isolate into pure culture (4). Here, we describe the development of a robust multiplex real-time PCR assay for detection and differentiation of members within Elizabethkingia. This assay was designed to rapidly detect all validated species of Elizabethkingia, as well as to distinguish E. anophelis and E. meningoseptica in a single real-time PCR test. The sensitivity and specificity of this assay show promise as a powerful tool for detection of Elizabethkingia. However, due to the increase in sensitivity, this real-time PCR is not intended for use with high concentrations of DNA from pure culture, although dilution of the culture or extract circumvents this issue. Future plans include assessing the sensitivity of the assay in determining the presence of Elizabethkingia spp. in various primary specimens (e.g., blood and cerebral spinal fluid) and validation of this assay for clinical and environmental specimens, as well as adding the detection of E. miricola and other species to the multiplex assay.
Supplementary Material
ACKNOWLEDGMENT
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.01619-18.
REFERENCES
- 1.Kim KK, Kim MK, Lim JH, Park HY, Lee S-T. 2005. Transfer of Chryseobacterium meningosepticum and Chryseobacterium miricola to Elizabethkingia gen. nov. as Elizabethkingia meningoseptica comb. nov. and Elizabethkingia miricola comb. nov. Int J Syst Evol Microbiol 55:1287–1293. doi: 10.1099/ijs.0.63541-0. [DOI] [PubMed] [Google Scholar]
- 2.King EO. 1959. Studies on a group of previously unclassified bacteria associated with meningitis in infants. Am J Clin Pathol 31:241–247. [DOI] [PubMed] [Google Scholar]
- 3.Moore LS, Owens DS, Jepson A, Turton JF, Ashworth S, Donaldson H, Holmes AH. 2016. Waterborne Elizabethkingia meningoseptica in adult critical care. Emerg Infect Dis 22:9–17. doi: 10.3201/eid2201.150139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balm MN, Salmon S, Jureen R, Teo C, Mahdi R, Seetoh T, Teo JT, Lin RT, Fisher DA. 2013. Bad design, bad practices, bad bugs: frustrations in controlling an outbreak of Elizabethkingia meningoseptica in intensive care units. J Hosp Infect 85:134–140. doi: 10.1016/j.jhin.2013.05.012. [DOI] [PubMed] [Google Scholar]
- 5.Hoque SN, Graham J, Kaufmann ME, Tabaqchali S. 2001. Chryseobacterium (Flavobacterium) meningosepticum outbreak associated with colonization of water taps in a neonatal intensive care unit. J Hosp Infect 47:188–192. doi: 10.1053/jhin.2000.0908. [DOI] [PubMed] [Google Scholar]
- 6.Lau SK, Chow WN, Foo CH, Curreem SO, Lo GC, Teng JL, Chen JH, Ng RH, Wu AK, Cheung IY, Chau SK, Lung DC, Lee RA, Tse CW, Fung KS, Que TL, Woo PC. 2016. Elizabethkingia anophelis bacteremia is associated with clinically significant infections and high mortality. Sci Rep 6:26045. doi: 10.1038/srep26045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doijad S, Ghosh H, Glaeser S, Kampfer P, Chakraborty T. 2016. Taxonomic reassessment of the genus Elizabethkingia using whole-genome sequencing: Elizabethkingia endophytica Kampfer et al. 2015 is a later subjective synonym of Elizabethkingia anophelis Kampfer et al. 2011. Int J Syst Evol Microbiol 66:4555–4559. doi: 10.1099/ijsem.0.001390. [DOI] [PubMed] [Google Scholar]
- 8.Nicholson AC, Humrighouse BW, Graziano JC, Emery B, McQuiston JR. 2016. Draft genome sequences of strains representing each of the Elizabethkingia genomospecies previously determined by DNA-DNA hybridization. Genome Announc 4:e00045-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nicholson AC, Gulvik CA, Whitney AM, Humrighouse BW, Graziano J, Emery B, Bell M, Loparev V, Juieng P, Gartin J, Bizet C, Clermont D, Criscuolo A, Brisse S, McQuiston JR. 2018. Revisiting the taxonomy of the genus Elizabethkingia using whole-genome sequencing, optical mapping, and MALDI-TOF, along with proposal of three novel Elizabethkingia species: Elizabethkingia bruuniana sp. nov., Elizabethkingia ursingii sp. nov., and Elizabethkingia occulta sp. nov. Antonie Van Leeuwenhoek 111:55–72. doi: 10.1007/s10482-017-0926-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chew KL, Cheng B, Lin RTP, Teo JWP. 2018. Elizabethkingia anophelis is the dominant Elizabethkingia species found in blood cultures in Singapore. J Clin Microbiol 56:e01445-17. doi: 10.1128/JCM.01445-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kenna DTD, Fuller A, Martin K, Perry C, Pike R, Burns PJ, Narayan O, Wilkinson S, Hill R, Woodford N, Logan JMJ, Turton JF. 2018. rpoB gene sequencing highlights the prevalence of an Elizabethkingia miricola cluster over other Elizabethkingia species among UK cystic fibrosis patients. Diagn Microbiol Infect Dis 90:109–114. doi: 10.1016/j.diagmicrobio.2017.10.014. [DOI] [PubMed] [Google Scholar]
- 12.Frost F, Nazareth D. 2018. Case report: first report of Elizabethkingia miricola infection in a patient with cystic fibrosis. F1000Res 7:440. doi: 10.12688/f1000research.14441.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Navon L, Clegg WJ, Morgan J, Austin C, McQuiston JR, Blaney DD, Walters MS, Moulton-Meissner H, Nicholson A. 2016. Notes from the field: investigation of Elizabethkingia anophelis cluster—Illinois, 2014-2016. MMWR Morb Mortal Wkly Rep 65:1380–1381. doi: 10.15585/mmwr.mm6548a6. [DOI] [PubMed] [Google Scholar]
- 14.Elbadawi LI, Borlaug G, Gundlach K, Monson T, Noble-Wang J, Moulton-Meissner H, Ansari U, Yoder JS, Wise M, McQuiston JR, Kallen A, Davis JP, Walters MS. 2016. A large and primarily community associated outbreak of Elizabethkingia anophelis infections, Wisconsin, 2015–2016. Open Forum Infect Dis 3:LB-9-LB-9. [Google Scholar]
- 15.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. 2004. GenBank. Nucleic Acids Res 33:D34–D38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finn RD, Clements J, Eddy SR. 2011. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res 39:W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cock PJ, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B, de Hoon MJ. 2009. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25:1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 20.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boratyn G, Camacho C, Federhen S, Merezhuk Y, Madden T, Schoch C, Zaretskaya I. 2014. MOLE-BLAST a new tool to search and classify multiple sequences. NCBI, Bethesda, MD: https://blast.ncbi.nlm.nih.gov/blast/moleblast/moleblast.cgi. [Google Scholar]
- 22.Gulvik CA, Effler TC, Wilhelm SW, Buchan A. 2012. De-MetaST-BLAST: a tool for the validation of degenerate primer sets and data mining of publicly available metagenomes. PLoS One 7:e50362. doi: 10.1371/journal.pone.0050362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen CY, Nace GW, Irwin PL. 2003. A 6 × 6 drop plate method for simultaneous colony counting and MPN enumeration of Campylobacter jejuni, Listeria monocytogenes, and Escherichia coli. J Microbiol Methods 55:475–479. [DOI] [PubMed] [Google Scholar]
- 24.Green SL, Bouley DM, Tolwani RJ, Waggie KS, Lifland BD, Otto GM, Ferrell JE Jr. 1999. Identification and management of an outbreak of Flavobacterium meningosepticum infection in a colony of South African clawed frogs (Xenopus laevis). J Am Vet Med Assoc 214:1833–1838. [PubMed] [Google Scholar]
- 25.Vancanneyt M, Segers P, Hauben L, Hommez J, Devriese LA, Hoste B, Vandamme P, Kersters K. 1994. Flavobacterium meningosepticum, a pathogen in birds. J Clin Microbiol 32:2398–2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bernardet JF, Vancanneyt M, Matte-Tailliez O, Grisez L, Tailliez P, Bizet C, Nowakowski M, Kerouault B, Swings J. 2005. Polyphasic study of Chryseobacterium strains isolated from diseased aquatic animals. Syst Appl Microbiol 28:640–660. doi: 10.1016/j.syapm.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 27.Kyritsi MA, Mouchtouri VA, Pournaras S, Hadjichristodoulou C. 2018. First reported isolation of an emerging opportunistic pathogen (Elizabethkingia anophelis) from hospital water systems in Greece. J Water Health 16:164–170. doi: 10.2166/wh.2017.184. [DOI] [PubMed] [Google Scholar]
- 28.Lau SK, Wu AK, Teng JL, Tse H, Curreem SO, Tsui SK, Huang Y, Chen JH, Lee RA, Yuen KY, Woo PC, 2015. Evidence for Elizabethkingia anophelis transmission from mother to infant, Hong Kong. Emerg Infect Dis 21:232–241. doi: 10.3201/eid2102.140623. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.