Abstract
Widespread linkage disequilibrium and incomplete annotation of cell-to-cell state variation represent substantial challenges to elucidating mechanisms of trait-associated genetic variation. Here, we perform genetic fine-mapping for blood cell traits in the UK Biobank to identify putative causal variants. These variants are enriched in genes encoding for proteins in trait-relevant biological pathways and in accessible chromatin of hematopoietic progenitors. For regulatory variants, we explore patterns of developmental enhancer activity, predict molecular mechanisms, and identify likely target genes. In several instances, we localize multiple independent variants to the same regulatory element or gene. We further observe that variants with pleiotropic effects preferentially act in common progenitor populations to direct the production of distinct lineages. Finally, we leverage fine-mapped variants in conjunction with continuous epigenomic annotations to identify trait-cell type enrichments within closely related populations and in single cells. Our study provides a comprehensive framework for single-variant and single-cell analyses of genetic associations.
Editorial summary:
Fine mapping of blood cell traits in UK Biobank identifies putative causal variants and enrichment of fine-mapped variants in accessible chromatin of hematopoietic progenitor cells. The study provides an analytical framework for single-variant and single-cell analyses of genetic associations.
Hematopoiesis is a paradigm of cellular differentiation that is highly coordinated to ensure balanced proportions of mature blood cells1. Despite our sophisticated understanding gained primarily from model organisms, many aspects of this process remain poorly understood in humans. At the population level, there is substantial variation in commonly measured blood cell traits, such as hemoglobin levels and specific blood cell counts, which can manifest as diseases at extreme ends of the spectrum2. Identifying genetic variants that drive these differences in blood cell traits in human populations may reveal regulatory mechanisms and genes critical for blood cell production and hematologic diseases.
To these ends, genome-wide association studies (GWAS) have identified thousands of genomic loci linked to complex phenotypes including blood cell traits3, but a major challenge has been the identification of causal genetic variants and relevant cell types underlying the observed associations4. In particular, linkage disequilibrium (LD) has confounded the precise identification of functional variants. In an effort to address these issues, several analytical approaches have been developed. The first, termed genetic fine-mapping, attempts to resolve trait-associated loci to likely causal variants by modeling LD structure and the strength of associations. In practice, a major limitation has been the computational burden imposed when allowing for multiple causal variants, and thus methods that assume exactly one causal variant per locus are most commonly used5,6, despite strong evidence that many loci contain multiple independent associations7–10.
The second suite of approaches focus instead on identifying functional tissue enrichments. It has been well established that ~80–90% of associated loci do not tag coding variants and that ~40–80% of the narrow-sense heritability of many complex traits can be resolved to genomic regulatory regions11,12. Given this observation, tissue-specific measurements of regulatory element activity are often overlapped with significant loci (e.g. epigenomic fine-mapping) or with polygenic signal from millions of variants (e.g. partitioned heritability) in order to identify variants and cell types most likely to underlie the measured trait or disease11,13. These enrichment methods have revealed causal tissues for diseases, such as pancreatic islets in diabetes13 and central nervous system cells in schizophrenia11, but are only beginning to be applied to highly related traits and cell types within single systems, such as the hematopoietic hierarchy.
To gain insights into hematopoietic lineage commitment and differentiation, we performed GWASs and genetic fine-mapping for 16 blood cell traits on individuals from the UK Biobank3, identifying multiple likely causal variants in hundreds of individual regions. We comprehensively annotated fine-mapped variants and identified high confidence molecular mechanisms and putative target genes at scale. This allowed us to not only gain insights into patterns of developmental regulation, but to learn about the pleiotropic regulatory processes underlying blood cell production and maintenance. Finally, we describe and validate a novel method (g-chromVAR) to discriminate between closely related cell types in an effort to identify relevant stages of hematopoiesis that are affected by these common genetic variants. Applying g-chromVAR to single hematopoietic cell data revealed substantial heterogeneity of genetic enrichment within classically defined hematopoietic progenitor populations. Thus, we demonstrate that using a well-powered method to identify cell populations that are trait-relevant provides a critical step towards broadly deciphering causal mechanisms underlying phenotypic variation.
Results
Fine-mapping pinpoints hundreds of likely causal variants.
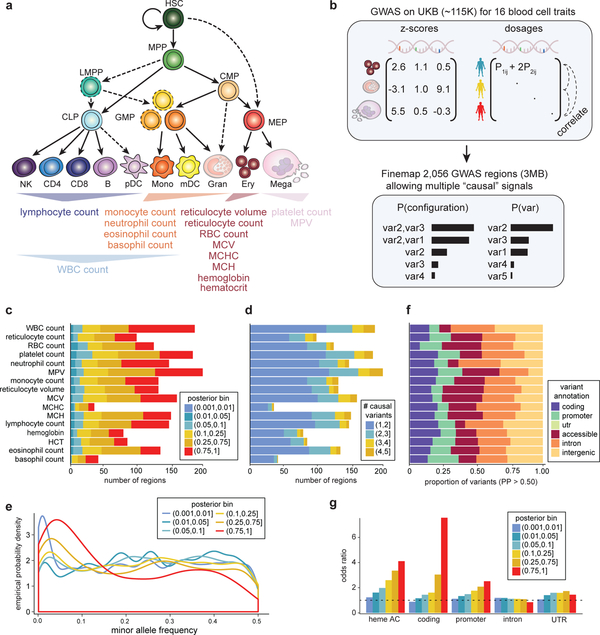
We performed GWASs on ~115,000 individuals from the UK Biobank (UKB) for 16 blood cell traits representing 7 distinct hematopoietic lineages (erythroid, platelet, lymphocyte, monocyte, and granulocyte (neutrophil, eosinophil, and basophil)) (Fig. 1a). Similar to previous reports, these traits were highly heritable, with common genetic variants explaining an average of 15.4% of narrow-sense heritability ()14 (Supplementary Fig. 1). Traits from the same lineage, such as red blood cell (RBC) count and hemoglobin (rg = 0.89, P = 7.1 × 10−25), typically had high genetic correlations, whereas traits from distinct lineages had low genetic correlations with some exceptions, such as platelet count and lymphocyte count (rg = 0.26, P = 3.8 × 10−18) (Supplementary Fig. 1). This suggests that the genetic regulation of blood production could potentially occur across various stages of hematopoiesis.
Figure 1 |. Overview of hematopoiesis, UKB GWAS, and fine-mapping.
(a) Schematic of the human hematopoietic hierarchy showing the primary cell types analyzed in this work. Colors used in this schematic are consistent throughout all figures. Mono, monocyte; gran, granulocyte; ery, erythroid; mega, megakaryocyte; CD4, CD4+ T cell; CD8, CD8+ T cell; B, B cell; NK, natural killer cell; mDC, myeloid dendritic cell; pDC, plasmacytoid dendritic cell. The 16 blood traits that were genetically fine-mapped are shown below the hierarchy. (b) Schematic of UKB GWAS and fine-mapping approach. Briefly, blood traits from ~115K individuals were fine-mapped allowing for multiple causal variants and using imputed genotype dosages as reference LD. (c) Number of fine-mapped regions for each trait with the highest posterior probability for a variant being causal indicated. (d) Breakdown of the number of causal variants (min = 1, max = 5) for all regions in each trait. (e) Empirical distribution of the minor allele frequency of variants in each posterior bin. (f) Proportion of fine-mapped variants within intronic, promoter, coding, UTR, and intergenic regions. (g) Local-shifting enrichments of fine-mapped variants across all traits for varying posterior probability bins.
To begin to dissect the nature and stage-specificity of these genetic effects, we performed genetic fine mapping to identify high confidence variants across 2,056 3-Mb regions containing a genome-wide significant association. Traditional fine-mapping approaches assume only one causal variant per locus and are either agnostic to LD or use small reference panels, which are inaccurate when scaled to large sample sizes15. To overcome these limitations, we calculated LD directly from the imputed genotype probabilities (dosages) for individuals in our GWASs, rather than from a hard-called reference panel (Fig. 1b).
Across all common variants (MAF > 0.1%, INFO16 > 0.6) in 2,056 regions, our method identified 38,654 variants with > 1% posterior probability (PP) of being causal for a trait association, comprising a significant proportion of narrow-sense heritability explained by all common variants (trait average of 24.9% of total for PP > 0.01) (Supplementary Fig. 1 and Supplementary Table 1). 993 regions (48%) contained at least one variant with PP > 0.50 (Fig. 1c), providing strong evidence that our approach was successful in pinpointing causal variants. The posterior expected number of independent causal variants was > 2 for 35% of regions and > 3 for 13% of regions (Fig. 1d). Given their increased complexity, regions with a greater expected number of causal variants had lower top configuration PPs (Supplementary Fig. 2 and Supplementary Table 2). The majority of variants (74%) with PP > 0.75 had MAF > 5% (Fig. 1e), consistent with the known polygenic nature of blood cell traits3. Fine-mapped variants had potentially diverse mechanisms, ranging from putative regulatory variants in accessible chromatin (AC) to coding variants, including 164 unique missense variants and 6 loss of function variants with PP > 0.10 (Fig. 1f, Supplementary Fig. 3 and Supplementary Table 3).
To validate our approach, we investigated the overlap of fine-mapped variants (binned by PP) with several annotations previously shown to be enriched for GWAS signals (Fig. 1g)11,12. To generate a null distribution, we locally shifted annotations within a 3-Mb window, similar to the method implemented in GoShifter17. We observed minimal enrichment for intronic and untranslated regions of genes, but found strong, focal, and stepwise enrichments across higher PP bins for hematopoietic AC, promoters, and coding regions (OR = 4.2, 2.9, and 8.5 for PP > 0.75, respectively) (Fig. 1f)11,12,17. Notably, strong enrichments persisted even after we excluded all variants with high correlation (R2 > 0.8) to the sentinel variants at each locus (Supplementary Fig. 3).
Dissecting mechanisms of core gene regulation in hematopoiesis.
We next sought to delineate the precise mechanisms underlying how fine-mapped genetic variants affect hematopoietic traits. For all 140,739 variants with PP > 0.001 we combined several lines of functional and predictive evidence to better understand the (i) cell populations, (ii) molecular mechanisms, and (iii) target genes involved in blood cell production (Supplementary Fig. 4). First, we identified fine-mapped (PP > 0.10) non-synonymous and loss of function coding variants in 77 RBC, 59 platelet, 20 monocyte, 28 lymphocyte, and 46 granulocyte (neutrophil, basophil, and eosinophil) trait genes (Supplementary Table 3). Within the set of genes identified from RBC trait variants, we identified both validated GWAS genes (SH2B318, TRIM5819) (Supplementary Fig. 5) and several Mendelian disease genes for diverse RBC disorders (HFE, TMPRSS6, PFKM, PKLR, PIEZO1, SPTA1, ANK1, RHD, GYPA, KLF1)20. Genes perturbed by fine-mapped coding variants were enriched for trait-relevant known and novel biological pathways. For example, RBC trait genes were involved in iron homeostasis, platelet trait genes in coagulation and wound healing, lymphocyte trait genes in T cell migration and activation, and monocyte and granulocyte trait genes in cytokine and inflammatory responses (Supplementary Fig. 6 and Supplementary Table 3). Of note, we identified several pathways corresponding to cholesterol and lipid regulation that were enriched in RBC trait genes (Supplementary Fig. 6), suggesting a connection between lipid metabolism and RBCs, which are major stores of cholesterol21.
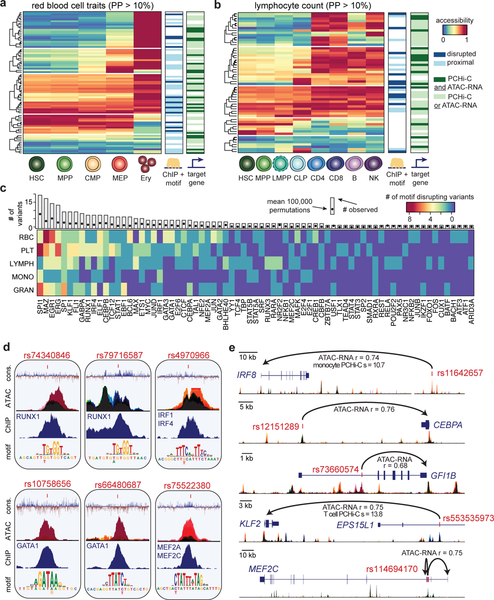
To investigate the exact stages of hematopoietic differentiation during which variants could regulate transcription, we overlapped fine-mapped variants (PP > 0.10) with chromatin accessibility profiles (ATAC-seq) of 18 hematopoietic progenitor, precursor, and differentiated cell populations primarily sorted from the bone marrow or blood of healthy donors (Fig. 1a, Supplementary Fig. 7 and Supplementary Table 4). Across traits representing the five major blood cell lineages, we used k-means clustering to categorize the developmental timing of AC peaks containing fine-mapped variants (Fig. 2A–B, Supplementary Fig. 8). For example, across RBC traits, we identified 80 fine-mapped regulatory variants, of which 26% (21/80) were restricted to erythroid progenitors, 18% (14/80) were restricted to megakaryocyte-erythroid progenitors (MEPs) and erythroid progenitors, and 29% (23/80) could regulate transcription across the entire erythroid lineage from hematopoietic stem cells (HSCs) to erythroid progenitors, whereas 14% (11/80) could only act in other hematopoietic lineages (Fig. 2a). In some cases, we identified small clusters of variants that followed slightly different regulatory programs, such as variants that could only regulate transcription in upstream multipotent progenitors and lymphocyte variants that could regulate transcription in T cell, but not B cell, subsets (Fig. 2a,b and Supplementary Fig. 8).
Figure 2 |. Mechanisms of core gene regulation in blood production.
(a,b) Heatmaps depicting red blood cell trait-associated variants (PP > 0.10) across the erythroid lineage (a) and lymphocyte count-associated variants (PP > 0.10) across the lymphoid lineage (b), clustered by chromatin accessibility. Each row marks a fine-mapped variant, each column denotes a cell type within the relevant lineage, and color denotes relative chromatin accessibility along the lineage at each variant (blue, least accessible chromatin; red, most accessible chromatin). Putative target genes (predicted by ATAC-RNA correlation and/or PCHi-C) and disrupted TFs (predicted by ChIP-seq occupancy and motif disruption) are indicated to the right. (c) Transcription factor motifs disrupted in lineage-specific hematopoietic traits. Each row represents a set of traits where variants disrupt specified TF motifs and are occupied by that TF in hematopoietic cells. The unique margin sums across each lineage are shown in the bar plot for each TF. The expected number of variants with ChIP + motif disruption across all PPs is estimated using 100,000 permutations and is shown as a single point. (d) Examples of molecular mechanisms from the analysis in c reveals putative causal variants that disrupt cis-binding of hematopoietic TFs known to be involved in regulating hematopoiesis for various blood cell traits: rs10758656 and rs66480687 are associated with red blood cell traits; rs75522380 and rs74340846 are associated with platelet traits; rs4970966 is associated with monocyte count; and rs79716587 is associated with lymphocyte count. Black color represents accessibility throughout hematopoiesis, whereas other stacked colors represent accessibility for the cell types shown in Figure 3d. (e) Examples of putative target genes from the analysis in a and b: rs11642657 and rs12151289 are associated with monocyte count; rs73660574 is associated with red blood cell traits; rs553535973 is associated with lymphocyte count; and rs114694170 is associated with platelet traits. Colors for accessible chromatin are the same as in d.
Next, we investigated the molecular mechanisms underlying fine-mapped regulatory variants. To nominate a high confidence molecular mechanism, we required that a variant (i) disrupt one of 426 motifs corresponding to known binding preferences of human TFs22 and (ii) show occupancy by that specific TF in a relevant hematopoietic primary tissue or cell line, based on 2,115 uniformly processed ChIP-seq profiles23. In total, we identified one or more such mechanisms for 145 distinct fine-mapped non-coding variants (Fig. 2c). Specifically, we identified 13 RBC, 28 platelet, 8 monocyte, 11 lymphocyte, and 18 granulocyte high confidence molecular mechanisms for variants also in primary hematopoietic AC (Fig. 2a,b, Supplementary Fig. 8 and Supplementary Table 5). These variants most commonly disrupted the binding sites of key transcriptional regulators of hematopoietic lineage commitment and differentiation (FDR < 10% for 33 TFs). For example, we observed 7 PU.1 (SPI1)24,25, 6 ERG26–28, 4 FLI128,29, 3 IRF430, and 3 RUNX131,32 binding site disrupting variants associated with platelet traits (Fig. 2c,d), in addition to many other compelling lineage-specific regulatory mechanisms for experimental follow-up (Supplementary Fig. 8 and Supplementary Note).
Finally, in order to identify high confidence target genes of fine-mapped regulatory variants, we built hematopoietic-specific enhancer-promoter maps using (i) measurements of physical DNA interactions in 15 primary hematopoietic cell populations from promoter capture Hi-C (PCHi-C)33 and (ii) the correlation between chromatin accessibility and cis gene expression across 16 primary hematopoietic populations34,35. Altogether, we identified one or more experimentally supported target genes for 415 variant-trait associations, providing testable biological hypotheses for 79% of fine-mapped regulatory variants (Fig. 2a,b, Supplementary Figs. 5 and 8, and Supplementary Tables 6 and 7). Interestingly, a number of variants were predicted to disrupt the transcription of hematopoietic TFs (Fig. 2d,e and Supplementary Fig. 8). For example, IRF8 and CEBPA, two essential TFs involved in monocyte differentiation36,37, are targets of fine-mapped monocyte count associated variants that fall within monocyte precursor AC (Fig. 2e). Similarly, we determined that GFI1B, KLF2, and MEF2C are targets of fine-mapped variants in progenitor-specific AC for mean reticulocyte volume, lymphocyte count, and platelet count, respectively (Fig. 2e). Overall, this functional analysis will likely facilitate experimental investigation into how common genetic variants regulate hematopoietic lineage commitment and differentiation.
Regions with multiple causal variants.
We next conducted a closer examination of the 785 trait-associated regions with multiple independent causal signals. Amongst proximal pairs of variants in which both variants had PP > 0.50, the majority were > 10 kb apart (76%), although 7 pairs were within fewer than 100 bp (Supplementary Fig. 9 and Supplementary Table 8). Across all pairs, 42% of the variants were of the same class (e.g. coding-coding), and pairs of variants in AC but in different regulatory regions within 1 Mb were typically lineage-specific (Supplementary Fig. 9). Examples of coding-coding pairs include hemoglobin-associated rs1800730 and rs1799945 (PP > 0.66; 4 bp apart) in HFE, the classic gene mutated in hereditary hemochromatosis, WBC count-associated rs146125856 and rs148783236 (PP > 0.98; 24 bp apart) in USP8, which encodes an immune-specific ubiquitin ligase and is mutated in Cushing’s disease38,39, and MPV-associated rs41303899 and rs415064 (PP > 0.76; 835 bp apart) in TUBB1, which encodes a β-tubulin protein important for pro-platelet formation that is mutated in monogenic forms of macrothrombocytopenia40.
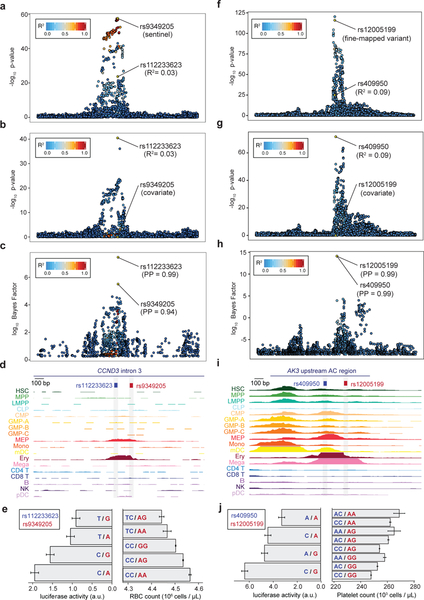
Although there were several other interesting pairs of variants in AC (see Supplementary Note and Supplementary Fig. 10), we specifically investigated the RBC count association at the CCND3 locus, in which we previously identified a causal variant and its target gene41. At this locus, our current approach correctly identified the known causal variant (rs9349205) as the primary association, as well as ~4 additional independent signals, including a secondary imputed variant (rs112233623) associated with decreased RBC count (Fig. 3a–c). Stepwise conditional analysis further validated these findings (Fig. 3b). Notably, these variants were missed by fine-mapping if we instead used LD estimated from either the UK10K whole genome sequencing (WGS) reference panel or hard-called variants from the UKB population (Supplementary Fig. 11), highlighting the importance of calculating LD using imputed genotype dosages from the GWAS population. Remarkably, rs112233623 is only 161 bp from rs9349205, and both lie within erythroid-specific AC (Fig. 3d). Luciferase reporter assays showed that each variant affected enhancer activity independently with minor allele effects in opposing directions, consistent with the genetic directionality (Fig. 3e). At a separate locus associated with platelet traits, we similarly observed a large number of independent signals (~8), allowing us to identify a variant pair (rs49950 and rs12005199; PP > 0.99; 123 bp apart) within a single AC region ~20 kb upstream of AK3, a gene whose zebrafish homolog is essential for platelet (thrombocyte) formation (Fig. 3f–i)42. Importantly, we again observed that each variant significantly affected enhancer activity additively and in concordance with population phenotypes (Fig. 3j).
Figure 3 |. Characterization and validation of CCND3 and AK3 regions with multiple causal variants.
(a,b) Regional association plots (n = 116,667 individuals; BOLT-LMM P-values) for RBC count in the CCND3 locus from the initial GWAS (a) and after conditioning on the sentinel variant rs9349205 (b). (c,d) Fine-mapping identifies two putative causal variants (rs9349205, PP = 0.94; rs112233623, PP = 0.99) located 161 bp apart (c), both of which lie within the same erythroid-specific accessible chromatin (AC) (d). (e) Luciferase reporter assays for four haplotypes (left) corroborate independent additive effects of rs9349205 (red; P = 1.78 × 10−3) and rs112233623 (blue; P = 2.86 × 10−6) on RBC count (right). (f,g) Regional association plots (n = 116,666 individuals, BOLT-LMM P-values) for platelet count in the AK3 locus from the initial GWAS (f) and after conditioning on sentinel variant rs12005199 (g). (h,i) Fine-mapping identifies two putative causal variants (rs12005199, PP = 0.99; rs409950, PP = 0.99) 123 bp apart (h), both located within a strong megakaryocyte AC region (i). (j) Luciferase reporter assays (n = 9 biological replicates) for four haplotypes (left) corroborate independent additive effects of rs12005199 (red; two-sided Wald test P = 5.19 × 10−4) and rs409950 (blue; two-sided Wald test P = 3.57 × 10−5) on platelet count (right). Mean and standard error are indicated for both phenotype and regulatory activity.
Mechanisms of pleiotropic variants across distinct blood cell lineages.
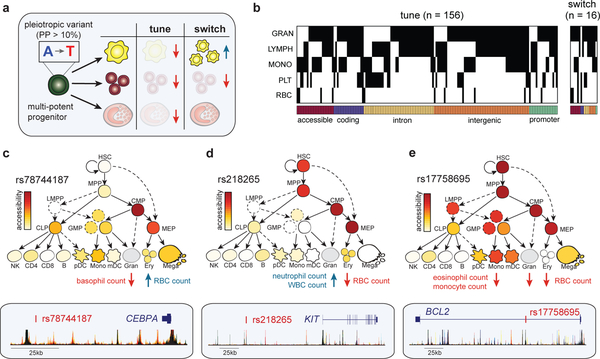
We next sought to examine the effects of variants associated with two or more of the seven distinct blood cell types for which phenotypes were available in the UK Biobank. We hypothesized that these pleiotropic variants could either (i) tune overall blood production by simultaneously increasing or decreasing the levels of terminal blood cells across multiple lineages or (ii) switch blood cell production such that one lineage is favored at the expense of others (Fig. 4a).
Figure 4 |. Dissecting mechanisms of pleiotropic variants across multiple blood cell lineages.
(a) Schematic that illustrates fine-mapped variants acting in multi-potential or heterogeneous progenitors on distinct hematopoietic lineages, either by tuning lineages in the same direction or switching the regulation in opposite directions. (b) A heatmap depicting 172 fine-mapped variants (PP > 0.10) with pleiotropic effects on cell counts in two or more hematopoietic lineages (eosinophil, neutrophil, basophil, lymphocyte, monocyte, platelet, RBC). Effects on eosinophil, neutrophil, and basophil counts are visualized together as a singular granulocyte lineage. Genomic annotations are indicated below each variant. (c) Pleiotropic variant rs78744187, located downstream of CEBPA, has high chromatin accessibility in CMP and MEP progenitors (top) and demonstrates a switch mechanism by downregulating basophil count while upregulating RBC count (bottom). (d) rs218265, located upstream of stem cell factor KIT, has high chromatin accessibility in several early progenitors (HSC, MPP, CMP, MEP) and demonstrates a switch mechanism by upregulating neutrophil and WBC count while downregulating RBC count. (e) rs17758695, located within an intron of anti-apoptotic factor BCL2, has high chromatin accessibility in several early progenitors (HSC, MPP, CMP, MEP) and exhibits a tuning mechanism, simultaneously downregulating eosinophil, monocyte, and RBC counts.
We restricted our analyses to quantified blood cell counts for interpretability and identified 172 pleiotropic variants that co-localized43 (PP > 0.10) across two or more traits (Fig. 4b–d, Supplementary Fig. 12, and Supplementary Table 9). Surprisingly, 91% (156/172) of these variants exhibited a tuning mechanism, modifying two or more lineages in the same direction, whereas the remaining 9% (16/172) favored one lineage at the expense of other lineages (P = 5.08 × 10−30; Binomial test). Regardless of direction, 88% of all pleiotropic variants were non-coding, and those in regions of AC had 60% more ATAC-seq reads in progenitors than terminal cell types (mean 4.01 vs. 2.44 counts per million; P = 0.025; Student’s t-test), consistent with the hypothesis that many of these variants act in common progenitor cell populations44,45.
One example of a variant exhibiting a switch mechanism is rs78744187 (PP = 0.99 and 0.99), which increases RBC count, while concomitantly decreasing basophil count (Fig. 4c). rs78744187 is located in an enhancer specific for CMPs, which encompasses a heterogeneous population containing progenitors for both basophils and RBCs, approximately 36 kb downstream of CEBPA, which encodes for a key myeloid TF46. We previously reported the association between rs78744187 and basophil count, but not RBC count, and showed that this variant switches the production of the closely related basophil and mast cell lineages45. A second switch variant, rs218265 (PP = 0.99 and 0.64), located within a gene desert 1.15 Mb upstream of KIT, increases neutrophil count but decreases RBC count. KIT encodes the receptor protein for stem cell factor, a growth stimulating cytokine involved in hematopoietic progenitor cell proliferation47. rs218265 falls within a region of AC that is exclusively open in multipotential and heterogenous populations (Fig. 4d), consistent with a role for this enhancer variant in regulating KIT expression in common upstream progenitors of neutrophils and RBCs. Taken together, our results suggest that tuning the dosage of key regulatory genes in upstream progenitors may switch the production of one lineage in favor of another during the early stages of lineage commitment.
As an example of a pleiotropic variant exhibiting the predominant tune mechanism, we found that rs17758695 (PP = 0.99, 0.99, and 0.99) is associated with decreases in eosinophil, monocyte, and RBC count (Fig. 4e). This variant is located within a progenitor-specific region of AC in the intron of BCL2, an anti-apoptotic protein known to regulate hematopoietic differentiation48. This is consistent with the idea that regulating a general cell death protein such as BCL2 in a common multipotential progenitor would tune the production of multiple cell types, in contrast to the switch variants proximal to key regulators of hematopoietic differentiation. An additional tune variant is the missense variant rs12459419 (PP = 0.30, 0.28, and 0.11) in the CD33 gene, which is associated with decreases in eosinophil, monocyte, and platelet counts. CD33 is broadly expressed in hematopoietic progenitors and is a surface marker of myeloid differentiation49 (Supplementary Fig. 12). In summary, our analyses support a prominent role for pleiotropy in hematopoietic differentiation, whereby individual variants can act in upstream progenitors to simultaneously tune or switch production and maintenance of multiple lineages.
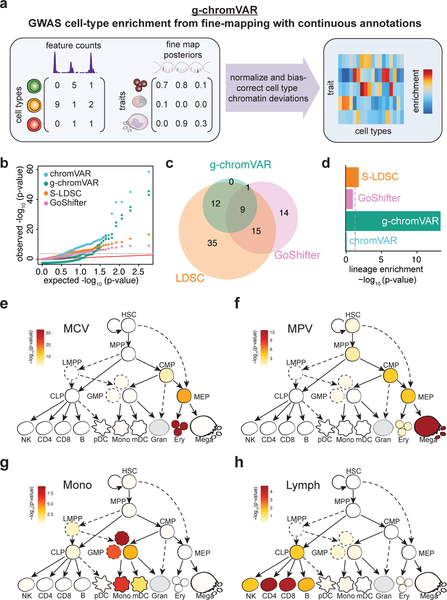
g-chromVAR, a novel method to measure fine-mapped GWAS enrichment amongst closely related tissues.
We next shifted our focus in the reciprocal direction – using fine-mapping to determine the exact stages of human hematopoiesis at which regulatory genetic variation underlying each blood cell trait is most likely acting. Although recent methods11,17 have been developed to calculate enrichment of genetic variation with genomic annotations, a method which accounts for both (i) the strength and specificity of the genomic annotation and (ii) the probability of variant causality, accounting for LD structure, is needed to resolve associations within the closely related, stepwise hierarchies that define hematopoiesis. To these ends, we developed a new approach called genetic-chromVAR (g-chromVAR), a generalization of the recently described chromVAR method,50 to measure the enrichment of regulatory variants in each cell state using fine-mapped variant PPs and quantitative measurements of regulatory activity (Fig. 5a; see Supplementary Note and Online Methods for details). We show that g-chromVAR is generally robust to variant PP thresholds and numbers of background peaks (Supplementary Fig. 13), captures true enrichments in a simulated setting (Supplementary Fig. 14), is robust to the choice of fine-mapping method (Supplementary Table 10), and can identify novel enrichments in large epigenomic datasets (Supplementary Table 11; see Supplementary Note for details).
Figure 5 |. Overview of g-chromVAR and application to hematopoietic cell types.
(a) Schematic showing inputs for continuous epigenomic data for each cell type and a matrix of fine-mapped variant posterior probabilities for GWAS traits. (b-d) Results from the application of g-chromVAR and three similar methods to 16 blood cell traits for 18 hematopoietic cell types. (b) Quantile-quantile representation of the P-values from each method. (c) Overlap between methods for Bonferroni-corrected trait enrichments. (d) Lineage enrichment of all trait-pairs (n = 288 pairs) for each method. A two-tailed Mann-Whitney rank-sum test was used to evaluate the relative enrichment of lineage-specific trait-cell type pairs (true positives). (e-h) Enrichments for four representative traits using g-chromVAR: mean corpuscular volume (e); mean platelet volume (f); monocyte count (g); lymphocyte count (h).
In order to validate g-chromVAR in a realistic setting, we used it along with seven other methods to estimate the enrichment of each of the 16 blood cell traits within the accessible chromatin of 18 hematopoietic progenitor and terminal cell populations (Figs. 1a and 5c, Supplementary Figs. 15 and 16, Supplementary Table 4)34,35. To compare g-chromVAR’s performance to other state-of-the-art enrichment tools, we leveraged our knowledge of the hematopoietic system and devised a lineage specificity test (see Supplementary Note), which is a nonparametric rank-sum test that compares the relative ranking of lineage specific and non-lineage specific enrichments for each of the compared methodologies. We found that g-chromVAR was the most specific of all tested methods, while still retaining sufficient power to identify 22 trait-cell type associations (Fig. 5d, and Supplementary Figs. 13a and16).
Having validated our approach, we investigated cell type enrichments for each of the 16 traits. We found that the most lineage-restricted or terminal populations were typically most strongly enriched for a corresponding trait association (Fig. 5e–h). For example, RBC count was most strongly enriched in erythroid precursors (Fig. 5e), and lymphocyte count was most strongly enriched in CD4+ and CD8+ T cells (Fig. 5h). In several instances, we observed significant enrichments for traits in earlier progenitor cells within each lineage, including enrichment for platelet traits in CMPs and enrichment for monocyte traits in a specific subpopulation of GMPs (Supplementary Fig. 13a). We sought to investigate these progenitor enrichments further at the single cell level.
GWAS enrichment in single-cell chromatin accessibility data.
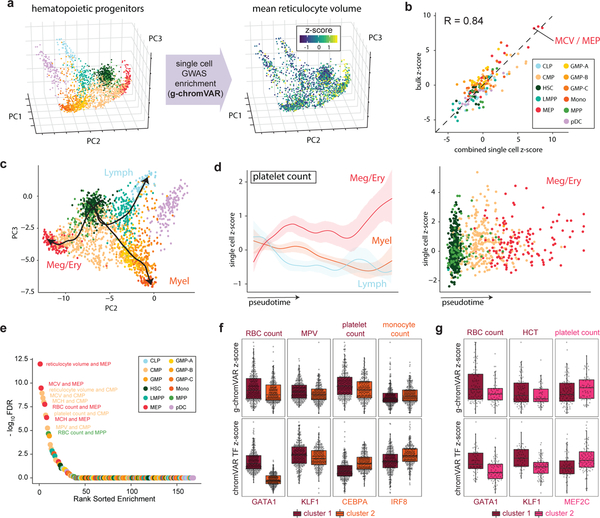
Although the strongest g-chromVAR enrichments for blood traits were in the most lineage restricted precursors, we reasoned that investigating progenitor populations that did have robust enrichment signals, such as CMPs and MEPs, could inform principles of the genetic regulation of terminal blood cell production51–54. To these ends, we scored 2,034 single bone-marrow derived hematopoietic stem and progenitor cells34 for GWAS enrichment using g-chromVAR (Fig. 6a). Single-cell composite and bulk cell type enrichments were highly correlated (r = 0.84) (Fig. 6b), and enrichments along inferred pseudotime trajectories of cellular differentiation mirrored our observations from bulk data, albeit with finer granularity (Fig. 6c,d). These results suggest that g-chromVAR is able to recover known biology from sparse single cell (sc-)ATAC-seq profiles.
Figure 6 |. Application of g-chromVAR to single-cell chromatin accessibility data.
(a) 2,034 hematopoietic single cells projected onto a three-dimensional principal components embedding. Single cells colored by g-chromVAR enrichment scores for mean reticulocyte volume reveal specific regulatory enrichment in the MEP population. (b) Validation of g-chromVAR enrichments using synthetic bulk populations from sums of single cells (n = 2,034 cells). Aggregated single-cell g-chromVAR z-scores across all trait-cell type pairs (individual points) strongly correlate (Pearson r = 0.84) with bulk population z-scores. (c) Inferred pseudotime trajectories of three hematopoietic lineages from scATAC-seq data. (d) Pseudotime trends (mean and 95% CIs) of g-chromVAR scores for platelet count across all single cells (n = 2,034 cells) corroborates regulatory dynamics of megakaryocyte/erythroid differentiation. (e) Rank order plot highlighting the trait-cell type pairs with the greatest variance over that of a χ2 distribution. (f) K-medoids partitioning of ATAC-seq counts in CMP cells (n = 502 cells) reveals two subpopulations: one that is enriched for monocyte genetic variants and one that is enriched for megakaryocyte/erythroid variants (RBC count, FDR = 1.28 × 10−4; MPV, FDR = 2.36 × 10−4; platelet count, FDR = 1.40 × 10−5; monocyte count, FDR = 2.21 × 10−2). ChromVAR scores for master transcription factors (TFs) of each blood cell type support biological hypotheses for genetic enrichments (GATA1, FDR = 1.76 × 10−82; KLF1, FDR = 4.33 × 10−3; CEBPA, FDR = 2.58 × 10−16; IRF8, FDR = 4.65 × 10−15). Two-tailed t-tests were used for each comparison; boxplots represent median and interquartile range. (g) Similar k-medoids partitioning of MEP cells (n = 138 cells) reveals two subpopulations with differential enrichments for megakaryocyte or erythroid associated genetic variants (RBC count, FDR = 0.155; HCT, FDR = 3.98 × 10−2; platelet count, FDR = 7.65 × 10−2), along with consistent differences in chromVAR TF-deviation scores for master TFs of each blood cell type (GATA1, FDR = 2.18 × 10−4; KLF1, FDR = 4.02 × 10−6; MEF2C, FDR = 2.52 × 10−3).
To explore potential heterogeneity within each of the 11 hematopoietic progenitor populations, we estimated the variation in regulatory genetic enrichments for each trait within populations. We found that classically defined CMP (n = 502 cells) and MEP (n = 138 cells) populations exhibited significant heterogeneity in g-chromVAR enrichments for both erythroid and megakaryocyte traits (Fig. 6e). We thus hypothesized that the CMP population could be subdivided into megakaryocyte/erythrocyte-primed and monocyte-primed subtypes, whereas the MEP population could be further subdivided into erythrocyte-primed and megakaryocyte-primed subtypes. To test this hypothesis, we performed unsupervised clustering on chromatin accessibility profiles for the CMP and MEP populations (Supplementary Fig. 17) and found that the (GWAS-naïve) subpopulations were indeed differentially enriched for the specific GWAS traits. In agreement with these genetic enrichments, we observed differential chromatin accessibility of motifs for lineage-specific master TFs between the subpopulations that corresponded to the trait enrichments, such as increased chromatin accessibility of GATA1 motifs within the clusters enriched for erythroid traits (Fig. 6f,g and Supplementary Table 12). Additional studies are needed to determine whether these differences are due to distinct lineage-biased subpopulations or whether they reflect gradations along a common axis of differentiation. Regardless, our findings demonstrate that genetic variation acts heterogeneously within classically defined progenitor populations.
Discussion
Two outstanding challenges in the post-GWAS era are (i) the precise identification of causal variants within associated loci and (ii) determination of the exact mechanisms by which these variants result in the observed phenotypes. To address (i), we used robust genetic fine-mapping to identify hundreds of putative causal variants for 16 blood cell traits, allowing for up to 5 causal variants in each locus. At PP > 0.10, we identified 240 fine-mapped coding variants as well as 647 regulatory variants in AC in at least one of 18 primary hematopoietic populations. Several compelling anecdotes, including a number of instances in which the activity of a single regulatory element is modulated by multiple functional variants, highlight the advantages of allowing for multiple causal variants when fine-mapping.
To address (ii), we compiled and derived functional annotations to nominate regulatory mechanisms and identify putative target genes. Overall, our comprehensive approach identified a high confidence regulatory mechanism for 145 variants and an experimentally supported target gene for 79% of variants in AC for distinct lineages. Our investigations into these fine-mapped pleiotropic variants revealed that ~90% of these variants act to tune total hematopoietic production, whereas the remaining ~10% favored production of one lineage at the expense of another (switch). To further improve causal cell type identification, we developed a novel enrichment method (g-chromVAR) that can discriminate between closely related cell types and applied it to directly probe the regulatory dynamics of hematopoiesis within classically defined progenitors in bulk and at the single cell level. Our “top loci” method is complementary to enrichment methods that investigate polygenic signals, such as S-LDSC.
Overall, our integrated approach is designed to sequentially identify causal genetic variants, their molecular mechanisms, their target genes, and the cell types in which they act. We expect that better-powered fine-mapping studies, more numerous and higher quality bulk and single-cell epigenomic datasets, and improved computational tools will extend the inferences discussed herein. Altogether, our study represents a paradigm for the comprehensive mapping of variant to function, which can be applied broadly to gain insights into the specific functions of variants associated with a range of human traits and diseases.
URLs.
A UCSC Genome Browser visualization hub for all bulk ATAC data is available with this hub URL: https://s3.amazonaws.com/atachematopoesis/hub.txt. The web app to visualize putative causal variants and corresponding annotations is available at http://molpath.shinyapps.io/ShinyHeme. Functional genomic annotations are available here: https://github.com/caleblareau/singlecell_bloodtraits/tree/master/data/annotations.
Online Methods
Genome-wide association studies.
Genome-wide association studies were carried out for 16 different blood cell indices in 114,910–116,667 “white British” individuals from UK Biobank. Imputation was performed using the combined 1000 Genomes Phase 3-UK10K panel (http://biobank.ctsu.ox.ac.uk/crystal/refer.cgi?id=157020). To account for population substructure in blood cell traits, we regressed each phenotype against the first 10 principal components of genetic ancestry, age, and sex. We then inverse normalized the residuals, which were used as the phenotype measurements for the genetic association tests. Specifically, we regressed each phenotype measurement against the probabilistic imputed allele dosage using a linear mixed model approach as implemented in BOLT-LMM v2.255. Genome-wide significance was defined as P < 5 × 10−8.
Linkage disequilibrium score regression.
We used LD score regression (LDSC) to compute the narrow-sense heritability estimates and genetic correlations of the 16 blood cell traits in the UKB. Reference LD scores were computed with a subset of unrelated European individuals from the UK10K cohort. To remove genetically related individuals, we first used PLINK to construct a filtered list of variants with MAF > 0.10 and no pair of variants with R2 > 0.10. These LD and MAF-pruned variants were then used to calculate an identity-by-descent (IBD) matrix, and one individual in each pair of samples with proportion IBD () > 0.125 were removed to produce a final subset of 3,677 unrelated individuals to serve as the reference panel for LDSC. After applying the recommended variant filtering, z-scores for an average of 6,655,000 variants per trait were used as input to LDSC. For heritability estimates for variants identified by fine-mapping or by linkage to the sentinel, we note that these estimates may be either over-estimated or under-estimated from the reported values as previously noted56.
Fine-mapping.
Sentinel association regions were constructed as follows: first, all variants were ranked by decreasing χ2statistics. Next, we derived 3-Mb regions centered at the top variant; each region is ~3 cMs, so all relevant LD structure should be fully captured for nearly every region (Yu et al.57 reported that 95% of region recombination rates fall within 3 Mb). This process was repeated for each top association variant that did not overlap any 3-Mb regions created thus far until there were no genome-wide significant variants remaining in undefined regions. Within each region, we identified all imputed variants with MAF > 0.1% and imputation quality (INFO) > 0.6 and extracted z-scores from the summary statistics for each. We next derived dosage LD matrices for each region using LDstore15 on the genotype probability files (.bgen) used for the association studies. To be exact, we computed LD matrices from 120,086 individuals who had a phenotype for at least one of the 16 blood cell traits.
Fine-mapping was performed on genome-wide significant GWAS regions using FINEMAP v1.1 software with the z-score and LD matrices as input16. The output from FINEMAP is (i) a list of potential causal configurations together with their posterior probabilities and Bayes Factors, (ii) the posterior probability marginalized over the causal configurations that individual variants are causal, and (iii) the posterior probabilities that there are a specific number (between 1 and 5) of statistically independent associations in each region. Default FINEMAP settings were used and all variants with posterior probabilities > 0.1% were retained for downstream analyses. For the CCND3 and AK3 regions in which follow-up luciferase reporters were performed, we reran FINEMAP allowing for up to 10 causal variants, confirming ~4 independent effects in the CCND3 locus (60.6% posterior probability) but revealing ~8 independent effects for the AK3 locus (59.9% posterior probability).
To confirm select regions with multiple putative causal variants, we performed conditional analysis using BOLT-LMM by first conditioning on the variant with the lowest P-value in the region and then stepwise adding to the model the variant with the lowest conditional P-value until no additional variant reached the genome-wide significance threshold of 5 × 10−8 in the combined model.
Local annotation shifting.
We implemented a slightly modified version of GoShifter to calculate the enrichment between fine-mapped variants with PP > 0.01 for every trait and 5 different genomic annotations (see Supplementary Note for details). To obtain the annotation for hematopoietic AC, we used the consensus peak set for all blood cell types, performed row and column quantile normalization on the counts matrix, and kept only peaks that had a maximum count in the top 80% for at least one of the 18 cell types. The coding, intron, promoter, and 5’ untranslated region annotations were obtained from the UCSC Genome Browser as previously processed (see URLs)12.
Variant classification and annotation.
To partition fine-mapped variants into bins of non-overlapping annotations (Figs. 1f,g and 4b), we overlapped variant positions with genomic intervals and then classified each variant based on the following hierarchy: (i) coding; (ii) promoters; (iii) UTRs; (iv) hematopoietic chromatin accessible; (v) intronic; and (vi) intergenic. For example, for a variant falling in an AC region that is an annotated promoter, this variant was assigned to the “promoter” class. Variant effect predictor (VEP) was used to further annotate the functions of coding variants58.
To define pleiotropic variants and relative effect directions, we considered a subset of 7 of the 16 total traits that were defined “count” traits for distinct cell types: basophil, eosinophil, neutrophil, platelet, red blood cell, monocyte, and lymphoid count were the traits used for their respective lineages. Note that basophil, eosinophil, and neutrophil count were represented together as granulocytes for visualization purposes (Fig. 4b), but still considered as distinct cell types. “Tune” variants were defined as those that exhibited the same direction of effect for the minor allele across all lineages. Conversely, “switch” variants were designated when the minor allele had differing effect directions for two or more lineages.
Gene set enrichment analysis.
Gene set enrichments of fine-mapped coding variants with PP > 0.10 were calculated using Functional Mapping and Annotation of Genome-Wide Association Studies (FUMA)59, using all protein-coding genes as the background model and requiring a minimum overlap of two genes and an FDR-adjusted P < 0.01 for each gene set. Only Gene Ontology (GO) biological processes were considered.
ATAC and scATAC sequencing and data preprocessing.
Chromatin accessibility profiles for a total of 18 populations, including 16 previously reported, were assayed using FastATAC, an optimized ATAC-seq protocol optimized for primary blood cells, as previously described35,60. Sequencing data for each of the 18 populations was uniformly processed using a custom pipeline that includes sequencing adaptor removal, alignment using Bowtie261, and PCR duplicate removal with Picard RemoveDups command.
Accessible chromatin peaks were called from the 18 sorted populations of blood cells using MACS262. To derive a consensus set of loci for downstream analysis, individual peaks were resized to a uniform width of 500 bp, centered at the summit from the MACS2 call as previously described35. To derive a consensus peak set for the blood cell types, peaks were combined by removing any other peak overlapping with a peak with greater signal at the summit within a particular cell type. A total of 451,283 peaks representing a consensus set across these 18 sorted bulk populations were called. The average number of fragments in this consensus peak set ranged from 4.4 million (pDCs) to 37.1 million (CMPs) for a mean of 19.3 million reads in peaks per sorted cell type (Supplementary Table 4).
FACS-sorted cells from 9 distinct cellular populations from CD34+ human bone marrow, which included cell types spanning the myeloid, erythroid, and lymphoid lineages, were additionally profiled as previously described34,60. Single-cells were sorted then assayed using scATAC-seq34,63 across a total of 30 independent single-cell experiments representing 6 human donors, with each population assayed from two or more distinct donors. In total, our raw data set comprised 3,072 single-cell chromatin accessibility landscapes with 2,034 cells passing stringent quality filtering. These cells yielding a median of 8,268 fragments per cell with 76% of those fragments mapping to peaks, resulting in a median of 6,442 fragments in peaks per cell again using a consensus peak set that was inferred for these specific progenitor populations34.
To infer dynamic GWAS enrichments across hematopoietic differentiation, pseudotime orderings of single cells across three lineages (erythroid, lymphoid, and myeloid) were estimated using an adaptation of the Waterfall algorithm64 as previously described. In brief, this supervised approach fits a regression line through the relevant cluster centroids (k total = 14) in principal component space. The pseudo-time values then represent the Euclidean distance along the interpolated lines. Lines are scaled such that the center of the HSC cluster is 0 in all trajectories. Further details and diagnostics of this approach are discussed in a previous work64.
To assess regulatory heterogeneity of single cells, we computed a chi-squared statistic for each trait/cell type’s z-scores to test whether the observed variance was greater than expected. Under the null, the variance of z-scores is 1 from the definition of our statistic (see g-chromVAR methods below), and we observed greater variation than expected only for traits within the CMP and MEP populations. Within CMP and MEP populations, we applied k-medoids clustering on the first 5 principal components within each sorted population from global chromatin accessibility profiles for each cell34. For both the CMPs and MEPs, the optimal cluster number was determined by maximum average silhouette width. Post-hoc analyses of heterogeneity within the partitioned clusters of the erythroid-enriched CMPs confirmed that megakaryocyte-erythroid enrichment was not distinct within CMPs.
Isolation of mDCs and pDCs.
Peripheral blood cells from healthy volunteers were enriched for cell surface markers using the strategy shown in Supplementary Figure 7. 55,000 cells from two healthy volunteers (two replicates total) were sorted into RPMI1640 medium supplemented with 10% FBS, washed with PBS and immediately transposed as previously above. Post-sort purities of > 95% were confirmed by flow cytometry for all of the samples.
Target gene identification.
Raw sequencing reads from sorted populations were obtained from bulk RNA-seq experiments previously described34,35 and were aligned to the hg19 reference genome using STAR version 2.5.1b65 with default parameters. Per-gene transcript quantifications were summed over biological and technical replicates to provide a single transcript count per sorted cell type for 16 total populations matching the analogous bulk ATAC profiles (RNA for megakaryocytes and mDCs was absent). To determine empirical peak-gene associations, Pearson correlation was computed for each peak within a 1-Mb window of the transcription start site per gene using the log counts per million value for each feature.
PCHi-C for 15 terminal hematopoietic datasets as well as for CD34+ hematopoietic stem and progenitor cells were processed as previously reported33,66. Specifically, variants in AC regions were only considered to physically interact with a gene’s promoter when the CHiCAGO score was > 5.
Transcription factor motif analysis.
Prediction of the effects of fine-mapped variants on transcription factor binding sites (TFBS) was performed using the motifbreakR package67 and a comprehensive collection of human TFBS models (HOCOMOCO22). For all fine-mapped variants with PP > 0.1%, we applied the “information content” scoring algorithm and used a P-value cutoff at 5 × 10–4 for a TFBS match; all other parameters were kept at default settings.
To identify recurrent motifs that were disrupted by fine-mapped variants or were spatially proximal to these motifs, we used the findOverlaps() function from the GenomicRanges package68. To identify variants near motifs (Supplementary Fig. 8d), we extended the range of the motif 20 bp in both directions. For either motif breaking variants or motif proximal variants, variant/motif pairs were filtered such that they intersected a relevant factor in hematopoietic tissue from 2,115 uniformly processed datasets in ChIP-Atlas. Relevant TFs were defined by “bagging” motifs based upon the similarity of their position weight matrices (Pearson r > 0.7). A match was determined when the exact name of the TF from the ChIP-Atlas dataset exactly matched the name of the motif or any motif in the same “bag”. Conservation profiles for example motif disrupting variants were obtained are PhyloP estimates69.
To determine whether specific TFs were disrupted or proximal to variants more than expected by chance, we performed 100,000 permutations where we sampled the same number of unique variants with PP > 0.10 from across all variants in the 2,054 investigated regions. The expected number of TFs that were disrupted or proximal to variants was taken to be the mean across all permutations and significance was determined as one over the number of times that the number of overlaps was greater for PP > 0.10 than for the random sample.
Luciferase reporter analysis.
Firefly luciferase reporter constructs (pGL4.24) were generated by cloning the variant(s) of interest centered in 300–400 nucleotides (AK3 325 bp; CCND3 363 bp) of genomic context upstream of the minimal promoter using BglII and XhoI sites. The Firefly constructs (500 ng) were co-transfected with a pRL-SV40 Renilla luciferase construct (50 ng) into 100,000 K562 cells using Lipofectamine LTX (Invitrogen) according to manufacturer’s protocol. After 48 h, luciferase activity was measured by Dual-Glo Luciferase assay system (Promega) according to manufacturer’s protocol. For each sample, the ratio of Firefly to Renilla luminescence was measured and normalized to the empty pGL4.24 construct.
A total of four haplotypes were constructed per locus to examine the effects of two fine-mapped putative causal variants. For the CCND3 locus, we examined the effects of rs112233623 (ref: C, alt: T) and rs9349205 (ref: G, alt: A), which are 161 bp apart. For the AK3 locus, we examined rs409950 (ref: A; alt: C) and rs12005199 (ref: A, alt: G), which are separated by 123 bp. A total of nine (n = 9) experimental replicates per haplotype (four haplotypes per locus), including the empty pGL construct, were measured across two experimental batches.
To compute the additive and multiplicative effects of each variant, we used a generalized linear model of the following form for both of the AK3 and CCND3 loci separately:
Here, the luciferase intensity is defined as the ratio of Firefly to Renilla luminescence normalized to the empty vector for each experimental replicate. The additive effects of the two SNPs were estimated using β1 and β2 whereas the multiplicative effect of both SNPs on the same haplotype was computed using an interaction term, β3. We encoded each variable such that the reference allele was a 0 whereas the alternate allele was a 1 for each experimental sample. Finally, we adjusted for variable infection efficiency between the experimental batches using a fixed effect variable B (B ∈ {0,1}). To increase power, point estimates and standard errors were realized directly from the linear model using the β coefficients from each reporter set rather than the mean of the specific haplotype.
g-chromVAR methodology.
The bias-corrected enrichment statistics for T traits and a set of S samples (chromatin cell type profiles) with P peaks computed by g-chromVAR is a generalization of the chromVAR method50. Intuitively, our implementation of g-chromVAR relaxes the requirement in chromVAR that trait-peak annotations be binary, allowing for uncertainty in annotations such as transcription factor binding or in our case, localization of GWAS variants (see Supplementary Note for details). Briefly, we use a matrix of variant posterior probabilities G, where gikis the sum of the posterior probabilities of the variants contained in the genomic coordinates of peak i for each trait k. Using the matrix of fragment counts in peaks X, where xij represents the number of fragments from peak i in sample j, a matrix multiplicationXT·G yields the total number of fragments weighted by the fine-mapped variant posterior probabilities for S samples (rows) and T traits (columns). To compute a raw weighted accessibility deviation, we compute the expected number of fragments per peak per sample in E, where eij is computed as the proportion of all fragments across all samples mapping to the specific peak multiplied by the total number of fragments in peaks for that sample:
Analogously, XT·E ields the expected number of fragments weighted by the fine-mapped variant posterior probabilities for S samples (rows) and T traits (columns). Using the G, X, and E matrices, we then compute the raw weighted accessibility deviation matrix Y for each sample j and trait k (yj,k) as follows:
To correct for technical confounders present in assays (differential PCR amplification or variable Tn5 tagmentation conditions), each peak is assigned a background set of peaks that are matched in mean nucleotide GC content and average fragment accessibility between the sums of the cell types. An inverse Cholesky transformation is applied to a P by 2 matrix containing these variables to generate two uncorrelated dimensions describing the per-peak confounding. The matrix B(b) encodes this background peak mapping where is 1 if peak i has peak j as its background peak in the b background set (b ∈ {1,2,…,50})and 0 otherwise. The matrices B(b) ·X and B(b) ·E thus give an intermediate for the observed and expected counts also of dimension P by S. For each background set b, sample j, and trait k, the elements of the background weighted accessibility deviations matrix Y(b)are computed as follows:
After the background deviations are computed over the 50 sets, the bias-corrected matrix Z for sample j and trait k (zj,k) can be computed as follows:
where the mean and variance of is taken over all values of b (b ∈ {1,2,…,50}). Sample-trait P-values can then be computed from the one-tailed normal distribution of these z-scores using the pnorm function in R. Our implementation of g-chromVAR utilizes efficient matrix operations for each step and can compute pair-wise trait-cell type enrichments in ~1 minute on a standard laptop computer.
Other cell-type enrichment methods.
To estimate cell type enrichments for each trait using stratified LDSC (S-LDSC), we partitioned each trait’s heritability into the baseline model of 53 annotations, as well as each of the 18 hematopoietic ATAC-seq annotations (one at a time). Similarly, GREGOR70, GPA71, and fGWAS72 were run using the same 18 hematopoietic ATAC-seq annotations (one at a time) using default parameters for single trait and single annotation enrichments. P-values for cell-type enrichment were required to meet a stringent Bonferroni threshold of 0.00017 (corrected for 16 traits and 18 cell types).
Reporting Summary.
Further information on research design is available in the Life Sciences Reporting Summary linked to this article.
Supplementary Material
Acknowledgements
We thank members of the Sankaran, Buenrostro, and Finucane labs for their helpful discussions. This work was supported by National Institutes of Health (NIH) Grants R01 DK103794 and R33 HL120791 (to V.G.S.), the New York Stem Cell Foundation (NYSCF, to V.G.S.), and by the Harvard Society and Broad Institute Fellows programs (to J.D.B.). J.C.U. is supported by an NIH training grant (5T32 GM007226–43). C.A.L. is supported by an NIH predoctoral fellowship (F31 CA232670). E.L.B. is supported by the Howard Hughes Medical Institute Medical Research Fellows Program. V.G.S. is a NYSCF-Robertson Investigator. This research was conducted using the UK Biobank Resource under projects 11898 and 31063.
Footnotes
Competing interests
The authors declare no competing interests.
Data availability. All processed data are available on GitHub (https://github.com/caleblareau/singlecell_bloodtraits). ATAC-seq profiles are available from NCBI GEO GSE119453 and SRA PRJNA491478.
Code availability. g-chromVAR is available as an open source R-package distributed freely at http://caleblareau.github.io/gchromVAR. All code required for reproducing results discussed herein is made available at http://github.com/caleblareau/singlecell_bloodtraits.
References
- 1.Doulatov S, Notta F, Laurenti E & Dick JE Hematopoiesis: a human perspective. Cell Stem Cell 10, 120–36 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Sankaran VG & Orkin SH Genome-wide association studies of hematologic phenotypes: a window into human hematopoiesis. Curr Opin Genet Dev 23, 339–44 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Astle WJ et al. The Allelic Landscape of Human Blood Cell Trait Variation and Links to Common Complex Disease. Cell 167, 1415–1429 e19 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyle EA, Li YI & Pritchard JK An Expanded View of Complex Traits: From Polygenic to Omnigenic. Cell 169, 1177–1186 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farh KK et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 518, 337–43 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wellcome Trust Case Control, C. et al. Bayesian refinement of association signals for 14 loci in 3 common diseases. Nat Genet 44, 1294–301 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lango Allen H et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature 467, 832–8 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flister MJ et al. Identifying multiple causative genes at a single GWAS locus. Genome Res 23, 1996–2002 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galarneau G et al. Fine-mapping at three loci known to affect fetal hemoglobin levels explains additional genetic variation. Nat Genet 42, 1049–51 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung CC et al. Fine mapping of a region of chromosome 11q13 reveals multiple independent loci associated with risk of prostate cancer. Hum Mol Genet 20, 2869–78 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finucane HK et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet 47, 1228–35 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gusev A et al. Partitioning heritability of regulatory and cell-type-specific variants across 11 common diseases. Am J Hum Genet 95, 535–52 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thurner M et al. Integration of human pancreatic islet genomic data refines regulatory mechanisms at Type 2 Diabetes susceptibility loci. Elife 7(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bulik-Sullivan BK et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 47, 291–5 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benner C et al. Prospects of Fine-Mapping Trait-Associated Genomic Regions by Using Summary Statistics from Genome-wide Association Studies. Am J Hum Genet 101, 539–551 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benner C et al. FINEMAP: efficient variable selection using summary data from genome-wide association studies. Bioinformatics 32, 1493–501 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trynka G et al. Disentangling the Effects of Colocalizing Genomic Annotations to Functionally Prioritize Non-coding Variants within Complex-Trait Loci. Am J Hum Genet 97, 139–52 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giani FC et al. Targeted Application of Human Genetic Variation Can Improve Red Blood Cell Production from Stem Cells. Cell Stem Cell 18, 73–78 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thom CS et al. Trim58 degrades Dynein and regulates terminal erythropoiesis. Dev Cell 30, 688–700 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wakabayashi A et al. Insight into GATA1 transcriptional activity through interrogation of cis elements disrupted in human erythroid disorders. Proc Natl Acad Sci U S A 113, 4434–9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sidore C et al. Genome sequencing elucidates Sardinian genetic architecture and augments association analyses for lipid and blood inflammatory markers. Nat Genet 47, 1272–1281 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kulakovskiy IV et al. HOCOMOCO: expansion and enhancement of the collection of transcription factor binding sites models. Nucleic Acids Res 44, D116–25 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oki S et al. ChIP-Atlas: a data-mining suite powered by full integration of public ChIP-seq data. EMBO Rep 19(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arinobu Y et al. Reciprocal activation of GATA-1 and PU.1 marks initial specification of hematopoietic stem cells into myeloerythroid and myelolymphoid lineages. Cell Stem Cell 1, 416–27 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Hoppe PS et al. Early myeloid lineage choice is not initiated by random PU.1 to GATA1 protein ratios. Nature 535, 299–302 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Loughran SJ et al. The transcription factor Erg is essential for definitive hematopoiesis and the function of adult hematopoietic stem cells. Nat Immunol 9, 810–9 (2008). [DOI] [PubMed] [Google Scholar]
- 27.Carmichael CL et al. Hematopoietic overexpression of the transcription factor Erg induces lymphoid and erythro-megakaryocytic leukemia. Proc Natl Acad Sci U S A 109, 15437–42 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kruse EA et al. Dual requirement for the ETS transcription factors Fli-1 and Erg in hematopoietic stem cells and the megakaryocyte lineage. Proc Natl Acad Sci U S A 106, 13814–9 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vo KK et al. FLI1 level during megakaryopoiesis affects thrombopoiesis and platelet biology. Blood 129, 3486–3494 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang S, He Q, Ma D, Xue Y & Liu F Irf4 Regulates the Choice between T Lymphoid-Primed Progenitor and Myeloid Lineage Fates during Embryogenesis. Dev Cell 34, 621–31 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Elagib KE et al. RUNX1 and GATA-1 coexpression and cooperation in megakaryocytic differentiation. Blood 101, 4333–41 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Blyth K et al. Runx1 promotes B-cell survival and lymphoma development. Blood Cells Mol Dis 43, 12–9 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Javierre BM et al. Lineage-Specific Genome Architecture Links Enhancers and Non-coding Disease Variants to Target Gene Promoters. Cell 167, 1369–1384 e19 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buenrostro JD et al. Integrated Single-Cell Analysis Maps the Continuous Regulatory Landscape of Human Hematopoietic Differentiation. Cell 173, 1535–1548 e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Corces MR et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat Genet 48, 1193–203 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li P et al. IRF8 and IRF3 cooperatively regulate rapid interferon-beta induction in human blood monocytes. Blood 117, 2847–54 (2011). [DOI] [PubMed] [Google Scholar]
- 37.Hohaus S et al. PU.1 (Spi-1) and C/EBP alpha regulate expression of the granulocyte-macrophage colony-stimulating factor receptor alpha gene. Mol Cell Biol 15, 5830–45 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dufner A et al. The ubiquitin-specific protease USP8 is critical for the development and homeostasis of T cells. Nat Immunol 16, 950–60 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Reincke M et al. Mutations in the deubiquitinase gene USP8 cause Cushing’s disease. Nat Genet 47, 31–8 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Burley K, Westbury SK & Mumford AD TUBB1 variants and human platelet traits. Platelets 29, 209–211 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Sankaran VG et al. Cyclin D3 coordinates the cell cycle during differentiation to regulate erythrocyte size and number. Genes Dev 26, 2075–87 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gieger C et al. New gene functions in megakaryopoiesis and platelet formation. Nature 480, 201–8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gusev A et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat Genet 48, 245–52 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Giladi A et al. Single-cell characterization of haematopoietic progenitors and their trajectories in homeostasis and perturbed haematopoiesis. Nature Cell Biology 20, 836–846 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Guo MH et al. Comprehensive population-based genome sequencing provides insight into hematopoietic regulatory mechanisms. Proc Natl Acad Sci U S A 114, E327–E336 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang D-E et al. Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein α-deficient mice. Proceedings of the National Academy of Sciences 94, 569 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Edling CE & Hallberg B c-Kit—A hematopoietic cell essential receptor tyrosine kinase. The International Journal of Biochemistry & Cell Biology 39, 1995–1998 (2007). [DOI] [PubMed] [Google Scholar]
- 48.Opferman JT & Kothari A Anti-apoptotic BCL-2 family members in development. Cell Death And Differentiation 25, 37 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paul SP, Taylor LS, Stansbury EK & McVicar DW Myeloid specific human CD33 is an inhibitory receptor with differential ITIM function in recruiting the phosphatases SHP-1 and SHP-2. Blood 96, 483 (2000). [PubMed] [Google Scholar]
- 50.Schep AN, Wu B, Buenrostro JD & Greenleaf WJ chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat Methods 14, 975–978 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Drissen R et al. Distinct myeloid progenitor-differentiation pathways identified through single-cell RNA sequencing. Nat Immunol 17, 666–676 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee J et al. Lineage specification of human dendritic cells is marked by IRF8 expression in hematopoietic stem cells and multipotent progenitors. Nat Immunol 18, 877–888 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Notta F et al. Distinct routes of lineage development reshape the human blood hierarchy across ontogeny. Science 351, aab2116 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Paul F et al. Transcriptional Heterogeneity and Lineage Commitment in Myeloid Progenitors. Cell 163, 1663–77 (2015). [DOI] [PubMed] [Google Scholar]
Methods-Only References
- 55.Loh PR et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat Genet 47, 284–90 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hormozdiari F et al. Leveraging molecular quantitative trait loci to understand the genetic architecture of diseases and complex traits. Nat Genet 50, 1041–1047 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu A et al. Comparison of human genetic and sequence-based physical maps. Nature 409, 951–3 (2001). [DOI] [PubMed] [Google Scholar]
- 58.McLaren W et al. The Ensembl Variant Effect Predictor. Genome Biol 17, 122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Watanabe K, Taskesen E, van Bochoven A & Posthuma D Functional mapping and annotation of genetic associations with FUMA. Nat Commun 8, 1826 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Buenrostro JD, Wu B, Chang HY & Greenleaf WJ ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr Protoc Mol Biol 109, 21 29 1–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Langmead B & Salzberg SL Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Y et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Buenrostro JD et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523, 486–90 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shin J et al. Single-Cell RNA-Seq with Waterfall Reveals Molecular Cascades underlying Adult Neurogenesis. Cell Stem Cell 17, 360–72 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dobin A et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mifsud B et al. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat Genet 47, 598–606 (2015). [DOI] [PubMed] [Google Scholar]
- 67.Coetzee SG, Coetzee GA & Hazelett DJ motifbreakR: an R/Bioconductor package for predicting variant effects at transcription factor binding sites. Bioinformatics 31, 3847–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lawrence M et al. Software for computing and annotating genomic ranges. PLoS Comput Biol 9, e1003118 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pollard KS, Hubisz MJ, Rosenbloom KR & Siepel A Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res 20, 110–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schmidt EM et al. GREGOR: evaluating global enrichment of trait-associated variants in epigenomic features using a systematic, data-driven approach. Bioinformatics 31, 2601–6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chung D, Yang C, Li C, Gelernter J & Zhao H GPA: a statistical approach to prioritizing GWAS results by integrating pleiotropy and annotation. PLoS Genet 10, e1004787 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pickrell JK Joint analysis of functional genomic data and genome-wide association studies of 18 human traits. Am J Hum Genet 94, 559–73 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.