Abstract
Gene-diet interaction studies have reported that individual variations in phenotypic traits may be due to variations in individual diet. Our study aimed to evaluate (i) the association of ADRB2 rs1042713 with obesity and obesity-related metabolic parameters and (ii) the effect of dietary nutrients on these associations in Malaysian adults. ADRB2 genotyping, dietary, physical activity, anthropometric, and biochemical data were collected from 79 obese and 99 nonobese individuals. Logistic regression revealed no association between ADRB2 rs1042713 and obesity (p=0.725). However, the carriers of G allele (AG + GG genotypes) of rs1042713 were associated with increased odds of insulin resistance, 2.83 (CI = 1.04–7.70, adjusted p=0.042), in the dominant model, even after adjusting for potential confounders. Obese individuals carrying the G allele were associated with higher total cholesterol (p=0.011), LDL cholesterol levels (p=0.008), and total cholesterol/HDL cholesterol ratio (p=0.048), compared to the noncarriers (AA), even after adjusting for potential confounders. Irrespective of obesity, the carriers of GG genotype had significantly lower fasting glucose levels with low saturated fatty acid intake (<7.3% of TE/day) (4.92 ± 0.1 mmol/L vs 5.80 ± 0.3 mmol/L, p=0.011) and high intake of polyunsaturated fatty acid:saturated fatty acid ratio (≥0.8/day) (4.83 ± 0.1 mmol/L vs 5.93 ± 0.4 mmol/L, p=0.006). Moreover, the carriers of GG genotype with high polyunsaturated fatty acid intake (≥6% of TE/day) had significantly lower HOMA-IR (1.5 ± 0.3 vs 3.0 ± 0.7, p=0.026) and fasting insulin levels (6.8 ± 1.6 µU/mL vs 11.4 ± 2.1 µU/mL, p=0.036). These effects were not found in the noncarriers (AA). In conclusion, G allele carriers of ADRB2 rs1042713 were associated with increased odds of insulin resistance. Obese individuals carrying G allele were compromised with higher blood lipid levels. Although it is premature to report gene-diet interaction on the regulation of glucose and insulin levels in Malaysians, we suggest that higher quantity of PUFA-rich food sources in regular diet may benefit overweight and obese Malaysian adults metabolically. Large-scale studies are required to replicate and confirm the current findings in the Malaysian population.
1. Background
Obesity and related chronic diseases have become the leading causes of morbidity and mortality worldwide. Long-term consumption of energy dense food coupled with sedentary lifestyle are the main contributors to the development of obesity and related complications [1]. Multiple genetic loci determined by genome-wide association studies (GWAS) have been found to be associated with increased susceptibility to obesity, diabetes, and dyslipidemia [2]. Earlier studies have reported on the adrenergic receptor system for its role in the stimulation of thermogenesis and in activating lipid mobilization from the fat stores. With respect to this, the beta-2 adrenoceptor gene is a candidate gene since it is the dominating lipolytic receptor in the human white adipose tissue [3]. It stimulates lipid mobilization through lipolysis in adipocytes and regulates body fat accumulation and energy expenditure [4].
A strong association between obesity and a single nucleotide polymorphism located at codon 16 substituting arginine for glycine (rs1042713/Arg16Gly) has been reported by Large et al. [5]. Masuo et al. reported that insulin-resistant subjects had higher frequencies of the G allele of rs1042713 [6]. Total body fat mass and blood pressure levels were higher in nonobese and nonhypertensive men with G allele in the Japanese population. The authors speculated that insulin resistance could, in part, be determined by the genetic variant of the beta-2 adrenoceptor gene and that polymorphism and higher plasma adrenaline could increase insulin resistance, adiposity, and high blood pressure in their subjects. Thus, the G allele could lead to heightened sympathetic nerve activity, insulin resistance, and higher blood pressure and adiposity in nonobese and nonhypertensive individuals. Other studies have adequately reported that insulin resistance was strongly associated with heightened sympathetic nerve activity [7]. In other words, adrenergic receptor defects lead to the sympathetic nervous system over activity that may play a role in the development of insulin resistance, hypertension, and obesity [8].
A study on the Swedish population reported that ADRB2 rs1042713 was significantly associated with elevated central body fat, systolic blood pressure, serum leptin, and triglyceride levels but not with obesity [9]. Studies from Saudi Arabia reported significant association between ADRB2 rs1042713 polymorphism and the development of insulin resistance, dyslipidemia, overweight, and obesity [10, 11]. However, findings from Asian populations (Japanese and Korean) reported negative association between obesity and ADRB2 gene polymorphisms. Moreover, these studies did not investigate interaction of gene variants with dietary nutrients [12, 13].
Individual or population differences in the development of obesity-related metabolic diseases may result not only from genetic variation but may also be the modulatory effect of dietary nutrients on gene and gene variants [14]. In the Malaysian population, relatively little is known with respect to the interaction between dietary nutrients and ADRB2 gene variations on obesity, insulin resistance, and glucose homeostasis. It is believed that early identification of the candidate gene variants and their interaction with diet may allow for the provision of good quality personalised dietary recommendation to achieve effective weight loss and reduction in metabolic risk factors [15]. Although a couple of intervention studies have reported lipid outcomes associated with ADRB2 rs1042713, there is no such study done in Malaysian adults. Since there has been conflicting results with respect to ADRB2 rs1042713 between Asian, Caucasian, and Arabic populations, the current study on the Malaysian population is valuable and will shed light and add to the existing evidence on gene variants and phenotypic outcomes and influence of diet on the latter. To the best of our knowledge, this is the first study in Malaysian adults that investigates the interaction between ADRB2 rs1042713 and dietary nutrients on obesity-related metabolic traits. This study is nested in a broader study investigating the association of single nucleotide polymorphisms in genes that have widely been reported to influence obesity and obesity-related metabolic disorders in human individuals. In an earlier publication, we have reported that FTO rs9930506 may interact with dietary protein and Vitamin E intake and modulate hsCRP levels in our Malaysian participants [16]. The aim of the current study was to evaluate (i) the effect of ADRB2 rs1042713 on obesity and obesity-related anthropometric and blood biochemical parameters and (ii) the influence of diet on the association between ADRB2 rs1042713 and obesity phenotypes, in Malaysian adults.
2. Methods
2.1. Ethical Approval
This study was reviewed and approved by the University of Nottingham Malaysia Campus (UNMC) Science and Engineering Research Ethics Committee and was registered under Medical Research and Ethics Committee (MREC) of National Medical Research Registry (Research ID: 25110), Ministry of Health Malaysia (MOH). Written informed consent was obtained from all participants.
2.2. Study Design
This cross-sectional study was conducted from 2014–2017 on Malaysian adults aged between 18 and 74 years. The study investigated (i) the association between ADRB2 rs1042713 with obesity and insulin resistance; (ii) the association between ADRB2 rs1042713 and phenotypes in obese and nonobese individuals; and (iii) the interaction between ADRB2 rs1042713 and dietary nutrients on phenotypic traits.
2.3. Participant Selection
Detailed information on the study design and methods can be found in our earlier publication [16]. Therefore, with respect to assessment of anthropometric parameters, dietary nutrients analysis, and physical activity level assessment, we refer the readers to our earlier publication [16].
2.4. Blood Collection and Biochemical Analysis
10–12 hour fasting blood was collected into vacutainer tubes containing fluoride oxalate for plasma glucose analysis and vacutainer tubes with clot activator and gel (Becton Dickinson, Oxford, United Kingdom) for serum lipid profile (total cholesterol, triglyceride, and HDL cholesterol levels), insulin, and high-sensitivity C-reactive protein (hsCRP) analysis. The analysis for above biochemical parameters were assessed using Abbott Architect CI8200 Automatic System following the manufacturer's instructions. Homeostatic model assessment to estimate insulin resistance (HOMA-IR) was calculated by multiplying fasting plasma glucose (mmol/L) by fasting serum insulin (µU/ml) and divided by 22.5 [17]. Low-density lipoprotein (LDL) cholesterol was calculated using the Friedewald formula: LDL cholesterol = total cholesterol − ((triglyceride/5) + HDL cholesterol) [18]. This information has been reported in our earlier publication [16]. This study investigates ADRB2 rs1042713 gene polymorphism in the same population with the aim to evaluate (i) the effect of ADRB2 rs1042713 on obesity and obesity-related anthropometric and blood biochemical parameters and (ii) the influence of diet on the association between ADRB2 rs1042713 and obesity phenotypes.
2.5. Genotyping of ADRB2 rs1042713 Gene Polymorphism
Five milliliters of whole blood was drawn from an antecubital vein into vacutainer tubes (Becton, Dickinson and Co., Franklin Lakes, NJ) containing EDTA. The genomic DNA was extracted from leukocytes using the MasterPure DNA Purification kit (Lucigen Corporation Middleton, WI, USA) according to the manufacturer's instructions. DNA samples were stored at −20°C until use. A DNA fragment of 310 bp containing rs1042713 was amplified by using polymerase chain reaction (PCR) for the identification of ADRB2 rs1042713 gene polymorphism with specific primers (forward primer: 5′-CCGCCGTGGGTCCGCC-3′ and reverse primer: 5′-CCATGACCAGATCAGCAC-3′) derived from an earlier study [19]. PCR was performed using 5 µl of genomic DNA (∼1 ng/µl), 0.2 µM forward primer, and reverse primer with 5 µl of Taq 5X Master Mix. Thermal cycling was performed as follows: initial denaturing at 95°C for 5 min; 35 cycles of denaturation at 94°C for 45 s, annealing at 64°C for 40 s and extension at 72°C for 45 sec, and then a final extension at 72°C for 5 min. The amplicons were verified by using electrophoresis on 2% agarose gel and visualized under ultraviolet illumination after staining by ethidium bromide. The verified amplicons were then sequenced by using BigDye® terminator v3.1 cycle sequencing kit chemistry.
2.6. Power and Sample Size Calculation
We performed power calculation using software QUANTO, Version 1.2.4, to find the minimum detectable effect for a given sample size. This calculation takes into account the type 1 error rate of 0.05, and the population prevalence of insulin resistance (using a cut off of HOMA-IR ≥ 1.7) of 45%, as reported in the present study. Given that the minor allele frequency (A) of ADRB2 rs1042713 in our study was 0.49 with 57 insulin-resistant and 69 non-insulin-resistant participants, we had 68.2% power to detect an effect of 2.83 (odds ratio) (dominant model).
With regard to the gene-diet interaction, given that the mean of fasting glucose levels in our population was 5.2 mmol/L, with a SD of 2.0, environmental effect (differences of fasting glucose levels between high and low PUFA : SFA ratio) of −0.4 (5.0 mmol/L–5.4 mmol/L), genetic effect (differences of fasting glucose levels between the carriers of G allele of ADRB2 rs1042713 and the noncarriers (A)) of 0.3 (5.4 mmol/L − 5.1 mmol/L), and interaction effect of −1.1 (4.83 mmol/L–5.93 mmol/L), a power of 61% for the gene-diet interaction was computed.
2.7. Statistical Analysis
Statistical analysis was performed using the statistical package for social sciences (IBM SPSS statistic, Chicago, IL, USA, version 22). Data were expressed as mean ± standard error (SE) or number (percentage). Log transformation was performed to transform nonnormally distributed data into normally distributed data. Independent t-test and chi-squared test were performed to assess the differences between the two genotype groups (AA vs AG + GG) on baseline continuous variables and categorical variables, respectively. Allele frequency was estimated by gene counting and chi-squared test was used to assess deviation from Hardy–Weinberg equilibrium (HWE) [20]. To study the effect of ADRB2 rs1042713 on obesity, data were dichotomised into obese and nonobese groups (obesity was defined by BMI ≥ 27.5 kg/m2) [21]. The AA genotype of ADRB2 rs1042713 was used as the reference group in both codominant and dominant models, whereas the combination of AG and GG genotypes was used as the reference group in recessive model. Odds ratios (ORs) with 95% confidence intervals (95% CIs) were estimated for each genotype by logistic regression to determine the odds of obesity associated with gene variants, after adjusting for covariates age, gender, physical activity status, smoking status, and alcohol consumption. Same analysis was performed to study the odds of ADRB2 gene variants on insulin resistance. Data were dichotomised into (i) non-insulin-resistant and insulin-resistant groups (using a cutoff of HOMA-IR ≥ 1.7 [22]).
Differences in means between gene variants in anthropometric, blood biochemical, and dietary parameters in obese and nonobese groups were assessed by using one-way analysis of covariance (ANCOVA). Adjustment for covariates such as age, gender, physical activity status, smoking status, alcohol consumption, BMI, WC, fat mass, body fat percent, and total energy intake were applied where appropriate. The intake of macronutrients (energy-adjusted) was dichotomised into two groups based on the median intake of the population. A multivariate general linear model (GLM) was used to investigate the effect of the interaction between dietary macronutrients and ADRB2 rs1042713 on obesity-related metabolic traits, after adjusting for potential confounders (age, gender, physical activity status, smoking status, alcohol consumption, BMI, and total energy intake). A statistical probability level of p < 0.05 (two-sided) was considered significant. No significant association was found between rs1042713 and protein and carbohydrate intake on obesity-related traits. Therefore, the results were not reported.
3. Result
3.1. Baseline Characteristics
In total, 178 Malaysian adults (female = 154; male = 24) were recruited for anthropometric measurement and genetic analysis. For biochemical and dietary analysis, 126 participants (female = 106, male = 20) were available. General characteristics of the study participants are reported in Table 1. The ages between the two groups (AA vs AG + GG) did not differ significantly (43.4 ± 1.8 y vs 41.1 ± 1.0 y; p=0.286). The gender distribution was not significantly different between the gene variants in the genotype groups (AA vs AG + GG), females (84.1% vs 87.3%), and males (15.9% vs 12.7%) (p=0.587). Ethnicity distribution was not significantly different between the two genotype groups (p=0.556). No significant difference was found in physical activity (p=0.256), smoking (p=0.606), and alcohol consumption (p=0.717) between the two genotype groups. No significant difference was found in height (p=0.907), body weight (p=0.505), and BMI (p=0.427) between the two genotype groups (p > 0.05). Due to the higher number of female participants, all data analyses were adjusted for by gender.
Table 1.
General characteristics of the study participants; differences between ADRB2 rs1042713 gene variants.
| Genotype | ADRB2 rs1042713 (dominant model) | ||
|---|---|---|---|
| AA (n=44) | AG + GG (n=134) | p value | |
| Age (years) | 43.4 ± 1.8 | 41.1 ± 1.0 | 0.286 |
| Gender | |||
| Female (n) | 37 (84.1%) | 117 (87.3%) | 0.587 |
| Male (n) | 7 (15.9%) | 17 (12.7%) | |
| Ethnicity | |||
| Malays | 20 (23.3%) | 66 (46.7%) | 0.556 |
| Chinese | 13 (31%) | 29 (69%) | |
| Indians | 11 (22%) | 39 (78%) | |
| Physical activity status | |||
| Physically active | 41 (24%) | 130 (76%) | 0.256 |
| Physically inactive | 3 (42.9%) | 4 (57.1%) | |
| Smoking status | |||
| Never | 44 (25%) | 132 (75%) | 0.606 |
| Former | 0 | 1 (100%) | |
| Current | 0 | 2 (100%) | |
| Alcohol consumption status | |||
| Never | 44 (25.1%) | 131 (74.9%) | 0.717 |
| Former | 0 | 1 (100%) | |
| Current | 0 | 1 (100%) | |
| Weight (kg) | 65.7 ± 1.1 | 68.1 ± 1.6 | 0.505 |
| Height (cm) | 157.9 ± 1.1 | 157.8 ± 0.6 | 0.907 |
| BMI (kg/m2) | 26.3 ± 0.7 | 27.2 ± 0.5 | 0.427 |
a p value based on the independent t-test. b p value based on the chi-squared test. p < 0.05 was considered as significant. Physically active was defined as accumulation of at least 150 minutes/week of moderate intensity activity (3–6 METs) or 60 minutes/week of vigorous physical activity (>6 METs) as defined by the Ministry of Health Malaysia [23].
3.2. Allele Frequencies of the Gene Variants of ADRB2 rs1042713
G allele was the most frequent variant in our study population (51.1%). The allele frequency for the minor allele of rs1042713 (A allele) was 0.49, which did not deviate from Hardy–Weinberg equilibrium as tested by the chi-squared test, χ 2 = 0.36 (χ 2 < 3.841) (Table 2).
Table 2.
Genotype distribution and allele frequency of ADRB2 rs1042713 in obese and nonobese groups.
| Genotype | Overall n=178 | Χ 2 (Hardy–Weinberg) | Obese BMI ≥ 27.5 n=79 | Nonobese BMI < 27.5 n=99 | Unadjusted OR (95% CI) | p value | Adjusted OR (95% CI)∗ | p value∗ |
|---|---|---|---|---|---|---|---|---|
| Codominant model | ||||||||
| AA | 44 (24.7%) | 0.36 | 19 (24.0%) | 25 (25.2%) | 1 | — | 1 | — |
| AG | 86 (48.3%) | 41 (51.9%) | 45 (45.5%) | 1.20 (0.58–2.49) | 0.627 | 1.26 (0.59–2.71) | 0.548 | |
| GG | 48 (27.0%) | 19 (24.1%) | 29 (29.3%) | 0.86 (0.38–1.98) | 0.726 | 0.94 (0.40–2.23) | 0.884 | |
| Dominant model | ||||||||
| AA | 44 (24.7%) | 19 (24.0%) | 25 (25.2%) | 1 | — | 1 | — | |
| AG + GG | 134 (75.3%) | 60 (76.0%) | 74 (74.8%) | 1.07 (0.54–2.12) | 0.853 | 1.14 (0.56–2.33) | 0.725 | |
| Recessive model | ||||||||
| AA + AG | 130 (73.0%) | 60 (75.9%) | 70 (70.7%) | 1 | — | 1 | — | |
| GG | 48 (27.0%) | 19 (24.1%) | 29 (29.3%) | 0.76 (0.39–1.50) | 0.434 | 0.80 (0.40–1.61) | 0.538 | |
| Allele frequency | ||||||||
| A | 174 (48.9%) | 79 (50.0%) | 95 (48.0%) | |||||
| G | 182 (51.1%) | 79 (50.0%) | 103 (52.0%) |
The agreement of genotype frequencies with Hardy–Weinberg equilibrium was tested by using the chi-squared test, with χ 2 < 3.841 considered as no deviation from Hardy–Weinberg equilibrium. Logistic regression was conducted to determine the risk of obesity associated with gene variants. Odds ratios (ORs) with 95% confidence intervals (95% CIs) were estimated for each genotype. ∗Adjusted for age, gender, physical activity status, smoking status, and alcohol consumption. p < 0.05 was considered significant.
3.3. Association between ADRB2 rs1042713 and Obesity and Insulin Resistance
Logistic regression was performed to examine the independent effect of ADRB2 rs1042713 on the odds of obesity and insulin resistance. We found no significant association between ADRB2 rs1042713 and obesity (obesity as defined by BMI ≥ 27.5 kg/m2) under codominant (AG p=0.548 and GG, p=0.884), dominant (p=0.725), and recessive (p=0.538) models, after adjusting for covariates age, gender, physical activity status, smoking status, and alcohol consumption (Table 2).
However, our results revealed significant association between ADRB2 rs1042713 and insulin resistance (using a cutoff of HOMA-IR ≥ 1.7) (Table 3). The carriers of GG genotype of rs1042713 had increased odds of insulin resistance, compared to AA genotype in both codominant and dominant models, 4.43 (CI = 1.31–15.0, adjusted p=0.016) and 2.83 (CI = 1.04–7.70, adjusted p=0.042), respectively, even after adjusting for covariates age, gender, BMI, physical activity status, smoking status, and alcohol consumption. No significant association was found in the recessive model (adjusted p=0.060).
Table 3.
Genotype distribution and allele frequency of ADRB2 rs1042713 in insulin-resistant and non-insulin-resistant groups.
| Genotype | HOMA < 1.7 n=69 | HOMA ≥ 1.7 n=57 | Unadjusted OR (95% CI) | p value | Adjusted OR (95% CI)∗ | p value∗ |
|---|---|---|---|---|---|---|
| Codominant model | ||||||
| AA | 21 (30.5%) | 9 (15.8%) | 1 | — | 1 | — |
| AG | 33 (47.8%) | 30 (52.6%) | 2.12 (0.84–5.35) | 0.111 | 2.31 (0.81–6.61) | 0.118 |
| GG | 15 (21.7%) | 18 (31.6%) | 2.80 (1.0–7.91) | 0.052 | 4.43 (1.31–15.0) | 0.016 |
| Dominant model | ||||||
| AA | 21 (30.5%) | 9 (15.8%) | 1 | — | 1 | — |
| AG + GG | 48 (69.5%) | 48 (84.2%) | 2.33 (0.97–5.61) | 0.058 | 2.83 (1.04–7.70) | 0.042 |
| Recessive model | ||||||
| AA + AG | 54 (78.3%) | 39 (68.4%) | 1 | — | 1 | — |
| GG | 15 (21.7%) | 18 (31.6%) | 1.66 (0.75–3.70) | 0.213 | 2.46 (0.96–6.27) | 0.060 |
| Allele frequency | ||||||
| A | 75 (54.3%) | 48 (42.1%) | ||||
| G | 63 (45.7%) | 66 (57.9%) |
Odds ratios (ORs) with 95% confidence intervals (95% CIs) were estimated for each genotype by logistic regression to determine the risk of insulin resistance associated with ADRB2 rs1042713. p ≤ 0.05 was considered as significant. ∗Adjusted for age, BMI, gender, physical activity status, smoking status, and alcohol consumption. Non-insulin-resistant and insulin-resistant groups were dichotomised using a cutoff of HOMA-IR ≥ 1.7 [22].
3.4. Differences in Means between ADRB2 rs1042713 Gene Variants in Anthropometric, Biochemical, and Dietary Parameters in Obese and Nonobese Groups
The age, anthropometric, biochemical, and dietary parameters of the study participants between gene variants in obese and nonobese groups have been reported in Table 4. In obese participants, we found that the carriers of G allele of rs1042713 had significantly higher total cholesterol (p=0.011), LDL cholesterol levels (p=0.008), and total cholesterol per HDL cholesterol ratio (p=0.048), compared to the noncarriers (AA), even after adjusting for covariates age, gender, BMI, WC, fat mass, body fat percent, physical activity status, smoking status, and alcohol consumption. Interestingly, such differences in blood biochemical parameters within genotypes were not observed in the nonobese group. With respect to dietary parameters, we found that the carriers of the G allele had significantly lower consumption of PUFA compared to the noncarriers (AA), even after adjusting for covariates (p=0.036). No significance association was found between ADRB2 rs1042713 and others dietary parameters.
Table 4.
Differences in means (±SE) between ADRB2 rs1042713 gene variants in anthropometric, biochemical, and dietary parameters in obese and nonobese groups.
| ADRB2 rs1042713 (dominant model) | ||||||
|---|---|---|---|---|---|---|
| Obese (n=79) | Nonobese (n=99) | |||||
| General characteristics | AA (n=19) | AG + GG (n=60) | p value | AA (n=25) | AG + GG (n=74) | p value |
| Age (years) | 47.8 ± 2.0 | 42.8 ± 1.5 | 0.038 ∗ | 40.0 ± 2.7 | 39.7 ± 1.3 | 0.958 |
| 1Weight (kg) | 76.0 ± 2.0 | 81.2 ± 2.3 | 0.097 | 57.9 ± 1.9 | 57.4 ± 1.0 | 0.957 |
| 1Height (cm) | 157.7 ± 1.7 | 157.7 ± 1.0 | 0.688 | 158.1 ± 1.5 | 157.9 ± 0.8 | 0.871 |
| 1BMI (kg/m2) | 30.5 ± 0.5 | 32.4 ± 0.6 | 0.086 | 23.0 ± 0.6 | 23.0 ± 0.3 | 0.936 |
| 1WC (cm) | 97.6 ± 2.2 | 99.4 ± 1.6 | 0.173 | 68.6 ± 3.7 | 68.6 ± 2.0 | 0.899 |
| 1WHR | 0.90 ± 0.01 | 0.93 ± 0.01 | 0.117 | 0.87 ± 0.01 | 0.87 ± 0.01 | 0.728 |
| 1Muscle mass (kg) | 24.0 ± 0.9 | 24.4 ± 0.7 | 0.525 | 20.9 ± 1.0 | 20.9 ± 0.6 | 0.941 |
| 1Fat mass (kg) | 32.3 ± 1.2 | 36.6 ± 1.4 | 0.065 | 19.2 ± 1.0 | 19.5 ± 0.6 | 0.887 |
| 1Fat-free mass (kg) | 43.8 ± 1.6 | 44.6 ± 1.2 | 0.475 | 38.6 ± 1.6 | 37.9 ± 0.7 | 0.678 |
| 1Percent body fat (%) | 42.5 ± 1.2 | 44.8 ± 0.7 | 0.110 | 33.3 ± 1.3 | 33.8 ± 0.8 | 0.683 |
| 1Systolic BP (mmHg) | 124.0 ± 3.6 | 124.0 ± 2.0 | 0.712 | 122.6 ± 5.0 | 119.6 ± 2.3 | 0.587 |
| 1Diastolic BP (mmHg) | 81.0 ± 2.3 | 81.5 ± 1.3 | 0.535 | 81.9 ± 2.7 | 79.2 ± 1.8 | 0.588 |
| 1Pulse rate (bpm) | 75.7 ± 2.5 | 76.1 ± 1.3 | 0.948 | 75.1 ± 4.5 | 77.9 ± 2.0 | 0.556 |
|
| ||||||
| Blood biochemical parameters | AA (n=19) | AG + GG (n=60) | p value | AA (n=11) | AG + GG (n=36) | p value |
| 2Fasting glucose (mmol/L) | 5.1 ± 0.2 | 5.3 ± 0.3 | 0.509 | 5.0 ± 0.3 | 5.2 ± 0.3 | 0.561 |
| 2Fasting insulin (uU/mL) | 7.74 ± 1.6 | 11.2 ± 1.3 | 0.209 | 11.7 ± 3.7 | 7.5 ± 1.4 | 0.361 |
| 2HOMA-IR | 1.9 ± 0.5 | 2.8 ± 0.3 | 0.176 | 2.9 ± 1.0 | 2.2 ± 0.7 | 0.494 |
| 2Total cholesterol (mmol/L) | 5.0 ± 0.2 | 5.7 ± 0.1 | 0.011 ∗ | 5.4 ± 0.3 | 5.6 ± 0.2 | 0.942 |
| 2Triglyceride (mmol/L) | 1.2 ± 0.2 | 1.4 ± 0.1 | 0.083 | 1.7 ± 0.4 | 1.4 ± 0.1 | 0.983 |
| 2HDL cholesterol (mmol/L) | 1.6 ± 0.1 | 1.5 ± 0.1 | 0.791 | 1.5 ± 0.1 | 1.6 ± 0.1 | 0.636 |
| 2LDL cholesterol (mmol/L) | 2.9 ± 0.2 | 3.6 ± 0.1 | 0.008 ∗ | 3.3 ± 0.4 | 3.3 ± 0.2 | 0.650 |
| 2Total cholesterol/HDL cholesterol | 3.4 ± 0.2 | 3.9 ± 0.1 | 0.048 ∗ | 3.7 ± 0.3 | 3.7 ± 0.2 | 0.650 |
| 2hsCRP (mg/L) | 5.2 ± 1.5 | 8.8 ± 1.4 | 0.059 | 2.2 ± 0.5 | 2.9 ± 0.5 | 0.750 |
|
| ||||||
| Dietary parameters | AA (n=19) | AG + GG (n=60) | p value | AA (n=11) | AG + GG (n=36) | p value |
| 3Total energy intake (kcal) | 2145.5 ± 108.0 | 2065.6 ± 41.7 | 0.766 | 1897.5 ± 53.6 | 1867.5 ± 47.4 | 0.836 |
| 4Actual total carbohydrate intake (g) | 272.3 ± 20.7 | 259.4 ± 7.2 | 0.967 | 237.8 ± 13.1 | 234.0 ± 11.0 | 0.625 |
| 4Actual total protein intake (g) | 75.1 ± 5.3 | 77.1 ± 3.1 | 0.982 | 72.8 ± 5.3 | 70.0 ± 3.2 | 0.276 |
| 4Actual total dietary fat intake (g) | 90.2 ± 5.5 | 84.7 ± 3.1 | 0.810 | 77.1 ± 4.1 | 77.7 ± 2.6 | 0.817 |
| 4Percentage energy from carbohydrate (%) | 46.9 ± 1.8 | 46.9 ± 1.2 | 0.995 | 46.8 ± 2.1 | 47.7 ± 1.3 | 0.520 |
| 4Percentage energy from protein (%) | 13.8 ± 0.6 | 14.9 ± 0.5 | 0.799 | 15.5 ± 1.2 | 14.4 ± 0.7 | 0.181 |
| 4Percentage energy from dietary fat (%) | 37.9 ± 1.9 | 36.4 ± 1.0 | 0.744 | 36.4 ± 1.5 | 36.8 ± 1.1 | 0.968 |
| 4SFA (%TE) | 7.0 ± 1.0 | 7.6 ± 0.5 | 0.514 | 7.0 ± 0.8 | 8.7 ± 0.6 | 0.307 |
| 4MUFA (%TE) | 11.8 ± 1.1 | 10.3 ± 0.6 | 0.339 | 11.8 ± 1.1 | 12.1 ± 0.6 | 0.895 |
| 4PUFA (%TE) | 8.0 ± 0.8 | 5.6 ± 0.4 | 0.036 ∗ | 6.5 ± 1.1 | 7.0 ± 0.6 | 0.523 |
| 4Trans fat (%TE) | 0.1 ± 0.01 | 0.1 ± 0.04 | 0.109 | 0.1 ± 0.03 | 0.1 ± 0.02 | 0.497 |
| 4Dietary cholesterol (%TE) | 88.8 ± 14.0 | 108.3 ± 10.5 | 0.987 | 111.4 ± 22.9 | 109.7 ± 16.1 | 0.471 |
One-way analysis of covariance was performed to determine the differences between means between gene variants in anthropometric, biochemical, and dietary parameters in obese and nonobese participants, after adjusting for covariates in different model 1age, gender, physical activity status, smoking status, and alcohol consumption; 2model1 + BMI, WC, body fat mass, and body fat percent; 3model1 + BMI; and 4model3 + total energy intake. ∗ p < 0.05 was considered significant. HOMA-IR: homeostatic model assessment-insulin resistance; HDL: high-density lipoprotein; LDL: low-density lipoprotein; hsCRP: high-sensitivity C-reactive protein; SFA: saturated fatty acid; MUFA: monounsaturated fatty acid; PUFA: polyunsaturated fatty acid; % TE: percentage of total energy intake.
3.5. Differences between Blood Biochemical Parameters and Respective Diagnostic Cutoffs in Obese Individuals Carrying ADRB2 rs1042713 G Allele
In obese individuals carrying the G allele of ADRB2 rs1042713, HOMA-IR was significantly higher than the diagnostic cut off (2.8 ± 0.3) (Table 5). Although HDL cholesterol levels (1.5 ± 0.1 mmol/L) were positively, significantly higher than 1 mmol/L (diagnostic cutoff), total cholesterol (5.7 ± 0.1 mmol/L) and LDL cholesterol levels (3.6 ± 0.1 mmol/L) were significantly higher than the respective cutoffs, indicating metabolic risk (Table 5).
Table 5.
Differences between blood biochemical parameters and respective diagnostic cutoffs in obese individuals carrying ADRB2 rs1042713 G allele.
| Blood biochemical parameters | Mean (±SE) | Diagnostic cutoff | p value |
|---|---|---|---|
| Fasting glucose (mmol/L) | 5.3 ± 0.3 | 5.6 [24] | 0.327 |
| Fasting insulin (µU/mL) | 11.2 ± 1.3 | 10.6 [25] | 0.330 |
| HOMA-IR | 2.8 ± 0.3 | 1.7 [22] | 0.003 ∗ |
| Total cholesterol (mmol/L) | 5.7 ± 0.1 | 5.2 [26] | 0.001 ∗ |
| Triglyceride (mmol/L) | 1.4 ± 0.1 | 1.7 [24] | <0.001 ∗ |
| HDL cholesterol (mmol/L) | 1.5 ± 0.1 | 1.0 [24] | <0.001 ∗ |
| LDL cholesterol (mmol/L) | 3.6 ± 0.1 | 2.6 [26] | <0.001 ∗ |
| Total cholesterol/HDL cholesterol | 3.9 ± 0.1 | — | — |
The one-sample t-test was performed to assess the difference between blood biochemical parameters and respective diagnostic cutoffs ∗ p < 0.05 was considered significant. HOMA-IR: homeostatic model assessment-insulin resistance; HDL: high-density lipoprotein; LDL, low-density lipoprotein.
3.6. Interaction between Dietary Fat Intake and ADRB2 rs1042713 on Fasting Glucose Levels, Insulin Levels, and HOMA-IR
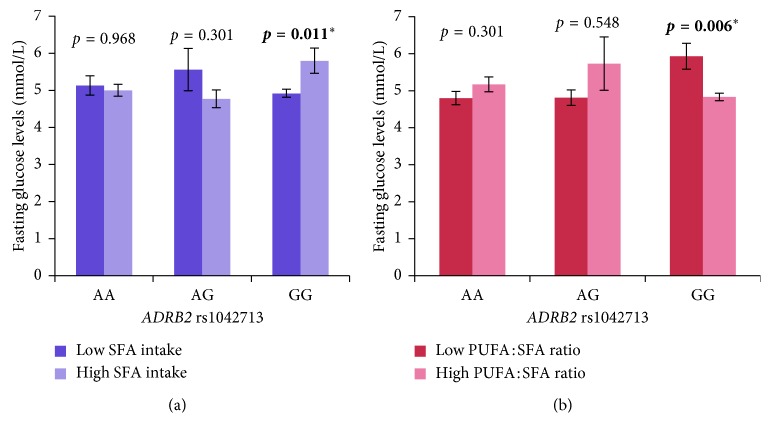
The multivariate general linear model was performed to investigate the effect of ADRB2 rs1042713 and dietary macronutrients on phenotypic variations. In our study, our results revealed that, irrespective of obesity, the carriers of GG genotype of rs1042713 had significantly lower fasting glucose levels with low SFA intake (<7.3% of TE/day) (FBG: 4.92 ± 0.1 mmol/L vs 5.80 ± 0.3 mmol/L, p=0.011) (Figure 1(a)) and high PUFA : SFA ratio (≥0.8/day) (FBG: 4.83 ± 0.1 mmol/L vs 5.93 ± 0.4 mmol/L, p=0.006) (Figure 1(b)), even after adjusting for covariates age, gender, BMI, physical activity status, smoking status, alcohol consumption, and total energy intake.
Figure 1.
The effect of the interaction between ADRB2 rs1042713 and (a) SFA intake and (b) PUFA : SFA ratio. SFA and PUFA : SFA ratios were dichotomised into two groups by using the median value of 7.3% of TE/day and 0.8/day (for all participants), respectively, for analysis. Gene-diet interaction was evaluated by using the multivariate general linear model after adjusting for age, gender, BMI, physical activity status, smoking status, alcohol consumption, and total energy intake. ∗ p < 0.05 was considered statistically significant. SFA: saturated fatty acid; PUFA: polyunsaturated fatty acid.
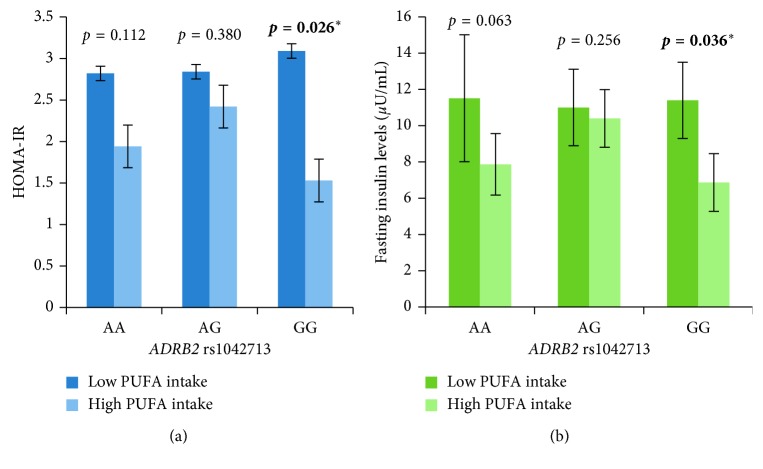
Moreover, the carriers of GG genotype of rs1042713 with high PUFA intake (≥6% of TE/day) had significantly lower HOMA-IR (1.5 ± 0.3 vs 3.0 ± 0.7, p=0.026) (Figure 2(a)) and fasting insulin levels (6.8 ± 1.6 µU/mL vs 11.4 ± 2.1 µU/mL, p=0.036) (Figure 2(b)) compared to low intake.
Figure 2.
The effect of the interaction between ADRB2 rs1042713 and PUFA intake on (a) HOMA-IR and (b) fasting insulin levels. PUFA intake was dichotomised into two groups by using the median value of 6% of TE/day (for all participants) for analysis. Gene-diet interaction was evaluated by using the multivariate general linear model after adjusting for age, gender, BMI, physical activity status, smoking status, alcohol consumption, and total energy intake. ∗ p < 0.05 was considered statistically significant. PUFA: polyunsaturated fatty acid; HOMA-IR: homeostatic model assessment-insulin resistance.
4. Discussion
Single-nucleotide polymorphism (SNP) in ADRB2 rs1042713 causes alterations in the structural conformation of the receptor which eventually affect the function of β-adrenergic receptors (ADRB) [27]. This may influence the binding of catecholamines to the beta-2 adrenoceptors and hence alter lipolysis. A meta-analysis involving 18 published articles revealed that there was no association between rs1042713 and obesity [28]. Contrary to the latter, studies on the Saudi population reported significant association between ADRB2 rs1042713 polymorphism and the development of obesity, as also with insulin resistance and dyslipidemia [10, 11]. However, findings from Asian populations (Japanese and Korean) reported negative association between obesity and ADRB2 gene polymorphisms [12, 13]. Moreover, these studies did not investigate interaction of gene variants, phenotypes, and dietary nutrients. To date, there are no data reported on ADRB2 rs1042713 in the Malaysian population. To the best of our knowledge, this is the first study that investigates the effect of ADRB2 rs1042713 on obesity and obesity-related metabolic parameters and its interaction with dietary nutrients in Malaysian adults.
4.1. Association between ADRB2 rs1042713 and Obesity and Insulin Resistance
In our study, we found no association between ADRB2 rs1042713 and odds of obesity (obesity was defined as BMI ≥ 27.5 kg/m2). However, ADRB2 rs1042713 was associated with insulin resistance (using a cutoff of HOMA-IR ≥ 1.7). We found that the carriers of G allele of rs1042713 had increased odds of insulin resistance compared to the noncarriers (AA), in the dominant model, even after adjusting for potential confounders. These findings suggest that variations in ADRB2 rs1042713 may interfere with glucose homeostasis and cause insulin resistance. Prior et al. reported that ADRB2 Arg16Gly–Gln27Glu haplotype was associated with glucose intolerance and insulin resistance in obese postmenopausal women [29]. A possible explanation for this observation could be the alteration in the structural conformation of the receptor, which may have enhanced sympathetic stimulation leading to increased lipolysis [8]. This overstimulation of ADRB2 is found to be associated with the pathogenesis of insulin resistance as it inhibits the insulin-induced translocation of GLUT4 and reduces glucose uptake via the cAMP-dependent protein kinase A-dependent pathways [30].
We found that HOMA-IR in the obese individuals carrying G allele of ADRB2 rs1042713 was above the diagnostic cutoff (1.7). It is now an established fact that higher levels of nonesterified fatty acids (NEFA) in the blood can induce preferential use of free fatty acids over glucose to generate ATP even in the presence of insulin in muscle and adipose tissue resulting in hyperglycaemia [31, 32]. Stimulation by noradrenaline of adipose tissue ADRB2 increases the release of NEFA. In addition, free fatty acids (FFAs) can stimulate hepatic gluconeogenesis and alter pancreatic insulin release and subsequent metabolism in individuals with impaired glucose metabolism [33].
4.2. Association between ADRB2 rs1042713 and Blood Lipid Levels
In the present study, we report that obese individuals carrying the G allele of rs1042713 had significantly higher total cholesterol, LDL cholesterol levels, and total cholesterol per HDL cholesterol ratio compared to the noncarriers (AA). Total cholesterol (5.7 ± 0.1 mmol/L) and LDL cholesterol levels (3.6 ± 0.1 mmol/L) in obese individuals carrying the G allele were above the diagnostic cutoffs (5.2 mmol/L and 2.6 mmol/L, respectively [26]). However, these differences in biochemical parameters were not observed in nonobese individuals. In our participants, excess FFAs in circulation in obese individuals carrying the gene variants of ADRB2 may have driven dyslipidemia. Increased lipolysis due to the polymorphisms of ADRB2 gene may have caused increased levels of NEFA induced hepatic production of VLDL and hence higher LDL levels in our participants [34].
4.3. Interaction between Dietary Fats and ADRB2 rs1042713 on Glycaemic Indices
We report that the level of fasting glucose was modulated by the types of dietary fatty acids such as SFA and PUFA in the carriers of GG genotype of ADRB2 rs1042713. The carriers of GG genotype of rs1042713 had significantly lower fasting glucose levels with intake of relatively higher PUFA:SFA ratio (≥0.8/day) (Figure 1(b)) and lower SFA intake (<7.3% of TE/day) (Figure 1(a)). Moreover, the carriers of GG genotype of rs1042713 had significantly lower fasting insulin levels and HOMA-IR when consuming higher PUFA intake (≥6% of TE/day).
These findings suggest that carriers of GG genotype of ADRB2 rs1042713 consuming higher percentage of PUFA demonstrated better homeostatic control of fasting blood glucose and insulin sensitivity. A meta-analysis of 102 randomised controlled feeding trials with 4,200 subjects has reported that PUFA had the most beneficial effects in improving glycaemia, insulin resistance, and insulin secretion in comparison to dietary carbohydrate, SFA, and MUFA [35]. The anti-inflammatory properties of PUFA increase the production of adiponectin via PPARα activation and alleviate adipose tissue inflammation via GPR120 and resolvins/protectins, which favour insulin sensitivity. It also suppresses oxidative stress and pancreatic lipotoxicity, reduces toxicity of tissue free fatty acids, and increases membrane fluidity [36]. This body of evidence indicates that the composition of dietary fatty acid intake plays an important role in affecting glucose metabolism and insulin sensitivity.
As per Malaysian recommendations, 10% of total energy should come from SFA and 3 to 8% of total energy should come from PUFA [23]. On an average, dietary intake of our participants with respect to SFA and PUFA were within these ranges. However, individuals with G allele of rs1042713 may need to consume higher quantity of PUFA to combat diet-related noncommunicable diseases. Replacing isocaloric quantity of foods rich in SFA with PUFA may improve homeostatic control of blood glucose and enhance insulin sensitivity (HOMA-IR) in such individuals.
5. Limitations
A major limitation of our study is the small sample size. We acknowledge that the current study is exploratory and is underpowered to detect the gene-diet interaction between ADRB2 rs1042713 and dietary nutrients on phenotypic and metabolic alterations in our population. However, with association and interaction analysis in the current study, we have generated a hypothesis. In future, large-scale studies are required to confirm such findings in the Malaysian population. In this study, we did not stratify our participants by gender for analysis due to the small sample size of male participants. To account for this, we have adjusted for gender statistically in all our data analysis to eliminate Type 1 error.
6. Conclusion
In conclusion, our study revealed that there was no association between ADRB2 rs1042713 and obesity. However, ADRB2 rs1042713 was associated with insulin resistance in Malaysian adults. The carriers of G allele of rs1042713 had increased odds of insulin resistance compared to noncarriers (AA). Obese individuals carrying the G allele of rs1042713 had significantly higher total cholesterol, LDL cholesterol levels, and total cholesterol per HDL cholesterol ratio compared to the noncarriers (AA). These differences were not observed in nonobese individuals. There is evidence from earlier studies that high PUFA intake is associated with favourable effects on glycaemia and insulin resistance. Over and above the latter, we found that higher PUFA intake was beneficial in individuals carrying the G allele with respect to glycaemic indices compared to the noncarriers. Although it is premature to report gene-diet interaction on the regulation of glucose and insulin levels in Malaysians, we suggest that higher quantity of PUFA-rich food sources in regular diet may benefit overweight and obese Malaysian adults metabolically. Large-scale studies are required to replicate and confirm the current findings in the Malaysian population.
Acknowledgments
The authors would like to express their gratitude to the participants of the study for their time and cooperation. The authors wish to thank the staff of the diagnostic center for their support and assistance with the blood analysis. The authors extend their appreciation to Leh Hui Eng for assistance on data collection. This study was funded by an internal grant from University of Nottingham Malaysia Campus (UNMC) (UNHB0008) and internal grants from University College Sedaya International (UCSI).
Abbreviations
- ADRB2:
Beta-2 adrenergic receptor
- AHA:
American Heart Association
- ANCOVA:
One-way analysis of covariance
- Arg:
Arginine
- BMI:
Body mass index
- BMR:
Basal metabolic rate
- cAMP:
Cyclic adenosine monophosphate
- CI:
Confidence interval
- DNA:
Deoxyribonucleic acid
- DSM-BIA:
Direct segmental multifrequency-bioelectrical impedance analysis method
- EDTA:
Ethylenediaminetetraacetic acid
- FFAs:
Free fatty acids
- GLM:
General linear model
- Gln:
Glutamine
- Glu:
Glutamic acid
- GLUT4:
Glucose transporter type 4
- Gly:
Glycine
- GPR120:
G-protein coupled receptor 120
- GWAS:
Genome-wide association studies
- HDL:
High-density lipoproteins
- HOMA:
Homeostatic model assessment
- HOMA-IR:
Homeostatic model assessment-insulin resistance
- hsCRP:
High-sensitivity C-reactive protein
- HWE:
Handy–Weinberg equilibrium
- LDL:
Low-density lipoproteins
- MET:
Metabolic equivalent
- MOH:
Ministry of health Malaysia
- MREC:
Medical Research and Ethics Committee
- MUFA:
Monounsaturated fatty acid
- NEFA:
Nonesterified fatty acids
- OR:
Odds ratio
- PCR:
Polymerase chain reaction
- PPARα:
Peroxisome proliferator-activated receptor alpha
- PUFA:
Polyunsaturated fatty acid
- SFA:
Saturated fatty acid
- SE:
Standard error of mean
- SNPs:
Single-nucleotide polymorphisms
- TE:
Total energy intake
- UCP-1:
Uncoupling protein 1
- UCSI:
University College Sedaya International
- UNMC:
University of Nottingham Malaysia Campus
- VLDL:
Very-low-density lipoproteins
- vs:
versus
- WC:
Waist circumference
- WHO:
World Health Organization
- WHR:
Waist hip ratio
- χ2:
Chi-square test.
Data Availability
The datasets generated and/or analysed during the present study are not publicly available, since ethical approval and participants' consent do not allow public sharing of data, but are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare that there are no conflicts of interest regarding the publication of this article.
Authors' Contributions
SRM designed the study; SRM and FA supervised and conducted data collection; PYT collected data and performed genotype analyses; PYT captured data and performed statistical analysis on the data under the supervision of SRM; SRM and PYT wrote the paper; all authors read and approved the final manuscript.
Supplementary Materials
Supplementary Table 1: physical activity and lifestyle of study participants.
References
- 1.Spiegelman B. M., Flier J. S. Obesity and the regulation of energy balance. Cell. 2001;104(4):531–543. doi: 10.1016/s0092-8674(01)00240-9. [DOI] [PubMed] [Google Scholar]
- 2.Speliotes E. K., Willer C. J., Berndt S. I., et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nature Genetics. 2010;42(11):937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arner P., Hoffstedt J. Adrenoceptor genes in human obesity. Journal of Internal Medicine. 1999;245(6):667–672. doi: 10.1046/j.1365-2796.1999.00495.x. [DOI] [PubMed] [Google Scholar]
- 4.Taylor M. R. Pharmacogenetics of the human beta-adrenergic receptors. Pharmacogenomics Journal. 2007;7(1):29–37. doi: 10.1038/sj.tpj.6500393. [DOI] [PubMed] [Google Scholar]
- 5.Large V., Hellström L., Reynisdottir S., et al. Human beta-2 adrenoceptor gene polymorphisms are highly frequent in obesity and associate with altered adipocyte beta-2 adrenoceptor function. Journal of Clinical Investigation. 1997;100(12):3005–3013. doi: 10.1172/jci119854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Masuo K., Katsuya T., Fu Y., Rakugi H., Ogihara T., Tuck M. L. β 2-adrenoceptor polymorphisms relate to insulin resistance and sympathetic overactivity as early markers of metabolic disease in nonobese, normotensive individuals. American Journal of Hypertension. 2005;18(7):1009–1014. doi: 10.1016/j.amjhyper.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 7.Masuo K., Mikami H., Ogihara T., Tuck M. L. Sympathetic nerve hyperactivity precedes hyperinsulinemia and blood pressure elevation in a young, nonobese Japanese population. American Journal of Hypertension. 1997;10(1):77–83. doi: 10.1016/s0895-7061(96)00303-2. [DOI] [PubMed] [Google Scholar]
- 8.Masuo K., Lambert G. W. Relationships of adrenoceptor polymorphisms with obesity. Journal of Obesity. 2011;2011:10. doi: 10.1155/2011/609485.609485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosmond R., Ukkola O., Chagnon M., Bouchard C., Bjorntorp P. Polymorphisms of the β 2-adrenergic receptor gene (ADRB2) in relation to cardiovascular risk factors in men. Journal of Internal Medicine. 2000;248(3):239–244. doi: 10.1046/j.1365-2796.2000.00721.x. [DOI] [PubMed] [Google Scholar]
- 10.Daghestani M. H., Warsy A., Daghestani M. H., et al. Arginine 16 glycine polymorphism in β 2-adrenergic receptor gene is associated with obesity, hyperlipidemia, hyperleptinemia, and insulin resistance in saudis. International Journal of Endocrinology. 2012;2012:8. doi: 10.1155/2012/945608.945608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daghestani M. H., Warsy A. A., Daghestani M. H., et al. The Gln27Glu polymorphism in β 2-adrenergic receptor gene is linked to hypertriglyceridemia, hyperinsulinemia and hyperleptinemia in Saudis. Lipids in Health and Disease. 2010;9(1):p. 90. doi: 10.1186/1476-511X-9-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayakawa T., Nagai Y., Kahara T., et al. Gln27Glu and Arg16Gly polymorphisms of the β 2-adrenergic receptor gene are not associated with obesity in Japanese men. Metabolism. 2000;49(9):1215–1218. doi: 10.1053/meta.2000.8622. [DOI] [PubMed] [Google Scholar]
- 13.Kim S.-H., Kim D.-J., Seo I. A., et al. Significance of β 2-adrenergic receptor gene polymorphism in obesity and type 2 diabetes mellitus in Korean subjects. Metabolism. 2002;51(7):833–837. doi: 10.1053/meta.2002.33347. [DOI] [PubMed] [Google Scholar]
- 14.Heianza Y., Qi L. Gene-diet interaction and precision nutrition in obesity. International Journal of Molecular Sciences. 2017;18(4):p. 787. doi: 10.3390/ijms18040787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fenech M., El-Sohemy A., Cahill L., et al. Nutrigenetics and nutrigenomics: viewpoints on the current status and applications in nutrition research and practice. Journal of Nutrigenetics and Nutrigenomics. 2011;4(2):69–89. doi: 10.1159/000327772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitra S. R., Tan P. Y., Amini F. Effect of FTO rs9930506 on obesity and interaction of the gene variants with dietary protein and vitamin E on C-reactive protein levels in multi-ethnic Malaysian adults. Journal of Human Nutrition and Dietetics. 2018;31(6):758–777. doi: 10.1111/jhn.12593. [DOI] [PubMed] [Google Scholar]
- 17.Wallace T. M., Levy J. C., Matthews D. R. Use and abuse of HOMA modeling. Diabetes Care. 2004;27(6):1487–1495. doi: 10.2337/diacare.27.6.1487. [DOI] [PubMed] [Google Scholar]
- 18.Friedewald W. T., Levy R. I., Fredrickson D. S. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clinical Chemistry. 1972;18(6):499–502. [PubMed] [Google Scholar]
- 19.Macho-Azcarate T., Marti A., González A., Martinez J. A., Ibañez J. Gln27Glu polymorphism in the beta2 adrenergic receptor gene and lipid metabolism during exercise in obese women. International Journal of Obesity. 2002;26(11):1434–1441. doi: 10.1038/sj.ijo.0802129. [DOI] [PubMed] [Google Scholar]
- 20.Namipashaki A., Razaghi-Moghadam Z., Ansari-Pour N. The essentiality of reporting hardy-weinberg equilibrium calculations in population-based genetic association studies. Cell Journal. 2015;17(2):187–192. doi: 10.22074/cellj.2016.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.WHO. Appropriate body-mass index for Asian populations and its implications for policy and intervention strategies. The Lancet. 2004;363(9403):157–163. doi: 10.1016/s0140-6736(03)15268-3. [DOI] [PubMed] [Google Scholar]
- 22.Yamada C., Moriyama K., Takahashi E. Optimal cut-off point for homeostasis model assessment of insulin resistance to discriminate metabolic syndrome in non-diabetic Japanese subjects. Journal of Diabetes Investigation. 2012;3(4):384–387. doi: 10.1111/j.2040-1124.2012.00194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.National Coordinating Committee on Food and Nutrition. Malaysian Dietary Guidelines 2010. Putrajaya, Malaysia: Ministry of Health Malaysia; 2010. [Google Scholar]
- 24.IDF. The IDF Consensus Worldwide Definition of the Metabolic Syndrome. Watermael-Boitsfort, Belgium: International Diabetes Federation; 2006. [Google Scholar]
- 25.Lee S., Choi S., Kim H. J., et al. Cutoff values of surrogate measures of insulin resistance for metabolic syndrome in Korean non-diabetic adults. Journal of Korean Medical Science. 2006;21(4):695–700. doi: 10.3346/jkms.2006.21.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.NCEP. Third report of the national cholesterol education program (NCEP) expert panel on detection, evaluation, and treatment of high blood cholesterol in adults (adult treatment panel III) final report. Circulation. 2002;106(25):3143–3421. doi: 10.1161/circ.106.25.3143. [DOI] [PubMed] [Google Scholar]
- 27.Green S. A., Turki J., Liggett S. B., Innis M. Amino-terminal polymorphisms of the human β 2-adrenergic receptor impart distinct agonist-promoted regulatory properties. Biochemistry. 1994;33(32):9414–9419. doi: 10.1021/bi00198a006. [DOI] [PubMed] [Google Scholar]
- 28.Zhang H., Wu J., Yu L. Association of Gln27Glu and Arg16Gly polymorphisms in beta2-adrenergic receptor gene with obesity susceptibility: a meta-analysis. PLoS One. 2014;9(6) doi: 10.1371/journal.pone.0100489.e100489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prior S. J., Goldberg A. P., Ryan A. S. ADRB2 haplotype is associated with glucose tolerance and insulin sensitivity in obese postmenopausal women. Obesity. 2011;19(2):396–401. doi: 10.1038/oby.2010.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mangmool S., Denkaew T., Phosri S., et al. Sustained βAR stimulation mediates cardiac insulin resistance in a PKA-dependent manner. Molecular Endocrinology. 2016;30(1):118–132. doi: 10.1210/me.2015-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koves T. R., Ussher J. R., Noland R. C., et al. Mitochondrial overload and incomplete fatty acid oxidation contribute to skeletal muscle insulin resistance. Cell Metabolism. 2008;7(1):45–56. doi: 10.1016/j.cmet.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 32.Zhang L., Keung W., Samokhvalov V., Wang W., Lopaschuk G. D. Role of fatty acid uptake and fatty acid beta-oxidation in mediating insulin resistance in heart and skeletal muscle. Biochimica et Biophysica Acta (BBA)-Molecular and Cell Biology of Lipids. 2010;1801(1):1–22. doi: 10.1016/j.bbalip.2009.09.014. [DOI] [PubMed] [Google Scholar]
- 33.Boden G., Shulman G. I. Free fatty acids in obesity and type 2 diabetes: defining their role in the development of insulin resistance and beta-cell dysfunction. European Journal of Clinical Investigation. 2002;32(S3):14–23. doi: 10.1046/j.1365-2362.32.s3.3.x. [DOI] [PubMed] [Google Scholar]
- 34.Jung U. J., Choi M.-S. Obesity and its metabolic complications: the role of adipokines and the relationship between obesity, inflammation, insulin resistance, dyslipidemia and nonalcoholic fatty liver disease. International Journal of Molecular Sciences. 2014;15(4):6184–6223. doi: 10.3390/ijms15046184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Imamura F., Micha R., Wu J. H. Y., et al. Effects of saturated fat, polyunsaturated fat, monounsaturated fat, and carbohydrate on glucose-insulin homeostasis: a systematic review and meta-analysis of randomised controlled feeding trials. PLOS Medicine. 2016;13(7) doi: 10.1371/journal.pmed.1002087.e1002087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coelho O. G. L., da Silva B. P., Rocha D. M. U. P., Lopes L. L., Alfenas R. C. G. Polyunsaturated fatty acids and type 2 diabetes: impact on the glycemic control mechanism. Critical Reviews in Food Science and Nutrition. 2015;57(17):3614–3619. doi: 10.1080/10408398.2015.1130016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1: physical activity and lifestyle of study participants.
Data Availability Statement
The datasets generated and/or analysed during the present study are not publicly available, since ethical approval and participants' consent do not allow public sharing of data, but are available from the corresponding author upon reasonable request.