Abstract
Cholangiocarcinoma (CCA) is a malignant cancer with an unknown etiology and an unfavorable prognosis. Most patients are diagnosed at an advanced stage, thus making it essential to find novel curative targets for CCA. Metabolic reprogramming of the tumor cells includes metabolic abnormalities in glucose (known as the Warburg effect) and other substances such as amino acids and fats. Metabolic reprogramming produces anti-oxidant substances, reduces tumor oxidative stress, and finally promotes the proliferation of tumors. There is increasing evidence to imply that SIRT2, a histone deacetylase, and its downstream target cMYC, play metabolic regulatory roles in tumor cells. However, the role of the SIRT2/cMYC pathway in CCA is unclear. To assess the metabolic reprogramming function of the SIRT2/cMYC pathway in CCA and to determine the downstream targets as well as evaluate the therapeutic effect, the CCA RNA-Seq data were downloaded from the TCGA database. Differentially expressed genes were confirmed and KEGG pathway enrichment analysis was performed. Overall, 48 paired CCA samples were collected and subjected to immunohistochemical detection, and the clinical characteristics of participants were summarized. The CCA cells were suppressed or overexpressed with different downstream targets of SIRT2 and then subjected to apoptosis, immunoblotting, seahorse, and metabolites tracing analysis. In vivo experiments were also performed. We found that the SIRT2/cMYC pathway contributed to the proliferation of CCA cells and confirmed that the downstream target is PHDA1 and the serine synthesis pathway. The up-regulated SIRT2 and cMYC levels resulted in low levels of mitochondrial oxidative phosphorylation and increased conversion of glucose to serine and led to poor patient survival. The highly active SIRT2/cMYC pathway up-regulated the serine synthesis pathway pyruvate and increased antioxidant production, thus consequently protecting the CCA cells from oxidative stress-induced apoptosis. Our data revealed that the SIRT2/cMYC pathway plays a critical role in transforming glucose oxidative metabolism to serine anabolic metabolism, thus providing antioxidants for stress resistance. SIRT2/cMYC-induced metabolic reprogramming may represent a new therapeutic target for treating CCA.
Introduction
Cholangiocarcinoma (CCA) is a malignant cancer with an unknown etiology and an unfavorable prognosis. The morbidity and mortality of CCA have increased year by year worldwide [1]. Because of limited diagnostic methodologies, the majority of patients are diagnosed at advanced stages and are not eligible for surgery, and many of these patients are further chemo-resistant to conventional chemotherapy [2]. Therefore, for the past three decades, the 5-year survival rates of CCA has remained only 10% [2]. Consequently, it is essential to find novel curative targets and therapeutic strategies for CCA.
To meet the high demand for rapid proliferation under different stressed conditions, cancer cells reprogram their metabolisms [3]. Known as the Warburg effect, this metabolic reprograming is marked by a conversion from oxidative phosphorylation to aerobic glycolysis [4], [5]. This metabolic reprogramming produces anti-oxidant substances, reduces tumor oxidative stress, thereby finally promoting the proliferation of tumors. These metabolic changes facilitate the accumulation of lactate and glycolytic intermediates to promote the growth and invasion of tumor. Hence, reversing the Warburg effect can be a potential mechanism for cancer therapy [6]. Changes in the expression of key enzymes that catalyze these biosynthesis pathways have been found in multiple tumor metabolic reprogramming [7]. Pyruvate dehydrogenase (PDH) complex is a multi-enzyme complex that regulates carbohydrate and fat metabolism, of which PDHA1 is the major component [8]. PDH can catalyze the irreversible decarboxylation of pyruvate into acetyl-CoA, which plays a crucial role in many biological reactions [9]. Abnormal expression of PDH has been found in a variety of tumors. Several studies have found that PDH activates the metabolism of cancer cells from glycolysis to glucose oxidation, which reduces the lactate production and increases reactive oxygen species (ROS), thus inducing the apoptosis and proliferation of tumor cells [10].
In the process of metabolic reprogramming, another important factor for cancer cell proliferation is the activation of the serine synthesis pathway (SSP) [11]. Serine is a non-essential amino acid in mammals, but plays an essential role in cancer progression [12]. Glycolytic metabolisms of glucose can be catalyzed by 3-phosphoglycerate dehydrogenase (PHGDH), phosphoserine aminotransferase (PSAT1), and phosphoserine phosphatase (PSPH) to produce serine. Therefore, PHGDH, PSAT1, and PSPH are considered important components of SSP. As a scavenger of ROS, serine contributes to the redox balance to promote the proliferation of cancer cells [13]. However, the specific role of SSP in cancer remains largely unexplored.
Post-translational modifications in metabolism regulation have attracted attention because of their ability to respond to changes in cellular metabolic status and regulate through upstream signal transduction pathways. Acetylation is a crucial post-translational modification found in the metabolic enzymes of cell regulation [14]. Deacetylases are classified into four groups (I-IV), wherein sirtuin (SIRTs) are NAD+-dependent class III histone deacetylases (HDACs). Among the seven SIRT homologues in mammals [15], SIRT2 catalyzes a variety of biological processes such as metabolism and gene expressions [16]. There is growing evidence that SIRT2 promotes proliferation in malignant tumors [16], [17]. Studies have demonstrated that SIRT2 inhibitors have anti-tumor, anti-diabetic, and anti-inflammatory effects [18]. Recent studies have shown that SIRT2 can inhibit the ubiquitin degradation of cMYC, thus contributing to the stability of cMYC and the proliferation of cancer. However, a SIRT2 inhibitor, thiomyristoyl (Tm), can inhibit the growth of cancer cells by degrading cMYC [19]. cMYC is an important transcription factor, which is up-regulated in numerous human tumors [20]. A recent research has further found that cMYC can stimulate SSP activation by transcriptionally up-regulating the expressions of SSP-related enzymes [21]. Together, these findings suggest that targeting the inhibition of SIRT2/cMYC may play a crucial role in cancer prevention and treatment. However, it remains unclear regarding whether the SIRT2/cMYC pathway has a synergistic effect on the metabolic reprogramming of CCA.
In the present study, we found that SIRT2/cMYC promoted metabolic reprogramming in CCA through two signaling pathways: 1) promoting PDHA1 phosphorylation (p-PDHA1) to inhibit oxidative phosphorylation; and 2) promoting serine synthesis by activating the SSP pathway to reduce apoptosis. The SIRT2/cMYC pathway plays a crucial role in transforming glucose oxidative metabolism to serine anabolic metabolism and thus provides antioxidants for stress resistance. SIRT2/cMYC-induced metabolic reprogramming may thus represent a new therapeutic target for treating CCA.
Materials and Methods
Ethics Approval and Consent to Participate
All experiments utilizing human samples and involving animals were reviewed and approved by the Ethical Committee and Animal Welfare Committee of Drum Tower Hospital, Nanjing University.
Cell Culture
The cells were maintained at 37°C with 5% CO2. Three human CCA cell lines were used: HIBEpic (ScienCell, Carlsbad, CA, USA), HuCCT1 (JCRB, Osaka, Japan) and RBE (The Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai, China). The cells were maintained in RPMI-1640 medium (Invitrogen, Waltham, MA, USA) containing 10% fetal bovine serum (Biological Industries, Cromwell, CT, USA), penicillin (Invitrogen) (100 U/ml), and streptomycin (Invitrogen) (100 U/ml). N-acetyl-L-cysteine (NAC, A7250) and CPI-613 (S2776) were purchased from Sigma-Aldrich (St. Louis, MO, USA) and Selleck (Houston, TX, USA), respectively. Tm (MCE, Monmouth Junction, NJ, USA) was commercially purchased.
Cell Transfection
SiRNAs targeting cMYC (5′-GUCAAGAGGCGAACACACA-3′), PDHA1 (5′-CCGAATGGAGTTGAAAGCAGAT-3′), and negative control siRNA were purchased from RiboBio (Guangzhou, China). The full-length cMYC and PDHA1 plasmids were gifts from the Zhao lab of Fudan University (Shanghai, China). HuCCT1 and RBE cells were transfected using Lipofectamine RNAiMax reagent (Invitrogen) according to the manufacturer’s instructions.
Cell Viability Assay
Cell viability was investigated by CCK-8 assay in 96-well plates (5×103 cells/well). Different concentrations of Tm or 0.1% dimethyl sulfoxide (DMSO) were added at indicated times. A total of 10 μl CCK-8 solutions (Dojindo, Minato-ku, Tokyo, Japan) was added to each well and they were incubated at 37 °C for 1 h. Absorbance was recorded at 450 nm. For each experimental trial, wells were conducted in triplicate and each well was assayed in triplicate.
Migration and Invasion Assays
Cellular motility and invasive abilities were determined using Transwell (Corning Life Sciences, Bedford, MA, USA) and Matrigel invasion (BD Biosciences, San Jose, CA, USA), respectively. For the transwell migration and invasion assay, 5 × 104 cells and 2 × 105 cells were seeded, respectively. The cells that migrated to the underside of the membrane were fixed and stained with 0.1% crystal violet, and 10 microscopic fields were counted. The average values of the migrating or invading cells were expressed as percentages. Each experiment was performed in replicate inserts, and the average value was calculated from three independent experiments.
Colony Formation Assay
CCA cells were counted and seeded in 6-well plates with 500 cells/well. After treatment with 25 μM Tm for 24 hours, the Tm was removed and normal medium was added for 14 days. The cells were fixed in methanol and colonies were counted after staining with 0.1% crystal violet in 25% methanol for 20 mins. The results were presented as the average number of counted colonies per well under each condition. All the experiments were performed in triplicate and repeated three times independently.
Apoptosis Assay
Apoptosis was detected by flow cytometry using the Annexin V-fluorescein isothiocyanate (FITC) Apoptosis Detection Kit (556, 547, BD Biosciences), following the manufacturer’s instructions.
Western Blotting
The cells were lysed using RIPA buffer (150 mM NaCl, 50 mM Tris-HCl at pH 7.4, 1 mM EDTA, 0.1% SDS, 1% Triton X-100, 1% sodium deoxycholate and 1% NP-40) mixed with a protease and phosphatase inhibitor cocktail (Roche Diagnostics GmbH, Mannheim, Germany) and phenylmethylsulfonyl fluoride (PMSF) (Biosharp, Hefei, China) for 15 min on ice. The proteins were subjected to western blotting according to standard protocols. Antibodies were as follows: SIRT2 (HPA011165,Sigma), SIRT2 (s8447, Sigma), cMYC (ab32072, Abcam, Cambridge, UK), p293-PDHA1 (ab92696, Abcam), GAPDH (ab128915, Abcam), cleaved-caspase3 (9664s, CST, Danvers, MA, USA), cleaved-PARP (5625s, CST), PDHA1 (ab110334, Abcam), PHGDH (14719-1-AP, Proteintech, Chicago, IL, USA), PSAT1 (10501-1-AP, Proteintech), PSPH (14513-1-AP, Proteintech), HRP-conjugated anti-rabbit (7074, CST), and anti-mouse antibodies (7076, CST).
Metabolite Analysis
Metabolism experiments were carried out as described previously [22]. Metabolite extracts were collected. A total of 2 μl of metabolite extracts were injected for gas chromatography-mass spectrometer (GC-MS) analysis using an Agilent 6980 GC coupled to an Agilent 5973 MS system. Relative metabolite abundances were investigated by comparing the abundance of each metabolite with internal standards and cell protein standards.
Mitochondrial Oxidative Phosphorylation Analysis
The real-time oxygen consumption rate (OCR) was detected using an XF96 Extracellular Flux Analyzer (Seahorse Bioscience, North Billerica, MA, USA). According to the manufacturer’s instructions, the cells were seeded at 1x104 cells/well in 96-well XF cell culture microplates and incubated at 37 °C for 24 h. A total of 25 μM Tm was added to each well of each plate. Before measurement, the medium was replaced with 175 μl/well XF-96 running medium (supplemented with RPMI-1640 without serum) and pre-incubated at 37°C without CO2 for 20 min. For each analysis, different compounds that modulate mitochondrial respiration were injected into each well, according to standard protocols: for the OCR – oligomycin, carbonylcyanide p-trifluoromethoxy-phenylhydrazone, rotenone and antimycin A; for the extracellular acidification rate (ECAR) – glucose, oligomycin and 2-deoxy-D-glucose. Cell number was used for data normalization. The OCR was expressed as picomoles/minute (pmol/min). The ECAR was expressed as milli-pH units per minute (mpH/min).
ROS and Glutathione Measurement.
For cellular ROS the analyses, 3 × 105 cells/well were seeded into a six-well plate and different concentrations of Tm were added at the indicated times. After treatment, the redox-sensitive probe DCF (5 μM) was incubated for 30 min. Fluorescence intensity was measured immediately by flow cytometry analysis in the six-well plate-treated cells. The levels of GSH and GSSG were measured in the cells or tumor lysates according to the GSH and GSSG Assay Kit instructions (S0053, Beyotime Biotechnology, Jiangsu, China). The experiments were conducted in triplicate and repeated three times independently.
Differentially expressed genes (DEGs) of Paired-CCA from the Cancer Genome Atlas (TCGA) Data
The CCA RNA-Seq data was downloaded from the TCGA database using the GDC Data Portal at https://gdc-portal.nci.nih.gov. The mRNA expression data included 18 samples consisting of 9 normal samples and 9 matched CCA samples. The sequencing data were public, and ethical issues were not relevant. The edgeR package in Bioconductor was used to screen the DEGs in the CCA and normal tissue samples. The edgeR package is based on a negative binomial (NB) distribution, which corrects the problem of overdispersion in RNA-Seq data using the Poisson model and the Bayes procedure. Data with a value of zero were deleted. If |FoldChange| >2, the genes were considered to be DEGs with a P value <.01 and a false discovery rate (FDR) <0.05.
Functional Annotation
The Database for Annotation Visualization and Integrated Discovery (DAVID) online tool (https://david.ncifcrf.gov/) was used to perform functional and pathway enrichment analyses in our study. GO and KEGG pathway enrichment assays were performed to detect the potential biological functions and pathways of the high- and low-expression genes in CCA.
Immunohistochemistry
Two to five human CCA specimens from one patient were used in the immunohistochemistry (IHC) study. Tumor specimens from humans and mice were fixed in 4% paraformaldehyde and embedded in paraffin. Except for target detection, the IHC followed standard procedures. The intensity of staining in each tissue was evaluated by multiplying the staining’s range and intensity [23].
CCA Cancer Xenograft Model
Female nude mice were subcutaneously injected in the flanks with 100 μL PBS or 2 × 106 HuCCT1 cells suspended in 100 μL Matrigel. The mice were allowed to recover after the injections and monitored for three weeks. Tumors were measured using a caliper and once the majority of tumors reached a threshold size of 200 mm3, intraperitoneal (IP) injections of DMSO or Tm were initiated. IP injections of 1.5 mg Tm in 50 μL DMSO were administered once a day. After one month of treatment, or if the mice met the humane endpoint criteria, they were euthanized via CO2 asphyxiation. Tissues were collected, fixed with 10% neutral formaldehyde, embedded in paraffin, sectioned, and stained with hematoxylin and eosin (HE). Serum, organs, and tumor tissues were frozen in liquid nitrogen and then stored at -80°C for subsequent analyses.
Statistics
All the data were presented as means ± SEM. The data were analyzed using one-way ANOVA followed by post hoc Duncan tests (GraphPad Prism version 5.0, GraphPad Software, La Jolla, CA, USA). P < .05 was considered significant.
Results
SIRT2, cMYC, and p-PDHA1 contributed to CCA metabolic reprogramming and were associated with poor prognosis
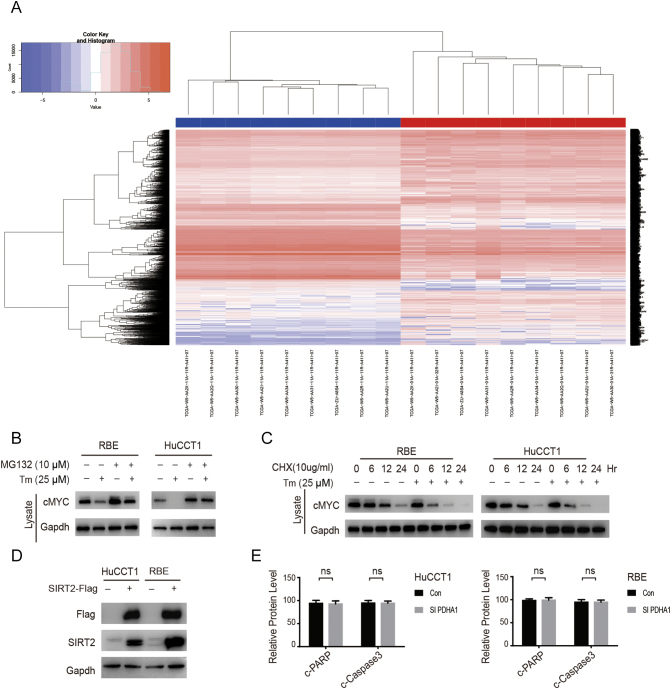
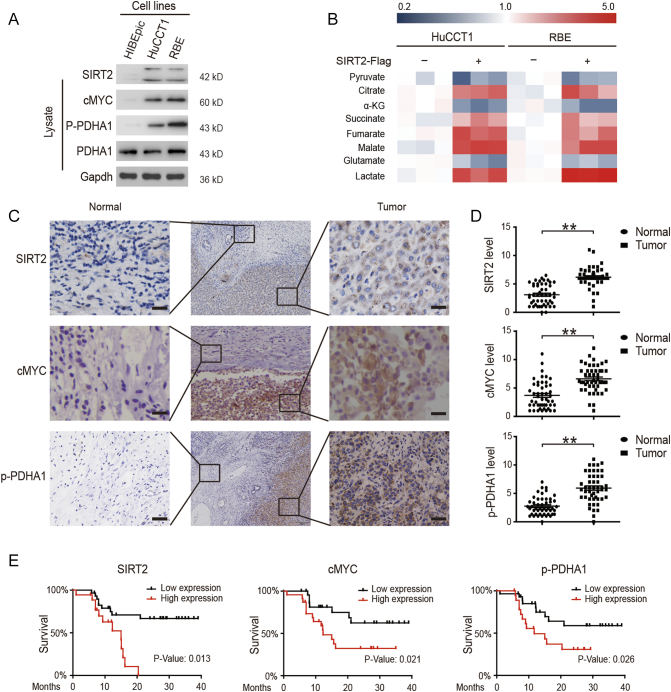
Metabolic reprogramming can be regulated through the expression of multiple genes to accelerate the malignant behavior of tumor cells [24]. Therefore, differentially expressed genes (DEGs) in CCA were analyzed using data from the Cancer Genome Atlas (TCGA) and the most relevant DEGs associated with metabolism and apoptosis were confirmed (sFigure 1A). As a transcription factor and downstream protein of SIRT2, cMYC has attracted extensive interest for its potential role in tumorigenesis in many different human cancers – especially for its role in regulating the expression of metabolic genes such as lactate dehydrogenase (LDH) and muscle-specific pyruvate kinase 2 (PKM2) [25], [26]. Furthermore, PDH is a gatekeeper of tricarboxylic acid cycle (TCA) and is always highly phosphorylated, thereby leading to an inactive state and contributing to the Warburg effect [27]. In order to identify the relevant protein targets of metabolic reprogramming, we first screened the cMYC-related targets in a normal bile duct epithelial cell line (Hibepicc) and two human CCA cell lines (HuCCT1 and RBE). We confirmed high levels of p-PDHA1, SIRT2, and its upstream target cMYC in CCA cells (Figure 1A). Because the Warburg effect is the key feature of tumor metabolism, the metabolic reprogramming changes were then evaluated by over-expressing SIRT2 in CCA cells. We found that while SIRT2 decreased the levels of TCA-related metabolites, it increased the level of lactate, thereby promoting the Warburg effect (Figure 1A and sFigure 1B). Overall, 48 CCA samples were used and the clinical characteristics were summarized in Table 1. We found that the expressions levels of SIRT2, cMYC, and p-PDHA1 were elevated in CCA tissues compared to those in adjacent tissues (Figure 1, C and D). In addition, these elevated expressions were associated with lower survival rates (Figure 1E). Collectively, these results revealed that high levels of SIRT2, cMYC, and p-PDHA1 contributed to CCA metabolic reprogramming and were associated with a poor prognosis.
Supplementary Figure 1.
(A) Differentially expressed genes (DEGs) in CCA were analyzed using data from The Cancer Genome Atlas (TCGA). (B & C) CCA cells were treated with or without indicated Tm, MG132 or CHX for 24 hours. Cells were harvested and lysed, and endogenous cMYC was visualized by Western blotting. Data represent mean ± SEM, n ≥ 3.
Figure 1.
High expression levels of SIRT2, cMYC, and p-PDHA1 were found in CCA and are associated with poor prognosis.
(A) The expression levels of SIRT2, cMYC, and p-PDHA1 in a normal bile duct epithelial cell line (HIBEpic) and two human CCA cell lines (HuCCT1 and RBE) were determined by western blotting. (B) CCA cells were overexpressed with either SIRT2 plasmids or control vectors; the cells were then collected and lysed and the concentrations of metabolites were measured by MS. (C & D) The SIRT2, cMYC, and p-PDHA1 protein levels in the tumors and adjacent tissues from 60 CCA patients were detected by IHC and quantified. The magnification is ×200. Scale bars, 100 μm. (E) The 3-year survival was evaluated in CCA patients with SIRT2, cMYC, and p-PDHA1 protein expressions, respectively. Data represent mean ± SEM, n ≥ 3. **P < .01.
Table 1.
Clinical characteristics of patients with cholangiocarcinoma
| n | Low (%) | High (%) | X2 | P value | |
|---|---|---|---|---|---|
| cMYC expression, n (%) | |||||
| Gender | 0.333 | .564 | |||
| Male | 24 | 11 (45.8) | 13 (54.2) | ||
| Female | 24 | 13 (54.2) | 11 (45.8) | ||
| Age | 1.371 | .242 | |||
| ≤60 | 20 | 8 (40.0) | 12 (60.0) | ||
| >60 | 28 | 16(57.1) | 12 (42.9) | ||
| Location | 0.785 | .675 | |||
| Intrahepatic | 19 | 8 (42.1) | 11 (57.9) | ||
| Hepatic portal | 9 | 5 (55.6) | 4 (44.4) | ||
| Extrahepatic | 20 | 11 (55.0) | 9 (45.0) | ||
| Size (mm) | 0.403 | .525 | |||
| ≥40 | 14 | 6 (42.9) | 8 (57.1) | ||
| <40 | 34 | 18 (52.9) | 16 (47.1) | ||
| Differentiation | 6.155 | .046 | |||
| Well | 16 | 4 (25.0) | 12 (75.0) | ||
| Medium | 28 | 18 (64.3) | 10 (35.7) | ||
| Poor | 4 | 2 (50.0) | 2 (50.0) | ||
| T Stage | 0.949 | .330 | |||
| T1~T2 | 35 | 19 (54.3) | 16 (45.7) | ||
| T3~T4 | 13 | 5 (38.5) | 8 (61.5) | ||
| Lymph node metastasis | 0.403 | .525 | |||
| Negative | 34 | 18 (52.9) | 16 (47.1) | ||
| Positive | 14 | 6 (42.9) | 8 (57.1) | ||
| Distant metastasis | 1.091 | .296 | |||
| Negative | 44 | 23 (50.0) | 21 (50.0) | ||
| Positive | 4 | 1 (50.0) | 3 (50.0) | ||
| Venous invasion | 0.085 | .771 | |||
| Negative | 27 | 14 (51.9) | 13 (48.1) | ||
| Positive | 21 | 10 (47.6) | 11 (52.4) | ||
| Nerve invasion | 1.613 | .204 | |||
| Negative | 14 | 5 (35.7) | 9 (64.3) | ||
| Positive | 34 | 19 (55.9) | 15 (44.1) | ||
| SIRT2 expression, n (%) | |||||
| Gender | 0.784 | .376 | |||
| Male | 24 | 13 (54.2) | 11 (45.8) | ||
| Female | 24 | 16 (66.7) | 8 (33.3) | ||
| Age | 0.002 | .960 | |||
| ≤60 | 20 | 12 (60.0) | 8 (40.0) | ||
| >60 | 28 | 17 (60.7) | 11 (39.3) | ||
| Location | 0.843 | .656 | |||
| Intrahepatic | 19 | 13 (68.4) | 6 (31.6) | ||
| Hepatic portal | 9 | 5 (55.6) | 4 (44.4) | ||
| Extrahepatic | 20 | 11 (63.6) | 9 (36.4) | ||
| Size (mm) | 0.089 | .766 | |||
| ≥40 | 14 | 8 (57.1) | 6 (42.9) | ||
| <40 | 34 | 21 (61.8) | 13 (38.2) | ||
| Differentiation | 0.463 | .793 | |||
| Well | 16 | 9 (56.3) | 7 (43.8) | ||
| Medium | 28 | 18 (64.3) | 10 (35.7) | ||
| Poor | 4 | 2 (50.0) | 2 (50.0) | ||
| T Stage | 6.553 | .010 | |||
| T1~T2 | 35 | 25(71.4) | 10 (28.6) | ||
| T3~T4 | 13 | 4 (30.8) | 9 (69.2) | ||
| Lymph node metastasis | 0.089 | .766 | |||
| Negative | 34 | 21 (61.8) | 13 (38.2) | ||
| Positive | 14 | 8 (57.1) | 6 (42.9) | ||
| Distant metastasis | 6.660 | .010 | |||
| Negative | 44 | 29 (65.9) | 15 (34.1) | ||
| Positive | 4 | 0 (0.0) | 4 (100.0) | ||
| Venous invasion | 0.167 | .683 | |||
| Negative | 27 | 17 (63.0) | 10 (37.0) | ||
| Positive | 21 | 12 (57.1) | 9 (42.9) | ||
| Nerve invasion | 1.002 | .317 | |||
| Negative | 14 | 10 (71.4) | 4 (28.6) | ||
| Positive | 34 | 19 (55.9) | 15 (44.1) | ||
| Gender | 4.269 | .039 | |||
| SIRT2 expression, n (%) | |||||
| Male | 24 | 11 (45.8) | 13 (54.2) | ||
| Female | 24 | 18 (75.0) | 6 (25) | ||
| Age | 3.407 | .065 | |||
| ≤60 | 20 | 9 (45.0) | 11 (55.0) | ||
| >60 | 28 | 20 (71.4) | 8 (28.6) | ||
| Location | 3.220 | .200 | |||
| Intrahepatic | 19 | 10 (52.6) | 9 (47.4) | ||
| Hepatic portal | 9 | 4 (44.4) | 5 (55.6) | ||
| Extrahepatic | 20 | 15 (75.0) | 5 (25.0) | ||
| Size (mm) | 0.897 | .344 | |||
| ≥40 | 14 | 7 (50.0) | 7 (50.0) | ||
| <40 | 34 | 22 (64.7) | 12 (35.3) | ||
| Differentiation | 3.388 | .184 | |||
| Well | 16 | 7 (43.8) | 9 (56.3) | ||
| Medium | 28 | 20 (71.4) | 8 (28.6) | ||
| Poor | 4 | 11(50.0) | 10 (50.0) | ||
| T Stage | 4.365 | .037 | |||
| T1~T2 | 35 | 18 (48.6) | 17 (51.4) | ||
| T3~T4 | 13 | 11 (84.6) | 2 (15.4) | ||
| Lymph node metastasis | 2.859 | .091 | |||
| Negative | 44 | 25 (56.8) | 19 (43.2) | ||
| Positive | 4 | 4 (100.0) | 0 (0.0) | ||
| Distant metastasis | 0 | 1 | |||
| Negative | 56 | 28 (50.0) | 28 (50.0) | ||
| Positive | 4 | 2 (50.0) | 2 (50.0) | ||
| Venous invasion | 0.167 | .683 | |||
| Negative | 27 | 17 (63.0) | 10 (37.0) | ||
| Positive | 21 | 12 (57.1) | 9 (42.9) | ||
| Nerve invasion | 5.043 | .025 | |||
| Negative | 14 | 5 (35.7) | 9 (64.3) | ||
| Positive | 34 | 24 (70.6) | 10 (29.4) | ||
PDHA1 is the Downstream Target of SIRT2/cMYC, But Is Unnecessary for CCA Apoptosis
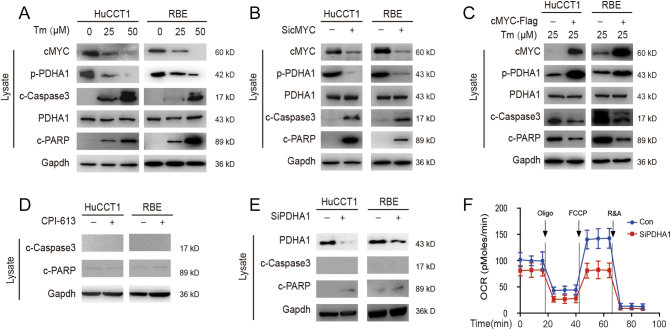
PDHA1 is the downstream target of SIRT2/cMYC, but is unnecessary for CCA apoptosis. As a well-known metabolic regulator, cMYC can be stabilized by the deacetylation activity of SIRT2 [19]. We confirmed that SIRT2 stabilized cMYC protein through post-translational modification in CCA (sFigure 1B and 1C). To further investigate how SIRT2 regulates metabolic reprogramming, Tm (a SIRT2 inhibitor) was added to CCA cells. After the inhibition of SIRT2, cMYC and p-PDHA1 decreased while apoptosis-related proteins (c-Caspase 3 and c-PARP) increased (Figure 2A). Furthermore, after knocking-down cMYC in CCA cells, we found that p-PDHA1 and apoptosis-related proteins decreased (Figure 2B). Also, rescue experiments showed over-expressing cMYC help CCA cells resist apoptosis in the presence of TM (Figure 2C). This suggests that PDHA1 is the downstream of the SIRT2/cMYC pathway. Conversely, neither CPI613 (a PDHA1 inhibitor) nor PDHA1 knockdown could significantly change the levels of apoptotic proteins in CCA cells (Figure 2, D and E). Additionally, the inhibition of PDHA1 led to a reduction of the OCR, which suggests a decrease in TCA (Figure 2F). The aforementioned results together suggest that PDHA1 is the downstream target of SIRT2/cMYC by regulating glucose metabolism in CCA. The inhibition of PDHA1 alone was insufficient to promote apoptosis in CCA.
Figure 2.
PDHA1 is the downstream target of SIRT2/cMYC.
(A) HuCCT1 and RBE cells were treated with 0, 25, or 50 μm Tm and subjected to western blotting. (B & C & D & E) HuCCT1 and RBE cells were transfected with siRNA of cMYC, plasmids of cMYC, CPI-613, Tm, or siRNA of PDHA1, respectively, and subjected to western blotting. (F) The OCRs were detected at different time points. The OCR was under oligomycin, carbonyl cyanide-m-chlorophenylhydrazone (FCCP), and antimycin A/rotenone treatments, respectively. Data represent mean ± SEM, n ≥ 3.
SIRT2 Inhibition Reduces CCA Biological Activity
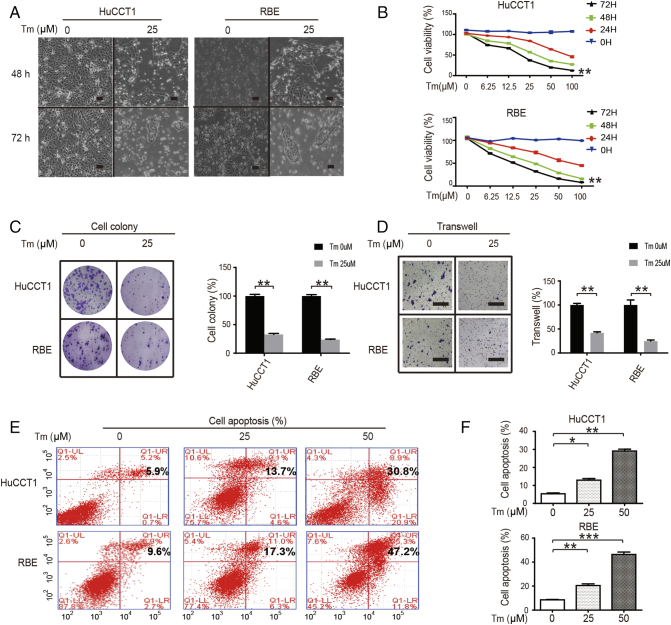
To observe the effects of SIRT2 inhibitor on the biological activity of CCA, HuCCT1 and RBE cells were treated with indicated Tm. After the inhibition of SIRT2, CCA cells showed marginal differences in phenotypes (Figure 3A). The cell growth was consistently affected after Tm treatment (Figure 3B). In accordance with these results, colony formation in CCA cells decreased when treated with Tm (Figure 3C). Additional transwell assays were then performed, and when SIRT2 was inhibited, a reduction in cell migration and invasion was observed (Figure 3D). Next, flow cytometry was used to study the mechanism by which Tm inhibited CCA cell proliferation. It was found that Tm enhanced apoptosis in CCA cells (Figure 3, E and F). This indicates that Tm, the SIRT2 inhibitor, can reduce the biological activity of CCA cells.
Figure 3.
SIRT2 inhibitor reduces CCA biological activity.
(A) HuCCT1 and RBE cells were treated with or without 25 μM Tm for 24 h or 72 h and the morphological changes were observed. Scale bars, 100 μm. (B) CCA cells were treated with 0-100 μM Tm for 0-72 h, and the cell proliferation was then quantified via CCK-8 assays. (C) CCA cells were treated with or without 25 μM Tm; colonies were stained with crystal violet (left) and quantified (right). Scale bars, 1 cm. (D) CCA cells were treated with or without 25 μM Tm; cell invasion was quantified. (E & F) CCA cells were treated with 0, 25, or 50 μM Tm; apoptotic cells were measured and quantified using flow cytometry. Data represent mean ± SEM, n ≥ 3. *P < .05, **P < .01, and ***P < .001.
The SIRT2/cMYC Pathway has Antioxidant Effects on CCA by Promoting the Downstream SSP Pathway
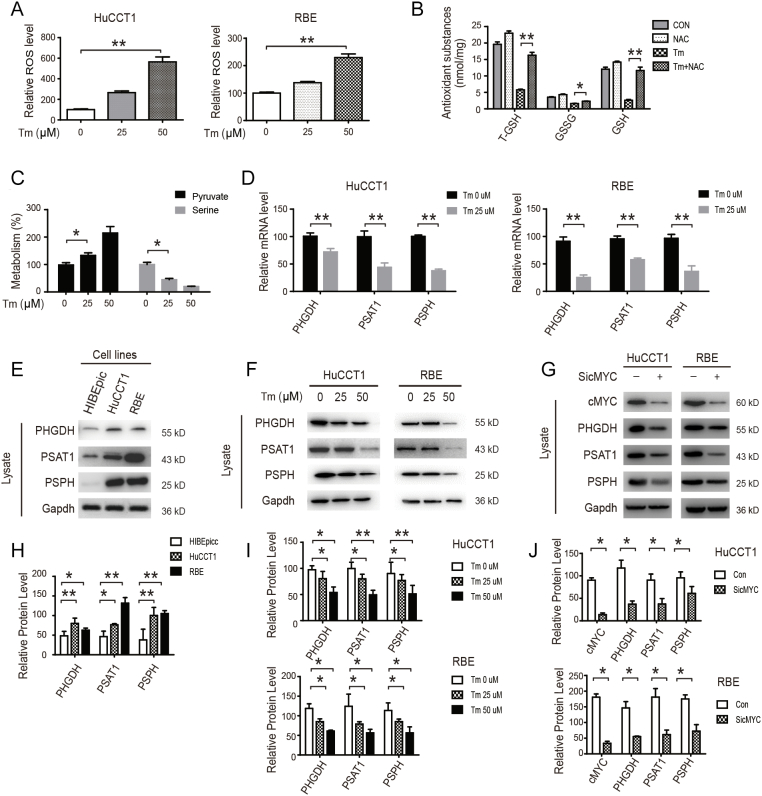
The imbalance between the antioxidants and pro-oxidants caused by superfluous reactive oxygen species (ROS) can promote apoptosis [28]. On the other hand, SSP plays a key role in antioxidant (T-GSH/GSSG/GSH) production and anti-apoptotic processes [29]. Consequently, we investigated whether Tm-induced apoptosis is related to ROS production. We found that Tm increased ROS level in a concentration-dependent manner (Figure 4A). More importantly, Tm decreased antioxidant (T-GSH/GSSG/GSH) levels, which can be reversed by NAC (acetylcysteine, a known antioxidant) (Figure 4B). We also found that Tm treatment decreased serine synthesis and the SSP pathway (Figure 4, C and D). Our results further revealed that Tm treatment inhibited the SSP pathway in both mRNA and protein levels (Figure 4, D–F, H and I). Similar to Tm treatment, cMYC knockdown also significantly decreased the levels of SSP pathway proteins in CCA cells (Figure 4, G and J). These results suggest that the SIRT2/cMYC pathway inhibited oxidative stress by promoting the SSP pathway and antioxidant production.
Figure 4.
SIRT2/cMYC has antioxidant effects on CCA by promoting the downstream SSP pathway.
(A) HuCCT1 and RBE cells were treated with 0, 25, or 50 μM Tm and the levels of ROS were detected. (B) CCA cells were treated with NAC, Tm, or a combination of both. The levels of T-GSH, GSSG, and GSH were detected. (C & D) CCA cells were treated with or without indicated Tm. The levels of pyruvate and serine were detected by MS. The relative mRNA levels of PHGDH, PSAT1, and PSPH were detected by qPCR. (E) The expressions of PHGDH, PSAT1, and PSPH in the HIBEpic and CCA cells were determined by western blotting. (F) CCA cells were treated with 0, 25, or 50 μM Tm and the levels of PHGDH, PSAT1, and PSPH were detected by Western blotting. (G) CCA cells were transfected with siRNA of cMYC and subjected to western blotting. Data represent mean ± SEM, n ≥ 3. *P < .05 and **P < .01
SIRT2/cMYC Pathway Inhibits Oxidative Stress-Related Apoptosis by Promoting Antioxidants Production
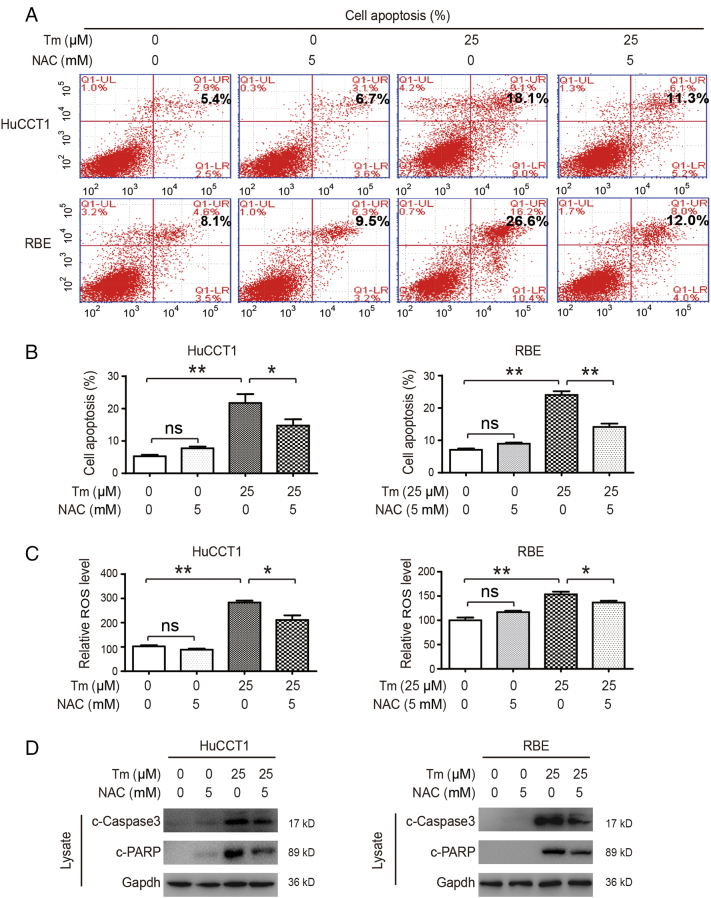
Studies have found that both lowering glycolytic metabolism and promoting antioxidant production can lead to tumor apoptosis [28], [30]. As previously demonstrated, the SIRT2/cMYC pathway promotes metabolic reprogramming in CCA through two signaling pathways: 1) promoting p-PDHA1 to inhibit oxidative phosphorylation and 2) promoting serine synthesis by activating the SSP pathway to increase antioxidant production. To evaluate whether the SSP pathway plays a key role in CCA cell apoptosis, NAC (an antioxidant) was used in combination with Tm. Our results revealed that NAC treatment alone did not affect apoptosis in CCA cells, though Tm treatment alone was able to promote apoptosis. The combination of Tm and NAC, however, led to a significant decrease in apoptosis as compared with Tm used alone (Figure 5, A and B). Accordingly, ROS levels did not change in the NAC-treated cells and increased in the Tm-treated cells, and the combination of Tm and NAC led to a decrease in ROS compared to Tm used alone (Figure 5C). Detection of apoptosis proteins showed similar results (Figure 5D), suggesting that the SIRT2/cMYC pathway reduced oxidative stress-induced apoptosis by promoting the downstream SSP pathway.
Figure 5.
The SIRT2/cMYC pathway inhibits oxidative stress-related apoptosis by promoting antioxidant production.
(A & B) After treatment with Tm, NAC, or a combination of both, CCA cells were analyzed using Annexin V/PI staining and apoptosis was measured by flow cytometry. Percentages of early and late apoptotic cells were quantified. (C & D) After treatment with Tm, NAC, or a combination of both, the levels of ROS and proteins were detected. Data represent mean ± SEM, n ≥ 3. *P < .05 and **P < .01.
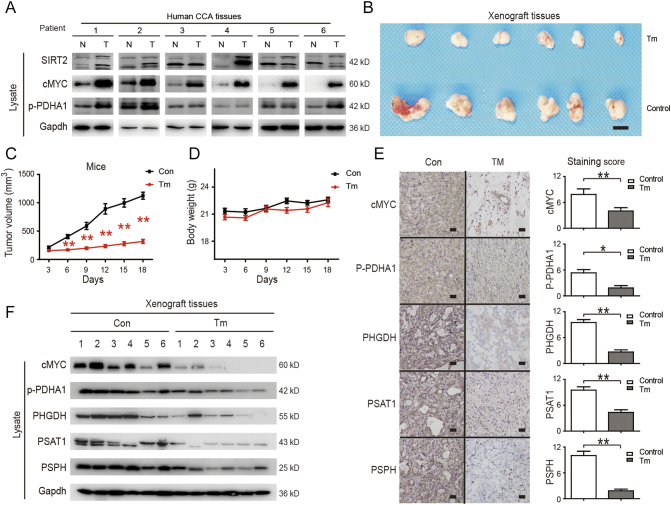
SIRT2 Inhibitor has an Antitumor Effect on CCA In Vivo
Finally, the expression levels of SIRT2, cMYC, and p-PDHA1 were assessed in fresh human CCA tissues. We found that these proteins significantly increased in tumor tissues as compared to that in adjacent tissues (Figure 6A). To further validate our findings, the anti-tumor effects of Tm were evaluated in vivo using a CCA cell tumor xenograft model. We observed that the Tm treatment group significantly inhibited tumor growth (Figure 6, B and C) without affecting mouse body weight (Figure 6D), suggesting that Tm treatment was safe in vivo. Lastly, the expression levels of these proteins were assessed in fresh xenograft tissues by both staining and western blotting, and we found that cMYC, p-PDHA1, PHGDH, PSAT1, and PSPH were markedly down-regulated following Tm treatment (Figure 6, E and F). These findings indicated that by inhibiting SIRT2 activity, Tm can degrade cMYC to decrease p-PDHA1 and SSP, thus exerting an anti-tumor effect in vivo.
Figure 6.
SIRT2 inhibitor has an anti-tumor effect on CCA in vivo.
(A) The levels of cMYC, p-PDHA1, and SIRT2 proteins in tumors and adjacent normal tissues from 6 donors were detected by western blotting. (B) HuCCT1 cells were injected subcutaneously into the flanks of nude mice. When tumors were palpable, DMSO (control) or Tm were administered. Tumors were collected and photographed. Scale bars, 1 cm. (C) Tumor volumes of mice were measured. (D) The body weights of mice were compared among the groups. (E & F) cMYC, p-PDHA1, PHGDH, PSAT1 and PSPH protein levels in xenograft tissues from control and Tm group were detected by staining and Western blotting. Data represent mean ± SEM, n ≥ 3. *P < .05 and **P < .01.
Discussion
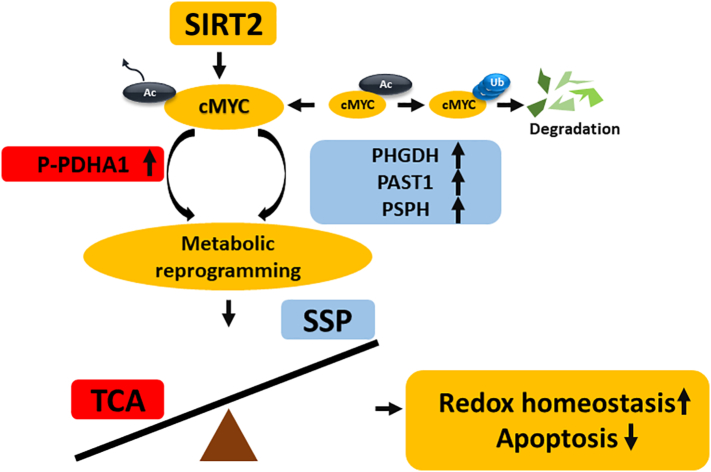
Metabolic reprogramming of tumor cells includes abnormal metabolism in glucose and other substances such as amino acids and fats. Among these, the metabolic reprogramming of glucose is called the Warburg effect. Nonetheless, the question of how metabolites in metabolic reprogramming help induce tumorigenesis and which proteins are key targets for this metabolism remains unanswered. In this study, we identified the promoting role of the SIRT2/cMYC pathway in CCA, identified its downstream targets, and evaluated its therapeutic effect (Figure 7).
Figure 7.
Schematic model of the effects of SIRT2/cMYC pathway in CCA cells.
SIRT2 and cMYC reprogrammed the metabolism of CCA cells by up-regulating downstream p-PHDA1 and PHGDH/PSAT1/PSPH. The metabolic reprogramming increased SSP and decreased mitochondrial oxidative phosphorylation, thus contributing to redox homeostasis, consequently protecting CCA cells from oxidative stress-induced apoptosis.
SIRT2 is a highly conserved enzyme that widely exists across species, from bacteria to humans. The removal of the acetyl group from lysine residues is coupled with the hydrolysis of NAD to generate nicotinamide, lysine, and O-acetyl-ADP-ribose. SIRT2 is responsible for catalyzing many biological processes, including genetic control, development and metabolism. Many previous studies have shown that SIRT2 increases the migration and invasion of cancer cells, demonstrating that it may play a role in promoting cancer proliferation and metastasis [31], [32]. However, some studies have shown that SIRT2 may be a tumor-suppressor gene in various other cancers [33]. Therefore, SIRT2 might promote tumor progression in certain cases, such as human gastric cancer and pancreatic cancer, while under other conditions it may inhibit tumor progression. It is worth noting that LDH-A [34], tubulin [35], and phosphoenolpyruvate carboxykinase (PEPCK) [36] are substrates of this deacetylase, suggesting that SIRT2 affects different metabolism in different tumors to promote the biological behaviors of various tumors. In fact, after inhibiting SIRT2 in CCA cells, two significant changes in different metabolic pathways are observed: one is glycolysis reversal by promoting PDHA1 phosphorylation (p-PDHA1) to inhibit oxidative phosphorylation and the other is promotion of the SSP pathway via aiding serine synthesis activity. Interestingly, we found that the reversal of PDHA1 phosphorylation alone was insufficient to induce significant apoptosis. This indicates that although glucose metabolism changes induced by SIRT2 are observed in CCA apoptosis, a high degree of serine anabolic metabolism provides an antioxidant effect against stress, which plays a more important role.
There are many kinds of SIRT2 inhibitors, mainly including nicotinamide, which inhibits NAD+-dependent reactions [37], and competitive inhibitors such as sirtinol, spitomicin, and cambinol [38]. Researchers have found that SIRT2 inhibitors have anti-tumor [25], anti-inflammatory [39], and anti-diabetic properties. SIRT2 shares a common structure with other members of the SIRT family, such as SIRT1, which is one of the most widely studied SIRTs and regulates cell migration, apoptosis, and proliferation [40]. Therefore, the off-target effects of inhibitors should not be ignored. Consequently, a specific inhibitor (Tm) was used. It inhibited SIRT2 with an IC50 value of 0.028 mM and SIRT1 with an IC50 value of 98 mM, but did not inhibit SIRT3, even at 200 mM [25]. Hence, the inhibition of SIRT2, rather than other SIRTs, may play a leading and more efficient role in reversing metabolic reprogramming. In addition, in the CCA tumorigenesis mouse model in which SIRT2 was inhibited by Tm, tumor growth was delayed without significant side effects.
As an oncogene, cMYC has received much attention as a result of its potential role in promoting tumorigenesis. There are many mechanisms by which cMYC induces tumorigenesis, and the enhancement of the Warburg effect is one method. This study found that cMYC is a crucial downstream target of SIRT2. Many studies have also elucidated that cMYC can elevate metabolic proteins such as LDHA and PKM2 [26], and is therefore considered a promising anti-cancer target. The present study demonstrates that cMYC has an elevated expression in CCA and predicts an unfavorable prognosis. In addition, cMYC is positively associated with p-PDHA1 and SSP-related enzymes and contributes to a high serine level. Our work has determined that the inhibition of cMYC can be used to reduce serine, thereby effectively promoting apoptosis in CCA cells.
Glucose is the main source for serine biosynthesis [41]. However, little is known about how glycolysis intertwines with SSP activation to coordinate cell survival and proliferation. Previous studies have reported that increased SSP activation was observed in breast cancer during serine starvation to maintain cell survival [42]. Herein we confirmed the importance of SSP activation in CCA proliferation. We also observed that by inhibiting the SIRT2/cMYC pathway, the reduced glycolytic metabolism changed to an elevated serine synthesis metabolism. These results suggest that high serine anabolic metabolism, which provides antioxidants for stress resistance, is essential for preventing apoptosis in CCA cells. We further observed that SIRT2/cMYC and its downstream targets may represent the intersecting points of metabolic reprogramming between glycolysis and SSP, highlighting the importance of SIRT2 for cell survival under redox imbalance conditions.
Our data show that the SIRT2/cMYC pathway plays a crucial role in the conversion of glucose oxidative metabolism to serine anabolic metabolism, thus providing antioxidants for stress resistance. SIRT2/cMYC-induced metabolic reprogramming may thus represent a new target for the treatment of CCA.
The following are the supplementary data related to this article.
Author contributions
Mingming Zhang, Lei Wang, and Xiaoping Zou designed the study. Lei Xu, Dehua Tang, and Yida Pan conducted the cell experiments. Yuming Wang and Lei Xu collected the tissue samples. Lei Xu and Mingming Zhang performed the protein analysis. Lixing Zhou, Yuming Wang, Mingming Zhang, and Dehua Tang drafted the manuscript and conducted the immunohistochemistry experiments. Lei Xu performed the metabolite analysis. Robert G. Dorfman and Mingming Zhang wrote the manuscript. Mingming Zhang and Xiaoping Zou supported the study. All of the authors read and approved the final manuscript.
Acknowledgments
Acknowledgments
We thank the Zhao lab for their assistance.
Grant support
This work was supported by grants from the National Natural Science Foundation of China (Nos. 81602076, 81672935, and 81472756), the Outstanding Youth Project of Nanjing City (No. JQX17002), the Jiangsu Clinical Medical Center of Digestive Disease (BL2012001), the Natural Science Foundation from the Department of Science & Technology of Jiangsu Province (BK20160113), the Fund of Jiangsu Provincial Commission of Health and Family Planning (No. Q201611) and the Fundamental Research Funds for the Central Universities (No. 021414380244).
Conflicts of interest
The authors declare no conflicts of interest.
Contributor Information
Xiaoping Zou, Email: 13770771661@163.com.
Jianlin Wu, Email: jlwu@must.edu.mo.
Mingming Zhang, Email: doczmm@126.com.
References
- 1.Razumilava N, Gores GJ. Cholangiocarcinoma. Lancet. 2014;383:2168–2179. doi: 10.1016/S0140-6736(13)61903-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rizvi S, Gores GJ. Pathogenesis, diagnosis, and management of cholangiocarcinoma. Gastroenterology. 2013;145:1215–1229. doi: 10.1053/j.gastro.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Warburg O. On the Origin of Cancer Cell. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 5.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg Effect: The Metabolic Requirements of Cell Proliferation. Science. 2009;324:1029. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Green DR, Galluzzi L, Kroemer G. Cell biology. Metabolic control of cell death. Science. 2014;345 doi: 10.1126/science.1250256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pinweha P, Rattanapornsompong K, Charoensawan V, Jitrapakdee S. MicroRNAs and oncogenic transcriptional regulatory networks controlling metabolic reprogramming in cancers. Comput Struct Biotechnol J. 2016;14:223–233. doi: 10.1016/j.csbj.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pilegaard H, Birk JB, Sacchetti M, Mourtzakis M, Hardie DG, Stewart G, Neufer PD, Saltin B, Van HG, Wojtaszewski JF. PDH-E1alpha dephosphorylation and activation in human skeletal muscle during exercise: effect of intralipid infusion. Diabetes. 2006;55:3020–3027. doi: 10.2337/db06-0152. [DOI] [PubMed] [Google Scholar]
- 9.Linn TC, Pettit FH, Reed LJ. Alpha keto acid dehydrogenase complexes. X. Regulation of the activity of PDC from beef kidney mitochondria by phosphorylation and dephosphorylation. Proc Natl Acad Sci U S A. 1969;62:234–241. doi: 10.1073/pnas.62.1.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun RC, Board PG, Blackburn AC. Targeting metabolism with arsenic trioxide and dichloroacetate in breast cancer cells. Mol Cancer. 2011;10 doi: 10.1186/1476-4598-10-142. [1(2011-11-18) 10, 1-15] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalhan SC, Hanson RW. Resurgence of serine: an often neglected but indispensable amino Acid. J Biol Chem. 2012;287:19786–19791. doi: 10.1074/jbc.R112.357194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vazquez A, Tedeschi PM, Bertino JR. Overexpression of the mitochondrial folate and glycine–serine pathway: a new determinant of methotrexate selectivity in tumors. Cancer Res. 2013;73:478–482. doi: 10.1158/0008-5472.CAN-12-3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Circu ML, Aw TY. Reactive oxygen species, cellular redox systems and apoptosis. Free Radic Biol Med. 2010;48:749–762. doi: 10.1016/j.freeradbiomed.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu Rev Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- 16.Harting K, Knoell B. SIRT2-mediated protein deacetylation: An emerging key regulator in brain physiology and pathology. Eur J Cell Biol. 2010;89:262–269. doi: 10.1016/j.ejcb.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 17.Chen J, Chan AWH, To KF, Chen W, Zhang Z, Ren J, Song C, Cheung YS, PBS Lai, Cheng SH. SIRT2 overexpression in hepatocellular carcinoma mediates epithelial to mesenchymal transition via akt/GSK-3β/β-catenin signaling (revised version) Hepatology. 2013;57 doi: 10.1002/hep.26278. [DOI] [PubMed] [Google Scholar]
- 18.Heltweg B, Gatbonton T, Schuler AD, Posakony J, Li H, Goehle S, Kollipara R, Depinho RA, Gu Y, Simon JA. Antitumor activity of a small-molecule inhibitor of human silent information regulator 2 enzymes. Cancer Res. 2006;66:4368–4377. doi: 10.1158/0008-5472.CAN-05-3617. [DOI] [PubMed] [Google Scholar]
- 19.Liu PY, Xu N, Malyukova A, Scarlett CJ, Sun YT, Zhang XD, Ling D, Su SP, Nelson C, Chang DK. The histone deacetylase SIRT2 stabilizes Myc oncoproteins. Cell Death Differ. 2013;20:503–514. doi: 10.1038/cdd.2012.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dang CV. Glutaminolysis: Supplying carbon or nitrogen or both for cancer cells? Cell Cycle. 2010;9:3884. doi: 10.4161/cc.9.19.13302. [DOI] [PubMed] [Google Scholar]
- 21.Sun L, Song L, Wan Q, Wu G, Li X, Wang Y, Wang J, Liu Z, Zhong X, He X. cMyc-mediated activation of serine biosynthesis pathway is critical for cancer progression under nutrient deprivation conditions. Cell Res. 2015;25:429. doi: 10.1038/cr.2015.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frezza C, Zheng L, Folger O, Rajagopalan KN, Mackenzie ED, Jerby L, Micaroni M, Chaneton B, Adam J, Hedley A. Haem oxygenase is synthetically lethal with the tumour suppressor fumarate hydratase. Nature. 2011;477:225. doi: 10.1038/nature10363. [DOI] [PubMed] [Google Scholar]
- 23.Xiao Y, Wang J, Qin Y, Xuan Y, Jia Y, Hu W, Yu W, Dai M, Li Z, Yi C. Ku80 cooperates with CBP to promote COX-2 expression and tumor growth. Oncotarget. 2015;6:8046. doi: 10.18632/oncotarget.3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shibata T, Aburatani H. Exploration of liver cancer genomes. Nat Rev Gastroenterol Hepatol. 2014;11:340. doi: 10.1038/nrgastro.2014.6. [DOI] [PubMed] [Google Scholar]
- 25.Jing H, Hu J, He B, Negron Abril YL, Stupinski J, Weiser K, Carbonaro M, Chiang YL, Southard T, Giannakakou P. A SIRT2-selective inhibitor promotes c-Myc oncoprotein degradation and exhibits broad anticancer activity. Cancer Cell. 2016;29:767–768. doi: 10.1016/j.ccell.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 26.David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463:364–368. doi: 10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McFate T, Mohyeldin A, Lu H, Thakar J, Henriques J, Halim ND, Wu H, Schell MJ, Tsang TM, Teahan O. Pyruvate dehydrogenase complex activity controls metabolic and malignant phenotype in cancer cells. J Biol Chem. 2008;283:22700–22708. doi: 10.1074/jbc.M801765200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacquemin G, Margiotta D, Kasahara A, Bassoy EY, Walch M, Thiery J, Lieberman J, Martinvalet D. Granzyme B-induced mitochondrial ROS are required for apoptosis. Cell Death Differ. 2015;22:862–874. doi: 10.1038/cdd.2014.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Samanta D, Semenza GL. Serine synthesis helps hypoxic cancer stem cells regulate redox. Cancer Res. 2016;76:6458–6462. doi: 10.1158/0008-5472.CAN-16-1730. [DOI] [PubMed] [Google Scholar]
- 30.Niu W, Luo Y, Wang X, Zhou Y, Li H, Wang H, Fu Y, Liu S, Yin S, Li J. BRD7 inhibits the Warburg effect and tumor progression through inactivation of HIF1alpha/LDHA axis in breast cancer. Cell Death Dis. 2018;9:519. doi: 10.1038/s41419-018-0536-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Li Y, Zhang M, Dorfman RG, Pan Y, Tang D, Xu L, Zhao Z, Zhou Q, Zhou L, Wang Y. SIRT2 promotes the migration and invasion of gastric cancer through RAS/ERK/JNK/MMP-9 pathway by increasing PEPCK1-related metabolism. Neoplasia. 2018;20:745–756. doi: 10.1016/j.neo.2018.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao D, Zou SW, Liu Y, Zhou X, Mo Y, Wang P, Xu YH, Dong B, Xiong Y, Lei QY. Lysine-5 acetylation negatively regulates lactate dehydrogenase A and is decreased in pancreatic cancer. Cancer Cell. 2013;23:464–476. doi: 10.1016/j.ccr.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harting K, Knoll B. SIRT2-mediated protein deacetylation: An emerging key regulator in brain physiology and pathology. Eur J Cell Biol. 2010;89:262–269. doi: 10.1016/j.ejcb.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 34.Huang S, Zhao Z, Tang D, Zhou Q, Li Y, Zhou L, Yin Y, Wang Y, Pan Y, Dorfman RG. Downregulation of SIRT2 inhibits invasion of hepatocellular carcinoma by inhibiting energy metabolism. Transl Oncol. 2017;10:917–927. doi: 10.1016/j.tranon.2017.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee JK, Lee J, Go H, Lee CG, Kim S, Kim HS, Cho H, Choi KS, Ha GH, Lee CW. Oncogenic microtubule hyperacetylation through BEX4-mediated sirtuin 2 inhibition. Cell Death Dis. 2016;7:e2336. doi: 10.1038/cddis.2016.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Avalos JL, Bever KM, Wolberger C. Mechanism of sirtuin inhibition by nicotinamide: altering the NAD(+) cosubstrate specificity of a Sir2 enzyme. Mol Cell. 2005;17:855–868. doi: 10.1016/j.molcel.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 38.Heltweg B, Gatbonton T, Schuler AD, Posakony J, Li H, Goehle S, Kollipara R, Depinho RA, Gu Y, Simon JA. Antitumor activity of a small-molecule inhibitor of human silent information regulator 2 enzymes. Cancer Res. 2006;66:4368–4377. doi: 10.1158/0008-5472.CAN-05-3617. [DOI] [PubMed] [Google Scholar]
- 39.Kanda T, Sasaki R, Nakamoto S, Haga Y, Nakamura M, Shirasawa H, Okamoto H, Yokosuka O. The sirtuin inhibitor sirtinol inhibits hepatitis A virus (HAV) replication by inhibiting HAV internal ribosomal entry site activity. Biochem Biophys Res Commun. 2015;466:567–571. doi: 10.1016/j.bbrc.2015.09.083. [DOI] [PubMed] [Google Scholar]
- 40.Liu T, Liu PY, Marshall GM. The critical role of the Class III histone deacetylase SIRT1 in cancer. Cancer Res. 2009;69:1702–1705. doi: 10.1158/0008-5472.CAN-08-3365. [DOI] [PubMed] [Google Scholar]
- 41.Sun L, Song L, Wan Q, Wu G, Li X, Wang Y, Wang J, Liu Z, Zhong X, He X. cMyc-mediated activation of serine biosynthesis pathway is critical for cancer progression under nutrient deprivation conditions. Cell Res. 2015;25:429–444. doi: 10.1038/cr.2015.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, Sethumadhavan S, Woo HK, Jang HG, Jha AK. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]