Abstract
BACKGROUND
Anti-tumor necrosis factor α (TNFα) represents the best therapeutic option to induce mucosal healing and clinical remission in patients with moderate-severe ulcerative colitis. On the other side gut microbiota plays a crucial role in pathogenesis of ulcerative colitis but few information exists on how microbiota changes following anti-TNFα therapy and on microbiota role in mucosal healing.
AIM
To elucidate whether gut microbiota and immune system changes appear following anti TNFα therapy during dextran sulfate sodium (DSS) colitis.
METHODS
Eighty C57BL/6 mice were divided into four groups: “No DSS”, “No DSS + anti-TNFα”, “DSS” and “DSS + anti-TNFα”. “DSS” and “DSS + anti-TNFα” were treated for 5 d with 3% DSS. At day 3, mice whithin “No DSS+anti-TNFα” and “DSS+anti-TNFα” group received 5 mg/kg of an anti-TNFα agent. Forty mice were sacrificed at day 5, forty at day 12, after one week of recovery post DSS. The severity of colitis was assessed by a clinical score (Disease Activity Index), colon length and histology. Bacteria such as Bacteroides, Clostridiaceae, Enterococcaceae and Fecalibacterium prausnitzii (F. prausnitzii) were evaluated by quantitative PCR. Type 1 helper T lymphocytes (Th1), type 17 helper T lymphocytes (Th17) and CD4+ regulatory T lymphocytes (Treg) distributions in the mesenteric lymph node (MLN) were studied by flow cytometry.
RESULTS
Bacteria associated with a healthy state (i.e., such as Bacteroides, Clostridiaceae and F. prausnitzii) decreased during colitis and increased in course of anti-TNFα treatment. Conversely, microorganisms belonging to Enterococcaceae genera, which are linked to inflammatory processes, showed an opposite trend. Furthermore, in colitic mice treated with anti-TNFα microbial changes were associated with an initial increase (day 5 of the colitis) in Treg cells and a consequent decrease (day 12 post DSS) in Th1 and Th17 frequency cells. Healthy mice treated with anti-TNFα showed the same histological, microbial and immune features of untreated colitic mice. “No DSS + anti-TNFα” group showed a lymphomononuclear infiltrate both at 5th and 12th d at hematoxylin and eosin staining, an increase of in Th1 and Th17 frequency at day 12, an increase of Enterococcaceae at day 5, a decrease of Bacteroides and Clostridiaceae at day 12.
CONCLUSION
Anti-TNFα treatment in experimental model of colitis improves disease activity but it is associated to an increase in Th17 pathway together with gut microbiota alteration.
Keywords: Gut microbiota, Dextran sodium sulphate colitis, Immune system, T cells, Mesenchymal lymphnode, Tumor necrosis factor α
Core tip: The effect of the gut microbiota on the immune responses is not limited to the gut. On the other hand, autoimmune diseases, such as inflammatory bowel disease, are associated to different degree of dysbiosis involving different bacterial genera. Using a colitic mouse model, we aimed to evaluate the impact of anti-tumor necrosis factor α (TNFα) therapy on the intestinal immune system and gut microbiota concurrently. Healthy mice treated with anti-TNFα showed similar histological, microbial and immune features of untreated colitic mice, a lymphomononuclear infiltrate both at day 5 and 12 at hematoxylin and eosin staining, an increase of type 1 helper T lymphocytes and type 17 helper T lymphocytes at day 12, and finally increase of Enterococcaceae at day 5, a decrease of Bacteroides and Clostridiaceae at day 12.
INTRODUCTION
Inflammatory bowel disease (IBD) is a chronic, relapsing, inflammatory disorder of the gastrointestinal tract due to a dysregulated immune response towards the microbiota[1]. Although many factors underlying IBD have been identified, the exact etiology is unknown, reflecting its complexity and the broad spectrum of its manifestations. IBD includes ulcerative colitis (UC) and Crohn’s disease (CD), which show differences in the pathology and clinical characteristics[2] and with an increasing incidence , especially in adolescence[3-6], becoming a global disease[7].
IBD is a multifactor disease where environment, genetics, hygiene and microbiota interact with host immune system in a dynamic equilibrium[8,9]. This equilibrium between gut microbiota and immune system is stable throughout adulthood and in health condition, while it is perturbed in diseases like IBD and in predisposing risk conditions[10-12]. Recent studies indicate that a healthy commensal gut microbiota stimulates CD4+ regulatory T lymphocytes (Tregs) that, in turn, are responsible for maintaining gut homeostasis[12]. Under homeostatic conditions, a balance exists between the production of pro-inflammatory cytokines by CD4+ T lymphocytes and anti-inflammatory cytokines [e.g., interleukin (IL)-10] by Tregs[13]. The phyla Bacteroidetes and Firmicutes dominate the gut microbiota of healthy humans[14].
In course of IBD a decrease of Firmicutes [and, among these, the species Fecalibacterium prausnitzii (F. prausnitzii)][15] and Bacteroidetes occurs with an increase in Actinobacteria and Proteobacteria[16,17]. Among genera a decrease of Bacteroidaceae and an increase of Enterobacteriaceae (specifically Escherichia coli) and Clostridiaceae[18-20] has been described, together with a reduced phylogenetic diversity associated to an increase of T lymphocyte differentiation toward type 1 helper T lymphocytes (Th1), type 17 helper T lymphocytes (Th17) subsets and a decrease of Treg[21-24].
How these conditions are modified in course of medical treatment aimed to control active and chronic gastrointestinal inflammation, and whether this could have a role in non-response rates to biologic agents[24], is not known. In a previous study we showed the importance of local immunity in course of anti-tumor necrosis factor α (TNFα) therapy using animal model of IBD[25]. In this study. we aim to understand whether anti-TNFα therapy could affect the gut microbiota and intestinal mucosa immune activation in experimental model of this disease.
MATERIAL AND METHODS
Experimental acute colitis
The animal protocol was designed to minimize pain or discomfort to the animals. The animals were acclimatized to laboratory conditions (23 °C, 12 h/12 h light/dark, 50% humidity, ad libitum access to food and water) for 2 wk prior to experimentation. C57BL/6 mice, 8 wk old, were fed for 5 d with 3% dextran sulfate sodium (DSS) polymers in drinking water (MP Biomedicals, Aurora, OH, United States) provided ad libitum. Mice were divided into four groups (Figure 1). Following three days from DSS treatment (day 3), two groups received an anti-TNFα drug iv [infliximab (IFX) 5 mg/kg]. Every day weight, fecal consistency, body weight of mice and the presence of occult fecal blood was monitored in mice as described[25,26], in order to calculate disease activity index (DAI). Mice were sacrificed at 12th d after starting DSS, it means 7 d after DSS stop to evaluate the recovery post colitis. Some animals were sacrificed following 5 d from DSS (day 5), for intermediate measures in the moment of acute colitis. Parallel experiments were undertaken during same period also on healthy mice receiving drinking water instead of DSS.
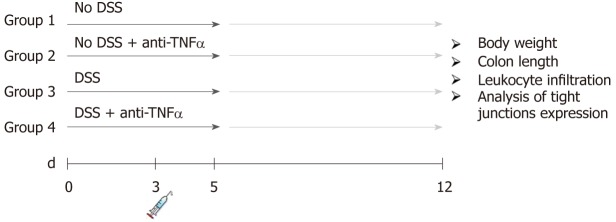
Figure 1.
Experimental design of the dextran sulfate sodium colitis model. Acute colitis was induced by administration of 3% dextran sulfate sodium (DSS) to drinking water (DSS) for 5 d. At 3rd d, one singular injections of an anti-tumor necrosis factor α was administrated. Each group counts 20 mice: 10 mice were sacrificed at day 5, 10 mice at day 12 after 1 wk of recovery post-DSS. DSS: Dextran sulfate sodium; TNFα: Tumor necrosis factor α.
Biological samples collection and analysis
At the sacrifice from each animal was collected the colon, the mesenchymal lymph nodes (MLN) and stool and stored at -80 °C. The colonic length was evaluated. The last part of the small intestine was divided in three parts: two were stored at -80 °C for RNA and protein expression studies, one was fixed in 4% formalin and embedded in paraffin. The stools of animals were harvested at day 5 and 12, subsequently weighed, and stored at -80 °C for further analysis.
General assessment of colitis
Colon length was measured as an indication of colonic inflammation. Briefly, the animals were anaesthetized and sacrified by cervical dislocation. The colon was resected between the ileo-caecal junction and the proximal rectum, close to its passage under the pelvis. The colon was placed onto a non-absorbent surface and measured with a ruler, taking care not to stretch the tissue.
Histological observation and scoring
Colon tissue was fixed in 4% formalin, embedded in paraffin and sliced into 3-mm-thick sections. Hematoxylin and eosin (HE)-stained sections were scored blindly on the basis of severity and extent of inflammation, as well as presence and extent of ulceration[27]. Scores ranged from 0 to 4 for each parameter; single values were summed up to obtain the overall histological score (maximum possible value equals 16). Photomicrographs were taken with 40 × magnification on a Nikon E400 Eclipse microscope.
RNA extraction and real time PCR
RNA was extracted from freeze colon using RNAqueous™ Total RNA Isolation Kit (Thermo-Fisher). cDNA was synthesized by High-Capacity cDNA Reverse Transcription kit (Applied Biosystems), following the manufacturer’s instructions. Quantitative real-time PCR (qPCR) was performed in duplicates using iQ SYBR Green Supermix (BioRad) on an iCycler iQ5 (BioRad) with the following scheme: an initial denaturation at 95 °C for 3 min, 40 cycles of PCR amplification, each consisting of a denaturing step of 95 °C for 10 s, annealing at 55 °C for 30 s, and a final step at 72 °C for 1 min. The primer pairs used were for CD3: forward 5’-ATGCGGTGGAACACTTTCTG-3’ and reverse 5’-GCACGTCAACTCTACACTGGT-3’; for β-actin: forward 5’-TGTTACCAACGTGGACGACA-3’ and reverse 5’-CTGGGTCATCTTTTCA-3’. β-actin was used as an endogenous reference gene to normalize the expression of CD3.
Western blot
Colon tissues were lysed in RIPA buffer [50 mmol/L Tris, pH 8, 150 mmol/L NaCl, 1 mmol/L EDTA, 1% NP-40, 0.05% sodium deoxycholate, 0.1% sodium dodecyl sulfate (SDS)] supplemented with protease inhibitors (10 μg/mL leupeptin, 20 μg/mL aprotinin, 1 mmol/L phenylmethanesulfonyl fluoride, 1 mmol/L NaVO4, 100 mmol/L NaF). Protein concentrations were determined using the Bradford protein assay (Bio-Rad). Protein extracts (50 μg) were resolved by 8% SDS-PAGE, transferred to PVDF membranes, and probed with rabbit polyclonal anti-ZO1 (1:500, Thermo Fisher Scientific) anti -Occludin (1:500, Thermo Fisher Scientific) anti-actin (1:1000, Sigma-Aldrich) primary antibodies. Horseradish peroxidase-conjugated secondary antibodies (GE Healthcare) were detected by use of the ECL Prime Western Blotting Detection Reagent (GE Healthcare) and the ChemiDoc XRS system (Bio-Rad). Densitometric analysis was performed by using the ImageJ software (http://imagej.nih.gov/ij/).
T lymphocyte activation and characterization
T lymphocytes were obtained from MLN of C57BL/6 mice, treated or not with DSS, using sterile strainers. MLN-derived T lymphocytes were then activated by plate-bound anti-CD3/anti-CD28 (10 and 4 μg/mL, respectively) in fully supplemented RPMI 1640 medium containing 10% fetal calf serum, 2 mmol/L glutamine, 100 IU/mL penicillin, 0.1 mg/mL streptomycin for 48 h. At the end of incubation, T lymphocytes were stimulated with phorbol myristate acetate (PMA) and ionomycin in the presence of brefeldin A for 4 h, fixed in 4% formyl saline and permeabilized with 0.1% saponin buffer prior to intracellular cytokine staining. Flow cytometry analysis was performed on an Epics XL Coulter instrument (Beckman Coulter). The following monoclonal antibodies (Mab) were used: PeCy5-labelled anti-CD4 Mab, PE-labelled anti-TNFα Mab, PE-labelled anti-IL6 Mab, PE-labelled anti-IL17A Mab, PE-labelled anti-FOXP3 Mab, PECF594-labelled anti-interferon (IFN)γ Mab, PE CF594-labelled anti-IL4 Mab, PE CF594 labelled anti-CD25 Mab (all from eBioscience, San Diego, CA, United States). Cell viability was assessed by Fixable Viability Dye eFluor 450 (eBioscience), according to the manufacturer’s protocol.
Fecal DNA extractions and qPCR
DNA from stool samples was extracted using the QIAamp DNA Stool Mini Kit (Qiagen, Inc., Valencia, CA, United States) according to manufacturer’s instructions and as described previously[28]. qPCR was performed in duplicates by using iQ SYBR Green Supermix (BioRad) on an CFX96 cycler (BioRad) with the following scheme: an initial denaturation at 95 °C for 3 min, 40 cycles of PCR amplification, each consisting of a denaturing step of 95 °C for 10 s, annealing at 55 °C for 15 s, and a final step at 60 °C for 1 min. The primer pairs used were: Fecalibacterium prausnitzii Forward 5'-AGA TGG CCT CGC GTC CGA-3', Reverse 5'- CCG AAG ACC TTC TTC CTC -3', Clostridiaceae Forward 5'-CGG TAC CTG ACT AAG AAG-3', Reverse 5'-AGT TTY ATT CTT GCG AAC-3', Enterococcaceae Forward 5'-CAT TGA CGT TAC CCG CAG AAG AAG C-3', Reverse 5'-CTC TAC GAG ACT CAA GCT TGC-3', Bacteroides Forward 5'- GAA GGT CCC CCA CAT TG -3', Reverse 5'-CAA TCG GAG TTC TTC GTG-3.
As standards for qPCR was used DNA extracted from Escherichia coli ATCC 25922 (LGC STANDARD) suspension. DNA was extracted starting from a suspension of 0,5 MacFarland (approximately 1.5 × 108 CFU/mL) using High pure PCR template preparation kit (Roche). Subsequently extracted DNA (approximately 108 copy/mL) was diluted two fold 1:100 to obtain 106 and 104 standards. The number of CT was normalized in number of genome copy using a standard curve. The results were divided between T0 and T2, where T0 represented the conditions before any treatment, and T2 represented the changes that appeared after 5 or 12 d of administration of DSS and anti-TNFα, alone or in association. The exact number of days was specified in the figures.
Bioinformatics analysis
Statistical analysis of bacterial genera and species were performed using R packages FactorMineR and factorextra[29], while ggplot2 for graphical rappresentations[30]. Principal component analysis (PCA) was computed in order to highlights differences in species relative abundances among samples. Finally, Mann-Whitney-Wilcoxon was performed for each bacterial species between T0 and T2 in every group considered in this study.
Statistical analysis
ANOVA and adjusted Bonferroni post-hoc analysis were performed in order to assess differences in DAI, protein and RNA expression and T lymphocyte subsets among treatment groups. A P value < 0.05 was considered statistically significant. Statistical analysis was performed using IC STATA 12 for Mac.
RESULTS
Anti-TNFα ameliorates DAI and histological scores in a murine model of colitis
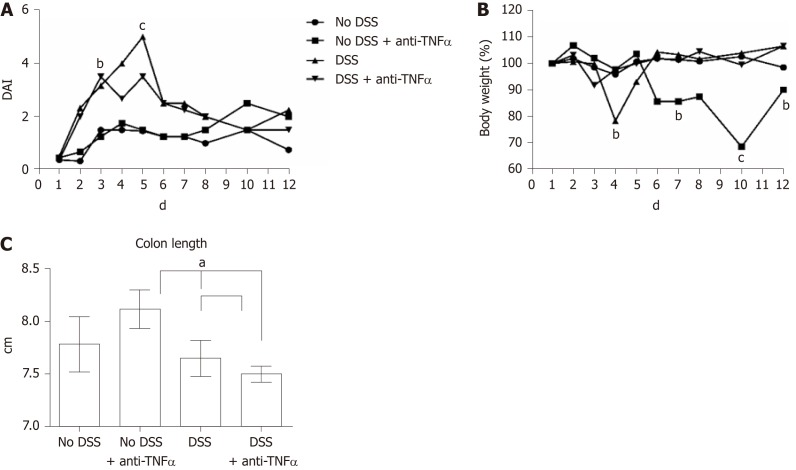
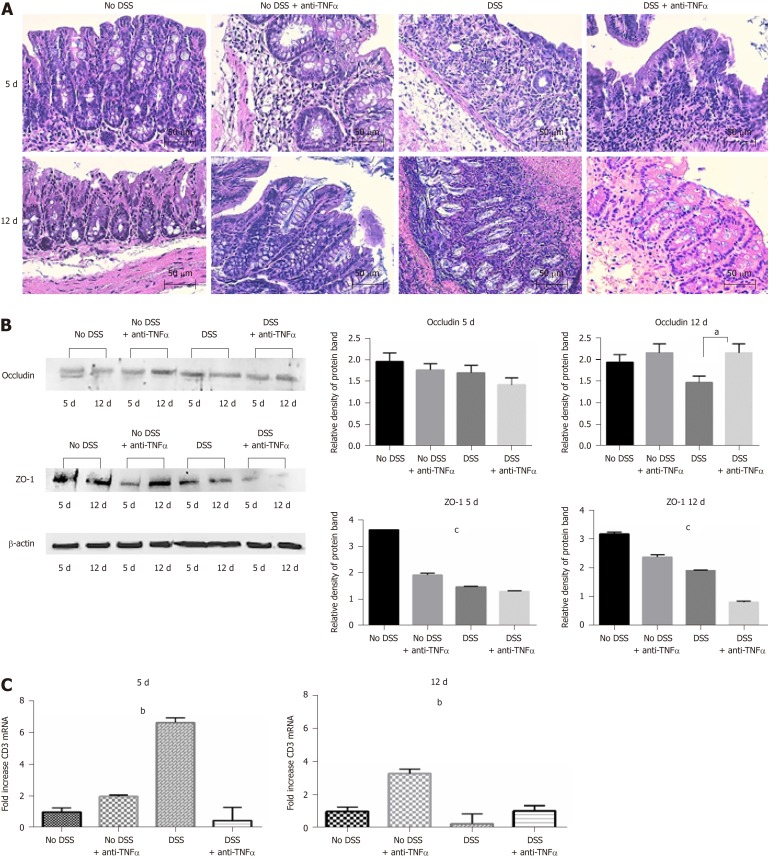
In the last 4 years, our group use a well-established model of colitis in mice, largely recognized in literature: the DSS model[26,31,32]. As shown in Figure 2A and B, during DSS treatment, particulary at the day 4 and 5, there was a significant increase of the DAI and a loss of body weight, that equates in an escalation of severity of the DSS-induced colitis. During the week of recovery, the values of body weight and DAI came back close to the controls, except in “No DSS + anti-TNFα group”, that showed gain of DAI score and an unexpected loss of body weight. Colon length was measured as an indication of colonic inflammation[33,34]. Bowels were resected from the 4 groups of mice and subjected to macroscopic and histopathological examination. The appearance of the organs from the DSS-treated mice showed obvious reddening and shortening of the colon, which are typical signs of acute intestinal inflammation (Figure 2C). Histological analysis, performed by HE staining, showed important modification on colonic mucosa, especially at day 5 not only in “DSS” group, but also in “No DSS + anti-TNFα” (Figure 3A). After 5 d of DSS treatment, intestinal villi were flatted, the glandular epithelium disappeared, the globet cells were no longer recognizable and there was an evident mononuclear cell infiltrate. In “DSS + anti-TNFα” group, few glands were preserved, lymphomononuclear cell infiltrate was present. Mononuclear inflammatory cells were present also in colon of mice treated only with anti-TNFα (“No DSS + anti-TNFα” group) at day 5 and 12. Following 7 d of recovery post-DSS, the “DSS” and “DSS + anti-TNFα” groups still displayed atrophic globet cells , but more preserved than day 5.
Figure 2.
Estimation of clinical and morphological changes of dextran sulfate sodium-induced colitis. A: The four-point disease activity index (DAI) was evaluated scoring daily body weight of mice, the percentage of body weight loss, the trait of the stool and the presence of occult blood in feces. B: Body weight, one of the DAI parameter, is also represented separately by percentage, where the mice weight at 1st d of experiment is equal to 100%. C: Colon length is another parameter to evaluate the healthy or disease state of the mice: the mean values in the 4 groups at day 5 are shown. aP < 0.05; bP < 0.01; cP < 0.0001. DSS: Dextran sulfate sodium; TNFα: Tumor necrosis factor α; DAI: Disease activity index.
Figure 3.
Histological analysis confirms the strength of the dextran sulfate sodium model. A: Hematoxylin and eosin staining of colon sections of mice from 4 groups of treatment at day 5 and 12. Forty-fold magnification was used. B: Protein expression of tight junctions occludin and ZO-1 in colon tissue. Antibody anti-β-actin was used as endogenous control. On the right, the histograms represent the mean of densitometry values from 3 different experiments. C: CD3 infiltration in colon mucosa was assessed by quantitative PCR of CD3 mRNA. One-way ANOVA (and non-parametric) was calculated; aP < 0.05; bP < 0.01; cP < 0.0001. DSS: Dextran sulfate sodium; TNFα: Tumor necrosis factor α.
Anti-TNFα has effects on tight junction expression and lymphocyte infiltration
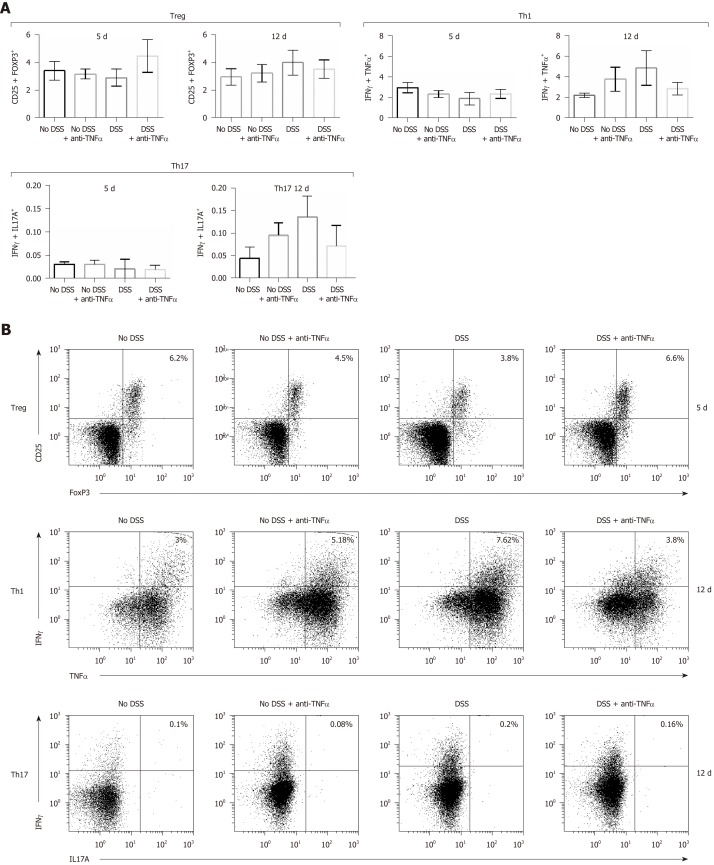
We analyzed the mononuclear cells infiltration in each condition (other than control group or “No DSS”) and the tight junction integrity of the colonic epithelium, and in particular occludin and ZO-1 (Figure 3B). Occludin expression decline was more evident at day 12 in “DSS”group (Figure 3B, right panel, P = 0.05); ZO-1 decreased at day 5 (also in “No DSS + anti-TNFα”group, P < 0.001) and its levels remained lower than control mice also at day 12 (Figure 3B, P < 0.001). CD3 expression was studied by qPCR in the last part of small intestine of mice from different groups. CD3 mRNA increased in “DSS group” during acute colitis at day 5 (Figure 3C, left panel). At day 12, after 7 d of recovery, CD3 mRNA level remained higher in samples from “No DSS + anti-TNFα” group while decreased in other mice (Figure 3C, right panel, P < 0.001): this finding correlates with the presence of lymphomononuclear cell infiltrate observed at day 5 and 12. The data in mucosa were confirmed by the data from MLN. In the presence of inflammatory processes, due in this case to DSS administration, Treg lymphocytes decreased early at day 5, except in “DSS + anti-TNFα” group (Figure 4A and B); Th1 and Th17 lymphocytes increased especially later at day 12 (Figure 4A, C and D); these modifications are less prominent in “DSS + anti-TNFα”. In “No DSS + anti-TNFα” group, several MLN-derived T lymphocyte subsets were found modified at day 12. In particular, Th1 and Th17 lymphocytes were found to be more frequent than in the “DSS + anti-TNFα” treatment group whereas they were found diminished as compared to the “DSS” treatment group.
Figure 4.
T cells subsets characterization in presence of an anti-tumor necrosis factor α agent. T cells were isolated from mesenteric lymphnode of mice and CD4+ cells were studied by flow cytometry. In particular, CD4+ regulatory T lymphocytes characterization was possible through the use of anti-CD25 and anti–FoxP3 antibodies (B); type 1 helper T lymphocytes expressed interferon (IFN)γ and tumor necrosis factor α (C); IFNγ+ and interleukin 17+ cells identified the type 17 helper T lymphocytes cluster (D). In each panel (B), (C) and (D) the cells in the upper right quadrant were the double staining cells. Treg: CD4+ regulatory T lymphocytes; Th1: Type 1 helper T lymphocytes; Th17: Type 17 helper T lymphocytes; DSS: Dextran sulfate sodium; TNFα: Tumor necrosis factor α; IFNγ: Interferon γ; IL: Interleukin.
The role of TNFα to maintain a healthy gut microbiota
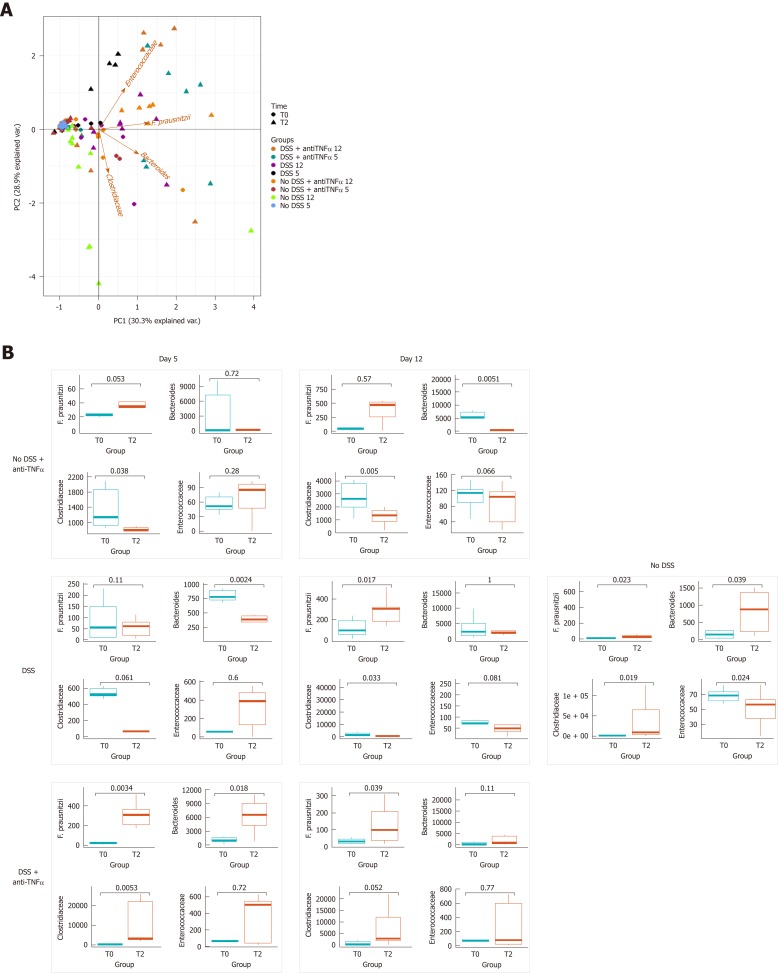
Increased epithelial tight junction permeability, with commensal bacteria, promotes intestinal CD4+ T cell expansion[31]. The distribution of specific genera and species of bacteria in the different groups of mice was initially evaluated, by PCA (Figure 5A). As we expect, the biplot at T0 shows the samples clustered together, and reflects the homogeneity in the abundance of genera and species in the microbiota of inbred mice.
Figure 5.
Bacterial genera and species characterization in presence of an anti- tumor necrosis factor α agent. A: Principal component analysis was represented by a biplot, where the variability among samples and bacterial weight in the variability were expressed. B: Dot plot represents the distribution of bacteria at the beginning of the experiment and at the moment of sacrifice; in the case of “No DSS” mice, the samples from day 5 and 12 are collected together, since no significant changes occurred between day 5 and 12. In left panel, the results were distinguished based on the day of the end of experiment. DSS: Dextran sulfate sodium; TNFα: Tumor necrosis factor α.
At T2, after 5th or 12th d of treatment, the samples from groups “DSS” or “DSS + anti-TNFα” diverged from other T2’s samples and from their respective T0. Moreover, it is possible to observe a gradient of distance starting with “DSS” group. Enterococcaceae weighted on this gradient: they were main factor that causes this distance of T2 from T0 in “DSS” and “DSS + TNFα” groups. We also compared, trough Mann-Whitney-Wilcoxon, the distribution of genera Bacteroides, Clostridaceae, Enterococcaceae, and of species F. prausnitzii in each group of mice. As we expected, in “No DSS” group no main changes occurred between T0 and T2, i.e., between the beginning of the experiment and the sacrifice (Figure 5B, right panel). As described eldewhere[35], animals with DSS induced acute colitis showed an increase of Enterococcaceae, and a decrease of Bacteroides and Clostridaceae, that is also statistically significant (P < 0.05, Figure 5B, left panel). The level of F. prasunitzii is not affected. After 7 d of recovery in “DSS” group all bacteria modifications reached the T0 level again.
In “DSS + anti-TNFα” group, the changes after 5 d were opposed to those seen in “DSS group”. In the “DSS + anti-TNFα” group there is no evident effect of the inflammation on Bacteroides, Clostridaceae and F. prausnitzii as their abundance increased while Enterococcaceae behaved as per “DSS” group. After 7 d of recovery the effects of anti-TNFα were maintained only by F. prausnitzii (Figure 5B, left panel). The increase of F. prausnitzii seems due the TNFα inhibition as in “No DSS + anti-TNFα”: after 2 d from one singular administration of anti-TNFα, the abundance of F. prausnitzii increased and remained stable 10 d after drug administration. In “No DSS + anti-TNFα”, the Clostridiaceae decreased at day 5 (P < 0.05), and Bacteroides fell down at day 12 too (P < 0.05). Enterococacceae show a moderate increase, but in this case the data were not significant (Figure 5B, left panel).
DISCUSSION
An unfavorable alteration of the commensal structure of gut microbiota is referred to as “dysbiosis”; this includes a reduction in the number of tolerogenic bacteria and an overgrowth of potentially pathogenic bacteria (“pathobionts”) that can penetrate the intestinal epithelium and induce disease in certain genetic or environmental contexts[32,33].
In the last decade a growing body of evidence suggests the crucial role of the gut microbiota in human health and studies are looking to define the link between “dysbiosis” and several pathologies[19,34,36].
In this paper, we use an animal model of experimental colitis to study the behavior of few bacterial genera and species during inflammation and anti-TNFα therapy. A model of severe murine colitis, which most closely resembles human UC, results from administration of 40-50 kDa DSS in drinking water[37]. The mechanism by which DSS induces intestinal inflammation is due to the damage to the epithelial monolayer lining in the large intestine allowing the dissemination of pro inflammatory intestinal contents (e.g., bacteria and their products) into underlying tissue. The DSS colitis model is very popular in IBD research due to its rapidity, simplicity, reproducibility and controllability[38,39].
DSS treatment induces changes in the abundance of few bacteria, whose dynamics shift toward an unhealthy state, confirmed by clinical data in mice (DAI, changes in body weight and histological score) (Figure 2). We have previously described the efficacy of IFX in neutralizing also murine TNFα[25,40]. The aim of this study was to evaluate the effects of a selective immunosuppression on the gut microbiota, in particular on few genera and species involved in T lymphocytes expansion. To differentiate between the effects of DSS and of IFX, we used two control groups of mice: “No DSS” and “NO DSS + anti-TNFα”. During the first evaluations, we noticed that the mice from “No DSS + anti-TNFα” group were characterized by a raise of DAI (Figure 2A), due to the loss of body weight (Figure 2B). “No DSS + anti-TNFα” group showed an increased mononuclear cell infiltrate compared to the control mice (Figure 3A), epithelial tight junction permeability (Figure 3B) and an increased expression of CD3 at qPCR (Figure 3C). In the first lymphoid station that drains the microbial products from intestine (MLN) Th1 and Th17 level increased at day 12 both in colitic mice and in control mice treated only with IFX (Figure 4A-D). Next, we looked at changes in Bacteroides, Clostridiaceae, Enterococcaceae and F. prausnitzii abundance. We have previously shown[15,16,18] that this mouse model mimics the changes that happens in the intestine of IBD patients . While in “No DSS” mice no main changes occurred between T0 and T2 (both at day 5 and at day 12, Figure 5B, right panel), in “No DSS + anti-TNFα” “DSS + anti-TNFα” and “DSS” groups there was an increase of Enterococcaceae abundance at day 5, a decrease of Bacteroides and Clostridiaceae at day 5 and day 12. This suggests that either DSS or anti-TNFα may cause a shift in the abundance of Enterococcaceae, Bacteroides and Clostridiaceae in the gut. The level of Enterococcaceae, Bacteroides and Clostridiaceae might have a role in the increase of CD3+ cells in colon mucosa, in the loss of tight junction expression and the raise of Th1 and Th17 in MLN observed in mice treated only with IFX.
On the other hand, a positive effect due to IFX was the augmented abundance of F. prausntizii at day 5 (after only 2 d from the i.v.) that remained also at day 12. The use of an anti-TNFα in healthy conditions resulted in dramatic changes in intestinal mucosa inflammation, modulation of tight junction permeability, and to an interesting shift in the overall intestinal mucosa T cell immune response at cytometry analysis, in parallel to variation of Enterococcaceae, Bacteroides and Clostridiaceae abundance. This observation, if confirmed in humans, has several potential implications. First, the dysbiotic effect of anti-TNFα must be considered in patients not responding to this drug. New and emerging data show potential positive effect of modulating gut microbiota composition in UC, including fecal microbiota transplantation[41,42].
Furthermore, the shift toward Th17 pathway could be considered when deciding to swap therapy in non-responder colitis patients: indeed emerging data suggest the positive effect of blocking the Th17/IL23 pathway also in UC, as shown by phase 3 induction trial of ustekinumab in UC patients[43].
A direct translation to human disease cannot be done. Furthermore, no direct correlation has been shown between anti-TNFα use and change in microbiota composition. It can be argued that microbial change could arise indirectly following the immunological changes due to IFX treatment. Finally, other factors, like the mucus layer, are also implied into the pathogenesis of DSS colitis, could exert an important role in these findings.
Overall our data suggest a deep knowledge of the interaction between drug treatment, immune system and microbiota is key to better understand IBD and its response to therapy.
ARTICLE HIGHLIGHTS
Research background
Anti-tumor necrosis factor α (TNFα) represents the best therapeutic option to induce mucosal healing and clinical remission in patients with moderate-severe ulcerative colitis. On the other side gut microbiota plays a crucial role in pathogenesis of ulcerative colitis but few information exists on how microbiota changes following anti-TNFα therapy and on microbiota role in mucosal healing.
Research motivation
The hypothesis behind this study is that anti-TNFα could induce a dysbiosis sustained by immunological changes: dysbiosis could represent one of the reasons for loss of response to anti-TNFα therapy or primary failure.
Research objectives
With this manuscript we aimed to evaluate, in colitic mice as well as healthy mice, intestinal immune system status and gut microbiota modulation induced by anti-TNFα therapy. A more comprehensive approach including gut microbiota modulation, if clarified, could be assessed by dedicated studies on active gut microbiota modulation during biologic therapy in inflammatory bowel disease (IBD).
Research methods
Healthy mice treated with anti-TNFα showed similar histological, microbial and immune features of untreated colitic mice, in particular a lymphomononuclear infiltrate both at 5th and 12th d at hematoxylin and eosin staining, an increase of type 1 helper T lymphocytes (Th1) and type 17 helper T lymphocytes (Th17) at day 12, and finally increase of Enterococcaceae at day 5, a decrease of Bacteroides and Clostridiaceae at day 12.
These findings are particularly relevant to understand the role that anti-TNFα modulation plays on gut microbiota (and vice-versa) also in humans: this finding could be of major interest in order to give more lights on mechanisms of loss of response to anti-TNFα, mechanisms of immunological shift with towards other pathways, like Th17 pathways, as well as novel potential therapeutic targets in IBD.
Research results
Gut microbiota contributes to immune system priming, development and activation. In course of IBD a decrease of Firmicutes (and, among these, the species Faecalibacterium prausnitzii) together with an increase in Proteobacteria has been demonstrated, associated to an increase of T lymphocyte differentiation toward Th1, Th17 subsets and a decrease of CD4+ regulatory T lymphocytes (Tregs). How these conditions are modified in course of medical treatment is not well clarified, particularly when a powerful modulator of the immune system, like anti-TNFα, is utilized.
Research conclusions
Our study confirmed data from the literature. The same features were confirmed in the experimental model of colitis presented in this paper. Furthermore, it was shown that higher representation of Bacteroides, Clostridiaceae and Faecalibacterium prausnitzii and lower representation of Enterococcaceae are associated to a healthy state while the opposite happens during colitis. Anti-TNFα is able to induce changing in gut microbiota toward a health state in course of active colitis, but a more dysbiotic effect when utilized in healthy controls. Healthy related-microbial assessment is associated to higher Treg cells count and lower Th1 and Th17 frequency, condition modified in course of active colitis or use of anti-TNFα in course of healthy condition.
The study conclusion demonstrates a clear relationship between microbial changes and immunological changes in healthy as well as in controls. However, whether these findings correlate to human disease needs to be assessed in a dedicated study as well the need of checking and correction of dysbiosis in patients exposed to anti-TNFα.
Research perspectives
This study strongly suggests a complete assessment of immune system and microbiota in course of powerful immunomodulatory therapy like anti-TNFα and opens new and important considerations. In order to personalize the therapeutic approach to an IBD patient, clinical studies assessing gut microbiota composition before starting an immunomodulatory therapy and in course of therapy are needed. Once an alteration has been demonstrated, the potential role of concomitant microbiota modulation could also be considered for newer and more personalized therapeutic approaches.
ACKNOWLEDGEMENTS
Dr. Alessandro Rettino, from West Midlands Regional Genetics Laboratory, Birmingham Women’s and Children’s NHS Foundation Trust, for language editing.
Footnotes
Institutional animal care and use committee statement: This manuscript was approved by the institutional animal care and use committee, No. NN42 (25/11/2013-25/11/2016).
Conflict-of-interest statement: Authors disclose no any conflict of interest.
Data sharing statement: No additional data are available.
ARRIVE guidelines statement: it was uploaded on Novembre 11th, 2018
Manuscript source: Invited manuscript
Peer-review started: November 14, 2018
First decision: January 6, 2019
Article in press: February 23, 2019
Specialty type: Gastroenterology and hepatology
Country of origin: Italy
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Chiba T, Sitkin S S-Editor: Ma RY L-Editor: A E-Editor: Song H
Contributor Information
Valentina Petito, Istituto di Patologia Speciale Medica, Università Cattolica del Sacro Cuore, Roma 00168, Italy.
Cristina Graziani, UOC di Medicina Interna e Gastroenterologia, Area di Gastroenterologia e Oncologia Medica, Dipartimento di Scienze Gastroenterologiche, Endocrino-Metaboliche e Nefro-Urologiche, Fondazione Policlinico Universitario “A. Gemelli” IRCCS, Roma 00168, Italy.
Loris R Lopetuso, Istituto di Patologia Speciale Medica, Università Cattolica del Sacro Cuore, Rome 00168, Italy; UOC di Medicina Interna e Gastroenterologia, Area di Gastroenterologia e Oncologia Medica, Dipartimento di Scienze Gastroenterologiche, Endocrino-Metaboliche e Nefro-Urologiche, Fondazione Policlinico Universitario “A. Gemelli” IRCCS, Roma 00168, Italy. lopetusoloris@libero.it.
Marco Fossati, Dipartimento delle Scienze della Salute della Donna e del Bambino, Fondazione Policlinico Universitario “A. Gemelli” IRCCS, Roma 00168, Italy.
Alessandra Battaglia, Istituto di Clinica Ostetrica e Ginecologica, Università Cattolica del Sacro Cuore, Roma 00168, Italy.
Vincenzo Arena, U.O.S.A. Gineco-Patologia e Patologia Mammaria, Fondazione Policlinico Universitario “A. Gemelli” IRCCS, Roma 00168, Italy; Istituto di Anatomia Patologica, Università Cattolica del Sacro Cuore, Roma 00168, Italy.
Domenico Scannone, Dipartimento di Anatomia Patologica e Istologia, Fondazione Policlinico Universitario “A. Gemelli” IRCCS, Roma 00168, Italy.
Gianluca Quaranta, Dipartimento di Microbiologia, Fondazione Policlinico Universitario “A. Gemelli” IRCCS, Roma 00168, Italy; Istituto di Microbiologia, Università Cattolica del Sacro Cuore, Roma 00168, Italy.
Andrea Quagliariello, Unità di Microbioma Umano, Ospedale Pediatrico Bambino Gesù IRCCS, Roma 00146, Italy.
Federica Del Chierico, Unità di Microbioma Umano, Ospedale Pediatrico Bambino Gesù IRCCS, Roma 00146, Italy.
Lorenza Putignani, Unità di Microbioma Umano, Ospedale Pediatrico Bambino Gesù IRCCS, Roma 00146, Italy; Unità di Parassitologia, Ospedale Pediatrico Bambino Gesù IRCCS, Roma 00146, Italy.
Luca Masucci, Dipartimento di Microbiologia, Fondazione Policlinico Universitario “A. Gemelli” IRCCS, Roma 00168, Italy; Istituto di Microbiologia, Università Cattolica del Sacro Cuore, Roma 00168, Italy.
Maurizio Sanguinetti, Dipartimento di Microbiologia, Fondazione Policlinico Universitario “A. Gemelli” IRCCS, Roma 00168, Italy; Istituto di Microbiologia, Università Cattolica del Sacro Cuore, Roma 00168, Italy.
Alessandro Sgambato, Istituto di Patologia Generale, Università Cattolica del Sacro Cuore, Roma 00168, Italy.
Antonio Gasbarrini, Istituto di Patologia Speciale Medica, Università Cattolica del Sacro Cuore, Roma 00168, Italy; UOC di Medicina Interna e Gastroenterologia, Area di Gastroenterologia e Oncologia Medica, Dipartimento di Scienze Gastroenterologiche, Endocrino-Metaboliche e Nefro-Urologiche, Fondazione Policlinico Universitario “A. Gemelli” IRCCS, Roma 00168, Italy.
Franco Scaldaferri, Istituto di Patologia Speciale Medica, Università Cattolica del Sacro Cuore, Roma 00168, Italy; UOC di Medicina Interna e Gastroenterologia, Area di Gastroenterologia e Oncologia Medica, Dipartimento di Scienze Gastroenterologiche, Endocrino-Metaboliche e Nefro-Urologiche, Fondazione Policlinico Universitario “A. Gemelli” IRCCS, Roma 00168, Italy.
References
- 1.Pott J, Kabat AM, Maloy KJ. Intestinal Epithelial Cell Autophagy Is Required to Protect against TNF-Induced Apoptosis during Chronic Colitis in Mice. Cell Host Microbe. 2018;23:191–202.e4. doi: 10.1016/j.chom.2017.12.017. [DOI] [PubMed] [Google Scholar]
- 2.Scaldaferri F, Fiocchi C. Inflammatory bowel disease: progress and current concepts of etiopathogenesis. J Dig Dis. 2007;8:171–178. doi: 10.1111/j.1751-2980.2007.00310.x. [DOI] [PubMed] [Google Scholar]
- 3.Benchimol EI, Fortinsky KJ, Gozdyra P, Van den Heuvel M, Van Limbergen J, Griffiths AM. Epidemiology of pediatric inflammatory bowel disease: a systematic review of international trends. Inflamm Bowel Dis. 2011;17:423–439. doi: 10.1002/ibd.21349. [DOI] [PubMed] [Google Scholar]
- 4.Mir-Madjlessi SH, Michener WM, Farmer RG. Course and prognosis of idiopathic ulcerative proctosigmoiditis in young patients. J Pediatr Gastroenterol Nutr. 1986;5:571–575. [PubMed] [Google Scholar]
- 5.Molodecky NA, Soon IS, Rabi DM, Ghali WA, Ferris M, Chernoff G, Benchimol EI, Panaccione R, Ghosh S, Barkema HW, Kaplan GG. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology. 2012;142:46–54.e42; quiz e30. doi: 10.1053/j.gastro.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 6.Ye Y, Pang Z, Chen W, Ju S, Zhou C. The epidemiology and risk factors of inflammatory bowel disease. Int J Clin Exp Med. 2015;8:22529–22542. [PMC free article] [PubMed] [Google Scholar]
- 7.Ng SC, Shi HY, Hamidi N, Underwood FE, Tang W, Benchimol EI, Panaccione R, Ghosh S, Wu JCY, Chan FKL, Sung JJY, Kaplan GG. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet. 2018;390:2769–2778. doi: 10.1016/S0140-6736(17)32448-0. [DOI] [PubMed] [Google Scholar]
- 8.Yang L, Liu C, Zhao W, He C, Ding J, Dai R, Xu K, Xiao L, Luo L, Liu S, Li W, Meng H. Impaired Autophagy in Intestinal Epithelial Cells Alters Gut Microbiota and Host Immune Responses. Appl Environ Microbiol. 2018:84. doi: 10.1128/AEM.00880-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 10.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, Essers J, Mitrovic M, Ning K, Cleynen I, Theatre E, Spain SL, Raychaudhuri S, Goyette P, Wei Z, Abraham C, Achkar JP, Ahmad T, Amininejad L, Ananthakrishnan AN, Andersen V, Andrews JM, Baidoo L, Balschun T, Bampton PA, Bitton A, Boucher G, Brand S, Büning C, Cohain A, Cichon S, D'Amato M, De Jong D, Devaney KL, Dubinsky M, Edwards C, Ellinghaus D, Ferguson LR, Franchimont D, Fransen K, Gearry R, Georges M, Gieger C, Glas J, Haritunians T, Hart A, Hawkey C, Hedl M, Hu X, Karlsen TH, Kupcinskas L, Kugathasan S, Latiano A, Laukens D, Lawrance IC, Lees CW, Louis E, Mahy G, Mansfield J, Morgan AR, Mowat C, Newman W, Palmieri O, Ponsioen CY, Potocnik U, Prescott NJ, Regueiro M, Rotter JI, Russell RK, Sanderson JD, Sans M, Satsangi J, Schreiber S, Simms LA, Sventoraityte J, Targan SR, Taylor KD, Tremelling M, Verspaget HW, De Vos M, Wijmenga C, Wilson DC, Winkelmann J, Xavier RJ, Zeissig S, Zhang B, Zhang CK, Zhao H, Silverberg MS, Annese V, Hakonarson H, Brant SR, Radford-Smith G, Mathew CG, Rioux JD, Schadt EE, Daly MJ, Franke A, Parkes M, Vermeire S, Barrett JC, Cho JH International IBD Genetics Consortium (IIBDGC) Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neurath MF. Cytokines in inflammatory bowel disease. Nat Rev Immunol. 2014;14:329–342. doi: 10.1038/nri3661. [DOI] [PubMed] [Google Scholar]
- 12.Jones-Hall YL, Nakatsu CH. The Intersection of TNF, IBD and the Microbiome. Gut Microbes. 2016;7:58–62. doi: 10.1080/19490976.2015.1121364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Himmel ME, Yao Y, Orban PC, Steiner TS, Levings MK. Regulatory T-cell therapy for inflammatory bowel disease: more questions than answers. Immunology. 2012;136:115–122. doi: 10.1111/j.1365-2567.2012.03572.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sokol H, Seksik P, Furet JP, Firmesse O, Nion-Larmurier I, Beaugerie L, Cosnes J, Corthier G, Marteau P, Doré J. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis. 2009;15:1183–1189. doi: 10.1002/ibd.20903. [DOI] [PubMed] [Google Scholar]
- 16.Bernstein CN, Forbes JD. Gut Microbiome in Inflammatory Bowel Disease and Other Chronic Immune-Mediated Inflammatory Diseases. Inflamm Intest Dis. 2017;2:116–123. doi: 10.1159/000481401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sartor RB, Wu GD. Roles for Intestinal Bacteria, Viruses, and Fungi in Pathogenesis of Inflammatory Bowel Diseases and Therapeutic Approaches. Gastroenterology. 2017;152:327–339.e4. doi: 10.1053/j.gastro.2016.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walters WA, Xu Z, Knight R. Meta-analyses of human gut microbes associated with obesity and IBD. FEBS Lett. 2014;588:4223–4233. doi: 10.1016/j.febslet.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lopetuso LR, Petito V, Graziani C, Schiavoni E, Paroni Sterbini F, Poscia A, Gaetani E, Franceschi F, Cammarota G, Sanguinetti M, Masucci L, Scaldaferri F, Gasbarrini A. Gut Microbiota in Health, Diverticular Disease, Irritable Bowel Syndrome, and Inflammatory Bowel Diseases: Time for Microbial Marker of Gastrointestinal Disorders. Dig Dis. 2018;36:56–65. doi: 10.1159/000477205. [DOI] [PubMed] [Google Scholar]
- 20.Wright EK, Kamm MA, Teo SM, Inouye M, Wagner J, Kirkwood CD. Recent advances in characterizing the gastrointestinal microbiome in Crohn's disease: a systematic review. Inflamm Bowel Dis. 2015;21:1219–1228. doi: 10.1097/MIB.0000000000000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larmonier CB, Shehab KW, Ghishan FK, Kiela PR. T Lymphocyte Dynamics in Inflammatory Bowel Diseases: Role of the Microbiome. Biomed Res Int. 2015;2015:504638. doi: 10.1155/2015/504638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ivanov II, Frutos Rde L, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, Finlay BB, Littman DR. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4:337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Atarashi K, Nishimura J, Shima T, Umesaki Y, Yamamoto M, Onoue M, Yagita H, Ishii N, Evans R, Honda K, Takeda K. ATP drives lamina propria T(H)17 cell differentiation. Nature. 2008;455:808–812. doi: 10.1038/nature07240. [DOI] [PubMed] [Google Scholar]
- 24.Holleran G, Lopetuso L, Petito V, Graziani C, Ianiro G, McNamara D, Gasbarrini A, Scaldaferri F. The Innate and Adaptive Immune System as Targets for Biologic Therapies in Inflammatory Bowel Disease. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18102020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lopetuso LR, Petito V, Cufino V, Arena V, Stigliano E, Gerardi V, Gaetani E, Poscia A, Amato A, Cammarota G, Papa A, Sgambato A, Gasbarrini A, Scaldaferri F. Locally injected Infliximab ameliorates murine DSS colitis: differences in serum and intestinal levels of drug between healthy and colitic mice. Dig Liver Dis. 2013;45:1017–1021. doi: 10.1016/j.dld.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 26.Ito R, Shin-Ya M, Kishida T, Urano A, Takada R, Sakagami J, Imanishi J, Kita M, Ueda Y, Iwakura Y, Kataoka K, Okanoue T, Mazda O. Interferon-gamma is causatively involved in experimental inflammatory bowel disease in mice. Clin Exp Immunol. 2006;146:330–338. doi: 10.1111/j.1365-2249.2006.03214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stalnikowicz R, Pollak D, Eliakim A, Wengrower D, Fich A, Goldin E, Ligumsky M, Rachmilewitz D. Cimetidine decreases indomethacin induced duodenal mucosal damage in patients with acute musculoskeletal disorders. Gut. 1988;29:1578–1582. doi: 10.1136/gut.29.11.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Papa E, Docktor M, Smillie C, Weber S, Preheim SP, Gevers D, Giannoukos G, Ciulla D, Tabbaa D, Ingram J, Schauer DB, Ward DV, Korzenik JR, Xavier RJ, Bousvaros A, Alm EJ. Non-invasive mapping of the gastrointestinal microbiota identifies children with inflammatory bowel disease. PLoS One. 2012;7:e39242. doi: 10.1371/journal.pone.0039242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lê S, Josse J, Husson F. FactoMineR: An R Package for Multivariate Analysis. J Statistical Software. 2008:25. [Google Scholar]
- 30.Wickham H. New York: Springer; 2009. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
- 31.Edelblum KL, Sharon G, Singh G, Odenwald MA, Sailer A, Cao S, Ravens S, Thomsen I, El Bissati K, McLeod R, Dong C, Gurbuxani S, Prinz I, Mazmanian SK, Turner JR. The Microbiome Activates CD4 T-cell-mediated Immunity to Compensate for Increased Intestinal Permeability. Cell Mol Gastroenterol Hepatol. 2017;4:285–297. doi: 10.1016/j.jcmgh.2017.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanoue T, Honda K. Induction of Treg cells in the mouse colonic mucosa: a central mechanism to maintain host-microbiota homeostasis. Semin Immunol. 2012;24:50–57. doi: 10.1016/j.smim.2011.11.009. [DOI] [PubMed] [Google Scholar]
- 33.Chow J, Mazmanian SK. A pathobiont of the microbiota balances host colonization and intestinal inflammation. Cell Host Microbe. 2010;7:265–276. doi: 10.1016/j.chom.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Natividad JM, Verdu EF. Modulation of intestinal barrier by intestinal microbiota: pathological and therapeutic implications. Pharmacol Res. 2013;69:42–51. doi: 10.1016/j.phrs.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 35.Wang K, Jin X, You M, Tian W, Le Leu RK, Topping DL, Conlon MA, Wu L, Hu F. Dietary Propolis Ameliorates Dextran Sulfate Sodium-Induced Colitis and Modulates the Gut Microbiota in Rats Fed a Western Diet. Nutrients. 2017;9 doi: 10.3390/nu9080875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sartor RB. Mechanisms of disease: pathogenesis of Crohn's disease and ulcerative colitis. Nat Clin Pract Gastroenterol Hepatol. 2006;3:390–407. doi: 10.1038/ncpgasthep0528. [DOI] [PubMed] [Google Scholar]
- 37.Okayasu I, Hatakeyama S, Yamada M, Ohkusa T, Inagaki Y, Nakaya R. A novel method in the induction of reliable experimental acute and chronic ulcerative colitis in mice. Gastroenterology. 1990;98:694–702. doi: 10.1016/0016-5085(90)90290-h. [DOI] [PubMed] [Google Scholar]
- 38.Berry D, Kuzyk O, Rauch I, Heider S, Schwab C, Hainzl E, Decker T, Müller M, Strobl B, Schleper C, Urich T, Wagner M, Kenner L, Loy A. Intestinal Microbiota Signatures Associated with Inflammation History in Mice Experiencing Recurring Colitis. Front Microbiol. 2015;6:1408. doi: 10.3389/fmicb.2015.01408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Munyaka PM, Rabbi MF, Khafipour E, Ghia JE. Acute dextran sulfate sodium (DSS)-induced colitis promotes gut microbial dysbiosis in mice. J Basic Microbiol. 2016;56:986–998. doi: 10.1002/jobm.201500726. [DOI] [PubMed] [Google Scholar]
- 40.Bossen C, Ingold K, Tardivel A, Bodmer JL, Gaide O, Hertig S, Ambrose C, Tschopp J, Schneider P. Interactions of tumor necrosis factor (TNF) and TNF receptor family members in the mouse and human. J Biol Chem. 2006;281:13964–13971. doi: 10.1074/jbc.M601553200. [DOI] [PubMed] [Google Scholar]
- 41.Mikami S, Nakase H, Yamamoto S, Takeda Y, Yoshino T, Kasahara K, Ueno S, Uza N, Oishi S, Fujii N, Nagasawa T, Chiba T. Blockade of CXCL12/CXCR4 axis ameliorates murine experimental colitis. J Pharmacol Exp Ther. 2008;327:383–392. doi: 10.1124/jpet.108.141085. [DOI] [PubMed] [Google Scholar]
- 42.Scaldaferri F, Pecere S, Petito V, Zambrano D, Fiore L, Lopetuso LR, Schiavoni E, Bruno G, Gerardi V, Laterza L, Pizzoferrato M, Ianiro G, Stojanovic J, Poscia A, Papa A, Paroni Sterbini F, Sanguinetti M, Masucci L, Cammarota G, Gasbarrini A. Efficacy and Mechanisms of Action of Fecal Microbiota Transplantation in Ulcerative Colitis: Pitfalls and Promises From a First Meta-Analysis. Transplant Proc. 2016;48:402–407. doi: 10.1016/j.transproceed.2015.12.040. [DOI] [PubMed] [Google Scholar]
- 43.Sands BE, Sandborn WJ, Panaccione R, O'Brien CD, Zhang H, Johanns J, Peyrin-Biroulet L, van Assche G, Danese S, Targan S, Abreu MT, Hisamatsu T, Szapary P, Marano C. Safety and Efficacy of Ustekinumab Induction Therapy in Patients With Moderate to Severe Ulcerative Colitis: Results From the Phase 3 UNIFI Study (late-breaking abstract at UEGW 2018) Available from: URL: https://www.ueg.eu/education/document/safety-and-efficacy-of-ustekinumab-induction-therapy-in-patients-with-moderate-to-severe-ulcerative-colitis-results-from-the-phase-3-unifi-study/180031/