Abstract
Glycosylphosphatidylinositols (GPIs) are linked to many cell-surface proteins, anchor these proteins in the membrane, and are well characterized. However, GPIs that exist in the free form on the mammalian cell surface remain largely unexplored. To investigate free GPIs in cultured cell lines and mouse tissues, here we used the T5-4E10 mAb (T5 mAb), which recognizes unlinked GPIs having an N-acetylgalactosamine (GalNAc) side chain linked to the first mannose at the nonreducing terminus. We detected free GPIs bearing the GalNAc side chain on the surface of Neuro2a and CHO, but not of HEK293, K562, and C2C12 cells. Furthermore, free GPIs were present in mouse pons, medulla oblongata, spinal cord, testis, epididymis, and kidney. Using a panel of Chinese hamster ovary cells defective in both GPI-transamidase and GPI remodeling pathway, we demonstrate that free GPIs follow the same structural remodeling pathway during passage from the endoplasmic reticulum to the plasma membrane as do protein-linked GPI. Specifically, free GPIs underwent post-GPI attachment to protein 1 (PGAP1)-mediated inositol deacylation, PGAP5-mediated removal of the ethanolamine phosphate from the second mannose, and PGAP3- and PGAP2-mediated fatty acid remodeling. Moreover, T5 mAb recognized free GPIs even if the inositol-linked acyl chain or ethanolamine-phosphate side chain linked to the second mannose is not removed. In contrast, addition of a fourth mannose by phosphatidylinositol glycan anchor biosynthesis class Z (PIGZ) inhibited T5 mAb-mediated detection of free GPIs. Our results indicate that free GPIs are normal components of the plasma membrane in some tissues and further characterize free GPIs in mammalian cells.
Keywords: glycosylphosphatidylinositol (GPI anchor), glycolipid, galactose, membrane lipid, glycosylation, post-translational modification (PTM), fatty acid remodeling, galactose, glycan biology, GPI transamidase, N-acetylgalactosamine
Introduction
Glycosylphosphatidylinositols (GPIs)3 act as membrane anchors of many cell-surface proteins in eukaryotic cells. GPI-anchoring is a ubiquitous mode of post-translational modification of proteins used for their cell-surface localization (1). In humans, at least 150 different proteins are GPI-anchored (2). Mammalian GPI-anchored proteins (GPI-APs) have various functions, such as enzymes, receptors, adhesion molecules, and complement regulatory proteins, and play critical roles in processes such as embryogenesis, neurogenesis, the immune system, and fertilization (3).
GPI is synthesized in the endoplasmic reticulum (ER) and preassembled GPI is transferred en bloc to precursor proteins that have a GPI-attachment signal peptide at the C terminus. The GPI-attachment signal peptide is cleaved off and replaced with GPI by GPI transamidase (GPI-Tase). Nascent GPI-APs undergo GPI maturation reactions in the ER and the Golgi apparatus. The mature GPI-APs are displayed on the cell surface. The structure of the core of GPI is protein-EtNP-6Manα-2Manα-6Manα-4GlcNα-6inositol phospholipid (where EtNP, Man, and GlcN are ethanolamine phosphate, mannose, and glucosamine, respectively) that is conserved among various organisms from protozoa to mammals (Fig. 1A). Man linked to GlcN is termed the first Man (Man1); Man linked to Man1 is termed the second Man (Man2), and Man linked to Man2 is termed the third Man (Man3). The core glycan moiety is variously modified by side chains in different organisms and in different proteins.
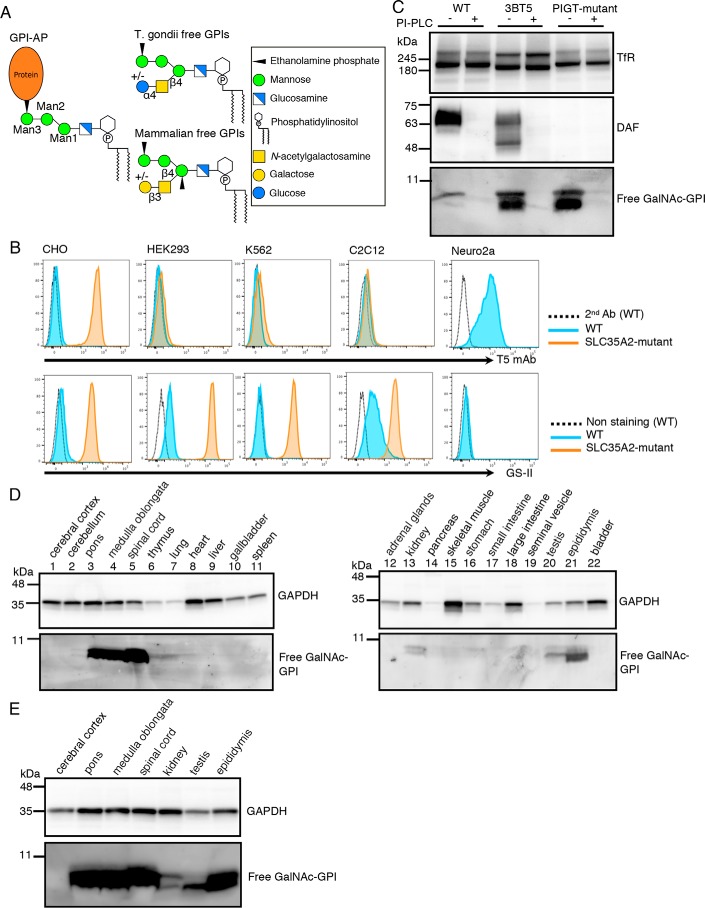
Figure 1.
Detection of free GPIs in some cultured cell lines and mouse tissues using T5 mAb. A, common structure of GPI-AP and structures of free GPIs from T. gondii and mammals. Monosaccharide symbols are according to the Symbol Nomenclature for Glycans (40). B, flow cytometric analysis of wildtype (WT) and SLC35A2 (UDP-Gal transporter)-deficient cell lines. WT (blue) or SLC35A2-mutant (orange) CHO, HEK293, K562, C2C12, and Neuro2a cells were stained with T5 mAb plus secondary antibody for free GPI (top) or with lectin GS-II for nonreducing terminal GlcNAc (bottom). Dotted lines, WT cells stained by secondary antibody only. C, Western blotting of free GPIs of CHO cells. Lysates of WT (3B2A), SLC35A2-mutant (3BT5), and PIGT-mutant cells treated with or without PI-PLC were analyzed by Western blotting. DAF, a GPI-AP; TfR, a loading control. D and E, Western blot analysis of various mouse tissue lysates using T5 mAb. GAPDH was used as a loading control.
The protozoan parasite Toxoplasma gondii expresses nonprotein-linked GPI as free GPI, as well as various GPI-APs. T. gondii-free GPI has a disaccharide side chain linked to Man1 (Fig. 1A). The disaccharide consists of glucose α1,4-linked to GalNAc that, in turn, is linked to Man1 via a β1,4-bond (4). The toxoplasma-free GPI is highly immunogenic and causes IgM response in infected mice (5, 6). A number of mAbs against free GPI have been established and their epitopes analyzed. One of them, named T5-4E10 mAb (T5 mAb) recognized free GPI only when the glucose capping was absent, indicating that the epitope for T5 mAb includes GalNAc exposed as the nonreducing end of the side chain. It was also shown that T5 mAb recognized free GPI but not GPI-APs (7, 8), suggesting that the terminal EtNP must be free for recognition by T5 mAb.
Earlier studies demonstrated the presence of free, nonprotein-linked GPIs on the cell surface of several mammalian cultured cell lines (9–11). Characteristics of the reported free GPIs are similar to those of fully or almost fully assembled GPI that are competent for attachment to proteins. However, whether they have undergone maturation steps, including side chain modifications that occur in protein-linked GPI-anchors, has been unclear. Many mammalian GPI-APs have a GalNAc side chain β1,4-linked to Man1, similar to Toxoplasma-free GPI. The mammalian GalNAc side chain can be elongated by β1,3-linked galactose (Gal) (12) and further by sialic acid (13). We recently found that Chinese hamster ovary (CHO) cells defective in SLC35A2 gene encoding a UDP-Gal transporter were strongly stained by T5 mAb as analyzed by flow cytometry, whereas WT CHO cells were negative or only weakly stained (14). It was suggested, therefore, that WT CHO cells express free GPIs bearing a Gal-capped GalNAc side chain and that SLC35A2-defective CHO cells express GalNAc-exposed free GPIs (Fig. 1A). It is unknown how widely such free GPIs are present on various cultured cell lines and cells in various mammalian tissues. Physiological roles of free GPIs in mammals are currently also unclear.
Although structural and functional information on mammalian-free GPIs is limited, a possible pathological role of free GPIs present in GPI-Tase-defective cells is being studied. Paroxysmal nocturnal hemoglobinuria (PNH) is an acquired hemolytic disease caused by somatic mutations of the PIGA gene in hematopoietic stem cells. Because PIGA is essential for the initial step in GPI biosynthesis, no GPI (or its biosynthetic intermediates) are generated in PIGA-defective cells and precursors of GPI-APs are degraded, resulting in GPI-AP deficiency. Affected red blood cells are highly sensitive to complement due to a lack of GPI-anchored complement regulatory proteins CD59 and DAF, leading to complement-mediated hemolysis (15). In contrast, patients with atypical PNH, caused by mutations in the PIGT gene, which encodes a component of GPI-Tase, have various autoinflammatory symptoms, such as urticaria, joint pain, fever, and noninfectious meningitis, in addition to hemolysis (16, 17). GPI is assembled, but not used for protein membrane anchoring, in PIGT-defective cells. It is therefore likely that nonprotein-linked free GPI is causally related to the autoinflammatory symptoms seen in PNH caused by PIGT mutations. How the nonprotein anchor GPIs are involved in autoinflammatory symptoms is a current focus of investigation.
Here, we report detection of free GPIs using T5 mAb in both cultured cell lines and mouse tissues, indicating that free GPIs are membrane components of normal mammalian cells. To further characterize structures of free GPIs, we used mutant CHO cells simultaneously defective in GPI-Tase and one of the genes in the GPI maturation pathway, and tested the binding of T5 mAb to the affected free GPIs. Our results indicate that free GPIs undergo similar structural remodeling to GPI-APs.
Results
Free, nonprotein-anchor GPIs are cell membrane glycolipids of some cultured cell lines and mouse tissues
T5 mAb is the only currently available probe to specifically detect free, nonprotein-anchor GPI in mammalian cells. The binding specificity of T5 mAb was partially determined using mutant CHO cells (14). T5 mAb bound to SLC35A2-defective CHO cells, whereas knockout (KO) of GalNAc transferase PGAP4 (also known as TMEM246 or C9orf125) in SLC35A2-defective CHO cells caused complete loss of T5 mAb binding. Therefore, T5 mAb binds to free GPI only when a GalNAc side chain is linked to Man1 and is not capped by Gal.
To determine whether free GPIs are widely expressed cell membrane components in cultured cell lines, we analyzed HEK293 (human embryonic kidney), K562 (human erythroleukemia), C2C12 (mouse myoblast), and Neuro2a (mouse neuroblastoma) cells by flow cytometry after staining with T5 mAb. Neuro2a cells, but not the others, were positively stained by T5 mAb (Fig. 1B, top panels), suggesting the expression of free GPI with exposed GalNAc on Neuro2a cells. To remove potential Gal-capping of GalNAc, we generated SLC35A2 KO cells from HEK293, K562, and C2C12. KO of SLC35A2 was confirmed by GS-II lectin staining. Consistent with defective galactosylation of glycoproteins, these SLC35A2 KO cells were strongly stained by GS-II (recognizing nonreducing terminal GlcNAc) (Fig. 1B, bottom panels). Nevertheless, these SLC35A2 KO cells were almost negative for staining by T5 mAb (Fig. 1B, top panels). These results indicate that only some cultured cell lines express free GPI detectable by T5 mAb, and that the free GPIs have structural variation. Specifically, CHO cells express free GPI bearing a galactosylated GalNAc side chain; Neuro2a cells express free GPI bearing a nongalactosylated GalNAc side chain; and HEK293, K562, and C2C12 cells rarely express GalNAc-bearing free GPI. The presence of free GPI lacking a GalNAc side chain cannot be excluded because T5 mAb recognition is GalNAc-dependent (14).
We then used T5 mAb in Western blotting. Free GPI in CHO 3BT5 (SLC35A2-mutant) cell extracts appeared as two closely adjacent bands below the 11-kDa marker, suggesting some structural heterogeneity of the GalNAc-exposed free GPI (Fig. 1C). Only a weak upper band appeared in WT CHO cell extracts. We reported that T5 mAb staining of CHO 3BT5 cells was completely eliminated by treatment with phosphatidylinositol-specific phospholipase C (PI-PLC) (14). Western blot analysis of the detergent extracts prepared from PI-PLC-treated cells did not detect any T5 mAb-reactive band, indicating almost all free GPI detected by T5 mAb was on the cell surface (Fig. 1C). No specific bands corresponding to GPI-APs were seen in the 180- to 11-kDa region, confirming that T5 mAb is specific for free GPI, as reported for T. gondii-free GPI (Fig. S1).
To test the presence and distribution of free GPIs in mammalian tissues, we analyzed detergent extracts of various tissues from adult male mice by Western blotting. Free GPIs of molecular sizes similar to those from CHO 3BT5 cells were strongly detected by T5 mAb in pons, medulla oblongata, spinal cord, and epididymis, and weakly in kidney and testis (Fig. 1, D and E). Other tissues tested (cerebral cortex, cerebellum, thymus, lung, heart, liver, gallbladder, spleen, adrenal gland, pancreas, skeletal muscle, stomach, small and large intestines, seminal vesicles, and bladder) did not have, or had only a trace amount of, free GPI detectable by T5 mAb (Fig. 1D). Distribution of free GPI bearing a Gal-capped GalNAc side chain cannot be evaluated because Gal transfer defective mice to eliminate Gal-capping or probes for Gal-capped free GPI are not available at the moment. Furthermore, free GPI lacking a GalNAc side chain cannot be studied either, because proper probes have not yet been developed. Nevertheless, these results indicate that cell-surface free GPIs are cell membrane glycolipids of some cultured cell lines and mouse tissues and they may be regulated in a cell/tissue-specific way.
Cell-surface appearance of free, nonprotein-anchor GPI when GPI-Tase is defective
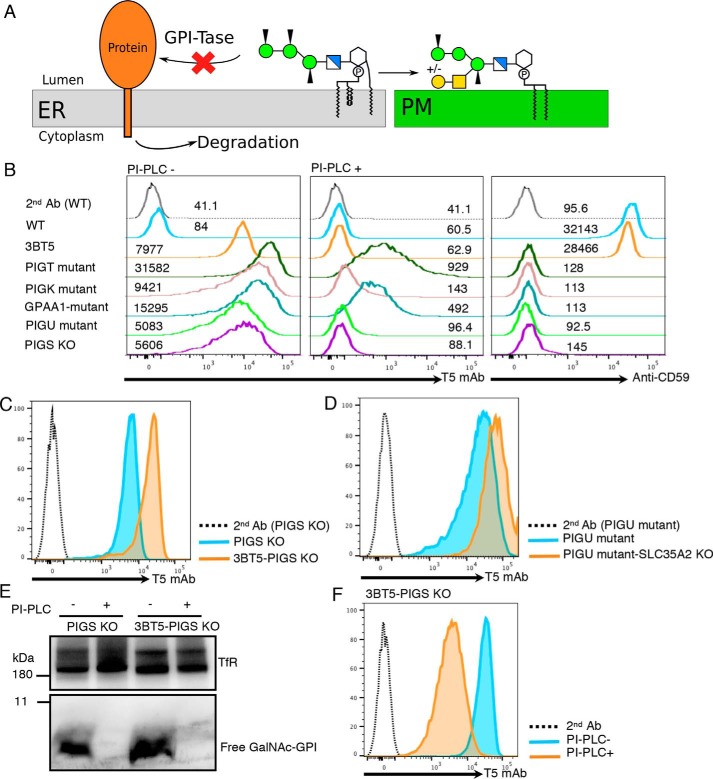
We next followed the fate of GPIs when they were not attached to proteins (schematic in Fig. 2A). Mammalian GPI-Tase is a complex of five proteins PIGK, GPAA1, PIGS, PIGT, and PIGU. We used mutant CHO cells defective in PIGT, PIGK, GPAA1, or PIGU, and PIGS KO CHO cells (see Table 1 for KO cells used in this study). All these cells lost GPI-AP (CD59) as expected (Fig. 2B, right panel) and were stained by T5 mAb (Fig. 2B, left panel). Although the staining intensity varies among these GPI-Tase–defective cells, the result indicates that GPIs that failed to attach to proteins in the ER were transported to the cell surface after modification by GalNAc side chain, and some of them were displayed on the cell surface without modification by Gal. This is in contrast to free GPIs on GPI-Tase–sufficient CHO cells because their GalNAc side chains were fully galactosylated and detectable by T5 mAb only when SLC35A2 is defective (WT versus 3BT5 in Fig. 2B, left panel). The variation in T5 staining intensities of GPI-Tase–defective cells might be caused by variation in Gal modification. Because the T5 mAb staining levels of PIGU-mutant and PIGS-KO CHO cells were lowest among the GPI-Tase–deficient cells (Fig. 2B, left panel), we tested this possibility by knocking out PIGS in SLC35A2-defective CHO-3BT5 cells (Fig. S2A). Mean fluorescence intensity of T5 mAb staining was 3.6 times that of PIGS KO cells derived from WT CHO-3B2A cells (17,483 versus 4,853) (Fig. 2C). Therefore, non-Gal–capped GPI accounted for only 27.8% of GalNAc side chain-bearing free GPI and the majority (72.2%) had a Gal–capped GalNAc side chain in PIGS KO cells. Similarly, T5 mAb staining mean fluorescence intensity of SLC35A2 KO cells derived from PIGU-mutant CHO cells (Fig. S2B) was 4.1 times that of PIGU-mutant CHO cells (57,014 versus 17,391) (Fig. 2D), indicating only 22.8% of the free GPI had a GalNAc-exposed side chain in PIGU-mutant cells. Collectively, free GPIs in PIGU-mutant cells and PIGS-KO CHO cells are modified in the Golgi during the transport to the cell surface at the GalNAc side chain, which is mainly elongated by Gal.
Figure 2.
Appearance of free GPIs on the cell surfaces of GPI-Tase–deficient cells. A, fate of GPI in GPI-Tase–deficient cells. GPI, which is not transferred to a precursor protein in the ER because of defective GPI-Tase, is transported to the plasma membrane (PM) as free GPI, whereas the precursor protein is degraded. B, flow cytometric analysis of GPI-Tase–defective CHO cells. 3B2A (WT), 3BT5 (SLC35A2-mutant), PIGT-, PIGK-, GPAA1-, and PIGU-mutant and PIGS KO CHO cells were stained with T5 mAb before (PI-PLC −) and after (PI-PLC +) treatment with PI-PLC. Mean fluorescence intensities are given above each line. C, flow cytometric analysis of 3B2A-PIGS KO (blue) and 3BT5-PIGS KO (orange) cells stained with T5 mAb and Alexa Fluor 488 secondary antibody. Dotted line, 3BT5-PIGS KO cells stained with secondary antibody alone. D, flow cytometric analysis of PIGU-mutant (blue) and PIGU-mutant SLC35A2 KO (orange) CHO cells. Cells were stained with T5 mAb and Alexa Fluor 488 secondary antibody. Dotted line, PIGU-mutant CHO cells stained with secondary antibody alone. E, Western blotting of free GPIs of 3B2A-PIGS KO (left) and 3BT5-PIGS KO (right) cells. Cells were treated with or without PI-PLC, lysed, and analyzed by Western blotting using T5 mAb. TfR, a loading control. F, PI-PLC sensitivity of free GPIs of 3BT5-PIGS KO cells. 3BT5-PIGS KO cells were treated with (orange) or without (blue) PI-PLC and stained with T5 mAb and Alexa Fluor 488 secondary antibody. C–F, results were reproducible in at least two independent experiments.
Table 1.
CHO cell lines used in this study
| Cells | Parent cells | Phenotype | Reference |
|---|---|---|---|
| 3B2A | CHO-K1 | Wildtype expressing human CD59 and DAF | 33 |
| PIGS KO | 3B2A | PIGS-deficient | This study |
| 3BT5 | 3B2A | SLC35A2-deficient (Lec8 phenotype) | 14 |
| 3BT5-PIGS KO | 3BT5 | SLC35A2 and PIGS-deficient | This study |
| C19 | 3B2A-FF8 | PGAP5-deficient | 19 |
| C19-PIGS KO | C19 | PGAP5 and PIGS-deficient | This study |
| C19-PIGS-SLC35A2 DKO | C19-PIGS KO | PGAP5, PIGS, and SLC35A2-deficient | This study |
| C10 | 3B2A | PGAP1-deficient | 18 |
| C10-PIGS KO | C10 | PGAP1 and PIGS-deficient | This study |
| C10-PIGS-SLC35A2 DKO | C10-PIGS KO | PGAP1, PIGS, and SLC35A2-deficient | This study |
| 3BT5-PGAP3 KO | 3BT5 | PGAP3 & SLC35A2-deficient | 14 |
| 3BT5-PGAP3-PIGS DKO | 3BT5-PGAP3 KO | PGAP3, SLC35A2, and PIGS-deficient | This study |
| 3BT5-PIGS-PGAP2 DKO | 3BT5-PIGS KO | PGAP2, SLC35A2, and PIGS-deficient | This study |
| 3BT5-PIGZ KO | 3BT5 | PIGZ and SLC35A2-deficient | This study |
More than 97% of T5 mAb-reactive free GPIs on the GPI-Tase–defective CHO cells were removed from the cell surface by treatment with PI-PLC (Fig. 2B, middle panel). The T5 mAb-reactive free GPI detected by Western blotting disappeared almost completely after PI-PLC treatment of PIGT-mutant cells (Fig. 1C, right two lanes) and PIGS KO cells (Fig. 2E, left two lanes). Therefore, almost all the T5 mAb–reactive free GPI was present on the cell surface in the GPI-Tase–defective cells.
Free GPIs undergo maturation reactions in the ER of GPI-Tase–defective cells
We next asked whether free GPIs in GPI-Tase–defective cells undergo maturation steps similar to protein-linked GPI during transport from the ER to the Golgi and the plasma membrane. To gain insight into any difference in free GPI maturation in WT cells, we compared free GPIs in GPI-Tase–defective and -sufficient CHO cells whenever possible.
The fully assembled GPI, which is competent for GPI-Tase–mediated attachment to proteins, has a fatty acyl chain linked to the 2-position of inositol and an EtNP group linked to Man1, Man2, and Man3, respectively (Fig. 2A). The first maturation reaction in the ER is inositol deacylation mediated by PGAP1 (18). The GPI moiety is resistant to PI-PLC when inositol is acylated and is sensitive after deacylation. As already shown in Fig. 2B (middle panel), T5 mAb-detectable free GPIs on GPI-Tase–defective CHO cells, including PIGS KO cells, were almost completely sensitive to PI-PLC treatment, indicating that inositol deacylation occurs in free GPI. In contrast to PIGS KO cells, free GPIs on 3BT5-PIGS KO cells contained a population resistant to PI-PLC as shown by a partial reduction in T5 mAb staining (86.1 ± 6.9% (mean ± S.D., n = 3) reduction) after PI-PLC treatment (Fig. 2F). As shown above in Fig. 2C, PIGS KO cells should also have Gal-capped free GPIs, which are not detected by T5 mAb. Therefore, these results suggest that a fraction of free GPIs in PIGS KO cells left the ER without inositol deacylation and were modified by GalNAc and Gal, whereas the majority of free GPIs were inositol deacylated before the ER exit, were modified by GalNAc, and remained either as GalNAc-exposed free GPIs or were expressed as Gal-capped free GPIs. As reported previously (14) and also shown in Fig. 1C (middle), both GPI-APs (DAF in Fig. 1C) and free GPI in GPI-Tase–sufficient CHO cells are completely sensitive to PI-PLC. It seems possible that inositol deacylation is less efficient in GPI-Tase–defective CHO cells.
The second maturation reaction in the ER is removal of the EtNP side chain from Man2, mediated by PGAP5 (19). To determine whether free GPIs undergo PGAP5-mediated reaction in GPI-Tase–defective CHO cells, we knocked out PIGS in PGAP5-defective CHO-C19 cells (termed C19-PIGS KO cells, Fig. S2C). In contrast to PIGS KO cells, which were stained strongly by T5 mAb, C19-PIGS KO cells were only slightly stained by T5 mAb (Fig. 3B). Because PGAP5-sufficient PIGS KO cells were strongly stained, this result indicates that when PGAP5 is not functional, the GalNAc side chain of free GPI is affected, which in turn indicates that free GPIs undergo PGAP5-mediated maturation reaction.
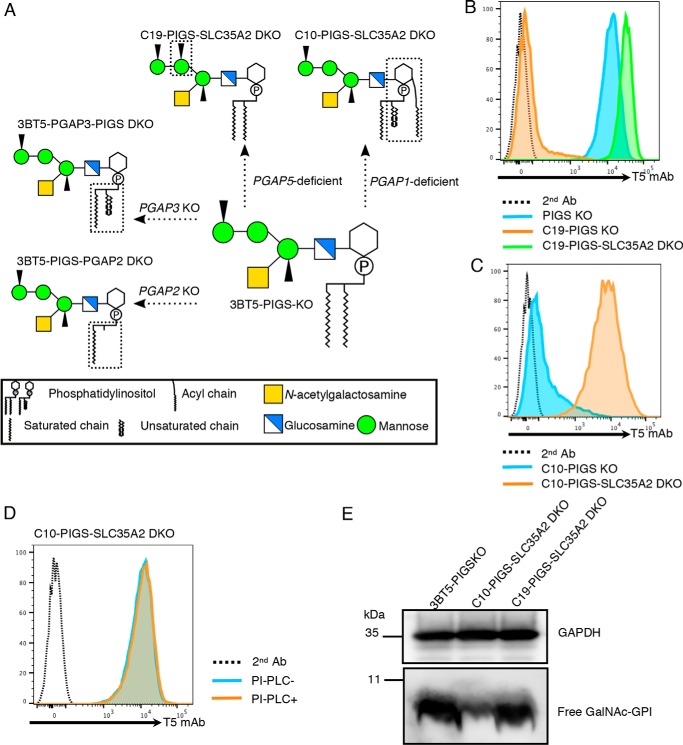
Figure 3.
Structural maturation of free GPI in the ER. A, schematic presentation of the free GPI structures in CHO cells defective in one of the GPI remodeling steps. 3BT5-PIGS KO cells express free GPI bearing the GalNAc side chain, C10-PIGS-SLC35A2 DKO cells express free GPI with an inositol-linked acyl chain, C19-PIGS-SLC35A2 DKO cells express free GPI with Man2-linked EtNP, 3BT5-PGAP3-PIGS DKO cells express free GPI bearing unremodeled fatty acid, and 3BT5-PIGS-PGAP2 DKO cells express lyso-form free GPI. Structures different between 3BT5-PIGS KO cells and other mutant cells are surrounded by broken lines. B, PIGS KO, C19-PIGS KO, and C19-PIGS-SLC35A2 DKO cells were stained with T5 mAb and Alexa Fluor 488 secondary antibody. C, C10-PIGS KO and C10-PIGS-SLC35A2 DKO cells were stained with T5 mAb and Alexa Fluor 488 secondary antibody. D, PI-PLC sensitivity of free GPIs of PGAP1-deficient cells. C10-PIGS-SLC35A2 DKO cells were treated with or without PI-PLC and stained with T5 mAb and Alexa Fluor 488 secondary antibody. E, Western blotting of free GPIs with inositol-linked acyl chain or Man2-linked EtNP. Lysates of 3BT5-PIGS KO, C10-PIGS-SLC35A2 DKO, and C19-PIGS-SLC35A2 DKO cells were analyzed by Western blotting with T5 mAb. GAPDH, a loading control. Similar results were obtained in at least two independent experiments.
We then assessed the basis of only slight staining by T5 mAb of C19-PIGS KO cells. This could be caused either by inefficient addition of the GalNAc side chain or efficient Gal-capping of the GalNAc side chain when Man2-linked EtNP remains. To test these possibilities, we further knocked out SLC35A2 in C19-PIGS KO cells (termed C19-PIGS-SLC35A2 DKO cells, Fig. 3A; see Fig. S2E for validation of SLC35A2 KO). C19-PIGS-SLC35A2 DKO cells were strongly stained by T5 mAb, indicating that the GalNAc side chain is added and more efficiently capped by Gal than in GPI-Tase–sufficient cells (Fig. 3B). We also found that PGAP1-defective PIGS KO cells are only weakly stained by T5 mAb (Fig. 3C) and that strong T5 mAb staining appeared when SLC35A2 was knocked out (C10-PIGS-SLC35A2 DKO cells, Fig. 3A; see Fig. S2D for validation of SLC35A2 KO). Free GPI on C10-PIGS-SLC35A2 DKO cells was, as expected, resistant to PI-PLC (Fig. 3D). Taken together, these results indicate that when the inositol-linked acyl chain remains in free GPI, its GalNAc side chain is almost completely Gal-capped. Therefore, whereas a sizable fraction of the GalNAc side chain of free GPI remains uncapped in PIGS KO cells, either PGAP1 defect or PGAP5 defect in PIGS KO cells result in almost complete Gal-capping. The reason for the latter phenomenon is yet to be investigated.
Free GPIs in GPI-Tase defective CHO cells undergo fatty acid remodeling in the Golgi
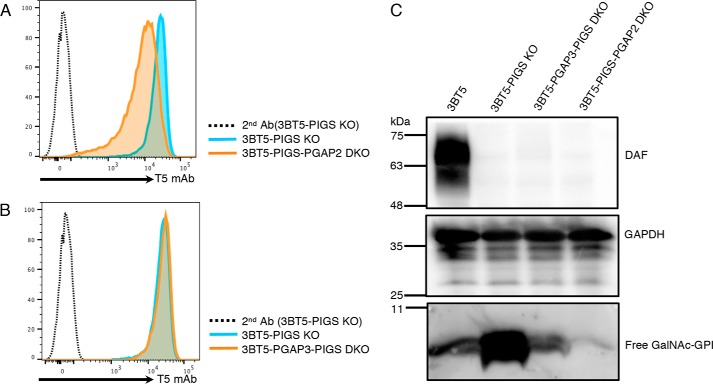
After transport to the Golgi, GPI-APs undergo fatty acid remodeling by PGAP3 and PGAP2, which is essential for lipid raft association of GPI-APs (20–22). PGAP3, a Golgi-resident GPI-specific phospholipase A2, removes an unsaturated fatty acid from the sn-2 position of PI and then the sn-2 position is reacylated with stearic acid, a saturated fatty acid, by PGAP2-dependent reaction. When PGAP2 is defective, fatty acid remodeling is terminated at the lyso-form intermediate and GPI-APs bearing the lyso form GPI, which has only one hydrocarbon chain (Fig. 3A), are transported to the cell surface. The lyso-forms of GPI-APs are released to medium because of unstable association with the plasma membrane or due to cleavage by phospholipase D, resulting in significant reduction of the levels of GPI-APs in PGAP2-KO cells (21). To determine whether free GPIs of 3BT5-PIGS KO cells undergo fatty acid remodeling, we knocked out PGAP2 (Fig. 3A). PGAP2 KO in 3BT5-PIGS KO cells resulted in a reduced level of T5 mAb-detectable free GPIs as assessed by flow cytometry (Fig. 4A), and a more strongly reduced level as assessed by Western blotting (Fig. 4C, right end lane), suggesting fatty acid remodeling of free GPIs occurred in the Golgi.
Figure 4.
Fatty acid remodeling occurs in free GPI. A, staining of 3BT5-PIGS KO and 3BT5-PIGS-PGAP2 DKO cells with T5 mAb and Alexa Fluor 488 secondary antibody. Dotted line, 3BT5-PIGS KO cells stained by secondary antibody only. B, staining of 3BT5-PIGS KO and 3BT5-PGAP3-PIGS DKO cells with T5 mAb and Alexa Fluor 488 secondary antibody. Dotted line, 3BT5-PIGS KO cells stained by secondary antibody only. C, Western blotting of free GPIs of fatty acid remodeling-deficient cells. Lysates of 3BT5, 3BT5-PIGS KO, 3BT5-PGAP3-PIGS DKO, and 3BT5-PIGS-PGAP2 DKO cells were analyzed by Western blotting. DAF, a GPI-AP; GAPDH, a loading control. Similar results were obtained in at least two independent experiments.
Further characterization of free GPI detected by T5 mAb
We next analyzed the importance of the lipid moiety of free GPI for binding of T5 mAb. Because an unsaturated fatty acid remains at the sn-2 position of PI moiety in PGAP3-defective cells (20), we generated CHO-3BT5-PGAP3-defective PIGS KO cells (Fig. S2F) to determine involvement of the sn-2–linked fatty acid in recognition by T5 mAb. PGAP3 defect did not affect T5 mAb binding to the cell-surface free GPI, as shown by flow cytometry (Fig. 4B). As Fig. 3D shows, T5 mAb recognized cell-surface free GPI bearing an inositol-linked acyl chain in flow cytometry. These results together indicate that the PI moiety is not involved in T5 mAb binding to free GPI when assessed by flow cytometry. In Western blotting against T5 mAb, however, levels of free GPIs bearing the inositol-linked acyl chain (PGAP1 KO cells) (Fig. 3E, middle) and those bearing an unsaturated fatty acid at the sn-2 position (3BT5-PGAP3-PIGS KO cells) (Fig. 4C) were very low compared with those from 3BT5-PIGS KO cells. It seems therefore that binding of T5 mAb to its epitope on a PVDF membrane is affected by the structure of the lipid moiety in GPI. A similar phenomenon was reported for GPI-APs from PGAP3-defective cells (23).
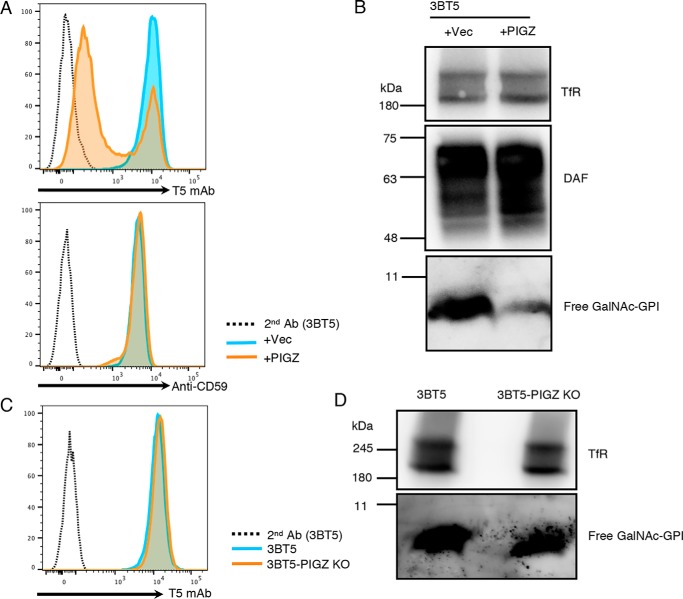
In some cell types, a fourth mannose (Man4) is transferred by PIGZ to Man3 as a side chain before attachment to proteins (23). The GalNAc side chain can be transferred to Man4-bearing GPI-APs (24). We asked whether the Man4 addition affects T5 mAb recognition of free GPIs. After transfection of PIGZ, T5 staining of CHO-3BT5 cells were greatly reduced (Fig. 5A) and detection by Western blotting became inefficient (Fig. 5B). These results indicate that T5 mAb binding is severely affected by the presence of Man4 and that free GPIs on CHO cells detected by T5 mAb do not have the Man4 side chain. The fact that Man4 inhibits T5 mAb binding allowed us to determine whether CHO cells have any Man4-bearing free GPI. KO of PIGZ in CHO-3BT5 cells (Fig. S3) did not increase, or only slightly increased, cell-surface T5 mAb staining (Fig. 5C) and the free GPI band on Western blots (Fig. 5D), indicating that CHO cells express a very low level of PIGZ and mainly have free GPI lacking Man4.
Figure 5.
Man4 inhibits detection of free GPIs with T5 mAb. A, flow cytometry of 3BT5 cells transiently transfected with empty vector (blue) or PIGZ cDNA (orange). Top, staining with T5 mAb; bottom, staining with anti-CD59. Dotted lines, secondary antibody only. B, Western blotting of free GPI in the lysates of 3BT5 cells transiently transfected with empty vector (left) or PIGZ cDNA (right). DAF, a GPI-AP; TfR, a loading control. C, flow cytometry of free GPIs on 3BT5-PIGZ KO (orange) and 3BT5 (blue) cells. Cells were stained with T5 mAb and Alexa Fluor 488 secondary antibody. Dotted line, 3BT5 stained by secondary antibody only. D, Western blotting of free GPIs in the lysates of 3BT5 (left) and 3BT5-PIGZ KO (right) cells. Similar results were obtained in at least two independent experiments.
Discussion
A major finding in this study is that free, nonprotein-linked GPIs undergo maturation reactions in the ER and Golgi, and exist as glycolipids in some mouse tissues and cultured mammalian cell lines. Using T5 mAb that recognizes free GPI bearing a Man1-linked GalNAc side chain, we show its presence on the cell surface of some cultured cell lines by flow cytometry and in the lysate of mouse pons, medulla oblongata, spinal cord, kidney, testis, and epididymis by Western blotting. Taking advantage of various mutant CHO cells defective in GPI maturation and based on the binding specificity of T5 mAb, known structures of protein GPI-anchors and sensitivity to PI-PLC, we conclude that free GPIs take basically the same maturation pathway as GPI-APs. Namely, the inositol-linked acyl chain is removed from or remains in free GPIs dependent upon the efficiency of PGAP1; free GPIs undergo fatty acid remodeling; and free GPIs are modified by GalNAc side chains with or without elongation by Gal (Fig. 1A). This finding advances previous works demonstrating the presence of nonprotein-linked GPIs on mammalian cultured cell lines (9–11) and emphasizes a view that free GPIs are normal components of the plasma membrane in at least some tissues.
T5 mAb-detectable free GPIs were found only in some tissues in mice and other tissues were negative (Fig. 1, D and E). Tissues positive for T5 mAb-detectable free GPIs express PGAP4 mRNA according to BioGPS (25–27). However, according to BioGPS, cerebral cortex and cerebellum have PGAP4 mRNA, whereas T5 mAb showed no free GPI band in these tissues. The negative results indicate: 1) that free GPIs are not expressed in these tissues at all, or 2) that they are expressed but all of them are Gal-capped, or 3) that free GPIs in these tissues, if expressed, do not have the GalNAc side chain. Because T5 mAb requires the presence of the uncapped GalNAc side chain and is the only currently available probe for free GPI, we cannot determine whether cells have free GPI lacking the GalNAc side chain, or carry it, but it is elongated by Gal and further by sialic acid.
Because the GalNAc side chain is transferred by PGAP4 in the Golgi to the fully assembled GPI, we cannot determine whether GPI biosynthetic intermediates are transported to the cell surface. This point is important for characterizing cells from patients with inherited GPI deficiency caused by mutations in genes involved in GPI biosynthesis (28–31). Because GPI biosynthetic intermediates are expected to accumulate in the cells of those patients, development of probes for various GPI intermediates is necessary to clarify this point.
When the inositol-linked acyl chain and the Man2-linked EtNP were normally removed from free GPI, a sizable fraction of the GalNAc side chain was not capped by Gal (Fig. 2C). In contrast, when the inositol-linked acyl chain or the Man2-linked EtNP was not removed from free GPI, its GalNAc side chain was very efficiently capped by Gal (Fig. 3, B and C). We previously reported that ER to Golgi transport of GPI-APs in PGAP1- or PGAP5-defective CHO cells is much slower than in the WT cells (32). It seems likely that the ER to Golgi transport of free GPI is similarly slowed because binding of GPI to its cargo receptor at the ER exit site is inefficient when the inositol-linked acyl chain or the Man2-linked EtNP remained. Under such conditions, a ratio of Gal transferase to free GPI may be higher, which in turn may result in efficient Gal-capping in the Golgi.
By using CHO cells defective in different GPI structural maturation steps, we further characterized the binding specificity of T5 mAb. T5 mAb bound free GPI bearing an inositol-linked acyl chain (Fig. 3D) and free GPI in which fatty acid remodeling did not occur (Fig. 4B) (14), indicating that the lipid moiety in GPI does not contribute to recognition by T5 mAb. Concerning the core glycan moiety, the 2-position of Man1 is not involved in recognition because the corresponding position in T. gondii GPI is open, whereas mammalian GPI has an EtNP modification (Fig. 1A). T5 mAb bound free GPI bearing EtNP linked to Man2 (Fig. 3B), indicating that the 6-position of Man2 is not included in the epitope. In contrast, the 2-position of Man3 must be open, because Man4 inhibited T5 mAb binding (Fig. 5A). Because EtNP at the 6-position of Man3 must be exposed for T5 mAb binding, the presence of Man4 at the 2-position of Man3 might sterically hinder access by T5 mAb.
We showed in this study that T5 mAb recognized free GPIs from Chinese hamster and mouse (Fig. 1, B–E). It is known that some GPI-APs from human, rat, and pig bear non-Gal-capped GalNAc side chains (1). Torpedo acetylcholinesterase, a GPI-AP, also has non-Gal–capped GalNAc side chain (1). If these organisms express free GPIs, T5 mAb should recognize them. Therefore, it is likely that T5 mAb is widely useful in studying free GPIs in humans, other mammals, and at least some non-mammalian vertebrates.
We showed that free GPIs detectable by T5 mAb on CHO cells almost completely lack Man4. Consistent with this, our previous study of the GPI-anchor of CD59 from CHO cells showed almost no Man4 in its GPI (14). There is no established functional assay for PIGZ. However, based on the result indicating that attachment of Man4 inhibits binding of T5 mAb (Fig. 5A, top panel), mannosyltransferase activity of PIGZ can be easily assessed by transfection of PIGZ cDNA into CHO 3BT5 cells. The assay should be useful to determine functional activities of PIGZ variants that may be found by whole exome sequencing being applied to healthy individuals and individuals suffering from various diseases.
Experimental procedures
Cells
CHO-K1, C2C12, Neuro2a, and K562 cells were obtained from ATCC. The 3B2A cell was established by stably expressing human DAF (CD55) and human CD59 in CHO-K1 cells, and selecting by cell sorting a clone highly expressing DAF and CD59 (33). The 3BT5 cell defective in SLC35A2 was established by several rounds of cell sorting from 3B2A cells to obtain a clone highly stained by T5 mAb (14). The FF8 cells are 3B2A cells stably transfected with pTRE2puro-VSVGex-FF-mEGFP-GPI in conjunction with pUHrT62-1, an expression plasmid for reverse tetracycline-controlled transactivators (34, 35). The CHO F21 cells stably express human CD59 and DAF and 12 PIG genes (36). The C19 mutant cells defective in PGAP5 are derivatives of FF8 cells (19). C10 mutant cells derived from 3B2A cells are defective in PGAP1 (18). CHO PA16.1 cells defective in PIGU were derived from 3B2A cells (37). PIGK-, GPAA1- and PIGT-deficient cells were isolated from mutagenized CHO F21 cells as described before (36). 3BT5-PGAP3 KO cells were generated previously by using the CRISPR-Cas9 system (14). In this study, SLC35A2 KO, PIGZ KO, PIGS KO, and PGAP2 KO cells were generated by knockout of SLC35A2, PIGZ, PIGS, and PGAP2, respectively. KO cells used in this study are listed in Table 1.
Animals
The experiments described here were performed in compliance with the regulations of the Review Committee for Animal Experimentation of Osaka University. Nine-week-old male C57BL/6 mice were used for Western blot analysis of free GPIs with T5 mAb.
Cell culture
CHO cells and Neuro2a cells were cultured in Dulbecco's modified Eagle's medium/Ham's F-12 medium (Nacalai Tesque, Japan) containing 10% fetal bovine serum (FBS, Sigma). K562 cells were cultured in RPMI 1640 medium (Life Technologies) containing 10% FBS. HEK293 and C2C12 cells were cultured in high glucose Dulbecco's modified Eagle's medium (Nacalai Tesque) containing 10% FBS. All cells were maintained at 37 °C in 5% CO2 atmosphere.
Antibodies and reagents
T5-4E10 mAb against free GPI-GalNAc (mouse IgM) was a gift from Dr. Jean François Dubremetz (Montpellier University, France). T5-4E10 mAb is available from BEI Resources, NIAID, National Institutes of Health. Other antibodies used were mouse mAb against human CD59 (clone 5H8, unconjugated or biotinylated), human DAF (clone IA10, BD Biosciences), GAPDH (Thermo Fisher), and transferrin receptor (Thermo Fisher). The secondary antibodies/reagents used for Western blotting were horseradish peroxidase-conjugated goat anti-mouse IgG (GE Healthcare) and goat anti-mouse IgM (Thermo Fisher), and for FACS analysis were phycoerythrin-conjugated goat anti-mouse IgG (Biolegend), allophycocyanin-conjugated streptavidin (Biolegend), and Alexa Fluor 488-conjugated goat anti-mouse IgM (Thermo Fisher). Lectins used for FACS analysis were Alexa Fluor 647-conjugated Griffonia simplicifolia lectin II (GS-II, Thermo Fisher) and Alexa Fluor 488-conjugated Helix pomatia-agglutinin lectin (HPA, Thermo Fisher). Phospholipase C, phosphatidylinositol-specific from Bacillus cereus (PI-PLC, Thermo Fisher) was used. Transfection reagents, including Polyethylenimine “MAX” (PEI-MAX, Polysciences) and Lipofectamine 2000 (Thermo Fisher), were used.
Generations of knockout cell lines
The SLC35A2, PIGS, PGAP2, and PIGZ genes were knocked out by the CRISPR/Cas9 system (38). pX330-mEGFP plasmid generated from pX330 were used to generate single guide RNA-Cas9 expression plasmids (39). The targeting sequences were cloned into pX330 or pX330-mEGFP plasmid digested with BbsI. The target sequences are listed in Table S1. The sequences were ligated into digested pX330 or pX330-EGFP. All the targeting sequences were confirmed by Sanger sequencing. After transfection of cells with knockout constructs, GFP-positive cells were sorted using a FACS Aria II cell sorter (BD Biosciences). Sorted cells were further cultured for more than 10 days and analyzed. Clonal cell lines were obtained by limiting dilution. For genotyping 3BT5-PIGZ KO cells (Fig. S3), PIGZ exon2 was amplified by primers 5′-ATGTAGATCTCCAGAGTGGCACC (CHO-PIGZ-exon2-F) and 5′-CTGCCATGACCTCAGGTGACTG (CHO-PIGZ-exon2-R) followed by Sanger sequencing.
Plasmid construction
To construct pME-PIGZ-3HA, human PIGZ cDNA amplified by PCR from a human brain cDNA library using primers (5′-aaaaGAATTCCACCATGCAGATCTGTGGATCCAGC and 5′-aaaaACGCGTGGTTTCTTCCCCCAGCTCC) was digested with EcoRI and MluI and cloned into the same site of pME-3HA plasmid.
Plasmid DNA transfection
Plasmids were transfected to CHO cells by electroporation. Around 5 × 106 cells were suspended in 400 μl of Opti-MEM (Life Technologies) with 10 μg of plasmid DNA, then electroporated by a Gene Pulser (Bio-Rad) at 260 V and 1000 microfarads. According to the manufacturers' instructions, HEK293 cells were transfected by PEI-MAX, C2C12 cells and K562 cells were transfected by Lipofectamine 2000.
Flow cytometry assay
Cells were stained with T5 mAb (1:100 ascites) and biotinylated, or unlabeled mouse anti-CD59 (10 μg ml−1) in FACS buffer (PBS containing 1% BSA, 0.1% NaN3) on ice for 25 min. Cells were then washed twice in FACS buffer followed by staining with Alexa Fluor 488-conjugated goat anti-mouse IgM (1:250) for T5 mAb, phycoerythrin-conjugated goat anti-mouse IgG (1:100) for anti-CD59, and allophycocyanin-conjugated streptavidin (1:100) for biotin-labeled anti-CD59 in FACS buffer. To analyze the glycosylation profiles of surface proteins, cells were stained by Alexa Fluor 488-conjugated and Alexa Fluor 647-conjugated lectins (1:100) in FACS buffer containing 1 mm CaCl2, 1 mm MnCl2, and 1 mm MgCl2 on ice for 15 min. After washing twice by FACS buffer, cells were analyzed by the BD FACS Canto II. Data were analyzed by the FlowJo software (FlowJo LLC).
PI-PLC sensitivity assay
Cells were detached from culture plates by treatment with detaching buffer, PBS containing 5 mm EDTA and 0.5% BSA (Nacalai Tesque). Cells were washed once in detaching buffer, around 5 × 105 cells were then treated with 1 unit/ml or without PI-PLC in 50 μl of reaction buffer (4 volumes of Opti-MEM and 1 volume of detaching buffer) at 37 °C for 1.5–2 h. The cells were then stained by T5 mAb, anti-CD59 followed by FACS analysis with the BD FACS Canto II. In some experiments, PI-PLC–treated cells were lysed and analyzed by Western blotting using anti-DAF and T5 mAb.
Preparation of protein extracts from mouse tissues
Mouse tissues were washed with cold PBS and homogenized in 25 volumes of lysis buffer (20 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm EDTA, 60 mm n-octyl-β-d-glucoside, 1× protease inhibitor mixture) using a handy microhomogenizer (Microtec Co. Ltd., Funabashi, Japan) and rotated at 4 °C for 2 h, and followed by centrifugation at 17,900 × g, 4 °C for 15 min. The protein concentrations of homogenates of each sample were measured using a Pierce BCA Protein Assay Kit (Thermo Fisher) and adjusted appropriately. Samples were mixed with 4× SDS sample buffer and boiled at 95 °C for 5 min, 5 μg of each sample was then loaded to SDS-PAGE for Western blotting analyses.
Western blotting
Cell lysates were prepared on ice in lysis buffer, followed by centrifugation at 17,900 × g for 15 min. Supernatants were collected, then mixed with 4× SDS sample buffer with 5% β-mercaptoethanol and boiled at 95 °C for 5 min. Samples were run on 10–20% SDS-PAGE gels and transferred to PVDF membranes. The PVDF membranes were blocked at 4 °C in TBST containing 5% nonfat milk overnight. Free GPIs were then detected by T5 mAb (1:1000 ascites). Other antibodies used were mouse anti-TfR (1:1000), anti-GAPDH (1:1000), and anti-DAF (0.5 μg ml−1) mAbs.
Author contributions
Y. W. and T. K. conceptualization; Y. W. investigation; Y. W. and T. K. writing-original draft; T. H., Y. Maeda, Y. Murakami, and M. F. resources; T. H., Y. Murakami, and M. F. methodology; T. H., Y. Maeda, Y. Murakami, M. F., and T. K. writing-review and editing; T. K. supervision; T. K. funding acquisition; T. K. project administration.
Supplementary Material
Acknowledgments
We thank Dr. Jean François Dubremetz (Montpellier University, France) for T5-4E10 antibody, Dr. Y. Tashima and Dr. Gun-Hee Lee for discussion, K. Uchikawa and Y. Kabumoto for cell sorting, and K. Kinoshita, S. Umeshita, Y. Onoe, K. Miyanagi, and Dr. N. Kanzawa for technical help.
This work was supported by grants-in aid Grant Number JP16H04753 from MEXT Japan Society for the Promotion of Science (JSPS) (to T. K). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Table S1 and Figs. S1–S3.
- GPI
- glycosylphosphatidylinositol
- T5 mAb
- T5-4E10 monoclonal antibody
- GPI-AP
- GPI-anchored proteins
- CHO
- Chinese hamster ovary
- ER
- endoplasmic reticulum
- GPI-Tase
- GPI transamidase
- PNH
- paroxysmal nocturnal hemoglobinuria
- EtNP
- ethanolamine phosphate
- PI-PLC
- phosphatidylinositol-specific phospholipase C
- KO
- knockout
- PVDF
- polyvinylidene difluoride
- TfR
- transferrin receptor
- PIGZ
- phosphatidylinositol glycan anchor biosynthesis class Z
- FBS
- fetal bovine serum
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase.
References
- 1. Ferguson M. A. J., Hart G. W., and Kinoshita T. (2017) Glycosylphosphatidylinositol Anchors, 3rd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York: [PubMed] [Google Scholar]
- 2. UniProt Consortium. (2015) UniProt: a hub for protein information. Nucleic Acids Res. 43, D204–D212 10.1093/nar/gku989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kinoshita T., and Fujita M. (2016) Biosynthesis of GPI-anchored proteins: special emphasis on GPI lipid remodeling. J. Lipid Res. 57, 6–24 10.1194/jlr.R063313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gas-Pascual E., Ichikawa H. T., Sheikh M. O., Serji M. I., Deng B., Mandalasi M., Bandini G., Samuelson J., Wells L., and West C. M. (2019) CRISPR/Cas9 and glycomics tools for toxoplasma glycobiology. J. Biol. Chem. 294, 1104–1125 10.1074/jbc.RA118.006072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Götze S., Azzouz N., Tsai Y. H., Groβ U., Reinhardt A., Anish C., Seeberger P. H., and Silva D. V. (2014) Diagnosis of toxoplasmosis using a synthetic glycosylphosphatidylinositol glycan. Angew. Chemie Int. Ed. 53, 13701–13705 10.1002/anie.201406706 [DOI] [PubMed] [Google Scholar]
- 6. Sharma S. D., Mullenax J., Araujo F. G., Erlich H. A., and Remington J. S. (1983) Western blot analysis of the antigens of Toxoplasma gondii recognized by human IgM and IgG antibodies. J. Immunol. 131, 977–983 [PubMed] [Google Scholar]
- 7. Tomavo S., Couvreur G., Leriche M. A., Sadak A., Achbarou A., Fortier B., and Dubremetz J. F. (1994) Immunolocalization and characterization of the low molecular weight antigen (4–5 kDa) of Toxoplasma gondii that elicits an early IgM response upon primary infection. Parasitology 108, 139–145 10.1017/S0031182000068220 [DOI] [PubMed] [Google Scholar]
- 8. Striepen B., Zinecker C. F., Damm J. B., Melgers P. A., Gerwig G. J., Koolen M., Vliegenthart J. F., Dubremetz J. F., and Schwarz R. T. (1997) Molecular structure of the “low molecular weight antigen” of Toxoplasma gondii: a glucose α1–4 N-acetylgalactosamine makes free glycosyl-phosphatidylinositols highly immunogenic. J. Mol. Biol. 266, 797–813 10.1006/jmbi.1996.0806 [DOI] [PubMed] [Google Scholar]
- 9. Singh N., Liang L. N., Tykocinski M. L., and Tartakoff A. M. (1996) A novel class of cell surface glycolipids of mammalian cells: free glycosyl phosphatidylinositols. J. Biol. Chem. 271, 12879–12884 10.1074/jbc.271.22.12879 [DOI] [PubMed] [Google Scholar]
- 10. Baumann N. A., Vidugiriene J., Machamer C. E., and Menon A. K. (2000) Cell surface display and intracellular trafficking of free glycosylphosphatidylinositols in mammalian cells. J. Biol. Chem. 275, 7378–7389 10.1074/jbc.275.10.7378 [DOI] [PubMed] [Google Scholar]
- 11. van't Hof W., Rodriguez-Boulan E., and Menon A. K. (1995) Nonpolarized distribution of glycosylphosphatidylinositols in the plasma membrane of polarized Madin-Darby canine kidney cells. J. Biol. Chem. 270, 24150–24155 10.1074/jbc.270.41.24150 [DOI] [PubMed] [Google Scholar]
- 12. Baldwin M. A., Stahl N., Reinders L. G., Gibson B. W., Prusiner S. B., and Burlingame A. L. (1990) Permethylation and tandem mass spectrometry of oligosaccharides having free hexosamine: analysis of the glycoinositol phospholipid anchor glycan from the scrapie prion protein. Anal. Biochem. 191, 174–182 10.1016/0003-2697(90)90405-X [DOI] [PubMed] [Google Scholar]
- 13. Stahl N., Baldwin M. A., Hecker R., Pan K. M., Burlingame A. L., and Prusiner S. B. (1992) Glycosylinositol phospholipid anchors of the scrapie and cellular prion proteins contain sialic acid. Biochemistry 31, 5043–5053 10.1021/bi00136a600 [DOI] [PubMed] [Google Scholar]
- 14. Hirata T., Mishra S. K., Nakamura S., Saito K., Motooka D., Takada Y., Kanzawa N., Murakami Y., Maeda Y., Fujita M., Yamaguchi Y., and Kinoshita T. (2018) Identification of a Golgi GPI-N-acetylgalactosamine transferase with tandem transmembrane regions in the catalytic domain. Nat. Commun. 9, 405 10.1038/s41467-017-02799-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hill A., DeZern A. E., Kinoshita T., and Brodsky R. A. (2017) Paroxysmal nocturnal haemoglobinuria. Nat. Rev. Dis. Prim. 3, 17028 10.1038/nrdp.2017.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Krawitz P. M., Höchsmann B., Murakami Y., Teubner B., Krüger U., Klopocki E., Neitzel H., Hoellein A., Schneider C., Parkhomchuk D., Hecht J., Robinson P. N., Mundlos S., Kinoshita T., and Schrezenmeier H. (2013) A case of paroxysmal nocturnal hemoglobinuria caused by a germline mutation and a somatic mutation in PIGT. Blood 122, 1312–1315 10.1182/blood-2013-01-481499 [DOI] [PubMed] [Google Scholar]
- 17. Kawamoto M., Murakami Y., Kinoshita T., and Kohara N. (2018) Recurrent aseptic meningitis with PIGT mutations: a novel pathogenesis of recurrent meningitis successfully treated by eculizumab. BMJ Case Rep. 2018, bcr-2018–225910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tanaka S., Maeda Y., Tashima Y., and Kinoshita T. (2004) Inositol deacylation of glycosylphosphatidylinositol-anchored proteins is mediated by mammalian PGAP1 and yeast Bst1p. J. Biol. Chem. 279, 14256–14263 10.1074/jbc.M313755200 [DOI] [PubMed] [Google Scholar]
- 19. Fujita M., Maeda Y., Ra M., Yamaguchi Y., Taguchi R., and Kinoshita T. (2009) GPI glycan remodeling by PGAP5 regulates transport of GPI-anchored proteins from the ER to the Golgi. Cell 139, 352–365 10.1016/j.cell.2009.08.040 [DOI] [PubMed] [Google Scholar]
- 20. Maeda Y., Tashima Y., Houjou T., Fujita M., Yoko-o T., Jigami Y., Taguchi R., and Kinoshita T. (2007) Fatty acid remodeling of GPI-anchored proteins is required for their raft association. Mol. Biol. Cell 18, 1497–1506 10.1091/mbc.e06-10-0885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tashima Y., Taguchi R., Murata C., Ashida H., Kinoshita T., and Maeda Y. (2006) PGAP2 is essential for correct processing and stable expression of GPI-anchored proteins. Mol. Biol. Cell 17, 1410–1420 10.1091/mbc.e05-11-1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Seong J., Wang Y., Kinoshita T., and Maeda Y. (2013) Implications of lipid moiety in oligomerization and immunoreactivities of GPI-anchored proteins. J. Lipid Res. 54, 1077–1091 10.1194/jlr.M034421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Taron B. W., Colussi P. A., Wiedman J. M., Orlean P., and Taron C. H. (2004) Human Smp3p adds a fourth mannose to yeast and human glycosylphosphatidylinositol precursors in vivo. J. Biol. Chem. 279, 36083–36092 10.1074/jbc.M405081200 [DOI] [PubMed] [Google Scholar]
- 24. Homans S. W., Ferguson M. A., Dwek R. A., Rademacher T. W., Anand R., and Williams A. F. (1988) Complete structure of the glycosyl phosphatidylinositol membrane anchor of rat brain Thy-1 glycoprotein. Nature 333, 269–272 10.1038/333269a0 [DOI] [PubMed] [Google Scholar]
- 25. Wu C., Jin X., Tsueng G., Afrasiabi C., and Su A. I. (2016) BioGPS: building your own mash-up of gene annotations and expression profiles. Nucleic Acids Res. 44, D313 10.1093/nar/gkv1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wu C., MacLeod I., and Su A. I. (2013) BioGPS and MyGene.info: organizing online, gene-centric information. Nucleic Acids Res. 41, D561–D565 10.1093/nar/gks1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wu C., Orozco C., Boyer J., Leglise M., Goodale J., Batalov S., Hodge C. L., Haase J., Janes J., Huss J. W., and Su A. I. (2009) BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 10.1186/gb-2009-10-11-r130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Almeida A. M., Murakami Y., Layton D. M., Hillmen P., Sellick G. S., Maeda Y., Richards S., Patterson S., Kotsianidis I., Mollica L., et al. (2006) Hypomorphic promoter mutation in PIGM causes inherited glycosylphosphatidylinositol deficiency. Nat. Med. 12, 846–851 10.1038/nm1410 [DOI] [PubMed] [Google Scholar]
- 29. Chiyonobu T., Inoue N., Morimoto M., Kinoshita T., and Murakami Y. (2014) Glycosylphosphatidylinositol (GPI) anchor deficiency caused by mutations in PIGW is associated with West syndrome and hyperphosphatasia with mental retardation syndrome. J. Med. Genet. 51, 203–207 10.1136/jmedgenet-2013-102156 [DOI] [PubMed] [Google Scholar]
- 30. Krawitz P. M., Schweiger M. R., Rödelsperger C., Marcelis C., Kölsch U., Meisel C., Stephani F., Kinoshita T., Murakami Y., Bauer S., et al. (2010) Identity-by-descent filtering of exome sequence data identifies PIGV mutations in hyperphosphatasia mental retardation syndrome. Nat. Genet. 42, 827–829 10.1038/ng.653 [DOI] [PubMed] [Google Scholar]
- 31. Krawitz P. M., Murakami Y., Hecht J., Krüger U., Holder S. E., Mortier G. R., Delle Chiaie B., De Baere E., Thompson M. D., Roscioli T., et al. (2012) Mutations in PIGO, a member of the GPI-anchor-synthesis pathway, cause hyperphosphatasia with mental retardation. Am. J. Hum. Genet. 91, 146–151 10.1016/j.ajhg.2012.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fujita M., Watanabe R., Jaensch N., Romanova-Michaelides M., Satoh T., Kato M., Riezman H., Yamaguchi Y., Maeda Y., and Kinoshita T. (2011) Sorting of GPI-anchored proteins into ER exit sites by p24 proteins is dependent on remodeled GPI. J. Cell Biol. 194, 61–75 10.1083/jcb.201012074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nakamura N., Inoue N., Watanabe R., Takahashi M., Takeda J., Stevens V. L., and Kinoshita T. (1997) Expression cloning of PIG-L, a candidate N-acetylglucosaminyl-phosphatidylinositol deacetylase. J. Biol. Chem. 272, 15834–15840 10.1074/jbc.272.25.15834 [DOI] [PubMed] [Google Scholar]
- 34. Takida S., Maeda Y., and Kinoshita T. (2008) Mammalian GPI-anchored proteins require p24 proteins for their efficient transport from the ER to the plasma membrane. Biochem. J. 409, 555–562 10.1042/BJ20070234 [DOI] [PubMed] [Google Scholar]
- 35. Maeda Y., Ide T., Koike M., Uchiyama Y., and Kinoshita T. (2008) GPHR is a novel anion channel critical for acidification and functions of the Golgi apparatus. Nat. Cell Biol. 10, 1135–1145 10.1038/ncb1773 [DOI] [PubMed] [Google Scholar]
- 36. Ashida H., Hong Y., Murakami Y., Shishioh N., Sugimoto N., Kim Y. U., Maeda Y., and Kinoshita T. (2005) Mammalian PIG-X and yeast Pbn1p are the essential components of glycosylphosphatidylinositol-mannosyltransferase I. Mol. Biol. Cell 16, 1439–1448 10.1091/mbc.e04-09-0802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hong Y., Ohishi K., Kang J. Y., Tanaka S., Inoue N., Nishimura J., Maeda Y., and Kinoshita T. (2003) Human PIG-U and yeast Cdc91p are the fifth subunit of GPI transamidase that attaches GPI-anchors to proteins. Mol. Biol. Cell 14, 1780–1789 10.1091/mbc.e02-12-0794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cong L., Ran F. A., Cox D., Lin S., Barretto R., Habib N., Hsu P. D., Wu X., Jiang W., and Marraffini L. A. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hirata T., Fujita M., Nakamura S., Gotoh K., Motooka D., Murakami Y., Maeda Y., and Kinoshita T. (2015) Post-Golgi anterograde transport requires GARP-dependent endosome-to-TGN retrograde transport. Mol. Biol. Cell 26, 3071–3084 10.1091/mbc.E14-11-1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Varki A., Cummings R. D., Aebi M., Packer N. H., Seeberger P. H., Esko J. D., Stanley P., Hart G., Darvill A., Kinoshita T., Prestegard J. J., Schnaar R. L., Freeze H. H., Marth J. D., Bertozzi C. R., et al. (2015) Symbol nomenclature for graphical representations of glycans. Glycobiology 25, 1323–1324 10.1093/glycob/cwv091 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.