Significance
Most natural proteins fold into a native folded conformation stabilized by noncovalent interactions. The free-energy difference between the folded and unfolded conformations of a protein (“thermodynamic folding stability”) establishes the fraction of its population that is in a folded conformation at equilibrium. Here, we used a mass spectrometry-based approach to measure the stabilities of ∼10,000 unique domains within ∼3,000 proteins in human cell extracts. Our data provide a global census of the magnitudes of folding stabilities within the human proteome and identify specific subsets of the proteome that are enriched in stable and unstable domains. We also show that the stability of proteins impacts their tendency to become oxidized and is globally altered by the chemical chaperone trimethylamine N-oxide (TMAO).
Keywords: protein stability, protein folding, methionine oxidation, intrinsically disordered proteins, osmolytes
Abstract
The stability of proteins influences their tendency to aggregate, undergo degradation, or become modified in cells. Despite their significance to understanding protein folding and function, quantitative analyses of thermodynamic stabilities have been mostly limited to soluble proteins in purified systems. We have used a highly multiplexed proteomics approach, based on analyses of methionine oxidation rates, to quantify stabilities of ∼10,000 unique regions within ∼3,000 proteins in human cell extracts. The data identify lysosomal and extracellular proteins as the most stable ontological subsets of the proteome. We show that the stability of proteins impacts their tendency to become oxidized and is globally altered by the osmolyte trimethylamine N-oxide (TMAO). We also show that most proteins designated as intrinsically disordered retain their unfolded structure in the complex environment of the cell. Together, the data provide a census of the stability of the human proteome and validate a methodology for global quantitation of folding thermodynamics.
Within a cell, most proteins exist in a dynamic equilibrium between folded and unfolded conformations (1, 2). The free-energy difference between these two states (ΔGfolding or “thermodynamic folding stability”) establishes the fraction of its population that is in a folded conformation at equilibrium (3). Folding stabilities can impact the tendency of proteins to aggregate, oxidize, or undergo degradation, and alterations in protein stabilities play a role in diverse biological processes and physiological disorders (e.g., aging and neurodegenerative proteinopathies) (4–8). Despite their fundamental importance to protein function, the magnitude and span of folding stabilities within the proteome remain unclear. For example, estimations of the fraction of the proteome that harbors “intrinsically disordered” domains are largely based on theoretical predictions and, with a few exceptions (9, 10), have yet to be verified by proteome-wide experimental data (11).
Historically, quantitative measurements of protein folding stabilities have been conducted for proteins in purified systems using in vitro denaturation experiments that employ global folding probes such as circular dichroism or isothermal calorimetry (12). Although these studies have made significant contributions to our understanding of folding thermodynamics, they have inherent limitations that mitigate their in vivo relevance. First, these studies have been biased toward well-folded stable protein model systems that can be easily expressed and purified. Hence, the thermodynamic folding parameters measured for well-studied folding models may not be representative of the proteome as a whole. Second, experiments conducted in purified systems typically do not account for the effects of interacting partners and other cellular factors that may influence protein stabilities in vivo. Third, these methodologies are inherently low throughput and cannot be easily used to investigate protein stabilities on proteome-wide scales.
To address these shortcomings, a number of recently developed techniques have investigated protein folding patterns using probes that can be analyzed by mass spectrometry-based proteomics. Hydrogen/deuterium exchange (HDX) (13, 14), hydroxyl radical footprinting (15), and limited proteolysis (9, 16–19) have been coupled with liquid chromatography–tandem mass spectrometry (LC-MS/MS) to provide proteome-wide measures of protein structure. These global studies have successfully investigated thermostabilities (melting temperatures), ligand binding interactions, and conformational changes induced by environmental alterations. However, due in part to technical limitations inherent in each of these approaches, it has proven difficult to obtain measurements of thermodynamic folding stabilities (ΔGfolding) on proteome-wide scales.
In this study, we globally measured protein folding stabilities by using methionine oxidation as a probe. This approach, referred to as stability of proteins from rates of oxidation (SPROX), was initially described by West et al. (20) and is conceptually similar to previously described probing methodologies based on thiol labeling (21). SPROX offers a number of advantages that are particularly conducive to proteomic workflows. Methionine oxidation is a small, nonlabile modification that can be readily coupled with chemical denaturation and is retained during the course of LC-MS/MS analyses. Selective oxidation of methionines allows for straightforward database searches, identification of modification sites, and interpretation of oxidation kinetics. Furthermore, the use of chemical denaturants prevents the irreversible aggregation of proteins during the course of denaturation—a phenomenon that typically confounds thermal unfolding experiments. To date, SPROX has been used to analyze the stability of individual intact proteins and to examine proteome-wide alterations in denaturation patterns induced by ligand binding (20, 22, 23). Here, we have coupled SPROX with highly multiplexed tandem mass tagging (TMT) to obtain high-resolution denaturation curves—an approach we have termed high-resolution SPROX (HR-SPROX). By analyzing HR-SPROX data in the context of thermodynamic protein folding models, we were able to measure localized ΔGfolding values on a proteome-wide scale.
Results
HR-SPROX Workflow.
Fig. 1 outlines the premise and workflow of HR-SPROX analyses, and detailed methods are provided in SI Appendix. Methionine is a readily oxidizable amino acid and can be converted to methionine sulfoxide by the oxidizing agent hydrogen peroxide (H2O2) (24). The oxidation rate of a methionine residue within a protein is contingent on its local structural environment. Exposed methionines in unfolded regions oxidize faster than protected methionines in folded regions (25–27). Thus, the energy difference between the folded and unfolded conformations (ΔGfolding) can modulate the folding equilibrium and rate of methionine oxidation. The progressive addition of a chemical denaturant results in the unfolding of the protein (as ΔGfolding increases) and escalates the rate of oxidation. Monitoring the extent of oxidation (at a constant oxidation time) in the presence of increasing concentrations of a chemical denaturant produces a denaturation curve (20).
Fig. 1.
HR-SPROX as a tool for measuring protein stability. (A) Theoretical basis of HR-SPROX. Methionine residues (white squares) are oxidized by H2O2 to form methionine sulfoxides (red squares). Methionine oxidation rates differ between folded and unfolded states. Thus, protein denaturation can be quantified by measuring the extent of oxidation across different denaturant concentrations at a constant oxidation time (dashed line). The resulting denaturation curves can be used to measure four types of information (numbered 1–4): baseline oxidation, midpoint of denaturation ([denaturant]1/2), m value, and free energy of folding (ΔGfolding or ΔGf). For two-state proteins, the latter two parameters can be determined by fitting the denaturation curve to an equation derived from the linear extrapolation model (LEM) as described in the text. (B) HR-SPROX workflow. Cells are lysed under native conditions, and folded proteins (black ovals) are unfolded with increasing concentrations of GdmCl. Methionines are converted to methionine sulfoxides (red squares) by addition of H2O2. A control experiment lacking H2O2 is included and used as a normalization point. Extracts are digested into peptides, and each sample (corresponding to a different denaturant concentration) is labeled with a unique tandem mass tag (TMT) and subsequently combined and analyzed by LC-MS/MS. Reporter ion intensities at the MS2 level are internally normalized to create denaturation curves, monitoring either the increase in methionine sulfoxide-containing peptides or the decrease in unoxidized methionine-containing peptides.
The SPROX denaturation curve can potentially provide four types of information: baseline oxidation, [denaturant]1/2, ΔGfolding, and m value (Fig. 1A). The level of baseline oxidation (the extent of oxidation induced by H2O2 in the absence of denaturant) provides a measure of the susceptibility of the methionine to oxidation in the context of the native protein. As we show below, this parameter is largely influenced by the level of solvent exposure of the methionine sidechain. The midpoint of denaturation ([denaturant]1/2) is a model-independent parameter that provides the denaturant concentration required to unfold one-half of the protein population. For proteins that conform to a two-state folding model, where only the fully folded and fully unfolded conformations are significantly populated at all denaturant concentrations, the resulting sigmoidal denaturation curve can be interpreted in terms of the energy difference between the two conformations using the following equation (see SI Appendix for formal derivation and assumptions):
| [1] |
where ΔGfolding is the folding stability in the absence of denaturant, and the m value is the slope of the linear relationship between ΔGfolding and [denaturant] in accordance to the linear extrapolation model of two-state folding (LEM) (28). It has been shown that the m value is correlated with the size and accessible surface area of the folding unit (29). Thus, for proteins or domains that unfold in a two-state fashion, least-squares fitting the SPROX curve with the above equation can provide ΔGfolding measurements for the folding unit that encompasses the methionine.
Accurate determination of ΔGfolding values from HR-SPROX curves using LEM is contingent on the availability of a significant number of data points in the transition regions of the sigmoidal curves (28). To obtain sufficient resolution required for analysis of a large number of polypeptides with diverse stabilities, we obtained data points at numerous denaturation concentrations (see below for exact statistics) and multiplexed the samples using TMTs (Fig. 1B). Each methionine can potentially produce two HR-SPROX curves based on the disappearance of methionine or appearance of methionine sulfoxide as a function of denaturant concentration. For a given methionine, these two curves are expected to be mirror images of one another, providing two independent datasets for analysis of its localized stability.
It should be noted that methionine residues have a tendency to become oxidized in the cell or during mass-spectrometric analysis, even in the absence of H2O2. However, the level of such spurious oxidation is minimal relative to the forced oxidation induced by H2O2. Furthermore, this background oxidation is expected to be constant at all denaturant concentrations and does not influence the measurements of normalized fractional oxidation.
Analysis of Purified Lysozyme Validates the Accuracy of ΔGfolding Measurements by HR-SPROX.
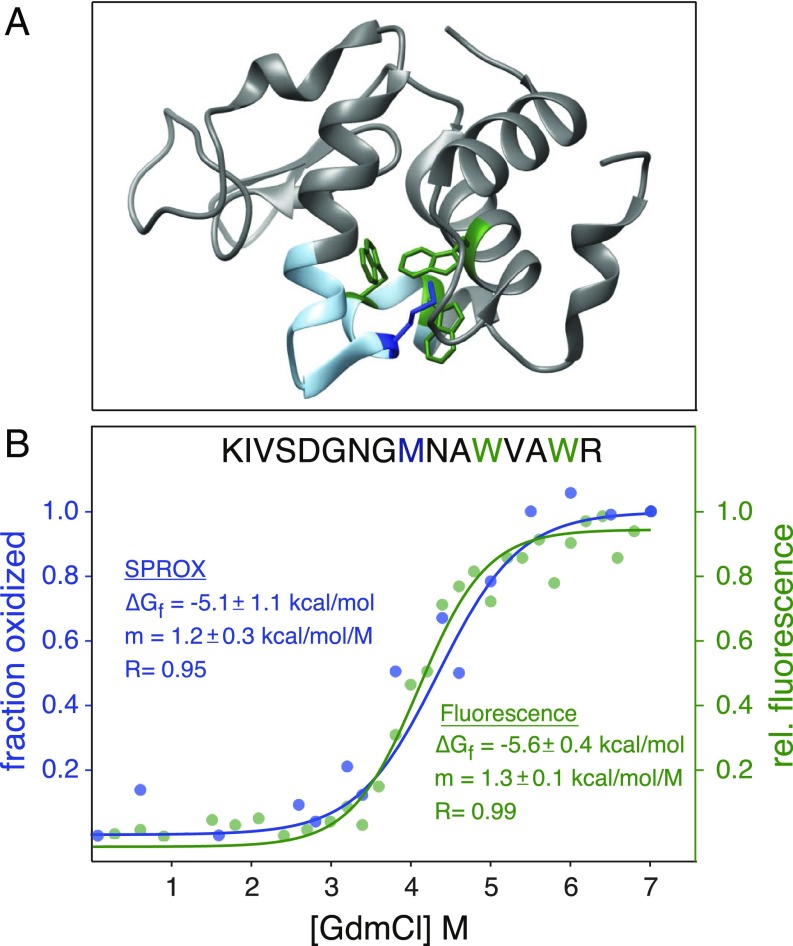
We first validated the HR-SPROX approach by analyzing purified lysozyme, a well-studied two-state protein folding model. Lysozyme has three buried tryptophan residues whose fluorescence emission spectra change upon denaturation (Fig. 2A). A protected methionine resides in the same region of the protein, enabling parallel analysis of stability by HR-SPROX. We obtained fluorescence and HR-SPROX denaturation curves using guanidine hydrochloride (GdmCl) as a denaturant. The experiments were carried out at room temperature at pH 7.4. Fig. 2B compares the denaturation curves obtained by fluorescence and HR-SPROX for a methionine-containing tryptic peptide that also encompasses two of the buried tryptophan residues. Analysis of the two denaturation curves with the two-state folding model provides similar measurements for [GdmCl]1/2, m value, and ΔGfolding. These values are also consistent with previous stability measurements obtained for lysozyme under similar conditions (30).
Fig. 2.
Validation of HR-SPROX by analysis of purified lysozyme. (A) The structure of hen lysozyme (1DPX). Buried tryptophan residues analyzed by fluorescence are colored in green, and the buried methionine residue analyzed by SPROX is colored in blue. (B) Denaturation curves obtained by intrinsic fluorescence (green) and SPROX (blue). The data were fit with Eq. 1 to determine ∆Gfolding and m values.
HR-SPROX Enables Proteome-Wide Analysis of Folding Thermodynamics in Human Fibroblast Extracts.
We next extended our analysis to the proteome-wide investigation of complex extracts. Human diploid fibroblasts expressing the catalytic component of human telomerase (HCA2-hTert) (31) were grown to a contact-inhibited quiescent state in culture and extracts were obtained under nondenaturing conditions. A series of HR-SPROX analyses were carried out with GdmCl concentrations ranging from 0 to 3 M (SI Appendix, Fig. S1). The experiments included five technical replicates obtained from two biological replicates, with each technical replicate being exposed to 30 denaturant concentrations and tagged with three different sets of 10-plex TMTs before LC-MS/MS analysis. Together, the data provided quantitative information for 10,412 unique methionine-containing peptides (in unoxidized and/or oxidized forms) mapped to 3,158 protein groups (Table 1 and Datasets S1–S3). All raw and processed data have been deposited to the ProteomeXchange Consortium via the PRIDE (32) partner repository with the dataset identifier PXD011456.
Table 1.
Combined quantified proteome coverage of HR-SPROX analyses (-TMAO)
| Incorporated residue | Unique peptides | Unique proteins |
| Met(unmodified) | 5,719 | 1,977 |
| Met(ox) | 9,350 | 2,977 |
| Met(unmodified) and Met(ox) | 4,565 | 1,745 |
| Met(unmodified) or Met(ox) | 10,412 | 3,158 |
The H2O2 concentrations and oxidation times used in our experiments were sufficient to oxidize exposed methionines to methionine sulfoxides (Met-O) but did not lead to the formation of significant levels of methionine sulfones (Met-O2) (SI Appendix, Fig. S2). The only other oxidized residue that was detectable at appreciable levels was sulfonic acid (Cys-O3). However, levels of sulfonic acid-containing peptides were significantly lower than methionine sulfoxide-containing peptides and their formation did not result in any calculation bias in measurements of methionine oxidation (SI Appendix, Fig. S2).
We initially analyzed methionine oxidation levels in individual experiments and showed that there was a significant correlation between replicate experiments (SI Appendix, Fig. S3). Additionally, there was a significant correlation between denaturation curves of individual peptides independently obtained from disappearance of methionines and appearance of methionine sulfoxides (SI Appendix, Fig. S3). Given that replicate experiments were in general agreement with each other, we combined all available data (all replicates, encompassing measurements of oxidized and/or unoxidized peptides based on availability) to obtain high-resolution denaturation curves for individual peptides. After merging datasets, the number of denaturant concentration points analyzed for individual methionine-containing peptides ranged from 10 to 300, with a median of 50 (SI Appendix, Fig. S4).
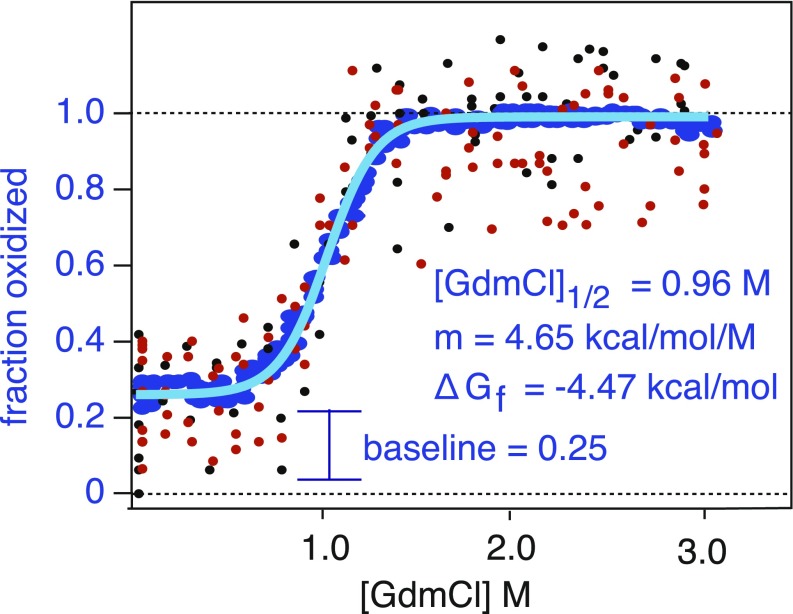
As an example, Fig. 3 illustrates a HR-SPROX denaturation curve for a protected methionine residue in ADP ribosylhydrolase. We noticed that, although individual TMT data points for a given peptide generally overlapped and had consistent trends, outliers were typically present and heavily biased subsequent least-squares regression analyses of the denaturation curves. We therefore obtained moving averages of the denaturation data and used the resulting smoothed plots to conduct least-squares regression analyses (Fig. 3). The baseline oxidation, [GdmCl]1/2, m value, and ΔGfolding measurements for individual methionine-containing peptides are tabulated in Dataset S4.
Fig. 3.
Example data for proteome-wide HR-SPROX analysis. Example HR-SPROX data for a protected methionine within a tryptic peptide (EAFDEVDMAHR) in ADP ribosylhydrolase. By combining a number of replicate measurements, detailed denaturation curves were obtained. Small black and red points indicate data obtained from methionine and methionine sulfoxide-containing peptides, respectively. The plots were smoothed by obtaining moving averages of the data (blue points) and fit by a two-state model (light blue line) to determine ΔGfolding, m values, [GdmCl]1/2, and baseline oxidation values.
Baseline Methionine Oxidation Levels Correlate with Secondary Structure and Solvent Accessibilities.
We first focused on analysis of baseline oxidation levels in the absence of denaturant. The data indicated that the distribution of baseline oxidation levels within the proteome is remarkably bimodal, with approximately one-half of the methionines forming a relatively well-protected population and one-half forming a poorly protected population (Fig. 4A). Based on these measurements, we set out to identify sequence and structural parameters that correlate with methionines’ susceptibility to oxidation.
Fig. 4.
Global analysis of methionine baseline oxidation. (A) The distribution of fractional methionine baseline oxidation levels within the analyzed proteome. Note that, due to experimental noise, it is possible for +H2O2/0 M GdmCl reporter ions to have a higher intensity than +H2O2/3 M GdmCl reporter ions, resulting in measured oxidation levels greater than 1. (B) Relative baseline oxidation levels of peptides with different methionine-neighboring residues. The columns are rank-ordered from low to high oxidation levels using the median value of the six relative positions. The most hydrophobic residues are highlighted in blue. Note that because the data consist of tryptic peptides, sequences having lysine and arginine residues on the N-side of methionines are rare and not included in the analysis. (C–F) Comparison of baseline oxidation levels of methionines contained within different DSSP secondary-structure elements (C), having different SASAs (D), having different disordered designations in accordance with MobiDB (E), and contained within proteins mapped to different GO terms (F). For all box plots, the medians are represented by yellow lines, and blue boxes denote interquartile ranges. The distributions being compared in box plots significantly differ from one another with values of P < 0.001 using the Mann–Whitney U test, with the exception of those grouped together under bars labeled “N.S.” The histograms indicate the distributions of selected categories. Cross-hatching indicates overlapping bars.
We did not find strong correlations between methionine oxidation and identities of neighboring residues in the primary sequence, beyond slightly decreased oxidation levels near hydrophobic residues (Fig. 4B and SI Appendix, Fig. S5). As discussed below, we can rationalize this trend by considering that hydrophobic residues are more likely to be in the protein core where methionines would lack exposure to solvent and thus be less susceptible to oxidation. Alternatively, as has been noted previously, the direct interaction of the methionine side chain with aromatic residues may decrease the reactivity of the sulfur atom with oxygen (33).
We next cross-referenced baseline oxidation levels to the secondary structure context of the methionine residues using the Define Secondary Structure of Proteins (DSSP) analysis of corresponding Protein Data Bank (PDB) structures (34). Structural data mapped to 1,016 proteins in our dataset indicated that highly oxidizable methionines tend to be contained in turns, bends, and unstructured coils, whereas more protected methionines tended to be in rigid secondary-structure elements such as β-strands and α-helices (Fig. 4C and Dataset S5). We subsequently used PDB structures to measure the solvent-accessible surface area (SASA) of methionines using the VMD molecular visualization program (35). We observed a very strong positive correlation between the SASA of a methionine residue and its susceptibility to oxidation (Fig. 4D and Dataset S5). Together, the data provide proteome-wide evidence that secondary-structure context and solvent accessibilities of methionine residues are the primary determinants of their susceptibility to oxidation. This trend had been previously noted for a limited number of individual proteins (27).
Intrinsically Disordered Proteins Are Highly Prone to Oxidation in Cell Extracts.
A large number of protein regions and domains have been classified as “intrinsically disordered,” primarily based on lack of evidence for structure in crystallography or NMR studies, or low complexity patterns of amino acid usage in their primary sequence (36). However, it is unclear what fraction of these sites retain their disorder in the complex intracellular environment of the cell where they could potentially be interacting with binding partners and other stabilizing factors. Given the strong correlation between localized structure and methionine oxidation established above, we assessed the baseline oxidation levels of methionines within regions classified as “disordered” in MobiDB, a curated database of intrinsically disordered proteins (IDPs) (37). Our analysis indicated that methionines within regions classified as disordered are significantly more prone to oxidation than those classified as “structured” (Fig. 4E and Dataset S5). This suggests that as a group, proteins that have been classified as intrinsically disordered mostly retain this property in crude extracts where they could potentially be interacting with stabilizing factors and binding partners. Thus, methionine oxidation rates can potentially help resolve ambiguities where conflicting evidence exists regarding the structured/disordered designation of a protein region.
Susceptibility to Oxidation Is Enriched in Specific Gene Ontologies.
We conducted gene ontology (GO) enrichment analyses to identify GO categories that were statistically enriched in protected or exposed methionines (Fig. 4F, SI Appendix, Fig. S6, and Dataset S6). GO categories most enriched in protected methionines included mitochondrial proteins, as well as proteins involved in oxidation–reduction reactions and ATP synthesis. It is notable that these categories contain proteins that are particularly exposed to reactive oxygen species (ROS) within the cell. It is possible that the prevalence of protected methionines in these proteins is an adaptive response to safeguard against ROS-induced oxidative damage. Among the GO categories most enriched in exposed methionines were cytoskeletal proteins. It has been shown that enzyme-mediated methionine oxidation is a regulatory mechanism for cytoskeletal dynamics. MICAL proteins are F-actin disassembling factors that modify the actin cytoskeleton by oxidizing two conserved methionine residues in actin (38, 39). Our data indicate that not only are MICAL-targeted methionines highly prone to oxidation (Fig. 5A) but that, as a group, cytoskeletal proteins contain an atypically large number of exposed methionines that could potentially be targetable by other oxygenases (40).
Fig. 5.
Global analysis of thermodynamic folding parameters. (A) Quantified methionine-containing peptides were separated into one of three categories based on the observed characteristics of their denaturation curves: solvent-exposed methionines (baseline oxidation, >0.5), protected two-state proteins (baseline oxidation, <0.5; goodness-of-fit r2 to two-state model, >0.9), and protected non-two-state proteins (baseline oxidation, <0.5; r2 < 0.9). The scatter plots show averaged denaturation curves for example peptides in each category. The peptide used as an example of an exposed methionine is the MICAL oxidation site on actin, as discussed in text. The pie chart indicates the number of peptides and proteins in each category. (B) Distributions of [GdmCl]1/2, m values, and ∆Gfolding measurements for two-state peptides. (C and D) Distributions of [GdmCl]1/2 and ΔGfolding measurements for example GO terms enriched in stable or unstable methionine-containing peptides. (E) The relationship between protein stability (∆Gfolding) and baseline methionine oxidation levels. See Fig. 4 for description of box and bar plots.
HR-SPROX Uncovers Proteome-Wide Distributions of [GdmCl]1/2, ΔGfolding, and m Values.
We next focused on analyzing thermodynamic folding parameters based on peptide HR-SPROX denaturation curves. We limited our subsequent analyses to peptides that contained protected methionines (conservatively defined as having a baseline oxidation levels less than 0.5) and were well fit by a two-state folding model (r2 > 0.9). This “two-state” subset consisted of 1,798 peptides mapped to 991 proteins (mean peptide to protein ratio of 1.7) (Fig. 5A). Peptides that contained protected methionines but were poorly fit by a two-state folding model (r2 < 0.9) were designated as “non–two-state.” These peptides fell into two categories. The first were peptides that showed poor fit to the two-state model due to low signal to noise (e.g., low intensity peptides). The second were peptides that displayed multiphasic unfolding patterns, perhaps due to the presence of partially folded intermediates populated during the course of denaturation. However, the resolution and signal-to-noise level of our data were not sufficient to accurately quantify the thermodynamic properties of folding intermediates in the latter category of peptides.
For “two-state” peptides, the distributions of ΔGfolding, m values, and [GdmCl]1/2 measurements are plotted in Fig. 5B. The median values for these three parameters were −2.6 kcal/mol, 2.8 kcal⋅mol−1⋅M−1, and 0.91 M, respectively. As a whole, our measured stabilities and m values appear lower than the majority of protein folding models studied to date and theoretical proteome-wide predictions for global protein stabilities (29, 41). It is therefore likely that, for most proteins, HR-SPROX measurements do not reflect the global stability of the protein, but rather subglobal stabilities of localized cooperative unfolding units encompassing the methionines. In this way, the exposure of methionines to oxidation through subglobal unfolding may be similar in nature to native HDX experiments (42).
Protein Stability Is Elevated in Specific GOs.
We conducted GO analyses to identify protein categories that are statistically enriched in highly stable or highly unstable regions as determined by [GdmCl]1/2 and ΔGfolding measurements of their methionine residues (Fig. 5 C and D, SI Appendix, Figs. S7 and S8, and Dataset S6). Among the most stable subsets of the proteome were proteins localized to the lysosomal lumen and extracellular space. Unfolding of target proteins in the denaturing environment of the lysosome is known to be required for their hydrolysis (43). It is therefore likely that resident proteins of the lysosome have evolved to be resistant to denaturation. Similarly, proteins localized to the oxidizing environment of the extracellular space are enriched in stabilizing disulfide bonds.
Among the least stable subsets of the proteome were ATP-binding proteins and ribosomal proteins. At ATP concentrations present in cell extracts, many ATP-binding proteins may exist in ATP-unbound forms that are predicted to be less stable than the ATP-bound forms (44). At higher ATP concentrations, the binding equilibrium may shift to the bound form, resulting in ligand-induced stabilization. Similarly, ribosomal proteins are stabilized by stoichiometric association to form the intact small and large subunits of the ribosome. During the course of chemical denaturation, as the higher-order structure of the ribosome is disrupted, orphaned ribosomal proteins appear to be relatively thermodynamically unstable.
Protein Stability Impacts the Rate of Oxidation for a Subset of the Proteome.
We next analyzed the relationship between folding stability and baseline methionine oxidation (Fig. 5E). The data indicated that ΔGfolding is not a strong predictor of baseline susceptibility to oxidation for two-state proteins that are relatively stable (ΔGfolding less than −1.5 kcal/mol). However, once protein stability declines beyond a minimum threshold, baseline methionine oxidation rates begin to rise. Thus, in unstable protein regions, oxidation from the unfolded state is the dominant pathway for oxidation, and the rate of oxidation is contingent on the folding equilibrium. Conversely, in stable protein regions, where the folding equilibrium is heavily shifted toward the native state, oxidation occurs primarily from the folded structure. In this regime, the rate of oxidation is independent of the folding equilibrium and instead appears to be limited by the solvent accessibility of the methionine in the native state.
HR-SPROX Enables Analysis of Interdomain and Intradomain Variability in Stability.
An important feature of the HR-SPROX analysis is that it can potentially provide localized stability information within proteins. In theory, different methionines within the same protein provide independent and localized stability probes. In some multidomain proteins, each domain may act as an independent cooperative unfolding unit (45). For these proteins, different methionines within the same domain are expected to report similar stabilities. To assess this feature of our data, we examined intradomain variability in our stability measurements. In general, we found that different methionines within the same domain have similar stabilities (Fig. 6).
Fig. 6.
Interdomain and intradomain comparisons of thermodynamic folding parameters. (A) Talin is presented as an example of an analyzed multidomain protein. The depicted structure is a composite of available structures for individual domains. The color coding indicates “weak” (blue), “intermediate” (pink), and “strong” (red) mechanical stabilities designated by Haining et al. (46) based on unfolding force magnitudes of domains in smAFM and steered molecular-dynamics studies. Denaturation curves represent exposed (red) or protected (blue) methionine-containing peptides in different domains. For protected methionines, fractional oxidation measurements were normalized with respect to the native and denatured baselines. The reported ΔGfolding is the median of all protected methionines. The Inset numbers indicate the number of peptide denaturation curves contained within plots for each domain. (B–D) Global analysis of intradomain variation in measured folding parameters. Using boundaries defined by the Pfam database, [GdmCl]1/2 (B), ΔGfolding (C), and m values (D) were assigned to specific domains. Measurements within 221 domains that contained more than one methionine were rank ordered based on their median values. Peptide measurements mapped to a given domain are plotted on the y axis for each rank-ordered domain represented on the x axis. (E) The distributions of coefficients of variation (CVs) of [GdmCl]1/2, ΔGfolding, and m-value measurements for different peptides within individual domains (intradomain variation, blue box plots), within individual proteins (intraprotein variation, orange box plots), and CV of the measurements in the entire dataset (intraproteome variation, black lines). See Fig. 4 for description of box plots.
An example of this trend is shown for the cytoskeletal protein talin (Fig. 6A). Talin is a large, amphipathic multidomain protein that forms a mechanical linkage between integrins and the cellular cytoskeleton. The interaction between talin domains and vinculin in focal adhesions is force dependent and is thought to occur through mechanical unfolding of specific talin domains that expose cryptic vinculin binding sites. To identify potential binding sites, mechanical stabilities of individual talin domains have been previously investigated by single-molecule atomic force microscopy (smAFM) (46–48). Here, we were able to track the stability and solvent exposure of 16 of the 18 domains in talin. Our data are largely consistent with the smAFM findings. We found that domains shown to have low mechanical stability by smAFM, including the linker region, R3 and R4, are solely composed of highly exposed methionines. Conversely, domains that have been shown to be highly stable, such as R5 and R9, contained protected and stable methionines. Additionally, our analysis provided thermodynamic data on domains that have not been previously studied by smAFM. For example, we identified R13 as another highly stable talin domain. These results highlight the ability of HR-SPROX to analyze localized thermodynamic stability data for large multidomain proteins that are difficult to study by traditional protein denaturation experiments.
Within individual talin domains where multiple methionine-containing peptides could be identified, the stability trends were in good agreement with each other (see overlap between protected methionine curves in Fig. 6A). To examine the generality of this trend, we used the Pfam database of protein domains (49) to globally define domain boundaries within our dataset and analyzed intradomain variability in our measurements. The analysis indicated that variabilities in [GdmCl]1/2, ΔGfolding, and m values were significantly lower within domains in comparison with variabilities of the parameters within the overall proteome (Fig. 6 B–E). The results are consistent with the idea that many domains behave as independent cooperative unfolding units.
The Chemical Chaperone Trimethylamine N-Oxide Has Variable Stabilizing Effects on the Proteome.
To determine whether HR-SPROX is capable of monitoring changes in protein stability induced by environmental alterations, we repeated our analyses in the presence of the chemical chaperone trimethylamine N-oxide (TMAO). TMAO is an amine-oxide osmolyte (50). It is found in the tissues of a variety of marine organisms where it is thought to counteract the destabilizing effects of urea and pressure on proteins. It has been postulated that TMAO stabilizes proteins by increasing the free energy of the unfolded state through solvent exclusion effects, thus shifting the folding equilibrium toward the native state (51, 52). Although the stabilizing effects of TMAO had previously been observed for a number of proteins (53, 54), the selectivity of its stabilizing effects have not been investigated on a proteome-wide scale. We conducted a limited HR-SPROX analysis in the presence of 1 M TMAO (SI Appendix, Fig. S9) and compared the baseline oxidation levels, [GdmCl]1/2, ΔGfolding, and m values of methionine-containing peptides to untreated controls (Fig. 7 and SI Appendix, Fig. S9). Overall, we observed that TMAO decreased baseline oxidation levels and increased the stability of the proteome, although the magnitudes of these effects were variable between proteins (Fig. 7A).
Fig. 7.
The effect of the chemical chaperone TMAO on proteome stability. (A) The effects of TMAO on denaturation curves of three example peptides mapped to different proteins. The data indicate that TMAO has variable effects on baseline oxidation levels and stabilities of different protein regions. (B) The distribution of relative methionine baseline oxidation levels within the analyzed proteome in the absence (blue lines, reproduced from Fig. 4) and presence of 1 M TMAO (orange bars). (C) Impact of TMAO on baseline oxidation levels of exposed and protected proteins with different stabilities. (D–F) Impact of TMAO on distributions of [GdmCl]1/2 (D), m values (E), and ΔGfolding (F). The distributions of the measurements are compared in the absence (blue lines, reproduced from Fig. 5) and presence of 1 M TMAO (orange bars). See Fig. 4 for description of box plots.
Our data indicated that TMAO decreased baseline oxidation levels of protected methionines within unstable proteins (ΔGfolding greater than −1.0 kcal/mol) but that this effect was diminished for more stable proteins (Fig. 7 B and C). This trend is consistent with the idea that, unlike stable protein regions, methionine oxidation in unstable protein regions occurs primarily from the unfolded state. Hence, modulation of the folding equilibrium by a chemical chaperone can effectively decrease oxidation rates in unstable protein regions (see above and Fig. 5E). However, TMAO did not significantly decrease the oxidation rates of most highly exposed methionines, indicating that most unstructured regions within proteins (including regions with nonrigid secondary structures, high solvent accessibility, and those characterized as “intrinsically disordered”) do not transform to stably folded structures in the presence of 1 M TMAO (Fig. 7C and SI Appendix, Fig. S9). Although our experiments did not investigate the efficacy of TMAO at higher concentrations, the results indicate that, at levels where it has a significant impact on the stability of structured proteins (Fig. 7 D–F), TMAO is not able to impose structure on the majority of disordered regions within the proteome.
Discussion
In this study, we investigated the thermodynamic folding stability of the human proteome using methionine oxidation as a probe. Our data indicate that rates of methionine oxidation are correlated to localized structure and amenable to detection and quantitation by bottom-up proteomics. However, this approach has at least two important limitations. First, the analysis is limited to tryptic peptides that contain methionines, which is one of the least abundant residues in natural proteins. For example, in our analyses, although we were able to detect a total of 60,776 unique peptides in our combined experiments, only 19% contained methionines and could be analyzed by HR-SPROX. Second, methionine oxidation is not a benign modification and may in itself impact folding stability. Thus, oxidation of one methionine could artifactually alter the localized stability of a second methionine located elsewhere in the same protein. This effect could potentially skew ΔGfolding and m-value measurements and mask multiphasic unfolding behavior. Nonetheless, methionine oxidation likely represents a significantly less disruptive modification in comparison with alternative proteomic approaches for analysis of protein structure based on limited proteolysis or indiscriminate radical footprinting.
Using HR-SPROX, we were able to quantify the susceptibility of a large number of methionines to oxidation by H2O2. Our results indicate that the distribution of methionine oxidation susceptibilities is strikingly bimodal. Methionines within the human proteome appear to be divided into two roughly equal-sized groups: solvent-exposed methionines that are highly prone to oxidation, and buried methionines that are protected from oxidation. Methionines are hydrophobic residues that would be expected to reside primarily in the cores of proteins based solely on their chemical properties. However, it has been suggested that exposed surface methionines can play a protective role by acting as sinks for ROS (55). Within the cell, the oxidation of methionines is enzymatically reversible and their reduction by methionine sulfoxide reductases, and subsequent formation of NADPH, forms a complete redox cycle (56, 57). Thus, reversible oxidation of surface methionines can act as a removal mechanism for cellular ROS and prevent the irreversible oxidation of other biomolecules (55). In support of this idea, our data provide global experimental evidence for the high prevalence of exposed oxidation-prone methionines in the human proteome.
Our analysis was able to measure thermodynamic folding stabilities (ΔGfolding) for ∼1,800 protein regions that contain highly protected methionines and unfold in accordance with a two-state model. To the best of our knowledge, this analysis represents the largest survey of thermodynamic protein stabilities conducted to date. The data highlight the remarkable range of stabilities within the proteome and suggest that stability is a physical property that has adapted to meet the specific functional needs of proteins. For example, our data highlight lysosomal and extracellular proteins as two subsets of the proteome that have evolved high stabilities, likely due to the physical demands of their local environments.
Our proteome-wide data also uncovered an important relationship between folding stability and methionine oxidation. We showed that, in general, there are two oxidation pathways for buried methionine residues. Oxidation can occur rapidly from the denatured state or more slowly from the folded state. Most buried and protected methionines reside in structured regions with stabilities that are high enough to heavily favor oxidation through the native state. However, ∼10% of protected methionines analyzed in our study were located in regions that were sufficiently unstable (ΔGfolding greater than −1.5 kcal/mol) to favor oxidation through the denatured state. The oxidation rate of this subset of the proteome is contingent on folding stability and can be diminished by stabilizing factors such as chemical chaperones (e.g., TMAO).
It has long been known that TMAO stabilizes proteins in saltwater fish, sharks, crustaceans, and molluscs (58). It is thought that this activity may counteract the destabilizing effects of urea and pressure in deep-sea animals. However, the exact physical mechanism of the stabilizing effects of TMAO remains under active investigation. It has been shown that interactions between TMAO and the peptide backbone raise the energy of unfolded structures and energetically favors the folded state (51, 59). Here, we have shown that TMAO does indeed stabilize much of the proteome. However, the magnitude of its effect is variable between proteins and domains. Our experiments have quantified the effects of TMAO on diverse protein substrates, and the results may provide a useful resource for evaluating putative mechanisms of stabilization by this osmolyte. More generally, this study demonstrates that HR-SPROX can be used to identify client sets of chaperones by globally quantifying their effects on protein folding stabilities.
It has been predicted that ∼30% of the human proteome contains regions that can be classified as intrinsically disordered (IDPs) (11). However, investigating the prevalence and properties of these regions within the cellular environment has proven challenging. In theory, the presence of interacting partners and molecular chaperones in cell extracts may impose structure on regions that otherwise appear disordered in purified systems. Here, we used HR-SPROX to probe the structure and stability of regions classified as IDPs in crude extracts. Our data indicate that, in comparison with the rest of the proteome, methionines in IDPs are highly prone to oxidation. These results suggest that most proteins designated as IDPs remain relatively unstructured even in crude extracts.
Our data also indicate that the addition of TMAO does not alter the oxidation properties of most IDPs, suggesting that most disordered regions cannot be significantly folded by the addition of 1 M concentrations of this osmolyte. As described above, TMAO is thought to modulate the equilibrium constant between the unfolded and the native state of stably folded proteins. However, this mechanism of action is contingent on the existence of localized energy minima representative of denatured and native states. Our data suggest that the folding energy landscapes of most IDPs may not contain localized minima representative of unique native states. Thus, IDPs may represent fundamentally different structures in comparison with “unstable proteins.” While the folding equilibrium of the latter can be altered in favor of a native state by the addition of osmolytes such as TMAO, the former inherently lack a native structure and are incapable of achieving a singular folded state even in the presence of stabilizing agents.
Together, our data validate a global methodology for quantitative analysis of protein stability and provide a proteome-wide census of ΔGfolding for the human proteome. This approach may prove useful in investigating proteome-wide trends in folding stabilities and can be employed in future studies to investigate the stabilizing effects of molecular chaperones, characterize the impact of environmental stresses on protein folding, and examine cross-species conservation of folding thermodynamics.
Materials and Methods
For more information on HR-SPROX protocol, details of bioinformatic analyses, reagents, and chemicals, please consult SI Appendix. SI Appendix also includes a detailed description of the protein folding thermodynamic model used in our analyses and its assumptions. SI Appendix also includes datasets that tabulate the proteomic search parameters, raw and processed proteomic data, and bioinformatic analyses.
Supplementary Material
Acknowledgments
We thank Dr. Michael Fitzgerald, Dr. Mark Dumont, and the members of the S.G. and Dr. Dragony Fu laboratories for helpful comments on the manuscript. This work was supported by grants from the National Science Foundation (MCB-1350165 CAREER) and National Institutes of Health (R35 GM119502-01, T32 GM068411, 1S100D021486-01, and 1S10OD025242-01).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited with the ProteomeXchange Consortium via the PRIDE partner repository (dataset identifier PXD011456).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1819851116/-/DCSupplemental.
References
- 1.Henzler-Wildman K, Kern D. Dynamic personalities of proteins. Nature. 2007;450:964–972. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- 2.Bryngelson JD, Onuchic JN, Socci ND, Wolynes PG. Funnels, pathways, and the energy landscape of protein folding: A synthesis. Proteins. 1995;21:167–195. doi: 10.1002/prot.340210302. [DOI] [PubMed] [Google Scholar]
- 3.Schellman JA. The thermodynamic stability of proteins. Annu Rev Biophys Biophys Chem. 1987;16:115–137. doi: 10.1146/annurev.bb.16.060187.000555. [DOI] [PubMed] [Google Scholar]
- 4.Berlett BS, Stadtman ER. Protein oxidation in aging, disease, and oxidative stress. J Biol Chem. 1997;272:20313–20316. doi: 10.1074/jbc.272.33.20313. [DOI] [PubMed] [Google Scholar]
- 5.Redler RL, Das J, Diaz JR, Dokholyan NV. Protein destabilization as a common factor in diverse inherited disorders. J Mol Evol. 2016;82:11–16. doi: 10.1007/s00239-015-9717-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boopathy S, et al. Structural basis for mutation-induced destabilization of profilin 1 in ALS. Proc Natl Acad Sci USA. 2015;112:7984–7989. doi: 10.1073/pnas.1424108112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith CD, et al. Excess brain protein oxidation and enzyme dysfunction in normal aging and in Alzheimer disease. Proc Natl Acad Sci USA. 1991;88:10540–10543. doi: 10.1073/pnas.88.23.10540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dobson CM. Protein folding and misfolding. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 9.Leuenberger P, et al. Cell-wide analysis of protein thermal unfolding reveals determinants of thermostability. Science. 2017;355:eaai7825. doi: 10.1126/science.aai7825. [DOI] [PubMed] [Google Scholar]
- 10.Minde D-P, Ramakrishna M, Lilley KS. 2018. Cellular labelling favours unfolded proteins. bioRxiv:10.1101/274761. Preprint, posted September 13, 2018. [DOI] [PMC free article] [PubMed]
- 11.Oldfield CJ, Dunker AK. Intrinsically disordered proteins and intrinsically disordered protein regions. Annu Rev Biochem. 2014;83:553–584. doi: 10.1146/annurev-biochem-072711-164947. [DOI] [PubMed] [Google Scholar]
- 12.Pace CN, Scholtz JM. 1997. Measuring the conformational stability of a protein. Protein Structure: A Practical Approach, The Practical Approach Series, ed Creighton TE (Oxford Univ Press, Oxford), Vol 2, pp 299–321.
- 13.Krishna MM, Hoang L, Lin Y, Englander SW. Hydrogen exchange methods to study protein folding. Methods. 2004;34:51–64. doi: 10.1016/j.ymeth.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 14.Ghaemmaghami S, Fitzgerald MC, Oas TG. A quantitative, high-throughput screen for protein stability. Proc Natl Acad Sci USA. 2000;97:8296–8301. doi: 10.1073/pnas.140111397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kiselar J, Chance MR. High-resolution hydroxyl radical protein footprinting: Biophysics tool for drug discovery. Annu Rev Biophys. March 14, 2018 doi: 10.1146/annurev-biophys-070317-033123. [DOI] [PubMed] [Google Scholar]
- 16.Park C, Marqusee S. Pulse proteolysis: A simple method for quantitative determination of protein stability and ligand binding. Nat Methods. 2005;2:207–212. doi: 10.1038/nmeth740. [DOI] [PubMed] [Google Scholar]
- 17.Feng Y, et al. Global analysis of protein structural changes in complex proteomes. Nat Biotechnol. 2014;32:1036–1044. doi: 10.1038/nbt.2999. [DOI] [PubMed] [Google Scholar]
- 18.Savitski MM, et al. Tracking cancer drugs in living cells by thermal profiling of the proteome. Science. 2014;346:1255784. doi: 10.1126/science.1255784. [DOI] [PubMed] [Google Scholar]
- 19.Becher I, et al. Pervasive protein thermal stability variation during the cell cycle. Cell. 2018;173:1495–1507.e18. doi: 10.1016/j.cell.2018.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.West GM, Tang L, Fitzgerald MC. Thermodynamic analysis of protein stability and ligand binding using a chemical modification- and mass spectrometry-based strategy. Anal Chem. 2008;80:4175–4185. doi: 10.1021/ac702610a. [DOI] [PubMed] [Google Scholar]
- 21.Sridevi K, Udgaonkar JB. Unfolding rates of barstar determined in native and low denaturant conditions indicate the presence of intermediates. Biochemistry. 2002;41:1568–1578. doi: 10.1021/bi011494v. [DOI] [PubMed] [Google Scholar]
- 22.West GM, et al. Quantitative proteomics approach for identifying protein-drug interactions in complex mixtures using protein stability measurements. Proc Natl Acad Sci USA. 2010;107:9078–9082. doi: 10.1073/pnas.1000148107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu Y, Wallace MA, Fitzgerald MC. Thermodynamic analysis of the geldanamycin-Hsp90 interaction in a whole cell lysate using a mass spectrometry-based proteomics approach. J Am Soc Mass Spectrom. 2016;27:1670–1676. doi: 10.1007/s13361-016-1457-2. [DOI] [PubMed] [Google Scholar]
- 24.Reeg S, Grune T. Protein oxidation in aging: Does it play a role in aging progression? Antioxid Redox Signal. 2015;23:239–255. doi: 10.1089/ars.2014.6062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zull JE, Chuang J. Characterization of parathyroid hormone fragments produced by cathepsin D. J Biol Chem. 1985;260:1608–1613. [PubMed] [Google Scholar]
- 26.Lam XM, Yang JY, Cleland JL. Antioxidants for prevention of methionine oxidation in recombinant monoclonal antibody HER2. J Pharm Sci. 1997;86:1250–1255. doi: 10.1021/js970143s. [DOI] [PubMed] [Google Scholar]
- 27.Xu K, Uversky VN, Xue B. Local flexibility facilitates oxidization of buried methionine residues. Protein Pept Lett. 2012;19:688–697. doi: 10.2174/092986612800494084. [DOI] [PubMed] [Google Scholar]
- 28.Pace CN. Determination and analysis of urea and guanidine hydrochloride denaturation curves. Methods Enzymol. 1986;131:266–280. doi: 10.1016/0076-6879(86)31045-0. [DOI] [PubMed] [Google Scholar]
- 29.Myers JK, Pace CN, Scholtz JM. Denaturant m values and heat capacity changes: Relation to changes in accessible surface areas of protein unfolding. Protein Sci. 1995;4:2138–2148. doi: 10.1002/pro.5560041020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kurtin WE, Lee JM. The free energy of denaturation of lysozyme: An undergraduate experiment in biophysical chemistry. Biochem Mol Biol Educ. 2002;30:244–247. [Google Scholar]
- 31.Bodnar AG, et al. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 32.Deutsch EW, et al. The ProteomeXchange Consortium in 2017: Supporting the cultural change in proteomics public data deposition. Nucleic Acids Res. 2017;45:D1100–D1106. doi: 10.1093/nar/gkw936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aledo JC, Cantón FR, Veredas FJ. Sulphur atoms from methionines interacting with aromatic residues are less prone to oxidation. Sci Rep. 2015;5:16955. doi: 10.1038/srep16955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kabsch W, Sander C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 35.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 36.Romero P, et al. Sequence complexity of disordered protein. Proteins. 2001;42:38–48. doi: 10.1002/1097-0134(20010101)42:1<38::aid-prot50>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 37.Piovesan D, et al. MobiDB 3.0: More annotations for intrinsic disorder, conformational diversity and interactions in proteins. Nucleic Acids Res. 2018;46:D471–D476. doi: 10.1093/nar/gkx1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hung RJ, et al. Mical links semaphorins to F-actin disassembly. Nature. 2010;463:823–827. doi: 10.1038/nature08724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hung RJ, Pak CW, Terman JR. Direct redox regulation of F-actin assembly and disassembly by Mical. Science. 2011;334:1710–1713. doi: 10.1126/science.1211956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manta B, Gladyshev VN. Regulated methionine oxidation by monooxygenases. Free Radic Biol Med. 2017;109:141–155. doi: 10.1016/j.freeradbiomed.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ghosh K, Dill K. Cellular proteomes have broad distributions of protein stability. Biophys J. 2010;99:3996–4002. doi: 10.1016/j.bpj.2010.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Englander SW, Sosnick TR, Englander JJ, Mayne L. Mechanisms and uses of hydrogen exchange. Curr Opin Struct Biol. 1996;6:18–23. doi: 10.1016/s0959-440x(96)80090-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bohley P, Seglen PO. Proteases and proteolysis in the lysosome. Experientia. 1992;48:151–157. doi: 10.1007/BF01923508. [DOI] [PubMed] [Google Scholar]
- 44.Patel A, et al. ATP as a biological hydrotrope. Science. 2017;356:753–756. doi: 10.1126/science.aaf6846. [DOI] [PubMed] [Google Scholar]
- 45.Batey S, Nickson AA, Clarke J. Studying the folding of multidomain proteins. HFSP J. 2008;2:365–377. doi: 10.2976/1.2991513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haining AW, von Essen M, Attwood SJ, Hytönen VP, Del Río Hernández A. All subdomains of the talin rod are mechanically vulnerable and may contribute to cellular mechanosensing. ACS Nano. 2016;10:6648–6658. doi: 10.1021/acsnano.6b01658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.del Rio A, et al. Stretching single talin rod molecules activates vinculin binding. Science. 2009;323:638–641. doi: 10.1126/science.1162912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yao M, et al. Mechanical activation of vinculin binding to talin locks talin in an unfolded conformation. Sci Rep. 2014;4:4610. doi: 10.1038/srep04610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Finn RD, et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yancey PH, Clark ME, Hand SC, Bowlus RD, Somero GN. Living with water stress: Evolution of osmolyte systems. Science. 1982;217:1214–1222. doi: 10.1126/science.7112124. [DOI] [PubMed] [Google Scholar]
- 51.Ma J, Pazos IM, Gai F. Microscopic insights into the protein-stabilizing effect of trimethylamine N-oxide (TMAO) Proc Natl Acad Sci USA. 2014;111:8476–8481. doi: 10.1073/pnas.1403224111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baskakov I, Bolen DW. Forcing thermodynamically unfolded proteins to fold. J Biol Chem. 1998;273:4831–4834. doi: 10.1074/jbc.273.9.4831. [DOI] [PubMed] [Google Scholar]
- 53.Mello CC, Barrick D. Measuring the stability of partly folded proteins using TMAO. Protein Sci. 2003;12:1522–1529. doi: 10.1110/ps.0372903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ganguly P, Boserman P, van der Vegt NFA, Shea JE. Trimethylamine N-oxide counteracts urea denaturation by inhibiting protein-urea preferential interaction. J Am Chem Soc. 2018;140:483–492. doi: 10.1021/jacs.7b11695. [DOI] [PubMed] [Google Scholar]
- 55.Levine RL, Mosoni L, Berlett BS, Stadtman ER. Methionine residues as endogenous antioxidants in proteins. Proc Natl Acad Sci USA. 1996;93:15036–15040. doi: 10.1073/pnas.93.26.15036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cabreiro F, Picot CR, Friguet B, Petropoulos I. Methionine sulfoxide reductases: Relevance to aging and protection against oxidative stress. Ann N Y Acad Sci. 2006;1067:37–44. doi: 10.1196/annals.1354.006. [DOI] [PubMed] [Google Scholar]
- 57.Weissbach H, et al. Peptide methionine sulfoxide reductase: Structure, mechanism of action, and biological function. Arch Biochem Biophys. 2002;397:172–178. doi: 10.1006/abbi.2001.2664. [DOI] [PubMed] [Google Scholar]
- 58.Yancey PH, Blake WR, Conley J. Unusual organic osmolytes in deep-sea animals: Adaptations to hydrostatic pressure and other perturbants. Comp Biochem Physiol A Mol Integr Physiol. 2002;133:667–676. doi: 10.1016/s1095-6433(02)00182-4. [DOI] [PubMed] [Google Scholar]
- 59.Zou Q, Bennion BJ, Daggett V, Murphy KP. The molecular mechanism of stabilization of proteins by TMAO and its ability to counteract the effects of urea. J Am Chem Soc. 2002;124:1192–1202. doi: 10.1021/ja004206b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.