Significance
We detected cfDNA somatic mutations in combination with protein markers to efficiently identify early stage HCC from asymptomatic HBsAg-seropositive individuals in a community population. We designed an assay to profile multiple types of genetic variations in parallel so that 2 mL of plasma were sufficient to achieve high sensitivity in the detection of cfDNA mutations. The assay successfully identified four early-stage HCC cases (<3 cm) from 331 HBsAg (+) individuals who were negative based on screening with serum AFP and ultrasonography. The positive predictive value was 17%, which was much higher than in previous studies. This paper shows that the combination of cfDNA and serum protein markers has significant promise for the early detection of HCC in the community population.
Keywords: cell free DNA, hepatocellular carcinoma, early detection of cancer, HBsAg-seropositive
Abstract
Liquid biopsies, based on cell free DNA (cfDNA) and proteins, have shown the potential to detect early stage cancers of diverse tissue types. However, most of these studies were retrospective, using individuals previously diagnosed with cancer as cases and healthy individuals as controls. Here, we developed a liquid biopsy assay, named the hepatocellular carcinoma screen (HCCscreen), to identify HCC from the surface antigen of hepatitis B virus (HBsAg) positive asymptomatic individuals in the community population. The training cohort consisted of individuals who had liver nodules and/or elevated serum α-fetoprotein (AFP) levels, and the assay robustly separated those with HCC from those who were non-HCC with a sensitivity of 85% and a specificity of 93%. We further applied this assay to 331 individuals with normal liver ultrasonography and serum AFP levels. A total of 24 positive cases were identified, and a clinical follow-up for 6–8 mo confirmed four had developed HCC. No HCC cases were diagnosed from the 307 test-negative individuals in the follow-up during the same timescale. Thus, the assay showed 100% sensitivity, 94% specificity, and 17% positive predictive value in the validation cohort. Notably, each of the four HCC cases was at the early stage (<3 cm) when diagnosed. Our study provides evidence that the use of combined detection of cfDNA alterations and protein markers is a feasible approach to identify early stage HCC from asymptomatic community populations with unknown HCC status.
The worldwide incidence of liver cancer in 2018 was 841,080, and it is the third leading cause of cancer death in the world (1). Hepatocellular carcinoma (HCC) represents the major histological type of liver cancer, accounting for ∼85–90% of cases (2). Moreover, there is no effective therapy for advanced stage HCC. Screening is recommended for patients with cirrhosis who have a high risk of developing HCC (3, 4). In China, early screening for HCC has been performed in several cohorts following the Asian-Pacific Association for the Study of the Liver guidelines, which recommends that HCC surveillance should be performed for individuals with cirrhosis and those that are surface antigen of hepatitis B virus (HBsAg) positive, using ultrasonography (US) and serum α-fetoprotein (AFP) tests every 6 mo (3). Although these modalities have demonstrated significant improvements in overall survival due to early detection and receipt of curative therapies in previous studies (5), precise detection of HCC requires experienced specialists, limiting their widespread application in all HBsAg-positive individuals. Furthermore, biannual screening is also associated with follow-up appointments and anxiety-producing procedures. Currently, most HCC cases in China are detected on the basis of clinical symptoms rather than by HCC screenings and are at an advanced stage when diagnosed in hospitals.
In recent studies, liquid biopsies based on genetic alterations in cell free DNA (cfDNA) have shown promising results in the early detection of cancer (6, 7). In combination with protein markers, the sensitivity and specificity might be further improved, and multiple tumor types might be screened in one test (8–10). Among the tumor types studied in this way, screening for HCC had among the highest sensitivities, possibly due to the abundant blood supply in the liver (9). However, these studies were conducted mainly on hospitalized HCC patients and healthy individuals without hepatitis B virus (HBV) infection (9). The performance of the liquid biopsy assays could be compromised in high-risk populations with chronic HBV infection because some precancerous lesions, such as cirrhosis, might also harbor driver mutations prevalent in HCC. Profiling hepatitis, cirrhosis, and noncancer liver nodules might be necessary to draw a baseline to precisely identify HCC that can be clinically validated by imaging or histology. Here, we report a method, the hepatocellular carcinoma screen (HCCscreen), that is based on detection of both serum protein markers and cfDNA alterations, and we demonstrate its utility when applied to early detection of HCC in the multicenter community population with chronic HBV infection.
Results
Clinical Parameters of Participants at Baseline in the Four Screening Centers and the Follow-Up of HCC Outcome.
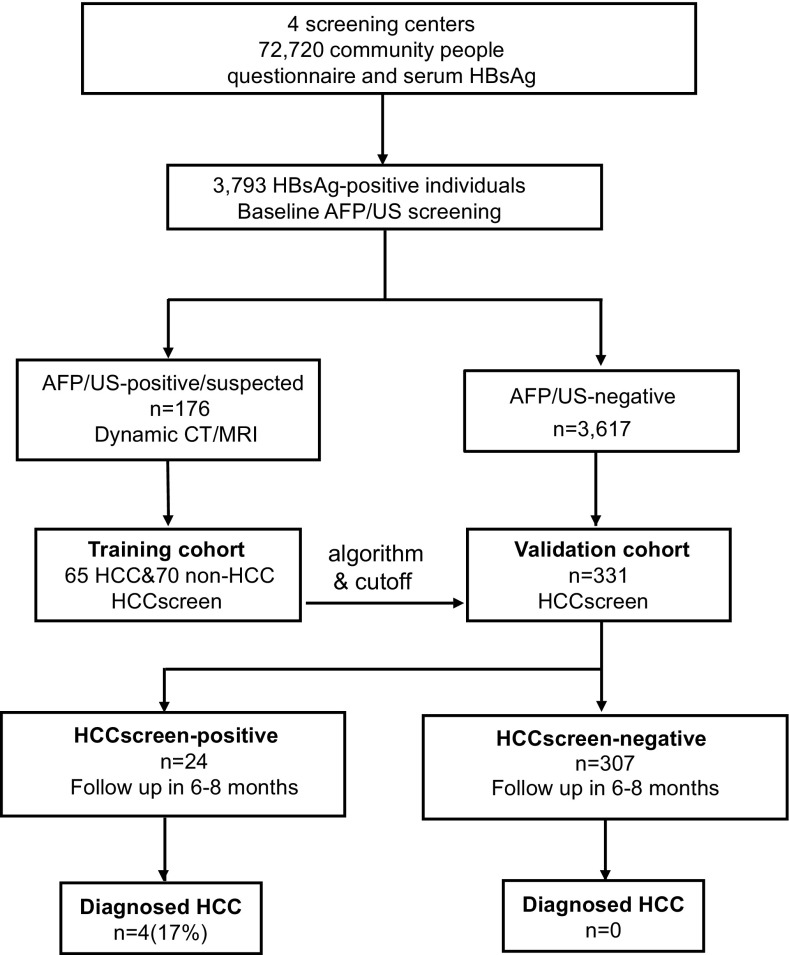
Community individuals (n = 72,720) in four screening centers were screened with blood HBsAg tests followed by questionnaire surveys. HBsAg-positive individuals (n = 3,793) were invited to participate in AFP/US screening. Of these HBsAg-positive individuals, 176 had concerning AFP/US results (named the AFP/US-positive/suspected group), whereas the remaining HBsAg-positive patients made up the AFP/US-negative group (n = 3,617) (Fig. 1 and SI Appendix, Table S1). To determine their HCC status, all AFP/US-positive/suspected individuals were recommended to undergo dynamic CT/MRI within 2 mo of the initial screening. Those with a reliable diagnosis of HCC status were included as a training cohort in this study, and we performed the HCCscreen test on the baseline AFP/US screening blood samples from these individuals (Fig. 1).
Fig. 1.
Study design. The enrollment, training of the HCCscreen model, and validation in sampled AFP/US-negative individuals.
Of the 3,617 AFP/US-negative individuals, about 60% had submitted to AFP/US screens before the baseline screen in this study (SI Appendix, Fig. S1 and Table S1). To reduce the anxiety and noncompliance in the follow-up procedures, we mainly chose the individuals who had undergone AFP/US screens during the previous 1–3 y for the validation cohort (n = 331). The distribution of the sampled AFP/US-negative participants was comparable to the total HBsAg-positive participants based on gender, proportion of US-detected cirrhosis, and serum albumin levels (SI Appendix, Table S1). We performed the HCC liquid biopsy testing (HCCscreen) on the blood samples collected from the validation cohort at the baseline AFP/US screening and followed up on HCC status 6–8 mo after the baseline screening. We also performed the HCCscreen on 70 healthy individuals without HBV infection.
Selection and Detection of Markers of HCC Using HCCscreen.
We developed the HCCscreen assay with two classes of biomarkers: (i) genetic alterations that are highly prevalent in HCC and can be detected in cfDNA; and (ii) the serum protein markers AFP and des γ carboxy prothrombin (DCP). In previous cancer genomic studies, most of the HBV-associated HCCs harbor, at least, one mutation in the following genes/locations: TP53, CTNNB1, AXIN1, or the TERT promoter (11, 12). We also considered the HBV integration breakpoint as a potential biomarker of HCC. Since the HBV integration site should be unique in each individual cell, the detection of multiple copies (>2) of a specific integration site from plasma (2 to 3 mL) could be an indication of clonal expansion of a single cell harboring HBV integration. Only in this case does the resultant neoplasm release multiple copies of the same genomic DNA into the blood. We designed an assay that can profile the genetic variations in parallel. The extracted cfDNA was ligated to a customized adapter with a DNA barcode and then amplified to generate a whole genome library. We enriched the targets of point mutations and HBV integrations with a method similar to rapid amplification of cDNA ends (RACE) using multiple primers covering the coding regions of TP53, CTNNB1, and AXIN1, the promoter region of TERT, and the HBV sequence (SI Appendix, Fig. S2) (7, 13). The reads from next generation sequencing can be tracked to the original cfDNA molecule with the DNA barcode to filter false positive single nucleotide variants (SNVs) from sequencing/amplification errors (14).
Based on our previous findings and other reports conducted on hospitalized patients affected by HCC, liver cirrhosis, and chronic hepatitis, the combination of serum protein levels of AFP and DCP displayed significant sensitivity and specificity in discriminating early-stage HCC and decompensated liver cirrhosis (15). Thus, we included these two serum protein markers in combination with cfDNA alterations to investigate whether this combination of liquid biopsy-based assays, including AFP, DCP, and cfDNA, would be an effective assay to screen for early-stage HCC.
Consistency of Clinical Diagnoses with the HCCscreen Assay.
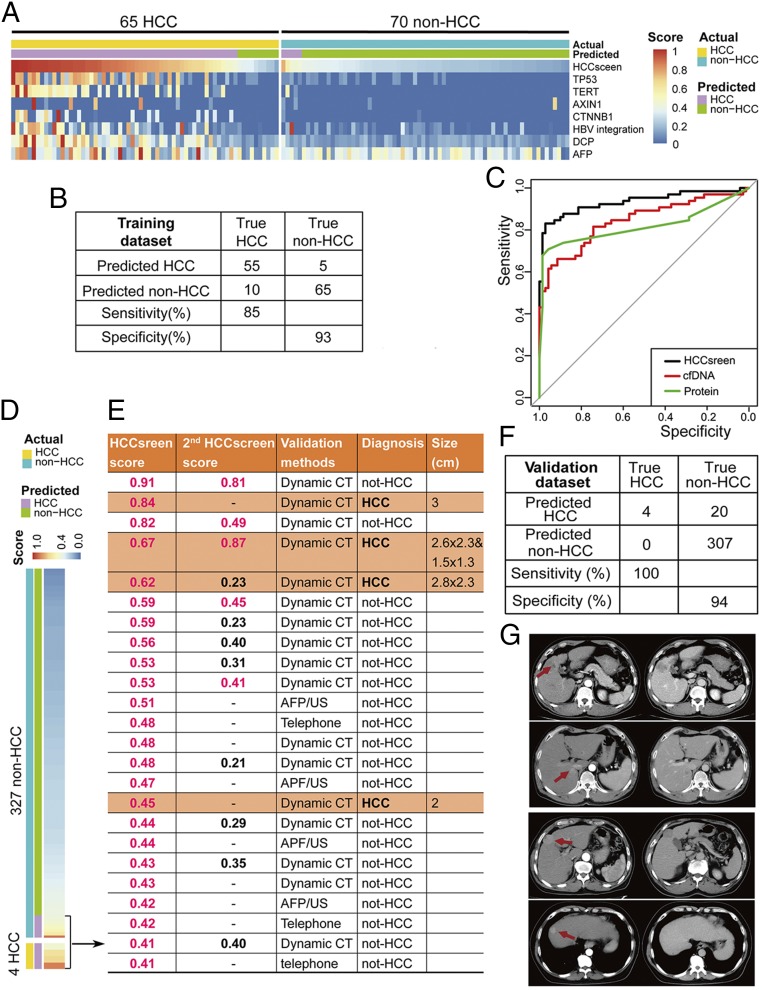
To determine its utility in the detection of HCC, we assessed the HCCscreen assay in the individuals who have a known diagnosis of HCC or have been excluded (non-HCC). We obtained 65 HCC cases and 70 non-HCC cases from the AFP/US-positive/suspected individuals. This HCC-positive or HCC-negative status was based on dynamic CT/MRI imaging and histologic confirmation. These 135 cases were used as the training cohort, and the HCCscreen results were compared with the clinical diagnosis. To build a classifier integrating different types of biomarkers in the assay, we first collapsed the different types of cfDNA mutations into a region of interest (ROI) score for each gene or locus (see Methods). The ROI score was a weighted sum of the damaging effects and frequency for each point mutation within a ROI. Besides ROI scores for SNV/indel mutations in the genes, we added two structural variant features (HBV integration in the TERT promoter region and other HBV integrations), one experimental feature (the cfDNA concentration), two protein markers (AFP and DCP), and two clinical features (age and gender) as the final features to build a diagnostic classifier to predict HCC status (Table 1). Using a penalized logistic regression algorithm with these markers, the HCCscreen model robustly differentiated HCC cases from non-HCC cases (Fig. 2A). With 100 iterations of leave-one-out cross validation on the training data set of 65 HCC and 70 non-HCC cases, we found that the assay would yield sensitivity of 85% and specificity of 93% in the diagnosis of HCC (area under the curve = 0.928) (Fig. 2 B and C). The HCCscreen score cutoff was 0.4 for the highest Youden index score (SI Appendix, Fig. S3B and Table S2). Both cfDNA and protein markers showed significant contributions to the identification of HCC (Fig. 2C and SI Appendix, Table S3).
Table 1.
Characteristics of HCCscreen features and their coefficients
| Feature class | Features | Coefficients |
| cfDNA | TP53 other than R249S | 2.02 |
| TP53 R249S | 0.21 | |
| TERT | 1.37 | |
| SV_TERT | 0.70 | |
| CTNNB1 | 1.20 | |
| AXIN1 | 0.01 | |
| HBV integrations | 0.82 | |
| cfDNA concentration | −0.27 | |
| Protein | AFP | 0.21 |
| DCP | 1.59 | |
| Clinical information | gender | 0.66 |
| age | 1.69 | |
| (Intercept) | −2.68 |
Penalized logistic regression: λ = 0.14; α = 0.
Fig. 2.
The performance of HCCscreen in the training and validation cohorts. (A) The HCCscreen scores and contributions of the cfDNA and protein biomarkers in the diagnostic model in the training cohort. (B) Binary results of the diagnostic model in the training cohort. (C) ROC of the diagnostic model of the HCCscreen in the training cohort. (D) The HCCscreen performance of the diagnostic model in the validation cohort. (E) Follow-up and diagnosis of the HCCscreen-positive cases in the validation cohort. (F) Binary results of the diagnostic model in the validation cohort. (G) Dynamic CT imaging of the four HCC cases detected with the HCCscreen among the AFP/US-negative individuals.
Predictive Value of the HCCscreen Assay for Early Stage HCC Among the AFP/US-Negative Individuals.
We further tested whether the HCCscreen could detect HCC from HBsAg-positive individuals who were AFP/US-negative and exhibited no clinical symptoms. We tested 331 AFP/US-negative individuals with HCCscreen and identified 24 positive cases (termed HCCscreen positive) based on the algorithm derived from the training cohort (Fig. 2D).
Follow-up procedures were performed on the 24 HCCscreen-positive individuals in 6–8 mo for the HCC clinical outcome. Of these individuals, 17 were examined by dynamic CT, 4 by AFP/US, and 3 were followed by telephone interviews. Four of the 24 HCCscreen-positive individuals were eventually diagnosed with HCC, and the positive predictive value of HCC detection was 17% (Fig. 2E). In addition, a group of HCCscreen-negative participants (n = 70) agreed to take the dynamic CT examination in 6–8 mo, and none were diagnosed with HCC. We also tracked 172 HCCscreen-negative participants by AFP/US 6–8 mo after the baseline AFP/US screen, and no cases of HCC were diagnosed. No HCC was found in the 65 participants followed up with telephone interviews (SI Appendix, Fig. S1). Altogether, no HCC cases were found in these HCCscreen-negative cases. Taken together, the HCCscreen assay yielded a 17% positive predictive value, 100% (4/4) sensitivity, and 94% specificity (307/327) in AFP/US-negative individuals (Fig. 2F). All four HCC tumors identified were <3cm when diagnosed by dynamic CT (Fig. 2G), and the four patients did not have liver cirrhosis according to the US result at the baseline.
We offered the AFP/US examination 6–8 mo after the baseline to the 944 participants who were AFP/US-negative at the baseline and did not take the HCCscreen test. Four HCC cases (0.4%, 4/944) were detected and further confirmed. Cancer registry records indicated that by June 30, 2018 no liver cancer outcome (ICD-10 code C22) had been identified among the 337 participants who were AFP/US-negative in the baseline screening and did not take the HCCscreen or any further AFP/US screen (SI Appendix, Fig. S1).
Blood was drawn a second time from 13 out of the 24 HCCscreen-positive cases for performing a repeated HCCscreen assay 6–8 mo after the first blood draw at the baseline. One HCC case available for the second HCCscreen test continued to be positive, and the score was higher than that obtained 6 mo previously. Another HCC case had already had the tumor surgically resected by the time of the second blood draw, and the HCCscreen turned out negative, consistent with this status. Seven out of the 11 HCCscreen-positive non-HCC cases (64%) were negative in the second HCCscreen test, although two of them were near to the threshold (0.40). The remaining four non-HCC cases were still positive in the second HCCscreen (Fig. 2E). These results indicated that the positive predictive value could be further improved by repeat testing at a second time point. All of them are currently followed to further validate the assay.
Training the Liquid Biopsy Assay with Healthy Individuals.
The HCCscreen assay showed a robust ability to identify HCC in a high-risk population. Previous studies predicted that the sensitivity and specificity in such a high-risk cohort would be lower than when comparing the cancer patients with healthy individuals without HBV infection or other risk factors (9). To test this hypothesis, we performed the HCCscreen assay on 70 healthy individuals without HBV infection (HBsAg negative) and used these data to replace the 70 HBsAg-positive non-HCC cases in the training cohort. By profiling the cfDNA and protein markers, the HCCscreen assay robustly identified the HCC cases from the healthy individuals, leading to 98% sensitivity and 100% specificity (SI Appendix, Fig. S3A). However, the algorithm derived from this training cohort (HCC and healthy individuals) performed poorly in the HBsAg-positive non-HCC cases. Most of the non-HCC cases were classified as positive, and the HCC and non-HCC cases were highly overlapping according to the algorithm (SI Appendix, Fig. S3B). Furthermore, the performance on the validation cohort was poor. Although all four HCC cases were positive in the test, many of the HBsAg (+) individuals were classified as positive, yielding specificity and positive predictive values of only 58% and 2.8%, respectively (SI Appendix, Fig. S3B). On the other hand, the algorithm derived from HCC vs. non-HCC cases correctly classified all healthy individuals (100%) as negative in addition to its performance on the HBsAg-positive validation cohort (SI Appendix, Fig. S3B).
Discussion
Early detection of cancer is the most effective way to reduce deaths from cancer. In recent studies, liquid biopsy assays based on cfDNA and/or proteins showed promise for detecting early-stage cancers of diverse tissue types (9). However, most of these studies were conducted on individuals previously diagnosed with cancer or as healthy, and no high-risk individuals were used as controls (9). In this study, we developed and tested a liquid biopsy assay to identify HCC cases in a high-risk asymptomatic community population. The HCC and non-HCC cases in the training cohort were identified from the same population with the same method AFP/US. Such non-HCC cases, especially those with liver nodules, were useful for the identification of HCC-specific biomarkers and thresholds to precisely detect HCC cases. In the selection of biomarkers, we focused on frequently altered genetic biomarkers with a clear oncogenic mechanism, such as the TERT promoter mutations and protein markers that are of clear diagnostic value, such as DCP (16). We included a limited number of candidate biomarkers with a clear link to HCC to avoid the overfitting effect when studying numerous candidate biomarkers on a limited number of tumor/normal cases. With the combination of population and biomarker selections, we aimed to set up an HCC early detection model that would not only show appropriate sensitivity and specificity in the training cohort, but also have comparable diagnostic value of early-stage HCC from high-risk individuals in different populations.
Using this HCCscreen assay, we found it possible to identify individuals with early-stage HCC and to discriminate them from those non-HCC individuals with chronic liver disease, including cirrhosis. The assay yielded 85% sensitivity and 93% specificity in the diagnosis of HCC from the individuals with US-detected liver nodules and/or increased serum AFP. More importantly, the performance was also maintained with the AFP/US-negative validation cohort where the sensitivity and specificity were 100% and 94%, respectively. The current sensitivity is based on a limited number of HCC cases. It might change with long term follow up or dynamic CT/MRI on all individuals, if additional HCC cases are identified. In this case, a prospective and large-scale clinical trial would be necessary to determine the sensitivity and specificity according to the follow-up time. However, the current positive predictive value (PPV) of 17% from the validation cohort was significantly higher than previously obtained with screening for AFP levels alone (17). The PPV could be further improved if a second HCCscreen test is provided to the cases positive in the first test. A high PPV would be very helpful for routine application in the clinic as it would reduce unnecessary anxiety and follow-up examinations of the non-HCC individuals.
To confirm the advantage of studying a high-risk population as a control, we also trained the model by comparison of HCC cases to healthy individuals without HBV infection. The assay can clearly separate the cases of HCC from the healthy individuals with 98% sensitivity and 100% specificity. However, the algorithm derived from this training cohort (HCC and healthy individuals) performed poorly on a validation cohort of all HBsAg-positive individuals. Although all four HCC cases were positive in the test, 189 non-HCC cases were classified as positive as well, yielding specificity and PPVs of only 58% and 2.8%, respectively. These results indicate the importance of the high-risk cases in defining the baseline to detect HCCs with high specificity. High-risk non-HCC cases in the training cohort are important to set up an algorithm and threshold with favorable performance in the screening of early HCC to be developed as the screening of HCC most likely happens in the context of high-risk individuals in clinical practice.
Tumor size at the time of diagnosis is an important clinical parameter affecting the survival of HCC patients. Unlike protein- or RNA-based biomarkers, a tumor cell generally harbors only one copy of mutant DNA in most cases. A fundamental question for cfDNA-based early detection screens is whether an early-stage tumor releases enough copies of mutant DNA for detection in the circulation. Among all of the HCCs identified by HCCscreen in this study, 85% and 68% of the cases were <5 and <3 cm, respectively. HCC tumors of <5 cm are early stage and amenable to curative surgery. The patients with tumors of <3 cm could have an even better outcome, thus underscoring the value of HCCscreen for reducing HCC morbidity and mortality. In the validation cohort, we identified four HCCs that were 2 to 3 cm in size from the AFP/US-negative population. These results clearly show that the sensitivity of the HCCscreen has promise for the detection of early-stage HCCs.
In both training and validation cohorts, there were some non-HCC cases that were positive in the HCCscreen test. These false positive results might not be due to technical errors as the genetic alterations, such as TP53 R249S, were validated with a second method (digital PCR). It is possible that the signals released for some minor lesions were from those that were undetectable by dynamic CT. Such lesions could be eliminated by the immune system or persist for the long term as the second HCCscreen test might become negative or continue to be positive. We are tracking the individuals with HCCscreen-positive results to determine whether they have an increased risk of developing HCC in the future. If this were true, the HCCscreen assay could potentially help to predict HCC long term outcome and lead to preventive chemo- or immunotherapeutic measures for some patients before diagnosis by dynamic CT.
An ideal tumor screening method should have high sensitivity and specificity. It should also be easy to perform in clinical practice. The HCCscreen assay detects mutations in coding regions and translocations/HBV integrations with unknown breakpoints at a cost of <150 US dollars. In addition, the liquid biopsy assay enables centralized and standard processing and requires minimal expertise and equipment in local hospitals/clinics. Taken together, the assay is highly suitable for HCC screening as a routine test in at-risk individuals.
There were also several limitations in this study. First, the follow-up period of 6–8 mo was relatively short. Current false positive cases could develop HCC in the future, and the PPV could be increased. All of the cases are being followed. Second, the training cohort was relatively small as the percentage of AFP/US-positive cases in the community HBsAg(+) population was not high. Additional early-stage HCC and non-HCC cases would be helpful to improve the algorithm of the assay. Third, the healthy control group in this study has a median age of 32, which is lower than the non-HCC group. The decreased performance of the algorithm trained by this cohort could be due to the loss of two risk factors, HBV infection and age. An age-matched HBsAg(−) group would further clarify the importance of HBV infected controls in the training of HCC screening assays.
Our study provides evidence that screening based on cfDNA mutations and protein markers in a high-risk population has efficacy in the identification of patients with HCC. It is noninvasive and can detect early-stage as well as late-stage tumors. More importantly, as somatic mutations in driver genes are common in the development of most cancers, the strategy could be modified for use in the early screening of other tumor types or of multiple tumor types from a single tube of blood.
Materials and Methods
We established the community-based cohort study on population with high risk of liver cancer (the CCOP-LC cohort; Chinese Clinical Registry, ChiCTR-EOC-17012853) in 2017, based on the early HCC screening program conducted in the community population. The study protocol (NCC201709011) was approved by the Institutional Review Board (IRB) of National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital, Chinese Academy of Medical Sciences in Beijing, China following the guidelines issued by the Ministry of Science and Technology of the People’s Republic of China. Human samples were collected and anonymously coded. All participants provided written informed consent before any protocol-directed procedures were provided. Detailed Materials and Methods are included in the SI Appendix.
Supplementary Material
Acknowledgments
This study was supported by State Key Projects Specialized on Infectious Diseases (2017ZX10201201-006), National Key R&D Program of China (2018YFC1312100), Key Research Projects for Precision Medicine (2017YFC0908103 and 2012ZX10002008), the Chinese Academy of Medical Sciences (CAMS) Innovation Fund for Medical Sciences (CIFMS) (Grants 2016-I2M-1-001 and 2016-I2M-1-007), the National Key Basic Research Program of China (Grant 2015CB553902), and the National Natural Science Foundation Fund (81472559).
Footnotes
Conflict of interest statement: S.W. and H. Yan are the founders of Genetron Health (Beijing) Co. Ltd. Y.J. is one of the cofounders of Genetron Health (Beijing) Co. Ltd. B.H.D. is a consultant for Genetron Health (Beijing) Co. Ltd. Y.J., Q.C., P.W., Yuting Wang, K.C., Q.S., S.W., and H. Yan have filed patents/patent applications based on the data generated from this work. Other authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1819799116/-/DCSupplemental.
References
- 1.Bray F, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Wong MCS, et al. The changing epidemiology of liver diseases in the Asia-Pacific region. Nat Rev Gastroenterol Hepatol. 2018;16:57–73. doi: 10.1038/s41575-018-0055-0. [DOI] [PubMed] [Google Scholar]
- 3.Omata M, et al. Asia-pacific clinical practice guidelines on the management of hepatocellular carcinoma: A 2017 update. Hepatol Int. 2017;11:317–370. doi: 10.1007/s12072-017-9799-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marrero JA, et al. Diagnosis, staging and management of hepatocellular carcinoma: 2018 practice guidance by the American association for the study of liver diseases. Hepatology. 2018;68:723–750. doi: 10.1002/hep.29913. [DOI] [PubMed] [Google Scholar]
- 5.Singal AG, Pillai A, Tiro J. Early detection, curative treatment, and survival rates for hepatocellular carcinoma surveillance in patients with cirrhosis: A meta-analysis. PLoS Med. 2014;11:e1001624. doi: 10.1371/journal.pmed.1001624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bettegowda C, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaudhuri AA, et al. Early detection of molecular residual disease in localized lung cancer by circulating tumor DNA profiling. Cancer Discov. 2017;7:1394–1403. doi: 10.1158/2159-8290.CD-17-0716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Springer S, et al. A combination of molecular markers and clinical features improve the classification of pancreatic cysts. Gastroenterology. 2015;149:1501–1510. doi: 10.1053/j.gastro.2015.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen JD, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018;359:926–930. doi: 10.1126/science.aar3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen JD, et al. Combined circulating tumor DNA and protein biomarker-based liquid biopsy for the earlier detection of pancreatic cancers. Proc Natl Acad Sci USA. 2017;114:10202–10207. doi: 10.1073/pnas.1704961114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Totoki Y, et al. Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nat Genet. 2014;46:1267–1273. doi: 10.1038/ng.3126. [DOI] [PubMed] [Google Scholar]
- 12.Zhang W, et al. Genetic features of aflatoxin-associated hepatocellular carcinomas. Gastroenterology. 2017;153:249–262. doi: 10.1053/j.gastro.2017.03.024. [DOI] [PubMed] [Google Scholar]
- 13.Waltari E, et al. 5′ rapid amplification of cDNA ends and illumina MiSeq reveals B cell receptor features in healthy adults, adults with chronic HIV-1 infection, cord blood, and humanized mice. Front Immunol. 2018;9:628. doi: 10.3389/fimmu.2018.00628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci USA. 2011;108:9530–9535. doi: 10.1073/pnas.1105422108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen H, et al. Direct comparison of five serum biomarkers in early diagnosis of hepatocellular carcinoma. Cancer Manag Res. 2018;10:1947–1958. doi: 10.2147/CMAR.S167036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lok AS, et al. HALT-C Trial Group Des-gamma-carboxy prothrombin and alpha-fetoprotein as biomarkers for the early detection of hepatocellular carcinoma. Gastroenterology. 2010;138:493–502. doi: 10.1053/j.gastro.2009.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chun S, Rhie SY, Ki CS, Kim JE, Park HD. Evaluation of alpha-fetoprotein as a screening marker for hepatocellular carcinoma in hepatitis prevalent areas. Ann Hepatol. 2015;14:882–888. doi: 10.5604/16652681.1171776. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.