Abstract
Tau proteins are proteins that stabilize micro-tubules, but their hyperphosphorylation can result in the formation of protein aggregates and, over time, neuro-degeneration. This phenomenon, termed tauopathy, is pathologically involved in several neurodegenerative disorders. DNA aptamers are single-stranded oligonucleotides capable of specific binding to target molecules. Using tau epitopes predisposed for phosphorylation, we identified six distinct aptamers that bind to tau at two phosphorylatable epitopes (Thr-231 and Ser-202) and to full-length Tau441 proteins with nanomolar affinity. In addition, several of these aptamers also inhibit tau phosphorylation (IT4, IT5, IT6) and tau oligomerization (IT3, IT4, IT5, IT6). This is the first report to identify tau epitope-specific aptamers. Such tau aptamers can be used to detect tau in biofluids and uncover the mechanism of tauopathy. They can be further developed into novel therapeutic agents in mitigating tauopathy-associated neurodegenerative disorders.
Graphical Abstract
INTRODUCTION
Tau proteins are microtubule-associated proteins known to promote the assembly of microtubules and to maintain microtubule integrity, which is essential for axonal transport and morphogenesis.1,2 Normal tau proteins bind with micro-tubules and prevent these track-like structures from breaking apart, allowing nutrients and molecules to be transported along the cells. However, tau is also found to be pathologically involved in several neurological disorders, termed tauopathies, in which aggregations of tau are deposited in brain neurons.3 In the case of Alzheimer’s disease, pathological tau proteins self-assemble into paired helical filaments, which later aggregate into insoluble neurofibrillary tangles.4 The transport system for neurons is disrupted along the process, causing nutrients and other essential supplies to cease moving along the cells. Neurons with tangles and nonfunctioning micro-tubules consequently undergo apoptosis and eventually cell death. Such a phenomenon is also observed in a range of other neurodegenerative diseases.5,6 Various forms of insoluble abnormal tau aggregates are involved in tauopathies, but they share a common composition of hyperphosphorylated tau.
Aptamers are nucleic acid probes capable of specific binding to defined targets.7 They are selected through an amplification-evolution process termed systematic evolution of ligands by exponential enrichment (SELEX).8 Aptamers have been selected against a variety of targets, including metal ions,9 fluorescent dyes,10 amino acids,11 nucleotides,12 antibiotics,13 metabolites,14 peptides,15 proteins,16 viruses,17 organelles,18 or even whole cells.19 As such, aptamers have shown remarkable specificity in discriminating targets from their analogue counterparts, such as differentiating among homologous proteins differing by only a few amino acids20 or one single amino acid,21 or even between enantiomers.22 Because molecular recognition is essential in many biological processes, aptamers can potentially inhibit the functions of their targets. For instance, binding of thrombin with its aptamers has been shown to undermine its activity and decrease the rate of blood clotting.23
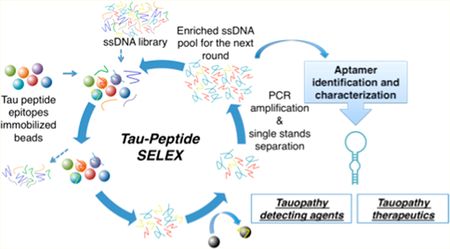
The most fundamental difference between healthy tau and pathological tau can be ascribed to the level of phosphor-ylation. Abnormally phosphorylated tau lesion causes death of neuron cells, resulting in irreversible and progressive neuro-degeneration. Understanding the origin and mechanism of tauopathy is a key step toward developing a means to delay or even fight against it. Since pathological tau loses its affinity for microtubules due to an abnormally high degree of phosphor-ylation, we hypothesized that aptamers binding to phosphor-ylatable regions on tau protein might be useful in studying the molecular mechanism(s) underlying tauopathy. It was also anticipated that aptamers binding to the phosphorylated sites on tau could be exploited to investigate the formation of hyperphosphorylated tau aggregates and detect the phosphor-ylation level. Therefore, we herein describe a SELEX process using fragments of phosphorylatable regions from tau protein and their corresponding phosphorylated forms as targets to search for site-specific tau aptamers for potential further use as tauopathy-detecting agents and possible therapeutic agents.
RESULTS AND DISCUSSION
Tau Epitope-Specific Aptamer Discovery and Selection.
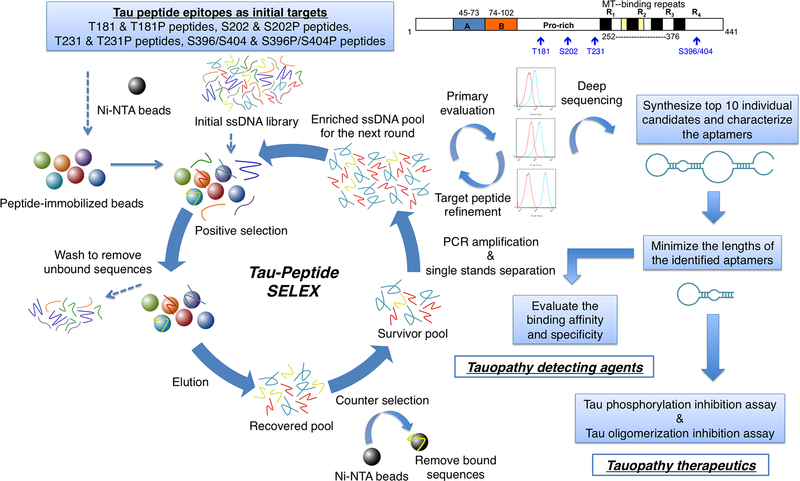
The selection began with a library containing 20 nmol of primer-flanked, 66-nucleotide-long, single-stranded DNA. Four phosphorylatable peptide epitopes from tau and their corresponding phosphorylated peptides (Table S1) were used as putative targets (Figure 1). The detailed screening strategy is described in Supporting Information.
Figure 1.
Tau aptamer discovery by SELEX and characterization workflow. Four peptide epitopes from tau and their corresponding phosphorylated peptides were used as putative targets. The enrichment was monitored using flow cytometry. The most abundant 10 candidates were characterized after deep sequencing. The binding affinity/specificity and the inhibitory effects with the identified aptamers were then verified.
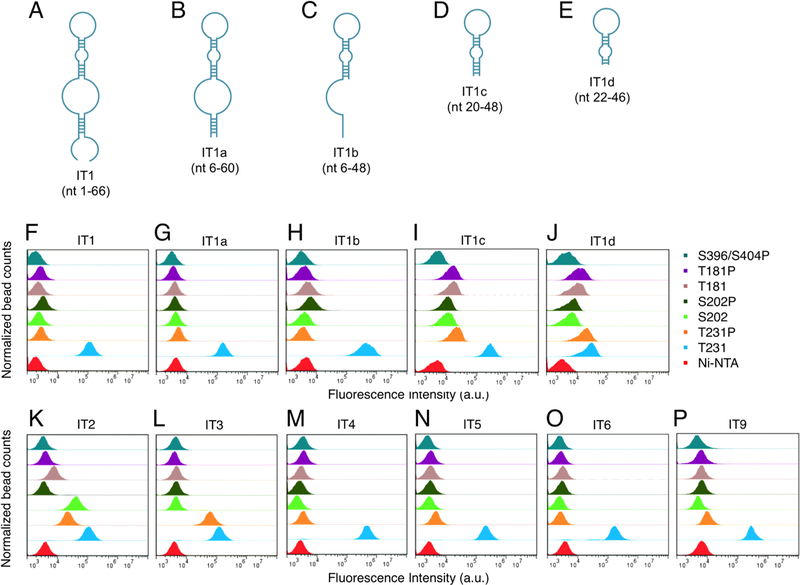
The 10 most abundant sequences found in pool #17 and their population percentage in each of the sequenced pools are listed in Table S2. They are denoted as IT1 through IT10 and were chemically synthesized and labeled with FAM at the 5’-end, followed by HPLC purification. Primary binding analysis was examined by flow cytometry. Seven out of the 10 candidates are confirmed aptamers (Figure 2F, K–P). Five of them (IT1, IT4, IT5, IT6, and IT9) possess strong and specific binding to T231 peptide, while IT3 bypassed the phosphorylated site and bound to both T231 and T231P peptides. Aptamer IT2, on the other hand, recognized not only both T231 and T231P, but it also bound to S202. It is noteworthy that IT8, IT9, and IT10 bore a high level of sequence homology to IT3, IT5, and IT2, respectively. In fact, the discrepancy only occurs at the 19th nucleotide (Table S3). IT9 and its predecessor IT5 displayed similar binding strength toward T231, suggesting that the nucleotide at position 19 for these two similar sequences had no significant impact on their binding abilities to T231. However, IT8 and IT10 both lost their binding abilities to the prospective targets found with IT3 and IT2, proving that the binding strength of an aptamer could be greatly compromised by merely altering one nucleotide within the crucial binding region. Based on the fact that IT8 and IT10 manifested no binding preference to either of the peptides, they are likely the byproducts that resulted from PCR amplification with edge effect.
Figure 2.
Primary binding analysis between aptamer candidates and their putative peptide targets. (A-E) Representative predicted secondary structures of aptamer IT1 and its subsequent truncated forms, IT1a, IT1b, IT1c, and IT1d. (F and M–P) Full-length sequences IT1, IT4, IT5, IT6, and IT9 are aptamers that specifically bind to T231 peptide. (F–J) The binding abilities of truncated aptamers IT1a, IT1b, and IT1c to T231 epitope are not seriously compromised when compared to the full-length aptamer IT1. However, binding strength is lost when further truncating the stem of IT1c into IT1d. (K) Sequence IT2 demonstrates a unique binding profile on three of the peptides (T231, T231P, and S202). (L) Sequence IT3 bypasses the phosphorylated site and recognizes both T231 and T231P.
The initial binding tests were carried out at 4 °C to ensure having the optimal secondary structures for aptamers. The binding abilities of the selected aptamers were then examined at room temperature and at 37 °C. None of the aptamers lost binding ability at room temperature or 37 °C (data not shown), suggesting that these aptamers are suitable for future in vivo studies.
Sequence Truncation of the Selected Aptamers.
The aptamers identified are full-length sequences evolved from the initial library, which contains the fixed primer binding regions on both ends to serve the PCR amplification process. However, some full-length aptamers can be shortened into a minimally functional sequence without compromising direct interaction with the target.24 Herein we synthesized such truncated versions of aptamers IT1, IT2, IT3, IT4, IT5, and IT6 based on secondary structures predicted by Integrated DNA Technologies OligoAnalyzer Tool (Figure 2A–J, Table S4). Two to three T-bases were also added as a spacer next to fluorescein if the truncated sequence ended with G-base or GC pair in the stem.
We successfully identified the binding motif from aptamer IT1 as sequence IT1c, which is less than half the length of its original IT1 sequence. IT2 was at first truncated to IT2a based on an evident stem–loop motif observed in the predicted secondary structure, and IT2a demonstrated binding ability equal to IT2 for targets T231, T231P, and S202. Still, an even more stringent truncation of IT2, candidate IT2b, failed to maintain the properties of the original aptamer. A less exacting approach was then implemented to further shorten aptamer IT2a. Sequence IT2c presented binding ability similar to IT2a, but a gradual loss of binding ability was observed by further truncating the stem of IT2c into IT2d and IT2e, suggesting the importance of a stable stem for binding of aptamer IT2c to its targets. A less restrictive truncation of aptamer IT3 into sequence IT3c resulted in partial binding to T231, but it caused a complete loss of binding to T231P. For IT4 and IT5, the complete sequences are required for target recognition. Finally, a branched hairpin structure, IT6a, retained selective binding ability to T231 as aptamer IT6.
Determination of Binding Kinetics/Affinities of the Selected Aptamers.
In this kinetics study, his-tag peptides, as well as his-tag Tau441 protein, were immobilized on the surface of the biosensor, while aptamer candidates were the analytes in the solution phase. The binding interaction of analyte to immobilized ligand was measured in real time as the change in the number of molecules bound to the biosensor that caused a shift in the interference pattern reflected from sensor surfaces. The kinetic on-rates (kon), off-rates (koff), and equilibrium dissociation constants (Kd) measured for all aptamer-target pairs are summarized in Table S5.
All aptamers reported here displayed high binding affinities toward Tau441 with Kd values ranging from 5.5 nM to 68 nM. The observed on-rates (kon) ranged between 104 M−1 s−1 and ~106 M−1 s−1. Aptamer IT1 presented the fastest on-rate ((9.3 ± 1.9) × 105 M−1 s−1) toward Tau441 protein, while the slowest on-rate ((1.067 ± 0.018) × 104 M−1 s−1) was detected for IT2 binding to Tau441. However, IT2 also demonstrated an extremely slow off-rate (koff) ((5.9 ± 1.2) × 10−5 s−1) for Tau441, exhibiting the lowest Kd (5.5 ± 1.1 nM) for Tau441 protein among all aptamers. The lowest Kd toward peptides, on the other hand, was observed between IT5 and T231 (5.0 ± 0.3 nM). Meanwhile, IT5 also exhibited the second lowest Kd (7.6 ± 0.6 nM) for Tau441 protein among all aptamers, indicating the high binding affinities of IT5 toward both T231 peptide and Tau441 protein. Although aptamer IT9 differs by only one base from IT5, as shown in Table S3, it showed appreciably slower kon and, therefore, higher equilibrium dissociation constants compared to IT5.
While IT1 showed similar binding kinetics toward T231 and Tau441, the truncated aptamer IT1c exhibited a much slower off-rate for Tau441 protein than for T231 peptide. On the other hand, truncation of IT2 did not affect the association of the aptamers to T231 peptide, but the shorter aptamer IT2a did display a faster dissociation rate for T231 compared to IT2. In fact, the dissociation rates with any of the targets were increased with IT2a compared to the off-rates with IT2. In addition, a faster on-rate for T231P peptide was found with either IT2 or IT2a in comparison to that of T231. With IT2a, especially, the on-rate toward T231P was almost 5 times faster than its on-rate toward T231, and this on-rate was also 2.5-fold faster than the association of IT2 to T231P. However, the association between IT2a and S202 peptide showed an opposite tendency. The on-rate of IT2a for S202 was found to be more than 6 times slower than its IT2 counterpart. Therefore, the truncation of IT2 appears to benefit its recognition of T231P, while, at the same time, losing its binding affinity toward the S202 site. Moreover, while the association of IT2a to Tau441 is almost 50 times faster than IT2, its off-rate is also 60 times faster than that of IT2. Overall, the binding affinity of IT2a was weakened by the truncation. Unlike IT1c and IT2a, the shorter version of IT6 did not affect the binding kinetics and affinity. IT6a still behaved much like IT6. Finally, the on-rates of IT3 to both T231 and T231P peptides were found to be quite similar, but the off-rate with T231P was almost 2-fold faster than that with T231.
Specificity of the Selected Tau Aptamers against Tau441 Protein.
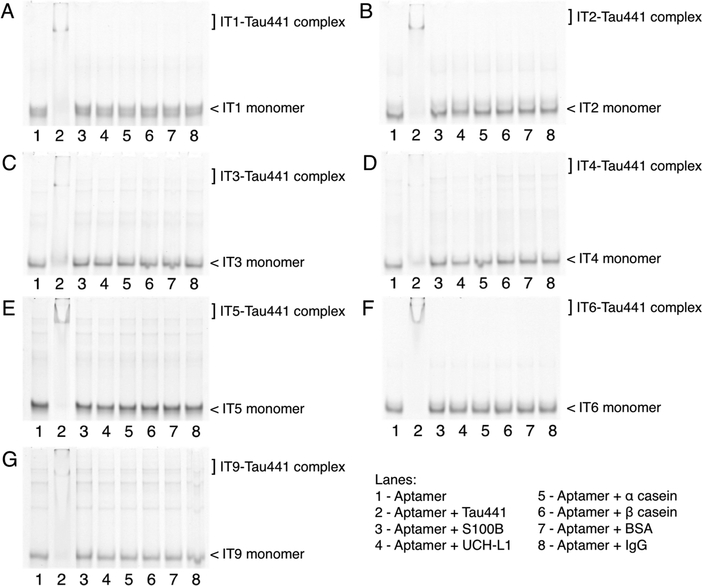
The binding specificity of the selected tau aptamers to full-length Tau441 protein was confirmed by nondenaturing gel electrophoresis after incubating each of the FITC-labeled aptamers with either target Tau441 protein or the individual nontarget proteins separately. Each of the tau aptamers (IT1-IT6 and IT9) alone displayed one main band at the lower part of the gel (Figure 3, lane 1). Some minor upper bands observed could have resulted from dimeric or multimeric forms of the oligonucleotides. The monomeric aptamers disappeared in the presence of the target Tau441 protein due to the formation of aptamer-Tau441 complexes (Figure 3, lane 2). No cross-reactivity was observed between the aptamers and the nontarget proteins, including S100B (S100 calcium-binding protein B) (11 kDa), UCH-L1 (ubiquitin carboxyl-terminal hydrolase L1) (25 kDa), α casein (23 kDa), β casein (24 kDa), BSA (bovine serum albumin) (66 kDa), and IgG (immunoglobin G) (150 kDa) (Figure 3, lane 3–8). In particular, S100B25 and UCH-L126 are important references because they are also brain-associated protein and traumatic brain injury (TBI) biofluid biomarker proteins.27
Figure 3.
Binding specificity of the tau aptamers toward tau protein. Gel electrophoresis of each FITC-labeled tau aptamer, (A) IT1, (B) IT2, (C) IT3, (D) IT4, (E) IT5, (F) IT6, and (G) IT9, after incubation with the protein of interest. Aptamers form binding complexes with tau and therefore show a smaller migration in the presence of tau. S100B, UCH-L1, α casein, β casein, BSA, and IgG are nontarget reference proteins and have no retention effect on the migration of aptamers under electrophoresis.
Aptamer-Based Sandwich ELISA for Detection of Tau.
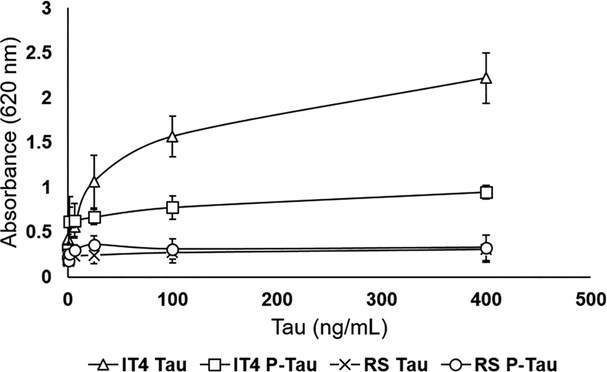
To further study the feasibility of using tau aptamers for tau protein detection, a sandwich enzyme-linked immunosorbent-assay (ELISA) was performed in triplicate. Tau protein was first captured by DAKO antibody. The biotin-labeled IT4 aptamer was used as the detection probe to specifically recognize tau protein. The 620 nm absorbance was recorded after incubating samples with streptavidin-labeled polyHRP enzyme and TMB substrate. As shown in Figure 4, IT4 aptamer strongly binds to tau protein with a dose-dependent increase in signal, as opposed to nontarget phosphorylated tau, indicating the possibility of utilizing IT4 aptamer to specifically quantify the tau level in biological samples.
Figure 4.
Aptamer-antibody sandwich ELISA for detection of tau. Both tau and phosphorylated tau can be captured by total tau antibody. However, aptamer IT4 only detects the presence of nonphosphorylated T231 residue. The scrambled random sequence (RS) and phosphorylated tau protein are included in this test as negative controls. All the sequences are labeled with biotin, which later reacts with streptavidin-labeled polyHRP enzyme and TMB substrate to reveal the colorimetric reaction products.
Inhibitory Effects of Tau Aptamers on Tau Phosphor-ylation.
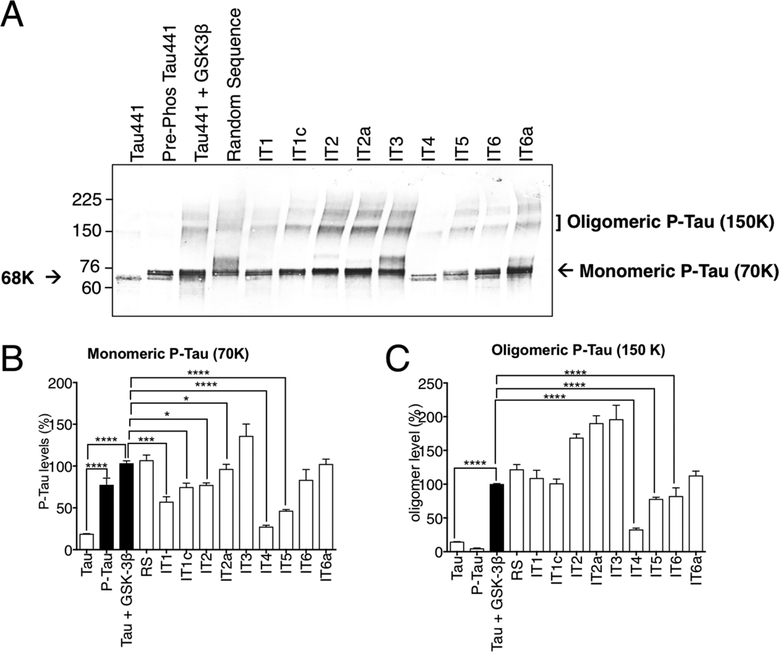
To analyze the ability of tau aptamers to inhibit tau phosphorylation, tau was phosphorylated by kinase glycogen synthase kinase-3β (GSK3β) for 24 h at 37 °C in the presence or absence of tau aptamers. We used phospho-T231 tau monoclonal antibody (RZ3) to probe the phosphorylated tau. A faint 68K band was observed with control Tau441 protein (Figure 5A, lane 1), while an intense band showing a molecular shift from 68K to 70K was detected with the commercially available positive control GSK3β prephosphorylated Tau441 (Figure 5A, lane 2). Lane 3 shows the experimentally GSK3β phosphorylated Tau441 at both the monomeric 70K band and oligomeric 150 K p-tau band. The presence of random oligonucleotide had no effect on the levels of phosphorylated monomeric tau or oligomeric p-tau (Figure 5, lane 4). IT1 and IT1c showed minor reduction of monomeric p-tau, but they had no effect on oligomeric p-tau (Figure 5, lanes 5 and 6). The presence of IT2, IT2a and IT3 somehow promoted phosphorylated tau toward the oligomeric form (Figure 5, lanes 7–9). The reason behind this result is not fully understood yet, but we suspect this is due to the fact that these aptamers bind to more than one epitope (Figure 2K,L). Among all aptamers, IT4 eliminated both monomeric p-tau and oligomeric p-tau most dramatically (Figure 5, lane 10). IT5 also showed some inhibition on monomeric p-tau, but IT6 and IT6a had no significant effects (Figure 5, lanes 11–13).
Figure 5.
Inhibitory effects of tau-binding aptamers on phosphorylation of Tau441 in vitro. (A) In vitro tau phosphorylation and oligomerization assay was performed by incubating Tau441 (1 μg) with aptamers (50 μM) for 1 h, followed by incubation with GSK3β (200 ng) for 16 h. Samples were analyzed by SDS-PAGE, followed by Western blotting with phospho-tau antibody (RZ3). (B) Quantification of monomeric p-tau (70K) and (C) oligomeric p-tau (150 K) bands of tau protein. The monomeric p-tau (70K) and oligomeric tau (150 K) bands were normalized by the levels of GSK3β-phosphorylated Tau441 and are shown as percentage. *p< 0.05, **p< 0.01, ***p< 0.001 and ****p< 0.0001 (n = 3, two-way ANOVA).
Heparin-Induced Aggregation of Tau441 and the Inhibitory Effects of Tau Aptamers.
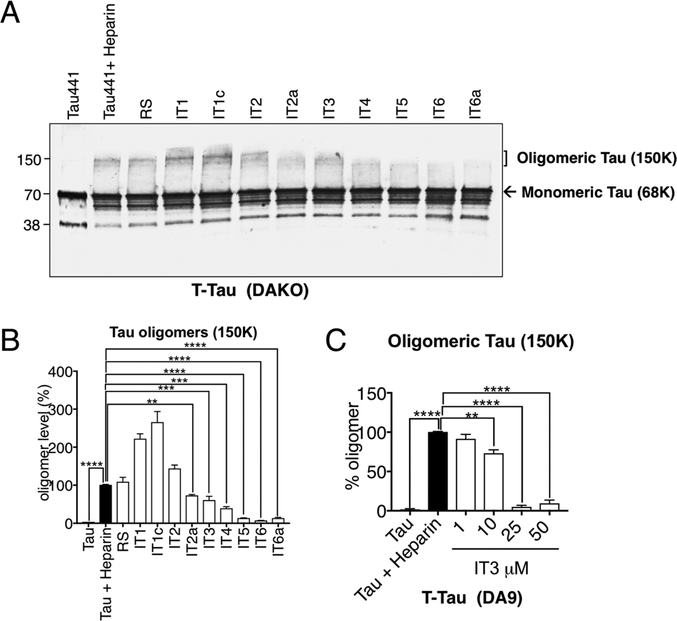
To examine the inhibitory effects of tau aptamer on the formation of tau oligomers in vitro, heparin was used as an oligomerization inducer.28 In brief, tau protein was incubated with either the aptamer or a random sequence for 1 h followed by treatment with heparin for 16 h to form tau oligomers. The reaction products were then analyzed by SDS-PAGE and immune-blotted with polyclonal total tau antibody. Tau441 was detected at 68K in monomeric form (Figure 6A, lane 1). Heparin treatment caused the formation of high molecular weight (~150 K oligomeric tau) (Figure 6A, lane 2). When random sequence oligonucleotide was preincubated with tau, the oligomeric assembly of tau induced was virtually unaffected. In contrast, IT4, IT5, IT6, and the truncated IT6a showed the most robust inhibition of oligomeric tau formation among the tested aptamers. Truncated IT2a and IT3 also showed partial inhibition of tau oligomerization. However, the remaining aptamers did not show any statistically significant effects on tau oligomerization (Figure 6A,B).
Figure 6.
In vitro inhibitory effects of aptamers on oligomerization of Tau441. (A) In vitro tau oligomerization assay was performed using Tau441 (10 μM), heparin (5 mg/mL) and aptamers (50 μM) for 16 h. Samples were analyzed by SDS-PAGE, followed by Western blotting with total tau antibody (DAKO). (B) Quantification of oligomeric tau (150 K). (C) In vitro dose-response effect of IT3 on heparin-induced tau oligomerization assay as detected by DA9 antibody. The reaction was performed in the presence of IT3 aptamer (1, 10, 25, 50 μM) for 24 h. The oligomeric tau bands (150 K) were normalized by the level of heparin-induced tau oligomers only and are shown as percentage. *p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001 (n = 3, two-way ANOVA).
In a separate experiment, an oligomerization study was performed with 1, 10, 25, and 50 μM of IT3, followed by probing with a monoclonal tau antibody (DA9). A dose-dependent effect to reduce the formation of tau oligomers was confirmed (≥90% inhibition of tau oligomers was observed at 50 μM of IT3) (Figure 6C).
Investigation of Tau Binding to Microtubules in the Presence of Aptamers.
A microtubule binding protein spin-down assay was used to investigate the binding of tau to microtubules in the presence of aptamers. The assay was first validated according to the manufacturer’s instructions (Figure S7). Briefly, microtubules were collected in the pellet fraction along with any proteins bound on MTs. Proteins that have no affinity to MTs would remain in the supernatant. The existence of each protein in the supernatant and pellet fractions was analyzed through gel electrophoresis and protein staining. We then confirmed that Tau441 was only found in the supernatant fraction when there were no MTs. Conversely, in the presence of MTs, Tau441 predominantly bound to MTs and was pulled down together with MTs during centrifugation (Figure S8A). A much reduced amount of tau stayed in the supernatant. To study the effects of aptamers to the tau-bound and preformed MT, the two most promising aptamers from the inhibition study (IT4 and IT5) were added separately into the mixture of Tau441/MTs for 30 min before centrifugation. The results indicated that IT4 and IT5 could detach partially of the bound Tau441 from MTs. This outcome was not unanticipated though, as it has been reported that the T231 site on Tau441, the target epitope of IT4 and IT5, has a pronounced influence on the binding affinity of tau to microtubules, although it is distinct from the well-recognized microtubule-binding domain.29
CONCLUSIONS
Although the exact causes of tauopathy and the related neurodegeneration remain unclear, evidence has shown that pathological tau always appears in the hyperphosphorylated and oligomeric form. To provide a specific tool for further investigating the relationship between tauopathy and tau phosphorylation/oligomerization, we herein propose a peptide-based selection process to identify site-specific tau-binding aptamers. Even though some previous studies have found tau-binding DNA sequences30 and a tau-binding RNA aptamer,31 none of these probes has a confirmed binding region. One essential advantage of using peptides instead of the whole protein to carry out the aptamer selection for tau is that the binding sites have been predetermined and are confined to the regions that are pathologically relevant.
Four phosphorylatable regions (T181, S202, T231, S396/S404) from tau protein and the corresponding phosphorylated epitopes from pathological tau are used here as putative targets to demonstrate the peptide-based tau aptamer selection method. Aptamer IT3 bypasses phosphorylation on T231 and recognizes both T231 and T231P. Aptamer IT2 could bind to T231, T231P, and S202, but not S202P. The remaining selected aptamers are highly specific to the T231 site. Additionally, aptamers IT1, IT2, and IT6 were structurally minimized into shorter versions (IT1c, IT2c, IT6a) without appreciably compromising their binding abilities. However, aptamers IT3, IT4, and IT5 were extremely susceptible to any attempts at sequence truncation. All tau aptamers reported show high affinity and specificity to tau protein. The dissociation constants of these aptamers against Tau441 protein range from 5.5 nM to 68 nM.
We then successfully demonstrated the feasibility of using tau aptamers as a detection tool by performing an aptamer–antibody sandwich ELISA using total tau antibody and IT4 aptamer. Phosphorylated tau was used as a negative control here, and our results showed that this assay is highly specific to the desired target.
Next, since our tau aptamers preferentially bind to nonphosphorylated tau, we reasoned that they might prevent tau from being phosphorylated at the T231 site by blocking substrate site access to protein kinases. Our results did, indeed, confirm this by showing that aptamers IT4 and IT5 strongly inhibited tau phosphorylation by GSK3β at T231. In our results, the GSK3β-phosphorylated Tau441 showed a high molecular weight form detected at 150 K by RZ3 antibody, while this effect was not seen with the purified prephosphorylated Tau441. It has been reported previously that high concentration of tau in solution causes it to aggregate when incubated at 37 °C.28 Thus, the likely explanation is that the hyperphosphorylated Tau441 by GSK3β had a higher tendency to oligomerize in solution. Indeed, we confirmed that IT4, IT5, and IT6 also reduced the levels of oligomeric tau. In parallel, we also hypothesized that tau aptamers might interfere with tau oligomerization, and such hypothesis was tested with an oligomerization study. It was observed that aptamers IT3, IT4, IT5, IT6, and IT6a could partially, or completely, inhibit tau oligomerization in vitro. Taken together, IT4 and IT5 showed the most robust effects on the reduction of tau phosphorylation and oligomerization, suggesting that they are therapeutic candidates for future animal-based models and possible clinical studies. Though we later found that IT4 and IT5 have moderate influence on the binding affinity of tau to microtubules, this result does not prevent us from utilizing these aptamers for detection in biofluids. Besides, this result can be attributed to the fact that the T231 epitope has been identified as a critical site in regulating tau’s ability to bind microtubules.29 This does not eliminate the possibility of using our platform to find aptamers that target other epitopes and have no impacts on tau’s binding ability to microtubules. A separate study reporting an RNA aptamer targeting tau also showed that the RNA-based tau aptamer could alleviate synthetic tau oligomer-mediated neurotoxicity in primary rat hippocampal neurons in culture.31
However, it is worth noting that RNA aptamers are not as stable as DNA-based aptamers in biological conditions. Furthermore, unlike our study, the reported RNA aptamer has undefined tau binding sites, and its specificity, affinity, and on- and off-rates for tau protein were not defined.
In summary, our novel findings demonstrate, for the first time, the feasibility of identifying tau epitope-specific aptamers. When tau DNA aptamers are further optimized, we envision that they might have multiple utility in terms of uncovering the mechanism of tauopathy or in diagnosing and treating tauopathy-associated disorders. In terms of diagnosis of tauopathies, tau and p-tau antibodies exist, but the advantages of using tau aptamers include their stability, ease of storage, and thus longer shelf life when being used in a point-of-care device. Aside from tau phosphorylation, tau acetylation has also been identified as a dominant post-translational modification in tauopathies. A recent study revealed that tau acetylation at residues K280/K281 impairs tau-mediated microtubule stabilization and enhances the formation of tau aggregates.32 Hence, our peptide-based aptamer selection process could also be applied to identify probes that recognize specific tau acetylation sites, thus providing new tools to investigate the role of tau acetylation in tau pathology, or even new antagonists to convey therapeutic approaches by binding to critical acetylation sites. In terms of therapeutic treatments for tauopathy, immunotherapy with tau/p-tau antibodies for tauopathy is a current research area.33 However, significant shortcomings have been assigned to immunotherapy. In particular, antibodies are biological drugs, which are expensive to produce because biological purity, activity qualification, batch-to-batch consistency, removal of toxic contaminants, and antibody humanization must be achieved. However, on the basis of the present study, we conclude that tau aptamers can be a practical alternative to antibodies, as tau aptamer synthesis can be readily scaled up for human trials. Importantly, nonphosphorylated tau was identified to be a major component of tau aggregate in mouse and human tauopathy brain.34 Thus, our nonphosphorylated tau-preferring aptamers are ideally positioned as a therapeutic for tauopathy diseases. In addition, aptamers have already been used in human as FDA-approved therapeutics.35–37 To realize the potential of tau aptamers as therapeutics, future studies should focus on further optimizing aptamer properties (such as reducing their interference of tau–MT interaction), optimizing their plasma half-life, improving blood–brain barrier penetration, and conducting proof-of principle studies in animal models of tauopathies.
EXPERIMENTAL SECTION
Buffers and Peptides.
The buffer used to disperse peptides and peptide-beads was Dulbecco’s phosphate-buffered saline (PBS) with calcium chloride and magnesium chloride (Sigma-Aldrich). The buffer used to prepare DNA solutions, as well as the buffer for all binding reactions, was PBS supplemented with additional 5 mM MgCl2. The sequences of the peptides used in this study are listed in Table S1. All peptides were purchased from GenScript; they all came with N-terminal acetylation and 2 units of 6-aminocaproic acid (Ahx) linker followed by six histidine residues (his-tag) at their C-terminus.
Immobilization of Tau/p-Tau Peptides.
Twenty microliters of stock Ni Sepharose high performance beads (~7 × 105 Ni-NTA beads, GE Healthcare Life Sciences) was equilibrated with 500 μL of PBS three times. After quick spin-down with a minicentrifuge (Fisher Scientific), the supernatant was discarded. The remaining pellet was suspended in 100 μL of PBS containing either 400 μg/mL or 800 μg/mL of the designated peptide and left overnight at 4 °C on a shaker. The prepared peptide beads were kept in the peptide solution and left unwashed at 4 °C until use. Beads used in counter selections or negative controls in binding tests were prepared as mentioned above but suspended in PBS as opposed to peptide solution.
DNA Library and Primers.
The forward primer and the reverse primer were labeled with FAM and biotin, respectively, at their 5’-ends. The sequence of the forward primer was 5’-FAM-CAG CAC CGT CAA CTG AAT-3’; the sequence of the reverse primer was 5’-biotin-ACA TCT CCA TCG CAT CAC-3’. The DNA library consisted of a randomized 30-nt region flanked by primer binding sites: 5’-CAG CAC CGT CAA CTG AAT-(N30)-GTG ATG CGA TGG AGA TGT-3’. All library and primer sequences were purchased from Integrated DNA Technologies and purified by reverse phase HPLC.
Polymerase Chain Reaction (PCR).
PCR parameters were optimized before the selection process. All PCR mixtures contained 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, 0.2 mM each dNTP, 0.5 μM each primer, and Hot Start Taq DNA polymerase (0.015 units/μL). PCR was performed on a BioRad C1000 Thermo Cycler, and all PCR reagents were purchased from Takara. The amplification began with a hot start at 95 °C for 90 s to activate Taq DNA polymerase. Then each of the repeated amplification cycles was performed at 95 °C for 30 s, 57 °C for 30 s, and 72 °C for 60 s, followed by a one-time final extension at 72 °C for 3 min.
SELEX Procedures.
The SELEX was performed by binding the DNA library to the peptide-immobilized beads and eluting DNA survivors from the washed peptide beads, followed by PCR amplification in a reciprocal manner. In the first round, an initial DNA library consisting of 20 nmol of the randomized oligonucleo-tides was used as a starting pool. For later rounds, 150 pmol to 20 pmol of samples amplified from previously recovered survivors were used. In each round, the DNA pool was heated at 95 °C for 3 min, followed by rapid cooling on ice for 5 min before starting incubation, allowing the DNA sequences to form the most favorable secondary structures. Beads were washed with 1 mL of PBS three times before use. Counter selection with bare Ni-NTA beads was introduced in round #6 and was included in all subsequent rounds. Sandwiched counter selections were used in round #8 and all later rounds. All incubations were performed at 4 °C. After each cycle of negative incubation, supernatant was collected, while the undesired sequences bound on Ni-NTA beads were discarded. For positive incubations, peptide beads were washed with buffer to remove unbound sequences. The washing stringency was enhanced as the selection progressed to remove weaker candidates. The washed peptide beads were heated at 95 °C for 7 min, followed by quick spin-down with a minicentrifuge. The surviving candidates in the supernatant were collected and were ready for PCR amplification during the first five rounds when negative selections were not included. For later rounds, the sequences recovered from peptide beads were treated with counter selection before PCR amplification. The number of optimized PCR amplification cycles for each round was confirmed with agarose gel electrophoresis. Streptavidin Sepharose high performance beads (GE Healthcare Life Sciences) were used to isolate the PCR products from the reaction mixture. The fluorophore-labeled amplicons were then separated from the biotinylated antisense DNA by eluting with 20 mM NaOH. Finally, the ssDNA was desalted with a NAP-5 column (GE Healthcare Life Sciences). The entire process is summarized in Table S6.
Flow Cytometric Analysis of Enriched Pools or Aptamer Candidates.
BD Accuri C6 flow cytometry (BD Immunocytometry Systems) was used to monitor the enrichment of candidates during the selection process and evaluate the binding specificity of the selected candidates. For each sample, 20 μL of peptide beads or bare beads (~1.4 × 105 beads) was washed with 500 μL of PBS three times. After removal of the supernatant, beads were suspended in 80 μL of buffer containing either a DNA pool or an aptamer candidate to be tested at 250 nM for 30 min. When examining the saturation of fluorescent signal with the selection pools, the DNA concentration used was decreased to 100 nM instead. Incubations were carried out at 4 °C unless stated otherwise. Afterward, beads were washed against 500 μL of buffer twice and then suspended in 80 μL of buffer for flow cytometric analysis.
Next-Generation Deep Sequencing of DNA Survivors.
Enriched DNA pools from rounds #15–17 were submitted for sequencing. Primers without FAM or biotin modification were used to amplify the DNA pools to be sequenced. The amplicons from each pool were then barcoded separately using the TruSeq DNA library preparation kit (Illumina). The samples were submitted to Illumina next-generation DNA sequencing at the University of Florida, ICBR Sequencing Core Facility. The sequencing results were analyzed using in-house software.
Chemical Synthesis and Purification of Aptamer Candidates.
The 10 most abundant sequences from round #17 were synthesized by the standard phosphoramidite method using a 3400 DNA synthesizer (Applied Biosystems). Reagents for synthesis were purchased from Glen Research. The synthesized candidates were then purified by reversed phase HPLC (Varian Prostar), using a C18 column and triethylammonium acetate (TEAA, 0.1M)/acetonitrile (Fisher Scientific) as the mobile phase.
Sequence Truncation of the Selected Aptamers.
The selected aptamers were truncated based on their secondary structures, as predicted by Integrated DNA Technologies OligoAnalyzer Tool. Shorter versions of each of the selected aptamers were synthesized and purified as described above. Their binding abilities were evaluated by flow cytometry as described previously for full-length candidates.
Measurement of Binding Kinetics/Affinities of the Selected Aptamers.
The binding affinities and binding kinetics of the selected aptamers were determined on an Octet RED384 instrument (Pall Fortebio). All measurements were performed on 384-well plates that were agitated at 1000 rpm at 30 °C. The his-tag peptides were immobilized at a concentration of 5 μg/mL on Ni-NTA sensors for 10 min, followed by rinsing the sensors in PBS for 10 min. Then the sensors were brought into fresh association buffer (PBS with 5 mM Mg2+) to establish a baseline for another 10 min. For kinetics analysis, the peptide-immobilized sensors were transferred to the wells containing aptamer dilutions (1 μM, 1.5 μM, 2 μM, 3 μM, 4 μM) for the association step (10 min) and then moved to the wells with fresh buffer for the dissociation step (10 min). A reference sensor was used without the treatment with aptamer solution, but other steps remained the same. When measuring the binding of aptamers with his-tag Tau441 protein (SignalChem), the experimental layout was the same as that described above, except that the loading concentration of his-tag Tau441 protein was 1 μg/mL supplemented with 0.75 mM imidazole, and the concentrations for aptamer dilutions were 0.75 μM, 1.5 μM, 3 μM, 6 μM, and 12 μM. Association rate constants (kon), dissociation rate constants (koff), and equilibrium dissociation constants (Kd) for each aptamer binding to its target peptide were calculated using the ForteBio data analysis software 10.0.
Specificity of the Selected Tau Aptamers against Tau441 Protein.
Gel electrophoresis was carried out to confirm the binding specificity between the aptamers against Tau441 protein. S100B, UCH-L1, α casein, β casein, BSA, and IgG were used as nontarget proteins for reference. A 10-μL mixture containing FITC-labeled aptamer at 200 nM and designated protein at 0.05 mg/mL were incubated at 4 °C for 30 min before loading onto an 8% nondenaturing polyacrylamide gel. Electrophoresis was initially carried out at 70 V for 10 min after introduction of the samples, and then the voltage was increased to 150 V for 45 min in 1x TBE buffer. Finally, gels were scanned using a Typhoon Imaging System (Amersham Biosciences).
Aptamer–Antibody Sandwich ELISA.
The coating buffer containing 1 μg/mL of total tau antibody (DAKO) in carbonate-bicarbonate buffer (pH 9.6) was pipetted into a 96-well plate (100 μL per well) and left overnight at 4 °C. The wells were washed three times with TBST (Tris-buffered saline, with Tween 20, pH 8.0, Sigma) and incubated with StartingBlock Blocking Buffers (Thermo Scientific) for 1 h at room temperature. After removal of the blocking buffer and washing three times with TBST, different concentrations of Tau441 protein were added into each well (100 μL). The plate was stored at 4 °C overnight.
On the next day, the plate was washed three times with TBST and then treated with 500 nM detecting aptamer or randomly scrambled aptamer sequence (RS) in 100 μL of PVP buffer (20 mg/mL polyvinylpyrrolidone and 100 μg/mL salmon sperm in TBST). The reaction was left on a shaker for 1 h. Afterward, the wells were washed with TBST three times and 100 μL of 1:10K diluted Pierce Streptavidin Poly-HRP (Thermo Scientific) solution was added to each well and incubated for 1 h at room temperature. Finally, 100 μL of TMB solution was added to each well, and the 620 nm absorbance was recorded by a plate reader.
Inhibition of Tau441 Phosphorylation by Aptamers.
Purified Tau441 (R-peptide) was used to perform phosphorylation by GSK3β Tau441 (1 μg) was first preincubated with the aptamers (50 μM) for 1 h before adding the enzyme GSK3β (200 ng) to the mixture of kinase assay buffer. The reaction buffer contained 20 mM Tris-HCl, 50 mM KCl, 10 mM MgCl2, pH 7.5. The assay was initiated by the addition of6 mM ATP. The reaction was carried out at 37 °C for 16 h on a shaker. Prephosphorylated GSK3β Tau441 (400 ng) was used as a positive control. Total reaction volume was 25 μL. Five microliters of the reaction sample was diluted with 15 μL of 4X Laemmli sample buffer, followed by SDS-PAGE and Western blotting with RZ3 antibody.
Assay of in Vitro Oligomerization of Tau441 Induced by Heparin.
To examine the inhibitory effect of tau aptamers on heparin-induced tau oligomerization, we preincubated different aptamers (50 μM) with Tau441 (10 μM) for 1 h at room temperature in the oligomerization buffer (20 mM Tris HCL, pH 7.4, 100 mM NaCl, 1 mM DTT, and 1 mM EDTA), followed by the addition of heparin (5 mg/mL) for 16 h incubation at 37 °C. The aptamers were prepared in 1X PBS with 5 mM MgCl2. The reaction was stopped by addition of Laemmli sample buffer. The reaction products were analyzed by SDS-PAGE, followed by immunoblotting with antitau antibody (DAKO).
SDS–PAGE Electrotransfer and Immunoblot Analysis.
Equal protein amount was prepared for sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) in 8X loading buffer containing 0.25 M Tris (pH 6.8), 2 mM DTT, 8% SDS, and 0.02% bromophenol blue. Each sample was subjected to SDS-PAGE on 4–20% precast gels and then transferred onto PVDF membranes. The membranes were blocked in 5% milk for 1 h and then incubated with primary antibodies RZ3 (pT231; 1:1000) or DAKO (total tau; 1:3000) that were detected using goat antimouse or goat antirabbit IgG conjugated to alkaline phosphatase (Amersham, Piscataway, NJ) for 1 h at room temperature. Immunoreactive bands were detected by developing with nitro blue tetrazolium and 5-bromo-4-chloro-3’-indolylphosphate (NBT/BCIP) (KPL). Quantitative evaluation of protein levels was performed via computer-assisted densitometric scanning (NIH ImageJ, version 1.6).
Statistical Analysis.
Monomeric p-tau (70K) and oligomeric tau (150 K) were normalized by the levels of GSK3β phosphorylated Tau441 or heparin-induced tau oligomers and shown as percentage. Data were presented as mean ± SEM for n = 3. Statistical analysis was performed with two-way ANOVA Tukey’s Test. For multiple comparisons, one-way ANOVA, followed by the Bonferroni’s post hoc test, was performed. *p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001, ns: nonsignificant. GraphPad Prism 7.0 (GraphPad).
Investigation of Tau Binding to Microtubules in the Presence of Aptamers.
The Microtubule Binding Protein Spin-Down Assay Kit (Cytoskeleton Inc., Denver, CO) was used to investigate the binding of tau to microtubules in the presence of aptamers. To validate the assay, microtubules (MTs) were first polymerized in the presence of GTP from tubulin proteins (mixture of α- and β subunits), and then microtubule-associated protein fraction (MAP2, 16 μg) and BSA protein (7.5 μg) were incubated with MTs (8 nM) as positive and negative protein controls, respectively. Mixtures were centrifuged at 100 000g at 4 °C for 40 min. The collected supernatant and pellet fractions were then mixed with Novex 4X LDS sample buffer (Thermo Scientific) and analyzed via a 4–20% SDS-PAGE gel (Thermo Scientific). The gel was stained by GelCode Blue Safe Protein Stain (Coomassie blue) (Thermo Scientific) and scanned by Typhoon using Cy5 channel. To study the interference of tau aptamers in the binding between Tau441 and MTs, Tau411 (5 μg) was incubated with tubulin proteins, which later polymerized into MTs accordingly to the manufacturer’s instructions as above. In a separate experiment, 20 μM FITC-labeled IT4 or IT5 was added to the mixture of preformed MT in the presence of tau for 30 min of incubation. The samples were centrifuged and analyzed as described above. The gel was scanned using both Cy5 and Cy2 channel for protein and aptamer imaging, respectively.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by National Institute of Health (GM R35 127130) and NSF 1645215 as well as NSFC grants (NSFC 21521063) (W.H.T.), and by University of Florida College of Medicine funding, Department of Veterans Affairs (Merit Award Veterans Affairs, (I01 RX001859 A02), Rehabilitation Research and Development Office, BRRC award no. B9256-C and BRRC Innovation Award no. 0118BRRC-01 (K.K.W).
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.8b08645.
Additional experimental details, additional characterization figures and data, peptide sequences, and DNA sequences (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Weingarten MD; Lockwood AH; Hwo SY; Kirschner MW A protein factor essential for microtubule assembly. Proc. Natl. Acad. Sci. U. S. A 1975, 72 (5), 1858–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Mandelkow EM; Biernat J; Drewes G; Gustke N; Trinczek B; Mandelkow E Tau domains, phosphorylation, and interactions with microtubules. Neurobiol. Aging 1995, 16 (3), 355–362. [DOI] [PubMed] [Google Scholar]
- (3).Sergeant N; Delacourte A; Buee L Tau protein as a differential biomarker of tauopathies. Biochim. Biophys. Acta, Mol. Basis Dis 2005, 1739 (2–3), 179–197. [DOI] [PubMed] [Google Scholar]
- (4).Brunden KR; Trojanowski JQ; Lee VMY Advances in tau-focused drug discovery for Alzheimer’s disease and related tauopathies. Nat. Rev. Drug Discovery 2009, 8 (10), 783–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Mandelkow E Alzheimer’s disease: The tangled tale of tau. Nature 1999, 402 (6762), 588–589. [DOI] [PubMed] [Google Scholar]
- (6).Schraen-Maschke S; Sergeant N; Dhaenens CM; Bombois S; Deramecourt V; Caillet-Boudin ML; Pasquier F; Maurage CA; Sablonniere B; Vanmechelen E; Buee L Tau as a biomarker of neurodegenerative diseases. Biomarkers Med. 2008, 2 (4), 363–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Ellington AD; Szostak JW In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346 (6287), 818–822. [DOI] [PubMed] [Google Scholar]
- (8).Tuerk C; Gold L Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249 (4968), 505–510. [DOI] [PubMed] [Google Scholar]
- (9).Qu H; Csordas AT; Wang J; Oh SS; Eisenstein MS; Soh HT Rapid and label-free strategy to isolate aptamers for metal ions. ACS Nano 2016, 10 (8), 7558–7565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Wilson C; Szostak JW Isolation of a fluorophore-specific DNA aptamer with weak redox activity. Chem. Biol 1998, 5 (11), 609–617. [DOI] [PubMed] [Google Scholar]
- (11).Famulok M Molecular recognition of amino acids by RNA-aptamers: An L-citrulline binding RNA motif and its evolution into an L-arginine binder. J. Am. Chem. Soc 1994, 116 (5), 1698–1706. [Google Scholar]
- (12).Lauhon CT; Szostak JW RNA aptamers that bind flavin and nicotinamide redox cofactors. J. Am. Chem. Soc 1995, 117 (4), 1246–1257. [DOI] [PubMed] [Google Scholar]
- (13).Niazi JH; Lee SJ; Gu MB Single-stranded DNA aptamers specific for antibiotics tetracyclines. Bioorg. Med. Chem 2008, 16 (15), 7245–7253. [DOI] [PubMed] [Google Scholar]
- (14).Paige JS; Nguyen-Duc T; Song W; Jaffrey SR Fluorescence imaging of cellular metabolites with RNA. Science 2012, 335 (6073), 1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Ferreira CSM; Matthews CS; Missailidis S DNA aptamers that bind to MUC1 tumour marker: design and characterization of MUC1-binding single-stranded DNA sptamers. Tumor Biol. 2006, 27 (6), 289–301. [DOI] [PubMed] [Google Scholar]
- (16).Bock LC; Griffin LC; Latham JA; Vermaas EH; Toole JJ Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature 1992, 355 (6360), 564–566. [DOI] [PubMed] [Google Scholar]
- (17).Balogh Z; Lautner G; Bardóczy V; Komorowska B; Gyurcsányi RE; Mészáros T Selection and versatile application of virus-specific aptamers. FASEB J. 2010, 24 (11), 4187–4195. [DOI] [PubMed] [Google Scholar]
- (18).Tawaraya Y; Hyodo M; Ara MN; Yamada Y; Harashima H RNA aptamers for targeting mitochondria using a mitochondria-based SELEX method. Biol. Pharm. Bull 2014, 37 (8), 1411–1415. [DOI] [PubMed] [Google Scholar]
- (19).Shangguan D; Li Y; Tang Z; Cao ZC; Chen HW; Mallikaratchy P; Sefah K; Yang CJ; Tan W Aptamers evolved from live cells as effective molecular probes for cancer study. Proc. Natl. Acad. Sci. U. S. A 2006, 103 (32), 11838–11843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Conrad R; Keranen LM; Ellington AD; Newton AC Isozyme-specific inhibition of protein kinase C by RNA aptamers. J. Biol. Chem 1994, 269 (51), 32051–32054. [PubMed] [Google Scholar]
- (21).Chen L; Rashid F; Shah A; Awan HM; Wu M; Liu A; Wang J; Zhu T; Luo Z; Shan G The isolation of an RNA aptamer targeting to p53 protein with single amino acid mutation. Proc. Natl. Acad. Sci. U. S. A 2015, 112 (32), 10002–10007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Shoji A; Kuwahara M; Ozaki H; Sawai H Modified DNA aptamer that binds the (R)-isomer of a thalidomide derivative with high enantioselectivity. J. Am. Chem. Soc 2007, 129 (5), 1456–1464. [DOI] [PubMed] [Google Scholar]
- (23).Griffin LC; Tidmarsh GF; Bock LC; Toole JJ; Leung LL In vivo anticoagulant properties of a novel nucleotide-based thrombin inhibitor and demonstration of regional anticoagulation in extracorporeal circuits. Blood 1993, 81 (12), 3271–3276. [PubMed] [Google Scholar]
- (24).Bing T; Yang X; Mei H; Cao Z; Shangguan D Conservative secondary structure motif of streptavidin-binding aptamers generated by different laboratories. Bioorg. Med. Chem 2010, 18 (5), 1798–1805. [DOI] [PubMed] [Google Scholar]
- (25).Selinfreund RH; Barger SW; Pledger WJ; Van Eldik LJ Neurotrophic protein S100 beta stimulates glial cell proliferation. Proc. Natl. Acad. Sci. U. S. A 1991, 88 (9), 3554–3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Bishop P; Rocca D; Henley JM Ubiquitin C-terminal hydrolase L1 (UCH-L1): Structure, distribution and roles in brain function and dysfunction. Biochem. J 2016, 473 (16), 2453–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Wang KK; Yang Z; Zhu T; Shi Y; Rubenstein R; Tyndall JA; Manley GT An update on diagnostic and prognostic biomarkers for traumatic brain injury. Expert Rev. Mol. Diagn 2018, 18 (2), 165–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Lim S; Haque MM; Kim D; Kim DJ; Kim YK Cell-based models to investigate tau aggregation. Comput. Struct. Biotechnol J. 2014, 12 (20–21), 7–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Cho JH; Johnson GV Primed phosphorylation of tau at Thr231 by glycogen synthase kinase 3beta (GSK3beta) plays a critical role in regulating tau’s ability to bind and stabilize microtubules. J. Neurochem 2004, 88 (2), 349–358. [DOI] [PubMed] [Google Scholar]
- (30).Krylova SM; Musheev M; Nutiu R; Li Y; Lee G; Krylov SN Tau protein binds single-stranded DNA sequence specifically – the proof obtained in vitro with non-equilibrium capillary electrophoresis of equilibrium mixtures. FEBS Lett. 2005, 579 (6), 1371–1375. [DOI] [PubMed] [Google Scholar]
- (31).Kim JH; Kim E; Choi WH; Lee J; Lee JH; Lee H; Kim DE; Suh YH; Lee MJ Inhibitory RNA aptamers of tau oligomerization and their neuroprotective roles against proteotoxic stress. Mol. Pharmaceutics 2016, 13 (6), 2039–2048. [DOI] [PubMed] [Google Scholar]
- (32).Trzeciakiewicz H; Tseng JH; Wander CM; Madden V; Tripathy A; Yuan CX; Cohen TJ A dual pathogenic mechanism links tau acetylation to sporadic tauopathy. Sci. Rep 2017, 7, 44102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Li C; Götz J Tau-based therapies in neurodegeneration: opportunities and challenges. Nat. Rev. Drug Discovery 2017, 16 (12), 863–883. [DOI] [PubMed] [Google Scholar]
- (34).Kimura T; Hatsuta H; Masuda-Suzukake M; Hosokawa M; Ishiguro K; Akiyama H; Murayama S; Hasegawa M; Hisanaga S The abundance of nonphosphorylated tau in mouse and human tauopathy brains revealed by the use of phos-tag method. Am. J. Pathol 2016, 186 (2), 398–409. [DOI] [PubMed] [Google Scholar]
- (35).Stein CA; Castanotto D FDA-approved oligonucleotide therapies in 2017. Mol. Ther 2017, 25 (5), 1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Dassie JP; Giangrande PH Current progress on aptamer-targeted oligonucleotide therapeutics. Ther. Delivery 2013, 4 (12), 1527–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Röthlisberger P; Gasse C; Hollenstein M Nucleic acid aptamers: emerging applications in medical imaging, nanotechnology, neurosciences, and drug delivery. Int. J. Mol. Sci 2017, 18 (11), 2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.