Abstract
Background
Endoplasmic reticulum (ER) oxidoreductin-1α (Ero1α) and protein disulfide isomerase (PDI) constitute the pivotal pathway of oxidative protein folding, and are highly expressed in many cancers. However, whether targeting the functional interplay between Ero1α and PDI could be a new approach for cancer therapy remains unknown.
Methods
We performed wound healing assays, transwell migration and invasion assays and xenograft assays to assess cell migration, invasion and tumorigenesis; gel filtration chromatography, oxygen consumption assay and in cells folding assays were used to detect Ero1α-PDI interaction and Ero1α oxidase activity.
Findings
Here, we report that elevated expression of Ero1α is correlated with poor prognosis in human cervical cancer. Knockout of ERO1A decreases the growth, migration and tumorigenesis of cervical cancer cells, through downregulation of the H2O2-correlated epithelial-mesenchymal transition. We identify that the conserved valine (Val) 101 of Ero1α is critical for Ero1α-PDI complex formation and Ero1α oxidase activity. Val101 of Ero1α is specifically involved in the recognition of PDI catalytic domain. Mutation of Val101 results in a reduced ER, retarded oxidative protein folding and decreased H2O2 levels in the ER of cervical cancer cells and further impairs cell migration, invasion, and tumor growth.
Interpretation
Our study identifies the critical residue of Ero1α for recognizing PDI, which underlines the molecular mechanism of oxidative protein folding for tumorigenesis and provides a proof-of-concept for cancer therapy by targeting Ero1α-PDI interaction.
Fund
This work was supported by National Key R&D Program of China, National Natural Science Foundation of China, and Youth Innovation Promotion Association, CAS.
Keywords: Cervical cancer, Ero1α, PDI, Protein interaction, Redox
Research in context.
Evidence before this study
Cervical cancer is one of the leading causes of cancer-associated deaths among women, and the 5-year survival rate of these patients is unsatisfying. Under the progression of tumorigenesis, protein synthesis and processing are of vital importance for rapid proliferation of cancer cells. Ero1α-PDI pathway in the endoplasmic reticulum is responsible for protein disulfide formation of membrane proteins and secretary proteins. Although Ero1α has been reported to be highly expressed in breast cancer and colon cancer as a poor prognosis marker, the key role of Ero1α in cervical cancer progression is largely unknown. Moreover, whether targeting the functional interplay between Ero1α and PDI could be a new approach for cancer therapy is an open question.
Added value of this study
This study firstly identifies high Ero1α expression as a marker for poor prognosis in cervical cancer. We also show that valine 101 in Ero1α is a critical residue involved in Ero1α-PDI functional interplay, ER redox balance and cervical cancer cell migration, invasion and tumorigenesis. Our study sheds light on not only new intervention strategy in cervical cancer, but also the molecular mechanism of oxidative protein folding.
Implications of all the available evidence
Further investigations based on our study could lead to the development of small chemicals or peptides that target the Ero1α-PDI interface around valine 101, so as to disrupt the interaction between Ero1α and PDI and prevent cancer progression. As Ero1α and PDI proteins are upregulated in many types of cancers, the strategy of disruption of Ero1α-PDI interaction may be applied not only in cervical cancer, but also in many other cancers.
Alt-text: Unlabelled Box
1. Introduction
Cervical cancer is one of the most common malignancies and the second leading cause of cancer-related deaths among women aged 20–39 years old [1]. Although concomitant chemoradiotherapy has been recommended as the standard treatment for cervical cancer, the 5-year survival rate of these patients still needs to be improved. Therefore, new therapeutic targets are urgently needed.
In the process of tumorigenesis, cells often encounter hypoxia, nutrient deprivation, proteasome dysfunction, and protein mutations. Due to uncontrolled proliferation and growth, cancer cells require a great capacity for folding and assembly of proteins in the endoplasmic reticulum (ER), which could lead to an accumulation of misfolded proteins and trigger ER stress [2]. Recent studies have increased our knowledge about the relationships between ER stress and cancer biology by demonstrating the impacts of the unfolded protein response (UPR) on cell transformation, inflammation, tumor angiogenesis, tumor aggressiveness, tumor invasion and metastasis [3]. Proteostasis in the ER has been highlighted as one of the key players in cancer development and aggressiveness, and some UPR components such as GRP78 [4] and CHOP [5] have been reported as independent prognostic biomarkers.
Biosynthesis of all membrane proteins and secretary proteins, which typically contain disulfide bonds, is performed in the ER lumen and occurs through the secretory pathway [6,7]. Sulfhydryl oxidase ER oxidoreductin-1α (Ero1α) and protein disulfide isomerase (PDI) constitute the pivotal pathway for catalyzing oxidative protein folding and are responsible for ER redox homeostasis [8]. Ero1α utilizes oxygen to oxidize PDI with hydrogen peroxide (H2O2) as a byproduct; oxidized PDI catalyzes the formation of disulfides in reduced substrates [9]. Ero1α is constitutively expressed in all tissues [10] and upregulated during cellular hypoxia [11] and UPR [12]. Notably, Ero1α is highly expressed in breast cancer [13] and colon cancer [14] and has been suggested to be a predictor for poor prognosis in breast cancer [15]. Moreover, PDI participates in the proliferation, survival, and metastasis of several types of cancer cells [16]. Nevertheless, little is known about the correlation of Ero1α-PDI pathway with cervical carcinogenesis.
Here, we propose that specifically disrupting the binding interaction between Ero1α and PDI could be an effective method to suppress tumorigenesis. PDI consists of four thioredoxin (Trx)-like domains arranged as a-b-b'-a'. Specifically, a and a' are catalytic domains and b and b' are noncatalytic domains. We previously reported that Ero1α binds to the b'xa' fragment and preferentially oxidizes the a' domain of PDI [17]. However, the details of the interaction between Ero1α and PDI are still not very clear. In this study, we aim to further characterize the molecular mechanism of Ero1α-PDI interaction and investigate its role in tumorigenesis.
We found that Ero1α was upregulated in cervical cancer, and increased Ero1α expression correlated with poor prognosis. Knockout (KO) of ERO1A impaired cervical cancer cell growth, migration and tumorigenesis. We further determined that Valine (Val) 101, a conserved hydrophobic residue located in the active site-containing loop of Ero1α, played a critical role in Ero1α-PDI interaction by recognizing the a' domain of PDI. Mutation of Val101 abolished the oxidase activity of Ero1α, reduced ER redox states and retarded oxidative protein folding. Importantly, mutation of Val101 suppressed cervical cancer cell growth, migration and tumorigenesis. Our study provides new insights into the molecular mechanism of Ero1α-PDI oxidative protein folding machinery for tumorigenesis and may guide cancer therapy by targeting Ero1α-PDI pathway.
2. Materials and methods
2.1. Patients and tissues collection
The tissue samples were collected in the First Affiliated Hospital of Wenzhou Medical University, Wenzhou, China in 2013. The Board and Ethical Committee of Wenzhou Medical University approved this study. All patients participated in this study provided written informed consents in accordance with the Declaration of Helsinki. Each pair of normal and cancerous tissue was obtained from the same patient without radiotherapy or chemotherapy prior to the operation. Western blotting was performed to detect Ero1α and PDI expression in these tissues.
2.2. Tissue microarray and immunohistochemistry
A human uterine cervix tissue microarray (CR2082) containing 60 cases of malignant tissues and 9 cases of normal tissues was purchased from Biomax. The formalin-fixed, paraffin-embedded sections were stained using anti-Ero1α antibody (177156, 1:200; Abcam). The staining intensity was divided into four categories: negative, weak, moderate and strong staining, according to the weighted intensity and extension of cancerous area.
2.3. Plasmid construction and protein preparation
For protein expression in bacteria, pQE30 plasmids encoding PDI and E. coli thioredoxin 1 (Trx1) and pGEX-6P-1 plasmids encoding Ero1α and Ero1β were used [18]. pET28a-Ero1p, pET23b-Pdi1p and pET15b-ERp46 plasmids were previously described [19]. For expression in mammalian cells, pcDNA3.1-Ero1α-myc and pcDNA3.1-Ero1α-HA were used [20]. pcDNA3.1-Ero1α with a C-terminal FLAG tag and all point mutations of Ero1α, Ero1β, and Ero1p were generated by overlap extension PCR and verified by DNA sequencing.
Recombinant Ero1α, Ero1p, PDI, Pdi1p, ERp46 and Trx1 proteins were purified as described [19]. Ero1β was purified as previously described [18]. PDI, Pdi1p, ERp46 and Trx1 protein concentrations were determined by absorbance at 280 nm, and Ero1 protein concentrations were determined by Bradford method.
For reduced protein preparation, PDI at 100 μM, Trx1 at 100 μM or Ero1α at 10 μM were incubated with 100 mM DTT in buffer A (50 mM Tris-HCl, pH 7.6, 150 mM NaCl, 2 mM EDTA) for 1 h at 25 °C. Excess DTT was removed using a HiTrap desalting column (GE Healthcare) pre-equilibrated with buffer A, and the reduced proteins were stored on ice for use only in the same day. For oxidized protein preparation, PDI at 100 μM or Ero1α at 50 μM was incubated with 50 mM potassium ferricyanide in buffer A for 1 h at 25 °C and then chromatographed through a Superdex-200 10/300 GL column (GE Healthcare) pre-equilibrated with buffer A. The monomer fraction was collected, concentrated and stored at −80 °C in aliquots.
2.4. Cell culture and transfection
HeLa and HEK293T cells were obtained from ATCC (Manassas, VA, USA) and were cultured in Dulbecco's Modified Eagle's Medium (DMEM, HyClone) containing 10% fetal bovine serum (Gibco), 100 units/ml penicillin and 100 μg/ml streptomycin (Gibco) at 5% CO2 and 37 °C. Plasmids were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. The transfected cells were harvested after 24–48 h.
2.5. Lentivirus preparation
The cDNA of HA-tagged Ero1α and its mutants were cloned into a pLE4 lentiviral vector kindly provided by Dr. Guang-Hui Liu (Institute of Biophysics, Chinese Academy of Sciences). Lentivirus particles were generated from HEK293T cells by cotransfecting lentiviral vectors with the packaging plasmids psPAX2 (Addgene) and pMD2.G (Addgene) and used for transduction in the presence of 4 μg/ml polybrene.
2.6. Generation of ERO1A KO HeLa cell lines by CRISPR/Cas9 genome editing
The 20-nt guide sequences targeting human ERO1A were designed and cloned into the expression vector pSpCas9(BB)-2A-GFP (PX458) containing human codon optimized Cas9, the RNA components and GFP (a gift from Dr. Feng Zhang, Addgene plasmid # 48138). The guide sequence targeting exon 1 of human ERO1A is:
5′-CGGCTGGGGATTCTTGTTTG- 3′ (clone 10 and 13).
HeLa cells were transfected with pSpCas9(BB)-2A-GFP vector containing the single-guide RNAs (sgRNAs) by Viafect (Promega) for 48 h, and GFP-positive cells were single-cell-sorted and screened. Genomic DNA (gDNA) was purified from clones using the TransDirect Animal Tissue PCR Kit (TransGen), and the region surrounding the protospacer adjacent motif (PAM) was amplified using the following primers:
Forward: 5′- GATCGCTGAGAGGCAGGA- 3′.
Reverse: 5′- AGAAGCACCTCTGTGCCG- 3′.
PCR products were cloned into pEASY-T5 Zero Cloning Vector (TransGen). The InDels of individual alleles were determined by plasmid DNA purification and sequencing (Invitrogen).
2.7. Western blotting
Cells were lysed in radio immunoprecipitation assay (RIPA) lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.25% deoxycholic acid, 1% NP-40, 1 mM EDTA, Millipore) containing protease inhibitor cocktail (Roche) for 30 min and then centrifuged at 13,000 ×g for 20 min to remove cell debris. The proteins in supernatant were quantified using a BCA Kit (Beyotime) and analyzed by SDS-PAGE. The gel was then transferred to a polyvinylidene difluoride membrane (Millipore) using a semi-dry transfer apparatus or a wet transfer apparatus. The membrane was blocked in 5% milk, incubated with antibodies and visualized by using a Chemi-Scope Mini imaging system (Clinx Science) with enhanced chemiluminescence (Thermo Fisher Scientific). Band intensity was quantified using ImageJ software (National Institutes of Health).
The following antibodies were used for immunoblotting: mouse monoclonal anti-Ero1α (2G4, 1:1000; Millipore), anti-PDI (RL90, 1:2000; Abcam), anti-Flag (M2, 1:2000; Sigma-Aldrich), anti-myc (9E10, 1:5000; Sigma-Aldrich), anti-HA (HA-7, 1:5000; Sigma-Aldrich), anti-E-cadherin (4A2, 1:1000; Cell Signaling Technology), anti-GAPDH (GAPDH-71.1, 1:50000; Sigma-Aldrich); rabbit monoclonal anti-Vimentin (D21H3, 1:1000, Cell Signaling Technology), anti-Snail (C15D3, 1:1000, Cell Signaling Technology), anti-Slug (C19G7, 1:1000, Cell Signaling Technology), goat anti-mouse IgG (1:10000; Sigma-Aldrich), and goat anti-rabbit IgG (1:10000, Sigma-Aldrich). The PDI and ERp46 antisera were generated by immunizing rabbits with purified recombinant proteins (Animal Facility, Institute of Genetics and Developmental Biology, CAS).
2.8. Immunoprecipitation
HEK293T cells transfected with pcDNA3.1-Ero1α-FLAG for 24 h were harvested and lysed in RIPA buffer containing protease inhibitor cocktail, and the supernatants were incubated with anti-FLAG M2-agarose beads (A2220, Sigma-Aldrich) overnight at 4 °C. The beads were washed three times with phosphate-buffered saline (PBS), resuspended in reducing loading buffer, and analyzed by western blotting.
2.9. Cell cycle analysis
HeLa cells of 2 × 106 were trypsinized, washed twice with PBS and fixed with 70% cold ethanol at -20 °C overnight. Fixed cells were washed with PBS and stained with 200 μl propidium iodide (Invitrogen) for 30 min in the dark. The cell cycle was analyzed by a flow cytometry.
2.10. Wound healing assays
HeLa cells were seeded in an insert of Culture-Insert 2 Well μ-Dish (Ibidi). Once cells grew to confluent monolayers, the insert was removed to introduce a uniform wound, and the medium was changed to DMEM containing 1% FBS. The migration of cells into the wound was monitored under a microscope for up to 24 h.
2.11. Transwell migration and invasion assays
Transwell migration assays were performed in a 24-well Transwell Inserts (Corning). HeLa cells (5 × 104) suspended in 200 μl serum-free medium were seeded in the upper chamber, and 600 μl DMEM containing 1% FBS was placed in the lower chamber. After incubation for 24 h at 37 °C, the cells in the upper chamber were carefully removed with a cotton swab, and the cells that had migrated to the reverse face of the membrane were fixed in methanol and stained with 0.1% crystal violet. Five fields were randomly selected from the central and surrounding parts of the membrane under a microscope, and cells were counted and normalized as indicated for statistical analysis. Transwell invasion assays were performed similarly as the migration assay, except the membrane of the upper chamber was Matrigel (BD Biosciences) precoated to mimic extracellular matrix.
2.12. Xenograft assays
For xenograft assays, HeLa cells (1 × 106 cells/mice) in 100 μl PBS were injected subcutaneously into the right and left flanks of 6–8 w female BALB/c nude mice (Vital River). After 36–42 days, the mice were sacrificed by cervical dislocation, and tumor tissues were excised, imaged, and weighed. Animal experiments were conducted with the approval of the Institutional Biomedical Research Ethics Committee of the Institute of Biophysics, Chinese Academy of Science.
2.13. Determination of ER redox state with superfolded-roGFP-iEER
HeLa cells transfected with pcDNA3.1-superfolded-roGFP-iEER alone or cotransfected with pcDNA3.1-Ero1α-myc for 48 h were harvested, washed twice with Hanks' Balanced Salt Solution (HBSS, Gibco), and then seeded onto a flat-bottom 96-well plate. The fluorescence intensities were measured at 525 nm with excitation at 390 and 465 nm by using an EnSpire Multimode Plate Reader (Perkin Elmer), and the fluorescence intensity ratio at 390/465 nm excitation was calculated.
2.14. In vivo oxidative folding assays
The in vivo myc-tagged Ig-J chains (JcM) folding assays were performed as described [20]. Briefly, HeLa cells cotransfected with pcDNA3.1-JcM and pcDNA3.1-Ero1α-HA for 48 h were harvested and incubated in DMEM containing 5 mM DTT for 5 min at 37 °C. After two washes in ice-cold PBS, cells were resuspended in fresh DMEM at 20 °C. Aliquots were taken at different time points, quenched by 20 mM NEM, and lysed in RIPA buffer containing protease inhibitor cocktail and 20 mM NEM. The supernatants were resolved by nonreducing SDS-15% PAGE and analyzed by western blotting.
2.15. Determination of H2O2 in the ER with HyPerER probe
Determination of H2O2 in the ER with HyPerER probe were performed according to previous work [21]. HeLa cells transfected with HyPerER [22] for 24 h were harvested, washed twice with HBSS, and then seeded onto a flat-bottom 96-well plate. After the addition of 0.5 mM DTT for 5 min, the fluorescence intensities were measured at 530 nm with excitation at 405 and 488 nm using an EnSpire Multimode Plate Reader (Perkin Elmer). The fluorescence intensity ratio at 488/405 nm excitation was calculated.
2.16. Circular dichroism analysis
The circular dichroism spectra of proteins at concentration of 0.2 mg/ml in 50 mM Na-Pi buffer (pH 7.0) were recorded from 195 nm to 260 nm in 1 nm step at 25 °C on a Chirascan Plus instrument (Applied Photophysics).
2.17. Oxygen consumption assays
Oxygen consumption was measured at 25 °C using an Oxygraph Clark-type oxygen electrode (Hansatech Instruments) as previously described [18]. Briefly, reactions were initiated by injecting Ero1 proteins into buffer B (100 mM Tris-HAc, 50 mM NaCl, 2 mM EDTA, pH 8.0) containing PDI/GSH or reduced Trx1 with or without FAD.
2.18. Analysis of Ero1α-PDI complex formation by gel filtration chromatography
Gel filtration chromatography of PDI, Ero1α and its mutants was performed on a Superdex-200 10/300GL column (GE Healthcare) pre-equilibrated with buffer A. Absorbance at 280 nm was recorded, and the monomer fraction was collected. Ero1α (or its mutants) and PDI monomers both at 5 μM were then mixed for incubation in buffer A for 30 min at 25 °C, and the complex was analyzed using the Superdex-200 10/300GL column.
2.19. Determination of Ero1α redox state in vitro
For the activation of Ero1α, oxidized Ero1α proteins were incubated with 10-fold reduced PDI in buffer A at 25 °C. For the inactivation of Ero1α, freshly prepared reduced Ero1α proteins were incubated alone in buffer A at 25 °C. Aliquots were taken at different times for immediate quenching with 20 mM N-ethylmaleimide (NEM, Sigma-Aldrich). The samples were then analyzed by nonreducing SDS-9% PAGE followed by western blotting.
2.20. Statistical analysis
The data were presented as the mean ± SEM from three or more independent experiments. The Student's t-test assumed two-tailed distributions to calculate statistical significance between groups. Differences were analyzed by GraphPad Prism 5, and p-values <.05 were considered to reach statistical significance. For the correlation between ERO1A mRNA expression and survival probability, the difference was evaluated by Kaplan–Meier analysis using the log-rank test. ERO1A expression greater than the third tercile of all patients was defined as high, and expression lower than the first tercile was defined as low. P-values of the log-rank test <0.05 indicates statistical significance.
3. Results
3.1. Increased Ero1α expression in human cervical cancer is associated with malignancy extent and poor prognosis
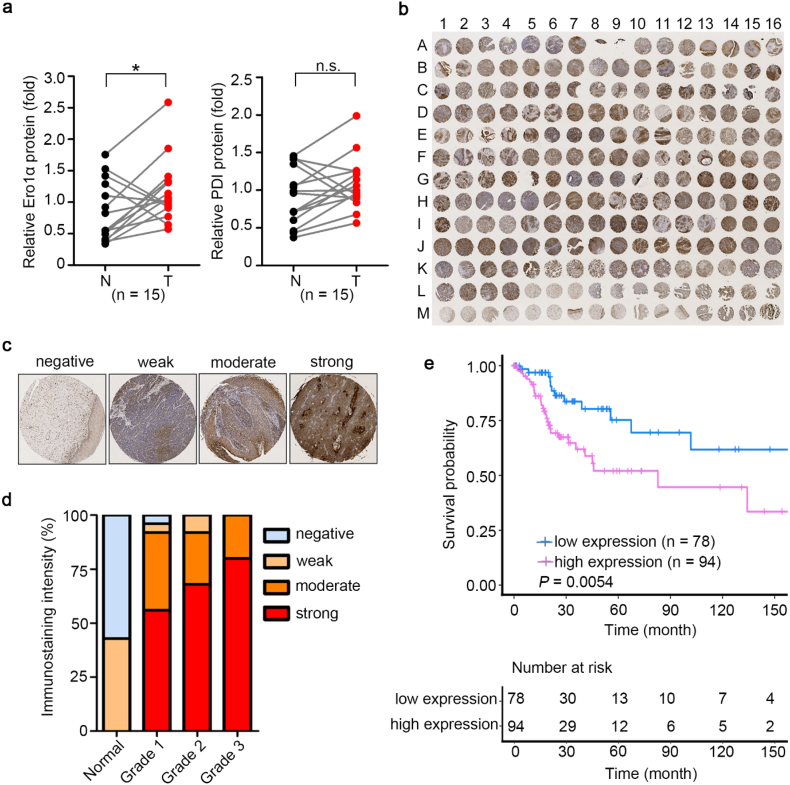
We analyzed Ero1α and PDI protein levels in cervical cancer tissue and adjacent normal cervical tissue of the same patient. Ero1α expression in cancer samples was aberrantly upregulated compared with normal tissues in 12 out of 15 patients, suggesting a positive correlation between Ero1α levels and cervical cancer. Interestingly, there seems to be a trend for samples with high Ero1α expression to also exhibit high PDI expression; however, the finding was not statistically significant between cancer and normal tissues for PDI expression (Fig. 1a and S1). By immunohistochemistry staining, we analyzed Ero1α expression patterns in a tissue microarray containing 60 cases of cervical cancer tissues with 9 cases of normal cervical tissues (Fig. 1b and Table S1). Ero1α expression levels in the array were classified into four categories, i.e., negative, weak, moderate, and strong immunohistochemistry staining (Fig. 1c), and clustered into four grades of cervical cancer tissues (Fig. 1d and Table S1). As shown in Fig. 1d, normal tissues exhibited negative or weak Ero1α expression, whereas cancer tissues mainly exhibited moderate or strong Ero1α staining. Notably, Ero1α expression was evidently elevated as the grade of cancer malignancy increased. Kaplan–Meier analysis from TCGA database showed that cervical cancer patients with high Ero1α expression had drastically shorter overall survival compared with patients with low Ero1α expression (Fig. 1e). Thus, Ero1α might be a biomarker for poor prognosis in cervical cancer.
Fig. 1.
Expression of Ero1α in human cervical cancer is upregulated with malignancy extent and poor prognosis. (a) Relative expression of Ero1α and PDI proteins in cervical tumor tissues (T) and adjacent normal cervical tissues (N) using western blotting. Band intensity was quantified and normalized to the mean value of expression level of the 15 patients. Each line segment between N (black) and T (red) represented one individual, * P < 0.05 (two-tailed, paired Student's t-test). (b) Tissue microarray containing 60 cases (from A1 to L4, 3 samples for each case) of cervical cancer with 9 normal cervical tissues (from L5 to M15, 3 samples for each case) was subjected to immunohistochemistry with antibody against Ero1α. The sample M16 was from malignant melanoma of skin as a positive marker. (c) Representative images of immunohistochemical staining intensities for Ero1α expression in cervical cancer tissue array. The images of negative, weak, moderate, and strong staining were from sample M1, H5, D1, G14 in (b), respectively. (d) Proportions of Ero1α expression levels in the four malignant grades of cervical tissue of (b). (e) Kaplan–Meier analysis of overall survival in 172 CESC (cervical squamous cell carcinoma and endocervical adenocarcinoma) patients from TCGA database expressing high (greater than the third tercile) and low (lower than the first tercile) levels of ERO1A. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. ERO1A KO impairs HeLa cell growth, migration, and tumorigenesis
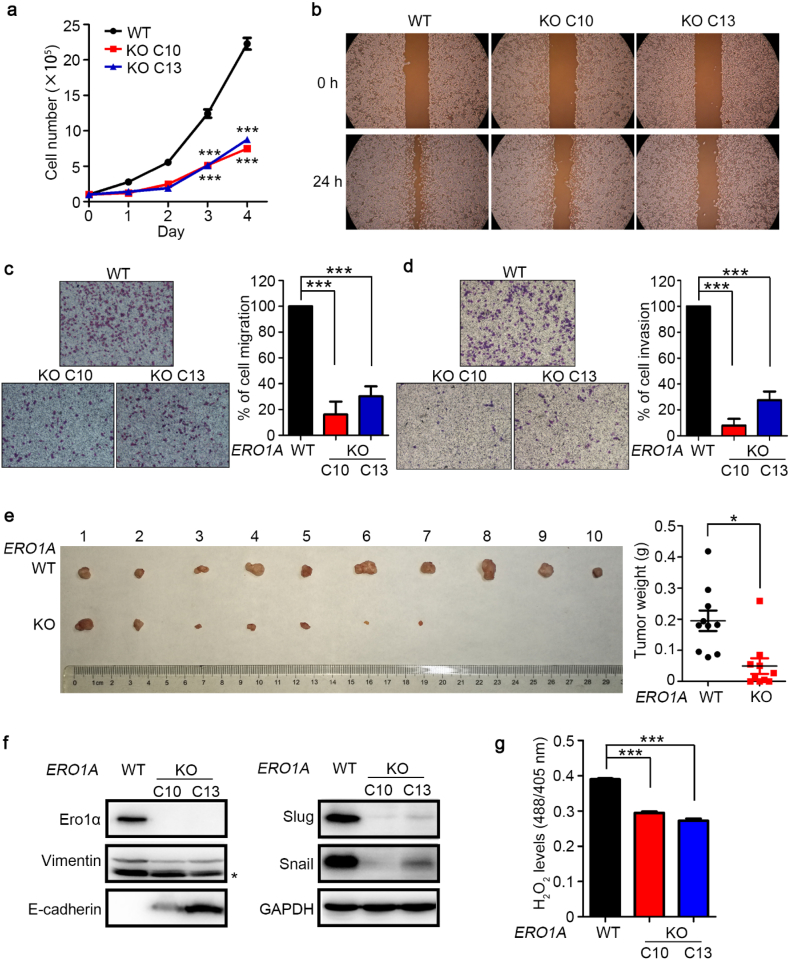
To investigate the role of Ero1α in cervical tumorigenesis, we generated ERO1A KO HeLa cervical cancer cells by means of CRISPR/Cas9 genome editing (Fig. S2). Growth curves of wild-type (WT) cells and two clones (C10 and C13) of ERO1A KO HeLa cells revealed that depletion of ERO1A impeded cancer cell growth (Fig. 2a), and cell cycle analysis demonstrated that the ERO1A KO HeLa cells were more arrested in G1 phase compared with WT cells (Fig. S3). Wound healing assay indicated that the migration ability of ERO1A KO cells was impaired (Fig. 2b). In transwell migration assays, the numbers of ERO1A KO cells that migrated to the underside of filter membrane were dramatically reduced to approximately 20–30% of migrated WT cells (Fig. 2c). The invasion ability of ERO1A KO cells was again significantly impaired (Fig. 2d). These results indicated the importance of Ero1α in cervical cancer cell migration and invasion. In BALB/c nude mice injected with WT or ERO1A KO HeLa cells, the tumors derived from ERO1A KO cells showed much smaller sizes and lower weights compared with those derived from WT cells (Fig. 2e). The above results suggest that Ero1α is important for cervical cancer progression, and ERO1A KO impedes cancer cell proliferation and tumor growth.
Fig. 2.
ERO1A knockout (KO) impairs HeLa cell growth, migration, invasion, and tumorigenesis. (a) Growth curves of wild-type (WT) and two clones (C10 and C13) of ERO1A KO cells. Data were shown as mean ± SEM of three biological replicates; *** P < 0.001 (two-tailed Student's t-test). (b) Wound healing assays for WT and ERO1A KO cells. (c) Transwell migration assays and (d) transwell invasion assays. Migrated cells to the underside of filter membrane were counted and normalized to WT. Data were shown as mean ± SEM from three independent experiments performed in two technical replicates; *** P < 0.001 (two-tailed Student's t-test). (e) Xenograft assay of WT and ERO1A KO (C10) cells. WT and ERO1A KO cells were respectively injected subcutaneously into the right and left flanks of a female BALB/c nude mouse. Images of xenograft tumors at 38–42 days after inoculation were shown and the tumor weights were shown as mean ± SEM of ten biological replicates; * P < 0.05 (two-tailed, paired Student's t-test). (f) Expression of EMT-associated molecules in WT and ERO1A KO HeLa cells analyzed by western blotting. (g) H2O2 levels in the ER of WT and ERO1A KO cells were determined by the fluorescence intensity ratio at 488/405 nm excitation of the HyPerER probe, after the addition of 0.5 mM DTT for 5 min. Data were shown as mean ± SEM from four technical replicates; *** P < 0.001 (two-tailed Student's t-test). Results are representative of two independent experiments.
Epithelial-mesenchymal transition (EMT) is a common mechanism for promoting tumor cell metastasis [23], by which epithelial cells lose their cell polarity and cell-cell adhesion and gain migratory and invasive properties to become mesenchymal stem cells. The expression of EMT-promoting molecules Slug, Snail, and Vimentin in ERO1A KO cells was greatly downregulated, and cellular adherence molecule E-cadherin was upregulated (Fig. 2f). Ero1α is responsible for oxidative protein folding in the ER by generating disulfides with H2O2 as a byproduct [10], which was reported to enhance EMT [24,25]. To detect H2O2 levels in the ER, an ER lumen-localized H2O2 probe (HyPerER) was used by monitoring the fluorescence intensity ratio at 488/405 nm excitation [22,26]. Because in the ER, HyPerER can be oxidized either by H2O2 or by PDI, the reductant dithiothreitol (DTT) was typically included to maintain PDI in a reduced state [21,27]. Accordingly, in DTT-flooded cells, we found that the H2O2 levels in the ER lumen of ERO1A KO cells were lower than that in WT cells (Fig. 2g). Addition of H2O2 to the cells restored the signal to a similar level, validating the usage of HyPerER probe (Fig. S4). To further confirm the role of Ero1α in EMT signaling, we treated cells with N-acetylcysteine (NAC), a cell-permeable glutathione precursor and a widely used reactive oxygen species (ROS) scavenger [28]. Indeed, NAC treatment suppressed EMT signaling triggered by Ero1α-overexpression (Fig. S5), in line with the observation that NAC could revert Ero1α-induced hyperoxidation in the ER [29]. Altogether, Ero1α promotes cervical cancer cell proliferation, migration, invasion and tumorigenesis, at least partially through H2O2-dependent EMT pathways.
3.3. The conserved Val101 is critical for Ero1α-PDI complex formation and Ero1α activity
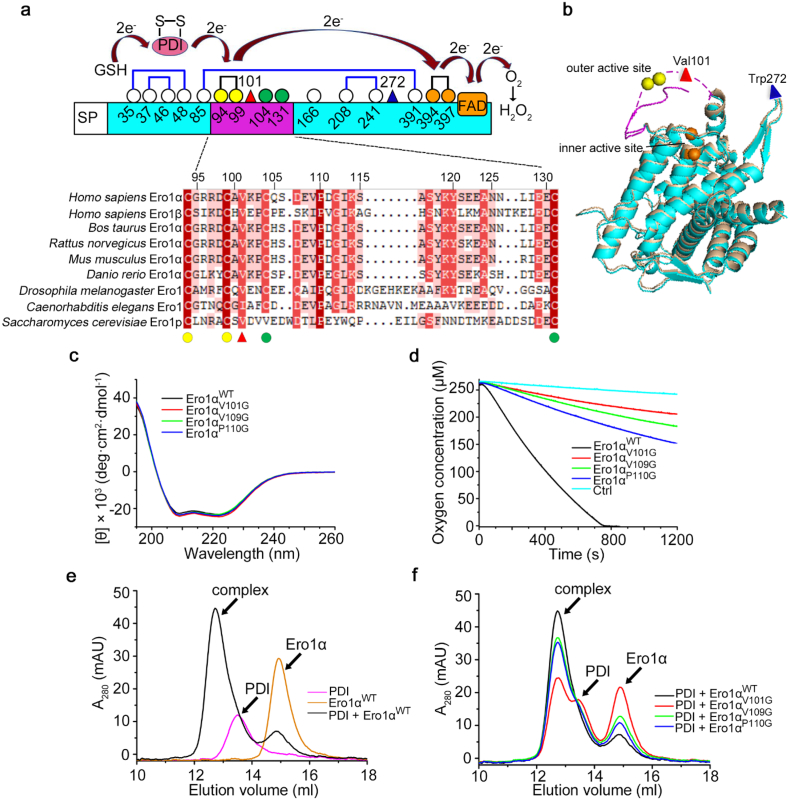
Ero1α catalyzes oxidative protein folding through the oxidation of its downstream oxidoreductase PDI. The interaction between Ero1α and PDI is a prerequisite for their functional interplay [8]. Therefore, the disruption of the interaction between Ero1α and PDI could represent an effective strategy to suppress tumorigenesis. Previous studies have confirmed that functional electron transfer occurs through the binding of Ero1α to the b'xa' fragment of PDI [17], and the Trp272 residue in a protruding β-hairpin of Ero1α is responsible for recognizing the hydrophobic pocket in the b' domain of PDI [30]. The binding affinity of PDI with active Ero1α was significantly higher than that of PDI with inactive Ero1α [19,31]; however, no substantial conformation change was observed between active and inactive forms of Ero1α based on structure superposition, including the protruding β-hairpin region. A flexible loop in Ero1α, which contains the outer active site (Cys94-Cys99) and two regulatory cysteines (Cys104 and Cys131), enables electron shuttling from PDI to the FAD-proximal inner active site (Cys394-Cys397) of Ero1α and finally to oxygen (Fig. 3a and b). Unfortunately, structural information of this loop is missing in both active and inactive Ero1α structures due to its intrinsic flexibility. Sequence alignment of this loop region from different species reveals that Val101, Val109 and Pro110 are the most conserved hydrophobic residues (Fig. 3a). To verify their potential roles in recognizing PDI, we first mutated these residues to glycine, and the mutations did not change the overall secondary structures as shown by circular dichroism spectrum (Fig. 3c). In a reconstituted system, the three Ero1α mutants showed decreased activities towards the oxidation of physiological substrate PDI reduced by glutathione (GSH), and Ero1αV101G is the least active (Fig. 3d). Analysis of Ero1α-PDI complex formation by gel-filtration chromatography showed that V101G mutation drastically destroyed the interaction between Ero1α and PDI; however, the other two mutations exhibited less effects (Fig. 3e and f). Given that Ero1α activity is allosterically controlled by PDI [19], we also assessed whether the V101G mutation affects these regulatory processes. Neither the activation nor the inactivation process of Ero1α was affected by the mutation of Val101 (Fig. S6). Taken together, we have identified that Val101 is a critical residue for Ero1α-PDI physical binding and complex formation, which is necessary for Ero1α oxidase activity.
Fig. 3.
The conserved Val101 located in the outer active site-containing loop is critical for the activity of Ero1α. (a) Schematic representation of the cysteine pattern of Ero1α in the active form. The cysteine residues were shown as white (non-catalytic), green (regulatory), yellow (outer active site) and orange (inner active site) circles with numbering. Val101 and Trp272 were shown as a red and a blue triangle, respectively. The electron transfer pathway between the active sites of Ero1α and its substrates, PDI and reduced glutathione (GSH), were indicated. Multiple sequence alignment of the flexible loops containing the outer active sites in Ero1 proteins from different species was performed by DNAman. Identical (100%), highly conserved (75–99%), semi-conserved (50–74%) and variable (<50%) residues were shaded in red, magenta, pink, and white, respectively. (b) Structural superposition of the active form (cyan, PDB: 3AHQ) and the inactive form (tint, PDB: 3AHR) of human Ero1α. The missing outer active site-containing loop in the active form was shown in lilac dashed line with the Val101 residue as a red triangle. Trp272 in the protruding β-hairpin was shown as a blue triangle. (c) Far-UV circular dichroism analysis of Ero1α proteins. (d) Oxygen consumption catalyzed by 2 μM Ero1α proteins in the presence of 20 μM PDI and 10 mM GSH. (e, f) Gel-filtration analysis of the interactions between PDI and Ero1α proteins. Ero1αWT monomer, PDI monomer, and their 1:1 mixture were analyzed individually (e). Complexes formed between PDI and Ero1αWT, Ero1αV101G, Ero1αV109G or Ero1αP110G were shown as in (f). The peaks for Ero1α, PDI, and Ero1α-PDI complex were indicated. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.4. Val101 of Ero1α is specifically involved in the recognition of the catalytic domain of PDI
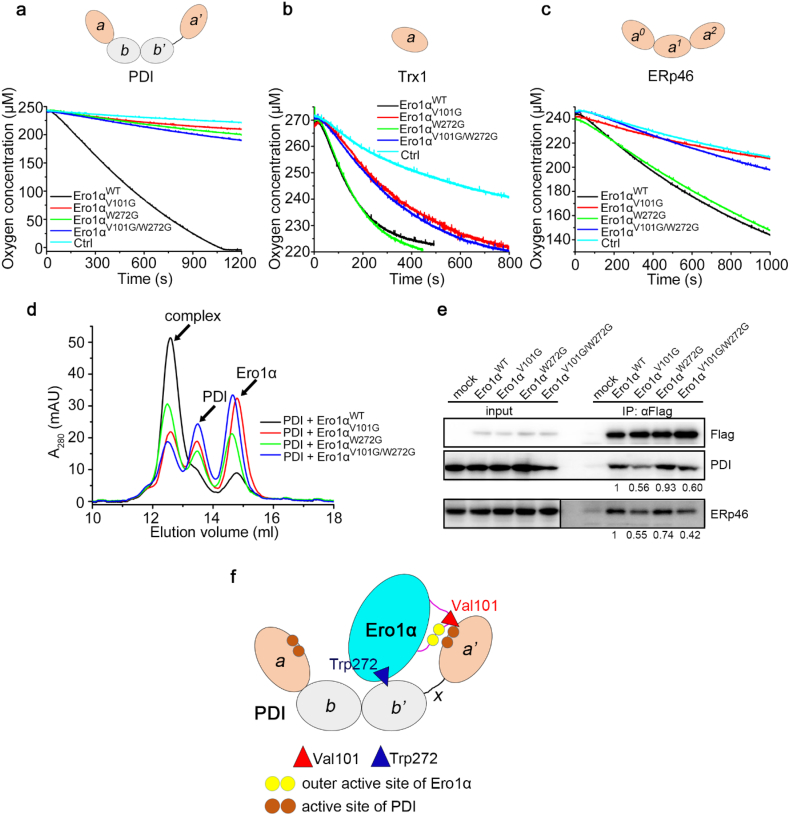
Next, we employed PDI, Trx1 and ERp46 to investigate which part of PDI is recognized by Val101 of Ero1α. PDI consists of four Trx-like domains. Among these domains, two are catalytic domains (a and a'), and two are noncatalytic domains (b and b'). Trx1 contains only one catalytic domain. ERp46 is a PDI family member containing three a-type catalytic domains [32]. Oxygen consumption assays showed that mutation of either Val101 or Trp272 resulted in the disability of Ero1α to oxidize PDI (Fig. 4a). In contrast, Ero1αW272G exhibited the same activity as Ero1αWT towards the oxidation of reduced Trx1 or ERp46, whereas both Ero1αV101G and Ero1αV101G/W272G were incapable to do so (Fig. 4b and c). These results suggest that Val101 of Ero1α interacts with the catalytic domain rather than the noncatalytic domain of PDI. Moreover, mutation of Val101 drastically impeded the oxidation of PDI a' domain and minimally affected the a domain (Fig. S7), in accordance with our previous finding that Ero1α prefers to oxidize the a' domain of PDI [17]. Gel-filtration analyses demonstrated that Val101 played a more powerful role than Trp272 in Ero1α-PDI complex formation (Fig. 4d). In accordance, co-immunoprecipitation analyses showed that mutation of Val101 markedly reduced Ero1α-PDI and Ero1α-ERp46 complexes in cells, whereas mutation of Trp272 caused less changes in complex formation (Fig. 4e). The conserved role of this valine residue was also verified in human Ero1β and yeast Ero1p (Fig. S8). Altogether, Val101 and Trp272 recognizes the a' and b' domains of PDI, respectively, and the former plays a dominant role in Ero1α-PDI complex formation (Fig. 4f).
Fig. 4.
Val101 of Ero1α is specifically involved in the recognition of the catalytic domain of PDI. (a) Oxygen consumption catalyzed by 2 μM Ero1αWT or its mutants in the presence of 10 mM GSH and 20 μM PDI. (b) Oxygen consumption catalyzed by 1 μM Ero1αWT or the mutants in the presence of 50 μM reduced Trx1. (c) Oxygen consumption catalyzed by 2 μM Ero1αWT or the mutants in the presence of 10 mM GSH and 60 μM ERp46. The domain organizations of PDI, Trx1 and ERp46 were shown on the top of each panel in (a-c). (d) Gel filtration analysis of the interaction between PDI and Ero1αWT, Ero1αV101G, Ero1αW272G or Ero1αV101G/W272G as in Fig. 3f. (e) Co-immunoprecipitation of Ero1α and PDI or ERp46 in HEK 293 T cells expressing FLAG-tagged Ero1α or the mutants. (f) Schematic model illustrating the roles of Val101 and Trp272 of Ero1α in the interaction with PDI. Trp272 in the protruding β-hairpin binds to the hydrophobic pocket in the b' domain of PDI, while Val101 in the outer active site-containing loop recognizes the catalytic a' domain of PDI.
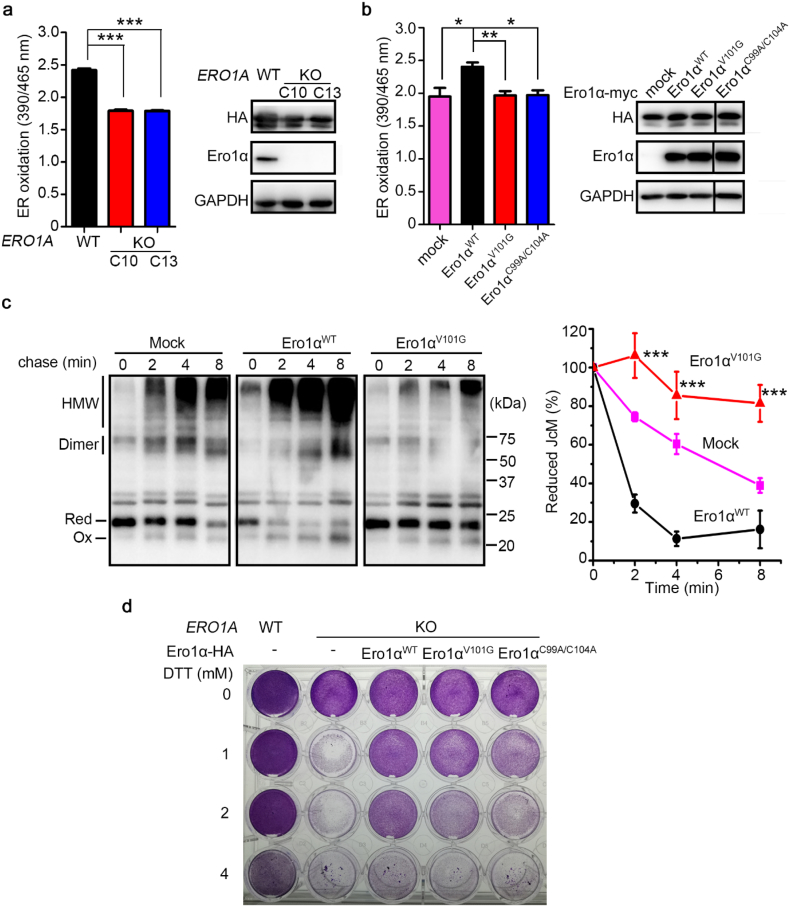
3.5. Mutation of Val101 triggers a reduced ER and inhibits the oxidase activity of Ero1α in cells
The ER lumen is significantly more oxidized compared with the cytosol with a lower GSH/GSSG ratio [33,34]. To study whether Val101 of Ero1α functions in ER GSH/GSSG maintenance, we employed an ER-localized superfolded redox-sensitive green fluorescent protein (superfolded-roGFP-iEER, hereafter referred to as roGFPER) as previously described [35]. roGFPER displays ratiometric fluorescence, and the fluorescence intensity ratio at 390/465 nm excitation increases under oxidizing conditions and decreases when the ER becomes reducing. Using this probe, we observed a more reduced ER in the two clones of ERO1A KO HeLa cells (Fig. 5a). Next, we stably expressed Ero1αWT, Ero1αV101G or Ero1αC99A/C104A (an inactive Ero1α for its outer active site was ruined, used as a negative control, also see Fig. S6) in ERO1A KO HeLa cells. Flow cytometry analyses showed that apoptosis was not induced in any of these cells (Fig. S9). The redox state in the ER was considerably recovered with the replenishment of Ero1αWT, whereas Ero1αV101G and Ero1αC99A/C104A did not restore the ER redox balance (Fig. 5b).
Fig. 5.
Mutation of Val101 generates a reduced ER and impairs the oxidase activity of Ero1α in HeLa cells. (a) ER oxidation determined by fluorescence intensity ratios at 390/465 nm excitation of HA-tagged roGFPER in WT and the two ERO1A KO cells (left). Protein immunoblotting of roGFPER (HA) and Ero1α in WT and the two ERO1A KO cells (right). (b) ER oxidation determined by fluorescence intensity ratios at 390/465 nm excitation of roGFPER in ERO1A KO (C10) cells expressing pcDNA3.1 (mock), Ero1αWT, Ero1αV101G or Ero1αC99A/C104A (left). Protein immunoblotting of roGFPER (HA) and Ero1α (right). Data in (a) and (b) were shown as mean ± SEM from three independent experiments performed in three technical replicates; * P < 0.05, ** P < 0.01, *** P < 0.001 (two-tailed Student's t-test). (c) Oxidative folding of Myc-tagged J-chain (JcM). WT cells co-transfected with JcM and empty vector (mock) or pcDNA3.1-Ero1α-HA were pulsed by DTT and chased at indicated times after DTT removal. Cell lysates were analyzed by non-reducing SDS-15% PAGE and α-myc western blotting. JcM in reduced (Red) and oxidized (Ox) monomer, dimer and high-molecular-weight species (HMW) were indicated on the left margin (left). The disappearance of fully reduced JcM monomers was quantified by densitometry and plotted as the percent remaining at each time point relative to 0 min (right). Data were shown as mean ± SEM from five independent experiments; *** P < 0.001, between Ero1αWT and Ero1αV101G (two-tailed Student's t-test). (d) Cell viabilities of WT and ERO1A KO (C10) cells stably expressing Ero1αWT and its mutants by lentivirus. Cells were treated by DTT at indicated concentrations for 24 h and visualized by crystal violet staining. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
We next explored whether mutation of Val101 in Ero1α affected oxidative protein folding in cells by monitoring the folding of immunoglobulin J-chain, a model substrate of Ero1α [36]. When HeLa transfectants expressing Myc-tagged J-chain (JcM) were exposed to DTT, most of the JcM was forced to be a reduced monomer. After DTT removal, the reduced monomers gradually disappeared, and oxidized monomers, dimers and high-molecular-weight (HMW) species appeared (Fig. 5c). Cells co-expressing Ero1αWT drastically accelerated the refolding kinetics of JcM. However, Ero1αV101G did not promote and even slowed down the JcM refolding compared to that in mock cells (Fig. 5c). The importance of Ero1α activity on cell survival was further evaluated using DTT as a reductive stress inducer. As shown in Fig. 5d, cell viability was reduced as the concentration of DTT increased, and depletion of Ero1α markedly decreased cell viability. Replenishment of Ero1αWT largely restored cell viability under reductive stress; however, Ero1αV101G and Ero1αC99A/C104A showed insufficient effects (Fig. 5d). Therefore, Val101 of Ero1α is essential for Ero1α oxidase activity in cells and is important for ER redox homeostasis maintenance.
3.6. Val101 of Ero1α is critical for cell migration, invasion, and tumor growth
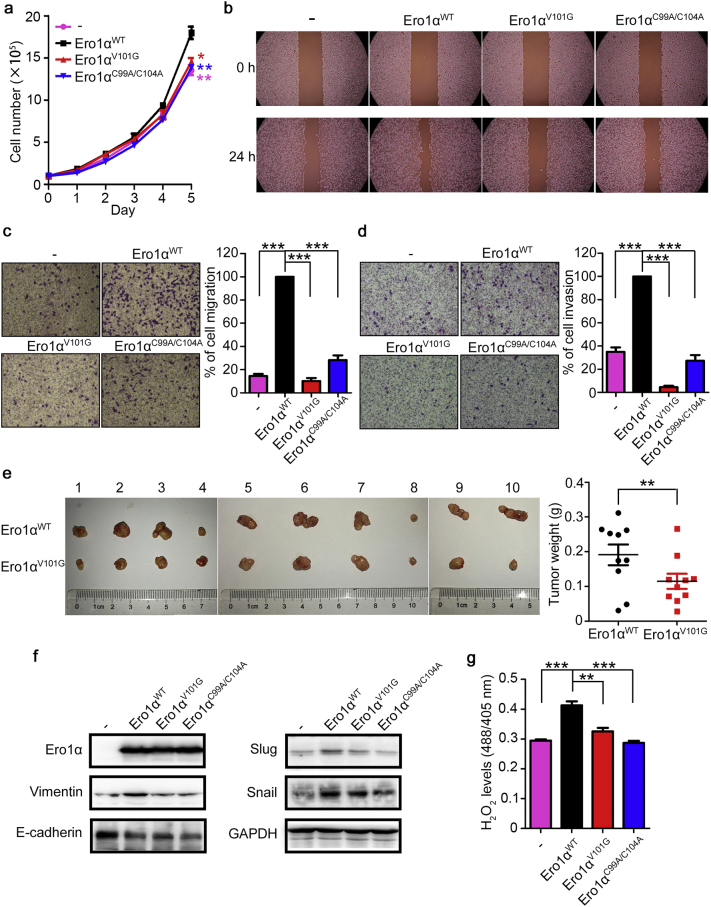
To determine the effects of mutation of Val101 on tumorigenesis, we first depicted the growth curves of ERO1A KO cells stably expressing Ero1αWT, Ero1αV101G, and Ero1αC99A/C104A. Mutation of Val101 caused marked growth retardation (Fig. 6a). In addition, when cells expressing Ero1αWT stopped growing with contact inhibition, cells expressing Ero1αV101G were still not confluent (Fig. S10). Cells expressing Ero1αV101G also exhibited delayed wound healing (Fig. 6b), impaired cell migration (Fig. 6c) and invasion (Fig. 6d) compared with cells expressing Ero1αWT. These results indicated that mutation of Val101 prevented cervical cell migration and invasion. Moreover, xenograft assays showed that the tumors derived from ERO1A KO HeLa cells stably expressing Ero1αV101G were significantly smaller than those from cells expressing Ero1αWT (Fig. 6e). Further, we found that replenishment of Ero1αWT induced the expression of mesenchymal markers, including Vimentin, Slug, Snail, while suppressed the epithelial marker E-cadherin; however, Ero1αV101G was incapable to promote efficient EMT signaling in ERO1A KO HeLa cells compared with Ero1αWT (Fig. 6f). Consistent with the role of H2O2 in EMT signaling, the H2O2 levels in the ER of Ero1αV101G-expressing cells were lower than that in Ero1αWT-expressing cells (Fig. 6g and Fig. S4). Taken together, mutation of Val101 in Ero1α impairs cervical cell migration, invasion and tumor growth.
Fig. 6.
The Val101 residue of Ero1α is critical for cell migration, invasion, and tumor growth in HeLa cells. (a) Growth curves of ERO1A KO (C10) cells stably expressing Ero1αWT and its mutants. Data were shown as mean ± SEM of three biological replicates; * P < 0.05, ** P < 0.01 (two-tailed Student's t-test). (b) Wound healing assays for ERO1A KO (C10) cells stably expressing Ero1αWT and its mutants. (c, d) Transwell migration assays (c) and transwell invasion assays (d) for ERO1A KO (C10) cells stably expressing Ero1αWT and its mutants. Migrated cells to the underside of filter membrane were counted and normalized to Ero1αWT. Data were shown as mean ± SEM from three independent experiments performed in two technical replicates; *** P < 0.001 (two-tailed Student's t-test). (e) Xenograft assays of ERO1A KO (C10) cells stably expressing Ero1αWT or Ero1αV101G. Cells were injected subcutaneously into the right (Ero1αWT) or left (Ero1αV101G) flanks of female BALB/c nude mice. Images of xenograft tumors at 36–41 days after inoculation were shown. Weights of the xenograft tumors were shown as mean ± SEM of ten biological replicates; ** P < 0.01 (two-tailed, paired Student's t-test). (f) Expression of EMT-associated molecules in ERO1A KO (C10) cells stably expressing Ero1αWT and its mutants analyzed by western blotting. (g) H2O2 levels in the ER of ERO1A KO (C10) cells stably expressing Ero1αWT and its mutants were determined by HyPerER probe, after the addition of 0.5 mM DTT for 5 min. Data were shown as mean ± SEM from four technical replicates; ** P < 0.01, *** P < 0.001 (two-tailed Student's t-test). Results are representative of two independent experiments.
4. Discussion
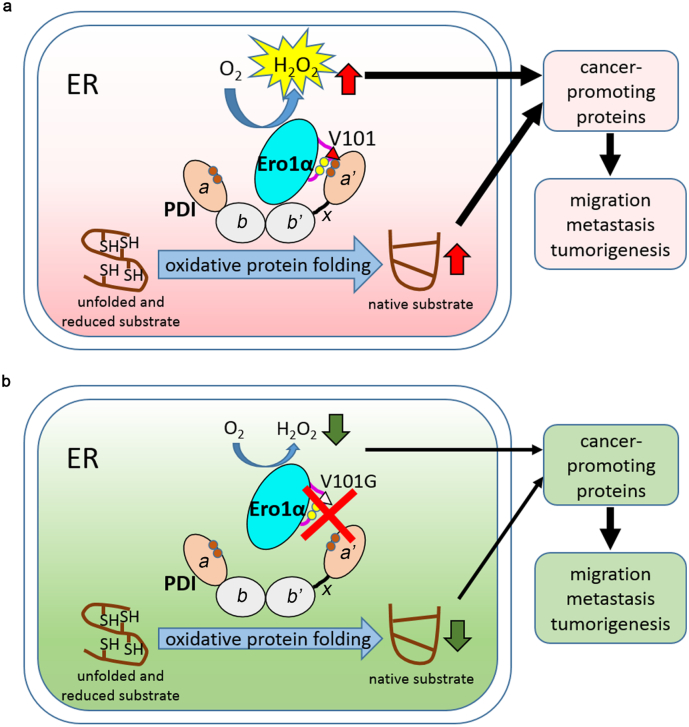
Ero1α, a sulfhydryl oxidase responsible for oxidative protein folding [8], exhibits highly upregulated expression in a variety of cancers [13,14,[37], [38], [39]] and thus represents a potential target for tumor intervention. Cervical cancer is one of the most common cancers in women, but little is known about the correlation of Ero1α with cervical carcinogenesis. In this study, we focused on the oxidative protein folding pathway and established the link between Ero1α-PDI interaction and tumorigenesis in cervical cancer. Based on the analyses of Ero1α expression levels in cervical cancer patients and immunohistochemistry staining of cervical cancer tissue microarray, we identified markedly increased Ero1α expression in tumor tissues compared with in normal tissues. In particular, the expression of Ero1α increased with the malignant degree of cervical cancer and poor prognosis (Fig. 1), indicating that Ero1α-mediated oxidative protein folding pathway is hijacked for the survival and proliferation of cancer cells. Knockout of ERO1A diminished tumor weight, providing direct evidence for the importance of Ero1α in tumorigenesis (Fig. 2e). Based on sequence alignment and biochemical analysis, we revealed that Val101 of Ero1α is a critical residue mediating Ero1α-PDI interaction (Fig. 3). Mutation of Val101 generated a reduced ER, retarded the oxidative folding of substrates (Fig. 5), and further impaired cervical cancer cell migration, invasion, and tumor growth (Fig. 7). Therefore, we provide the first evidence that disruption of Ero1α-PDI functional complex is an efficient strategy for prevention of tumorigenesis.
Fig. 7.
Role of Ero1α-PDI functional interplay in tumorigenesis. Ero1α coordinates with its partner PDI to catalyze oxidative protein folding in the ER with H2O2 as a byproduct. Ero1α binds to the b'xa' fragment of PDI through Val101 recognizing the a' domain of PDI. (a) In tumor cells, Ero1α and/or PDI are highly expressed to facilitate protein folding for tumor cell growth and proliferation. Ero1α generates more H2O2 and the ER becomes more oxidizing in tumor cells than in normal cells. Increased H2O2 and oxidative protein folding generate more cancer-promoting proteins which endow cells with higher ability of migration, metastasis and tumorigenesis. (b) Mutation of Val101 disrupts the interaction between Ero1α and PDI, decreases Ero1α-catalyzed oxidative folding and H2O2 production, and suppresses cell migration, metastasis and tumorigenesis.
Previous studies have reported that the expression and folding of several tumor-associated factors are promoted by Ero1α, such as growth factor VEGF, the master regulator for angiogenesis and tumor metastasis [38]. Ero1α promoted colon cancer progression through modulation of integrin-β1 glycosylation and membrane transport [39]. It will be interesting to know how the overall profile of proteins that drive metastasis and tumorigenesis changes in Ero1α KO cells or Ero1αV101G knock-in cells. Here, we report that in addition to oxidative protein folding catalyzed by Ero1α/PDI, H2O2, the byproduct of Ero1α oxidase activity, could also play an important role in tumorigenesis. H2O2 functions as a two-edged sword. It participates in many cellular pathways as a signaling molecule, but it also acts as an ROS to promote EMT, which is a notable mechanism involved in the progression of tumors [24]. Our results demonstrated that ERO1A KO or alternatively mutation of Val101 indeed decreased the production of H2O2 in the ER (Fig. 2g and Fig. 6g) and suppressed tumorigenesis. Thus, elimination of excess H2O2 generated by Ero1α might be another strategy for prevention of tumorigenesis. There are three peroxidases (Prx4, GPx7 and GPx8) located in the ER lumen, which utilize H2O2 for further oxidative protein folding by catalyzing PDI oxidation and concomitantly limit oxidative damage [40]. Notably, these peroxidases are connected to various diseases, underscoring the importance of ER redox homeostasis mediated by efficient H2O2 scavenging. For example, Prx4 has recently been proposed as a biomarker for type 2 diabetes, cancer, and sepsis [41] and GPx7 has tumor suppressor functions in esophageal adenocarcinoma [42] as well as geroprotective functions [43,44]. Interestingly, Ero1α is also upregulated in esophageal and gastric cancer cells [45], which is in contrast to the expression variation of GPx7. The decrease in GPx7 and increase in Ero1α in the development of these cancers may also imply the potential relationship between H2O2 and cancers.
Previous attempts for cancer therapy by developing small molecular inhibitors for either PDI or Ero1α have been reported. For example, 16F16 inhibits PDI activity by irreversible binding to the active-site cysteines of PDI [46], and EN460 competes with FAD cofactor of Ero1α to prevent its re-oxidation [47]. Nevertheless, 16F16 shows cytotoxicity given its PDI inhibition as well as potential off-target effects, and 16F16 could bind to other proteins that containing free cysteines within a similar binding cavity as in PDI [46]. Although EN460 exhibits high specificity towards Ero1α, it has low potency in vivo and exhibits a toxicity profile due to promiscuous interactions with free thiols [47]. Therefore, these inhibitors are far from clinic usage and awaiting further evaluation of their specificity and off-target toxicity. Here, we found that mutation of the critical Val101 residue of Ero1α led to the disruption of Ero1α-PDI functional complex and efficiently inhibited cervical cell migration, invasion, and tumorigenesis. Because the hydrophobic residue Val101 is very close to the outer active site (Cys94-Cys99) of Ero1α, we thus proposed that Val101 very possibly affects the docking between Ero1α outer active site and the -Cys-Gly-His-Cys- active site in the a' domain of PDI, which is also located in a hydrophobic patch [48] (Fig. 7). Although the structure of this loop is missing due to its intrinsic flexibility, we speculate that this flexible loop can be recognized by PDI as an unfolded peptide, and Val101 plays a dominant role in the recognition. Future structural study on Ero1α-PDI complex should provide more detailed information about how the Val101 of Ero1α is involved in the binding with PDI. In addition to PDI, Ero1α also oxidizes other PDI family members, such as ERp46 and P5 [49]. ERp46 promotes tumor growth in prostate carcinoma cells [50]; P5 promotes the proliferation of HeLa cells through activating the Wnt/β-catenin signaling pathway [51]. All these PDI homologues contain the a-type catalytic domains; thus, the strategy of disruption of Ero1α-PDI interaction might also be useful to inhibit cancer progression mediated by PDI homologues. Indeed, mutation of Val101 also destroys the functional interplay between Ero1α and ERp46 (Fig. 4c and e).
The current therapies for cervical cancer include surgery, radiation therapy and chemotherapy. However, patients receiving these treatments often suffer from poor quality of life. Based on our findings, we propose a new strategy for intervention of cervical cancer by targeting the Ero1α-PDI interface formed by Val101 and corresponding residues in PDI. Small chemicals or peptides that mimic the conformation around Val101 of Ero1α should disrupt the interaction between Ero1α and PDI to prevent cancer progression. To this end, efficient chemical or peptide discovery is expected for the realization of this therapeutic concept. Given that Ero1α and PDI proteins are upregulated in many types of cancers, the strategy of disruption of Ero1α-PDI interaction may be applied not only in cervical cancer but also in many other cancers. Collectively, our study offers mechanistic insights into the oncogenic roles of Ero1α and reveals Ero1α as a potential biomarker and therapeutic target for cancers.
Acknowledgments
Acknowledgments
We acknowledge Sai Yang and Jingjing Huang for subcutaneous injection in xenograft assays, Junying Jia, Shuang Sun, Jianhui Li for technical assistance, Lu Wang and Qianqian Chen for invaluable help.
Funding sources
This work was supported by National Key R&D Program of China (2017YFA0504000, 2016YFA0500200, 2017YFA0205501); National Natural Science Foundation of China (31771261, 31571163, 31870761, 31470814, 31671175, 31171345, and 31570772); and Youth Innovation Promotion Association, CAS, to LW and XW.
Declaration of interests
The authors have no financial conflicts to disclose.
Author contributions
LW conceived the project. YZ, TW, CCW, and LW designed research; YZ, TL, LZ, FS, GS, XW, XEW, XW, YL and BL performed research; YZ, GS, YC, TW, and LW analyzed data; YZ, CCW, and LW wrote the paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.02.041.
Appendix A. Supplementary data
Supplementary figures
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Oakes S.A. Endoplasmic reticulum proteostasis: a key checkpoint in cancer. Am J Physiol Cell Physiol. 2017;312(2):C93–C102. doi: 10.1152/ajpcell.00266.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galmiche A., Sauzay C., Chevet E., Pluquet O. Role of the unfolded protein response in tumor cell characteristics and cancer outcome. Curr Opin Oncol. 2017;29(1):41–47. doi: 10.1097/CCO.0000000000000339. [DOI] [PubMed] [Google Scholar]
- 4.Thornton M., Aslam M.A., Tweedle E.M. The unfolded protein response regulator GRP78 is a novel predictive biomarker in colorectal cancer. Int J Cancer - J Int du cancer. 2013;133(6):1408–1418. doi: 10.1002/ijc.28137. [DOI] [PubMed] [Google Scholar]
- 5.Dalton L.E., Clarke H.J., Knight J. The endoplasmic reticulum stress marker CHOP predicts survival in malignant mesothelioma. Br J Cancer. 2013;108(6):1340–1347. doi: 10.1038/bjc.2013.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hatahet F., Ruddock L. Protein disulfide isomerase: a critical evaluation of its function in disulfide bond formation. Antioxid Redox Signal. 2009;11(11):2807–2850. doi: 10.1089/ars.2009.2466. [DOI] [PubMed] [Google Scholar]
- 7.Bulleid N.J., Ellgaard L. Multiple ways to make disulfides. Trends Biochem Sci. 2011;36(9):485–492. doi: 10.1016/j.tibs.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 8.Wang L., Wang X., Wang C.C. Protein disulfide-isomerase, a folding catalyst and a redox-regulated chaperone. Free Radic Biol Med. 2015;83:305–313. doi: 10.1016/j.freeradbiomed.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 9.Sevier C.S., Kaiser C.A. Ero1 and redox homeostasis in the endoplasmic reticulum. Biochim Biophys Acta. 2008;1783(4):549–556. doi: 10.1016/j.bbamcr.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 10.Cabibbo A., Pagani M., Fabbri M. ERO1-L, a human protein that favors disulfide bond formation in the endoplasmic reticulum. J Biol Chem. 2000;275(7):4827–4833. doi: 10.1074/jbc.275.7.4827. [DOI] [PubMed] [Google Scholar]
- 11.Gess B., Hofbauer K.-H., Wenger R.H., Lohaus C., Meyer H.E., Kurtz A. The cellular oxygen tension regulates expression of the endoplasmic oxidoreductase ERO1-Lα. Eur J Biochem. 2003;270(10):2228–2235. doi: 10.1046/j.1432-1033.2003.03590.x. [DOI] [PubMed] [Google Scholar]
- 12.Rao J., Zhang C., Wang P. C/EBP homologous protein (CHOP) contributes to hepatocyte death via the promotion of ERO1alpha signalling in acute liver failure. Biochem J. 2015;466(2):369–378. doi: 10.1042/BJ20140412. [DOI] [PubMed] [Google Scholar]
- 13.Tanaka T., Kajiwara T., Torigoe T., Okamoto Y., Sato N., Tamura Y. Cancer-associated oxidoreductase ERO1-alpha drives the production of tumor-promoting myeloid-derived suppressor cells via oxidative protein folding. J Immunol. 2015;194(4):2004–2010. doi: 10.4049/jimmunol.1402538. [DOI] [PubMed] [Google Scholar]
- 14.Kukita K., Tamura Y., Tanaka T. Cancer-associated oxidase ERO1-alpha regulates the expression of MHC class I molecule via oxidative folding. J Immunol. 2015;194(10):4988–4996. doi: 10.4049/jimmunol.1303228. [DOI] [PubMed] [Google Scholar]
- 15.Kutomi G., Tamura Y., Tanaka T. Human endoplasmic reticulum oxidoreductin 1-alpha is a novel predictor for poor prognosis of breast cancer. Cancer Sci. 2013;104(8):1091–1096. doi: 10.1111/cas.12177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee E., Lee D.H. Emerging roles of protein disulfide isomerase in cancer. BMB Rep. 2017;50(8):401–410. doi: 10.5483/BMBRep.2017.50.8.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang L., Li S.J., Sidhu A. Reconstitution of human Ero1-Lalpha/protein-disulfide isomerase oxidative folding pathway in vitro. Position-dependent differences in role between the a and a' domains of protein-disulfide isomerase. J Biol Chem. 2009;284(1):199–206. doi: 10.1074/jbc.M806645200. [DOI] [PubMed] [Google Scholar]
- 18.Wang L., Zhu L., Wang C.C. The endoplasmic reticulum sulfhydryl oxidase Ero1beta drives efficient oxidative protein folding with loose regulation. Biochem J. 2011;434(1):113–121. doi: 10.1042/BJ20101357. [DOI] [PubMed] [Google Scholar]
- 19.Zhang L., Niu Y., Zhu L. Different interaction modes for protein-disulfide isomerase (PDI) as an efficient regulator and a specific substrate of endoplasmic reticulum Oxidoreductin-1alpha (Ero1alpha) J Biol Chem. 2014;289(45):31188–31199. doi: 10.1074/jbc.M114.602961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang L., Zhang L., Niu Y., Sitia R., Wang C.C. Glutathione peroxidase 7 utilizes hydrogen peroxide generated by Ero1alpha to promote oxidative protein folding. Antioxid Redox Signal. 2014;20(4):545–556. doi: 10.1089/ars.2013.5236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramming T., Hansen H.G., Nagata K., Ellgaard L., Appenzeller-Herzog C. GPx8 peroxidase prevents leakage of H2O2 from the endoplasmic reticulum. Free Radic Biol Med. 2014;70:106–116. doi: 10.1016/j.freeradbiomed.2014.01.018. [DOI] [PubMed] [Google Scholar]
- 22.Enyedi B., Varnai P., Geiszt M. Redox state of the endoplasmic reticulum is controlled by Ero1L-alpha and intraluminal calcium. Antioxid Redox Signal. 2010;13(6):721–729. doi: 10.1089/ars.2009.2880. [DOI] [PubMed] [Google Scholar]
- 23.Brabletz T., Kalluri R., Nieto M.A., Weinberg R.A. EMT in cancer. Nat Rev Cancer. 2018;18(2):128–134. doi: 10.1038/nrc.2017.118. [DOI] [PubMed] [Google Scholar]
- 24.Li W., Zhang L., Chen X., Jiang Z., Zong L., Ma Q. Hyperglycemia promotes the epithelial-mesenchymal transition of pancreatic Cancer via hydrogen peroxide. Oxid Med Cell Longev. 2016;2016:5190314. doi: 10.1155/2016/5190314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim Y.M., Cho M. Activation of NADPH oxidase subunit NCF4 induces ROS-mediated EMT signaling in HeLa cells. Cell Signal. 2014;26(4):784–796. doi: 10.1016/j.cellsig.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 26.Belousov V.V., Fradkov A.F., Lukyanov K.A. Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nat Methods. 2006;3(4):281–286. doi: 10.1038/nmeth866. [DOI] [PubMed] [Google Scholar]
- 27.Konno T., Pinho Melo E., Lopes C. ERO1-independent production of H2O2 within the endoplasmic reticulum fuels Prdx4-mediated oxidative protein folding. J Cell Biol. 2015;211(2):253–259. doi: 10.1083/jcb.201506123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zafarullah M., Li W.Q., Sylvester J., Ahmad M. Molecular mechanisms of N-acetylcysteine actions. Cell Mol Life Sci CMLS. 2003;60(1):6. doi: 10.1007/s000180300001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hansen H.G., Schmidt J.D., Soltoft C.L. Hyperactivity of the Ero1alpha oxidase elicits endoplasmic reticulum stress but no broad antioxidant response. J Biol Chem. 2012;287(47):39513–39523. doi: 10.1074/jbc.M112.405050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Masui S., Vavassori S., Fagioli C., Sitia R., Inaba K. Molecular bases of cyclic and specific disulfide interchange between human ERO1alpha protein and protein-disulfide isomerase (PDI) J Biol Chem. 2011;286(18):16261–16271. doi: 10.1074/jbc.M111.231357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Inaba K., Masui S., Iida H., Vavassori S., Sitia R., Suzuki M. Crystal structures of human Ero1alpha reveal the mechanisms of regulated and targeted oxidation of PDI. EMBO J. 2010;29(19):3330–3343. doi: 10.1038/emboj.2010.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ellgaard L., Ruddock L.W. The human protein disulphide isomerase family: substrate interactions and functional properties. EMBO Rep. 2005;6(1):28–32. doi: 10.1038/sj.embor.7400311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hwang C., Sinskey A.J., Lodish H.F. Oxidized redox state of glutathione in the endoplasmic reticulum. Science. 1992;257(5076):1496–1502. doi: 10.1126/science.1523409. [DOI] [PubMed] [Google Scholar]
- 34.Montero D., Tachibana C., Winther J.R., Appenzeller-Herzog C. Intracellular glutathione pools are heterogeneously concentrated. Redox Biol. 2013;1(1):508–513. doi: 10.1016/j.redox.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J., Zhu Q., Wang X. Secretory kinase Fam20C tunes endoplasmic reticulum redox state via phosphorylation of Ero1alpha. EMBO J. 2018;37(14) doi: 10.15252/embj.201798699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mezghrani A., Fassio A., Benham A., Simmen T., Braakman I., Sitia R. Manipulation of oxidative protein folding and PDI redox state in mammalian cells. EMBO J. 2001;20(22):6288–6296. doi: 10.1093/emboj/20.22.6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanaka T., Kutomi G., Kajiwara T. Cancer-associated oxidoreductase ERO1-α promotes immune escape through up-regulation of PD-L1 in human breast cancer. Oncotarget. 2017;8(15):24706–24718. doi: 10.18632/oncotarget.14960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tanaka T., Kutomi G., Kajiwara T. Cancer-associated oxidoreductase ERO1-α drives the production of VEGF via oxidative protein folding and regulating the mRNA level. Br J Cancer. 2016;114(11):1227–1234. doi: 10.1038/bjc.2016.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takei N., Yoneda A., Sakai-Sawada K., Kosaka M., Minomi K., Tamura Y. Hypoxia-inducible ERO1alpha promotes cancer progression through modulation of integrin-beta1 modification and signalling in HCT116 colorectal cancer cells. Sci Rep. 2017;7(1):9389. doi: 10.1038/s41598-017-09976-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kakihana T., Nagata K., Sitia R. Peroxides and peroxidases in the endoplasmic reticulum: integrating redox homeostasis and oxidative folding. Antioxid Redox Signal. 2012;16(8):763–771. doi: 10.1089/ars.2011.4238. [DOI] [PubMed] [Google Scholar]
- 41.Schulte J. Peroxiredoxin 4: a multifunctional biomarker worthy of further exploration. BMC Med. 2011;9:137. doi: 10.1186/1741-7015-9-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peng D., Belkhiri A., Hu T. Glutathione peroxidase 7 protects against oxidative DNA damage in oesophageal cells. Gut. 2012;61(9):1250–1260. doi: 10.1136/gutjnl-2011-301078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wei P.C., Hsieh Y.H., Su M.I. Loss of the oxidative stress sensor NPGPx compromises GRP78 chaperone activity and induces systemic disease. Mol Cell. 2012;48(5):747–759. doi: 10.1016/j.molcel.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fang J., Yang J., Wu X. Metformin alleviates human cellular aging by upregulating the endoplasmic reticulum glutathione peroxidase 7. Aging Cell. 2018;17:e12765. doi: 10.1111/acel.12765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Battle D.M., Gunasekara S.D., Watson G.R. Expression of the endoplasmic reticulum oxidoreductase Ero1α in gastro-intestinal cancer reveals a link between homocysteine and oxidative protein folding. Antioxid Redox Signal. 2013;19(1):24. doi: 10.1089/ars.2012.4651. [DOI] [PubMed] [Google Scholar]
- 46.Xu S., Sankar S., Neamati N. Protein disulfide isomerase: a promising target for cancer therapy. Drug Discov Today. 2014;19(3):222–240. doi: 10.1016/j.drudis.2013.10.017. [DOI] [PubMed] [Google Scholar]
- 47.Blais J.D., Chin K.T., Zito E. A small molecule inhibitor of endoplasmic reticulum oxidation 1 (ERO1) with selectively reversible thiol reactivity. J Biol Chem. 2010;285(27):20993–21003. doi: 10.1074/jbc.M110.126599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chao W., Wei L., Jinqi R. Structural insights into the redox-regulated dynamic conformations of human protein disulfide isomerase. Antioxid Redox Signal. 2013;19(1):44–53. doi: 10.1089/ars.2012.4630. [DOI] [PubMed] [Google Scholar]
- 49.Araki K., Iemura S., Kamiya Y. Ero1-alpha and PDIs constitute a hierarchical electron transfer network of endoplasmic reticulum oxidoreductases. J Cell Biol. 2013;202(6):861–874. doi: 10.1083/jcb.201303027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Duivenvoorden W.C.M., Hopmans S.N., Austin R.C., Pinthus J.H. Endoplasmic reticulum protein ERp46 in prostate adenocarcinoma. Oncol Lett. 2017;13(5):3624–3630. doi: 10.3892/ol.2017.5908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gao H., Sun B., Fu H. PDIA6 promotes the proliferation of HeLa cells through activating the Wnt/β-catenin signaling pathway. Oncotarget. 2016;7(33):53289–53298. doi: 10.18632/oncotarget.10795. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures