Abstract
Plant range expansion is occurring at a rapid pace, largely in response to human-induced climate warming. While the movement of plants along latitudinal and altitudinal gradients is well documented, effects on the belowground microbial communities remains largely unknown. Further, in range expansion not all plant species are equal: in a new range the relatedness between range-expanding plant species and native flora can influence plant-microbe interactions. Here we used a latitudinal gradient across Europe to examine bacterial and fungal communities in the rhizosphere and surrounding soils of range-expanding plant species. We selected range expanders with and without congeneric natives in the new range, and as a control, the congeneric natives, totaling 382 plant individuals collected across Europe. In general, a plant’s status as range expander was a weak predictor of bacterial and fungal community composition. However, microbial communities of range-expanding plant species became more similar to each other farther from their original range. Range expanders unrelated to the native community also experienced a decrease in the ratio of plant pathogens to symbionts, giving weak support to the enemy release hypothesis. Even at a continental scale the effects of plant range expansion on the belowground microbiome are detectable, though changes to specific taxa remain difficult to decipher.
Keywords: bacteria, fungi, plant-soil interactions, climate change, microbial ecology
Species range expansion in response to climate change is recognized as a major uncertainty in predicting consequences of global warming for biodiversity and functioning of ecosystems1,2. Initially, attention was given to the ability of species to keep up with their shifting climate envelope; now research questions have expanded to include the consequences of range shifts for community interactions3. The disruption of plant range expansions on aboveground interactions have been well documented4–6, including on aboveground herbivores and higher tropic levels7,8. While evidence suggests that introduced invasive species can alter soil communities9–11, the effects of plant range expansion on belowground microbial communities remain ambiguous.
The relationships between plants and their associated microbes can influence plant establishment, fitness, and community assembly12–14. It has been proposed that plant range expanders will be successful in their new range because they lose their specialized soil pathogens15–17. At the same time, range expanders may also lose specialized mutualistic microbes18–20. Results of these studies lead to the similar expectation that the plant-associated microbial community in the rhizosphere and surrounding soil (named here the belowground plant microbiome) of range expanding plant species will associate less with the belowground microbiome in their new range than in their native range, and also when compared to native plant species. However, few studies have characterized or compared the microbiome community structure and diversity of range expanding plant species (however see21) and no study has made a direct comparison with related native plant species at a continental scale.
The soil and rhizosphere microbiome, made up largely of bacteria and fungi, is taxonomically and functionally diverse22. The community composition of the belowground microbiome is broadly structured by abiotic factors, yet effects differ between bacteria and fungi23,24. For example, while at large spatial scales bacterial communities are strongly influenced by soil pH25,26, the composition of fungal communities are simultaneously affected by climate and nutrients27–29. At the same time, both the soil and rhizosphere microbiome are strongly controlled by biotic factors, including the composition of root exudates, plant species identities and plant traits30–32. Through these properties plant species can assemble species-specific microbiomes where microbial taxa are enriched or suppressed under some plants and not under others14,33–36. At the same time, phylogenetic relatedness of range-expanding plants with native flora can represent another potential effect of range expansion on microbial communities - where some research suggests that closely related plant species can harbor similar microbial taxa, especially pathogens37,38. Finally, plant-microbe interactions evolve over time, changing over years and even decades39,40; thus during range expansion, both distance from the original range and evolutionary history between plants and microbes41 have potential to influence the belowground plant microbiome.
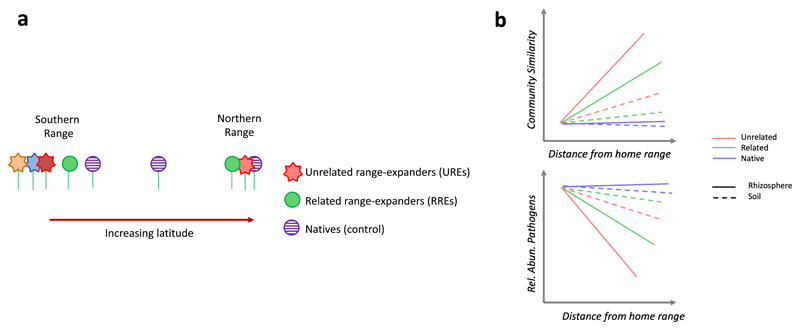
Here we analyze the microbiome of intra-continental range-expanding plant species along a latitudinal gradient to explore key hypotheses that have been previously proposed for exotic and invasive plants but may also apply to climate warming-induced range-expansions. To test for the influence of plant phylogeny on the belowground microbiome during range expansion, we selected range expanders that are either related or unrelated to the native flora (Fig. 1A). To test for the effects of range expansion on the belowground plant microbiome compared changes in community composition and pathogen relative abundance across the range expansion gradient (Fig.1B). We hypothesize that if plant range expansion influences the belowground plant microbiome, observed patterns will be stronger in the rhizosphere42 than in the bulk soil. Further, if range-expanding plants farther from their original range either lose the ability to interact with certain microbial taxa or preferentially promote the growth of a beneficial community, in the new range the microbiome or range-expanders will become more similar and alpha diversity with decrease. However, because plants more closely related to the native community may share microbes, this change will be less pronounced for range expanders that encounter congeneric natives in the new habitat. Finally, if the enemy release hypothesis common to invasive plant species is also applicable to range-expanders, we expect fewer belowground pathogens to be associated with range expanders unrelated to the native flora compared to related expanders and native.
Figure 1. Changes in microbial community during plant range-expansions.
A. When plants move from the southern range to a new range, range expanders can be either related to the native flora (circles) or unrelated (stars). B. Hypothesized responses of microbial community similarity and pathogen relative abundance to range-expansion; where we expect observed patterns to be stronger in the rhizosphere (solid lines) than in the bulk soil (dashed lines), and that the relatedness of the range-expander to the native flora will affect the strength of the response.
In Europe, climate change induced range expansion is well documented; many plant species are expanding their range into higher latitudes and altitudes2,43. Here we used high-throughput Illumina sequencing to explore how the belowground microbiome of plant species changes when plants expand from their original range (in lower latitudes) to new ranges (in higher latitudes). We targeted the microbiome of three plant groups: unrelated range-expanders - plant species without native species from the same genus in their new range; related range-expanders - plant species that have native species from the same genus in their new range (Supplementary Table 1 and Supplementary Figure 1); and native plant species, which are congeneric to the related range expanders and native throughout the entire gradient. All range expanding plants either arrived to or had greatly expanded within the Netherlands in the late 20th and early 21st centuries44. In an effort to minimize variation in abiotic factors, we selected 11 plant species grown on similar parent soil (see methods). For each species, we sampled the microbiome in the rhizosphere and surrounding (bulk) soil of up to 9 plant individuals collected from up to 6 countries, spanning from Greece to the Netherlands, totaling 382 plant individuals (Figure 1 and Table 1). While some species were cosmopolitan45, others were quite rare and more difficult to find. Regardless of plant species dominance, here we have replicates not just in plant species, but in plant types (native, related and unrelated range expanders) and 382 bulk soil and rhizosphere samples in order to obtain a number that should be sufficient to capture large scale patterns in the microbial community26,28.
Results & Discussion
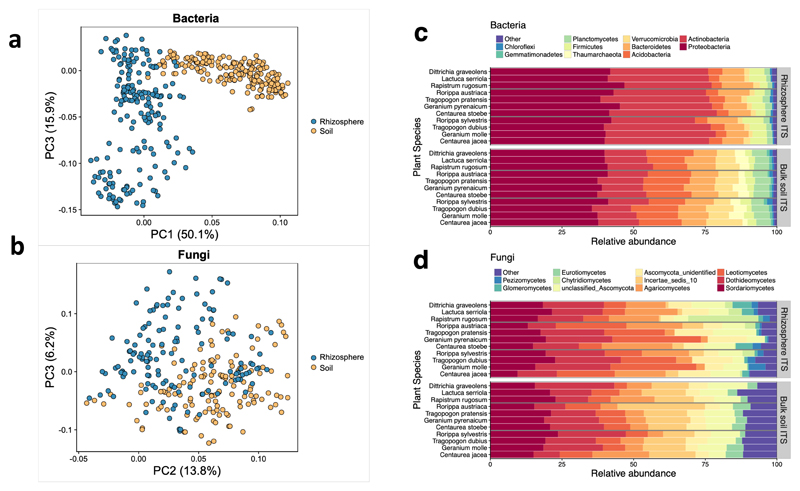
Overall, rhizosphere and bulk soil communities were significantly different from each other, both in community overlap as visualized by a PCA (p < 0.001 for both bacteria and fungi; Figure 2A and 2B), and in taxa overlap (Figure 2C and 2D). We found 47,704 bacterial phylotypes and 9,374 fungal phylotypes in soils, and 33,939 bacterial phylotypes and 6,438 fungal phylotypes in the rhizosphere. Further, there was low community overlap among plant individuals in both soil (averaging 4,092 (8%) unique bacterial taxa and 523 (5.5%) unique fungal phylotypes per sample) and the rhizosphere (averaging 1,932 (5.6%) unique bacterial phylotypes and 257 (4%) unique fungal phylotypes per sample). High microbiome diversity among 11 plant species is not a surprise, especially because the selected plants represent a range of phylogenetically and ecologically distinct species36,46,47.
Figure 2. Rhizosphere and soils harbor different microbial communities.
Differences in bulk soil (yellow) and rhizosphere (blue) of A bacterial and B fungal communities, visualized by PCoA and differences determined by NMDS of Bray-Curtis differences (PERMANOVA: p < 0.001 for both). Relative abundance of C bacterial and D fungal taxa from rhizosphere and soils.
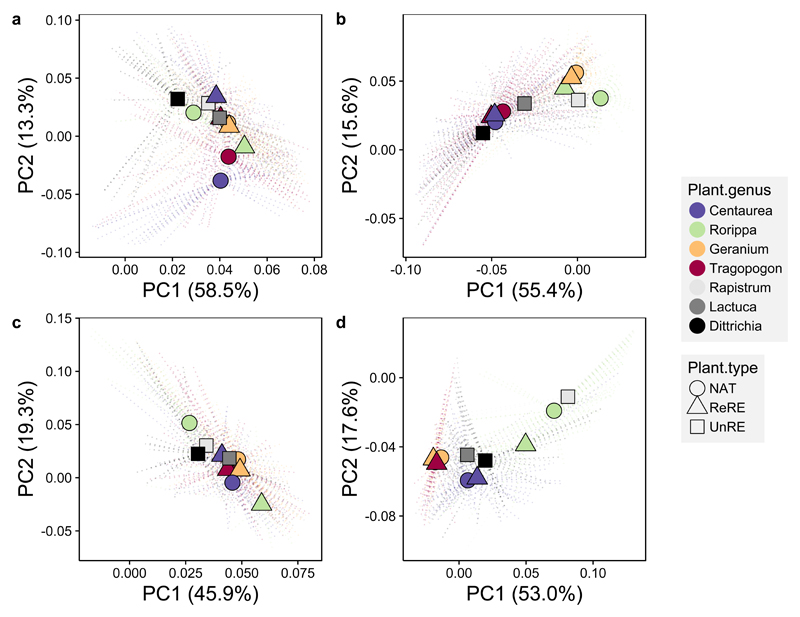
Across the gradient, plant species was the strongest predictor of bacterial and fungal community composition in both soil and rhizosphere environments, explaining 7 to 14% of the variation (Figure 3; Supplementary Table 2), and plant genus a proxy of phylogenetic relatedness (Supplementary Figure 1) provided no additional prediction power. Conversely, the effects of plant grouping (unrelated range-expander, related range-expander and native) and latitude had a much smaller effect on microbial composition and explained a maximum of 2% of the variation in all cases. In general, soil abiotic factors also had a minor influence on variation, accounting for less than 1% of the variation for all factors (e.g. pH, N, C), except for soil bacterial communities where pH explained approximately 5% of the variation. The relatively minor effect of soil abiotics on microbial communities - compared to previous studies25 - can be explained by the small variation in soil factors across the gradient and between plants (Supplementary Figure 2), as was the goal of choosing plant species growing on the same parent soil material. In comparison, other studies have been more focused on elucidating patterns in microbial community composition relative to changes in abiotic factors26,28,48. Thus, the differences observed here are more likely due to plant species effects sensu47, such as plant ecology, relatedness with native flora, and life history traits45,49,50.
Figure 3. Plant species was the strongest predictor of bacterial (top) and fungal (bottom) community structure in both the soil (left) and the rhizosphere (right).
PCoA ordinations show the centroid of all individuals for each plant species with lines representing connections to individual samples (not plotted). Plant group (native, related range expander and unrelated range expander) is represented by shape, and plant genus by color.
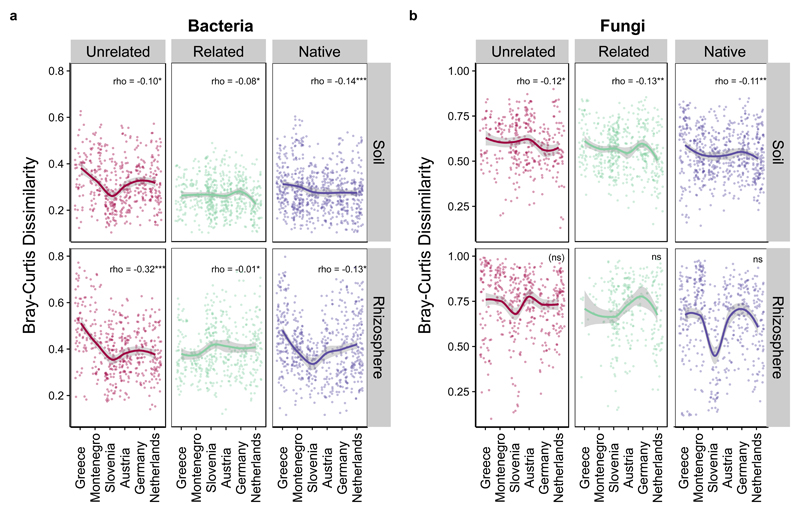
In support of our hypothesis, we found that range-expanders farther from their original range had more similar microbial communities to other plant individuals. Put another way, the variation in community composition decreased among individuals in the new range. Further, there were negative correlations between “range” (country samples were collected from) and community dissimilarity for all plant groups (Figure 4 and Supplementary Table 3); latitude and distance, gave equivalent results. This pattern was significant for bacterial communities in the soil and rhizosphere of all plant types (rho varied between -0.08 and -0.32 and p < 0.05 for all). However, for fungal communities, correlations were only observed in soils (rho varied from -0.10 to -0.13, p < 0.05 for all) and not in the rhizosphere. The negative correlation between range and community dissimilarity was strongest in unrelated range-expander species (Supplementary Table 3). We also found a significant difference in the degree of microbial community similarity by plant group yet there was an interaction of country in two scenarios (p< 0.0001 in all cases) (Supplementary Table 4). This suggests that controls on native and range-expanding plant microbiome community composition differs across the gradient. For instance, native plants microbiomes (and to a lesser extent related range-expanders) may be more influenced by a long-term co-evolutionary history that would be consistent across this latitudinal gradient51,52, while unrelated range expander microbiome patterns might be more determined by more recent spatial effects and the native (neighbor) plant community53. Because we used a survey to explore changes to the belowground microbiome across a natural range expansion transect, we were unable to test for co-evolutionary history between microbes and plants. Still, our results suggest that future studies should be designed with this process in mind, particularly to identify the role of the microbial community for plant adaptions during climate change39,54.
Figure 4. Changes in microbial community dissimilarly across the range expansion gradient.
A Both soil and rhizosphere bacterial communities become more similar under unrelated range-expanders (red) and to some extent native plants (purple) farther from the original range. Similar but weaker patterns were observed in related range-expanders (green). B Fungal communities showed a weaker response, and significant decreases were only observed in soils. (Spearman’s Rank Correlation Coefficient: P < 0.05*; p <0.01**; p<<0.001***, SE shown in grey)
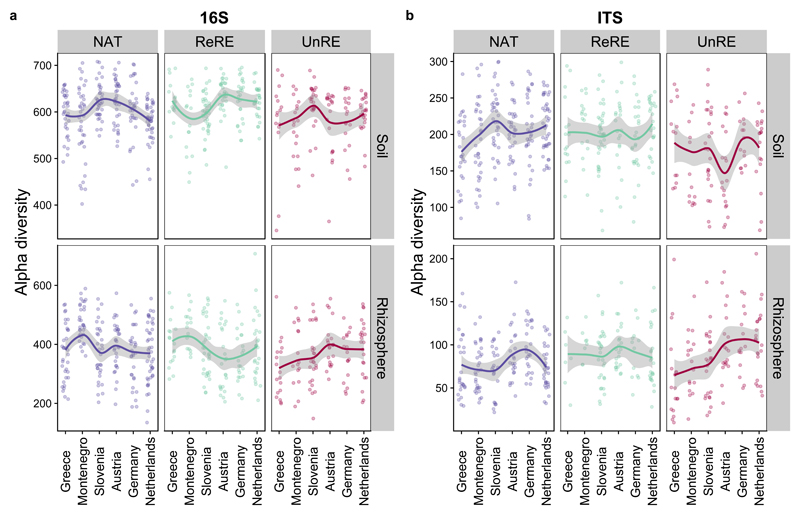
While community structure became more similar across the gradient, changes in bacterial richness and fungal richness was much more variable (Figure 5; Supplementary Table 5). Under unrelated range-expanders, fungal alpha diversity in the rhizosphere of significantly increased with distance from the original range (rho = 0.36 p <0.001 in the rhizosphere, p>0.05 in soil). However, related range-expanders showed no relationship between fungal diversity and distance from original range (p > 0.05 for both soil and rhizosphere) in comparison to native plants which where fungal alpha diversity increased with latitude in both the rhizosphere (rho = 0.20, p < 0.05) and in the bulk soil (rho = 0.23 p < 0.05). The mechanisms behind increased fungal diversity in the rhizosphere of unrelated range-expanders remains in question. It could be that if range-expanding plants do not need to invest in belowground defense55,56 the rhizosphere becomes accessible for a larger proportion of microbes, though this varies by plant species57. Alternatively, it has been proposed that exotics and range-expanders promote high microbial diversity as part of a defense mechanism53,57. The later proposition, that range-expanding plants enrich their rhizosphere, is congruent with our findings that community composition becomes more similar among individuals in the northern part of the range (Figure 4), and that unrelated range-expanders had higher fungal and bacterial diversity in their rhizosphere and lower diversity in the associated soils (p < 0.0001 in all cases) (Supplementary Table 6). Overall, the inconsistency between the responses of the two expanders suggests that related and unrelated range-expanders have different controls on microbial diversity. Further, the variability in alpha diversity patterns indicates that alpha diversity and community similarity are affected by different mechanisms.
Figure 5. Changes in alpha diversity across the latitudinal gradient of range expansion differed between bacterial and fungal communities.
While A bacterial alpha diversity did not significantly change (ns in all cases), B fungal diversity increased in the rhizosphere of unrelated range expanders (red), to some extent native plant (purple), and no pattern was seen in related range expanders (green). (Spearman’s Rank Correlation Coefficient: P < 0.05*; p <0.01**; p<<0.001***, SE shown in grey)
It has been proposed that in novel ecosystems plant success or failure is based on reduced exposure to soil-borne pathogens combined with continued association with symbionts58,59. We applied this concept here and used FunGuild60 to test how the abundance of potential fungal functional groups change as range-expanding plants move farther from their original range. Specifically, we examined potential plant pathogens and arbuscular mycorrhizal fungi (AMF), as these are the relevant mutualistic symbionts for most of our plant species, except for the crucifers. However, we could detect no significant change in the relative abundance in either of these groups under range expanding plant species (Supplementary Figure 3). Though there was a significant positive correlation in the ratio of plant pathogens to symbionts across the transect (rho = 0.31 p< 0.001) (Supplementary Table 7). On the other hand, under native plants the relative abundance of plant pathogens increased in both the soil and rhizosphere from south to north (rho = 0.23 for both). Contrary to previous studies, these results do not directly verify that range-expanders lose their specialist microbes58 or are released from specialist enemies61. Instead, the results suggest that compared to natives, range expanders are exposed to fewer potential pathogens and symbionts in the new range, which has been predicted for range-expanding plant species62 and demonstrated for introduced exotics in their new range63,64. At the same time, recent studies of plant succession 65,66 clearly demonstrate that plant success and nutrient cycling is tied to the microbial communities. Yet it remains unclear if the mechanisms underlying plant range-expansion are the same as those observed elsewhere.
Still, these results are not without caveats. The first being that the molecular methods used are not infallible- the DNA community analysis does not assess the active microbial community nor the true functional capabilities. Thus, potential functional groupings and relative abundances of taxa cannot indicate the expected pathogenicity of these fungi in the host plant’s rhizospheres. Equally important is that for all plant groups the relative abundance of these functional groupings make up approximately 5% of the fungal community. Meaning, any changes in composition or diversity may overinflate or obscure true changes in these low abundance groups67 and specific primers or culture work is necessary to explore functional changes more thoroughly. Our study exemplifies that high-throughput sequence data can be used to assess large-scale patterns in plant-soil associations, but future functional analyses (e.g. metagenomics and metatranscriptomics approaches) and experimental studies must be designed to take the low abundance of pathogen sequences into account.
Our study contributes initial steps of identifying the patterns of changes in the plant microbiome during plant range expansion. While, microbial community and diversity dynamics change across a range expansion gradient, clarifying the mechanisms behind the observed changes would require further experimental study. In the present study, we attempted to link the concepts from plant ecology to the microbiome by assuming that plant establishment outside the native range will result in altered exposure for soil microbes. Our results suggest that while terms like ‘exotic’, ‘range-expander’ and ‘native’ are helpful descriptors in plant ecology, it should not be assumed that these labels are equally relevant to describe the belowground microbial community of such plant species. Future research will require consideration of the ecological roles of both plants and microbes26,36 but currently, the ecological roles on many microbial taxa still remain unknown. At the same time, we think that this large-scale biogeographical studies of plant-soil-microbe associations of native, related and unrelated range expanders along a latitudinal gradient is an essential step to understand how climate warming-induced range-expanding plant species may assemble a new microbiome in their novel range. This approach may also stand as a model for processes that take place belowground upon the introduction of exotic plant species in a new continent. Subsequent experimental work is needed in order to understand functional consequences for invasiveness and naturalization.
Almost 4% of extant global vascular flora have established outside their native range68, and climate change induced range expansion is not expected to slow down69. Though soil microbes exert strong selective pressures on plant species and communities70,71 our understanding of microbial community dynamics during range expansion remains limited. Range expansion offers an opportunity to explore how global change may alter the relationship between plants and their microbiome, but also how the belowground microbiome changes across large geographic scales. Understanding the effect of range expansion on the belowground plant microbiome can provide baseline knowledge for predicting ecological consequences of current rapid climate warming, and it may also be used to enhance understanding of community responses to invasion scenarios for introduced exotic species.
Methods
Plant species and soil collection
In central Europe, rivers flow to the south and north away from the Alps, resulting in habitats with sediments from similar parent materials and soils that spread across a latitudinal gradient. Within these well-connected river habitats, and in response to climate change, many plant species are expanding their range with much more movement expected in the coming decades1,74,75. Within this latitudinal gradient, spanning from Greece in the south to the Netherlands in the north, we identified 7 range-expander species whose range has expanded north into Austria, Germany and the Netherlands over the last 50 years, approximately76. Range-expanders without native congenerics in the northern sites (here named unrelated range-expanders) include: Dittrichia graveolens, Lactuca serriola and Rapistrum rugosum. Range-expanders with native congenerics (related range-expanders) include: Centaurea stoebe, Geranium pyrenaicum, Tragopogon pratensis and Rorippa austriaca. As a control, we also included 4 native plant species that are congeneric with the related range-expanders: Centaurea jacea, Geranium molle, Tragopogon dubious and Rorippa sylvestris. C. stoebe and R. austriaca originated from Central and Eastern Europe, while all other range-expanders originated from southern Europe (www.gbif.org). Plant populations were sampled from 6 countries in Europe – Greece, Montenegro, Slovenia, Austria, Germany and the Netherlands - in the summer growing seasons of 2013 and 2014. All plants were flowering at the time of sampling. At each sampling site, environmental parameters, including weather conditions at sampling dates, were recorded. For each sampling location of a single species, 3 individuals of 3 distinct populations (in most cases at least 400m separation) were chosen, totaling 9 plant individuals for each location (see Supplementary Figure 1 for sample numbers). For collection of all samples, permissions were obtained from both nature reserves and government agencies responsible for the land.
To assess the soil and rhizosphere microbiomes of native and range-expanding plant species, soils and roots plus rhizosphere were collected from under individual plants. Briefly, the entire plant was dug up to a 10 cm radius around the plant and soil was shaken off the plant roots. Bulk soil was homogenized and 10 g was collected for microbial and chemical analyses. The fine plant roots plus rhizosphere soil were collected separately, hereon referred to as the rhizosphere community. All rhizosphere and soil samples were stored at 4°C until shipped, within 1 week, to the Netherlands Institute of Ecology (NIOO). At the NIOO soil and rhizosphere samples for DNA extraction were frozen at -80°C. A subset of soil was stored in the fridge at 4°C for chemical analyses.
Soil chemical analyses
For all soil samples collected in 2014 nutrients and pH were measured on fresh soil stored at 4°C (Supplementary data; Supplementary Figure 2). Gravimetric moisture (% water) was determined on soils oven dried at 105°C. Total soil C and N content was determined from these dried soils on an elemental analyzer (LECO, St Joseph, MI, USA). Extractable NO3 and NH4 were measure using the KCl extraction protocol. Briefly, soils were dried at 4°C, 10 g dry soil were then mixed with 1M potassium chloride (KCl) solution, shaken, and then the supernatant is used for analyses of NO3 and NH4. Soil pH was measured in an H20 slurry solution using a bench-top pH meter following the ISO 10309 standard procedure.
Community level sequence analysis
To identify the bulk soil and rhizosphere microbiomes from native and range-expanding plants, DNA was extracted from 0.25 g of ground bulk soil and 0.35 g of ground rhizosphere material using the PowerSoil-htp 96 Well Soil DNA isolation kit (MO BIO Laboratories, Inc., California, USA) according to the manufacturer’s instructions. Bacterial community composition was determined by targeting 16S rRNA amplicons using 515F/806R primers77, and the fungal community composition by targeting the ITS region using primers ITS4/fITS978. To prevent the amplification of plant material79, PNA Clamps© (PCR Blockers) (CGACACTGACACTGA-KK) were added at the PCR step for rhizosphere bacterial DNA. For all samples, DNA was PCR amplified in duplicate using barcoded primers77. PCR products were purified using the Agencourt AMPure XP magnetic bead system (Beckman Coulter Life Sciences, Indianapolis, Indiana, USA) and analyzed via the Standard Sensitivity NGS Fragment Analysis kit (1bp-6000bp). Pooled PCR amplicons were sequenced with the Illumina MiSeq platform at BGI Tech Solutions (HongKong) Co Limited.
MiSeq paired-end reads targeting 16s rRNA amplicon were merged and only reads which had minimum overlap of 150bp with PHRED score of 25 (estimated by RDP extension of PANDASeq80). Primer sequences were stripped using Flexbar version 2.581. Sequences were then clustered to OTUs with VSEARCH v1.0.1082, using the UPARSE strategy of de-replication, sorting by abundance and clustering using the UCLUST smallmem algorithm83. All singletons were removed, and potential chimeric sequences were removed using the UCHIME algorithm84. Taxonomic classification for each OTU was obtained by using the RDP Classifier version 2.1085.
Likewise, MiSeq paired-end reads targeting ITS region were treated as described above with following adjustments: ITS primer sequences were stripped using ITSx 1.0.1186 before clustering, and sequences were classified using the UNITE database87. All bioinformatics steps were implemented with a publicly available workflow made with Snakemake88. After samples were removed due to sampling error or falling below the rarified threshhold, 382 samples were included in downstream analyses of plant soil and rhizosphere microbiomes.
Community similarity was visualized with a PCA of the dissimilarity matrix based on Bray-Curtis distances. Plotted in Figure 1 is the centroid of each plant species community with lines representing connections to all other samples of that species. We quantified phylogenetic distances between all plant species used, but did not make a full analysis of these distances with differences in microbiome composition, as plant genus or family-specific issues might interfere with pure phylogenetic distances (Supplementary Figure 1). To investigate how distance from the original range influences the microbiome for each plant species we tested within country dissimilarity of bacterial and fungal communities both in the rhizosphere and in soil. Briefly, pairwise Bray-Curtis dissimilarity was estimated between samples of each plant species within each country. Diversity of soil communities were analyzed using “vegan” package89 using the PERMANOVA test and visualized with “ggplot2” package. Correlation patterns were visualized with the LOESS smoothing function90. Because within country distance was so much smaller than between country distance diversity patterns were the same whether plotted by latitude, country or geographic distance, which here we refer to as “range”. Spearman’s Rank Correlations were run on latitude and plots show country name for clarity. FUNGUILD analyses were generated using the web interface and only taxa that received a ‘highly probably’ classification were included. When all taxa were included results remained the same. All other analyses were performed using R programming language (R Development Core Team 2008).
Supplementary Material
Acknowledgements
We are grateful to the support of Željka Modrić-Surina (Natural History Museum, Croatia), Snežana Dragićević (Natural History Museum of Montenegro), Dr Indra Starke (Senckenberg, Frankfurt, Germany) and Dr Michael Hohla (Austria) who all helped in the extensive sampling. This work was supported in large part by the European Research Council (ERC advanced grant ERC-Adv 323020 (SPECIALS) to WHvdP. Additional support came from the Estonian Research Council (grant PUTJD78) (KK), and the Slovenian Research Agency (research core funding No. P1-0236) (BV, TČ).
Footnotes
Data Availability: The authors declare that the data supporting the findings of this study are available within the paper and its supplementary information files. Sequences have been deposited under PRJEB25697, PRJEB25694, PRJEB25693, PRJEB25692.
Author Contributions WvdP conceived the idea of this study. Sample collection was completed WvdP, KSR, KK, SG, LJB, FtH, OK, NK, MM, DC, MAT, BV, TC, CW, RAW. Soil analyses and sequencing was completed by LJB, FtH, CW, DvR, KSR. Data analyses were completed by LBS and KSR. The manuscript was written by KSR, with contribution from all co-authors.
The authors declare no competing interests.
References
- 1.Pecl GT, et al. Biodiversity redistribution under climate change: Impacts on ecosystems and human well-being. Science (80-. ) 2017;355 doi: 10.1126/science.aai9214. [DOI] [PubMed] [Google Scholar]
- 2.Parmesan C, Yohe G. A globally coherent fingerprint of climate change impacts across natural systems. Nature. 2003;421:37–42. doi: 10.1038/nature01286. [DOI] [PubMed] [Google Scholar]
- 3.Classen AT, et al. Direct and indirect effects of climate change on soil microbial and soil microbial-plant interactions: What lies ahead? Ecosphere. 2015;6 art130. [Google Scholar]
- 4.Meisner A, De Deyn GB, de Boer W, van der Putten WH. Soil biotic legacy effects of extreme weather events influence plant invasiveness. Proc Natl Acad Sci U S A. 2013;110:9835–8. doi: 10.1073/pnas.1300922110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Engelkes T, et al. Successful range-expanding plants experience less above-ground and below-ground enemy impact. Nature. 2008;456:946–948. doi: 10.1038/nature07474. [DOI] [PubMed] [Google Scholar]
- 6.van der Putten WH, Bradford MA, Brinkman EP, van de Voorde TFJ, Veen GF. Where, when and how plant-soil feedback matters in a changing world. Funct Ecol. 2016;30:1109–1121. [Google Scholar]
- 7.Gonzalez-Megias A, Menendez R. Climate change effects on above- and below-ground interactions in a dryland ecosystem. Philos Trans R Soc B Biol Sci. 2012;367:3115–3124. doi: 10.1098/rstb.2011.0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tylianakis JM, Didham RK, Bascompte J, Wardle DA. Global change and species interactions in terrestrial ecosystems. Ecol Lett. 2008;11:1351–1363. doi: 10.1111/j.1461-0248.2008.01250.x. [DOI] [PubMed] [Google Scholar]
- 9.Kourtev PS, Ehrenfeld JG, Häggblom M. Exotic Plant Species Alter the Microbial Community Structure and Function in the Soil. Ecology. 2002;83:3152–3166. [Google Scholar]
- 10.McLeod ML, et al. Exotic invasive plants increase productivity, abundance of ammonia-oxidizing bacteria and nitrogen availability in intermountain grasslands. J Ecol. 2016;104:994–1002. [Google Scholar]
- 11.Coats VC, Rumpho ME. The rhizosphere microbiota of plant invaders: an overview of recent advances in the microbiomics of invasive plants. Front Microbiol. 2014;5:368. doi: 10.3389/fmicb.2014.00368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klironomos JN. Feedback with soil biota contributes to plant rarity and invasiveness in communities. Nature. 2002;417:67–70. doi: 10.1038/417067a. [DOI] [PubMed] [Google Scholar]
- 13.Kardol P, Wardle DA. How understanding aboveground-belowground linkages can assist restoration ecology. Trends Ecol Evol. 2010;25:670–9. doi: 10.1016/j.tree.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 14.Van Nuland ME, Bailey JK, Schweitzer JA. Divergent plant–soil feedbacks could alter future elevation ranges and ecosystem dynamics. Nat Ecol Evol. 2017;1:0150. doi: 10.1038/s41559-017-0150. [DOI] [PubMed] [Google Scholar]
- 15.Engelkes T, et al. Successful range-expanding plants experience less above-ground and below-ground enemy impact. Nature. 2008;456:946–8. doi: 10.1038/nature07474. [DOI] [PubMed] [Google Scholar]
- 16.Van Grunsven RHA, et al. Reduced plant-soil feedback of plant species expanding their range as compared to natives. J Ecol. 2007;95:1050–1057. [Google Scholar]
- 17.Dostálek T, Münzbergová Z, Kladivová A, Macel M. Plant–soil feedback in native vs. invasive populations of a range expanding plant. Plant and Soil. 2015;399 [Google Scholar]
- 18.Berg G, Smalla K. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol Ecol. 2009;68:1–13. doi: 10.1111/j.1574-6941.2009.00654.x. [DOI] [PubMed] [Google Scholar]
- 19.De Frenne P, et al. Plant movements and climate warming: Intraspecific variation in growth responses to nonlocal soils. New Phytol. 2014;202:431–441. doi: 10.1111/nph.12672. [DOI] [PubMed] [Google Scholar]
- 20.Van GRUNSVEN RHA, Van Der PUTTEN WH, MARTIJN BEZEMER T, BERENDSE F, VEENENDAAL EM. Plant–soil interactions in the expansion and native range of a poleward shifting plant species. Glob Chang Biol. 2010;16:380–385. [Google Scholar]
- 21.Collins CG, Carey CJ, Aronson EL, Kopp CW, Diez JM. Direct and indirect effects of native range expansion on soil microbial community structure and function. J Ecol. 2016;104:1271–1283. [Google Scholar]
- 22.Fierer N. Embracing the unknown: disentangling the complexities of the soil microbiome. Nat Rev Microbiol. 2017;15:579–590. doi: 10.1038/nrmicro.2017.87. [DOI] [PubMed] [Google Scholar]
- 23.Kuramae E, Gamper H, van Veen J, Kowalchuk G. Soil and plant factors driving the community of soil-borne microorganisms across chronosequences of secondary succession of chalk grasslands with a neutral pH. FEMS Microbiol Ecol. 2011;77:285–294. doi: 10.1111/j.1574-6941.2011.01110.x. [DOI] [PubMed] [Google Scholar]
- 24.Tecon R, Or D. Biophysical processes supporting the diversity of microbial life in soil. FEMS Microbiol Rev. 2017;41:599–623. doi: 10.1093/femsre/fux039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lauber CL, Hamady M, Knight R, Fierer N. Pyrosequencing-Based Assessment of Soil pH as a Predictor of Soil Bacterial Community Structure at the Continental Scale. Appl Environ Microbiol. 2009;75:5111–5120. doi: 10.1128/AEM.00335-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delgado-Baquerizo M, et al. A global atlas of the dominant bacteria found in soil. Science (80-. ) 2018;359:320–325. doi: 10.1126/science.aap9516. [DOI] [PubMed] [Google Scholar]
- 27.Talbot JM, et al. Endemism and functional convergence across the North American soil mycobiome. Proc Natl Acad Sci U S A. 2014;111:6341–6. doi: 10.1073/pnas.1402584111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tedersoo L, et al. Global diversity and geography of soil fungi. Science (80-. ) 2014;346 doi: 10.1126/science.1256688. [DOI] [PubMed] [Google Scholar]
- 29.Barberán A, et al. Why are some microbes more ubiquitous than others? Predicting the habitat breadth of soil bacteria. Ecol Lett. 2014;17 doi: 10.1111/ele.12282. [DOI] [PubMed] [Google Scholar]
- 30.Lekberg Y, Rosendahl S, Olsson PA. The fungal perspective of arbuscular mycorrhizal colonization in ‘nonmycorrhizal’ plants. New Phytol. 2015;205:1399–1403. doi: 10.1111/nph.13118. [DOI] [PubMed] [Google Scholar]
- 31.Lau JA, Lennon JT. Rapid responses of soil microorganisms improve plant fitness in novel environments. Proc Natl Acad Sci U S A. 2012;109:14058–62. doi: 10.1073/pnas.1202319109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Vries FT, et al. Land use alters the resistance and resilience of soil food webs to drought. Nat Clim Chang. 2012;2:276–280. [Google Scholar]
- 33.Peay KG. Back to the future: natural history and the way forward in modern fungal ecology. Fungal Ecology. 2014;12:4–9. [Google Scholar]
- 34.Edwards J, et al. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc Natl Acad Sci U S A. 2015;112:E911–20. doi: 10.1073/pnas.1414592112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pieterse CMJ, de Jonge R, Berendsen RL. The Soil-Borne Supremacy. Trends Plant Sci. 2016;21:171–173. doi: 10.1016/j.tplants.2016.01.018. [DOI] [PubMed] [Google Scholar]
- 36.Prober SM, et al. Plant diversity predicts beta but not alpha diversity of soil microbes across grasslands worldwide. Ecol Lett. 2015;18:85–95. doi: 10.1111/ele.12381. [DOI] [PubMed] [Google Scholar]
- 37.Gilbert GS, Webb CO. Phylogenetic signal in plant pathogen-host range. Proc Natl Acad Sci. 2007;104:4979–4983. doi: 10.1073/pnas.0607968104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parker IM, et al. Phylogenetic structure and host abundance drive disease pressure in communities. Nature. 2015;520:542–544. doi: 10.1038/nature14372. [DOI] [PubMed] [Google Scholar]
- 39.Lankau RA. Coevolution between invasive and native plants driven by chemical competition and soil biota. Proc Natl Acad Sci. 2012;109:11240–11245. doi: 10.1073/pnas.1201343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morriën E, et al. Soil networks become more connected and take up more carbon as nature restoration progresses. Nat Commun. 2017;8 doi: 10.1038/ncomms14349. 14349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keymer DP, Lankau RA. Disruption of plant-soil-microbial relationships influences plant growth. J Ecol. 2017;105:816–827. [Google Scholar]
- 42.Leach JE, Triplett LR, Argueso CT, Trivedi P. Communication in the Phytobiome. Cell. 2017;169:587–596. doi: 10.1016/j.cell.2017.04.025. [DOI] [PubMed] [Google Scholar]
- 43.Bakkenes M, Alkemade JRM, Ihle F, Leemans R, Latour JB. Assessing effects of forecasted climate change on the diversity and distribution of European higher plants for 2050. Glob Chang Biol. 2002;8:390–407. [Google Scholar]
- 44.Wilschut RA, Kostenko O, Koorem K, van der Putten WH. Nematode community responses to range-expanding and native plant communities in original and new range soils. Ecol Evol. 2018 doi: 10.1002/ece3.4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koorem K, et al. Relatedness with plant species in native community influences ecological consequences of range expansions. Oikos. 2018 doi: 10.1111/oik.04817. [DOI] [Google Scholar]
- 46.van der Heijden MGA, Hartmann M. Networking in the Plant Microbiome. PLoS Biology. 2016;14 doi: 10.1371/journal.pbio.1002378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leff JW, et al. Predicting the structure of soil communities from plant community taxonomy, phylogeny, and traits. ISME J. 2018 doi: 10.1038/s41396-018-0089-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fierer N, et al. Reconstructing the Microbial Diversity and Function of Pre-Agricultural Tallgrass Prairie Soils in the United States. Science (80-. ) 2013;342:621–624. doi: 10.1126/science.1243768. [DOI] [PubMed] [Google Scholar]
- 48.Emmett BD, Youngblut ND, Buckley DH, Drinkwater LE. Plant Phylogeny and Life History Shape Rhizosphere Bacterial Microbiome of Summer Annuals in an Agricultural Field. Front Microbiol. 2017;8:2414. doi: 10.3389/fmicb.2017.02414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goberna M, Navarro-Cano JA, Verdú M. Opposing phylogenetic diversity gradients of plant and soil bacterial communities. Proc R Soc B Biol Sci. 2016;283 doi: 10.1098/rspb.2015.3003. 20153003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reynolds HL, Packer A, Bever JD, Clay K. Grassroots ecology: Plant-microbe-soil interactions as drivers of plant community structure and dynamics. Ecology. 2003;84:2281–2291. [Google Scholar]
- 52.Bennett JA, et al. Plant-soil feedbacks and mycorrhizal type influence temperate forest population dynamics. Science (80-. ) 2017;355 doi: 10.1126/science.aai8212. [DOI] [PubMed] [Google Scholar]
- 53.Golivets M, Wallin KF. Neighbour tolerance, not suppression, provides competitive advantage to non-native plants. Ecol Lett. doi: 10.1111/ele.12934. n/a--n/a. [DOI] [PubMed] [Google Scholar]
- 54.Geml J, Wagner MR. Out of sight, but no longer out of mind - towards an increased recognition of the role of soil microbes in plant speciation. New Phytol. 2018;217:965–967. doi: 10.1111/nph.14979. [DOI] [PubMed] [Google Scholar]
- 55.Dawson W. Release from belowground enemies and shifts in root traits as interrelated drivers of alien plant invasion success: A hypothesis. Ethol Ecol Evol. 2015;5:4505–4516. doi: 10.1002/ece3.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Blumenthal D, Mitchell CE, Pysek P, Jarosik V. Synergy between pathogen release and resource availability in plant invasion. PNAS. 2009;106:7899–7904. doi: 10.1073/pnas.0812607106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wilschut RA, Silva JCP, Garbeva P, van der Putten WH. Belowground Plant–Herbivore Interactions Vary among Climate-Driven Range-Expanding Plant Species with Different Degrees of Novel Chemistry. Front Plant Sci. 2017;8:1861. doi: 10.3389/fpls.2017.01861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Inderjit, van der Putten WH. Impacts of soil microbial communities on exotic plant invasions. Trends Ecol Evol. 2010;25:512–519. doi: 10.1016/j.tree.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 59.Rout ME, Callaway RM. Interactions between exotic invasive plants and soil microbes in the rhizosphere suggest that ‘everything is not everywhere’. Ann Bot. 2012;110:213–222. doi: 10.1093/aob/mcs061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nguyen NH, et al. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016;20:241–248. [Google Scholar]
- 61.Blumenthal D, Mitchell CE, Pysek P, Jarosík V. Synergy between pathogen release and resource availability in plant invasion. Proc Natl Acad Sci U S A. 2009;106:7899–904. doi: 10.1073/pnas.0812607106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Van der Putten WH. Climate Change, Aboveground-Belowground Interactions, and Species’ Range Shifts. 2012 [Google Scholar]
- 63.Mitchell CE, Power AG. Release of invasive plants from fungal and viral pathogens. Nature. 2003;421:625. doi: 10.1038/nature01317. [DOI] [PubMed] [Google Scholar]
- 64.Bever JD, Mangan SA, Alexander HM. Maintenance of Plant Species Diversity by Pathogens. Annu Rev Ecol Evol Syst. 2015;46:305–325. [Google Scholar]
- 65.Morriën E, et al. Soil networks become more connected and take up more carbon as nature restoration progresses. Nat Commun. 2017;8:14349. doi: 10.1038/ncomms14349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hannula SE, et al. Shifts in rhizosphere fungal community during secondary succession following abandonment from agriculture. ISME J. 2017;11:2294–2304. doi: 10.1038/ismej.2017.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jousset A, et al. Where less may be more: how the rare biosphere pulls ecosystems strings. ISME J. 2017;11:853–862. doi: 10.1038/ismej.2016.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van Kleunen M, Dawson W, Maurel N. Characteristics of successful alien plants. Mol Ecol. 2015;24:1954–1968. doi: 10.1111/mec.13013. [DOI] [PubMed] [Google Scholar]
- 69.Chen IC, Hill JK, Ohlemuller R, Roy DB, Thomas CD. Rapid Range Shifts of Species Associated with High Levels of Climate Warming. Science (80-. ) 2011;333:1024–1026. doi: 10.1126/science.1206432. [DOI] [PubMed] [Google Scholar]
- 70.Bever J, Platt T, Morton E. Microbial population and community dynamics on plant roots and their feedbacks on plant communities. Annu Rev Microbiol. 2012;66:265–283. doi: 10.1146/annurev-micro-092611-150107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wubs ERJ, van der Putten WH, Bosch M, Bezemer TM. Soil inoculation steers restoration of terrestrial ecosystems. Nat Plants. 2016;2 doi: 10.1038/nplants.2016.107. 16107. [DOI] [PubMed] [Google Scholar]
- 72.Morriën E, van der Putten WH. Soil microbial community structure of range-expanding plant species differs from co-occurring natives. J Ecol. 2013;101:1093–1102. [Google Scholar]
- 73.Wall DH, Nielsen UN, Six J. Soil biodiversity and human health. Nature. 2015;528:69–76. doi: 10.1038/nature15744. [DOI] [PubMed] [Google Scholar]
- 74.Alexander JM, Diez JM, Levine JM. Novel competitors shape species’ responses to climate change. Nature. 2015;525:515–518. doi: 10.1038/nature14952. [DOI] [PubMed] [Google Scholar]
- 75.Fordham DA, et al. Plant extinction risk under climate change: are forecast range shifts alone a good indicator of species vulnerability to global warming? Glob Chang Biol. 2012;18:1357–1371. [Google Scholar]
- 76.Tamis WLM, Zelfde MV, Der Meijden R, Van De Haes HAU. Changes in Vascular Plant Biodiversity in the Netherlands in the 20th Century Explained by their Climatic and other Environmental Characteristics. Clim Change. 2005;72:37–56. [Google Scholar]
- 77.Caporaso JG, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–4. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ihrmark K, et al. New primers to amplify the fungal ITS2 region--evaluation by 454-sequencing of artificial and natural communities. FEMS Microbiol Ecol. 2012;82:666–77. doi: 10.1111/j.1574-6941.2012.01437.x. [DOI] [PubMed] [Google Scholar]
- 79.Lundberg DS, Yourstone S, Mieczkowski P, Jones CD, Dangl JL. Practical innovations for high-throughput amplicon sequencing. Nat Methods. 2013;10:999–1002. doi: 10.1038/nmeth.2634. [DOI] [PubMed] [Google Scholar]
- 80.Masella AP, Bartram AK, Truszkowski JM, Brown DG, Neufeld JD. PANDAseq: paired-end assembler for illumina sequences. BMC Bioinformatics. 2012;13:31. doi: 10.1186/1471-2105-13-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dodt M, Roehr J, Ahmed R, Dieterich C. FLEXBAR—Flexible Barcode and Adapter Processing for Next-Generation Sequencing Platforms. Biology (Basel) 2012;1:895–905. doi: 10.3390/biology1030895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rognes T, Flouri T, Nichols B, Quince C, Mahé F. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–1. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 84.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cole JR, et al. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014;42:D633–42. doi: 10.1093/nar/gkt1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bengtsson-Palme J, et al. Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol Evol. 2013;4:914–919. [Google Scholar]
- 87.Kõljalg U, et al. Towards a unified paradigm for sequence-based identification of fungi. Mol Ecol. 2013;22:5271–5277. doi: 10.1111/mec.12481. [DOI] [PubMed] [Google Scholar]
- 88.Koster J, Rahmann S. Snakemake--a scalable bioinformatics workflow engine. Bioinformatics. 2012;28:2520–2522. doi: 10.1093/bioinformatics/bts480. [DOI] [PubMed] [Google Scholar]
- 89.Oksanen J, et al. Vegan: Community Ecology Package. R Package Version. 2.0-10. CRAN. 2013 [Google Scholar]
- 90.Wickham H. Ggplot2: Elegant Graphics for Data Analysis. Springer Publishing Company, Incorporated; 2009. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.