Abstract
Prophages are commonly found in Listeria genomes, potentially enhancing survival or fitness of Listeria spp. Currently, there is still limited information on the distribution of prophages among Listeria isolates of different allelic types and from various sources. In this study, by using mitomycin C induction, prophages were found in 23/144 isolates (16.0%), including 13 L. monocytogenes and 10 Listeria spp. isolates, resulting in 28 and 11 induced phages, respectively. These prophage-carrying isolates (lysogens) were obtained from foods and food-related environments presenting 3 common allelic types (ATs) of L. monocytogenes (lineage I, II and IV), 4 ATs of L. innocua and 1 AT of L. welshimeri. The likelihood of prophage-carrying isolates of L. monocytogenes was 14.4 (95% CI: 4.9–35.4), and 18.5 (95% CI: 4.8–50.2) for Listeria spp. The 39 induced phages were classified into 3 lysis groups by the host range test against 9 major serotypes of L. monocytogenes and 5 species of Listeria. Most phages were host-specific with higher ability to lyse L. monocytogenes serotype 4 than other serotypes. The genome size of phages ranged from 35±2 kb to 50±2 kb and belonged to two common phage families, Myoviridae and Siphoviridae. Restriction analysis classified 19 selected phages into 16 restriction profiles, suggesting highly diverse prophages with at least 16 types. This may contribute to the variation in the genomes of Listeria. Information obtained here provides basic knowledge for further study to understand the overall role of prophages in Listeria, including roles in survival or fitness in foods and food processing environments.
Introduction
Listeria monocytogenes is an important foodborne pathogen that can cause listeriosis-a serious foodborne illness with mortality rate up to 30% [1]. The genus Listeria comprises 17 species. Several new species have been discovered in the past decade, for example, L. marthii from soil [2]; L. fleischmannii and L. weihenstephanensis in cheese and water, respectively [3,4]. Previous studies have reported that various types of foods and food processing environments can be contaminated with Listeria spp., including L. monocytogenes [5,6]. L. monocytogenes showed good survival in specific environments and they were resistant to the deleterious effects of freezing, drying, and heat [7–9]. There could be some potential factors such as gene transfer, gene gain or loss that may facilitate survival, evolution and speciation of this pathogen.
Bacteriophages (phages) are viruses of bacteria, and are the most common component in the biosphere [10]. Phages can be classified into two groups based on their life cycles [11]. Phages from lytic cycle, called lytic phages, replicate using bacterial machinery and then destroy the host. Phages from lysogenic cycle, called lysogenic phages, have the same lytic capacity but in addition, they can also integrate their DNA into the bacterial chromosome to establish a prophage. The interests of prophage and the host are partly aligned because the lysogen will prolong prophage status [12]. Prophages have several important roles in facilitating the lysogen’s survival [13,14], virulence [15] and phage resistance [16]. Therefore, studying the diversity of prophages and their characteristics (e.g., host specificity) is useful for further studies to understand prophage’s contribution to overall life of the bacteria.
Previous studies have revealed the pervasiveness of prophages in Listeria genomes, and multiple prophages were found in a single strain [17,18]. For example, L. innocua CLIP11262 harbored up to six prophage-like elements, including 5 prophages and 1 monocin [19]. A recent study reported that various types of prophage inserted into the genomes of L. monocytogenes ST121 [20]. However, these studies mostly have applied bioinformatics analyses to search for prophage regions in Listeria genomes. Alternatively, if no sequencing is performed on Listeria isolates, induction is an effective approach to examine the presence of prophages. Among the reported inducing agents such as antibiotics, UV radiation, sunlight, temperature, or pressure [21–23], mitomycin C has been reported as the most effective in prophage induction [16,24].
In this study, mitomycin C induction was performed to investigate the distribution of prophage among Listeria isolates obtained from various foods and food-processing environments. These isolates were classified based on partial SigB sequences, which is useful for the prediction of prophage in each allelic type representing different lineages/ species. Characterization of the induced phages phenotypically and genotypically allows us to better understand prophage diversity, which may contribute to the variation in Listeria genomes or to Listeria survival and fitness in foods and food-related environments. Moreover, host range data obtained here could allow predicting particular subtypes of L. monocytogenes or Listeria spp. in which gene transfer may occur upon phage infection, leading higher survival of the pathogen in the food production chain.
Materials and methods
L. monocytogenes and Listeria spp. isolates used in this study
A total of 144 isolates of L. monocytogenes and other Listeria spp. (non- monocytogenes) were used for prophage induction in this study (Table 1). These isolates were previously obtained from multiple sources [5,6,25] and are maintained at the Department of Food Technology, Prince of Songkla University (PSU): animal origin products (n = 38), seafood/ aquatic products (n = 53), vegetable products (n = 8), food contact surfaces (n = 22) and non-food contact surfaces (n = 23). Four L. monocytogenes reference strains, including F2365, Mack, FSL F2-695 and FSL J1-208 were used as propagating hosts for prophage induction and phage lysate preparation (Table 2). A total of 31 Listeria strains/ isolates were used for host range determination. These included 19 reference strains [25,26] obtained from the Food Safety Lab (FSL), Cornell University, and from the Department of Medical Science Thailand (DMST), Ministry of Health, Thailand. Additional 12 isolates of L. monocytogenes and Listeria spp. were selected from the collection of 144 isolates used in this study [5,6,25]. Of these, eight L. monocytogenes hosts were subjected to serotype classification by multiplex PCR following the protocol of Doumith et al. [27] combined with the classification of sigB allelic types.
Table 1. Source of L. monocytogenes and Listeria spp. isolates used in this study.
| Source of isolates | Number of isolates | ||
|---|---|---|---|
| L. monocytogenes | Listeria spp. | Total | |
| Animal origin products (Ani) | 18 | 20 | 38 |
| Seafood/ aquatic products (Sea) | 27 | 26 | 53 |
| Vegetable products (Veg) | 3 | 5 | 8 |
| Food contact surfaces (FCS) | 19 | 3 | 22 |
| Non-food contact surfaces (NFCS) | 23 | 0 | 23 |
| Total | 90 | 54 | 144 |
Table 2. L. monocytogenes and Listeria spp. strains/ isolates subjected to mitomycin C induction and host range determination.
| Strain/ isolate ID | Lineage/ species | Serotype | Source |
|---|---|---|---|
| L. monocytogenesa and other Listeria spp. reference strains | |||
| FSL J1-175 | I | 1/2b | Water |
| FSL J1-194 | I | 1/2b | Human |
| FSL J1-169 | I | 3b | Human |
| FSL J1-049 | I | 3c | Human |
| FSL R2-574 (F2365)* | I | 4b | Food |
| FSL F6-367(Mack)* | II | 1/2a | Lab strain |
| FSL R2-0559 | II | 1/2a | Food |
| FSL J1-094 | II | 1/2c | Human |
| FSL C1-115 | II | 3a | Human |
| FSL F2-695* | IIIA | 4a | Human |
| FSL F2-501 | IIIA | 4b | Human |
| FSL J2-071 | IIIA | 4c | Animal |
| FSLW1-110 | IIIC | 4b | Unknown |
| FSL J1-208* | IV | 4a | Animal |
| FSL J1-158 | IV | 4b | Animal |
| DMST-9011 | L. innocua | Unknown | |
| DMST-9012 | L. ivanovii | Unknown | |
| FSL C7-0084 | L. marthii | Soil, forest | |
| FSL C7-0015 | L. seeligeri | Soil | |
| L. monocytogenes and other Listeria spp. isolates | |||
| PSU-KV-032LM | I | 1/2b, 3b | Environmental-FCS |
| PSU-KV-042LM | I | 1/2b, 3b | Environmental-NFCS |
| PSU-KV-038LM | I | 4b | Environmental-FCS |
| PSU-KV-105LM | I | 4b | Seafood/aquatic product |
| PSU-KV-148LM | II | 1/2c | Animal origin product |
| PSU-KV-159LM | II | 1/2c | Animal origin product |
| PSU-KV-108LM | IV | 4a, 4c | Seafood/aquatic product |
| PSU-KV-120LM | IV | 4a, 4c | Animal origin product |
| PSU-KV-114LS | L. innocua | Animal origin product | |
| PSU-KV-146LS | L. innocua | Animal origin product | |
| PSU-KV-131LS | L. welshimeri | Seafood/aquatic product | |
| PSU-KV-181LS | L. welshimeri | Vegetable product | |
aFour reference strains with ‘*’ were used as the propagating hosts for mitomycin C induction and phage lysate preparation.
SigB allelic typing
Typing of a partial sigB gene has been used for allelic type classification and species confirmation of Listeria isolates in previous studies [5,28,29]. Of the 144 Listeria isolates used for prophage induction in this study, 60 have been assigned an allelic type (AT) previously [5,6]. SigB allelic typing was performed to 84 Listeria isolates in this study. The protocol of Nightingale et al. [28] was followed for PCR amplification of a partial sigB gene (780 bp) using the same primers. The purified PCR products were sequenced at Macrogen Inc. (Seoul, South Korea). Allelic type classification was performed using the SigB allelic type database of all species of Listeria (kindly provided by Prof. Martin Wiedmann and Dr. Renato H Orsi, Cornell University, Ithaca, New York). A phylogenetic tree was generated by MEGA5 program [30] using sequences of the allelic types found in this study with some closely related allelic types in the database mentioned above. The maximum likelihood method with gamma distribution was used for constructing the tree with 1,000 bootstrap replications [29,31].
Induction of Listeria prophage by mitomycin C and phage lysate preparation
The culture of an isolate was prepared by inoculating an isolated colony in 5 ml of Luria Bertani (LB) broth (Oxoid, UK) supplemented with 50 mM morpholinepropanesulfonic acid (MOPS), 1% (wt/vol) glucose, 10 mM CaCl2, and 10 mM MgCl2 (LB-MOPS-Glu-salts) [26]. The culture was incubated at 30°C (220 rpm) to reach an optical density (at 600 nm) of 0.4 to 0.5, and a 1 ml-aliquot was mixed with mitomycin C (Sigma-Aldrich, St Louis, USA) to a final concentration of 1 μg/ml (modified from [32]). The mixture was subsequently incubated for 7 h. Then, 200 μl of this mixture was later mixed with 100 μl of a given propagating host in a total volume of 2 ml of LB MOPS, followed by an incubation for 18 h at 30°C (220 rpm). The double layer technique was applied with the filtered lysate of the overnight co-culture following the procedure described by Vongkamjan et al. [26]. Plaque formation was observed as appearance of induced phage. An isolated plaque representing a distinct plaque morphology type (A: translucent plaque, rather round shape, Φ ≥ 1 mm; B: turbid at the edge, star-shape, Φ = 0.5–1 mm; C: turbid zone, Φ ≤ 0.5 mm; D: clear zone, round, Φ = 1 mm; E: turbid zone, tiny, Φ ≤ 0.2 mm) was selected for three-time-purification, followed by phage lysate preparation by the double layer method [26]. Titers of induced phages were determined by spotting 5μl of ten-fold serial dilutions of phage on the propagating host lawn. High titer of phage lysate was kept at 4°C for further analysis. The likelihood of prophage-carrying isolates among L. monocytogenes and Listeria spp. was determined as odd ratio with 95% confidence interval (95% CI) using a generalized linear model in R program version 3.1.2 (https://cran.r-project.org).
Host range determination of the induced phages
Phage host range determination was performed by spotting 5 μl of diluted phage representing 100×RTD (routine test dilution) [26], approximately 106–107 PFU/ml, on the 31 Listeria hosts mentioned above. Each spot on the lawn was examined and recorded for lysing (+) or no lysing (-) after overnight incubation at 30°C [26]. The experiment was carried out in triplicate. Then, a clustering analysis based on lysis ability of the induced phages against the tested hosts was performed using R program.
Estimation of phage genome size
Pulsed-Field Gel Electrophoresis (PFGE) analysis was used to estimate genome size of the phages as described previously [25,26,33]. Briefly, high-titer lysate of a given phage (107–109 PFU/ml) was used to prepare a plug with 1.3% low-melting-point agarose. PFGE analysis was performed on a CHEF-DR III system (Bio-Rad, Hercules, CA, USA) in 0.5×TBE buffer (1 M Tris, 0.5M EDTA and boric acid, pH 8.0) for 22 h with a 0.5 s to 5.0 s switch time, 6 V/cm, and an included angel of 120° [26]. Genome estimation was performed using Uvitec UVI-1D software (Uvitec Limited Co., Cambridge, UK) with the tool for molecular weight estimation.
Restriction enzyme analysis
A total of 19 phages representing each genome size group, but from different lysis groups and sources were selected for restriction enzyme analysis. DNA of the induced phages was extracted by phenol/chloroform as described previously [26]. Restriction analysis was performed using Fast digest HindIII (Thermo Fisher Scientific, MA, USA) and EcoRI (Vivantis, Selangor Darul Ehsan, Malaysia), following the manufacturers’ instructions. Two restriction profiles were considered different when at least one distinguishing band was present [32].
Transmission electron microscopy (TEM)
Examination of the phage morphology and family was performed with four induced phages selected to represent a given genome size group. A 3 μl-drop of freshly prepared lysate of a given phage (108 PFU/ml) was deposited onto carbon-coated copper grid. The grid was left to dry for 15 s, then slowly stained with 20 μl of uranyl acetate (2%, pH 4.5) [32,34]. The imaging was done at 160 kV with a transmission electron microscope JEM-2010, JEOL (Japan) at the Scientific Equipment Center, Prince of Songkla University, Hat Yai, Thailand.
Results
Classification of allelic types based on Listeria partial sigB sequences
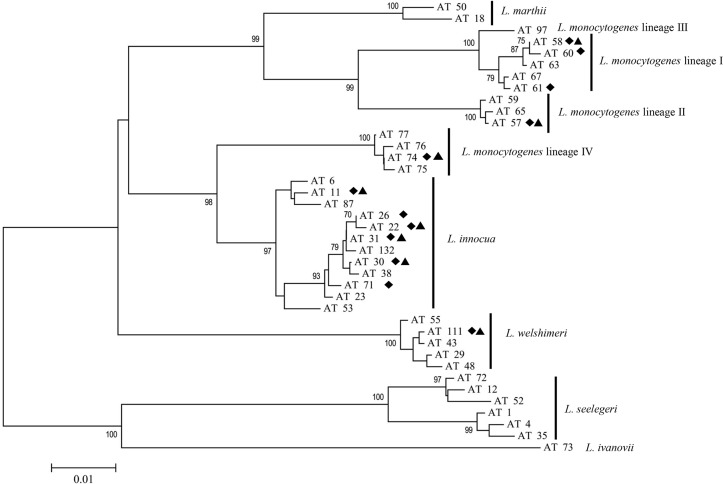
Overall, a total of 144 L. monocytogenes and Listeria spp. isolates were subjected to partial sigB sequencing analysis to classify them into allelic types (Table 3 and Fig 1). Classification of these isolates showed 12 allelic types, which represented lineages I, II or IV of L. monocytogenes, or other species, including L. innocua and L. welshimeri. In the 90 L. monocytogenes isolates used in this study, five allelic types were observed. Of which, AT 58, AT 60 and AT 74 were commonly found in the majority of isolates. Allelic types 58 and 60 represented L. monocytogenes lineage I while AT 74 represented L. monocytogenes lineage IV. In the 54 Listeria spp. isolates, AT 111 and AT 30 were common allelic types found in this study; these isolates could be classified as L. welshimeri and L. innocua, respectively.
Table 3. Presence of prophages in the isolates of L. monocytogenes and Listeria spp.
| Lineage/ species | SigB allelic type (AT) | No. of isolates | No. of lysogens | The likelihood of prophage-carrying isolates (95% Confidence interval) | |
|---|---|---|---|---|---|
| L. monocytogenes | 14.4 (4.9–35.4) | ||||
| Lineage I (n = 60) | AT 58 | 24 | 8 | ||
| AT 60 | 22 | - | |||
| AT 61 | 14 | - | |||
| Lineage II (n = 4) | AT 57 | 4 | 4 | ||
| Lineage IV (n = 26) | AT 74 | 26 | 1 | ||
| Listeria spp. | 18.5 (4.8–50.2) | ||||
| L. innocua (n = 27) | AT 11 | 6 | 1 | ||
| AT 22 | 3 | 1 | |||
| AT 26 | 1 | - | |||
| AT 30 | 13 | 2 | |||
| AT 31 | 3 | 2 | |||
| AT 71 | 1 | - | |||
| L. welshimeri (n = 27) | AT 111 | 27 | 4 | ||
Fig 1. Neighbor-joining tree describes the sigB allelic type (AT) of 144 isolates included in this study (♦) and some closely related allelic types from the database.
The maximum likelihood method and a gamma distribution were used to construct the tree with 1,000 bootstrap replications. Only bootstrap values ≥70% are presented on the tree. Triangles (▲) indicates that the allelic type contained prophage.
Distribution of prophages in Listeria isolates obtained from foods and food-related environments
Mitomycin C could induce the prophages in 23/144 (16.0%) of L. monocytogenes and Listeria spp. isolates (Table 3). These prophage-carrying isolates belonged to 8 out of 12 allelic types found among the tested isolates (Table 3 and Fig 1). The eight common allelic types associated with the occurrence of prophages included 3 ATs of L. monocytogenes [AT 58 (lineage I), AT 57 (lineage II), and AT 74 (lineage IV)] and 5 ATs of Listeria spp. [AT 11, AT 22, AT30, and AT 31 (L. innocua); and AT 111 (L. welshimeri)]. Overall, the likelihood of prophage-carrying isolates of L. monocytogenes was 14.4 (95% CI: 4.9–35.4) and 18.5 (95% CI: 4.8–50.2) for Listeria spp.
Based on the presence of distinct types of plaque morphology based on the size, shape and the turbidity of each examined plaque, 39 inducible phages were obtained from 23 lysogenic isolates (Table 4). The majority of phages (23 phages) were obtained from eight lysogens of L. monocytogenes lineage I. Other four phages were from four lysogens of lineage II and only one phage was from lysogens of lineage IV. Ten lysogens of Listeria spp. yielded 11 phages, including six phages from L. innocua and five phages from L. welshimeri. Among four L. monocytogenes hosts used as propagating hosts, two L. monocytogenes strains of serotype 4a were common hosts for phage propagation, while the strain FSL F2-695 was propagating host for phages from the lysogens of L. monocytogenes lineage I and L. innocua. The strain FSL J1-208 was suitable as the propagating host for phages from the lysogens of L. monocytogenes lineage I and L. welshimeri. The strain L. monocytogenes serotype 1/2a (Mack) was a suitable host for phages from the lysogens of L. monocytogenes lineage II. F2365 (serotype 4b) could be propagating host for phages from the lysogens of L. monocytogenes lineage I and IV. While one lysogen of lineage IV could yield a phage on only F2365 host, some lysogens such as PSU-KV-165LM and PSU-KV-167LM provided up to five phages in three propagating hosts.
Table 4. Distribution of induced Listeria phages on different propagating hosts.
| Lineage/ species | Lysogen IDa | Allelic type (AT) of lysogen | Induced phages IDb on each propagating host (genome size group) | |||
|---|---|---|---|---|---|---|
| Mack (1/2a) | FSL F2-695 (4a) | FSL J1-208 (4a) | F2365 (4b) | |||
| L. monocytogenes lineage I | ||||||
| 112LM | AT 58 | - | LP013 (4) | LP012 (1) | LP010/011 (1) | |
| 133LM | AT 58 | - | - | LP017 (2) | LP016 (2) | |
| 134LM | AT 58 | - | - | LP019 (2) | LP018 (2) | |
| 160LM | AT 58 | - | - | LP027/028 (1) | - | |
| 165LM | AT 58 | - | LP034 (3) | LP032/033 (1) | LP030/031 (1) | |
| 167LM | AT 58 | - | LP039 (4) | LP037/038 (1) | LP035/036 (1) | |
| 036LM | AT 58 | - | LP041 (4) | - | LP040 (2) | |
| 038LM | AT 58 | - | - | - | LP042 (2) | |
| L. monocytogenes lineage II | ||||||
| 148LM | AT 57 | LP022 (2) | - | - | - | |
| 149LM | AT 57 | LP023 (2) | - | - | - | |
| 150LM | AT 57 | LP024 (2) | - | - | - | |
| 159LM | AT 57 | LP026 (2) | - | - | - | |
| L. monocytogenes lineage IV | ||||||
| 218LM | AT 74 | - | - | - | LP047 (1) | |
| L. innocua | ||||||
| 143LS | AT 31 | - | LP020 (3) | - | - | |
| 145LS | AT 31 | - | LP021 (3) | - | - | |
| 152LS | AT 30 | - | LP025 (3) | - | - | |
| 199LS | AT 30 | - | LP045 (2) | - | - | |
| 192LS | AT 11 | - | LP044 (4) | - | - | |
| 200LS | AT 22 | - | LP046 (4) | - | - | |
| L. welshimeri | ||||||
| 104LS | AT 111 | - | - | LP009 (2) | - | |
| 130LS | AT 111 | LP014/015 (2) | - | - | - | |
| 164LS | AT 111 | - | - | LP029 (2) | - | |
| 181LS | AT 111 | - | - | LP043 (2) | - | |
aID of Listeria lysogen has a prefix of “PSU-KV-”.
bID of induced Listeria phages has a prefix of “PSU-VKH-”. Multiple induced phages listed are separated by “/”. Phages in bold indicates that were selected for restriction enzyme analysis.
Lysis ability of the induced phages against 31 Listeria hosts
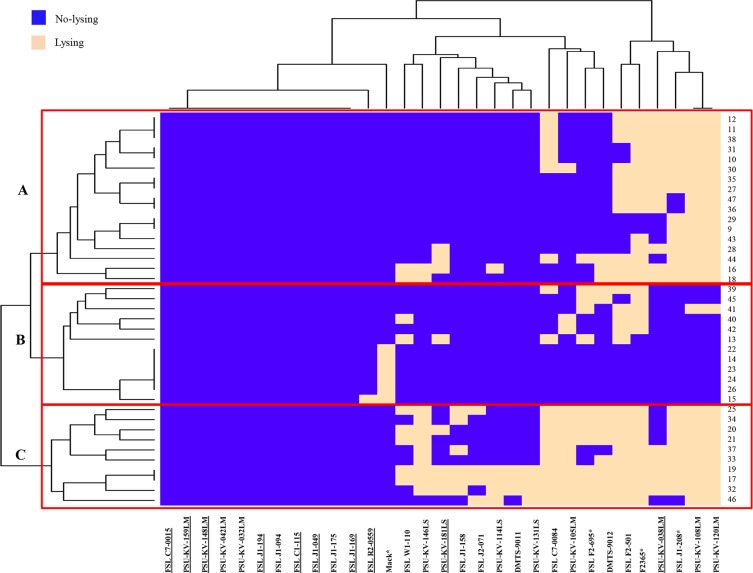
The host range of 39 induced phages was evaluated on 19 Listeria reference strains and 12 Listeria isolates from the collection of those isolates used in this study. These hosts were selected to represent 9 major L. monocytogenes serotypes and 5 distinct Listeria species. Clustering analysis based on the lysis similarity classified these 39 phages into 28 lysis profiles, presenting 3 major lysis groups (A, B and C) (Fig 2). Each lysis group included 10–17 induced phages. Group B contained those phages (n = 12) that were host-specific with the ability to lyse only 1–5 host strains (<16%) of L. monocytogenes serotype 4 and L. marthii. Phages (n = 17) in group A showed similar host range as those in group B (as host-specific phages), however, they could also lyse Mack (1/2a) and L. welshimeri. In comparison, phages (n = 10) in group C had a broader host range than those in groups A and B. These phages could lyse 10–18 hosts (32–58%) of L. monocytogenes serotype 4 and other Listeria species used as hosts, except L. seeligeri. Overall, the majority of phages (30/39 phages) could lyse hosts of L. monocytogenes serotype 4. Interestingly, 10/31 hosts resistant to the induced phages were lysogens.
Fig 2. Clustering analysis shows lysis capabilities of 39 induced Listeria phages against 31 tested hosts.
Blue represents no-lysing and beige represents lysing of a given host. Host strains are shown on the x-axis, while induced phages are shown in the y-axis. Clusters of the induced phages are designated A to C based on similarities of the lysis profiles on 31 hosts. The clustering was done using R-program v.3.1.2. Underlined hosts represent lysogenic hosts.
Estimated genome size of the induced phages
Genome estimation of the induced phages by PFGE classified them into four groups (Table 4), including group 1 (35±2 kb, 14 phages), group 2 (40±2 kb, 16 phages), group 3 (45±2 kb, 4 phages), and group 4 (50±2 kb, 5 phages). Most phages were in groups 1 and 2. Group 1 contained phages induced from lysogens of L. monocytogenes lineage I and the only lysogen of L. monocytogenes lineage IV, while group 2 contained phages induced from lysogens of all L. monocytogenes lineages and Listeria species found in this study (except L. monocytogenes lineage IV). In comparison, groups 3 and 4 included phages induced from lysogens of L. monocytogenes lineage I and L. innocua. Interestingly, many Listeria lysogens harbored different induced phages with different genome size representing different prophage types in their genomes.
Restriction analysis of the induced phages
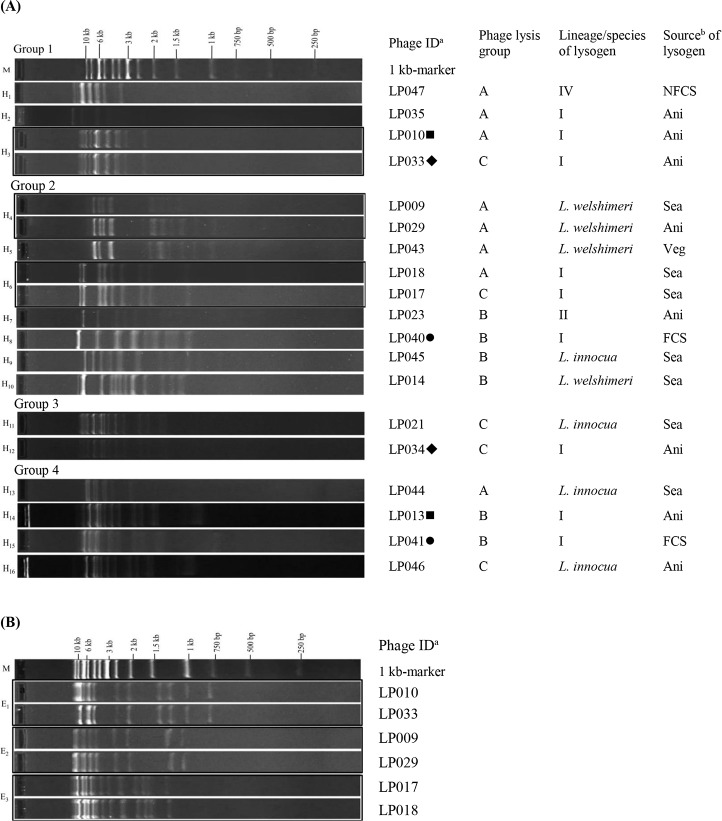
Restriction enzyme analysis using HindIII was performed with 19 representative phages, resulting in 16 restriction profiles (H1 to H16) (Fig 3). In each genome size group, 2–7 different restriction profiles were obtained. Seven different profiles were obtained in phage genome size group 2 (40±2 kb). The profiles H3 (group 1) and H6 (group 2) were both observed in two phages from the lysogens of L. monocytogenes lineage I obtained from the same source. For group 2, profile H4 was found in two phages (LP009 and LP029) from the lysogens of L. welshimeri obtained from different sources. Two and four distinct restriction profiles were observed in phages of genome groups 3 and 4, respectively.
Fig 3. Restriction analysis of induced Listeria phages with enzymes HindIII and EcoRI.
HindIII-restriction profiles of selected induced phages grouping by genome size (group 1 to 4) (Fig 3A). EcoRI-restriction profiles of induced phages that showed similar restriction profiles by HindIII (Fig 3B). a“M” is a 1-kb molecular maker. Induced phages obtained from a single lysogen are marked with the same symbol next to the phage ID. Phages within a box had the same restriction pattern. bRefer to Table 1 for abbreviations of the lysogen sources.
Phages with the same HindIII restriction profiles (H3, H4, or H6) were further analyzed by enzyme EcoRI. Results showed that two phages with identical HindIII profiles had identical EcoRI profiles (E1, E2 or E3). In summary, by using two restriction enzymes, these 19 phages could be classified into 16 restriction profiles, suggesting 16 different prophage types. Interestingly, up to seven prophage types were obtained from the lysogens of L. monocytogenes lineage I (AT 58). In addition, three and four prophage types were obtained from the lysogens of L. welshimeri (AT 111) and L. innocua (AT 11, AT 22, AT 30, AT 31), respectively.
TEM analysis of the induced phages
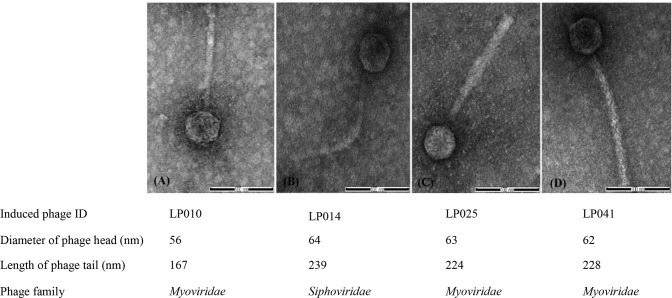
Morphology of four phages selected to represent the four genome size groups was examined by TEM (Fig 4). Morphology observation showed that three phages belonged to the Myoviridae family with an isometric head (diameter 56 to 63 nm) with long contractile tail (167 to 228 nm) with a sheath. These phages were from the lysogens of L. monocytogenes lineage I or L. innocua with the genome sizes 35±2 kb, 45±2 kb and 50±2 kb. Another phage was classified to the Siphoviridae family with a hexagonal head (diameter of 64 nm) and longer non-contractile tail (239 nm). This phage was from the lysogen of L. welshimeri and had genome size 40±2 kb.
Fig 4. Morphology of induced Listeria phages by TEM analysis.
Induced phages were stained with 2% uranyl acetate (pH 4.5) and visualized at a final magnification of 100kx. All panels are shown at the same scale with the scale bar indicating 100 nm.
Discussion
Distribution of prophages among L. monocytogenes, L. innocua, and L. welshimeri obtained from foods and food-related environments
A total of 39 induced phages were obtained from the 23/144 isolates of L. monocytogenes and Listeria spp. tested (16%). Prophages were detected in most allelic types found in this study (8/12) of L. monocytogenes lineage I, II and IV, L. innocua, and L. welshimeri. No isolates of L. monocytogenes in the tested collection belonged to lineage III, therefore the results lack information on the prophage distribution from isolates of this lineage.
Prophages were found to be absent or rare in five specific allelic types representing L. monocytogenes lineages I, IV and L. innocua. There is still limited information to explain why those allelic types are lack of presence of prophage. However, we speculate that the isolates of these lineages are particularly resistant to uptake of extraneous DNA as suggested previously [35]. Another possible reason could be the lack of prophage insertion sites for these lineages as opposed to L. monocytogenes lineage I and II [17,18]. Interestingly, finding here may link to the host range data; L. monocytogenes serotype 1/2 and 3 (mostly belonged to lineages I, II) are resistant to phages as those hosts could be lysogens containing prophage sequences that are homologous to the induced phages [36]. Another possibility could be appropriated propagating host in the induction as mentioned in previous studies [14,37]. This hypothesis was supported when different sets of prophages were obtained using different propagating hosts, even if those hosts represented serotype 4a (FSL F2-695 and FSL J1-208).
In addition, our study showed the likelihood of having prophages in L. monocytogenes isolates as 14.4 (95% CI: 4.9–35.4) and 18.5 (95% CI: 4.8–50.2) for the isolates of Listeria spp. (including L. innocua and L. welshimeri). It seems reasonable as L. innocua and L. welshimeri were reported that derived from L. monocytogenes through early evolutionary events of gene acquisition by phage transduction [38]. Therefore, isolates of these Listeria species may have more insertion site for prophages to incorporate into the host chromosomes. In addition, in this study, different prophage types were found among Listeria lysogens or even within a single lysogen. Similarly, genome analysis has previously revealed multiple prophages in Listeria genomes, especially in L. monocytogenes and L. innocua [17–19]
Induced Listeria phages appear to be host-specific with higher ability to lyse L. monocytogenes serotype 4 than other serotypes
Induced Listeria phages in this study showed different lysis profiles with 74% of these represented host-specific phages. A previous study revealed that lysogenic Listeria phages have narrower host range than isolated Listeria phages [39]. The majority of Listeria phages isolated from a turkey processing plant showed broad host ranges [40]. In related bacteria, phages obtained from lysogenic strains of Streptococcus iniae were reported to have narrow lytic spectrum [41]. This can be explained by the unique characteristic of induced phages as they have the ability to incorporate their genome in the host chromosome instead of lysing the host.
In this study, most induced phages could lyse the hosts of L. monocytogenes serotype 4. Similarly, Listeria phages from silage or turkey processing environments were highly susceptible to L. monocytogenes serotype 4 strains [26,40]. Moreover, the induced phages could not lyse L. monocytogenes serotype 1/2 and serotype 3 hosts. This is of interest since serotype 1/2a is linked to the increasing cases of listeriosis in the last decade [42,43]. Therefore, we speculated that prophages may facilitate the survival of Listeria hosts. Another potential support is that the differences in phage susceptibility between serotypes of L. monocytogenes can be explained by the different in structure of cell wall teichoic acids (WTA). This is because L. monocytogenes serotype 4 contains WTA with terminal glucose and galactose residues, which is important for phage adsorption [44,45] and further facilitation for phages to lyse the host.
Most Listeria species used as hosts for the host range determination were sensitive to the induced phages, except L. seeligeri. This may be because the L. seeligeri strain is a lysogen. We also observed that the induced phages were likely to be resistant to 10/31 lysogenic Listeria hosts. This supports the hypothesis that lysogens could have phage resistance in certain bacterial hosts, because the bacteria may harbor specific (pro)phage sequences that could increase their survival without being affected by phage with homologous sequence as reported in Oenococcus oeni [36]. However, sequencing analysis of the induced phage and its host is still needed to elucidate the phage resistance characteristics.
Induced phages show highly similar genome size as previously reported temperate Listeria phages, but rather high genetic diversity and belong to two common phage families
The induced phages in this study showed genome size ranged from 35±2 kb to 50±2 kb and were classified into four groups. Similarly, genomes of previously reported temperate Listeria phages also had sizes from 35 kb to 48 kb [46–48]. Temperate phages typically have smaller genomes, that may contain only the necessary sequences for their replication and encapsulation [49,50]. Only the basic, important genes coding for six main functional modules were present in the genomes of temperate phages [47,51]. However, genomes of lytic phage contained a number of additional gene coding sequences with no function [34].
Restriction analysis of 19 selected phages using enzymes HindIII and EcoRI resulted in 16 restriction profiles, suggesting considerable diversity of prophages in the genome of Listeria lysogens. The diversity of prophages may contribute to the variation of host genomes by prophage incorporation as mentioned in a previous study [18]. Another study has also shown that the differences in a 42-kb-prophage sequence could differentiate four examined L. monocytogenes strains [52]. In this current study, seven and three prophage types were found in a single allelic type (AT 58 and AT 111, respectively). This suggests the usefulness of prophage to the classification of Listeria isolates of the same allelic types.
Two common families Myoviridae and Siphoviridae were observed among the induced phages in this study. LP014 belonged to Siphoviridae family with long, non-contractile tails, which is the most common among phages (60%) [53]. The morphological characteristics revealed that our Siphoviridae phage was similar to the Listeria phages LP-032-2 and LP-032-3 with a head diameter of 53–55 nm and a tail length of 160–297 nm reported previously [34].
In summary, this is the first study that investigated the distribution of phages induced from Listeria isolates of different allelic types of distinct L. monocytogenes lineages or Listeria species. Characterization of induced phages allows us to better understand their diversity and lysis ability against Listeria hosts representing different L. monocytogenes serotypes and distinct Listeria species. Diversity of prophage may have contributed to the genetic diversity of Listeria spp. isolated from foods and food-related environments in Thailand. Recombination and mosaicism caused by prophages in Listeria genomes may occur and gene transfer may be affected and could later drive host survival and fitness in foods or food-associated environments.
Acknowledgments
We would like to acknowledge Prof. Martin Wiedmann and Dr. Renato H Orsi (Cornell University, Ithaca, New York) for providing the database of Listeria SigB allelic types.
Data Availability
All relevant data are within the paper.
Funding Statement
This project is financially supported by the Higher Education Research Promotion and the Thailand’s Education Hub for Southern Region of ASEAN Countries Project Office of the Higher Education Commission (to HTKV). Funding from Prince of Songkla University (AGR600584S; to KV) and the TRF Distinguished Research Professor Grant (to SB) is also acknowledged.
References
- 1.Swaminathan B, Gerner-Smidt P. The epidemiology of human listeriosis. Microbes Infect. 2007;9: 1236–1243. 10.1016/j.micinf.2007.05.011 [DOI] [PubMed] [Google Scholar]
- 2.Graves LM, Helsel LO, Steigerwalt AG, Morey RE, Daneshvar MI, Roof SE, et al. Listeria marthii sp. nov., isolated from the natural environment, Finger Lakes National Forest. Int J Syst Evol Microbiol. 2010;60: 1280–1288. 10.1099/ijs.0.014118-0 [DOI] [PubMed] [Google Scholar]
- 3.Bertsch D, Rau J, Eugster MR, Haug MC, Lawson PA, Lacroix C, et al. Listeria fleischmannii sp. nov., isolated from cheese. Int J Syst Evol Microbiol. 2013;63: 526–532. 10.1099/ijs.0.036947-0 [DOI] [PubMed] [Google Scholar]
- 4.Halter EL, Neuhaus K, Scherer S. Listeria weihenstephanensis sp. nov., isolated from the water plant Lemna trisulca taken from a freshwater pond. Int J Syst Evol Microbiol. 2013;63: 641–647. 10.1099/ijs.0.036830-0 [DOI] [PubMed] [Google Scholar]
- 5.Vongkamjan K, Fuangpaiboon J, Jirachotrapee S, Turner MP. Occurrence and diversity of Listeria spp. in seafood processing plant environments. Food Control. Elsevier Ltd; 2015;50: 265–272. 10.1016/j.foodcont.2014.09.001 [DOI] [Google Scholar]
- 6.Vongkamjan K, Fuangpaiboon J, Turner MP, Vuddhakul V. Various ready-to-eat products from retail stores linked to occurrence of diverse Listeria monocytogenes and Listeria spp. isolates. J Food Prot. 2016;79: 239–245. 10.4315/0362-028X.JFP-15-361 [DOI] [PubMed] [Google Scholar]
- 7.Tolvanen R, Hellström S, Elsser D, Morgenstern H, Björkroth J, Korkeala H. Survival of Listeria monocytogenes strains in a dry sausage model. J Food Prot. 2008;71: 1550–1555. 10.4315/0362-028X-71.8.1550 [DOI] [PubMed] [Google Scholar]
- 8.Burgess CM, Gianotti A, Gruzdev N, Holah J, Knøchel S, Lehner A, et al. The response of foodborne pathogens to osmotic and desiccation stresses in the food chain. Int J Food Microbiol. The Authors; 2016;221: 37–53. 10.1016/j.ijfoodmicro.2015.12.014 [DOI] [PubMed] [Google Scholar]
- 9.Palumbo SA, Williams AC. Resistance of Listeria monocytogenes to freezing in foods. Food Microbiol. 1991;8: 63–68. 10.1016/0740-0020(91)90017-V [DOI] [Google Scholar]
- 10.Kutter E, Sulakvelidze A. Bacteriophages: Biology and applications. 1st ed Boca Raton: CRC Press; 2005. [Google Scholar]
- 11.Fortier L-C, Sekulovic O. Importance of prophages to evolution and virulence of bacterial pathogens. Virulence. 2013;4: 354–365. 10.4161/viru.24498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Casjens S. Prophages and bacterial genomics: What have we learned so far? Mol Microbiol. 2003;49: 277–300. 10.1046/j.1365-2958.2003.03580.x [DOI] [PubMed] [Google Scholar]
- 13.Bondy-denomy J, Qian J, Westra ER, Buckling A, Guttman DS, Davidson AR, et al. Prophages mediate defense against phage infection through diverse mechanisms. ISME J. Nature Publishing Group; 2016;10: 2854–2866. 10.1038/ismej.2016.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferreira V, Barbosa J, Stasiewicz M, Vongkamjan K, Moreno Switt A, Hogg T, et al. Diverse geno- and phenotypes of persistent Listeria monocytogenes isolates from fermented meat sausage production facilities in Portugal. Appl Environ Microbiol. 2011;77: 2701–2715. 10.1128/AEM.02553-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rabinovich L, Sigal N, Borovok I, Nir-Paz R, Herskovits A a. Prophage excision activates Listeria competence genes that promote phagosomal escape and virulence. Cell. 2012;150: 792–802. 10.1016/j.cell.2012.06.036 [DOI] [PubMed] [Google Scholar]
- 16.Verghese B, Lok M, Wen J, Alessandria V, Chen Y, Kathariou S, et al. comK prophage junction fragments as markers for Listeria monocytogenes genotypes unique to individual meat and poultry processing plants and a model for rapid niche-specific adaptation, biofilm formation, and persistence. Appl Environ Microbiol. 2011;77: 3279–3292. 10.1128/AEM.00546-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuenne C, Billion A, Mraheil MA, Strittmatter A, Daniel R, Goesmann A, et al. Reassessment of the Listeria monocytogenes pan-genome reveals dynamic integration hotspots and mobile genetic elements as major components of the accessory genome. BMC Genomics. 2013;14: 47 10.1186/1471-2164-14-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.den Bakker HC, Desjardins CA, Griggs AD, Peters JE, Zeng Q, Young SK, et al. Evolutionary dynamics of the accessory genome of Listeria monocytogenes. Brissette CA, editor. PLoS One. 2013;8: e67511 10.1371/journal.pone.0067511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nelson KE, Fouts DE, Mongodin EF, Ravel J, DeBoy RT, Kolonay JF, et al. Whole genome comparisons of serotype 4b and 1/2a strains of the food-borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Res. 2004;32: 2386–2395. 10.1093/nar/gkh562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rychli K, Wagner EM, Ciolacu L, Zaiser A, Tasara T, Wagner M, et al. Comparative genomics of human and non-human Listeria monocytogenes sequence type 121 strains. PLoS One. 2017;12: e0176857 10.1371/journal.pone.0176857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lemaiıtre J-P, Delcourt A, Rousset A. Optimization of the detection of bacteriophages induced from Listeria sp. Lett Appl Microbiol. 1997;24: 51–54. 10.1046/j.1472-765X.1997.00346.x [DOI] [PubMed] [Google Scholar]
- 22.Jiang S, Paul J. Occurrence of lysogenic bacteria in marine microbial communities as determined by prophage induction. Mar Ecol Prog Ser. 1996;142: 27–38. 10.3354/meps142027 [DOI] [Google Scholar]
- 23.López E, Domenech A, Ferrándiz M-J, Frias MJ, Ardanuy C, Ramirez M, et al. Induction of prophages by fluoroquinolones in Streptococcus pneumoniae: implications for emergence of resistance in genetically-related clones. PLoS One. 2014;9: e94358 10.1371/journal.pone.0094358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loessner MJ, Goeppl S, Busse M. Comparative inducibility of bacteriophage in naturally lysogenic and lysogenized strains of Listeria spp. by UV light and Mitomycin C. Lett Appl Microbiol. 1991;12: 196–199. 10.1111/j.1472-765X.1991.tb00538.x [DOI] [Google Scholar]
- 25.Vongkamjan K, Benjakul S, Vu HTK, Vuddhakul V. Longitudinal monitoring of Listeria monocytogenes and Listeria phages in seafood processing environments in Thailand. Food Microbiol. Elsevier Ltd; 2017;66: 11–19. 10.1016/j.fm.2017.03.014 [DOI] [PubMed] [Google Scholar]
- 26.Vongkamjan K, Moreno Switt A, den Bakker HC, Fortes ED, Wiedmann M. Silage collected from dairy farms harbors an abundance of listeriaphages with considerable host range and genome size diversity. Appl Environ Microbiol. 2012;78: 8666–8675. 10.1128/AEM.01859-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doumith M, Buchrieser C, Glaser P, Jacquet C, Martin P. Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J Clin Microbiol. 2004;42: 3819–3822. 10.1128/JCM.42.8.3819-3822.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nightingale KK, Windham K, Wiedmann M. Evolution and molecular phylogeny of Listeria monocytogenes isolated from human and animal listeriosis cases and foods. J Bacteriol. 2005;187: 5537–5551. 10.1128/JB.187.16.5537-5551.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liao J, Wiedmann M, Kovac J. Genetic stability and evolution of the sigB allele, used for Listeria sensu stricto subtyping and phylogenetic inference. Elkins CA, editor. Appl Environ Microbiol. 2017;83: e00306–17. 10.1128/AEM.00306-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28: 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nale JY, Shan J, Hickenbotham PT, Fawley WN, Wilcox MH, Clokie MRJ. Diverse temperate bacteriophage carriage in Clostridium difficile 027 strains. PLoS One. 2012;7: e37263 10.1371/journal.pone.0037263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fortier L-C, Moineau S. Morphological and genetic diversity of temperate phages in Clostridium difficile. Appl Environ Microbiol. 2007;73: 7358–7366. 10.1128/AEM.00582-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moreno Switt AI, den Bakker HC, Vongkamjan K, Hoelzer K, Warnick LD, Cummings KJ, et al. Salmonella bacteriophage diversity reflects host diversity on dairy farms. Food Microbiol. Elsevier Ltd; 2013;36: 275–285. 10.1016/j.fm.2013.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Denes T, Vongkamjan K, Ackermann HW, Moreno Switt AI, Wiedmann M, den Bakker HC. Comparative genomic and morphological analyses of Listeria phages isolated from farm environments. Appl Environ Microbiol. 2014;80: 4616–4625. 10.1128/AEM.00720-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goerke C, Pantucek R, Holtfreter S, Schulte B, Zink M, Grumann D, et al. Diversity of prophages in dominant Staphylococcus aureus clonal lineages. J Bacteriol. 2009;191: 3462–3468. 10.1128/JB.01804-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poblet-Icart M, Bordons A, Lonvaud-Funel A. Lysogeny of Oenococcus oeni (syn. Leuconostoc oenos) and study of their induced bacteriophages. Curr Microbiol. 1998;36: 365–369. 10.1007/s002849900324 [DOI] [PubMed] [Google Scholar]
- 37.Domelier A-S, van der Mee-Marquet N, Sizaret P-Y, Hery-Arnaud G, Lartigue M-F, Mereghetti L, et al. Molecular characterization and lytic activities of Streptococcus agalactiae bacteriophages and determination of lysogenic-strain features. J Bacteriol. 2009;191: 4776–4785. 10.1128/JB.00426-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hain T, Steinweg C, Kuenne CT, Billion A, Ghai R, Chatterjee SS, et al. Whole-genome sequence of Listeria welshimeri reveals common steps in genome reduction with Listeria innocua as compared to Listeria monocytogenes. J Bacteriol. 2006;188: 7405–7415. 10.1128/JB.00758-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loessner MJ, Busse M. Bacteriophage typing of Listeria species. Appl Environ Microbiol. 1990;56: 1912–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim J-W, Siletzky RM, Kathariou S. Host ranges of Listeria-specific bacteriophages from the turkey processing plant environment in the United States. Appl Environ Microbiol. 2008;74: 6623–6630. 10.1128/AEM.01282-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wright EE, Elliman JR, Owens L. Induction and characterization of lysogenic bacteriophages from Streptococcus iniae. J Appl Microbiol. 2013;114: 1616–1624. 10.1111/jam.12192 [DOI] [PubMed] [Google Scholar]
- 42.Mammina C, Parisi A, Guaita A, Aleo A, Bonura C, Nastasi A, et al. Enhanced surveillance of invasive listeriosis in the Lombardy region, Italy, in the years 2006–2010 reveals major clones and an increase in serotype 1/2a. BMC Infect Dis. 2013;13: 152 10.1186/1471-2334-13-152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marini E, Magi G, Vincenzi C, Manso E, Facinelli B. Ongoing outbreak of invasive listeriosis due to serotype 1/2a Listeria monocytogenes, Ancona province, Italy, January 2015 to February 2016. Eurosurveillance. 2016;21: 30217 10.2807/1560-7917.ES.2016.21.17.30217 [DOI] [PubMed] [Google Scholar]
- 44.Eugster MR, Loessner MJ. Rapid analysis of Listeria monocytogenes cell wall teichoic acid carbohydrates by ESI-MS/MS. Riggs PD, editor. PLoS One. 2011;6: e21500 10.1371/journal.pone.0021500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wendlinger G, Loessner MJ, Scherer S. Bacteriophage receptors on Listeria monocytogenes cells are the N-acetylglucosamine and rhamnose substituents of teichoic acids or the peptidoglycan itself. Microbiology. 1996;142: 985–992. 10.1099/00221287-142-4-985 [DOI] [PubMed] [Google Scholar]
- 46.Dorscht J, Klumpp J, Bielmann R, Schmelcher M, Born Y, Zimmer M, et al. Comparative genome analysis of Listeria bacteriophages reveals extensive mosaicism, programmed translational frameshifting, and a novel prophage insertion site. J Bacteriol. 2009;191: 7206–7215. 10.1128/JB.01041-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Loessner MJ, Inman RB, Lauer P, Calendar R. Complete nucleotide sequence, molecular analysis and genome structure of bacteriophage A118 of Listeria monocytogenes: Implications for phage evolution. Mol Microbiol. 2000;35: 324–340. 10.1046/j.1365-2958.2000.01720.x [DOI] [PubMed] [Google Scholar]
- 48.Zimmer M, Sattelberger E, Inman RB, Calendar R, Loessner MJ. Genome and proteome of Listeria monocytogenes phage PSA: An unusual case for programmed + 1 translational frameshifting in structural protein synthesis. Mol Microbiol. 2003;50: 303–317. 10.1046/j.1365-2958.2003.03684.x [DOI] [PubMed] [Google Scholar]
- 49.Desiere F, Lucchini S, Canchaya C, Ventura M, Brüssow H. Comparative genomics of phages and prophages in lactic acid bacteria. Antonie Van Leeuwenhoek. 2002;82: 73–91. 10.1023/A:1020676825358 [DOI] [PubMed] [Google Scholar]
- 50.Klumpp J, Loessner MJ. Listeria phages: Genomes, evolution, and application. Bacteriophage. 2013;3: e26861 10.4161/bact.26861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goh S, Ong PF, Song KP, Riley T V, Chang BJ. The complete genome sequence of Clostridium difficile phage ΦC2 and comparisons to ΦCD119 and inducible prophages of CD630. Microbiology. 2007;153: 676–685. 10.1099/mic.0.2006/002436-0 [DOI] [PubMed] [Google Scholar]
- 52.Orsi RH, Borowsky ML, Lauer P, Young SK, Nusbaum C, Galagan JE, et al. Short-term genome evolution of Listeria monocytogenes in a non-controlled environment. BMC Genomics. 2008;9: 539 10.1186/1471-2164-9-539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guttman B, Raya R, Kutter E. Basic phage biology In: Kutter E, Sulakvelidze A, editors. Bacteriophages: Biology and applications. 1st ed Florida, USA: CRC Press; 2005. pp. 29–66. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.