Abstract
Background
Hypoxia suppresses global protein production, yet certain essential proteins are translated through alternative pathways to survive under hypoxic stress. Translation via the internal ribosome entry site (IRES) is a means to produce proteins under stress conditions such as hypoxia; however, the underlying mechanism remains largely uncharacterized.
Methods
Proteomic and bioinformatic analyses were employed to identify hnRNPM as an IRES interacting factor. Clinical specimens and mouse model of tumorigenesis were used for determining the expression and correlation of hnRNPM and its target gene. Transcriptomic and translatomic analyses were performed to profile target genes regulated by hnRNPM.
Findings
Hypoxia increases cytosolic hnRNPM binding onto its target mRNAs and promotes translation initiation. Clinical colon cancer specimens and mouse carcinogenesis model showed that hnRNPM is elevated during the development of colorectal cancer, and is associated with poor prognosis. Genome-wide transcriptomics and translatomics analyses revealed a unique set of hnRNPM-targeted genes involved in metabolic processes and cancer neoplasia are selectively translated under hypoxia.
Interpretation
These data highlight the critical role of hnRNPM-IRES-mediated translation in transforming hypoxia-induced proteome toward malignancy.
Fund
This work was supported by the Ministry of Science and Technology, Taiwan (MOST 104–2320-B-006-042 to HSS and MOST 105–2628-B-001-MY3 to TMC).
Keywords: Hypoxia, Colon cancer, IRES, Translational reprogramming, Translatomics, Tumorigenesis
Research in Context.
Evidence before this study
Hypoxia prevents cap-dependent translation initiation and leads to a decrease in global translation rates. However, essential proteins with important physiological functions need to be translated to ensure cell survival and growth in unfavorable conditions. Translation via the internal ribosome entry site has been discovered under hypoxia; however, the underlying mechanism remains largely uncharacterized.
Added value of this study
The finding that hnRNPM promotes IRES-containing mRNA translation under hypoxia fills a critical missing piece of the fact that critical genes are actively translated under low oxygen condition. A unique set of genes involved in metabolic processes and cancer neoplasia are selectively translated under hypoxia via hnRNPM-dependent mechanism.
Implications of all the available evidence
Genome-wide profiling of the translational signature of hnRNPM demonstrates that the hnRNPM-IRES-mediated protein production is involved in cancer development, which provides a new direction in searching for anti-tumor therapies.
Alt-text: Unlabelled Box
1. Introduction
Because fast expanding tumor cells outgrow their vasculatures and cause regional low oxygen tension, hypoxia is a commonly known pathophysiological feature in many solid tumors [1]. As hypoxia inhibited mTORC1 activity results in increases the association of dephosphorylated eukaryotic translation initiation factor 4E (eIF4E)-binding protein 1 (4E-BP1) with the cap-binding protein eIF4E, the low oxygen condition often prevents cap-dependent translation initiation and leads to a decrease in global translation rates [1]. On the other hand, certain essential proteins, with important physiological functions which allow cells to adapt to hypoxic conditions and to acquire sufficient oxygen and energy, are translated through IRES- [2] or an eIF4E2-dependent [3] manner. This translational switch is an essential mechanism for cancer cell survival and growth in unfavorable conditions. In contrast to the well documented mechanism that triggers translation through the use of cap-binding protein eIF4E2 and the O2-regulated hypoxia-inducible factor (HIF)-2α [4,5], the molecular details of how hypoxia activates IRES-mediated translation is largely unknown.
The molecular mechanisms of ribosome recruitment and translation initiation on cellular IRES are still unclear. A growing list of IRES trans-acting factors (ITAFs) has been identified to control IRES-mediated translation in response to different stress conditions in a target-specific manner [6,7]. For example, the polypyrimidine tract-binding protein 1 (PTBP1) is known to stimulate IRES-mediated translation of HIF-1α when oxygen supply is limited and the heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1) is an ITAF which controls cell type or cell status-specific expression of fibroblast growth factor 2 (FGF2) isoforms [8], resulting in different subcellular localizations and functions of FGF2. It has been suggested that these cellular IRESs also rely on regional RNA organization and the ITAFs to recruit translation machinery [9]. The characterization at the molecular level of IRES-mediated translation machinery will open new opportunities to better control protein production for mRNAs using this specialized mechanism of translation initiation under various stress conditions such as ER stress, viral infection, and hypoxia.
Human fibroblast growth factor 9 (FGF9) is a potent mitogen involved in many physiological processes including endometriosis [10], neuronal degeneration [11], and various cancers including prostate, brain, lung, and colorectal cancers [[12], [13], [14], [15]]. This study aims to investigate the mechanism underlying the hypoxia-induced, IRES-mediated translational switch using FGF9 as a model. Herein, we identified hnRNPM as a novel and pivotal ITAF to regulated translational switch under hypoxia. The novel hnRNPM-IRES-mediated translation activates a specific set of mRNAs that are involved in promoting tumorigenesis, which demonstrates a dynamic and multi-dimensional mechanism of mRNA translation is utilized by hypoxic cancer cells.
2. Materials and methods
2.1. Clinical samples and cell lines
Paired normal and tumor specimens from 29 patients with colorectal cancer (CRC) employed in our previous study [12] as well as tissue slides from 220 paired CRC cases arranged into 11 tissue arrays were used to study the control of hnRNPM on FGF9 expression. This study was approved by the Clinical Research Ethics Committee at National Cheng Kung University Medical Center, and informed consent was obtained from all subjects who participated in this study.
HEK293 and colon cancer cell lines used in this study were routinely maintained according to the guidelines from ATCC and the detailed information is shown in supplementary Table S2. For hypoxia treatment, the cells were incubated for 9 h at 37 °C in a hypoxic chamber with 1% O2.
2.2. In vitro transcription
All constructs were made based on the human cDNA sequence of FGF9 (D14838.1) and FGF2 (NM_002006.2). The cloning strategy was described previously [12]. DNA templates for the synthesis of the FGF9 and FGF2 RNA probes were generated. Plasmid DNAs were linearized by NcoI restriction enzyme (NEB, Massachusetts, UK). Two microgram of linearized DNA was used as the template for RNA probe synthesis. For biotinylated RNA, the Riboprobe systems (Promega, Madison, WI, USA) were used and mixed with the biotin-labeled dUTP (Roche Diagnostics, Indianapolis, IN, USA). The reaction mixture was incubated at 37 °C for 1.5 h, DNA template was digested by RQ1 DNase (Promega, Madison, WI, USA) for 15 min. The RNA probes were purified by phenochloroform precipitation.
2.3. RNA pull-down and protein identification
Biotinylated RNAs containing full length (FL) IRES only (IRES), or 5’UTR sequences without IRES (ΔIRES) sequences were in vitro transcribed (Riboprobe systems, Promega) and used in pull-down assays. The differentially stained bands were analyzed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometer (MALDI-TOF MS). The Mass Spec data were uploaded to ProteomeXchange (http://www.proteomexchange.org/) under the accession number of PXD012892.
In parallel, the protein tryptic digests were fractionated using a nano-UPLC system (nanoACQUITY UPLC, Waters, Milford, MA) coupled to an ion trap mass spectrometer (LTQ Orbitrap Discovery Hybrid FTMS, Thermo Fisher Scientific) equipped with an electrospray ionization source. The raw data were searched against the NCBI-nr database (18132328 sequences, 6219145704 residues) using the Mascot search engine (Matrix Science) with the Proteome Discoverer 1.4 interface (Thermo Fisher Scientific).
2.4. Transient transfection and luciferase reporter assay
HEK293 cells were transfected with each plasmid construct (0.5 μg) plus plasmids carrying Renilla luciferase or β-galactosidase as an internal control. The cells were incubated for 24 h and harvested by adding 100 μl of reporter lysis buffer (Luciferase Assay System; Promega). The activity of firefly and Renilla luciferase in cell lysates was measured using a dual-luciferase reporter assay system (Promega) and TD20/20 luminometer from Turner BioSystems. β-galactosidase activity was assayed using a β-galactosidase enzyme assay system (Promega).
2.5. Overexpression and knockdown assays
The hnRNPM expression vector was purchased from Origene and the pCMV5 empty vector was used as negative control. In addition, Pseudotyped lentiviruses containing short hairpin RNA (shRNA) sequence against hnRNPM (TRCN0000001246, TRCN0000350395) were obtained from the RNAi core facility at the Academia Sinica (http://rnai.genmed.sinica.edu.tw/about) and were used to knockdown hnRNPM expression. Lentiviruses containing shRNA against luciferase (TRCN0000231693) was used as control.
2.6. Quantitative real-time PCR (RT-qPCR)
RNA was isolated and reverse-transcribed into cDNA using the High-Capacity cDNA Reverse Transcription Kits (Applied Biosystems). The cDNA was subjected to conventional PCR or quantitative real-time PCR amplification. The TaqMan assays for FGF9, PLOD2, and ACTB (Applied Biosystems) were performed and the levels of FGF9 and PLOD2 mRNA were measured using the 2-ΔΔCt method, and normalized to the expression levels of ACTB. In addition, SYBR qPCR assays were developed to measure relative amount of CYR61 mRNA and 18S rRNA. Information for the primers and probes designed for these assays is given in Supplementary Table S4.
2.7. RNA immunoprecipitation (RIP) and RIP sequencing
RNA immunoprecipitation (RIP) assay was performed using EZ-Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Millipore). The isolated RNAs were reverse-transcribed to cDNA for RT-PCR or high-throughput sequencing (RIP-Seq). The RIP was conducted under the use of RNase inhibitor to protect RNA from RNase degradation (Recombinant RNasin® Ribonuclease Inhibitor, Promega Cat. No. N2515). In brief, cells were lysed and incubated with hnRNPM antibody (sc-20,002; 5 μg) or control IgG (SC-2025; 5 μg) conjugated with magnetic beads (50 μl) for 4 h. The protein-RNA complexes were immunoprecipitated and magnetically separated. The RNAs were isolated by RNeasy mini kit (QIAGEN, Hilden, Germany), and treated with a DNA-free kit (Ambion) to eliminate any DNA contamination.
2.8. Polysome profiling and polysome-bound RNA sequencing
Polysomal fraction assay was performed according to our pervious study [12]. In parallel, puromycin treatment was performed with 2 μg/ml puromycin for 4 h in HEK293 cells prior to sucrose gradient sedimentation. For the genome-wide translatomic analysis, RNAs isolated from the fractions 8–11 were pooled to form the polysome-bound samples. The pooled polysome samples from normoxic and hypoxic conditions were used for the preparation of fragment libraries, followed by high-throughput sequencing using a SOLiD™ 5500xl sequencer (Applied Biosystems, Foster City, CA, USA). The raw reads were further analyzed using LifeScope™ Genomic Analysis Software (version 2.5) and mapped to human genome (hg19) released from UCSC database. To identify the differentially polysome-bound mRNAs, the mapped profiles from duplicated experiments were analyzed using the Partek Genomics Suite (version 4.9, http://www.partek.com/).
2.9. Western blotting, immunohistochemistry, immunofluorescence staining and enzyme-linked immunosorbent assay
Procedures of Western blotting, immunohistochemistry, immunofluorescence staining and Enzyme-linked immunosorbent assay were performed routinely in the lab using standard protocols. Antibodies used in this study are listed in Supplementary Table S3.
2.10. RNA-protein network analyses
RNA-Protein Network Analyses were performed on translationally upregulated, hnRNPM-target transcripts using the Database for Annotation, Visualization and Integrated Discovery (DAVID,v6.7, https://david.ncifcrf.gov/tools.jsp) web-accessible programs [16]. Only enriched fold changes >2 and P < .05 were considered as significant pathway/process presented in the dataset.
2.11. In vivo AOM/DSS-induced colon carcinogenesis mouse model
On day 1, the male B6 mice in the treatment group (n = 6 mice/time point) were injected with azoxymethane (AOM) working solution (10 mg/kg body weight) while the control mice (n = 6 mice/time point) received sterile isotonic saline injection. The dextran sodium sulfate (DSS, MP Biomedicals, Santa Ana, CA, USA)-containing water (2%) was given at weeks 1, 4, and 7 to mice in the AOM group. Mice were sacrificed at 1, 2, 3, 4, 8, 12, 16, and 20 weeks after AOM injection. The study was approved by the Animal Ethics Committee at National Cheng Kung University and all experiments applied to the animals have followed the relevant regulatory standards.
2.12. Statistical analysis
Data were analyzed with Student's t-test or one-way ANOVA followed by Tukey's multiple test using GraphPad Prism 5.0 (GraphPad Software). Kaplan-Meier estimator and Mann-Whitney test were used for survival and non-parametric analysis, respectively. All data are presented as the means ± SEM and the significance was set at P < .05.
3. Results
3.1. hnRNPM is a novel IRES-binding protein
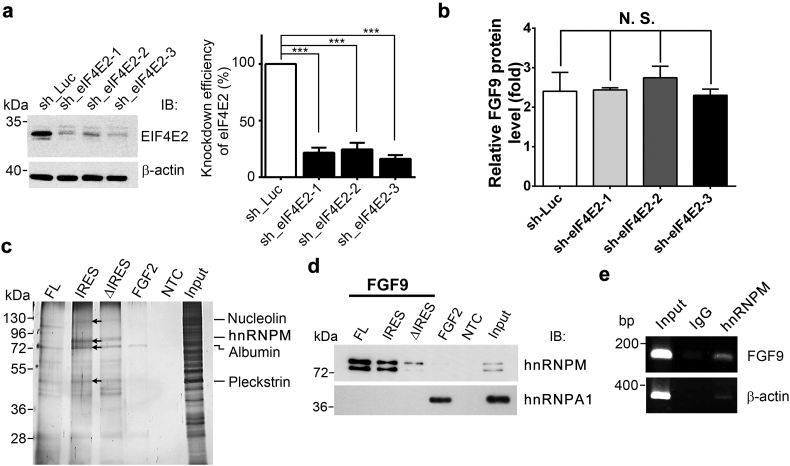
We have previously shown that FGF9 is overexpressed in colon cancer and contributes to treatment failure of patients [12]. To determine the relative contributions of cap-dependent and -independent pathways to FGF9 protein production, we first knocked down eIF4E2, a key factor that regulates cap-dependent translation, in the HEK293 cells and measured the amount of FGF9 protein under hypoxia (Fig. 1a and Fig. S1a). Knockdown of eIF4E2 did not affect the FGF9 protein levels (Fig. 1b), indicating that cap-dependent translation contributes very little to the translational activation of FGF9 protein in low oxygen conditions. We then sought to identify the protein that binds to FGF9 IRES and controls FGF9 protein synthesis in hypoxia. RNA pulled-down followed by immunoblotting results showed no interactions between FGF9 5’UTR (FL)/IRES RNAs and the known hypoxia-responsible ITAFs, such as PTBP1 and hnRNPA1 (Fig. S1b), suggesting unidentified ITAFs may interact with FGF9-IRES under hypoxia. To identify the proteins that bind to FGF9-IRES, the RNA-protein complexes were visualized by sliver staining (Fig. 1c), followed by cutting specific bands for protein identification using matrix-assisted laser desorption/ionization time-of-flight mass spectrometer (MALDI-TOF MS). Four proteins, nucleolin (~110KD), hnRNPM (~80kD), albumin (~72kD), and pleckstrin (~45kD) were identified (Table S1). In parallel, the digested protein fragments were analyzed using an ion trap mass spectrometer to obtain matched peptide sequences. Results from the ion trap mass spectrometry showed a match of 5 peptides to just one protein, the human hnRNPM. As only the hnRNPM was confirmed with Linear Trap Quadropole (LTQ)-Orbitrap analysis, we then focused on the hnRNPM for further investigation.
Fig. 1.
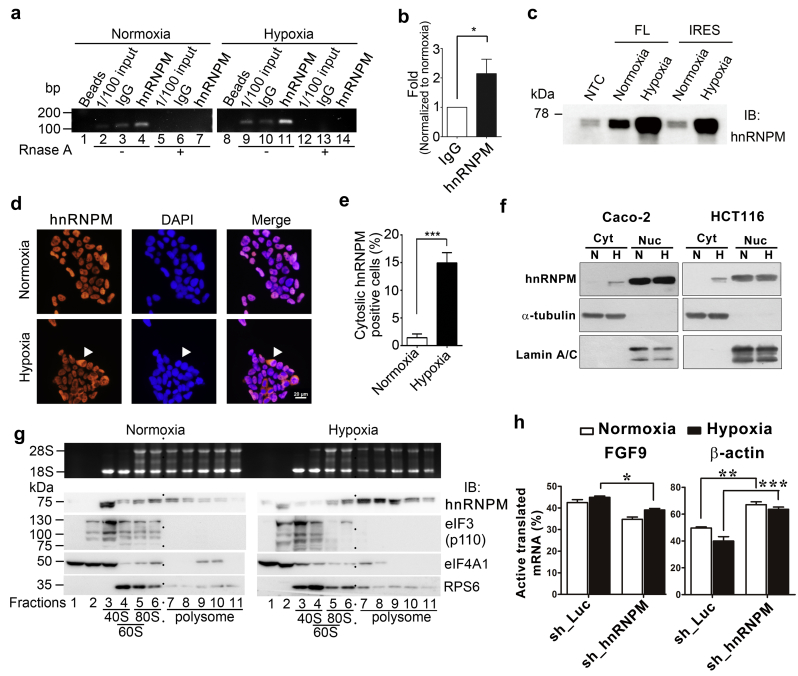
hnRNPM is a novel FGF9 IRES-binding protein. (a) Immunoblot of HEK293 cells expressing control shRNA (sh_Luc) or three eIF4E2-specific shRNAs (sh_eIF4E2–1–3, left). Relative knockdown efficiency of each shRNA compared to sh_Luc was shown (right). The bars represent the mean ± SEM (n = 3). ⁎⁎⁎P < .001 (ANOVA followed by Tukey's multiple test). (b) FGF9 ELISA shows that all three eIF4E2-specific shRNAs knockdown do not change the FGF9 protein production during hypoxia (n = 3). (c) A silver stain image representing unique proteins identified by MALDI-TOF mass spectrometry (indicated by arrows). The Input and NTC were used to show the target proteins in the cell lysates and a negative control for the pull-down assay. (d) Alignment of peptide sequences from LTQ-Orbitrap analysis matched to the human hnRNPM protein sequences. (e) Immunoblot of hnRNPM and hnRNPA1 that were pulled down by indicated biotinylated probes. The FGF2/hnRNPA1 and NTC were used as positive and negative controls for the pull-down assay. Input was used to show the target proteins in the cell lysates. (f) Representative gel image shows that FGF9 mRNAs specifically interact with hnRNPM. β-actin and normal mouse IgG were used as a control transcript and a negative control, respectively for RNA immunoprecipitation assay.
To confirm the binding of hnRNPM and FGF9-IRES, biotinylated FGF9 5’UTR RNA fragments were incubated with HEK293 whole cell lysates to pull down interacting proteins. Using a pan-antibody which recognizes different hnRNPM isoforms [17], an immunoblot showed that the upper band of hnRNPM interacts with all in vitro transcribed FGF9 RNAs (Fig. 1d). However, the low molecular weight isoform of hnRNPM was found to specifically bind to RNAs containing IRES (Fig. 1d). Result of RNA immunoprecipitation (RIP) assay showed that the FGF9 signal was enriched in hnRNPM precipitated RNAs in comparison to the IgG control (Fig. 1e). Taken together, these data suggest that hnRNPM binds to FGF9 IRES and may control FGF9 protein synthesis under hypoxia.
3.2. hnRNPM controls IRES-mediated FGF9 overexpression in hypoxia
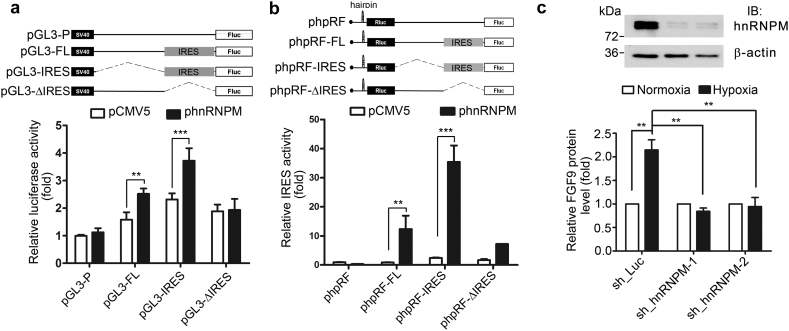
We then determined if hnRNPM regulates FGF9 expression. Forced expression of hnRNPM induced a significant increase of luciferase activity in FL and IRES reporter constructs, respectively, while the luciferase activity of construct without IRES sequences (∆IRES) was not changed (Fig. 2a and S2a). To further verify that hnRNPM-induced luciferase activity is through FGF9 IRES, we co-transfected hnRNPM expression vector and various FGF9 5’UTR bicistronic constructs in HEK293 cells. Overexpression of hnRNPM induced ~15- and ~35-folds increases of the luciferase activity in phpFL and phpIRES, respectively (Fig. 2b). In contrast, the ∆IRES construct only exerted a minor increase (Fig. 2b). Similar result was also obtained in a colon cancer cell line, indicating that overexpression of hnRNPM significantly elevates IRES activity (Fig. S2b). Next, we determined whether hnRNPM is required for hypoxia-induced FGF9 protein expression. As shown in Fig. 2C, level of FGF9 protein was increased ~2·2 folds by hypoxia, which was abolished by knockdown of hnRNPM (Fig. 2c). Furthermore, the drop of FGF9 protein expression was not resulted from a decrease of FGF9 mRNA level or a reduction of FGF9 protein stability (Fig. S2c, d). Collectively, these data support the role of hnRNPM in controlling hypoxia-induced, IRES-mediated upregulation of FGF9 protein expression.
Fig. 2.
hnRNPM controls hypoxia-induced FGF9 overexpression. (a-b) Relative luciferase activities of indicated monocistronic (a) or bicistronic (b) reporter constructs (upper panel) co-transfected with pCMV5 or phnRNPM. Data show fold-increase of pGL3-P of pCMV5. (c) Representative Western blot (upper panel) of hnRNPM knockdown (sh_hnRNPM-1&2) in HEK293 cells. The detection of endogenous FGF9 protein productions in sh_Luc- or hnRNPM-knockdown HEK293 cells exposed to hypoxia or normoxia (lower panel). Data were normalized to respective normoxia group. All data in this figure represent the mean ± SEM (n = 3). ⁎⁎P < .01; ⁎⁎⁎P < .001 (Student's t-test).
3.3. hnRNPM-mediated translational activation is involved in colon cancer development and prognosis
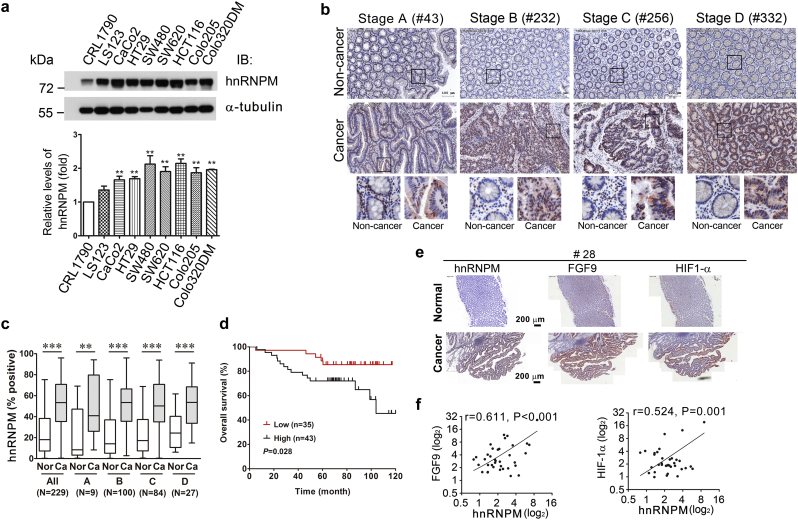
As the results suggest that hnRNPM is a critical ITAF for IRES-mediated translational activation under hypoxia, we speculated that hnRNPM may play a vital role in aberrant expression of FGF9 in colon cancer cells. To test this hypothesis, we first evaluated the expression of hnRNPM in one normal and 8 colorectal cancer (CRC) cell lines. As shown in Fig. 3a, only a basal level of hnRNPM was detected in the normal human colon epithelial cell, CRL1790, while all CRC cell lines showed overexpression of hnRNPM (Fig. 3a).
Fig. 3.
hnRNPM expression is associated with colon cancer (CRC) development and prognosis. (a) Immunoblots (upper panel) and quantified results (lower panel panel) of levels of hnRNPM in normal colon cells (CRL1790) and 8 CRC cell lines (n = 3). α–tubulin served as a loading control. ⁎⁎P < .01; ⁎⁎⁎P < .001 (Student's t-test). (b) Representative images of immunochemical (IHC) staining of hnRNPM in paired CRC tumor and noncancerous tissues with indicated Duke's stage A to D (number indicates patient ID used for the IHC). Lower panel under each tissue sample indicates amplified images from the box marked region. (c) A Box and Whisker plot shows the quantified IHC intensity of hnRNPM in paired noncancerous (Nor) and cancer (Ca) tissues. The number (N) of involved cases for each indicated stage is shown. ⁎⁎P < .01; ⁎⁎⁎P < .001 (Student's t-test). The ends of the box are the upper and lower quartiles while the Whisker shows standard deviation. (d) Kaplan-Meier survival curve shows the overall survival probability between CRCs with high (top 30%, n = 43) and low (bottom 30%, n = 35) hnRNPM expressions. (e) Representative images (case number #28) show the IHC staining of hnRNPM, FGF9 and HIF-1α in paired noncancerous (Normal) and cancer tissues. (f) The correlations between hnRNPM- and FGF9-positive cells (left), and between hnRNPM- and HIF1-α-positive cells (right) in paired non-cancerous and CRC cases (n = 40) were plotted (Spearman's correlation test).
Next, the expression of hnRNPM protein was examined in 229 pairs of colon cancer and adjacent normal tissues using tissue arrays. An elevated level of hnRNPM was found in cancer tissues when compared to the adjacent non-cancerous counterparts (Fig. 3b, c) and in metastatic liver nodules (Fig. S3a, b). Interestingly, the presence of hnRNPM protein in cytosolic fraction was more evident in the cancer cells as compared to that in non-cancer counterparts (Fig. 3b, lowest panel). Moreover, the overall survival rate of patients with high hnRNPM levels was significantly lower than those with low expression levels (Fig. 3d).
To illustrate the control of hnRNPM on FGF9 translation, immunostaining was performed using antibodies specific against hnRNPM, HIF1-α, and FGF9 in serial sections of 49 pairs of CRC tissues. Our data revealed the upregulation of all three proteins in the tumor part when compared to the non-tumor part of CRC tissues (Fig. 3e). Consistent with its role in controlling FGF9 protein translation, we found the levels of hnRNPM and FGF9 were positively correlated (Fig. 3f, left). In addition, the concordant expression of hnRNPM and HIF1-α indicates that a hypoxic microenvironment triggers hnRNPM expression (Fig. 3f, right). Together with the in vitro promoter activity assay, our data demonstrate that hnRNPM activates the translation of FGF9 through binding to the IRES in the 5’UTR, which is correlated with poor prognosis.
3.4. Expression of hnRNPM is associated with colon cancer development in vivo
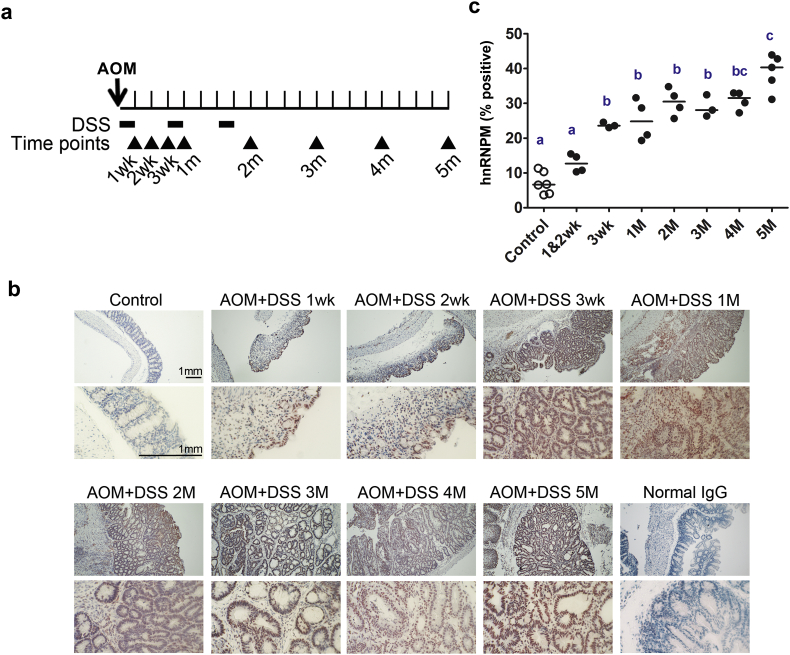
To investigate when hnRNPM is expressed during cancer development, an azoxymethane/dextran sodium sulfate (AOM/DSS)-induced CRC mouse model was used (Fig. 4a). Using immunohistochemical staining, the expression of hnRNPM was detected at the nucleus of pre-neoplastic colitis in AOM/DSS mice after 1 week treatment. Notably, hnRNPM-positive cells were abundant in the area surrounding the aberrant crypt foci in colon tumors from AOM/DSS mice at 3 weeks, and their expression was observed in the nucleus and cytoplasm of tumor cells (Fig. 4b). Our data demonstrated that the expression levels of hnRNPM in colon tissues of the AOM/DSS mice increased as the cancer progressed (Fig. 4c). These data clearly showed that overexpression of hnRNPM occurs at an early stage of colon cancer development and throughout the course of CRC progression.
Fig. 4.
Expression of hnRNPM is positively correlated with cancer progression in the mouse model of AOM-induced tumorigenesis. (a), Schematic representation of the experimental protocol of AOM/DSS murine model to induce CRC. The AOM was injected at Day 1 (arrow), followed by giving DSS at weeks 1, 4, and 7 (Bold line). The arrowhead indicates time points when the tissues were collected. (b) IHC images shows the IHC staining of hnRNPM in the colon sections of AOM/DSS-treated mice. Saline-injected and age-matched mice were used as control for AOM/DSS treatment; normal rabbit IgG was used as negative control for IHC. (c), The plot shows the comparison of hnRNPM-positive cells from IHC staining in indicated AOM/DSS-treated mice groups (n = 3–6). ⁎⁎⁎P < .001 (ANOVA followed by Tukey's multiple test).
3.5. Hypoxia increases binding of cytosolic hnRNPM onto its target and promotes translation
Using an hnRNPM-IP followed by reverse transcriptase quantitative PCR (RT-qPCR), our data showed a 2-fold enrichment of FGF9 mRNA under hypoxia (Fig. 5a, b). Similarly, marked increase of hnRNPM proteins were pulled down by FGF9 FL and IRES RNAs under hypoxic conditions compared to those under normoxia (Fig. 5c). These data indicate that hypoxia enhances the interaction between hnRNPM and IRES-containing target transcripts (i.e., FGF9 mRNA).
Fig. 5.
Hypoxia enhances interaction between hnRNPM and FGF9 mRNA and promotes translation. (a) Gel image of FGF9 RT-PCR from hnRNPM-immunoprecipitation assay. RNase treatment was performed to indicate that PCR products were amplified from RNA origin. (b) The quantitative measurement of FGF9 TaqMan assays from hnRNPM-immunoprecipitation assay. The relative FGF9 mRNA level was shown as the amount measured in hypoxia normalized to the amount in normoxia. (c) Representative Western blot image shows levels of hnRNPM pulldown by full length FGF9 RNA 5’UTR (FL) or just IRES sequences from cells cultured under normoxia or hypoxia conditions. (d) Immunofluorescent images showed cellular distribution of hnRNPM (red) in HEK293 cells cultured in normoxia or hypoxia conditions. The arrowheads indicate cytosolic hnRNPM. Nuclei were marked with DAPI (blue). (e) Number of cytosolic hnRNPM positive cells in normoxia or hypoxia (A total of 100 cells per independent experiment were counted). (f) Representative Western blot image shows the expression of hnRNPM in the cytosolic and nuclear fractions of indicated cells treated with normoxia or hypoxia for 9 h (n = 3 independent experiments). The levels of α-tubulin and lamin A/C are used as markers for cytosolic and nuclear fractions, respectively. (g) The 18S and 28S rRNAs were resolved on a 1% formaldehyde/agarose gel and visualized by ethidium bromide staining (Upper panel). Immunoblotting analysis of gradient fractions was performed using antibodies against hnRNPM, PTBP1, eIF3, eIF4A1, and ribosomal protein RPS6 (Lower panel). (h) The quantitative measurement of mRNAs from hnRNPM-knockdown cytoplasmic extracts. Bar graphs represent the proportion of actively translated mRNA of FGF9 (left panel) and β-actin (right panel) from 3 independent experiments. ⁎P < .05; ⁎⁎P < .01; ⁎⁎⁎P < .001 (Student's t-test). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
As hnRNPM is a known RNA binding protein that is involved in splicing and mRNA processing [18], shuttling between nucleus and cytoplasm would be predicted in order for hnRNPM to exert its biological function. To examine whether hypoxia changes the cellular distribution of hnRNPM, we performed immunofluorescent staining to detect cellular distribution of hnRNPM. As shown in Fig. 5d, hnRNPM is predominantly co-localized with DAPI in the nucleus under both normoxic and hypoxic conditions. Notably, an increase in cytosolic hnRNPM was detected when cells were cultured under hypoxia (Fig. 5e). Both the number of cells with cytosolic hnRNPM staining and the fluorescent intensity of cytosolic hnRNPM were significantly increased under hypoxia (Fig. 5e and Fig. S4a). In parallel, we performed Western blot on fractionated cell lysates in Caco2 and HCT116 colon cancer cells cultured under normoxia or hypoxia to support the observation. As shown in the Fig. 5f, hnRNPM was predominantly present in the nuclear fraction in both normoxic and hypoxic conditions. While the levels of hnRNPM in nuclear fraction showed no differences between normoxia and hypoxia, a significant increase of hnRNPM expression in cytosolic fraction were observed in hypoxic condition (Fig. 5f and S4b).
To examine whether cytosolic hnRNPM would interact with IRES-containing mRNA to promote translation, we investigated whether hnRNPM is associated with translation machinery to activate translation. Cytoplasmic fractions of HEK293 in normoxia and hypoxia were subjected to sucrose density gradient centrifugation. The distribution of 28S and 18S rRNA in all fractions was analyzed by denaturing agarose gel electrophoresis (Fig. 5g, upper). Results showed that hnRNPM was largely associated with fractions 3–6 under normoxia, while the distribution of hnRNPM was mostly found in fractions 7–11 (polysome) under hypoxia, indicating a switch to active translation under low oxygen conditions (Fig. 5g, lower). On the other hand, such hypoxia-induced polysome association shift was not observed in eIF3 and eIF4A1, two well-known translation initiation factors, in cells treated with hypoxia. To conclude whether the sedimentation of the hypoxic hnRNPM reflected their association with polysomes, cells were treated with puromycin prior to sucrose gradient sedimentation. As expected, puromycin treatment caused the relocation of hnRNPM, and RPS6 to lower sedimenting fractions in the gradient (Fig. S4c). The relocation of hnRNPM toward the less dense fractions in the gradient after puromycin treatment suggests that a portion of hnRNPM was associated with ribosomes in hypoxia. Furthermore, we observed that hnRNPM distributed together with several translation initiation factors (eIF3 and eIF4A1) in fractions enriched with the 40S ribosome subunit (Fig. 5g, right lower panel, fractions 1–2), implying that hnRNPM may be associated with the 43S preinitiation complex and participate in translational control during hypoxia. Nevertheless, further in-depth investigation is needed to verify whether hnRNPM can be involved in the translation initiation to regulate protein synthesis under hypoxia.
Next, we examined the role of hnRNPM in controlling hypoxia-induced FGF9 translation. Knockdown of hnRNPM significantly decreased the actively translated FGF9 mRNA under hypoxia but not in normoxia as compared to the Luc knockdown control (Fig. 5h, left panel). It is noticed that the proportion of actively translated ACTB mRNA is elevated in hnRNPM-knockdown cells under both hypoxia and normoxia (Fig. 5h, right panel). Whether the results suggest that hnRNPM may function as a negative regulator for ACTB mRNA translation remains to be determined. Nevertheless, altogether these data strongly support the novel role of hnRNPM in controlling FGF9 mRNA translation under low oxygen condition.
3.6. hnRMPM activates translation in a specific set of IRES-containing transcripts and promotes colon cancer tumorigenesis in response to hypoxia
To study the global impact of hnRNPM regulation, we performed RIP followed by deep sequencing (RIP-seq) to identify hnRNPM-bound RNA targets in low oxygen condition (SRA submission #: SUB2903395). The RIP-seq results revealed that the global RNA-hnRNPM interaction changed dramatically from normoxia to hypoxia (Fig. 6a). In parallel, we combined polysome-bound RNA fractions (fractions 7–11) and ran deep sequencing to identify genes that are regulated through hypoxia-induced translational reprograming (SRA submission #: SUB2978848). The integration of results from RIP-seq (12,495 transcripts with overall read counts >10) and translatomics (11,997 transcripts with overall read counts >10) revealed that over 87% of polysome-bound mRNAs are also targeted by hnRNPM. The ribosome occupancy was compared between non-targets (absent from hnRNPM-RIP, 1502 transcripts) and common targets (present in hnRNPM-RIP with >40% of changing reads, 7353 transcripts). The results revealed that the numbers of reads in association with ribosome-bound mRNA for non-targets were significantly more than those for common targets (Fig. S5a), suggesting that the function of hnRNPM binding onto its target mRNA is not to change translation efficiency but to select specific mRNAs for translation under hypoxia.
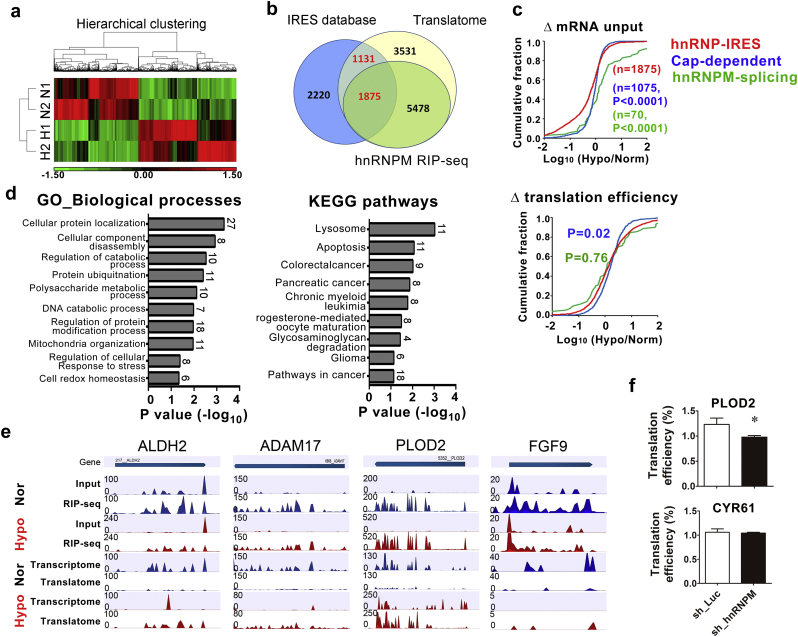
Fig. 6.
hnRMPM selectively controls IRES-mediated translation in response to hypoxia. (a) Hierarchical clustering of duplicated hnRNPM-IP-seq from cells exposed to normoxia (N1 and N2) or hypoxia (H1 and H2). Each column represents individual transcripts with significant (P < .05) up- (red) or down-expression (green). (b) Venn diagram shows overlapped targets from hnRNPMRIP-Seq (HEK293), translatome (HT-29), and predicted IRES-containing gene datasets. (c) Cumulative distribution log10-fold changes of mRNA input (upper) and translation efficiency (lower) among hnRNPM-IRES targets (red), hnRNPM-splicing targets (green), and cap-dependent translational targets (blue). P values were calculated using Mann-Whitney test (two-sided). (d) Gene ontology (biological processes, left) and KEGG pathways (right) analysis of 546 translation efficiency (TE) up-regulated targets. The number showing with each bar represents targets identified in each designated category. P value was presented as transformed –log10 value. (e) Genome browser view of the distribution of the RIP-seq and Ribo-Seq reads of represented genes. The upper panels show the hnRNPM-binding targets identified by RIP-seq (input, input RNA; RIP-Seq, hnRNPM immuno-precipitated RNAs), and the lower panels depict the Ribosome profiling (Ribo-Seq) reads of the represented genes (Transcriptome, input RNAs; Translatome, polyribome-bound RNAs). Normalized read densities of indicated dataset were shown for normoxia (blue) and hypoxia (red) (f) The quantitative measurement of mRNAs from hnRNPM-knockdown cytoplasmic extracts. Bar graphs represent the proportion of actively translated mRNA of PLOD2 (upper panel) and CYR61 (lower panel) from 3 independent experiments. ⁎P < .05 (Student's t-test). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
As our previous study suggested that cellular IRES may function as an RNA switch to turn protein synthesis “on” during hypoxia and/or in cancer cells [12], we then cross-referenced the hnRNPM common targets identified in this study to the dataset of 5226 IRES-containing genes obtained previously to identify those transcripts that are potentially controlled by the hnRNPM-IRES-mediated translational switch. We found that 1875 hnRNPM-target, polysome-bound mRNAs contain the predicted IRES motif (Fig. 6b), indicating the protein synthesis of these mRNAs may be controlled by hypoxia-induced, hnRNPM-IRES-mediation. The possibilities that the identified hnRNPM-bound transcripts may represent either the hnRNPM splicing targets [19] or that the observed polysome-association may result from cap-dependent translation in hypoxia [5] were then considered. The comparison of three datasets revealed that only a very small proportion of hnRNPM-IRES-target genes overlap with hnRNPM-splicing targets (8 genes, 0.43%) or cap-dependent translated targets (175 genes, 9.3%, Fig. S5b). In addition, while the amounts of mRNA input in the hnRNPM-IRES targets are much less than in hnRNPM-splicing targets (Fig. 6c, upper), the overall translation efficiencies (TE, defined by normalized ribosome-bound reads with its mRNA input) are similar between the two groups (Fig. 6c, lower). We also found a higher amount of input mRNA (P < .001, Fig. 6c, upper) and better TE (P < .05, Fig. 6c, lower) in association with cap-dependent translated targets than the hnRNPM-IRES targets. These results confirmed the notion that cap-dependent translation is still the most efficient way to generate proteins even under stress conditions. Nevertheless, our data also demonstrated that the hnRNPM-IRES-transcripts represent a unique mRNA population that is regulated by cap-independent translation in hypoxia.
The biological significance of hypoxia-induced, hnRNPM-IRES-mediated translational activation was explored using 546 TE up-regulated targets with enhanced binding of hnRNPM. GO terms enrichment analysis revealed that many of these IRES-containing mRNAs translationally activated by hnRNPM either participated in the mitochondrial process, a well-known role of mitochondrial function in cellular energy and reactive oxygen species homeostasis in hypoxia (Fig. S5c, left), or functioned in the regulation of transcriptional activity (Fig. S5c, right). In addition, these hnRNPM-IRES translated targets are likely to be involved in the processes including cellular protein localization, regulation of protein modification, regulation of cellular response to stress and cell redox homeostasis (Fig. 6d, left). Furthermore, the hypoxia-induced, hnRNPM-IRES targets were highly enriched in various pathways including lysosome, apoptosis, and various types of cancers (Fig. 6d, right). Detailed read distribution of RIP-seq and polysome-bound RNA-seq analyses showed that the genomic regions of transcripts functioning in the response to oxygen levels, such as ALDH2 [20], ADAM17 [21] and P4HA1 [22], or which are involved in cancer/ catabolic pathways, such as Procollagen-Lysine,2-Oxoglutarate 5-Dioxygenase 1 (PLOD2 [23], FGF9 [12], DCUN1D5 [24], CKS1B [25] and PSMD13 [26], were bound by hnRNPM and that their polysome-bound reads were up-regulated by hypoxia treatment (Fig. 6e and Fig. S5d).
To validate the role of hnRNPM as a master regulator for controlling IRES-mediated translational activation in hypoxia, hnRNPM was knocked down in cells and the proportion of actively translated mRNA for the predicted hnRNPM target gene, PLOD2, was quantified. Result showed that hypoxia significantly increased hnRNPM-bound PLOD2 mRNA (Fig. S5e, left) while knockdown of hnRNPM drastically repressed hypoxia-induced PLOD2 mRNA translation (Fig. 6f, upper). On the other hand, neither oxygen levels nor hnRNPM-knockdown affected the interaction of CYR61 mRNA, an IRES-containing non-hnRNPM-target gene, with hnRNPM (Fig. S5e, right) or changed the translation efficiency (Fig. 6f, lower). Collectively, these results provide strong evidence to support the notion that hnRNPM activates translation in a specific subset of mRNAs that are involved in hypoxic responses to promoting tumorigenesis in cancer cells.
4. Discussion
Translation of cellular mRNAs via initiation at IRESs has received increasing attention in recent years because it controls important cell-fate decisions under stress, leading to either survival or cell death [27,28]. While ITAFs are believed to play a pivotal role in IRES-mediated translation and function in a target-specific manner [7], the key player(s) that guide cellular IRES-mediated expression under hypoxic stress remain largely unknown. Herein, we provide compelling evidence to show that hnRNPM binds to IRES-containing mRNAs and activates their translation in hypoxia. Overexpression of hnRNPM increased FGF9 protein production in hypoxic conditions, while knockdown of hnRNPM abolished hypoxia-induced FGF9 protein synthesis. Since FGF9 has been shown to play critical roles in tumorigenesis and cancer malignancy [12,13,15], these results, for the first time, demonstrate that hnRNPM is a novel hypoxia-induced ITAF that activates IRES-mediated translation in stress conditions and offer a new avenue in studying aberrant gene expression during cancer progression. Nevertheless, it is of notice that we focused on selected bands for proteomic analysis. Thus the approach limited the protein identified in the current study restrict to the IRES sequences but not a complete list of interacting proteins for FGF9 5’UTR. To reconstruct the complete RNA-protein interaction of FGF9 5’UTR under hypoxia, a comprehensive approach will be employed in the future.
Prior to this study, hnRNPM was a protein known for its regulation of pre-mRNA splicing [29], alternative splicing site selection [30], and RNA quality control [31]. Hypoxia has been shown to have a significant effect on the alternative splicing of genes in various conditions including normal endothelial cells [32] and human breast cancer cells [33], perhaps through the upregulation of many splice factors and splice factor kinases that helps cells adapt through and/or survive under hypoxia [34]. In supporting previous findings, hnRNPM has been suggested to promote breast cancer metastasis via regulating alternative splicing [35]. Notably, the hnRNPM-IRES-target genes identified in this study only overlapped 8 genes with previously discovered hnRNPM-splicing targets [19] (0.43%), demonstrating that the hnRNPM-IRES targets are a novel population of mRNA that is controlled by hypoxia-induced translation machinery. Taken together, these results showed that hnRNPM is a versatile molecule to promote cancer development through multiple mechanisms.
It is known that cells activate alternative translations, including cap-dependent and IRES-dependent pathways, when eIF4F activity is inhibited by hypoxia. While the cap-dependent protein synthesis pathway is well documented [5], the control of IRES-dependent pathway remains unclear [36]. Our experimental evidence supports a role of hnRNPM in binding IRES-containing cellular mRNAs and directly facilitating translation. In addition to the significant elevation of hnRNPM-bound mRNAs associated with polysomes when oxygen is limited, hnRNPM-bound mRNAs were found to uniquely co-exist with eIF3 and eIF4A1 in free mRNA and pre-40S ribosomal particles in hypoxia-treated fractions. Taken together, these data imply that hnRNPM participates in the formation of a specific translational initiation complex, and promotes protein synthesis for hnRNPM-target mRNAs under hypoxia (Fig. 7). Recent studies have shown that an eIF4G homolog, DAP5, associates with eIF2β and eIF4A1 to stimulate IRES-dependent translation of some cellular mRNAs [37], and XIAP IRES directly recruited eIF3 to the precise structural conformation near the initiating AUG in a poly(A)- and PABP-dependent manner [38]. Our findings that hnRNPM facilitates the formation of initiation complex provide further insights into the machinery of IRES-mediated translation under stress conditions, and suggest that hnRNPM may represent the key regulator to control cap-independent translation in hypoxia. Whether hnRNPM would partner with DAP5 and/or PABP to drive IRES-dependent translation in hypoxia is unclear and warrants further investigation.
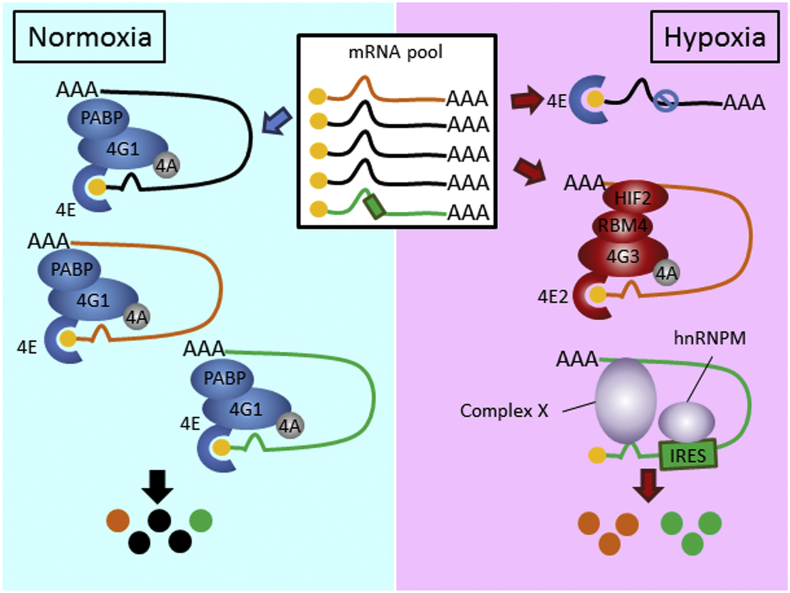
Fig. 7.
A working model of hnRNPM-IRES-mediated translational switch in hypoxia. There are 3 types of cytoplasmic mRNAs (labeled with different colors) in the mRNA pool in the cells. Cap-dependent translation carried out by eIF4F (eIF4E-eIF4A-eIF4G1) is predominately used for protein synthesis in normoxia (left). When eIF4F-dependent translation is inhibited during hypoxia (transcripts in black), mRNA translation is then carried out selectively by the hypoxic eIF4F (eIF4E2-eIF4A-eIF4G3)-dependent (transcripts in orange) pathway [5] (right). Alternatively, a subset of IRES-containing mRNAs (transcripts in green) undergoes conformational change in hypoxia and is recognized by the novel hypoxia-induced ITAF hnRNPM. Together with the help from complex X (other RBPs like hnRNPC1/C2 and HuR) and the interaction of hnRNPM-IRES trigger the formation of a new translation initiation complex and prime cap-independent translation to take place (right). Overall, protein synthesis through co-regulated, co-occupied IRES-containing mRNAs by hnRNPM can achieve vital physiological responses to combat environmental hypoxic stress. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The translational reprogramming is known to be an essential mechanism for cancer cell growth, progression, and survival in unfavorable conditions. The hypoxia-induced translational switch through the use of the eIF4E2-dependent pathway has been well documented [4,39]; however, the key regulators of hypoxic IRES-mediated translation and their contribution to tumorigenesis remain a missing piece of the cancer genome. By cross-referencing predicted IRES-containing genes with our transcriptomic and translatomic datasets, we identified a subset of hnRNPM-targeted mRNAs actively translated in low oxygen conditions through an IRES-mediated mechanism. <10% of genes on the hnRNPM-IRES-targeted mRNA list are overlapped with previous identified genes that are translated through cap-dependent translation [5], indicating that the hnRNPM-IRES targets represent a unique population of mRNA that is regulated by different translation machinery in hypoxia. By analyzing the ontology of these genes, we found that a large number of up-regulated genes participate in the mitochondrial process, in which their functions concerning cellular energy and reactive oxygen species homeostasis in hypoxia are well recognized. Furthermore, these hypoxia-induced, hnRNPM-IRES targets are highly enriched in various pathways, such as lysosome and apoptosis, and are involved in the tumorigenesis for several different types of cancers. In supporting this notion, our recent study also found that hypoxia induces autophagy through translational up-regulation of lysosomal proteins in human colon cancer cells [40]. In addition, hnRNPM has been suggested as a novel biomarker for colorectal carcinoma [41]. Collectively, these results suggest the possibility of using hnRNPM as an anticancer target for future research.
In summary, the role of hnRNPM in promoting IRES-containing mRNA translation fills a critical missing piece of the hypoxia-induced gene activation. This hnRNPM-IRES-dependent function complements the mRNA translation pathway mediated through eIF4E2-HIF2α to implement dynamic gene regulation and offers an alternative path for translation recovery from stress-mediated global translational suppression. Our study provides the first genome-wide profile of the translational signature of hnRNPM, and demonstrates that the hnRNPM-IRES-mediated protein production is involved in cancer development and treatment outcomes. These findings not only support the notion that IRES-mediated translation acts at an important node in tumorigenesis [27], but also provide a new direction in searching for anti-tumor therapies.
Acknowledgments
Acknowledgements
The authors would like to thank the Bioinformaticians in the Bioinformatics core at National Cheng Kung University for providing excellent assistance in computational analyses. We are also grateful to Dr. Laising Yen from the Baylor College of Medicine for the critical reading and comments on the manuscript.
Funding sources
This work was supported by the Ministry of Science and Technology, Taiwan (MOST 104-2320-B-006-042 to HSS and MOST 105-2628-B-001-MY3 to TMC).
Declaration of interests
The authors declare no competing interests.
Conflict of interest
All authors declare no conflict of interest.
Author contributions
Conceptualization, S.J.T. and H.S.S.; Methodology, M.C.L., and S.Y.H.; Investigation, T.M.C., M.C.L., Y.H.L., Y.L.C., C.H.W., Y.M.W., and C.W.C.; Writing - Original Draft, T.M.C., H.S.S., and S.J.T.; Writing - Review & Editing, H.S.S., and S.J.T.; Funding Acquisition, T.M.C., and H.S.S.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.02.059.
Contributor Information
H. Sunny Sun, Email: hssun@mail.ncku.edu.tw.
Shaw-Jenq Tsai, Email: seantsai@mail.ncku.edu.tw.
Supplementary data
Supplementary material
References
- 1.Wouters B.G., Koritzinsky M. Hypoxia signalling through mTOR and the unfolded protein response in cancer. Nat Rev Cancer. 2008;8:851–864. doi: 10.1038/nrc2501. [DOI] [PubMed] [Google Scholar]
- 2.Silvera D., Schneider R.J. Inflammatory breast cancer cells are constitutively adapted to hypoxia. Cell Cycle. 2009;8:3091–3096. doi: 10.4161/cc.8.19.9637. [DOI] [PubMed] [Google Scholar]
- 3.Koumenis C. Wouters BG. "translating" tumor hypoxia: unfolded protein response (UPR)-dependent and UPR-independent pathways. Mol Cancer Res. 2006;4:423–436. doi: 10.1158/1541-7786.MCR-06-0150. [DOI] [PubMed] [Google Scholar]
- 4.Uniacke J., Holterman C.E., Lachance G. An oxygen-regulated switch in the protein synthesis machinery. Nature. 2012;486:126–129. doi: 10.1038/nature11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ho J.J., Wang M., Audas T.E. Systemic reprogramming of translation efficiencies on oxygen stimulus. Cell Rep. 2016;14:1293–1300. doi: 10.1016/j.celrep.2016.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.King H.A., Cobbold L.C., Willis A.E. The role of IRES trans-acting factors in regulating translation initiation. Biochem Soc Trans. 2010;38:1581–1586. doi: 10.1042/BST0381581. [DOI] [PubMed] [Google Scholar]
- 7.Komar A.A., Hatzoglou M. Cellular IRES-mediated translation: the war of ITAFs in pathophysiological states. Cell Cycle. 2011;10:229–240. doi: 10.4161/cc.10.2.14472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonnal S., Pileur F., Orsini C. Heterogeneous nuclear ribonucleoprotein A1 is a novel internal ribosome entry site trans-acting factor that modulates alternative initiation of translation of the fibroblast growth factor 2 mRNA. J Biol Chem. 2005;280:4144–4153. doi: 10.1074/jbc.M411492200. [DOI] [PubMed] [Google Scholar]
- 9.Martinez-Salas E., Lozano G., Fernandez-Chamorro J., Francisco-Velilla R., Galan A., Diaz R. RNA-binding proteins impacting on internal initiation of translation. Int J Mol Sci. 2013;14:21705–21726. doi: 10.3390/ijms141121705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wing L.Y., Chuang P.C., Wu M.H., Chen H.M., Tsai S.J. Expression and mitogenic effect of fibroblast growth factor-9 in human endometriotic implant is regulated by aberrant production of estrogen. J Clin Endocrinol Metab. 2003;88:5547–5554. doi: 10.1210/jc.2003-030597. [DOI] [PubMed] [Google Scholar]
- 11.Nakamura S., Arima K., Haga S. Fibroblast growth factor (FGF)-9 immunoreactivity in senile plaques. Brain Res. 1998;814:222–225. doi: 10.1016/s0006-8993(98)01042-7. [DOI] [PubMed] [Google Scholar]
- 12.Chen T.M., Shih Y.H., Tseng J.T. Overexpression of FGF9 in colon cancer cells is mediated by hypoxia-induced translational activation. Nucleic Acids Res. 2014;42:2932–2944. doi: 10.1093/nar/gkt1286. [Epub 2013 Dec 10] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giri D., Ropiquet F., Ittmann M. FGF9 is an autocrine and paracrine prostatic growth factor expressed by prostatic stromal cells. J Cell Physiol. 1999;180:53–60. doi: 10.1002/(SICI)1097-4652(199907)180:1<53::AID-JCP6>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 14.Ohgino K., Soejima K., Yasuda H. Expression of fibroblast growth factor 9 is associated with poor prognosis in patients with resected non-small cell lung cancer. Lung Cancer. 2014;83:90–96. doi: 10.1016/j.lungcan.2013.10.016. [DOI] [PubMed] [Google Scholar]
- 15.Todo T., Kondo T., Kirino T. Expression and growth stimulatory effect of fibroblast growth factor 9 in human brain tumors. Neurosurgery. 1998;43:337–346. doi: 10.1097/00006123-199808000-00098. [DOI] [PubMed] [Google Scholar]
- 16.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 17.Datar K.V., Dreyfuss G., Swanson M.S. The human hnRNP M proteins: identification of a methionine/arginine-rich repeat motif in ribonucleoproteins. Nucleic Acids Res. 1993;21:439–446. doi: 10.1093/nar/21.3.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hovhannisyan R.H., Carstens R.P. Heterogeneous ribonucleoprotein m is a splicing regulatory protein that can enhance or silence splicing of alternatively spliced exons. J Biol Chem. 2007;282:36265–36274. doi: 10.1074/jbc.M704188200. [DOI] [PubMed] [Google Scholar]
- 19.Huelga S.C., Vu A.Q., Arnold J.D. Integrative genome-wide analysis reveals cooperative regulation of alternative splicing by hnRNP proteins. Cell Rep. 2012;1:167–178. doi: 10.1016/j.celrep.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun A., Zou Y., Wang P. Mitochondrial aldehyde dehydrogenase 2 plays protective roles in heart failure after myocardial infarction via suppression of the cytosolic JNK/p53 pathway in mice. J Am Heart Assoc. 2014;3 doi: 10.1161/JAHA.113.000779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X.J., Feng C.W., Li M. ADAM17 mediates hypoxia-induced drug resistance in hepatocellular carcinoma cells through activation of EGFR/PI3K/Akt pathway. Mol Cell Biochem. 2013;380:57–66. doi: 10.1007/s11010-013-1657-z. [DOI] [PubMed] [Google Scholar]
- 22.Gilkes D.M., Bajpai S., Chaturvedi P., Wirtz D., Semenza G.L. Hypoxia-inducible factor 1 (HIF-1) promotes extracellular matrix remodeling under hypoxic conditions by inducing P4HA1, P4HA2, and PLOD2 expression in fibroblasts. J Biol Chem. 2013;288:10819–10829. doi: 10.1074/jbc.M112.442939. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Noda T., Yamamoto H., Takemasa I. PLOD2 induced under hypoxia is a novel prognostic factor for hepatocellular carcinoma after curative resection. Liver Int. 2012;32:110–118. doi: 10.1111/j.1478-3231.2011.02619.x. [DOI] [PubMed] [Google Scholar]
- 24.Bommelje C.C., Weeda V.B., Huang G. Oncogenic function of SCCRO5/DCUN1D5 requires its Neddylation E3 activity and nuclear localization. Clin Cancer Res. 2014;20:372–381. doi: 10.1158/1078-0432.CCR-13-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Black J.C., Atabakhsh E., Kim J. Hypoxia drives transient site-specific copy gain and drug-resistant gene expression. Genes Dev. 2015;29:1018–1031. doi: 10.1101/gad.259796.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li S., Hu J., Jin R.J. Temperature-sensitive retinoid isomerase activity of RPE65 mutants associated with Leber congenital Amaurosis. J Biochem. 2015;158:115–125. doi: 10.1093/jb/mvv028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walters B., Thompson S.R. Cap-independent translational control of carcinogenesis. Front Oncol. 2016;6:128. doi: 10.3389/fonc.2016.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.El-Naggar A.M., Sorensen P.H. Translational control of aberrant stress responses as a hallmark of cancer. J Pathol. 2018;244:650–666. doi: 10.1002/path.5030. [DOI] [PubMed] [Google Scholar]
- 29.Kafasla P., Patrinou-Georgoula M., Lewis J.D., Guialis A. Association of the 72/74-kDa proteins, members of the heterogeneous nuclear ribonucleoprotein M group, with the pre-mRNA at early stages of spliceosome assembly. Biochem J. 2002;363:793–799. doi: 10.1042/0264-6021:3630793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lleres D., Denegri M., Biggiogera M., Ajuh P., Lamond A.I. Direct interaction between hnRNP-M and CDC5L/PLRG1 proteins affects alternative splice site choice. EMBO Rep. 2010;11:445–451. doi: 10.1038/embor.2010.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hessle V., Bjork P., Sokolowski M. The exosome associates cotranscriptionally with the nascent pre-mRNP through interactions with heterogeneous nuclear ribonucleoproteins. Mol Biol Cell. 2009;20:3459–3470. doi: 10.1091/mbc.E09-01-0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weigand J.E., Boeckel J.N., Gellert P., Dimmeler S. Hypoxia-induced alternative splicing in endothelial cells. PLoS One. 2012;7 doi: 10.1371/journal.pone.0042697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han J., Li J., Ho J.C. Hypoxia is a key driver of alternative splicing in human breast Cancer cells. Sci Rep. 2017;7:4108. doi: 10.1038/s41598-017-04333-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bowler E., Porazinski S., Uzor S. Hypoxia leads to significant changes in alternative splicing and elevated expression of CLK splice factor kinases in PC3 prostate cancer cells. BMC Cancer. 2018;18:355. doi: 10.1186/s12885-018-4227-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu Y., Gao X.D., Lee J.H. Cell type-restricted activity of hnRNPM promotes breast cancer metastasis via regulating alternative splicing. Genes Dev. 2014;28:1191–1203. doi: 10.1101/gad.241968.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xia X., Holcik M. Strong eukaryotic IRESs have weak secondary structure. PLoS One. 2009;4 doi: 10.1371/journal.pone.0004136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liberman N., Gandin V., Svitkin Y.V. DAP5 associates with eIF2beta and eIF4AI to promote internal ribosome entry site driven translation. Nucleic Acids Res. 2015;43:3764–3775. doi: 10.1093/nar/gkv205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thakor N., Smith M.D., Roberts L. Cellular mRNA recruits the ribosome via eIF3-PABP bridge to initiate internal translation. RNA Biol. 2016:1–15. doi: 10.1080/15476286.2015.1137419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uniacke J., Perera J.K., Lachance G., Francisco C.B., Lee S. Cancer cells exploit eIF4E2-directed synthesis of hypoxia response proteins to drive tumor progression. Cancer Res. 2014;74:1379–1389. doi: 10.1158/0008-5472.CAN-13-2278. [DOI] [PubMed] [Google Scholar]
- 40.Lai M.C., Chang C.M., Sun H.S. Hypoxia induces autophagy through translational up-regulation of lysosomal proteins in human Colon Cancer cells. PLoS One. 2016;11 doi: 10.1371/journal.pone.0153627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen S., Zhang J., Duan L. Identification of HnRNP M as a novel biomarker for colorectal carcinoma by quantitative proteomics. Am J Physiol Gastrointest Liver Physiol. 2014;306:G394–G403. doi: 10.1152/ajpgi.00328.2013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material