Abstract
The human hCLCA1 gene is a member of the CLCA gene family that has a well-documented role in inflammatory airway diseases. Previously, we demonstrated that secreted hCLCA1 plays a role in regulating the innate immune response by activating airway macrophages. However, the mechanism of this regulation remains unclear. In this present study, recombinant proteins containing different hCLCA1 domains are expressed to determine the specific hCLCA1 domain(s) responsible for macrophage activation. Specifically, hCLCA1 constructs containing the hydrolase domain (HYD), the von Willebrand Factor Type A (VWA) domain, and the fibronectin type III (FN3) domain were heterologously expressed and affinity purified through fast protein liquid chromatography. Circular dichroism spectroscopy revealed that the purified hCLCA1 constructs exhibited secondary structure consistent with folded proteins. The VWA domain clearly demonstrated an ability to activate macrophages, inducing an increase in both IL-1β mRNA and protein expression. This activation was associated with the activation of MAPKs and NF-κB pathways, identifying potential mechanistic pathways by which hCLCA1's VWA domain exerts its signaling effect. Altogether, this work identifies a domain with signaling function within hCLCA1, providing a specific target to one of the most highly induced gene products of airway inflammatory disease.
Highlights
-
•
hCLCA1 is an immune modulator upregulated in inflammatory airway diseases.
-
•
hCLCA1's VWA domain upregulated IL-1β mRNA and protein in U-937 macrophages.
-
•
Macrophage activation by the VWA domain correlated with MAPK and NF-κB activation.
-
•
The VWA domain thus represents an attractive target for pharmacological treatment.
1. Introduction
The human hCLCA1 protein is a member of the CLCA (chloride-channel modulating, calcium-activated) gene family with a well-established role in inflammatory airway diseases such as asthma, cystic fibrosis, and chronic obstructive pulmonary disease [[1], [2], [3], [4]]. Although hCLCA1 was originally identified as the pore subunit of calcium activated chloride channels, later studies have shown that it is a chloride channel accessory protein [[5], [6], [7], [8], [9]]. Studies have reported that hCLCA1 is induced in inflamed airway epithelial cells, and this induced expression often exceeds that of most other inflammatory mediators [10,11]. We have previously identified the novel role of hCLCA1 as a signaling molecule [12], suggesting a possible mechanism by which hCLCA1 expression contributes to inflammatory airway disease.
In our previous study, we have demonstrated hCLCA1 acts as a signaling molecule and modulates innate immune response in macrophages [12]. A recent study also demonstrated that mCLCA1 (formerly mCLCA3), the murine ortholog of hCLCA1, modulates leukocyte recruitment via IL-17 and CXCL-1 in bacterial pneumonia [13], which is in line with our newly identified function of hCLCA1 as an immune modulator. However, the mechanism regarding this modulation remains unclear. In addition to the previously mentioned hydrolase domain (HYD), hCLCA1 also contains both von Willebrand factor A (VWA) and Fibronectin type III (FN3) domains. Such domains have been shown to play a role in different aspects of the inflammatory response. The hydrolase domain of hCLCA1 is similar to the protease domain of matrix metalloproteases, which are known to play a role in activation of pro-inflammatory cytokines such as IL-8 [14,15]. The VWA domain is a ligand binding motif first described in the hemostasis protein Von Willebrand Factor and has been known to contribute to the inflammatory response through leukocyte recruitment [[16], [17], [18]]. Finally, the FN3 domain has been shown to increase inflammatory cytokine expression in fibroblast cells [19,20]. In this study, we expressed proteins containing different hCLCA1 domains to determine the specific domain(s) and associated pathways that are involved in hCLCA1 macrophage activation. The results demonstrate that the von Willebrand factor type A (VWA) domain of hCLCA1 can induce a modest up-regulatation of both mRNA and protein expression of IL-1β. Additionally, both the NF-κB and MAPK signaling pathways were activated when macrophages were exposed to VWA domain.
2. Materials and methods
2.1. Cell culture
Human monocytes (U-937 cell line; CRL1593.2; ATCC) were grown in RPMI-1640 medium supplemented with 10% heat inactivated Fetal bovine serum and 1% penicillin-streptomycin 37 °C in a humidified atmosphere with 5% CO2.
2.2. Cloning, expression, purification, and endotoxin removal of hCLCA1 domain containing proteins
DNA corresponding to the hydrolase domain (amino acids 1–273), von Willebrand Factor Type A (VWA) domain (amino acids 240–544), Hydrolase-VWA domain (amino acids 1–544), and Fibronectin type III (FN3) domain (amino acids 643–915) were amplified by PCR utilizing primers containing ligation-independent cloning tags (Table 1). PCR products were then cloned into a modified pET28 vector encoding a hexahistidine tag (His-tag) and maltose binding protein tag on the N-terminus using ligation independent cloning method [21]. Success of cloning was confirmed by PCR and sequencing (Table 2).
Table 1.
Primers used for cloning.
| Human CLCA1 PCR Primers for cloning | |
|---|---|
| Primer Name | Primer Sequence (5′ → 3′) |
| F.P. LIC-N1-hCLCA1 | TAC TTC CAA TCC AAT GCA ATG GGC CCC TTC AAG |
| F.P. LIC-N240-hCLCA1 | TAC TTC CAA TCC AAT GCA GCC CAG CAC GTG GAC |
| F.P. LIC-N643-hCLCA1 | TAC TTC CAA TCC AAT GCA CTG GAC AAT GGA GCC |
| R.P. LIC-C544-hCLCA1 | TTA TCC ACT TCC AAT GGC CTA CTT ATC GAC CAC AAA |
| R.P. LIC-C915-hCLCA1 | TTA TCC ACT TCC AAT GGC TCA GGC GAT ACT CAG |
Table 2.
Primers used for sequencing.
| Human CLCA1 PCR Primers for sequencing | |
|---|---|
| Primer Name | Primer Sequence (5′ → 3′) |
| F.P. 306-hCLCA1 | ATA TAA AAA TGC AGA CGT GCT GGT CGC CGA AA |
| R.P. 475-hCLCA1 | GCA GAG TAT GGG CCC CAG GGC CGA GCT TTT G |
| F.P. 900-hCLCA1 | CTT CAG CCT GCT GCA GAT CGG GCA GCG GAT C |
| F.P. 1522-hCLCA1 | GGC ACA GTG ATC GTC GAC TCA ACT GTG GGA |
| F.P. 2215-hCLCA1 | GGG TCC TTT GTG GCT TCT GAC GTG CCA AAC GC |
For protein expression E. coli Rosetta (DE3) cells carrying the plasmid of interest were grown in 2 YT broth containing antibiotic at 37 °C to an OD600 of ∼0.6 and induced with 0.2 mM IPTG, after which point the cultures were grown for 18–22 h at 18 °C. The cells were then harvested by centrifugation, resuspended in buffer A (250 mM KCl, 10 mM HEPES, 25 μg/mL DNase I, 25 μg/mL lysozyme, 1 mM PMSF and 10 mM βME, pH 7.4) and lysed by sonication. The soluble fraction was loaded onto a Ni2+-NTA (Bio-Rad Profinity IMAC resin. 1560123; Biorad) affinity column and eluted with a gradient of 20–300 mM imidazole. Eluted fractions were pooled and loaded onto an amylose (E8021S; New England Biolabs) column, with pure protein being eluted with 10 mM maltose. The eluted proteins were dialyzed against Buffer D (10 mM KCl and 20 mM HEPES pH 7.4) at 4 °C overnight and loaded onto a Resource Q anion exchange column (17117901; General Electric). Bound proteins were eluted with a gradient of 10–1000 mM KCl. The eluted proteins were then concentrated with Amicon ultra-15 centrifugal filter units (30 kDa MWCO, UFC903008; Fisher Scientific)and polished by gel-filtration chromatography on a Hiload 16/60 Superdex 200 column (28989335; General Electric) by running 1 column volume of Buffer A through the column. Eluted proteins were routinely assessed to be ≥ 95% pure utilizing SDS-PAGE and Coomassie staining. Eluted proteins were then concentrated to a final volume of ∼1.5 mL and treated with 1% triton-X114 for 20 min at 4 °C, 10 min at 37 °C, and centrifuged for 10 min at 20,000g. This process was repeated 3× to completely remove endotoxin residuals. Endotoxin contamination was assessed to be ≤ 0.01 EU/mL using a quantitative Limulus Amebocyte Lysate assay (L00350; Genscript). Protein concentration was estimated using the BCA assay (23225; Thermofisher).
2.3. Circular dichroism spectroscopy
Proteins (500 μg/mL) were dialyzed into 25 mM sodium phosphate buffer (pH 7.4) containing 250 mM NaF and CD spectra were collected at 20 °C utilizing a PiStar-180 circular dichroism spectrometer with a 0.05 cm path-length cuvette. Four spectra from 260 to 190 nm were recorded with 0.5 nm resolution for each protein, averaged, and buffer subtracted.
2.4. Monocyte differentiation and activation
Monocyte cells were seeded in each well (1.3 × 106 to 1.5 × 106 cells/well) in a 6-well plate and differentiated into macrophages with 0.1 nM phorbol-12-myristate-13-acetate (PMA) in supplemented FBS-free RPMI-1640 medium for 18 h. The cells were washed 2 times with FBS-free RPMI-1640 medium and incubated in supplemented RPMI-1640 medium containing 6% FBS. For the IL-1β experiment, 1 μg/mL or 5 μg/mL of different purified hCLCA1 domain proteins were added to macrophages for 48 h. For the cell signaling experiment, 5 μg of His-MBP or His-MBP-VWA proteins were added to macrophages for 2, 4, 6, or 12 h.
2.5. RNA isolation and real-time quantitative PCR
RNA was extracted using TRIzol reagent (15596026; Thermofisher) according to manufacturers’ protocols. The collected RNA was analyzed with a GoTaq 2-Step RT-qPCR system and Mx3005P real-time PCR machine cDNA of each sample was measured in duplicates in Mx3005P real-time qPCR machine, and the average CT (cycle threshold) value was used to calculate the fold difference of each gene. Primers were designed for GAPDH, TNF-α, IL-12a, IL-8, IL-1β, IL-6 and IL-10 (Table 3).
Table 3.
Primers used in RT-qPCR experiments.
| Human qPCR Primers | ||
|---|---|---|
| Gene Name | Forward Primers (5′ → 3′) | Reverse Primers (5′ → 3′) |
| GAPDH | CAAGGTCATCCATGACAACTTTG | GGGCCATCCACAGTCTTCTG |
| TNF-α | TGCTGCACTTTGGAGTGATCG | TGCTACAACATGGGCTACAGG |
| IL-12a | CAGTGGAGGCCTGTTTACCATTG | TACTACTAAGGCACAGGGCCATC |
| IL-8 | TCTCTTGGCAGCCTTCCTGATTTC | ATTTCTGTGTTGGCGCAGTGTG |
| IL-1β | GCTGATGGCCCTAAACAGATG | TGTAGTGGTGGTCGGAGATTC |
| IL-6 | AGCCACTCACCTCTTCAGAAC | GTGCCTCTTTGCTGCTTTCAC |
| IL-10 | AAGCTGAGAACCAAGACCCAGACA | AAAGGCATTCTTCACCTGCTCCAC |
2.6. Efficiency and fold difference calculations
Dilution series from 1 × 100-fold to 1 × 10−5-fold of cDNA were used to determine the primer efficiency. The CT value obtained in each dilution was used to generate a linear plot of CT vs. log copies. The efficiency of the primer set was determined with the equation Eff = 10(−1/slope). Primer efficiencies were within the range 1.9–2. The fold difference between hCLCA1's domains-activated and unstimulated samples was determined using an efficiency-corrected calculation with unstimulated macrophage served as control and GAPDH was served as reference gene [22]:
2.7. SDS-PAGE and western blot analysis
For IL-1β experiment, cell lysates were collected using M-PER mammalian protein extraction reagent (78501; Thermofisher) with the addition of Halt protease and phosphatase inhibitor cocktail (78440; Thermofisher). The cell lysates were resolved by SDS-PAGE and electroblotted onto a PVDF membrane. The membranes were probed overnight at 4 °C with primary antibodies GAPDH (1:1000) or IL-1β (1:500), then incubated for 1 h at room temperature with secondary antibodies (DyLight 488 conjugate Goat anti-Rabbit IgG antibody (35552; Thermo Scientific) and ECL Plex Goat anti-Mouse IgG-Cy5 antibody (PA45009; Amersham Biosciences). Proteins were detected and analyzed using Typhoon Trio and ImageQuant TL system.
For cell signaling experiment, stimulated macrophages were extracted using Pierce NE-PER nuclear and cytoplasmic extraction kit (78833; Pierce) with the addition of Halt protease and phosphatase inhibitor cocktail. The nuclear fractions were then resolved by SDS-PAGE and electroblotted onto PVDF membrane. After protein detection, the antibodies were removed from membrane by incubating with Amresco gentle review stripping buffer (N552; Amresco) for 30 min at room temperature, and the membranes were probed against primary β-actin antibody (C-4; sc-47778; Santa Cruz) overnight at 4 °C. The membranes were then incubated with secondary antibodies for 1 h at room temperature. Proteins were detected and analyzed using Typhoon Trio and ImageQuant TL system. Densitometry analysis of phosphorylated proteins was normalized to β-actin in each sample. Background noise is automatically calculated by ImageQuant analysis software, based on the most common pixel values around the target bands (‘rolling ball’ algorithm). The primary antibodies used were phospho-p44/42 MAPK (Thr202/Tyr204)), phospho-p38 MAPK (Thr180/Tyr182), phospho-SAPK/JNK (Thr183/Tyr185), and phospho-IKappaB-alpha (Ser32). The secondary antibodies used were DyLight 488 conjugate Goat anti-Rabbit IgG antibody and ECL Plex Goat anti-Mouse IgG-Cy5 antibody.
2.8. Statistics
All data are expressed as means ± standard error of the mean (SEM). The normality and variance tests were done using Shaprio-Wilk test and Levene's test respectively, and the fold difference values of RT-qPCR and IL-1β western blot densitometry were analyzed using ANOVA following Tukey's Honestly-Significant-Difference Test [23]. For IL-1β expression western blot densitometry, the fold difference of hCLCA1 protein domain treated macrophages was compared to the media only negative control macrophages. For cell signaling western blot densitometry, the fold difference of His-MBP-VWA treated macrophages at given time points was compared to that of His-MBP treated macrophages at the same time points utilizing the Mann-Whitney U test. Each western blot was a result of an individual biological replicate. Significance was determined at p < 0.05.
3. Results and discussion
3.1. Purification of hCLCA1 domain proteins
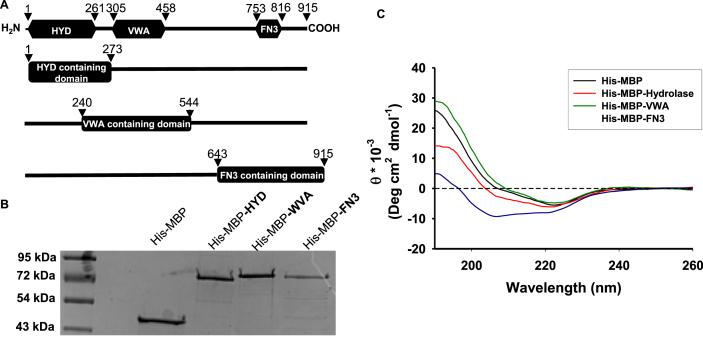
hCLCA1 cDNAs corresponding to the hydrolase domain (HYD) (amino acids 1–273), von Willebrand factor type A (VWA) domain (amino acids 240–544), and fibronectin type III (FN3) domain (amino acids 643–915) were ligated into a pET-28 expression vector containing an N-terminal hexahistidine-tag and maltose binding protein (Fig. 1-A). This created the His-MBP-HYD, His-MBP-VWA, and His-MBP-FN3 constructs. These constructs were designed to comprise both the aforementioned protein domains and the flanking regions of each domain when possible. This was done in order to ensure proper folding of the functional hCLCA1 domains and to limit the size of the fusion proteins to ∼72 kDa. An additional construct comprising all 915 amino acids of hCLCA1 was also prepared, however the large size of this fusion protein negatively affected protein stability and this protein was unable to be successfully purified (data not shown). These constructs were initially purified by Ni2+ and amylose affinity chromatography, after which point anion exchange and size exclusion chromatography were utilized to remove contaminants from the final protein preparations. All proteins were determined to be >95% pure by SDS-PAGE and Coomassie staining (Fig. 1-B). To verify folding of the maltose-binding protein-hCLCA1 domain fusion proteins far UV (260-190 nm) circular dichroism spectra of the three hCLCA1 constructs (His-MBP-HYD, His-MBP-VWA, and His-MBP-FN3) and the His-MBP protein were obtained (Fig. 1-C). Deconvolution of these spectra revealed all the protein constructs exhibited a mixture of α-helix and β-sheet secondary structure (Table 4). Of note was the dramatic increase in β-sheet secondary structure in His-MBP-HYD and His-MBP-VWA, with both constructs exhibiting ∼48% β-sheets (parallel and antiparallel combined), in contrast to the 21.3% β-sheet content calculated for His-MBP. While minima at 208 and 220 nm were apparent for the His-MBP-FN3 spectrum (Fig. 1c), these did not translate into increased α-helical structure, as His-MBP-FN3 possessed 30.1% β-sheet and only 14.7% α-helix. However, it is important to note that protein secondary structures calculated utilizing UV circular dichroism spectra are not definitive and may often differ greatly from data obtained by X-ray diffraction. This discrepancy is illustrated in the deconvolution of His-MBP protein, with the CDNN algorithm estimating 26.2% α-helix and 21.3% β-sheet (antiparallel and parallel combined) versus 43% α-helix and 20% β-sheet determined by X-ray diffraction [24]. Nevertheless, the CD spectra of the His-MBP-hCLCA1 constructs differing from the His-MBP control, with regards to an increase in beta sheet content, indicates that our protein constructs are folded. While the protein preparations were relatively pure after Ni2+ and amylose affinity chromatography, anion exchange and size exclusion chromatography steps were required to remove small amounts of contaminants from the final preparations. Comparison of the size exclusion chromatography elution profiles of the hCLCA1 protein constructs (Fig. 1s) to a set of gel filtration standards indicated that the final purified proteins eluted at volumes consistent with monomeric (His-MBP-HYD and His-MBP-FN3) or dimeric (His-MBP-VWA) proteins.
Fig. 1.
hCLCA1 domain construct design, purification, and circular dichroism spectroscopy: hCLCA1 is comprised of three functional domains (A, top): hydrolase (HYD), VWA, and FN3. Four constructs correlating to these domains were designed: amino acids 1–273 containing the hydrolase domain (His-MBP-HYD), amino acids 240–544 containing the VWA domain (His-MBP-WVA), and amino acids 643–915 containing the FN3 domain (His-MBP-FN3). His-MBP-hCLCA1 domain fusion proteins were obtained from the soluble fraction by affinity, ion exchange, and size exclusion chromatography and were assessed by SDS-PAGE (B). Structural integrity of purified His-MBP and His-MBP-hCLCA1 fusion proteins was assessed by circular dichroism spectroscopy in the far UV region from 260 to 190 nm (C).
Table 4.
Estimated Secondary Structure of Protein Constructs Used in this study. The CD spectra from Fig. 1-C were deconvoluted with CDNN software utilizing the “complex” sample dataset [54].
| His-MBP | His-MBP-HYD | His-MBP-VWA | His-MBP-FN3 | |
|---|---|---|---|---|
| α-Helix | 26.20% | 18.00% | 24.80% | 14.70% |
| Antiparallel β-sheet | 13.40% | 40.00% | 37.90% | 24.60% |
| Parallel β-sheet | 8.30% | 7.00% | 11.00% | 4.50% |
| β-Turn | 14.80% | 16.10% | 11.80% | 23.60% |
| Random Coil | 29.10% | 25.60% | 20.10% | 35.20% |
Escherichia coli based expression systems are widely used for heterologous expression and purification of proteins, including immune modulators such as interferon-α [25]. The maltose binding protein tag has found widespread use in E. coli based expression systems in recent years due to its ability to assist protein folding and improve solubility [26,27]. While the use of the MBP tag in our expression system allowed us to express and purify large amounts of the above-mentioned protein constructs, removal of the His-MBP tags by TEV protease cleavage resulted in a dramatic loss of protein stability. As a result, the His-MBP affinity tags were not cleaved off our protein constructs in subsequent experiments.
3.2. Activation of U-937 macrophages with hCLCA1 domains
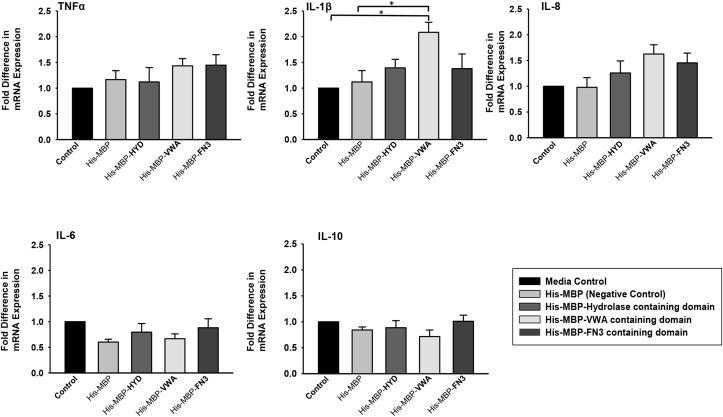
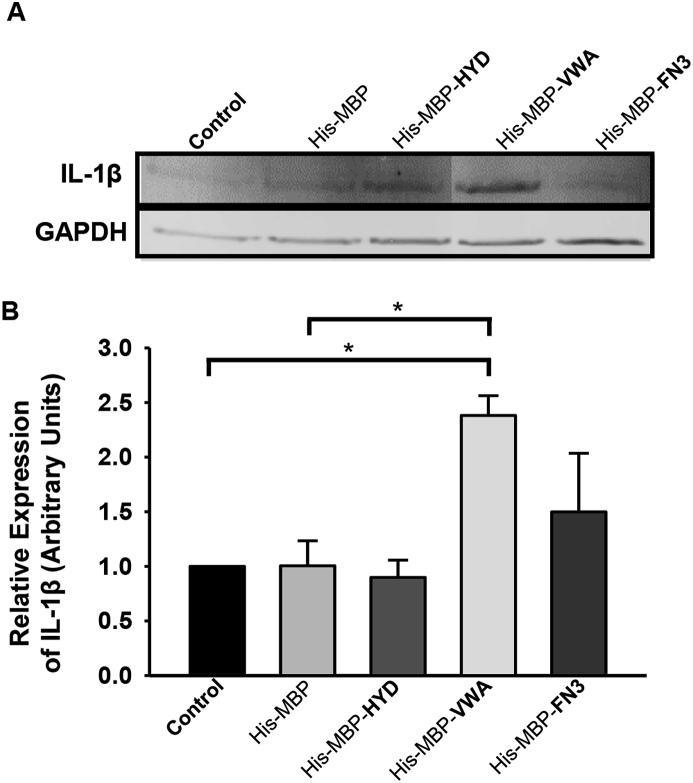
Macrophage activation by hCLCA1 was investigated using the U-937 macrophage cell line treated with 1 or 5 μg/mL of purified hCLCA1 domain proteins for 48 h; macrophage activation was assessed by expression of the pro-inflammatory cytokines TNF-α, IL-8, IL-1β, IL-6, and IL-10. Treatment with 1 μg/mL hCLCA1 constructs did not elicit an inflammatory response (data not shown), while treatment with 5 μg/mL His-MBP-VWA induced modest induction of IL-1β mRNA, with a 2.08 ± 0.19 fold increase over control observed (Fig. 2). The increase in IL-1β mRNA expression was translated into an elevated IL-1β protein expression, with western blot analysis again showing a modeset 2.38 ± 0.18 fold increase in IL-1β protein expression in His-MBP-VWA treated macrophages over the control (Fig. 3). In our previous study IL-1β was the most induced cytokine activated by immune-purified hCLCA1 [12], and was used as a marker of macrophage activation in the current work. Conversely, TNF-α, IL-8, IL-6, and IL-10 mRNA levels were not increased to a statistically significant level upon treatment with any of the hCLCA1 protein constructs, although His-MBP-VWA was capable of upregulating IL-6 transcripts 1.63 ± 0.18 fold over the media control.
Fig. 2.
Effect on cytokine expression of macrophages exposed to 5 μg/mL purified hCLCA1 domains. U-937 macrophages were treated for 48 h using 5 μg/mL of different purified hCLCA1 domain proteins. TNF-α, IL-8, IL-1β, IL-6, and IL-10 mRNA expression was quantified using RT-qPCR. The fold difference of each sample was compared against the control. Results were the means of 5 samples ± SEM. Significant fold differences from corresponding control values are indicated by * (p < 0.05).
Fig. 3.
VWA containing domain (amino acids 240–544) of hCLCA1 increased IL-1β protein expression level. U-937 macrophages were treated for 48 h using 5 μg/mL of different purified hCLCA1 domain proteins. IL-1β protein levels were then assessed by western blotting. The fold difference of each sample was compared against the negative control (media only). Results were the means of 3 samples ± SEM. Significant fold differences from corresponding control values are indicated by * (p < 0.05).
In our previous study, ∼150 pg/mL of immuno-purified hCCLA1 was sufficient to induce macrophage activation [12], while another study reported a physiological hCLCA1 concentration of ∼0.23 μg/mL in IL-13 induced NHBE cells [28], values that are several orders of magnitude lower than the 5 μg/mL doses used in this experiment. One possible explanation for this discrepancy is the inability of our E. coli based expression system to post-translationally glycosylate expressed proteins [29]. Secreted hCLCA1 has been shown to be a highly glycosylated protein [30,31], and glycosylation is known to modulate the structure and function of signaling molecules [[32], [33], [34]]. Additionally, while prior studies have reported that the MBP tag does not affect bioactivity of purified proteins [35,36], we cannot preclude the possibility that the MBP tag may impair macrophage activation by His-MBP-VWA due to conformational hindrance. Finally, a 3 step Triton X-114 phase separation protocol was required to reduce endotoxin levels of purified hCLCA1 constructs below 0.01 EU/mL. Given that Adam et al., reported a 50% loss of bioactivity utilizing a 2-step Triton X-114 endotoxin removal protocol [37] it is possible that the high dose of His-MBP-VWA required for macrophage activation in this experiment was a result of reduced protein activity.
The VWA domain was first identified in von Willebrand factor, a protein responsible for platelet and collagen binding during hemostasis [38]. Since this initial discovery VWA domains have been identified in a variety of proteins such as integrins, extracellular matrix proteins, collagens, where they primarily function as adhesion molecules [[39], [40], [41], [42]]. This adhesion functionality plays a crucial role in the inflammatory response, as von Willebrand factor has been shown to facilitate leukocyte and neutrophil recruitment and adhesion during inflammation [[16], [17], [18]]. In some cases, this adhesion to von Willebrand factor by leukocytes was shown to be mediated by β2 integrins [17]. Interestingly, several other VWA domain containing proteins are known to adhere to integrins. In particular, matrilins 1, 2, and 3 interact with integrin α1β1 [43,44], while the basement membrane protein AMACO promotes cell attachment to β1 integrins [45]. β1 and αVβ3 integrins have been shown to induce activation of both NF-κB and MAPK pathways in response to ligand binding [46,47]. This activation resulted in increased expression of the pro-inflammatory cytokines TNF-α, IL-1β, IL-6, and IL-8 in macrophages [47]. Thus macrophage activation induced by the VWA domain of hCLCA1 could be due to integrin binding and subsequent downstream signaling effects.
3.3. Phosphorylation of MAPKs and NF-κB pathways by VWA domain
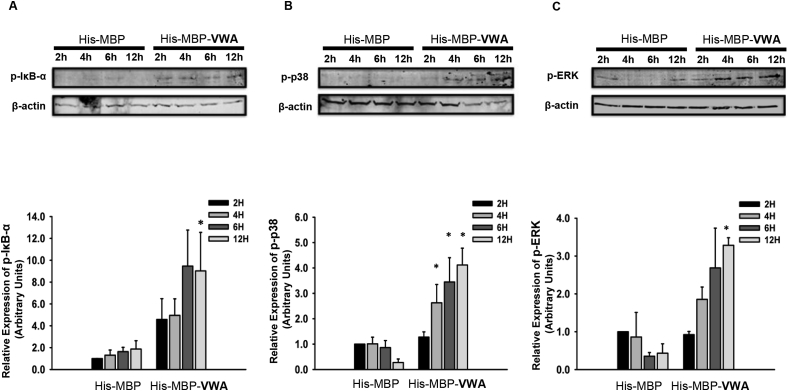
Given the precedence for VWA domain induced integrin binding, as well as the role of the MAPK and NF-κB pathways induced by integrin activation in IL-1β expression [[48], [49], [50], [51], [52], [53]], we sought to investigate potential activation of the MAPK and NF-κB pathways by the hCLCA1 VWA domain. For this experiment macrophages were induced with 5 μg/mL His-MBP-VWA for 2, 4, 6, or 12 h and activation of the MAPK pathway was then assessed by quantification of phosphorylated p38, ERK, and JNK by western blotting, while quantification of phosphorylated IκB-α was used to examine activation of the NF-κB pathway.
Western blot and densitometry analysis showed that His-MBP-VWA significantly increased levels of phosphorylated IκB-α (Fig. 4A), p38 (Fig. 4B), and ERK (Fig. 4C) in macrophages over the His-MBP activated macrophages. While IκB-α phosphorylation was only significantly greater than control at 12 h, large (4.58 and 4.95 fold) increases in phosphorylation were apparent at 2 and 4 h, while the level of phosphorylation plateaued after 6 h, suggesting an early activation of the NF-κB pathway. Conversely, increased levels of phosphorylation of p38 and ERK were only apparent after 4 h of incubation and appeared to increase in a linear fashion until the end of the 12 h timepoint. Macrophage treatment with His-MBP-VWA did not induce detectable levels of phosphorylated JNK, while p-JNK was detected in LPS stimulated macrophages (Fig. s2), suggesting that the signaling response observed was solely due to macrophage activation by the hCLCA1 VWA domain and not due to residual endotoxin contamination.
Fig. 4.
Levels of phosphorylated IκB-α, p38 and ERK with VWA domain treatment. U-937 macrophages were treated for 2, 4, 6, or 12 h with 5 μg/mL of His-MBP or His-MBP-VWA protein. The levels of phosphorylated IκB-α (A), p38 (B) and ERK (C) was then assessed by western blotting. The fold difference of each sample was compared against His-MBP treated macrophage at 1 h. Results were the means of 3 samples ± SEM. Significant fold differences from corresponding control values at the given timepoints are indicated by * (p < 0.05).
In the present study we have identified the VWA domain of hCLCA1 to be sufficient for macrophage activation, inducing modest expression of IL-1β in U-937 macrophages. This activation was shown to correlate with the activation of NF-κB and MAPK pathways, both of which have been implicated in inflammatory cytokine expression. The activation of macrophages by hCLCA1 would modify the inflammatory response, mucus secretion, and airway higher responsiveness. Thus, identification of the protein domain within hCLCA1 with signaling function is of great therapeutic interest, as it provides a specific target to one of the most highly induced gene products of airway inflammatory disease.
Acknowledgements
The work was supported by a Natural Sciences and Engineering Research Council of Canada Discovery Grant 371364-2010 (MEL).
Footnotes
Transparency document related to this article can be found online at https://doi.org/10.1016/j.bbrep.2019.100630.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2019.100630.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Representative size exclusion elution profiles of molecular size standards, His-MBP-HYD, His-MBP-VWA, and His-MBP-FN3. The elution profile of the Hiload 16/60 Superdex 200 column was first established with a set of gel filtration standards (1511901; Biorad) of sizes 670 kDa (thyroglobin, A), 150 kDa (immunoglobulin gamma, B), 44 kDa (ovalbumin, C), 17 kDa (myoglobin, D), and 1.35 kDa (vitamin B12, E). Representative chromatograms of the purification of His-MBP-HYD, His-MBP-VWA, and His-MBP-FN3 are attached. High molecular weight aggregates (>600 kDa), dimeric protein (∼150 kDa calculated MW), monomeric protein (∼72 kDa calculated MW), and small impurities (≤45 kDa calculated MW) are annotated on each chromatogram. In all cases monomeric His-MBP-hCLCA1 domain proteins were isolated and used for downstream applications, with the exception of His-MBP-VWA, which could only be isolated as a dimer.
Validation of phospho-specific antibodies with LPS-induced macrophages. Macrophages were activated with 1 μg/mL LPS for 4 h before protein isolation. Nuclear fraction proteins were analyzed with western blot against different phosphor-specific antibodies: p-IκB-α, p-p38, p-ERK, and p-JNK. A representative blot showing no detectable p-JNK from macrophages stimulated with His-MBP-VWA for 12 h is also shown.
Transparency document
References
- 1.Kamada F., Suzuki Y., Shao C., Tamari M., Hasegawa K., Hirota T., Shimizu M., Takahashi N., Mao X.Q., Doi S., Fujiwara H., Miyatake A., Fujita K., Chiba Y., Aoki Y., Kure S., Tamura G., Shirakawa T., Matsubara Y. Association of the hCLCA1 gene with childhood and adult asthma. Genes Immun. 2004;5:540–547. doi: 10.1038/sj.gene.6364124. [DOI] [PubMed] [Google Scholar]
- 2.Brouillard F., Bensalem N., Hinzpeter A., Tondelier D., Trudel S., Gruber A.D., Ollero M., Edelman A. Blue native/SDS-PAGE analysis reveals reduced expression of the mClCA3 protein in cystic fibrosis knock-out mice. Mol. Cell. Proteomics : MCP. 2005;4:1762–1775. doi: 10.1074/mcp.M500098-MCP200. [DOI] [PubMed] [Google Scholar]
- 3.Hegab A.E., Sakamoto T., Uchida Y., Nomura A., Ishii Y., Morishima Y., Mochizuki M., Kimura T., Saitoh W., Massoud H.H., Massoud H.M., Hassanein K.M., Sekizawa K. CLCA1 gene polymorphisms in chronic obstructive pulmonary disease. J. Med. Genet. 2004;41:e27. doi: 10.1136/jmg.2003.012484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hauber H.P., Lavigne F., Hung H.L., Levitt R.C., Hamid Q. Effect of Th2 type cytokines on hCLCA1 and mucus expression in cystic fibrosis airways. J. Cyst. Fibros. : Off. J. Eur. Cyst. Fibros. Soc. 2010;9:277–279. doi: 10.1016/j.jcf.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 5.Cunningham S.A., Awayda M.S., Bubien J.K., Ismailov I.I., Arrate M.P., Berdiev B.K., Benos D.J., Fuller C.M. Cloning of an epithelial chloride channel from bovine trachea. J. Biol. Chem. 1995;270:31016–31026. doi: 10.1074/jbc.270.52.31016. [DOI] [PubMed] [Google Scholar]
- 6.Gaspar K.J., Racette K.J., Gordon J.R., Loewen M.E., Forsyth G.W. Cloning a chloride conductance mediator from the apical membrane of porcine ileal enterocytes. Physiol. Genom. 2000;3:101–111. doi: 10.1152/physiolgenomics.2000.3.2.101. [DOI] [PubMed] [Google Scholar]
- 7.Hamann M., Gibson A., Davies N., Jowett A., Walhin J.P., Partington L., Affleck K., Trezise D., Main M. Human ClCa1 modulates anionic conduction of calcium-dependent chloride currents. J. Physiol. 2009;587:2255–2274. doi: 10.1113/jphysiol.2009.170159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loewen M.E., Bekar L.K., Gabriel S.E., Walz W., Forsyth G.W. pCLCA1 becomes a cAMP-dependent chloride conductance mediator in Caco-2 cells. Biochem. Biophys. Res. Commun. 2002;298:531–536. doi: 10.1016/s0006-291x(02)02498-1. [DOI] [PubMed] [Google Scholar]
- 9.Loewen M.E., Bekar L.K., Walz W., Forsyth G.W., Gabriel S.E. pCLCA1 lacks inherent chloride channel activity in an epithelial colon carcinoma cell line. Am. J. Physiol. Gastrointest. Liver Physiol. 2004;287:G33–G41. doi: 10.1152/ajpgi.00023.2004. [DOI] [PubMed] [Google Scholar]
- 10.Loewen M.E., Forsyth G.W. Structure and function of CLCA proteins. Physiol. Rev. 2005;85:1061–1092. doi: 10.1152/physrev.00016.2004. [DOI] [PubMed] [Google Scholar]
- 11.Sabo-Attwood T., Ramos-Nino M., Bond J., Butnor K.J., Heintz N., Gruber A.D., Steele C., Taatjes D.J., Vacek P., Mossman B.T. Gene expression profiles reveal increased mClca3 (Gob5) expression and mucin production in a murine model of asbestos-induced fibrogenesis. Am. J. Pathol. 2005;167:1243–1256. doi: 10.1016/S0002-9440(10)61212-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ching J.C., Lobanova L., Loewen M.E. Secreted hCLCA1 is a signaling molecule that activates airway macrophages. PLoS One. 2013;8 doi: 10.1371/journal.pone.0083130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dietert K., Reppe K., Mundhenk L., Witzenrath M., Gruber A.D. mCLCA3 modulates IL-17 and CXCL-1 induction and leukocyte recruitment in murine Staphylococcus aureus pneumonia. PLoS One. 2014;9 doi: 10.1371/journal.pone.0102606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parks W.C., Wilson C.L., López-Boado Y.S. Matrix metalloproteinases as modulators of inflammation and innate immunity. Nat. Rev. Immunol. 2004;4:617. doi: 10.1038/nri1418. [DOI] [PubMed] [Google Scholar]
- 15.Van den Steen P.E., Proost P., Wuyts A., Van Damme J., Opdenakker G. Neutrophil gelatinase B potentiates interleukin-8 tenfold by aminoterminal processing, whereas it degrades CTAP-III, PF-4, and GRO-α and leaves RANTES and MCP-2 intact. Blood. 2000;96:2673–2681. [PubMed] [Google Scholar]
- 16.Bernardo A., Ball C., Nolasco L., Choi H., Moake J.L., Dong J.F. Platelets adhered to endothelial cell-bound ultra-large von Willebrand factor strings support leukocyte tethering and rolling under high shear stress. J. Thromb. Haemost. 2005;3:562–570. doi: 10.1111/j.1538-7836.2005.01122.x. [DOI] [PubMed] [Google Scholar]
- 17.Pendu R., Terraube V., Christophe O.D., Gahmberg C.G., de Groot P.G., Lenting P.J., Denis C.V. P-selectin glycoprotein ligand 1 and β2-integrins cooperate in the adhesion of leukocytes to von Willebrand factor. Blood. 2006;108:3746–3752. doi: 10.1182/blood-2006-03-010322. [DOI] [PubMed] [Google Scholar]
- 18.Denis C.V., André P., Saffaripour S., Wagner D.D. Defect in regulated secretion of P-selectin affects leukocyte recruitment in von Willebrand factor-deficient mice. Proc. Natl. Acad. Sci. U.S.A. 2001;98:4072–4077. doi: 10.1073/pnas.061307098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolachala V.L., Bajaj R., Wang L., Yan Y., Ritzenthaler J.D., Gewirtz A.T., Roman J., Merlin D., Sitaraman S.V. Epithelial-derived fibronectin expression, signaling, and function in intestinal inflammation. J. Biol. Chem. 2007;282:32965–32973. doi: 10.1074/jbc.M704388200. [DOI] [PubMed] [Google Scholar]
- 20.Kelsh R., You R., Horzempa C., Zheng M., McKeown-Longo P.J. Regulation of the innate immune response by fibronectin: synergism between the III-1 and EDA domains. PLoS One. 2014;9 doi: 10.1371/journal.pone.0102974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lobo P.A., Van Petegem F. Crystal structures of the N-terminal domains of cardiac and skeletal muscle ryanodine receptors: insights into disease mutations. Structure. 2009;17:1505–1514. doi: 10.1016/j.str.2009.08.016. [DOI] [PubMed] [Google Scholar]
- 22.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stahlberg A., Rusnakova V., Forootan A., Anderova M., Kubista M. RT-qPCR work-flow for single-cell data analysis. Methods. 2013;59:80–88. doi: 10.1016/j.ymeth.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 24.Quiocho F.A., Spurlin J.C., Rodseth L.E. Extensive features of tight oligosaccharide binding revealed in high-resolution structures of the maltodextrin transport/chemosensory receptor. Structure. 1997;5:997–1015. doi: 10.1016/s0969-2126(97)00253-0. [DOI] [PubMed] [Google Scholar]
- 25.EL-Baky N., Linjawi M.H., Redwan E.M. Auto-induction expression of human consensus interferon-alpha in Escherichia coli. BMC Biotechnol. 2015;15 doi: 10.1186/s12896-015-0128-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bach H., Mazor Y., Shaky S., Shoham-Lev A., Berdichevsky Y., Gutnick D.L., Benhar I. Escherichia coli maltose-binding protein as a molecular chaperone for recombinant intracellular cytoplasmic single-chain antibodies. J. Mol. Biol. 2001;312:79–93. doi: 10.1006/jmbi.2001.4914. [DOI] [PubMed] [Google Scholar]
- 27.Kapust R.B., Waugh D.S. Escherichia coli maltose-binding protein is uncommonly effective at promoting the solubility of polypeptides to which it is fused. Protein Sci. : Publ. Protein Soc. 1999;8:1668–1674. doi: 10.1110/ps.8.8.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alevy Y.G., Patel A.C., Romero A.G., Patel D.A., Tucker J., Roswit W.T., Miller C.A., Heier R.F., Byers D.E., Brett T.J., Holtzman M.J. IL-13-induced airway mucus production is attenuated by MAPK13 inhibition. J. Clin. Investig. 2012;122:4555–4568. doi: 10.1172/JCI64896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sahdev S., Khattar S.K., Saini K.S. Production of active eukaryotic proteins through bacterial expression systems: a review of the existing biotechnology strategies. Mol. Cell. Biochem. 2008;307:249–264. doi: 10.1007/s11010-007-9603-6. [DOI] [PubMed] [Google Scholar]
- 30.Gibson A., Lewis A.P., Affleck K., Aitken A.J., Meldrum E., Thompson N. hCLCA1 and mCLCA3 are secreted non-integral membrane proteins and therefore are not ion channels. J. Biol. Chem. 2005;280:27205–27212. doi: 10.1074/jbc.M504654200. [DOI] [PubMed] [Google Scholar]
- 31.Gruber A.D., Gandhi R., Pauli B.U. The murine calcium-sensitive chloride channel (mCaCC) is widely expressed in secretory epithelia and in other select tissues. Histochem. Cell Biol. 1998;110:43–49. doi: 10.1007/s004180050263. [DOI] [PubMed] [Google Scholar]
- 32.Ruddon R.W., Bedows E. Assisted protein folding. J. Biol. Chem. 1997;272:3125–3128. doi: 10.1074/jbc.272.6.3125. [DOI] [PubMed] [Google Scholar]
- 33.Shental-Bechor D., Levy Y. Effect of glycosylation on protein folding: a close look at thermodynamic stabilization. Proc. Natl. Acad. Sci. U.S.A. 2008;105:8256–8261. doi: 10.1073/pnas.0801340105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shental-Bechor D., Levy Y. Folding of glycoproteins: toward understanding the biophysics of the glycosylation code. Curr. Opin. Struct. Biol. 2009;19:524–533. doi: 10.1016/j.sbi.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 35.Pennati A., Deng J., Galipeau J. Maltose-binding protein fusion allows for high level bacterial expression and purification of bioactive mammalian cytokine derivatives. PLoS One. 2014;9 doi: 10.1371/journal.pone.0106724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Do B.H., Ryu H.B., Hoang P., Koo B.K., Choe H. Soluble prokaryotic overexpression and purification of bioactive human granulocyte colony-stimulating factor by maltose binding protein and protein disulfide isomerase. PLoS One. 2014;9 doi: 10.1371/journal.pone.0089906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adam O., Vercellone A., Paul F., Monsan P.F., Puzo G. A nondegradative route for the removal of endotoxin from exopolysaccharides. Anal. Biochem. 1995;225:321–327. doi: 10.1006/abio.1995.1161. [DOI] [PubMed] [Google Scholar]
- 38.Sadler J.E. Biochemistry and genetics of von Willebrand factor. Annu. Rev. Biochem. 1998;67:395–424. doi: 10.1146/annurev.biochem.67.1.395. [DOI] [PubMed] [Google Scholar]
- 39.Tuckwell D. Evolution of von Willebrand factor A (VWA) domains. Biochem. Soc. Trans. 1999;27:835–840. doi: 10.1042/bst0270835. [DOI] [PubMed] [Google Scholar]
- 40.Hynes R.O., Zhao Q. The evolution of cell adhesion. J. Cell Biol. 2000;150:F89–F96. doi: 10.1083/jcb.150.2.f89. [DOI] [PubMed] [Google Scholar]
- 41.Colombatti A., Bonaldo P., Doliana R. Type A modules: interacting domains found in several non-fibrillar collagens and in other extracellular matrix proteins. Matrix. 1993;13:297–306. doi: 10.1016/s0934-8832(11)80025-9. [DOI] [PubMed] [Google Scholar]
- 42.Whittaker C.A., Hynes R.O. Distribution and evolution of von Willebrand/integrin a domains: widely dispersed domains with roles in cell adhesion and elsewhere. Mol. Biol. Cell. 2002;13:3369–3387. doi: 10.1091/mbc.E02-05-0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Makihira S., Yan W., Ohno S., Kawamoto T., Fujimoto K., Okimura A., Yoshida E., Noshiro M., Hamada T., Kato Y. Enhancement of cell adhesion and spreading by a cartilage-specific noncollagenous protein, cartilage matrix protein (CMP/Matrilin-1), via integrin α1β1. J. Biol. Chem. 1999;274:11417–11423. doi: 10.1074/jbc.274.16.11417. [DOI] [PubMed] [Google Scholar]
- 44.Mann H.H., Sengle G., Gebauer J.M., Eble J.A., Paulsson M., Wagener R. Matrilins mediate weak cell attachment without promoting focal adhesion formation. Matrix Biol. 2007;26:167–174. doi: 10.1016/j.matbio.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 45.Gebauer J.M., Keene D.R., Olsen B.R., Sorokin L.M., Paulsson M., Wagener R. Mouse AMACO, a kidney and skin basement membrane associated molecule that mediates RGD-dependent cell attachment. Matrix Biol. 2009;28:456–462. doi: 10.1016/j.matbio.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 46.Reyes-Reyes M., Mora N., Zentella A., Rosales C. Phosphatidylinositol 3-kinase mediates integrin-dependent NF-(κ)B and MAPK activation through separate signaling pathways. J. Cell Sci. 2001;114:1579–1589. doi: 10.1242/jcs.114.8.1579. [DOI] [PubMed] [Google Scholar]
- 47.Antonov A.S., Antonova G.N., Munn D.H., Mivechi N., Lucas R., Catravas J.D., Verin A.D. αVβ3 integrin regulates macrophage inflammatory responses via PI3 kinase/akt-dependent NF-κB activation. J. Cell. Physiol. 2011;226:469–476. doi: 10.1002/jcp.22356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hiscott J., Marois J., Garoufalis J., D'Addario M., Roulston A., Kwan I., Pepin N., Lacoste J., Nguyen H., Bensi G. Characterization of a functional NF-kappa B site in the human interleukin 1 beta promoter: evidence for a positive autoregulatory loop. Mol. Cell Biol. 1993;13:6231–6240. doi: 10.1128/mcb.13.10.6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Risbud M.V., Shapiro I.M. Role of cytokines in intervertebral disc degeneration: pain and disc content, Nature reviews. Rheumatology. 2014;10:44–56. doi: 10.1038/nrrheum.2013.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Singer C.A., Lontay B., Unruh H., Halayko A.J., Gerthoffer W.T. Src mediates cytokine-stimulated gene expression in airway myocytes through ERK MAPK, Cell communication and signaling. CCS. 2011;9:14. doi: 10.1186/1478-811X-9-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen B.C., Lin W.W. PKC- and ERK-dependent activation of I kappa B kinase by lipopolysaccharide in macrophages: enhancement by P2Y receptor-mediated CaMK activation. Br. J. Pharmacol. 2001;134:1055–1065. doi: 10.1038/sj.bjp.0704334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goebeler M., Gillitzer R., Kilian K., Utzel K., Brocker E.B., Rapp U.R., Ludwig S. Multiple signaling pathways regulate NF-kappaB-dependent transcription of the monocyte chemoattractant protein-1 gene in primary endothelial cells. Blood. 2001;97:46–55. doi: 10.1182/blood.v97.1.46. [DOI] [PubMed] [Google Scholar]
- 53.Tamura D.Y., Moore E.E., Johnson J.L., Zallen G., Aiboshi J., Silliman C.C. p38 mitogen-activated protein kinase inhibition attenuates intercellular adhesion molecule-1 up-regulation on human pulmonary microvascular endothelial cells. Surgery. 1998;124:403–407. discussion 408. [PubMed] [Google Scholar]
- 54.Böhm G., Muhr R., Jaenicke R. Quantitative analysis of protein far UV circular dichroism spectra by neural networks. Protein Eng. Des. Sel. 1992;5:191–195. doi: 10.1093/protein/5.3.191. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Representative size exclusion elution profiles of molecular size standards, His-MBP-HYD, His-MBP-VWA, and His-MBP-FN3. The elution profile of the Hiload 16/60 Superdex 200 column was first established with a set of gel filtration standards (1511901; Biorad) of sizes 670 kDa (thyroglobin, A), 150 kDa (immunoglobulin gamma, B), 44 kDa (ovalbumin, C), 17 kDa (myoglobin, D), and 1.35 kDa (vitamin B12, E). Representative chromatograms of the purification of His-MBP-HYD, His-MBP-VWA, and His-MBP-FN3 are attached. High molecular weight aggregates (>600 kDa), dimeric protein (∼150 kDa calculated MW), monomeric protein (∼72 kDa calculated MW), and small impurities (≤45 kDa calculated MW) are annotated on each chromatogram. In all cases monomeric His-MBP-hCLCA1 domain proteins were isolated and used for downstream applications, with the exception of His-MBP-VWA, which could only be isolated as a dimer.
Validation of phospho-specific antibodies with LPS-induced macrophages. Macrophages were activated with 1 μg/mL LPS for 4 h before protein isolation. Nuclear fraction proteins were analyzed with western blot against different phosphor-specific antibodies: p-IκB-α, p-p38, p-ERK, and p-JNK. A representative blot showing no detectable p-JNK from macrophages stimulated with His-MBP-VWA for 12 h is also shown.