Abstract
Resolution of the Holliday junction (HJ) is essential for homologous recombination and DNA repair. In Saccharomyces cerevisiae, HJ resolvase Yen1 and the Mus81-Mms4 complex are redundant in DNA damage repair. In cultured mammalian cells, such redundancy also exists between Yen1 ortholog GEN1 and the Mus81-Mms1 ortholog MUS81-EME1. In this report, we further tested if GEN1 and EME1 redundantly affect HJ-related physiological processes in mice. We found that combined homozygous mutations of Gen1 and Eme1 led to synthetic lethality during early embryonic stages. Homozygous Gen1 mutations did not cause DNA repair deficiency in mouse embryonic fibroblast (MEF) cells, but made heterozygous Eme1 mutant MEFs more sensitive to various DNA-damaging reagents. Gen1 mutations also reduced the meiotic recombination efficiency in Eme1 mutant mice. These results suggest that Gen1 and Eme1 play redundant roles in DNA repair and meiotic recombination in vivo.
Keywords: : Gen1, Eme1, DNA repair, meiotic recombination
Introduction
A Holliday junction (HJ) is a branched intermediate structure that usually exists in meiosis and DNA recombination repair (Matos and West, 2014). HJs connect two double-stranded DNA molecules together so that they must be resolved to allow the segregation of both molecules (West, 2009). Defective HJ resolution may lead to sterility or increased susceptibility to cancer (Holloway et al., 2008; Matos et al., 2011). Several evolutionary conserved pathways are involved in HJ resolution in eukaryotic organisms.
In Saccharomyces cerevisiae, the Yen1 nuclease can symmetrically cut HJ in a manner analogous to the Escherichia coli HJ resolvase RuvC (Ip et al., 2008; Rass et al., 2010), whereas the XPF-family heterodimeric endonuclease complex Mus81-Mms4 can asymmetrically cleave HJ (Boddy et al., 2001; Chen et al., 2001). Slx1, another structure-selective endonuclease, is also capable of processing HJs (Fricke and Brill, 2003; Fekairi et al., 2009; Munoz et al., 2009). In addition to these structure-specific nucleases, HJs can be dissolved by the Sgs1-Top3-Rmi1 (STR) complex to produce noncrossover recombinants (Ellis et al., 1995; Wu and Hickson, 2003; Cejka et al., 2010).
In human cells, the Yen1 ortholog GEN1, Mus81-Mms4 ortholgs MUS81-EME1, and the Slx1 ortholog SLX1 can process HJs in similar ways as they do in yeast cells (Ip et al., 2008; Matos and West, 2014). It has been shown that GEN1 forms a dimer that juxtaposes two products in a substrate-like complex, and the chromodomain of GEN1 is indispensable for its DNA recognition and cleavage activities (Lee et al., 2015; Liu et al., 2015). Both MUS81 and SLX1 can be activated by interacting with SLX4, a scaffold protein that serves as an essential docking platform to cooperate with multiple structure-specific endonucleases (Fekairi et al., 2009; Munoz et al., 2009; Svendsen et al., 2009). HJs could also be dissolved by the Bloom's syndrome complex (BLM helicase-topoisomerase IIIα-RMI1/2) to produce noncrossover recombinants (Ellis et al., 1995; Wu and Hickson, 2003; Cejka et al., 2010).
Yen1 and the Mus81-Mms4 complex play redundant roles in HJ resolution in yeasts. Deletion of yen1 in S. cerevisiae does not affect DNA repair, whereas disruption of both yen1 and mus81 caused hypersensitivity to various DNA-damaging reagents (Blanco et al., 2010; Ho et al., 2010; Tay and Wu, 2010). During meiosis, deletion of both yen1 and mms4 also caused more severe decrease in chromosome crossover than the single mutations did (Zakharyevich et al., 2012). In addition, ectopic expression of human GEN1 could overcome HJ defects caused by mus81 deletion in fission yeasts, resulting in the formation of chromosome crossover (Lorenz et al., 2010). Recent studies in mitotic human cells have shown sequential activation of GEN1 and MUS81-EME1 to eliminate persistently jointed DNA molecules (Matos et al., 2011; Matos and West, 2014). Depletion of both GEN1 and MUS81 in cultured cells from Bloom's syndrome patients resulted in elongated and segmented chromosomes, as well as high levels of cell mortality (Wechsler et al., 2011; Wyatt et al., 2013). These results indicate that the functional redundancy between GEN1 and MUS81-EME1 also exists in mammalian cells. However, little evidence has been provided for the functional redundancy between GEN1 and EME1, making this conclusion remain to be confirmed in mammals.
We have generated Gen1 and Eme1 mutations in mice by insertional mutagenesis with the piggyBac (PB) transposon. We report synthetic lethality, reduced DNA repair, and decreased meiotic recombination efficiency in mice and cells carrying both mutations. These results suggest a redundant role between Gen1 and Eme1 in DNA recombination in mice.
Materials and Methods
Mice
All animal experiments were performed in accordance with protocols approved by the Animal Care and Use Committee of the Institute of Developmental Biology and Molecular Medicine (IDM), Fudan University. Both Gen1 (081125049-HLA) and Eme1 (081028120-HLA) mutants were generated by inserting a PB transposon in target genes during the process of a large-scale insertional mutagenesis project on the FVB/NJ background (Ding et al., 2005; Sun et al., 2008). In the Gen1PB allele, the PB insertion was mapped in the second intron (Chr:12.11268138, Ensembl release 54). In the Eme1PB allele, the PB inserts into the third intron (Chr: 11.94510958, Ensembl release 54).
Cell culture and DNA damage assay
Mouse embryonic fibroblasts (MEFs) were isolated from E14.5 embryos and cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum, 1% l-glutamine, and 1% penicillin–streptomycin. The assay for sensitivity to DNA damage was performed as previously described (Lei et al., 2012). In brief, MEFs of indicated genotype were plated in three wells of a six-well plate at a density of 5 × 104 per well for 12 h, then treated with CPT (264933; Jingke Chem), Etoposide (E1383; Sigma), MMC (M4287; Sigma), MMS (129925; Sigma), or HU (H8627; Sigma) at indicated doses for 24 h. Surviving cells in each well were counted by FACS after 1-week incubation. Cell survival rate was calculated by dividing survival cell number in each well by mean survival cell number in control group without drug treatment. Statistical analysis was performed by unpaired t-test.
Polymerase chain reaction
Genotyping polymerase chain reaction (PCR) was performed with a PB-specific primer (LB2) and two flanking genomic primers (Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/dna). PCR and reverse transcription polymerase chain reaction (RT-PCR) primers are listed in Supplementary Table S1.
Measurement of meiotic recombination efficiency
Male Gen1 mutant mice with mixed FVB/NJ and C57BL/6J background were bred in two steps: Gen1PB/+ (FVB/NJ) mice were mated with C57BL/6J wild-type mice to generate Gen1PB/+ (FVB/C57); Gen1PB/+ (FVB/C57) mice were then mated with Gen1PB/+ (FVB/NJ) mice to generate wild-type and Gen1PB/PB mice with mixed background. Combined Gen1 and Eme1 mutant mice with mixed background were generated by mating Gen1PB/+ mice (FVB/C57) with Gen1PB/+; Eme1PB/PB mice (FVB/NJ). Heterozygosity of the single nucleotide polymorphisms (SNPs) in these mice was confirmed using Sanger sequencing after genotyping PCR (Supplementary Table S1). The meiotic recombination efficiency was measured in the offspring of male mice with indicated genotypes and FVB/NJ females by genotyping PCR and Sanger sequencing. Meiotic recombination efficiency = [No. (rs32032816G/G; rs32330931A/G) + No. (rs32032816C/G; rs32330931G/G)]/No. (all offspring).
Statistics
Statistical methods were indicated in the figure legend. The threshold for significance was set at p < 0.05.
Results
Disruption of Gen1 and Eme1 causes synthetic lethality
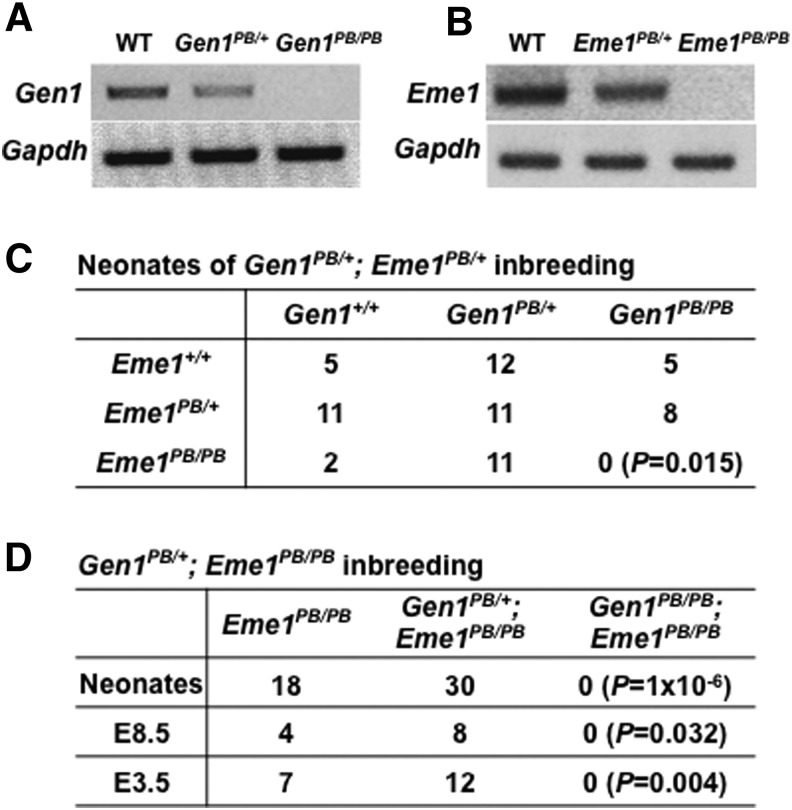
We have isolated Gen1 and Eme1 mutants from a PB insertional mutagenesis project in mice (Sun et al., 2008). The PB transposon was inserted into the second and third intron of Gen1 and Eme1, respectively. Both mutants were viable and fertile. RT-PCR analysis of individual mutant alleles showed that gene expression was reduced or extinguished in MEF cells isolated from heterozygous or homozygous mutants, respectively (Fig. 1A, B). To test if Gen1 and Eme1 play redundant roles in mice, we first tried to generate mice carrying both mutations. We have successfully obtained double heterozygous mice (Gen1PB/+; Eme1PB/+) and Gen1PB/+; Eme1PB/PB mutants. However, inconsistent with expected Mendelian distribution, double homozygotes (Gen1PB/PB; Eme1PB/PB) were observed neither among 65 neonates generated from inbreeding of Gen1PB/+; Eme1PB/+ mice (Fig. 1C, p = 0.015), nor among 48 progenies born by Gen1PB/+; Eme1PB/PB inbreeding (Fig. 1D, p = 1 × 10−6), indicating prenatal lethality of double homozygotes. To precisely pinpoint the time of embryonic death, embryos from Gen1PB/+; Eme1PB/PB intercrosses were dissected at different stages and genotyped by PCR (Fig. 1D). We failed to identify double homozygotes at embryonic day 8.5 (E8.5) (0/12, p = 0.032), or even at E3.5 (0/19 blastocysts, p = 0.004), indicating that Gen1PB/PB; Eme1PB/PB embryos could not survive through early embryonic development. The synthetic lethality caused by Gen1 and Eme1 mutations indicates that both genes are required for embryonic development.
FIG. 1.
Disruption of Gen1 and Eme1 caused synthetic lethality. RT-PCR showed disrupted expression of Gen1 (A) and Eme1 (B) in MEFs bearing homozygous mutations, respectively. Genotyping PCR of neonates (C, D) and embryonic day 8.5 (E8.5) offspring (D), and E3.5 blastocysts (D) from indicated mice inbreeding detected no double homozygous mutant (Gen1PB/PB; Eme1PB/PB), respectively. p-Value of Fisher's exact test was labeled in the figure. MEFs, mouse embryonic fibroblasts; PB, piggyBac; RT-PCR, reverse transcription polymerase chain reaction; WT, wild-type.
Gen1 and Eme1 play redundant roles in cellular resistance to DNA damage-induced cell death
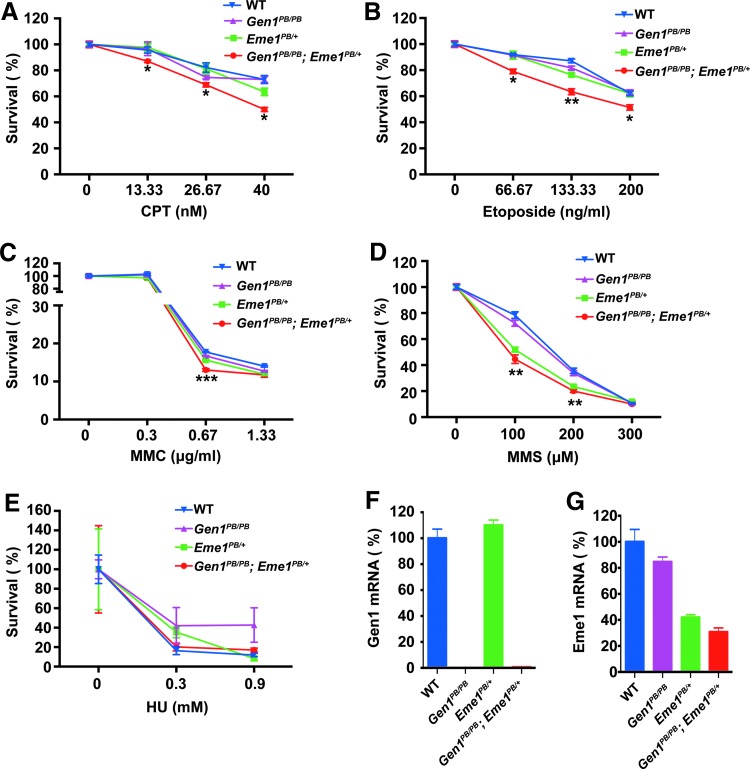
We speculate that the lethality of Gen1 and Eme1 double mutants may be due to the redundant roles of GEN1 and the MUS81-EME1 complex in DNA repair in mice. To confirm this hypothesis, we prepared primary MEFs carrying different combinations of Gen1 and Eme1 mutations to test their sensitivities to various DNA damaging reagents, such as mitomycin C (MMC), methyl methanesulfonate (MMS), camptothecin (CPT), and Etoposide. We did not observe any significant differences in the survival rates between the wild-type and Gen1PB/PB MEFs (Fig. 2A–D).
FIG. 2.
Gen1 mutation increased the sensitivity of Eme1PB/+ MEFs to DNA-damaging reagents. WT, Gen1PB/PB, Eme1PB/+, and Gen1PB/PB; Eme1PB/+ MEF cells were subjected to doses of CPT (A), Etoposide (B), MMC (C), MMS (D), and HU (E) as indicated. The survival rate of the cells was quantified 1 week later. Compared with Eme1PB/+ MEF cells, Gen1PB/PB; Eme1PB/+ MEF cells were more sensitive to CPT (A), Etoposide (B), and MMC (C). (F) Real-time RT PCR results of Gen1 in the four cell lines. (G) Real-time RT-PCR results of Gen1 in the four cell lines. Data are presented as mean ± SEM. *, p < 0.05; **, p < 0.01, ***, p < 0.001; unpaired t test. CPT, camptothecin; HU, hydroxyurea; MMC, mitomycin C; MMS, methyl methanesulfonate.
However, Gen1PB/PB mutations significantly increased the sensitivity of Eme1PB/+ MEFs to DNA-damaging reagents. Compared with that of Eme1PB/+ MEFs, the survival rate of Gen1PB/PB; Eme1PB/+ MEFs dropped by ∼12% under the treatment with three different concentrations of CPT or Etoposide (Fig. 2A, B). Although low and high concentrations of MMC treatment did not cause any differences, medial level of MMC (0.67 μg/mL) killed slightly more Gen1PB/PB; Eme1PB/+ MEFs than Eme1PB/+ MEFs (Fig. 2C). The survival rates of Gen1PB/PB; Eme1PB/+ and Eme1PB/+ MEFs are comparable with each other after MMS treatment, both lower than those of the wild-type or Gen1PB/PB MEFs (Fig. 2D). Meanwhile, DNA replication stress by hydroxyurea (HU) treatment did not bring any differences between the wild-type and Gen1PB/PB; Eme1PB/+ MEFs (Fig. 2E).
Real-time RT PCR showed that Gen1 mRNA was undetectable in homozygous mutant MEFs (Fig. 2F), whereas ∼31% of Eme1 mRNA still remained in Gen1PB/PB; Eme1PB/+ MEFs (Fig. 2G).
These results indicate that Gen1 and Eme1 are functionally redundant in the resistance to DNA damage-induced cell death in mammalian cells.
Gen1 and Eme1 play redundant roles in meiotic recombination
As the intermediate structure of DNA recombination, HJs exist not only in DNA repair, but also in meiosis. We thus checked if Gen1 and Eme1 also play redundant roles in meiotic recombination in mice.
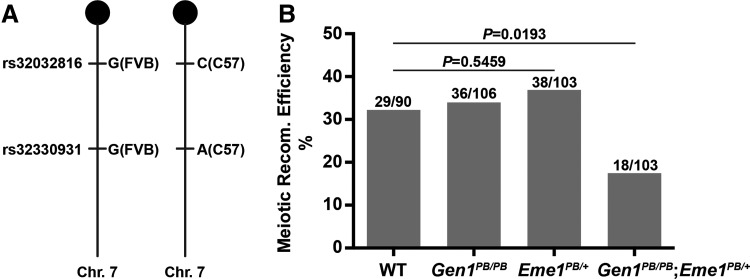
We first introduced Gen1 and Eme1 mutations into the mixed genetic background of FVB/NJ and C57BL/6J by breeding, then collected ∼100 offspring of each of the four genotypes (WT, Gen1PB/PB, Eme1PB/+, and Gen1PB/PB; Eme1PB/+) to measure the meiotic recombination efficiency by scoring the segregation of SNPs from both ancestral genetic background. After screening >40 SNPs that are not only different in FVB/NJ and C57BL/6J, but also mapped ∼25–50 Mbps apart from each other (Supplementary Table S2), we found a pair of Chr. Seven SNPs (rs32032816 and rs32330931) that were both heterozygous in all of the four genotypes so that they could be used to calculate the recombination efficiency (Fig. 3A).
FIG. 3.
Decreased meiotic recombination efficiency in Gen1PB/PB; Eme1PB/+ mice. (A) Brief illustration shows the genotype of two SNPs on FVB/NJ and C57BL/6J background, respectively. (B) Meiotic recombination efficiency of WT, Gen1PB/PB, Eme1PB/+, and Gen1PB/PB; Eme1PB/+ mice. Recombinant pups/total pups is labeled above each column. p Value was calculated by Fisher's exact test. SNPs, single nucleotide polymorphisms.
The calculated meiotic recombination efficiency in the offspring of wild-type male mice was 32.22% (29/90). Similar results were obtained in the offspring of Gen1PB/PB (33.96%, n = 106) and Eme1PB/+ (36.89%, n = 103) males. However, the rate was significantly reduced in the offspring of Gen1PB/PB; Eme1PB/+ mice. Only 17.48% (18/103, p = 0.0193) of the progeny were identified as recombinants. Thus, disruption of Gen1 affects meiotic recombination not on the wild type, but on the Eme1 mutant background (Fig. 3B). Other than this, we did not find obvious differences in the size and gross morphology of the testis in Gen1PB/PB; Eme1PB/+ and other mutants (Supplementary Fig. S1A–D). The mating between male mice of each genotype and WT FVB/NJ females resulted in similar litter sizes as well (Supplementary Fig. S1E). Taken together, these results suggest that Gen1 and Eme1 also play redundant roles in meiotic recombination.
Discussion
GEN1, MUS81-EME1, and SLX1 are three HJ resolvases that could process persistent HJs in mammalian cells (Boddy et al., 2001; Chen et al., 2001; Fricke and Brill, 2003; Ip et al., 2008; Fekairi et al., 2009; Munoz et al., 2009; Rass et al., 2010). However, their contributions to HJ-mediated processes in vivo have not been well defined. In this study, we found that a Gen1 mutation alone did not affect DNA repair or meiotic recombination in mice. It caused synthetic lethality, nevertheless, when combined with Eme1 mutations at an early embryonic stage. In addition, the combination of Gen1PB/PB; Eme1PB/+ mutations makes MEFs more sensitive to DNA damage and mice less capable of meiotic recombination. These results indicate that Gen1 and Eme1 are functionally redundant in DNA repair and meiotic recombination in mice.
Compared with those of the Mus81 and Eme1 mutants, the phenotypes related to DNA recombination are milder in Gen1 mutants. Homozygous deletion of Eme1 caused hypersensitivity to MMC and other DNA-damaging reagents in embryonic stem cells and MEFs (Abraham et al., 2003). Mus81−/− mice exhibited significant meiotic defects such as the depletion of mature epididymal sperm and partial failure of DNA double-strand breaks repair (Holloway et al., 2008). Thus, GEN1 likely plays a supporting role in DNA repair and meiosis recombination in mice. This may also explain why Gen1PB/PB; Eme1PB/+ MEFs showed only a mild additional effect on drug sensitivity when compared with Eme1PB/+ MEFs, in which substantial amount of Eme1 expression could still be detected (Fig. 2G).
Increased sensitivity of Gen1 and Eme1 double mutants to DNA damaging reagents may be a result of multiple cellular events. It has been shown that codepletion of MUS81 and GEN1 in human fibroblasts led to severe chromosome segregation defects (Wyatt et al., 2013). MUS81- and GEN1-codepleted HeLa cells also showed genome instability, which is exemplified by impaired replication fork movement and S-phase progression, endogenous checkpoint activation, chromosome segmentation, and multinucleation (Sarbajna et al., 2014). These events may also contribute to the synthetic lethality observed in Gen1PB/PB; Eme1PB/PB embryos, which may experience DNA damages caused by environment during development.
Meanwhile, mutant embryos may die of HJ processing deficiency during mitosis. DNA replication is known to produce HJs (Petermann and Helleday, 2010). Failure of the resolution of replication-induced HJs could cause delayed or arrested mitosis in mammalian cells, leading to cell death in the absence of exogenous DNA damage (Garner et al., 2013). Depletion of GEN1 in human cells expressing mutant SLX4 that no longer binds and activates MUS81 would result in dysfunctional mitosis and cell death under normal culturing conditions (Garner et al., 2013; Sarbajna et al., 2014). Double mutations of GEN1 and EME1 may cause cell death through similar mitotic defects.
Conclusion
In summary, GEN1 and MUS81-EME1 are functionally redundant in mice to ensure the elimination of HJs during both mitotic DNA damage repair and meiotic recombination. Given that both MUS81-EME1 and SLX1 interact with the scaffold protein SLX4 to resolve HJs together in mammalian cells, it would be reasonable to speculate that GEN1 and SLX1 are also functionally redundant in these processes in vivo.
Supplementary Material
Acknowledgments
The authors thank Dr. Kai Lei, Zhisheng Ye, and Xiaoqiang Zhu for helpful discussion, and Guicheng Wang, Yanyan Nie, Xiaorong Huang, and Zhiyan Xia for technical assistance. This study was supported by grants from the National Natural Science Foundation of China (81270763), Chinese Hi-Tech Research and Development Project (2014AA021104), Chinese Key Projects for Basic Research (2011CB944001), Shanghai Municipal Government (15XD1500500 and 12431900100), and the Research Fund of the State Key Laboratory of Genetic Engineering, Fudan University.
Disclosure Statement
No competing financial interests exist.
References
- Abraham J., Lemmers B., Hande M.P., Moynahan M.E., Chahwan C., Ciccia A., Essers J., Hanada K., Chahwan R., Khaw A.K., et al. (2003). Eme1 is involved in DNA damage processing and maintenance of genomic stability in mammalian cells. EMBO J 22, 6137–6147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco M.G., Matos J., Rass U., Ip S.C., and West S.C. (2010). Functional overlap between the structure-specific nucleases Yen1 and Mus81-Mms4 for DNA-damage repair in S. cerevisiae. DNA Repair (Amst) 9, 394–402 [DOI] [PubMed] [Google Scholar]
- Boddy M.N., Gaillard P.H., McDonald W.H., Shanahan P., Yates J.R., 3rd and Russell P. (2001). Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell 107, 537–548 [DOI] [PubMed] [Google Scholar]
- Cejka P., Plank J.L., Bachrati C.Z., Hickson I.D., and Kowalczykowski S.C. (2010). Rmi1 stimulates decatenation of double Holliday junctions during dissolution by Sgs1-Top3. Nat Struct Mol Biol 17, 1377–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.B., Melchionna R., Denis C.M., Gaillard P.H., Blasina A., Van de Weyer I., Boddy M.N., Russell P., Vialard J., and McGowan C.H. (2001). Human Mus81-associated endonuclease cleaves Holliday junctions in vitro. Mol Cell 8, 1117–1127 [DOI] [PubMed] [Google Scholar]
- Ding S., Wu X., Li G., Han M., Zhuang Y., and Xu T. (2005). Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell 122, 473–483 [DOI] [PubMed] [Google Scholar]
- Ellis N.A., Groden J., Ye T.Z., Straughen J., Lennon D.J., Ciocci S., Proytcheva M., and German J. (1995). The Bloom's syndrome gene product is homologous to RecQ helicases. Cell 83, 655–666 [DOI] [PubMed] [Google Scholar]
- Fekairi S., Scaglione S., Chahwan C., Taylor E.R., Tissier A., Coulon S., Dong M.Q., Ruse C., Yates J.R., 3rd, Russell P. et al (2009). Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell 138, 78–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricke W.M., and Brill S.J. (2003). Slx1-Slx4 is a second structure-specific endonuclease functionally redundant with Sgs1-Top3. Genes Dev 17, 1768–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner E., Kim Y., Lach F.P., Kottemann M.C., and Smogorzewska A. (2013). Human GEN1 and the SLX4-associated nucleases MUS81 and SLX1 are essential for the resolution of replication-induced holliday junctions. Cell Rep 5, 207–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho C.K., Mazon G., Lam A.F., and Symington L.S. (2010). Mus81 and Yen1 promote reciprocal exchange during mitotic recombination to maintain genome integrity in budding yeast. Mol Cell 40, 988–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway J.K., Booth J., Edelmann W., McGowan C.H., and Cohen P.E. (2008). MUS81 generates a subset of MLH1-MLH3-independent crossovers in mammalian meiosis. PLoS Genet 4, e1000186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip S.C., Rass U., Blanco M.G., Flynn H.R., Skehel J.M., and West S.C. (2008). Identification of Holliday junction resolvases from humans and yeast. Nature 456, 357–361 [DOI] [PubMed] [Google Scholar]
- Lee S.H., Princz L.N., Klugel M.F., Habermann B., Pfander B., and Biertumpfel C. (2015). Human Holliday junction resolvase GEN1 uses a chromodomain for efficient DNA recognition and cleavage. Elife 4, e12256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei K., Zhu X., Xu R., Shao C., Xu T., Zhuang Y., and Han M. (2012). Inner nuclear envelope proteins SUN1 and SUN2 play a prominent role in the DNA damage response. Curr Biol 22, 1609–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Freeman A.D., Declais A.C., Wilson T.J., Gartner A., and Lilley D.M. (2015). Crystal structure of a eukaryotic GEN1 resolving enzyme bound to DNA. Cell Rep 13, 2565–2575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz A., West S.C., and Whitby M.C. (2010). The human Holliday junction resolvase GEN1 rescues the meiotic phenotype of a Schizosaccharomyces pombe mus81 mutant. Nucleic Acids Res 38, 1866–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos J., Blanco M.G., Maslen S., Skehel J.M., and West S.C. (2011). Regulatory control of the resolution of DNA recombination intermediates during meiosis and mitosis. Cell 147, 158–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos J., and West S.C. (2014). Holliday junction resolution: regulation in space and time. DNA Repair (Amst) 19, 176–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz I.M., Hain K., Declais A.C., Gardiner M., Toh G.W., Sanchez-Pulido L., Heuckmann J.M., Toth R., Macartney T., Eppink B., et al. (2009). Coordination of structure-specific nucleases by human SLX4/BTBD12 is required for DNA repair. Mol Cell 35, 116–127 [DOI] [PubMed] [Google Scholar]
- Petermann E., and Helleday T. (2010). Pathways of mammalian replication fork restart. Nat Rev Mol Cell Biol 11, 683–687 [DOI] [PubMed] [Google Scholar]
- Rass U., Compton S.A., Matos J., Singleton M.R., Ip S.C., Blanco M.G., Griffith J.D., and West S.C. (2010). Mechanism of Holliday junction resolution by the human GEN1 protein. Genes Dev 24, 1559–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbajna S., Davies D., and West S.C. (2014). Roles of SLX1-SLX4, MUS81-EME1, and GEN1 in avoiding genome instability and mitotic catastrophe. Genes Dev 28, 1124–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L.V., Jin K., Liu Y., Yang W., Xie X., Ye L., Wang L., Zhu L., Ding S., Su Y., et al. (2008). PBmice: an integrated database system of piggyBac (PB) insertional mutations and their characterizations in mice. Nucleic Acids Res 36(Database issue), D729–D734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svendsen J.M., Smogorzewska A., Sowa M.E., O'Connell B.C., Gygi S.P., Elledge S.J., and Harper J.W. (2009). Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell 138, 63–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay Y.D., and Wu L. (2010). Overlapping roles for Yen1 and Mus81 in cellular Holliday junction processing. J Biol Chem 285, 11427–11432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler T., Newman S., and West S.C. (2011). Aberrant chromosome morphology in human cells defective for Holliday junction resolution. Nature 471, 642–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S.C. (2009). The search for a human Holliday junction resolvase. Biochem Soc Trans 37, 519–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., and Hickson I.D. (2003). The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature 426, 870–874 [DOI] [PubMed] [Google Scholar]
- Wyatt H.D., Sarbajna S., Matos J., and West S.C. (2013). Coordinated actions of SLX1-SLX4 and MUS81-EME1 for holliday junction resolution in human cells. Mol Cell 52, 234–247 [DOI] [PubMed] [Google Scholar]
- Zakharyevich K., Tang S., Ma Y., and Hunter N. (2012). Delineation of joint molecule resolution pathways in meiosis identifies a crossover-specific resolvase. Cell 149, 334–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.